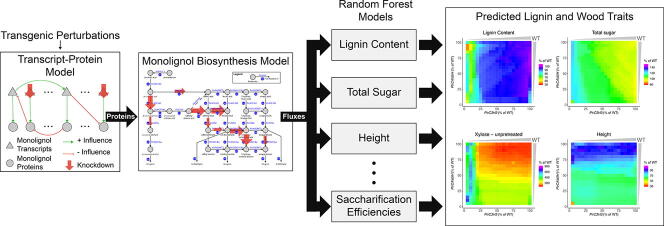
Graphical abstract
Keywords: Lignin biosynthesis, Multiscale modeling, Cross-regulatory influences, Random forests
Abstract
Understanding the mechanisms behind lignin formation is an important research area with significant implications for the bioenergy and biomaterial industries. Computational models are indispensable tools for understanding this complex process. Models of the monolignol pathway in Populus trichocarpa and other plants have been developed to explore how transgenic modifications affect important bioenergy traits. Many of these models, however, only capture one level of biological organization and are unable to capture regulation across multiple biological scales. This limits their ability to predict how gene modification strategies will impact lignin and other wood properties. While the first multiscale model of lignin biosynthesis in P. trichocarpa spanned the transcript, protein, metabolic, and phenotypic layers, it did not account for cross-regulatory influences that could impact abundances of untargeted monolignol transcripts and proteins. Here, we present a multiscale model incorporating these cross-regulatory influences for predicting lignin and wood traits from transgenic knockdowns of the monolignol genes. The three main components of this multiscale model are (1) a transcript-protein model capturing cross-regulatory influences, (2) a kinetic-based metabolic model, and (3) random forest models relating the steady state metabolic fluxes to 25 physical traits. We demonstrate that including the cross-regulatory behavior results in smaller predictive error for 23 of the 25 traits. We use this multiscale model to explore the predicted impact of novel combinatorial knockdowns on key bioenergy traits, and identify the perturbation of PtrC3H3 and PtrCAld5H1&2 monolignol genes as a candidate strategy for increasing saccharification efficiencies while reducing negative impacts on wood density and height.
1. Introduction
Due to its recalcitrant chemical and physical nature, lignin is a key barrier to sustainable biofuel and biomaterial production [1], [2], [3], [4]. A phenylpropanoid polymer found in secondary plant cell walls, lignin is entangled with cellulosic biomass making their conversion to biofuel difficult and expensive [2], [5], [6]. Lignin is composed of three main subunits, the H, G, and S monolignols that are synthesized through a series of enzymatic reactions and polymerized with phenolic aldehydes, phenolic alcohols, and unusual metabolites, which are integrated using traditional and nontraditional linkages through radical coupling reactions [7], [8]. The amount and ratio of these monolignols and other components define the content, composition, and structure of lignin [5], [6], [7], [8]. Modifying the expression of the monolignol specific genes associated with the enzymes in the biosynthetic pathway has been shown to alter lignin content and composition as well as associated wood properties, enabling opportunities for increased efficiency of biofuel production [9], [10]. However, lignin plays an important role in plant growth and adaptation [6], [11], and many attempts to alter the structure and composition of lignin have resulted in plants with other unfavorable phenotypes such as dwarfism [10], [12], [13], [14], [15], [16], [17]. Researchers have turned to computational models to explore modifications to the lignin pathway, which will result in favorable lignin and bioenergy traits while avoiding unfavorable phenotypic properties.
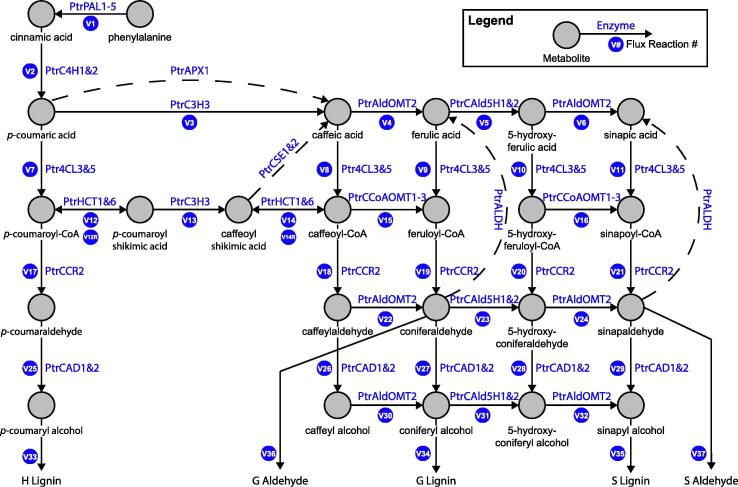
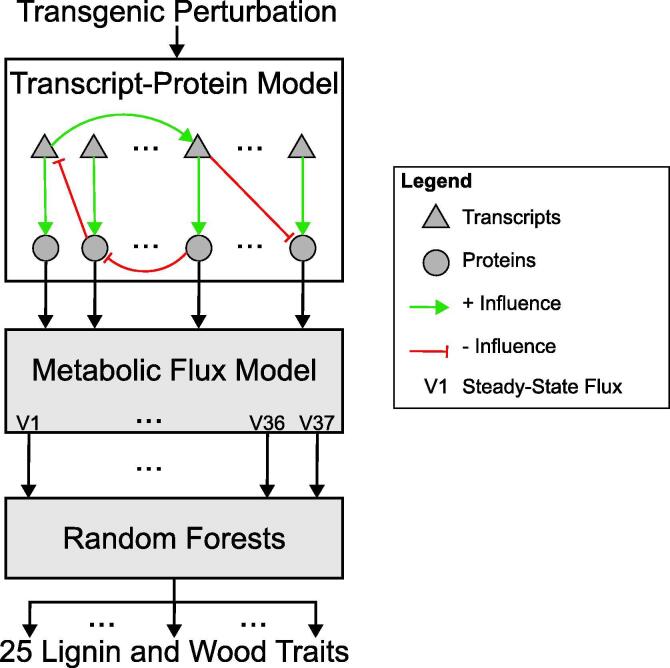
Computational models of the monolignol biosynthesis pathway (Fig. 1) have been developed for several bioenergy crops and trees to explore how gene modifications can alter lignin content and composition [18], [19], [20], [21], [22], [23], [24]. While these models have provided insight into previous unexplained behavior of the metabolic pathway, they lack the ability to fully predict how transgenic modifications impact lignin biosynthesis. These models do not account for the emergent properties that occur across biological levels of organization (e.g., transcriptional, post-transcriptional, and post-translational regulations). The vertical integration of these levels through multiscale modeling approaches is needed to explore what single or combinatorial gene modifications can lead to desirable lignin and wood traits while reducing unfavorable phenotypic properties [9]. The first multiscale model of lignin biosynthesis was developed for Populus trichocarpa to explore how modifying transcript abundance through gene modification strategies impacted 25 lignin and wood traits [9]. This model made the simplifying assumption that the enzyme abundances were dependent only on their associated transcript abundance, ignoring any epistatic cross-regulatory influences. Subsequent work developed a transcript-protein model to capture the effects of these cross-regulatory influences on the monolignol transcript and protein abundances under transgenic knockdowns in P. trichocarpa [25]. The incorporation of such a transcript-protein model that captures the cross-influences observed in the transgenic data into a multiscale model of lignin biosynthesis is expected to improve our ability to predict how different gene modification strategies impact lignin properties and wood traits.
Fig. 1.
Monolignol biosynthetic pathway in P. trichocarpa. The PtrCSE1&2, PtrAPX1, and PtrALDH reactions (dashed lines) are not included in the model as the reactions were discovered after the onset of the study or had not been confirmed in poplar species.
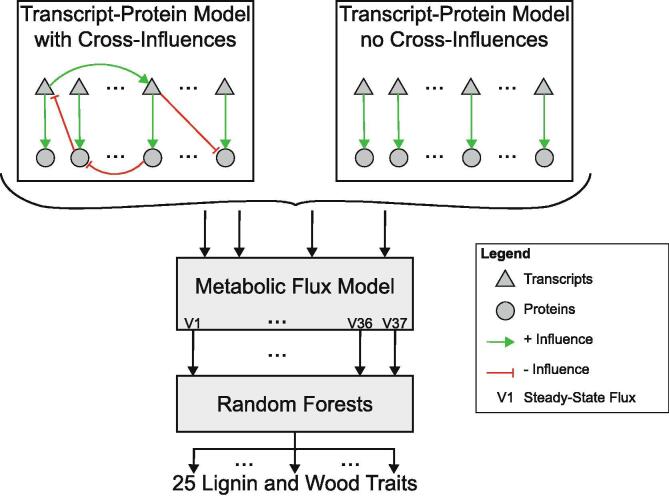
In this paper we present a multiscale model of lignin biosynthesis in P. trichocarpa that connects (1) the transcript-protein model capturing cross-regulatory influences [25]; (2) the kinetic monolignol biosynthesis model [9], [19]; and (3) 25 new random forest models that relate the steady state flux outputs from the kinetic model to 25 lignin and wood traits (Fig. 2)) to predict how these lignin and wood traits are altered by single and combinatorial knockdowns of the monolignol specific genes. We used random forest models to capture the relationships between the steady state fluxes and wood traits due to their ability to capture complex relationships [26]. We used the Minimally Biased Variable Selection in R (MUVR) algorithm [27] to train the 25 random forest models on the steady state fluxes that resulted from the kinetic model when it was simulated using the experimental protein abundances [9]. The MUVR algorithm simultaneously performs variable selection and validation by implementing recursive variable elimination within a repeated double cross-validation (rdCV) procedure. Incorporating the cross-influences at the transcript and protein level improved our ability to predict 23 of the 25 lignin and wood traits, and showed specific improvements of the predictions in the PtrCAld5H1&2, Ptr4CL3&5, PtrHCT1&6, and PtrC3H3 simulated single and family knockdowns. The improved overall prediction of the phenotypes due to the systemic integration of the models across different biological scales is a demonstration of (1) the functionality of the individual models and (2) our ability to integrate these models into a functioning multiscale system.
Fig. 2.
Multiscale model of lignin biosynthesis in Populus trichocarpa.
We used the multiscale model to explore the predicted impact of five novel combinatorial knockdowns on six key lignin and wood bioenergy traits: lignin content, plant height, relative wood density, total sugar content, and the wood saccharification efficiencies for glucose and xylose productions from unpretreated samples. The multiscale model under these novel combinatorial knockdowns predicted favorable changes to the wood bioenergy and plant growth traits not observed in the single gene and gene family knockdowns. Further, we identified the PtrC3H3 and PtrCAld5H1&2 combinatorial knockdown as a candidate for increasing total sugar content and the saccharification efficiencies of glucose and xylose, while mitigating negative impacts of relative wood density and height. The multiscale model presented in this work is a useful tool for exploring the space of combinatorial gene perturbation strategies that achieve increased saccharificaition efficiencies while mitigating negative impacts on plant growth and adaptation.
2. Material and methods
2.1. Experimental data
Wang et al. performed a series of systematic experimental transgenic knockdowns of 21 of the 25 monolignol genes and gene families in P. trichocarpa [9]. PtrCSE1&2, PtrAPX1, and PtrALDH (Fig. 1, dashed lines) were not included in these experiments or the subsequent models [9], [19], [25] that compose the multiscale model presented here, as the reactions were discovered after the onset of their study or had not been confirmed in poplar species. The experiments were divided into six batches and phenomic and proteomic measurements were taken after 6 months of growth in a greenhouse. Multiple independent lines were grown for each transgenic, and up to three of those lines were chosen to represent a range of the expression of the knocked out genes.
2.1.1. Transcriptomics and proteomics data
The absolute abundances for the 21 monolignol transcripts and proteins were measured using RNAseq and protein cleavage-isotope dilution mass spectrometry (PC-IDMS) respectively [9], [28]. To account for batch effects, the transcript and protein abundances were normalized to the to the wildtype of each batch [9], and missing protein abundance values were imputed as described in [25]. The RNAseq libraries are available under GEO accession number GSE78953 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78953), and the proteomics data set is available on CyVerse(https://datacommons.cyverse.org/browse/iplant/home/shared/LigninSystemsDB). Plots comparing the transcript and protein abundances and their log fold-changes can be found in [9], [25].
2.1.2. Phenomic data
Twenty-five phenomic characteristics describing lignin and other wood chemical properties, wood physical properties, and saccharification (extraction of sugar) efficiencies were obtained from subsets of these transgenic lines (Table 1).
Table 1.
The 25 experimentally measured lignin and wood traits.
| Lignin and wood traits |
|---|
| Lignin content |
| Glucose content |
| Xylose content |
| Total sugar content |
| C:L ratio |
| S-subunit abundance |
| G-subunit abundance |
| H-subunit abundance |
| S/G ratio |
| -O-4 linkages |
| -1 linkages |
| -5 linkages |
| - linkages |
| End-group linkages |
| p-hydroxybenzoate |
| hydroxycinnamaldehydes |
| Tree height |
| Tree diameter |
| Stem volume |
| Modulus of elasticity (MOE) |
| Relative wood density |
| Saccharification efficiency of glucose from unpretreated samples |
| Saccharification efficiency of xylose from unpretreated samples |
| Saccharification efficiency of glucose from pretreated samples |
| Saccharification efficiency of xylose from pretreated samples |
The wood chemical properties obtained include lignin content, glucose content, xylose content, total sugar content, and total sugar to lignin (C:L) ratio, which were measured from 181 of the transgenics and 18 of the wildtypes. The abundances of lignin subunit composition (S-subunits, G-subunits, H-subunits, and S/G ratio), interunit linkages (-O-4, -1, -5, -, and end-groups), and two non-monolignol phenolics (p-hydroxybenzoate and hydroxycinnamaldehydes) were further analyzed in 56 transgenics and 8 wildtypes using semi-quantitative two-dimensional nuclear magnetic resonance (2D NMR). NMR data was not measured for any of the Ptr4CL and PtrAldOMT2 knockdowns.
The wood physical properties that were obtained relate to plant growth, wood mechanical strength, and wood density. Tree height, diameter, and stem volume were measured in 171 of the transgenics and 15 wildtypes. The modulus of elasticity (MOE) is a quantitative indication of wood mechanical strength, where a larger MOE indicates a stiffer wood that is less likely to become deformed. The MOE was measured in 108 of the transgenics and 12 of the wildtype trees. Relative wood density was measured in 67 of the transgenics and 9 of the wildtypes.
The last four phenotypic properties that were measured relate to how well enzymes are able to break down the cellulose to glucose, which we refer to as the saccharifciation efficiency. Wood is a promising resource for sustainable biofuel and biomaterial production. However, lignin, which is embedded with the celluloses and hemicelluloses, impedes enzyme saccharification. Acid pre-treatments are often used to facilitate this process, but they are costly and produce enzyme inhibitors [1]. Lowering lignin content has been shown to reduce the need for chemical pre-treatments [29]. The saccharification efficiency was calculated from the amounts of glucose and xylose that were released from pretreated and unpretreated wood samples from 180 of the transgenics and 17 of the wildtypes.
Following the same procedure as the transcripts and protein measurements [9], [25], the measurements from each trait were normalized to the mean wildtype in each of the six batches to remove batch effects. Distributions of the phenomic data are shown in Fig. S1. The phenomic data sets are available on CyVerse (https://datacommons.cyverse.org/browse/iplant/home/shared/LigninSystemsDB).
2.2. Multiscale lignin model
We constructed the multiscale lignin biosynthesis model (Fig. 2) to predict how single and combinatorial transgenic monolignol gene knockdowns impact lignin and other wood traits. The first two components of this model, the transcript-protein model [25] and the kinetic monolignol biosynthesis model [9], [19], were implemented as described in their respective publications. The development of the third component, the random forest models, is described in Section 2.2.1. To simulate transgenic knockdowns, the abundances of the monolignol transcripts being targeted for knockdown were used as inputs to the transcript-protein model. The transcript-protein model predicted the abundance of the other monolignol transcripts and proteins. Those protein abundances were used as inputs to the kinetic metabolic model, which was then run until each of the fluxes reached a steady state. The values of the parameters and initial conditions for metabolite concentrations used here are listed in [9]. The steady state fluxes were then used as inputs to the random forest models, which predicted the twenty-five lignin and wood traits.
2.2.1. Random forest models
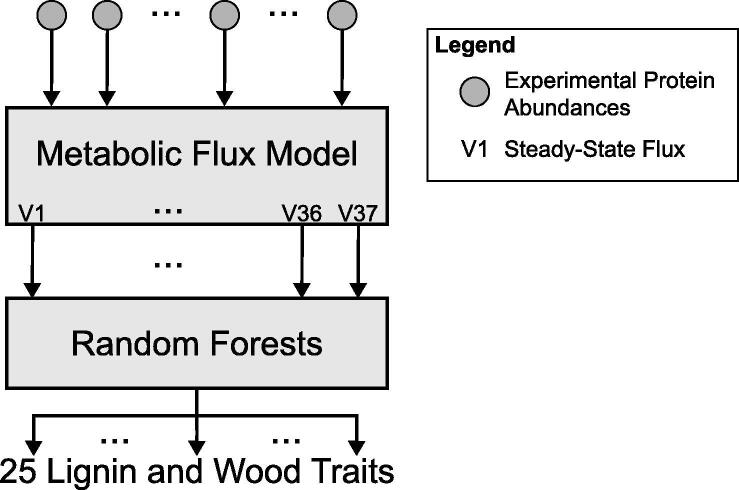
Random forest models were created for each of the 25 lignin and wood traits using the MUVR algorithm [27]. The MUVR algorithm simultaneously performs variable selection and validation by implementing recursive variable elimination within a repeated double cross-validation (rdCV). The models were trained using experimentally measured lignin and wood physical traits from a series of systematic transgenic knockdown experiments [9] (see Section 2.1.2) and the steady state fluxes obtained from the kinetic monolignol pathway model when run with the experimental protein abundances from the same transgenic experiments (Fig. 3, see Section 2.1.1). Table 2 contains the parameters used when calling the MUVR algorithm.
Fig. 3.
Block diagram describing the training of the random forest models.
Table 2.
Parameters used to call the MUVR algorithm.
| MUVR Parameter | Value | Description |
|---|---|---|
| nrep | 50 | Number of MUVR repetitions |
| nOuter | 8 | Number of outer cross-validation segments |
| varRatio | 0.9 | Proportion of variables kept per iteration |
| method | Model type: Random forests | |
| fitness | Fitness metric: root mean square error of predicted test data |
3. Results and discussion
3.1. Random forest model training
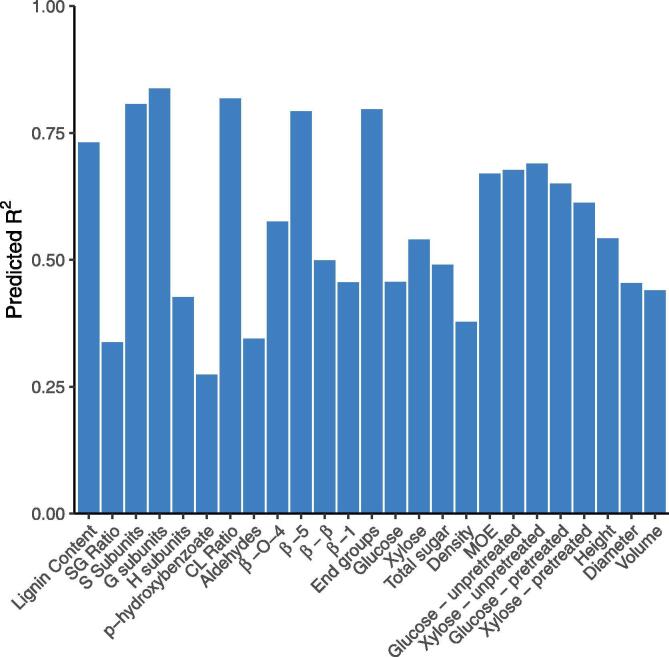
We trained a random forest model for each of the 25 lignin and wood traits using the MUVR algorithm [27] and the steady state fluxes output from the kinetic monolignol biosynthesis model [9], [19] simulated with the experimentally measured protein abundances (Fig. 3, see Section 2.2.1). The trained random forest models range from two to 24 steady state fluxes as predictor variables out of a total 39 fluxes (Table S1). The random forest models captured the variation in the physical traits with an R2 for 11 out of the 25 traits including lignin content, C:L ratio, S subunits, G subunits, linkages, end groups, modulus of elasticity (MOE), glucose saccharification yield from unpretreated samples, xylose saccharification yield from unpretreated samples, glucose saccharification yield from pretreated samples, and xylose saccharification yield from pretreated samples (Fig. 4). The variances of 11 of the 25 predicted phenotypic traits were explained moderately well with R2 values between 0.35 and 0.6, including H subunits, linkages, linkages, linkages, glucose, xylose, total sugar content, relative wood density, height, diameter, and volume. The remaining 3 traits, S/G ratio, p-hydroxybenzoate, and aldehyde content, had prediction R2 values less than 0.35. The low R2 value for S/G ratio can be explained by the poor prediction of one data point that was much higher than any other S/G values (Fig. S2). Since we predict S/G ratio separate from the S and G monolignol predictions, there can be cases where the predicted values are inconsistent due to limited amounts of training data (e.g., Fig. S8). The low R2 values for p-hydroxybenzoate, and aldehyde content could also be due to insufficient training data for these traits, or that factors other than the steady state fluxes are needed to predict these traits.
Fig. 4.
Predicted R2 MUVR model metric for the 25 lignin and wood traits.
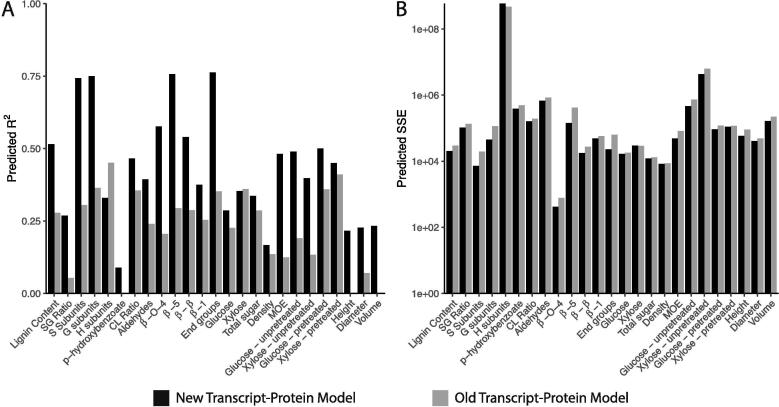
3.2. Impact of cross-influences between the lignin transcripts and proteins on the predicted lignin and wood traits
To evaluate the impact of including the cross-influences between monolignol transcripts and proteins, we simulated the transgenic experiments using the multiscale model with the new transcript-protein model, which captures these cross influences [25], and the old transcript-protein model [9] that assumes each monolignol protein abundance is dependent only on its associated transcript abundance (Fig. 5). For 23 of the lignin and wood traits, the predictions when using the new transcript-protein model had a higher R2 (Fig. 6A) and a lower SSE (Fig. 6B) than predictions obtained using the old transcript-protein model, when compared to the experimentally measured lignin and wood trait values. The exceptions included the H subunits and xylose where the old transcript-protein model had better R2 and SSE metrics. Further, H subunits and the saccharification efficiency of xylose from pretreated samples were the only traits to have an R2 when using the old transcript-protein model, while the new transcript-protein model had 12 traits with a predicted R2 .
Fig. 5.
Block diagram describing how the new transcript-protein model that includes cross-regulatory influences [25] and the old transcript-protein model [9] are connected to the other parts of the multiscale model to evaluate how incorporating the cross-regulatory influences impact lignin and wood trait prediction.
Fig. 6.
(A) R2 and (B) SSE of predicted lignin and wood traits using multiscale model with the new transcript-protein model (black) and the old transcript-protein model (gray).
Additionally, we simulated knockdowns for each monolignol gene and gene family from wildtype levels to a complete knockout at 1% decrements using the new transcript-protein model and the old transcript-protein model. The simulated knockdowns that showed the most differences in the predicted phenotypes between the two transcript-protein models are discussed below. The simulation results for all 25 traits for each individual and family knockdown are located in the Supplemental Figures.
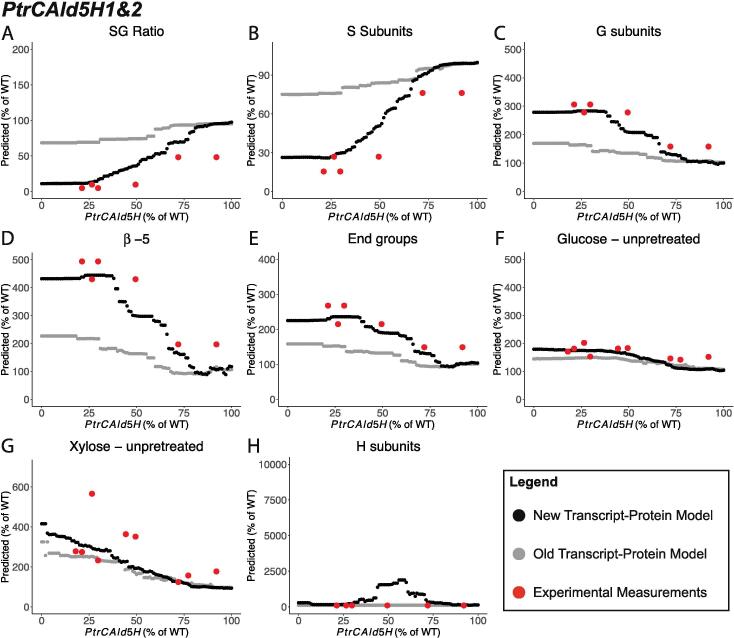
3.2.1. PtrCAld5H1&2 knockdown simulations
We observed the most differences between the predicted phenotypes when using the old transcript-protein model versus the new transcript-protein model in the PtrCAld5H family knockdowns. The predicted results for the two models started to diverge once PtrCAld5H1&2 were knocked down below ~75% of their wildtype levels for eight of the 25 traits: S/G ratio, S subunits, G subunits, H subunits, linkages, end-groups, and the saccharification efficiencies of glucose and xylose from unpretreated samples (Fig. 7). Below 75% of the PtrCAld5H1&2 wildtype levels, the new transcript-protein model led to predictions that were closer to the experimental measurements in seven of those eight traits (Fig. 7A–G). H subunits were the only trait that was better predicted using the old transcript-protein model (Fig. 7H). Our random forest model had the highest prediction SSE (Fig. 6B) for the H subunits, suggesting that a different approach is needed to predict how the H subunits are altered. The predictions from both models were similar to each other for the other 17 lignin and wood traits.
Fig. 7.
Selected predicted traits from simulated knockdowns of PtrCAld5H1&2 using the multiscale model with the new transcript-protein model (black) and the old transcript-protein model (gray); (A) SG ratio, (B) S subunits, (C) G subunits, (D) -5 linkages, (E) End groups, (F) Saccharification efficiency of glucose from unpretreated samples, (G) Saccharification efficiency of xylose from unpretreated samples, and (H) H subunits.
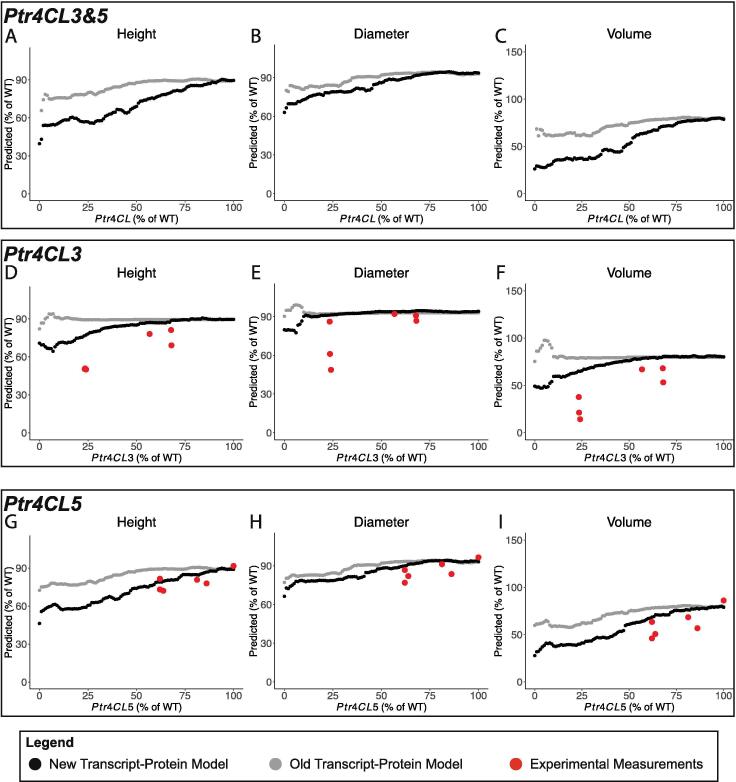
3.2.2. Ptr4CL3&5, Ptr4CL3, and Ptr4CL5 knockdown simulations
Differences in predicted height, diameter, and volume were observed between the new and old transcript-protein models when knockdowns in Ptr4CL3&5, Ptr4CL3, and Ptr4CL5 were simulated (Fig. 8). In all three knockdown scenarios the new transcript-protein model predicted a larger decrease in each of the three traits. We do not have experimental measurements for these three traits when Ptr4CL3&5 were both knocked down (Fig. 8A–C), however, the reduction in the experimentally measured height, diameter, and volume for the single knockdowns, Ptr4CL3 (Fig. 8D–F) and Ptr4CL5 (Fig. 8G–I), support this larger decrease. With an ~25% knockdown of Ptr4CL3, the average height, diameter, and volume were measured to be ~50%, ~65%, and ~25% of their wildtype levels respectively (Fig. 8D–F). At a simulated 25% knockdown of both Ptr4CL3&5, the new transcript-protein model predicted reductions in height, diameter, and volume to ~55%, ~80%, and ~40% of their respective wildtype levels, while the old transcript-protein model only predicted reductions to ~80%, ~85%, and ~65% of their respective wildtype levels (Fig. 8A–C).
Fig. 8.
Predicted tree height, diameter, and stem volume from simulated knockdowns of (A–C) Ptr4CL3&5, (D–F) Ptr4CL3, and (G–I) Ptr4CL5 using the multiscale model with the new transcript-protein model (black) and the old transcript-protein model (gray).
When Ptr4CL5 was knocked down, the predicted height, diameter and volume resulting from the new transcript-protein model were more consistent with the Ptr4CL5 experimental results than the predicted results from the old transcript-protein model were (Fig. 8G–I). The measured height, diameter, and volume from the Ptr4CL3 knockdowns were more severe than the decreases predicted by either model, however, the new transcript-protein model predictions were more similar (Fig. 8D–F).
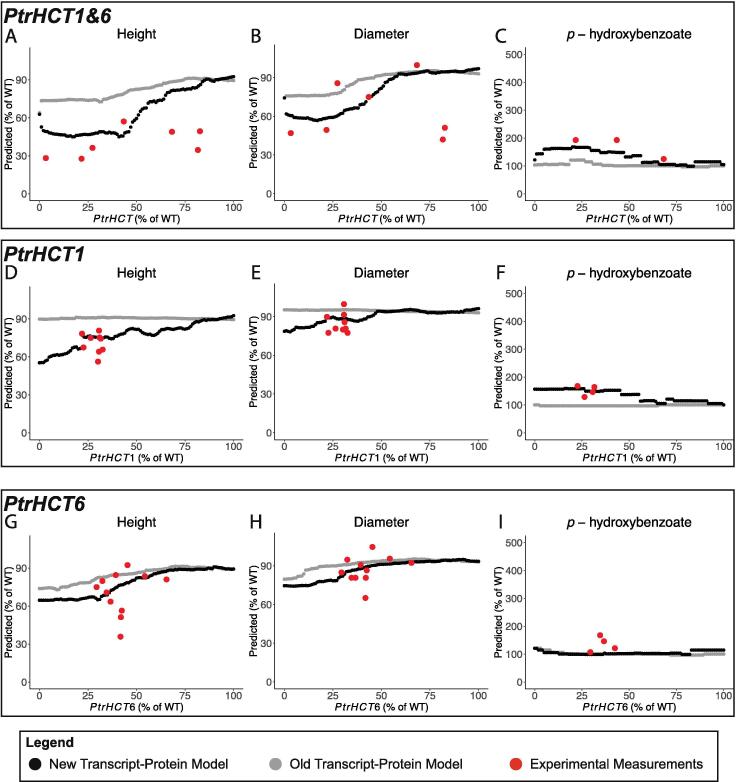
3.2.3. PtrHCT1&6, PtrHCT1, and PtrHCT6 knockdown simulations
The predicted height and diameter traits differed between the new and old transcript-protein models when PtrHCT1&6, PtrHCT1, andPtrHCT6 were knocked down. When PtrHCT1&6 were knocked down the new transcript-protein model predicted a larger decrease in the height and diameter, more closely matching the experimental measurements (Fig. 9A, B). The predicted height and diameter, however, did not reach the experimental levels until the PtrHCT transcripts were knocked down to ~50% of their wildtype levels, despite experimentally observing these values when the transcripts were at ~75% of their wildtype levels. When PtrHCT1 was knocked down (Fig. 9D, E), the predicted height and diameter using the new transcript-protein model matched the decreases observed in the experimental measurements, while no change from wildtype was predicted when the old transcript-protein model was used. The predicted height and diameters from both transcript-protein models when PtrHCT6 was knocked were in line with the experimentally measured heights and diameters (Fig. 9G, H).
Fig. 9.
Predicted tree height, diameter, and p-hydroxybenzoate from simulated knockdowns of (A–C) PtrHCT1&6, (D–F) PtrHCT1, and (G–I) PtrHCT6 using the multiscale model with the new transcript-protein model (black) and the old transcript-protein model (gray).
In the PtrHCT1&6 and PtrHCT1 simulated knockdowns, p-hydroxybenzoate was also different between the two transcript-protein models. The new transcript-protein model predicted an increase in p-hydroxybenzoate, matching the experimental measurements, where the predictions from the old transcript-protein model remained at wildtype levels (Fig. 9C, F). Neither model predicted an increase in p-hydroxybenzoate when PtrHCT6 was knocked down (Fig. 9I), though experimentally an increase was measured similar to PtrHCT1&6 and PtrHCT1 knockdowns.
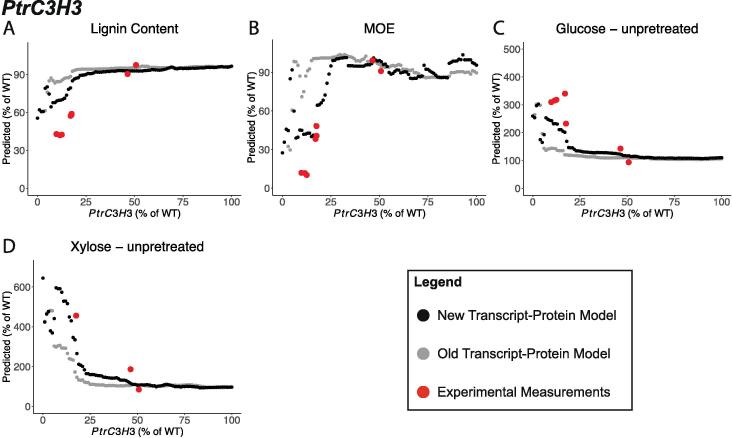
3.2.4. PtrC3H3 knockdown simulations
The predictions for lignin content, MOE, and saccharification efficiencies of glucose and xylose from unpretreated samples differed between the new and old transcript-protein models when PtrC3H3 was knocked down (Fig. 10). The predictions from the two models were largely consistent until the PtrC3H3 was knocked down to ~25% of its wildtype abundance. At this point, the new transcript-protein model predicted a larger decrease in lignin content (Fig. 10A) and MOE (Fig. 10B), and a larger increase in the saccharification efficiencies of glucose and xylose from unpretreated samples (Fig. 10C, D). For all four of these traits, the new transcript-protein model’s predictions were more consistent with the experimental measurements.
Fig. 10.
Selected predicted traits from simulated knockdowns of PtrC3H3 using the multiscale model with the new transcript-protein model (black) and the old transcript-protein model (gray); (A) Lignin Content, (B) MOE, (C) Saccharification efficiency of glucose from unpretreated samples, and (D) Saccharification efficiency of xylose from unpretreated samples.
3.2.5. Capturing the regulatory cross-influences among the lignin specific transcripts and proteins improves lignin and wood trait prediction
Overall, for many of the targeted knockdowns and lignin and wood traits, both transcript-protein models resulted in similar predictions. However, the multiscale model using the new transcript-protein model better estimated changes in S/G ratio, S subunits, G subunits, linkages, and end-groups in the PtrCAld5H1&2 knockdowns; height, volume and diameter in the Ptr4CL3&5, Ptr4CL3, and Ptr4CL5 knockdowns; height, diameter, and p-hydroxybenzoate in the PtrHCT1&6, PtrHCT1, and PtrHCT6 knockdowns; and the saccharification efficiencies of glucose and xylose from unpretreated samples in the PtrCAld5H1&2 and the PtrC3H3 knockdowns.
3.3. Exploring the impact of combinatorial knockdowns on key lignin and wood bioenergy traits
An intended use of this multiscale model is to explore novel combinatorial perturbations of the monolignol genes and gene families to identify potential gene perturbation strategies that yield improved lignin and wood traits. This involves balancing the changes to different physical traits such as aiming to improve saccharification efficiencies while maintaining growth traits like height and relative wood density similar to wildtype. In the following sections we explore how our multiscale model, using the new transcript-protein model, predicted changes in lignin content, height, relative wood density, total sugar content, and the saccharification efficiencies of glucose and xylose from unpretreated samples under five different combinatorial gene perturbations. As the multiscale model was developed with data from transgenic knockdown experiments only, we limited our simulations to combinatorial knockdowns of the monolignol genes. These knockdown combinations were heuristically chosen based on the predicted results from the single and family knockdowns.
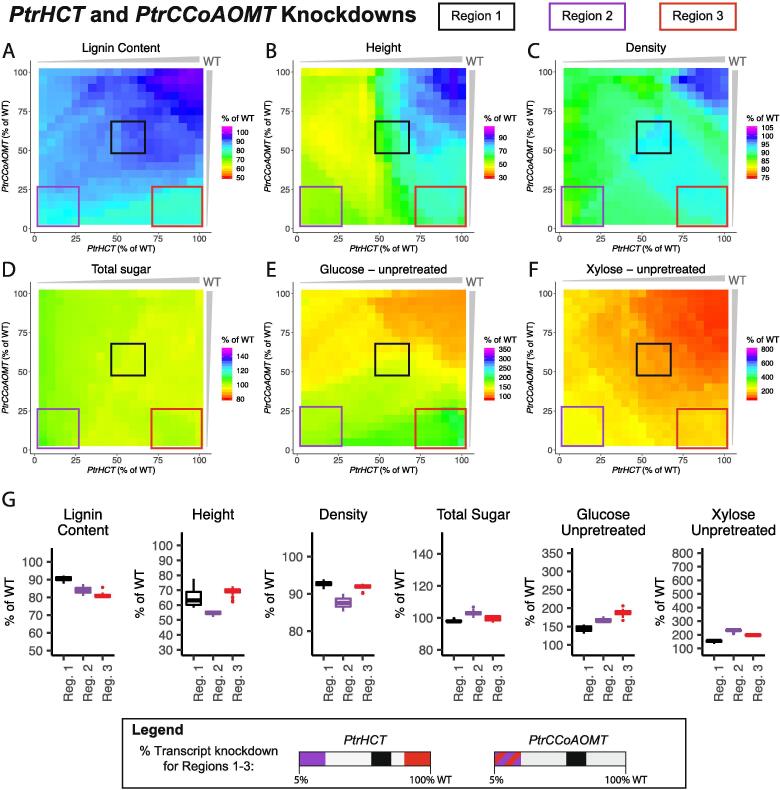
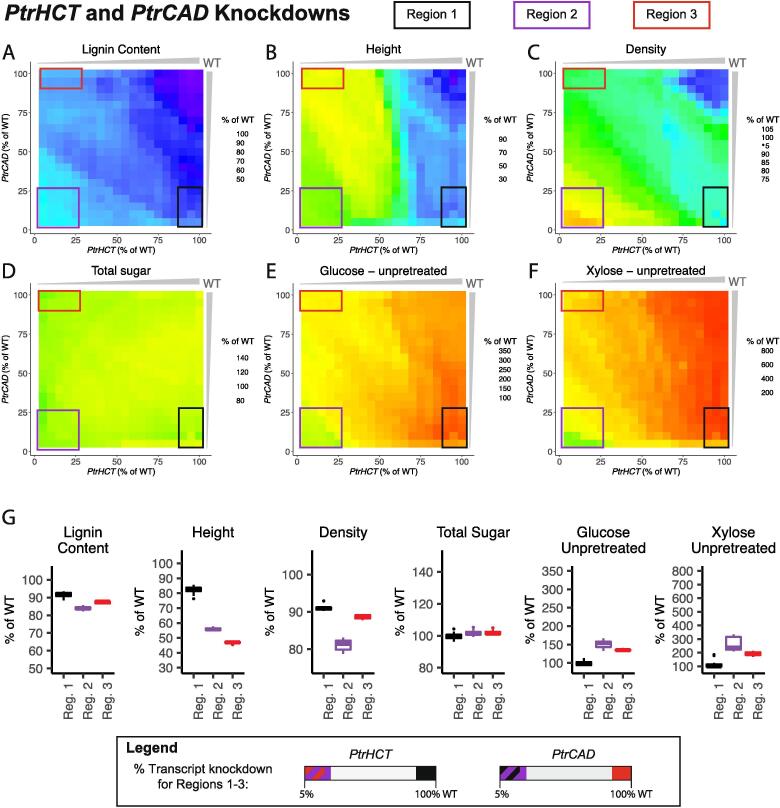
3.3.1. PtrHCT and PtrCCoAOMT combinatorial knockdowns
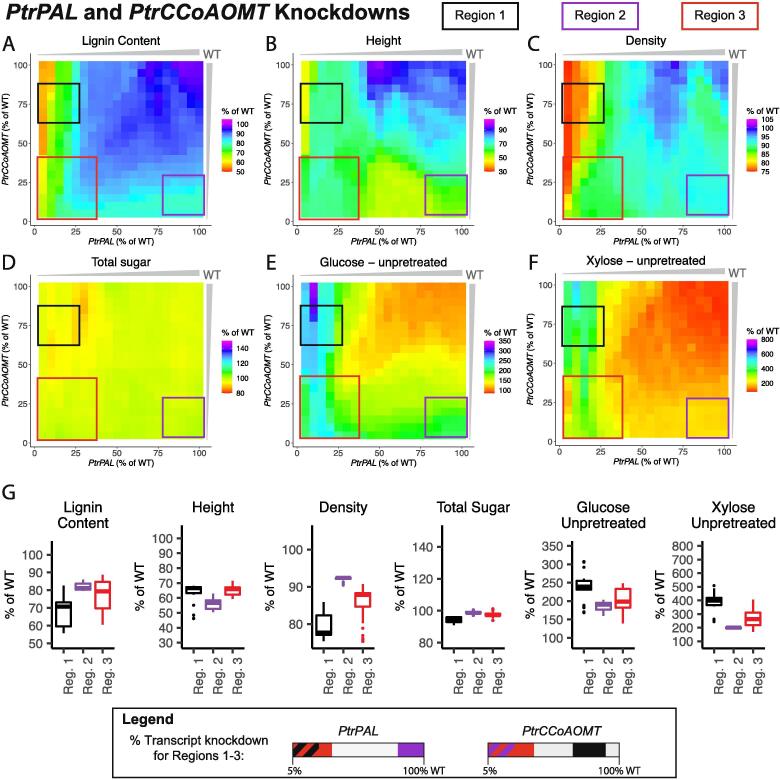
We simulated combinatorially knocking down the PtrHCT and PtrCCoAOMT gene families from 100% to 5% of wildtype levels at 5% decrements, and highlighted three regions of interest in the six lignin and wood traits (Fig. 11A–F). In all three regions, our model predicted an increase in the saccharification efficiencies of glucose and xylose from unpretreated samples ranging from ~144–187% increase for glucose and ~154–231% increase for xylose (Fig. 11G). The largest increase in the saccharification efficiency of xylose from unpretreated samples was found in Region 2 (purple box, Fig. 11), where both gene families were knocked down to low levels. However, this region also had the largest negative predicted impact to height and relative wood density (Fig. 11G). The largest predicted increase in glucose saccharification efficiency was in Region 3 (red box, Fig. 11), where the PtrCCoAOMTgenes were knocked down to low levels, but the PtrHCT genes were only knocked down between 75% and 100% of their wildtype levels. Of the three highlighted areas, this region had the least predicted negative impact on height and the largest decrease in lignin content (Fig. 11G). Region 1 (black box, Fig. 11), where both gene families were knocked down to around half of their wildtype abundances had the smallest increase in the saccharification efficiencies of the three regions, but the predicted relative wood density was the highest at ~93% of its wildtype levels (Fig. 11G).
Fig. 11.
Lignin and wood trait predictions under combinatorial knockdowns of the PtrHCT and PtrCCoAOMT monolignol gene families; (A) Lignin Content, (B) Tree height, (C) Relative wood density, (D) Total sugar content, (E) Saccharification efficiency of glucose from unpretreated samples, and (F) Saccharification efficiency of xylose from unpretreated samples. (G) Boxplots of the predicted values of the six traits in Region 1 (black), Region 2 (purple), and Region 3 (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3.2. PtrPAL and PtrCCoAOMT combinatorial knockdowns
We simulated knocking down the PtrPAL and PtrCCoAOMT families from 100% to 5% of their wildtype levels, and highlighted three regions of interest in the six lignin and wood traits (Fig. 12A–F). In all three of these regions, the multiscale model predicted an increase in the saccharification efficiencies of glucose and xylose from unpretreated samples, ranging from ~150–300% and ~200–500% of their wildtype levels respectively (Fig. 12G). Region 1 (black box, Fig. 12), where the PtrPAL genes were knocked down below 25% of their wildtype levels and the PtrCCoAOMT genes were knocked down to ~75% of their wildtype levels, had the largest increase in the saccharification efficiencies. However, its predicted relative wood density was also the most negatively impacted of the three regions. Region 3 (red box, Fig. 12), where both the PtrPAL and PtrCCoAOMT gene families were knocked down below 40% and 35% of their wildtype levels respectively, had the second largest increase in the saccharification efficiencies, and the second highest predicted relative wood density of the three regions. Region 2 (purple box, Fig. 12), where the PtrCCoAOMT genes were knocked down below 25% of their wildtype levels, and the PtrPAL genes remained around their wildtype levels, showed the least increase in the saccharificaiton efficiencies and the smallest predicted decrease in relative wood density of the three regions. However, Region 2 had the largest predicted decrease in height, while Regions 1 and 3 had similar predicted decreases in tree height (Fig. 12G).
Fig. 12.
Lignin and wood trait predictions under combinatorial knockdowns of the PtrPAL and PtrCCoAOMT monolignol gene families. (A) Lignin Content, (B) Tree height, (C) Relative wood density, (D) Total sugar content, (E) Saccharification efficiency of glucose from unpretreated samples, and (F) Saccharification efficiency of xylose from unpretreated samples. (G) Boxplots of the predicted values of the six traits in Region 1 (black), Region 2 (purple), and Region 3 (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
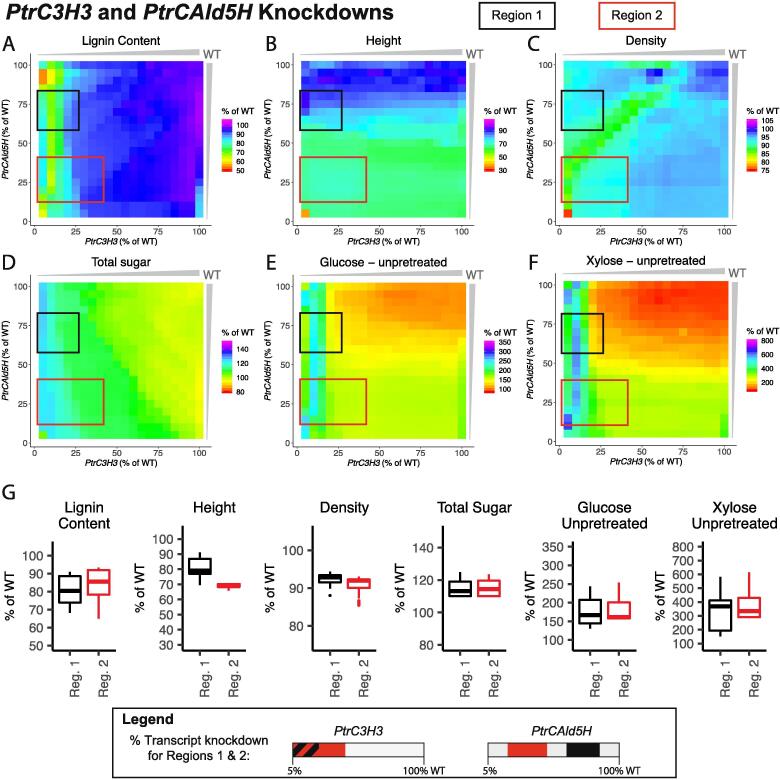
3.3.3. PtrC3H3 and PtrCAld5H combinatorial knockdowns
We simulated knocking down PtrC3H3 and the PtrCAld5H family from 100% to 5% of their wildtype levels, and highlighted two regions of interest in the six lignin and wood traits (Fig. 13A–F). In these combinatorial knockdowns, lignin content (Fig. 13A), height (Fig. 13B), and the saccharification efficiencies (Fig. 13E, F) follow the trends for single PtrC3H3 or PtrCAld5H family knockdowns. When PtrC3H3 was less than 25% of its wildtype levels, lignin content and the saccharification efficiencies were predicted to be similar values regardless of how much the PtrCAld5H genes were knocked down (Fig. 13A, E, F). Similarly, height and the saccharification efficiencies were predicted to change similar to the PtrCAld5H family knockdown for any knockdown level of PtrC3H3 (height) or when PtrC3H3 was greater than 25% of its wildtype (saccharification efficiencies) (Fig. 13B, E, F). Relative wood density (Fig. 13C) and total sugar content (Fig. 13D) were predicted to have a combinatorial effect as PtrC3H3 and the PtrCAld5H genes were knocked down.
Fig. 13.
Lignin and wood trait predictions under combinatorial knockdowns of the PtrC3H3 and PtrCAld5H monolignol gene and gene family. (A) Lignin Content, (B) Tree height, (C) Relative wood density, (D) Total sugar content, (E) Saccharification efficiency of glucose from unpretreated samples, and (F) Saccharification efficiency of xylose from unpretreated samples. (G) Boxplots of the predicted values of the six traits in Region 1 (black) and Region 2 (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The two highlighted regions had similar predicted changes in 5 of the 6 traits, with the exception of height, where Region 2 (red box, Fig. 13) was predicted to have a more negative impact (Fig. 13G). Region 2, however, achieved similar levels of improvement over a larger range of knockdown of PtrC3H3 than Region 1 (black box, Fig. 13). In both regions, the saccharification efficiencies of glucose and xylose from unpretreated samples were predicted to range from ~150–250% and ~200–600% respectively (Fig. 13G).
3.3.4. PtrHCT and PtrCAD combinatorial knockdowns
We simulated knocking down the PtrHCT and PtrCAD families from 100% to 5% of wildtype levels, and highlight three regions in the six lignin and wood traits (Fig. 14A–F). These regions correspond to knocking down the PtrCAD genes to below 25% their wildtype levels while keeping the PtrHCT genes around their wildtype levels (Region 1, black box, Fig. 14), knocking down both the PtrHCT and PtrCAD genes below 25% their wildtype levels (Region 2, purple box, Fig. 14), and knocking down the PtrHCT genes below 25% their wildtype levels while keeping the PtrCAD genes around their wildtype levels (Region 3, red box, Fig. 14). Our multiscale model predicted that the combinatorial effect of knocking down both gene families will result in higher sacharification efficiencies for glucose and xylose from unpretreated samples than knocking down only one of the gene families (Fig. 14G). Relative wood density was predicted to be lowest in this region, however, the predicted height in this region is higher than the predicted height when only the PtrHCT genes are knocked down (Fig. 14G).
Fig. 14.
Lignin and wood trait predictions under combinatorial knockdowns of the PtrHCT and PtrCAD monolignol gene families. (A) Lignin Content, (B) Tree height, (C) Relative wood density, (D) Total sugar content, (E) Saccharification efficiency of glucose from unpretreated samples, and (F) Saccharification efficiency of xylose from unpretreated samples. (G) Boxplots of the predicted values of the six traits in Region 1 (black), Region 2 (purple), and Region 3 (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
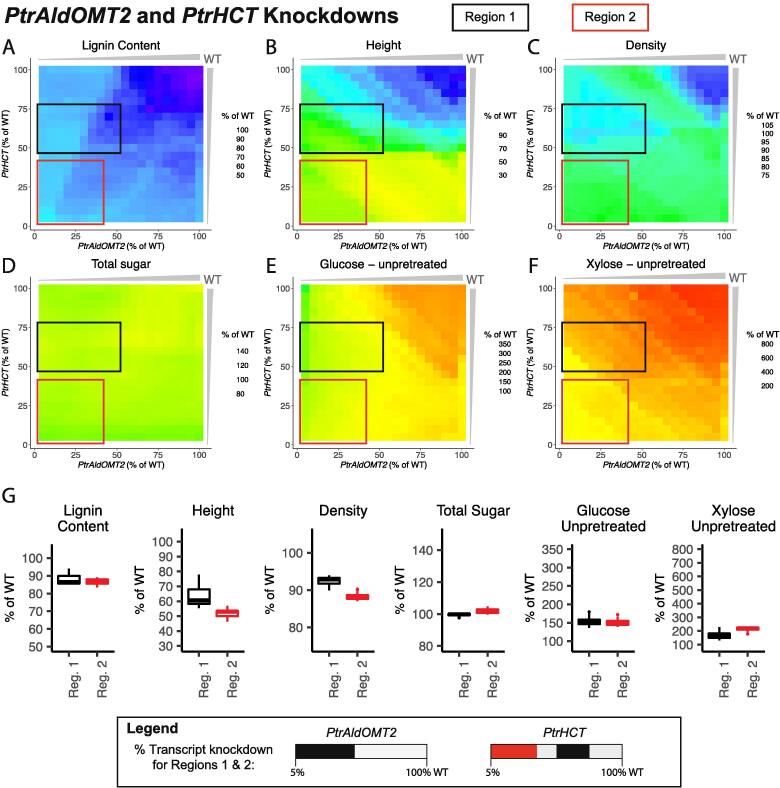
3.3.5. PtrAldOMT2 and PtrHCT combinatorial knockdowns
We simulated knocking down PtrAldOMT2 and the PtrHCT family from 100% to 5% of wildtype levels, and highlight two regions of interest in the six lignin and wood traits (Fig. 15A–F). In both of these regions, the multiscale model predicted a slight reduction in lignin content to ~87% of its wildtype levels and increased predicted saccharification efficiency of glucose from unpretreated samples to ~150% of its wildtype levels. Region 2 (red box, Fig. 15), where the PtrHCT genes were knocked down to below 40% of their wildtype levels and PtrAldOMT2 was knocked down below 40% of its wildtype levels, had a larger predicted increase in the saccharification efficiency of xylose from unpretreated samples to ~215% of its wildtype levels. However this region also had a lower predicted height and relative wood density, ~50% and ~88% of their wildtype levels respectively (Fig. 15G). Region 1 (black box, Fig. 15), where PtrAldOMT2 was knocked down below 50% and the PtrHCT genes were knocked down to between 50–75% of their wildtype levels, only saw a predicted increase in saccharification efficiency of xylose from unpretreated samples to ~175% of its wildtype levels. Height and density, however, were predicted to be around 60% and 93% of their wildtype levels respectively (Fig. 15G).
Fig. 15.
Lignin and wood trait predictions under combinatorial knockdowns of the PtrAldOMT2 and PtrHCT monolignol gene and gene family. (A) Lignin Content, (B) Tree height, (C) Relative wood density, (D) Total sugar content, (E) Saccharification efficiency of glucose from unpretreated samples, and (F) Saccharification efficiency of xylose from unpretreated samples. (G) Boxplots of the predicted values of the six traits in Region 1 (black) and Region 2 (red). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3.6. Combinatorial knockdowns of the lignin genes and gene families could lead to improved bioenergy traits
These five examples demonstrate that combinatorial knockdowns of the monolignol gene and gene families could lead to improved lignin and wood traits beyond what has been observed in the single gene or gene families knockdowns. Further, combinatorial knockdowns could improve our ability to identify gene perturbation strategies that improve bioenergy traits while mitigating negative impacts to plant growth and adaptation. Previously, Wang et al., identified the combinatorial knockdown of the PtrPAL and PtrCCoAOMT monolignol gene families as a possible combination for maximizing wood density, saccharification efficiencies, and C:L ratio [9]. This knockdown consists of 8 genes, PtrPAL1-5 and PtrCCoAOMT1-3, which is an impractical number of genes to simultaneously silence. Our model predicted similar or greater increases in the saccharification efficiencies of glucose and xylose from unpretreated samples when only PtrC3H3 and PtrCAld5H1&2 were knocked down (Fig. 13E–G) versus predictions obtained when PtrPAL1-5 and PtrCCoAOMT1-3 were knocked down (Fig. 12E–G). Our model also predicted smaller negative impacts on relative wood density and height, as well as a larger increase in total sugar content when PtrC3H3 and PtrCAld5H1&2 were knocked down (Fig. 12, Fig. 13). These results suggest that knocking down 3 genes (PtrC3H3 and PtrCAld5H1&2) could achieve similar traits as knocking down 8 genes, and is more experimentally feasible. Combinatorial knockdown simulations of our model have not been validated as there are currently no published combinatorial knockdown studies for P. trichocarpa. Comparisons to single and combinatorial knockdowns in hybrid poplar and tobacco were assessed (Supplemental Text 1), and showed some consistency.
4. Conclusion
We developed a multiscale model capturing transcript, protein, metabolic, and phenotypic layers of lignin biosynthesis in P. trichocarpa. This multiscale model is composed of three components (1) a transcript-protein model that includes cross-regulatory influences [25], (2) a kinetic monolignol biosynthesis model [9], [19] that uses the predicted protein abundances to predict the steady state fluxes in the monolignol biosynthesis pathway, and (3) 25 random forest models that relate the steady state monolignol fluxes to lignin and other wood traits of interest to the bioenergy and biomaterials industries. Incorporating the regulatory cross-influences between the monolignol transcripts and proteins improved prediction of 23 of the 25 lignin and wood traits. Further, when including the regulatory cross-influences, our multiscale model better estimated the changes in S/G ratio, S subunits, G subunits, linkages, and end-groups in simulated knockdowns of the PtrCAld5H1&2; height, volume and diameter in simulated single and family knockdowns of Ptr4CL3, Ptr4CL5, PtrHCT1, PtrHCT6, and PtrHCT1&6; p-hydroxybenzoate in the PtrHCT1 and PtrHCT1&6 simulated knockdowns; and the saccharification efficiencies of glucose and xylose productions in the simulated single and family knockdowns of PtrC3H3 and PtrCAld5H1&2. We used the multiscale model to explore the predicted impact of five novel combinatorial knockdowns, on six bioenergy and plant growth traits. Our model predicted that through combinatorial knockdowns we can alter the lignin and wood traits in ways not seen in the single gene or gene family knockdowns, such as increasing saccharification efficiencies in a combined knockdown of the PtrHCT and PtrCAD gene families. We further identified the combinatorial knockdown of the PtrC3H3 and PtrCAld5H1&2 genes as a candidate for increasing the saccharification efficiencies of glucose and xylose, and total sugar content, while mitigating negative impacts of relative wood density and height.
By exploring combinatorial knockdowns, gene perturbation strategies can be identified that increase these saccharification efficiencies, or other bioenergy traits, while reducing negative impacts to plant growth and adaptation. Future work will involve experimentally testing and validation of the combinatorial knockdown multiscale model predictions, and developing a systematic multi-objective optimization for exploring the space of these knockdowns for user-defined objectives. Beyond optimizing for set traits, these objectives could include constraints on the number of genes that would have to be perturbed, or the size of the perturbation range, to predict a desired set of traits.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Megan L. Matthews: Conceptualization, Methodology, Investigation, Formal analysis, Software, Writing - original draft, Writing - review & editing, Visualization. Jack P. Wang: Conceptualization, Methodology, Writing - review & editing. Ronald Sederoff: Conceptualization, Writing - review & editing, Funding acquisition. Vincent L. Chiang: Conceptualization, Methodology, Writing - review & editing, Project administration, Funding acquisition. Cranos M. Williams: Conceptualization, Methodology, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgements
We thank David C. Muddimann for his work quantifying the proteomics and John Ralph for his work quantifying the lignin structures used in this manuscript. This work was supported in part by the Innovation Project of State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University, Grant No. A01), the Fundamental Research Funds for the Central Universities of China grant 2572018CL01, Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team), National Science Foundation, Grant DBI-0922391, and by the National Physical Science Consortium Graduate Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.csbj.2020.11.046.
Supplementary data
The following are the Supplementary data to this article:
References
- 1.Yang B., Wyman C.E. Pretreatment: the key to unlocking low-cost cellulosic ethanol. Biofuels Bioprod Biorefin. 2008;2(1):26–40. doi: 10.1002/bbb.49. [DOI] [Google Scholar]
- 2.Valdivia M., Galan J.L., Laffarga J., Ramos J.-L. Biofuels 2020: biorefineries based on lignocellulosic materials. Microb Biotechnol. 2016;9(5):585–594. doi: 10.1111/1751-7915.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang V.L. From rags to riches. Nat Biotechnol. 2002;20(6):557–558. doi: 10.1038/nbt0602-557. [DOI] [PubMed] [Google Scholar]
- 4.Chen F., Dixon R.A. Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol. 2007;25(7):759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- 5.Freudenberg K. Lignin: its constitution and formation from p-hydroxycinnamyl alcohols. Science. 1965;148(3670):595–600. doi: 10.1126/science.148.3670.595. [DOI] [PubMed] [Google Scholar]
- 6.Higuchi T. Biochemistry and molecular biology of wood. Springer; Berlin, Heidelberg: 1997. Biosynthesis of wood components; pp. 93–262. [DOI] [Google Scholar]
- 7.Wilkerson C.G., Mansfield S.D., Lu F., Withers S., Park J.-Y., Karlen S.D., Gonzales-Vigil E., Padmakshan D., Unda F., Rencoret J., Ralph J. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science. 2014;344(6179):90–93. doi: 10.1126/science.1250161. [DOI] [PubMed] [Google Scholar]
- 8.del Río J.C., Rencoret J., Gutiérrez A., Elder T., Kim H., Ralph J. Lignin monomers from beyond the canonical monolignol biosynthetic pathway: another brick in the wall. ACS Sustain Chem Eng. 2020;8(13):4997–5012. doi: 10.1021/acssuschemeng.0c01109. [DOI] [Google Scholar]
- 9.Wang J.P., Matthews M.L., Williams C.M., Shi R., Yang C., Tunlaya-anukit S., Chen H.-C., Li Q., Liu J., Lin C.-Y., Naik P., Sun Y.-H., Loziuk P.L., Yeh T.-F., Kim H., Gjersing E., Shollenberger T., Shuford C.M., Song J., Miller Z., Huang Y.-Y., Edmunds C.W., Liu B., Sun Y., Lin Y.-C.J., Li W., Chen H., Peszlen I., Ducoste J.J., Ralph J., Chang H.-M., Muddiman D.C., Davis M.F., Smith C., Isik F., Sederoff R., Chiang V.L. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat Commun. 2018;9(1):1579. doi: 10.1038/s41467-018-03863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon RA, Reddy MSS, Gallego-Giraldo L. Monolignol biosynthesis and its genetic manipulation: the good, the bad, and the ugly. In: Recent advances in polyphenol research. John Wiley & Sons Ltd; 2014. Ch. 1, p. 1–38. doi:10.1002/9781118329634.ch1.
- 11.Vance C.P., Kirk T.K., Sherwood R.T. Lignification as a mechanism of disease resistance. Annu Rev Phytopathol. 1980;18(1):259–288. doi: 10.1146/annurev.py.18.090180.001355. [DOI] [Google Scholar]
- 12.Boerjan W., Ralph J., Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54(1):519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 13.Umezawa T. Lignin modification in planta for valorization. Phytochem Rev. 2018;17(6):1305–1327. doi: 10.1007/s11101-017-9545-x. [DOI] [Google Scholar]
- 14.Sederoff R.R., MacKay J.J., Ralph J., Hatfield R.D. Unexpected variation in lignin. Curr Opin Plant Biol. 1999;2(2):145–152. doi: 10.1016/S1369-5266(99)80029-6. [DOI] [PubMed] [Google Scholar]
- 15.Elkind Y., Edwards R., Mavandad M., Hedrick S.A., Ribak O., Dixon R.A., Lamb C.J. Abnormal plant development and down-regulation of phenylpropanoid biosynthesis in transgenic tobacco containing a heterologous phenylalanine ammonia-lyase gene. Proc Nat Acad Sci. 1990;87(22):9057–9061. doi: 10.1073/pnas.87.22.9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu W.-J., Harding S.A., Lung J., Popko J.L., Ralph J., Stokke D.D., Tsai C.-J., Chiang V.L. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol. 1999;17(8):808–812. doi: 10.1038/11758. [DOI] [PubMed] [Google Scholar]
- 17.Li L., Zhou Y., Cheng X., Sun J., Marita J.M., Ralph J., Chiang V.L. Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Nat Acad Sci USA. 2003;100(8):4939–4944. doi: 10.1073/pnas.0831166100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J.P., Matthews M.L., Naik P.P., Williams C.M., Ducoste J.J., Sederoff R.R., Chiang V.L. Flux modeling for monolignol biosynthesis. Curr Opin Biotechnol. 2019;56:187–192. doi: 10.1016/j.copbio.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Wang J.P., Naik P.P., Chen H.-C., Shi R., Lin C.-Y., Liu J., Shuford C.M., Li Q., Sun Y.-H., Tunlaya-Anukit S., Williams C.M., Muddiman D.C., Ducoste J.J., Sederoff R.R., Chiang V.L. Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell. 2014;26(3):894–914. doi: 10.1105/TPC.113.120881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y., Escamilla-Treviño L., Dixon R.A., Voit E.O. Functional analysis of metabolic channeling and regulation in lignin biosynthesis: a computational approach. PLOS Comput Biol. 2012;8(11) doi: 10.1371/journal.pcbi.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee Y., Chen F., Gallego-Giraldo L., Dixon R.A., Voit E.O. Integrative analysis of transgenic alfalfa (Medicago sativa L.) suggests new metabolic control mechanisms for monolignol biosynthesis. PLOS Comput Biol. 2011;7(5) doi: 10.1371/journal.pcbi.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y., Voit E.O. Mathematical modeling of monolignol biosynthesis in Populus xylem. Math Biosci. 2010;228(1):78–89. doi: 10.1016/J.MBS.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Faraji M., Fonseca L.L., Escamilla-Treviño L., Dixon R.A., Voit E.O. Computational inference of the structure and regulation of the lignin pathway in Panicum virgatum. Biotechnol Biofuels. 2015;8(1):151. doi: 10.1186/s13068-015-0334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faraji M., Voit E.O. Improving bioenergy crops through dynamic metabolic modeling. Processes. 2017;5(4):61. doi: 10.3390/pr5040061. [DOI] [Google Scholar]
- 25.Matthews M.L., Wang J.P., Sederoff R., Chiang V.L., Williams C.M. Modeling cross-regulatory influences on monolignol transcripts and proteins under single and combinatorial gene knockdowns in Populus trichocarpa. PLOS Comput Biol. 2020;16(4) doi: 10.1371/journal.pcbi.1007197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 27.Shi L., Westerhuis J.A., Rosén J., Landberg R., Brunius C. Variable selection and validation in multivariate modelling. Bioinformatics. 2019;35(6):972–980. doi: 10.1093/bioinformatics/bty710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuford C.M., Li Q., Sun Y.-H., Chen H.-C., Wang J., Shi R., Sederoff R.R., Chiang V.L., Muddiman D.C. Comprehensive quantification of monolignol-pathway enzymes in Populus trichocarpa by protein cleavage isotope dilution mass spectrometry. J Proteome Res. 2012;11(6):3390–3404. doi: 10.1021/pr300205a. [DOI] [PubMed] [Google Scholar]
- 29.Min D., Li Q., Jameel H., Chiang V., Chang H.-M. The cellulase-mediated saccharification on wood derived from transgenic low-lignin lines of black cottonwood (Populus trichocarpa) Appl Biochem Biotechnol. 2012;168(4):947–955. doi: 10.1007/s12010-012-9833-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.