Abstract
Background
Exosomes derived from stem cells have been demonstrated to be good candidates for the treatment of osteochondral injury. Our previous studies have demonstrated that mechanical stimulation could be crucial for the secretion of exosomes derived from umbilical cord mesenchymal stem cells (U-MSCs). Therefore, we explore whether mechanical stimulation caused by a rotary cell culture system (RCCS) has a beneficial effect on exosome yield and biological function.
Methods
U-MSCs were subjected to an RCCS at different rotational speeds and exosomes were characterised by transmission electron microscopy, nanoparticle tracking analysis and western blotting. small-interfering RNAs of Rab27a (siRNA-Rab27a) was used to reduce exosome production. Quantitative real-time PCR (qRT-PCR) was used to detect the expression of mechanically sensitive long non-coding RNA H19 (LncRNA H19). The effects of exosomes on chondrocyte proliferation were examined using cell counting kit-8 (CCK-8), toluidine blue staining and a series of related genes. Annexin V-FITC and PI (V-FITC/PI) flow cytometry was used to detect the effect of exosomes on the inhibition of chondrocyte apoptosis. Macroscopic evaluation, MRI quantification and immunohistochemical staining were conducted to investigate the in vivo effects of exosomal LncRNA H19 through SD rat cartilage defect models.
Results
RCCS significantly promoted exosome production at 36 rpm/min within 196 h. Mechanical stimulation was able to increase the expression level of exosomes. The exosomal LncRNA H19 was found to promote chondrocyte proliferation and matrix synthesis and inhibit apoptosis in vitro. Chondral regeneration activity was lost in LncRNA H19-defective exosomes. The injection of exosomal LncRNA H19 in vivo resulted in improved macroscopic assessment, MRI quantification and histological analysis. Moreover, exosomal LncRNA H19 was able to relieve pain levels during the early stages of cartilage repair in an animal experiment.
Conclusion
Our findings confirmed that mechanical stimulation can enhance exosome yield as well as biological function for the repair of cartilage defects. The underlying mechanism may be related to the high expression of LncRNA H19 in exosomes. The translational potential of this article: This study provides a theoretical support of optimizing exosome production. It advances the yield of mesenchymal stem cell exosome and facilitate the clinical application to repair of osteochondral damage.
Keywords: Chondrocytes, Exosomes, LncRNA H19, Mechanical stimulation, Mesenchymal stem cells
Introduction
Mesenchymal stem cells (MSCs) have been proven to have a therapeutic effect on cartilage defects or osteoarthritis [1]. It has been shown through in vivo experiments that MSCs could differentiate into chondrocytes. Therefore, the goal of those studies was to better promote MSC differentiation into chondrocytes. Our research group previously found that mechanical stimulation is a positive physical regulator of chondrogenic differentiation [2]. Both the generation of mechanical stimulation with a special instrument [3] and the transfection of mechanically sensitive genes [4] promoted increased chondrogenic capacity and matrix deposition in MSCs. In in vivo experiments, MSCs that were mechanically prestimulated improved the early stages of cartilage defect repair [4,5].
Although the use of MSCs to repair cartilage is based on the hypothesis that MSCs act as regenerative cells to stimulate endogenous repairs, our research group found that only a small number of transplanted MSCs actually survive in the body [5]. Consequently, only a small number of MSCs ultimately differentiate into chondrocytes. The reparative effect of MSCs is now thought to be due to a paracrine effect via the secretion of a variety of nutrient factors, rather than through a cell replacement effect [6]. Our previous study demonstrated that exosomes derived from U-MSCs regulate chondrocyte homeostasis and coordinate subsequent regeneration processes such as cell proliferation, migration, differentiation and matrix synthesis [7]. Therefore, we speculate that mechanical stimulation may not only enhance the therapeutic effect of MSCs, but may also optimise the biological function of their paracrine factors in cartilage defect repair.
Exosomes are nanosized (50–150 nm) extracellular vesicles that participate in numerous physiological and pathological phenomena [8,9]. Exosomes derived from stem cells have been demonstrated to be good candidates for the treatment of injured tissues or to act as natural carriers of therapeutic molecules [10]. Exosomes have been shown to mediate the communication between cells by transporting noncoding RNAs [11]. Long noncoding RNAs (LncRNAs), a class of non-protein coding transcribed RNA molecules >200 nucleotides in size, are involved in a variety of biological processes at both the transcriptional and post-transcriptional levels [12]. Studies have confirmed that LncRNAs can be enriched in many kinds of cells and that these LncRNAs are released through exosomes [13].
In this study, we used a rotary cell culture system (RCCS) to simulate a mechanical environment. We first explored the optimal mechanical stimulation for the generation of exosomes from U-MSCs by RCCS. To our surprise, RCCS significantly promoted exosome production at 36 rpm/min within 196 h. RCCS was able to attenuate the downregulation of exosome production caused by inhibiting Rab27a. More importantly, we found that the expression of LncRNA H19 was highly upregulated in exosomes produced in a mechanical environment. Mechanical stimulation promoted the chondral regeneration activity of exosomes when they were co-cultured with human chondrocytes. These beneficial effects were suppressed in LncRNA H19-deficient U-MSCs both in vivo and in vitro. In summary, this study demonstrated that mechanical stimulation not only enhances exosome yield but also promotes chondral damage repair by upregulating LncRNA H19 in exosomes derived from U-MSCs.
Method
Cell culture in RCCS
In this study, we used the RCCS system to simulate mechanical stimulation. The culture container was invented by NASA to simulate microgravity. In the RCCS instrument, the cells–carrier complex is suspended in the medium while the container is rotated, which is similar to the homogeneous liquid suspension track [14]. The distribution of cells will be random because of the surface gravity vector. The combination of simultaneous hydrostatic pressure, shear stress and buoyancy force generates a microgravity environment beneficial to cell adhesion, aggregation and proliferation throughout the duration of incubation [15].
The protocol for cell culture was based on our previous studies [3,5]. Briefly, Cytodex 3 microcarriers (GE Healthcare Life Sciences, Little Chalfont, UK) were used to provide a stable but non-rigid surface that allows the adhesion of U-MSCs. Primary umbilical cord mesenchymal stem cells (U-MSCs) (Shanghai Wonderful Medical Science Co. Ltd, UC-MSC-7530) were mixed thoroughly in exosome-free conditioned medium (Dulbecco's Modified Eagle Media: Nutrient Mixture F-12 (DMEM-F12) containing 10% foetal bovine serum and 1% penicillin/streptomycin, 4 × 105 cells/mL). The rotational speed of RCCS was initially set to 10 rpm/min to allow cells to fully make contact with the microcarriers. Then, the rotational speed was increased to 15–48 rpm/min. Culture medium was collected every 48 h for exosome extraction.
Isolation of U-MSCs-Exos
We extracted three kinds of exosomes: (1) exosomes from U-MSCs cultured normally without RCCS mechanical stimulation (N-Exos); (2) exosomes from U-MSCs transfected with siRNA H19 in a mechanical environment stimulated by RCCS (si-Exos) and (3) exosomes from U-MSCs with mechanical stimulation in RCCS (S-Exos). The procedure for the isolation of exosomes was adapted from Thery et al. [8]. First, supernatants collected from the RCCS were centrifuged at 3000 g for 15 min and then at 20,000 g for 45 min. Supernatants were passed through a 0.22-μm filter and then centrifuged at 110,000 g for 70 min to pellet exosomes. 5 ml To resuspend the pellets, PBS was used and the suspension was then centrifuged at 110,000 g for 70 min. All operations were performed at 4 °C. The final exosome pellets were stored at −80 °C. The Bradford method (Beyotime, Shanghai, China) was used to quantify the protein concentration of the exosomes.
Transmission electron microscopy
Exosomes were suspended in buffer and dropped on the copper grid. After drying, they were fixed with 3% (w/v) glutaraldehyde for 2 h and then negatively stained with 2% uranyl acetate for 30 s. Transmission electron microscopy was used to observe the samples.
Nanoparticle tracking analysis
The ZetaView instrument was used to measure the size distribution of exosomes. Exosomes were diluted 500-fold with buffer and added to the nanopores for measurement. Readings were recorded using a particle analyser.
Transfection assay
The specific small-interfering RNAs (siRNA) targeting Rab27a and LncRNA H19 were purchased from Invitrogen (Gene, Shanghai, China). siRNA transfection was performed using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. The sequences of siRNAs for gene silencing are listed in Supplemental Table 1.
Chondrocyte toluidine blue staining
Chondrocytes (BNCC339995, Beina Biology Research Institute, China) of passage 3 were treated with three types of exosomes at a concentration of 10 μg/ml for three days. Chondrocytes were digested and smeared. Toluidine blue staining solution (0.1%) was used to dye digested cells for 2 min. Then, the cell smear was gently washed with distilled water for 3 min. After dyeing, the cells were observed through microscopic examination. Image J software was used to calculate the average optical density, which represents the intensity of staining.
Chondrocyte proliferation assay (Cell counting Kit-8)
The Cell Counting Kit-8 assay (Beyotime, Shanghai, China) was used to determine the proliferative effect of exosomes on chondrocytes. Chondrocytes (2 × 103 cells/well) were seeded on 96-well cell plates and co-cultured with exosomes (10 μg/ml) for four days. The media was changed daily. The standard curves were made by measuring the optical density value at 450 nm.
Cell apoptosis assay
Chondrocyte apoptosis was detected using V-FITC/PI according to the manufacturer's instructions (Beyotime, Shanghai, China). Chondrocytes (5 × × 105 cells/mL) were pretreated with IL-1β (10 ng/mL) for one day and co-cultured with different exosomes (10 μg/ml). Chondrocytes were harvested and washed after three days. A binding buffer was used to resuspend the chondrocytes. Annexin V-FITC (5 μl) and PI (5 μl) were both added to the samples. After incubation avoiding light for 20 min, samples were analysed using flow cytometry (BD Biosciences).
Western blot
Specific markers of exosomes and chondrocytes were tested using western blotting. Chondrocytes were co-cultured with exosome (10 μg/ml) for three days. Chondrocytes and exosomes were all lysed using radio immunoprecipitation assay (RIPA) and then mixed with a protein loading buffer boiled for five min. The samples were subjected to 6–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) polyacrylamide gels and transferred to a polyvinylidene fluoride (PVDF) membrane for 60 min. The PVDF membranes were blocked with 5% bovine serum albumin (BSA) and then incubated with primary antibodies at 4 °C for 12 h. Membranes were washed for three times with hosphate buffered saline-Tween (PBST) and incubated with the corresponding secondary antibody for 2 h. Membranes were prepared by an enhanced chemiluminescence substrate. Glyceraldehyde 3-phosphate dehydrogenase was used as the internal reference.
Quantitative real-time PCR analysis
For exosomal RNA detection, total RNA was isolated from U-MSC-derived exosomes using the PrimeScript RT-PCR kit (TaKaRa, China). RT-PCR reactions were carried out under the following conditions: 95 °C for 3 min, 40 cycles of 95 °C for 3 s and 60 °C for 30 s. The synthetic miRNA Caenorhabditis elegans miR-39 (cel-miR-39: 5′-UCACCGGGUGUAAAUCAGCUUG-3′) was used as an exogenous reference to standardise the expression of exosomal LncRNAs. Beta-2-microglobulin was used as an internal reference to standardise the expression of other mRNAs. The results were calculated using the 2–ΔΔCT method, expressed as the mean of three samples and presented as fold increases relative to the negative control. The primer sequences are listed in Supplemental Table 2.
In vivo SD (Sprague Dawley) rat cartilage defect model and histological evaluation
All procedures were performed according to the animal experimental ethical inspection of laboratory animal centre, Shanghai Tenth People's Hospital. Eighteen SD rats (190–210 g) were divided into three groups randomly. A drill bit (1.5 mm diameter) was used to make cartilage defects on the distal femurs. Every week, they were intra-articularly injected with: (1) 100 μL PBS, (2) 100 μL si-Exos suspension (1 mg/mL) or (3) 100 μL S-Exos suspension (1 mg/mL). Four and eight weeks later, all the animals were sacrificed with a lethal dose of anaesthesia.
Animal behaviour measurement
The protocol for animal behaviour measurement was based on the method of Ren et al. [16]. Briefly, the rat was kept in a standing position and laid on the experimenter's hand. Von Fery microfilament procedures (Aesthesio, USA) were used to stimulate the knee joint of the hind leg using the lowest force possible. Each procedure was tested five times at various intervals. The test was considered positive when the leg withdrawal occurred at least three times. The leg withdrawal threshold (LWT) represented the responsiveness of the defective knee to pain. The measurement was carried out on the third day after weekly injection of exosomes. Normal rats without surgery served as the control group.
Magnetic resonance imaging analysis
T2 mapping techniques have been shown to reflect cartilage structure using the relaxation constant [17]. Samples were soaked in Fomblin to avoid the tissue––air junction, and then MRI imaging analysis was performed on a 3.0 T MRI scanner (MR750, GE Healthcare, Waukesha, WI, USA). The T2 mapping sequence was as follows: time of repetition = 1285 ms, time of echo = 13.8 ms, scan time = 2:05 min, field of view = 80 mm, acquisition matrix = 512 × 256 pixels and slice thickness = 1 mm. The T2 relaxation times of the centre of the cartilage repair site were obtained from a GE workstation using T2 imaging software.
Gross and histological analysis
The joints were harvested for magnetic resonance imaging analysis and gross appearance according to the International Cartilage Repair Society (ICRS) macroscopic assessment (Supplemental Table 3) by three independent blinded observers. The ICRS was firstly reported by Mainil [18]. Its proposal was to use visual patterns for the histological assessment of cartilage repair.
Haematoxylin and eosin (HE), toluidine blue (TB) and safranin-O/fast green (Saf-O) were used for histochemical staining. Immunohistochemical staining was performed for type II collagens. Therapeutic effects on cartilage defect repair were assessed via the histologic grading scale by three independent blinded observers. The histologic grading scale was firstly reported by Wakitani [19]. It was used to evaluate the quality of the repaired tissue.
Statistical analysis
The data are all shown as the mean ± standard deviation (SD) and were used for comparisons among groups. Comparisons of difference between two groups were performed using the unpaired Student t test. Comparisons of differences among multiple groups were performed using one-way analysis of variance (ANOVA). GraphPad Prism version 8.0 was used to perform the statistical analysis. A p value <0.05 was considered statistically significant.
Result
Characterisation of U-MSCs and exosomes
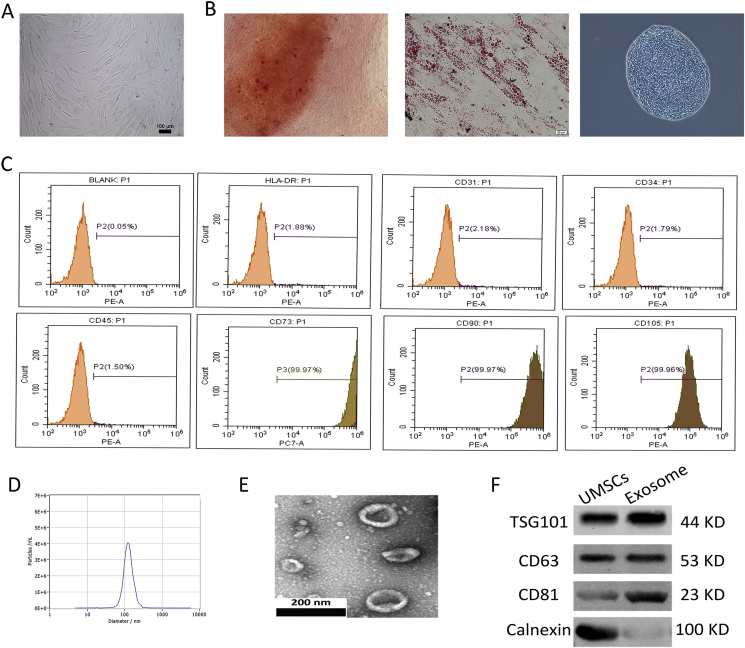
U-MSCs were successfully obtained from umbilical cord Wharton's jelly. More than 95% of U-MSCs exhibited homogeneous fibroblastic morphology after three propagations (Fig. 1A). U-MSCs exhibited a multi-differentiation capacity for osteogenesis, adipogenesis and chondrogenesis (Fig. 1B). Flow cytometric analysis revealed that a majority of U-MSCs expressed CD105, CD73 and CD90 and were negative for CD31, CD34, CD45 and human leukocyte antigen DR (HLA-DR) (Fig. 1C). Nanosight analysis demonstrated that the diameter of exosomes was approximately 120 nm (Fig. 1D). Transmission electron microscopy revealed a cup-shaped exosome morphology (Fig. 1E). Western blotting revealed that the exosomes express exosome-associated proteins (CD63, CD81 and TSG101) as well as a negative protein (Calnexin) (Fig. 1F).
Figure 1.
Characterisation of U-MSCs and exosomes. (A) Morphological observation of U-MSCs (×100). (B) U-MSCs exhibited multi-differentiation capacity for osteogenesis, adipogenesis and chondrogenesis. (C) Flow cytometric analysis of umbilical cord mesenchymal positive markers, such as CD105, CD73 and CD90, and negative markers, such as CD31, CD34, CD45 and HLA-DR. (D) The concentration and size distribution of exosomes by Nanosight. (E) Morphology of exosomes under transmission electron microscopy, scale bar: 200nm. (F) Western blot analysis of exosome surface markers (TSG101, CD63, CD81 and Calnexin).
The most optimal conditions for enhancing exosome yield
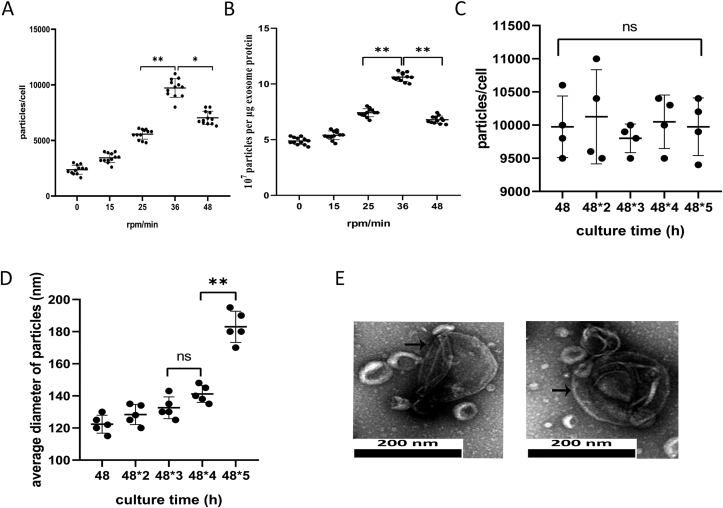
To confirm the most optimal conditions for exosome production, we compared exosome yields derived from different rotational speeds: 0, 15, 25, 36 and 48 rpm/min. As shown in Fig. 2A, when the rotational speed increased gradually, the number of exosome particles secreted per cell reached its peak at 36 rpm/min (9717 ± 834, p < 0.01). However, as the rotational speed continued to increase to 48 rpm/min, the yield of exosome production decreased (7033 ± 594, p < 0.01). Moreover, the purity of exosomes (the ratio of particles to proteins) was also highest at 36 rpm/min (10.62 ± 0.38, p < 0.01, Fig. 2B). Based on the high yield and purity, we determined 36 rpm/min to be the optimal rotation rate for the development of a scalable exosome acquisition method.
Figure 2.
High-yield exosome production from RCCS. (A) Yield of exosomes isolated by RCCS at different rotational speeds (n = 12). Exosome yield = the number of exosome measured by Nanosight/the number of cells. (B) Particle purity of exosomes at different rotational speeds ( = 12). Particle purity = the number of particles/exosomal protein (μg). (C) Yield of exosomes isolated by RCCS at 36 rpm/min from different culture times (n = 4). (D) Average diameter of particles isolated by RCCS at 36 rpm/min from different culture times (n = 5). (E) Morphology of exosomes under transmission electron microscopy after 48∗5 h, scale bar: 200nm. Plots show yield for each method and the mean ± SD of all measurements. ∗p < 0.05, ∗∗p < 0.01, n = 3.
Under the 36 rpm/min culture conditions, supernatant was extracted every 48 h and exosomes were centrifuged at the same speeds. With increasing culture time, there was no difference in exosome production (p > 0.05, Fig. 2C). The diameter of exosomes increased slightly, but there was no statistical difference in the first four times examined (p > 0.5). However, at 48∗5 h, the diameter of exosomes increased to 183 ± 9.7 nm (p < 0.01, Fig. 2D). As shown in Fig. 2E, some cell vesicles appeared very large under transmission electron microscopy. This suggests that U-MSCs secrete normal-sized exosomes under short-term mechanical stimulation (<48∗4 h), but exosome homogeneity changes under long-term stress (>48∗5 h). Therefore, U-MSCs used to derive exosomes by RCCS should be replaced after 196 h of culture.
RCCS attenuates the reduction in exosome production caused by inhibition of Rab27a
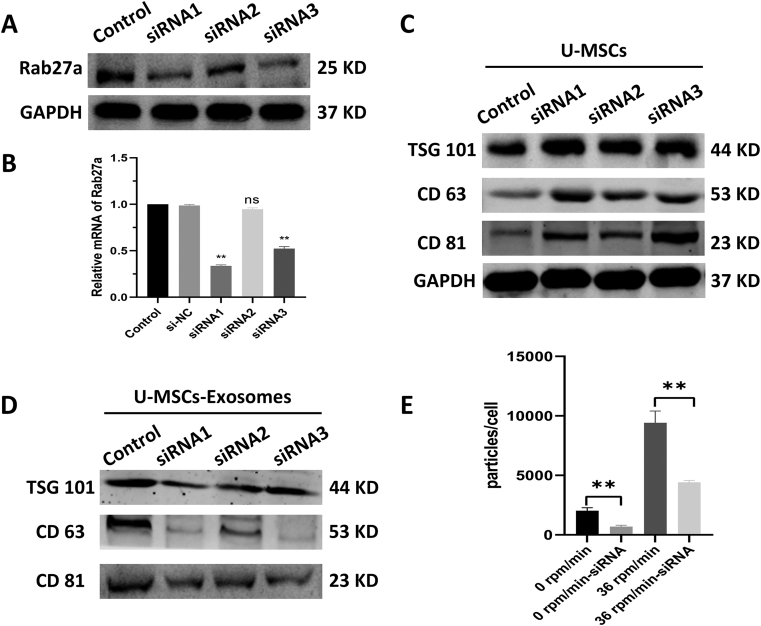
To further determine the increase in exosome yield due to mechanical stimulation caused by RCCS rather than 3D culture, U-MSCs were transfected with siRNA-Rab27a to reduce exosome production. Rab27a, a type of small GTPase, has been demonstrated to be important for the secretion of exosomes [20].
As shown in Fig. 3A–B, the protein and mRNA level of Rab27a in U-MSCs was significantly decreased by siRNAs (p < 0.01). Exosomal markers (TSG101, CD63 and CD81) were higher in the siRNA-Rab27a group of U-MSCs compared to the control group (Fig. 3C). However, the exosomal markers (TSG101, CD63 and CD81) in exosomes showed the opposite trend (Fig. 3D). These results suggested that siRNA-Rab27a was an effective method for reducing exosome production. Among three kinds of siRNAs, the knockdown effect of siRNA1 was strongest (p < 0.01). Therefore, siRNA1 was used for the following interference assay. We cultured U-MSCs transfected with siRNA1 at two RCCS speeds: one completely static (0 rpm/min) and the other at the optimal rotational speed (36 rpm/min). Without mechanical stimulation (0 rpm/min), the exosome yield in the siRNA group was reduced approximately three fold (p < 0.01). However, the yield only dropped 1.5-fold in the mechanical environment (p < 0.01, Fig. 3E). These results demonstrated that mechanical stimulation can attenuate the reduction in exosome production caused by siRNA interference.
Figure 3.
Effect of Rab27a inhibition on U-MSCs exosomes secretion and yield. (A-B) The protein and mRNA level of Rab27a was decreased in the Rab27a-siRNAsgroup. (B) The expression of exosomal marker (TSG101, CD63 and CD81) in U-MSCs by western blot. (C) The expression of exosomal marker (TSG101, CD63 and CD81) in exosome derived from U-MSCs by western blot. (D) The yield of exosome at static and optimal speed with or without interference. Plots show yield for each method and the mean ± SD of all measurements (∗p < 0.05; ∗∗p < 0.01, n = 3). (E) Therefore, siRNA1 was used for the following interference assay. We cultured U-MSCs transfected with siRNA1 at two RCCS speeds: one completely static (0 rpm/min) and the other at the optimal rotational speed (36 rpm/min). Without mechanical stimulation (0 rpm/min), the exosome yield in the siRNA group was reduced approximately three fold (p<0.01). However, the yield only dropped 1.5-fold in the mechanical environment (p<0.01, Fig. 3E). These results demonstrated that mechanical stimulation can attenuate the reduction in exosome production caused by siRNA interference.
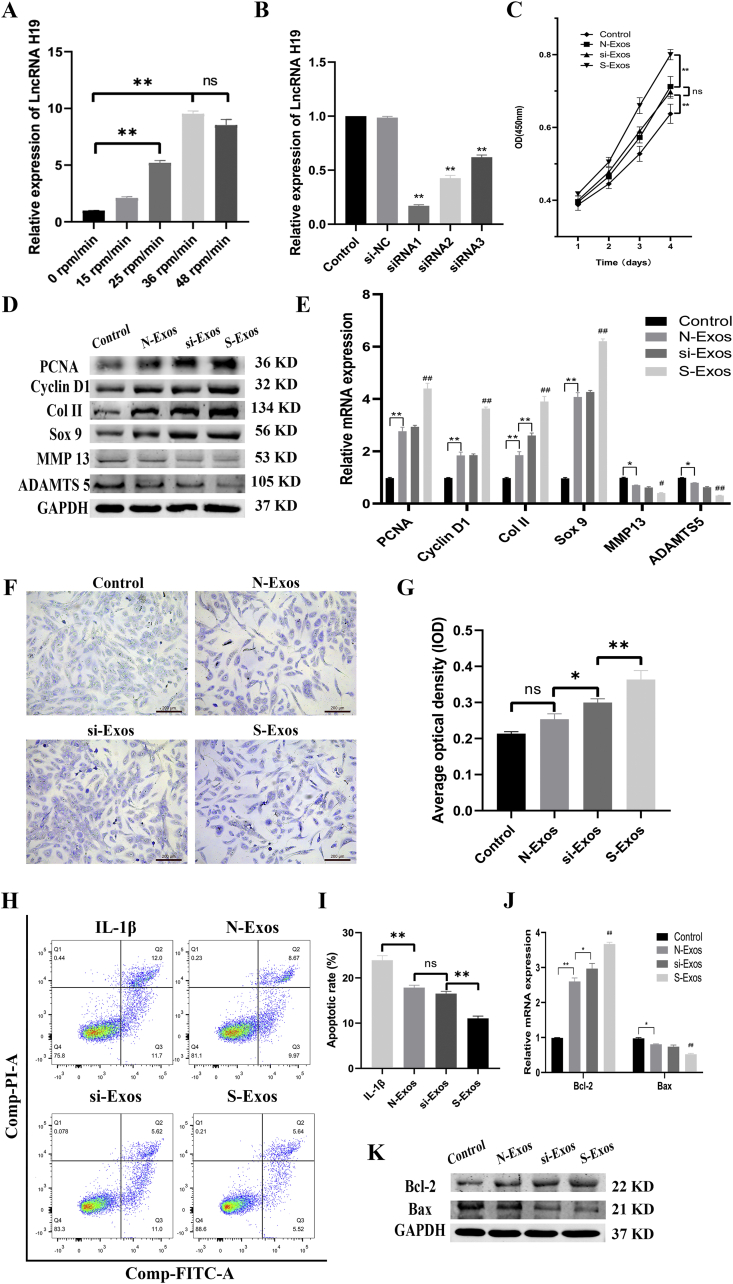
Exosomal LncRNA H19 in a mechanical environment enhances proliferation and inhibits chondrocyte apoptosis
LncRNA H19, which has been previously reported to be mechanically sensitive [21], was found via qRT-PCR to increase nearly 10-fold when chondrocytes were stimulated at 36 rpm/min (p < 0.01, Fig. 4A). In order to determine whether high levels of exosomal H19 affect the activity of chondrocytes, we silenced H19 in U-MSCs via siRNA. Among three different siRNAs, the knockdown effect of siRNA1 was the strongest (p < 0.01, Fig. 4B). Therefore, siRNA1 was used for the following interference assay.
Figure 4.
Exosomal LncRNA H19 in mechanical environment enhances proliferation and inhibits apoptosis of chondrocytes. (A) The expression level of LncRNA H19 in exosomes under mechanical environment detected by qRT-PCR. (B) The expression level of LncRNA H19 in U-MSCs treated with different siRNAs. (C) The proliferation was assessed by CCK-8 assay. (D–E) The protein and mRNA level of genes associated with proliferation (PCNA, Cyclin D1) and matrix synthesis (Col II, Sox 9, MMP 13, ADMST5). (F) Toluidine blue staining and the quantification of chondrocytes treated with different kinds of exosomes. (G) The quantification of toluidine blue staining. (H) Chondrocytes were pretreated with si-Exos and S-Exos followed by IL-1β (10 ng/mL) challenge for 24 h. Apoptosis was assessed by flow cytometry. (I) Apoptotic rate of chondrocytes. (J–K) The protein and mRNA level of genes associated with anti-apoptosis (Bcl-2, Bax). Data represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, n = 3; #p < 0.05, ##p < 0.01 compared to each other group, n = 3.
Chondrocytes were co-cultured with 10 μg/ml of each exosomes. As shown in Fig.4C, S-Exos exhibited the highest cell proliferation, as assessed by CCK-8 after four days (p < 0.01). There was no statistical difference between N-Exos and si-Exos (p = 0.77). The control group had the least proliferative activity. Proliferating cell nuclear antigen (PCNA) and Cyclin D1 were genes associated with cell proliferation. The protein and mRNA levels of PCNA and Cyclin D1 were enhanced after treatment with exosomes (Fig. 4D–E). S-Exos showed a stronger effect on proliferation than si-Exos and N-Exos.
Matrix synthesis is closely linked to proliferation. Anabolic markers, such as Col II and Sox9, were upregulated by S-Exos. Moreover, catabolic markers, such as matrix metalloproteinase 13 (MMP 13) and A disintegrin and metalloproteinase with thrombospondin motifs 5 (ADAMTS 5), were downregulated by S-Exos (Fig. 4D–E). The effects of N-Exos and si-Exos were inferior to that of S-Exos, but superior to that of the control group. Toluidine blue staining generally identifies the cartilaginous matrix. As shown in Fig. 4F–G, the sulfate groups of proteoglycans in chondrocytes treated with S-Exos showed the strongest effect of metachromatic staining (p < 0.01).
Inhibition of apoptosis is an important factor in promoting cell proliferation. We used V-FITC/PI flow cytometry to detect the effect of exosomes on the inhibition of chondrocyte apoptosis (Fig. 4F). IL-1β was used to induce apoptosis (23.9 ± 1%). Co-culture with all three exosome types decreased the apoptosis rates of chondrocytes. There was no difference between N-Exos (17.9 ± 0.5%) and si-Exos (16.5 ± 0.6%) (p = 0.15). The anti-apoptotic activity of S-Exos (11.1 ± 0.5%) was the highest (p < 0.01, Fig. 4I). Proteins and mRNA level associated with apoptosis, such as Bcl-2 and Bax, were consistent with the above results (Fig. 4J–K).
These results clearly demonstrate the superior efficacy of S-Exos in promoting cell proliferation, matrix synthesis and inhibiting cell apoptosis. Altogether, our results show that interference against H19 in U-MSCs significantly weakens the efficacy of exosomes derived from U-MSCs.
The effect of exosomes on cartilage defect repair
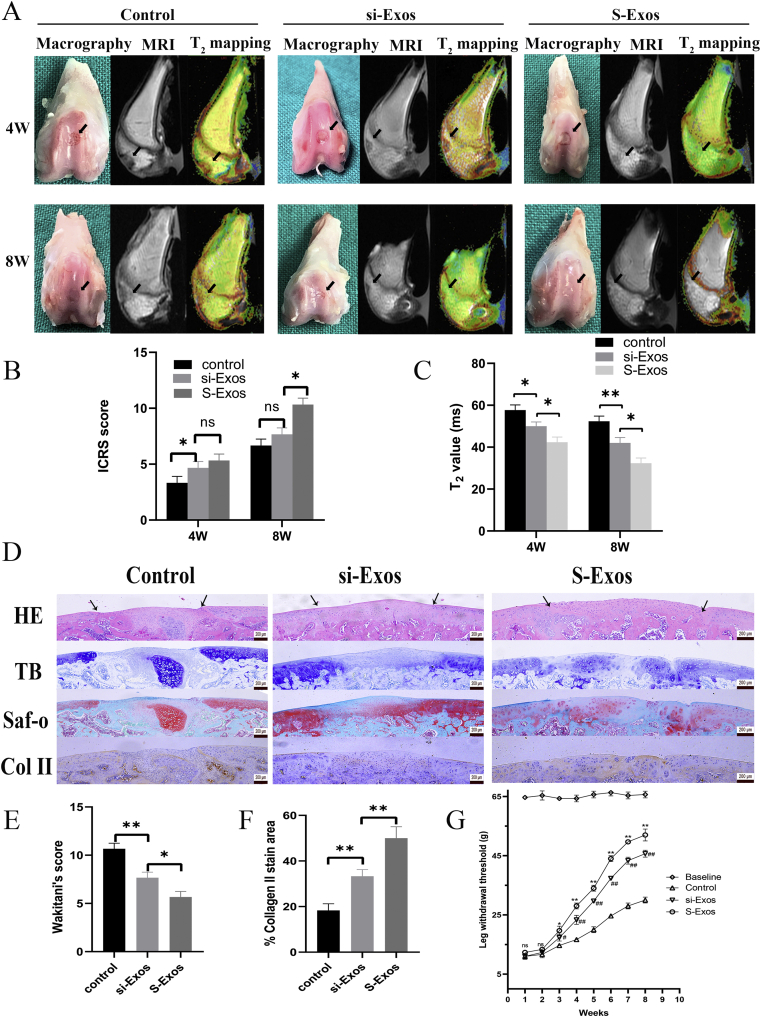
Articular joint samples were harvested from SD rats after 4 and 8 weeks for macroscopic evaluation, MRI quantification and immunohistochemical staining. Because we found that there was no significant difference between N-Exos and si-Exos in vitro, we only compared si-Exos and S-Exos in vivo.
As shown in Fig. 5A, an obvious defect was present in the control group after 4 weeks. The defects in si-Exos and S-Exos were mostly covered by new tissue; however, there was a clear boundary between the new and old surrounding cartilage in si-Exos. After 8 weeks, si-Exos-treated defects exhibited better surface regularity and integration with the surrounding cartilage. The cartilage defects in the S-Exos group were filled with complete and uniform tissue, with obscured boundaries. The result of ICRS scoring indicated that there was no difference in early repair between S-Exos (5.3 ± 0.58) and si-Exos (4.7 ± 0.58) at 4 weeks (p = 0.39, Fig. 5B). At 8 weeks, S-Exos (10.3 ± 0.58) showed its superiority in defect repair over si-Exos (7.7 ± 0.58) and the control group (6.7 ± 0.58) (p < 0.01, Fig. 5B). There was no significant difference between si-Exos and the control group (p = 0.16).
Figure 5.
Effect of exosomes on repair of cartilage defect. (A) Representative macroscopic, MRI, T2 mapping images of the regenerated tissues. (B) ICRS macroscopic scores. (C) T2 mapping scores. (D) Staining results of HE, TB, Saf-O and immunohistochemical staining for type II collagens. (E) Wakitani scores for the histological sections. (F) Percentage of collagen II stain area. Data represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, n = 3. (G) Time-dependent nociceptive responses after injection of different exosomes. Data represent mean ± SEM. ∗p < 0.05, ∗∗p < 0.01 compared to the si-Exos group; #p < 0.05, ##p < 0.01 compared to the control group, n = 3.
The MRI images showed that the cartilage defects in the control group were poorly filled at 4 and 8 weeks (Fig. 5A). The new tissues in the si-Exos group were partially filled until 8 weeks. Only the defects of the S-Exos group showed a similar intensity to that of surrounding cartilage at 8 weeks. T2 mapping values were used for quantitative analysis. As the tissue repair occurs, the T2 value decreases. The T2 values of the S-Exos group at 4 weeks (42.3 ± 2.5 vs. 57.7 ± 2.5/50 ± 2, p = 0.001/0.015) and 8 weeks (32.3 ± 2.5 vs. 52.3 ± 2.5/42 ± 2.6, p < 0.01) were both lower than that of the control/si-Exos groups (Fig. 5C).
Samples at 8 weeks were sliced for HE, toluidine blue and safranin O staining (Fig. 5D). HE and TB staining revealed that the defects treated with S-Exos showed the best surface regularity, the most ordered tissues and the most glycosaminoglycan deposition. In the si-Exos group, the defects were repaired to some extent, but were still worse than in the S-Exos group. In contrast, fibrous and ectopic cartilage tissues were found in the control group. The repair of the subchondral bone in both the si-Exos and S-Exos groups was better than in the control group, as determined via safranin O staining.
At 8 weeks, S-Exos-treated defects exhibited a significantly lower Wakitani score of 5.67 ± 0.58 compared to 7.66 ± 0.58 in the si-Exos group (p = 0.013). The control group had the highest Wakitani score, 10.77 ± 0.57 (Fig. 5E). Moreover, S-Exos was rich in Col II, similar to the adjacent cartilage (Fig. 5D). The Col II staining area in the S-Exos (51.7 ± 6.2%) was the largest compared to the si-Exos (33.3 ± 2.9%) and control groups (18.4 ± 3.2%), which indicates increased Col II secretion and matrix synthesis in vivo (Fig. 5F).
As shown in Fig. 5G, the surgery-induced pain persisted for at least 2 weeks, during which there were no differences among the three groups (p > 0.05). The pain gradually resolved at 3 weeks, and S-Exos rats (19.7 ± 1.5 g) had reduced pain with higher LWT compared to the si-Exos (17.3 ± 1.2 g) and PBS groups (14.7 ± 0.6 g). At 7 weeks, the pain in all three groups remained stable and above the control baseline (p > 0.05). This suggested that the cartilage defect had been repaired to varying degrees in the different groups, with the S-Exos group (52 ± 3 g) showing the greatest improvement, but still not reaching normal levels (65.7 ± 1.1 g).
Discussion
In our previous studies, we found that MSCs exposed to mechanical stimulation via RCCS showed improved therapeutic effects on osteochondral regeneration activity in vivo [3,5]. Thus, we were curious about whether mechanical stimulation has an impact on the quantity or quality of the MSC secretome. The former question has been partially investigated by several previous studies [22,23]. Jo et al. [24] used a microfluidic device to create shear stress and succeeded in increasing the yield of extracellular vesicles. Mechanical stimulation has been demonstrated to induce pro-angiogenic factor secretion by MSCs to enhance the tube formation of endothelial cells in vitro [25]. Our results confirmed that mechanical stimulation was crucial to the quantity and biological quality of MSC exosomes. We will discuss both of these aspects.
First, we detected whether mechanical stimulation could enhance exosome yield. Bradley et al. [15] found that the production of IL-2 by T cells in the RCCS environment was enhanced compared to the static group. They also investigated the relation between culture time and production. To their surprise, RCCS T cell production of IL-2 gradually rose to its peak within 72 h and then started to descend. In our study, the yield of exosomes via RCCS was rather consistent during extraction every 48 h. However, culture time was a real consideration, as the diameter of exosomes increased after long cultivation times (196 h). The specific reasons for exosomal changes require further investigation. Another important aspect of RCCS is the rotational speed. Previous studies have reported speeds ranging from 12 to 48 rpm/min, and no universal rotational speed has been identified [[26], [27], [28]]. Difference in rotational speed may lead to different mechanical stimulation, which can generate diverse cell states. We predicted that if the speed was too low, the mechanical stimulation could be too subtle, and if the speed was too high, abnormal stress could be generated. Therefore, we wanted to determine an optimal rotational speed at which the cells could produce substantial exosomes. The speed of 36 rpm/min that we used throughout may be the best model, and it has been shown that excessive mechanical stimulation can damage exosomal production. Ostrowski et al. [29] previously identified that silencing Rab27a was a way to block exosome secretion. Therefore, we used siRNA to inhibit Rab27a in U-MSCs, and the number of exosomes in the siRNA-Rab27a group was reduced approximately three-fold at the static state. However, the yield only dropped 1.5-fold in the mechanical environment. Our results showed that inhibition of Rab27a decreased the production of exosomes, but mechanical stimulation could attenuate this reduction in production.
Next, we detected the relationship between mechanical stimulation and osteochondral activity. We used the effects of exosomes on the biological activity of chondrocytes as a readout for exosome activity. We found that exosomes derived from U-MSCs in a mechanical environment significantly promoted the proliferation of chondrocytes. We predicted that certain mechanically sensitive exosomal components might be upregulated. Thus, several mechanically sensitive miRNAs or LncRNAs, which were reported by other studies, were detected by qRT-PCR. Through screening and literature reading, we preliminarily judged that LncRNA H19 could play an important role in osteochondral activity. Wu et al. [21] also showed that mechanical tension could increase the expression of LncRNA H19 in MSCs.
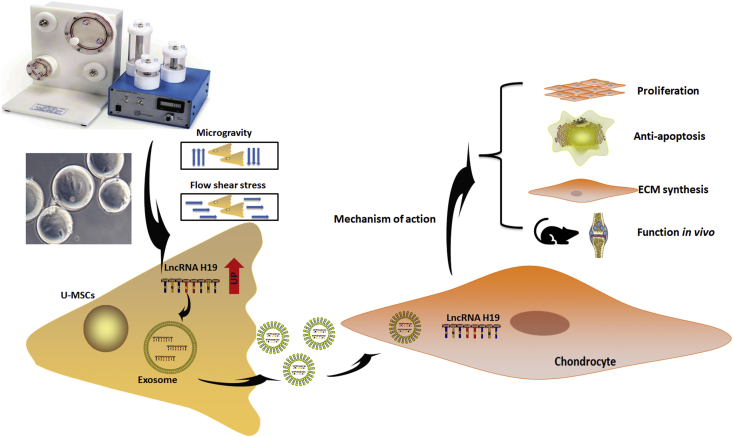
We found that exosomes released by U-MSCs in a mechanical environment (S-Exos) significantly promoted the proliferation of chondrocytes and prevented chondrocyte apoptosis when stimulated by IL-1β. Moreover, our study showed that downregulating LncRNA H19 in exosomes impaired the proliferative and anti-apoptotic effects of exosomes on chondrocytes. These results suggest that mechanical stimulation increases the expression of H19 in exosomes derived from U-MSCs and that exosomal H19 plays a key role in promoting proliferation and preventing apoptosis in chondrocytes (Fig. 6).
Figure 6.
Schematic presentation involved in the proliferation and anti-apoptosis of chondrocytes by exosomal LncRNA H19 derived from U-MSCs in mechanical environment. Mechanical stimulation caused by RCCS could enhance exosome biological function for repair of cartilage defect. The underlying mechanism may be through high expression of LncRNA H19 in exosomes.
The LncRNA H19 is a highly conserved sequence, about 2.3 kb in length, that is involved in stem cell differentiation, embryonic growth and tumourigenesis [30]. LncRNA H19 has been found to be upregulated in embryonic tissues and downregulated in most adult tissues, except for articular cartilage and skeletal muscle [31,32]. Dudek et al. [32] found that H19 was highly expressed in healthy young human articular chondrocytes. Even more interestingly, the level of LncRNA H19 was comparable to the most abundant matrix genes, i.e., collagen II and aggrecan. However, the specific mechanism of action of LncRNA H19 on osteochondral activity is still controversial and contradictory. Some studies have shown that LncRNA H19 has a positive regulatory effect on chondrocytes. For example, Dudek et al. [32] reported that H19 could be transcribed into miRNA-675, which upregulates collagen II expression in chondrocytes via a de-repression mechanism.
Zou et al. [33] used C28/I2 cell lines as normal chondrocytes and found that LncRNA H19 was dose-dependently elevated when treated with lipopolysaccharide (LPS). Knockdown of H19 in C28/I2 cells could attenuate the decreased viability and increased inflammatory factors caused by LPS. These results were different from our results as well as those of Dudek. One reason may be that Zou used a cell line other than primary human chondrocytes. Moreover, LncRNA H19 may not be the sole LncRNA in exosomes that plays a role in osteochondral activity. Exosomes derived from mechanical conditions may differ in a myriad of components. Thus, subsequent studies should focus on RNA-sequence analyses to identify the different miRNAs, LncRNAs and circRNAs present in exosomes.
Conclusion
Our findings confirmed that mechanical stimulation could enhance the exosome yield, as well as the biological function for cartilage defect repair. The underlying mechanism may be high expression of the LncRNA H19 in exosomes.
Funding
This project is supported by the National Natural Science Foundation of China (grant 81772324).
CRediT authorship contribution statement
Xing Wu: Funding acquisition, Conceptualisation, Writing—Review & Editing, Supervision.
Litao Yan: Investigation, Writing—original draft.
Gejun Liu: Investigation, Resources.
Ethics approval and consent to participate
I will conscientiously abide by the ethical principles of animal welfare, accept the supervision and inspection of committee at any time, and voluntarily accept the punishment if any infringement.
Availability of data and materials
The datasets in this study are available.
Consent for publication
Not applicable.
Conflicts of Interest Statement
The authors declare that they have no competing interests.
Acknowledgement
We appreciate our members for their valuable efforts on this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jot.2020.03.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figs1Anterior view of the RCCS system.
Figs2.
Lateral view of the RCCS system.
References
- 1.Zhang S., Hu B., Liu W., Wang P., Lv X., Chen S. Articular cartilage regeneration: the role of endogenous mesenchymal stem/progenitor cell recruitment and migration. Semin Arthritis Rheum. 2020;50(2):198–208. doi: 10.1016/j.semarthrit.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Liu P.C., Liu K., Liu J.F., Xia K., Chen L.Y., Wu X. Transfection of the IHH gene into rabbit BMSCs in a simulated microgravity environment promotes chondrogenic differentiation and inhibits cartilage aging. Oncotarget. 2016;7(39):62873–62885. doi: 10.18632/oncotarget.11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L., Liu G., Li W., Wu X. Sonic hedgehog promotes chondrogenesis of rabbit bone marrow stem cells in a rotary cell culture system. BMC Dev Biol. 2019;19(1):18. doi: 10.1186/s12861-019-0198-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J., Wu X. Cyclic tensile strain promotes chondrogenesis of bone marrow-derived mesenchymal stem cells by increasing miR-365 expression. Life Sci. 2019;232:116625. doi: 10.1016/j.lfs.2019.116625. [DOI] [PubMed] [Google Scholar]
- 5.Chen L., Liu G., Li W., Wu X. Chondrogenic differentiation of bone marrow-derived mesenchymal stem cells following transfection with Indian hedgehog and sonic hedgehog using a rotary cell culture system. Cell Mol Biol Lett. 2019;24:16. doi: 10.1186/s11658-019-0144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcaraz M.J., Compañ A., Guillén M.I. Extracellular vesicles from mesenchymal stem cells as novel treatments for musculoskeletal diseases. Cells. 2019;9(1) doi: 10.3390/cells9010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan L., Wu X. Exosomes produced from 3D cultures of umbilical cord mesenchymal stem cells in a hollow-fiber bioreactor show improved osteochondral regeneration activity. Cell Biol Toxicol. 2019 doi: 10.1007/s10565-019-09504-5. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao X., Wei W., Wang X., Chenglin L., Björklund M., Ouyang H. Stem cell derived exosomes: microRNA therapy for age-related musculoskeletal disorders. Biomaterials. 2019;224:119492. doi: 10.1016/j.biomaterials.2019.119492. [DOI] [PubMed] [Google Scholar]
- 10.Ullah M., Ng N.N., Concepcion W., Thakor A.S. Emerging role of stem cell-derived extravesicular MicroRNAs in age-associated human diseases and in different therapies of longevity. Ageing Res Rev. 2019:100979. doi: 10.1016/j.arr.2019.100979. [DOI] [PubMed] [Google Scholar]
- 11.Nawaz M. Extracellular vesicle-mediated transport of non-coding RNAs between stem cells and cancer cells: implications in tumor progression and therapeutic resistance. Stem Cell Investig. 2017;4:83. doi: 10.21037/sci.2017.10.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J., Yu W., Wang Y., Xia K., Huang Y., Xu A. lncRNAs: function and mechanism in cartilage development, degeneration, and regeneration. Stem Cell Res Ther. 2019;10(1):344. doi: 10.1186/s13287-019-1458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conigliaro A., Costa V., Lo Dico A., Saieva L., Buccheri S., Dieli F. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morabito C., Steimberg N., Mazzoleni G., Guarnieri S., Fanò-Illic G., Mariggiò M.A. RCCS bioreactor-based modelled microgravity induces significant changes on in vitro 3D neuroglial cell cultures. Biomed Res Int. 2015;2015:754283. doi: 10.1155/2015/754283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley J.H., Stein R., Randolph B., Molina E., Arnold J.P., Gregg R.K. T cell resistance to activation by dendritic cells requires long-term culture in simulated microgravity. Life Sci Space Res (Amst) 2017;15:55–61. doi: 10.1016/j.lssr.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Ren K. An improved method for assessing mechanical allodynia in the rat. Physiol Behav. 1999;67(5):711–716. doi: 10.1016/s0031-9384(99)00136-5. [DOI] [PubMed] [Google Scholar]
- 17.Mosher T.J., Dardzinski B.J. Cartilage MRI T2 relaxation time mapping: overview and applications. Semin Musculoskelet Radiol. 2004;8(4):355–368. doi: 10.1055/s-2004-861764. [DOI] [PubMed] [Google Scholar]
- 18.Mainil-Varlet P., Aigner T., Brittberg M., Bullough P., Hollander A., Hunziker E. Histological assessment of cartilage repair: a report by the histology endpoint committee of the international cartilage repair society (ICRS) J Bone Joint Surg Am. 2003;85–A(Suppl 2):45–57. [PubMed] [Google Scholar]
- 19.Wakitani S., Goto T., Pineda S.J., Young R.G., Mansour J.M., Caplan A.I. Mesenchymal cell-based repair of large, full-thickness defects of articular cartilage. J Bone Joint Surg Am. 1994;76(4):579–592. doi: 10.2106/00004623-199404000-00013. [DOI] [PubMed] [Google Scholar]
- 20.Li X., Zheng Y., Hou L., Zhou Z., Huang Y., Zhang Y. Exosomes derived from maxillary BMSCs enhanced the osteogenesis in iliac BMSCs. Oral Dis. 2020;26(1):131–144. doi: 10.1111/odi.13202. [DOI] [PubMed] [Google Scholar]
- 21.Wu J., Zhao J., Sun L., Pan Y., Wang H., Zhang W.B. Long non-coding RNA H19 mediates mechanical tension-induced osteogenesis of bone marrow mesenchymal stem cells via FAK by sponging miR-138. Bone. 2018;108:62–70. doi: 10.1016/j.bone.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Shah N., Morsi Y., Manasseh R. From mechanical stimulation to biological pathways in the regulation of stem cell fate. Cell Biochem Funct. 2014;32(4):309–325. doi: 10.1002/cbf.3027. [DOI] [PubMed] [Google Scholar]
- 23.Fahy N., Alini M., Stoddart M.J. Mechanical stimulation of mesenchymal stem cells: implications for cartilage tissue engineering. J Orthop Res. 2018;36(1):52–63. doi: 10.1002/jor.23670. [DOI] [PubMed] [Google Scholar]
- 24.Jo W., Jeong D., Kim J., Cho S., Jang S.C., Han C. Microfluidic fabrication of cell-derived nanovesicles as endogenous RNA carriers. Lab Chip. 2014;14(7):1261–1269. doi: 10.1039/c3lc50993a. [DOI] [PubMed] [Google Scholar]
- 25.Kasper G., Dankert N., Tuischer J., Hoeft M., Gaber T., Glaeser J.D. Mesenchymal stem cells regulate angiogenesis according to their mechanical environment. Stem Cell. 2007;25(4):903–910. doi: 10.1634/stemcells.2006-0432. [DOI] [PubMed] [Google Scholar]
- 26.Imura T., Otsuka T., Kawahara Y., Yuge L. "Microgravity" as a unique and useful stem cell culture environment for cell-based therapy. Regen Ther. 2019;12:2–5. doi: 10.1016/j.reth.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tackett N., Bradley J.H., Moore E.K., Baker S.H., Minter S.L., DiGiacinto B. Prolonged exposure to simulated microgravity diminishes dendritic cell immunogenicity. Sci Rep. 2019;9(1):13825. doi: 10.1038/s41598-019-50311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arun R.P., Sivanesan D., Patra B., Varadaraj S., Verma R.S. Simulated microgravity increases polyploid giant cancer cells and nuclear localization of YAP. Sci Rep. 2019;9(1):10684. doi: 10.1038/s41598-019-47116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. doi: 10.1038/ncb2000. sup pp. 1–13. [DOI] [PubMed] [Google Scholar]
- 30.Chen C., Liu M., Tang Y., Sun H., Lin X., Liang P. LncRNA H19 is involved in myocardial ischemic preconditioning via increasing the stability of nucleolin protein. J Cell Physiol. 2020 doi: 10.1002/jcp.29524. In press. [DOI] [PubMed] [Google Scholar]
- 31.Steck E., Boeuf S., Gabler J., Werth N., Schnatzer P., Diederichs S. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med (Berl) 2012;90(10):1185–1195. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
- 32.Dudek K.A., Lafont J.E., Martinez-Sanchez A., Murphy C.L. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285(32):24381–24387. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y., Li S., Zou Y. Knockdown of LncRNA H19 relieves LPS-induced damage by modulating miR-130a in osteoarthritis. Yonsei Med J. 2019;60(4):381–388. doi: 10.3349/ymj.2019.60.4.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets in this study are available.