Abstract
In this paper, two novel, powerful, and robust convolutional neural network (CNN) architectures are designed and proposed for two different classification tasks using publicly available data sets. The first architecture is able to decide whether a given chest X-ray image of a patient contains COVID-19 or not with 98.92% average accuracy. The second CNN architecture is able to divide a given chest X-ray image of a patient into three classes (COVID-19 versus normal versus pneumonia) with 98.27% average accuracy. The hyperparameters of both CNN models are automatically determined using Grid Search. Experimental results on large clinical data sets show the effectiveness of the proposed architectures and demonstrate that the proposed algorithms can overcome the disadvantages mentioned above. Moreover, the proposed CNN models are fully automatic in terms of not requiring the extraction of diseased tissue, which is a great improvement of available automatic methods in the literature. To the best of the author’s knowledge, this study is the first study to detect COVID-19 disease from given chest X-ray images, using CNN, whose hyperparameters are automatically determined by the Grid Search. Another important contribution of this study is that it is the first CNN-based COVID-19 chest X-ray image classification study that uses the largest possible clinical data set. A total of 1,524 COVID-19, 1,527 pneumonia, and 1524 normal X-ray images are collected. It is aimed to collect the largest number of COVID-19 X-ray images that exist in the literature until the writing of this research paper.
Keywords: convolutional neural network, COVID-19 detection, deep learning, image classification, medical image processing
INTRODUCTION
Research on the diagnosis and treatment of the new type of coronavirus (COVID-19), which first appeared in Wuhan Province of China in December 2019 and brought life to almost a standstill all over the world, has gained great momentum. The main reason why this dangerous virus brings life to a halt in the world is its very high contagious feature. Deaths occur when the disease turns into pneumonia (10). People can be contagious before they develop symptoms, making it difficult to control the spread of the virus. Development of any vaccine can take at least 12 months according to the research conducted until the writing of this paper (27). Currently, there are no effective antiviral drugs (29). COVID-19 disease caused by coronavirus has been declared a pandemic by the World Health Organization (WHO) as of March 11. According to the Johns Hopkins Coronavirus Research Center, as of July 10, 2020, the total number of confirmed COVID-19 cases worldwide was 12,294,117 and the total number of active cases 4,976,653. As of the same date, the number of deaths due to COVID-19 disease was 555,531. These statistics reveal that this novel coronavirus can be deadly, with a 4.94% case fatality rate. Distribution of COVID-19 deaths reported worldwide, as of July 10, 2020 by the European Centre for Disease Prevention and Control can be seen in Supplemental Fig. S1; all Supplemental material is available at (https://doi.org/10.6084/m9.figshare.12957488.v1).
Looking at the current medical technological advances, COVID-19 disease diagnosis is typically based on swabs from the nose and throat (22). The definitive diagnosis is made after pathological examinations. The major disadvantages of this procedure are that it is time consuming and susceptible to sampling error and therefore inefficient. These tests are known as reverse-transcription polymerase chain reactions (RT-PCR), and it is confirmed that the sensitivity of the tests is not high enough for early detection (19). It is possible to increase the diagnostic capabilities of physicians and reduce the time spent for accurate diagnosis with the aid of computer-assisted automatic detection and diagnosis systems. The purpose of these systems is to help experts make quick and accurate decisions. Automatic detection of COVID-19 disease from medical images is a critical component of the new generation of computer-assisted diagnostic (CAD) technologies and has emerged as an important area in recent years. X-rays are a widely used imaging method for the detection, classification, and analysis of diseases caused by viruses. The motivation of this study is the early diagnosis of COVID-19 disease by using a fully automatic, deep convolutional neural network (CNN), whose hyperparameters are determined using Grid Search. The design of a reliable and robust CAD system is proposed using the highest possible number of COVID-19 X-ray images in the literature.
There are quite a few studies about the detection of COVID-19 disease using CNN because it is a new type of disease. Ozturk et al. (22) used a deep CNN model for binary classification [COVID-19(+) versus COVID-19(−)(−)] and multiclassification (COVID-19(+) vs. normal vs, pneumonia). With their 17 convolutional layered CNN architecture, they achieved 98.08% success for binary classification and 87.02% success for multiple classification. Although the accuracy rate they obtained is not bad, the number of images they used is not high enough, namely 127 X-ray images for COVID-19, 500 X-ray images for normal, and 500 X-ray images for pneumonia. Another study about detection of COVID-19 disease is by Loey et al. (20), who proposed the deep transfer learning-based generative adversarial network (GAN) model. Their objective was to collect all possible COVID-19 X-ray images. They achieved collection of 307 X-ray images for each of four different classes. They obtained an overall accuracy rate of 85.2% for multiclassification [COVID-19(+) vs. normal vs. pneumonia]. Although they tried to collect the highest number of COVID-19 X-ray images, they were able to collect only 307 images. Another study that is worth examining is in Ref. 19. In this study, the researchers proposed a fully automatic CNN framework to detect COVID-19 disease. They obtained a 96% overall accuracy rate for the detection COVID-19 disease using 1,296 COVID-19 CT images from six different hospitals. Togacar et al. (29) presented a COVID-19 disease detection study using the MobileNetV2 deep learning model. They obtained a 99.27% overall accuracy rate for the COVID-19(+), normal, and pneumonia multiclassification. They used 295 COVID-19, 65 normal, and 458 pneumonia X-ray images. They achieved a satisfactory result, but the number of images they used was still not high enough. There are other studies about usage of deep learning techniques for COVID-19 disease detection (2, 4). Researchers who are interested in more studies in the literature can investigate those papers.
The goal of this study is to present two novel and fully automatic deep CNN models whose hyperparameters are automatically determined by Grid Search for COVID-19 detection and virus classification using the largest possible number of X-ray images of COVID-19 that exist in the literature. The rest of this paper is organized as follows. The following section, proposed cnn models, presents the proposed method. All the steps about the proposed architectures for each task can be found in this section. In experimental results and discussion, experimental results are discussed, and these results are compared with the state-of-art methods The final section offers conclusions of the paper.
PROPOSED CNN MODELS
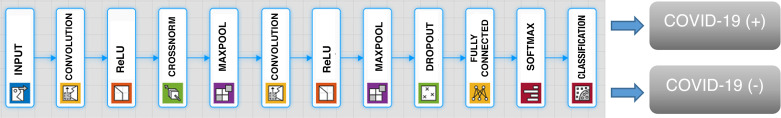
In this paper, two novel, powerful, and robust CNN architectures are designed and proposed for two different classification tasks using publicly available data sets. The hyperparameters of these CNN architectures are automatically tuned by Grid Search. The first of these architectures is used to decide whether a given chest X-ray image of a patient has COVID-19 or not. We can call this Task 1. The proposed CNN architecture for Task 1 consists of 12 weighted layers, in which there are two convolutional layers and one fully connected layer, as shown in Fig. 1. The convolutional layer is the most important CNN layer and is also known as the transformation layer. This transformation is performed by moving a particular filter over the whole input image (30, 31). The convolutional layers are followed by the rectified linear units layer (ReLU layer) and maxpooling layers. The ReLU layer follows the convolution layers and is used as a rectifier unit. The RELU layer’s effect on the input data is that it takes negative values to zero. In all architectures, ReLU is used as an activation function, since it is already a standard activation function in image classification tasks. A pooling layer is generally placed after the ReLU layer, and its main task is to decrease input size (width × height) for the subsequent convolution layer. In the CNN architecture, the consecutive convolution, ReLU, and pooling layers are followed by a fully connected layer (30, 31). This layer is connected to all neurons of the previous layer. The fully connected layer, resulting in a two-dimensional feature vector, is fed as an input to Softmax classifier, which makes the final prediction whether there is the novel coronavirus or not. There are two neurons in the output layer, as this model tries to classifies an image into two classes: COVID-19(+) or COVID-19(−). The first convolutional layer has 96 kernels of size 7 × 7 with stride 4 and padded with [0 0 0 0], whereas the second convolutional layer has two groups of 128 kernels of size 5 × 5 with stride 1 and padded with [2 2 2 2]. The size of the input images that are forwarded into the whole architectures is 227 × 227 × 3. More information about the layer information, their activations, and the number of total learnable parameters is shown in Table 1.
Fig. 1.
Proposed convolutional neural network architecture for Task 1. ReLU, rectified linear units layer.
Table 1.
CNN architecture details for Task 1
| Name | Type | Activations | Learnable Parameters | Total Learnables | |
|---|---|---|---|---|---|
| 1 | 227×227×3 images | Input image | 227×227×3 | - | 0 |
| 2 | 96 7×7×3 convolutions with stride [4 4] and padding [0 0 0 0] | Convolution | 56×56×96 | Weights: 7×7×3×96 Bias: 1×1×96 |
14,208 |
| 3 | ReLU-1 | ReLU | 56×56×96 | - | 0 |
| 4 | Cross channel normalization with 5 channels per element | Cross-channel normalization | 56×56×96 | - | 0 |
| 5 | 3×3 maxpooling with stride [2 2] and padding [0 0 0 0] | Maxpooling | 27×27×96 | - | 0 |
| 6 | 2 groups of 128 5×5×48 convolutions with stride [1 1] and padding [2 2 2 2] | Grouped convolution | 27×27×256 | Weights: 5×5×48×128×2 Bias: 1×1×12×2 |
307,456 |
| 7 | ReLU-2 | ReLU | 27×27×256 | - | 0 |
| 8 | 3×3 maxpooling with stride [2 2] and padding [0 0 0 0] | Maxpooling | 13×13×256 | - | 0 |
| 9 | 50% dropout | Dropout | 13×13×256 | - | 0 |
| 10 | 2 fully connected layer | Fully connected | 1×1×2 | Weights: 2×43,264 Bias: 2×1 |
86,530 |
| 11 | Softmax | Softmax | 1×1×2 | - | 0 |
| 12 | Crossentropyex with classes COVID-19(+) and COVID-19(−) |
classification output | - | - | 0 |
The number of the layers for both tasks is determined by intuition, depending on the experiences and current trends obtained from literature survey. While deciding on the number of layers, I have tried to minimize the number of layers so that the model is not as complex as possible, and the model has a compact and simple structure while the exhaustive capability and the best accuracy rate are still preserved. In deep learning studies while designing CNN architecture, it is still not easy to choose a theoretically optimal CNN architecture for a targeted image classification task. Therefore, implementing and reviewing several CNN architectures with different number of layers should be a logical procedure for that targeted classification task. In this paper, we adopted this procedure while deciding the number of layers and layer architectures. Different network architectures are compared to reach the desired target. For example: Sultan et al. (28) presented CNN architecture to classify brain tumor types and tumor grades. The CNN architecture they proposed consists of 16 weighted layers, in which there are three convolutional layers and one fully connected layer. They achieved a 96.13% accuracy rate for tumor type classification and 98.7% for tumor grading. Anaraki et al. (16) proposed two CNN architectures optimized for using Genetic Algorithm for multiclassification of brain tumors. The first architecture consists of 15 weighted layers, in which there are five convolutional layers and one fully connected layer. They achieved 90.9% accuracy for tumor type classification. The second architecture consists of 17 weighted layers, in which there are five convolutional layers and one fully connected layer. They achieved 94.2% for tumor grading. Abiwinanda et al. (1) suggested an optimal CNN architecture consisting of 12 weighted layers, in which there are two layers of convolution, ReLU, and maxpooling, followed by two fully connected ones. They obtained 98.51% for brain tumor detection. Ertosun et al. (12) proposed a CNN model for grading of gliomas. The architecture they used for low-grade glioma (LGG) versus high-grade glioma (HGG) consisted of 10 weighted layers, in which there are four convolutional layers and one fully connected layer, and they obtained 96% for LGG versus HGG classification. Readers interested in more similar studies that underpin the CNN architectures and the number of layers in this study can read the following articles (9, 24, 37).
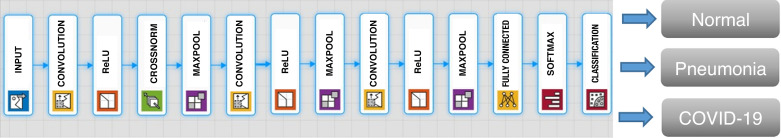
The second CNN architecture classifies the X-ray image into three classes, i.e., normal, pneumonia, and COVID-19. We can call this Task 2. The proposed CNN architecture for Task 2 consists of 14 weighted layers, in which there are three convolutional layers and one fully connected layer, as shown in Fig. 2. The convolutional layers are followed by ReLU and maxpooling layers. The fully connected layer, resulting in a three-dimensional feature vector is fed as an input to Softmax classifier, which makes the final prediction of a chest X-ray image. There are three neurons in the output layer, as this model tries to classify an image into three classes: normal, pneumonia, and COVID-19. The first convolutional layer has 96 kernels of size 11 × 11 with stride 4 and padded with [0 0 0 0], whereas the second and third convolutional layers have two groups of 128 kernels of size 5 × 5 with stride 1 and padded with [2 2 2 2]. More information about the layers, their activations, and the number of total learnable parameters is shown in Table 2.
Fig. 2.
Proposed convolutional neural network architecture for Task 2.
Table 2.
CNN architecture details for Task 2
| Name | Type | Activations | Learnable Parameters | Total Learnables | |
|---|---|---|---|---|---|
| 1 | 227×227×3 input images | Input image | 227×227×3 | - | 0 |
| 2 | 96 11×11×3 convolutions with stride [4 4] and padding [0 0 0 0] | Convolution | 55×55×96 | Weights: 11×11×3×96 Bias: 1×1×96 |
34,944 |
| 3 | ReLU-1 | ReLU | 55×55×96 | - | 0 |
| 4 | Cross channel normalization with 5 channels per element | Cross-channel normalization | 55×55×96 | - | 0 |
| 5 | 3×3 maxpooling with stride [2 2] and padding [0 0 0 0] | Maxpooling | 27×27×96 | - | 0 |
| 6 | 2 groups of 128 5×5×48 convolutions with stride [1 1] and padding [2 2 2 2] | Grouped convolution | 27×27×256 | Weights: 5×5×48×128×2 Bias: 1×1×128×2 |
307,456 |
| 7 | ReLU-2 | ReLU | 27×27×256 | - | 0 |
| 8 | 3×3 maxpooling with stride [2 2] and padding [0 0 0 0] | Maxpooling | 13×13×256 | - | 0 |
| 9 | 2 groups of 128 5×5×48 convolutions with stride [1 1] and padding [2 2 2 2] | Grouped convolution | 27×27×256 | Weights: 5×5×48×128×128×2 Bias: 1×1×128×2 |
307,456 |
| 10 | ReLU-3 | ReLU | 13×13×256 | - | 0 |
| 11 | 3×3 maxpooling with stride [2 2] and padding [0 0 0 0] | Maxpooling | 6×6×256 | - | 0 |
| 12 | 3 fully connected layer | Fully connected | 1×1×3 | Weights: 3×9216 Bias: 3×1 |
27,651 |
| 13 | Softmax | Softmax | 1×1×2 | - | 0 |
| 14 | Crossentropyex with classes Normal, ‘Pneumonia, and COVID-19 | classification output | - | - | 0 |
Hyperparameter tuning using Grid Search.
Along with deep learning, studies in machine learning evolved from feature engineering to architectural engineering. In previous studies, researchers mostly created the best feature sets to represent a problem in order to solve the problem; that is, they were working on extracting attributes and selecting those with the highest representational capability among them (13, 36). The development of deep learning approaches included how to design a multilayered artificial neural network: how many layers it consists of, how many neurons it contains, which optimization algorithm or activation function will be more important in problem solving. Solving problems with deep learning has become equivalent to designing the multilayered network structure in the best and optimum way. In this architectural engineering, the most frequently used tools after the researcher's intuition were hyperparameters. While designing machine learning models from data, the algorithms or techniques used in the model bring along some parameters that the designer should decide on. The parameters that vary according to the problem and data set and which are left to the person who designed the model are called hyperparameters. Choosing the most appropriate hyperparameter group is one of the important problems to be tackled. The choice of hyperparameters generally varies depending on the intuition of the designer, the experiences obtained from previous problems, the reflection of the applications in different fields to our own problem, current trends, and the design dependency within the model (25). The selection of such hyperparameters is a tedious and time-consuming process. However, recently, different techniques have been introduced to choose the most suitable hyperparameter group for the solution of a problem (11, 14).
While making the model design, the first choices we make for hyperparameters generally do not lead us to the correct results. Hyperparameters are changed in an iterative way; the success of the model is observed, and the most suitable hyperparameter group is tried to be selected for the model. In addition, there are methods that automate this selection job. Some of the hyperparameters are in a position to take an infinite number of values. However, we can determine ranges for the values that hyperparameters can take by using the preliminary information we have about the problem. Value lists are created for hyperparameters by selecting certain key points from these ranges.
In the hyperparameter selection process with Grid Search, the network is trained for the combinations of all values in the specified range, and the best combination is selected as the hyperparameter group according to the observed conditions (25, 36). To decide the configuration values, trends in Grid Search should be observed. It should not focus only on the best-performing result; all results in the Grid Search should be reviewed. In other words, fixed parameters or intervals should be observed by observing the relationships and trends among the parameters. Since training deep networks is a lengthy process, a subset of the data set can be worked out for hyperparameter selection, thus saving time. The purpose of this process is not to find the most suitable values for hyperparameters but to create a general opinion. Thus, the range of hyperparameters is determined.
Grid Search is used to find the optimal hyperparameters of a model that result in the most “accurate” predictions. The optimal hyperparameter values are set and tuned before the learning process is started, since hyperparameters have big effects on the result of the learning process (5, 6). In all tasks of this study, a stochastic gradient descent momentum (SGDM) optimizer is used to update the network parameters (weights and biases) to minimize the loss function by taking small steps at each iteration in the direction of the negative gradient of the loss. CNN and Grid Search were implemented on Matlab’s 2019a software environment with the help of Machine Learning Toolbox. A grid search function for deep learning in Matlab has been created for four hyperparameters (learning, rate, minibatch size, momentum, and regularization). The created function has been designed considering “fitness function value,” which has the following formula in Eq. 1:
| (1) |
The hyperparameters that give the minimum fval value are accepted as the optimum hyperparameters and are given as output of the parameter optimization process. The algorithm of Grid Search optimizer used in the hyperparameter optimization of the CNN models in this study can be summarized as follows.
In this study, a limited range of hyperparameters has been chosen to optimize, because when choosing a grid search method for optimizing hyperparameters, the number of hyperparameters to be optimized should be taken into account, as well as the value ranges for these parameters. However, since the optimization of these hyperparameters requires high computation time, it is not practically meaningful to evaluate all possible probabilities specified for wide parameter value ranges in the experiments to be carried out and to use grid search method. That is why a limited range of each hyperparameter is adopted in this paper. If the selected hyperparameter subset and value ranges form a small search space, the grid search method can be used to obtain successful results. However, when the space begins to expand, random search, metaheuristic algorithms or reinforcement learning algorithms may be preferred. Therefore, using metaheuristic methods (such as genetic algorithms, particle swarm optimization, and differential evolution) for hyperparameter optimization will be more efficient in practical terms. Hyperparameters that needed to be optimized for CNN models in this study are learning rate, minibatch size, momentum, and (ridge regression) regularization, because these are among the most important fine-adjustment hyperparameters for the accuracy and success of the classification task. The optimal hyperparameters that were obtained for Task 1 and Task 2 using the Grid Search optimizer algorithm above are shown in Table 3 and Table 4, respectively.
Table 3.
Optimal hyperparameters obtained by Grid Search for Task 1
| Parameters | Default | Optimum |
|---|---|---|
| Learning rate | 0.01 | 0.0001 |
| Minibatch size | 128 | 32 |
| Momentum | 0.9 | 0.9 |
| Regularization | 0.0001 | 0.0001 |
Table 4.
Optimal hyperparameters obtained by Grid Search for Task 2
| Parameters | Default | Optimum |
|---|---|---|
| Learning rate | 0.01 | 0.0001 |
| Minibatch size | 128 | 64 |
| Momentum | 0.9 | 0.9 |
| Regularization | 0.0001 | 0.001 |
Performance evaluation metrics.
After the classification algorithm is performed, the performance of the classification must be evaluated. This is done in a variety of ways in previous studies in the literature (16). To check the performance of the classification algorithm, the confusion matrix is used in this paper. Confusion matrix provides helpful information regarding the actual image labels and predicted image labels proposed by the classification method. Different aspects of the classification performance can be assessed using this valuable information. Confusion matrix has diagonal values that show true positives (TP). TP is the number of samples classified as true when it is actually true. True false (TF) is the number of samples classified as false when it is actually false. False positives (FP) is the number of samples classified as positive when it is actually false. False negatives (FN) is the number of samples classified as negative when it is actually true. The most used performance evaluation metrics are accuracy, specifity, sensitivity, and area of receiver operation characteristic curve (curve (area under the curve, AUC). Accuracy is by far the most preferred one (16). In this study, training, validation, and test data sets are randomly assigned for multiple independent iterations to comprehensively test the CNN model. The performance measures, such as accuracy, AUC, sensitivity, specificity, and precision are evaluated on test data sets in this paper. These metrics can be computed using Eqs. 2–5, respectively.
| (2) |
| (3) |
| (4) |
| (5) |
EXPERIMENTAL RESULTS AND DISCUSSION
Data set.
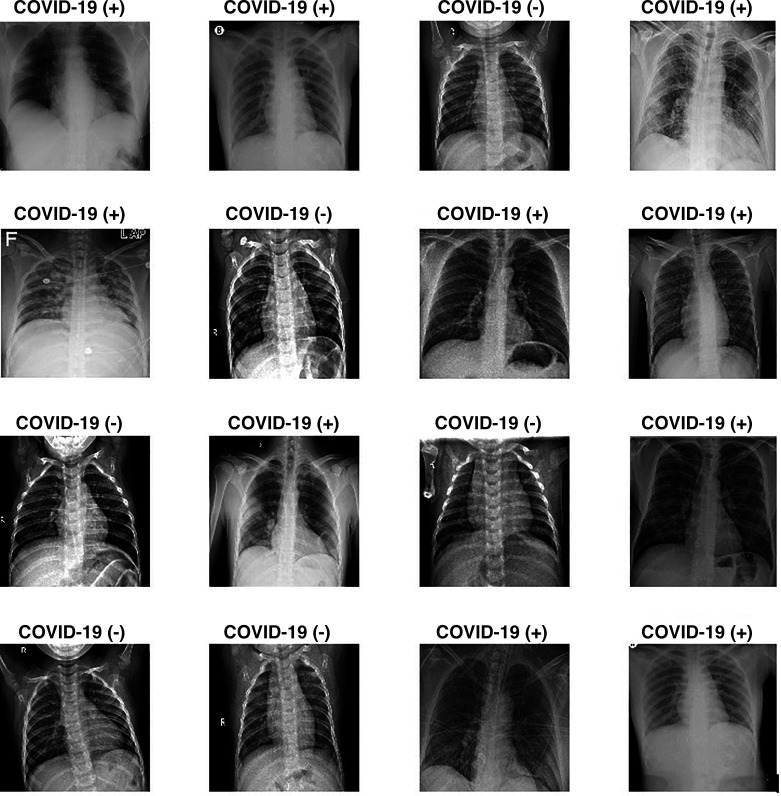
Since the COVID-19 virus is a new virus and appeared very recently, finding a data set is a huge problem. One of the main contributions of this study to the literature is that a wide variety of data sets have been used. Most data sets that are available in the literature have been found meticulously and used in this study. The first data set is called COVID-19 Image Data Collection by Joseph Paul Cohen, Paul Morrison, and Lan Dao and contains 542 frontal chest X-ray images from 262 people from 26 countries (8). This is a public open data set of chest X-ray images of patients who are positive or suspected of COVID-19 or other viral and bacterial pneumonias (MERS, SARS, and ARDS). The second data set that is used in this study has been created by Paul Money (21) and consists of 5,863 X-ray images and two categories: pneumonia and normal. The third data set used in this study is known as ChestX-ray8 by Wang et al. (35) and consists of 108,948 frontal view chest X-ray images of 32,717 patients. This data set is used to increase the number of pneumonia images. The fourth data set used in this study is the COVID-19 Radiography Database, created by a research team from Qatar University (23). This is a database of chest X-ray images for COVID-19-positive cases along with normal and viral pneumonia images; 219 COVID-19-positive images, 1,341 normal images, and 1,345 viral pneumonia images are currently available. The last data set is by Kermany et al. (18) and was added to the data set to increase the number of pneumonia and COVID-19 chest X-ray images. A total of 1,524 COVID-19 images, 1,527 pneumonia images, and 1,524 normal images were collected for this study. All the data sets used in this study are publicly available, and corresponding websites are given in this paper. Figure 3 shows some of the chest X-ray images with and without COVID-19 from the datastore prepared.
Fig. 3.
Some of the chest X-ray images with and without COVID-19 from data store.
Experiment platform and time consumption.
The hardware and software environments used in this study iares as follows:
-
•
Software environment: Windows 10 (64-bit) operating system, Matlab R2019a.
-
•
Hardware environment: NVIDIA GeForce GTX-850M GM107 GPU, Intel Core i7 5400 GPU 2.60 Ghz, 16.0 GB RAM.
-
•
Elapsed time for training the deep learning model in Task 1 (1,828 images) was 25 min and in Task 2 (2,745 images) was 49 min.
Results and discussion.
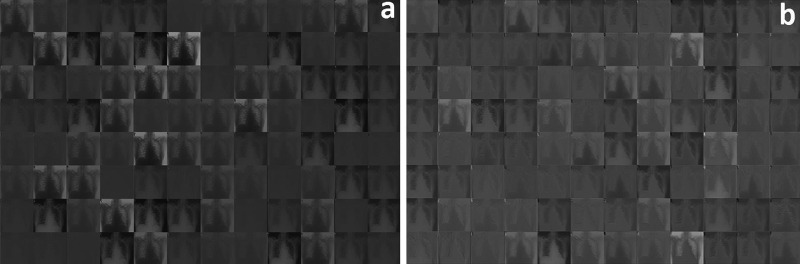
Each CNN model is trained separately by splitting the data into training, validation, and testing sets. The training set is used to train the network, and then the testing set is used for testing the model and parameter optimization processes. Training, validation, and testing data sets are randomly separated. A total of 3,048 images, with a training subset of 1,828, validation subset of 610, and testing subset of 610 images (60, 20, and 20%) is used for Task 1; 305 images are randomly excluded from the data set of each class, and they are used for test purposes to prevent a biased data set assignment effect on the CNN and to comprehensively test the CNN model. Figure 4 shows how to feed an image with COVID-19 to a CNN and demonstrates the activations of various layers of the network. After the CNN model is trained, displaying the activations of network layers gives a lot of clues about the features that the deep neural network learns. To discover the features and parameters that the network learns, these activations can be investigated by comparing areas of activation with the original image. CNN uses the first convolutional layer to learn how to detect features like color and edges. More complicated features are detected by deeper convolutional layers, and subsequent convolutional layers of the CNN build up their features by combining features learned by the earlier convolutional layers. For instance, activations of the first convolutional layer in Fig. 4A shows that simple features like color and edges are learned, since this is an earlier layer, whereas channels in the second convolutional layer (Fig. 4B), which is a deeper layer, learns complex features like patchy or ground-glass opacities due to COVID-19. Hence, researchers can find out what the network has learned by identifying features in this way. The proposed CNN architecture for Task 1 can be seen in Fig. 1. This architecture has two convolutional layers that perform convolutions with learnable layers. CNN learns important features, generally with one feature per channel. Every layer of CNN includes many two-dimensional arrays named “channels.” The first convolutional layer has 96 channels, as shown in Fig. 4A, while the second convolutional layer has 256 channels; 96 of these channels are shown in Fig. 4B.
Fig. 4.
Activations of first (A) and second (B) convolutional layer for Task 1.
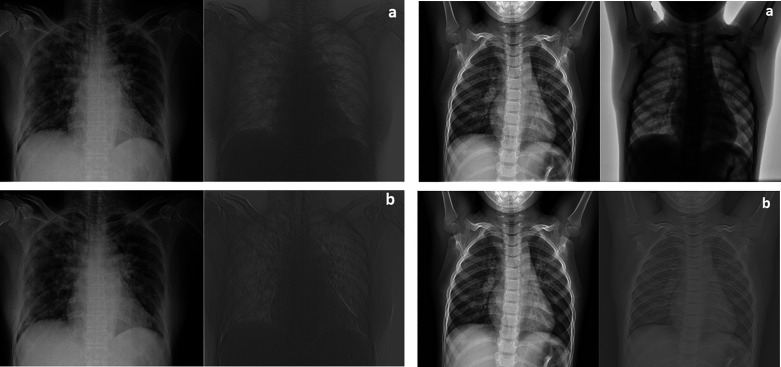
Every single image in the grid of activations (Fig. 4A) corresponds to the output of each channel in the first convolutional layer. White pixels display strong positive activations, while black pixels display strong negative activations. Gray pixels, on the other hand, represent a channel that is not activated as strongly on the input image. The position of a pixel in the activation of a channel corresponds to the same position in the original image. Figure 5 (left) is an image with COVID-19, whereas Fig. 5 (right) is an image with no COVID-19 and is added to compare with the COVID-19 image. Figure 5A (left) shows activation in a specific channel of first convolutional layer for Task 1. White pixels in the channel of Fig. 5B (left) show that this channel is strongly activated at COVID-19. It is seen from the activations that the first convolutional layer is able to activate on COVID-19. Even if the network has never been told to learn about COVID-19, it has learned that COVID-19 has useful features to distinguish between classes of images. Here, a very superior feature of the CNN emerges. In this study, learning to identify COVID-19 helps to distinguish between a COVID-19(+) and a COVID-19(−) image.
Fig. 5.
Investigation of activations in specific channels of first convolutional layer (A) and the strongest activations channels of first convolutional layer (B) for Task 1 (left: COVID-19(+) image; right: COVID-19(−) image).
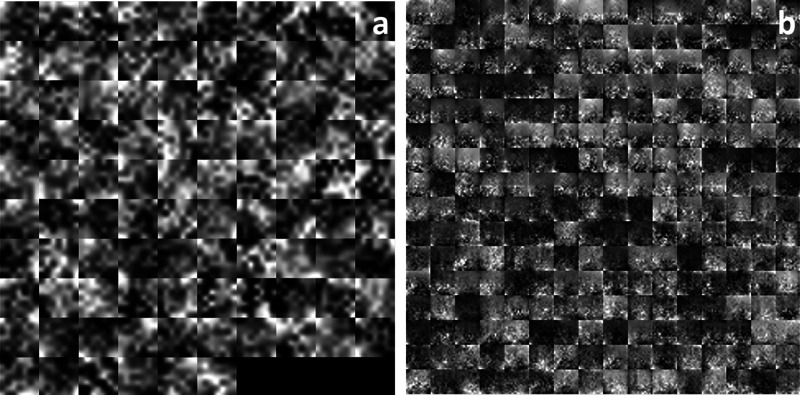
CNN in Task 1 classifies the images into COVID-19(+)and COVID-19(−) images using features that are learned by the network itself during the training process. What the CNN learns during training is sometimes unclear; however, the high-level combinations of the features can be visualized: 96 features learned by CNN in the first convolutional layer are visualized in Fig. 6A, and 256 features learned in the second convolutional layer (Fig. 6AB) are visualized in Fig. 6. CNN architecture for Task 1 has two 2-D convolutional layers. Low-level features are learned in the earlier convolutional layer (Fig. 6A), since they have small receptive field size, whereas deeper layers (Fig. 6B) of the network have larger receptive field size and learn more complicated features. By investigating the channels in Fig. 6A, it can be seen that the filters at this first convolutional layer activate on edges and color, which allows the network to construct more complex features in the second convolutional layer (Fig. 6B).
Fig. 6.
Features of first (A) and second (B) convolutional layer for Task 1.
CNN architecture for Task 1 has one fully connected layer. The fully connected layers are toward the end of the network and learn high-level combinations of the features learned by the earlier layers. The output image classes are the images generated from the final fully connected layer. Features of fully connected layer for Task 1 and detailed images of fully connected layers that strongly activate the classes for Task 1 are shown in Supplemental Fig. S2 (https://doi.org/10.6084/m9.figshare.12957482.v1).
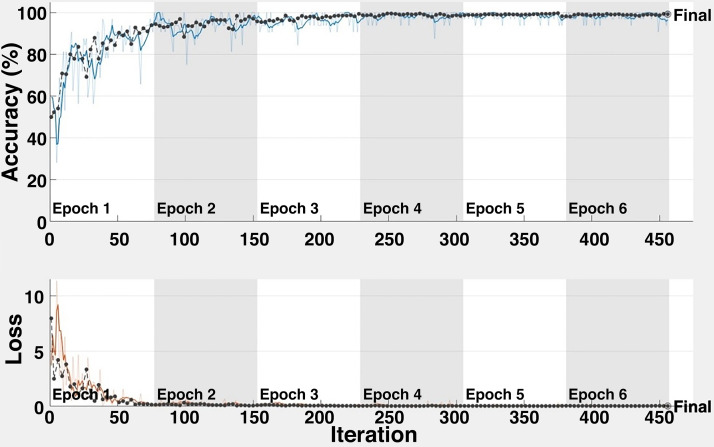
Average accuracy and loss plots for the task of COVID-19(+) image vs COVID-19(−) image (Task 1) are shown in Fig. 7. The proposed CNN model achieves an overall average classification accuracy of 98.92% after 456 iterations for Task 1. This result indicates the ability of the architecture for COVID-19 disease detection. As can clearly be seen from Fig. 7, almost 100% accuracy is obtained after 210 iterations.
Fig. 7.
Accuracy and loss for Task 1.
After the classification process, the performance of the models should be assessed in various ways. In this study, confusion matrix is used to evaluate the comparison performance. Confusion matrix provides precious information about the predicted and actual classes obtained by the proposed architectures. The performance of the study is assessed from different aspects. Performance of the architecture is evaluated using accuracy, specificity, sensitivity, and precision metrics using Eqs. 2–5. Five independent iterations are performed. Classification performance for the task is evaluated for each independent iteration, and the average classification performance of the model is calculated. Performance metrics are calculated using the results from the confusion matrix, and the corresponding results are shown in Table 5. Average accuracy of 98.92% is obtained to classify COVID-19(+) and COVID-19(−) in Task 1. The ROC curve is used as another method to quantify the performance of the architectures. The average value of the AUC of the ROC curve is found to be 0.9957 for Task 1. Confusion matrix results and ROC curves of each independent iteration for Task 1 are shown in Supplemental Fig. S3 (https://doi.org/10.6084/m9.figshare.13100048.v1). It is important to note that the performance measures, such as accuracy, AUC, sensitivity, specificity, and precision, are evaluated on test data sets.
Table 5.
Accuracy metrics in terms of accuracy, specificity, sensitivity, and precision
| Performance Metrics (%) | Iteration 1 | Iteration 2 | Iteration 3 | Iteration 4 | Iteration 5 | Average |
|---|---|---|---|---|---|---|
| Task 1 | ||||||
| Sensitivity | 99.15 | 99.88 | 98.86 | 98.14 | 97.71 | 98.72 |
| Specificity | 99.17 | 99.84 | 98.86 | 98.03 | 97.71 | 98.72 |
| Precision | 99.15 | 99.84 | 98.85 | 98.03 | 97.72 | 98.72 |
| Accuracy | 99.18 | 99.84 | 99.85 | 98.03 | 97.71 | 98.92 |
| AUC | 0.9996 | 0.9997 | 0.9997 | 98.67 | 99.28 | 0.9957 |
| Task 2 | ||||||
| Sensitivity | 96.77 | 98.58 | 98.03 | 98.80 | 98.25 | 98.09 |
| Specificity | 98.83 | 99.29 | 99.02 | 99.40 | 99.12 | 99.13 |
| Precision | 98.67 | 98.60 | 98.04 | 98.84 | 98.28 | 98.49 |
| Accuracy | 97.70 | 98.58 | 98.03 | 98.80 | 98.25 | 98.27 |
| AUC | 0.9992 | 0.9977 | 0.9992 | 0.9967 | 0.9969 | 0.9979 |
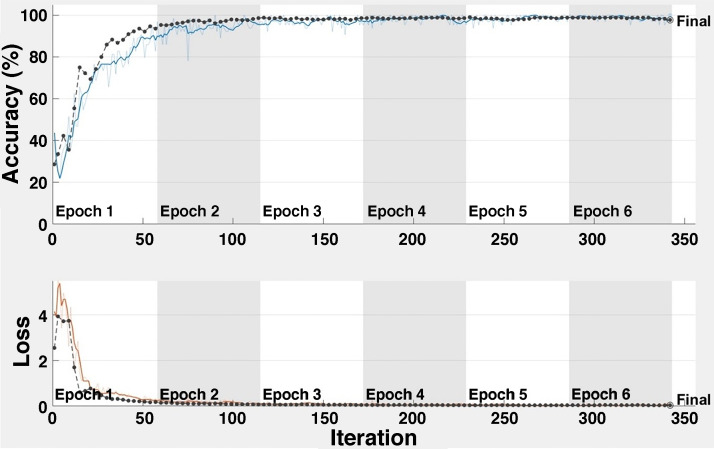
Four sample validation images with predicted labels and the predicted probabilities of the images having those labels for Task 1 are shown in Supplemental Fig. S4 (https://doi.org/10.6084/m9.figshare.12957485.v1). A total of 4,575 images, with a training subset of 2,745, validation subset of 915, and testing subset of 915 images (60, 20, and 20%, respectively) is used for Task 2; 305 images are randomly excluded from the data set of each class and used for test purposes to prevent a biased data set assignment effect on the CNN and to comprehensively test the CNN model. Five independent iterations are performed. Classification performance for the task is evaluated for each independent iteration, and the average classification performance of the model is calculated. Average accuracy and loss plots for Task 2 are shown in Fig. 8. The proposed CNN model achieves an overall average classification accuracy of 98.27% after 342 iterations for Task 2. This result indicates the ability of the architecture for COVID-19 disease classification. As can clearly be seen from Fig. 8, almost 100% accuracy is obtained after 100 iterations.
Fig. 8.
Accuracy and loss for Task 2.
Performance of the architecture is evaluated using accuracy, specificity, sensitivity, and precision metrics using Eqs. 2–5. These metrics are calculated using the results from the confusion matrix and are shown in Table 5. Confusion matrix results and ROC curves of each independent iteration for Task 2 are shown in Supplemental Fig. S5 (https://doi.org/10.6084/m9.figshare.13100084.v1). The average value of the AUC of the ROC curve is found to be 0.9979 for Task 2. It is important to note that the performance measures, such as accuracy, AUC, sensitivity, specificity, and precision, are evaluated on test data sets.
Comparison with state-of-the-art methods.
This section is devoted to the comparison of the proposed method with state-of-the-art methods. Table 6 shows a detailed comparison of the results found using the proposed method with the results of the state-of-the-art methods. The number of COVID-19 disease detection studies using deep learning methods is not high due to the lack of COVID-19 X-ray images in the literature. Togacar et al. (29) used existing deep learning models such as MobileNetV2 and SqueezeNet to detect COVID-19 disease. They created a combined data set comprised of three classes: normal, pneumonia, and COVID-19. They used 295 COVID-19 images, 65 normal X-ray images, and 458 pneumonia images in total to train their deep learning model, and that number of data set is obviously quite small to train a deep learning model successfully. With this data set, they obtained a classification accuracy of 99.27% for Task 2. There was no study for Task 1. Lin et al. (19) proposed a three-dimensional deep learning model for COVID-19 detection. They called their model COVID-19 Detection Model Neural Network (COVNet), which actually consists of ResNet50 as the backbone. They collected 1,296 COVID-19 CT images, 1,325 normal CT images, and 1,735 pneumonia CT images from six different hospitals. They obtained a classification accuracy of 96% for Task 1 and there was no study for Task 2. Ozturk et al. (22) used the DarkCovidnet deep learning model to classify X-ray images as COVID-19 and No Findings (Task 1) and as COVID-19, pneumonia, and normal (Task 2). They obtained an overall accuracy of 98.08% for Task 1 and 87.02% for Task 2. The number of images they used was 127 COVID-19 X-ray images, 500 normal X-ray images, and 500 pneumonia X-ray images. Loey et al. (20) aimed to create the largest possible number of X-ray images for COVID-19 that exists in the literature. They collected 307 images for four different types of classes. The classes are COVID-19, normal, pneumonia bacterial, and pneumonia viral. Loey et al. (20) used pretrained models (Alexnet, Googlenet, and Resnet18) with deep transfer learning to detect COVID-19 disease and to multiclassify between COVID-19, normal, pneumonia bacterial, and pneumonia viral. They obtained a classification accuracy of 85.2% for Task 2 and 99.9% for Task 1. Although the classification rate they obtained for Task 1 seems to be high, the number of images they used is still not enough for successful deep learning training. Apostolopoulos et al. (4) adopted a procedure based on transfer learning for detection COVID-19 detection from X-ray images. They collected 224 COVID-19 X-ray images, 504 normal X-rays, and 714 pneumonia X-rays. They obtained a classification accuracy of 96.78% for Task 2 and there was not a study for Task 1.
Table 6.
Detailed comparison of the proposed method with the-state-of-art methods
| Model | Overall Accuracy for Task 1 | Overall Accuracy for Task 2 | Classification Type | Number of Cases | Data Sets Used | Type of Images |
|---|---|---|---|---|---|---|
| Togacar et al. (29) | — | 99.27% | • Normal, Pneumonia and COVID-19 | 295 COVID-19(+) 458 Pneumonia 65 Normal |
COVID-19 image data collection (8) | Chest X-ray images |
| Lin et al. (19) | 96% | — | • Normal, Pneumonia and COVID-19 | 1296 COVID-19(+) 1735 Pneumonia 1325 Normal |
6 different hospitals (19) | Chest CT images |
| Ozturk et al. (22) | 98.08% | 87.02% | • Healthy, Not healthy • Normal, Pneumonia and COVID-19 |
127 COVID-19(+) 500 pneumonia 500 normal |
COVID-19 image data collection (8) and ChestX-ray8 (35) |
Chest X-ray images |
| Loey et al. (20) | 99.9% | 85.2% | • Healthy, Not healthy • Normal, Pneumonia and COVID-19 |
307 COVID-19(+) 307 Pneumonia 307 Normal |
COVID-19 image data collection (8) and Data set by Kermany et al. (18) | Chest X-ray images |
| Apostolopoulos et al. (4) | — | 96.78% | • Normal, Pneumonia and COVID-19 | 224 COVID-19(+) 714 Pneumonia 504 Normal |
COVID-19 image data collection (8) and COVID-19 X-rays (8a) | Chest X-ray images |
| Hemdan et al. (15) | — | 90% | • Normal, Pneumonia and COVID-19 | 25 COVID-19(+) 25 Normal |
COVID-19 image data collection (8) | Chest X-ray images |
| Wang and Wong (32) | — | 92.4% | • Normal, Pneumonia and COVID-19 | 53 Covıd-19(+) 5526 Covid-19(−) 8066 Healthy |
COVID-19 image data collection (8), COVID-19 Chest X-ray Data set initiative (34), RSNA Pneumonia Detection Challenge (22a), Actualmed COVID-19 Chest X-ray Data set Initiative (33) and (23) | Chest X-ray images |
| Proposed Method | 98.92% | 98.27% | • Healthy, Not healthy • Normal, Pneumonia and COVID-19 |
1524 COVID-19(+) 1524 Normal 1527 Pneumonia |
COVID-19 image Data Collection (8), Chest X-Ray images (21), ChestX-ray8 (23), COVID-19 Radiography Database (35), Data set by Kermany et al. (18) and COVID-19 X-rays (8a) | Chest X-ray images |
When the above-mentioned and existing studies in the literature are carefully examined, the contributions of the study proposed in this paper can be summarized as follows.
-
•
To the best of the author’s knowledge, this is the first study that detects the COVID-19 disease using CNN whose hyperparameters are automatically determined by Grid Search.
-
•
To the best of the author’s knowledge, COVID-19 disease was detected with the largest data set in the literature until the writing of this paper.
-
•
The CNN models used in the proposed method are novel, and the CNN architectures for both tasks are designed by the author.
-
•
By use of the proposed CNN model for Task 1, COVID-19 disease can be detected with a satisfactory rate such as 98.92%.
-
•
By use of the proposed CNN model for Task 2, COVID-19 disease, pneumonia disease and healthy X-ray images can be classified with a satisfactory rate such as 98.27%.
-
•
The proposed method not only detects COVID-19 disease but also detects pneumonia disease with high accuracy.
Hyperparameters are indispensable for deep learning algorithms and are highly effective on performance. The deep learning model is almost equivalent to choosing the most suitable hyperparameter group. However, pairs of parameters are not always sought for the best performance. In such cases, how to proceed, which hyperparameters will be changed, to what extent, which hyperparameters are in correlation with each other should be investigated, trends should be determined and presented in studies. Recently, hyperparameter groups that have given the best performance are given in studies involving deep learning studies. In some studies, even these parameter pairs are not fully given; the choice of hyperparameters and how they achieve success are not discussed. This weakens the analysis value and importance of the study. In the deep learning studies, we proposed the selection of hyperparameters at certain intervals instead of the intuitive hyperparameter selection method, and detailed analysis of how the model performance and working time are affected by the hyperparameter change in these intervals, and discussion of the hyperparameter groups that are correlated with each other, are analyzed in relation to the hyperparameters. It is imperative that a parameter analysis section be present in all studies. In our opinion, it is essential to establish such a standard in such an area where we can no longer keep up with the pace.
CONCLUSIONS
In this paper, two novel and fully automatic studies using deep convolutional neural networks are presented for COVID-19 detection and virus classification. Two novel, powerful, and robust CNN architectures are designed and proposed for two different classification tasks using publicly available data sets. The hyperparameters of both CNN architectures are automatically determined using the Grid Search Optimizer method. Detection of COVID-19 disease is achieved with high accuracy, such as 98.92%. Moreover, classification of chest X-ray images into normal, pneumonia, and COVID-19 is obtained with satisfying accuracy of 98.27%. Experimental results on large clinical data sets show the effectiveness of the proposed architectures. The results of Task 1 and Task 2 indicate that, to the best of the author’s knowledge, state-of-the-art classification performance is achieved using a large clinical data set without data augmentation. One of the main contributions of this study to the literature is that a wide variety of data sets have been used. It aims to create the largest possible number of X-ray images of COVID-19 that exist in the literature until the writing of this research. A total of 1,524 COVID-19 images, 1,527 pneumonia images, and 1,524 normal images were collected and used for this research. It is believed that, thanks to their simplicity and flexibility, the models proposed in this paper can be readily used in practice to help physicians in diagnosing the COVID-19 disease.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author.
AUTHOR CONTRIBUTIONS
E.I. conceived and designed research; performed experiments; analyzed data; interpreted results of experiments; prepared figures; drafted manuscript; edited and revised manuscript; approved final version of manuscript.
REFERENCES
- 1.Abiwinanda N, Hanif M, Hesaputra ST, Handayani A, Mengko TR. Brain tumor classification using convolutional neural network. IFMBE Proc 68: 183–189, 2019. doi: 10.1007/978-981-10-9035-6_33. [DOI] [Google Scholar]
- 2.Alimadadi A, Aryal S, Manandhar I, Munroe PB, Joe B, Cheng X. Artificial intelligence and machine learning to fight COVID-19. Physiol Genomics 52: 200–202, 2020. doi: 10.1152/physiolgenomics.00029.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med 43: 635–640, 2020. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badriyah T, Santoso DB, Syarif I. Deep learning algorithm for data classification with hyperparameter optimization method. J Phys Conf Ser 1193: 012033, 2019. doi: 10.1088/1742-6596/1193/1/012033. [DOI] [Google Scholar]
- 6.Bergstra JS, Bardenet R, Bengio Y, Kégl B. Algorithms for hyper-parameter optimization. Adv Neural Inf Process Syst 25: 2546–2554, 2011. [Google Scholar]
- 8.Cohen JP. 2020. COVID-19 Image Data Collection. https://github.com/ieee8023/covid-chestxray-dataset.
- 8a.Dadario AMV. COVID-19 X Rays. Kaggle; 2020. https://www.kaggle.com/andrewmvd/convid19-X-rays. [Google Scholar]
- 9.Decuyper M, Van Holen R. Fully automatic binary glioma grading based on pre-therapy MRI using 3D convolutional neural networks (Preprint). arXiv 1908.01506, 2019.
- 10.Di Gennaro F, Pizzol D, Marotta C, Antunes M, Racalbuto V, Veronese N, Smith L. Coronavirus diseases (COVID-19) current status and future perspectives: A narrative review. Int J Environ Res Public Health 17: 2690, 2020. doi: 10.3390/ijerph17082690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan H, Wang R, Liu X, Liu H. A method to determine the hyperparameter range for tuning RBF support vector machines. International Conference on E-Product E-Service and E-Entertainment, pp. 16–19, 2010. [Google Scholar]
- 12.Ertosun MG, Rubin DL. 2015. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. In AMIA Annual Symposium Proceedings, vol. 2015, American Medical Informatics Association, 2015. pp. 1899–1908. [PMC free article] [PubMed] [Google Scholar]
- 13.Ghawi R, Pfeffer J. Efficient hyperparameter tuning with Grid Search for text categorization using kNN approach with BM25 similarity. Open Comput Sci 9: 160–180, 2019. doi: 10.1515/comp-2019-0011. [DOI] [Google Scholar]
- 14.Gulcu A, Kus Z. Hyper-parameter selection in convolutional neural networks using microcanonical optimization algorithm. IEEE Access 8: 52528–52540, 2020. doi: 10.1109/ACCESS.2020.2981141. [DOI] [Google Scholar]
- 15.Hemdan EE-D, Shouman MA, Karar ME. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images (Preprint) arXiv 2003.11055, 2020.
- 16.Kabir Anaraki A, Ayati M, Kazemi F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern Biomed Eng 39: 63–74, 2019. doi: 10.1016/j.bbe.2018.10.004. [DOI] [Google Scholar]
- 18.Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha MK, Pei J, Ting MYL, Zhu J, Li C, Hewett S, Dong J, Ziyar I, Shi A, Zhang R, Zheng L, Hou R, Shi W, Fu X, Duan Y, Huu VAN, Wen C, Zhang ED, Zhang CL, Li O, Wang X, Singer MA, Sun X, Xu J, Tafreshi A, Lewis MA, Xia H, Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172: 1122–1131.e9, 2018. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K, Liu D, Wang G, Xu Q, Fang X, Zhang S, Xia J, Xia J. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology 296: E65–E71, 2020.doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loey M, Smarandache F, Khalifa NEM. Within the lack of chest COVID-19 X-ray dataset: A novel detection model based on GAN and deep transfer learning. Symmetry (Basel) 12: 651, 2020. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- 21.Money P. Chest X-Ray Images (Pneumonia). https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia, 2018.
- 22.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med 121: 103792, 2020. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Radiological Society of North America. RSNA Pneumonia Detection Challenge. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data. [DOI] [PubMed]
- 23.Rahman T, Chowdhury M, Khandakar A. COVID-19 Radiography Database. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database, 2020.
- 24.Sajjad M, Khan S, Muhammad K, Wu W, Ullah A, Baik SW. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J Comput Sci 30: 174–182, 2019. doi: 10.1016/j.jocs.2018.12.003. [DOI] [Google Scholar]
- 25.Shekar BH, Dagnew G. 2019. Grid search-based hyperparameter tuning and classification of microarray cancer data. 2019 Second International Conference on Advanced Computational and Communication Paradigms, pp. 1–8, 2019. [Google Scholar]
- 27.Singh D, Kumar V, Vaishali, Kaur M. Classification of COVID-19 patients from chest CT images using multiobjective differential evolution-based convolutional neural networks. Eur J Clin Microbiol Infect Dis 39: 1379–1389, 2020. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sultan HH, Salem NM, Al-Atabany W. Multi-classification of brain tumor images using deep neural network. IEEE Access 7: 69215–69225, 2019. doi: 10.1109/ACCESS.2019.2919122. [DOI] [Google Scholar]
- 29.Toğaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med 121: 103805, 2020. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toğaçar M, Ergen B, Cömert Z. Detection of lung cancer on chest CT images using minimum redundancy maximum relevance feature selection method with convolutional neural networks. Biocybern Biomed Eng 40: 23–39, 2020. doi: 10.1016/j.bbe.2019.11.004. [DOI] [Google Scholar]
- 31.Toğaçar M, Özkurt KB, Ergen B, Cömert Z. BreastNet: a novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Physica A 545: 123592, 2020. doi: 10.1016/j.physa.2019.123592. [DOI] [Google Scholar]
- 32.Wang L, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray image (Preprint). arXiv , 2003.09871, 2020. [DOI] [PMC free article] [PubMed]
- 33.Wang L, Wong A, Lin ZQ, McInnis P, Chung A, Gunraj H. Actualmed COVID-19 Chest X-ray Dataset Initiative. https://github.com/agchung/Actualmed-COVID-chestxray-dataset.
- 34.Wang L, Wong A, Lin ZQ, McInnis P, Chung A, Lee J. COVID-19 Chest X-Ray Dataset Initiative. https://github.com/agchung/Figure1-COVID-chestxray-dataset, 2020
- 35.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 3462–3471, 2017. [Google Scholar]
- 36.Wong J, Manderson T, Abrahamowicz M, Buckeridge DL, Tamblyn R. Can hyperparameter tuning improve the performance of a super learner?: a case study. Epidemiology 30: 521–531, 2019. doi: 10.1097/EDE.0000000000001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu H, Fang Q, He H, Hu J, Jiang D, Xu K. Automatic prediction of meningioma grade image based on data amplification and improved convolutional neural network. Comput Math Methods Med 2019: 7289273, 2019. doi: 10.1155/2019/7289273. [DOI] [PMC free article] [PubMed] [Google Scholar]