Abstract
Background
Early diagnosis of tumor metastasis is crucial for clinical treatment. Artificial intelligence (AI) has shown great promise in the field of medicine. We therefore aimed to evaluate the diagnostic accuracy of AI algorithms in detecting tumor metastasis using medical radiology imaging.
Methods
We searched PubMed and Web of Science for studies published from January 1, 1997, to January 30, 2020. Studies evaluating an AI model for the diagnosis of tumor metastasis from medical images were included. We excluded studies that used histopathology images or medical wave-form data and those focused on the region segmentation of interest. Studies providing enough information to construct contingency tables were included in a meta-analysis.
Findings
We identified 2620 studies, of which 69 were included. Among them, 34 studies were included in a meta-analysis with a pooled sensitivity of 82% (95% CI 79–84%), specificity of 84% (82–87%) and AUC of 0·90 (0·87–0·92). Analysis for different AI algorithms showed a pooled sensitivity of 87% (83–90%) for machine learning and 86% (82–89%) for deep learning, and a pooled specificity of 89% (82–93%) for machine learning, and 87% (82–91%) for deep learning.
Interpretation
AI algorithms may be used for the diagnosis of tumor metastasis using medical radiology imaging with equivalent or even better performance to health-care professionals, in terms of sensitivity and specificity. At the same time, rigorous reporting standards with external validation and comparison to health-care professionals are urgently needed for AI application in the medical field.
Funding
College students' innovative entrepreneurial training plan program .
Keywords: Tumor metastasis, Medical imaging, Artiificial intelligence, Deep learning, Diagnostic meta-analysis
Research in context.
Evidence before this study
The accurate diagnosis of tumor metastasis without misdiagnosis and missed diagnosis is a challenging task. Artificial intelligence (AI) has already shown great promise for automated diagnosis from medical imaging with rapid speed and high accuracy. There is an urgent need for the application of such diagnostic technologies for the detection of tumor metastasis from medical radiology imaging. We searched PubMed and Web of Science for studies published from Jan 1, 1997, to Jan 30, 2020, with no restrictions on regions, languages, or publication types. Studies were included if they evaluated an AI model for the diagnosis of tumor metastasis from medical images. We found one systematic review comparing performance of AI algorithms with health-care professionals for all diseases, but we did not find systematic reviews focusing on tumor metastasis.
Added value of this study
To the best of our knowledge, this systematic review and meta-analysis is the first to show that AI algorithms were beneficial for the diagnosis of tumor metastasis from medical radiology imaging across a broad range of primary tumors and metastasis sites. During the process, we also found several common methodological defects that should be considered by algorithm developers. High-quality evidence with externally validated results and comparison to health-care professionals are urgently needed for studies on AI application in the medical field.
Implications of all the available evidence
AI algorithms were beneficial for the diagnosis of tumor metastasis from medical radiology imaging. The methodology and reporting of studies on the AI application in the medical field is often flawed. Normative and rigorous reporting standards should be established to enable the results to be more credible.
Alt-text: Unlabelled box
1. Introduction
Tumor metastasis, including lymph node metastasis (LNM) and distant metastasis (DM), contributes to cancer-related death. Regarding tumor classification, N and M staging are essential for both the treatment strategy, like the plan for surgery and chemoradiotherapy, and prognosis prediction [1,2]. Thus, it is crucial to conduct a complete and accurate pre-operative clinical evaluation of tumor metastasis.
Medical imaging is commonly used to visualize tumor dissemination and quantify the severity, providing valuable information for diagnosis, staging and treatment decision [3] with satisfactory diagnostic accuracy. For example, the sensitivity and specificity of contrast-enhanced ultrasound (CEUS), multidetector computed tomography (MDCT), magnetic resonance imaging (MRI), and fluorodeoxyglucose (FDG) positron emission tomography (PET)/CT in the detection of colorectal cancer liver metastasis was 80–97% [4], which is similar in other diseases [5,6]. However, owing to the uncoordinated ratio of doctors to patients and the difficulty of radiological diagnosis, making a correct and timely diagnosis from medical imaging is challenging [7].
Artificial intelligence (AI) has already shown great promise to address this problem through automated diagnosis from medical imaging [8,9]. In the 1980s, artificial neural networks (ANNs) were developed [10], resulting in a surge of machine learning (ML) based on statistical models. In the 1990s, various ML models were successively proposed, such as support vector machines (SVM) [11] and random forests (RF) [12]. It is not until 2006 that deep learning (DL), a new branch of ML, gained great attention [13,14]. Since then, DL, such as convolutional neural networks (CNN) and deep neural networks (DNN), has been applied in many fields, including photo captioning, automatic speech recognition, image recognition, natural language processing, drug discovery and bioinformatics [15], [16], [17], [18], [19]. Over the past few decades, due to the progress of high-throughput technologies, biomedical data like genome sequences and medical images has experienced explosive growth [20]. With the promising performance of AI in big data and image processing [21,22], more and more people anticipate similar success in the medical field, especially in medical imaging. AI can automatically detect details in medical images, and thus make a quantitative assessment rather than the subjective visual assessment by clinicians. Moreover, human experts may leave out some small metastases, resulting in a missed diagnosis [23], [24], [25].
Considering high expectations and demands for AI diagnosis tools in the clinical practice, it is time to review the evidence supporting AI-based diagnosis systematically. In this systematic review and meta-analysis, we were the first to evaluate the diagnostic performance of AI algorithms in tumor metastasis from medical radiology imaging, aiming to guide clinical practice.
2. Methods
2.1. Search strategy and selection criteria
In this systematic review and meta-analysis, we searched for studies that developed or validated an AI model for the diagnosis of tumor metastasis (LNM and DM) from medical radiology imaging. We searched PubMed and Web of Science for studies published from January 1, 1997, to January 30, 2020, with no restrictions on regions, languages, or publication types. A major milestone that happened in 1997 may explain why this starting time was chosen. In 1997, IBM's "Deep Blue" computer defeated the world chess champion Kasparov. After that, artificial intelligence began its positive development. [26] Full search terms and search strategies are provided in the Appendix Section 1.
Reviewers (QZ and LY) screened titles and abstracts of the search results. Uncertainties about inclusion were resolved by the other reviewer (BZ). Studies were included if they evaluated an AI model for the diagnosis of tumor metastasis from medical images with all forms of diagnostic outcomes, such as accuracy, precision, Dice-ratio and recall, etc.. There were no limits on the participants, the type of tumor metastasis, or the intended context for using the model. For the study reference standard to identify whether there is the presence of metastasis, we accepted clinical notes, expert opinion or consensus, and histopathology or laboratory testing.
Giving for radiology images were most widely used in clinical practice to diagnose tumor metastasis, we excluded studies that used histopathology images or medical wave-form data and those focused on the region segmentation of interest to make our study more consistent. We extracted binary diagnosis accuracy data, so ternary diagnosis outcomes were excluded because it had some difference when constructing contingency tables by binary outcomes. Studies that used pre-treatment images to predict conditions of lymph nodes after treatment (e.g. radiotherapy and chemotherapy) were not included because our focus is “diagnosis” other than “prediction”. Studies based on animals or nonhuman samples or those presented duplicate data were also excluded.
This systematic review was done following the recommendations of the PRISMA statement [27]. The research question was formulated according to previously published recommendations for systematic reviews of prediction models (CHARMS checklist) [28].
2.2. Data collection
Three reviewers (QZ, LY and JL) extracted data independently using a predefined data extraction sheet, and uncertainties were resolved by another reviewer (BZ). We extracted binary diagnosis accuracy data and constructed contingency tables, which included true-positive (TP), false-positive (FP), true-negative (TN), and false-negative (FN) results if the study provided enough information. Sensitivity and specificity results were calculated from contingency tables.
To evaluate the performance of the AI model, we conducted a meta-analysis from studies providing enough information to construct contingency tables. If a study provided several contingency tables for different algorithms or primary tumors, we treated them as independent items.
The quality of the included studies was evaluated by the reviewers (QZ and KG) and conformed to the revised version of the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) [29].
2.3. Statistical analysis
Receiver operating characteristic (ROC) curves were constructed to evaluate the accuracy of the AI model. The ROC figures provide average sensitivity and specificity across included studies with a 95% confidence interval (CI) of the summary operating point. The ROC figures also provide the 95% prediction region representing the confidence intervals for forecasts of sensitivity and specificity in a future study. Areas under the ROC curve (AUCs) with 95% CI were also calculated. Odds ratio (OR) and 95% CI for each study was calculated to estimate the performance of the AI algorithms.
We calculated heterogeneity between studies using the χ² test (threshold P = 0·1), which was quantified using the I² statistic. We also conducted the subgroup analysis and regression analysis to identify the sources of heterogeneity. Random effects models were used during the process. P value of 0·05 or less was considered to indicate a statistically significant difference.
Two separate analyses were performed according to different algorithms and whether studies were externally validated. Following its development, we divided AI algorithms into ML algorithms (ANN, KNN, SVM, RF, logistic regression and decision tree) and DL algorithms (CNN, DNN and DCNN). External validation means studies were validated by out-of-sample dataset.
To compare diagnostic performance between AI algorithms and health-care professionals, we did another separate analysis for studies providing contingency tables for both health-care professionals and AI algorithm using the same sample. We evaluated the quality of included studies according to QUADAS-2 by RevMan (Version 5.3). Stata (Version 15.0) was used in the ROC curves, the calculation of AUC, subgroup analysis, Deeks’ Funnel Plot Asymmetry Test and forest plots. Data analysis was performed by BZ. This study is registered with PROSPERO, CRD42020172924.
2.4. Role of the funding source
Our study was funded by the College Students' Innovative Entrepreneurial Training Plan Program (No.201901249). The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
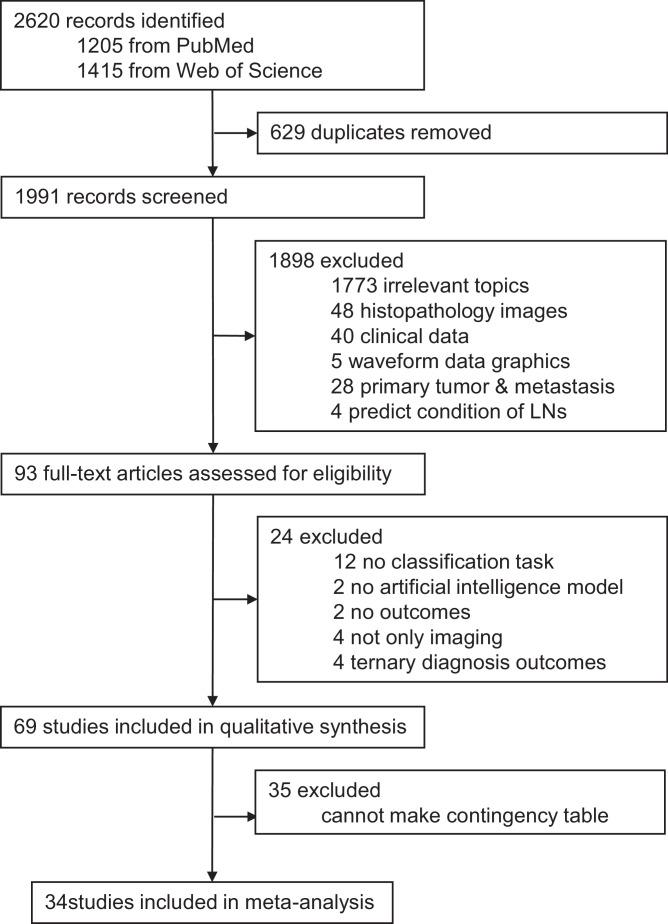
Fig. 1 summarized our literature search for eligible studies. Our search identified 2620 records, of which 1991 were screened after removing 629 duplicates. 1898 articles were excluded as they did not meet the inclusion criteria. 93 full-text articles were assessed for eligibility and 24 articles were excluded when scanning the full text. As a result, 69 studies were included in the systematic review. Among the 69 studies, 34 studies provided enough information to construct contingency tables and calculate test performance parameters, and were therefore included in the meta-analysis.
Fig. 1.
Study selection.
These 69 studies described 72 patient cohorts. In these studies, target conditions were divided into LNM (45 studies) and DM (26 studies) (2 studies involved both LNM and DM), which included bone metastasis (13 studies), brain metastasis (3 studies), liver metastasis (4 studies), lung metastasis (2 studies) and others (4 studies). Primary tumors comprised breast cancer (10 studies), head and neck cancer (9 studies), gastric cancer (7 studies), lung cancer (6 studies), colorectal cancer (5 studies), prostate cancer (3 studies) and other primary tumors (6 studies). Thirteen studies did not report this. In addition, 10 studies contained several different primary tumors. Study characteristics are shown in Tables 1, 2 and 3. All included studies used retrospective data and were not open-access. Seven studies excluded low-quality images which meant that the location and size of the lesion on the images did not match that seen at pathologic examination or one or two of the most representative images were selected for each patient, while 62 studies did not report this. Comparison between AI models and health-care professionals by the same test set was only provided in 8 studies. As for the verification of the model, 7 studies collected out-of-sample dataset to do an external validation, and the others were internally validated. Furthermore, different algorithms including DL (23 studies) and ML (34 studies) were included in the systematic review. Four studies used both DL and ML algorithms and 5 studies did not report the detailed types of algorithms.
Table 1.
Participant demographics for the 69 included studies.
| First author and year | Participants | |||||
|---|---|---|---|---|---|---|
| Inclusion criteria | Exclusion criteria | Patient/Sample | Positive Patients(samples)/Negative Patients(samples) | Mean age (SD; range), year | Percentage of male participants | |
| Mitsuru Koizumi et al. (2020) [40] | NR | Skeletal metastasis did not meet the criteria of the term ‘disseminated’; no skeletal metastasis | 54/54 | 54(NR)/0(NR) | NR | NR |
| Jing Li et al. (2020) [41] | Patients underwent gastrectomy plus lymph node dissection and were diagnosed gastric adenocarcinomas; patients were scanned with GSI mode; without any local or systematic treatment before CT scans and surgery; with definite postoperative pathologic data. | Invisible lesion on CT images; with a minimum diameter of tumor less than 5 mm insufficient to outline a valid ROI; insufficient stomach distension; poor image quality for post-processing. | 204/NR | 122(NR)/82(NR) | Training set:59(12;28–81) Test set:59(11;28–74) |
Training set:72% Test set:72% |
| L. Zhang et al. (2020) [42] | NR | NR | 51/NR | 32(NR)/19(NR) | NR | 47% |
| Li-Qiang Zhou et al. (2020) [43]* | Patients with histologically confirmed primary breast cancer who underwent surgery; T1 or T2 primary breast cancer with clinically negative LNs and no preoperative therapy; standard preoperative breast US | T3 or T4 stage; physically positive LNs; imaging positive LNs; physically and imaging positive LNs; preoperative therapy; low quality US images | Cohort1: 756/974 Cohort 2:78/81 |
Training set:343(441)/337(436) Testing set: internal validation:37(49)/39(48) external validation:40(43)/38(38) |
Training set:48(NR;24–81) Test set: internal validation:50(NR;25–82) external validation:46(NR;30–74) |
NR |
| Endre Grøvik et al. (2020) [44] | The presence of known or possible metastatic disease; no prior surgical or radiation therapy; the availability of all required MRI sequences; patients with ≥1 metastatic lesion | NR | 156/156 | 156(156)/0(0) | 63(12;29–92) | 33% |
| Yu Zhao et al. (2019) [45] | Patients with metastatic castration-resistant prostate cancer | NR | 193/NR | 193(NR)/0(NR) | 69.6(7.9;NR) | NR |
| Jie Xue et al. (2019) [46] | Definitely histopathological results of the primary tumor lesion; patients with only metastatic lesions in brain; with an age over 18 years old; 3D T1 MPRAGE sequence was acquired. | Unqualified imaging quality of 3D T1 MPRAGE; data missing; skull metastases and meningeal metastases | Dataset 1:1201/1201 Dataset 2:231/231 Dataset 3:220/220 |
Dataset 1:1201(1201)/0(0) Dataset 2:231(231)/0(0) Dataset 3:220(220)/0(0) |
Dataset 1:58(18;NR) Dataset 2:60(18;NR) Dataset 3:59(15;NR) |
Dataset 1:57% Dataset 2:53% Dataset 3:52% |
| Bettina Baessler et al. (2019) [47]* | Patients with retroperitoneally metastasized testicular germ cell tumors prior to post-chemotherapy LN dissection | Absence of contrast-enhanced CT imaging data after chemotherapy and prior to post-chemotherapy LN dissection; insufficient image quality; insufficient matching of histopathology to the individual LNs | 80/204 Training set:63/120 Testing set: internal validation:19/23 external validation:41/61 |
44(107)/36(97) Training set: NR(60)/NR(60) Testing set: NR(15)/NR(8) Validation set: NR(25)/NR(36) |
LNM:34(13;NR) N-LNM:36(10;NR) |
NR |
| Xiaojun Yang et al. (2019) [48]* | Preoperative contrast-enhanced CT images within 2 weeks before surgery; histologically confirmed primary invasive breast cancer; SLN biopsy (and ALND); pathologically results after operation confirmed SLN metastasis | Neoadjuvant therapy before CT examination and surgery; poor visualization of the tumor for segmentation due to serious artifacts caused by metallic foreign bodies on the breast; tumor was too small to be seen on CT images; incomplete clinicopathological data | 348/348 Training set:184/184 Testing set:164/164 |
Training set:71(71)/113(113) Testing set:63(63)/101(101) |
Training set: SLN-P:52(9;NR); SLN—N:50(11;NR) Testing set: SLN-P:50(10;NR); SLN—N:53(10;NR) |
NR |
| Yuan Gao et al. (2019) [49] | NR | No metastatic LNs revealed by CT; with preoperative neoadjuvant radio-chemotherapy; complicated with abdominal infection; pathological grouping different from CT grouping; LN adhesions | 602/38,495 | NR | 62(NR;20–91) | 72% |
| David Coronado-Gutierrez et al. (2019) [50]* | Positive metastatic nodes by ultrasound-guided FNA or CNB; Negative metastatic nodes determined by histopathology | Surgical biopsy showed positive result after not suspicious nodes in ultrasound exam or negative results of ultrasound-guided FNA or CNB; Patients refused to receive SLNB | 127/118 | NR(53)/NR(65) | 54.6 (NR;26~91) | NR |
| Yukinori Okada et al. (2019) [51] | NR | NR | 56/NR | 56(NR)/0(0) | 59 (12.7;NR) | 0 |
| Jeong Hoon Lee et al. (2019) [52]* | NR | NR | 202/995 | NR(348)/NR(647) | NR | NR |
| Jansen et al. (2019) [53] | NR | Based on visual evaluation, DW-MRI failed to register on the DCE-MR series | 111/111 | 72(NR)/39(NR) | NR | NR |
| Chuangming Li et al. (2019) [54]* | Patients had breast cancer confirmed by histology; underwent a DCE-MRI scan before tumor resection or biopsy; received tumor resection and SLNB within 1 week after MRI examination | MRI examination data were incomplete, or image quality was poor | 62/62 | 35(NR)/27(NR) | SLN-P:48.14 (8.35; NR) SLN—N:49.78 (12.53; NR) |
NR |
| M. Dohopolski et al. (2019) [55] | Patients with oropharyngeal squamous cell carcinoma; underwent neck dissections; had preoperative PET and CT imaging | NR | 129/543 | NR | NR | NR |
| Yige Peng et al. (2019) [56]* | NR | No detailed metastases information | 48/NR | 24(NR)/24(NR) | NR | NR |
| Qiuxia Feng MD et al. (2019) [57]* | Definitive diagnosis by histopathology | Neoadjuvant chemotherapy or radiotherapy or endoscopic resection; end-stage disease or severe complications precluding surgery; disease that could not be detected on imaging; poor imaging quality or poor gastric resection | 490/NR | 279(NR)/211(NR) | 61.8(10.4; NR) | Training and validation set: 73% Test set: 77% |
| Thoma Schnelldorfer et al. (2019) [58] | Underwent a laparoscopic operation with the initial intent for either resection or palliation of the underlying malignancy; Video recordings of the operation were available | Malignancy originating from esophageal, hepatic and colorectal malignancies | 35/35 | 20(20)/15(15) | 67 (NR;44~85) | 66% |
| Samir D. Mehta et al. (2019) [59]* | Underwent CT of the abdomen and pelvis or radiographs of the lumbar spine and DEXA studies; CT studies/ lumbar spine radiographs were performed not more than 1 year prior to the DEXA study | NR | 200/NR | 45(NR)/155(NR) | Case: 70.5 (NR;63.9~76.7) Control: 62 (NR;53.5~69) |
Case: 78% Control: 83% |
| Yoshiko Ariji, et al. (2019) [60]* | Underwent intravenous contrast enhanced CT and dissection of cervical lymph nodes | NR | 45/441 | NR(127(/NR(314) | 63 (NR;33~95) | 53% |
| Yunpeng Zhou et al. (2019) [61] | Definite lymph node metastasis reported by preoperative imaging | With a history of abdominal pelvic surgery, and pelvic radio-chemotherapy | 301/12,060 | 301(NR)/0(NR) | 59.5(NR; NR) | 75% |
| Yu Li et al. (2019) [62]* | Received radical colectomy with lymph node dissection; Patients with colon cancer diagnosis; Patients with no history of previous or coexisting other malignancies; Patients who underwent preoperative enhanced CT for local colon cancer staging and for liver metastasis diagnosis; | Patients who underwent treatment (radiotherapy, chemotherapy or chemoradiotherapy) before the baseline CT examination; Poor image quality; Patients with liver metastasis who did not receive synchronous resection of the primary tumor and liver metastasis | 48/NR | 24(NR)/24(NR) | LNM: 63.3 (11.21; NR) Non-LNM: 59.71 (13.86; NR) |
63% |
| Zhiguo Zhou et al. (2019) [63]* | NR | NR | 129/543 | Training set: NR (91)/NR (287) Test set: NR (39)/NR (126) |
NR | NR |
| eMine acar et al. (2019) [64] | Sclerotic lesions >2 cm in patients with at least three sclerotic metastatic lesions; sclerosis areas of the bones that located on the surface of the joint and/or on the surface of the other side of the joint; osteophytes not considered as metastasis. | No bone metastasis; <3 bone metastasis; no sclerotic metastasis; uptake<liver uptake | 75/257 | NR(153)/NR(104) | 69(9; NR) | NR |
| Fang Hou et al. (2019) [65]* | NR | NR | 28/573 | Training set: NR (21)/NR (293) Test set: NR (25)/NR (234) |
NR | NR |
| Yoshiko Ariji et al. (2019) [66]* | Oral squamous cell carcinoma; underwent neck dissection; pathology confirms cervical lymph node metastasis | NR | 54/143 (LN) 703 (image) | NR (33)/NR (110) | 64(NR; NR) | 52.94% |
| Xiaojuan Xu et al. (2019) [67] | Patients who received standard FIGO surgical staging for endometrial cancer between January 2011 and December 2017 | Patients without DCE-MRI 2 weeks before surgery; patients with serious MR artifacts and without uniform MR scanner; patients missing clinical characteristics data and endometrial biopsy histological information; patients with any preoperative therapy; patients suffering from other malignant tumor diseases concurrently | 200/NR | 67(NR)/133(NR) | Training cohort: pN(+):55.7(NR; NR) pN(-):55.7 (NR; NR) Test Cohort: pN(+):57.4(NR; NR) pN(-):51.7(NR’; NR) |
NR |
| Jiaxiu Luo et al. (2018) [68]* | NR | NR | 172/NR | 74(NR)/98(NR) | NR | NR |
| Richard Ha et al. (2018) [69] | NR | NR | 275/275 | 133(133)/142(142) | NR | NR |
| B.H. Kann et al. (2018) [70]* | NR | NR | 270/653 | NR (380)/NR (273) | NR | NR |
| Jeong Hoon Lee et al. (2018) [71]* | NR | NR | 804/812 cohort1:604/612 cohort2:200/200 |
Training set: NR (286)/NR (263) Validation set: NR (33)/NR (30) Test set: NR (100)/NR (100) |
Training & Validation set:44(NR;13–84) Test set:55(NR;10–81) |
Training & Validation set:30.6% Test set:27% |
| Yun Lu et al. (2018) [72] | NR | NR | Training set:351/28,080 Test set:414/36,000 |
Training set:351(28,080)/0(0) Test set: NR |
NR | NR |
| José Raniery Ferreira Junior et al. (2018) [73]* | NR | No standard contrast-enhanced CT protocol; did not present all clinical data; presented other opacities attached to the tumor | 68/NR | LNM: Test set:23(NR)/29(NR) Validation set:9(NR)/7(NR) DM: Test set:8(NR)/44(NR) Validation set:5(NR)/11(NR) |
Test set:66.6(9.1;41–85) Validation set:64.88(9.1;41–79) |
Test set:57.7% Validation set:62.5% |
| Tzu-Yun Lo et al. (2018) [74] | NR | NR | 70/75 | 70(75)/0(0) | NR | NR |
| Jin Li et al. (2018) [75] | NR | NR | NR/619 | Original data: NR(307)/NR(312) augmented data: NR(1535)/NR(1560) |
NR | NR |
| Mohamed Amine Larhmam et al. (2018) [76] | NR | NR | NR/153 | NR (87)/NR (66) | NR | NR |
| Yan Zhong et al. (2018) [77]* | Underwent surgical resection and systematic LN dissection according to the American Thoracic Society criteria; had no enlargement of the hilar or mediastinal LNs at CT (enlargement defined as short axis of a node ≥ 10 mm on axis images) and clinical N0; no distal metastasis | IV administration of contrast material; unsatisfactory image quality due to respiratory artifact during the examination that may have disturbed feature extraction; and surgical resection not performed within 90 days of thin-section CT | 492/492 | 78(78)/414(414) | 61.4(9.7; NR) N-LNM:61.28(9.8; NR) LNM:61.71(9.62; NR) |
35% N-LNM:32% LNM:50% |
| Wang, H et al. (2017) [78]* | NR | NR | 168/1397 | NR (127)/NR (1270) | 61(NR;38–81) | 54% |
| Mitsuru Koizumi et al. (2017) [79]* | NR | NR | 265/265 | 124(124)/101(101) | NR | NR |
| Juan Wang et al. (2017) [80] | NR | NR | 26/NR | 26(NR)/0(NR) | 58(14; NR) | 54% |
| Zhi-Long Wang et al. (2017) [81] | NR | Pathologically proven adenocarcinoma, small cell carcinoma, mixed cancer, or other diseases; other preoperative therapies simultaneously; esophageal multiple primary carcinoma; death within 30 days after surgery; enhanced CT data before preoperative chemotherapy not obtained or images not interpretable; non-suitability for radical esophagectomy | 131/NR | 51(NR)/80(NR) | 58(NR;42–75) | 77.90% |
| Tuan D. Pham et al. (2017) [82]* | Biopsy-proven primary lung malignancy with pathological mediastinal nodal staging; | Patients with nodal biopsy more than three months from CT | 148/NR | Test set: NR (133)/NR (138) | 69.4(NR;36–84) | 63% |
| Qi Zhang et al. (2017) [83]* | Underwent axilla conventional US and RTE simultaneously | Take neoadjuvant therapy before SLNB or ALND | 158/161 | NR (92)/NR (69) | 55.2(5.2;21–81) | NR |
| Yu-wen Wang et al. (2016) [84]* | NR | A relatively large (minimal axial diameter up to 10 mm) necrotic node, which did not promptly respond to RT | Stage I: 335/663 Stage II: 210/410 |
Stage I: NR (337)/NR (326); Stage II: NR (211)/NR (199) | NR | NR |
| Ali Aslantas et al. (2016) [85]* | NR | NR | 60/130 | 39(34)/21(96) | 57(NR;30–87) | 60% |
| Aneta Chmielewski et al. (2015) [86]* | Underwent surgical treatment for invasive breast cancer with axillary lymph node evaluation | NR | 77/105 | NR (24)/NR (81) | NR | 0 |
| Mitsuru Koizumi et al. (2015) [87]* | NR | NR | 426/NR | 152(NR)/274(NR) | NR | NR |
| Mitsuru Koizumi et al. (2015) [88] | NR | Patient showing segmentation error on BONENAVI version 2 | 394/NR | 142(NR)/252(NR) | NR | NR |
| Nesrine Trabelsi et al. (2015) [89] | NR | NR | 11/NR | 11(NR)/0(NR) | NR | NR |
| Xuan Gao et al. (2015) [90] | NR | NR | 132/768 | NR | NR | 60.60% |
| Osamu Tokuda, et al. (2014) [91],* | NR | Benign conditions; did not undergo follow-up examinations; younger than 20 years of age | 406/3248 | 90(235)/316(3013) Prostatic cancer: NR(104)/NR(464); Breast cancer: NR(42)/NR(830); Males with other cancer: NR(56)/NR(1168); Females with other cancers: NR(33)/NR(551) |
66(NR;27–92) | 55% |
| Ari Seff et al. (2014) [92] | NR | NR | Mediastinal LN:90/389(LN) Abdominal:86/595(LN) |
Mediastinal LN:NR(960Candidates)/NR(3208Candidates) Abdominal: NR(1005Candidates)/NR(3484Candidates) |
NR | NR |
| Zhi-Guo Zhou et al. (2013) [93]* | NR | NR | 175/175 | 134(NR)/41(NR) | 59.8(NR;30–85) | 71% |
| Seungwook Yang et al. (2013) [94]* | NR | Excessive motion artifacts | 26/90 | Test Set: black-blood:26(53)/0(443); MP-RAGE:26(53)/0(5788) | NR | NR |
| Jianfei Liu et al. (2013) [95] | NR | NR | 50/NR | Training set: NR; Test set:44(102)/NR | NR | NR |
| Yoshihiko Nakamura et al. (2013) [96] | NR | NR | 28/NR | 28(95)/0(NR)` | NR | NR |
| Chuan-Yu Chang et al. (2013) [97] | NR | NR | 6/177 | All positive | NR | NR |
| Johannes Feulner et al. (2013) [98] | NR | NR | 54/1086 | NR(289)/NR(NR) | NR | NR |
| Chao Li et al. (2012) [99] | NR | NR | 38/NR | 27(NR)/11(NR) | NR | NR |
| Hongmin Cai et al. (2012) [100] | NR | NR | 228/NR | NR | 58(NR;19–86) | 61% |
| Shao-Jer Chen et al. (2012) [101] | NR | NR | 37/149 | 13(55)/24(94) | LN:64(10;44–77) N-LN:47(13;15–68) |
LN:61.5% N-LN:41.7% |
| Xiao-Peng Zhang et al. (2011) [102]* | Patients received radical gastrectomy and D2 lymph nodes dissection; Preoperatively examined with multi-detector row CT; Confirmed as gastric cancer by postoperative histopathology | Received preoperative neoadjuvant therapy; Distant metastasis was found in the preoperative examination or in the operation | 175/NR | 134(NR)/41(NR) | 59.8 (NR;30~85) | 71% |
| Matthias Dietzel et al. (2010) [103] | Invasive breast lesions with histopathological verification after bMRI | With a history of breast biopsy/interventions (surgical or minimally invasive) and chemotherapy/radiation therapy up to 12 months before bMRI; Histopathological grading not possible | 194/NR | 97(NR)/97(NR) | 60.6 (12.1; 25~87) | NR |
| May Sadik et al. (2008) [104]* | Underwent whole-body bone scintigraphy with a dual-detector r-camera; Patients with a complete set of technically sufficient images; At least 1 yr follow-up bone scan | Patients with a urine catheter, large bladder, sternotomy or fracture that could be misleading for the CAD system | NR/869 | NR(297)/NR(572) | Training set: 66 (NR;25~92) Test set: 65 (NR;43~86) |
Training: 65% Test: 69% All: 62% |
| Junji Shiraishi et al. (2008) [105] | NR | NR | 97/103 | NR(26);NR(77) | NR | NR |
| Junhua Zhang et al. (2008) [106]* | NR | NR | 112/210 | NR(114)/NR(96) | 53 (17;17~81) | NR |
| Rie Tagaya et al. (2008) [107]* | NR | NR | 91/91 | Training set:6(6)/3(3) Test set:60(60)/22(22) |
NR | NR |
| K. Marten et al. (2004) [108] | Patients with pulmonary metastasis; undergoing clinical staging and follow-up CT examinations of the chest | NR | 20/135 | 20(NR)/0(NR) | 62.4(NR;NR) | NR |
Abbreviation: NR=not reported. CT=computed tomography. GSI=Gemstone spectral imaging. LN= Lymph node. US= ultrasound. 3D-T1-MPRAGE images=Three-dimensional T1 magnetization prepared rapid acquisition gradient echo. SLN= sentinel lymph node. ALND= axillary lymph node dissection. FDG-PET/CT= fluoro-deoxy glucose positron emission tomography with CT. MRI= magnetic resonance imaging. FNA= fine needle aspiration. CNB= core needle biopsy. DW-MRI= diffusion-weighted magnetic resonance imaging. DCE-MR= contrast-enhanced magnetic resonance imaging. OPSCC= oropharyngeal squamous cell carcinoma. DEXA=Dual-energy X-ray absorptiometry. HNC=head and neck cancer. DCE-MRI= dynamic contrast enhanced MRI. FIGO=International Federation of Gynecology and Obstetrics. RTE=real-time elastography. NPC=nasopharyngeal carcinoma. CAD=computer-assisted diagnosis.
34 studies included in the meta-analysis.
Table 2.
Model training and validation for the 69 included studies.
| First author and year | Metastasis type | Target condition | Primary tumor | Reference standard | Type of internal validation | External validation |
|---|---|---|---|---|---|---|
| Mitsuru Koizumi et al. (2020) [40] | DM | Disseminated skeletal metastasis | prostate cancer(n = 12), GC=(n = 12), breast cancers(n = 15), miscellaneous cancers (n = 10) | Expert consensus | NR | YES |
| Jing Li et al. (2020) [41] | LNM | LNM in GC | GC | Histopathology; follow up | Resampling method | NO |
| L. Zhang et al. (2020) [42] | DM | Lung metastasis in STS | STS | Histopathology | Random split sample validation | NO |
| Li-Qiang Zhou et al. (2020) [43]* | LNM | Clinically negative axillary lymph node metastasis in primary breast cancer | Breast cancer | Histopathology | NR | YES |
| Endre Grøvik et al. (2020) [44] |
DM | Detection and Segmentation of Brain Metastases | Lung (n = 99), breast (n = 33), melanoma (n = 7), genitourinary (n = 7), gastrointestinal (n = 5), and miscellaneouscancers (n = 5) | Expert consensus | NR | NO |
| Yu Zhao et al. (2019) [45] | DM& LNM | Bone metastasis, lymph node metastasis in prostate cancer | Metastatic castration-resistant prostate cancer | Expert consensus | NR | NO |
| Jie Xue et al. (2019) [46] | DM | Detection and Segmentation of Brain Metastases | Lung, Breast, Kidney, Other organs (rectum, colon, melanoma, ovary and liver) | Expert consensus | Resampling method | NO |
| Bettina Baessler et al. (2019) [47]* | LNM | LNM in NSTGCT patients | NSTGCT | Histopathology | Resampling method | NO |
| Xiaojun Yang et al. (2019) [48]* | LNM | SLNM in Breast Cancer | Breast cancer | Histopathology | Resampling method | NO |
| Yuan Gao et al. (2019) [49] | LNM | PGMLNs in GC | GC | Histopathology; expert consensus | Resampling method | NO |
| David Coronado-Gutierrez et al. (2019) [50]* | LNM | Metastasis in the axillary lymph node | Breast cancer | Histopathology | Resampling method | NO |
| Yukinori Okada et al. (2019) [51] | DM | Bone metastasis | Breast cancer | Based on CT, MRI and clinical findings: expert consensus | NR | NR |
| Jeong Hoon Lee et al. (2019) [52]* | LNM | Metastasis in the cervical lymph node | Thyroid cancer | Histopathology by FNA and/or surgery | Random split sample validation | NO |
| Jansen et al. (2019) [53] | DM | Liver metastasis | NR | Expert consensus | NR | NO |
| Chuangming Li et al. (2019) [54]* | LNM | Sentinel lymph node metastasis | Breast cancer | Histopathology; expert consensus | NR | NO |
| M. Dohopolski et al. (2019) [55] | LNM | Small Lymph node metastasis | Oropharyngeal squamous cell carcinoma | Histopathology | NR | NO |
| Yige Peng et al. (2019) [56]* | DM | Distant metastasis in STS | STS | Biopsy or CT and/or PET images | NR | NO |
| Qiuxia Feng MD et al. (2019) [57]* | LNM | LNM in GC | GC | Histopathology | NR | NO |
| Thoma Schnelldorfer et al. (2019) [58] | DM | Distinguish metastasis in the peritoneal from the benign lesions | Gastric adenocarcinoma: 19. Pancreatic adenocarcinoma: 11; Gallbladder carcinoma: 2. Metastatic pancreatic neuroendocrine tumor, jejunal adenocarcinoma, ampullary adenocarcinoma: 1 each | Histopathology | NR | NO |
| Samir D. Mehta et al. (2019) [59]* | DM | Osteoblastic metastases involving one or more vertebral bodies from L1 to L4 | NR | Clinical notes | Random split sample validation | NO |
| Yoshiko Ariji, et al. (2019) [60]* | LNM | Metastasis in the cervical lymph node | Oral cancer | Histopathology | Resampling method | NO |
| Yunpeng Zhou et al. (2019) [61] | LNM | LNM in rectal cancer | Rectal cancer | Expert consensus | NR | NO |
| Yu Li et al. (2019) [62]* | DM | Metastasis in the liver of the preoperative CT | Colon cancer | Histopathology | Resampling method | NO |
| Zhiguo Zhou et al. (2019) [63]* | LNM | LNM in HNC | HNC | Histopathology | NR | NO |
| eMine acar et al. (2019) [64] | DM | Differentiating metastatic and completely responded sclerotic bone lesion in prostate cancer |
Prostate cancer | Expert consensus | Resampling method | NO |
| Fang Hou et al. (2019) [65]* | LNM | LNM | NR | Histopathology | NR | NO |
| Yoshiko Ariji et al. (2019) [66]* | LNM | LNM in Oral squamous cell carcinoma | Oral squamous cell carcinoma | Histopathology | NR | NO |
| Xiaojuan Xu et al. (2019) [67] | LNM | LNM in EC | EC | Histopathology | NR | NO |
| Jiaxiu Luo et al. (2018) [68]* | LNM | SLNM in breast cancer | Breast cancer | Histopathology | NR | NO |
| Richard Ha et al. (2018) [69] | LNM | LNM in breast cancer | Breast cancer | Biopsy; follow up | Resampling method | NO |
| B.H. Kann et al. (2018) [70]* | LNM | LNM in HNC | HNC | Histopathology | Resampling method | NO |
| Jeong Hoon Lee et al. (2018) [71]* | LNM | LNM in thyroid tumor | Thyroid tumor | FNA and/or laboratory tests | Random split sample validation | NO |
| Yun Lu et al. (2018) [72] | LNM | Pelvis LNM in rectal cancer | Rectal cancer | Expert consensus | Random split sample validation | YES |
| José Raniery Ferreira Junior et al. (2018) [73]* | DM& LNM | LNM and distant metastasis in lung cancer | Lung cancer | Clinical notes | Resampling method | NO |
| Tzu-Yun Lo et al. (2018) [74] | LNM | LNM in HNC | HNC | Clinical notes | Resampling method | NO |
| Jin Li et al. (2018) [75] | LNM | LNM in Colorectal Cancer | Colorectal Cancer | Expert consensus | NR | NO |
| Mohamed Amine Larhmam et al. (2018) [76] | DM | Spine metastasis | NR | Single expert | Resampling method | NO |
| Yan Zhong et al. (2018) [77]* | LNM | Occult mediastinal LNM of lung adenocarcinoma | Lung adenocarcinoma | Histopathology | Resampling method | NO |
| Wang, H et al. (2017) [78]* | LNM | Mediastinal LNM of non-small cell lung cancer | Non-small cell lung cancer | Histopathology | Resampling method | NO |
| Mitsuru Koizumi et al. (2017) [79]* | DM | Skeletal metastasis in prostate cancer | Prostate cancer | BS&CT expert consensus; follow up; and/or biopsy | NR | YES |
| Juan Wang et al. (2017) [80] | DM | Spinal metastasis | 15 lung, 5 thyroid, two liver, 1 breast, 1 prostate, 1 esophagus, 1 urinary tract | Biopsy | Resampling method | NO |
| Zhi-Long Wang et al. (2017) [81] | LNM | LNM in esophageal cancer with preoperative chemotherapy | Esophageal cancer | Postoperative pathological results | Random split sample validation | NO |
| Tuan D. Pham et al. (2017) [82]* | LNM | Mediastinal lymph nodes in lung Cancer | Lung cancer | Histopathology | Resampling method | NO |
| Qi Zhang et al. (2017) [83]* | LNM | Axillary lymph node metastasis in breast cancer | Breast cancer | Histopathology | Resampling method | NO |
| Yu-wen Wang et al. (2016) [84]* | LNM | Metastasis in the retropharyngeal lymph nodes | NPC | MRI follow-up | Random split sample validation | NO |
| Ali Aslantas et al. (2016) [85]* | DM | Bone metastatic | Chest, prostate, lung cancers | Single expert (laboratory tests, and other accessible radiographic images) | Resampling method | NO |
| Aneta Chmielewski et al. (2015) [86]* | LNM | Axillary lymph node metastasis in breast cancer patients | Breast cancer | Imaging-pathology gold standards: FNA, biopsy, LND, normal image with long term follow-up | Resampling method | NO |
| Mitsuru Koizumi et al. (2015) [87]* | DM | Metastasis in bone | Prostate cancer, lung cancer, breast cancer, and other cancers | Radiology (CT, MR or PET/CT), follow-up scan and patients' clinical course | NR | YES |
| Mitsuru Koizumi et al. (2015) [88] | DM | Metastasis in bone | Prostate cancer, lung cancer, breast cancer, and other cancers | Radiology (CT, MR or PET/CT), follow-up scan and patients' clinical course | NR | YES |
| Nesrine Trabelsi et al. (2015) [89] | DM | Metastasis in liver | NR | NR | NR | NO |
| Xuan Gao et al. (2015) [90] | LNM | Mediastinal lymph nodes in lung cancer | Lung cancer | Histopathology | Random split sample validation | NO |
| Osamu Tokuda, et al. (2014) [91]* | DM | Bone metastasis | Prostatic cancer (N = 71), breast cancer (N = 109), other cancers (N = 226) | All bone-scan images, including the follow-up scans, expert consensus; laboratory tests;(OR) biopsy | NR | YES |
| Ari Seff et al. (2014) [92] | LNM | LNM | NR | Expert consensus | Resampling method | NO |
| Zhi-Guo Zhou et al. (2013) [93]* | LNM | LNM in GC | GC | Surgery and histopathology | Resampling method | NO |
| Seungwook Yang et al. (2013) [94]* | DM | Brain metastases | NR | Single expert | NR | NO |
| Jianfei Liu et al. (2013) [95] | DM | Ovarian Cancer Metastases | Ovarian Cancer | Single expert | NR | NO |
| Yoshihiko Nakamura et al. (2013) [96] | LNM | Abdominal Lymph Node | 5 colorectal; 23 stomach cancer | 26cases: single expert 2 cases: experts consensus using a particular medical image |
Resampling method | NO |
| Chuan-Yu Chang et al. (2013) [97] | LNM | LNM | NR | Histopathology | NR | NO |
| Johannes Feulner et al. (2013) [98] | LNM | Mediastinal lymph nodes | NR | Single expert | Resampling method | NO |
| Chao Li et al. (2012) [99] | LNM | LNM in GC | GC | Histopathology | NR | NO |
| Hongmin Cai et al. (2012) [100] | LNM | Regional LNM | Rectal cancer | Histopathology | Resampling method | NO |
| Shao-Jer Chen et al. (2012) [101] | LNM | LNM | NR | Histopathology; follow up | Resampling method | NO |
| Xiao-Peng Zhang et al. (2011) [102]* | LNM | LNM in GC | GC | Histopathology | Resampling method | NO |
| Matthias Dietzel et al. (2010) [103] | LNM | Metastasis to the ipsilateral axilla lymph node | Breast cancer | Surgicopathology | Random split sample validation | NO |
| May Sadik et al. (2008) [104]* | DM | Metastasis to bone | Testing: Breast/prostate cancer | Training: Clinical reports and the bone scan images Testing: Final clinical assessments made by the same experienced physician |
NR | NO |
| Junji Shiraishi et al. (2008) [105] | DM | Metastasis to the liver | NR | Biopsy or surgical specimens | NR | NO |
| Junhua Zhang et al. (2008) [106]* | LNM | Metastasis to the cervical lymph nodes | NR | Histopathology | Resampling method | NO |
| Rie Tagaya et al. (2008) [107]* | LNM | Diagnosis of LNM by B-Mode Images from Convex-Type Echobronchoscopy | 66 lung cancer,25sarcoidosis | Histopathology or cytologic testing | NR | NO |
| K. Marten et al. (2004) [108] | DM | Pulmonary nodules | NR | Expert consensus | NR | NO |
Characteristics only be described in 1 or 2 studies are classified to others.
Abbreviation: NR=not reported. LNM=Lymph node metastasis. DM= distant metastasis. BS=bone scintigraphy. GC=gastric cancers. STS=soft-tissue sarcoma. NSTGCT= Non-seminomatous testicular germ cell tumor. PGMLNs= perigastric metastatic lymph nodes. EC=Endometrial cancer. FNA=fine needle aspiration.
34 studies included in the meta-analysis.
Table 3.
Indicator, algorithm, and data source for the 69 included studies.
| First author and year | Indicator definition |
Algorithm |
Data source |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Method for predictor measurement | Exclusion of poor-quality imaging | Heatmap provided | Extracted features | Algorithm architecture name | Algorithm architecture | Transfer learning applied | Source of data | Number of images for training/testing) | Data range | Open access data | |
| Mitsuru Koizumi et al. (2020) [40] | BS | NR | NR | NO | NR | ANN | NR | Retrospective clinical data from cancer institute hospital, Tokyo, Japan | NR/54 | 2013.1–2019.8 | NO |
| Jing Li et al. (2020) [41] | dual-energy CT | YES | NR | YES | DCNNs; ANN; Ksvm | CNN; ANN; SVM | NR | Retrospective cohort | 136/68 | 2012.1–2018.11 | NO |
| L. Zhang et al. (2020) [42] | MRI, CT | NR | NR | NO | Inception V3 | CNN; Inception | YES | Data collected from Cancer Imaging Archive | 25/15 | NR | YES |
| Li-Qiang Zhou et al. (2020) [43]* | US image | YES | YES | NO | Inception V3; Inception-ResNet V2; ResNet-101 | CNN; Inception; Residual Network | NR | Cohort 1: retrospective cohort collected from Tongji Hospital; Cohort 2: retrospective cohort collected from Hubei Cancer Hospital (Hubei, China) | 877/97(internal test) +81(external test) | Cohort 1:2016.5–2018.10; Cohort 2:2018.10–2019.4 | NO |
| Endre Grøvik et al. (2020) [44] |
Multisequence MRI | NR | YES | NO | GoogLeNet | CNN | NR | Retrospective cohort | 100/51 | 2016.6–2018.6 | NO |
| Yu Zhao et al. (2019) [45] | PSMA PET/CT, CT | NR | NR | NR | triple combing 2.5D U-NET | CNN | NR | Retrospective cohort from medical centers of Technical University of Munich, University of Munich and University of Bern | 130/63 | NR | NR |
| Jie Xue et al. (2019) [46] | 3D-T1-MPRAGE images | YES | NR | NO | 3D CNN | CNN | NR | Dataset 1: Retrospective clinical data from the Shandong Provincial Hospital Affiliated to Shandong University; Dataset 2: Retrospective clinical data from the Affiliated Hospital of Qingdao University Medical College; Dataset 3: Retrospective clinical data from the Second Hospital of Shandong University | 1201/451 | Dataset 1:2016.10–2019.5 Dataset 2:2017.8–2019.3 Dataset 3:2017.4–2019.4 |
NO |
| Bettina Baessler et al. (2019) [47]* | CT | YES | NR | YES | logistic regression | logistic regression | NR | Retrospective cohort | 120/23(internal test)+61(external test) | 2008–2017 | NO |
| Xiaojun Yang et al. (2019) [48]* | CT | YES | NR | YES | CNN-F; multivariable logistic regression | CNN; logistic regression | YES | Retrospective cohort | 184/164 | 2016.1–2018.11 | NO |
| Yuan Gao et al. (2019) [49] | CT | YES | NR | YES | FR-CNN | CNN | NR | Cohort 1: retrospective cohort collected from Tongji Hospital Cohort 2: retrospective cohort collected from Hubei Cancer Hospital (Hubei, China) |
32,495/6000 | 2011.1–2018.5 | No |
| David Coronado-Gutierrez et al. (2019) [50]* | US | YES | NR | YES | CNN; VGG-M | VGG | NR | Retrospective cohort | NR/NR | 2015.4~2018.8 | NO |
| Yukinori Okada et al. (2019) [51] | BS | NR | NR | NO | NR | CNN | NR | Retrospective cohort | NR/NR | 2012.1~2014.11 | NO |
| Jeong Hoon Lee et al. (2019) [52]* | CT(Axial) | NR | YES | NO | VGG16; VGG19; Inception; Inception V3; InceptionResNetV2; D3nseNet121; DenseNet169; ResNet | CNN; VGG; Inception; Residual Network | NR | Retrospective cohort | 891/104 | 2017.7~2018.1 | NO |
| Jansen et al. (2019) [53] | Contrast-enhanced MRI, diffusion-weighted MRI | NR | NR | NR | NR | CNN-F | NR | Retrospective cohort from University Medical Center Utrecht, The Netherlands | 55 /17 | 2015.2–2018.2 | NO |
| Chuangming Li et al. (2019) [54]* | Contrast-enhanced MRI | YES | NR | YES | Logistic regression; SVM; XGBoost | NR | NR | Clinical data from the Second Affiliated Hospital of Chongqing Medical University, China | 49/13 | 2013.3–2018.12 | YES |
| M. Dohopolski et al. (2019) [55] | PET, CT | NR | NR | NR | AlexNet-like, UNET | CNN | NR | NR | 4074/54 | NR | NR |
| Yige Peng et al. (2019) [56]* | PET-CT | NR | NR | YES | 3D deep multi-modality collaborative learning | CNN | NR | Public PET-CT dataset of STS patients | NR/NR | NR | YES |
| Qiuxia Feng MD et al. (2019) [57]* | CT | YES | NR | YES | NR | NR | NR | Retrospective cohort from the First Affiliated Hospital with Nanjing Medical University, Nanjing, China | 326/164 | 2014.1–2016.12 | NO |
| Thoma Schnelldorfer et al. (2019) [58] | Laparoscopy | NR | NR | NR | DNN | Deep neural network | NR | Retrospective cohort | NR/NR | 2014.1.1~2017.9.30 | NO |
| Samir D. Mehta et al. (2019) [59]* | Dual X-ray absorptiometry | NR | NR | NR | Radom forest algorithm; SVM | Radom forest algorithm; SVM | NR | Retrospective cohort | 160/40 | 2010.1.1~2018.8.31 | NO |
| Yoshiko Ariji, et al. (2019) [60]* | CT (Contrast enhanced, axial) |
NR | NR | NR | AlexNet | AlexNet | NR | Retrospective cohort | 353/88 | 2007~2015 | NO |
| Yunpeng Zhou et al. (2019) [61] | High-resolution MRI | NR | NR | NR | Faster region-based CNN | FRCNN | NO | Retrospective cohort | Patients: 201/100 Images: 12,060/6030 |
2016.7~2017.12 | NO |
| Yu Li et al. (2019) [62]* | CT | YES | NR | YES | SVM | SVM | NR | Retrospective cohort | 240/240 | 2015.10~2018.7 | NO |
| Zhiguo Zhou et al. (2019) [63]* | CT; PET; PEC&CT | NR | NR | YES | MO; CNN; AutoMO | SVM; CNN | NR | Retrospective cohort from the University of Texas Southwestern Medical Center | 378/165 | 2009–2018 | NO |
| eMine acar et al. (2019) [64] | 68Ga-PSMA PET/CT |
NR | NR | YES | Decision tree; discriminant analysis; SVM; KNN; | Decision tree; discriminant analysis; SVM; KNN, | NR | Retrospective cohort | 153/104 | 2017.1–2018.11 | NO |
| Fang Hou et al. (2019) [65]* | OCT | NR | NR | YES | BP-ANN | ANN | NR | Retrospective cohort from Department of Head and neck Tumor, Tianjin Medical University Cancer Institute and Hospital, China | 314/259 | NR | NO |
| Yoshiko Ariji et al. (2019) [66]* | CT | NR | NR | NR | AlexNet | CNN | NR | Retrospective cohort from Aichi-Gakuin University School of Dentistry, Nagoya, Japan | 562/141 | 2017–2018 | NR |
| Xiaojuan Xu et al. (2019) [67] | Contrast-enhanced -MRI | NR | YES | YES | NR | NR | NR | Retrospective cohort from National Cancer Center, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China | 140/60 | 2011.1–2017.12 | NO |
| Jiaxiu Luo et al. (2018) [68]* | diffusion-weighted MRI | NR | NR | YES | CNN; SVM | SVM; CNN | NR | Retrospective cohort | 122/50 | 2014.3–2016.6 | NO |
| Richard Ha et al. (2018) [69] | MRI | NR | NO | NO | CNN; VGG-16 | CNN; VGG | NR | Retrospective cohort | NR/NR | 2013.1–2016.6 | NO |
| B.H. Kann et al. (2018) [70]* | CT | NR | NR | NO | DCNNs | CNN | NR | Retrospective cohort | 522/131 | 2013–2017 | NO |
| Jeong Hoon Lee et al. (2018) [71]* | US | NR | YES | NO | VGG-Class Activation Map;CNN-GAP | CNN; VGG | NR | Retrospective cohort | 612/200 | cohort1:2008.1–2015.11 cohort2:2016.1–2016.11 |
NO |
| Yun Lu et al. (2018) [72] | MRI | NR | NR | NO | FR-CNN; VGG16 | CNN; VGG | YES | Training set: Retrospective cohort from Affiliated Hospital of Qingdao University; Test set: Retrospective cohort from 6 Chinese Medical Centers | 28,080/36,000 | cohort1:2011.9–2018.10 cohort2:NR |
NO |
| José Raniery Ferreira Junior et al. (2018) [73]* | CT | YES | YES | YES | NB; KNN; RBF; ANN | KNN; ANN | NR | Retrospective cohort | 52/16 | NR | NO |
| Tzu-Yun Lo et al. (2018) [74] | CT | NR | NR | YES | SVM | SVM | NR | Retrospective cohort from Taipei Veterans General Hospital of Taiwan | NR/NR | NR | NO |
| Jin Li et al. (2018) [75] | MRI | NR | NR | NO | Inception-v3 | CNN | YES | Data collected from Harbin Medical University Cancer Hospital | NR/NR | NR | NO |
| Mohamed Amine Larhmam et al. (2018) [76] | MRI | NR | NR | YES | SVM | SVM | NR | NR | NR/NR | NR | NR |
| Yan Zhong et al. (2018) [77]* | CT | YES | NR | YES | RBF; SVM | SVM | NR | Retrospective cohort | NR/NR | 2013.1–2016.9 | NO |
| Wang, H et al. (2017) [78]* | 18F-FDG PET/CT |
NR | NO | YES | Random forest; AdaBoost; SVM; BP-ANN | Random forest; AdaBoost; SVM; BP-ANN | NR | Retrospective cohort from Cancer Hospital Affiliated to Harbin Medical University | 200/1197 | 2009.6–2014.9 | NO |
| Mitsuru Koizumi et al. (2017) [79]* | BS | NR | NR | NO | BONENAVI | ANN | NR | NR | NR/NR | 2013.2–2017.1 | NO |
| Juan Wang et al. (2017) [80] | MRI | NR | NR | NO | Siamese neural network | CNN | NR | Clinical data collected from the Peking University Third Hospital | 85,503/NR | NR | NO |
| Zhi-Long Wang et al. (2017) [81] | CT | YES | NR | YES | LS-SVM | SVM | NR | Clinical data collected from the Peking University Cancer Hospital & Institute, Beijing, China | 66/65 | 2006.1–2012.1 | NO |
| Tuan D. Pham et al. (2017) [82]* | CT | NR | NR | YES | Logistic regression; SVM; NBLDA | Logistic regression; SVM; NBLDA | NR | Retrospective cohort | NR/271 | 2010.4–2015.4 | NO |
| Qi Zhang et al. (2017) [83]* | Real-time elastography and B-mode ultrasound | NR | NR | YES | SVM | SVM | NR | Retrospective cohort | NR/NR | 2013.11–2014.11 | NO |
| Yu-wen Wang et al. (2016) [84]* | MRI | NR | NR | YES | Feed-forward back-propagation NN | ANN | NR | Retrospective cohort | Stage I: 331/332 Stage II: 410/205 |
NR | NO |
| Ali Aslantas et al. (2016) [85]* | BS | NR | NR | YES | ANN | ANN | NR | Retrospective cohort from Medical Faculty of Suleyman Demirel University, Konya Education and Research Hospital | NR/130 | 2003–2013 | NO |
| Aneta Chmielewski et al. (2015) [86]* | US | NR | NR | YES | SVM | SVM | NR | Retrospective cohort | 80/25 | NR | NO |
| Mitsuru Koizumi et al. (2015) [87]* | BS | NR | NR | YES | BONENAVI | ANN | NR | Retrospective cohort | NR/NR | 2013.1~2013.12 | NO |
| Mitsuru Koizumi et al. (2015) [88] | BS | NR | NR | YES | BONENAVI 2 | ANN | NR | Retrospective cohort | NR/NR | 2013.1~2013.12 | NO |
| Nesrine Trabelsi et al. (2015) [89] | CT | NR | NR | YES | Neural network | Neural network | NR | Retrospective cohort | 8/3 | NR | NO |
| Xuan Gao et al. (2015) [90] | 18F-FDG PET/CT | NR | NR | YES | RBF; SVM | SVM | NR | Retrospective cohort | 30/30 | 2009.6–2013.7 | NO |
| Osamu Tokuda, et al. (2014) [91]* | BS | NR | NR | NO | BONENAVI | ANN | NR | NR | NR/3248 | 2006.1–2011.5 | NO |
| Ari Seff et al. (2014) [92] | CT | NR | YES | YES | Random forest; SVM | Random forest; SVM | NR | NR | NR/984 | NR | NO |
| Zhi-Guo Zhou et al. (2013) [93]* | MDCT | YES | NR | YES | ER based model | ER | NR | Retrospective cohort from Peking University Cancer Hospital & Institute (Beijing, China P. R.) | NR/NR | 2006.4–2008.9 | NO |
| Seungwook Yang et al. (2013) [94]* | Magnetic resonance black-blood imaging | NR | NR | YES | Conjugate gradient BP-ANN | ANN | NR | Retrospective cohort | 37/53 | NR | NO |
| Jianfei Liu et al. (2013) [95] | Abdominal contrast-enhanced CT | NR | NR | NO | Joint framework | NR | NR | Retrospective cohort | 6/44 | NR | NO |
| Yoshihiko Nakamura et al. (2013) [96] | 3-D X-ray CT | NR | NR | YES | SVM | SVM | NR | Retrospective cohort | NR/NR | NR | NO |
| Chuan-Yu Chang et al. (2013) [97] | US | NR | NR | YES | PSONN; one-against-one multi-class SVM | SVM | NR | Retrospective cohort | 88/89 | 2005–2007 | NO |
| Johannes Feulner et al. (2013) [98] | CT | NR | NR | YES | Spatial prior; AdaBoost | Spatial prior; AdaBoost | NR | NR | 289/1086 | NR | NO |
| Chao Li et al. (2012) [99] | GSI-CT | NR | NR | YES | SFS-KNN; mRMR-KNN; Metric Learning | KNN | NR | Retrospective cohort from GE Healthcare equipment in Ruijin Hospital | NR/NR | 2010.4 | NO |
| Hongmin Cai et al. (2012) [100] | CT | NR | NR | YES | SVM | SVM | NR | Retrospective cohort | NR/228 | 2007.1–2008.11 | NO |
| Shao-Jer Chen et al. (2012) [101] | US | NR | NR | YES | SVM | SVM | NR | Retrospective cohort from Buddhist Dalin Tzu Chi General Hospital | NR/NR | NR | NO |
| Xiao-Peng Zhang et al. (2011) [102]* | Multi-detector row CT | NR | NR | YES | LibSVM 2.89 | SVM | NR | Retrospective cohort | NR/NR | 2006.4~2008.9 | NO |
| Matthias Dietzel et al. (2010) [103] | Breast MRI | NR | NR | YES | ANN | ANN | NR | Retrospective cohort | 123/71 | NR | NO |
| May Sadik et al. (2008) [104]* | BS | NR | NR | YES | ANN | ANN | NR | Retrospective cohort | 810/59 | Training: 1999.1~2002.6 Testing: 1999.8~2001.1 |
NO |
| Junji Shiraishi et al. (2008) [105] | Contrast-enhanced ultrasonography | NR | NR | YES | ANN | ANN | NR | Retrospective cohort | NR/NR | NR | NO |
| Junhua Zhang et al. (2008) [106]* | US | NR | NR | YES | v-SVM | SVM | NR | Retrospective cohort | NR/NR | 2005.7~2006.6 | NO |
| Rie Tagaya et al. (2008) [107]* | US from convex-type echobronchoscopy | NR | NR | NO | BP-ANN | ANN | NR | Retrospective cohort from St. Marianna University School of Medicine, Tokyo, Japan | 9/82 | 2005.4–2007.3 | NO |
| K. Marten et al. (2004) [108] | MSCT | NR | NR | NR | NR | NR | NR | Retrospective cohort from Klinikum rechts der Isar, Technical University Munich, Germany | NR/NR | NR | NR |
Abbreviation: NR=not reported. BS=bone scintigraphy. GC=gastric cancers. CT=computed tomography. MRI= magnetic resonance imaging. ANN= artificial neural network. SVM= support vector machine. NN= neural networks. CNN= convolutional neural networks. US= ultrasound. PSMA= Prostate specific-membrane antigen. 3D-T1-MPRAGE images=Three-dimensional T1 magnetization prepared rapid acquisition gradient echo. FR-CNN= fast region convolutional neural networks. CNN-F= CNN fast. PET: positron emission tomography. DNN= Deep neural network. MO= multi-objective model. KNN= k nearest neighbors. OCT= Optical coherence tomography. ANN= artificial neural network. BP-ANN= back-propagation artificial neural network. MSCT= multi-slice CT.
34 studies included in the meta-analysis.
We accepted all forms of the reference standard for the diagnosis of metastasis. Forty-three studies used histopathology; 21 studies used varying models of expert evaluation; 10 studies used other imaging types to confirm the diagnosis; 7 studies used existing clinical notes; 4 studies used clinical follow-up, and 1 study did not report this. A part of studies applied several different references.
A total of 34 studies and 123 contingency tables were included in the meta-analysis. In these studies, primary tumors included breast cancer (7 studies), head and neck cancer (7 studies), gastrointestinal cancer (4 studies), lung cancer (5 studies) and others (3 studies). 4 studies had several different primary tumors; 4 studies did not report this. There were 25 studies targeting LNM and 10 studies targeting DM (1 study related to both LNM and DM). None of the 8 studies included in the systematic review with comparison between AI models and health-care professionals were excluded in the meta-analysis. After removing 3 from the 7 studies included in the systematic review with external validation because of the lack of contingency tables, only 4 studies were used for the meta-analysis.
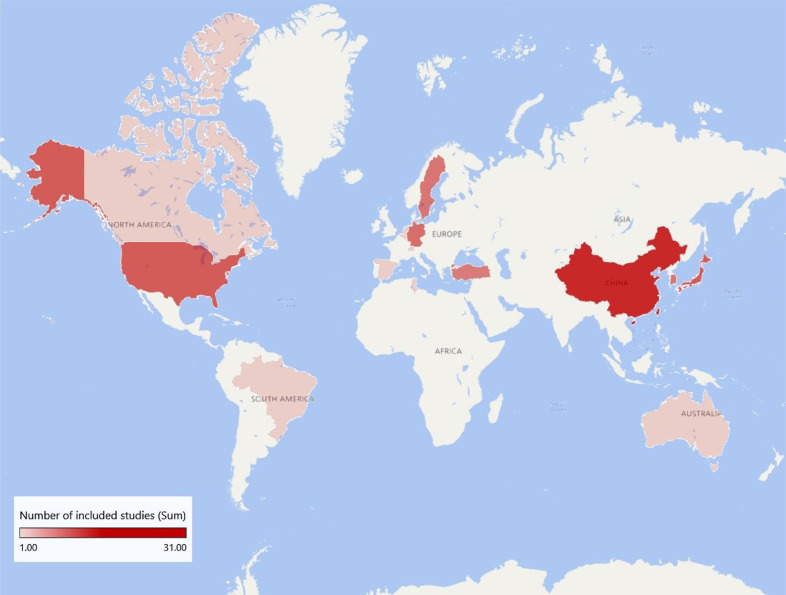
In addition, we investigated the international research situation of this subject, finding that the studies mostly concentrated on China, America and Japan, with 31, 11 and 11 studies respectively. Included studies were also widely distributed in South Korea and Europe. South America, Australia and the Middle east had some sporadic distribution as well (Fig. 2).
Fig. 2.
International research situation.
The quality of studies included in the meta-analysis was assessed by the QUADAS-2 score [29] (Supplementary figure 1). Three and 5 studies showed a high risk respectively for patient selections and reference standards because these studies did not clarify whether enrolled patients were consecutive or use non-histopathology methods as reference standard, which we think were acceptable. So, these studies were not excluded.
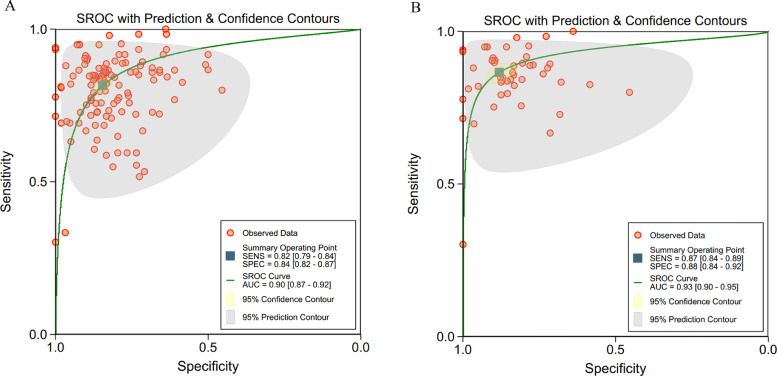
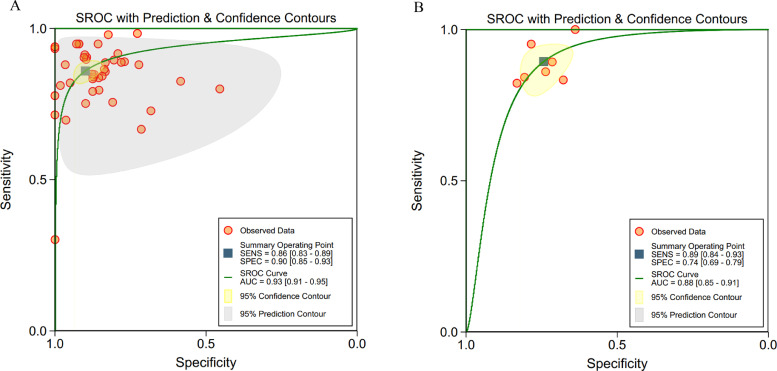
ROC curves of these 34 studies (123 contingency tables) are shown in Fig. 3a, in which the pooled sensitivity was 82% (95% CI 79–84%) for all studies, and the pooled specificity was 84% (82–87%), with AUC of 0·90 (0·87–0·92). Many studies used more than one algorithm with several different accuracy for each algorithm. So, when selecting the contingency tables reporting the highest accuracy for different algorithms in these 34 studies with 48 tables, the pooled sensitivity was 87% (95% CI 84–89%), and the pooled specificity was 88% (84–92%), with AUC of 0·93(0·90–0·95) (Fig. 3b).
Fig. 3.
(a, b). ROC curves of all studies included in the meta-analysis (34 studies)
a: ROC curves of all studies included in the meta-analysis (34 studies with 123 tables)
b: ROC curves of studies when selecting contingency tables reporting the highest accuracy (34 studies with 48 tables)
Abbreviations: ROC=receiver operating characteristic; SENS= sensitivity; SPEC= specificity.
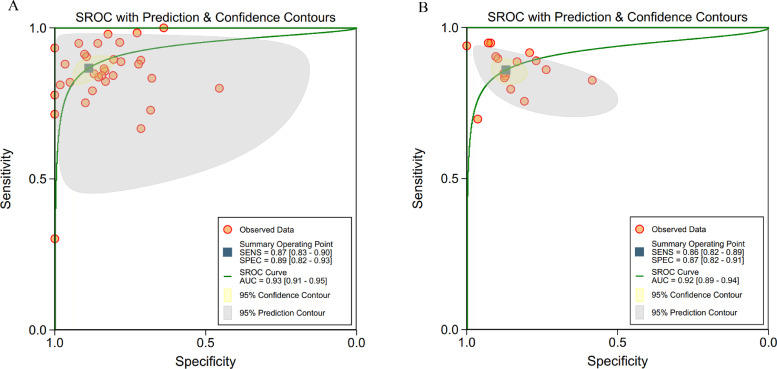
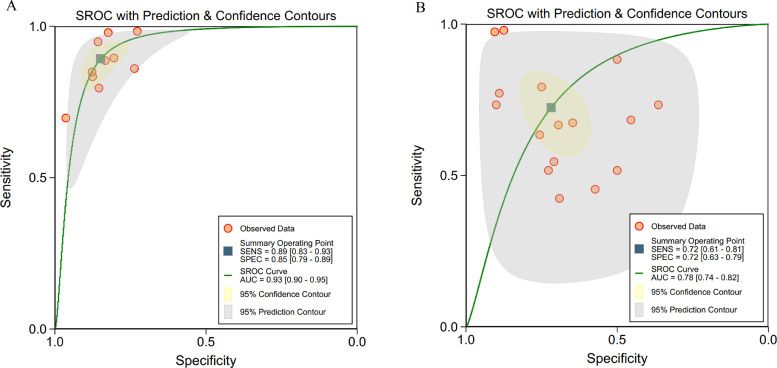
Considering different algorithms were used in the included studies, we divided them into ML algorithms (ANN, KNN, SVM, RF, logistic regression and decision tree) and DL algorithms (CNN, DNN and DCNN) and did separate analysis for them, which showed a pooled sensitivity of 87% (95% CI 83–90%) for ML and 86% (82–89%) for DL, and a pooled specificity of 89% (82–93%) for ML and 87% (82–91%) for DL (Fig. 4).
Fig. 4.
(a, b): ROC curves of studies using different algorithms
a: ROC curves of studies using machine learning algorithms (32 tables)
b: ROC curves of studies using deep learning algorithms (16 tables).
30 studies included in the meta-analysis were validated by in-sample dataset with a pooled sensitivity of 86% (95% CI 83–89%) and a pooled specificity of 90% (85–93%). Only 4 studies used out-of-sample dataset to perform an external validation, for which sensitivity was 89% (84–93%) and specificity was 74% (69–79%) (Fig. 5).
Fig. 5.
(a, b): ROC curves of studies with or without external validation
a: ROC curves of studies without external validation (41 tables)
b: ROC curves of studies with external validation (7 tables).
Of these 34 studies, 8 compared performance between AI algorithms and health-care professionals using the same sample, with 10 contingency tables for AI algorithm and 16 tables for health-care professionals (Fig. 6). The pooled sensitivity was 89% (95% CI 83–93%) for AI algorithms and 72% (61–81%) for health-care professionals. The pooled specificity was 85% (79–89%) for AI algorithms and 72% (63–79%) for health-care professionals. Only 1 of the 8 studies was validated by out-of-sample dataset, and therefore a comparison between the performance of AI and health-care professionals by the identical external sample could not be performed.
Fig. 6.
(a, b). ROC curves of studies using the same sample for comparing performance between health-care professionals and artificial intelligence algorithms (8 studies)
a: Artificial intelligence models (10 tables)
b: Health-care professionals (16 tables).
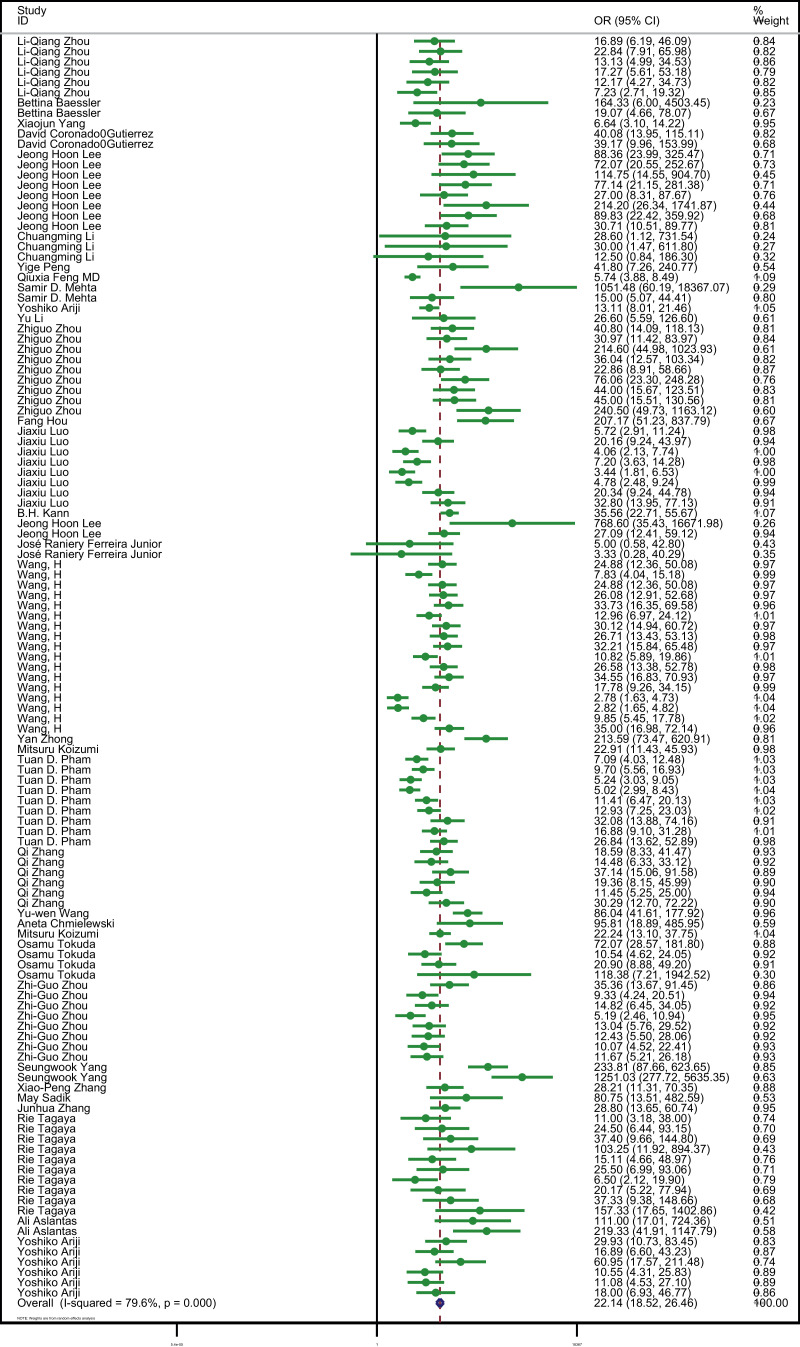
All studies showed that the AI algorithms were beneficial for the diagnosis of tumor metastasis from medical radiology imaging when compared to the reference standard used in each study (OR 22·14 [95% CI 18·52–26·46] P<0·001, I²=79·6%) (Fig. 7), from which we can also see high heterogeneity among these studies. Visual inspection of funnel plots suggested there was no publication bias (P = 0·19) (Supplementary figure 2).
Fig. 7.
Forest plot of studies included in the meta-analysis (34 studies).
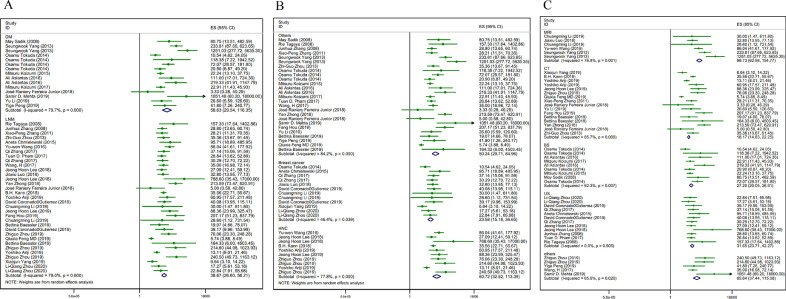
To determine the source of heterogeneity, we did several subgroup analyses. In terms of metastasis types, there were DM whose pooled sensitivity was 88% (95% CI 80–93%), pooled specificity was 90% (76–96%), and AUC was 0·94 (0·92–0·97) (n = 15, I²=79·7%, P<0·001) and LNM whose sensitivity was 86% (95% CI 83–88%), specificity was 87% (84–90%), and AUC was 0·93 (0·90–0·95) (n = 33, I²=79·0%, P<0·001) (Fig. 8a). The outcomes were similar regarding the primary tumor types and medical imaging types. When it comes to the primary tumor types, in the breast cancer group, the sensitivity was 85% (95% CI 81–87%), the specificity was 82% (75–87%), and AUC was 0·86 (0·83–0·89) (n = 12, I²=46.4·0%, P = 0·039). In the head and neck cancer group, the sensitivity was 87% (95% CI 81–91%), the specificity was 91% (87–94%), and AUC was 0·95 (0·92–0·96) (n = 10, I²=77·8%, P<0·001). Regarding the other primary tumor types, the sensitivity was 88% (95% CI 83–91%), the specificity was 89% (81–94%), and AUC was 0·94 (0·91–0·95) (n = 26, I²=84·2%, P<0·001) (Fig. 8b). As for medical imaging types, there were 16 contingency tables using CT (I²=85·7%, P<0·001), 12 tables using ultra sound (I²=0·0%, P = 0·505), 9 tables using bone scintigraphy (I²=62·3%, P = 0·007), 6 tables using MRI (I²=76·8%, P = 0·001) and 5 tables using other imaging types (I²=65·6%, P = 0·02) (Fig. 8c). Subgroup analysis for different AI algorithms contained ML (n = 32, I²=82·4%, P<0·001) and DL (n = 16, I²=70·8%, P<0·001). While in the studies were externally validated, heterogeneity was acceptable (n = 7, I²=45·1%, P = 0·091). We could not find a reasonable explanation for heterogeneity from subgroup analysis. We also did regression analysis to find the sources of heterogeneity. However, the results also could not make an explanation (regression analysis results are provided in Supplementary table).
Fig. 8.
(a, b, c). Forest plot of 3 subgroups
a: Subgroup 1. Different metastasis types
b: Subgroup 2. Different primary tumors
c: Subgroup 3. Different imaging types
Abbreviations: ES= estimate.
4. Discussion
With great attention to the development of AI, more and more people are curious about its performance in medicine. In this systematic review and meta-analysis, we found that AI algorithms may be used for the diagnosis of tumor metastasis from medical radiology imaging material with equivalent or even better performance to health-care professionals, in terms of sensitivity and specificity. Tumor metastasis, as one of the main reasons for tumor-induced death, has a great impact on the treatment plan and prognosis judgment. Tumor metastasis sites may involve lymph nodes and distant organs, such as liver, lung and brain, which may be difficult to diagnose in clinical examination. Medical imaging is an important tool to diagnose tumor metastasis. However, the accurate diagnosis of tumor metastasis without misdiagnosis and missed diagnosis is a challenging task. The excellent performance of AI in image identification with rapid speed, high accuracy and significant manpower reduction excited the public. In 2019, Liu XX, et al. [30]. conducted a systematic review and meta-analysis and found the diagnostic performance of deep learning models from medical imaging to be equivalent to that of health-care professionals in classifying diseases, with the sensitivity of 87·0% and specificity of 92·5%, which provided the basis for the clinical use of deep learning models. As for the diagnosis of tumor metastasis, there were no other meta-analyses focus on this subject to date, where we also reached a similar positive conclusion.
The first appearance of AI as a term can be dated back to a conference in 1956 [31]. As a branch of computer science, AI attempted to use computers to simulate the thought processes and intelligent behaviors of people, of which machine learning is an important part. The presence of ANN, SVM and other ML algorithms aroused people's enthusiasm towards ML. It is not until 2006 that Geoffrey Hinton [13] the greatness of ML, proposed the concept of DL, which was the further development of ML. Twenty-three of the included studies in 2018 and beyond witnessed an increase in DL, in contrast to that only 1 study before 2018 involved in DL. Taking into account the different development stages of AI, we did a separate analysis for studies using different algorithms, where no significant difference was observed. This may be attributed to the small dataset of included studies, most of which collected a few hundred data, limiting the advantages of DL.
In our research, we observed statistically significant heterogeneity among the included studies. So, we did several subgroup analyses and meta-regression for different algorithms, existence of external validation, the type of metastasis, primary tumors and medical imaging. The heterogeneity of studies validated by external sample was acceptable. 3 of the 4 studies with external validation based on the different version of the same computer assisted diagnosis system, which may contribute to the result. Generally, the results still cannot explain the source of heterogeneity, which may be contributed to the broad nature of the review (accepting any classification task using any imaging types for any metastasis types of any primary tumors).
Although the outcome of our research seems to bring light to the application of AI in detecting tumor metastasis from medical radiology imaging, several common methodological defects should be noted.
First, the design and practice of some included studies may make the research results out of clinical practice, among which the most common is the lack of comparison with health-care professionals in diagnostic accuracy. In the 69 included studies, only 8 studies made a comparison with health-care professionals. Assessing the performance of AI in insolation instead of comparing with the most common way in clinical practice (review the medical imaging by a radiologist) makes the outcomes unreliable when applied in the clinical setting. Even if some studies had the comparison, very few of them made it with humans using the same test dataset, resulting in a lack of comparability. Although we have reached the conclusion that AI models had the equivalent or even better diagnostic performance from medical imaging compared to health-care professionals, some factors still need to be considered. Only 8 studies using the same sample to compare health-care professionals and AI algorithms. Different studies recruited radiologists with different years of experience and different numbers. Some studies did not train radiologists in advance. All of above may influence the result. Furthermore, we included the studies that only used medical imaging to identify the presence of tumor metastasis, and excluded those that used other clinical materials, such as electronic medical record and clinical information of patients. It made our research topic more consistent. With the additional information available in the clinical practice, some prediction models can predict the possibility of metastasis based on the patient's gender, age and history to assist diagnosis [32], [33], [34], [35], [36].
Second, there were no prospective studies. All included studies were retrospective studies, whose participants were selected from hospital medical records. Some studies used online open-access datasets instead of being done in the real clinical environment. And some studies provided poor description of missing data. In terms of the standard to diagnose metastasis, some studies only used the opinion of a single radiologist as a standard, which may not be convincing.
Third, various indicators of diagnostic performance were used in the studies. The value of TP, TN, FP and FN at a specified threshold should at least be provided, but most studies did not give a threshold or explain the reason for choosing this threshold. Most studies set the threshold at the value of 0·5, which is a convention in machine learning development [37,38]. Indicators like the sensitivity, specificity and accuracy were used in most studies. When the number of patients with/without metastasis in the test dataset was reported, sensitivity and specificity can be used to calculate TP, TN, FP and FN for contingency tables construction. Other indicators such as precision, dice ratio, F1 score and recall, which are common in the field of computer science, also appeared as the only measure in some studies. However, these indicators are not comprehensive, only with which we cannot get enough information to construct contingency tables.
Last but not least, in the 69 included studies there were only 4 with external validation, which means testing the model with out-of-sample dataset from one or more other centers. Most studies split the dataset from one center into training set and test set randomly or according to different time periods. The performance was evaluated by the test set, which should be called internal validation. Since the goal of validation is to investigate the performance within patients from different population, it is appropriate to collect a new dataset from different center. The absence of external validation made it hard to ensure the generalizability of the model, leading to overestimated results [39]. In our research, studies with external validation had an expectedly worse performance than internally validated studies. It is understandable that better performance can be achieved with the less heterogeneous samples. Strict external validation in the development of diagnostic model is urgently needed.
During the research, we also found some common deficiencies in AI studies. The most obvious point is that some key terminology is not uniformly named. Different studies have different definitions of the same terminology. For instance, for one AI model, the dataset is usually divided into several different parts, including the initial training set and one or more testing sets used to evaluate model effectiveness. While the term “validation” is used causally, some authors used this word to indicate the dataset used to test the diagnostic performance of the final model. Others defined it as a dataset with tuning function during the development process. The naming confusion makes it difficult to judge whether the test set is independent. The independent dataset, which is never learned by the model, is crucial to the credibility of the final model. So, canonical naming is urgently needed. Some scholars [30] have put forward suggestions. They distinguished the dataset used for a model as training set (for training the model), tuning set (for tuning the parameters of the model) and validation test set (for evaluating the performance of the final model), which is also accepted by our article. As for different types of validation test set, Altman and Royston's suggestion [39] may be adopted. They named dataset for in-sample validation as internal validation, dataset for in-sample validation with a temporal split as temporal validation, and dataset for out of sample validation as external validation. Studies on the AI application in the medical field should strive to avoid problems mentioned above in the future.
Diagnosis of tumor metastasis using AI algorithms has great potential. From this meta-analysis, we conservatively draw a conclusion that the AI algorithms may be used for the diagnosis of tumor metastasis from medical radiology imaging with equivalent or even better performance to health-care professionals, in terms of sensitivity and specificity, providing a basis for its clinical application. Its widespread clinical application may alleviate the shortage of medical resources, improve the detection rate and accuracy of tumor metastasis and then the prognosis of patients. However, it should be acknowledged that more high-quality studies on the AI application in the medical field with adaption to the clinical practice and standardized research routines are needed. In this review, we also put forward some existing problems of design and reporting that the algorithm developers should consider. High-quality studies are always the cornerstone of evaluation for diagnostic performance by various algorithms, which will finally benefit patients and the health care system.
Contributors
YL and GL contributed to the conception and design of the study. QZ, LY, JL and BZ contributed to the literature search and data extraction. QZ and KG contributed to risk of bias evaluation. BZ contributed to data analysis and interpretation. QZ wrote the first draft of the report with input from LY. All authors contributed to critical revision of the manuscript. All authors approved the manuscript.
Declaration of Competing Interest
All authors declare no competing interests.
Acknowledgments
Acknowledgement
This work was supported by College Students' Innovative Entrepreneurial Training Plan Program.
Data sharing statement
The search strategy was shown in Appendix Section 1, and the contingency tables of 34 studies included in the meta-analysis were shown in Appendix Section 2. The results of risk of bias and publication bias were separately provided in the Supplementary Figure 1 and 2. Additional data are available on request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2020.100669.
Contributor Information
Yujie Liang, Email: yujie0350@126.com.
Guiqing Liao, Email: drliaoguiqing@hotmail.com.
Appendix. Supplementary materials
References
- 1.Yamashita K., Hosoda K., Ema A., Watanabe M. Lymph node ratio as a novel and simple prognostic factor in advanced gastric cancer. Eur J Surg Oncol. 2016;42(9):1253–1260. doi: 10.1016/j.ejso.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan B., Brierley J., Byrd D. The TNM classification of malignant tumours-towards common understanding and reasonable expectations. Lancet Oncol. 2017;18(7):849–851. doi: 10.1016/S1470-2045(17)30438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruytenberg T., Verbist B.M., Vonk-Van Oosten J., Astreinidou E., Sjögren E.V., Webb A.G. Improvements in high resolution laryngeal magnetic resonance imaging for preoperative transoral laser microsurgery and radiotherapy considerations in early lesions. Front Oncol. 2018;8:216. doi: 10.3389/fonc.2018.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsili A.C., Alexiou G., Naka C., Argyropoulou M.I. Imaging of colorectal cancer liver metastases using contrast-enhanced US, multidetector CT, MRI, and FDG PET/CT: a meta-analysis. Acta Radiol. 2020 doi: 10.1177/0284185120925481. 284185120925481. [DOI] [PubMed] [Google Scholar]
- 5.Zhen L., Liu X., Yegang C. Accuracy of multiparametric magnetic resonance imaging for diagnosing prostate Cancer: a systematic review and meta-analysis. BMC Cancer. 2019;19(1):1244. doi: 10.1186/s12885-019-6434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozturk M., Selcuk M.B., Polat A.V., Ozbalci A.B., Baris Y.S. The diagnostic value of ultrasound and shear wave elastography in the differentiation of benign and malignant soft tissue tumors. Skeletal Radiol. 2020 doi: 10.1007/s00256-020-03492-y. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Wang H., Li Q., Zhao M.-.H., Zhan Q.-.M. Big data and medical research in China. BMJ. 2018;360:j5910. doi: 10.1136/bmj.j5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King B.F. Artificial intelligence and radiology: what will the future hold? J Am Coll Radiol. 2018;15(3 Pt B):501–503. doi: 10.1016/j.jacr.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Schlemmer H.-.P., Bittencourt L.K., D'Anastasi M. Global challenges for cancer imaging. J Glob Oncol. 2018;4 doi: 10.1200/JGO.17.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukushima K. Neocognitron: a self organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern. 1980;36(4):193–202. doi: 10.1007/BF00344251. [DOI] [PubMed] [Google Scholar]
- 11.Cortes C., Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. [Google Scholar]
- 12.The random subspace method for constructing decision forests -IEEE J Magazine [Internet]. [cited 2018 Jul 10]. Availablefrom: https://ieeexplore.ieee.org/document/709601/.
- 13.Hinton G.E., Osindero S., Teh Y.-.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18(7):1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 14.Hinton G.E., Salakhutdinov R.R. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 15.Krizhevsky A., Sutskever I., Hinton G.J. Ainips. ImageNet classification with deep convolutional. Neural Netw. 2012;25(2) [Google Scholar]
- 16.Litjens G., Kooi T., Bejnordi B.E. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Rajkomar A., Lingam S., Taylor A.G., Blum M., Mongan J. High-throughput classification of radiographs using deep convolutional neural networks. J Digit Imaging. 2017;30(1) doi: 10.1007/s10278-016-9914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vamathevan J., Clark D., Czodrowski P. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18(6):463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinyals O., Toshev A., Bengio S., Erhan D. Show and tell: lessons learned from the 2015 MSCOCO image captioning challenge. IEEE Trans Pattern Anal Mach Intell. 2017;39(4):652–663. doi: 10.1109/TPAMI.2016.2587640. [DOI] [PubMed] [Google Scholar]
- 20.Cao C., Liu F., Tan H. Deep learning and its applications in biomedicine. Geno Proteomics Bioinform. 2018;16(1):17–32. doi: 10.1016/j.gpb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anthimopoulos M., Christodoulidis S., Ebner L., Christe A., Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016;35(5):1207–1216. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 22.Xu Y., Hosny A., Zeleznik R. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res. 2019;25(11):3266–3275. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kooi T., Litjens G., van Ginneken B. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal. 2017;35:303–312. doi: 10.1016/j.media.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Yang X., Kwitt R., Styner M., Niethammer M. Quicksilver: fast predictive image registration - a deep learning approach. Neuroimage. 2017;158:378–396. doi: 10.1016/j.neuroimage.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosny A., Parmar C., Quackenbush J., Schwartz L.H., Aerts H.J.W.L. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18(8):500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ensmenger N. Is chess the drosophila of artificial intelligence? A social history of an algorithm. Soc Stud Sci. 2012;42(1):5–30. doi: 10.1177/0306312711424596. [DOI] [PubMed] [Google Scholar]
- 27.Moher D., Liberati A., Tetzlaff J., Altman D.G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moons K.G., de Groot J.A., Bouwmeester W. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014;11(10) doi: 10.1371/journal.pmed.1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M., Sterne J.A., Bossuyt P.M., QUADAS-2 Group QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 30.Liu X.X., Faes L., Kale A.U. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health. 2019;1(6):E271–EE97. doi: 10.1016/S2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- 31.Miller R.A. Medical diagnostic decision support systems–past, present, and future: a threaded bibliography and brief commentary. J Am Med Inform Ass. 1994;1:8–27. doi: 10.1136/jamia.1994.95236141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu T., Ge X., Yu J. Comparison of the application of B-mode and strain elastography ultrasound in the estimation of lymph node metastasis of papillary thyroid carcinoma based on a radiomics approach. Int J Comput Assist Radiol Surg. 2018;13(10):1617–1627. doi: 10.1007/s11548-018-1796-5. [DOI] [PubMed] [Google Scholar]
- 33.Ozden S., Er S., Saylam B., Yildiz B.D., Senol K., Tez M. A comparison of logistic regression and artificial neural networks in predicting central lymph node metastases in papillary thyroid microcarcinoma. Ann Ital Chir. 2018;89:193–198. [PubMed] [Google Scholar]
- 34.Nowikiewicz T., Wnuk P., Małkowski B., Kurylcio A., Kowalewski J., Zegarski W. Application of artificial neural networks for predicting presence of non-sentinel lymph node metastases in breast cancer patients with positive sentinel lymph node biopsies. Arch Med Sci. 2017;13(6):1399–1407. doi: 10.5114/aoms.2016.57677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu C., Jiang L., Cao Y., Hu C., Yu Y., Zhang H.J.T.C.R. Factors associated with de novo metastatic disease in invasive breast cancer: comparison of artificial neural network and logistic regression models. 2019. 2019;8(1):77–86. doi: 10.21037/tcr.2019.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaquero-Garcia J., Lalonde E., Ewens K.G. PRiMeUM: a model for predicting risk of metastasis in uveal melanoma. Invest Ophthalmol Vis Sci. 2017;58(10):4096–4105. doi: 10.1167/iovs.17-22255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin H., Chen H., Graham S., Dou Q., Rajpoot N., Heng P.A. Fast ScanNet: fast and dense analysis of multi-gigapixel whole-slide images for cancer metastasis detection. IEEE Trans Med Imaging. 2019;38(8):1948–1958. doi: 10.1109/TMI.2019.2891305. [DOI] [PubMed] [Google Scholar]
- 38.Bhooshan N., Giger M.L., Jansen S.A., Li H., Lan L., Newstead G.M. Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology. 2010;254(3):680–690. doi: 10.1148/radiol.09090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altman D.G., Royston P. What do we mean by validating a prognostic model. Stat Med. 2000;19(4):453–473. doi: 10.1002/(sici)1097-0258(20000229)19:4<453::aid-sim350>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 40.Koizumi M., Motegi K., Koyama M. Diagnostic performance of a computer-assisted diagnostic system: sensitivity of BONENAVI for bone scintigraphy in patients with disseminated skeletal metastasis is not so high. Ann Nucl Med. 2020 doi: 10.1007/s12149-020-01435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Dong D., Fang M. Dual-energy CT-based deep learning radiomics can improve lymph node metastasis risk prediction for gastric cancer. Eur Radiol. 2020 doi: 10.1007/s00330-019-06621-x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L., Ren Z. Comparison of CT and MRI images for the prediction of soft-tissue sarcoma grading and lung metastasis via a convolutional neural networks model. Clin Radiol. 2020;75(1):64–69. doi: 10.1016/j.crad.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Zhou L Q, Wu X.L., Huang S.Y. Lymph node metastasis prediction from primary breast cancer US images using deep learning. Radiology. 2020;294(1):19–28. doi: 10.1148/radiol.2019190372. [DOI] [PubMed] [Google Scholar]
- 44.Grovik E., Yi D., Iv M., Tong E., Rubin D., Zaharchuk G. Deep learning enables automatic detection and segmentation of brain metastases on multisequence MRI. J Magnetic Resonance Img. 2020;51(1):175–182. doi: 10.1002/jmri.26766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y., Gafita A., Tetteh G. Conference proceedings: annual international conference of the IEEE engineering in medicine and biology society IEEE engineering in medicine and biology society annual conference. Vol. 2019. 2019. Deep neural network for automatic characterization of lesions on 68Ga-PSMA PET/CT Images; pp. 951–954. [DOI] [PubMed] [Google Scholar]
- 46.Xue J., Wang B., Ming Y. Deep-learning-based detection and segmentation-assisted management on brain metastases. Neuro-oncology. 2019 doi: 10.1093/neuonc/noz234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baessler B., Nestler T., Pinto Dos Santos D. Radiomics allows for detection of benign and malignant histopathology in patients with metastatic testicular germ cell tumors prior to post-chemotherapy retroperitoneal lymph node dissection. Eur Radiol. 2019 doi: 10.1007/s00330-019-06495-z. [DOI] [PubMed] [Google Scholar]
- 48.Yang X., Wu L., Ye W. Deep learning signature based on staging ct for preoperative prediction of sentinel lymph node metastasis in breast cancer. Acad Radiol. 2019 doi: 10.1016/j.acra.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Gao Y., Zhang Z.-.D., Li S. Deep neural network-assisted computed tomography diagnosis of metastatic lymph nodes from gastric cancer. Chin Med J. 2019;132(23):2804–2811. doi: 10.1097/CM9.0000000000000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coronado-Gutierrez D., Santamaria G., Ganau S. Quantitative ultrasound image analysis of axillary lymph nodes to diagnose metastatic involvement in breast cancer. Ultrasound Med Biol. 2019;45(11):2932–2941. doi: 10.1016/j.ultrasmedbio.2019.07.413. [DOI] [PubMed] [Google Scholar]
- 51.Okada Y., Matsushita S., Nakajima Y. Comparison of diagnostic precision for bone metastasis of primary breast cancer between BONENAVI version 1 and BONENAVI version 2. Nucl Med Commun. 2019;40(11):1148–1153. doi: 10.1097/MNM.0000000000001081. [DOI] [PubMed] [Google Scholar]
- 52.Lee J.H., Ha E.J., Kim J.H. Application of deep learning to the diagnosis of cervical lymph node metastasis from thyroid cancer with CT. Eur Radiol. 2019;29(10):5452–5457. doi: 10.1007/s00330-019-06098-8. [DOI] [PubMed] [Google Scholar]
- 53.Jansen M.J.A., Kuijf H.J., Niekel M. Liver segmentation and metastases detection in MR images using convolutional neural networks. J Med Img (Bellingham, Wash) 2019;6(4) doi: 10.1117/1.JMI.6.4.044003. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J., Sun D., Chen L. Radiomics analysis of dynamic contrast-enhanced magnetic resonance imaging for the prediction of sentinel lymph node metastasis in breast cancer. Front Oncol. 2019;9(980) doi: 10.3389/fonc.2019.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dohopolski M., Chen L., Sher D.J., Wang J. Predicting lymph node metastasis in patients with oropharyngeal cancer by convolutional neural networks with associated epistemic uncertainty. Int J Radiation Oncol Biol Phys. 2019;105S(1):S122. doi: 10.1088/1361-6560/abb71c. -S. [DOI] [PubMed] [Google Scholar]
- 56.Peng Y., Bi L., Guo Y., Feng D., Fulham M., Kim J. Conference proceedings: annual international conference of the IEEE engineering in medicine and biology society IEEE engineering in medicine and biology society annual conference. Vol. 2019. 2019. Deep multi-modality collaborative learning for distant metastases predication in PET-CT soft-tissue sarcoma studies; pp. 3658–3688. [DOI] [PubMed] [Google Scholar]
- 57.Feng Q.-.X., Liu C., Qi L. An intelligent clinical decision support system for preoperative prediction of lymph node metastasis in gastric cancer. J Am College Radiol. 2019;16(7):952–960. doi: 10.1016/j.jacr.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 58.Schnelldorfer T., Ware M.P., Liu L.P., Sarr M.G., Birkett D.H., Ruthazer R. Can we accurately identify peritoneal metastases based on their appearance? an assessment of the current practice of intraoperative gastrointestinal cancer staging. Ann Surg Oncol. 2019;26(6):1795–1804. doi: 10.1245/s10434-019-07292-0. [DOI] [PubMed] [Google Scholar]
- 59.Mehta S.D., Sebro R. Random forest classifiers aid in the detection of incidental osteoblastic osseous metastases in DEXA studies. Int J Comput Assist Radiol Surg. 2019;14(5):903–909. doi: 10.1007/s11548-019-01933-1. [DOI] [PubMed] [Google Scholar]
- 60.Ariji Y., Fukuda M., Kise Y. Contrast-enhanced computed tomography image assessment of cervical lymph node metastasis in patients with oral cancer by using a deep learning system of artificial intelligence. Oral Surg Oral Med Oral Pathol Oral Radiol. 2019;127(5):458–463. doi: 10.1016/j.oooo.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Zhou Y.P., Li S., Zhang X.X. High definition MRI rectal lymph node aided diagnostic system based on deep neural network. Zhonghua Wai Ke Za Zhi. 2019;57(2):108–113. doi: 10.3760/cma.j.issn.0529-5815.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 62.Li Y., Eresen A., Shangguan J. Establishment of a new non-invasive imaging prediction model for liver metastasis in colon cancer. Am J Cancer Res. 2019;9(11):2482–2492. [PMC free article] [PubMed] [Google Scholar]
- 63.Zhiguo Z, Dohopolski M, Liyuan C, et al. Reliable lymph node metastasis prediction in head & neck cancer through automated multi-objective model. 2019: 4. [DOI] [PMC free article] [PubMed]
- 64.Acar E., Leblebici A., Ellidokuz B.E., Basbinar Y., Kaya G.C. Machine learning for differentiating metastatic and completely responded sclerotic bone lesion in prostate cancer: a retrospective radiomics study. Br J Radiol. 2019;92(201902861101) doi: 10.1259/bjr.20190286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou F., Yang Z., Gu W., Yu Y., Liang Y. OCT Automatic identification of metastatic lymph nodes in OCT images. In: Fujimoto JG, Izatt JA, editors. Proceedings of SPIE. 2019. [Google Scholar]
- 66.Ariji YA-OH, Sugita Y, Nagao T, et al. CT evaluation of extranodal extension of cervical lymph node metastases in patients with oral squamous cell carcinoma using deep learning classification. doi: 10.1007/s11282-019-00391-4. [DOI] [PubMed]
- 67.Xu X, Li H, Wang S, et al. Multiplanar MRI-Based predictive model for preoperative assessment of lymph node metastasis in endometrial cancer. [DOI] [PMC free article] [PubMed]
- 68.Luo J., Ning Z., Zhang S., Feng Q., Zhang Y. Bag of deep features for preoperative prediction of sentinel lymph node metastasis in breast cancer. Phys Med Biol. 2018;63(24501424) doi: 10.1088/1361-6560/aaf241. [DOI] [PubMed] [Google Scholar]
- 69.Ha R., Chang P., Karcich J. Axillary lymph node evaluation utilizing convolutional neural networks using MRI dataset. J Digit Imaging. 2018;31(6):851–856. doi: 10.1007/s10278-018-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kann B.H., Aneja S., Loganadane G.V. Successful identification of head and neck cancer (HNC) nodal metastasis (NM) and extranodal extension (ENE) using deep learning neural networks. Int J Radiation Oncol Biol Phys. 2018;102S(3):S60. doi: 10.1038/s41598-018-32441-y. -S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee J.H., Baek J.H., Kim J.H. Deep learning-based computer-aided diagnosis system for localization and diagnosis of metastatic lymph nodes on ultrasound: a pilot study. THYROID. 2018;28(10):1332–1338. doi: 10.1089/thy.2018.0082. [DOI] [PubMed] [Google Scholar]
- 72.Lu Y., Yu Q., Gao Y. Identification of metastatic lymph nodes in MR imaging with faster region-based convolutional neural networks. Cancer Res. 2018;78(17):5135–5143. doi: 10.1158/0008-5472.CAN-18-0494. [DOI] [PubMed] [Google Scholar]
- 73.Ferreira Junior J.R., Koenigkam-Santos M., Garcia Cipriano F.E., Fabro A.T., de Azevedo-Marques P.M. Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput Methods Programs Biomed. 2018;159:23–30. doi: 10.1016/j.cmpb.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 74.Lo T-Y, Wei P-Y, Yen C-H, et al. Prediction of metastasis in head and neck cancer from computed tomography images. 2018: 18–23.
- 75.Jin L, Peng W, Yanzhao L, Yang Z, Xiaolong L, Kuan L. Transfer learning of pre- trained inception-v3 model for colorectal cancer lymph node metastasis classification. 2018: 1650–4.
- 76.Larhmam MA, Mahmoudi S, Drisis S, Benjelloun M. A Texture analysis approach for spine metastasis classification in T1 and T2 MRI. In: Rojas I, Ortuno F, editors; 2018: 198–211.
- 77.Zhong Y, Yuan M, Zhang T, Zhang YD, Li H, Yu TF. Radiomics approach to prediction of occult mediastinal lymph node metastasis of lung adenocarcinoma. [DOI] [PubMed]
- 78.Wang H., Zhou Z., Li Y. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res. 2017;7(1):11. doi: 10.1186/s13550-017-0260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koizumi M., Motegi K., Koyama M., Terauchi T., Yuasa T., Yonese J. Diagnostic performance of a computer-assisted diagnosis system for bone scintigraphy of newly developed skeletal metastasis in prostate cancer patients: search for low-sensitivity subgroups. Ann Nucl Med. 2017;31(7):521–528. doi: 10.1007/s12149-017-1175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J., Fang Z., Lang N., Yuan H., Su M.-.Y., Baldi P. A multi-resolution approach for spinal metastasis detection using deep Siamese neural networks. Comput. Biol. Med. 2017;84:137–146. doi: 10.1016/j.compbiomed.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z.-.L., Zhou Z.-.G., Chen Y., Li X.-.T., Sun Y.-.S. Support vector machines model of computed tomography for assessing lymph node metastasis in esophageal cancer with neoadjuvant chemotherapy. J Comput Assist Tomogr. 2017;41(3):455–460. doi: 10.1097/RCT.0000000000000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pham T.D., Watanabe Y., Higuchi M., Suzuki H. Texture analysis and synthesis of malignant and benign mediastinal lymph nodes in patients with lung cancer on computed tomography. Sci Rep. 2017;7(43209) doi: 10.1038/srep43209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Q, Suo J, Chang W, Shi J, Chen M. Dual-modal computer-assisted evaluation of axillary lymph node metastasis in breast cancer patients on both real-time elastography and B-mode ultrasound. [DOI] [PubMed]
- 84.Wang Y.-.W., Wu C.-.S., Zhang G.-.Y. Can parameters other than minimal axial diameter in mri and pet/ct further improve diagnostic accuracy for equivocal retropharyngeal lymph nodes in nasopharyngeal carcinoma. PLoS ONE. 2016;11(e016374110) doi: 10.1371/journal.pone.0163741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aslantas A, Dandil E, Saglam S, Cakiroglu M. CADBOSS: a computer-aided diagnosis system for whole-body bone scintigraphy scans. [DOI] [PubMed]
- 86.Chmielewski A., Dufort P., Scaranelo A.M. A computerized system to assess axillary lymph node malignancy from sonographic images. Ultrasound Med Biol. 2015;41(10):2690–2699. doi: 10.1016/j.ultrasmedbio.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 87.Koizumi M., Miyaji N., Murata T. Evaluation of a revised version of computer-assisted diagnosis system, BONENAVI version 2.1.7, for bone scintigraphy in cancer patients. Ann Nucl Med. 2015;29(8):659–665. doi: 10.1007/s12149-015-0988-0. [DOI] [PubMed] [Google Scholar]
- 88.Koizumi M., Wagatsuma K., Miyaji N. Evaluation of a computer-assisted diagnosis system, BONENAVI version 2, for bone scintigraphy in cancer patients in a routine clinical setting. Ann Nucl Med. 2015;29(2):138–148. doi: 10.1007/s12149-014-0921-y. [DOI] [PubMed] [Google Scholar]
- 89.Trabelsi N, Ben Sellem D. Comparison of supervised and unsupervised classifications in the detection of hepatic metastases. 2015.
- 90.Gao X, Chu C, Li Y, et al. The method and efficacy of support vector machine classifiers based on texture features and multi-resolution histogram from (18)F-FDG PET-CT images for the evaluation of mediastinal lymph nodes in patients with lung cancer. [DOI] [PubMed]
- 91.Tokuda O., Harada Y., Ohishi Y., Matsunaga N., Edenbrandt L. Investigation of computer-aided diagnosis system for bone scans: a retrospective analysis in 406 patients. Ann Nucl Med. 2014;28(4):329–339. doi: 10.1007/s12149-014-0819-8. [DOI] [PubMed] [Google Scholar]
- 92.Seff A., Lu L., Cherry K.M. 2D view aggregation for lymph node detection using a shallow hierarchy of linear classifiers. In: Golland P, Hata N, Barillot C, Hornegger J, Howe R, editors. Lecture notes in computer science. 2014. pp. 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou Z.-.G., Liu F., Jiao L C. An evidential reasoning based model for diagnosis of lymph node metastasis in gastric cancer. BMC Med Inform Decis Mak. 2013;13(123) doi: 10.1186/1472-6947-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang S., Nam Y., Kim M.-.O., Kim E.Y., Park J., Kim D H. Computer-aided detection of metastatic brain tumors using magnetic resonance black-blood imaging. Invest Radiol. 2013;48(2):113–119. doi: 10.1097/RLI.0b013e318277f078. [DOI] [PubMed] [Google Scholar]
- 95.Liu J., Wang S., Linguraru M.G., Yao J., Summers R.M. A Variational framework for joint detection and segmentation of ovarian cancer metastases. In: Sakuma I, Barillot C, Navab N, editors. Lecture notes in computer science. 2013. pp. 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakamura Y., Nimura Y., Kitasaka T. Automatic abdominal lymph node detection method based on local intensity structure analysis from 3D X-ray CT images. In: Novak CL, Aylward S, editors. Proceedings of SPIE. 2013. [Google Scholar]
- 97.Chang C.-.Y., Lai C.-.C., Lai C.-.T., Chen S.-.J. Integrating PSONN and Boltzmann function for feature selection and classification of lymph nodes in ultrasound images. J Vis Commun Image Represent. 2013;24(1):23–30. [Google Scholar]
- 98.Feulner J, Zhou Sk Fau Hammon M, Hammon M, Fau Hornegger J, Hornegger J, Fau Comaniciu D, Comaniciu D. Lymph node detection and segmentation in chest CT data using discriminative learning and a spatial prior. [DOI] [PubMed]
- 99.Li C., Zhang S., Zhang H. Using the K-nearest neighbor algorithm for the classification of lymph node metastasis in gastric cancer. Comput Math Methods Med. 2012;(876545) doi: 10.1155/2012/876545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cai H, Cui C Fau Tian H, Tian H Fau Zhang M, Zhang M Fau Li L, Li L. A novel approach to segment and classify regional lymph nodes on computed tomography images. [DOI] [PMC free article] [PubMed]
- 101.Chen SJ, Lin Ch Fau Chang C-Y, Chang Cy Fau Chang K-Y, et al. Characterizing the major sonographic textural difference between metastatic and common benign lymph nodes using support vector machine with histopathologic correlation. [DOI] [PubMed]
- 102.Zhang X.-.P., Wang Z.-.L., Tang L., Sun Y.-.S., Cao K., Gao Y. Support vector machine model for diagnosis of lymph node metastasis in gastric cancer with multidetector computed tomography: a preliminary study. BMC Cancer. 2011;11(10) doi: 10.1186/1471-2407-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dietzel M., Baltzer P.A.T., Dietzel A. Application of artificial neural networks for the prediction of lymph node metastases to the ipsilateral axilla - initial experience in 194 patients using magnetic resonance mammography. Acta radiol. 2010;51(8):851–858. doi: 10.3109/02841851.2010.498444. [DOI] [PubMed] [Google Scholar]
- 104.Sadik M., Hamadeh I., Nordblom P. Computer-assisted interpretation of planar whole-body bone scans. J Nuclear Med. 2008;49(12):1958–1965. doi: 10.2967/jnumed.108.055061. [DOI] [PubMed] [Google Scholar]
- 105.Shiraishi J., Sugimoto K., Moriyasu F., Kamiyama N., Doi K. Computer-aided diagnosis for the classification of focal liver lesions by use of contrast-enhanced ultrasonography. Med Phys. 2008;35(5):1734–1746. doi: 10.1118/1.2900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang J., Wang Y., Dong Y., Wang Y. Computer-aided diagnosis of cervical lymph nodes on ultrasonography. Comput. Biol. Med. 2008;38(2):234–243. doi: 10.1016/j.compbiomed.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 107.Tagaya R., Kurimoto N., Osada H., Kobayashi A. Automatic objective diagnosis of lymph nodal disease by B-mode images from convex-type echobronchoscopy. Chest. 2008;133(1):137–142. doi: 10.1378/chest.07-1497. [DOI] [PubMed] [Google Scholar]
- 108.Marten K, Grillhosl A, Fau Seyfarth T, Seyfarth T, Fau Obenauer S, Obenauer S, Fau Rummeny EJ, Rummeny Ej Fau Engelke C, Engelke C. Computer-assisted detection of pulmonary nodules: evaluation of diagnostic performance using an expert knowledge-based detection system with variable reconstruction slice thickness settings. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.