PKA is essential for the virulence of eukaryotic human pathogens. Understanding PKA signaling mechanisms is therefore fundamental to deciphering pathogenesis and developing novel therapies.
KEYWORDS: Aspergillus, filamentous fungi, fungal pathogen, protein kinase A, protein phosphorylation, proteomics
ABSTRACT
Protein kinase A (PKA) signaling plays a critical role in the growth and development of all eukaryotic microbes. However, few direct targets have been characterized in any organism. The fungus Aspergillus fumigatus is a leading infectious cause of death in immunocompromised patients, but the specific molecular mechanisms responsible for its pathogenesis are poorly understood. We used this important pathogen as a platform for a comprehensive and multifaceted interrogation of both the PKA-dependent whole proteome and phosphoproteome in order to elucidate the mechanisms through which PKA signaling regulates invasive microbial disease. Employing advanced quantitative whole-proteomic and phosphoproteomic approaches with two complementary phosphopeptide enrichment strategies, coupled to an independent PKA interactome analysis, we defined distinct PKA-regulated pathways and identified novel direct PKA targets contributing to pathogenesis. We discovered three previously uncharacterized virulence-associated PKA effectors, including an autophagy-related protein, Atg24; a CCAAT-binding transcriptional regulator, HapB; and a CCR4-NOT complex-associated ubiquitin ligase, Not4. Targeted mutagenesis, combined with in vitro kinase assays, multiple murine infection models, structural modeling, and molecular dynamics simulations, was employed to characterize the roles of these new PKA targets in growth, environmental and antimicrobial stress responses, and pathogenesis in a mammalian system. We also elucidated the molecular mechanisms of PKA regulation for these effectors by defining the functionality of phosphorylation at specific PKA target sites. We have comprehensively characterized the PKA-dependent phosphoproteome and validated PKA targets as direct regulators of infectious disease for the first time in any pathogen, providing new insights into PKA signaling and control over microbial pathogenesis.
INTRODUCTION
The cAMP/protein kinase A (PKA) signaling pathway is ubiquitous and essential among all eukaryotes. Despite its critical importance for the virulence of numerous microbial pathogens, the precise mechanisms of PKA-dependent control over infectious disease remain unknown. This is mainly due to the paucity of direct PKA targets that have been well characterized in any organism. Previous studies have identified only potential PKA targets in humans and model organisms by in vitro proteome chip assays (1), computational prediction of target motifs (2), and targeted phosphoproteomics using enrichment with anti-PKA target motif antibodies (3).
The complexity of eukaryotic signaling pathways represents an especially daunting obstacle to defining their roles in disease caused by eukaryotic pathogens, such as invasive fungal infections. In the human-pathogenic yeast Candida albicans and the plant fungal pathogen Magnaporthe oryzae, interrogation of the PKA-dependent phosphoproteome identified potential PKA targets based on phosphorylation at canonical PKA target motifs (4, 5). However, these studies experimentally validated very few PKA targets predicted as in vivo substrates, and in even fewer cases has PKA-dependent site-specific phosphoregulation been investigated. Here, we comprehensively characterize the PKA phosphoproteome and validate PKA phosphorylation targets as direct regulators of microbial disease for the first time in any pathogen.
To define the roles of specific PKA signaling effectors in microbial pathogenesis, we used the fungus Aspergillus fumigatus as a prototypical eukaryotic pathogen. A. fumigatus is the primary etiology of invasive aspergillosis (IA), a leading cause of infection-associated mortality in the many immunocompromised patients treated for cancer or undergoing hematopoietic stem cell or solid-organ transplantation (6–10). Despite mortality rates of 40 to 60%, the specific molecular effectors responsible for A. fumigatus virulence are poorly understood. PKA signaling is critical for the growth and virulence of A. fumigatus (11–13), and thus, elucidation of the myriad processes regulated by the PKA signaling network will significantly advance the mechanistic understanding of overall eukaryotic microbial pathogenesis.
We employed high-resolution and quantitative liquid chromatography-tandem mass spectroscopy (LC-MS/MS) to examine the global effects of PKA on both the total fungal proteome and the phosphoproteome. Multiple proteases and two different peptide enrichment approaches, including titanium dioxide (TiO2) phosphopeptide enrichment and antibody enrichment of peptides containing canonical PKA target motifs, were utilized to maximize the number of unique phosphopeptides discovered. Our phosphomotif analysis, in conjunction with an independent PKA interactome analysis, identified 127 potential direct PKA targets in A. fumigatus. We characterized three probable direct PKA target proteins not previously identified as PKA effectors in any organism that represent diverse PKA-regulated pathways, including an autophagy-related protein, Atg24; a CCAAT-binding transcriptional regulator, HapB; and a CCR4-NOT complex-associated ubiquitin ligase, Not4. Furthermore, we demonstrated their impacts on disease in multiple mammalian infection systems and used a site-directed mutagenesis approach, coupled with computational modeling and molecular dynamics simulations, to define the functional effects of phosphorylation at specific PKA target sites on these effectors. This multifaceted strategy represents the most comprehensive analysis of the PKA-dependent phosphoproteome of any human pathogen, enabling the discovery of specific PKA effectors contributing to virulence. These findings have broad implications for the overall understanding of cAMP/PKA signaling mechanisms and the vital role of PKA signaling in infectious diseases.
RESULTS
PKA signaling impacts the abundance of proteins associated with essential cellular processes.
In order to determine the global effects of PKA on protein levels across the A. fumigatus whole proteome, we compared the levels from triplicate biological replicates of a wild-type (WT) A. fumigatus strain and a strain deleted for the major PKA catalytic subunit (ΔpkaC1) using quantitative LC-MS/MS analysis (see Fig. S1A in the supplemental material). Through this approach, 3,212 unique proteins were identified, representing approximately one-third of the estimated 9,840 proteins expressed by A. fumigatus (Fig. 1A) (14). Of these, 1,073 proteins were significantly increased in the WT strain, and 530 proteins were significantly decreased, compared to the ΔpkaC1 strain (Fig. 1B and C; Fig. S1B). The sets of significantly upregulated or downregulated proteins were each analyzed for functional category enrichment using both the FungiFun database and the Database for Annotation, Visualization, and Integrated Discovery (DAVID). While the PKA-downregulated proteins were primarily involved in aerobic respiration (Fig. 1D; Fig. S1C and Table S1), the upregulated proteins were strongly enriched for transcriptional regulation, protein synthesis, as well as proteasomal and autophagic protein degradation in addition to numerous other functional categories (Fig. 1E; Fig. S1D and Table S2).
FIG 1.
Global A. fumigatus PKA proteomic analysis. (A) Numbers of unique proteins identified in open, TiO2-enriched, and PKA motif-enriched data sets as well as unique phosphoproteins identified in combined TiO2- and PKA motif-enriched data sets. Of the 3,212 proteins identified in the open analysis, 799 were also identified in the combined phosphoprotein analyses. (B) Heat map of open protein abundances of 3,212 unique proteins identified in WT and ΔpkaC1 samples. Red indicates a higher abundance, while green indicates a lower abundance. Brackets indicate the clustering of proteins with similar abundance profiles. (C) Fold changes in the abundances of 3,212 identified proteins in the open analysis (of 9,840 annotated A. fumigatus proteins), grouped by P value (increasing for negative fold changes and decreasing for positive fold changes). Proteins with statistically significant fold changes with P values of <0.01, P values of between 0.05 and 0.01, and statistically insignificant P values of 0.05 or higher are shown. (D) Enriched functional categories of proteins identified in the open proteomic analysis demonstrating a ≥2-fold-lower abundance in the WT genetic background than in the ΔpkaC1 background and statistical significance of a P value of <0.05 in t test comparisons. Pie charts show enriched functional categories for this set of proteins in comparison to the total set of annotated A. fumigatus proteins based on the Functional Category (FunCat) classification system, as analyzed using FungiFun 2.2.8 bioinformatic software. FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide. (E) FungiFun output of enriched functional categories of identified proteins with a ≥2-fold-higher abundance in the WT than in the ΔpkaC1 background at a P value of <0.05. Pie charts show enriched categories based on FunCat classifications.
Global A. fumigatus PKA proteomic analysis. (A) Diagram of the proteomics experiment workflow. Total protein extracts from triplicate samples of A. fumigatus wild-type (WT) and ΔpkaC1 strain mycelia were split, subjected to both LysC and GluC protease digestions, split again into three parts, and treated with either TiO2 for phosphopeptide enrichment or anti-PKA motif antibodies for PKA motif enrichment or unenriched for open protein analysis. LysC digests were further digested with trypsin either immediately following LysC digestion for phosphopeptide enrichment and open samples or after antibody treatment for PKA motif enrichment. (B) Volcano plot of open protein abundances of 3,212 unique proteins identified, plotted as log2 average fold changes in WT samples compared to the ΔpkaC1 average (x axis), against the −log10 P value in a t test comparison of protein abundances between strain types (y axis). (C) Enriched functional categories of proteins identified by open proteomic analysis demonstrating a ≥2-fold-lower abundance in the WT genetic background than in the ΔpkaC1 background and statistical significance of a P value of <0.05 in t test comparisons. Pie charts show enriched functional categories for this set of proteins in comparison to the total set of annotated A. fumigatus proteins based on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) classification systems, as analyzed using FungiFun 2.2.8 bioinformatic software. (D) FungiFun output of enriched functional categories of identified proteins with a ≥2-fold-higher abundance in the WT than in the ΔpkaC1 background at a P value of <0.05. Pie charts show enriched categories based on GO and KEGG classifications. Download FIG S1, JPG file, 0.5 MB (482.5KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteins with decreased abundance in the wild type versus the ΔpkaC1 strain in open proteomic analysis. Download Table S1, PDF file, 0.02 MB (26.3KB, pdf) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteins with increased abundance in the wild type versus the ΔpkaC1 strain in open proteomic analysis. Download Table S2, PDF file, 0.03 MB (30.8KB, pdf) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of the PKA-dependent whole phosphoproteome and PkaC1 interactome identifies direct PKA target candidate proteins.
We compared the phosphoenriched fractions from the WT and ΔpkaC1 strains to identify proteins that were specifically phosphorylated in a PKA-dependent manner. Enhanced coverage of phosphosites was obtained by digestion with both GluC and a combination of LysC and trypsin proteases followed by TiO2-based phosphopeptide enrichment (Fig. S1A). As a dual orthogonal approach, extracts were also enriched for peptides specifically phosphorylated at canonical PKA target motifs using targeted antibodies. The combination of these two data sets resulted in the identification of 1,225 uniquely phosphorylated proteins, highlighting the robustness in using the two distinct methodologies (Fig. 1A). To further identify likely direct PKA targets, we compared the abundances of each phosphopeptide between the WT and ΔpkaC1 strains. Proteins displaying statistically significant increases in phosphopeptide abundances of at least 2-fold between the ΔpkaC1 strain and the WT strain were prioritized (Fig. S2A). To select potential direct PKA targets from the pool of enriched phosphoproteins, the data set was further examined for phosphorylation specifically at canonical PKA target motifs, resulting in the identification of 127 unique direct PKA target candidates (Fig. S2A). These included A. fumigatus proteins previously characterized, others that are uncharacterized but homologous to proteins with known functions in other species, and also proteins with no predictable function (Table 1; Table S3).
TABLE 1.
Functional categories of direct PKA target candidate proteins
| Functional category | No. of proteins |
|---|---|
| Transcriptional regulation | 48 |
| Chromatin organization | 44 |
| Protein subcellular localization | 19 |
| Extracellular import/export | 19 |
| Cytoskeletal interaction | 18 |
| Translation | 9 |
| Lipid metabolism | 9 |
| Cellular component recycling | 8 |
| Endosomal trafficking | 7 |
| Transcription factors | 7 |
| RNA processing | 6 |
| Cell wall | 3 |
| Nuclear distribution | 3 |
| Unknown/no predicted function | 38 |
Identification of direct PKA target candidate proteins. (A) Strategy for discerning likely direct PKA targets among phosphoproteins identified by LC-MS/MS analysis. The total set of identified phosphoproteins (from both TiO2- and antibody-enriched data sets) was narrowed to those yielding a ≥2-fold-higher abundance in the WT than in the ΔpkaC1 genetic background with a statistical significance level of a P value of <0.05. This set was further narrowed to those proteins phosphorylated at canonical PKA target motifs, resulting in a set of 127 direct PKA target candidate proteins. (B) PKA target candidates were further prioritized by comparison of their PKA-dependent fold changes in phosphorylation abundance as determined by our PKA phosphoproteome analysis (y axis) to the strength of evidence for their interaction with the PKA catalytic subunit as estimated based on the number of coimmunoprecipitated peptide spectra identified for each protein in our PkaC1 interactome study (x axis). Proteins considered to be of particular interest based on a large PKA-dependent increase in phosphopeptide abundance and/or strong evidence of PKA interaction are labeled on the graph. Three proteins selected for in-depth analysis (Atg24, HapB, and Not4) are shown in color. Download FIG S2, JPG file, 0.3 MB (288.3KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Direct PKA target candidate proteins. Download Table S3, PDF file, 0.1 MB (66.5KB, pdf) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To more precisely prioritize candidate PKA target proteins, the whole proteome was also interrogated by identifying proteins coimmunoprecipitated with PkaC1 in an affinity purification procedure via LC-MS/MS analysis, yielding a PKA interactome data set. As enzyme-substrate interactions tend to be transient in nature, it is likely that interactants captured in this manner may represent more frequently targeted substrate proteins. The set of 127 potential PKA targets derived from the phosphoproteome was cross-referenced against the interactome data for prioritization (Fig. S2B). In selecting PKA target candidates to pursue for in-depth characterization, three primary criteria were considered: (i) the magnitude of the fold change in phosphorylation at PKA target motifs between the WT and ΔpkaC1 strains in the phosphoproteome data set, (ii) identification in the PKA interactome data set, and (iii) a demonstrated or predicted association with important PKA-regulated processes.
The A. fumigatus autophagy-associated protein Atg24 is a direct PKA target in vivo and in vitro.
Using the above-described stringent criteria, we identified a candidate protein (Atg24) uncharacterized among the Aspergilli. Atg24 is homologous to a sorting nexin essential for mitophagy, growth, conidiation, and oxidative stress tolerance in the rice blast fungus Magnaporthe oryzae (15). Interestingly, the Saccharomyces cerevisiae Atg24 homolog is involved in vacuolar protein retrieval (16), indicating potentially diverse roles for Atg24 among various eukaryotic species. A. fumigatus Atg24 was phosphorylated at two canonical PKA target motifs (Ser47 and Ser266), and phosphorylation at Ser47 was found to be 31.67-fold upregulated in the WT compared to the ΔpkaC1 strain (Table 2 and Fig. 2A). Atg24 also contains two additional predicted PKA target motifs, but these were not identified as phosphorylated. Further evidence in support of Atg24 as a direct PKA target was provided by the identification of Atg24 in our independent PkaC1 interactome study (Table 2; Fig. S2B). The Atg24 sequence exhibits two putative conserved functional domains typical of sorting nexins: the PX domain, containing a phosphoinositide-binding region facilitating membrane association, and the BAR domain, associated with homodimerization (Fig. 2A). Both domains are essential for mitochondrial association in M. oryzae (15). As our bioinformatic analysis of the whole proteome implicated protein degradation/autophagy as a major process regulated by PKA, Atg24 represents a PKA target of potentially strong significance.
TABLE 2.
Highlighted direct PKA target candidate proteinsa
| Parameter | Value | Phosphopeptide sequence | Phosphorylation site(s) |
Localization probability(ies) (%) |
PKA target motif |
Phosphorylation fold change (WT vs ΔpkaC1) |
Adjusted phosphorylation fold change |
P value |
|---|---|---|---|---|---|---|---|---|
| Protein name | Atg24 | RRM(Sp)SVHEDPPQAGPLADAVDLAGIGDGVLEC | S47 | 82.6 | Yes | 25.3 | 31.67 | 0.0020 |
| GenBank accession no. | XP_751536.1 | RRM(Sp)SVHEDPPQAGPLADAVDLA | S47 | 97.5 | Yes | −1.16 | 1.08 | 0.3630 |
| Description | Vacuolar targeting protein | RM(Sp)SVHEDPPQAGPL | S47 | 100 | Yes | −1.53 | −1.22 | 0.0179 |
| No. of interactome spectra | 2 | RRM(Sp)SVHEDPPQAGPL | S47 | 99.8 | Yes | −1.88 | −1.50 | 0.0039 |
| No. of PKA motifs | 4 | RE(Sp)DLEADYNDL | S266 | 100 | Yes | −1.78 | −1.42 | 0.0285 |
| Expression ratio (WT/ΔpkaC1) | 0.80 | VARRE(Sp)DLEADYNDL | S266 | 100 | Yes | −2.94 | −2.35 | 0.0088 |
| RE(Sp)DLEADYNDL | S266 | 100 | Yes | −3.5 | −2.80 | 0.0011 | ||
| MEGDLDNDPEM(Sp)EHGR | S487 | 100 | No | −1.64 | −1.31 | 0.0103 | ||
| Protein name | HapB | RM(Sp)HVGSPHVQ | S181 | 100 | Yes | −1.13 | 1.67 | 0.4860 |
| GenBank accession no. | XP_755804.1 | RM(Sp)HVG(Sp)PHVQ | S181; S185 | 100; 100 | Yes; no | 1.3 | 2.46 | 0.3060 |
| Description | CCAAT-binding transcription factor subunit | MSHVG(Sp)PHVQ | S185 | 100 | No | 1.97 | 3.73 | 0.0033 |
| No. of interactome spectra | 0 | NVDSGKPAEENPSSAPKRK(Sp)SEVNDDNANSVKKS | S330 | 73.3 | Yes | 1.16 | 2.19 | 0.5290 |
| No. of PKA motifs | 3 | RKS(Sp)EVNDDNANSVK | S331 | 99.2 | Yes | 82.2 | 155.53 | 0.0081 |
| Expression ratio (WT/ΔpkaC1) | 0.53 | RKS(Sp)EVNDDNANSVK | S331 | 100 | Yes | 1.94 | 3.67 | 0.0345 |
| KS(Sp)EVNDDNANSVK | S331 | 99.8 | Yes | −1.71 | 1.11 | 0.0029 | ||
| Protein name | Not4 | RA(Sp)LAGSQASQSPRPVHAT | S311 | 100 | Yes | 102 | 108.84 | 0.0059 |
| GenBank accession no. | XP_750771.1 | RA(Sp)LAGSQAS | S311 | 100 | Yes | 20.3 | 21.66 | 0.0471 |
| Description | CCR4-NOT core complex subunit | RA(Sp)LAGSQASQSPR | S311 | 100 | Yes | 15.6 | 16.65 | 0.0364 |
| No. of interactome spectra | 2 | RA(Sp)LAGSQ | S311 | 100 | Yes | −2.38 | −2.23 | 0.0094 |
| No. of PKA motifs | 6 | TRRA(Sp)LAG(Sp)QASQSPRPVHAT | S311; S315 | 97.3; 99.5 | Yes; no | 1.07 | 1.14 | 0.8000 |
| Expression ratio (WT/ΔpkaC1) | 0.94 | TRRA(Sp)LAG(Sp)QA(Sp)QSPRPVHAT | S311; S315; S318 | 100; 100; 99.8 | Yes; no; no | 4.24 | 4.52 | 0.1370 |
| RA(Sp)LAGSQA(Sp)QSPRPVHAT | S311; S318 | 100; 86.3 | Yes; no | 2.12 | 2.26 | 0.0325 | ||
| TAG(Tp)PPISGGGMFAQ | T638 | 99.7 | No | 1.18 | 1.26 | 0.3500 | ||
| NISEISLG(Sp)PLPK | S919 | 100 | No | 1.32 | 1.41 | 0.0879 | ||
Lowercase “p” following parenthetically enclosed amino acid residue abbreviations within peptide sequences indicates phosphorylation of corresponding residues. Bold text indicates phosphorylated amino acid residues identified on peptide fragments with no additional phosphorylation and showing significant enrichment (P < 0.05) of at least 2-fold (unadjusted) in the WT strain compared to ΔpkaC1.
FIG 2.
PKA-dependent phosphorylation and deletion phenotype characterization of the target proteins Atg24, HapB, and Not4. (A) Diagram of the Atg24 primary amino acid sequence indicating putative functional domains, locations of canonical PKA target motif serines (S) and threonines (T), and identified sites of phosphorylation (green boxes). (B) In vitro phosphorylation of recombinant Atg24 by PKA. Glutathione S-transferase (GST)-labeled Atg24 proteins with the native sequence and with S47 and S48 mutated to alanines were expressed in BL21 Escherichia coli cells and purified, followed by incubation either with or without the purified recombinant mammalian PKA catalytic subunit. Reaction mixture contents were separated by SDS-PAGE and either probed with antiphosphopeptide nanopolymer followed by chemiluminescent detection using X-ray film (top) or stained with Coomassie blue dye (bottom). Positions of molecular weight marker bands are indicated, and the expected size of dephosphorylated Atg24 is ∼80 kDa. An increased phosphopeptide signal was identified for native Atg24 in the presence of PKA, and a corresponding upward shift in protein migration was observed for this sample following Coomassie blue staining. (C) Diagram of the HapB primary amino acid sequence indicating the putative functional domain, locations of canonical PKA target motif serines (S), and identified sites of phosphorylation (green boxes). (D) Diagram of the Not4 primary amino acid sequence indicating putative functional domains, locations of conserved and nonconserved canonical PKA target motif serines (S) and threonines (T), the identified site of phosphorylation (green box), a region unique to filamentous fungi, a region conserved between yeast and filamentous fungi, and a region conserved between fungi and mammals. (E) Reduced-radial-growth phenotype of the Atg24 deletion strain (Δatg24) grown on GMM agar for 5 days at 37°C, compared to the wild-type (Atg24-WT) and complementation (Atg24-Comp) strains. (F) Reduced-radial-growth phenotype of the HapB deletion strain (ΔhapB) grown on GMM agar for 4 days at 37°C, compared to the wild-type (HapB-WT) and complementation (HapB-Comp) strains. (G) Reduced-radial-growth phenotype of the Not4 deletion strain (Δnot4) grown on GMM agar for 4 days at 37°C, compared to the wild-type (Not4-WT) and complementation (Not4-Comp) strains.
To next validate the direct phosphorylation of Atg24 by PKA, in vitro PKA phosphorylation assays were performed on purified recombinant Atg24 native and nonphosphorylatable alanine mutant (Atg24S47A;S48A) proteins. While both the native and mutant forms revealed protein bands of the expected size (∼80 kDa) in the absence of PKA, only the corresponding band for native Atg24 displayed a shift in gel migration (∼100 kDa) in the presence of PKA, indicating probable posttranslational modification at Ser47/Ser48 (Fig. 2B, bottom). Additionally, probing with an antiphosphopeptide nanopolymer confirmed the phosphorylation of WT Atg24 in the presence of PKA (Fig. 2B, top). To verify the specific site(s) of phosphorylation on Atg24, the PKA-phosphorylated native Atg24 band was analyzed via LC-MS/MS, which confirmed the phosphorylation at Ser47 with a 100% localization probability (Table S4).
Atg24 in vitro PKA phosphorylation assay MS results. Download Table S4, PDF file, 0.02 MB (17.7KB, pdf) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The CCAAT-binding transcription factor complex component HapB is a potential direct PKA target.
Our current whole-proteomic analysis and previous studies (17, 18) indicated major roles for PKA in transcriptional control and regulation of carbon and nitrogen metabolism. To further dissect this specific aspect of PKA regulation, we characterized a second direct PKA target candidate homologous to Aspergillus nidulans HapB, a member of the CCAAT-binding transcriptional regulatory complex (CBC), not previously examined in A. fumigatus. In A. nidulans, HapB controls the nuclear localization of the CBC (19), and its deletion results in a severe growth defect with a reduced ability to metabolize several alternative carbon and nitrogen sources, including γ-aminobutyric acid (GABA) and acetamide (20). In our phosphoproteomic study, A. fumigatus HapB was identified as phosphorylated at two canonical PKA target motifs (Ser181 and Ser331) with a 100% localization probability and at a third PKA target motif site (Ser330) with a 73.3% probability. Furthermore, phosphorylation at Ser331 was found to be 155-fold upregulated in the WT compared to the ΔpkaC1 strain when adjusted for the differential abundance of HapB in the two genetic backgrounds via open proteomic analysis (Table 2 and Fig. 2C). While the phosphorylation of Ser331 was found to be strongly PKA dependent in our phosphoproteomic analysis, a direct interaction of HapB and PKA was not identified in our PkaC1 interactome analysis (Table 2; Fig. S2B), leaving open the possibility that the phosphorylation of HapB may be regulated indirectly rather than directly by PKA. However, it is also possible that HapB was simply missed in our interactome analysis due to the transient nature of kinase-substrate interactions. The HapB sequence exhibits a conserved HAP2 domain typical of homologous CBC members. A putative nuclear localization signal (NLS) was also identified in A. fumigatus HapB based on homology to the experimentally validated NLS of A. nidulans (19) that is located immediately N terminally to the Ser330 and Ser331 phosphosites (Fig. 2C).
The CCR4-NOT regulatory complex component Not4 is another probable direct PKA target.
Another direct PKA target candidate protein that met our stringent proteomic selection criteria was identified as Not4, which is also uncharacterized among filamentous fungi but showed sequence homology to S. cerevisiae Not4p, a member of the CCR4-NOT regulatory complex. S. cerevisiae Not4 is associated with the regulation of a number of essential cellular processes, including translation, transcription, protein degradation, and the DNA damage response (21). Not4p has also been found to be both polysome and proteasome associated, and its deletion results in increased susceptibility to translation stress (hygromycin B [HYG]), replication stress (hydroxyurea), and heat stress (22–24). While the N-terminal region of A. fumigatus Not4 shows significant homology to yeast Not4p, A. fumigatus Not4 is approximately twice the length, with its C-terminal region being absent in yeast Not4p. Interestingly, homology searches across a wide range of fungal species, including the filamentous ascomycete clade as well as ascomycete and basidiomycete yeasts and zygomycetes, revealed that the C-terminal domain is unique to filamentous fungi, suggesting a possible role in polarized growth. Moreover, the alignment of the mammalian CNOT4 protein indicated that only the extreme N-terminal region shared any significant sequence similarity between fungi and mammals (Fig. 2D; Fig. S3A). The only conserved functional domains identifiable in A. fumigatus Not4 were located in this N-terminal conserved region and included a RING/Ubox E3 ubiquitin ligase domain and an RMM RNA-binding domain (Fig. 2D). This is consistent with the role of yeast Not4p as an E3 ubiquitin ligase, known to target stalled translating peptides and the nucleosomal histone protein H2B (22, 23, 25, 26). Not4p-dependent H2B ubiquitination has also been associated with increased histone H3K4 methylation (23, 26).
ClustalW alignment of the amino acid sequences of Not4 homologs from human and fungal species and growth phenotypes of Not4 truncation mutants. (A) The amino acid sequences of the proteins homologous to A. fumigatus Not4 as determined by a BLAST search for Homo sapiens as well as 10 other fungal species representing the filamentous ascomycetes (A. fumigatus, A. nidulans, Penicillium brasilianum, M. oryzae, Neurospora crassa, and Sordaria macrospora), the ascomycete yeasts (S. cerevisiae, C. albicans, and Schizosaccharomyces pombe), as well as the basidiomycota (C. neoformans) and zygomycota (Mucor ambiguus) were aligned via the ClustalW algorithm using MegAlign v15.2.0.130 software (DNAStar). (B) Phenotypes of the Not4-WT, Not41–693 amino acid truncation mutant, Not41–233 truncation mutant, and Δnot4 strains on GMM agar (5 days at 37°C). Details show an uneven leading growth edge of the Not41–693 truncation mutant compared to Not4-WT. Download FIG S3, JPG file, 0.5 MB (472.8KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Our phosphoproteomic data showed that A. fumigatus Not4 is phosphorylated at five sites with a >95% localization probability, including one canonical PKA target motif (Ser311) with a 100% localization probability. The phosphorylation of a peptide fragment at Ser311 was increased in abundance 108.84-fold in the WT background compared to the ΔpkaC1 background (Table 2 and Fig. 2D). Additionally, two Not4 peptides were also identified in our PkaC1 interactome analysis, in addition to three other putative members of the CCR4-NOT complex (Not1, Not2, and Not3), indicating a likely interaction between the CCR4-NOT complex and PKA. Based on both phosphoproteome and interactome data and our whole-proteomic data indicating a major role for PKA in regulating protein synthesis in A. fumigatus (a primary function of CCR4-NOT in yeast), we selected Not4 as a likely direct PKA target of potentially important function.
PKA target candidates Atg24, HapB, and Not4 are essential for hyphal growth of A. fumigatus.
To ascertain the overall importance of the probable PKA targets Atg24, HapB, and Not4 for A. fumigatus growth and development, the radial growth of deletion strains for each (Δatg24, ΔhapB, and Δnot4) was compared to those of the corresponding WT strain expressing green fluorescent protein (GFP)-labeled protein (Atg24-WT, HapB-WT, and Not4-WT) and a complemented strain (Atg24-Comp, HapB-Comp, and Not4-Comp). In each case, the deletion strains exhibited markedly reduced radial growth compared to the control strains (Fig. 2E to G). To examine the impact of PKA-dependent phosphorylation at specific target residues, we also generated Atg24, HapB, and Not4 phosphomutant strains in which each of the identified phosphorylated PKA target serines and threonines (Table 2 and Fig. 2A, C, and D) was mutated to nonphosphorylatable alanine residues as well as to negatively ionizing phosphomimetic aspartic acid residues. Because A. fumigatus Not4 showed differential degrees of sequence conservation between filamentous fungi and yeasts or mammals (Fig. 2D), we also generated two mutant strains, Not41–693 and Not41–233, expressing truncated forms of Not4. The Not41–693 mutant exhibited a slight but not statistically significant reduction in radial growth compared to Not4-WT, while Not41–233 exhibited a significant radial growth defect, although this was not as severe as that of the Δnot4 strain (Fig. S3B). A more detailed discussion of the phenotypic effects of Not4 truncation is provided in Appendix I at https://doi.org/10.6084/m9.figshare.13087418.
Roles in stress responses and subcellular localization patterns for Atg24, HapB, and Not4.
We also identified important roles for Atg24, HapB, and Not4 in the fungal response to adverse environmental conditions, including oxidative stress, nutrient deprivation, and temperature stress, as well as to antimicrobial agents, including the three major classes of antifungals: azoles (voriconazole [VCZ]), echinocandins (caspofungin [CSP]), and polyenes (amphotericin B [AMB]), discussed in Appendix II at the URL mentioned above. Additionally, to examine the subcellular localization of the PKA target proteins Atg24, HapB, and Not4 in A. fumigatus, hyphae of the Atg24-WT, HapB-WT, and Not4-WT strains as well as selected mutant strains expressing GFP-labeled proteins were observed via live-cell fluorescence microscopy. These observations are discussed in Appendix III at the URL mentioned above.
PKA-dependent phosphorylation of Atg24, HapB, and Not4 regulates multiple stress response pathways.
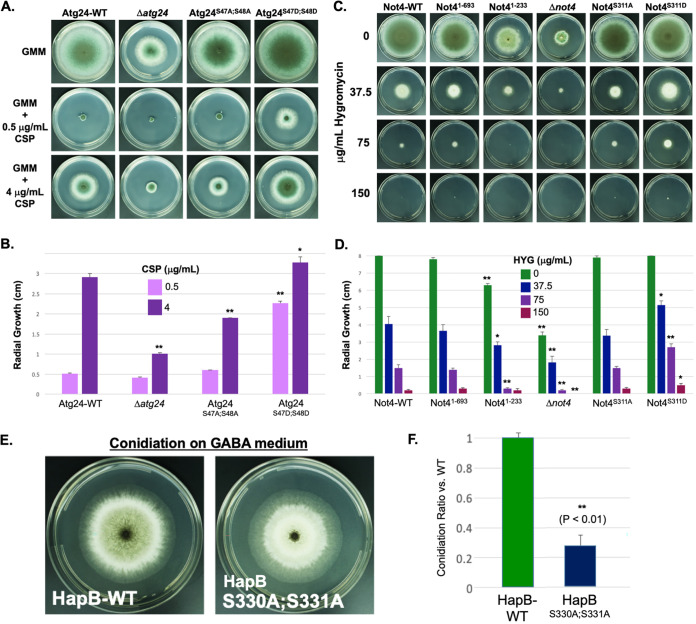
PKA deletion results in increased susceptibility to cell wall stress in A. fumigatus (12, 27), and autophagy has also been reported to play an important role in the fungal cell wall stress response (28). Therefore, we next examined whether the PKA-dependent phosphorylation of Atg24 impacts fungal cell wall stress tolerance (Fig. 3A and B). CSP, which inhibits fungal cell wall β-glucan synthesis, has been demonstrated to maximally inhibit A. fumigatus growth at lower concentrations (0.5 μg/ml), while higher concentrations (4 μg/ml) induce a “paradoxical effect” in which partial growth recovery occurs due to the activation of alternative stress response mechanisms (29). We found that the Δatg24 strain showed growth inhibition similar to that of Atg24-WT at 0.5 μg/ml CSP but had significantly reduced growth at 4 μg/ml. Interestingly, alanine substitution at Ser47 and Ser48 (Atg24S47A;S48A) also resulted in reduced paradoxical growth recovery compared to Atg24-WT at 4 μg/ml CSP despite showing growth inhibition similar to that of the control strain at 0.5 μg/ml CSP. Furthermore, the Ser47 and Ser48 phosphomimetic mutant strain (Atg24S47D;S48D) demonstrated markedly enhanced growth compared to Atg24-WT at 0.5 μg/ml as well as 4 μg/ml CSP. CSP tolerance was not affected by mutations at any of the other PKA target motifs. Taken together, these data indicate that the identified PKA-dependent phosphorylation of Atg24 at Ser47/Ser48 is an important regulator of the fungal cell wall stress response in A. fumigatus. However, expressing Atg24S47D;S48D in a ΔpkaC1 background failed to ameliorate the increased CSP sensitivity of the ΔpkaC1 strain, indicating that other factors besides Atg24 phosphorylation contribute to PKA-mediated cell wall stress tolerance (Fig. S4).
FIG 3.
Effects of PKA-dependent phosphorylation of target proteins on stress responses. (A) Comparison of radial growth of the Atg24-WT and Δatg24 strains as well as Ser27/Ser48 alanine (Atg24S47A;S48A) and aspartic acid (Atg24S47D;S48D) mutant strains on GMM agar infused with 0, 0.5, or 4 μg/ml caspofungin (CSP) for 5 days at 37°C. (B) Quantitative comparison of radial growth measurements from panel A. Growth of the Δatg24 and Atg24S47A;S48A strains was significantly more inhibited than that of Atg24-WT at 4 μg/ml CSP, while growth of the Atg24S47D;S48D strain was significantly less inhibited than that of Atg24-WT at both 0.5 and 4 μg/ml CSP. (C) Comparison of radial growth of Not4-WT, Not4 truncation, Δnot4, and Ser311 alanine (Not4S311A) and aspartic acid (Not4S311D) substitution mutant strains on GMM agar (5 days at 37°C) infused with the indicated concentrations of hygromycin (HYG). (D) Quantitative comparison of radial growth measurements from panel C. Growth of the Not41–233 and Δnot4 strains was significantly reduced compared to Not4-WT with or without HYG (*, P < 0.05; **, P < 0.01), while Not4S311D showed significantly increased growth compared to Not4-WT at all concentrations of HYG. (E) Growth (5 days at 37°C) and conidiation of HapB-WT and the HapBS330A;S331A alanine substitution mutant on solid agar medium using GABA as the sole carbon and nitrogen source. (F) Quantitative comparison of conidiation on GABA medium by the HapB-WT and HapBS330A;S331A strains from the experiment shown in panel E. Conidiation was significantly reduced (P < 0.01) in the alanine substitution mutant as determined by t test comparisons.
Cell wall stress sensitivity of Atg24 and PkaC1 mutant strains. Growth of the Atg24-WT, Atg24-S47D/S48D, ΔpkaC1, and ΔpkaC1; Atg24-S47D/S48D strains was observed on agar plates infused with the indicated concentrations of caspofungin (CSP). Both the ΔpkaC1 and ΔpkaC1; Atg24S47D/S48D strains showed patterns of growth inhibition similar to that of Atg24-WT, with some evidence of paradoxical growth recovery at CSP concentrations of 2 and 4 μg/ml. Download FIG S4, JPG file, 0.3 MB (349.9KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The yeast Not4 homolog has been associated with the regulation of protein translation and the response to the translation-inhibiting antibiotic hygromycin B (HYG). To determine whether PKA-dependent phosphorylation of Not4 plays a role in regulating the response of A. fumigatus to translation stress, the Not4-WT, Not41–694, Not41–233, Δnot4, Not4S311A, and Not4S311D strains were grown on media with concentrations of HYG ranging from 0 to 150 μg/ml (Fig. 3C and D). The growth of the Not41–693 and Not4S311A strains was not significantly different from that of Not4-WT at any HYG concentration, while the growth of the Not41–233 strain was significantly inhibited compared to Not4-WT at all HYG concentrations besides 150 μg/ml, and the Δnot4 strain was significantly inhibited compared to Not4-WT at all HYG concentrations. Interestingly, the Not4S311D mutant exhibited significantly increased growth compared to Not4-WT at 37.5, 75, and 150 μg/ml HYG, with the effect being most pronounced at 75 μg/ml. In order to further validate the contribution of Not4 to the translation stress response and determine whether these observed effects were specific to the mechanism of action of HYG, potentially through impacting glycosylation, which has been shown to contribute to HYG resistance in yeast (30), we additionally examined the growth of Not4 mutant strains under exposure to Geneticin (G418), an alternative aminoglycoside translation inhibitor (Fig. S5). While this compound was less inhibitory to A. fumigatus growth than HYG, we still found that growth reduction of the Δnot4 strain compared to Not4-WT was exacerbated by Geneticin treatment, while the Not4S311D mutant showed significantly increased growth compared to Not4-WT across a range of Geneticin concentrations. These observations suggest that Not4 plays an important role in the A. fumigatus response to translation stress and that PKA-dependent phosphorylation of Not4 at Ser311 is able to enhance the efficiency of this response. However, it remains possible that Not4 may regulate aminoglycoside tolerance through mechanisms indirectly associated with translation.
Effects of Not4 on Geneticin sensitivity. Quantitative comparison of the radial growth of Not4-WT, the Not4 Ser311-to-alanine (Not4S311A) and Ser311-to-aspartic acid (Not4S311D) substitution mutant strains, and the Δnot4 strain on GMM agar (4 days at 37°C) infused with the indicated concentrations of Geneticin (G418) was performed. Statistical comparisons of each strain to Not4-WT were made at the same Geneticin concentrations (*, P < 0.05; **, P < 0.01 [by Student’s t test]). Growth of the Δnot4 strain was significantly reduced compared to Not4-WT with or without Geneticin, but the ratio of Not4-WT to Δnot4 growth was increased under Geneticin treatment (2.03 at 0 mg/ml compared to 2.85 at 3.2 mg/ml), suggesting exacerbated growth inhibition. Not4S311D showed significantly increased growth compared to Not4-WT at all concentrations of geneticin examined. Download FIG S5, JPG file, 0.2 MB (216.6KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Previously, HapB was shown to regulate alternative carbon and nitrogen source utilization in A. nidulans (19, 20). To ascertain the functional importance of HapB phosphorylation by PKA, growth of the HapBS330A;S331A and HapBS330D;S331D phosphomutant strains was examined on standard medium, on medium containing GABA as the sole carbon and nitrogen source, and under nitrogen starvation conditions. While the phosphosite mutation did not affect radial growth under these conditions (see Appendix II at https://doi.org/10.6084/m9.figshare.13087418), when using GABA as the sole carbon and nitrogen source, conidiation of the HapBS330A;S331A strain was strongly suppressed compared to that of the HapB-WT strain (Fig. 3E and F). The ratio of ΔhapB to HapB-WT strain growth on GABA medium was higher than that observed for standard medium, indicating that the loss of HapB causes less severe relative growth inhibition when using GABA as the sole carbon and nitrogen source (see Appendix II at the URL mentioned above). This indicates a negative role for HapB in regulating GABA metabolism in A. fumigatus. Under such circumstances, the PKA-dependent phosphorylation of HapB at Ser330/Ser331 may have an inhibitory effect on the activity of the CBC.
Modeling and molecular dynamics simulations of Atg24 indicate significant phosphorylation-induced electrostatic effects.
Because nonphosphorylatable and phosphomimetic mutations of Ser47 and Ser48 residues in Atg24 produced significant effects on protein functionality as demonstrated by the altered cell wall stress responses of these phosphomutant strains, we sought to elucidate the possible biochemical basis for this altered functionality. As homodimerization has been established as a key functional feature of BAR domain-containing proteins in general and sorting nexins in particular (31, 32), we generated in silico structural models of dimerized Atg24 bound to phosphoinositide-3-phosphate [PI(3)P] in both dephosphorylated and Ser47/Ser48-phosphorylated forms. The structural model of Atg24 and the subsequent dimer model (Fig. 4A) are similar to that of the PI(3)P-bound PX-BAR membrane-remodeling unit of mammalian sorting nexin 9 (33). Molecular dynamics (MD) simulations of the subsequent models revealed stability in the last 90 ns of each simulation based on the average structures from Cα root mean square deviations (RMSD) and radius of gyration (Rg) (a measurement of overall compactness) (Fig. S6). Phosphorylated Ser47 and Ser48 residues are located in a poorly structurally defined region of Atg24 (Fig. 4A and B, gray). The phosphorylation sites reside ∼40 Å from the PI(3)P-binding site and ∼32 Å from the nearest residues involved in dimerization. Phosphorylation of residues Ser47 and Ser48 is predicted to have several structural consequences based on the 100-ns MD simulation. Phosphorylation results in a localized change in the electrostatic surface of Atg24 (Fig. 4C); however, there is a noticeable change in the negative surface electrostatics of the localized region (expanded red region in Fig. 4C). Phosphorylation of Ser47 and Ser48 results in increases (residues 1 and 2, 39 to 56, 59 to 64, 223 to 224, 379 to 393, 398 to 400, and 483 to 486) and decreases (residues 17 to 36 and 491) in the ability of certain residues to sample conformational space (Fig. 4D, ±1 Z-score). Of these, only residues Phe223 and Val24 are also involved in dimerization. The greatest significance in RMSD differences between the WT and the phosphorylated state occurred in the N terminus (residues 1 to 68) and a loop region between alpha helices (residues 375 to 400). The MD simulation of the WT Atg24 model has three short alpha helices in its N terminus that are not observed in the MD simulation of the phosphorylated model. These findings suggest that Ser47/Ser48 phosphorylation of Atg24 is unlikely to have a dramatic impact on either dimerization or membrane association via PI(3)P binding, but due to significant localized electrostatic surface alterations, it is likely that interactions with other as-yet-undetermined binding partners could be strongly affected by this modification.
FIG 4.
Structural models of Atg24-WT and phosphorylated Ser47 and Ser48. (A) Dimeric structure of Atg24 based on the phosphatidylinositol-3-phosphate [PI(3)P]-bound PX-BAR membrane remodeling unit of sorting nexin 9. The structure is shown in a cartoon format with the BAR domain in red, the PX domain in blue, and the sequence between the two domains in yellow. The second dimer component protein is made slightly transparent, and dimer component proteins A and B and Ser47 and Ser48, shown as spheres, are labeled for clarity. (B) Details of the site of phosphorylation of Ser47 and Ser48 as well as the PI(3)P-binding site with the same color coding as that noted above. (C) Electrostatic surface plots of the protein models created using the Adaptive Poisson-Boltzmann Solver in PyMOL for WT (left) and phosphorylated (right) proteins. The model is rotated 90° horizontally counterclockwise from the orientation shown in panel B. Blue denotes the positively charged electrostatic surface, while red denotes negatively charged areas. (D) Plot of the significance of observed root mean square fluctuation (RMSF) differences between WT and phosphorylated protein models, color coded as noted above. The secondary structure is plotted on top of the panel, dotted boxes denote residues involved in dimerization, and # denotes the site of phosphorylation for clarity. A positive Z-score suggests more significance for the phosphorylated protein, while a negative Z-score suggests more significance for the WT protein.
Visual inspection of the MD simulations of the A. fumigatus Atg24-WT and phosphorylated Ser47 and Ser48 models. The Cα RMSD (black and red lines) and Rg (radius of gyration) (overall compactness of structure) (green and blue lines) show stability after an initial period of equilibration (first 10 ns). Download FIG S6, JPG file, 0.3 MB (270.9KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Atg24, HapB, and Not4 are important virulence determinants in mammalian disease.
Given the strong impacts of Atg24, HapB, and Not4 on the hyphal growth and multiple stress responses of A. fumigatus, we next determined whether these PKA targets influence virulence in a mammalian host. For Atg24, immunosuppressed CD1 outbred mice were infected with conidia of the WT A. fumigatus strain (Ku80) as well as the Atg24 deletion and complementation strains via inhalation of aerosolized spores in an enclosed chamber. This experiment was repeated twice using two different levels of immunosuppression (Fig. 5A to C) for added validation. In both experiments, murine mortality was significantly reduced in the Δatg24 strain compared to the WT strain. In the first experiment, mortality from Δatg24 infection was also significantly reduced compared to Atg24-Comp, whereas in the second experiment, the difference between these strains was not statistically significant. This minor difference may be explained by the fact that the Atg24-Comp strain did not exhibit a completely recovered in vitro growth phenotype (Fig. S4A). Comparative histological analysis of murine lung tissues infected with the Ku80 and Δatg24 strains was also performed using tissue harvested at 3 days postinfection. Hematoxylin and eosin (HE) and silver staining revealed increased tissue damage and invasive hyphal growth, respectively, in the Ku80 strain-infected lung tissue compared to that infected with the Δatg24 strain (Fig. 5C).
FIG 5.
Effects of the PKA target proteins Atg24, HapB, and Not4 on virulence in a murine infection model. (A) Survival curve for Atg24 mutant strains in an inhalational murine model of invasive pulmonary aspergillosis. Fifteen mice immunosuppressed with 150 mg/kg cyclophosphamide or 40 mg/kg triamcinolone were exposed to 12 ml of aerosolized suspensions of 109 spores/ml of wild-type (Ku80), Δatg24, or Atg24-Comp strain conidia inside an inhalation chamber. Mice were monitored for survival for 14 days following infection. Mortality was significantly reduced for mice infected with the Δatg24 strain compared to the Ku80 (P = 0.0193) and Atg24-Comp (P = 0.0248) strains as determined by log rank comparisons. (B) Survival curve for repeat experiments of panel A, substituting 175 mg/kg cyclophosphamide for immunosuppression of mice. Mortality was significantly reduced (P = 0.0062) for mice infected with the Δatg24 strain compared to Ku80 but not compared to Atg24-Comp. (C) Histological analysis of mouse lung tissue 3 days after infection with Ku80 (top) or Δatg24 (bottom) from an experiment corresponding to the survival curve shown in panel A. Hematoxylin and eosin (HE) and silver staining were used to visualize tissue inflammation and invasive fungal growth, respectively. Examples of both are indicated by black arrows. (D) Survival curve of HapB mutant strains in the intranasal infection murine model. Ten mice immunosuppressed with 175 mg/kg cyclophosphamide or 40 mg/kg triamcinolone were inoculated with 40 μl of suspensions of 108 spores/ml of Ku80, ΔhapB, or HapB-Comp strain conidia. Mice were then monitored for survival for 14 days following infection. Mortality was significantly reduced for mice infected with the ΔhapB strain compared to the Ku80 (P = 0.0160) and HapB-Comp (P = 0.0036) strains as determined by log rank comparisons. (E) Survival curve for Not4 mutant strains in the intranasal infection murine model. Twenty-five mice were inoculated as described above for panel D with Ku80,Δnot4, or Not4-Compstrain conidia. Mortality was significantly reduced for mice infected with the Δnot4 strain compared to the Ku80 (P = 0.0295) and Not4-Comp (P = 0.0009) strains as determined by log rank comparisons. (F) Histological analysis of HE- and silver-stained mouse lung tissue 3 days after infection with Ku80 or ΔhapB conidia from the experiment shown in panel D. Arrows indicate inflammation and hyphal growth. (G) Histological analysis of HE- and silver-stained mouse lung tissue 3 days after infection with Ku80 or Δnot4 conidia from the experiment shown in panel E. Arrows indicate inflammation and hyphal growth.
To examine the contributions of HapB and Not4 to virulence, mice were infected intranasally with conidia of the Ku80, deletion, and complementation strains. Murine mortality was significantly reduced with the ΔhapB and Δnot4 strains compared to both Ku80 and the respective complementation strains, while the Ku80 and complementation strains did not produce significantly different rates of mortality from one another in either experiment (Fig. 5D and E). HE and silver staining of infected lung tissue revealed increased tissue damage and invasive hyphal growth in the Ku80-infected lung tissue compared to that infected with either the ΔhapB or the Δnot4 strain in each experiment (Fig. 5F and G). Overall, these results demonstrate that the activity of each of these three PKA targets contributes significantly to the disease severity of invasive aspergillosis in a mammalian host model, implicating manipulation of pathways regulated by these effectors as a potentially important mechanism for the control of pathogenesis by PKA.
Analysis of additional direct PKA target candidate proteins indicates minor effects on fungal physiology.
In addition to Atg24, HapB, and Not4, a number of other direct PKA targets with potential significance were identified in our phosphoproteomic analysis, including both previously characterized and uncharacterized proteins. Deletion and phosphomutational analyses were carried out for several of these selected proteins (Table 3), and a detailed discussion of this work is provided in Appendix IV at https://doi.org/10.6084/m9.figshare.13087418.
TABLE 3.
A. fumigatus strains used in this study
| Strain and/or GenBank accession no. | Genotype | Reference |
|---|---|---|
| Ku80 (WT) | akuB-Ku80 | 62 |
| Ku80; PyrG− | akuB-Ku80; ΔpyrG | 62 |
| ΔpkaC1 | pkaC1::pyrG | 27 |
| Δatg24 | atg24::pyrG | This study |
| Atg24-Comp | atg24::pyrG; atg24promo-atg24-hph | This study |
| Atg24-WT | Δatg24::atg24promo-atg24–egfp-pyrG | This study |
| Atg24S47A;S48A | Δatg24::atg24promo-atg24mt-S47A;S48A–egfp-pyrG | This study |
| Atg24T117A | Δatg24::atg24promo-atg24mt-T117A–egfp-pyrG | This study |
| Atg24T172A | Δatg24::atg24promo-atg24mt-T172A–egfp-pyrG | This study |
| Atg24T266A | Δatg24::atg24promo-atg24mt-T266A–egfp-pyrG | This study |
| Atg24S47D;S48D | Δatg24::atg24promo-atg24mt-S47D;S48D–egfp-pyrG | This study |
| ΔpkaC1; Atg24S47D;S48D | pkaC1::hph; Δatg24::atg24promo-atg24mt-S47D;S48D–egfp-pyrG | This study |
| ΔhapB | hapB::pyrG | This study |
| HapB-Comp | hapB::pyrG; hapBpromo-hapB-hph | This study |
| HapB-WT | ΔhapB::hapBpromo-hapB–egfp-hph | This study |
| HapBS181A;S185A | ΔhapB::hapBpromo-hapBmut-S181A;S185A–egfp-hph | This study |
| HapBS330A;S331A | ΔhapB::hapBpromo-hapBmut-S330A;S331A–egfp-hph | This study |
| HapBS330D;S331D | ΔhapB::hapBpromo-hapBmut-S330D;S331D–egfp-hph | This study |
| Δnot4 | not4::pyrG | This study |
| Not4-Comp | not4::pyrG; not4promo-not4trunc-1–693—egfp-hph | This study |
| Not4-WT | Δnot4::not4promo-not4–egfp-pyrG | This study |
| Not41–693 | Δnot4::not4promo-not4trunc-1–693—egfp-pyrG | This study |
| Not41–233 | Δnot4::not4promo-not4trunc-1–233—egfp-pyrG | This study |
| Not4S311A | Δnot4::not4promo-not4mut-S311A; not4trunc-1–693—egfp-hph | This study |
| Not4S311D | Δnot4::not4promo-not4mut-S311D; not4trunc-1–693—egfp-hph | This study |
| AtfA-GFP | ΔatfA::atfApromo-atfA–egfp-hph | This study |
| AtfAS234A;T379A;S433A;T441A | ΔatfA::atfApromo-atfAmut-S234A;T379A;S433A;T441A–egfp-hph | This study |
| AmdX-GFP | ΔamdX::amdXpromo-amdX–egfp-hph | This study |
| ΔpkaC1; AmdX-GFP | pkaC1::pyrG; ΔamdX::amdXpromo-amdX–egfp-hph | This study |
| Rum1-GFP | Δrum1::rum1promo-rum1–egfp-hph | This study |
| ΔpkaC1; Rum1-GFP | pkaC1::pyrG; Δrum1::rum1promo-rum1–egfp-hph | This study |
| Δrum1 | rum1::pyrG | This study |
| ΔSphK (XP_752463.1) | XP_752463.1::pyrG | This study |
| ΔPst1 (XP_750917.1) | XP_750917.1::pyrG | This study |
| ΔXP_750216.1 | XP_750216.1::pyrG | This study |
| ΔXP_751586.1 | XP_751586.1::pyrG | This study |
| ΔXP_749288.1 | XP_749288.1::pyrG | This study |
| ΔXP_001481443.1 | XP_001481443.1::pyrG | This study |
DISCUSSION
PKA regulation is critical for cellular processes, yet its direct effectors are not well defined in any human pathogen. We identified several major PKA-regulated pathways in a pathogenic fungus and characterized for the first time key effector proteins and their specific PKA-dependent phosphoregulatory mechanisms governing disease in multiple animal models of infection. Our data support a model for PKA signaling in which dramatic changes in overall physiology and pathogenesis are regulated by finely tuned and subtle modifications to the activity of target proteins associated with myriad diverse cellular processes.
The three major PKA-regulated pathways that we identified encompass fundamental cellular processes, including (i) the breakdown and recycling of cellular components via autophagy and proteasomal pathways, mediated in part through the sorting nexin Atg24; (ii) gene transcription via the regulation of chromatin remodeling and transcription factors such as the CBC component HapB; and (iii) protein biosynthesis, including amino acid and tRNA synthesis and translation, in part through the CCR4-NOT complex component Not4 (Fig. 6). Our identification of Atg24, HapB, and Not4 each as both probable direct PKA targets and important contributors to pathogenesis in a mammalian host demonstrates for the first time a direct link between PKA signaling and the respective pathways associated with each of these effectors in the virulence of a human pathogen. Importantly, neither Atg24 nor Not4 homologs have previously been associated with virulence in any pathogen. Another CBC component, HapC, was previously shown to be essential for the virulence of A. fumigatus (34), which together with our identification of HapB as an important virulence determinant strongly supports a role for the CBC in A. fumigatus pathogenesis. While an association with virulence has been previously reported for autophagy-associated proteins (35–37), CBC components (38), and CCR4-NOT complex members (39–41) in some pathogenic yeast species, our findings suggest a key role for PKA in regulating the impacts of these processes on pathogenesis. Considering the significant but relatively modest functional effects observed from individual phosphosite mutations in the PKA effectors, it is unlikely that the phosphorylation of any single target protein alone would have a major influence on pathogenesis. Instead, our results suggest that the overall dramatic impact of PKA signaling on fungal virulence is a result of the cumulative effects of regulatory modification of numerous effector proteins involved in diverse cellular pathways. Given the divergence of these pathways between mammals and microbial pathogens, elucidating their poorly defined mechanisms to identify novel pathogen-specific processes may lead to new targeted therapeutic strategies.
FIG 6.
Schematic model showing new PKA signaling effectors in A. fumigatus. Major pathways regulated by PKA as evidenced by whole-proteome and phosphoproteome analyses are depicted, as are the specific roles of the PKA effectors Atg24, HapB, and Not4 in these processes. “P” indicates phosphorylation of the effector proteins by PKA. Arrows indicate regulatory pathways. Areas of regulation that are suggested but not conclusively determined by existing evidence are denoted with question marks.
Such mechanistic insights were possible through a unique combination of robust quantitative whole-proteomic and phosphoproteomic approaches using dual-phosphopeptide enrichment strategies with three different proteases to maximize proteome coverage. Quantitative comparison between triplicate samples of each strain confirmed the identification of phosphorylated proteins statistically significantly enhanced exclusively by PKA activity. This quantitative approach is beneficial because many proteins may be phosphorylated by multiple kinases, even at the same target sites, and such proteins might thus be phosphorylated to some degree even in the absence of one targeting kinase and therefore overlooked in a qualitative screen. Integration of the PKA-dependent phosphoproteome data with PkaC1 interactome data enabled the most thorough interrogation of direct PKA target proteins that has been performed for any organism.
Of the 3,212 proteins quantified through open proteomic analysis, the abundances of approximately half were significantly altered by PKA, confirming its role as a master regulator of biological processes. Of the proteins upregulated by PKA, tandem bioinformatic approaches revealed that those involved in both protein expression and degradation were prominent, indicating that a primary mode of action for PKA is in the modulation of protein levels. This observation that PKA regulates protein homeostasis is in accordance with the PKA-dependent phosphoproteome of Cryptococcus neoformans, in which proteasomal regulation by PKA was found to influence capsule formation, and recent work in C. albicans, wherein proteasomal inhibition induced filamentation in a PKA-dependent manner (42, 43). The molecular chaperone Hsp90 also negatively regulates filamentation in C. albicans through suppression of PKA signaling (44), suggesting a major role for PKA in filamentous fungal growth, which may have implications for polarized cell growth among eukaryotes. We found that the most prominent functional category downregulated by PKA was proteins involved in aerobic respiration, illustrating the role of PKA as a regulatory check on the metabolic rate. The majority of the 127 strongly predicted PKA target proteins serve putative functions in transcriptional regulation, either as DNA-binding transcription factors or through chromatin remodeling, indicating this as a primary direct method by which PKA exerts its influence over cellular processes.
We validated the putative autophagy-associated protein Atg24 as an in vitro and in vivo target of PKA and demonstrated the functional significance of the PKA-dependent phosphorylation of Atg24. PKA has been previously linked to autophagic processes in S. cerevisiae, targeting the autophagy-associated proteins Atg1p and Atg13p (45, 46). In the plant pathogen M. oryzae, Atg24 may function similarly to yeast Atg32p, an essential component of the yeast mitophagy pathway (15, 47). A. fumigatus Atg24 also exhibits sequence similarity to the yeast sorting nexin Snx4p, involved in protein retrieval from vacuoles (16), and it is possible that A. fumigatus Atg24 is involved in either or both of these processes.
Molecular modeling and MD simulations of the nonphosphorylated and phosphorylated states of Atg24 provided a biochemical basis for the functional consequences of Atg24 phosphorylation by PKA, indicating that the phosphorylation of Ser47/Ser48 drastically altered the negative electrostatic character of the protein albeit in a localized manner. The Atg24 mammalian ortholog sorting nexin 9 contains an N-terminal amphipathic helix important for membrane tubulation/anchoring (33), and our MD simulation revealed a loss of three short alpha helices in the N terminus upon phosphorylation. This suggests that the phosphorylated state may still dimerize and bind PI(3)P; however, it may not associate efficiently with target membranes.
The effects of Atg24 PKA phosphosite mutations on the response to the antifungal caspofungin are of broad clinical relevance given the treatment resistance challenges of invasive fungal infections (48–51). Although an association between autophagy and the cell wall stress response has been reported (28), mechanisms by which the processes are related remain unclear. One possibility is that autophagy facilitates the fungal response to cell wall stress by freeing up resources from nonessential cellular components that may instead be redirected toward cell wall repair mechanisms. Our findings should facilitate investigation into new methods for enhancing the efficacy of cell wall-targeting antifungal agents.
We also present novel evidence of a specific mechanism for nutrient sensing regulation through the PKA-dependent phosphorylation of the CBC component HapB, the first demonstration of CBC regulation by PKA. The CBC is conserved among eukaryotes and has been found to regulate growth on nonfermentable carbon sources in S. cerevisiae (52) and to regulate both iron and redox homeostasis in A. nidulans (53, 54). It was also shown to regulate the expression of the acetamidase-encoding amdS gene of A. nidulans (55). Another transcription factor known to promote amdS expression in A. nidulans, AmdX (56), was also identified in our study as a direct PKA target, suggesting the possibility of coordinated regulation of metabolic pathways by PKA through multiple avenues. Additionally, the markedly reduced conidiation resulting from HapB phosphosite mutation during growth using GABA as the sole carbon and nitrogen source indicates an important role for PKA in modifying HapB function. Although HapB phosphosite mutation did not influence the nuclear localization of the protein, PKA-dependent phosphorylation of HapB may affect its association with other components of the CBC.
This work also provides the first identification of the E3 ubiquitin ligase CCR4-NOT complex component Not4 as a probable direct PKA target in any organism. A functional association between PKA signaling and CCR4-NOT activity has been reported in that both pathways appear to coordinate the repression of the stress response mediated by the Msn2/4p transcription factors in yeast (57). A PKA target site of Not4 regulating tolerance to HYG suggests an important role for PKA in regulating CCR4-NOT translational control. In addition, the identification of Not1, Not2, Not3, and Not4 in our PkaC1 interactome as well as a recent report on the identification of PKA-dependent Not3 phosphorylation in A. nidulans (58) together suggest a high degree of regulatory interaction between PKA and the CCR4-NOT complex. The identification of a filamentous fungus-specific C-terminal domain of Not4 is novel, as no component of the CCR4-NOT complex has previously been characterized in a filamentous fungus. Our observation of the dramatically increased abundance of truncated Not4 proteins (see Appendix III at https://doi.org/10.6084/m9.figshare.13087418) suggests a potential role for this C-terminal domain in regulating the expression and/or degradation of Not4. Additionally, the subcellular localization of the Not4 truncations indicates that the domain may be involved in nuclear import/export mechanisms.
These data comprise the most comprehensive analysis of direct and validated PKA targets in any organism and provide new insights into the specific ways in which PKA executes its role as a master regulator of biological signaling pathways governing overall microbial pathogenesis. Understanding how this ubiquitous signaling pathway controls virulence in all pathogens will allow the future development of targeted therapies for disease.
MATERIALS AND METHODS
Protein extraction and purification.
A. fumigatus strains were cultured in liquid glucose minimal medium (GMM) with shaking at 250 rpm for 24 h at 37°C. The total cell lysate was obtained by homogenizing mycelia (1 g [wet weight]) using a mortar and pestle as previously described (27, 59, 60). The total protein in the crude extracts was quantified by the Bradford method (61), and samples were normalized to contain 10 mg of protein each. GFP-Trap (ChromoTek, Planegg-Martinsried, Germany) affinity purifications were performed from crude extracts according to the manufacturer’s instructions, as previously described (59).
Quantitative LC-MS/MS analysis.
Triplicate liquid GMM cultures of WT and ΔpkaC1 strains were grown for 24 h at 37°C. The whole protein from each sample was extracted via bead homogenization with 8 M urea. Either LysC or GluC was added, and growth was allowed to proceed for 18 h. After digestion, the protein was subjected to antibody enrichment, trypsin digestion, and, finally, TiO2 enrichment, as applicable. Phosphopeptide enrichments were performed using TiO2 spin tips (GL Bioscience). For anti-PKA phosphopeptide antibody enrichment, resuspended peptides were then transferred in immunoaffinity purification buffer directly onto prealiquoted phospho-PKA substrate beads (Cell Signaling Technology). Immunoprecipitation was performed for 2 h at 4°C. Combined eluents were further enriched using TiO2 spin tips (GL Bioscience). Quantitative LC-MS/MS was performed using a nanoAcquity ultraperformance liquid chromatography (UPLC) system (Waters Corp.) coupled to a Thermo Fusion Lumos high-resolution accurate-mass tandem mass spectrometer via a nanoelectrospray ionization source. Analytical separation was performed using a 1.7-μm Acquity BEH130 C18 75-μm by 250-mm column (Waters Corp.) with a 90-min linear gradient of 3% to 30% acetonitrile (for open analysis, 5 to 30% acetonitrile). Data collection on the Fusion Lumos mass spectrometer was performed in the data-dependent acquisition (DDA) mode of acquisition. Following 9 total UPLC-MS/MS analyses for each open analysis set, data were imported into Proteome Discoverer 2.2 (Thermo Scientific Inc.). The relative peptide abundance was calculated based on the areas under the curve of the selected ion chromatograms. The MS/MS data were searched against the NCBI Aspergillus fumigatus database. Mascot Distiller and Mascot Server (v2.5; MatrixSciences) were utilized to produce fragment ion spectra and perform the database searches. To identify differences between the WT and ΔpkaC1 groups, a two-tailed t test was performed on log2 protein or phosphopeptide intensities. Fold changes were calculated by determining ratios of the average intensities. Considering something differentially expressed between the groups required a P value of <0.05 and a fold change of ≥2.
Bioinformatic analysis of protein functional cluster enrichment.
Sets of significantly upregulated or downregulated proteins from the open proteomic analysis were each analyzed for the enrichment of particular functional categories using both the FungiFun v2.2.8 (https://elbe.hki-jena.de/fungifun/) and DAVID v6.8 (https://david.ncifcrf.gov/) bioinformatic programs. Both programs compared sets of significantly upregulated or downregulated proteins to the complete set of annotated A. fumigatus (Af293) proteins to identify disproportionate enrichments of proteins associated with particular functions based on multiple categorization schemes.
Construction of gene deletions and mutations in A. fumigatus.
Genotypes of the strains used in this study are detailed in Table 3 (27, 62). Mutant strains were generated as described previously (27). Briefly, GFP labeling of proteins was accomplished by the insertion of the appropriate coding region 5′ to, and in frame with, the egfp coding region in a plasmid vector, followed by transformation of the akuBKU80 strain of A. fumigatus and screening via HYG or pyrG selection as described previously (60). Gene deletions were accomplished via the replacement of the open reading frame of interest with the Aspergillus parasiticus pyrG selectable marker. Deletions were verified via PCR and Southern blotting as described previously (63). Plasmid constructs for site-directed mutagenesis and gene truncations were generated via fusion PCR as described previously (27). All mutated strains were verified via DNA sequencing. The presence of GFP-labeled mutant protein forms was verified for key strains by Western blotting with anti-GFP antibodies as described previously (60). Phosphorylation of purified Escherichia coli-expressed recombinant Atg24 protein was detected via western blot probed with pIMAGO biotin-conjugated anti-phosphopeptide nanopolymers (Tymora Analytical Operations, West Lafayette, Indiana) followed by detection with avidin-conjugated horseradish peroxidase and chemiluminescent substrate according to manufacturer's instructions.
Assessment of radial growth, conidiation, and antifungal compound susceptibility.
For radial growth assays, conidia (104) were point inoculated onto GMM agar medium in petri plates and incubated for 4 or 5 days at 37°C. Growth media used included standard GMM (1% glucose, pH 6.5) as well as modified GMM containing either 50 mM H2O2, 0.5 μg/ml CSP, 4 μg/ml CSP, 3.75 μg/ml HYG, 75 μg/ml HYG, or 150 μg/ml HYG. Conidiation was quantified by collecting conidia from each plate in equal volumes of 0.05% Tween 20, followed by dilution of the collected spore stocks and counting of spores via a hemocytometer. The mean radial growth and conidiation rates for each of the strains were compared statistically by Student’s t test. For CSP, VCZ, and AMB sensitivity determinations, CLSI M38-A2 in vitro antifungal susceptibility standards were used (64).
Modeling and molecular dynamic simulations.
A structural model of monomeric Atg24 was created using iTASSER (65–67). The created structural model was then used to model the phosphorylated (Ser47 and Ser48) protein created through PyMOL (PyMOL Molecular Graphics System version 1.2r3pre; Schrödinger LLC). Molecular dynamics (MD) simulations of structural models were performed with the GROMACS 5.1 software package using the CHARMM force field and the flexible TIP3 water model individually (68). The initial structures were immersed in a periodic water box with a dodecahedron shape (1-nm thickness) and neutralized with counterions. The electrostatic energy was calculated using the particle mesh Ewald method with 0.9-nm cutoff distances for the Coulomb and van der Waals interactions. After energy minimization, the system was equilibrated to 300 K and normal pressure for 100 ps with position restraints for heavy atoms and Linear Constraint Solver (LINCS) constraints for all bonds. The system was coupled to the external bath by Parrinello-Rahman pressure and temperature coupling. The final MD calculations were performed under the same conditions except that the position restraints were removed and the simulation was run for 100 ns. The last frame of the 100-ns simulation was extracted for each model for electrostatic surface analysis. PyMOL Molecular Graphics System version 1.8 (Schrödinger) was used for structural analysis and image creation. The electrostatic surface plots of the protein models were created by using the Adaptive Poisson-Boltzmann Solver in PyMOL (69, 70). The PDB2PQR Web server (71) was used with the CHARMM force field and output naming schemes with a default pH of 7.5.
Murine invasive aspergillosis virulence assays and histopathological analysis.
Mice weighing 13 to 16 g (CD1; Charles River Laboratory, Raleigh, NC, USA) were immunosuppressed with cyclophosphamide (150 mg/kg of body weight for intranasal infection or 175 mg/kg for inhalational infection, intraperitoneally, on days −2 before infection and +3 postinfection) and triamcinolone acetonide (40 mg/kg, subcutaneously, on days −1 and +6). For intranasal infections, 40 μl of suspensions of 1 × 108 conidia/ml of the relevant strains was delivered intranasally following brief isoflurane anesthesia induction. For inhalational infections, mice were placed in an enclosed plexiglass chamber aerosolized with 12 ml of a spore suspension containing 109 conidia/ml for 1 h. Survival was plotted on a Kaplan-Meier curve and analyzed using log rank pairwise comparisons. P values of <0.05 were considered statistically significant. To characterize disease histopathology, additional mice were infected with each of the analyzed strains. Mice were euthanized on day +3 after infection, and lungs were harvested. Lung sections were stained with Gomori’s methenamine silver stain to visualize fungal hyphae and with hematoxylin and eosin stain to examine inflammation and tissue damage, as previously described (72). Animal studies at the Duke University Medical Center were performed in full compliance with all of the guidelines of the Duke University Medical Center Institutional Animal Care and Use Committee (IACUC) and in full compliance with the U.S. Animal Welfare Act (Public Law 98-198). The Duke University Medical Center IACUC approved all of the vertebrate studies under protocol number A-249-16-11. The studies were conducted in the Division of Laboratory Animal Resources (DLAR) facilities, which are accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Fluorescence microscopy.
Conidia (103) of the respective GFP-labeled strains were cultured in 35-mm cover-glass-bottom petri dishes containing 3 ml of liquid GMM and incubated for 16 h at 37°C prior to visualization. For mitochondrial staining, where noted, 3 μl of MitoTracker dye (Invitrogen) was added to the growth medium 15 min prior to visualization. Hyphae were visualized using an Axio Observer 3 microscope (Carl Zeiss, Oberkochen, Germany) equipped with ZEN Lite imaging software.
Data availability
Raw phosphoproteome data are accessible at the Duke Center for Genomic and Computational Biology Express Data Repository: https://discovery.genome.duke.edu/express/resources/5081/.
ACKNOWLEDGMENTS
Funding for this study was provided by National Institutes of Health-National Institute of Allergy and Infectious Diseases grant 5R21AI132911-02.
We thank D. Chris Cole for providing valuable technical laboratory support.
Footnotes
This article is a direct contribution from William J. Steinbach, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Axel Brakhage, Leibniz Institute for Natural Product Research and Infection Biology - Hans Knöll Institute (HKI), and David Askew, University of Cincinnati.
Citation Shwab EK, Juvvadi PR, Waitt G, Shaheen S, Allen J, IV, Soderblom EJ, Bobay BG, Asfaw YG, Moseley MA, Steinbach WJ. 2020. The protein kinase A-dependent phosphoproteome of the human pathogen Aspergillus fumigatus reveals diverse virulence-associated kinase targets. mBio 11:e02880-20. https://doi.org/10.1128/mBio.02880-20.
REFERENCES
- 1.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M. 2005. Global analysis of protein phosphorylation in yeast. Nature 438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 2.Gao X, Jin C, Ren J, Yao X, Xue Y. 2008. Proteome-wide prediction of PKA phosphorylation sites in eukaryotic kingdom. Genomics 92:457–463. doi: 10.1016/j.ygeno.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Giansanti P, Stokes MP, Silva JC, Scholten A, Heck AJ. 2013. Interrogating cAMP-dependent kinase signaling in Jurkat T cells via a protein kinase A targeted immune-precipitation phosphoproteomics approach. Mol Cell Proteomics 12:3350–3359. doi: 10.1074/mcp.O113.028456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao C, Wu M, Bing J, Tao L, Ding X, Liu X, Huang G. 2017. Global regulatory roles of the cAMP/PKA pathway revealed by phenotypic, transcriptomic and phosphoproteomic analyses in a null mutant of the PKA catalytic subunit in Candida albicans. Mol Microbiol 105:46–64. doi: 10.1111/mmi.13681. [DOI] [PubMed] [Google Scholar]
- 5.Franck WL, Gokce E, Randall SM, Oh Y, Eyre A, Muddiman DC, Dean RA. 2015. Phosphoproteome analysis links protein phosphorylation to cellular remodeling and metabolic adaptation during Magnaporthe oryzae appressorium development. J Proteome Res 14:2408–2424. doi: 10.1021/pr501064q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neofytos D, Treadway S, Ostrander D, Alonso CD, Dierberg KL, Nussenblatt V, Durand CM, Thompson CB, Marr KA. 2013. Epidemiology, outcomes, and mortality predictors of invasive mold infections among transplant recipients: a 10-year, single-center experience. Transpl Infect Dis 15:233–242. doi: 10.1111/tid.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pagano L, Caira M, Candoni A, Offidani M, Fianchi L, Martino B, Pastore D, Picardi M, Bonini A, Chierichini A, Fanci R, Caramatti C, Invernizzi R, Mattei D, Mitra ME, Melillo L, Aversa F, Van Lint MT, Falcucci P, Valentini CG, Girmenia C, Nosari A. 2006. The epidemiology of fungal infections in patients with hematologic malignancies: the SEIFEM-2004 study. Haematologica 91:1068–1075. [PubMed] [Google Scholar]
- 8.Pappas PG, Alexander BD, Andes DR, Hadley S, Kauffman CA, Freifeld A, Anaissie EJ, Brumble LM, Herwaldt L, Ito J, Kontoyiannis DP, Lyon GM, Marr KA, Morrison VA, Park BJ, Patterson TF, Perl TM, Oster RA, Schuster MG, Walker R, Walsh TJ, Wannemuehler KA, Chiller TM. 2010. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clin Infect Dis 50:1101–1111. doi: 10.1086/651262. [DOI] [PubMed] [Google Scholar]
- 9.Neofytos D, Horn D, Anaissie E, Steinbach W, Olyaei A, Fishman J, Pfaller M, Chang C, Webster K, Marr K. 2009. Epidemiology and outcome of invasive fungal infection in adult hematopoietic stem cell transplant recipients: analysis of Multicenter Prospective Antifungal Therapy (PATH) Alliance registry. Clin Infect Dis 48:265–273. doi: 10.1086/595846. [DOI] [PubMed] [Google Scholar]
- 10.van de Peppel RJ, Visser LG, Dekkers OM, de Boer MGJ. 2018. The burden of invasive aspergillosis in patients with haematological malignancy: a meta-analysis and systematic review. J Infect 76:550–562. doi: 10.1016/j.jinf.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Zhao W, Panepinto JC, Fortwendel JR, Fox L, Oliver BG, Askew DS, Rhodes JC. 2006. Deletion of the regulatory subunit of protein kinase A in Aspergillus fumigatus alters morphology, sensitivity to oxidative damage, and virulence. Infect Immun 74:4865–4874. doi: 10.1128/IAI.00565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller KK, Richie DL, Feng X, Krishnan K, Stephens TJ, Wikenheiser-Brokamp KA, Askew DS, Rhodes JC. 2011. Divergent protein kinase A isoforms co-ordinately regulate conidial germination, carbohydrate metabolism and virulence in Aspergillus fumigatus. Mol Microbiol 79:1045–1062. doi: 10.1111/j.1365-2958.2010.07509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebmann B, Muller M, Braun A, Brakhage AA. 2004. The cyclic AMP-dependent protein kinase a network regulates development and virulence in Aspergillus fumigatus. Infect Immun 72:5193–5203. doi: 10.1128/IAI.72.9.5193-5203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, Miyasato SR, Binkley J, Orvis J, Shah P, Wymore F, Sherlock G, Wortman JR. 2014. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res 42:D705–D710. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y, Deng YZ, Naqvi NI. 2013. Atg24-assisted mitophagy in the foot cells is necessary for proper asexual differentiation in Magnaporthe oryzae. Autophagy 9:1818–1827. doi: 10.4161/auto.26057. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki SW, Emr SD. 2018. Membrane protein recycling from the vacuole/lysosome membrane. J Cell Biol 217:1623–1632. doi: 10.1083/jcb.201709162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santangelo GM. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 70:253–282. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thevelein JM, Bonini BM, Castermans D, Haesendonckx S, Kriel J, Louwet W, Thayumanavan P, Popova Y, Rubio-Texeira M, Schepers W, Vandormael P, Van Zeebroeck G, Verhaert P, Versele M, Voordeckers K. 2008. Novel mechanisms in nutrient activation of the yeast protein kinase A pathway. Acta Microbiol Immunol Hung 55:75–89. doi: 10.1556/AMicr.55.2008.2.1. [DOI] [PubMed] [Google Scholar]
- 19.Steidl S, Tuncher A, Goda H, Guder C, Papadopoulou N, Kobayashi T, Tsukagoshi N, Kato M, Brakhage AA. 2004. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J Mol Biol 342:515–524. doi: 10.1016/j.jmb.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Steidl S, Papagiannopoulos P, Litzka O, Andrianopoulos A, Davis MA, Brakhage AA, Hynes MJ. 1999. AnCF, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol Cell Biol 19:99–106. doi: 10.1128/mcb.19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JE, Reese JC. 2012. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 47:315–333. doi: 10.3109/10409238.2012.667214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panasenko OO, Collart MA. 2012. Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol Microbiol 83:640–653. doi: 10.1111/j.1365-2958.2011.07957.x. [DOI] [PubMed] [Google Scholar]
- 23.Laribee RN, Shibata Y, Mersman DP, Collins SR, Kemmeren P, Roguev A, Weissman JS, Briggs SD, Krogan NJ, Strahl BD. 2007. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc Natl Acad Sci U S A 104:5836–5841. doi: 10.1073/pnas.0607996104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulder KW, Inagaki A, Cameroni E, Mousson F, Winkler GS, De Virgilio C, Collart MA, Timmers HT. 2007. Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics 176:181–192. doi: 10.1534/genetics.106.060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. 2009. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem 284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulder KW, Brenkman AB, Inagaki A, van den Broek NJ, Timmers HT. 2007. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res 35:2428–2439. doi: 10.1093/nar/gkm175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shwab EK, Juvvadi PR, Waitt G, Soderblom EJ, Moseley MA, Nicely NI, Asfaw YG, Steinbach WJ. 2017. A novel phosphoregulatory switch controls the activity and function of the major catalytic subunit of protein kinase A in Aspergillus fumigatus. mBio 8:e02319-16. doi: 10.1128/mBio.02319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin HY, Wang JJ, Feng MG, Ying SH. 2019. Autophagy-related gene ATG7 participates in the asexual development, stress response and virulence of filamentous insect pathogenic fungus Beauveria bassiana. Curr Genet 65:1015–1024. doi: 10.1007/s00294-019-00955-1. [DOI] [PubMed] [Google Scholar]
- 29.Wiederhold NP. 2007. Attenuation of echinocandin activity at elevated concentrations: a review of the paradoxical effect. Curr Opin Infect Dis 20:574–578. doi: 10.1097/QCO.0b013e3282f1be7f. [DOI] [PubMed] [Google Scholar]
- 30.Dean N. 1995. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc Natl Acad Sci U S A 92:1287–1291. doi: 10.1073/pnas.92.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carman PJ, Dominguez R. 2018. BAR domain proteins—a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys Rev 10:1587–1604. doi: 10.1007/s12551-018-0467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worby CA, Dixon JE. 2002. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol 3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- 33.Pylypenko O, Lundmark R, Rasmuson E, Carlsson SR, Rak A. 2007. The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J 26:4788–4800. doi: 10.1038/sj.emboj.7601889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gsaller F, Hortschansky P, Furukawa T, Carr PD, Rash B, Capilla J, Müller C, Bracher F, Bowyer P, Haas H, Brakhage AA, Bromley MJ. 2016. Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog 12:e1005775. doi: 10.1371/journal.ppat.1005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roetzer A, Gratz N, Kovarik P, Schuller C. 2010. Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol 12:199–216. doi: 10.1111/j.1462-5822.2009.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan W, Kraus PR, Boily MJ, Heitman J. 2005. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell 4:1420–1433. doi: 10.1128/EC.4.8.1420-1433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu G, Hacham M, Waterman SR, Panepinto J, Shin S, Liu X, Gibbons J, Valyi-Nagy T, Obara K, Jaffe HA, Ohsumi Y, Williamson PR. 2008. PI3K signaling of autophagy is required for starvation tolerance and virulence of Cryptococcus neoformans. J Clin Invest 118:1186–1197. doi: 10.1172/JCI32053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu PC, Chao CC, Yang CY, Ye YL, Liu FC, Chuang YJ, Lan CY. 2013. Diverse Hap43-independent functions of the Candida albicans CCAAT-binding complex. Eukaryot Cell 12:804–815. doi: 10.1128/EC.00014-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dagley MJ, Gentle IE, Beilharz TH, Pettolino FA, Djordjevic JT, Lo TL, Uwamahoro N, Rupasinghe T, Tull DL, McConville M, Beaurepaire C, Nantel A, Lithgow T, Mitchell AP, Traven A. 2011. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol Microbiol 79:968–989. doi: 10.1111/j.1365-2958.2010.07503.x. [DOI] [PubMed] [Google Scholar]
- 40.Panepinto JC, Heinz E, Traven A. 2013. The cellular roles of Ccr4-NOT in model and pathogenic fungi—implications for fungal virulence. Front Genet 4:302. doi: 10.3389/fgene.2013.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng S, Clancy CJ, Checkley MA, Zhang Z, Wozniak KL, Seshan KR, Jia HY, Fidel P, Jr, Cole G, Nguyen MH. 2005. The role of Candida albicans NOT5 in virulence depends upon diverse host factors in vivo. Infect Immun 73:7190–7197. doi: 10.1128/IAI.73.11.7190-7197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geddes JM, Caza M, Croll D, Stoynov N, Foster LJ, Kronstad JW. 2016. Analysis of the protein kinase A-regulated proteome of Cryptococcus neoformans identifies a role for the ubiquitin-proteasome pathway in capsule formation. mBio 7:e01862-15. doi: 10.1128/mBio.01862-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hossain S, Veri AO, Cowen LE. 2020. The proteasome governs fungal morphogenesis via functional connections with Hsp90 and cAMP-protein kinase A signaling. mBio 11:e00290-20. doi: 10.1128/mBio.00290-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, Heitman J, Cowen LE. 2009. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol 19:621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A 102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. 2009. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A 106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okamoto K, Kondo-Okamoto N, Ohsumi Y. 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell 17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 48.Clemons KV, Espiritu M, Parmar R, Stevens DA. 2006. Assessment of the paradoxical effect of caspofungin in therapy of candidiasis. Antimicrob Agents Chemother 50:1293–1297. doi: 10.1128/AAC.50.4.1293-1297.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens DA, Espiritu M, Parmar R. 2004. Paradoxical effect of caspofungin: reduced activity against Candida albicans at high drug concentrations. Antimicrob Agents Chemother 48:3407–3411. doi: 10.1128/AAC.48.9.3407-3411.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Astvad KMT, Johansen HK, Roder BL, Rosenvinge FS, Knudsen JD, Lemming L, Schonheyder HC, Hare RK, Kristensen L, Nielsen L, Gertsen JB, Dzajic E, Pedersen M, Ostergard C, Olesen B, Sondergaard TS, Arendrup MC. 2018. Update from a 12-year nationwide fungemia surveillance: increasing intrinsic and acquired resistance causes concern. J Clin Microbiol 56:e01564-17. doi: 10.1128/JCM.01564-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jimenez-Ortigosa C, Moore C, Denning DW, Perlin DS. 2017. Emergence of echinocandin resistance due to a point mutation in the fks1 gene of Aspergillus fumigatus in a patient with chronic pulmonary aspergillosis. Antimicrob Agents Chemother 61:e01277-17. doi: 10.1128/AAC.01277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinkham JL, Guarente L. 1985. Cloning and molecular analysis of the HAP2 locus: a global regulator of respiratory genes in Saccharomyces cerevisiae. Mol Cell Biol 5:3410–3416. doi: 10.1128/mcb.5.12.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hortschansky P, Eisendle M, Al-Abdallah Q, Schmidt AD, Bergmann S, Thon M, Kniemeyer O, Abt B, Seeber B, Werner ER, Kato M, Brakhage AA, Haas H. 2007. Interaction of HapX with the CCAAT-binding complex—a novel mechanism of gene regulation by iron. EMBO J 26:3157–3168. doi: 10.1038/sj.emboj.7601752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thon M, Al Abdallah Q, Hortschansky P, Scharf DH, Eisendle M, Haas H, Brakhage AA. 2010. The CCAAT-binding complex coordinates the oxidative stress response in eukaryotes. Nucleic Acids Res 38:1098–1113. doi: 10.1093/nar/gkp1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Heeswijck R, Hynes MJ. 1991. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res 19:2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy RL, Andrianopoulos A, Davis MA, Hynes MJ. 1997. Identification of amdX, a new Cys-2-His-2 (C2H2) zinc-finger gene involved in the regulation of the amdS gene of Aspergillus nidulans. Mol Microbiol 23:591–602. doi: 10.1046/j.1365-2958.1997.d01-1872.x. [DOI] [PubMed] [Google Scholar]
- 57.Lenssen E, Oberholzer U, Labarre J, De Virgilio C, Collart MA. 2002. Saccharomyces cerevisiae Ccr4-not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol Microbiol 43:1023–1037. doi: 10.1046/j.1365-2958.2002.02799.x. [DOI] [PubMed] [Google Scholar]
- 58.Ribeiro LFC, Chelius C, Boppidi KR, Naik NS, Hossain S, Ramsey JJJ, Kumar J, Ribeiro LF, Ostermeier M, Tran B, Ah Goo Y, de Assis LJ, Ulas M, Bayram O, Goldman GH, Lincoln S, Srivastava R, Harris SD, Marten MR. 2019. Comprehensive analysis of Aspergillus nidulans PKA phosphorylome identifies a novel mode of CreA regulation. mBio 10:e02825-18. doi: 10.1128/mBio.02825-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juvvadi PR, Ma Y, Richards AD, Soderblom EJ, Moseley MA, Lamoth F, Steinbach WJ. 2015. Identification and mutational analyses of phosphorylation sites of the calcineurin-binding protein CbpA and the identification of domains required for calcineurin binding in Aspergillus fumigatus. Front Microbiol 6:175. doi: 10.3389/fmicb.2015.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Juvvadi PR, Belina D, Soderblom EJ, Moseley MA, Steinbach WJ. 2013. Filamentous fungal-specific septin AspE is phosphorylated in vivo and interacts with actin, tubulin and other septins in the human pathogen Aspergillus fumigatus. Biochem Biophys Res Commun 431:547–553. doi: 10.1016/j.bbrc.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 62.da Silva Ferreira ME, Kress MRVZ, Savoldi M, Goldman MHS, Hartl A, Heinekamp T, Brakhage AA, Goldman GH. 2006. The akuBKU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cramer RAJ, Perfect BZ, Pinchai N, Park S, Perlin DS, Asfaw YG, Heitman J, Perfect JR, Steinbach WJ. 2008. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot Cell 7:1085–1097. doi: 10.1128/EC.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi, 2nd ed. CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 65.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. 2015. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 69.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lerner MG, Carlson HA. 2006. APBS plugin for PyMOL. University of Michigan, Ann Arbor, MI. [Google Scholar]
- 71.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res 32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steinbach WJ, Cramer RAJ, Perfect BZ, Asfaw YG, Sauer TC, Najvar LK, Kirkpatrick WR, Patterson TF, Benjamin DKJ, Heitman J, Perfect JR. 2006. Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:1091–1103. doi: 10.1128/EC.00139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global A. fumigatus PKA proteomic analysis. (A) Diagram of the proteomics experiment workflow. Total protein extracts from triplicate samples of A. fumigatus wild-type (WT) and ΔpkaC1 strain mycelia were split, subjected to both LysC and GluC protease digestions, split again into three parts, and treated with either TiO2 for phosphopeptide enrichment or anti-PKA motif antibodies for PKA motif enrichment or unenriched for open protein analysis. LysC digests were further digested with trypsin either immediately following LysC digestion for phosphopeptide enrichment and open samples or after antibody treatment for PKA motif enrichment. (B) Volcano plot of open protein abundances of 3,212 unique proteins identified, plotted as log2 average fold changes in WT samples compared to the ΔpkaC1 average (x axis), against the −log10 P value in a t test comparison of protein abundances between strain types (y axis). (C) Enriched functional categories of proteins identified by open proteomic analysis demonstrating a ≥2-fold-lower abundance in the WT genetic background than in the ΔpkaC1 background and statistical significance of a P value of <0.05 in t test comparisons. Pie charts show enriched functional categories for this set of proteins in comparison to the total set of annotated A. fumigatus proteins based on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) classification systems, as analyzed using FungiFun 2.2.8 bioinformatic software. (D) FungiFun output of enriched functional categories of identified proteins with a ≥2-fold-higher abundance in the WT than in the ΔpkaC1 background at a P value of <0.05. Pie charts show enriched categories based on GO and KEGG classifications. Download FIG S1, JPG file, 0.5 MB (482.5KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteins with decreased abundance in the wild type versus the ΔpkaC1 strain in open proteomic analysis. Download Table S1, PDF file, 0.02 MB (26.3KB, pdf) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proteins with increased abundance in the wild type versus the ΔpkaC1 strain in open proteomic analysis. Download Table S2, PDF file, 0.03 MB (30.8KB, pdf) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of direct PKA target candidate proteins. (A) Strategy for discerning likely direct PKA targets among phosphoproteins identified by LC-MS/MS analysis. The total set of identified phosphoproteins (from both TiO2- and antibody-enriched data sets) was narrowed to those yielding a ≥2-fold-higher abundance in the WT than in the ΔpkaC1 genetic background with a statistical significance level of a P value of <0.05. This set was further narrowed to those proteins phosphorylated at canonical PKA target motifs, resulting in a set of 127 direct PKA target candidate proteins. (B) PKA target candidates were further prioritized by comparison of their PKA-dependent fold changes in phosphorylation abundance as determined by our PKA phosphoproteome analysis (y axis) to the strength of evidence for their interaction with the PKA catalytic subunit as estimated based on the number of coimmunoprecipitated peptide spectra identified for each protein in our PkaC1 interactome study (x axis). Proteins considered to be of particular interest based on a large PKA-dependent increase in phosphopeptide abundance and/or strong evidence of PKA interaction are labeled on the graph. Three proteins selected for in-depth analysis (Atg24, HapB, and Not4) are shown in color. Download FIG S2, JPG file, 0.3 MB (288.3KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Direct PKA target candidate proteins. Download Table S3, PDF file, 0.1 MB (66.5KB, pdf) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Atg24 in vitro PKA phosphorylation assay MS results. Download Table S4, PDF file, 0.02 MB (17.7KB, pdf) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ClustalW alignment of the amino acid sequences of Not4 homologs from human and fungal species and growth phenotypes of Not4 truncation mutants. (A) The amino acid sequences of the proteins homologous to A. fumigatus Not4 as determined by a BLAST search for Homo sapiens as well as 10 other fungal species representing the filamentous ascomycetes (A. fumigatus, A. nidulans, Penicillium brasilianum, M. oryzae, Neurospora crassa, and Sordaria macrospora), the ascomycete yeasts (S. cerevisiae, C. albicans, and Schizosaccharomyces pombe), as well as the basidiomycota (C. neoformans) and zygomycota (Mucor ambiguus) were aligned via the ClustalW algorithm using MegAlign v15.2.0.130 software (DNAStar). (B) Phenotypes of the Not4-WT, Not41–693 amino acid truncation mutant, Not41–233 truncation mutant, and Δnot4 strains on GMM agar (5 days at 37°C). Details show an uneven leading growth edge of the Not41–693 truncation mutant compared to Not4-WT. Download FIG S3, JPG file, 0.5 MB (472.8KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cell wall stress sensitivity of Atg24 and PkaC1 mutant strains. Growth of the Atg24-WT, Atg24-S47D/S48D, ΔpkaC1, and ΔpkaC1; Atg24-S47D/S48D strains was observed on agar plates infused with the indicated concentrations of caspofungin (CSP). Both the ΔpkaC1 and ΔpkaC1; Atg24S47D/S48D strains showed patterns of growth inhibition similar to that of Atg24-WT, with some evidence of paradoxical growth recovery at CSP concentrations of 2 and 4 μg/ml. Download FIG S4, JPG file, 0.3 MB (349.9KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Effects of Not4 on Geneticin sensitivity. Quantitative comparison of the radial growth of Not4-WT, the Not4 Ser311-to-alanine (Not4S311A) and Ser311-to-aspartic acid (Not4S311D) substitution mutant strains, and the Δnot4 strain on GMM agar (4 days at 37°C) infused with the indicated concentrations of Geneticin (G418) was performed. Statistical comparisons of each strain to Not4-WT were made at the same Geneticin concentrations (*, P < 0.05; **, P < 0.01 [by Student’s t test]). Growth of the Δnot4 strain was significantly reduced compared to Not4-WT with or without Geneticin, but the ratio of Not4-WT to Δnot4 growth was increased under Geneticin treatment (2.03 at 0 mg/ml compared to 2.85 at 3.2 mg/ml), suggesting exacerbated growth inhibition. Not4S311D showed significantly increased growth compared to Not4-WT at all concentrations of geneticin examined. Download FIG S5, JPG file, 0.2 MB (216.6KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Visual inspection of the MD simulations of the A. fumigatus Atg24-WT and phosphorylated Ser47 and Ser48 models. The Cα RMSD (black and red lines) and Rg (radius of gyration) (overall compactness of structure) (green and blue lines) show stability after an initial period of equilibration (first 10 ns). Download FIG S6, JPG file, 0.3 MB (270.9KB, jpg) .
Copyright © 2020 Shwab et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Raw phosphoproteome data are accessible at the Duke Center for Genomic and Computational Biology Express Data Repository: https://discovery.genome.duke.edu/express/resources/5081/.