Abstract
Purpose
The purpose of this study was to perform a detailed longitudinal phenotyping of X-linked retinitis pigmentosa (RP) caused by mutations in the RPGR gene during a long follow-up period.
Methods
An Italian cohort of 48 male patients (from 31 unrelated families) with RPGR-associated RP was clinically assessed at a single center (mean follow-up = 6.5 years), including measurements of best-corrected visual acuity (BCVA), Goldmann visual field (GVF), optical coherence tomography (OCT), fundus autofluorescence (FAF), microperimetry, and full-field electroretinography (ERG).
Results
Patients (29.6 ± 15.2 years) showed a mean BCVA of 0.6 ± 0.7 logMAR, mostly with myopic refraction (79.2%). Thirty patients (62.5%) presented a typical RP fundus, while the remaining sine pigmento RP. Over the follow-up, BCVA significantly declined at a mean rate of 0.025 logMAR/year. Typical RP and high myopia were associated with a significantly faster decline of BCVA. Blindness was driven primarily by GVF loss. ERG responses with a rod-cone pattern of dysfunction were detectable in patients (50%) that were significantly younger and more frequently presented sine pigmento RP. Thirteen patients (27.1%) had macular abnormalities without cystoid macular edema. Patients (50%) with a perimacular hyper-FAF ring were significantly younger, had a higher BCVA and a better-preserved ellipsoid zone band than those with markedly decreased FAF. Patients harboring pathogenic variants in exons 1 to 14 showed a milder phenotype compared to those with ORF15 mutations.
Conclusions
Our monocentric, longitudinal retrospective study revealed a spectrum disease progression in male patients with RPGR-associated RP. Slow disease progression correlated with sine pigmento RP, absence of high myopia, and mutations in RPGR exons 1 to 14.
Keywords: RPGR, X-linked retinitis pigmentosa, sine pigmento retinitis pigmentosa, myopia, ORF15
Retinitis pigmentosa (RP) is the most common form of inherited retinal dystrophy with a prevalence of 1:3500 to 1:4000.1 The main cellular hallmark of RP is a progressive degeneration of photoreceptors that first leads to night blindness and loss of peripheral visual field, and subsequently may cause legal blindness.1 RP is more frequently observed as an isolated form (also called simplex RP), without extra-ocular involvement.2 It is characterized by high genetic heterogeneity, with over 130 genes reported to cause either isolated (approximately 90 genes) or syndromic forms (approximately 40 genes; http://www.sph.uth.tmc.edu/RetNet; accessed on April 2020). The majority of RP genes follow an autosomal recessive inheritance (AR). Nevertheless, autosomal dominant (AD) and X-linked (XL) patterns of inheritance (XLRP) are observed in a significant number of cases.2
Approximately 15% of RP cases are inherited as XL traits and are usually associated with both an earlier onset and a more severe phenotype.3–5 Male patients with XLRP usually present night blindness, myopia (often since childhood), progressive loss of peripheral visual fields, and subsequent loss of central vision in relation with severe rod and cone dysfunction.6 Disease progression in male patients with XLRP is one of the most rapid among all RP, reaching legal blindness by the third to the fourth decade.5,7 Other clinical hallmarks of XLRP include bone spicule pigmentary changes, attenuated retinal blood vessels, optic disc pallor, visual field loss, and diminished or non-recordable electroretinogram (ERG) responses.8 Female carriers show a spectrum of clinical manifestations that range from asymptomatic to severe, which can in some instances lead to the misdiagnosis of an AD mode of inheritance based on the evaluation of the family pedigree.9–12 The main genes associated with XLRP are the Retinitis Pigmentosa GTPase Regulator gene (RPGR; OMIM # 312610), accounting for up to 80% of XLRP cases,5,13,14 and the Retinitis Pigmentosa 2 (RP2; OMIM # 312600) responsible for approximately 5% to 15% of cases.15 Additional genes account only for a limited fraction of XLRP cases. For example, a severe form of XLRP (RP23) in a single family was associated with a deep intronic variant in the OFD1 Centriole and centriolar satellite protein (OFD1; OMIM # 300170), which is most commonly responsible for the X-linked dominant Oral-facial-digital syndrome type 1.16 Moreover, XLRP combined with variable degrees of neurological symptoms and hearing loss was reported in women heterozygous for a novel loss-of-function mutation in the phosphoribosyl pyrophosphate synthetase 1 gene (PRPS1; OMIM # 311850).17
RPGR was identified by positional cloning in the RP3 locus on chromosome Xp11.4 (OMIM # 300029).18,19 The RPGR gene has 19 exons and undergoes extensive alternative splicing that generates at least ten different isoforms.14,20–23 The two main RPGR isoforms are the constitutively expressed RPGR,1–19 composed of 19 exons and encoding an 815-aa protein, and the retina-specific RPGRORF15 isoform, composed of 15 exons and encoding a 1152-aa protein.18 Both isoforms share exons 1 to 14, encoding a regulator of chromosome condensation 1 (RCC1)-like domain essential to interact with binding partners,24 but diverge at their carboxyl-terminal ends. In particular, the terminal exon (open reading frame 15; ORF15) of the RPGRORF15 isoform contains a repetitive, purine-rich sequence coding for a low complexity stretch of 567-aa with a high glutamic acid and glycine (Glu/Gly) content.23 Until now, more than 350 sequence variants have been described in RPGR25 and almost 60% of them are found in ORF15.26 The high mutation frequency in ORF15 (mainly small deletions or duplications) is most likely due to the nucleotide composition and repetitive nature of its sequence, which might induce errors during DNA replication.27
To date, genotype-phenotype correlations have been investigated in several RPGR patient cohorts, yet a consensus remains to be reached. Studies suggested a more severe RP phenotype in patients with mutations in exons 1 to 14 compared to patients with variants in ORF15.7,8,28–30 However, other reports described a milder clinical presentation in patients carrying variants in exons 1 to 14 compared to those with ORF15 mutations.31,32 A common ground was found instead regarding RPGR variants located toward the 3′ end of ORF15, which were more frequently associated with milder XL Cone Rod Dystrophy (CRD) / XL Cone Dystrophy (CD) phenotypes.25,30,33–35 Finally, some RPGR variants in exons 1 to 14 were reported in patients with RP presenting progressive hearing loss, sinusitis, and chronic recurrent respiratory tract infections,36–38 in line with the more widespread expression of the RPGR1–19 isoform and its essential ciliary function in other organs apart from retina.39,40
In this study, we report the clinical and genetic findings in a cohort of 48 male patients of Italian origin with XLRP caused by variants in RPGR. We describe the spectrum of retinal phenotypes encountered in this cohort and the longitudinal progression of RPGR-associated RP. We also sought to identify clinical parameters that could reliably predict disease progression. In that respect, our observations gain strength by the fact that all subjects were diagnosed and followed up at a single center. Ultimately, these results can be useful for selecting and prioritizing patients in clinical trials, as well as for establishing effective read-outs to assess treatment outcomes.
Methods
Ethics Statement
All procedures adhered to the tenets of the Declaration of Helsinki and were approved by the Ethics Board of the Università degli Studi della Campania “Luigi Vanvitelli” (for adult protocol no. 8189/2015, 09.04.2015; for pediatric subjects’ protocol no. 500/2017, 12.09.2017). The cited protocols aimed to investigate genotype-phenotype correlations in inherited retinal dystrophies. Peripheral blood samples were collected upon written informed consent of the subjects for sample collection and genetic analysis. For minors, informed consent was obtained by the parents or legal guardians.
Patients’ Inclusion Criteria
In this study, we analyzed subjects who were recruited in the above-cited protocols and satisfied the following inclusion criteria: only male patients with a clinical diagnosis of RP and disease-causing variants in the RPGR gene. Therefore, we excluded female subjects (i.e. obligate female carriers, which usually show no or limited signs of the disease) and patients with RPGR-associated CD or CRD. Patients belonging to the same family were included in the study and assigned the same family identification number.
The clinical diagnosis of RP was formulated according to the criteria described by Hamel.1 Specifically, the diagnosis requires a dramatic reduction in a- and b-wave amplitudes at the ERG (with a rod-cone pattern of photoreceptor dysfunction at the early stages of the disease) associated with one or more of the following alterations: night blindness and/or photophobia even with preserved visual acuity in the early and mid-stages of the disease; visual field changes such as irregular loss of peripheral visual field evolving to ring scotoma, and eventually to tunnel vision; fundus abnormalities, such as pigmentary deposits resembling bone spicules, initially in the mid peripheral retina (hereafter referred to as typical RP fundus appearance), an attenuation of the retinal vessels, as well as waxy pallor of the optic disc accompanied by various degrees of retinal atrophy at the fundus examination. Based on the fundus appearance, RP is also defined as sine pigmento, sector, and pericentral on the basis of presence and localization of bone spicule pigmentary deposits.1,41
Disease onset was defined as the self-reported age of the first symptoms (i.e. night vision problems and/or visual abnormalities). The disease length was calculated over time between the self-reported onset of symptoms and the date of the examination. In case of incidental diagnosis, the age at diagnosis was considered as the disease onset.
Ophthalmological Examination
The medical records of all the included subjects seen at the Referral Center for Inherited Retinal Dystrophies of the Eye Clinic, University of Campania “Luigi Vanvitelli” were reviewed to extract the findings of the following ophthalmological examinations: best-corrected visual acuity (BCVA) measurements with the Snellen visual chart, slit lamp anterior segment examination, measurements of intraocular pressure, fundus examination, Goldmann visual field (GVF) examination, optical coherence tomography (OCT), Fundus Autofluorescence (FAF), Microperimetry (MP1), and standard full-field ERGs.
Spherical equivalent, defined as the sum of the sphere and one half the cylinder power and averaged between the two eyes, was used to calculate refractive error in diopters (D). Refractive errors were classified into high myopia (≤ −6 D), moderate myopia (> −6 D to ≤ −3 D), low myopia (< −3 D to ≤ −0.75 D), emmetropia (> −0.75 D to < 0.75 D), low hyperopia (≥ 0.75 D to < 3 D), medium hyperopia (≥ 3 D to < 6 D), and high hyperopia (≥ 6 D), using the criteria defined by the CREAM consortium (CREAM consortium meeting, 2012, Sardinia, Italy).
GVF was measured by moving the III4e and V4e stimulus target on a calibrated standard Goldmann perimeter by the same experienced ophthalmic technician. Legal blindness based on GVF was determined following the indications by the International Classification of Diseases (ICD-10 Version 2016; International Statistical Classification of Diseases and Related Health Problems 10th Revision [ICD-10]-World Health Organization [WHO] Version 2016; i.e. central III4e GVF area in the better-seeing eye no greater than 314 degrees,2 corresponding to an equivalent radius of 10 degrees).
OCT was performed by experienced operators either using the Heidelberg Spectralis OCTPlus with BluePeak (Heidelberg Engineering, Heidelberg, Germany) or the Cirrus HD-OCT (Carl Zeiss, Dublin, CA, USA). The macular abnormalities (MAs) revealed by OCT scans were classified for cystoid macular edema (CME), vitreo-macular traction syndrome (VMT), epiretinal membrane (ERM), full-thickness macular hole (FTMH), lamellar macular hole (LMH) and tractional macular edema (TME), as previously described.42 The mean macular thickness (MMT) and the length of the ellipsoid zone band (EZ)43,44 (previously known as the IS/OS line) were measured using SD-OCT performed on the Heidelberg Spectralis OCTPlus with the BluePeak imaging platform. The measurements were performed manually by two operators and revised by a third one in case of discrepancies. FAF imaging of the 30 degrees field was acquired with the Heidelberg Spectralis OCTPlus with BluePeak (Heidelberg Engineering, Heidelberg, Germany). Following previous studies,34,45 the FAFs were classified on the basis of the presence of a hyperautofluorescent ring or an abnormal central hypoautofluorescence.
MP1 was performed by an automatic fundus-related perimeter (MP1 Microperimeter, Nidek Technologies, Padova, Italy). The following parameters were used: a fixation target of 2 degrees in diameter consisting of a red ring; a white, monochromatic background with a luminance of 1.27 cd/m²; a Goldmann III-size stimulus with a projection time of 200 ms; and a predefined automatic test pattern (Humphrey 10-2) covering 10 degrees centered onto the gravitational center of the fixation points with 68 stimuli.
Full-field ERG was recorded using corneal contact lens electrodes with a Ganzfeld stimulator (Roland Consult, Brandenburg, Germany) according to the standards of the International Society for Clinical Electrophysiology of Vision.46
Next Generation Sequencing and Variant Interpretation
Genomic DNA was extracted from peripheral blood using the DNeasy Blood & Tissue Kit (QIAGEN) according to the manufacturer's instructions. Panel-based sequencing (RETplex) was performed as previously described.47 Libraries for clinical exome and whole-exome sequencing were prepared using the ClearSeq Inherited Disease Panel (Agilent Technologies) and the SureSelect Human All Exon V7 (Agilent Technologies), respectively. Targeted regions were enriched using the SureSelectQXT Target Enrichment system (Agilent Technologies). Libraries were run on a NextSeq500 sequencing platform (Illumina Inc., San Diego, CA, USA). Sequencing data were analyzed using an already reported pipeline.48 Only variants with a minor allele frequency (MAF) < 0.05 in the Genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org) were considered. Variant interpretation and analysis was performed as previously described.49 The identified variants were validated by Sanger sequencing of the corresponding genomic fragments. For the amplification of the exon sequence, PCR was performed on 20 ng of genomic DNA using AmpliTaq Gold polymerase (Thermo Fisher Scientific) according to standard protocols. Amplicons were Sanger sequenced and sequences were aligned to the reference genome (hg19) using the UCSC Genome Browser (http://genome.ucsc.edu/). Mutation detection was performed using the CodonCode Aligner software. The novel variants identified in this study have been deposited to the Leiden Open Variation Database (LOVD version 3.0; https://www.lovd.nl/).
ORF15 Amplification and Sanger Sequencing
The ORF15 sequence was analyzed similarly to previously described procedures.50 We amplified the ORF15 genomic region (NM_001034853.1) by PCR on 200 ng genomic DNA using the HOT FIREPol DNA Polymerase (Solis BioDyne) and the oligonucleotide primers R1Ampl and R2Ampl (Supplementary Table S1). Solution S (Solis BioDyne) was added in the PCR mix at a final concentration of 3X. The following cycling parameters were used: an initial denaturation step (95°C, 15 minutes) followed by 45 cycles at 95°C for 45 seconds, 60°C for 1 minute and 72°C for 3 minutes and 30 seconds, with a final extension of 10 minutes at 72°C. After a PCR clean-up step (ExoSAP-IT), amplicons were Sanger sequenced using the BigDye Terminator version 1.1 Cycle Sequencing Kit (Thermo Fisher Scientific) and the following primers: R7bSeq, R8bSeq, R9Seq, R5Seq, R4Seq, and R11Seq (Supplementary Table S1). Annealing and extension temperatures are reported in Supplementary Table S1.
Criteria for Genotype-Phenotype Correlation
To explore possible genotype-phenotype correlations, patients’ genotypes were stratified according to the position of the variants in the major RPGR isoform (i.e. variants in exons 1–14 versus variants in ORF15). Variants in exons 1 to 14 were further classified according to the mutation type (i.e. null variants, splice variants, missense variants). Regression models were applied as detailed below.
Statistical Analysis
Continuous variables are reported as mean ± standard deviation (SD) and categorical variables are reported as counts (percentage).
The natural history of disease was analyzed using previously applied methods.51,52 In order to include all the patients (i.e. also those with no longitudinal data), we performed a cross-sectional analysis on the data of the last visit for each of the selected outcome measures (e.g. BCVA, MMT, and EZ band width). In particular, linear regression models, estimated by a generalized estimating equation (GEE), were performed with each outcome measure as the dependent variable and age as the independent variable in order to estimate a slope (mean rate of change per year of age) with its relative standard error (SE).
Moreover, regression models were fitted with each outcome measure as the dependent variable and high myopia, sine pigmento fundus, gene region harboring the mutation (exons 1–14 versus ORF15), mutation type (i.e. null variants in exons 1–14, splice variants in exons 1–14, or variants in ORF15) as independent variable, in order to investigate whether these factors influenced the selected outcome. We also controlled for age (added as an additional independent variable) when a variable was shown to be age-related. To this regard, the Student's t-test was used to evaluate if the mean age of the patients, self-reported age of onset, and disease length were different between the following subgroups: patients with high myopia versus those without high myopia; patients with typical RP versus patients with sine pigmento; and patients with variants in exons 1 to 14 versus those with variants in ORF15. Analysis of variance (ANOVA) with Bonferroni correction for multiple comparisons performed with t-tests was adopted to evaluate differences in the age between the patients classified according to lens status and according to the mutation type. Fisher exact tests and Pearson χ2 tests were adopted to explore differences for dichotomous and categorical variables, respectively. For dichotomous or categorical variables related to age, logistic regression models were fitted including age as an additional independent variable.
Moreover, GEE were fitted on longitudinal data, using baseline values as offset and follow-up length expressed in years as an independent variable, to estimate the annual change of the BCVA, MMT, and the EZ band width over the follow-up period. The analysis of BCVA was conducted separately in the best-seeing eyes and in the worst-seeing eye in order to assess the symmetry in disease progression. Moreover, asymmetry in BCVA between the two eyes was defined as a difference of 0.3 logMAR (15 Early Treatment Diabetic Retinopathy Study [ETDRS] letters), which is the threshold for clinical significance of BCVA changes.53 Finally, regression models, including the interaction between follow-up length and the following factors: high myopia, sine pigmento fundus, gene region affected by the mutation, and mutation type, were fitted in order to investigate whether these factors influenced the decline of BCVA. We controlled for age (added as an additional independent variable) when a factor has been shown to be age-related.
GEE were applied because this method could accommodate the inter-eye correlation (i.e. between the two eyes of the same subject at a given visit) and longitudinal correlation (i.e. between values of the same eye followed over time) by adopting an appropriate covariance structure, as previously described.54
For the statistical analysis, BCVA were converted to logMAR using the values of 2.7 for hand motion, 2.8 for light perception, and 2.9 for no light perception. All the other measures (e.g. MMT and EZ band width) were also log-transformed to estimate the exponential rate of progression. Intercepts were included in all the models, except those regarding BCVA, because we assume that BCVA should start with the value of 0 logMAR (corresponding to healthy status).
Finally, a Kaplan-Meier survival analysis was performed to show the time to low vision, blindness based on visual acuity, and blindness based on visual field. We adopted the definition proposed by the ICD-10 version 2016: BCVA worse than 20/67 in the better-seeing eye for low vision; BCVA worse than 20/400 in the better-seeing eye for blindness based on visual acuity; III4e GVF area in the better-seeing eye no greater than 314 degrees2 (i.e. corresponding to an equivalent radius of 10 degrees) for blindness based on visual field. The relative distributions were compared with the Log Rank (Mantel-Cox) test.
For the analyses involving only clinical parameters, all the patients were included assuming that the observations could be considered as independent given the known intrafamiliar variability. On the other hand, the analyses involving genotype were performed only on unrelated patients. In other words, only one patient from each family was included (i.e. the first diagnosed patient; assigned the lowest ID number among the family components). Related patients were excluded to reduce the risk of findings biased toward families with higher numbers of affected subjects.
The P values (P) lower than 0.05 were considered statistically significant. The reported P values relative to the t-tests incorporate the Bonferroni correction in case of multiple comparisons. These statistical analyses were performed using the IBM SPSS Statistics platform (version 21.0.0.0).
Results
Clinical Characterization of the RPGR-RP Cohort
A total of 48 patients (mean age = 29.6 ± 15.2 years) from 31 families with a clinical diagnosis of RP and harboring a disease-causing mutation in the RPGR gene were involved in this clinical study. Two patients with disease-causing mutations in the RPGR gene but with a clinical diagnosis of CD were not included in the above cohort as they did not meet the inclusion criteria.
The main clinical findings of the cohort are summarized in Table 1. The age at onset of the first symptom was 6.9 ± 4.6 years (median = 5 years; range = 1–21 years; interquartile range = 2.5 years). Almost all patients (45; 93.8%) reported that night blindness was present at disease onset, whereas reduction of visual field (12; 25.0%) and decreased visual acuity (5; 10.4%) were less frequently observed at disease onset. A typical RP fundus was found in the majority of patients (30; 62.5%), whereas the sine pigmento RP form was detected in 37.5% of patients (n = 18; Fig. 1). Patients with sine pigmento RP were significantly younger compared to patients with a typical RP fundus (20.4 ± 6.9 years versus 35.1 ± 16.2 years; P = 0.001).
Table 1.
Clinical Findings in the Patients With RPGR-associated RP
| Parameters | Study Cohort (n = 48) | |
|---|---|---|
| Age, y | 29.6 ± 15.2 | |
| Self-reported age of onset, y | 6.9 ± 4.6 | |
| Disease length, y | 22.7 ± 15.4 | |
| Mean refractive error (D) | −4.0 ± 3.1 | |
| High myopia | 16 (33.3%) | |
| Typical RP | 30 (62.5%) | |
| Vitreomacular alteration | 13 (27.1%) | |
| Detectable photopic ERG | 24 (50.0%) | |
| Right Eye | Left Eye | |
| BCVA, logMAR | 0.63 ± 0.79 | 0.62 ± 0.74 |
| EZ-band width, µm | 1319.0 ± 685.9 | 1326.0 ± 571.9 |
| MMT, µm | 226.7 ± 43.1 | 224.6 ± 44.4 |
| Photopic ERG (b-wave amplitude), µV | 20.7 ± 14.0 | 24.0± 17.5 |
| Photopic ERG (b-wave implicit time), ms | 43.5 ± 2.8 | 43.1 ± 4.4 |
| 30 Hz Flicker ERG (trough-to-peak amplitude), µV | 11.8 ± 9.1 | 11.7 ± 10.7 |
| 30 Hz Flicker ERG (implicit time), ms | 46.6 ± 8.5 | 45.8± 8.0 |
| MS, dB | 2.6 ± 3.3 | 3.8 ± 4.4 |
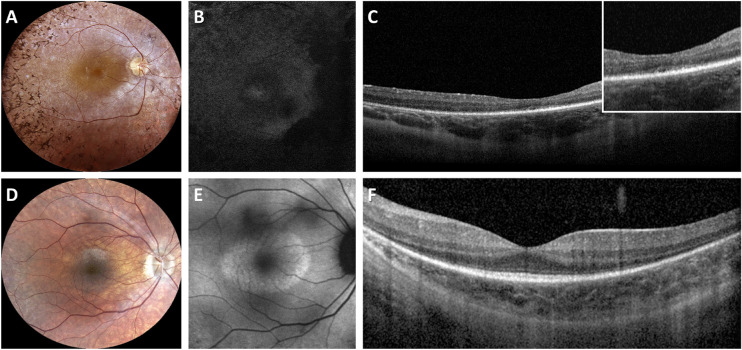
Figure 1.
Multimodal imaging findings in two representative patients with RPGR-associated RP. (A) Fundus photograph of a 31-year-old patient with typical RP (ID no. 13; BCVA 0,3; NM_001034853: c.1245+3A>T) showing bone-spicule pigment deposits in the mid peripheral retina, optic disc pallor and RPE atrophy. (B) Fundus autofluorescence (FAF) image showing a widespread hypoautofluorescence. (C) Spectral domain OCT revealing RPE atrophy. The inset shows the disruption of the ellipsoid zone (EZ) band. (D) Fundus photograph of a 25-year-old patient with sine pigmento RP (ID no. 37; BCVA 0,7; NM_001034853: c.1059+1G>A) showing absence of bone-spicule-like pigment migration, optic disc pallor, and mild RPE atrophy. (E) FAF image showing a hyperautofluorescent ring around the central macula. (F) Spectral domain OCT showing central sparing of the EZ band that is attenuated toward the peripheral macula.
The most recently measured mean spherical equivalent, averaged between eyes, was −4.0 ± 3.1 D (range = −12 D to +1.25 D). Most patients (39; 79.2%) were myopic. In particular, high, moderate, and low myopia were observed in 16 (33.3%), 12 (25.0%), and 11 (22.9%) patients, respectively. Eight patients (16.7%) were emmetropic and only one patient showed a low hyperopia (+1.25 D in both eyes). There was no statistically significant difference (P = 0.058) in the frequency of high myopia between typical RP and sine pigmento RP. Cataract or pseudophakia was found in 21 patients (43.7%) and we observed a significant association between cataract/pseudophakia and age (P < 0.001). Specifically, patients with clear lenses in both eyes were younger (20.1 ± 6.3 years) than those with cataract (34.1 ± 8.3; P < 0.001) or patients with pseudophakia (52.0 ± 15.3 years; P < 0.001).
In the study cohort, at the most recent visit, mean BCVA was 0.63 ± 0.79 logMAR in the right eyes and 0.62 ± 0.74 logMAR in the left eyes. The analysis of inter-eye asymmetry values showed a low asymmetry in the BCVA (median = 0.06 logMAR, equivalent to 3 ETDRS letters) with only eight patients (16.7%) recording higher values than the established threshold for clinically significant changes (i.e. 0.3 logMAR).
Longitudinal analysis of BCVA data collected in 41 patients over a mean follow-up period of 6.5 years (SD = 4.4; range = 1–18 years; median = 5 years; average number of visits = 5), reported in Table 2, showed a significant decline of 0.025 logMAR/year (SE = 0.012; P = 0.047) equivalent to about 1 ETDRS letter/year in the best-seeing eyes. A faster decline was observed in the worst-seeing eyes (i.e. 0.043 logMAR/year; P < 0.001), equivalent to two ETDRS letters. We observed a significantly faster decline of BVCA (P = 0.014) in patients with typical RP fundus appearance (0.060 logMAR/year; SE = 0.146) compared to patients with sine pigmento RP (0.011 logMAR/year; SE = 0.033). Moreover, BCVA declined significantly faster (P = 0.035) in patients with high myopia (0.081 logMAR/year; SE = 0.030) compared to those without high myopia (0.017 logMAR/year; SE = 0.039). Based on these observations, we then explored the combined effect of the sine pigmento RP and high myopia on BCVA. We found a significant slower BCVA decline (P < 0.05) in patients with sine pigmento RP and without high myopia (0.009 logMAR/year; SE = 0.004), followed by those with sine pigmento RP and high myopia (0.012 logMAR/year; SE = 0.0012). A faster BCVA decline was observed in patients that presented a typical RP fundus appearance in the absence of high myopia (0.020 logMAR/year; SE = 0.005), whereas the highest annual rate of BCVA change was observed in patients with a typical RP fundus and high myopia (0.098 logMAR/year; SE = 0.041). Finally, considering that patients with sine pigmento RP were significantly younger compared to patients with typical RP, we repeated the above analysis taking into account only observations within the same age range (i.e., 6 to 30 years) and we found a significantly (P < 0.001) faster decline of BCVA in patients with typical RP (0.024 logMAR/year; SE = 0.004) compared to patients with sine pigmento RP (0.007 logMAR/year; SE = 0.0006). Moreover, the regression model, including as covariate the age at the study baseline, confirmed a significantly faster progression (P = 0.017) in patients with typical RP compared to patients with sine pigmento RP.
Table 2.
Main Findings of the Longitudinal Analysis of BCVA in Patients With RPGR-RP
| Mean Annual Rate | P Value | |||
|---|---|---|---|---|
| (95% Confidence Interval) | Standard | (Comparison Between | ||
| Group | [logMAR/Year] | Error | P Value | Subgroups) |
| Overall cohort (best-seeing eye)* | 0.025 (0.0004 to 0.050) | 0.012 | 0.047 | n.a. |
| Overall cohort | 0.043 (0.023 to 0.063) | 0.010 | < 0.001 | n.a. |
| Patients with typical RP | 0.060 (0.031 to 0.088) | 0.146 | < 0.001 | 0.014 |
| Patients with sine pigmento RP | 0.011 (0.004 to 0.017) | 0.033 | 0.001 | |
| Patients with high myopia | 0.081 (0.022 to 0.141) | 0.030 | 0.007 | 0.035 |
| Patients without high myopia | 0.017 (0.009 to 0.025) | 0.039 | < 0.001 | |
| Patients with sine pigmento RP and without high myopia | 0.009 (0.008 to 0.010) | 0.004 | < 0.001 | 0.002 |
| Patients with sine pigmento RP and with high myopia | 0.012 (0.010 to 0.015) | 0.0012 | < 0.001 | |
| Patients with typical RP and without high myopia | 0.020 (0.010 to 0.030) | 0.005 | < 0.001 | |
| Patients with typical RP and with high myopia | 0.098 (0.017 to 0.179) | 0.041 | 0.017 | |
| Patients aged 6–30 years with typical RP | 0.024 (0.016 to 0.032) | 0.004 | < 0.001 | < 0.001 |
| Patients aged 6–30 years with sine pigmento RP | 0.007 (0.006 to 0.008) | 0.0006 | < 0.001 |
The progression is estimated on the worst-seeing eye unless otherwise specified.
n.a., non applicable.
To assess the risk of developing low vision and blindness with ageing, a Kaplan-Meier curve analysis (hereafter referred to as “survival analysis”) was performed in the entire cohort. This analysis indicated that the patients reached low vision and blindness based on BCVA at the median age of 48.2 years and 51.3 years, respectively (Fig. 2). Survival analysis of blindness based on GVF suggested that the development of blindness was driven primarily by GVF loss. Specifically, the survival curve based on a GVF not greater than 10 degrees (median age = 26.4 years; 95% confidence interval [CI] = 21.1–31.6 years) was significantly shifted (P < 0.001) to younger ages when compared with a survival curve based on BCVA (median age = 51.3 years; 95% CI = 49.1–53.4 years).
Figure 2.

Survival curves of low vision and legal blindness (based on visual acuity and visual field) for patients with RPGR - associated RP. Kaplan–Meier plot showing the risk of developing low vision (green) and legal blindness, based on visual field (blue) or on visual acuity (red), with age. Survival analyses showed that legal blindness based on visual field is reached at a median age of 26.4 years (blue circle), low vision at 48.2 years (green circle), and legal blindness based on visual acuity at 51.3 years (red circle).
Scotopic and photopic ERG responses were not detectable in 24 out of 48 patients. The remaining half showed a rod-cone pattern of photoreceptor dysfunction with undetectable scotopic and markedly abnormal photopic ERG responses. In particular, the mean b-wave amplitude of light-adapted 3.0 ERG was 20.7 ± 14.0 µV in right eyes and 24 ± 17.5 µV in left eyes, whereas the mean trough-to-peak amplitude of 30 Hz flicker ERG was 11.8 ± 9.1 µV in right eyes and 11.7 ± 10.7 µV in left eyes. Moreover, the light-adapted 3.0 ERG were delayed (normal range = 29–33 ms) with a mean implicit of 43.5 ± 2.8 ms in right eyes and 43.1 ± 4.4 ms in left eyes. Similarly, the 30 Hz flicker ERG showed longer implicit time than the normal range (46.6 ± 8.5 ms in right eyes and 45.8 ± 8.0 ms in left eyes; normal range = 25–31 ms).
The patients with a rod-cone pattern were significantly younger than those with undetectable ERG (24.8 ± 13.6 years versus 34.4 ± 15.5 years; P = 0.027). Moreover, a detectable photopic response was significantly associated with sine pigmento RP (13/18, 72.2% vs. 11/30, 36.7%; P = 0.018), also in the age-adjusted model (P = 0.032), whereas no association was found with high myopia (8/16, 50.0% vs. 16/32, 50.0%; P = 1.000).
Microperimetry examination, which was available for 24 patients (50.0%), showed a markedly reduced macular sensitivity (MS; 2.6 ± 3.3 dB in the right eyes; 3.8 ± 4.4 dB in the left eyes; normal value: 19.6 ± 0.4 dB). According to the evaluation in the eye with the best fixation stability, 17 patients (34.7%) showed stable fixation, 5 patients (10.2%) showed a relatively stable fixation, and 2 patients (4.1%) showed an unstable fixation. We did not identify any significant association of MS or fixation stability with age nor significant differences in their relationship with other clinical factors (i.e. high myopia and RP form).
Spectral domain OCT scans revealed MA in 27.1% of the patients analyzed (13/48). In particular, 12 patients (25.0%) had an ERM in at least one eye (7 bilateral; 5 monolateral) and one patient (2.1%) had a bilateral VMT. The patients with a MA were significantly older than patients without such alterations (39.5 ± 14.1 years vs. 25.9 ± 14.0 years; P = 0.005). The frequency of MA was similar in patients with and without high myopia (25.0% vs. 28.1%; P = 0.553) as well as in patients with typical RP and patients with sine pigmento RP (25.0% vs. 29.2%; P = 1.000). None of the patients presented a CME or had a history of CME (i.e. according to past medical records or in the OCT scans performed during the clinical follow-up).
Combined spectral domain OCT and fundus autofluorescence imaging were available for 28 patients (58.3%). The EZ band was detectable in all 28 patients and the mean width of the EZ band was 1319.0 ± 685.9 µm in the right eyes and 1326.0 ± 571.9 µm in the left eyes. The EZ band became significantly thinner with age at a linear rate of −27.5 µm/year (95% CI = −42.1 to −12.8 µm/year; P < 0.001) and at an exponential rate of −2.8%/year (95% CI = −4.4 to −1.1/year; P = 0.002). Spectral domain OCT scans acquired exclusively with the Heidelberg Spectralis OCTPlus with BluePeak showed that MMT, on average, was 226.7 ± 43.1 µm in the right eyes and 224.6 ± 44.4 µm in the left eyes. The progression of MMT with age did not reach statistical significance (P > 0.7) in both linear and exponential regression models. Analysis of follow-up data (available only in 8 patients) showed a loss of EZ band width at a linear rate of −183.7 µm/year (95% CI = −226.5 to −100.8 µm/year; P < 0.001) and at an exponential rate of −8.9%/year (95% CI = −12.8 to −4.8%/year; P < 0.001). Moreover, we observed a reduction of MMT at a linear rate of −5.8 µm/year (95% CI = −9.7 to −1.9 µm/year; P = 0.003) and at an exponential rate of −2.4%/year (95% CI = −4.2 to −0.6%/year; P = 0.011).
Based on the FAF imaging, we identified 14 patients (14/28; 50%) with a hyper-AF ring around the macula and no other significant FAF abnormalities. The remaining patients had a marked FAF decrease in the periphery with some degree of FAF in the posterior pole, whereas one patient showed an almost complete absence of FAF in the entire retina. We observed a significant difference when comparing the group of patients with a hyper-AF ring with those displaying a marked FAF decrease. The patients with a hyper-AF ring were younger than those with decreased FAF (17.5 ± 5.8 vs. 30.1 ± 14.6; P = 0.006; Table 3). They also had significantly better BCVA (β = −0.43 logMAR; P < 0.001) and a wider EZ band (β = 843 µm; P < 0.001). The frequency of MA was significantly lower in the patients with a hyper-AF ring compared to those with markedly decreased FAF. However, after correction for age, only the differences in EZ band width remained statistically significant (P < 0.001).
Table 3.
Comparison Between Different FAF Patterns in Patients With RPGR-RP
| Parameters | Hyper-AF Ring (n = 14) | Decreased FAF (n = 14) | P Value | |||
|---|---|---|---|---|---|---|
| Age, y | 17.5 ± 5.8 | 30.1 ± 14.6 | 0.006 | |||
| Self-reported age of onset, y | 5.5 ± 3.6 | 7.4 ± 4.9 | 0.247 | |||
| Disease length, y | 12.0 ± 5.8 | 22.7 ± 15.6 | 0.023 | |||
| Mean refractive error, D | −3.3 ± 0.7 | −4.7 ± 1.0 | 0.277 | |||
| High myopia | 4 (28.6%) | 7 (50.0%) | 0.220 | |||
| Typical RP | 7 (50.0%) | 7 (50.0%) | 1.000 | |||
| Vitreomacular alteration | 0 (0%) | 5 (35.7) | 0.020 | |||
| Detectable photopic ERG | 10 (71.4%) | 10 (71.4%) | 0.590 | |||
| Mean Difference | ||||||
| Estimation Beta (P Value) | ||||||
| Right Eye | Left Eye | Right Eye | Left Eye | Crude | Age-Adjusted | |
| BCVA (logMAR) | 0.15 ± 0.10 | 0.14 ± 0.09 | 0.41 ± 0.36 | 0.44 ± 0.34 | −0.43 (< 0.001) | −0.08 (0.126) |
| EZ-band width, µm | 1782.9 ± 556.9 | 1820.9 ± 702.6 | 855.1 ± 451.2 | 831.0 ± 390.9 | 843 (< 0.001) | 823 (< 0.001) |
| MMT, µm | 239.8 ± 39.6 | 237.4 ± 35.4 | 213.6 ± 43.8 | 211.9 ± 49.9 | 25.9 (0.089) | 28.9 (0.076) |
| Photopic ERG (b-wave amplitude), µV | 12.4 ± 17.3 | 15.3 ± 20.9 | 4.5 ± 8.1 | 4.3 ± 8.2 | 9.4 (0.067) | 6.9 (0.138) |
| Photopic ERG (b-wave implicit time), ms | 42.5 ± 3.1 | 42.6 ± 5.0 | 45.2 ± 1.7 | 44.7 ± 4.0 | −2.4 (0.194) | −3.5 (0.096) |
| 30 Hz Flicker ERG (trough-to-peak amplitude), µV | 13.8 ± 14.5 | 12.6 ± 9.8 | 5.9 ± 4.1 | 7.0 ± 5.3 | 6.7 (0.066) | 2.1 (0.455) |
| 30 Hz Flicker ERG (implicit time), ms | 44.2 ± 3.7 | 42.8 ± 2.4 | 50.9 ± 9.9 | 50.7 ± 9.8 | −7.2 (0.011) | −8.4 (0.037) |
| MS, dB | 4.8 ± 3.4 | 5.8 ± 3.6 | 2.2 ± 3.7 | 4.3 ± 5.6 | 2.0 (0.272) | 2.4 (0.111) |
Molecular Characterization of the RPGR-RP Cohort
We identified RPGR causative variants in 48 male patients (from 31 unrelated families) diagnosed with RP. Almost one third of the patients were part of larger pedigrees that showed an X-linked inheritance, whereas the rest were simplex cases. The patients were analyzed either by panel-based sequencing of known retinopathy genes or by more comprehensive clinical exome sequencing. One patient (no. 41 in Table 4) was analyzed by Whole Exome Sequencing (WES). Following a first-tier next-generation sequencing (NGS) analysis, likely pathogenic variants in the RPGR exons 1 to 14 were identified in 23 patients at the hemizygous state (Table 4). The remaining 25 male patients included in this cohort were unsolved after NGS-based screening. Given that standard NGS-based approaches fail to accurately probe the ORF15 mutational hot spot due to the presence of an extensive stretch of highly repetitive purine-rich sequence, we analyzed the latter subset of undiagnosed patients by direct PCR-based Sanger sequencing of the entire ORF15 optimized for purine-rich sequences. We identified likely pathogenic variants in all cases. All the RPGR identified variants segregated with the RP phenotype in available family members.
Table 4.
Variants Identified in the Patients With RPGR-RP
| Family | Patient | RefSeq | Nucleotide Change | Protein Change | Location | Mutation Type | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 1 | NM_001034853 | c.139deI | p.(Ser47Leufs*21) | Exon 2 | Frameshift | This study |
| 19 | 35 | NM_001034853 | c.154G>A | p.? | Exon 2 | Splice variant | 66 |
| 25 | 42, 43 | NM_001034853 | c.154G>A | p.? | Exon 2 | Splice variant | 66 |
| 18 | 33, 34 | NM_001034853 | c.155-2A>G | p.(?) | Intron 2 | Splice variant | 73 |
| 14 | 29 | NM_001034853 | c.202G>C | p.(Gly68Arg) | Exon 3 | Missense | 74 |
| 30 | 48 | NM_001034853 | c.237_238del | p.(Val81Glnfs*6) | Exon 3 | Frameshift | This study |
| 15 | 30 | NM_001034853 | c.247+1G>A | p.(?) | Intron 3 | Splice variant | This study |
| 23 | 40 | NM_001034853 | c.439G>C | p.(Ala147Pro) | Exon 5 | Missense | LOVD† |
| 12 | 25, 26 | NM_001034853 | c.619+5G>A | p.(?) | Intron 6 | Splice variant | 75 |
| 5 | 10 | NM_001034853 | c.706C>T | p.Gln236* | Exon 7 | Nonsense | 76 |
| 17 | 32 | NM_001034853 | c.807T>C | p.(Cys250Arg) | Exon 7 | Missense | 18 |
| 20 | 36, 37 | NM_000328 | c.1059+1G>A | p.(?) | Intron 9 | Splice variant | This study |
| 2 | 2 | NM_001034853 | c.1106_1115del | p.(Arg369Glnfs*9) | Exon 10 | Frameshift | This study |
| 13 | 27 | NM_001034853 | c.1243_1244del | p.(Arg415Glyfs*37) | Exon 10 | Frameshift | 77 |
| 7 | 13 | NM_001034853 | c.1245+3A>T | p.(?) | Intron 10 | Splice variant | 67 |
| 24 | 41 | NM_000328 | c.1246-3A>G | p.(?) | Intron 10 | Splice variant | This study |
| 4 | 9 | NM_001034853 | c.1283dup | p.(Pro429Thrfs*24) | Exon 11 | Frameshift | This study |
| 29 | 47 | NM_001034853 | c.1345C>T | p.(Arg449*) | Exon 11 | Nonsense | 78 |
| 10 | 23 | NM_001034853 | c.1582_1585del | p.(Thr528Leufs*4) | Exon 14 | Frameshift | 76 |
| 16 | 31 | NM_001034853 | c.2232_2235del | p.(Asp744Glufs*70) | ORF15 | Frameshift | This study |
| 11 | 24 | NM_001034853 | c.2236_2237delGA | p.(Glu746Argfs*23) | ORF15 | Frameshift | 23 |
| 6 | 11, 12 | NM_001034853 | c.2311delG | p.(Glu771Argfs*44) | ORF15 | Frameshift | This study |
| 8 | 14–16 | NM_001034853 | c.2405_2406del | p.(Glu802Glyfs*32) | ORF15 | Frameshift | 23 |
| 26 | 44 | NM_001034853 | c.2405_2406del | p.(Glu802Glyfs*32) | ORF15 | Frameshift | 23 |
| 28 | 46 | NM_001034853 | c.2405_2406del | p.(Glu802Glyfs*32) | ORF15 | Frameshift | 23 |
| 21 | 38 | NM_001034853 | c.2466_2467del | p.(Lys823Argfs*11) | ORF15 | Frameshift | 23 |
| 22 | 39 | NM_001034853 | c.2706_2707del | p.(Glu903Glyfs*175) | ORF15 | Frameshift | 79 |
| 9 | 17–22 | NM_001034853 | c.2760_2761del | p.(Glu922Glyfs*156) | ORF15 | Frameshift | 26 |
| 27 | 45 | NM_001034853 | c.2792del | p.(Glu931Glyfs*158) | ORF15 | Frameshift | 13 |
| 31 | 49 | NM_001034853 | c.2993_2997del | p.(Glu998Glyfs*79) | ORF15 | Frameshift | 23 |
| 3 | 3–8 | NM_001034853 | ORF15 del | p.(?) | ORF15 | Large deletion | This study |
†Leiden Open Variation Database, variant entry # 0000575845.
Half of our cohort harbored variants in ORF15 (n = 25; 12 families; 52%), while the remaining part had causative variants in exons 1 to 14 (n = 23; 19 families; 48%; Fig. 3). In terms of type of mutations, frameshifts were the most frequently observed variants in this cohort (n = 25; 17 families; 52%), followed by splice variants (n = 12; 8 families; 25%), missense (n = 3; 3 families; 6%), and nonsense mutations (n = 2; 2 families; 4%; Fig. 3). We also identified an extended deletion of ORF15 in 6 affected male subjects of a large pedigree (n = 6; 1 family; 13%). With the exception of this extended deletion, all variants detected in ORF15 were deletions of few nucleotides (1 to 5 bases) causing frameshifts predicted to introduce premature termination codons. Ten of the 28 putatively pathogenic variants identified in this study were novel (Table 4) and had not been previously reported neither in the Human Gene Mutation Database (HGMD) nor in the Leiden Open Variation Database (LOVD). The frequency of these variants in reference population databases (e.g. ExAC, 1000 Genomes,55 and gnomAD) is compatible with a causative role in conditions with X-linked inheritance. The missense variant c.439G>C; p.(Ala147Pro) was previously reported as VUS in LOVD (variant entry #0000575845) and its potential pathogenicity was corroborated by in silico predictions (“Disease causing” in Mutation-taster; “Probably damaging” according to PolyPhen-2; category: deleterious, score: 0.01 according to Sorting Intolerant from Tolerant; score 26.6. in Combined Annotation-Dependent Depletion). This variant is absent from control population datasets, such as gnomAD (http://gnomad.broadinstitute.org) and Bravo (http://bravo.sph.umich.edu/).
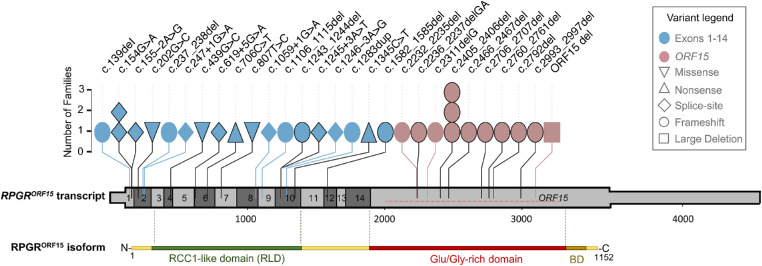
Figure 3.
Genetic variants detected in RPGR. Schematic drawing of the RPGRORF15 transcript showing the position and frequency of the variants identified in this cohort. The untranslated regions of the transcript are depicted as a thinner bar. Each symbol designates a family whose variant position is reported on the top. The symbol color indicates whether the variant is found in exons 1 to 14 or in the ORF15 region. The symbol shape indicates the mutation type. The large deletion (ORF15 del) is shown with a horizontal dotted line within the terminal exon of RPGRORF15. Already reported variants are denoted with a black border. Protein domains of the retina-specific isoform RPGRORF15 are shown below the transcript to indicate the RCC1-like domain (green), the Glu/Gly-rich region (red), and the basic domain (BD; brown; not to scale).
Genotype – Phenotype Correlation Analysis
To explore possible genotype-phenotype correlations, we stratified the unrelated patients according to the variant localization within the RPGRORF15 transcript (i.e. variants in exons 1–14 vs. variants in ORF15), as also implemented in other reports.28,32 We then assessed the disease severity of each patient group by considering the main clinical parameters of the cohort described herein (Table 5). We did not observe statistically significant differences in terms of patients’ age, self-reported age of onset, and disease length between patients with variants in exons 1 to 14 vs. those with variants in ORF15 (Table 5). Similarly, there were no significant differences in the frequency of high myopia, MA, detectable photopic ERG, and the presence of a hyperautofluorescent ring. In contrast, patients harboring ORF15 variants showed a faster loss of BCVA than patients with variants in exon 1 to 14 (P < 0.001). Specifically, in the former patient group, BCVA declined at a mean annual rate of 0.044 logMAR/year (equivalent to 2 ETDRS letters/year), whereas the mean rate of decline in the latter group was 0.011 logMAR/year (equivalent to 1/2 ETDRS letters/year). On average, patients with ORF15 variants showed lower values of EZ band width, MMT and MS compared to those with variants in exons 1 to 14, but these differences were not statistically significant (Table 5). None of the patients showed significant extra-ocular findings with the exception of five unrelated patients (nos. 2, 9, 32, 37, and 40) carrying mutations in the exons 1 to 14, who reported recurrent otitis during childhood.
Table 5.
Clinical Features of Patients With RPGR-RP Stratified According to the Localization of the Variant (Exons 1–14 vs. ORF15)
| Exon 1–14 | ORF15 (12 Unrelated | |||||
|---|---|---|---|---|---|---|
| Parameters | (19 Unrelated Patients) | Patients) | P Value | |||
| Age, y | 26.2 ± 12.5 | 33.3 ± 19.1 | 0.223 | |||
| Self-reported age of onset, y | 6.3 ± 5.0 | 7.6 ± 5.4 | 0.492 | |||
| Disease length, y | 20 ± 12.6 | 25.7 ± 18.9 | 0.321 | |||
| Mean refractive error, D | −3.9 ± 2.9 | −3.6 ± 3.4 | 0.774 | |||
| High myopia | 7 (36.8%) | 3 (25.0%) | 0.697 | |||
| Typical RP | 10 (52.6%) | 9 (75.0%) | 0.274 | |||
| Vitreomacular alteration | 4 (21.1%) | 2 (16.7%) | 1.000 | |||
| Detectable photopic ERG | 10 (52.6%) | 6 (50.0%) | 1.000 | |||
| Hyperautofluorescent ring | 6 of 12 (50.0%) | 3 of 7 (42.9%) | 1.000 | |||
| Annual BCVA decline (best-seeing eye) | 0.011 ± 0.004 (P = 0.003) |
0.044 ± 0.001 (P < 0.001) |
< 0.001 | |||
| Mean Difference | ||||||
| Estimation Beta (P Value) | ||||||
| Right Eye | Left Eye | Right Eye | Left Eye | Crude | Age-Adjusted | |
| BCVA, logMAR | 0.34 ± 0.42 | 0.34 ± 0.41 | 0.76 ± 0.79 | 0.86 ± 0.81 | 0.47 (0.044) | 0.32 (0.063) |
| EZ-band width, µm | 1492 ± 634 | 1518 ± 700 | 1184 ± 715 | 1224 ± 796 | −301.1 (0.345) | −216.2 (0.469) |
| MMT, µm | 239.6 ± 45.8 | 240.0 ± 44.1 | 205.7 ± 41.0 | 205 ± 44.7 | −34.4 (0.075) | −34.1 (0.095) |
| Photopic ERG (b-wave amplitude), µV | 17.8 ± 8.1 | 23.9 ± 16.4 | 28.3 ± 32.6 | 30.8 ± 33.7 | 8.68 (0.613) | −5.65 (0.706) |
| Photopic ERG (b-wave implicit time), ms | 43.5 ± 1.9 | 43.5 ± 3.0 | 44.0 ± 1.1 | 41.6 ± 2.5 | 1.6 (0.442) | 1.3 (0.571) |
| 30 Hz Flicker ERG (trough-to-peak amplitude), µV | 10.2 ± 8.5 | 10.1 ± 6.3 | 14.3 ± 17.4 | 15.7 ± 15.1 | 4.8 (0.451) | 4.42 (0.305) |
| 30 Hz Flicker ERG (implicit time), ms | 48.6 ± 10.5 | 48.7 ± 9.9 | 49.9 ± 5.5 | 37.2 ± 7.7 | −3.5 (0.245) | −3.1 (0.299) |
| MS, dB | 2.9 ± 3.5 | 4.3 ± 4.8 | 2.5 ± 4.1 | 2.4 ± 5.3 | −1.18 (0.58) | −0.87 (0.696) |
We then sought to further classify the variants in exons 1 to 14 into three groups according to the mutation types and their predicted consequence at the protein level (i.e. missense variants [M], splice-site variants [S], and null variants [N] that comprised nonsense and frameshift changes; Supplementary Table S2). Using this stratification, we found a differential effect of variants in exons 1 to 14. Patients bearing missense variants (n = 3) presented the mildest phenotypes, considering also their older mean age, the relatively preserved mean visual acuity, and the highest mean values of EZ band and MS (Supplementary Table S2). Moreover, the age-adjusted model revealed a reduction of flicker ERG responses in patients carrying null variants compared to the ORF15 group (P < 0.001), in spite of a better-preserved macular thickness (i.e. higher MMT values). The slower BCVA decline detected in patients harboring splice-site variants compared to patients with null variants (i.e. 0.008 logMAR/year vs. 0.011 logMAR/year) was not statistically significant (P = 0.468).
Overall, our data indicated that mutations in ORF15 are associated with a more severe phenotype compared to variants in exons 1 to 14.
Discussion
This study describes the natural history of RP due to RPGR mutations in a large Italian cohort (48 male patients from 31 different families) evaluated at a single center with a long-term follow-up (mean = 6.5 years). To our knowledge, this is the first report of a longitudinal natural history study in Italian patients with RPGR-associated RP that includes detailed morphological and functional assessments. The largest cohort of RP patients with RPGR mutations (113 patients) was characterized by Sandberg et al.29 focusing mainly on visual acuity, visual field, and ERG assessments in US patients. More recently, Talib et al. described a multicenter, retrospective cohort study involving 52 patients with RPGR-RP from various Dutch medical centers.32 However, as the authors also pointed out, the potential intercenter variability (which may not always be statistically accounted for in the retrospective study setting) may have complicated data interpretation.32 With respect to previous reports, our results extend the number of patients followed up at a single referral center and provide a comprehensive and homogeneous clinical description of RPGR-associated forms of RP. In this RP cohort, symptoms were first reported at 5 years of age, thus confirming the earlier onset of RPGR-RP, particularly compared to autosomal dominant RP forms.56,57 About 80% of patients presented variable degrees of myopia, and approximately one third of the cohort had high myopia, in line with previous observations in X-linked RP56,57 and in RPGR-related RP.32 Our data confirm a faster BCVA decline in patients with high myopia as opposed to those without high myopia.32 Moreover, the patients with typical RP and high myopia had a significantly faster progression of the disease compared to those with sine pigmento RP in the absence of high myopia. In terms of photoreceptor function, the sine pigmento RP forms were significantly associated with detectable photopic ERG responses. These findings suggest that the scarcity or absence of pigment deposits could be considered as a prognostic factor for a slower disease progression, particularly in the absence of high myopia and in the first three decades of life.
The survival analyses over an extended follow-up period showed that the BCVA decline led to blindness at a median age of 56 years. Blindness was mainly driven by visual field loss. In particular, patients reached blindness based on visual field at a significantly younger age (26.4 years; i.e. about 25 years earlier), than blindness based on BCVA. These findings are consistent with the report of Sandeberg et al.29 showing that blindness based on visual field was achieved about 10 years earlier than blindness based on BCVA. It should be noted though that the different thresholds in the definition of blindness by BCVA could impact the above comparison: specifically, Sandberg et al.29 adopted the value of 20/200, whereas we considered a visual acuity of 20/400 (according to the ICD-10 version 2016).
FAF imaging revealed the presence of a hyperautofluorescent ring in 14 patients (50%; 14/28) that were significantly younger and had a better preserved EZ band compared to patients displaying other FAF abnormalities (i.e. marked reduced macular FAF or absence of macular FAF). On this basis, it is tempting to hypothesize that FAF patterns could be indicative of different stages of disease progression. Specifically, the hyperautofluorecent ring may represent the front of advancing concentric photoreceptor cell loss, as previously shown in RP in general45 and in subjects with ORF15 variants.34
Finally, in terms of MA, we did not detect CME in any of our patients with RPGR-RP even though CME is the most frequently encountered MA in our general RP cohort, affecting 22.9% of patients.58 This finding is consistent with two previous studies that reported no case of CME in patients with RPGR-RP.9,35 Moreover, the lower frequency of CME observed in patients with X-linked RP (7.1%) compared to autosomal forms reported in our previous study58 strongly suggest that the incidence of CME in RP varies according to the mutated gene. On the contrary, the frequency of ERM and VMT in patients with RPGR-RP was similar to that observed in our overall RP cohort (ERM = 25% in RPGR-RP vs. 19.8% in RP; VMT = 2.5% in RPGR-RP vs. 5% in RP).58 The prevalence of ERM in our cohort was lower compared to that reported by Talib et al. (i.e. 40% in at least one eye).32 A possible explanation for this difference could be due to a selection bias, since only 29% (15/52) of their cohort underwent spectral domain OCT (compared to 100% in this study, 48/48). Moreover, even if longitudinal observations of spectral domain OCT scans acquired with the Heidelberg Spectralis and follow-up mode were available only for small subgroups of patients, the analysis showed a significant loss of the EZ band width at an annual rate of −183.7 µm/year and at an exponential rate of −8.9%/year. These estimates are comparable with the values reported in previous studies44,59 on X-linked patients, which range from −210 µm/year to −270 µm/year and from −7% to −9.6%/year. This evidence supports the adoption of the EZ band width as a relevant outcome measure for the evaluation of disease progression and of treatment efficacy in patients with RPGR-RP.
We identified 10 novel RPGR variants, expanding the list of RPGR disease-causing mutations (see Table 4). Among those, two novel single nucleotide variations (SNVs) and a large deletion were found within the ORF15. All ORF15 variants were identified by direct Sanger sequencing of this region in patients that remained undiagnosed after a panel-based NGS analysis, suggesting that alternative ad hoc methods (either Sanger sequencing34,50,60 or NGS-based61,62) should be deployed for accurate probing of ORF15 in unsolved cases after a first-tier NGS analysis (especially in men with XL or simplex RP/CD/CRD).
Several attempts have already been made to correlate the genotype with disease severity in patients carrying RPGR mutations, but conclusions are not always concordant. For example, a more severe clinical phenotype has been associated with variants in exon 1 to 14 in some cohorts.28,30,63,64 In contrast, other studies argue that variants in the ORF15 region of the RPGR gene present with a more deleterious phenotype.31,32 Cohort's size, selection bias, variability of patients' diagnoses (e.g. inclusion of both patients with RP and patients with CRD) as well as the potential nonuniformity of observations made in multicenter studies, represent some factors that may impact on the phenotypic-genotypic correlations inferred. In this study, we classified the variants according to their position within the RPGRORF15. We therefore explored possible correlations between the clinical parameters of patients with RP and the variant location in RPGR in this cohort. We found that variants in ORF15 correlated with a more severe phenotype in terms of BCVA loss. In particular, patients carrying ORF15 variants had a faster decline of BCVA compared with patients with mutations in exons 1 to 14. Moreover, these patients showed lower values of MMT compared with patients harboring variants in exons 1 to 14, albeit this was not statistically significant. These findings partially corroborate the recent observation that variants in ORF15 correlated with a more severe visual impairment, including higher myopia, a thinner central retina, and faster progression of visual field loss.32 The disease severity associated with ORF15 variants could be attributed to the presumed gain-of-function or dominant-negative effect of the resulting protein product, as previously suggested.28,30,35,40 Variants in ORF15 are mostly small deletions or duplications leading to frameshift changes. Because ORF15 is the terminal exon of the RPGRORF15 transcript, the stop or frame-shifting variants are, in principle, not subject to nonsense-mediated mRNA decay, but can be expected to lead to the synthesis of presumably dysfunctional protein products. Such proteins could severely perturb RPGR protein networks and exert a more deleterious effect compared to that of null variants in exons 1 to 14. Indeed, in vivo studies using transgenic mice demonstrated that certain truncated forms of RPGR behave as gain-of-function mutants and can be more deleterious than null alleles.65
By further stratifying patients with variants in exons 1 to 14 based on the mutation type, we noted that splice-site variants in this region were associated with milder phenotypes compared to null alleles (Supplementary Table S2). Given that the consequences of splice-site variations can vary greatly, we cannot exclude that this observation may be biased by the small number of splice variants in our cohort. In addition, two of the splice-site variants (c.154G>A and c.1245+3A>T) had already been shown to cause in-frame changes, namely, an in-frame deletion of 42 amino acids66 and in-frame skipping of exon 10.67 Therefore, to reliably interpret genotype-phenotype associations involving RPGR splice-altering variants, in vitro analyses of their consequence at the protein level should be pursued, as also performed for other genes (e.g. ABCA4).68 In this context, it is interesting that in patient-derived cell lines the consequence of the c.1245+3A>T splice site variant (exon 10 in-frame skipping) was shown to result in a strongly reduced localization of the RPGR protein along the cilium,39 suggesting a possible read-out assay to quantify the severity of RPGR mutations or efficacy of treatment approaches.
Considering the above points, further studies are required to establish clear correlations between RPGR variants and their impact on visual function and retinal morphology, especially given the phenotypic variability of RPGR patients. Some reports describe extensive phenotypic variability in families harboring the same variants in RPGR, as well as different diagnosis (i.e. XLRP and XLCRD)8,28,69,70 even in dizygotic twins within the same kindred.71 Clearly, much remains to be understood about the contribution of genetic modifiers, environmental effects, and their synergistic effects on the observed phenotypic variation. Therefore, any attempt to predict the clinical severity or disease progression on the basis of the RPGR mutation type or location should cautiously consider the complex interplay of different factors.
The current study has some limitations mainly related to its retrospective design. First, although the patients were invited to undergo yearly follow-up visits, only cross-sectional data are available for some patients and for some outcome measurements. Moreover, MP1 measurements are available for a subgroup of patients, because MP1 was introduced in our clinical practice only recently and some patients were not always willing or capable to perform the examination (e.g. because of poor visual acuity or young age). For similar reasons, also fundus autofluorescence was available only for a subset of patients. Finally, the follow-up length is not the same for all the patients and the relatively small sample size may hinder the detection of statistically significant associations in some comparisons due to the insufficient power of the test.
In conclusion, we present a single-center, retrospective, longitudinal study that comprehensively describes visual function and disease progression in subjects with RPGR-associated RP on the grounds of morphological and functional parameters. Based on the longitudinal analysis, we identified a subgroup of patients with RPGR-associated RP who showed a more severe phenotype and faster disease progression. These patients were older, had typical RP, elevated myopia, and harbored pathogenic variants in the ORF15 sequence. On the other hand, we defined a subgroup of patients presenting an overall milder phenotype with a better visual acuity, a better-preserved EZ band, the presence of an AF ring, fewer MA, and a slower disease progression. These were patients of younger age, with sine pigmento forms of RP, no myopia, and variants in exons 1 to 14. Taken together, these results suggest that the selection and prioritization of patients for clinical trials should consider the patient's age, the RP form according to the fundus appearance, the presence of high myopia, as well as the localization and type of causative variants in the RPGR gene. Finally, this study adds new knowledge on the disease and provides a novel resource to guide patient selection and the design of outcome measures in recently opened RPGR treatment trials.72
Supplementary Material
Acknowledgments
The authors thank the participating patients and the patient association Retina Italia Onlus. We are grateful to the European Reference Network dedicated to Rare Eye Diseases (ERN-EYE) for useful discussions. The authors also wish to thank Annalaura Torella and the TIGEM NGS facility for support in NGS analysis and Mariateresa Pizzo for technical support.
Supported by the Italian Fondazione Roma (to S.B. and F.S.), by the Ministero dell'Istruzione dell'Università e della Ricerca (MIUR) under PRIN 2015 (to S.B. and F.S.) and by the University of Campania ‘Luigi Vanvitelli’ under “VALERE: VAnviteLli pEr la RicErca” (to S.B. and F.S.). This study was also supported by the Foundation Fighting Blindness center grant (C-CMM-0907-0428-INSERM04; to I.A. and C.Z.), by the LABEX LIFESENSES (ANR-10-LABX-65) grant managed by the Agence Nationale de la Recherche within the Investissements d'Avenir program (ANR-11-IDEX-0004-0; to C.C., I.A., and C.Z.) and the IHU FOReSIGHT (ANR-18-IAHU-0001) grant supported by French state funds managed by the Agence Nationale de la Recherche within the Investissements d'Avenir program (to C.C., I.A., and C.Z.).
Disclosure: V. Di Iorio, None; M. Karali, None; P. Melillo, None; F. Testa, None; R. Brunetti-Pierri, None; F. Musacchia, None; C. Condroyer, None; J. Neidhardt, None; I. Audo, None; C. Zeitz, None; S. Banfi, None; F. Simonelli, None
References
- 1. Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006; 1: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang S, Vaccarella L, Olatunji S, Cebulla C, Christoforidis J.. Diagnostic challenges in retinitis pigmentosa: genotypic multiplicity and phenotypic variability. Curr Genomics. 2011; 12: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Branham K, Othman M, Brumm M, et al.. Mutations in RPGR and RP2 account for 15% of males with simplex retinal degenerative disease. Invest Ophthalmol Vis Sci. 2012; 53: 8232–8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lyraki R, Megaw R, Hurd T.. Disease mechanisms of X-linked retinitis pigmentosa due to RP2 and RPGR mutations. Biochem Soc Trans. 2016; 44: 1235–1244. [DOI] [PubMed] [Google Scholar]
- 5. Tee JJ, Smith AJ, Hardcastle AJ, Michaelides M.. RPGR-associated retinopathy: clinical features, molecular genetics, animal models and therapeutic options. Br J Ophthalmol. 2016; 100: 1022–1027. [DOI] [PubMed] [Google Scholar]
- 6. Fishman GA. Retinitis pigmentosa. Genetic percentages. Arch Ophthalmol. 1978; 96: 822–826. [DOI] [PubMed] [Google Scholar]
- 7. Tee JJL, Yang Y, Kalitzeos A, Webster A, Bainbridge J, Michaelides M.. Natural history study of retinal structure, progression, and symmetry using ellipzoid zone metrics in RPGR-associated retinopathy. Am J Ophthalmol. 2019; 198: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang L, Yin X, Feng L, et al.. Novel mutations of RPGR in Chinese retinitis pigmentosa patients and the genotype-phenotype correlation. PLoS One. 2014; 9: e85752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kurata K, Hosono K, Hayashi T, et al.. X-linked retinitis pigmentosa in Japan: clinical and genetic findings in male patients and female carriers. Int J Mol Sci. 2019; 20(6): 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nanda A, Salvetti AP, Clouston P, Downes SM, MacLaren RE.. Exploring the variable phenotypes of RPGR carrier females in assessing their potential for retinal gene therapy. Genes (Basel). 2018; 9(12): 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Talib M, van Schooneveld MJ, Van Cauwenbergh C, et al.. The spectrum of structural and functional abnormalities in female carriers of pathogenic variants in the RPGR gene. Invest Ophthalmol Vis Sci. 2018; 59: 4123–4133. [DOI] [PubMed] [Google Scholar]
- 12. Wu DM, Khanna H, Atmaca-Sonmez P, et al.. Long-term follow-up of a family with dominant X-linked retinitis pigmentosa. Eye (Lond). 2010; 24: 764–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pelletier V, Jambou M, Delphin N, et al.. Comprehensive survey of mutations in RP2 and RPGR in patients affected with distinct retinal dystrophies: genotype-phenotype correlations and impact on genetic counseling. Hum Mutat. 2007; 28: 81–91. [DOI] [PubMed] [Google Scholar]
- 14. Roepman R, Bernoud-Hubac N, Schick DE, et al.. The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum Mol Genet. 2000; 9: 2095–2105. [DOI] [PubMed] [Google Scholar]
- 15. Hardcastle AJ, Thiselton DL, Van Maldergem L, et al.. Mutations in the RP2 gene cause disease in 10% of families with familial X-linked retinitis pigmentosa assessed in this study. Am J Hum Genet. 1999; 64: 1210–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Webb TR, Parfitt DA, Gardner JC, et al.. Deep intronic mutation in OFD1, identified by targeted genomic next-generation sequencing, causes a severe form of X-linked retinitis pigmentosa (RP23). Hum Mol Genet. 2012; 21: 3647–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Almoguera B, He S, Corton M, et al.. Expanding the phenotype of PRPS1 syndromes in females: neuropathy, hearing loss and retinopathy. Orphanet J Rare Dis. 2014; 9: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meindl A, Dry K, Herrmann K, et al.. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat Genet. 1996; 13: 35–42. [DOI] [PubMed] [Google Scholar]
- 19. Roepman R, van Duijnhoven G, Rosenberg T, et al.. Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum Mol Genet. 1996; 5: 1035–1041. [DOI] [PubMed] [Google Scholar]
- 20. Hong DH, Li T.. Complex expression pattern of RPGR reveals a role for purine-rich exonic splicing enhancers. Invest Ophthalmol Vis Sci. 2002; 43: 3373–3382. [PubMed] [Google Scholar]
- 21. Neidhardt J, Glaus E, Barthelmes D, Zeitz C, Fleischhauer J, Berger W.. Identification and characterization of a novel RPGR isoform in human retina. Hum Mutat. 2007; 28: 797–807. [DOI] [PubMed] [Google Scholar]
- 22. Schmid F, Glaus E, Cremers FP, Kloeckener-Gruissem B, Berger W, Neidhardt J.. Mutation- and tissue-specific alterations of RPGR transcripts. Invest Ophthalmol Vis Sci. 2010; 51: 1628–1635. [DOI] [PubMed] [Google Scholar]
- 23. Vervoort R, Lennon A, Bird AC, et al.. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000; 25: 462–466. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Q, Giacalone JC, Searby C, Stone EM, Tucker BA, Sheffield VC.. Disruption of RPGR protein interaction network is the common feature of RPGR missense variations that cause XLRP. Proc Natl Acad Sci USA. 2019; 116: 1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mawatari G, Fujinami K, Liu X, et al.. Clinical and genetic characteristics of 14 patients from 13 Japanese families with RPGR-associated retinal disorder: report of eight novel variants. Hum Genome Var. 2019; 6: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shu X, Black GC, Rice JM, et al.. RPGR mutation analysis and disease: an update. Hum Mutat. 2007; 28: 322–328. [DOI] [PubMed] [Google Scholar]
- 27. Ball EV, Stenson PD, Abeysinghe SS, Krawczak M, Cooper DN, Chuzhanova NA.. Microdeletions and microinsertions causing human genetic disease: common mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum Mutat. 2005; 26: 205–213. [DOI] [PubMed] [Google Scholar]
- 28. Fahim AT, Bowne SJ, Sullivan LS, et al.. Allelic heterogeneity and genetic modifier loci contribute to clinical variation in males with X-linked retinitis pigmentosa due to RPGR mutations. PLoS One. 2011; 6: e23021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sandberg MA, Rosner B, Weigel-DiFranco C, Dryja TP, Berson EL.. Disease course of patients with X-linked retinitis pigmentosa due to RPGR gene mutations. Invest Ophthalmol Vis Sci. 2007; 48: 1298–1304. [DOI] [PubMed] [Google Scholar]
- 30. Sharon D, Sandberg MA, Rabe VW, Stillberger M, Dryja TP, Berson EL.. RP2 and RPGR mutations and clinical correlations in patients with X-linked retinitis pigmentosa. Am J Hum Genet. 2003; 73: 1131–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andreasson S, Breuer DK, Eksandh L, et al.. Clinical studies of X-linked retinitis pigmentosa in three Swedish families with newly identified mutations in the RP2 and RPGR-ORF15 genes. Ophthalmic Genet. 2003; 24: 215–223. [DOI] [PubMed] [Google Scholar]
- 32. Talib M, van Schooneveld MJ, Thiadens AA, et al.. Clinical and genetic characteristics of male patients with RPGR-associated retinal dystrophies: a long-term follow-up study. Retina. 2019; 39: 1186–1199. [DOI] [PubMed] [Google Scholar]
- 33. Demirci FY, Rigatti BW, Wen G, et al.. X-linked cone-rod dystrophy (locus COD1): identification of mutations in RPGR exon ORF15. Am J Hum Genet. 2002; 70: 1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ebenezer ND, Michaelides M, Jenkins SA, et al.. Identification of novel RPGR ORF15 mutations in X-linked progressive cone-rod dystrophy (XLCORD) families. Invest Ophthalmol Vis Sci. 2005; 46: 1891–1898. [DOI] [PubMed] [Google Scholar]
- 35. Nguyen XT, Talib M, van Schooneveld MJ, et al.. RPGR-associated dystrophies: clinical, genetic, and histopathological features. Int J Mol Sci. 2020; 21(3): 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iannaccone A, Breuer DK, Wang XF, et al.. Clinical and immunohistochemical evidence for an X linked retinitis pigmentosa syndrome with recurrent infections and hearing loss in association with an RPGR mutation. J Med Genet. 2003; 40: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Dorp DB, Wright AF, Carothers AD, Bleeker-Wagemakers EM.. A family with RP3 type of X-linked retinitis pigmentosa: an association with ciliary abnormalities. Hum Genet. 1992; 88: 331–334. [DOI] [PubMed] [Google Scholar]
- 38. Zito I, Downes SM, Patel RJ, et al.. RPGR mutation associated with retinitis pigmentosa, impaired hearing, and sinorespiratory infections. J Med Genet. 2003; 40: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Da Costa R, Glaus E, Tiwari A, Kloeckener-Gruissem B, Berger W, Neidhardt J. Localizing the RPGR protein along the cilium: a new method to determine efficacies to treat RPGR mutations. Gene Ther. 2015; 22: 413–420. [DOI] [PubMed] [Google Scholar]
- 40. Megaw RD, Soares DC, Wright AF.. RPGR: its role in photoreceptor physiology, human disease, and future therapies. Exp Eye Res. 2015; 138: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fahim AT, Daiger SP, Weleber RG.. Nonsyndromic retinitis pigmentosa overview. GeneReviews [Internet]: Seattle, WA: University of Washington; 2017. [Google Scholar]
- 42. Testa F, Melillo P, Rossi S, et al.. Prevalence of macular abnormalities assessed by optical coherence tomography in patients with Usher syndrome. Ophthalmic Genet. 2018; 39: 17–21. [DOI] [PubMed] [Google Scholar]
- 43. Hood DC, Zhang X, Ramachandran R, et al.. The inner segment/outer segment border seen on optical coherence tomography is less intense in patients with diminished cone function. Invest Ophthalmol Vis Sci. 2011; 52: 9703–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cai CX, Locke KG, Ramachandran R, Birch DG, Hood DC.. A comparison of progressive loss of the ellipsoid zone (EZ) band in autosomal dominant and x-linked retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014; 55: 7417–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Robson AG, El-Amir A, Bailey C, et al.. Pattern ERG correlates of abnormal fundus autofluorescence in patients with retinitis pigmentosa and normal visual acuity. Invest Ophthalmol Vis Sci. 2003; 44: 3544–3550. [DOI] [PubMed] [Google Scholar]
- 46. Marmor MF, Fulton AB, Holder GE, et al.. ISCEV Standard for full-field clinical electroretinography (2008 update). Doc Ophthalmol. 2009; 118: 69–77. [DOI] [PubMed] [Google Scholar]
- 47. Di Iorio V, Karali M, Brunetti-Pierri R, et al.. Clinical and genetic evaluation of a cohort of pediatric patients with severe inherited retinal dystrophies. Genes (Basel). 2017; 8(10): 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Musacchia F, Ciolfi A, Mutarelli M, et al.. VarGenius executes cohort-level DNA-seq variant calling and annotation and allows to manage the resulting data through a PostgreSQL database. BMC Bioinformatics. 2018; 19: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Karali M, Testa F, Brunetti-Pierri R, et al.. Clinical and genetic analysis of a european cohort with pericentral retinitis pigmentosa. Int J Mol Sci. 2019; 21(1): 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Neidhardt J, Glaus E, Lorenz B, et al.. Identification of novel mutations in X-linked retinitis pigmentosa families and implications for diagnostic testing. Mol Vis. 2008; 14: 1081–1093. [PMC free article] [PubMed] [Google Scholar]
- 51. Kong X, Strauss RW, Michaelides M, et al.. Visual acuity loss and associated risk factors in the retrospective progression of Stargardt Disease Study (ProgStar Report No. 2). Ophthalmology. 2016; 123: 1887–1897. [DOI] [PubMed] [Google Scholar]
- 52. Testa F, Melillo P, Bonnet C, et al.. Clinical presentation and disease course of Usher syndrome because of mutations in Myo7a or Ush2a. Retina. 2017; 37: 1581–1590. [DOI] [PubMed] [Google Scholar]
- 53. Csaky KG, Richman EA, Ferris FL 3rd. Report from the NEI/FDA Ophthalmic Clinical Trial Design and Endpoints Symposium. Invest Ophthalmol Vis Sci. 2008; 49: 479–489. [DOI] [PubMed] [Google Scholar]
- 54. Glynn RJ, Rosner B.. Regression methods when the eye is the unit of analysis. Ophthalmic Epidemiol. 2012; 19: 159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Genomes Project C, Abecasis GR, Altshuler D, et al.. A map of human genome variation from population-scale sequencing. Nature. 2010; 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grover S, Fishman GA, Alexander KR, Anderson RJ, Derlacki DJ.. Visual acuity impairment in patients with retinitis pigmentosa. Ophthalmology. 1996; 103: 1593–1600. [DOI] [PubMed] [Google Scholar]
- 57. Wright AF. Chapter 5 Towards the identification of genes in X-linked retinitis pigmentosa. Prog Retin Res. 1990; 9: 197–227. [Google Scholar]
- 58. Testa F, Rossi S, Colucci R, et al.. Macular abnormalities in Italian patients with retinitis pigmentosa. Br J Ophthalmol. 2014; 98: 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ramachandran R, Zhou L, Locke KG, Birch DG, Hood DC.. A comparison of methods for tracking progression in X-linked retinitis pigmentosa using frequency domain OCT. Transl Vis Sci Technol. 2013; 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sharon D, Bruns GA, McGee TL, Sandberg MA, Berson EL, Dryja TP.. X-linked retinitis pigmentosa: mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Invest Ophthalmol Vis Sci. 2000; 41: 2712–2721. [PubMed] [Google Scholar]
- 61. Chiang JPW, Lamey TM, Wang NK, et al.. Development of high-throughput clinical testing of RPGR ORF15 using a large inherited retinal dystrophy cohort. Invest Ophthalmol Vis Sci. 2018; 59: 4434–4440. [DOI] [PubMed] [Google Scholar]
- 62. Li J, Tang J, Feng Y, et al.. Improved diagnosis of inherited retinal dystrophies by high-fidelity PCR of ORF15 followed by next-generation sequencing. J Mol Diagn. 2016; 18: 817–824. [DOI] [PubMed] [Google Scholar]
- 63. Sandberg MA, Rosner B, Weigel-DiFranco C, McGee TL, Dryja TP, Berson EL.. Disease course in patients with autosomal recessive retinitis pigmentosa due to the USH2A gene. Invest Ophthalmol Vis Sci. 2008; 49: 5532–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tee JJL, Yang Y, Kalitzeos A, et al.. Characterization of visual function, interocular variability and progression using static perimetry-derived metrics in RPGR-associated retinopathy. Invest Ophthalmol Vis Sci. 2018; 59: 2422–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hong DH, Pawlyk BS, Adamian M, Li T.. Dominant, gain-of-function mutant produced by truncation of RPGR. Invest Ophthalmol Vis Sci. 2004; 45: 36–41. [DOI] [PubMed] [Google Scholar]
- 66. Demirci FY, Radak AL, Rigatti BW, Mah TS, Gorin MB.. A presumed missense mutation of RPGR causes abnormal RNA splicing with exon skipping. Am J Ophthalmol. 2004; 138: 504–505. [DOI] [PubMed] [Google Scholar]
- 67. Glaus E, Schmid F, Da Costa R, Berger W, Neidhardt J. Gene therapeutic approach using mutation-adapted U1 snRNA to correct a RPGR splice defect in patient-derived cells. Mol Ther. 2011; 19: 936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sangermano R, Khan M, Cornelis SS, et al.. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018; 28: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huang WC, Wright AF, Roman AJ, et al.. RPGR-associated retinal degeneration in human X-linked RP and a murine model. Invest Ophthalmol Vis Sci. 2012; 53: 5594–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zahid S, Khan N, Branham K, et al.. Phenotypic conservation in patients with X-linked retinitis pigmentosa caused by RPGR mutations. JAMA Ophthalmol. 2013; 131: 1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Walia S, Fishman GA, Swaroop A, et al.. Discordant phenotypes in fraternal twins having an identical mutation in exon ORF15 of the RPGR gene. Arch Ophthalmol. 2008; 126: 379–384. [DOI] [PubMed] [Google Scholar]
- 72. Cehajic-Kapetanovic J, Xue K, Martinez-Fernandez de la Camara C, et al.. Initial results from a first-in-human gene therapy trial on X-linked retinitis pigmentosa caused by mutations in RPGR. Nat Med. 2020; 26: 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Miano MG, Testa F, Strazzullo M, et al.. Mutation analysis of the RPGR gene reveals novel mutations in south European patients with X-linked retinitis pigmentosa. Eur J Hum Genet. 1999; 7: 687–694. [DOI] [PubMed] [Google Scholar]
- 74. Pyo Park S, Hwan Hong I, Tsang SH, Chang S. Cellular imaging demonstrates genetic mosaicism in heterozygous carriers of an X-linked ciliopathy gene. Eur J Hum Genet. 2013; 21: 1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zito I, Gorin MB, Plant C, Bird AC, Bhattacharya SS, Hardcastle AJ.. Novel mutations of the RPGR gene in RP3 families. Hum Mutat. 2000; 15: 386. [DOI] [PubMed] [Google Scholar]
- 76. Buraczynska M, Wu W, Fujita R, et al.. Spectrum of mutations in the RPGR gene that are identified in 20% of families with X-linked retinitis pigmentosa. Am J Hum Genet. 1997; 61: 1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Koenekoop RK, Loyer M, Hand CK, et al.. Novel RPGR mutations with distinct retinitis pigmentosa phenotypes in French-Canadian families. Am J Ophthalmol. 2003; 136: 678–687. [DOI] [PubMed] [Google Scholar]
- 78. Breuer DK, Yashar BM, Filippova E, et al.. A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet. 2002; 70: 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Birtel J, Gliem M, Mangold E, et al.. Next-generation sequencing identifies unexpected genotype-phenotype correlations in patients with retinitis pigmentosa. PLoS One. 2018; 13: e0207958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.