Abstract
Sucrose induces flavonoid accumulation in plants as a defense mechanism against various stresses. However, the relationship between the biosynthesis of flavonoids as secondary metabolites and sucrose levels remains unknown. To understand the change in flavonoid biosynthesis by sucrose, we conducted secondary metabolite profiling in Melissa officinalis treated with different levels of sucrose using ultraperformance liquid chromatography/quadrupole time-of-flight mass spectrometry. The partial least squares-discriminant and hierarchical clustering analyses showed significant differences in secondary metabolite profiles in M. officinalis at 50, 150, and 300 mM sucrose levels. The levels of 3 flavonoids such as quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside, 6-methoxyaromadendrin 3-O-acetate, and 3-hydroxycoumarin and 19 flavonoids including 6-methoxyaromadendrin 3-O-acetate, aureusidin, iridin, flavonol 3-O-(6-O-malonyl-β-d-glucoside) quercetin 3-O-glucoside, and rutin increased at 150 and 300 mM sucrose, respectively, compared to 50 mM sucrose, indicating that the flavonoids were accumulated in M. officinalis by a higher concentration of sucrose. This is the first investigation of the change in individual flavonoids as secondary metabolites in M. officinalis by varying sucrose levels, and the results demonstrate that the sucrose causes the accumulation of certain flavonoids as a defense mechanism against osmotic stress.
Introduction
Plants contain a variety of primary and secondary metabolites, which are the intermediate or end products of cellular processes.1 Secondary metabolites including flavonoids play an important role in various biochemistry and physiological processes in plants. Their levels are considered important because they are used in obtaining valuable information such as the physiological state; they reflect specific biochemical processes in plants as metabolite levels serve as the ultimate response of biological systems to various genetic or environmental changes.2
Metabolomics, the study of chemical processes involving the entire metabolome of an organism, is a useful tool in determining metabolites in response to such changes. Various analytical tools have been used for metabolite profiling of plants, including gas chromatography/mass spectrometry,3,4 liquid chromatography–mass spectrometry (LC–MS),5,6 and nuclear magnetic resonance.7,8 LC–MS is the most commonly used in secondary metabolite profiling of plants because it offers high selectivity and sensitivity and allows the analysis of nonvolatile, unstable, and high-molecular-weight compounds without derivatization.9,10
Melissa officinalis, a perennial herb distributed throughout East Asia, has been well known as a traditional medicine used in treating human disorders such as headache, digestion disorder, Alzheimer’s disease, and cancer.11,12 Various secondary metabolites in M. officinalis are known to be responsible for antioxidative, antibacterial, anti-inflammatory, antifungal, and antitumor activities.13−16 Thus, many studies have manipulated the metabolism of M. officinalis to produce target secondary metabolites that can be used as valuable substances.17,18
Sucrose can function as the hormone-like signaling molecule and control various metabolisms and growth in plants.19 It is an important factor affecting the synthesis of the secondary metabolites pathway including flavonoid.20,21 Secondary metabolites are well known to accumulate during stressful conditions because of defense mechanisms in plants.22 For example, flavonoids accumulate in the presence of sucrose as defense mechanisms against osmotic stress in plants.20,21,23 In these studies, analysis of gene expression or total flavonoid levels revealed that sucrose induces the upregulation of flavonoid biosynthesis. However, to our knowledge, there is no study on the relationship between flavonoid biosynthesis and sucrose levels through metabolite profiles, especially the individual levels of flavonoids.
In this study, the secondary metabolite profile changes in M. officinalis were analyzed in response to different levels of sucrose. To accomplish this, we used ultraperformance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF MS), and the metabolite profiles were statistically analyzed using partial least squares-discriminant analysis (PLS-DA) and hierarchical clustering analysis (HCA). These results can be used in understanding the alteration in metabolisms based on the sucrose level and give clues on the molecular breeding of plants for overproducing high-value metabolites.
Results and Discussion
Identification of Secondary Metabolites from M. officinalis
To analyze the changes in the profile of secondary metabolites of M. officinalis in response to sucrose, M. officinalis leaves treated with 50, 150, or 300 mM sucrose were extracted with MeOH and analyzed using UPLC-Q-TOF MS. More than 20,000 peaks of the negative electrospray ionization mode (ESI–) and positive electrospray ionization ions (ESI+) were detected, and 169 metabolites were identified using XCMS in all the 15 samples obtained from five biological replicates of each condition group (Table 1), indicating that our results were more accurate and less biased than the previous reports that showed metabolic changes under the stressful conditions with 6 unidentified secondary metabolites using LC–MS/MS,24 30 identified secondary metabolites using UPLC-Q-TOF MS,25 and 95 identified metabolites using LC–MS/MS.26 These metabolites were found to be major intermediates in the secondary metabolisms of plants, including the biosynthesis of carotenoids (e.g., (2′S)-deoxymyxol 2′-α-l-fucoside), phenylpropanoids (e.g., 1-O-galloyl-β-d-glucose, justicidin B, 1-acetoxypinoresinol, 3-hydroxycoumarin, cleistanthin A, and umbelliferone), flavones, and flavonols (e.g., scullcapflavone II, flavonol 3-O-(6-O-malonyl-β-d-glucoside), cyanidin 3-O-(6″-glucosyl-2″-xylosylgalactoside), iridin, isoorientin, kolaflavanone, thymonin, quercetin 3-O-glucoside, quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside, rutin, vitexin 2″-O-β-d-glucoside, malvidin, and petunidin).
Table 1. Identified Secondary Metabolites from M. officinalis with Retention Time and m/z.
| ESI | exact mass | mass error (ppm) | matched metabolite from database | PubChem ID |
|---|---|---|---|---|
| negative | 242.080 | 19.531 | lumichrome | 5326566 |
| negative | 174.100 | 11.809 | N5-ethyl-l-glutamine | 439378 |
| negative | 258.062 | 4.088 | streptamine phosphate | 439934 |
| negative | 276.025 | 2.137 | 2-carboxy-d-arabinitol 1-phosphate | 129417 |
| negative | 232.016 | 0.655 | N-phosphohypotaurocyamine | 16019959 |
| negative | 192.063 | 10.309 | valiolone | 443630 |
| negative | 634.132 | 2.383 | actinorhodine | 441143 |
| negative | 105.033 | 6.208 | cyanopyrazine | 73172 |
| negative | 588.127 | 5.164 | kolaflavanone | 155169 |
| negative | 184.999 | 15.551 | l-serine O-sulfate | 164701 |
| negative | 278.115 | 9.500 | isohelenol | 15558 |
| negative | 332.074 | 19.246 | 1-O-galloyl-β-d-glucose | 124021 |
| negative | 331.082 | 3.990 | malvidin | 159287 |
| negative | 504.169 | 7.928 | cellotriose | 5287993 |
| negative | 128.047 | 0.992 | 2-hydroxy-cis-hex-2,4-dienoate | 11953951 |
| negative | 317.066 | 7.140 | petunidin | 73386 |
| negative | 110.037 | 8.061 | catechol | 289 |
| negative | 514.115 | 1.270 | MK 571 | 5281888 |
| negative | 650.252 | 14.857 | BQ 518 | 443291 |
| negative | 292.121 | 17.599 | INF271 | 443080 |
| negative | 856.254 | 4.466 | 7-hydroxylpradimicin A | 441176 |
| negative | 342.110 | 14.691 | 3-(4-methoxyphenyl)-5,6,7-trimethoxy-4H-1-benzopyran-4-one | 248269 |
| negative | 674.221 | 18.819 | premithramycin A2′ | 443797 |
| negative | 326.121 | 8.104 | robinobiose | 441428 |
| negative | 490.171 | 19.730 | BMS-268770 | 56928083 |
| negative | 344.108 | 12.008 | TRAM-34 | 656734 |
| negative | 328.102 | 16.053 | 7-hydroxy-6-methyl-8-ribityl lumazine | 440869 |
| negative | 518.159 | 0.550 | esmeraldic acid | 443632 |
| negative | 216.027 | 4.060 | 5-carboxymethyl-2-hydroxymuconate | 54675765 |
| negative | 192.047 | 6.535 | 6-(allylthio)purine | 3633259 |
| negative | 133.038 | 11.861 | l-aspartate | 5960 |
| negative | 405.100 | 5.991 | cefaloglycin | 19150 |
| negative | 356.098 | 17.147 | Bay-K-8644 | 2303 |
| negative | 168.019 | 12.028 | butanoylphosphate | 266 |
| negative | 307.069 | 14.055 | narciclasine | 72376 |
| negative | 150.032 | 1.013 | α-oxo-benzeneacetic acid | 11915 |
| negative | 471.150 | 4.355 | 10-formyldihydrofolate | 135398690 |
| negative | 194.063 | 1.680 | 6-(isopropylthio)purine | 3698120 |
| negative | 273.086 | 10.110 | brugine | 442998 |
| negative | 296.105 | 18.929 | calophyllin B | 5281624 |
| negative | 109.900 | 6.524 | calcium chloride anhydrous | 5284359 |
| negative | 296.092 | 19.494 | 2,3,9,10-tetrahydroxyberberine | 443768 |
| negative | 332.069 | 10.829 | hypoxylone | 442747 |
| negative | 310.121 | 18.357 | 7-hydroxy-3-(4-methoxyphenyl)-4-propyl-2H-1-benzopyran-2-one | 5357444 |
| negative | 306.060 | 18.751 | isoprothiolane sulfoxide | 93275 |
| negative | 312.106 | 11.416 | 6-O-(β-d-xylopyranosyl)-β-d-glucopyranose | 443248 |
| negative | 814.211 | 3.276 | victorin C | 21549934 |
| negative | 506.100 | 7.917 | cassiamin C | 442728 |
| negative | 392.199 | 8.060 | abyssinone VI | 5281219 |
| negative | 610.153 | 1.052 | rutin | 5280805 |
| negative | 372.100 | 0.836 | ohioensin-A | 442531 |
| negative | 594.159 | 10.092 | vitexin 2″-O-β-d-glucoside | 5280641 |
| negative | 464.096 | 9.183 | quercetin 3-O-glucoside | 5280804 |
| negative | 294.021 | 15.387 | 4-[2,2-dichloro-1-(4-methoxyphenyl)ethenyl]phenol | 156639 |
| negative | 448.101 | 7.367 | isoorientin | 114776 |
| negative | 360.085 | 15.746 | thymonin | 442662 |
| negative | 575.058 | 16.657 | isopentenyladenosine-5′-triphosphate | 23724748 |
| negative | 522.137 | 7.259 | iridin | 5281777 |
| negative | 342.074 | 17.721 | dihydromethylsterigmatocystin | 5280636 |
| negative | 516.127 | 7.769 | 1,3-dicaffeoylquinic acid | 6474640 |
| negative | 580.312 | 11.523 | hordatine B | 72193633 |
| negative | 870.218 | 14.415 | iresinin I | 11953907 |
| negative | 714.486 | 11.184 | (2′S)-deoxymyxol 2′-α-l-fucoside | 23724611 |
| negative | 487.120 | 5.943 | luciferyl sulfate | 11953812 |
| negative | 486.116 | 6.751 | flavonol 3-O-(6-O-malonyl-β-d-glucoside) | 11953833 |
| negative | 463.074 | 3.945 | N6-(1,2-dicarboxyethyl)-AMP | 447145 |
| negative | 286.048 | 2.426 | aureusidin | 5281220 |
| negative | 136.039 | 17.392 | hypoxanthine | 135398638 |
| negative | 198.039 | 3.888 | 2,4-dinitrophenylhydrazine | 3772977 |
| negative | 719.446 | 19.427 | erythromycin C | 83933 |
| negative | 432.194 | 12.242 | aspulvinone H | 54675755 |
| negative | 743.204 | 15.328 | cyanidin 3-O-(6″-glucosyl-2″-xylosylgalactoside) | 441671 |
| negative | 181.038 | 19.968 | 2-methyl-3-hydroxy-5-formylpyridine-4-carboxylate | 440898 |
| negative | 374.058 | 7.988 | glucocochlearin | 5281135 |
| negative | 364.095 | 19.154 | justicidin B | 442882 |
| negative | 750.140 | 16.841 | UDP-N-acetylmuramoyl-l-alanine | 3037124 |
| negative | 560.081 | 17.449 | dTDP-4-oxo-5-C-methyl-l-rhamnose | 443215 |
| negative | 494.121 | 13.369 | 5′-methoxyhydnocarpin-D | 5281879 |
| negative | 538.111 | 9.840 | lithospermic acid | 6441498 |
| negative | 493.098 | 18.917 | MK826 | 443580 |
| negative | 664.382 | 2.599 | phytolaccoside B | 441939 |
| negative | 344.074 | 11.436 | theogallin | 442988 |
| negative | 374.100 | 17.102 | scullcapflavone II | 124211 |
| negative | 164.069 | 10.386 | β-d-fucose | 439650 |
| negative | 685.357 | 12.321 | avadharidine | 441710 |
| negative | 162.017 | 18.924 | allicin | 65036 |
| negative | 586.314 | 19.530 | 5-oxoavermectin “2b” aglycone | 11953969 |
| negative | 584.310 | 13.431 | lappaconitine | 5281279 |
| negative | 336.059 | 15.581 | 2,2-bis(4-hydroxyphenyl)hexafluoropropane | 73864 |
| negative | 652.315 | 11.849 | thalidasine | 159795 |
| negative | 608.289 | 19.119 | oxyacanthine | 442333 |
| negative | 380.216 | 5.889 | 4,4-difluoro-17-β-hydroxyandrost-5-en-3-one propionate | 253787 |
| negative | 568.305 | 5.251 | adouetine Y | 5281578 |
| negative | 626.148 | 4.216 | quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside | 5282166 |
| negative | 660.424 | 8.099 | 12-O-palmitoyl-16-hydroxyphorbol 13-acetate | 334044 |
| negative | 1051.605 | 14.892 | aculeacin A | 14315169 |
| negative | 137.905 | 1.711 | Ba2+ | 104810 |
| positive | 305.939 | 2.050 | 3-iodo-4-hydroxyphenylpyruvate | 440184 |
| positive | 542.121 | 11.040 | isochamaejasmin | 390361 |
| positive | 184.023 | 3.201 | 5-hydroxyisourate | 250388 |
| positive | 543.110 | 11.677 | CMP-3-deoxy-d-manno-octulosonate | 445888 |
| positive | 292.132 | 11.326 | SB 206553 | 5163 |
| positive | 146.069 | 6.737 | d-glutamine | 145815 |
| positive | 115.063 | 14.931 | proline | 145742 |
| positive | 129.043 | 1.863 | 5-oxoproline | 7405 |
| positive | 303.137 | 0.083 | evodiamine | 442088 |
| positive | 117.079 | 9.054 | l-valine | 6287 |
| positive | 324.106 | 4.923 | d-fructofuranose 1,2′:2,3′-dianhydride | 440332 |
| positive | 307.084 | 6.429 | glutathione | 124886 |
| positive | 334.057 | 1.241 | nicotinamide d-ribonucleotide | 14180 |
| positive | 450.116 | 4.305 | neoastilbin | 442437 |
| positive | 305.028 | 13.271 | 2,4-dinitro-1-(3-nitrophenoxy)benzene | 221812 |
| positive | 303.007 | 18.316 | 2-(((3,5-dichlorophenyl)carbamoyl)oxy)-2-methyl-3-butenoic acid | 119359 |
| positive | 900.168 | 18.543 | N-methylanthraniloyl-CoA | 24883420 |
| positive | 162.032 | 2.679 | umbelliferone | 5281426 |
| positive | 540.163 | 1.405 | cleistanthin A | 442833 |
| positive | 621.109 | 14.679 | cyanidin 3-O-3″,6″-O-dimalonylglucoside | 23724697 |
| positive | 360.085 | 3.860 | 6-methoxyaromadendrin 3-O-acetate | 442415 |
| positive | 522.110 | 1.595 | cefoselis | 5748845 |
| positive | 162.032 | 2.617 | 3-hydroxycoumarin | 13650 |
| positive | 610.132 | 0.389 | gallocatechin-(4α→8)-epigallocatechin | 442682 |
| positive | 341.054 | 18.994 | aristolochic acid | 2236 |
| positive | 492.090 | 14.101 | carmine | 14950 |
| positive | 249.173 | 4.366 | lophocerine | 442313 |
| positive | 338.045 | 7.327 | UK-47265 | 133777 |
| positive | 606.237 | 17.505 | cancentrine | 5462434 |
| positive | 418.324 | 9.894 | 8′-apo-β-carotenol | 5280991 |
| positive | 436.334 | 8.296 | 2-phytyl-1,4-naphthoquinone | 56927684 |
| positive | 300.063 | 2.857 | kaempferide | 5281666 |
| positive | 416.147 | 0.577 | 1-acetoxypinoresinol | 442831 |
| positive | 348.169 | 1.303 | enalaprilate | 5462501 |
| positive | 892.534 | 2.918 | zeaxanthin diglucoside | 10533723 |
| positive | 183.977 | 11.963 | 3-phosphonooxypyruvate | 105 |
| positive | 801.531 | 14.884 | PC(20:4(8Z,11Z,14Z,17Z)/18:4(6Z,9Z,12Z,15Z)/0:0) | none |
| positive | 803.547 | 9.142 | PC(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/16:1(9Z)/0:0) | none |
| positive | 260.116 | 10.860 | maculosin | 119404 |
| positive | 884.542 | 8.209 | PI(20:4(8Z,11Z,14Z,17Z)/18:1(11Z)) | 53480105 |
| positive | 743.547 | 10.424 | PC(15:0/18:2(9Z,12Z)/0:0) | none |
| positive | 777.531 | 14.567 | PC(22:5(7Z,10Z,13Z,16Z,19Z)/14:1(9Z)/0:0) | none |
| positive | 781.562 | 15.183 | PC(20:3(5Z,8Z,11Z)/16:1(9Z)/0:0) | none |
| positive | 755.547 | 15.502 | PC(18:3(6Z,9Z,12Z)/16:0/0:0) | none |
| positive | 779.547 | 14.305 | PC(16:1(9Z)/20:4(5Z,8Z,11Z,14Z)/0:0) | none |
| positive | 757.562 | 13.641 | PE(15:0/22:2(13Z,16Z)/0:0) | none |
| positive | 753.531 | 4.492 | PC(20:3(5Z,8Z,11Z)/14:1(9Z)/0:0) | none |
| positive | 774.528 | 16.736 | oligomycin C | 5281901 |
| positive | 596.459 | 0.803 | spirilloxanthin | 5366506 |
| positive | 612.475 | 17.956 | DG(14:1(9Z)/22:5(4Z,7Z,10Z,13Z,16Z)/0:0) | 53477996 |
| positive | 313.914 | 6.671 | tiron | 9001 |
| positive | 739.515 | 0.965 | PE(18:2(9Z,12Z)/18:2(9Z,12Z)/0:0) | none |
| positive | 741.531 | 5.923 | PE(18:1(11Z)/18:2(9Z,12Z)/0:0) | none |
| positive | 713.500 | 11.815 | PE(20:3(5Z,8Z,11Z)/14:0/0:0) | none |
| positive | 737.500 | 9.720 | PE(14:1(9Z)/22:4(7Z,10Z,13Z,16Z)/0:0) | none |
| positive | 206.006 | 0.849 | 3-oxalomalate | 5459790 |
| positive | 336.140 | 2.553 | steroid O-sulfate | 439761 |
| positive | 715.515 | 5.771 | PE(20:2(11Z,14Z)/14:0/0:0) | none |
| positive | 208.001 | 1.808 | stipitatonate | 54746226 |
| positive | 636.341 | 18.127 | ansatrienin A | 5282069 |
| positive | 329.116 | 5.177 | 2,2′-(1-phenyl-1H-1,2,4-triazole-3,5-diyl)bisphenol | 443276 |
| positive | 716.502 | 18.708 | 1′-hydroxy-γ-carotene glucoside | 23724600 |
| positive | 328.116 | 5.397 | anisatin | 115121 |
| positive | 332.142 | 3.872 | 1-dehydro-9-fluoro-11-oxotestololactone | 253326 |
| positive | 330.139 | 1.981 | 17α-chloroethynylestradiol | 245467 |
| positive | 499.297 | 1.360 | tauroursodeoxycholic acid | 9848818 |
| positive | 155.982 | 15.912 | 2-phosphoglycolate | 529 |
| positive | 477.316 | 0.573 | gentamicin C1 | 72395 |
| positive | 757.599 | 16.260 | PE(20:1(11Z)/dm18:0/0:0) | none |
| positive | 171.952 | 6.659 | 4-bromophenol | 7808 |
| positive | 168.972 | 12.949 | 2-aminoethylarsonate | 129501 |
| positive | 168.964 | 10.786 | l-selenocysteine | 6326983 |
The secondary metabolites identified in this study are well known to have beneficial health effects. For example, rutin, lithospermic acid, moxalactam, isoorientin, 5′-methoxyhydnocarpin-D, oxyacanthine, 1,3-dicaffeoylquinic acid, isohelenol, lappaconitine, phytolaccoside B, iridin, and scullcapflavone II are known to possess various physiological activities such as antioxidative,27 antibacterial,28 hepatoprotective,29 anti-HIV-1,30 antifungal,31 antimutagenic,32 and anti-inflammatory33 activities. Specifically, malvidin, a primary plant pigment, inhibits human leukemia cells by arresting the G2/M phase and then inducing apoptosis.34 Lithospermic acid can be used in diabetic retinopathy and mesenteric ischemia reperfusion injury because of its antioxidative, hepatoprotective, and anti-inflammatory effects.27,35
PLS-DA of the Sucrose Effect on Secondary Metabolite Profiles
To statistically compare changes in the profile of secondary metabolites of M. officinalis in response to different levels of sucrose, principal component analysis (PCA) was performed using SIMCA-P+. Because the metabolite profiles of the groups were slightly discriminated by PCA, with 0.52 of R2X and 0.33 of Q2 (data not shown), PLS-DA was employed to obtain better separations between the groups. Among the three groups treated with sucrose levels of 50, 150, and 300 mM, the metabolite profiles were clearly separated by partial least squares 1 (PLS1) and 2 (PLS2) in the score plot of PLS-DA (Figure 1). The model generated explained variation values, such as 0.52 of R2X and 0.97 of R2Y, and a predictive capability value, such as 0.87 of cumulative Q2, indicating a good model. Our previous study on the change in flavonoid levels in lemon balm by sucrose also showed that six metabolite profiles were significantly different between 50, 150, and 300 mM sucrose.24 However, the present results may be considered more accurate and reliable because only six secondary metabolites (i.e., 435.13, 523.129, 540.063, 573.200, 615.714, and 617.153) were used in the previous study without identification. In the permutation test, all points of permuted R2 and Q2 values to the left were located in the lower side contrary to the original points, and the regression line of Q2 had a negative intercept, indicating that the PLS-DA models were clearly validated without overfitting from the original model (Figure S2).36
Figure 1.
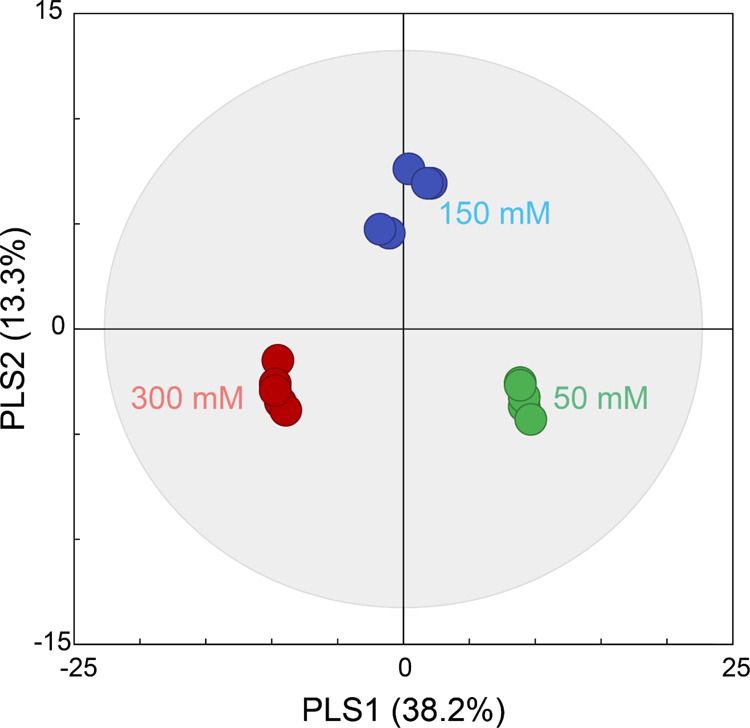
PLS-DA score plot of secondary metabolite profiles in M. officinalis treated with 50 (control; green), 150 (blue), and 300 mM (red).
The loading scores of the selected 20 metabolites, which represented the magnitude of the contribution of each metabolite to PLS, are listed in Table 2. Of the identified 169 metabolites in this study, 40 metabolites including anisatin, quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside, isohelenol, and l-aspartate contributed positively to PLS1. However, 129 metabolites such as lithospermic acid, iridin, 3-hydroxycoumarin, and 6-methoxyaromadendrin 3-O-acetate contributed negatively to PLS1. Seventy-nine metabolites including 10-formyldihydrofolate, 2,4-dinitrophenylhydrazine, and allicin contributed positively to PLS2, while 90 metabolites such as quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside, rutin, thalidasine, and victorin C contributed negatively to PLS2.
Table 2. Top 20 Identified Metabolites with High Absolute Loadings on PLS1 and PLS2 as Determined by PLS-DA.
| PLS1 |
PLS2 |
||
|---|---|---|---|
| metabolite | loading | metabolite | loading |
| anisatin | 0.113 | 10-formyldihydrofolate | 0.197 |
| 2,2′-(1-phenyl-1H-1,2,4-triazole-3,5-diyl)bisphenol | 0.107 | 2,4-dinitrophenylhydrazine | 0.159 |
| quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside | 0.107 | 2-carboxy-d-arabinitol 1-phosphate | 0.157 |
| isohelenol | 0.101 | 2-methyl-3-hydroxy-5-formylpyridine-4-carboxylate | 0.112 |
| l-aspartate | 0.092 | 5′-methoxyhydnocarpin-D | 0.138 |
| thalidasine | 0.091 | 7-hydroxy-6-methyl-8-ribityl lumazine | 0.134 |
| aspulvinone H | 0.091 | allicin | 0.132 |
| 2,2-bis(4-hydroxyphenyl)hexafluoropropane | 0.091 | α-oxo-benzeneacetic acid | 0.130 |
| allicin | 0.090 | Ba2+ | 0.120 |
| 6-O-(β-d-xylopyranosyl)-β-d-glucopyranose | 0.090 | butanoylphosphate | 0.115 |
| isopentenyladenosine-5′-triphosphate | –0.115 | CMP-3-deoxy-d-manno-octulosonate | –0.107 |
| 6-methoxyaromadendrin 3-O-acetate | –0.115 | DG(14:1(9Z)/22:5(4Z,7Z,10Z,13Z,16Z)/0:0) | –0.107 |
| BMS-268770 | –0.115 | 5-oxoproline | –0.109 |
| 2,4-dinitrophenylhydrazine | –0.115 | 1-dehydro-9-fluoro-11-oxotestololactone | –0.115 |
| gentamicin C1 | –0.115 | cassiamin C | –0.121 |
| iridin | –0.118 | cellotriose | –0.121 |
| 3-hydroxycoumarin | –0.118 | victorin C | –0.130 |
| cefoselis | –0.119 | rutin | –0.132 |
| lithospermic acid | –0.119 | thalidasine | –0.132 |
| dihydromethylsterigmatocystin | –0.123 | quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside | –0.141 |
In variable importance in projection (VIP) analysis, VIP values greater than 1 are considered important.36 In this study, 78 metabolites such as quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside, rutin, umbelliferone, and cleistanthin A were shown to have VIP values greater than 1, of which 16 metabolites belong to flavonoid classes (Table 3). These results suggested that the flavonoids were critical metabolites for discriminating between the groups.
Table 3. VIP Scores of the 78 Metabolites with a VIP >1.0 That Strongly Contributed to the PLS-DA Model.
| metabolite | VIP |
|---|---|
| 2-methyl-3-hydroxy-5-formylpyridine-4-carboxylate | 1.772 |
| narciclasine | 1.598 |
| glutathione | 1.588 |
| allicin | 1.500 |
| α-oxo-benzeneacetic acid | 1.417 |
| quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside | 1.400 |
| PE(15:0/22:2(13Z,16Z)/0:0) | 1.400 |
| phytolaccoside B | 1.378 |
| N6-(1,2-dicarboxyethyl)-AMP | 1.377 |
| iresinin I | 1.375 |
| rutin | 1.366 |
| 10-formyldihydrofolate | 1.363 |
| umbelliferone | 1.360 |
| cassiamin C | 1.359 |
| PC(18:3(6Z,9Z,12Z)/16:0/0:0) | 1.355 |
| victorin C | 1.350 |
| 2-carboxy-d-arabinitol 1-phosphate | 1.343 |
| cleistanthin A | 1.338 |
| 2,4-dinitrophenylhydrazine | 1.332 |
| 5′-methoxyhydnocarpin-D | 1.330 |
| gentamicin C1 | 1.318 |
| 6-methoxyaromadendrin 3-O-acetate | 1.317 |
| flavonol 3-O-(6-O-malonyl-β-d-glucoside) | 1.316 |
| luciferyl sulfate | 1.311 |
| erythromycin C | 1.302 |
| 5-oxoproline | 1.297 |
| 1-dehydro-9-fluoro-11-oxotestololactone | 1.290 |
| 3-hydroxycoumarin | 1.279 |
| proline | 1.274 |
| thymonin | 1.271 |
| l-selenocysteine | 1.252 |
| cefoselis | 1.250 |
| PC(22:5(7Z,10Z,13Z,16Z,19Z)/14:1(9Z)/0:0) | 1.233 |
| thalidasine | 1.226 |
| l-aspartate | 1.214 |
| 7-hydroxy-6-methyl-8-ribityl lumazine | 1.211 |
| aspulvinone H | 1.198 |
| 2,2-bis(4-hydroxyphenyl)hexafluoropropane | 1.197 |
| Ba2+ | 1.192 |
| catechol | 1.190 |
| scullcapflavone II | 1.182 |
| lithospermic acid | 1.173 |
| N-methylanthraniloyl-CoA | 1.170 |
| iridin | 1.168 |
| gallocatechin-(4α→8)-epigallocatechin | 1.168 |
| 2-(((3,5-dichlorophenyl)carbamoyl)oxy)-2-methyl-3-butenoic acid | 1.158 |
| PC(22:6(4Z,7Z,10Z,13Z,16Z,19Z)/16:1(9Z)/0:0) | 1.156 |
| dihydromethylsterigmatocystin | 1.151 |
| vitexin 2″-O-β-d-glucoside | 1.149 |
| evodiamine | 1.143 |
| lophocerine | 1.142 |
| CMP-3-deoxy-d-manno-octulosonate | 1.138 |
| aureusidin | 1.116 |
| INF271 | 1.115 |
| BQ 518 | 1.113 |
| tauroursodeoxycholic acid | 1.110 |
| isohelenol | 1.104 |
| hypoxanthine | 1.101 |
| isopentenyladenosine-5′-triphosphate | 1.093 |
| 2,2′-(1-phenyl-1H-1,2,4-triazole-3,5-diyl)bisphenol | 1.082 |
| PC(20:3(5Z,8Z,11Z)/16:1(9Z)/0:0) | 1.075 |
| UDP-N-acetylmuramoyl-l-alanine | 1.065 |
| PE(20:3(5Z,8Z,11Z)/14:0/0:0) | 1.060 |
| cellotriose | 1.057 |
| BMS-268770 | 1.056 |
| 3-iodo-4-hydroxyphenylpyruvate | 1.056 |
| 6-(isopropylthio)purine | 1.053 |
| carmine | 1.053 |
| butanoylphosphate | 1.046 |
| DG(14:1(9Z)/22:5(4Z,7Z,10Z,13Z,16Z)/0:0) | 1.045 |
| anisatin | 1.043 |
| 4-[2,2-dichloro-1-(4-methoxyphenyl)ethenyl]phenol | 1.042 |
| PI(20:4(8Z,11Z,14Z,17Z)/18:1(11Z)) | 1.040 |
| isoorientin | 1.034 |
| quercetin 3-O-glucoside | 1.017 |
| cefaloglycin | 1.015 |
| avadharidine | 1.013 |
| justicidin B | 1.009 |
HCA of the Sucrose Effect on Secondary Metabolite Profiles
To cluster and visualize the discrimination of secondary metabolite profiles with 50, 150, and 300 mM sucrose, HCA with the Euclidean distance coefficient and average linkage was performed using MeV software. After normalization using the sum of identified metabolites and then transformation using unit variance scaling, data composed of identified metabolites and groups (50, 150, and 300 mM sucrose) were exported into the heat map.
In the heat map, five biological replicates at each group had similar metabolite profiles (Figure 2). However, the metabolite profiles were significantly different depending on different sucrose levels, 50, 150, and 300 mM. The secondary metabolite profile of 150 mM sucrose was closer to that of 300 mM sucrose than to that of 50 mM sucrose. These results are similar to those obtained in a previous study on primary metabolite profiles in M. officinalis with 64 metabolites.4 This comparison indicates that the effect of sucrose level on primary metabolite profiles may be associated with the secondary metabolite profiles in M. officinalis. Moreover, the clustering of secondary metabolite profiles between sucrose levels was enabled by certain individual metabolites. For example, l-serine O-sulfate, thalidasin, spirilloxanthin, and quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside increased in 50 mM. However, the levels of proline, glutathione, isoorientin scullcapflavone II, flavonol 3-O-(6-O-malonyl-β-d-glucoside), luciferyl sulfate, cassiamin C, and rutin were much higher in 300 mM sucrose than in 50 and 150 mM sucrose.
Figure 2.
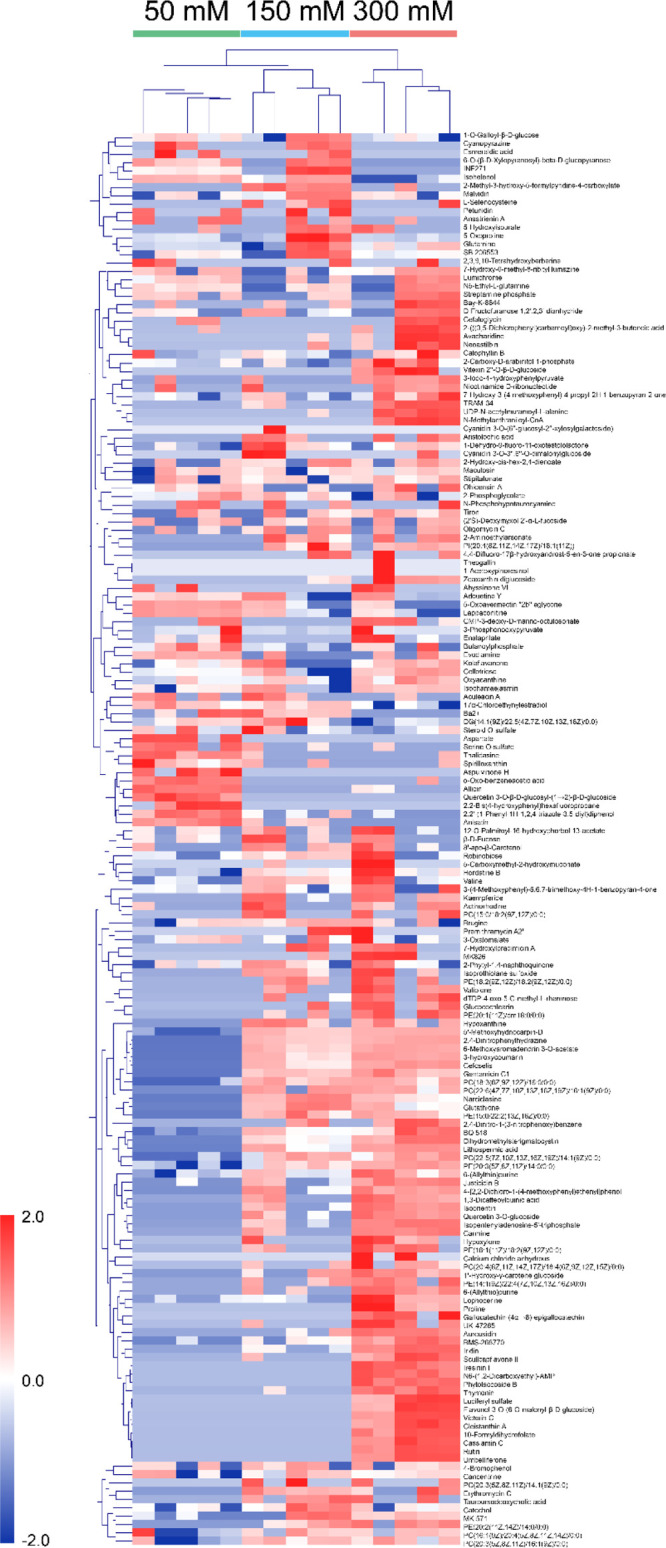
Clustered heat map of 169 secondary metabolites of M. officinalis treated with 50 (control; green), 150 (blue), and 300 mM (red) sucrose. Similarity assessment of clustering based on the Euclidean distance coefficient and average linkage method. Each column and each row represent different concentrations of sucrose and individual metabolite, respectively.
Comparison of Individual Flavonoid Levels with 50, 150, and 300 mM Sucrose
Most studies have reported only total flavonoid abundances to reveal the relationship between sucrose levels and contents of total flavonoids20,24,37,38 or the phenylpropanoid pathway39,40 without identifying or comparing individual flavonoid abundances. In this study, we identified individual secondary metabolites and determined the changes in each flavonoid, anthocyanindin, and phenlypropanoid levels depending on sucrose levels.
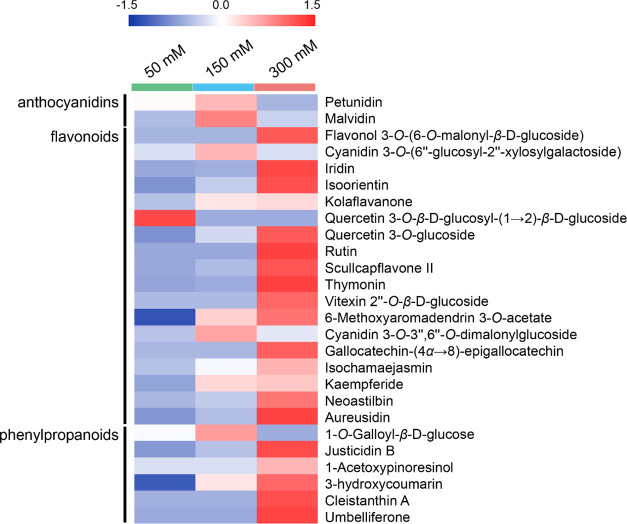
To compare the changes in flavonoid level between the groups, one-way analysis of variance with the post hoc Tukey’s honestly significant difference test was conducted using Statistica (p > 0.05). The abundance of three flavonoids such as quercetin 3-O-β-d-glucosyl-(1→2)-β-d-glucoside, 6-methoxyaromadendrin 3-O-acetate, and 3-hydroxycoumarin increased with 150 mM sucrose compared to those with 50 mM sucrose. However, compared to those with 50 mM sucrose, the abundances of most flavonoids such as 6-methoxyaromadendrin 3-O-acetate, 3-hydroxycoumarin, aureusidin, thymonin, rutin, justicidin B, isoorientin, quercetin 3-O-glucoside, umbelliferone, iridin, scullcapflavone II, cleistanthin A, flavonol 3-O-(6-O-malonyl-β-glucoside), isochamaejasmin, gallocatechin-(α→8)-epigallocatechin, vitexin 2″-O-β-glucoside, kolaflavanone, kaempferide, and neoastilbin were significantly increased with 300 mM (Figure 3). These results showed that flavonoids accumulated depending on the sucrose level, indicating that sucrose induced the production of more flavonoids via the phenylpropanoid pathway.
Figure 3.
Heat map of 26 flavonoids in M. officinalis treated with 50 (control; green), 150 (blue), and 300 mM (red). Each row represents individual flavonoids.
Similar to these results, previous studies have reported that rutin accumulates in Fagopyrum esculentum Moench in response to sucrose41 and quercetin 3-O-glucoside accumulates in Arabidopsis under abiotic and oxidative stress.42 Our results showed that the types of accumulating flavonoids in M. officinalis differed depending on sucrose levels, and active flavonoid biosynthesis served as a defense mechanism against osmotic stress, suggesting that the biosynthetic pathway of flavonoids was regulated by the sucrose signaling pathway. Previously, it was observed at the messenger RNA level that sucrose caused the accumulation of anthocyanins and the upregulation of anthocyanin synthesis.20 However, our results showed that anthocyanins (e.g., malvidin and petunidin) did not accumulate under a high sucrose level at both 150 and 300 mM. This is possibly because anthocyanins other than malvidin and petunidin were not identified in this study, and malvidin and petunidin could not represent the behaviors of all other anthocyanins under a high sucrose level. The precise prediction and speculation of the secondary metabolism and change in individual secondary metabolites of M. officinalis in response to the concentrations of sucrose should be supported and verified by further experiments.
Conclusions
This is the first report to investigate the change in secondary metabolite profiles in M. officinalis depending on the sucrose level using UPLC-Q-TOF MS. One hundred and sixty-nine metabolites were identified using XCMS; these metabolites were major intermediates in the secondary metabolism of plants such as the biosynthesis of carotenoids, phenylpropanoids, flavones, and flavonols, which serves as a defense mechanism against stress in plants. PLS-DA and HCA results showed a significant difference in secondary metabolite profiles in M. officinalis between 50, 150, and 300 mM sucrose. In contrast to that with 50 mM sucrose, 32 secondary metabolites such as 6-methoxyaromadendrin 3-O-acetate and 3-hydroxycoumarin accumulated in 150 mM, and 76 metabolites such as aureusidin, thymonin, quercetin 3-O-glucoside, and rutin increased in 300 mM. Accumulation of different types of flavonoids was observed depending on the sucrose level, suggesting that the accumulation of these flavonoids acts as a defense mechanism against osmotic stress. This study demonstrated that secondary metabolite profiles could be a useful tool for investigating the change in certain secondary metabolites and secondary metabolism in plants under osmotic stress and provide clues for manipulating plant metabolisms to produce target flavonoids, which have various properties such as antitumoral, antioxidant, antifungal, and antibacterial activities.
Materials and Methods
Plant Growth Conditions
M. officinalis was prepared as previously described.4 Briefly, M. officinalis was cultivated in 4 g/L Murashige and Skoog medium (0.025 mg/L of CoCl2·6H2O, 0.025 mg/L of CuSO4·5H2O, 36.70 mg/L of FeNaEDTA, 6.20 mg/L of H3BO3, 0.83 mg/L of KI, 16.9 mg/L of MnSO4·H2O, 0.25 mg/L of Na2MoO4·2H2O, 8.60 mg/L of ZnSO4·7H2O, 332.02 mg/L of CaCl2, 170.00 mg/L of KH2PO4, 1900.00 mg/L of KNO3, 180.54 mg/L of MgSO4, and 1650.00 mg/L of NH4NO3) containing 50 mM sucrose and 7 g/L agar at pH 5.7 after 2 cm-long explants with two leaves were transferred to the culture and test media with three different concentrations of sucrose, 50 (control), 150, or 300 mM, for examining the effects of sucrose concentration on flavonoid accumulation in M. officinalis.4,24 The leaves were incubated at 25 °C for 20 days (15:9 h light–dark cycle). The leaves of M. officinalis were harvested and quickly frozen in liquid nitrogen to quench cellular metabolism, and the frozen samples were stored at −80 °C.
Metabolite Extraction and UPLC-Q-TOF MS Analysis
Fifty milligrams of ground M. officinalis leaves were extracted with 0.5 mL of cold methanol (high-performance liquid chromatography grade, Merck, Darmstadt, Germany). The methanol extract was diluted with 50 μL and was thoroughly vortexed, after which it was centrifuged at 14,000g for 5 min. The supernatant was filtered using a 0.45 μm syringe filter (hydrophilic poly(tetrafluoroethylene), Advantec, Dublin, OH). The metabolite extract was stored at −20 °C before UPLC-Q-TOF MS analysis.
Metabolite extract was analyzed by UPLC-Q-TOF MS. The UPLC analysis was performed using a Waters ACQUITY UPLC system (Waters, Milford, MA) equipped with a Waters ACQUITY BEH C18 column (100 × 2.1 mm, 1.7 μm). The mobile phase consisted of solvent A, 0.1% (w/v) formic acid in distilled water, and solvent B, 0.1% (w/v) formic acid in acetonitrile. The UPLC was eluted first with a linear gradient from 10 to 100% of solvent B (0–7.0 min) and then eluted isocratically with 100% of solvent B (7.0–8.0 min). The flow rate was 0.3 mL/min, and the injection volume was 5 μL. The column and autosampler were maintained at 35 and 15 °C, respectively. Mass spectrometry was performed using a Q-TOF micromass detector (Waters, Manchester, UK). The conditions of the Q-TOF mass spectrometer in the negative electrospray ionization (ESI) mode were 2800 V of capillary voltage, 35 V of sample cone voltage, 1.0 V of extraction cone voltage, 250 °C of desolvation temperature, 100 °C of source temperature, and 500 L/h of desolvation gas flow rate. The positive ESI was under the same conditions, expect for an extraction cone voltage of 2.0 V. The ESI mass spectra were acquired over m/z 100–1500. Leucine-enkephalin was used as a reference ion by the LockSpray interface to measure mass more accurately and reproducibly.
Data Processing and Statistical Analysis
Acquired data were analyzed using Waters MassLynx (version 4.1). The noise elimination level was set at 6.0 with 10 masses per retention time being collected. Before further processing, lock spray scans were removed because lock spray peaks disrupted the detection and analysis of actual signals from samples (Figure S1A,B). UPLC-Q-TOF MS data were preprocessed using XCMS with signal-to-noise ratios as described in the literature (Table S1).43,44 Mass and retention time windows were set at 0.05 Da and 0.20 min, respectively. After normalization by log transformation, the processed data were further analyzed using PLS-DA and HCA with the Euclidean distance coefficient and average linkage methods. SIMCA-P+ (version 14.1, Umetrics AB, Umea, Sweden) was used for PLS-DA,36 and MeV (MultiExperiment Viewer; Dana-Farber Cancer Institute, Boston, MA) was used for HCA.45 Statistica (version 7.1; StatSoft, Tulsa, OK) was used for the univariate analysis.46
Acknowledgments
This work was supported by the Mid-career Researcher Program and Young Researcher Program from the National Research Foundation of Korea (NRF-2020R1A2B5B02002631 and NRF-2020R1G1A100826811, respectively). S.K. acknowledges the support of the Research Grant of Jeonju University in 2019. Facility support at the Korea University Food Safety Hall by the Institute of Biomedical Science and Food Safety is also acknowledged.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c04745.
Chromatograms for the same samples before and after removal of lock spray scans as examples; validation of the PLS-DA model using the 100 permutation test; and number of peaks detected, peak groups, IP clusters, and predictions in negative and positive modes (PDF)
Author Present Address
⊥ Forest Medicinal Resources Research Center, National Institute of Forest Science, Yongju 36040, South Korea.
Author Contributions
K.H.K. and D.L. conceived and designed the project. S.K. and H.L. collected the samples. S.K., J.K., and N.K. performed the experiments. S.K., J.K., N.K., D.-Y.L., and D.L. acquired the metabolomics data. S.K., D.L., H.L., D.-Y.L., and K.H.K. analyzed the data. S.K., D.-Y.L., and K.H.K. wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. 10.1007/978-94-010-0448-0_11. [DOI] [PubMed] [Google Scholar]
- Weckwerth W. Metabolomics in systems biology. Annu. Rev. Plant Biol. 2003, 54, 669–689. 10.1146/annurev.arplant.54.031902.135014. [DOI] [PubMed] [Google Scholar]
- Fiehn O.; Kopka J.; Trethewey R. N.; Willmitzer L. Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 2000, 72, 3573–3580. 10.1021/ac991142i. [DOI] [PubMed] [Google Scholar]
- Kim S.; Shin M. H.; Hossain M. A.; Yun E. J.; Lee H.; Kim K. H. Metabolite profiling of sucrose effect on the metabolism of Melissa officinalis by gas chromatography-mass spectrometry. Anal. Bioanal. Chem. 2011, 399, 3519–3528. 10.1007/s00216-011-4693-0. [DOI] [PubMed] [Google Scholar]
- Theodoridis G.; Gika H.; Franceschi P.; Caputi L.; Arapitsas P.; Scholz M.; Masuero D.; Wehrens R.; Vrhovsek U.; Mattivi F. LC-MS based global metabolite profiling of grapes: solvent extraction protocol optimisation. Metabolomics 2012, 8, 175–185. 10.1007/s11306-011-0298-z. [DOI] [Google Scholar]
- Matsuda F.; Yonekura-Sakakibara K.; Niida R.; Kuromori T.; Shinozaki K.; Saito K. MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 2009, 57, 555–577. 10.1111/j.1365-313x.2008.03705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan P.; Kruger N. J.; Ratcliffe R. G. Metabolite fingerprinting and profiling in plants using NMR. J. Exp. Bot. 2005, 56, 255–265. 10.1093/jxb/eri010. [DOI] [PubMed] [Google Scholar]
- Bundy J. G.; Spurgeon D. J.; Svendsen C.; Hankard P. K.; Osborn D.; Lindon J. C.; Nicholson J. K. Earthworm species of the genus Eisenia can be phenotypically differentiated by metabolic profiling. FEBS Lett. 2002, 521, 115–120. 10.1016/s0014-5793(02)02854-5. [DOI] [PubMed] [Google Scholar]
- Fraser P. D.; Pinto M. E. S.; Holloway D. E.; Bramley P. M. Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J. 2000, 24, 551–558. 10.1046/j.1365-313x.2000.00896.x. [DOI] [PubMed] [Google Scholar]
- Sumner L. W.; Mendes P.; Dixon R. A. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 2003, 62, 817–836. 10.1016/s0031-9422(02)00708-2. [DOI] [PubMed] [Google Scholar]
- Tagashira M.; Ohtake Y. A new antioxidative 1,3-benzodioxole from Melissa officinalis. Planta Med. 1998, 64, 555–558. 10.1055/s-2006-957513. [DOI] [Google Scholar]
- de Sousa A. C.; Gattass C. R.; Alviano D. S.; Alviano C. S.; Blank A. F.; Alves P. B. Melissa officinalis L. essential oil: antitumoral and antioxidant activities. J. Pharm. Pharmacol. 2004, 56, 677–681. 10.1211/0022357023321. [DOI] [PubMed] [Google Scholar]
- Kim S.; Yun E. J.; Bak J. S.; Lee H.; Lee S. J.; Kim C. T.; Lee J.-H.; Kim K. H. Response surface optimised extraction and chromatographic purification of rosmarinic acid from Melissa officinalis leaves. Food Chem. 2010, 121, 521–526. 10.1016/j.foodchem.2009.12.040. [DOI] [Google Scholar]
- Nikitina V. S.; Kuz’mina L. Y.; Melent’ev A. I.; Shendel’ G. V. Antibacterial activity of polyphenolic compounds isolated from plants of geraniaceae and rosaceae families. Appl. Biochem. Microbiol. 2007, 43, 629–634. 10.1134/s0003683807060117. [DOI] [PubMed] [Google Scholar]
- Viollon C.; Chaumont J.-P. Antifungal properties of essential oils and their main components upon Cryptococcus neoformans. Mycopathologia 1994, 128, 151–153. 10.1007/bf01138476. [DOI] [PubMed] [Google Scholar]
- Miron T. L.; Herrero M.; Ibáñez E. Enrichment of antioxidant compounds from lemon balm (Melissa officinalis) by pressurized liquid extraction and enzyme-assisted extraction. J. Chromatogr. A 2013, 1288, 1–9. 10.1016/j.chroma.2013.02.075. [DOI] [PubMed] [Google Scholar]
- Weitzel C.; Petersen M. Enzymes of phenylpropanoid metabolism in the important medicinal plant Melissa officinalis L. Planta 2010, 232, 731–742. 10.1007/s00425-010-1206-x. [DOI] [PubMed] [Google Scholar]
- Kong J.-Q. Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering. RSC Adv. 2015, 5, 62587–62603. 10.1039/c5ra08196c. [DOI] [Google Scholar]
- Smeekens S.; Hellmann H. A. Sugar sensing and signaling in plants. Front. Plant Sci. 2014, 5, 113. 10.3389/fpls.2014.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C.; Poggi A.; Loreti E.; Alpi A.; Perata P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006, 140, 637–646. 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.; Tian L.; Liu H.; Pan Q.; Zhan J.; Huang W. Sugars induce anthocyanin accumulation and flavanone 3-hydroxylase expression in grape berries. Plant Growth Regul. 2009, 58, 251–260. 10.1007/s10725-009-9373-0. [DOI] [Google Scholar]
- Bennett R. N.; Wallsgrove R. M. Secondary metabolites in plant defence mechanisms. New Phytol. 1994, 127, 617–633. 10.1111/j.1469-8137.1994.tb02968.x. [DOI] [PubMed] [Google Scholar]
- Lloyd J. C.; Zakhleniuk O. V. Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J. Exp. Bot. 2004, 55, 1221–1230. 10.1093/jxb/erh143. [DOI] [PubMed] [Google Scholar]
- Hossain M. A.; Kim S.; Kim K. H.; Lee S.-J.; Lee H. Flavonoid compounds are enriched in lemon balm (Melissa officinalis) leaves by a high level of sucrose and confer increased antioxidant activity. Hortscience 2009, 44, 1907–1913. 10.21273/hortsci.44.7.1907. [DOI] [Google Scholar]
- Pant B.-D.; Pant P.; Erban A.; Huhman D.; Kopka J.; Scheible W.-R. Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant, Cell Environ. 2015, 38, 172–187. 10.1111/pce.12378. [DOI] [PubMed] [Google Scholar]
- Matsuda F.; Yonekura-Sakakibara K.; Niida R.; Kuromori T.; Shinozaki K.; Saito K. MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 2009, 57, 555–577. 10.1111/j.1365-313x.2008.03705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C. J.; Yu S. H.; Wang X.-M.; Woo S. J.; Park H. J.; Lee H. C.; Choi S. H.; Kim K. M.; Kim J. H.; Park K. S.; Jang H. C.; Lim S. The effect of lithospermic acid, an antioxidant, on development of diabetic retinopathy in spontaneously obese diabetic rats. PLoS One 2014, 9, e98232 10.1371/journal.pone.0098232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.; Zheng B. W.; Yu W.; Niu T. S.; Xiao T. T.; Zhang J.; Xiao Y. H. Antibacterial effect evaluation of moxalactam against extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae with in vitro pharmacokinetics/pharmacodynamics simulation. Infect. Drug Resist. 2018, 11, 103–112. 10.2147/idr.s150431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q. F.; Zhang S. J.; Zheng L.; Liao M.; He M.; Huang R. B.; Zhuo L.; Lin X. Protective effect of isoorientin-2 ’’-O-alpha-L-arabinopyranosyl isolated from Gypsophila elegans on alcohol induced hepatic fibrosis in rats. Food Chem. Toxicol. 2012, 50, 1992–2001. 10.1016/j.fct.2012.03.044. [DOI] [PubMed] [Google Scholar]
- Varadaraju T. G.; Hwu J. R. Synthesis of anti-HIV lithospermic acid by two diverse strategies. Org. Biomol. Chem. 2012, 10, 5456–5465. 10.1039/c2ob25575h. [DOI] [PubMed] [Google Scholar]
- Escalante A.; Gattuso M.; Pérez P.; Zacchino S. Evidence for the mechanism of action of the antifungal phytolaccoside B isolated from phytolacca tetramera hauman. J. Nat. Prod. 2008, 71, 1720–1725. 10.1021/np070660i. [DOI] [PubMed] [Google Scholar]
- Wozniak D.; Janda B.; Kapusta I.; Oleszek W.; Matkowski A. Antimutagenic and anti-oxidant activities of isoflavonoids from Belamcanda chinensis (L.) DC. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2010, 696, 148–153. 10.1016/j.mrgentox.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Jang H.-Y.; Ahn K.-S.; Park M.-J.; Kwon O.-K.; Lee H.-K.; Oh S.-R. Skullcapflavone II inhibits ovalbumin-induced airway inflammation in a mouse model of asthma. Int. Immunopharmacol. 2012, 12, 666–674. 10.1016/j.intimp.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Hyun J. W.; Chung H. S. Cyanidin and malvidin from Oryza sativa cv. Heugjinjubyeo mediate cytotoxicity against human monocytic leukemia cells by arrest of G(2)/M phase and induction of apoptosis. J. Agric. Food Chem. 2004, 52, 2213–2217. 10.1021/jf030370h. [DOI] [PubMed] [Google Scholar]
- Chan K. W. K.; Ho W. S. Anti-oxidative and hepatoprotective effects of lithospermic acid against carbon tetrachloride-induced liver oxidative damage in vitro and in vivo. Oncol. Rep. 2015, 34, 673–680. 10.3892/or.2015.4068. [DOI] [PubMed] [Google Scholar]
- Umetrics A. B.User’s Guide to SIMCA-P, SIMCA-P+ version 11.0; Umetrics AB: UmeÅ, Sweden, 2005. [Google Scholar]
- Gollop R.; Even S.; Colova-Tsolova V.; Perl A. Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J. Exp. Bot. 2002, 53, 1397–1409. 10.1093/jxb/53.373.1397. [DOI] [PubMed] [Google Scholar]
- Pasqua G.; Monacelli B.; Mulinacci N.; Rinaldi S.; Giaccherini C.; Innocenti M.; Vinceri F. F. The effect of growth regulators and sucrose on anthocyanin production in Camptotheca acuminata cell cultures. Plant Physiol. Biochem. 2005, 43, 293–298. 10.1016/j.plaphy.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Li Y.; Van den Ende W.; Rolland F. Sucrose induction of anthocyanin biosynthesis Is bediated by DELLA. Mol. Plant 2014, 7, 570–572. 10.1093/mp/sst161. [DOI] [PubMed] [Google Scholar]
- Payyavula R. S.; Singh R. K.; Navarre D. A. Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 2013, 64, 5115–5131. 10.1093/jxb/ert303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Park N. I.; Park C. H.; Kim S. G.; Lee S. Y.; Park S. U. Influence of sucrose on rutin content and flavonoid biosynthetic gene expression in seedlings of common buckwheat (Fagopyrum esculentum Moench). Plant Omics 2011, 4, 215–219. [Google Scholar]
- Nakabayashi R.; Yonekura-Sakakibara K.; Urano K.; Suzuki M.; Yamada Y.; Nishizawa T.; Matsuda F.; Kojima M.; Sakakibara H.; Shinozaki K.; Michael A. J.; Tohge T.; Yamazaki M.; Saito K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. S.; Ho Y. S.; Yeo H. C.; Lin J. P. Y.; Lee D.-Y. Precursor mass prediction by clustering ionization products in LC-MS-based metabolomics. Metabolomics 2013, 9, 1301–1310. 10.1007/s11306-013-0539-4. [DOI] [Google Scholar]
- Huan T.; Forsberg E. M.; Rinehart D.; Johnson C. H.; Ivanisevic J.; Benton H. P.; Fang M.; Aisporna A.; Hilmers B.; Poole F. L.; Thorgersen M. P.; Adams M. W. W.; Krantz G.; Fields M. W.; Robbins P. D.; Niedernhofer L. J.; Ideker T.; Majumder E. L.; Wall J. D.; Rattray N. J. W.; Goodacre R.; Lairson L. L.; Siuzdak G. Systems biology guided by XCMS Online metabolomics. Nat. Methods 2017, 14, 461–462. 10.1038/nmeth.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A. I.; Bhagabati N. K.; Braisted J. C.; Liang W.; Sharov V.; Howe E. A.; Li J.; Thiagarajan M.; White J. A.; Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. 10.1016/s0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Lee D. Y.; Fiehn O. High quality metabolomic data for Chlamydomonas reinhardtii. Plant Methods 2008, 4, 7. 10.1186/1746-4811-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.