Abstract
Introduction:
Imprinted genes are preferentially expressed from one parentally inherited allele, and many are crucial to the regulation of placental function and fetal growth. Murine Krüppel-like factor 14 (Klf14) is a maternally expressed imprinted transcription factor that is a component of the Mest imprinted gene cluster on mouse chromosome 6. We sought to determine if loss of Klf14 expression alters the course of normal mouse extraembryonic development. We also used high-throughput RNA sequencing (RNAseq) to identify a set of differentially expressed genes (DEGs) in placentas with loss of Klf14.
Methods:
We generated a Klf14 knockout (Klf14null) mouse using recombineering and transgenic approaches. To identify DEGs in the mouse placenta we compared mRNA transcriptomes derived from 17.5dpc Klf14matKO and wild-type littermate placentas by RNAseq. Candidate DEGs were confirmed with quantitative reverse transcription PCR (qPCR) on an independent cohort of male and female gestational age matched Klf14matKO placentas.
Results:
We found that 17.5dpc placentas inheriting a maternal null allele (Klf14matKO) had a modest overgrowth phenotype and a near complete ablation of Klf14 expression. However, there was no effect on fetal growth. We identified 20 DEGs differentially expressed in Klf14matKO placentas by RNAseq, and subsequently validated five that are highly upregulated (Begain, Col26a1, Fbln5, Gdf10, and Nell1) by qPCR. The most enriched functional gene-networks included those classified as regulating cellular development and metabolism.
Conclusion:
These results suggest that loss of the maternal Klf14 locus in the mouse placenta acts results in changes in gene expression patterns that modulate placental growth.
Keywords: Imprinting, Mouse Model, Transcription, KLF14, RNAseq
Introduction
The metabolic and developmental processes controlling fetal and placental growth are strongly influenced by imprinted genes [1]. This remarkable set of genes is expressed exclusively or preferentially from one parentally inherited allele based on heritable DNA-methylation at gametic differentially methylated domains (DMDs). The widely accredited kinship theory postulates that misalignment of maternal and paternal selfish genetic reproductive interests contributes to a division in pro-growth paternally expressed and growth-limiting maternally expressed imprinted genes [2]. Murine Krüppel-like factor 14 (Klf14) is a maternally expressed imprinted gene encoding a transcription factor robustly expressed in the placental labyrinth [3].
Preferential expression of maternally inherited Klf14 is dependent on maternal parent-of-origin specific DNA methylation at the Mest DMD, which exerts transcriptional control of the paternally expressed Mest as well as a cluster of maternally expressed genes across a 400-kb region that includes Copg2, Cpa4 , and Klf14 [4, 5]. Disruption of either the establishment or inheritance of imprinted DNA methylation results in loss of Mest DMD methylation in utero and a lack of Klf14 expression within extraembryonic tissues [3, 6]. Our previous work revealed that loss of imprinting at the Mest locus in late gestation is associated with reduced fetal-placental weight ratio [6], increased spongiotrophoblast central volume and elevated triglyceride levels [7, 8]. We have interpreted these findings as strong evidence of a role for the Mest imprinting cluster in regulating placental growth and metabolism. This work is also in accordance with other studies linking the Mest imprinted locus to in utero placental and fetal development.
In late gestation (16.5-18.5dpc) in the mouse, an intrauterine and post-natal growth restriction phenotype is observed in fetuses with bimaternal inheritance of the subproximal 6q region encompassing the Mest and Nap1l5 imprinting clusters, whereas fetal growth enhancement that resolved by term pregnancy is observed with bipaternal inheritance [9, 10]. In humans, matUPD7 and Mest DMD hypermethylation are causes of intrauterine growth restriction in a minority of Silver-Russell syndrome cases [11, 12]. Together, this evidence suggests the imprinted (methylated) Mest/MEST maternal allele is integrated as a net growth restrictor during pre-natal development either through silencing of pro-growth paternally-expressed genes or maternal expression of growth restricting genes.
Multiple lines of evidence support a role for Klf14 as a key metabolic transcriptional regulator. In humans, KLF14 is a maternal-specific expressed quantitative trait locus (eQTL) associated with type-2 diabetes risk and HDL levels; variant polymorphisms rs731702 and rs972283 within the intergenic region upstream of KLF14 result in diminished KLF14 expression in cis, and altered downstream metabolic transcriptional networks in trans within adipocytes [13-15]. Recent work describing the function of Klf14 in the mouse liver suggest a conserved role for KLF14 in transcriptional regulation of metabolic homeostasis [16-18]. These effects are mediated by binding of KLF14 to SP-1 like GC rich motifs in target gene promoters, where it acts predominately as a transcriptional repressor [16-18]. While these studies in adipose and liver are informative, the tissue with the highest RNA expression of human KLF14 is the placenta, corroborated by strong immunohistochemical staining in chorionic villi syncytiotrophoblast [19].
In this study, we sought to determine whether loss of Klf14 expression alters the course of normal mouse extraembryonic development by examining placentas derived from a Klf14 knockout mouse. Furthermore, because Klf14 is a transcription factor, we pursued the identification of placenta-specific transcriptional changes by high-throughput RNA sequencing (RNAseq). We analyzed the differentially expressed gene sets to understand the pathways altered by loss of Klf14. Our work suggests that within the mouse placenta Klf14 is an imprinted transcription factor that modulates expression of a defined set of genes that may have a role in placental growth.
Materials and methods:
Generation of a Klf14 null mouse
We employed recombineering technology to construct a targeting vector, and generate Klf14flox and Klf14null alleles (Figure S1, Table S3). Recombineering utilizes bacterial strains with inducible gap-repair recombination enzymes that catalyze recombination between homologous 200-500 bp sequences (homology boxes) enabling efficient exchange of sequences from one vector to another (for reviews see [20, 21]). A 13.5kb region encompassing Klf14 and surrounding sequences was retrieved from bMQ6044J02 bacterial artificial chromosome, obtained from Source Bioscience, and cloned into a plasmid adjacent to a diphtheria toxin gene. We then engineered plasmids to insert LNL and LFNTF cassettes to generate a targeting allele. Standard methods of ES cell transfection, homologous recombination, Flp electroporation, Neomycin/Ganciclovir positive/negative selection and blastocyst injection were used to generate a transgenic Klf14flox mouse line [22]. The loxP sites were thereby added approximately 1.6-kb upstream of the Klf14 transcriptional start site and 300-bp downstream of the Klf14 3′-UTR separated by a total of 4.9-kb. This line was then crossed with Sox2:CRE transgenic females to generate a constitutive null allele (Klf14null).
Animal husbandry
Animals were cared for in accordance with the University of Pittsburgh Institutional Animal Care and Use Committee. Heterozygous Klf14null mice were backcrossed 5 generations to the 129sv Taconic strain prior to setting up timed matings. We bred Klf14hetKO female mice to 129sv inbred wild-type mice to generate Klf14matKO and control offspring for phenotypic and transcriptional analysis. Additionally, control (Klf14wt), heterozygous (Klf14hetKO), and homozygous (Klf14homKO) derived from heterozygous intercrosses were studied for late gestation placental abnormalities.
Genotyping
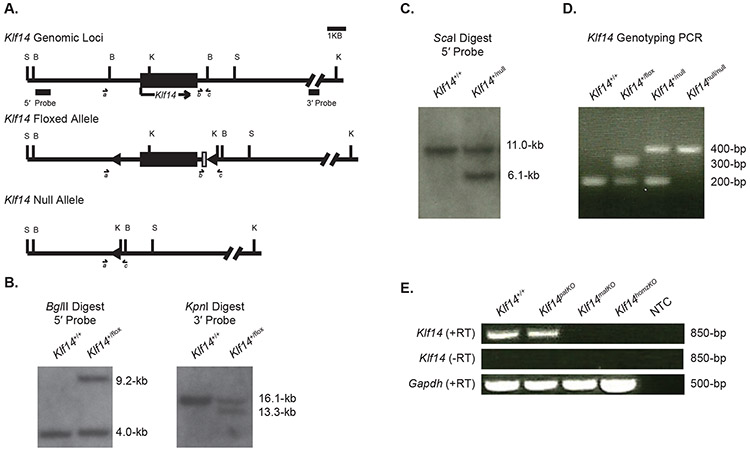
Floxed alleles were confirmed by Southern blotting of BglII digests with an external 5′ probe (Klf14wt band of 4.0-kb and Klf14flox band of 9.2-kb). Null alleles were distinguished from wild-type and floxed alleles by Southern blot of ScaI digested genomic DNA with an external 5′ probe (Klf14wt and Klf14flox bands of 11.0-kb and Klf14null band of 6.1-kb). Klf14 floxed, null and wild-type alleles were also genotyped by semi-nested PCR by the presence of 200-bp (Klf14wt), 300-bp (Klf14flox) and 400-bp (Klf14null) alleles (Table S3). Sox2:CRE transgene genotyping and Zfy sex-typing were also determined by PCR. Placental genotypes were ascertained from associated yolk-sac or fetal tissues.
Placental measurements
Late gestation (17.5dpc) mouse placentas were collected from crosses of heterozygous Klf14null females to either wild-type or heterozygous males. Placenta and fetal wet weights were recorded. Half of each placenta was cryo-preserved for histology. Nucleic acids were isolated from the remaining half for gene expression and methylation analysis. Placental layer fractions were determined using the ratio of the average spongiotrophoblast, and the labyrinth area determined by random grid sampling of a single central placental section for each sample.
Klf14 expression analysis
Nucleic acids were isolated using All-Prep or RNeasy kits (Qiagen) per manufacturer’s instructions from fresh placental tissues, and other fetal and adult tissues isolated by microdissection. Prior to reverse transcription, RNA was treated with DNaseI for 1h to remove genomic contaminants. First-strand cDNA synthesis was carried out with MMLV-RT (Promega) from 500 ng of RNA template using oligo(dT)18 primers. Amplification of an 800-bp RT-PCR of Klf14 cDNA product was performed using primers previously reported by Parker-Katiraee et al. [3].
Klf14 methylation analysis
The methylation status of the Mest DMD in wild-type and null placentas was examined by bisulfite genomic sequencing and combined bisulfite restriction analysis assays. Bisulfite genome conversion, PCR and Sanger sequencing of wild-type and Klf14null placental DNA was carried out as previously described using the same reagents and nested primers [6]. The BstBI restriction enzyme was utilized to cut 1ug of bisulfite converted Mest DMD PCR amplicon in order to examine for the presence of unconverted (methylated) sites within the Mest DMD.
Transcriptome analysis
Library preparation and high-throughput sequencing were performed at the Children’s Hospital of Pittsburgh Genomics core. An Illumina TruSeq mRNA library preparation kit was used with indexing i5 and i7 adapters. Four control and four mutant samples were sequenced to a depth of 40 million reads on a NextSeq500 (Illumina) instrument. Computational analysis was performed on the high throughput computing cluster of the University of Pittsburgh Center for Research Computing. Quality control and trimming of sequencing adapters was performed with Trimgalore [23]. The reads were aligned to the ensembl v96 mouse genome and annotation using STAR v2.7 and gene counts tabulated using HTSeq [24, 25]. Differential expression analysis was performed using the DESeq2 Bioconductor package [26]. Genome-wide transcription data can be found in the GEO Repository, Geo accession number GSE137918.
Quantitative RT-PCR
1μg RNA isolated from placental tissues was subjected to DNaseI treatment (Ambion) and then used as template for first strand cDNA synthesis using superscript IV (Invitrogen). Amplification of target genes was performed using SYBR green mastermix on a 7900HT real time PCR machine (Applied biosystems). Primer sequences are given as supplemental (Table S3) and for lineage markers partly drawn from Tunster et al. [27, 28].
Statistics
Morphometric comparisons were made between wild-type, heterozygous and null placentas by analysis of variance (ANOVA) followed by post-hoc Tukey-HSD, or Student’s T-test where appropriate. For qPCR data technical replicates were averaged and the ddCt method utilized to calculate the log2 fold change between mutant and control groups separated by sex or aggregated as well as the standard deviation for each. Comparison across groups was made by either one-way (for DEGs) or two-way (for lineage markers) Welch’s T-test with p value cutoffs at 0.05, 0.005 and 0.0005 for significance. A Benjamini-Hochberg false discovery rate (FDR) p-adjusted (q value) cutoff of q <0.10 was used to identify candidate DEGs from RNAseq transcriptome analysis. A less stringent p value <0.1 was used for a broader supplementary gene expression and gene network analysis via Ingenuity Pathway Analysis (Qiagen).
Results
Generation of a Klf14 maternal knockout
We generated a floxed Klf14 allele using homology directed mouse embryonic stem cell genome editing approaches to add loxP flanking the Klf14 single exon open reading frame (Figures 1A and S1). Analysis of transgenic mouse genomic DNA by Southern blotting confirmed the expected deletion of a Bgl2 site in the targeted floxed allele at the Klf14 upstream loxP site by the presence of a 5.2-kb larger BglII fragment detected by a dsDNA probe 5′ of Klf14 and external to the targeting construct (Figure 1B). Floxed male mice were crossed with Sox2:CRE mice to generate constitutive Klf14 null offspring. The deletion of a 4.9-kb region encompassing the single Klf14 open reading frame and promoter sequence was distinguished by Southern blot of ScaI digested DNA, probed with the same 5′ external probe (Figure 1C). The wild-type, floxed and null alleles can also readily be distinguished by semi-nested genomic PCR (Figure 1D).
Figure 1:
Genetic characterization of a novel Klf14 floxed and constitutive null allele. A) Schematic of the native Klf14 locus and genome edited floxed and null alleles. B) BglII restriction digest Southern blot of wild-type and heterozygous floxed mice. C) ScaI restriction digest and southern blot of wildtype and heterozygous null mice. D) Genomic genotyping semi-nested PCR identifies specific bands for wild-type (200-bp), floxed (300-bp) and null (400-bp) alleles. E) RT-PCR of an 850-bp Klf14 cDNA amplicon in wild-type, Klf14patKO, Klf14matKO and Klf14homzKO placentas. Key: Filled rectangle, Klf14 with arrow of 5′ to 3′ transcription; directional triangles, loxP elements; grey rectangle, residual LFNTF vector sequence; Vertical lines indicate restriction sites (S) ScaI, (B)-BglII, (K) KpnI, (N) NotI; and a, b and c semi-nested PCR primer s. Scale bar is 1-kb.
A survey of Klf14 mRNA expression in extraembryonic, fetal and adult wild-type tissues by reverse-transcription PCR (RT-PCR) revealed robust expression in yolk sac, placenta and fetal intestine with lesser expression in fetal brain, fetal liver, adult kidney and adult heart (Figure S2). We then tested the degree of preferential maternal Klf14 expression in placenta with inherited paternal null (Klf14patKO), maternal null (Klf14matKO) and homozygous null (Klf14homzKO) alleles. The placentas of Klf14patKO fetuses maintained wild-type levels of transcription. In contrast, expression of Klf14 in Klf14matKO and Klf14homzKO placentas was undetectable (Figure 1E). This result confirms strict maternal expression of Klf14 in the mouse placenta and complete loss of expression when inheriting a maternal null allele. Additionally, we found there was no change in imprinted DNA methylation at the Mest DMD associated with inheritance of the Klf14null allele, suggesting that there would be no in cis effects on the expression of neighboring imprinted genes within the subproximal 6q imprinted cluster (Figure S3).
Placentas of Klf14matKO offspring are modestly overgrown in late gestation without overt morphological changes in placental layers.
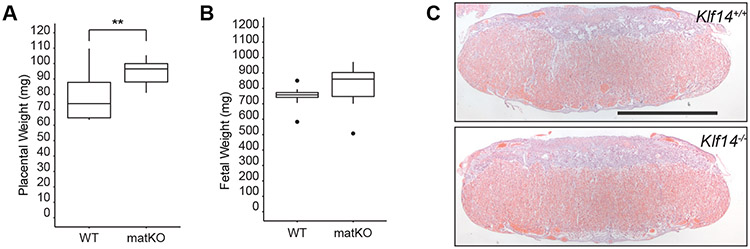
Consistent with other reports of Klf14 constitutive null alleles, we found that both heterozygous (Klf14hetKO) and Klf14homzKO mice were viable and fertile [16, 29]. Heterozygous intercrosses resulted in offspring that inherited the Klf14null allele at expected mendelian frequencies on both B6N and 129sv backgrounds (Chi square p > 0.05, Tables S1 and S2). A placental overgrowth phenotype was observed in late gestation placentas from Klf14matKO offspring compared to those of wild-type littermates (Figure 2A; Tukey-HSD p = 0.0076). However, although fetal weights in Klf14matKO offspring remained unchanged (Figure 2B), there was no difference in fetal-placenta weight ratio. Similarly, placental but not fetal overgrowth was observed across genotypes of late gestation mouse placentas derived from Klf14hetKO 129sv intercrosses (Figure S4; ANOVA p = 0.05). Representative images of a Klf14homzKO and control littermate H&E and Periodic acid-Schiff (PAS) histological staining demonstrate a moderate overall increase in size in all layers with lack of any structural abnormalities and consistent glycogen content (Figures 2C and S5). Likewise, there were no aberrations in the labyrinth to spongiotrophoblast placental layer fraction in Klf14homzKO placentas (Figure S4). The expression of lineage markers was similar among the placental layers in Klf14matKO placentas, except for a slight reduction of the labyrinth marker Gcm1 (p < 0.05; Figure S6).
Figure 2:
Placental overgrowth in Klf14matKO offspring at 17.5dpc. A) Placental weights are increased in Klf14matKO placentas compared to wild-type littermates. B) Fetal weights remain unchanged between Klf14matKO and wild-type littermates. C) Representative H&E images of wild-type and homozygous 17.5dpc placentas (scale, 2mm). WT, wild-type (n=9); matKO, Klf14matKO (n=10). Statistical comparisons by ANOVA and Tukey post hoc test.
Klf14 regulates a placental transcriptional network including Begain, Col26a1, Fbln5, Gdf10, and Nell1.
We sought to use Klf14matKO placentas to identify differentially expressed genes (DEGs) that could be mediating the overgrowth phenotype. Transcriptome wide gene expression profiling by RNAseq was performed on four Klf14matKO and four control 17.5dpc male placentas. The top 100 highest expressed genes in our RNAseq dataset (Table S4) overlapped considerably with known mouse placental labyrinth and spongiotrophoblast lineage markers and those highly expressed in rat placental compartments and were consistent across genotypes with abundant labyrinth and spongiotrophoblast transcripts and few from maternal decidual endometrium [30-32]. Expression of placental lineage markers used to examine layer composition by qPCR (Figure S6), revealed similar consistent expression across control and Klf14matKO placentas with modest variability in labyrinth markers (Table S5) [27, 28]. Not surprisingly, Klf14matKO placentas had a greater than 6 log2 fold decrease in Klf14 expression (Table 1), but the expression of other imprinted and non-imprinted genes within the Mest imprinting cluster as well as the Napl15 and Peg10 imprinting clusters on chromosome 6 were unchanged (Table S6). These results confirm strong, near absolute maternal parent-of-origin specific expression of Klf14 in the mouse placenta.
Table 1:
Table of differentially expressed genes identified in Klf14matKO placentas. Differential gene-level expression analysis of a comparison of four male matKO and four wild-type littermates, filtered (FDR q value <0.10) and arranged in ascending q value order. RPKM, averaged normalized gene counts as RPKM and log2 fold change (FC) for comparison of control and matKO.
| Gene | Ensembl ID | Control RPKM | matKO RPKM | log2 FC | FDR q value |
|---|---|---|---|---|---|
| Klf14 | ENSMUSG00000073209 | 431.6 | 5.3 | −6.34 | 7.04E-39 |
| Col26a1 | ENSMUSG00000004415 | 20.2 | 87.6 | 2.12 | 2.65E-10 |
| Nell1 | ENSMUSG00000055409 | 24.7 | 106.5 | 2.11 | 1.16E-04 |
| Slit2 | ENSMUSG00000031558 | 124.1 | 260.9 | 1.07 | 1.36E-03 |
| Adcy2 | ENSMUSG00000021536 | 53.6 | 136.8 | 1.36 | 1.46E-03 |
| Gdf10 | ENSMUSG00000021943 | 15.7 | 94.9 | 2.60 | 4.29E-03 |
| Pkia | ENSMUSG00000027499 | 45.8 | 103.2 | 1.18 | 4.29E-03 |
| Gm4779 | ENSMUSG00000045010 | 85.9 | 34.9 | −1.30 | 1.19E-02 |
| Piezo2 | ENSMUSG00000041482 | 461.9 | 319.3 | −0.53 | 1.19E-02 |
| Dkk2 | ENSMUSG00000028031 | 54.7 | 137.1 | 1.33 | 2.53E-02 |
| Naalad2 | ENSMUSG00000043943 | 48.2 | 91.0 | 0.92 | 4.10E-02 |
| Smarca1 | ENSMUSG00000031099 | 171.3 | 266.5 | 0.64 | 4.27E-02 |
| Cd163 | ENSMUSG00000008845 | 85.5 | 44.4 | −0.96 | 4.95E-02 |
| Fbln5 | ENSMUSG00000021186 | 465.4 | 901.1 | 0.95 | 4.95E-02 |
| Matn2 | ENSMUSG00000022324 | 421.5 | 633.8 | 0.59 | 4.95E-02 |
| Begain | ENSMUSG00000040867 | 13.0 | 66.4 | 2.36 | 4.95E-02 |
| Col8a1 | ENSMUSG00000068196 | 126.5 | 248.5 | 0.98 | 4.95E-02 |
| Raver2 | ENSMUSG00000035275 | 45.0 | 83.9 | 0.91 | 7.74E-02 |
| Tmem200a | ENSMUSG00000049420 | 10.4 | 28.1 | 1.43 | 7.74E-02 |
| Igdcc4 | ENSMUSG00000032816 | 105.7 | 211.4 | 1.00 | 7.82E-02 |
| Olfm2 | ENSMUSG00000032172 | 67.5 | 141.5 | 1.07 | 8.62E-02 |
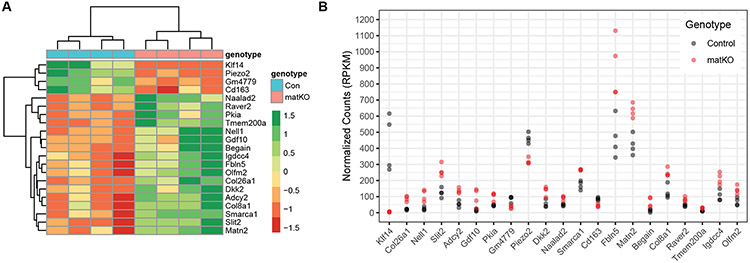
We identified 21 DEGs with an FDR q value less than 0.10 in Klf14matKO placentas (Table 1). The distribution of significant DEGs as shown by a volcano plot was skewed toward elevated expression, indicative of transcriptional de-repression (Figure S8). Hierarchical clustering of the DEGs revealed two sets of transcripts (Figure 3A). The first set was a small group of 4 genes with diminished expression including Klf14. The second set was a larger group of 17 genes with elevated expression. The normalized RNAseq read counts were consistent across samples of the same genotype for each identified DEG (Figure 3B). These results identify a possible network of trans regulated genes repressed by Klf14.
Figure 3:
Differential RNA-Seq expression analysis of 17.5dpc Klf14matKO placentas compared with wild-type littermates. Gene level RNA-Seq analysis of four KLF14matKO and four wild-type littermates. A) Heatmap clustergram of 21 DEGs including Klf14 in Klf14matKO placentas. Green = upregulated, Red = downregulated. B) Normalized RNAseq read counts for each of 21 DEGs including Klf14 . Grey circle are WT samples and red circles are Klf14matKO samples. RPKM, reads per kilobase of transcript per million reads.
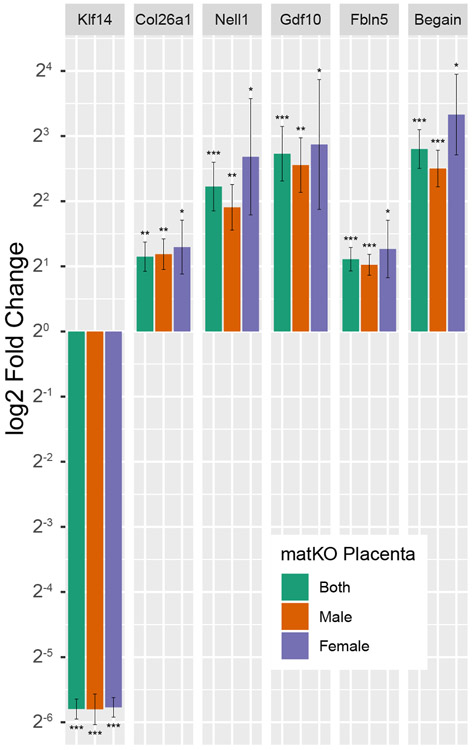
To confirm changes in gene expression identified by RNAseq we analyzed additional cohorts of both male and female Klf14matKO 17.5dpc placentas and wild-type littermates by quantitative RT-PCR (qPCR). This analysis confirmed the elevated expression of Begain, Col26a1 Fbln5, Gdf10, and Nell1 in Klf14matKO 17.5dpc placentas (Figure 4). Results were largely concordant between male, female and combined cohorts with lesser statistical significance in female cohorts due to lower numbers of samples assayed. The discovery of Gm14296, encoding a putative zinc finger protein, as differentially expressed in the RNASeq data set was refuted by qPCR, although it was impossible to distinguish it from segmentally duplicated isoforms (Figure S7). We were also able to verify changes in expression of Col9a2, Fgg, Tlr12, Enpp2, and Ltbp2 but not Cp which were identified using a less stringent unadjusted p value cutoff of less than 0.10 (Figure S7, Table S7). Additionally, we tested the known hepatic Klf14 targets Sphk1 and Pgc1a but found no difference in expression (Figure S7).
Figure 4:
qPCR validation of transcriptional changes in Klf14 and DEGs. Log2 fold change of six transcripts in independent control and matKO samples. Fold change was calculated by the ddCT method using Gapdh as the reference and wt sex-matched cohorts as 1.0 fold baseline. Male-wt n=5, Male-matKO n=7, Female-wt n=5, Female-mat KO n=4. Mean fold change and SEM are shown. * p <0.05, ** p <0.005 and *** p <0.0005.
To better understand the network of DEGs in Klf14matKO placentas we performed Ingenuity Pathway Analysis (IPA). We discovered two enriched functional groups that included cellular development and growth as facets (Table 2). By superimposing these two overlapping networks we exposed a putative placenta cellular developmental regulome controlled by Klf14 (Figure S9). Additionally, we performed IPA functional analysis with a larger list of 525 genes identified using a less stringent non-adjusted p value cutoff of 0.1 (Table S7-10). The larger analysis identified networks involved in cellular and organismal development, cardiovascular disease and metabolism among others (Table S8). We further identified additional canonical pathways and upstream regulators (Tables S9 and S10). These functional gene categories provide a potential molecular circuit that influences placental growth and development.
Table 2:
Functional enrichment of differentially expressed genes in Klf14matKO placentas. The highest enriched pathways suggest an overlap in cellular growth and development.
| Functional Category | Score | DEGs in Network |
|---|---|---|
| Cellular Development, Cellular Growth and Proliferation, Cellular Movement | 28 | Begain, Cd163, Col8a1, Fbln5, Gdf10, Nell1, Olfm2, Pkia, Smarca1, Tmem200a |
| Cell Morphology, Cellular Assembly and Organization, Cellular Development | 22 | Adcy2,Col26a1, Gm4479, Dkk2, Idgcc4, Naalad2, Piezo2, Raver2, Slit2 |
Discussion
KLF14 is a negative regulator of placental growth
Our results demonstrate that Klf14matKO placentas exhibit a modest placental overgrowth, which is reciprocal and in similar magnitude to the placental growth restriction seen in MestpatKO mice [33]. These findings suggest that Mest and Klf14 influence placental growth in an antagonistic manner, congruent with the Kinship hypothesis with the paternally expressed Mest functioning to enhance placental growth and Klf14 acting to limit it [2]. Our failure to observe an overt abnormal lipid accumulation in Klf14matKO placentas compared to the effect of concurrent loss of Klf14 expression and gain of Mest expression [7] suggests the importance of combined imprinting of both genes and/or expression of other components of the Mest cluster. The overall impact of maternal Klf14 deletion on placental growth was on par with maternal deletion of another imprinted gene, Grb10 [34, 35]. However, the Klf14matKO growth effect did not affect layer fractions, whereas Grb10matKO placentas had an increase in spongiotrophoblast at the expense of labyrinth [34, 35]. Other imprinted genes, notably Igf2, have a greater effect on placental development with paternal deletion of placental Igf2 (Igf2p0), somatic Igf2 or the H19 DMD having disrupted spongiotrophoblast and glycogen cell layers and pronounced placental growth effects [36]. Our work supports the hypothesis that imprinted genes have an integrated role in placental growth and metabolism with different clusters regulating specific processes.
Loss of KLF14 modulates expression of a network of genes
Our findings indicate that loss of Klf14 activates expression of multiple placental genes. These results are consistent with reported studies on KLF14 and its orthologs as repressors in both vertebrate and invertebrate species [37]. Our data are also consistent with KLF14’s role in human adipocytes, where it has been implicated primarily as a transcriptional repressor, with 9/10 trans-eQTL genes upregulated concomitant with diminished KLF14 expression associated with maternal inheritance of rs4731702 [13, 15]. More recently, the Klf14 eQTL studies have been expanded on to show that while the phenotypic associations with metabolic syndrome were female-specific the changes in gene expression were irrespective of sex [38]. However, none of the 20 DEGs identified in Klf14matKO placentas overlap the trans-eQTL targets nor those previously studied in targeted Klf14 deletion mice [16-18, 38, 39]. However we did find an overlap of 9 genes from an expanded 385 adipocyte trans-eQTL loci linked to rs4731702 [38], and our 525 less stringent cutoff DEG list (Table 7; data not shown). These discrepancies between studies identifying Klf14 expression networks may be explained by differences in adipocyte, liver and placental transcriptional regulation.
Our IPA analysis indicated functional enrichment for genes involved in cellular growth and development (Table 2 and S8). Canonical pathways identified from our larger list of candidate DEGs (Table S9) revealed links to sphingosine kinase-1 and polo-like kinases that have previously been identified as associated with Klf14 [16, 29], as well as activation of the ligand-activated nuclear receptors LXR and RXR, known components of placental developmental and transcriptional metabolic networks [40-42]. Intriguingly TGFB is one of the highest scoring upstream regulatory node identified by IPA in our study (Table S10). This corroborates studies showing Klf14 transcriptional activation by TGFB and subsequent attenuation of TGFBRII activation via repression in Panc-1 cells, and TGFB/progesterone induced Klf14 activation in Leydig cells [39, 43]. The associated gene networks we have identified point towards an interplay between multiple pathways influence by Klf14.
The set of DEGs identified herein is smaller than the 87 DEGs identified in Klf14 adipose-specific knockout mice [38] and could be explained by the diverse tissue lineages in the placenta. Thus, we suggest that RNAseq of Klf14matKO of specific placental compartments, namely the labyrinth, may yield a larger group of trans-regulated genes. Additionally, within the placenta there is expression of redundant KLF paralogs that also bind with similar affinities to SP1-like motifs. For example, the binding site for KLF6, the most highly expressed KLF in the mouse placenta, has been described in vivo as the SP1-like core GGCG motif at the promoter of genes coregulated by the placental developmental transcriptional regulators PPARG and LCoR [44]. This motif is encompassed by the predicted KLF14 binding site [45]. Moreover, the mouse Klf6 KO leads to severe placental anatomical abnormalities. A combinatorial approach to deletions of Klf paralogs in addition to Klf14 may lead to more severe morphological defects and transcriptional changes, and a larger DEG set.
In summary, our work shows that Klf14 is an imprinted gene in the mouse placenta that when ablated results in placental overgrowth and transcriptional changes. Based on our findings of a unique network of disrupted genes in Klf14matKO placentas we suggest that Klf14 has a discrete function in the placenta as a transcriptional regulator, and it influences the intrauterine environment by modulating pathways involved in placental growth.
Supplementary Material
Highlights:
Late gestation placental growth enhancement in a novel Klf14matKO mouse model
Identification of differentially expressed genes in Klf14matKO placentas by RNA-seq
Validation of Begain, Col26a1, Fbln5, Gdf10, and Nell1 as upregulated by qPCR
Acknowledgements
The project was supported by NIH P01-HD069316 (to YB, YS and RC).
Abbreviations:
- DEG
Differentially expressed gene
- DMD
Differentially methylated domain
- eQTL
expressed quantitative trait locus
- FDR
false discovery rate
- Klf14
Krüppel-like factor 14
- PAS
Periodic acid-Schiff RNAseq, High-throughput RNA sequencing
- RT-PCR
Reverse transcription PCR
- qPCR
Quantitative RT-PCR
References
- [1].Frost JM, Moore GE, The importance of imprinting in the human placenta, PLoS Genet 6(7) (2010) e1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Moore T, Haig D, Genomic imprinting in mammalian development: a parental tug-of-war, Trends Genet 7(2) (1991) 45–9. [DOI] [PubMed] [Google Scholar]
- [3].Parker-Katiraee L, Carson AR, Yamada T, Arnaud P, Feil R, Abu-Amero SN, Moore GE, Kaneda M, Perry GH, Stone AC, Lee C, Meguro-Horike M, Sasaki H, Kobayashi K, Nakabayashi K, Scherer SW, Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution, PLoS Genet 3(5) (2007) e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lefebvre L, Viville S, Barton SC, Ishino F, Surani MA, Genomic structure and parent-of-origin-specific methylation of Peg1, Hum Mol Genet 6(11) (1997) 1907–15. [DOI] [PubMed] [Google Scholar]
- [5].McMinn J, Wei M, Sadovsky Y, Thaker HM, Tycko B, Imprinting of PEG1/MEST isoform 2 in human placenta, Placenta 27(2-3) (2006) 119–26. [DOI] [PubMed] [Google Scholar]
- [6].Himes KP, Koppes E, Chaillet JR, Generalized disruption of inherited genomic imprints leads to wide-ranging placental defects and dysregulated fetal growth, Dev Biol 373(1) (2013) 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Himes KP, Young A, Koppes E, Stolz D, Barak Y, Sadovsky Y, Chaillet JR, Loss of inherited genomic imprints in mice leads to severe disruption in placental lipid metabolism, Placenta 36(4) (2015) 389–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Koppes E, Himes KP, Chaillet JR, Partial Loss of Genomic Imprinting Reveals Important Roles for Kcnq1 and Peg10 Imprinted Domains in Placental Development, PLoS One 10(8) (2015) e0135202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Beechey CV, Peg1/Mest locates distal to the currently defined imprinting region on mouse proximal chromosome 6 and identifies a new imprinting region affecting growth, Cytogenet Cell Genet 90(3-4) (2000) 309–14. [DOI] [PubMed] [Google Scholar]
- [10].Beechey CV, A reassessment of imprinting regions and phenotypes on mouse chromosome 6: Nap1l5 locates within the currently defined sub-proximal imprinting region, Cytogenet Genome Res 107(1-2) (2004) 108–14. [DOI] [PubMed] [Google Scholar]
- [11].Abu-Amero S, Monk D, Frost J, Preece M, Stanier P, Moore GE, The genetic aetiology of Silver-Russell syndrome, J Med Genet 45(4) (2008) 193–9. [DOI] [PubMed] [Google Scholar]
- [12].Hannula-Jouppi K, Muurinen M, Lipsanen-Nyman M, Reinius LE, Ezer S, Greco D, Kere J, Differentially methylated regions in maternal and paternal uniparental disomy for chromosome 7, Epigenetics : official journal of the DNA Methylation Society 9(3) (2014) 351–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Civelek M, Lusis AJ, Conducting the metabolic syndrome orchestra, Nat Genet 43(6) (2011) 506–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Langenberg C, Hofmann OM, Dupuis J, Qi L, Segre AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bengtsson Bostrom K, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Couper DJ, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jorgensen T, Kao WH, Klopp N, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Perry JR, Petersen AK, Platou C, Proenca C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparso T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, van Vliet-Ostaptchouk JV, Walters GB, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Mohlke KL, Morris AD, Palmer CN, Pramstaller PP, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Wareham NJ, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Hu FB, Meigs JB, Pankow JS, Pedersen O, Wichmann HE, Barroso I, Florez JC, Frayling TM, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI, M. investigators, G. Consortium, Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis, Nat Genet 42(7) (2010) 579–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Small KS, Hedman AK, Grundberg E, Nica AC, Thorleifsson G, Kong A, Thorsteindottir U, Shin SY, Richards HB, Consortium G, Investigators M, Consortium D, Soranzo N, Ahmadi KR, Lindgren CM, Stefansson K, Dermitzakis ET, Deloukas P, Spector TD, McCarthy MI, Mu TC, Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes, Nat Genet 43(6) (2011) 561–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].de Assuncao TM, Lomberk G, Cao S, Yaqoob U, Mathison A, Simonetto DA, Huebert RC, Urrutia RA, Shah VH, New role for Kruppel-like factor 14 as a transcriptional activator involved in the generation of signaling lipids, J Biol Chem 289(22) (2014) 15798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo Y, Fan Y, Zhang J, Lomberk GA, Zhou Z, Sun L, Mathison AJ, Garcia-Barrio MT, Zhang J, Zeng L, Li L, Pennathur S, Willer CJ, Rader DJ, Urrutia R, Chen YE, Perhexiline activates KLF14 and reduces atherosclerosis by modulating ApoA-I production, J Clin Invest 125(10) (2015) 3819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang L, Tong X, Gu F, Zhang L, Chen W, Cheng X, Xie L, Chang Y, Zhang H, The KLF14 transcription factor regulates hepatic gluconeogenesis in mice, J Biol Chem 292(52) (2017) 21631–21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, Proteomics. Tissue-based map of the human proteome, Science 347(6220) (2015) 1260419. [DOI] [PubMed] [Google Scholar]
- [20].Copeland NG, Jenkins NA, Court DL, Recombineering: a powerful new tool for mouse functional genomics, Nat Rev Genet 2(10) (2001) 769–79. [DOI] [PubMed] [Google Scholar]
- [21].Liu P, Jenkins NA, Copeland NG, A highly efficient recombineering-based method for generating conditional knockout mutations, Genome Res 13(3) (2003) 476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Behringer R, Manipulating the mouse embryo : a laboratory manual, Fourth edition. ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2014. [Google Scholar]
- [23].Kreueger F, Trim Galore: A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files, (2018). [Google Scholar]
- [24].Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data, Bioinformatics 31(2) (2015) 166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner, Bioinformatics 29(1) (2013) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol 15(12) (2014) 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tunster SJ, Tycko B, John RM, The imprinted Phlda2 gene regulates extraembryonic energy stores, Mol Cell Biol 30(1) (2010) 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tunster SJ, Van de Pette M, John RM, Impact of genetic background on placental glycogen storage in mice, Placenta 33(2) (2012) 124–7. [DOI] [PubMed] [Google Scholar]
- [29].Fan G, Sun L, Shan P, Zhang X, Huan J, Zhang X, Li D, Wang T, Wei T, Zhang X, Gu X, Yao L, Xuan Y, Hou Z, Cui Y, Cao L, Li X, Zhang S, Wang C, Loss of KLF14 triggers centrosome amplification and tumorigenesis, Nat Commun 6 (2015) 8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Shankar K, Zhong Y, Kang P, Blackburn ML, Soares MJ, Badger TM, Gomez-Acevedo H, RNA-seq analysis of the functional compartments within the rat placentation site, Endocrinology 153(4) (2012) 1999–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rawn SM, Cross JC, The evolution, regulation, and function of placenta-specific genes, Annu Rev Cell Dev Biol 24 (2008) 159–81. [DOI] [PubMed] [Google Scholar]
- [32].Simmons DG, Cross JC, Determinants of trophoblast lineage and cell subtype specification in the mouse placenta, Dev Biol 284(1) (2005) 12–24. [DOI] [PubMed] [Google Scholar]
- [33].Lefebvre L, Viville S, Barton SC, Ishino F, Keverne EB, Surani MA, Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest, Nat Genet 20(2) (1998) 163–9. [DOI] [PubMed] [Google Scholar]
- [34].Charalambous M, Cowley M, Geoghegan F, Smith FM, Radford EJ, Marlow BP, Graham CF, Hurst LD, Ward A, Maternally-inherited Grb10 reduces placental size and efficiency, Dev Biol 337(1) (2010) 1–8. [DOI] [PubMed] [Google Scholar]
- [35].Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A, Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism, Proc Natl Acad Sci U S A 100(14) (2003) 8292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Coan PM, Fowden AL, Constancia M, Ferguson-Smith AC, Burton GJ, Sibley CP, Disproportional effects of Igf2 knockout on placental morphology and diffusional exchange characteristics in the mouse, J Physiol 586(20) (2008) 5023–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lomberk G, Urrutia R, The family feud: turning off Sp1 by Sp1-like KLF proteins, Biochem J 392(Pt 1) (2005) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Small KS, Todorcevic M, Civelek M, El-Sayed Moustafa JS, Wang X, Simon MM, Fernandez-Tajes J, Mahajan A, Horikoshi M, Hugill A, Glastonbury CA, Quaye L, Neville MJ, Sethi S, Yon M, Pan C, Che N, Vinuela A, Tsai PC, Nag A, Buil A, Thorleifsson G, Raghavan A, Ding Q, Morris AP, Bell JT, Thorsteinsdottir U, Stefansson K, Laakso M, Dahlman I, Arner P, Gloyn AL, Musunuru K, Lusis AJ, Cox RD, Karpe F, McCarthy MI, Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition, Nat Genet 50(4) (2018) 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Truty MJ, Lomberk G, Fernandez-Zapico ME, Urrutia R, Silencing of the transforming growth factor-beta (TGFbeta) receptor II by Kruppel-like factor 14 underscores the importance of a negative feedback mechanism in TGFbeta signaling, J Biol Chem 284(10) (2009) 6291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Barak Y, Sadovsky Y, Shalom-Barak T, PPAR Signaling in Placental Development and Function, PPAR Res 2008 (2008) 142082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Larkin JC, Sears SB, Sadovsky Y, The influence of ligand-activated LXR on primary human trophoblasts, Placenta 35(11) (2014) 919–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Plosch T, van Straten EM, Kuipers F, Cholesterol transport by the placenta: placental liver X receptor activity as a modulator of fetal cholesterol metabolism?, Placenta 28(7) (2007) 604–10. [DOI] [PubMed] [Google Scholar]
- [43].Gonzalez CR, Vallcaneras SS, Calandra RS, Gonzalez Calvar SI, Involvement of KLF14 and egr-1 in the TGF-beta1 action on Leydig cell proliferation, Cytokine 61(2) (2013) 670–5. [DOI] [PubMed] [Google Scholar]
- [44].Shalom-Barak T, Liersemann J, Memari B, Flechner L, Devor CE, Bernardo TM, Kim S, Matsumoto N, Friedman SL, Evans RM, White JH, Barak Y, Ligand-Dependent Corepressor (LCoR) Is a Rexinoid-Inhibited Peroxisome Proliferator-Activated Receptor gamma-Retinoid X Receptor alpha Coactivator, Mol Cell Biol 38(9) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, Greenblatt J, Frey BJ, Hughes TR, C2H2 zinc finger proteins greatly expand the human regulatory lexicon, Nat Biotechnol 33(5) (2015) 555–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.