Abstract
Phytophthora species are notorious pathogens of several economically important crop plants. Several general elicitors, commonly referred to as Pathogen-Associated Molecular Patterns (PAMPs), from Phytophthora spp. have been identified that are recognized by the plant receptors to trigger induced defense responses in a process termed PAMP-triggered Immunity (PTI). Adapted Phytophthora pathogens have evolved multiple strategies to evade PTI. They can either modify or suppress their elicitors to avoid recognition by host and modulate host defense responses by deploying hundreds of effectors, which suppress host defense and physiological processes by modulating components involved in calcium and MAPK signaling, alternative splicing, RNA interference, vesicle trafficking, cell-to-cell trafficking, proteolysis and phytohormone signaling pathways. In incompatible interactions, resistant host plants perceive effector-induced modulations through resistance proteins and activate downstream components of defense responses in a quicker and more robust manner called effector-triggered-immunity (ETI). When pathogens overcome PTI—usually through effectors in the absence of R proteins—effectors-triggered susceptibility (ETS) ensues. Qualitatively, many of the downstream defense responses overlap between PTI and ETI. In general, these multiple phases of Phytophthora-plant interactions follow the PTI-ETS-ETI paradigm, initially proposed in the zigzag model of plant immunity. However, based on several examples, in Phytophthora-plant interactions, boundaries between these phases are not distinct but are rather blended pointing to a PTI-ETI continuum.
Keywords: Phytophthora, plant defense, plant immunity, zigzag model, PAMPS, effectors, RXLR, susceptibility genes
Introduction
Oomycetes are a unique class of eukaryotes classified in the kingdom Protoctista along with algae, diatoms, and other planktons under the sub kingdom Stramenopila (Kamoun et al., 2015). Morphologically they are quite similar to filamentous fungi but physiologically, genetically, and biochemically they are very different from fungi. Phytopathogenic oomycetes from the genus Phytophthora cause destructive diseases in many economically important crops and forest ecosystems (Hansen, 2015). Control strategies for Phytophthora diseases are very limited (Attard et al., 2014). Despite their differences, fungi and oomycetes use quite similar strategies to infect and colonize their hosts, but many chemicals that target sterol synthesis and chitin in fungal cell walls, which are absent from oomycetes, do not control diseases caused by oomycetes (Latijnhouwers et al., 2003; Attard et al., 2014). Deployment of host genetic resistance is considered the most effective, eco-friendly and cost-effective management strategy. To develop sustainable resistance against oomycetes, a thorough understanding of the molecular basis of Phytophthora-plant interactions is very important (Anderson et al., 2010).
Plant defense comprises both constitutive preformed and induced defense responses that offers several layers of protection against pathogen invasion (Anderson et al., 2010; Doughari, 2015). Constitutive defense consisted of physical or chemical barriers that restrict the attachment and entry of the most microbes into the plants (Osbourn, 1996; Chassot et al., 2008; Underwood, 2012). According to the zigzag model of plant-pathogen interaction, induced defense consists of two layers, the first one is known as pathogen associated molecular pattern (PAMP) triggered immunity (PTI). In PTI, conserved molecules or structures of pathogens are perceived by plant pattern recognition receptors (PRRs), followed by activation of defense responses. To circumvent PTI, pathogens deliver effector proteins inside host cells, where they interfere with defense responses. Plants perceive effectors through resistance (R) genes and activate a more robust and faster defense response, termed as effector-triggered immunity (ETI). When one or more pathogen effectors suppress PTI, pathogens successfully infect susceptible hosts and in the absence of effective R proteins, ETI is overcome, eventually leading to effector triggered susceptibility (ETS). Multiple shifts between ETS and ETI occur because of coevolution of effectors in pathogens and corresponding R genes in plant hosts (Jones and Dangl, 2006). Although the real-world applicability of this model has been challenged (Thomma et al., 2011; Pritchard and Birch, 2014), this is so far considered the most concise model of plant immunity for pedagogical purposes. The popularity of zigzag model could be attributed to the fact that it elegantly contrives the three principal outcomes of plant-pathogen interactions: the non-race-specific elicitor-induced defense (PTI), the race or cultivar-specific gene-for-gene resistance (ETI), and race-specific pathogen virulence and host susceptibility (ETS).
In the zigzag model, PTI and ETI are viewed as two separate sequential branches of plant immunity (Jones and Dangl, 2006). However, there are several examples where effectors fulfill the criteria to be designated as PAMPs (Thomma et al., 2011) and PAMPs as effectors. Moreover, in the zigzag model the term R gene was exclusively reserved for intracellular receptors harboring nucleotide binding (NB) and leucine rich repeat (LRR) domains. However, in the original gene-for gene resistance concept, Flor described both transmembrane as well as intracellular pathogen receptors as R proteins (Thomma et al., 2011). Accordingly, PRRs, at least some of them, have been classified as a subclass of R genes (Sanseverino et al., 2012). Based on several plant-pathogen interaction examples, Thomma et al., proposed a continuum between PTI and ETI, in which the strict boundaries of the zigzag model are blurred (Thomma et al., 2011). In this review, we have discussed the current state of Phytophthora-plant interactions in terms of zigzag model as well as deviations from this model. These discussions suggest that the PTI-ETI continuum concept also applies to Phytophthora-plant interactions. The application of current knowledge about Phytophthora-plant interactions to develop broad-spectrum long-lasting resistance in plants against Phytophthora spp. has also been discussed.
Pamp Triggered Immunity (PTI)
Plants recognize Phytophthora spp. by sensing a wide variety of elicitors (Table 1). The term “elicitor” was initially used to refer to both the exogenous and endogenous signaling molecules that can induce any defense response in plants (Eder and Cosio, 1994; Hahn, 1996). Exogenous elicitors could be either the components of pathogen's cell wall or membranes, released upon action of host enzymes or secreted by the pathogen during host-pathogen interaction to subvert the host defense and/or facilitate nutrient acquisition (Raaymakers and Van Den Ackerveken, 2016). Whereas, the endogenous elicitors were referred to components that originated from host plants, usually resulting from damage caused by the action of pathogen enzymes (Eder and Cosio, 1994). In the zigzag model, the terms exogenous and endogenous elicitors were replaced by PAMPs and DAMPs (Damage Associated Molecular Patterns), respectively and the induced defense upon recognition of either PAMP or DAMP was termed as PTI. PAMPs fall into a broad spectrum of chemistries ranging from carbohydrates to proteins as is discussed below.
Table 1.
Phytophthora spp. elicitors and associated PTI components.
| Name | Pathogen groups carrying PAMP | Chemical nature | Cognate PRR | PAMP perception model | Signaling | References |
|---|---|---|---|---|---|---|
| Elicitins | Unique to oomycete genera; Phytophthora and Pythium | Protein | ELR (Reported for Cryptogein, INF1 and ParA1) | BAK1/SERK3 dependent (Reported for INF1 and ParA1) | SA (Cryptogein) JA and ET (INF1) Calcium and MAPK (Cryptogein and INF1) | Kamoun et al., 1998; Lebrun-Garcia et al., 1998; Kawamura et al., 2009; Amelot et al., 2011; Du Y. et al., 2015; Peng et al., 2015; Derevnina et al., 2016 |
| OPEL | Oomycetes | Protein | Unknown | Unknown | SA | Chang et al., 2015 |
| Pep-13 | Phytophthora spp. | Protein/peptide | Unknown | BAK1/SERK3 independent | SA and JA Calcium and MAPK | Nürnberger et al., 1994; Blume et al., 2000; Halim et al., 2009; Wang H. et al., 2019 |
| β-glucans | Oomycetes and fungi | Carbohydrate | Unknown | Unknown | SA | Kopp et al., 1989; Klarzynski et al., 2000; Fesel and Zuccaro, 2016 |
| Eicosapolyenoic acids (EPs) | Oomycetes, primitive fungi and nematodes | Lipids | Unknown | Unknown | JA | Preisig and Kuć, 1985; Savchenko et al., 2010 |
| Elicitors with dual, PAMP and effector status | ||||||
| XEG1-GH12 | Fungi and oomycetes | Protein-CWDEs | Unknown | BAK1/SERK3 dependent | Unknown | Ma et al., 2015, 2017 |
| CBEL-CBM1 | Fungi and oomycetes | Protein-CWDEs | Unknown | BAK1/SERK3 dependent | SA, JA, and ET | Khatib et al., 2004; Larroque et al., 2013 |
| nlp20-NLPs | Bacteria, fungi and oomycetes | Protein | RLP23 | BAK1/SERK3 dependent | SA | Böhm et al., 2014; Albert et al., 2015 |
Proteinaceous PAMPs of Phytophthora spp.
Elicitins are extracellular structurally conserved cysteine-rich proteins secreted by species in the genus Pythium and Phytophthora. They do not display sequence similarity to plant proteins, and thus are recognized as non-self-molecules by the host plants leading to the induction of an array of defense responses (Derevnina et al., 2016). Elicitins are reported to bind sterols and lipids. Sterols are certain types of steroid alcohols, which are essential structural and functional components of eukaryotes. Pythium and Phytophthora lack sterol synthesis ability, and, therefore, rely on their host's sterols. They have adapted efficient mechanisms of sterol scavenging from the host cell membranes, possibly through elicitins (Mikes et al., 1998). Elicitins are designated as oomycetes PAMPs that can induce PTI. In non-host plants, pathogens are not capable of overcoming this elicitin-induced PTI, rendering pathogens as non-adapted; these interactions are called non-host/non-pathogen interactions.
Non-host resistance has long been studied using the tobacco-Phytophthora model because most of the Phytophthora spp. are non-adapted pathogens of tobacco. For example, cryptogein isolated from P. cryptogea is one of the earliest identified sterol-binding elicitins, which has been extensively used to explore defense mechanisms underlying non-host resistance. The sterol-cryptogein complex is recognized by the tobacco PRRs and triggers a strong defense response (Lochman et al., 2005), likely making P. cryptogea, a non-adapted pathogen of tobacco (Wendehenne et al., 2002). A number of elicitins from different Phytophthora spp. have been identified and they are being used to identify host factors that can offer broad spectrum non-race specific resistance to diverse phytopathogens (Du J. et al., 2015). These include INF1 from P. infestans, ParA1 from P. parasitica, CAP1 from P. capsici, PAL1 from P. palmivora and Quercinin from P. quercina (Kamoun et al., 1998; Fawke et al., 2015; Derevnina et al., 2016). The INF1 elicitin shares 79% amino acid sequence identity with cryptogein. INF1 is shown to bind phytosterols but its essentiality as a sterol carrier for pathogen is not confirmed because the INF1 deficient P. infestans can still survive (Kamoun et al., 1998; Fawke et al., 2015). This finding partially conflicts with the traditional definition, where PAMP is defined as an essential contributor to pathogen's fitness (Jones and Dangl, 2006). So far, INF1 is among the most widely used elicitin for understanding molecular mechanisms for the induction of PTI, and for the suppression of PTI by effectors (Kanzaki et al., 2003; Bos et al., 2006; Kawamura et al., 2009; Gilroy et al., 2011b; Cheng et al., 2012; King et al., 2014; Liu et al., 2015; Turnbull et al., 2017; He et al., 2018; Murphy et al., 2018).
OPEL is another unique oomycete-specific PAMP, which was identified in P. parasitica and has homologs in several Phytophthora spp. and other oomycetes such as Hyaloperonospora arabidopsidis, Pythium ultimum, and Albugo laibachii (Chang et al., 2015). So far, there is no evidence of its presence in fungi or any other phytopathogens. OPEL is reported to induce strong PTI in tobacco, making it resistant to subsequent infection by P. parasitica as well as other diverse pathogens like Tobacco mosaic virus (TMV) and Ralstonia solanacearum (Chang et al., 2015). OPEL is a 556 amino acid large modular protein consisting of a signal peptide and three conserved domains: a glycine-rich protein domain, a thaumatin-like domain, and a glycosyl hydrolase domain harboring a laminarinase active site. This laminarinase active site is associated with the elicitor activity of OPEL, which is recognized by a PRR either directly or indirectly through DAMPs generated through its enzymatic activity (Chang et al., 2015).
Pep-13 is another PAMP that is apparently unique to Phytophthora spp. (Nürnberger et al., 1994). Pep-13 is a highly conserved 13 amino acid peptide found in the cell-wall associated transglutaminases (TGases) of many Phytophthora spp. TGases are R-glutaminyl-peptide:amine-γ-glutamyl transferases involved in specific protein cross linking, which are associated with multiple physiological activities in animals (Brunner et al., 2002). No known biological function of TGases in Phytophthora spp. has been reported yet other than the induction of PTI in the plants (Reiss et al., 2011; Severino et al., 2014). The P. sojae's Pep-13 was found to induce PTI, strong enough to make parsley a non-host for the pathogen.
Non-proteinaceous PAMPs of Phytophthora spp.
In addition to proteinaceous PAMPs, components of the pathogen's cell wall or membranes that are generated by host enzymes also display PAMP activity. For example, β-glucans are cell wall polymers, which are released from a pathogen's cell wall by host glucanases (Umemoto et al., 1997). β-glucans are common PAMPs of filamentous pathogens (Fesel and Zuccaro, 2016). Different β-glucan oligosaccharides possess conserved characteristics patterns that are recognized by specific hosts. For example, β-1,6-glucans from P. sojae can be perceived as PAMPs by soybean but not by tobacco. Phytoalexin biosynthesis in rice and soybean is initiated by different β-1,3- and β-1,6-glucans. Also, different kinds of defense responses are induced by different β-glucans. These characteristics make β-glucans excellent candidates to dissect non-host resistance (Fesel and Zuccaro, 2016; Mélida et al., 2018).
Eicosapolyenoic acids (EPs) are polyunsaturated fatty acids including eicosapentaenoic acid (EPAs), and arachidonic acid (AA). EPs isolated from P. infestans were reported to function as PAMPs in a wide variety of plants. Application of low concentration of AAs (isolated from Phytophthora and fungi) on potato, tomato, sugar beet, and vine plants enhanced their resistance against a variety of diseases caused by filamentous pathogens (Dedyukhina et al., 2014). A 20-carbon chain and cis-unsaturation at their 5-position in EPs has been reported as the minimum structural features required for their elicitor activity (Preisig and Kuć, 1985). Among plant pathogens only oomycetes, primitive fungi and nematodes have been reported to synthesize these fatty acids. In mammals, EPAs and AAs have been reported to play an important role in mediating inflammatory responses, function of the central nervous system and immune signaling. Their association with both plant and animal immune systems and their presence in multiple pathogens make them putative evolutionary conserved cross-kingdom immune signaling molecules that perhaps evolved by convergent evolution. Another interesting fact about EPs is their potential interaction with β-1,3-glucans, which alone do not induce any defense in potatoes but remarkably enhance the sensitivity of potato tissues to EPs (Bostock et al., 2011).
Phytophthora spp. Elicitors With Conflicted PAMP or Effector Status
In addition to the well-defined oomycete PAMPs, there are some elicitors that qualify to be designated PAMPs as well as effectors. Cell wall degrading enzymes (CWDEs) are one such example. CWDEs have been reported as common elicitors of filamentous pathogens and they contribute to pathogen's virulence.
Plant cell wall is not only an important component of preformed defense that serves as a barrier to prevent pathogen penetration but also as an essential element of plant's surveillance system. PRRs monitor the apoplastic environment and perceive any intrusion in the cell wall integrity by sensing peptides or wall glucans, derived as a result of cell wall damage caused by the pathogen's CWDEs (Bacete et al., 2018). Cell wall damage facilitates pathogen's penetration and DAMPs also serve as nutrients for the pathogen (Ma et al., 2015; Gui et al., 2017). An endoglucanase type CWDEs, XEG1, which is a member of glycoside hydrolase family 12 (GH12), is identified in Phytophthora spp. It is defined as PAMP because the GH12 family is widely distributed among prokaryotic and eukaryotic microbial taxa and it is recognized by the host through the PTI recognition machinery. It is also described as an effector because it is an important virulence factor that is secreted in the host's apoplast to damage the host cell wall by hydrolyzing xyloglucan and β-1,4-glucan to reducing sugars (Ma et al., 2015, 2017). The role of CWDEs in the induction of plant defense response has long been established in case of oomycetes but the PAMP status of a fungal CWDE was recently established. The GH12 proteins of Verticillium dahlia fungus act as PAMPs as it can trigger PTI in Nicotiana benthamiana, independently of their enzymatic activity (Gui et al., 2017).
Carbohydrate-binding module family 1 (CBM1) domain-containing proteins are another type of CWDEs, common among fungi and oomycetes. Although structurally similar, CBM1 proteins of oomycetes and fungi have been reported to play different biological roles during their interaction with the plant cell walls. Cellulose binding elicitor lectin (CBEL), a CBM1-containing protein, first identified in the cell wall of P. parasitica is now a well-defined PAMP of Phytophthora spp. (Larroque et al., 2013). It can induce PTI in a limited number of species across diverse plant families (Khatib et al., 2004). In addition to triggering PTI, CBEL was found to be involved in maintaining cell wall integrity and pathogen adhesion to the host cell surface but devoid of any enzymatic activity (Séjalon-Delmas et al., 1997). Whereas, in fungi CBM1 containing proteins are enzymatically active and have been shown to be involved in cellulose degradation (Larroque et al., 2012). Interestingly, a fungal CBM1 was recently shown to suppress PTI induced by the GH12 fungal PAMP (Gui et al., 2017), indicating their effector activity in fungi.
Necrosis and ethylene-inducing peptide 1-like proteins (NLPs) are another example of PAMP effector overlap and have been found as common elicitors among the microbes of prokaryotic (bacteria) and eukaryotic (fungi and oomycetes) lineages (Gijzen and Nürnberger, 2006). NLPs were discovered as cytotoxic proteins secreted by necrotrophic and hemi-biotrophic pathogens to initiate necrosis in their dicotyledonous hosts. Later on, non-cytotoxic forms of NLPs were found, secreted by the pathogens during biotrophy (Böhm et al., 2014). NLPs were initially proposed as dual function effector toxins, acting both as virulence factors as well as trigger for ETI (Kamoun, 2006; Böhm et al., 2014). Recently, a highly conserved 20 amino acid fragment referred to as nlp20 found in both cytotoxic and non-cytotoxic NLPs of all three microbial taxa has been shown to incite PTI in various plants. Like XEG1, nlp20 is also recognized through the PTI recognition system. These findings suggest NLPs as unique widespread virulence factors, harboring a highly conserved PAMP motif and have the ability to incite both PTI and ETI (Böhm et al., 2014).
Like CWDEs, NLPs are also mentioned both as PAMPs and effectors in literature without clarification on their specific characteristics behind their respective designation (Haas et al., 2009; Hardham and Cahill, 2010; Böhm et al., 2014; Fawke et al., 2015; Raaymakers and Van Den Ackerveken, 2016). As discussed above, CWDEs and NLPs qualify both as PAMP and effector status and thus, are excellent examples of the PTI-ETI continuum in diverse pathosystems, including Phytophthora spp.
Cognate PRRs and PAMPs Perception Models
Based on bacterial, fungal, and oomycete PAMPs there are two PAMP perception models (Figure 1). The most well-described one is the BAK1/SERK3 dependent perception. BAK1/SERK3 is an LRR repeat receptor-like kinase (LRR-RLK) type cell surface receptor, that was first identified as an interactor of the brassinosteroid receptor 1 (BRI1). Later, it was identified as an essential component of PTI against diverse PAMPs. Induction of HR by INF1 in tobacco and N. benthamiana is BAK1/SERK3-mediated (Chaparro-Garcia et al., 2011). Recognition of INF1 by RLP or RLK type PRR and location of INF1-PRR interaction was not clear, but recently it was shown that multiple INFs are perceived by the extracellular domain of a wild potato RLP, elicitin response receptor (ELR), which associates with BAK1 to modulate localized cell death in potato (reviewed in Fawke et al., 2015). The suppressor of BIR1 (SOBIR1) was identified as another required component in ParA1-induced ELR-BAK1 mediated non-host resistance against P. parasitica in tomato and N. benthamiana (Peng et al., 2015). Wild potato ELR is also reported to recognize cryptogein (Du J. et al., 2015; Derevnina et al., 2016) but association of BAK1 and SOBIR1 with cryptogein induced HR is not clear yet.
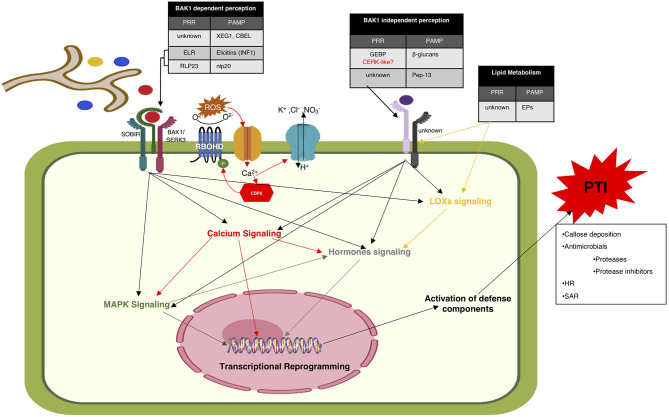
Figure 1.
Proposed model depicting the components and events of PAMP triggered immunity. According to the to date literature, there are two PAMPs perception models. One is BAK1/SERK3 dependent (left) and the other one is BAK1/SERK3 independent perception (right). All PAMPs shown in the leftmost table are perceived in BAK1/SERK3 dependent manner. Ca2+ ion influx is among the earliest events of PAMP perception followed by opening up the membrane transporters for the influx of H+ and efflux of K+, Cl−, and that leads to changes in extracellular pH and depolarization of plasma membrane. ROS burst is reported to occur within minutes of PAMP perception. Ca2+-dependent or Ca2+independent phosphorylated RBOHD generates ROS that leads to further increase in cytosolic Ca2+ concentrations. Calcium, MAPK, and hormonal signaling are all reported to be involved in receptor mediated perception of Phytophthora spp. PAMPs. PAMPs induce defense responses include; ROS burst, callose deposition, hypersensitive response (HR), expression of defense genes, accumulation of antimicrobial secondary metabolites and systemic acquired resistance (SAR).
The nlp20 PAMP of NLPs has been shown to be recognized by an Arabidopsis PRR, RLP23 that interacts with SOBIR1 and BAK1 to trigger various PTI responses (Albert et al., 2015). Both the cell wall associated glycoprotein CBEL and the endoglucanase XEG1 have also been found to require BAK1/SERK3 for PTI induction but their cognate PRRs are not yet known (Larroque et al., 2013; Ma et al., 2015). Evidence of involvement of these PAMP recognition components to induce defense in response to nlp20, XEG1, and CBEL is another solid reason in favor of their dual PAMP/effector designation (Ma et al., 2015).
All the above mentioned proteinaceous PAMPs of Phytophthora spp. are perceived in BAK1/SERK3 dependent manner. However, recently it was reported that unlike all known oomycetes PAMPs, perception of Pep-13 is not implicated through BAK1/SERK3 (Wang H. et al., 2019). Pep-13 has been shown to bind to a 100 kD monomeric integral plasma membrane receptor of parsley (Nennstiel et al., 1998; Blume et al., 2000) but its cognate PRR in other plant species is not yet known.
Although known for a long time, the polysaccharide PAMP, β-glucans are far less explored in terms of their host recognition. The PRRs binding β-glucans and the signaling mechanisms involved in β-glucans incited PTI are still unknown. A putative PRR named, GEBP (glucan elicitor binding protein) present on the plasma membrane of soybean was shown to bind P. sojae-derived β-1,6-glucan and transduce signals that results in accumulation of phytoalexins (Umemoto et al., 1997). After this finding, many studies were conducted to find β-glucans receptors in other plants. However, these findings are considered controversial because these studies were conducted on β-glucans derived through acidic hydrolysis in vitro and currently there is no evidence regarding the binding of plant derived β-glucans to these receptors (Fesel and Zuccaro, 2016). Recently, CERK1, an Arabidopsis fungal chitin receptor was reported to bind with synthetic β-1,3-glucan oligosaccharides that are present in both oomycete and fungi. Just like chitin perception via CERK1 (Wang H. et al., 2019), BAK1 was not found involved in this β-1,3-glucan-CERK1 induced PTI responses. Also, the cerk1 mutants showed higher susceptibility to a biotrophic oomycete H. arabidopsidis (Mélida et al., 2018). Thus, providing some bases that CERK1 could be a putative PRR for Phytophthora spp. derived β-glucans and their recognition could be independent of BAK1.
EPs are among the earliest discovered elicitors, defined almost 40 years ago. Until now, nothing is known about the cognate PRRs and role of any other member of the PAMP perception model in the recognition of EPs in the host.
Signaling Mechanisms Involved in Shaping PTI
Upon initial perception of a PAMP by PRR, stereotypic signaling events are initiated, leading to the activation of an array of defense components that collectively formulate PTI (Figure 1). Generally characterized PAMPs induced defense responses include reactive oxygen species (ROS) burst, callose deposition, expression of defense genes and accumulation of antimicrobial secondary metabolites (Larroque et al., 2013). Most of this information is based on the poster child pathosystem, P. syringae-Arabidopsis interaction. Signaling pathways involved in PTI defense responses induced by Phytophthora spp. pathogens are highly fragmented. In the following paragraphs, we have compiled the available information, consistencies and deviations from the general signaling mechanisms considered to be involved in PTI.
Calcium Signaling
Calcium signaling is considered as an integral part of receptor mediated perception of most PAMPs of diverse pathogens. Recent studies provide evidence of a complex cross talk between calcium and MAPK signaling pathways. Ca2+ ion influx is one of the earliest events that has been reported to trigger within 30 s of PAMP perception. This Ca2+ influx opens up the membrane transporters for the influx of H+ and efflux of K+, Cl−, and that leads to changes in extracellular pH and depolarization of plasma membrane within 1 to 3 min of PAMP perception (Bigeard et al., 2015; Westphal et al., 2019).
PAMP induced Ca2+ ion homeostasis in the host has been tested for only a few Phytophthora spp. PAMPs mainly because of the experimental limitations. Pep-13 application on transgenic parsley lines expressing aequorin (a Ca2+ sensor) represented a biphasic response (Blume et al., 2000). Cytosolic Ca2+ concentration was observed to increase after 30 to 40 s of Pep-13 treatment that rapidly peaked at 2 min, and subsequently started decreasing slowly during the next 10 to 40 min to a stable concentration that was almost three times more than the basal levels. The extent of induction in Ca2+ influx was found positively correlated with the Pep-13 concentration. Decreased Pep-13 concentration preferentially reduced the transient Ca2+ peak, but the sustained increases of cytosolic Ca2+ was elicited even at very low Pep-13 concentrations (Blume et al., 2000). Pep-13 has also been reported to induce K+ and Cl− efflux, extracellular alkalinization, activation of MAPK and ultimately generation of antimicrobial phytoalexins (Nennstiel et al., 1998; Blume et al., 2000). Elicitins have been reported to trigger calcium and protein kinase mediated signaling leading to the depolarization of plasma membrane, cell wall modifications, protein phosphorylation, potassium (K+) and chloride (Cl−) efflux and phenylpropanoid metabolism (Bourque et al., 1998; Lochman et al., 2005; Amelot et al., 2011). Induction in nitrate () efflux through anion channels regulated by Ca2+ influx and phosphorylation events upstream to cryptogein induced oxidative burst is also reported during cryptogein-tobacco interactions (Wendehenne et al., 2002).
Reactive Oxygen Species (ROS)
Generation of reactive oxygen species (ROS) is incited by all discovered Phytophthora spp. PAMPs. Extracellular ROS burst is reported to occur within 2 to 3 min after PAMP perception. The respiratory burst oxidase homolog D (RBOHD), a plasma membrane localized NADPH oxidase, mediates ROS induction upon PAMP perception (Bigeard et al., 2015). Regulation of RBOHD during PTI responses has been reported to occur in both Ca2+-dependent and Ca2+independent manner (Saijo et al., 2018). RBOHD has been reported as an integral component of plasma membrane immune receptor complex involved in antibacterial immunity. It has been shown to exist in complex with both FLS2 and EFR, the most well-studied PRRs of bacterial PAMPs, flagellin, and elongation factor, respectively. The plasma membrane associated kinase BIK1 is a direct substrate of this PRR complex. Upon perception of PAMPs, BIK1 is activated to phosphorylate RBOHD to initiate ROS production in a Ca2+-independent manner. Ca2+ can directly regulate RBOHD by binding to its N-terminal EF-hand motifs to stimulate ROS production. ROS has been shown to increase cytosolic Ca2+ concentrations as well as Ca2+-dependent protein kinases (CDPKs) dependent phosphorylation of RBOHD (Kadota et al., 2014; Bigeard et al., 2015). RBOHD association with known PRRs of oomycete and fungal PAMPs has not been reported yet. However, RBOHD has been shown to be involved in CBEL induced PTI responses (except necrosis) that are regulated in BAK1-dependent manner. Both RBOHD and BAK1 have been shown essential in developing Arabidopsis root resistance against P. parasitica (Larroque et al., 2013). On the other hand, BAK1-independent PTI responses induced upon fungal β-1,3-glucan perception by CERK1 did not require RBOHD to induce PTI responses including ROS burst (Mélida et al., 2018), and the same could be speculated for oomycetes β-glucans. In N. benthamiana RBOHA and RBOHB were reported to participate in resistance against P. infestans (Yoshioka et al., 2003). Silencing of these genes resulted in enhanced susceptibility of N. benthamiana to non-virulent P. infestans.
PAMP-Induced Hypersensitive Response (HR)
Activation of pathogenesis-related (PR) proteins and other defense components leading to the induction of HR has also been reported. PR proteins respond to cryptogein either in calcium dependent or independent manner (Lebrun-Garcia et al., 1998; Lochman et al., 2005; Dahan et al., 2009). Hoeberichts et al. investigated the effect of light during cryptogein-tobacco interaction and reported that cryptogein induced transcriptional changes are immensely affected by photoperiod and that the HR response is significantly delayed under light (Hoeberichts et al., 2013). Just like cryptogein, INF1 is shown to induce HR through calcium and protein kinase signaling mechanisms. Induction of INF1 induced HR is shown to be independently regulated by calcium and protein kinase signaling pathways. Oxidative burst is incited but not necessarily required for HR. However, it is reported to be involved in both calcium and protein kinase signaling pathways (Sasabe et al., 2000). MAPK interacting cytosolic proteins HSP70 and HSP90 are reported to be the essential components of INF1 induced HR (Kanzaki et al., 2003).
Plant lipoxygenases (LOXs) have also been reported to regulate PAMP induced HR particularly during incompatible host-pathogen interactions. LOXs are the enzymes that catalyze the oxygenation of polyunsaturated fatty acids to produce hydroperoxides that serve as precursors of different biologically active compounds with distinct physiological functions as well as in plant defense responses. Plant LOX precursors can be divided into two major classes; 9-LOXs and 13-LOXs according to the position on the carbon chain at which oxygen is incorporated into linoleic acid or linolenic acid, the lipid substrates for LOX catalysis in plants (Ricker and Bostock, 1994; Kolomiets et al., 2001). Distinct LOX mediated products have been suggested to be involved in plant-pathogen interactions in different ways; 9-LOXs are speculated to cause oxidative damage to plant membranes during HR and serve as direct antimicrobial agents whereas, 13-LOXs are reported to act as signaling molecules in Jasmonic acid (JA) metabolism (Slusarenko et al., 1993; Hwang and Hwang, 2010).
Discovery of lipid PAMPs, EPs as Phytophthora spp. elicitors first led to the hypothesis about the role of lipid metabolism in early stages of plant-pathogens interactions (Véronési et al., 1996). Since then, LOX activity has been widely monitored during plant-pathogen interactions and is reported to be positively correlated with plant resistance against pathogens of different lineages (Véronési et al., 1996; Kolomiets et al., 2001; Hwang and Hwang, 2010; Afroz et al., 2013). Phytophthora spp. pathogens and even their non-lipid PAMPs have been reported to enhance LOX activity in the host cells (Véronési et al., 1996). The Ca2+dependent TGase PAMP, Pep-13 showed enhanced LOX expression in potato cells. CBEL treated tobacco cell suspension and the whole plant, both showed up to 2.5-folds increase in LOX activity (Séjalon et al., 1995). Although known for a long time the pathway behind LOX related plant defense responses is not defined, except some fragmented clues generated from different pathosystem studies. The 9-LOX hydroperoxides generation upon cryptogein treatment has been reported as a crucial element for HR in tobacco (Rustérucci et al., 1999).
PAMP-Induced Systemic Acquired Resistance (SAR)
In addition to localized resistance, various Phytophthora spp. PAMPs are shown to trigger the transport of defense signals throughout the plant that results in broad-spectrum disease resistance against secondary infections. This spread of resistance across the plant is known as systemic acquired resistance (SAR) (Gao et al., 2015). For example, EPs can induce SAR in different plant species, which is effective against subsequent infection by diverse pathogens (Savchenko et al., 2010; Robinson and Bostock, 2015). In tobacco, treatment of lower leaves with AA was shown to induce SAR against TMV in the upper leaves of the plants. Transgenic Arabidopsis expressing EPs showed broad spectrum resistance against diverse pathogens including P. capsici (oomycete), B. cinerea (fungi), and aphids (insect), but were highly susceptible to Pseudomonas syringae pv tomato (bacteria). Increased transcription of JA related genes as well as enhanced JA levels were observed in these EP plants, whereas salicylic acid (SA) mediated signaling was found compromised (Savchenko et al., 2010). These findings are consistent with the general assumption that SA and JA signaling pathways are antagonistic to each other. In contrast, both SA and JA pathways were found to be involved in the induction of Pep-13 mediated SAR in potato (Halim et al., 2009). In tomato, INF1 infiltrated in the middle of fully expanded leaves was found to activate SAR against Ralstonia solanacearum in the distal parts of the challenged leaves (Kawamura et al., 2009). Jasmonic acid (JA) and ethylene (ET) signaling (but not SA) pathways were found in this SAR development. Conversely, cryptogein induced SAR in tobacco was SA mediated, which was found effective against P. parasitica var nicotianae (Wendehenne et al., 2002). OPEL-induced SAR in systemic leaves was found effective against cross kingdom pathogens like, TMV (virus), Ralstonia solanacearum (bacteria), and P. parasitica. Enhanced expression of several SA-responsive genes and lipid transfer protein DIR1 was observed in tobacco plants in response to OPEL treatment that suggests SA and/or DIR1-mediated SAR. Further investigation is required to confirm the involvement of these signaling molecules in OPEL-induced SAR (Chang et al., 2015). XEG1 induced SAR in soybean and N. benthamiana was found effective against P. sojae and P. parasitica, respectively (Ma et al., 2015). The signaling mechanisms behind the development of this SAR are not investigated yet. NLPs are generally characterized as ethylene (ET) inducing proteins (Böhm et al., 2014). Treatment of Arabidopsis with nlp20 protected the plant from subsequent infection with virulent isolates of Pseudomonas syringae pv. tomato, Botrytis cinerea, or H. arabidopsidis (Albert et al., 2015). Enhanced expression of SA related genes has been found in Arabidopsis upon treatment with HaNLP3 that is nlp20 homolog in a non-Phytophthora oomycete H. arabidopsidis (Oome et al., 2014). Specific signaling mechanisms behind nlp20 induced SAR needs further investigations. CBEL induced SAR was not specifically tested but unlike other PAMPs that activate selective phytohormones, CBEL has been shown to induce all three hormone signaling pathways; SA, JA, and ET that are differentially involved in the induction of necrosis and other defense responses in Arabidopsis (Khatib et al., 2004). Despite being one of the earliest discovered elicitors, signaling mechanisms behind β-glucan induced PTI are still not known. Crude β-glucan (Kopp et al., 1989) preparations from P. sojae and P. infestans, elicited systemic protection against different viruses in soybean and various Nicotianae spp. (Kopp et al., 1989). Application of different β-glucan preparations from P. sojae and brown alga Laminaria digitate showed accumulation of PR genes and SA in tobacco leaves. SAR was only tested for laminarin from L. digitate, which was found effective against soft rot bacterium Erwinia carotovora (Klarzynski et al., 2000).
Conclusively, upon recognition by cognate plant PRR, Phytophthora spp. PAMPs/elicitors can trigger PTI through crosstalk among several interlinked and independent defense signaling components. Point to be noted is, HR and SAR are generally considered as ETI responses but as discussed above Phytophthora PAMPs are reported to induce both localized (HR) and systemic (SAR) resistance that can protect the plant against diverse range of pathogens.
Effector Triggered Susceptibility (ETS)
In order to establish infection, successful Phytophthora pathogens have devised ways to either escape recognition or circumvent host immunity (Figure 2). In accordance with the zigzag model, Phytophthora spp. carry myriads of effector proteins that are secreted inside the host cells to modulate the PTI and to trigger ETS (Jones and Dangl, 2006). P. parasitica carries INF1 like elicitin called ParA1 that is recognized by tobacco to induce defense (Kamoun et al., 1993). It has been found that virulent strains of P. parasitica can bypass this recognition by suppressing ParA1 expression during their interaction with compatible hosts, and this adaptation enables it to cause black shank disease in tobacco (Colas et al., 2001). Since sequence divergence was not observed in ParA1 gene, this expression modulation could be the result of the effector activities during infection. The role of P. parasitica effectors in ParA1 downregulation is not investigated, but several RXLR effectors from different Phytophthora spp. had been reported to suppress INF1-induced cell death (Bos et al., 2006; Gilroy et al., 2011b; Wang et al., 2011; He et al., 2018; Murphy et al., 2018).
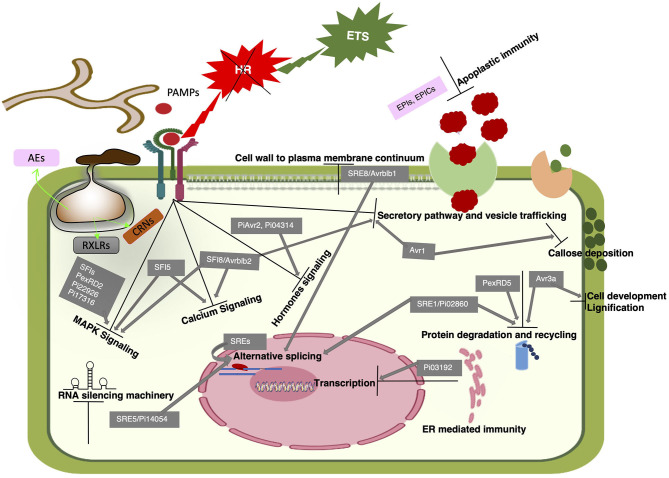
Figure 2.
A proposed model of PTI to ETS transition. Phytophthora spp. deploy multiple effectors to target various components of the host's signaling pathways, metabolic and physiological machinery to promote infection. Here we have only shown P. infestans effectors known to modulate defense responses in favor of infection. An immune component is intervened at different levels by multiple effectors acting at different locations. For example, apoplastic immunity is modulated by the action of pathogen's protease inhibitors (EPIs, EPICs) against host's proteases in the apoplast and RXLR (Avr1, Avrblb2) effectors operate intracellularly to prevent the secretion of host's proteases into the apoplast. Some effectors target multiple host proteins to interfere with different signaling pathways and also one signaling component is targeted by several effectors. For example, SFI8 interferes with calcium signaling, inhibits protease secretion into the apoplast and interferes with MAPK signaling. MAPK signaling is targeted by 11 RXLR effectors including eight SFIs. Alternative splicing is modulated by nine SREs and, SRE 1, 5, and 8 are known to target proteolysis, RNAi and cell wall to plasma membrane continuum, respectively. Most of these effectors (details in Table 2) have shown to overcome INF1 induced HR thus, indicate PTI suppression in various ways to establish ETS.
Phytophthora spp. effectors can be broadly categorized into two classes with respect to their localization: apoplastic effectors that are secreted in the apoplast of the host cells and cytoplasmic effectors that are translocated inside the host cells. Apoplastic effectors are diverse hydrolytic enzymes like glycoside hydrolases (GHs), pectinases, proteases, and protease inhibitors (Mcgowan and Fitzpatrick, 2017). Based on conserved motifs, cytoplasmic effectors of Phytophthora spp. are categorized in two groups, the RXLR effectors with RxLR dEER (Arg, any amino acid, Leu, Arg) motif and crinklers (CRNs) having LxFLAK motifs. Phytophthora spp. carry 300-700 RXLR effectors with almost no sequence similarity to known proteins (Guo et al., 2019). The characteristic N-terminal RXLR motif is implicated in the translocation of effectors inside host cells.The C-terminal portion of RxLR effectors, which has different amino acid sequences, carry the PTI-suppressing and disease promoting effector activity in susceptible plants and the R-activating avirulence activity in resistant plants (Figures 2, 3 and Table 2) (Birch et al., 2008; Bozkurt et al., 2012; Anderson et al., 2015).
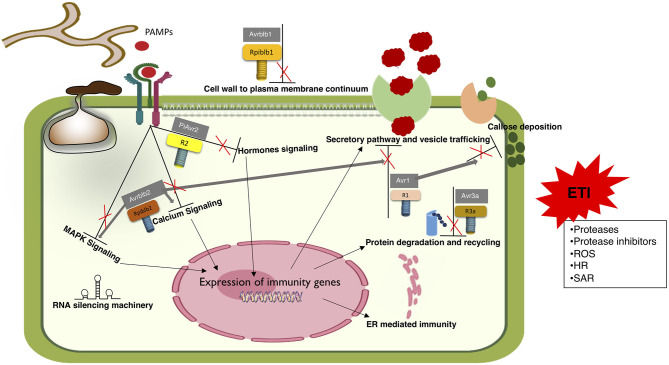
Figure 3.
R genes mediated transition of ETS to ETI. Proposed model depicting the activation of ETI through recognition of effectors by cognate R genes. Although numerous effectors are known to establish ETS by modulating all known defense responses, little is known in terms of ETI. Very few R genes are known to identify their cognate effectors. Mechanisms behind this recognition are also scarcely known. In agreement to the zigzag model, ETI responses are just prolonged and robust PTI responses.
Table 2.
Phytophthora spp. effectors that modulate host defense responses to establish ETS or trigger ETI.
| Phytophthora spp. | Host species | Effector | Host target | Specific elicitor induced cell death suppression | Virulence function to establish ETS | ETI and R gene | References |
|---|---|---|---|---|---|---|---|
| P. infestans | N. benthamiana | Avr1 | Sec5 Exocyst subunit | CRN2 | Prevent PR-1 secretion and callose deposition | YesR1 | Du Y. et al., 2015; Du et al., 2018 |
| Avrblb2/SFI8 | C14 Protease | Prevent C14 secretion | YesRpiblb2 | Bozkurt et al., 2011 | |||
| Tomato CaMs tested in N. benthamiana | CaMs | Interferes with calcium signaling to regulate ETI | Naveed et al., 2019 | ||||
| Tomato Arabidopsis | Unknown | Interferes with MAPK signaling | Zheng et al., 2014 | ||||
| SFI1, SFI2, SFI5, SFI6, SFI7 | |||||||
| Potato | SFI3 | UBK | He et al., 2019 | ||||
| SFI5 | CaMs | Interferes with calcium signaling to suppress PTI | Zheng et al., 2018 | ||||
| Potato N. benthamiana | PexRD2 | MAP3Kε | INF1 | Interferes with MAPK signalig | King et al., 2014 | ||
| Pi22926 | MAP3Kβ2 | Avr4/Cf4 and AvrPto/Pto associated HR | Ren et al., 2019 | ||||
| Arabidopsis N. benthamiana | Avrblb1/IPI-O1/SRE8 | LecRK-1.9 Lectin receptor kinase | Disrupt cell wall to plasma membrane continuum | YesRpi-blb1 | Bouwmeester et al., 2011 | ||
| Potato N. benthamiana | Pi03192 | NAC transcription factors | Prevent NAC localization from ER to nucleus | No | Mclellan et al., 2013 | ||
| Tobacco N. benthamiana | Avr3a | CMPG1 E3 ligase | INF1 | Prevent ubiquitin-dependent proteolysis | YesR3a | Bos et al., 2006; Gilroy et al., 2011b | |
| PexRD54 | ATG8CL autophagy protein | Modulate autophagy mediated defense | Dagdas et al., 2016, 2018 | ||||
| N. benthamiana | SRE3 | U1-70K | Modulate Alternative splicing | Huang et al., 2020 | |||
| Pi14054/SRE5 | Unknown | Suppressor of RNA silencing | Vetukuri et al., 2017 | ||||
| Potato | PITG_15718.2 | Unknown | Attenuate plant defense and growth | Wang J. et al., 2019 | |||
| S-factors | |||||||
| Arabidopsis N. benthamiana | PiAvr3a | CAD7 | Suppress lignification | Li T. et al., 2019 | |||
| Potato N. benthamiana | Pi02860 | NRL1 E3 ligase | INF1 | Enhance proteasome-mediated degradation of SWAP70 | He et al., 2018 | ||
| N. benthamiana | Pi04089/SRE2 | KRBP1 RNA-binding protein | Possibly affects post translational modifications | Wang et al., 2015 | |||
| Potato N. benthamiana | Pi17316 | StVIK (MAP3K) | INF1 | Interferes with MAPK signaling | Murphy et al., 2018 | ||
| Potato N. benthamiana | PiAvr2 | BSL family | INF1 | Interferes with brassinosteroid (BR) hormone signaling | YesR2 | Gilroy et al., 2011a; Turnbull et al., 2017, 2019 | |
| Potato N. benthamiana | Pi04314 | PP1cs | Modulate JA and SA signaling | Boevink et al., 2016b | |||
| P. sojae | soybean | Avh240 | AP1 Protease | Prevent AP1 secretion | Guo et al., 2019 | ||
| PsAvh238 | ACSs | Suppress ET biosynthesis | Yang et al., 2019 | ||||
| PSR1 and PSR2 | PINP1 RNA helicase | Suppress RNA silencing | Qiao et al., 2013, 2015 | ||||
| Arabidopsis and N. benthamiana | Avh331 | INF1 | Interferes with MAPK signaling | Cheng et al., 2012 | |||
| Arabidopsis | PsCRN63 (crinkler) | flg22 | Li et al., 2016 | ||||
| N. benthamiana | PsAvh262 | BiPs (S-factors) | Suppress ER mediated immunity | Jing et al., 2016 | |||
| P. capsici | Arabidopsis | Avr3a12 | FKBP15-2 | Inhibits ER stress sensing | Fan et al., 2018 | ||
| PcAvh103 | EDS1 | Suppress SA mediated defense | Li et al., 2020 | ||||
| RxLR48 | NPR1 | Li T. et al., 2019 | |||||
| PSR2 | RNA binding protein DRB4 | Modulates RNA silencing | Hou et al., 2019 | ||||
| RxLR207 | ACD11, BPA1 and BPLs (S-factors) | Supress ROS-mediated defense | Li T. et al., 2019 | ||||
| N. benthamiana | Avh1 | PP2Aa scaffolding subunit | Attenuate plant defense and growth | Chen et al., 2019 | |||
The blurriness of effector vs. PAMP terms is found more in case of apoplastic effectors (described in further sections). Cytoplasmic effectors on the other hand show less conflict of being designated effectors. In general, in contrast to cytoplasmic effectors, apoplastic effectors tend to behave either like PAMPs or effectors. In literature, some authors referred to CRNs as PAMPs, whereas others as effectors (Torto et al., 2003). Torto et al. discussed the possibility of CRNs as Phytophthora spp. PAMPs based on some observations pointing toward the similarities of CRNs with the then well-known PAMP, INF1 but the authors have clearly stated, “whether the CRN proteins function as PAMPs remains to be determined” (Torto et al., 2003). Based upon the existing evidence, CRNs fit well in the effector category (Wang et al., 2018; Maximo et al., 2019).
Identification of the host's targets hijacked by the pathogen's effectors and characterizing their role in the infection development and defense manipulation has immensely contributed to our understanding of the molecular basis of plant-pathogen interactions. Based on current research, it has been found that effectors from diverse pathogens target conserved cellular processes across diverse plant families. Following the norm, Phytophthora spp. effectors have also been found to target cellular processes targeted by bacterial and fungal pathogens across different plant hosts (Toruño et al., 2016). Immune suppression by Phytophthora pathogens can be achieved in two ways: (1) by inhibiting the activity of positive regulators of immunity or (2) by supporting the function of negative immune regulators. Till now, all the apoplastic effectors have been found targeting the positive immune regulators, whereas cytoplasmic effectors particularly, the RXLR effectors have been found targeting both the positive and negative regulators of defense involved in different cellular processes at various subcellular locations.
Phytophthora spp. Effectors Targeting Positive Immune Regulators
Modulation of Apoplastic Immunity by the Apoplastic Effectors
Plant's apoplast serves as a battleground for plant-pathogen warfare. Either as preformed defense or upon PTI activation, plants secret several diverse catalytic proteins to the apoplast against attackers. Pathogens deploy a wide variety of apoplastic effectors that compromise host defense proteins as well as cellular components to facilitate pathogen colonization and penetration into the host cells. As described above, many of these Phytophthora spp. apoplastic effectors are considered as PAMPs because they are conserved across the genus. For example, PsXEG1, a glycoside hydrolase of P. sojae, is secreted in the host's apoplast to damage the cell wall by reacting with specific glucans. A very interesting defense and counter defense strategy in the soybean-P.sojae system was discovered recently. A soybean glucanase inhibitor protein, the GmGIP1, has been shown to bind PsXEG1 in the apoplast and block its virulence activity. To counter this defense, P. sojae deploys a paralog of PsXEG1 named as PsXLP1(PsXEG1-like protein). PsXLP1 lacks endoglucanase activity but it can bind to GmGIP1 with far more affinity than PsXEG1, thus protecting PsXEG1 from inhibitory effect of GmGIP1 and freeing it for its virulence activity. Similar role of XEG1-XLP1 has been found to contribute to P. parasitica virulence in N. benthamiana. Many other Phytophthora spp. have been reported to carry this gene pair suggesting that XLP1 deployment to guard XEG1 against apoplastic glucanase inhibitors could be a conserved counter defense strategy across the genera (Ma et al., 2017). On the contrary to the above example where soybean produces glucanase inhibitor (GmGIP1) specifically against P. sojae's glucanase (XEG1), P. sojae was also found to secrete a specific glucanase inhibitor effector, PsGIP1, to inhibit host's endo-β-1,3-glucanases to prevent the degradation of β-1,3/1,6-glucans in the pathogen's cell wall and thus avert the release of defense-eliciting oligosaccharides (Damasceno et al., 2008).
Phytophthora spp. have also been reported to carry specific extracellular protease inhibitors (EPIs) against host's protective proteases in the apoplast. Different Phytophthora spp. protease inhibitors have also been found targeting different classes of secreted host proteases primarily subtilisin-like serine proteases (Figueiredo et al., 2018) and papain-like cysteine proteases (PLCPs) (Misas-Villamil et al., 2016). P. infestans secrets Kazal family serine protease inhibitors, EPI1 and EPI10, into the host's apoplasts where they interact with P69B, a tomato subtilisin-like serine protease (PR-7 family) secreted to the host's apoplast (Tian et al., 2004, 2005). Another class of P. infestans effectors is cystatin-like protease inhibitors, EPIC1 to EPIC4. Among these, EPIC1 and EPIC12B can interact and interfere with different host papain-like cysteine proteases (PLCPs). Tomato plants carry seven PLCPs, three out of these seven; C14, and Phytophthora-inhibited protease 1 (Pip1) that is also classified as PR protein closely related to Rcr3pim (Kaschani et al., 2010) have been found to interact and inhibited by both EPIC1 and EPIC12B in the apoplast of the host. These three PLCPs, C14, Rcr3pim, and Pip1 (3-PLCPs) are also targeted by fungal and bacterial effectors (Kaschani et al., 2010; Shindo et al., 2016), and thus can be speculated as hubs of apoplastic immunity targeted by pathogens of diverse lineages.
Modulation of Vesicle Trafficking and Secretion
In addition to modulation of apoplastic immunity by the apoplastic protease inhibitor effectors in the apoplast, some RXLR effectors are reported to interfere with plant secretory pathway and vesicle-trafficking intracellularly, to suppress the secretion of proteases and other antimicrobial compounds to the apoplast. For example, a P. infestans RXLR effector, Avr1 is reported to interact with Sec5, a subunit of exocyst complex to hijack exocytosis. Avr1 was found to be localized with Sec5-associated mobile bodies around perihaustorial membrane in the host cell (Wang et al., 2018), most probably to stop it from becoming a part of the exocyst complex. Sec5 has been found as a required component for PTI, and Avr1-Sec5 interaction is suggested to interfere with PR-1 secretion as well as callose deposition, thus attenuating host defense against P. infestans (Du Y. et al., 2015). Another highly conserved effector, RxLR24 is reported to interact with the members of RABA GTPase family putatively, involved in vesicular secretion of core antimicrobials like PR-1 and defensin (PDF1.2) to promote infection (Tomczynska et al., 2018). The P. infestans RXLR effector, PiAvrblb2 (PITG_04090), interacts with C14 protease at the plasma membrane and blocks its secretion into the apoplast of host leaves (Bozkurt et al., 2011). Similarly, another RXLR effector, PsAvh240, has been shown to interact with an aspartic protease, GmAP1, at the soybean plasma membrane to inhibit its secretion into the apoplast and thereby suppressing the apoplastic defense against P. sojae (Guo et al., 2019).
Modulation of Cell-to-Cell Trafficking
In higher plants, cells are connected through symplastic cell-to-cell channels called plasmodesmata (PD) that enable intercellular trafficking of nutrients and signaling molecules. This intercellular transport is regulated by adjusting the size exclusion limit (SEL) of PD that mainly depends upon callose depositions around the PD openings (Tomczynska et al., 2020). Callose deposition upon PAMP perception is crucial for host defense against Phytophthora pathogens (Chang et al., 2015; Du Y. et al., 2015; van den Berg et al., 2018). PD mediated cell-to-cell movement of P. ramorum hyphae was reported previously (Giesbrecht et al., 2011). A P. brassicae RXLR effector, RxLR3 is shown to inhibit callose deposition at PD by targeting host's callose synthases (CalS) to promote cell-to-cell conductivity that resulted in enhanced infection (Tomczynska et al., 2020). These findings suggest that Phytophthora spp. can modulate PD openings to physically spread across neighboring cells during infection.
Modulation of Host's Cell Wall to Plasma Membrane (CW-PM) Continuum
Another strategy used by filamentous pathogens to interfere with plant defense is the disruption of cell wall to plasma membrane (CW-PM) continuum in hosts. Adhesion of plant cell wall and plasma membrane is critical for cell wall sensory signaling and induction of several PTI responses like callose deposition, ROS burst and cell death. Peptides containing RGD (R: arginine; G: glycine; D: aspartic acid) motif are well-known to disrupt CW-PM adhesions in plants (Mellersh and Heath, 2001). Avrblb1 (IPI-O1), a P. infestans RXLR effector that also contain an RGD motif has been reported to disrupt CW-PM adhesion by targeting a plasma membrane associated lectin receptor kinase, LecRK-I.9. Loss of function mutants of LecRK-I.9 showed enhanced susceptibility to different Phytophthora pathogens (Bouwmeester et al., 2011).
Modulation of Endoplasmic Reticulum (ER)Mediated Immunity
Endoplasmic reticulum (ER) quality control system is the critical component involved in shaping PTI responses. Several RXLR effectors have been found to suppress this ER mediated immunity in multiple ways. For example, a P. infestans RXLR effector Pi03192 targets two N. benthamiana NAC transcription factors at ER membrane and prevent their localization to the nucleus thus, resulting in increased host susceptibility to P. infestans by preventing transcriptional regulation of defense components. These NAC TFs are induced by P. infestans but not in response to bacterial flg22 indicating their specificity in PTI induced by PAMP of diverse pathogens (Mclellan et al., 2013). ER stress sensing is a crucial component of ER-mediated plant immunity. Recently, a P. capsici RXLR effector PcAvr3a12 (Avr3a class), was found to interact with an ER stress sensing protein FKBP15-2 encoding a peptidyl-prolyl cis-trans isomerase (PPIase). This PcAvr3a12- FKBP15-2 interaction inhibits FKBP15-2 stress sensing activity thus, attenuating ER mediated defense response against P. capsici (Fan et al., 2018).
Modulation of Host's Alternative pre-mRNA Splicing by Phytophthora Pathogens
Alternative splicing (AS) of pre-mRNA to generate multiple transcripts of a gene is an essential post-transcriptional process that regulates numerous physiological processes including plant immunity. Recently, Huang et al. have put forth AS as an independent layer of host gene regulation in response to pathogens, and P. infestans has been shown to reprogram the AS of host's pre-mRNAs in favor of infection, putatively, by enhancing the production of host's transcripts that promote disease as well as by antagonizing the generation of core host defense transcripts (Huang et al., 2020). For example, PKFP gene that encodes a protein kinase, undergoes AS to generate PKFP.1, a functional isoform that positively regulates plant immunity and PKFP.2, a non-function isoform. P. infestans has been found to suppress the generation of PKFP.1 to enhance host's susceptibility. Like other host's physiological processes, modulation in AS by P. infestans is implemented by deploying effectors and nine RXLR effectors are presented as splicing regulatory effectors (SREs 1-9). Three of these SREs (3,6, and 7) were shown to bind with core host splicing factors (SFs). SRE3 was shown to modulate host immunity by physically interacting with U1-70K, a core spliceosomal component (Huang et al., 2020). Some of these SREs have been previously reported to target different host's proteins to modulate other physiological processes. For example, SRE1 (Pi02860) was reported to interact with NRL1 to modulate ubiquitin-mediated proteolysis (He et al., 2018), SRE2 (Pi04089) binds with KRBP1, which is presumed to contribute to AS, SRE5 (Pi14054) is a suppressor of host's RNA silencing (Vetukuri et al., 2017) and SRE8 (Pi21388/IPI-O1/Avrblb1) is reported to disrupt host's CW-PM continuum (Bouwmeester et al., 2011). Since, AS regulates almost all physiological responses, deployment of multiple effectors might be the pathogen's strategy to reprogram the host's protein profiling in pathogen's favor.
Modulation of RNA Interference Mediated Immunity
Host's RNA interference (RNAi) is well-known for antiviral defense, but its implications in defense against eukaryotic phytopathogens have just begun to unfold. Viral pathogens carry suppressors of RNA silencing to counter host's RNA silencing for successful infection. In 2013, Qiao et al. reported two P. sojae RXLR effectors, termed as Phytophthora Suppressor of RNA Silencing 1 and 2 (PSR1 and PSR2) that enhanced plant susceptibility by suppressing RNA silencing machinery (Qiao et al., 2013). PSR1 is reported to target a host RNA helicase PINP1 (PSR1-Interacting Protein 1) that regulates the accumulation of endogenous small interfering RNAs (siRNAs) and microRNAs in Arabidopsis (Qiao et al., 2015). Recently, Hou et al. (2019) presented an example of trans-kingdom RNAi during Phytophthora-Arabidopsis interaction. They have shown the induction of endogenous secondary siRNAs in Arabidopsis in response to P. capsici. Some of these siRNAs were found to target P. capsici transcripts for potential gene silencing to mitigate infection. PSR2 was found to counter this host induced pathogen's gene silencing by associating with a core component of host's RNA silencing machinery, a double stranded RNA binding protein (DRB4) that is known to be involved in the biogenesis of secondary siRNAs (Hou et al., 2019). A P. infestans effector, Pi14054, has been shown to act as a suppressor of RNA silencing in N. benthamiana (Vetukuri et al., 2017). These findings suggest that Phytophthora spp. deploy effectors to modulate cross-kingdom RNAi mechanisms, which play a role in host defense.
Modulation of Proteolysis and Autophagy Mediated Defense
Protein degradation and recycling either through proteasome or autophagy has been shown to play key roles in plant immunity (Üstün et al., 2016; Hofius et al., 2017). E3 ligases are known to ubiquitylate protein substrates for ubiquitin-dependent proteolysis by the 26S proteasome (Furniss et al., 2018; Serrano et al., 2018). A P. infestans RXLR effector Avr3a suppresses INF1 induced PTI by interacting and stabilizing the U-box E3 ligase CMPG1 that functions in protein degradation and is essential component of INF1 induced cell death (Bos et al., 2006; Gilroy et al., 2011b). Recently, P. infestans is reported to influence the alternative splicing of some core components of ubiquitin-dependent proteolysis. SP1 gene encodes an E3 ubiquitin protein ligase that undergoes AS to generate SP1.1, a functional isoform that is a positive regulator of plant immunity, and SP1.2, a non-function isoform. P. infestans was found to suppress the generation of SP1.1 to enhance host's susceptibility (Huang et al., 2020).
Autophagy has been shown to play a crucial role in shaping HR (Hofius et al., 2017). A P. infestans RXLR effector, PexRD54 binds to host autophagy protein ATG8CL to interfere with the formation of autophagosomes. This PexRD54-ATG8CL binding outcompetes ATG8CL binding with an autophagy cargo receptor Joka2 that plays a positive role in defense (Dagdas et al., 2016). They have shown that during P. infestans infection, defense-related autophagosomes labeled by ATG8CL-Joka2 are diverted toward perihaustorial membranes, where PexRD54 also localize with these autophagosomes. Moreover, overexpression of ATG9, the protein involved in early autophagosome biogenesis, enhances plant defense (Dagdas et al., 2018). These findings reveal complex modulations in the host's proteolysis and recycling mechanisms during the Phytophthora-plant interactions.
Modulation of MAPK and Calcium Signaling Pathways by RXLR Effectors
Recent investigations have shown that Phytophthora spp. modulate MAPK and calcium signaling pathways by deploying specific effectors that target core components of these signaling pathways (Figure 2). The MAPK signaling plays important roles in both PTI as well as ETI. The MAPK signaling includes three protein kinases: MAP kinase (MAPK), MAP kinase kinase (MAP2K) and MAP kinase kinase kinase (MAP3K). In a typical MAPK signaling cascade, the MAPKs are phosphorylated by MAP2Ks, which themselves are phosphorylated by MAP3Ks (Saijo et al., 2018). Recently, multiple P. infestans RXLR effectors have been shown to modulate immunity by suppressing MAPK signaling either by directly interacting with different MAP3Ks components or by targeting the upstream players of MAPK pathways (Ren et al., 2019).
For example, a P. infestans RXLR effector PexRD2 binds with the kinase domain of MAP3Kε in the cytosol to interrupt the associated MAPK signaling (King et al., 2014). Overexpression of MAP3Kε results in enhanced phosphorylation of MAPKs and also induce cell death in the plant tissues. PexRD2 is shown to inhibit both MAP3Kε triggered cell death and MAPKs phosphorylation, whereas it was unable to suppress INF1 mediated HR. The HR associated with the recognition of Cladosporium fulvum effector Avr4 mediated by the tomato Cf4 protein and the P. syringe effector AvrPto by Pto/Prf proteins are known to be MAP3Kε dependent. PexRD2 suppressed both these Avr4/Cf4 and AvrPto/Pto associated HR. On the contrary, it was unable to suppress the R proteins mediated HR triggered by diverse P. infestans effectors, AVR3a (RXLR) and CRN8 (crinkler), providing evidence that HR associated with AVR3a/R3a and CRN8/R protein is independent of MAP3Kε (King et al., 2014). Recently, Ren et al. reported another P. infestans RXLR effector, Pi22926 that interacts with a different MAP3K of potato, StMAP3Kβ2 in the nucleoplasm. Just like PexRD2, Pi22926 also inhibited Avr4/Cf4 and AvrPto/Pto associated HR but not the INF1 and AVR3a/R3a induced HR, indicating that Pi22926 and PexRD2 may suppress the same specific MAP3K dependent signaling pathway to promote disease. But further investigations revealed a further specification in MAPK signaling modulated by these two effectors. It was found that cell death responses triggered by NbMAP3Kβ2 and NbMAP3Kε are independent of each other and are specifically suppressed by Pi22926 and PexRD2, respectively. These findings suggest that P. infestans deploy PexRD2 and Pi22926 effectors at two different subcellular locations (cytoplasm and nucleus, to target two different MAP3Ks that act in parallel in MAP3K-dependent defense signaling induced upon the recognition of unidentified PAMP/PRR or effector/R protein pairs (King et al., 2014; Ren et al., 2019).
Another group of P. infestans RXLR effectors called suppressors of the early f lg22-induced immune response (SFIs) were found to suppress flg22 mediated PTI by targeting multiple steps of MAPK signaling (Zheng et al., 2014). All eight SFIs significantly suppressed activation of a flg22 induced PTI reporter gene having a PAMP-inducible promoter (pFRK1-Luc) in tomato cells. Out of eight, three SFIs, SFI5, SFI6, and SFI7 were found interrupting MAPK signaling at or upstream of the MAPK cascade in tomato (host of P. infestans), but not in Arabidopsis (non-host of P. infestans). SFI3 was found to suppress reported gene activity downstream of MAP kinase activation, which is defined as post-translational MAP Kinase modification by phosphorylation, in tomato only (Zheng et al., 2014). Whereas SFI1, SFI2, and SFI8/Avrblb2 were found functionally redundant in both tomato and Arabidopsis as they suppressed transcriptional activation of flg22-induced marker genes. Interestingly, none of the 8 SFIs attenuated flg22-dependent post-translational MAP kinase activation in Arabidopsis. Thus, SFI1, SFI2 and SFI8 putatively act downstream of post-translational MAP kinase activation (Zheng et al., 2014). In a recent study, SFI3 was shown to be localized in the host's nucleus and interacted with a protein UBK that contains both U-box and kinase domains. Importantly, this SFI3-UBK interaction is found responsible for flg22-induced transcription responses (He et al., 2019). A P. sojae RXLR effector, Avh331 is reported to suppress INF1 induced PTI by significantly suppressing transcriptional activation of resistance marker genes downstream of the MAPK signaling, thus leading to decreased H2O2 accumulation and callose deposition to promote Phytophthora infection of Arabidopsis and N. benthamiana (Cheng et al., 2012). Another P. sojae effector, PsCRN63 (crinkler) is reported to suppress flg22 induced PTI through MAPK signaling (Li et al., 2016).
SFI8/Avrblb2 is a core P. infestans RXLR effector, found to have multiple host targets associated with diverse defense mechanisms. In addition to targeting MAPK signaling, SFI8/Avrblb2 and its other homologs interact with diverse calmodulins (CaMs) (Naveed et al., 2019). CaMs are ubiquitous Ca2+sensors in eukaryotes that play a major role in calcium signaling by interacting with numerous downstream targets. We have found that CaM binding to Avrblb2 is required for its recognition by the cognate R protein, Rpi-blb2 (Naveed et al., 2019). Like SFI8/Avrblb2, SFI5 is also shown to interact with CaMs to suppress PTI (Zheng et al., 2018). CaM binding to SFI5 was found essential for MAPK suppression (Zheng et al., 2018). Targeting of both MAPK and calcium signaling components by Avrblb2 and SFI5 indicates highly complex cross talk between these signaling pathways during all PTI, ETS and ETI responses (Oh et al., 2014b).
Modulation of Defense Hormones Signaling
As discussed above, plant hormone signaling plays a crucial role in cell death and SAR responses. It is generalized that SA mediates resistance to biotrophic pathogens, whereas JA and ET signaling has been associated with resistance to necrotrophs. Since most of the Phytophthora pathogens are hemibiotrophs, SA, JA, and ET were all found involved in contriving defense against Phytophthora pathogens (Halim et al., 2009; Kawamura et al., 2009; Savchenko et al., 2010; Chang et al., 2015).
Recently, Yang et al. reported ET biosynthesis pathway as an essential defense component against P. sojae infection in soybean. To counter ET mediated defense, P. sojae deploys PsAvh238, a RXLR effector that can interact with host's ACSs (aminocyclopropane carboxylic acid synthases). ACSs contribute to plant defense by catalyzing a critical step in the ET biosynthesis pathway. PsAvh238 is shown to destabilize ACSs to suppress ET production by the host and thus, promote infection (Yang et al., 2019). Two P. capsici RXLR effectors were shown to target different components of SA signaling. Enhanced Disease Susceptibility 1 (EDS1) was first discovered as eds1 mutation in Arabidopsis that enhanced disease susceptibility against a biotrophic oomycete pathogen Peronospora parasitica. Further research revealed EDS1 as a positive regulator of plant immunity that interacts with Phytoalexin Deficient 4 (PAD4) and this EDS1-PAD4 complex plays a key role in SA accumulation and also regulate SA mediated defense (Wiermer et al., 2005). PcAvh103 is reported to promote P. capsici infection by binding with EDS1 and disrupting EDS1-PAD4 (Li et al., 2020). EDS1-PAD4 operates upstream of the SA pathway, whereas NPR1 (non-expressor of pathogenesis related1) is a SA receptor that operates downstream signaling. RxLR48 is found to associate with NPR1 to facilitate its nuclear localization and inhibits its proteasome-mediated degradation to suppress SA mediated defense and promote P. capsici infection (Li Q. et al., 2019b).
RXLR Effectors Target Negative Defense Regulators Involved in Diverse Mechanisms
In addition to the above-mentioned examples, in which positive regulators of plant immunity are modulated by Phytophthora spp. effectors, there are numerous examples in which negative regulators of plant immunity are also affected by Phytophthora spp. effectors. These negative immune regulators are termed as susceptibility factors (S-factors). The best studied S-factor with practical application in agriculture is the barley MLO gene and its homologs in diverse plants. Plants harboring dominant MLO alleles are highly susceptible to powdery mildew pathogens, whereas plants carrying recessive mlo alleles are immune (Bai et al., 2008). Similarly, there are numerous other examples of diverse S-factors. Some Phytophthora effectors targeting the host S-factors to promote ETS have been discovered (Boevink et al., 2016a). P. infestans has been shown to promote AS of two S-factors, a receptor like kinase (RLPK) and a bHLH transcription factor (bHLH025), to promote infection (Huang et al., 2020). Some splicing regulatory effectors (SREs) reported in this study were previously shown to interact with host's S-factors. SRE2 (Pi04089) was reported to interact with K-homology RNA-binding protein (KRBP1), which is an S-factor because its overexpression significantly increased P. infestans colonization (Wang et al., 2015).
As is described above, ubiquitin mediated protein degradation is an important component of plant immunity and thus, is targeted by multiple RXLR effectors. Unlike, above mentioned examples of targeting positive immune regulators, Pi02860 (SRE1) targets an S-factor involved in host's protein degradation machinery. NRL1 (non-phototrophic hypocotyl 3/root phototropism 2 (NPH3/RPT2)-like protein), which encodes a substrate adaptor component of a CULLIN3-associated ubiquitin E3 ligase in plants (He et al., 2018), is reported as S-factor because it favors the colonization of P. infestans in potato (Yang et al., 2016). A potential substrate of NRL1, SWAP70 (a guanine nucleotide exchange factor), was found as a positive regulator of immunity against P. infestans because it promotes INF1-mediated PTI. SRE1 was reported to attenuate this INF1-induced PTI responses through its interaction with NRL1 to enhance proteasome-mediated degradation of StSWAP70 (He et al., 2018).
In a recent report, a P. capsici effector, RxLR207, is shown to promote the 26S proteasome-dependent degradation of multiple proteins involved in ROS homeostasis (Li Q. et al., 2019a). RxLR207 is found to interact with BPA1 (binding partner of ACD11 [Accelerated cell Death11]) and four other members of BPLs (BPA1-Like proteins). ACD11 and BPAs are negative regulators of cell death as mutations in these genes show constitutive cell death. Since P. capsici and other Phytophthora spp. transition from a biotrophic to necrotrophic phase, in which plant cells and tissues die, and since RxLR207 is involved in this transition (Li Q. et al., 2019a), it can be speculated that RxLR207 promotes cell death by destabilizing BPAs. Unlike the above examples of RXLR promoted protein degradations, P. sojae effector PsAvh262 is reported to stabilize ER luminal binding immunoglobulin proteins (BiPs), as they are shown to act as S-factors because they suppress host's ER stress-triggered cell death and thus promote Phytophthora infection (Jing et al., 2016), perhaps at the biotrophic phase.
Another RXLR effector of P. infestans, Pi17316 is shown to target a MAP3K, a vascular highway1-interacting kinase (StVIK) to suppress INF1 induced PTI (Murphy et al., 2018). The interaction of Pi17316-StVIK (MAP3K) is different from other effector-MAPK interaction in a way that StVIK is a S-factor. Enhanced expression of both Pi17316 and StVIK is shown to suppress INF1 induced defense and promote P. infestans colonization. Silencing of NbVIK attenuates the ability of Pi17316 to suppress INF1 induced cell death and to promote disease. These results clearly showed the potential of StVIK and NbVIK as S-factors that promote late blight disease (Murphy et al., 2018).
Recently, Avr3a like effectors of three Phytophthora spp. were reported to interact with specific cinnamyl alcohol dehydrogenases (CADs) from Arabidopsis (AtCAD7) and N. benthamiana (NbCAD7). CADs are reported to contribute to structural lignification during plant cell development and some of them are specifically induced upon pathogen invasion to increase localized lignification that acts as a barrier against infection. However, both AtCAD7 and NbCAD7 are reported as S-factors against Phytophthora infection as their silencing resulted in reduced host susceptibility. Moreover, NbCAD7 was found to amplify the suppression of INF1 induced cell death by PiAvr3a and AtCAD7 suppressed core immune responses like ROS burst, callose deposition and WRKY33 expression (Li T. et al., 2019).
RXLR Effectors Target S-Factors and Positive Defense Regulators to Modulate Growth Hormone Signaling
Extensive research in diverse plant-pathogen systems, including Phytophthora, reveals a complex crosstalk—exhibiting both complementary and antagonizing effects- among growth- and defense-related phytohormone signaling (Turnbull et al., 2019). RXLR effectors have also been found to interfere with plant growth hormone signaling by targeting different signaling components that play a positive or negative role in host defense.
PcAvh1, a P. capsici RXLR effector, targets the scaffolding subunit of the protein phosphatase 2A (PP2Aa) involved in positively regulating plant immunity and growth. PP2A is involved in auxin signaling and silencing of PP2Aa in N. benthamiana resulted in attenuation of resistance to P. capsici as well as dwarfism of the plant (Michniewicz et al., 2007; Chen et al., 2019). The P. infestans RXLR effector, PiAvr2, has been shown to highly induce brassinosteroid (BR) hormone signaling. The BRI1-SUPPRESSOR1-like (BSL1) potato phosphatases are the components of BR signaling pathway crucial for plant growth. PiAvr2 was found to interact with all three BSL family members, StBSL1, StBSL2, and StBSL3 to attenuate immunity and enhance host susceptibility to P. infestans. Both BSL1 and BSL3 are shown to suppress INF1 induced cell death, whereas BSL2 and BSL3 are required for BSL1 stability. Moreover, PiAvr2 also caused the constitutive overexpression of bHLH transcription factor StCHL1, which is known to regulate antagonism between growth and immunity. It has also been found that suppression of INF1 induced cell death by StBSL1 and StBSL3 requires StCHL1. Since all these BR signaling components, BSLs and the TF CHL1 are negative regulators of host immunity, they all serve as S-factors (Turnbull et al., 2017, 2019). Another RXLR effector, Pi04314 has been shown to enhance leaf colonization of P. infestans by modulating JA and SA signaling (Boevink et al., 2016b). Pi04314 translocates to the host nucleus where it interacts with three isoforms of PP1c (protein phosphatase 1 catalytic) proteins to re-localize them from nucleolus to nulcleoplasm and also attenuate the induction of JA and SA responsive genes. PP1cs serve as S-factors because Pi04314-PPIcs interaction has shown to increase host susceptibility to P. infestans (Boevink et al., 2016b). As PiAvr2 acts to suppress INF1 induced immunity by switching host hormonal machinery toward growth enhancement and Pi04314 do so by suppressing the host defense (other than INF1 induced) related hormone signaling, it can be concluded that P. infestans is capable of differentially manipulating different types of hormone signaling pathways to attenuate PTI induced by different elicitors to promote late blight disease.
Effector Triggered Immunity (ETI) and ETS2
Effectors work as a double edge sword, which on one side suppress host defense responses, but on the other side act as avirulence (Avr) factors inducing R protein-mediated defenses in plants (Figure 3). This dual capability of effectors is well-represented in the ETS and ETI phases of the zigzag model (Jones and Dangl, 2006). All known Phytophthora R genes in potatoes are activated by effectors from Phytophthora spp. Introgression of R genes effective against Phytophthora spp. from wild Solanaceous plants into cultivated crop plants is a viable method against late blight disease, and more than a dozen R genes effective against various Phytophthora spp. have been identified. However, deployment of these single genes does not provide stable resistance against the rapidly evolving populations of P. infestans (Armstrong et al., 2005; Lokossou et al., 2009). R1, R3a, and R4 are among the 11 major dominant R genes introgressed from Solanum demissum into different potato cultivars. R1 recognizes P. infestans RXLR effector Avr1 to induce ETI. As mentioned above, Avr1 circumvents PTI by suppressing callose deposition (Du Y. et al., 2015). P. infestans isolates virulent on R1 containing potato plants lack Avr1 but carry a homologous gene named Avr1-like (Avr1-L) effector, which, due to a deletion of potent domains at the C terminus, is not recognized by R1 (Du et al., 2018). Similarly, P. infestans isolates virulent on potato cultivars carrying R4 gene carry truncated and non-functional cognate avr4 alleles (Van Poppel et al., 2008). The P. infestans RXLR effector, Avr3a is recognized by R3a. Interestingly, two P. sojae RXLR effectors, Avr1b and Avh1b have sequence similarity to Avr3a but only Avh1b is found to be weakly recognized by R3a whereas Avr1b like virulent alleles of Avr3a evade recognition by potato R3a. Avr1b is recognized by a soybean R protein Rpi1b and virulent isolates avoid recognition by either suppressing the expression of functional Avr1b or carry allele having multiple amino acid substitutions (Armstrong et al., 2005).
According to the zigzag model, perception of Avr effectors by the cognate R proteins can be either through their direct physical interaction or indirectly through recognizing modifications in host proteins targeted by these effectors. In case of Phytophthora-plant interactions, there is only one evidence of direct physical interaction-based recognition of class I variants of IPI-O (e.g., IPI-O1/Avrblb1, IPI-O2) family of P. infestans RXLR effectors by Rpi-blb1 that is a coiled-coil type NB-LRR (CC-NB-LRR) type R protein. Whereas class III variants of IPI-O family (i.e., IPI-O4) have also been found to physically bind with Rpi-blb1 but this interaction doesn't lead to induction of defense responses. Binding of IPI-O4 with Rpi-blb1 blocks recognition of IPI-O1 and also leads to the suppression of IPI-O1 elicited HR response (Chen et al., 2012). P. infestans lineages carrying IPI-O4 cause more severe disease in hosts harboring Rpi-blb1 and recently, evidence of suppression of Rpi-blb1 associated resistance is provided by in planta over-expression of IPI-O4 (Chen and Halterman, 2017). This IPI-O4 mediated suppression of IPI-O1 and Rpi-blb1 mediated ETI align well with the ETS2 phase of zigzag model.
Another CC-NB-LRR type R gene identified from S. bulbocastanum is Rpi-blb2 that can recognize PiAvrblb2, originally discovered as Avr factor. Transformation of this R gene into potato conferred broad-spectrum resistance against all the then known races of P. infestans (Song et al., 2003). Rpi-blb2 is one of the most well-studied R gene in terms of Phytophthora-plant interactions. The detailed mechanism behind this resistance is not fully explored but it seems to be involved in calcium and SA signaling in Rpi-blb2-mediated ETI. Transgenic N. benthamiana harboring Rpi-blb2 gene displayed strong HR in response to transient expression of the PiAvrblb2, through SGT1 and SA mediated pathways (Oh et al., 2014a,b). SGT1 is a eukaryotic ubiquitin ligase-associated co-chaperone that is widely reported to be involved in both PTI and ETI associated HR responses (Austin et al., 2002; Li et al., 2016). The P. capsici and P. infestans INF1 (PAMPs) and PexRD2 (RXLR effector) induced-HR is also shown to require SGT1 (Oh et al., 2009; Liu et al., 2016). Cross talk between SGT1 and calcium signaling pathways has been reported to regulate immune responses (Nowotny et al., 2003; Liu et al., 2016). In our laboratory, we showed that physical interaction of Avrblb2 with calmodulin is required for its recognition by Rpi-blb2 (Naveed et al., 2019). Based upon all these evidences, we suggest that calmodulin serves as a guardee, which is monitored by Rpi-blb2 to induce resistance response.
Other classes of four P. infestans resistant R proteins, Rpi-blb3, Rpi-abpt, R2, and R2-like harboring a characteristic leucine zipper nucleotide binding site leucine-rich repeat (LZ-NB-LRR) domain were cloned from different wild potato species. All these Rpi genes recognize PiAvr2 and trigger strong HR response (Lokossou et al., 2009).
Avr2 is a multiallelic highly divergent class of RXLR effectors that are recognized by the NB-LRR type R protein, R2 (Yang et al., 2020). As described above, PiAvr2 promotes ETS by modulating BR hormone signaling. Interestingly, both virulent and avirulent Avr2 alleles interact with the host phosphatase BSL1 but only the avirulent variants mediate activation of R2 (Saunders et al., 2012). This and additional studies suggest that BSL family proteins act as guardees and contribute toAvr2-mediated virulence of P. infestans (Turnbull et al., 2019). Many different R2 alleles and orthologs from six diverse Solanum spp. have been reported to recognize different Avr2 variants. P. infestans strains, virulent on R2 containing potatoes were found to either suppress Avr2 expression or carry a distinct variant of Avr2 that eludes R2 mediated perception (Gilroy et al., 2011a). Recently, Yang et al. (2020) discovered that virulent and avirulent Avr2 variants have different protein structures and virulent Avr2 are in fact intrinsically disordered proteins (IDPs). IDPs carry intrinsically disordered regions (IDRs) that may endow these proteins to have functional and evolutionary advantages over structured proteins. IDP- type Avr2 variants are predicted to be unstable and have a short protein half-life. The authors have suggested that these properties enable virulent forms of Avr2 effectors to elude recognition by R2 (Yang et al., 2020). This is an excellent example of effector evolution to circumvent ETI and cause ETS.
All these examples indicate that Phytophthora spp. carry multiple rapidly evolving effectors that aid them to overcome ETI and enter into the second phase of ETS. On the basis of extensive genome analysis of different Phytophthora spp. Haas et al. (2009) reported extensive sequence diversity among both inter and intraspecies RXLR and CRN effectors with high degree of expansion and pseudogene formation. Both RXLRs and CRNs are modular proteins with highly conserved N terminal domains and highly diverse C termini. Moreover, these effector genes typically reside in the gene sparse and repeat-rich genomic environment. Mobile elements in these repetitive regions contribute to the dynamic nature and high rates of gene gain and gene loss observed for these effectors (Haas et al., 2009). Although, both CRNs and RXLRs are shown to have the potential to contribute to the plant-pathogen evolutionary arm's race, currently, all known effectors with avirulence functions and cognate R genes belong to RXLR effectors.
Perspective on Developing Phytophthora-Resistant Plants
As discussed above, plant defense against Phytophthora spp. is a complex multilayered, interlinked phenomenon where each defense layer is intervened by effectors that have been rapidly coevolving during host-Phytophthora interactions. In order to develop sustainable resistance in crop plants against these noxious pathogens, a thorough understanding of the molecular basis of Phytophthora-plant interactions and discovery of multiple sources of genetic resistance are very important. In the past, several P. infestans R genes have been identified in wild potato, tomato, and other plants and are being used to develop Phytophthora-resistant plants (Zhu et al., 2012; Rodewald and Trognitz, 2013). But resistance was broken down because most of these R genes are very specific in recognizing their Phytophthora counterparts (avr genes) that are diverse not only among different Phytophthora spp. but also among different races and strains of the same species (Rodewald and Trognitz, 2013). Thus, the R gene-mediated resistance is not sufficient alone to provide broad-spectrum and durable resistance.
Apart from this effector-R gene model, recent discoveries of host's S-factors being targeted by multiple RXLR effectors is opening up new horizons for developing Phytophthora resistant crops. Genome editing through CRISPR/Cas-9 technology provides an excellent tool to knock out the negative immune regulators in plants. Efforts to implement these latest developments have already started and are showing promising results. For example, silencing of six S-genes in potato resulted in enhanced resistance against P. infestans (Sun et al., 2016).
In this era of high throughput technology, Phytophthora-plant interactions are being widely studied using different “omics” (genomics, transcriptomics, proteomics, metabolomics, and effectromics) approaches to identify novel components of plant defense. For example, comparative transcriptome studies of P. infestans-resistant wild tomato enabled the identification of P. infestans resistant transcription factors SpWRKY3 (Cui et al., 2018), and a long non-coding RNA (lncRNA16397) along with glutaredoxin (SpGRX) gene. Plants transformed with these non-race specific genes conferred broad spectrum resistance against P. infestans (Cui et al., 2017).
PAMP initiated non-host resistance have also been explored widely in an effort to identify non-race-specific sources of resistance. Recently, ELR that can recognize elicitins from diverse Phytophthora spp. was identified from a wild potato species S. microdontum. Transfer of ELR in cultivated potatoes exhibit enhanced resistance to diverse strains of P. infestans (Du J. et al., 2015). Identification of such non-race-specific sources of resistance and their pyramiding with multiple R genes and S-factors can offer stable broad-spectrum resistance against these rapidly evolving Phytophthora spp.
Summary and Conclusion
Most of the findings about Phytophthora-plant interactions described above are well-aligned with the basic zigzag model. However, there are some conflicts among the basic designation of components and the defense responses that contribute in PTI and ETI (Thomma et al., 2011). Based on wide distribution among diverse pathogen groups, and contribution in pathogen fitness and virulence as well as evasion of plant defense in multiple ways, some apoplastic effectors fulfill the criteria to be designated as PAMPs as well as effectors. In the zigzag model, the term R gene is confined to intracellular NB-LRR domain containing receptors that recognize effectors in the cytoplasm of the host cells to initiate quick defense responses up to a level where the threshold to trigger HR and SAR is attained (Jones and Dangl, 2006). Both known PRRs for Phytophthora spp. PAMPs, ELR, and RLP23 belong to RLPs that are currently considered a subclass of R proteins. Moreover, the timing, magnitude and the outcome of the defense responses cannot be strictly restricted to either PTI or ETI (Thomma et al., 2011). For example, as described above almost all Phytophthora spp. PAMPs have been found to induce HR and SAR upon their recognition and also the defense responses induced by CRN effectors have been found to develop slowly as compared to INF1 induced responses (Torto et al., 2003).
As mentioned above Phytophthora spp. PAMPs belong to diverse classes of biological molecules i.e., carbohydrates, proteins, and lipids. The pathogenic and evolutionary success of Phytophthora spp. can be ascribed to their diverse and rapidly evolving effector gene complements. Particularly, the RXLR effectors target diverse host proteins involved in different cellular processes to intervene host defense in multiple ways. Broadly, RXLR effectors can either suppress the positive regulators or support the negative regulators of plant defense to establish infection. Although evidence of defense modulation through intervention of diverse signaling pathways and physiological mechanisms have been found, compared to the 100s of Phytophthora effectors, these examples are just the tip of an iceberg. Moreover, unlike PAMPs, effector biology research is mainly limited to two pathosystems; P. sojae-soybean and P. infestans-tomato/potato/tobacco. In order to thoroughly understand the Phytophthora-plant interactions, there is dire need to expand the research beyond these model pathosystems.
Author Contributions
ZN, GA, and JC conceived the idea. ZN designed the layout of the article and wrote the paper. ZN and HM contributed to the figures. XW and GA revised and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
GA was employed by EukaryoTech LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Fulbright, Institute of international education (IIE) and United States education foundation of Pakistan (USEFP) for partially sponsoring graduate studies of ZN.
References
- Afroz A., Zahur M., Zeeshan N., Komatsu S. (2013). Plant-bacterium interactions analyzed by proteomics. Front. Plant Sci. 4:21. 10.3389/fpls.2013.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert I., Böhm H., Albert M., Feiler C. E., Imkampe J., Wallmeroth N., et al. (2015). An RLP23–SOBIR1–BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1:15140. 10.1038/nplants.2015.140 [DOI] [PubMed] [Google Scholar]
- Amelot N., Carrouche A., Danoun S., Bourque S., Haiech J., Pugin A., et al. (2011). Cryptogein, a fungal elicitor, remodels the phenylpropanoid metabolism of tobacco cell suspension cultures in a calcium-dependent manner. Plant Cell Environ. 34, 149–161. 10.1111/j.1365-3040.2010.02233.x [DOI] [PubMed] [Google Scholar]
- Anderson J. P., Gleason C. A., Foley R. C., Thrall P. H., Burdon J. B., Singh K. B. (2010). Plants versus pathogens: an evolutionary arms race. Funct. Plant Biol. 37, 499–512. 10.1071/FP09304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G., Deb D., Fedkenheuer K., Mcdowell J. M. (2015). Recent progress in RXLR effector research. Mol. Plant Microbe Interact. 28, 1063–1072. 10.1094/MPMI-01-15-0022-CR [DOI] [PubMed] [Google Scholar]
- Armstrong M. R., Whisson S. C., Pritchard L., Bos J. I., Venter E., Avrova A. O., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 102, 7766–7771. 10.1073/pnas.0500113102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard A. S., Evangelisti E., Kebdani-Minet N. M., PanabiãRes F., Deleury E., Maggio C., et al. (2014). Transcriptome dynamics of Arabidopsis thaliana root penetration by the oomycete pathogen Phytophthora parasitica. BMC Genomics 15:538. 10.1186/1471-2164-15-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M. J., Muskett P., Kahn K., Feys B. J., Jones J. D., Parker J. E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. 10.1126/science.1067747 [DOI] [PubMed] [Google Scholar]
- Bacete L., Mélida H., Miedes E., Molina A. (2018). Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 93, 614–636. 10.1111/tpj.13807 [DOI] [PubMed] [Google Scholar]
- Bai Y., Pavan S., Zheng Z., Zappel N. F., Reinstã¤Dler A., Lotti C., et al. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol. Plant Microbe Interact. 21, 30–39. 10.1094/MPMI-21-1-0030 [DOI] [PubMed] [Google Scholar]
- Bigeard J., Colcombet J., Hirt H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. 10.1016/j.molp.2014.12.022 [DOI] [PubMed] [Google Scholar]
- Birch P. R., Boevink P. C., Gilroy E. M., Hein I., Pritchard L., Whisson S. C. (2008). Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11, 373–379. 10.1016/j.pbi.2008.04.005 [DOI] [PubMed] [Google Scholar]
- Blume B., Nurnberger T., Nass N., Scheel D. (2000). Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell 12, 1425–1440. 10.1105/tpc.12.8.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P. C., Mclellan H., Gilroy E. M., Naqvi S., He Q., Yang L., et al. (2016a). Oomycetes seek help from the plant: Phytophthora infestans effectors target host susceptibility factors. Mol. Plant 9, 636–638. 10.1016/j.molp.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Boevink P. C., Wang X., Mclellan H., He Q., Naqvi S., Armstrong M. R., et al. (2016b). A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat. Commun. 7:10311. 10.1038/ncomms10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm H., Albert I., Oome S., Raaymakers T. M., Van Den Ackerveken G., Nürnberger T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10:e1004491. 10.1371/journal.ppat.1004491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. I., Kanneganti T. D., Young C., Cakir C., Huitema E., Win J., et al. (2006). The C terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48, 165–176. 10.1111/j.1365-313X.2006.02866.x [DOI] [PubMed] [Google Scholar]
- Bostock R. M., Savchenko T., Lazarus C., Dehesh K. (2011). Eicosapolyenoic acids: novel MAMPs with reciprocal effect on oomycete-plant defense signaling networks. Plant Signal. Behav. 6, 531–533. 10.4161/psb.6.4.14782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque S., Ponchet M., Binet M.-N., Ricci P., Pugin A., Lebrun-Garcia A. (1998). Comparison of binding properties and early biological effects of elicitins in tobacco cells. Plant Physiol. 118, 1317–1326. 10.1104/pp.118.4.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester K., De Sain M., Weide R., Gouget A., Klamer S., Canut H., et al. (2011). The lectin receptor kinase LecRK-I. 9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 7:e1001327. 10.1371/journal.ppat.1001327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt T. O., Schornack S., Banfield M. J., Kamoun S. (2012). Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483–492. 10.1016/j.pbi.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Bozkurt T. O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., et al. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. U.S.A. 108, 20832–20837. 10.1073/pnas.1112708109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F., Rosahl S., Lee J., Rudd J. J., Geiler C., Kauppinen S., et al. (2002). Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J 21, 6681–6688. 10.1093/emboj/cdf667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. H., Yan H. Z., Liou R. F. (2015). A novel elicitor protein from Phytophthora parasitica induces plant basal immunity and systemic acquired resistance. Mol. Plant Pathol. 16, 123–136. 10.1111/mpp.12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro-Garcia A., Wilkinson R. C., Gimenez-Ibanez S., Findlay K., Coffey M. D., Zipfel C., et al. (2011). The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS ONE 6:e16608. 10.1371/journal.pone.0016608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot C., Nawrath C., Métraux J.-P. (2008). The cuticle: not only a barrier for plant defense, a novel defense syndrome in plants with cuticular defects. Plant Signal. Behav. 3, 142–144. 10.4161/psb.3.2.5071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-R., Zhang Y., Li H., Zhang Z.-H., Sheng G.-L., Li Y.-P., et al. (2019). The RXLR effector PcAvh1 is required for full virulence of Phytophthora capsici. Mol. Plant Microbe Interact 32, 986–1000. 10.1094/MPMI-09-18-0251-R [DOI] [PubMed] [Google Scholar]
- Chen Y., Halterman D. (2017). Determination of virulence contribution from Phytophthora infestans effector IPI-O4 in a resistant potato host containing the RB gene. Physiol. Mol. Plant Pathol. 100, 30–34. 10.1016/j.pmpp.2017.05.00622438813 [DOI] [Google Scholar]
- Chen Y., Liu Z., Halterman D. A. (2012). Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLoS Pathog. 8:e1002595. 10.1371/annotation/75775518-f06e-4148-a639-31cfc6972b2e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B., Yu X., Ma Z., Dong S., Dou D., Wang Y., et al. (2012). Phytophthora sojae effector Avh331 suppresses the plant defence response by disturbing the MAPK signalling pathway. Physiol. Mol. Plant Pathol. 77, 1–9. 10.1016/j.pmpp.2011.10.002 [DOI] [Google Scholar]
- Colas V., Conrod S., Venard P., Keller H., Ricci P., Panabières F. (2001). Elicitin genes expressed in vitro by certain tobacco isolates of Phytophthora parasitica are down regulated during compatible interactions. Mol. Plant Microbe Interact. 14, 326–335. 10.1094/MPMI.2001.14.3.326 [DOI] [PubMed] [Google Scholar]
- Cui J., Luan Y., Jiang N., Bao H., Meng J. (2017). Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lnc RNA 16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 89, 577–589. 10.1111/tpj.13408 [DOI] [PubMed] [Google Scholar]
- Cui J., Xu P., Meng J., Li J., Jiang N., Luan Y. (2018). Transcriptome signatures of tomato leaf induced by Phytophthora infestans and functional identification of transcription factor SpWRKY3. Theor. Appl. Genet. 131, 787–800. 10.1007/s00122-017-3035-9 [DOI] [PubMed] [Google Scholar]
- Dagdas Y. F., Belhaj K., Maqbool A., Chaparro-Garcia A., Pandey P., Petre B., et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 5:e10856. 10.7554/eLife.10856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas Y. F., Pandey P., Tumtas Y., Sanguankiattichai N., Belhaj K., Duggan C., et al. (2018). Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. Elife 7:e37476. 10.7554/eLife.37476.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan J., Pichereaux C., Rossignol M., Blanc S., Wendehenne D., Pugin A., et al. (2009). Activation of a nuclear-localized SIPK in tobacco cells challenged by cryptogein, an elicitor of plant defence reactions. Biochem. J 418, 191–200. 10.1042/BJ20081465 [DOI] [PubMed] [Google Scholar]
- Damasceno C. M., Bishop J. G., Ripoll D. R., Win J., Kamoun S., Rose J. K. (2008). Structure of the glucanase inhibitor protein (GIP) family from Phytophthora species suggests coevolution with plant endo-β-1,3-glucanases. Mol. Plant Microbe Interact. 21, 820–830. 10.1094/MPMI-21-6-0820 [DOI] [PubMed] [Google Scholar]
- Dedyukhina E. G., Kamzolova S. V., Vainshtein M. B. (2014). Arachidonic acid as an elicitor of the plant defense response to phytopathogens. Chem. Biol. Technol. Agric. 1:18 10.1186/s40538-014-0018-9 [DOI] [Google Scholar]
- Derevnina L., Dagdas Y. F., De La Concepcion J. C., Bialas A., Kellner R., Petre B., et al. (2016). Nine things to know about elicitins. New Phytol. 212, 888–895. 10.1111/nph.14137 [DOI] [PubMed] [Google Scholar]
- Doughari J. (2015). An overview of plant immunity. J. Plant Pathol. Microbiol. 2015, 6–11. 10.4172/2157-7471.1000322 [DOI] [Google Scholar]
- Du J., Verzaux E., Chaparro-Garcia A., Bijsterbosch G., Keizer L. C., Zhou J., et al. (2015). Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 1:15034. 10.1038/nplants.2015.34 [DOI] [PubMed] [Google Scholar]
- Du Y., Mpina M. H., Birch P. R., Bouwmeester K., Govers F. (2015). Phytophthora infestans RXLR effector AVR1 interacts with exocyst component Sec5 to manipulate plant immunity. Plant Physiol. 169, 1975–1990. 10.1104/pp.15.01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Weide R., Zhao Z., Msimuko P., Govers F., Bouwmeester K. (2018). RXLR effector diversity in Phytophthora infestans isolates determines recognition by potato resistance proteins; the case study AVR1 and R1. Stud. Mycol. 89, 85–93. 10.1016/j.simyco.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder J., Cosio E. G. (1994). Elicitors of plant defense responses. Int. Rev. Cytol. 148, 1–36. 10.1016/S0074-7696(08)62404-3 [DOI] [Google Scholar]
- Fan G., Yang Y., Li T., Lu W., Du Y., Qiang X., et al. (2018). A Phytophthora capsici RXLR effector targets and inhibits a plant PPIase to suppress endoplasmic reticulum-mediated immunity. Mol. Plant 11, 1067–1083. 10.1016/j.molp.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Fawke S., Doumane M., Schornack S. (2015). Oomycete interactions with plants: infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 79, 263–280. 10.1128/MMBR.00010-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesel P. H., Zuccaro A. (2016). β-glucan: crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet. Biol. 90, 53–60. 10.1016/j.fgb.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Figueiredo J., Sousa Silva M., Figueiredo A. (2018). Subtilisin-like proteases in plant defence: the past, the present and beyond. Mol. Plant Pathol. 19, 1017–1028. 10.1111/mpp.12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furniss J. J., Grey H., Wang Z., Nomoto M., Jackson L., Tada Y., et al. (2018). Proteasome-associated HECT-type ubiquitin ligase activity is required for plant immunity. PLoS Pathog. 14:e1007447. 10.1371/journal.ppat.1007447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q. M., Zhu S., Kachroo P., Kachroo A. (2015). Signal regulators of systemic acquired resistance. Front. Plant Sci. 6:228 10.3389/fpls.2015.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesbrecht M. B., Hansen E. M., Kitin P. (2011). Histology of Phytophthora ramorum in Notholithocarpus densiflorus bark tissues. New Zealand J. For. Sci. 41, 89–100. [Google Scholar]
- Gijzen M., Nürnberger T. (2006). Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67, 1800–1807. 10.1016/j.phytochem.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Gilroy E. M., Breen S., Whisson S. C., Squires J., Hein I., Kaczmarek M., et al. (2011a). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191, 763–776. 10.1111/j.1469-8137.2011.03736.x [DOI] [PubMed] [Google Scholar]
- Gilroy E. M., Taylor R. M., Hein I., Boevink P., Sadanandom A., Birch P. R. (2011b). CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 190, 653–666. 10.1111/j.1469-8137.2011.03643.x [DOI] [PubMed] [Google Scholar]
- Gui Y. J., Chen J. Y., Zhang D. D., Li N. Y., Li T. G., Zhang W. Q., et al. (2017). Verticillium dahliae manipulates plant immunity by glycoside hydrolase 12 proteins in conjunction with carbohydrate-binding module 1. Environ. Microbiol. 19, 1914–1932. 10.1111/1462-2920.13695 [DOI] [PubMed] [Google Scholar]
- Guo B., Wang H., Yang B., Jiang W., Jing M., Li H., et al. (2019). Phytophthora sojae effector PsAvh240 inhibits host aspartic protease secretion to promote infection. Mol. Plant 12, 552–564. 10.1016/j.molp.2019.01.017 [DOI] [PubMed] [Google Scholar]
- Haas B. J., Kamoun S., Zody M. C., Jiang R. H., Handsaker R. E., Cano L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398. 10.1038/nature08358 [DOI] [PubMed] [Google Scholar]
- Hahn M. G. (1996). Microbial elicitors and their receptors in plants. Annu. Rev. Phytopathol. 34, 387–412. 10.1146/annurev.phyto.34.1.387 [DOI] [PubMed] [Google Scholar]
- Halim V. A., Altmann S., Ellinger D., Eschen-Lippold L., Miersch O., Scheel D., et al. (2009). PAMP-induced defense responses in potato require both salicylic acid and jasmonic acid. Plant J. 57, 230–242. 10.1111/j.1365-313X.2008.03688.x [DOI] [PubMed] [Google Scholar]
- Hansen E. M. (2015). Phytophthora species emerging as pathogens of forest trees. Curr. For. Rep. 1, 16–24. 10.1007/s40725-015-0007-7 [DOI] [Google Scholar]
- Hardham A. R., Cahill D. M. (2010). The role of oomycete effectors in plant–pathogen interactions. Funct. Plant Biol. 37, 919–925. 10.1071/FP10073 [DOI] [Google Scholar]
- He Q., McLellan H., Hughes R. K., Boevink P. C., Armstrong M., Lu Y., et al. (2019). Phytophthora infestans effector SFI 3 targets potato UBK to suppress early immune transcriptional responses. New Phytol. 222, 438–454. 10.1111/nph.15635 [DOI] [PubMed] [Google Scholar]
- He Q., Naqvi S., Mclellan H., Boevink P. C., Champouret N., Hein I., et al. (2018). Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proc. Natl. Acad. Sci. U.S.A. 115, E7834–E7843. 10.1073/pnas.1808585115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeberichts F. A., Davoine C., Vandorpe M., Morsa S., Ksas B., Stassen C., et al. (2013). Cryptogein-induced transcriptional reprogramming in tobacco is light dependent. Plant Physiol. 163, 263–275. 10.1104/pp.113.217240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Li L., Hafrén A., Coll N. S. (2017). Autophagy as an emerging arena for plant–pathogen interactions. Curr. Opin. Plant Biol. 38, 117–123. 10.1016/j.pbi.2017.04.017 [DOI] [PubMed] [Google Scholar]
- Hou Y., Zhai Y., Feng L., Karimi H. Z., Rutter B. D., Zeng L., et al. (2019). A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 25, 153–165. 10.1016/j.chom.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Lu X., Wu H., Xie Y., Peng Q., Gu L., et al. (2020). Phytophthora effectors modulate genome-wide alternative splicing of host mRNAs to reprogram plant immunity. Mol. Plant. 13, 1470–1484. 10.1016/j.molp.2020.07.007 [DOI] [PubMed] [Google Scholar]
- Hwang I. S., Hwang B. K. (2010). The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 152, 948–967. 10.1104/pp.109.147827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M., Guo B., Li H., Yang B., Wang H., Kong G., et al. (2016). A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant Binding immunoglobulin Proteins. Nat. Commun. 7:11685. 10.1038/ncomms11685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444, 323. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. 10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Kamoun S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. 10.1146/annurev.phyto.44.070505.143436 [DOI] [PubMed] [Google Scholar]
- Kamoun S., Furzer O., Jones J. D., Judelson H. S., Ali G. S., Dalio R. J., et al. (2015). The Top 10 oomycete pathogens in molecular plant pathology. Mol. Plant Pathol. 16, 413–434. 10.1111/mpp.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S., Klucher K. M., Coffey M. D., Tyler B. M. (1993). A gene encoding a host-specific elicitor protein of Phytophthora parasitica. Mol. Plant Microbe Interact. 6, 573–581. 10.1094/MPMI-6-573 [DOI] [PubMed] [Google Scholar]
- Kamoun S., Van West P., Vleeshouwers V. G., De Groot K.e, Govers, F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1426. 10.1105/tpc.10.9.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzaki H., Saitoh H., Ito A., Fujisawa S., Kamoun S., Katou S., et al. (2003). Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol. Plant Pathol. 4, 383–391. 10.1046/j.1364-3703.2003.00186.x [DOI] [PubMed] [Google Scholar]
- Kaschani F., Shabab M., Bozkurt T., Shindo T., Schornack S., Gu C., et al. (2010). An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 154, 1794–1804. 10.1104/pp.110.158030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y., Hase S., Takenaka S., Kanayama Y., Yoshioka H., Kamoun S., et al. (2009). INF1 elicitin activates jasmonic acid- and ethylene-mediated signalling pathways and induces resistance to bacterial wilt disease in tomato. J. Phytopathol. 157, 287–297. 10.1111/j.1439-0434.2008.01489.x [DOI] [Google Scholar]
- Khatib M., Lafitte C., Esquerré-Tugayé M. T., Bottin A., Rickauer M. (2004). The CBEL elicitor of Phytophthora parasitica var. nicotianae activates defence in Arabidopsis thaliana via three different signalling pathways. New Phytol. 162, 501–510. 10.1111/j.1469-8137.2004.01043.x [DOI] [Google Scholar]
- King S. R., Mclellan H., Boevink P. C., Armstrong M. R., Bukharova T., Sukarta O., et al. (2014). Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. Plant Cell 26, 1345–1359. 10.1105/tpc.113.120055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarzynski O., Plesse B., Joubert J.-M., Yvin J.-C., Kopp M., Kloareg B., et al. (2000). Linear β-1,3 glucans are elicitors of defense responses in tobacco. Plant Physiol. 124, 1027–1038. 10.1104/pp.124.3.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolomiets M. V., Hannapel D. J., Chen H., Tymeson M., Gladon R. J. (2001). Lipoxygenase is involved in the control of potato tuber development. Plant Cell 13, 613–626. 10.1105/tpc.13.3.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M., Rouster J., Fritig B., Darvill A., Albersheim P. (1989). Host-pathogen interactions: XXXII. A fungal glucan preparation protects Nicotianae against infection by viruses. Plant Physiol. 90, 208–216. 10.1104/pp.90.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroque M., Barriot R., Bottin A., Barre A., Roug,é P., Dumas B., et al. (2012). The unique architecture and function of cellulose-interacting proteins in oomycetes revealed by genomic and structural analyses. BMC Genomics 13:605. 10.1186/1471-2164-13-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larroque M., Belmas E., Martinez T., Vergnes S., Ladouce N., Lafitte C., et al. (2013). Pathogen-associated molecular pattern-triggered immunity and resistance to the root pathogen Phytophthora parasitica in Arabidopsis. J. Exp. Bot. 64, 3615–3625. 10.1093/jxb/ert195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers M., De Wit P. J., Govers F. (2003). Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 11, 462–469. 10.1016/j.tim.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Lebrun-Garcia A., Ouaked F., Chiltz A., Pugin A. (1998). Activation of MAPK homologues by elicitors in tobacco cells. Plant J. 15, 773–781. 10.1046/j.1365-313X.1998.00269.x [DOI] [PubMed] [Google Scholar]
- Li Q., Ai G., Shen D., Zou F., Wang J., Bai T., et al. (2019a). A Phytophthora capsici effector targets ACD11 binding partners that regulate ROS-mediated defense response in arabidopsis. Mol. Plant 12, 565–581. 10.1016/j.molp.2019.01.018 [DOI] [PubMed] [Google Scholar]
- Li Q., Chen Y., Wang J., Zou F., Jia Y., Shen D., et al. (2019b). A Phytophthora capsici virulence effector associates with NPR1 and suppresses plant immune responses. Phytopathol. Res. 1:6 10.1186/s42483-019-0013-y [DOI] [Google Scholar]
- Li Q., Wang J., Bai T., Zhang M., Jia Y., Shen D., et al. (2020). A Phytophthora capsici effector suppresses plant immunity via interaction with EDS1. Mol. Plant Pathol. 21, 502–511. 10.1111/mpp.12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhang M., Shen D., Liu T., Chen Y., Zhou J.-M., et al. (2016). A Phytophthora sojae effector PsCRN63 forms homo-/hetero-dimers to suppress plant immunity via an inverted association manner. Sci. Rep. 6:26951. 10.1038/srep26951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wang Q., Feng R., Li L., Ding L., Fan G., et al. (2019). Negative regulators of plant immunity derived from cinnamyl alcohol dehydrogenases are targeted by multiple Phytophthora Avr3a-like effectors. New Phytol. 10.1111/nph.16139. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Liu Z., Qiu A., Shi L., Cai J., Huang X., Yang S., et al. (2015). SRC2-1 is required in PcINF1-induced pepper immunity by acting as an interacting partner of PcINF1. J. Exp. Bot. 66, 3683–3698. 10.1093/jxb/erv161 [DOI] [PubMed] [Google Scholar]
- Liu Z.-Q., Liu Y.-Y., Shi L.-P., Yang S., Shen L., Yu H.-X., et al. (2016). SGT1 is required in PcINF1/SRC2-1 induced pepper defense response by interacting with SRC2-1. Sci. Rep. 6:21651. 10.1038/srep21651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochman J., Kasparovsky T., Damborsky J., Osman H., Marais A., Chaloupkova R., et al. (2005). Construction of cryptogein mutants, a proteinaceous elicitor from Phytophthora, with altered abilities to induce a defense reaction in tobacco cells. Biochemistry 44, 6565–6572. 10.1021/bi0502285 [DOI] [PubMed] [Google Scholar]
- Lokossou A. A., Park T.-H., Van Arkel G., Arens M., Ruyter-Spira C., Morales J., et al. (2009). Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant Microbe Interact. 22, 630–641. 10.1094/MPMI-22-6-0630 [DOI] [PubMed] [Google Scholar]
- Ma Z., Song T., Zhu L., Ye W., Wang Y., Shao Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. 10.1105/tpc.15.00390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Zhu L., Song T., Wang Y., Zhang Q., Xia Y., et al. (2017). A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 355, 710–714. 10.1126/science.aai7919 [DOI] [PubMed] [Google Scholar]
- Maximo H. J., Dalio R. J., Dias R. O., Litholdo C. G., Felizatti H. L., Machado M. A. (2019). PpCRN7 and PpCRN20 of Phythophthora parasitica regulate plant cell death leading to enhancement of host susceptibility. BMC Plant Biol. 19:544. 10.1186/s12870-019-2129-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgowan J., Fitzpatrick D. A. (2017). Genomic, network, and phylogenetic analysis of the oomycete effector arsenal. mSphere 2, e00408–e00417. 10.1128/mSphere.00408-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclellan H., Boevink P. C., Armstrong M. R., Pritchard L., Gomez S., Morales J., et al. (2013). An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog 9:e1003670. 10.1371/journal.ppat.1003670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélida H., Sopeña-Torres S., Bacete L., Garrido-Arandia M., Jordá L., López G., et al. (2018). Non-branched β-1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J 93, 34–49. 10.1111/tpj.13755 [DOI] [PubMed] [Google Scholar]
- Mellersh D. G., Heath M. C. (2001). Plasma membrane–cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 13, 413–424. 10.1105/tpc.13.2.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M., Zago M. K., Abas L., Weijers D., Schweighofer A., Meskiene I., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130, 1044–1056. 10.1016/j.cell.2007.07.033 [DOI] [PubMed] [Google Scholar]
- Mikes V., Milat M. L., Ponchet M., Panabières F., Ricci P., Blein J. P. (1998). Elicitins, proteinaceous elicitors of plant defense, are a new class of sterol carrier proteins. Biochem. Biophys. Res. Commun. 245, 133–139. 10.1006/bbrc.1998.8341 [DOI] [PubMed] [Google Scholar]
- Misas-Villamil J. C., van der Hoorn R. A., Doehlemann G. (2016). Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 212, 902–907. 10.1111/nph.14117 [DOI] [PubMed] [Google Scholar]
- Murphy F., He Q., Armstrong M., Giuliani L. M., Boevink P. C., Zhang W., et al. (2018). The potato MAP3K StVIK is required for the Phytophthora infestans RXLR effector Pi17316 to promote disease. Plant Physiol. 177, 398–410. 10.1104/pp.18.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed Z. A., Bibi S., Ali G. S. (2019). The Phytophthora RXLR effector Avrblb2 modulates plant immunity by interfering with Ca2+ signaling pathway. Front. Plant Sci. 10:374. 10.3389/fpls.2019.00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nennstiel D., Scheel D., Nürnberger T. (1998). Characterization and partial purification of an oligopeptide elicitor receptor from parsley (Petroselinum crispum). FEBS Lett. 431, 405–410. 10.1016/S0014-5793(98)00800-X [DOI] [PubMed] [Google Scholar]
- Nowotny M., Spiechowicz M., Jastrzebska B., Filipek A., Kitagawa K., Kuznicki J. (2003). Calcium-regulated interaction of Sgt1 with S100A6 (calcyclin) and other S100 proteins. J. Biol. Chem. 278, 26923–26928. 10.1074/jbc.M211518200 [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Nennstiel D., Jabs T., Sacks W. R., Hahlbrock K., Scheel D. (1994). High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78, 449–460. 10.1016/0092-8674(94)90423-5 [DOI] [PubMed] [Google Scholar]
- Oh S.-K., Kim H., Choi D. (2014a). Rpi-blb2-mediated late blight resistance in Nicotiana benthamiana requires SGT1 and salicylic acid-mediated signaling but not RAR1 or HSP90. FEBS Lett. 588, 1109–1115. 10.1016/j.febslet.2014.02.028 [DOI] [PubMed] [Google Scholar]
- Oh S.-K., Kwon S.-Y., Choi D. (2014b). Rpi-blb2-mediated hypersensitive cell death caused by Phytophthora infestans AVRblb2 requires SGT1, but not EDS1, NDR1, salicylic acid-, jasmonic acid-, or ethylene-mediated signaling. Plant Pathol. J. 30, 254 10.5423/PPJ.OA.03.2014.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.-K., Young C., Lee M., Oliva R., Bozkurt T. O., Cano L. M., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21, 2928–2947. 10.1105/tpc.109.068247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oome S., Raaymakers T. M., Cabral A., Samwel S., Böhm H., Albert I., et al. (2014). Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 16955–16960. 10.1073/pnas.1410031111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A. E. (1996). Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 8, 1821. 10.2307/3870232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K. C., Wang C. W., Wu C. H., Huang C. T., Liou R. F. (2015). Tomato SOBIR1/EVR homologs are involved in elicitin perception and plant defense against the oomycete pathogen Phytophthora parasitica. Mol. Plant Microbe Interact. 28, 913–926. 10.1094/MPMI-12-14-0405-R [DOI] [PubMed] [Google Scholar]
- Preisig C. L., Kuć J. A. (1985). Arachidonic acid-related elicitors of the hypersensitive response in potato and enhancement of their activities by glucans from Phytophthora infestans (Mont.) deBary. Arch. Biochem. Biophys. 236, 379–389. 10.1016/0003-9861(85)90638-1 [DOI] [PubMed] [Google Scholar]
- Pritchard L., Birch P. (2014). The zigzag model of plant-microbe interactions: is it time to move on? Mol. Plant Pathol. 15, 865–870. 10.1111/mpp.12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Liu L., Xiong Q., Flores C., Wong J., Shi J., et al. (2013). Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 45, 330–333. 10.1038/ng.2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Shi J., Zhai Y., Hou Y., Ma W. (2015). Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl. Acad. Sci. U.S.A. 112, 5850–5855. 10.1073/pnas.1421475112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaymakers T. M., Van Den Ackerveken G. (2016). Extracellular recognition of oomycetes during biotrophic infection of plants. Front. Plant Sci. 7:906. 10.3389/fpls.2016.00906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K., Kirchner E., Gijzen M., Zocher G., Löffelhardt B., Nürnberger T., et al. (2011). Structural and phylogenetic analyses of the GP42 transglutaminase from Phytophthora sojae reveal an evolutionary relationship between oomycetes and marine Vibrio bacteria. J. Biol. Chem. 286, 42585–42593. 10.1074/jbc.M111.290544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Armstrong M., Qi Y., Mclellan H., Zhong C., Du B., et al. (2019). Phytophthora infestans RXLR effectors target parallel steps in an immune signal transduction pathway. Plant Physiol. 180, 2227–2239. 10.1104/pp.18.00625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker K., Bostock R. (1994). Eicosanoids in the Phytophthora infestans-potato interaction: lipoxygenase metabolism of arachidonic acid and biological activities of selected lipoxygenase products. Physiol. Mol. Plant Pathol. 44, 65–80. 10.1016/S0885-5765(05)80095-5 [DOI] [Google Scholar]
- Robinson S. M., Bostock R. M. (2015). β-glucans and eicosapolyenoic acids as MAMPs in plant-oomycete interactions: past and present. Front. Plant Sci. 5:797. 10.3389/fpls.2014.00797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodewald J., Trognitz B. (2013). Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol. Plant Pathol. 14, 740–757. 10.1111/mpp.12036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C., Montillet J.-L., Agnel J.-P., Battesti C., Alonso B., Knoll A., et al. (1999). Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J. Biol. Chem. 274, 36446–36455. 10.1074/jbc.274.51.36446 [DOI] [PubMed] [Google Scholar]
- Saijo Y., Loo E. P. I., Yasuda S. (2018). Pattern recognition receptors and signaling in plant–microbe interactions. Plant J. 93, 592–613. 10.1111/tpj.13808 [DOI] [PubMed] [Google Scholar]
- Sanseverino W., Hermoso A., D' Alessandro R., Vlasova A., Andolfo G., Frusciante L., et al. (2012). PRGdb 2.0: towards a community-based database model for the analysis of R-genes in plants. Nucleic Acids Res. 41, D1167–D1171. 10.1093/nar/gks1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M., Takeuchi K., Kamoun S., Ichinose Y., Govers F., Toyoda K., et al. (2000). Independent pathways leading to apoptotic cell death, oxidative burst and defense gene expression in response to elicitin in tobacco cell suspension culture. Eur. J. Biochem. 267, 5005–5013. 10.1046/j.1432-1327.2000.01553.x [DOI] [PubMed] [Google Scholar]
- Saunders D. G., Breen S., Win J., Schornack S., Hein I., Bozkurt T. O., et al. (2012). Host protein BSL1 associates with Phytophthora infestans RXLR effector AVR2 and the Solanum demissum immune receptor R2 to mediate disease resistance. Plant Cell 24, 3420–3434. 10.1105/tpc.112.099861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko T., Walley J. W., Chehab E. W., Xiao Y., Kaspi R., Pye M. F., et al. (2010). Arachidonic acid: an evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 22, 3193–3205. 10.1105/tpc.110.073858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séjalon N., Dargent R., Villalba F., Bottin A., Rickauer M., Esquerré-Tugayé M. (1995). Characterization of a cell-surface antigen isolated from the plant pathogen Phytophthora parasitica var. nicotianae. Can. J. Bot. 73, 1104–1108. 10.1139/b95-365 [DOI] [Google Scholar]
- Séjalon-Delmas N., Mateos F. V., Bottin A., Rickauer M., Dargent R., Esquerré-Tugayé M. (1997). Purification, elicitor activity, and cell wall localization of a glycoprotein from Phytophthora parasitica var. nicotianae, a fungal pathogen of tobacco. Phytopathology 87, 899–909. 10.1094/PHYTO.1997.87.9.899 [DOI] [PubMed] [Google Scholar]
- Serrano I., Campos L., Rivas S. (2018). Roles of E3 ubiquitin-ligases in nuclear protein homeostasis during plant stress responses. Front. Plant Sci. 9:139. 10.3389/fpls.2018.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severino V., Farina A., Fleischmann F., Dalio R. J., Di Maro A., Scognamiglio M., et al. (2014). Molecular profiling of the Phytophthora plurivora secretome: a step towards understanding the cross-talk between plant pathogenic oomycetes and their hosts. PLoS ONE 9:e112317. 10.1371/journal.pone.0112317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T., Kaschani F., Yang F., Kovács J., Tian F., Kourelis J., et al. (2016). Screen of non-annotated small secreted proteins of Pseudomonas syringae reveals a virulence factor that inhibits tomato immune proteases. PLoS Pathog. 12:e1005874. 10.1371/journal.ppat.1005874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarenko A., Meier B. M., Croft K., Eiben H. (1993). “Lipoxygenase in plant disease,” in Mechanisms of Plant Defense Responses, eds P. Spencer-Phillips, and M. Jeger (Dordrecht: Springer; ), 211–220. 10.1007/978-94-011-1737-1_63 [DOI] [Google Scholar]
- Song J., Bradeen J. M., Naess S. K., Raasch J. A., Wielgus S. M., Haberlach G. T., et al. (2003). Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. U.S.A. 100, 9128–9133. 10.1073/pnas.1533501100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Wolters A.-M. A., Vossen J. H., Rouwet M. E., Loonen A. E., Jacobsen E., et al. (2016). Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 25, 731–742. 10.1007/s11248-016-9964-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B. P., Nurnberger T., Joosten M. H. (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell Online 23, 4–15. 10.1105/tpc.110.082602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Benedetti B., Kamoun S. (2005). A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 138, 1785–1793. 10.1104/pp.105.061226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Huitema E., Da Cunha L., Torto-Alalibo T., Kamoun S. (2004). A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 279, 26370–26377. 10.1074/jbc.M400941200 [DOI] [PubMed] [Google Scholar]
- Tomczynska I., Stumpe M., Doan T. G., Mauch F. (2020). A Phytophthora effector protein promotes symplastic cell-to-cell trafficking by physical interaction with plasmodesmata-localised callose synthases. New Phytol. 227, 1467–1478. 10.1111/nph.16653 [DOI] [PubMed] [Google Scholar]
- Tomczynska I., Stumpe M., Mauch F. (2018). A conserved Rx LR effector interacts with host RABA-type GTP ases to inhibit vesicle-mediated secretion of antimicrobial proteins. Plant J. 95, 187–203. 10.1111/tpj.13928 [DOI] [PubMed] [Google Scholar]
- Torto T. A., Li S., Styer A., Huitema E., Testa A., Gow N. A., et al. (2003). EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 13, 1675–1685. 10.1101/gr.910003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño T. Y., Stergiopoulos I., Coaker G. (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. 10.1146/annurev-phyto-080615-100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull D., Wang H., Breen S., Malec M., Naqvi S., Yang L., et al. (2019). AVR2 targets BSL family members, which act as susceptibility factors to suppress host immunity. Plant Physiol. 180, 571–581. 10.1104/pp.18.01143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull D., Yang L., Naqvi S., Breen S., Welsh L., Stephens J., et al. (2017). RXLR Effector AVR2 up-regulates a brassinosteroid-responsive bHLH transcription factor to suppress immunity. Plant Physiol. 174, 356–369. 10.1104/pp.16.01804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto N., Kakitani M., Iwamatsu A., Yoshikawa M., Yamaoka N., Ishida I. (1997). The structure and function of a soybean β-glucan-elicitor-binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 1029–1034. 10.1073/pnas.94.3.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood W. (2012). The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3:85. 10.3389/fpls.2012.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S., Sheikh A., Gimenez-Ibanez S., Jones A., Ntoukakis V., Börnke F. (2016). The proteasome acts as a hub for plant immunity and is targeted by Pseudomonas type III effectors. Plant Physiol. 172, 1941–1958. 10.1104/pp.16.00808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg N., Christie J., Aveling T., Engelbrecht J. (2018). Callose and β-1,3-glucanase inhibit Phytophthora cinnamomi in a resistant avocado rootstock. Plant Pathol. 67, 1150–1160. 10.1111/ppa.1281930332458 [DOI] [Google Scholar]
- Van Poppel P. M., Guo J., Van De Vondervoort P. J., Jung M. W., Birch P. R., Whisson S. C., et al. (2008). The Phytophthora infestans avirulence gene Avr4 encodes an RXLR-dEER effector. Mol. Plant Microbe Interact. 21, 1460–1470. 10.1094/MPMI-21-11-1460 [DOI] [PubMed] [Google Scholar]
- Véronési C., Rickauer M., Fournier J., Pouénat M.-L., Esquerré-Tugayé M.-T. (1996). Lipoxygenase gene expression in the tobacco-Phytophthora parasitica nicotianae interaction. Plant Physiol. 112, 997–1004. 10.1104/pp.112.3.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetukuri R. R., Whisson S. C., Grenville-Briggs L. J. (2017). Phytophthora infestans effector Pi14054 is a novel candidate suppressor of host silencing mechanisms. Eur. J. Plant Pathol. 149, 771–777. 10.1007/s10658-017-1222-9 [DOI] [Google Scholar]
- Wang H., He H., Qi Y., Mclellan H., Tian Z., Birch P. R., et al. (2019). The oomycete microbe-associated molecular pattern Pep-13 triggers SERK3/BAK1-independent plant immunity. Plant Cell Rep. 38, 173–182. 10.1007/s00299-018-2359-5 [DOI] [PubMed] [Google Scholar]
- Wang J., Gao C., Li L., Cao W., Dong R., Ding X., et al. (2019). Transgenic RXLR effector PITG_15718.2 suppresses immunity and reduces vegetative growth in potato. Int. J. Mol. Sci. 20:3031. 10.3390/ijms20123031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Han C., Ferreira A.O., Yu X., Ye W., Tripathy S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell. 23, 2064–2086. 10.1105/tpc.111.086082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Mclellan H., Bukharova T., He Q., Murphy F., Shi J., et al. (2018). Phytophthora infestans RXLR effectors act in concert at diverse subcellular locations to enhance host colonization. J. Exp. Bot. 70, 343–356. 10.1093/jxb/ery360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Boevink P., Mclellan H., Armstrong M., Bukharova T., Qin Z., et al. (2015). A host KH RNA-binding protein is a susceptibility factor targeted by an RXLR effector to promote late blight disease. Mol. Plant 8, 1385–1395. 10.1016/j.molp.2015.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendehenne D., Lamotte O., Frachisse J. M., Barbier-Brygoo H., Pugin A. (2002). Nitrate efflux is an essential component of the cryptogein signaling pathway leading to defense responses and hypersensitive cell death in tobacco. Plant Cell 14, 1937–1951. 10.1105/tpc.002295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal L., Strehmel N., Eschen-Lippold L., Bauer N., Westermann B., Rosahl S., et al. (2019). pH effects on plant calcium fluxes: lessons from acidification-mediated calcium elevation induced by the γ-glutamyl-leucine dipeptide identified from Phytophthora infestans. Sci. Rep. 9:4733. 10.1038/s41598-019-41276-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Feys B. J., Parker J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. 10.1016/j.pbi.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Yang B., Wang Y., Guo B., Jing M., Zhou H., Li Y., et al. (2019). The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 Gm ACS s to suppress ethylene biosynthesis and promote infection. New Phytol. 222, 425–437. 10.1111/nph.15581 [DOI] [PubMed] [Google Scholar]
- Yang L., Liu H., Duan G., Huang Y., Liu S., Fang Z., et al. (2020). Phytophtora infestans AVR2 effector escapes R2 recognition through effector disordering. Mol. Plant Microbe Interact. 33, 921–931. 10.1094/MPMI-07-19-0179-R [DOI] [PubMed] [Google Scholar]
- Yang L., Mclellan H., Naqvi S., He Q., Boevink P. C., Armstrong M., et al. (2016). Potato NPH3/RPT2-Like Protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiol. 171, 645–657. 10.1104/pp.16.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H., Numata N., Nakajima K., Katou S., Kawakita K., Rowland O., et al. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15, 706–718. 10.1105/tpc.008680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Mclellan H., Fraiture M., Liu X., Boevink P. C., Gilroy E. M., et al. (2014). Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathog. 10:e1004057. 10.1371/journal.ppat.1004057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Wagener N., Mclellan H., Boevink P. C., Hua C., Birch P. R. J., et al. (2018). Phytophthora infestans RXLR effector SFI5 requires association with calmodulin for PTI/MTI suppressing activity. New Phytol. 219, 1433–1446. 10.1111/nph.15250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Li Y., Vossen J. H., Visser R. G., Jacobsen E. (2012). Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 21, 89–99. 10.1007/s11248-011-9510-1 [DOI] [PMC free article] [PubMed] [Google Scholar]