Abstract
Many acute viral infections target tissue macrophages, yet the mechanisms of macrophage-mediated control of viruses are poorly understood. Here, we report that CD40 expressed by peritoneal macrophages restricts early infection of a broad range of RNA viruses. Loss of CD40 expression enhanced virus replication as early as 12–24 hours of infection and, conversely, stimulation of CD40 signaling with an agonistic antibody blocked infection. With peritoneal cell populations infected with the filovirus, wild-type (WT) Ebola virus (EBOV), or a BSL2 model virus, recombinant vesicular stomatitis virus encoding Ebola virus glycoprotein (rVSV/EBOV GP), we examined the mechanism conferring protection. Here we demonstrate that restricted virus replication in macrophages required CD154/CD40 interactions which stimulated IL-12 production through TRAF6-dependent signaling. In turn, IL-12 production resulted in interferon gamma (IFN-γ) production, which induced proinflammatory polarization of macrophages, protecting the cells from infection. These CD40-dependent events protected mice against virus challenge. CD40−/− mice were exquisitely sensitive to intraperitoneal challenge with a dose of rVSV/EBOV GP that was sublethal to CD40+/+ mice, exhibiting viremia within 12 hours of infection and rapidly succumbing to infection. This study identifies a previously unappreciated role for macrophage-intrinsic CD40 signaling in controlling acute virus infection.
Keywords: Interferon gamma, IL-12, Ebola virus, filovirus, innate immunity, virus restriction, CD40, CD40 signaling, peritoneum, macrophage, TRAF6, RNA virus
Summary sentence:
CD40 signaling in macrophages inhibits acute virus replication at early times during infection.
Introduction:
CD40 is a member of the tumor necrosis factor receptor superfamily expressed by antigen presenting cells (APCs) such as B cells, dendritic cells (DCs), macrophages as well as some non-immune cells (1). CD40 serves as a costimulatory molecule for adaptive immunity that interacts with CD154 (CD40 ligand or CD40L) expressed primarily on activated T cells (2). Both humoral and cellular immunity are critically dependent on CD40/CD154 interactions (1). These interactions lead to B cell maturation, immunoglobulin and cytokine production and generation of plasma cells as well as memory B cells. In addition, CD40 signaling in APCs results in proinflammatory cytokine production, upregulation of costimulatory molecules and enhanced expression of MHC class II molecules, which enhances T cell immunity (3–6).
CD40 signaling is well-established to stimulate proinflammatory events that promote and sustain adaptive immunity as well as promote autoimmunity (7–9). CD40/CD154 interactions or agonistic CD40 antibodies elicit signaling pathways, such as activation of NF-κB, mitogen-activated protein kinases and phosphoinositide 3-kinase, in multiple cell types (10). These signaling pathways stimulate the production and secretion of pro-inflammatory cytokines such as IL-12, leading to IFN-γ production (11–15). The activation of CD40-dependent signaling facilitates control of chronic parasitic and bacterial infections, including some Leishmania and Mycobacterium spp. (9, 16–20).
However, the ability of CD40 signaling to rapidly alter innate immune responses in vivo has had limited study. One such study of acute CD40-dependent innate immune responses by Gold, et al. showed that CD40-deficient (CD40−/−) mice have significantly reduced proinflammatory cytokine profiles and associated morbidity and mortality in a cecal ligation and puncture model of acute sepsis (21). The authors concluded enhanced survival was due to the reduced proinflammatory cytokine production in CD40−/− animals. Surprisingly, stimulation of CD40 signaling in this model was mediated through interactions between bacterial heat shock protein 70 and CD40, rather than CD40/CD154 interactions (22). Nonetheless, this study highlights the potential for CD40 signaling to induce rapid innate immune responses.
Here, we examine the role of CD40 in controlling acute virus infection of resident macrophages enriched from the peritoneum. Loss of CD40 signaling in peritoneal cell populations increased replication of a broad group of RNA viruses at early times of infection. Protection required CD40 signaling, generation of IL-12 and stimulation of IFN-γ production. These events resulted in proinflammatory polarization of macrophages that blocked acute virus infection. Our studies highlight the importance of this pathway for control of filovirus infection and have implications for a wide range of RNA viruses that target myeloid cells early during infection.
Materials and Methods:
Ethics statement:
This study was conducted in strict accordance with the Animal Welfare Act and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (University of Iowa (UI) Institutional Assurance Number: #A3021–01). All animal procedures were approved by the UI Institutional Animal Care and Use Committee (IACUC) which oversees the administration of the IACUC protocols and the study was performed in accordance with the IACUC guidelines (Protocol #8011280, Filovirus glycoprotein/cellular protein interactions).
Mice:
C57BL/6 CD40−/− Ifnar−/− mice were generated by crossing C57BL/6 Ifnar−/− (kind gift of Dr. John Harty, University of Iowa, Iowa City, IA) and C57BL/6 CD40−/− mice (Cd40tm1Kik Jackson Labs stock #002928 (23)). C57BL/6 Ifnar−/− Ifngr1−/− mice were generated by crossing C57BL/6 Ifnar−/− and C57BL/6 Ifngr1−/− mice (Ifngr1tm1Agt/J Jackson Labs stock #003288). MaFIA mice were purchased from Jackson Labs (C57BL/6-Tg(Csf1r-EGFP-NGFR/FKBP1A/TNFRSF6)2Bck/J, stock #005070). Genotyping was performed using primers and standard PCR conditions from Jackson labs. Mice were further validated by phenotyping peritoneal macrophages by flow cytometry using fluorescent antibodies to either CD40 or IFNγR. WT C57BL/6 mice were a kind gift of Dr. John Harty, University of Iowa.
Peritoneal cell isolation:
To isolate peritoneal cells, mice were euthanized and the peritoneal cavity was immediately lavaged with 10 mL of ice cold RPMI1640 + 10%FBS + 1% pen/strep. Cells were then washed and resuspended in fresh medium supplemented with 50 ng/mL murine M-CSF. Forty-eight hours after plating, non-adherent cells were removed by washing with PBS to obtained enriched peritoneal macrophage (pmac) cultures.
rVSV/EBOV GP production:
Recombinant vesicular stomatitis virus expressing the glycoprotein from EBOV (Mayinga) and a GFP reporter was generated as previously described (24). Virus was propagated by infecting Vero cells at low MOI (~0.1) and collecting supernatants at 48hpi. The resulting supernatants were filtered through a 0.45 μm filter and purified by ultra-centrifugation (28,000g, 4°C, 2 hr) through a sucrose cushion. The resulting stocks were resuspended in a small volume of PBS and further purified by treatment with an endotoxin removal kit (Detoxi-Gel Endotoxin Removing Gel, ThermoFisher Scientific 20339) before being aliquoted and stored at −80°C until use.
rVSV/EBOV GP TCID50/ml:
To determine titers, serum samples or tissue culture supernatants were filtered through 0.45 μm filters and serially diluted before being added onto Vero cells. Titers were determined by the number of wells expressing GFP at day 5 of infection. Titers were calculated as 50% tissue culture infectious dose per milliliter of serum (TCID50/mL) according to the Reed-Meunch method (25).
rVSV/EBOV GP infections:
Ex vivo:
All ex vivo studies used rVSV/EBOV GP that expresses GFP. Plated cells were infected with virus such that approximately 20% of cells were infected (GFP positive) at 24 hours. Peritoneal cells infected with rVSV/EBOV GP were lifted with Versene and samples fixed in 3.7% formalin for 15 minutes or unfixed cells were evaluated for GFP expression on a FACSCaliber. Data were analyzed by FlowJo software (Tree Star, Inc., Ashland, OR).
In vivo:
WT and Ifnar−/− C57BL/6 mice were used in these studies. For WT mice, 1 μg of anti-IFNAR antibody, MAR-1, was given one day prior to challenge. For in vivo infections, two separate stocks of virus were used over the course of the project and the LD50/TCID50 differed between the two stocks as previously described for stocks of wild-type EBOV (26). For each stock, the LD50 was determined by preliminary survival curves administering serial dilutions of the stock intraperitoneally (i.p.). For specific experiments, the dose administered was chosen based on the desired lethality given the questions asked in individual experiments. Thus, we use the term “lethal dose” to describe a dose of virus that kills greater than 80% of our Ifnar−/− mice and “sublethal dose” to describe a dose of virus that kills 0% - 50% of our Ifnar−/− mice. For Stock#1 a lethal dose in female mice was 1×103 TCID50 whereas a lethal dose of Stock#2 in the same mouse population was 5×102 TCID50. The TCID50 of the stocks was determined in Vero cells.
Six to eight week old mice of both genders as noted in the figure legends were used for these experiments. The rVSV/EBOV GP LD50 differs for males and females. Consequently, the amount of virus administered was adjusted accordingly and noted in figure legends to be either a sublethal or lethal dose. In vivo ligation of CD40 was performed by administering a single 250 μg dose of an agonistic rat anti-mouse CD40 mAb (Clone FGK4.5, BioXCell, West Lebanon, NH) or rat IgG2a control (Clone 2A3, BioXCell) 24 hours prior to viral challenge. In vivo blockage of CD40 signaling was performed by administering 250 μg of inhibitory hamster anti-mouse CD154 Ab (Clone MR-1, BioXCell) or polyclonal Armenian hamster IgG (BioXCell). Depletion of T-cells was performed by administering 200 μg of rat anti-mouse CD8 mAb (Clone 2.43, BioXCell) in combination with rat anti-mouse CD4 mAb (Clone GK1.5, BioXCell) or 400 μg of rat IgG2b isotype control mAb (Clone LTF-2, BioXCell).
EBOV infection:
Experiments with replication-competent EBOV variant Mayinga (NCBI accession number NC_002549.1) were performed in a biosafety level 4 laboratory (BSL-4) at the Texas Biomedical Research Institute (San Antonio, TX). The viral stock was amplified and characterized as described previously (27).
Murine Cells:
To test whether CD40 signaling restricts EBOV infection, 125,000 peritoneal cells were seeded into wells of a 96-well plate and treated as described in the figure legends (concentrations detailed below). All treatments were performed in quintuplicate. Twenty-four h later, cells were washed to remove non-adherent population and challenged with EBOV at MOI=0.0015. After another 24 h, some infected cells were collected in TRIzol (Thermo Fisher Scientific, Waltham, MA) and removed from the BSL-4 laboratory according to approved standard operating procedures. Subsequently, RNA from cells treated with TRIzol was extracted and qRT-PCR was performed. The results were quantified by delta Ct method comparing EBOV NP and HPRT transcripts (28). In parallel, supernatants were collected and cells were fixed after 24 hours. The supernatants were titrated on Vero cell monolayers for 24 hours. Subsequently, fixed cells were stained with anti-VP40 antibody (generous gift from Dr. Ricardo Carrion, Jr., Texas Biomedical Research Institute) to identify infected cells and Hoescht dye to stain nuclei. Imaging and analysis was performed as described previously (29).
Human Cells:
Monocyte derived macrophages were isolated as described previously (30, 31).Briefly, human peripheral blood was collected from healthy donors according to University of Texas Health approved IRB protocol #20180013HU. Heparinized blood was layered on a Ficoll-Paque cushion (GE Healthcare, Uppsala, Sweden) to allow for collection of peripheral blood mononuclear cells (PBMCs). PBMCs were cultured in Teflon wells (Savillex, Eden Prairie, MN) RPMI (Life Technologies, Carlsbad, CA) with 20% autologous serum for 5 days at 37°C/5% CO2. PBMCs were pelleted, resuspended in RPMI with 10% autologous serum, and plated into wells of a 96-well plate overnight. Subsequently, cells were treated as described in the figure legends for 24 hours. Stimulated MDMs were incubated with EBOV at MOI=0.1 for 30 min to allow binding, then washed, and overlaid with fresh medium. The cells were fixed after 24 hours. The supernatants were then titrated on Vero cell monolayers for 24 hours. Subsequently, all samples were treated with anti-VP40 antibody to identify infected cells and Hoescht dye to stain nuclei. Imaging and analysis were performed as described above.
Additional virus infections:
Vesicular stomatitis virus (Indiana) was obtained from Dr. Stanley Perlman (Univ. Iowa, Iowa City, IA), infections were performed at MOI=0.5. Ross river virus was obtained from Dr. David Sander (Purdue University, West Lafayette, IN) and infections were performed at an MOI=1. Sindbis virus (strain AR86) was obtained from Dr. Suchetana Mukhopadhyay at Indiana University (Bloomington, IN) and infections were performed at an MOI=1. Influenza virus (A/Puerto Rico/8/34) (32) was obtained from Dr, Balaji Manicassamy at The University of Iowa, and cells were infected at a MOI=1.
Phenotyping:
Peritoneal cells were stained extracellularly with monoclonal antibodies specific to CD45 (clone 30-F11), Siglec F (BD Biosciences, San Jose, CA; clone E50–2440), CD11c (clone N418), F4/80 (clone BM8), Ly6G (clone 1A8), Ly6C (clone HK1.4), MHC-II (clone M5/114.15.2), CD19 (clone 1D3), CD8 (clone 53–6.7), CD4 (clone RM4–5), CD3 (clone 145–2C11), NK1.1 (clone PK136), and NKp46 (clone 29A1.4) in FACS buffer (PBS, 2% FCS, 0.02% sodium azide) for 30 minutes at 4°C and fixed using 1-step Fix/Lyse solution (eBioscience, San Diego, CA) for 10 minutes at room temperature. All antibodies were purchased from BioLegend (San Diego, CA) unless otherwise indicated. Samples were run on a BD LSRFortessa and analyzed using FlowJo software (Tree Star, Inc., Ashland, OR). Cells were gated on live cells, singlets, and CD45+ cells, and cell populations were phenotyped as follows: eosinophils (SiglecF+CD11c+), macrophages (SiglecF−F4/80+), large pmacs (F4/80+ CD11c-high MHCII-low), small pmacs (F4/80+ CD11c-low MHCII-high), neutrophils (SiglecF−F4/80−Ly6G+Ly6C+), dendritic cells (SiglecF−F4/80−Ly6G−MHC-II+CD11c+), B cells (CD3−CD19+), CD8 T cells (CD3+CD8+), CD4 T cells (CD3+CD4+), and NK cells (CD3−NK1.1+NKp46+).
For phenotyping of infected peritoneal cells ex vivo: male C57 BL/6 mice (WT, Ifnar−/− or Ifnar/CD40−/−) were infected i.p. with 2×106 iu of rVSV/EBOV GP. Peritoneal cells were isolated at 24 hours post infection and stained and analyzed as reported above. Gating was performed as indicated on Figure S2A.
Organ harvest:
Mice were anesthetized using isoflurane and perfused through the left ventricle with 5 mL cold sterile PBS prior to being euthanized by rapid cervical dislocation in accordance to our IACUC protocol. Organs were harvested and snap frozen in liquid nitrogen to preserve virus. RNA from organs was isolated as described.
siRNA knockdown of TRAF6:
TriFECTa DsiRNA Kit (mm.Ri.Traf6.13) was purchased from Integrated DNA Technologies, Coralville, IA. Transfection of three siRNAs against TRAF6 was achieved with the transfection reagent Viromer Blue (Origene Technologies, Rockville, MD) according to manufacturer’s instructions. Twenty-four and forty-eight hours following transfection, knockdown was validated by immunoblotting. TRAF6 expression was normalized for GAPDH expression and transfection of all three siRNAs resulted in ~80–93% knock down. At 48 hours, cells were exposed to anti-CD40 antibody or IL-12 for 24 hours and infected as described below.
Virus infection in the presence of inhibitor, mAbs, cytokines and/or cell membranes:
Agonistic mouse anti-CD40 antibody (clone FGK4.5/ FGK45, BioXCell) was used at a final concentration of 1μg/mL for ex vivo experiments. Agonistic human anti-CD40 antibody (clone G28.5, BioXCell) was used at a final concentration of 1 μg/mL. Insect cell membranes expressing CD154 (CD40L) were generated as described (33) and added to cells at a 3:1 ratio (membranes/cells). TRAF6 inhibitors (ChemBridge ID #s 7651589 and 6872674 (34)) were obtained from Hit2Lead (San Diego, CA) and resuspended in DMSO. Compounds were diluted such that the final DMSO concentration was less than 0.01% in tissue culture. Apilimod was used at a concentration of 100 nM for all experiments. IL-12 (StemCell Technologies, Vancouver, CA) (murine: Catalog # 78028.1; human: Catalog # 78027.1) was used at 20 ng/μl. IFN-γ was purchased from StemCell Technologies (Catalog # 78020.1) and used at 20 ng/μl.
Magnetic bead separation:
Removal of individual cell populations from peritoneal cultures was performed using biotinylated antibodies (Tonbo Biosciences, San Diego, CA) against CD3ξ (145–2C11), Ly-6G (RB6–8C5) and CD19 (1D3). Separation was performed using MojoSort Streptavadin Nanobeads (Biolegend, catalog #480016) in accordance to manufacturer’s protocol. Validation of depletion was performed using antibodies described in the “phenotyping” section.
RNA isolation and qRT-PCR:
RNA was isolated using the TRIzol reagent from Thermo Fisher Scientific, Waltham, MA). All steps were performed according to the manufacturer’s specifications. For RNA isolation from whole organs we used the gentleMACS tissue dissociation system with associated gentleMACS M tubes (Miltenyi Biotec, Bergishch Gladbach, Germany). Organs were placed in 1mL of TRIzol in the M tube and dissociated for 1 minute. RNA was subsequently converted to cDNA with the High Capacity cDNA RevTrans Kit (#4368814) from Thermo Fisher Scientific. A total of 1 microgram of RNA was used as input for each reaction. Quantitative PCR was performed using POWER SYBR Green Master Mix (#4367659) (Thermo Fisher Scientific) according to the manufacturer’s instructions and utilizing a 7300 real time PCR machine (Thermo Fisher Scientific). Twenty nanograms of cDNA were used in each well. All primers used were generated using NCBI’s “pick primers” tool. Primers used are as follows: HPRT forward 5’-GCGTCGTGATTAGCGATGATG-3’, HPRT reverse 5’-CTCGAGCAAGTCTTTCAGTCC-3’, VSV-M forward 5’-CCTGGATTCTATCAGCCACTTC-3’, VSV-M reverse 5’-TTGTTCGAGAGGCTGGAATTAG-3’, EBOV NP forward 5’-CAGTGCGCCACTCACGGACA-3’, EBOV NP reverse 5’-TGGTGTCAGCATGCGAGGGC-3’, IRF-1 forward 5’-TCACACAGGCCGATACAAAG-3’, IRF-1 reverse 5’-GATCCTTCACTTCCTCGATGTC-3’, GBP5 forward 5’-CCCAGGAAGAGGCTGATAG-3’, GBP5 reverse 5’-TCTACGGTGGTGGTTCATTT-3’, IL-12p35 forward 5’-ATCACAACCATCAGCAGATCA-3’, IL-12p35 reverse 5’-CGCCATTATGATTCAGAGACTG-3’, RRV forward 5’-TAC AAG CAC GAC CCA TTG CCG-3’, RRV reverse 5’-GAT AGT CCT GCC GCC TGC TGT-3’, IAV forward 5’-CTT CTA ACC GAG GTC GAA AC-3’, IAV reverse 5’-CGT CTA CGC TGC AGT CCT C-3’, SINV forward 5’-CCA CTG GTC TCA ACA GTC AAA-3’ and SINV reverse 5’-CTC CTT TCT CCA GGA CAT GAA C-3’.
Statistical analysis
All ex vivo experiments were performed a minimum of three independent times and are shown as mean with error expressed as standard error of the mean (when pooling experiments) or standard deviation (when a single experiment is shown). Statistical significance was determined by two tailed Student’s T-test (alpha=0.05). In vivo experiments were performed a minimum of three times, unless otherwise noted in the figure legend. Statistical significance was determined using Log-rank (Mantel-Cox) test. All statistics were calculated using GraphPad Prism software (GraphPad Software, Inc.).
Results:
CD40 expression restricts infection of a wide range of RNA viruses in peritoneal cells and macrophages enriched from the peritoneal compartment.
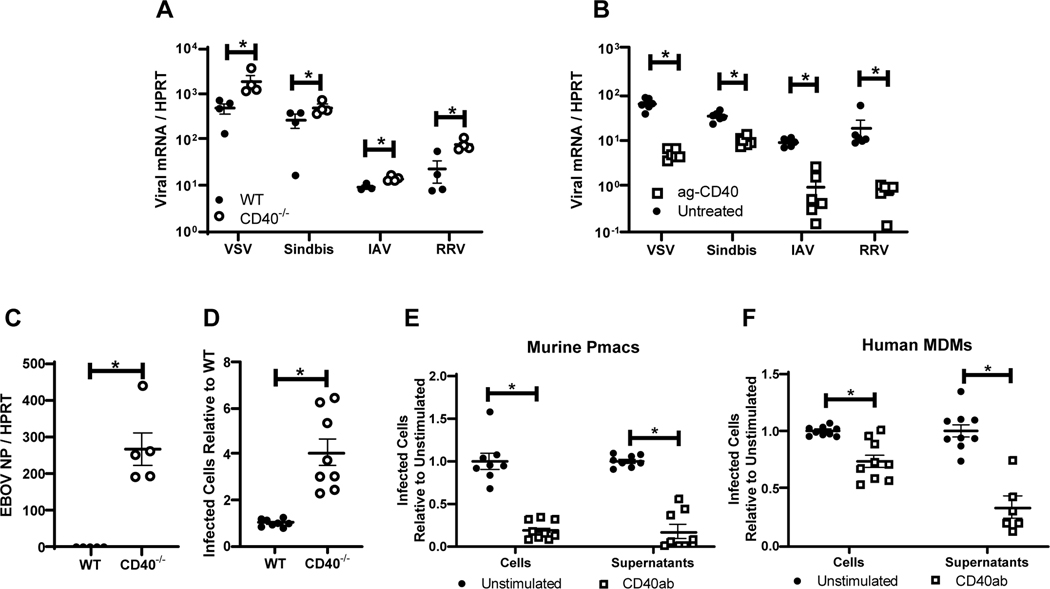
The infectivity of a series of enveloped RNA viruses were assessed on resident peritoneal cells from WT versus CD40−/− mice. Ex vivo experiments were performed with murine peritoneal cells, composed of ~20% macrophages, and/or enriched peritoneal macrophages (pmacs), that consisted of 80–90% macrophages (Figures S1 and S2). These cells were chosen as they are permissive to a wide range of viruses. Further, while pmacs and peritoneal B cells are CD40+, other peritoneal populations express the CD40 ligand, CD154 (35). Viruses tested included the plus and minus strand viruses: Sindbis virus (AR86) (SINV), Ross River virus (RRV), vesicular stomatitis virus (Indiana) (VSV), influenza A virus (A/Puerto Rico/8/34) (IAV) and WT Ebola virus (Mayinga) (EBOV). At 24 hours, virus load of all virus challenges was elevated in CD40−/− peritoneal cells compared to WT cells, as assessed by qRT-PCR (Figure 1A and C). In EBOV studies, the number of CD40−/− cells that were positive for EBOV VP40 antigen was ~4-fold greater at this time relative to infection of WT cells (Figure 1D).
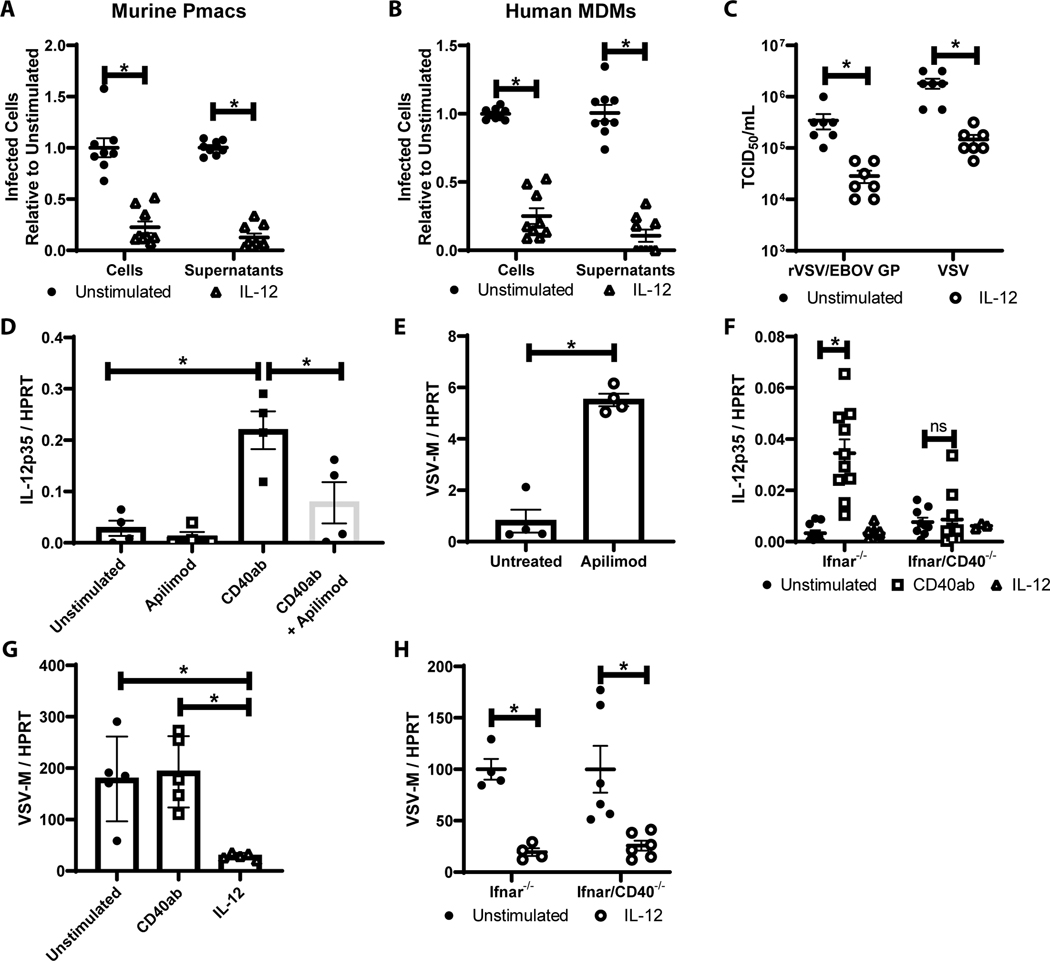
Figure 1. CD40 restricts replication of a variety of RNA viruses in cells from the peritoneal compartment.
A) Peritoneal cells from WT or CD40−/− male mice were infected with the indicated viruses (MOI=1 for all viruses except VSV where MOI = 0.5). Twenty-four hours following infection, RNA was extracted and viral load was quantified by qRT-PCR. B) Peritoneal cells from WT male mice were stimulated with an agonistic CD40 antibody, FGK4.5/FGK45, for 24 hours prior to infection and quantitation. C) Peritoneal cells from WT or CD40−/− mice were infected with EBOV (Mayinga) (MOI = 0.0015) and evaluated for virus load at 24 hours by qRT-PCR. D) Peritoneal cells from WT or CD40−/− mice were infected with by EBOV (Mayinga) (MOI = 0.0015) and number of infected cells were assessed at 24 hours by fixation followed by staining with anti-VP40 antibody and Hoescht dye. Infected cells were quantified by microscopy. E-F) Peritoneal cells from WT mice (E) or 6 day matured human MDMs (hMDMs) (F) were stimulated with an agonistic CD40 antibody (mouse CD40 antibody, FGK4.5/FGK45, or human CD40 antibody, G28.5) or left untreated (MOI = 0.0015 pmacs; MOI = 0.1 MDMs). After 24 hours, non-adherent cells were removed from the cultures and enriched pmacs or hMDMs were infected with EBOV(Mayinga). At 24 hours, cells were processed and stained as described in D. In parallel, supernatants from infected cells were titered on Vero cells and quantified using the same microscopic method. All experiments were performed at least three independent times. For all experiments, * indicates p<0.05. All qRT-PCR is quantified by delta Ct method comparing viral RNA to HPRT.
To determine if CD40 signaling was required for the protection observed in WT cells, an agonistic mouse CD40 monoclonal antibody (mAb), FGK4.5/ FGK45, was incubated with CD40+ cells for 24 hours followed by a 24-hour virus infection. Treatment of peritoneal cells or enriched pmacs with agonistic anti-CD40 mAb reduced virus loads (Figure 1B and E). Supernatants from EBOV-infected pmacs also demonstrated reduced production of infectious virus in the presence of agonistic anti-CD40 mAb (Figure 1E). Similar findings were observed with EBOV infection of agonistic anti-CD40 mAb treated human monocyte derived macrophages (hMDMs) (Figure 1F). These observations indicate that CD40 signaling in resident peritoneal cells, and specifically pmacs, restricts infection of a broad range of RNA viruses and implicates CD40 signaling as an important, but unappreciated, mechanism of innate immune control of RNA viruses over the first 24 hours of infection.
A BSL-2 model virus of EBOV, rVSV/EBOV GP, recapitulates findings with WT EBOV (Mayinga).
To study the mechanism by which CD40 restricts viral replication, we focused on EBOV infection as the virus is well established to initially target macrophages and dendritic cells (36). Peritoneal delivery of EBOV causes significant pathology in a variety of different animal models (37–39), with pmacs as potential early targets of infection (40, 41). In these studies, we also extensively utilized a BSL-2 model virus of EBOV, recombinant vesicular stomatitis virus encoding EBOV glycoprotein and GFP in place of the native G glycoprotein (rVSV/EBOV GP). Our lab has previously found that rVSV/EBOV GP recapitulates many aspects of EBOV infection both in vitro and in vivo, including virus tropism and responsiveness to proinflammatory and anti-inflammatory cytokines during macrophage infection (40–42). Use of this chimeric virus allowed us to perform studies in a more cost-effective manner, while maintaining the tropism of EBOV GP.
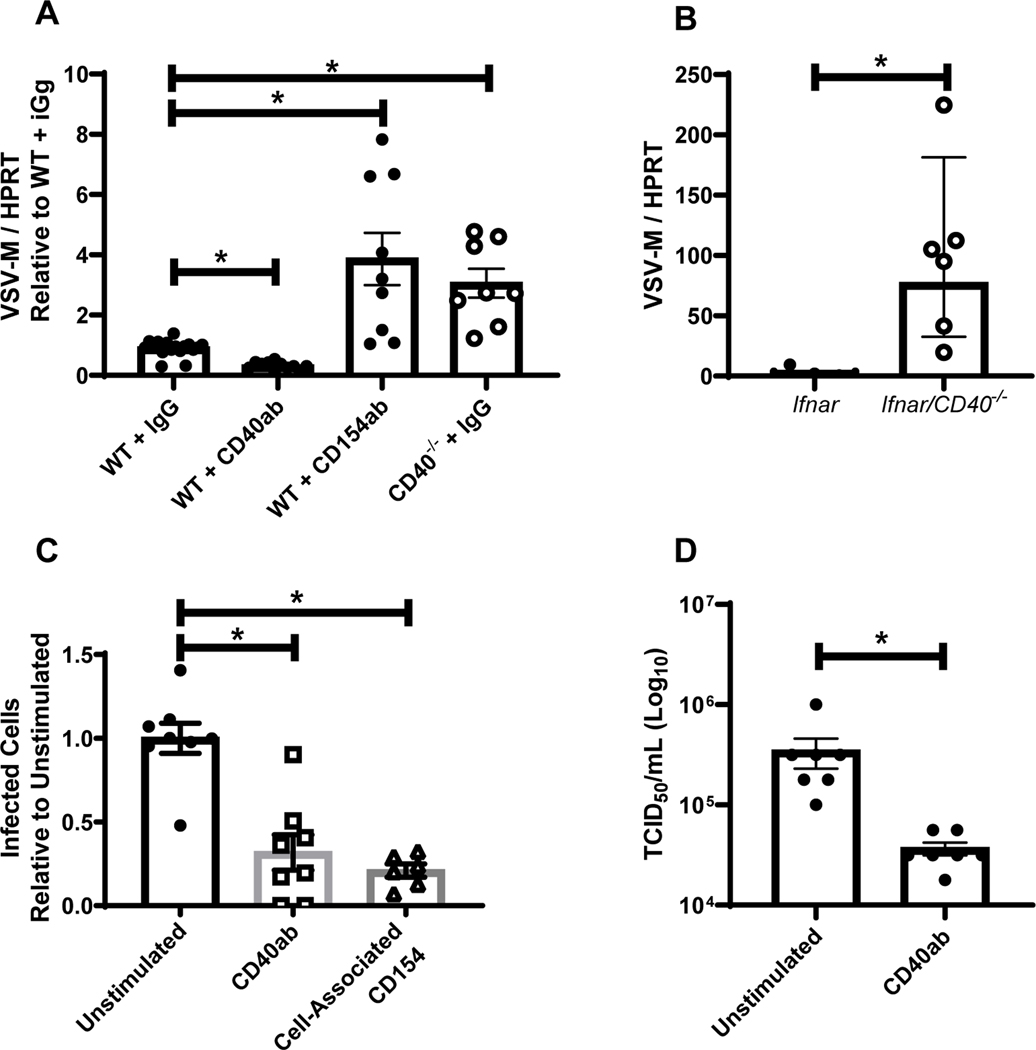
To evaluate the utility of rVSV/EBOV GP, WT or CD40−/− mice were left untreated, stimulated with agonistic anti-CD40 mAb, or treated with a blocking antibody against CD154 to inhibit CD40 signaling. After 24 hours of treatment, mice were infected with rVSV/EBOV GP for 24 hours and resident peritoneal cells were isolated and assessed for virus load by qRT-PCR (Figure 2A). Stimulation of CD40 signaling reduced virus load, whereas elevated levels of virus in the peritoneal cells were observed in CD40−/− mice. Blockade of CD154/CD40 interactions also enhanced virus load. These studies were consistent with our findings with WT EBOV and other RNA virus infections investigated, indicating that rVSV/EBOV GP can serve to model aspects of EBOV infection. However, VSV replication is strongly suppressed by type 1 interferon (43), so limited virus replication occurs and no rVSV/EBOV GP associated pathology is observed in WT mice. Thus, we assessed the effect of CD40 signaling on rVSV/EBOV GP replication in a model system where we had previously shown significant morbidity and mortality, interferon αβ receptor knock out (Ifnar−/−) mice (40). Ifnar−/− and Ifnar/CD40−/− mice were infected with rVSV/EBOV GP and virus loads were assessed by qRT-PCR from 24 hpi peritoneal cell RNA (Figure 2B). Cells from the Ifnar/CD40−/− mice were highly susceptible to rVSV/EBOV GP, with virus loads almost 90-fold higher than that observed in cells from CD40-competent mice. Thus, peritoneal cells from CD40-null mice on either a WT or Ifnar−/− background infected with either EBOV or rVSV/EBOV GP contained significantly higher levels of viral RNA than cells from CD40-expressing mice.
Figure 2. A BSL-2 model virus of EBOV, rVSV/EBOV GP, recapitulates EBOV findings.
A) WT or CD40−/− female mice were injected i.p. with 200 μg of agonistic CD40 antibody, blocking CD154 or IgG control as noted. Twenty-four hours later, mice were infected i.p. with a lethal dose of rVSV/EBOV GP. Peritoneal cells were isolated, RNA was harvested, and viral RNA was quantified by qRT-PCR at 24 hours of infection. B) Female Ifnar−/− and Ifnar/CD40−/− mice were infected i.p. with a dose of rVSV/EBOV GP that was lethal to Ifnar−/− mice. Twenty-four hours after infection, peritoneal cells were harvested, RNA was isolated and qRT-PCR analysis of viral RNA was performed. C-D) Enriched pmacs from male Ifnar−/− mice were incubated for 24 hours with agonistic CD40-specific mAb (C and D) or insect cell membranes expressing CD154 (C only). Treatments were removed and cells infected with rVSV/EBOV GP (MOI=1). Infection was quantified 24 hours following infection by flow cytometry and data are expressed as a percent change in the number of GFP+ cells relative to the unstimulated controls (C). Supernatants were titered on Vero cells (D). All experiments were performed three times. For all experiments, * indicates p<0.05.
The effect of CD40 signaling was also assessed ex vivo in peritoneal cells from Ifnar−/− mice. Cells were plated and stimulated with either agonistic anti-CD40 mAb or insect cell membrane fragments that express CD154 for 24 hours and subsequently infected with rVSV/EBOV GP. Infection was quantified by flow cytometry at 24 hours. Similar to our EBOV findings, stimulation of CD40 signaling through either method reduced virus load in peritoneal cells (Figure 2C) and production of infectious virus (Figure 2D), consistent with the broad restrictive effect of CD40 signaling on RNA virus replication. Together these experiments supported use of both our BSL2 model system and Ifnar−/− mice to explore and mechanistically characterize the role of CD40 in this acute virus infection.
Identification of peritoneal cell populations
To understand the CD40-dependent events within the peritoneum needed to control virus infection, we characterized the cells resident within the peritoneal cavity. For the mouse strains used in these studies, relative peritoneal cell numbers and absolute numbers were determined using the gating strategy shown (Figure S2A). Two CD40+ cell populations, SiglecF−, F4/80+ macrophages and CD19+, CD3− B cells, composed the largest numbers of cells of all mice regardless of genotype, with smaller populations of T cells, NK cells, neutrophils, eosinophils and DCs present (Figure S2B). Eosinophils and macrophages were increased as a percent of total cells in Ifnar−/− mice and DCs were elevated in Ifnar/CD40−/− mice; however, the absolute numbers of these populations were unchanged between strains (Figure S2C). As previously observed in spleen, B cells composed a smaller percentage and a lower absolute number of cells within the peritoneal cavity in Ifnar−/− and Ifnar/CD40−/− mice compared to their WT counterparts (44).
To assess the relative percent of different peritoneal cell populations targeted by rVSV/EBOV GP, WT and Ifnar−/− male mice were infected with 2 × 106 iu of rVSV/EBOV GP and GFP+ cells present within the peritoneum at 24 hours post infection were characterized. We found that, at this early time during infection of Ifnar−/− mice, pmacs are highly permissive for this virus (Figure S3D). Other cell populations had appreciable levels of GFP positivity, including DCs and, somewhat surprisingly, neutrophils. However, we do not believe that this short lived, EBOV antigen-expressing cell population contributes significantly to virus load as Mahamadzadel et al. has shown (45). Given the relative abundance of macrophages, they served as a predominant population of productively infected cells in the peritoneal cavity. Numbers of GFP+ cells were notably lower in the WT mice due to the limited infection and lack of spread by rVSV/EBOV GP.
CD154+ lymphocytes resident within the peritoneum initiate CD40 signaling
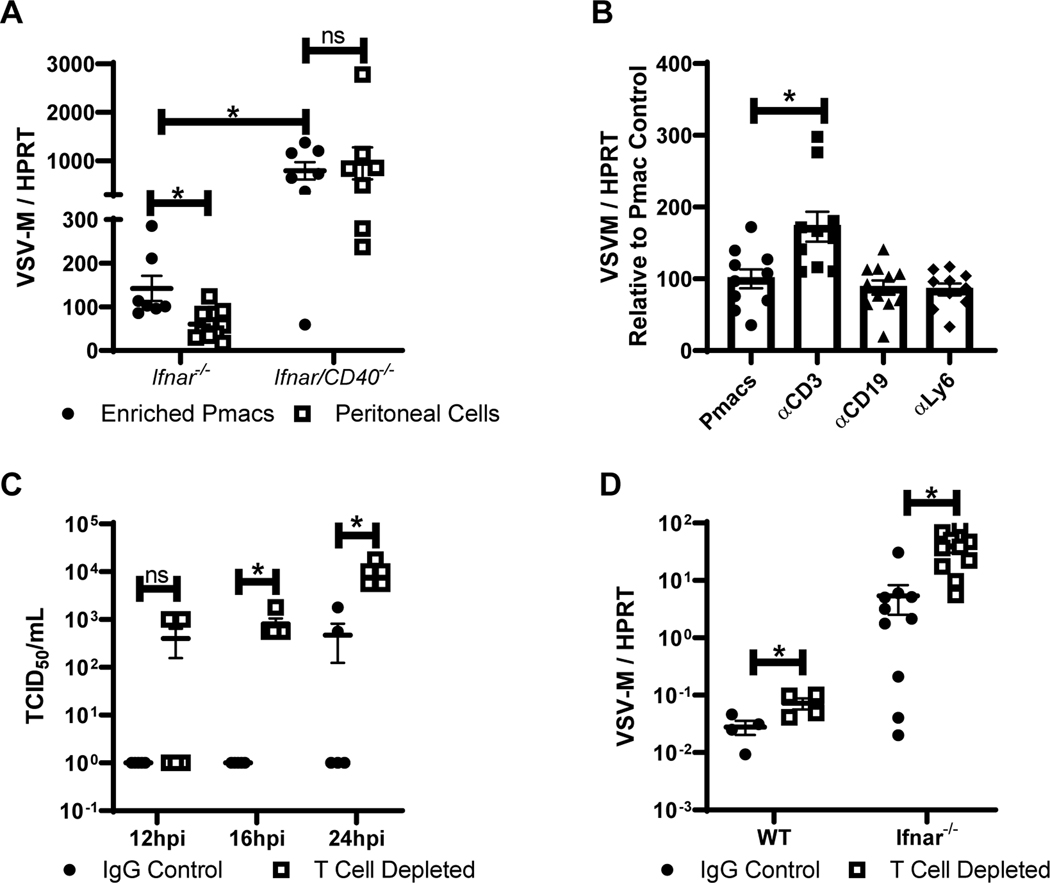
Our studies suggested that CD40-dependent restriction of virus infection requires CD40 signaling. To elicit CD40 signaling within the peritoneal cavity, the CD40 ligand, CD154 would be required either as a soluble or plasma membrane-associated protein. To determine the location of CD154 within the peritoneum, resident cells and fluid were obtained from naïve Ifnar−/− mice. Resident peritoneal cells were adhered overnight and the non-adherent cells separated by several washes. Cell lysates were generated and assessed along with peritoneal fluid for CD154 expression. Immunoblot analysis demonstrated that CD154 was expressed by non-adherent cells, with low-to-undetectable CD154 on adherent cells and in peritoneal fluids (Figure S3A). CD154 was also detected on the surface of peritoneal lymphocytes from naïve Ifnar−/− mice by flow cytometry (Figure S3B). Consistent with a role for non-adherent peritoneal cells in controlling virus infection, we observed that bulk peritoneal cells from CD40+ mice supported significantly lower levels of virus replication than peritoneal cultures where non-adherent cell populations were washed away, leaving an enriched pmac culture with many fewer lymphocytes present (Figure 3A). In CD40−/− peritoneal cells, virus loads were notably higher and the presence of non-adherent cells did not inhibit viral loads. In combination, these findings suggested that CD40 on pmacs regulated virus replication and that CD154 on non-adherent cells contributed to CD40-dependent control.
Figure 3. CD40 restriction of EBOV and rVSV/EBOV GP infection in peritoneal cells requires CD3+ T cells.
A) Peritoneal cells from male Ifnar−/− and Ifnar/CD40−/− mice were isolated. Forty-eight hours after isolation, cells were either washed to remove non-adherent cells, or incubated in the presence of non-adherent cells and infected with rVSV/EBOV GP (MOI=0.1). RNA was isolated 24 hours following infection and viral RNA was quantified by qRT-PCR. B) Peritoneal cells were isolated from male Ifnar−/− mice and cells expressing the indicated surface protein were removed via magnetic bead separation. The remaining cells were infected with rVSV/EBOV GP (MOI=0.1). RNA was isolated at 24 hours and quantified by qRT-PCR. C-D) Female C57BL/6 Ifnar−/− mice were injected i.p. with antibodies against CD4 and CD8 (200 μg each) or appropriate IgG controls. Twenty-four hours later, mice were infected i.p. with a sublethal dose of rVSV/EBOV GP. At 24 hours, serum was collected and titers were quantified by endpoint dilution on Vero cells (C) or peritoneal cell RNA was harvested, and viral RNA was quantified by qRT-PCR (D). All experiments were performed three times. All qRT-PCR is quantified by delta Ct method comparing VSV-M gene to HPRT. * indicates p<0.05.
To further examine the role of non-adherent cells in reducing virus load, we used magnetic nanobeads to deplete different non-adherent cell populations from the overall CD40+ peritoneal cells and subsequently infected the remaining cells. Targeted cell populations were reduced by at least 50% as confirmed by flow cytometry. While the overall depletion was incomplete, we found that removal of CD3+ cells, but not B cells or neutrophils, resulted in increased levels of infection (Figure 3B), suggesting that resident CD3+ cells present in the peritoneal cavity were involved in controlling virus infection. Naïve resident CD3+ cells are reported to express low constitutive levels of CD154, although CD154 is robustly upregulated upon CD3+ cell activation (46).
The effect of CD3+ T cells was also examined in vivo by systemic CD3+ cell depletion in Ifnar−/− mice using a combination of CD4- and CD8-specific mAbs as previously described (Figure S4) (47). T cell-depleted and isotype control Ab-treated mice were infected with a low dose of rVSV/EBOV GP and bled at 12, 16 and 24 hours. Whereas viremia was detected in the isotype control Ab-treated mice at 24 hours of infection, viremia was evident in some T cell-depleted mice as early as 12 hours with significantly higher viremia in these mice at 16–24h of infection (Figure 3C). Virus load within the peritoneum of Ifnar−/− or WT mice depleted of T cells was also elevated at 24 hours post infection (Figure 3D), indicating that T cells within the peritoneum during the initial 24h of infection were required for control of virus infection. Since the timing of these experiments precluded the generation of adaptive immune responses (9, 48, 49), we concluded that peritoneal T cells were contributing to innate control of virus infection.
CD40 restriction is TRAF6 dependent
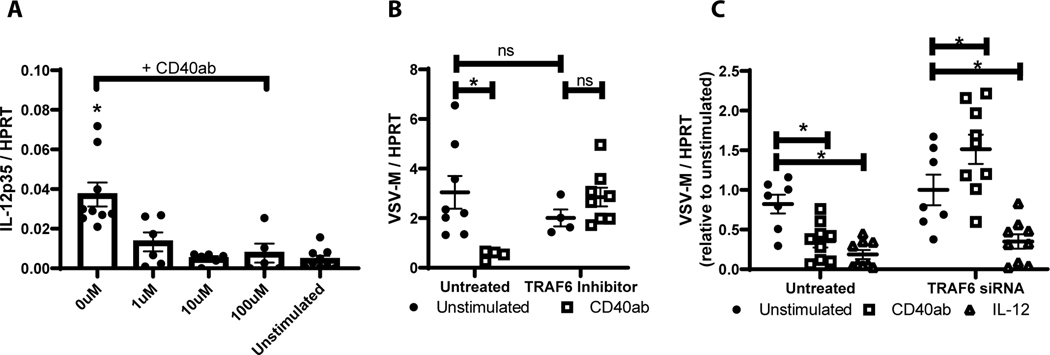
CD40 signaling in macrophages requiresTRAF6 recruitment to the intracellular cytoplasmic tail of CD40 (50). To evaluate the role of TRAF6 in CD40 control of rVSV/EBOV GP infection, the effect of two recently described inhibitors of TRAF6/CD40 interaction (34) was assessed. To verify that that inhibitor 7651589 inhibits CD40 signaling, increasing concentrations of inhibitor were added to enriched pmacs from Ifnar−/− mice in the presence of agonistic CD40 antibody and the production of IL-12p35 RNA was assessed, as it is well established that CD40 signaling in macrophages enhances IL12p35 transcription (51, 52). The TRAF6 inhibitor 7651589 blocked IL-12p35 transcription in a dose dependent manner (Figure 4A). Similar findings were observed with a second TRAF6 inhibitor, 6872674 (data not shown). Addition of 10 μM of the inhibitors blocked the ability of agonistic anti-CD40 mAb to restrict rVSV/EBOV GP infection (Figure 4B and data not shown). siRNA against TRAF6 was also used to evaluate the role of TRAF6 in virus infection. Knockdown of TRAF6 in enriched pmacs (Figure S5) reversed the protection conferred by CD40 ligation in peritoneal cells (Figure 4C). However, we observed that the addition of exogenous IL-12 to either untreated or TRAF6 siRNA-treated cells suppressed virus load, suggesting that CD40 restriction of virus infection may be mediated through IL-12 production and that IL-12 acted downstream from TRAF6. In total, these findings indicated that CD40 signaling through TRAF6 controls rVSV/EBOV GP replication, potentially through an IL-12-dependent pathway.
Figure 4. CD40 signaling through TRAF6 is required for CD40 restriction.
A-B) Pmacs were harvested from male Ifnar−/− mice and stimulated with agonistic CD40 antibody in the presence of varying concentrations of a TRAF6 inhibitor (7651589) for 7 hours. Following stimulation, RNA was isolated and IL-12p35 mRNA was quantified by qRT-PCR (A). In parallel, cells were stimulated for 24 hours with CD40 agonist (+/− 10 μM TRAF6 inhibitor 7651589) and subsequently challenged with rVSV/EBOV GP (MOI=1). RNA was harvested and viral RNA was assessed by qRT-PCR (B). C) Peritoneal cells were harvested from male Ifnar−/− mice and transfected with 3 TRAF6 siRNAs (25 nM each). At 48 hours, cells were stimulated with agonistic CD40 antibody or IL-12. After 24 hours cells were washed to remove non-adherent cells, media was refreshed, and cells were infected with rVSV/EBOV GP (MOI=1). RNA was harvested 24hpi and virus was quantified by qRT-PCR.
CD40 restriction requires IL-12 production
Inhibition of EBOV infection by IL-12-mediated mechanisms has not previously been reported. Thus, we sought to examine in more detail the effect of IL-12 on EBOV (Mayinga) infection and replication in WT pmacs and human MDMs (Figure 5A and B). IL-12 strongly inhibited infection in both primary cell populations and resulted in a reduction in supernatant-associated infectious EBOV as assessed in Vero cells. Representative images of these infections are shown in Figure S6. In a similar manner, exogenous IL-12 decreased both rVSV/EBOV GP and VSV production from Ifnar−/− pmacs (Figure 5C). IL-12 had previously been reported to reduce VSV replication and pathology in the CNS (53–55).
Figure 5. IL-12 production is a critical for CD40 restriction.
A/B) Peritoneal cells from male mice (A) or human monocyte derived macrophages (B) were untreated or treated with IL-12. After incubation, non-adherent cells were removed and enriched pmacs were infected for 24 hours with WT EBOV (MOI = 0.0015 pmacs; MOI = 0.1 MDMs) under BSL-4 containment. Cells were subsequently fixed, stained with anti-VP40 antibody and Hoescht dye, and infected cells were quantified by microscopy. Supernatants from infected cells were titered on Vero cells and quantified by the same microscopic method. C) Enriched pmacs from male Ifnar−/− mice were stimulated with IL-12 and at 24 hours infected with rVSV/EBOV GP (MOI=1) or VSV (MOI=0.5). Twenty-four hour following infection, supernatants were collected, filtered and TCID50s were determined on Vero cells. D/E) Pmacs were harvested from male Ifnar−/− mice and treated with agonistic CD40-specific mAb with or without the IL-12 inhibitor, apilimod, for 8 hours. RNA was harvested and qRT-PCR was performed to quantify IL-12p35 expression (D). In parallel, cells were stimulated with apilimod for 8 hours and infected with rVSV/EBOV GP (MOI=1). Twenty-four hours following infection, RNA was isolated and viral RNA was assessed by qRT-PCR (E). F) Enriched pmacs were obtained from Ifnar−/− and Ifnar/CD40−/− mice. Cells were stimulated with CD40 agonistic mAb or IL-12 and IL-12p35 gene expression was quantified by qRT-PCR. G) Pmacs from IL-12p40−/− mice were harvested and stimulated with agonistic CD40-specific antibody or IL-12 for 24h prior to infection with rVSV/EBOV GP (MOI=5). Twenty-four hours post infection, RNA was extracted and viral RNA was quantified by qRT-PCR. H) Enriched pmacs were obtained from Ifnar−/− and Ifnar/CD40−/− mice. Cells were stimulated with IL-12 and 24 hours later infected with rVSV/EBOV GP. Infections were evaluated for virus load by qRT-PCR at 24 hours. All experiments were performed three times. For all experiments, * indicates p<0.05.
Next, we sought to more closely examine the direct involvement of IL-12 in CD40 restriction of virus infection, using three independent approaches. In a first set of studies, we used the IL-12 inhibitor, apilimod, that selectively blocks IL-12 p40 and p35 production by inhibiting the lipid kinase, PIKfyve (56–58). Apilimod pre-treatment decreased CD40-elicited IL-12p35 expression in Ifnar−/− pmacs (Figure 5D). Concomitantly, apilimod enhanced rVSV/EBOV GP virus loads in these cells (Figure 5E), consistent with CD40 signaling eliciting IL-12 production that decreased virus infection. Our studies in the Ifnar−/− pmacs indicate that this effect is IFN-I-independent. In a second set of experiments, agonistic anti-CD40 mAb stimulated IL-12p35 transcription in Ifnar−/− pmacs; however, this was not observed in Ifnar/CD40−/− pmacs (Figure 5F). Finally, CD40 restriction of rVSV/EBOV GP infection was abolished in pmacs from IL-12p40−/− mice, but addition of IL-12 to those cultures profoundly inhibited virus infection (Figure 5G). In total, our findings indicate that CD40 restriction of virus infection is mediated through IL-12 production. Consistent with a critical role for IL-12, administration of IL-12 protected Ifnar−/− and Ifnar/CD40−/− pmacs from rVSV/EBOV GP infection (Figures 5H).
IL-12 dependent production of IFN-γ is critical for CD40-mediated protection from rVSV/EBOV GP challenge
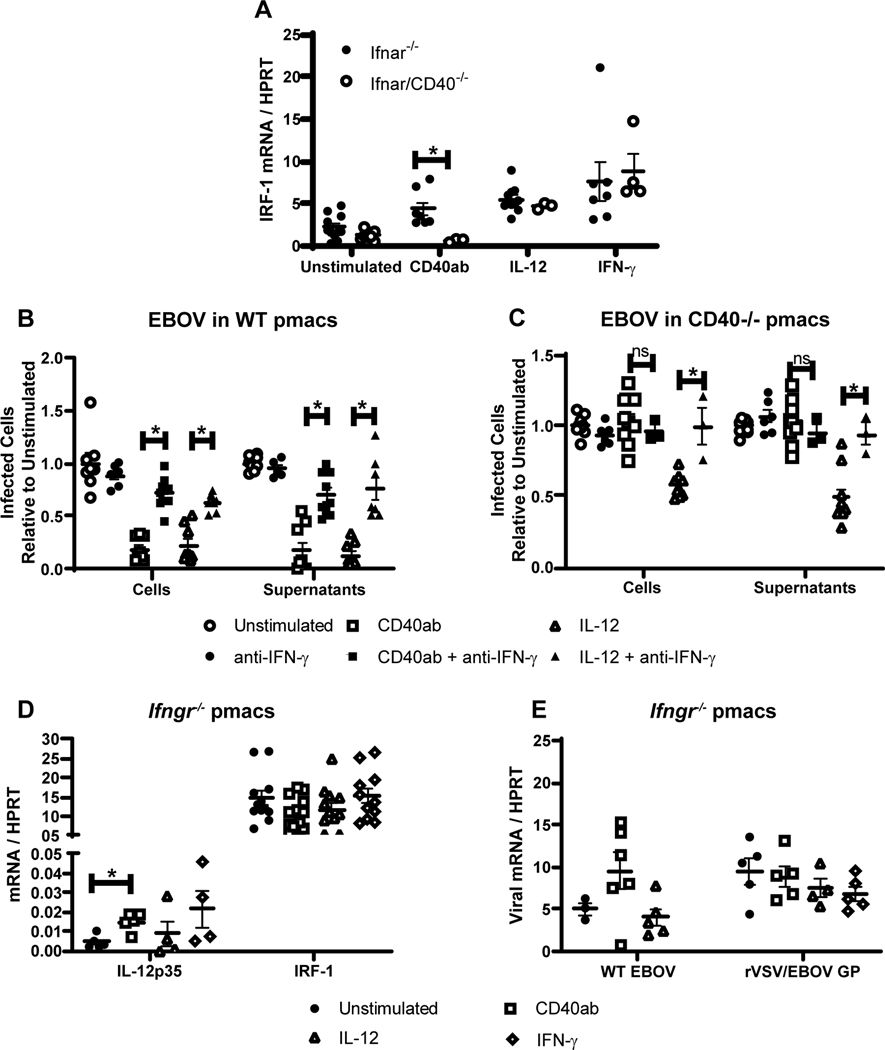
IL-12 stimulates the production of IFN-γ (59–61) and we have previously shown that IFN-γ plays a critical role in early control of EBOV (Mayinga) or rVSV/EBOV GP infection (40). Thus, we investigated if CD40-elicited IFN-γ production mediated protection of pmacs. The ability of CD40 ligation, exogenous IL-12 or IFN-γ to stimulate IFN-γ responses was evaluated in Ifnar−/− or Ifnar/CD40−/− pmacs. Expression of the transcription factor IRF-1 serves as a marker of IFN-γ signaling and accurately predicts whether pmacs are polarized to a proinflammatory (M1) phenotype, protecting against EBOV or rVSV/EBOV GP (40). IRF-1 expression was upregulated in Ifnar−/− pmacs in response to these stimuli, whereas pmacs from CD40-deficient mice were only responsive to IL-12 and IFN-γ (Figure 6A). Addition of blocking IFN-γ mAb reversed the protective effects of CD40 signaling or IL-12 against EBOV (Mayinga) infection in WT pmacs under BSL-4 conditions (Figure 6B, left). In a similar manner, EBOV present in the supernatant of cells at 24 hours was reduced by agonistic CD40 antibody or IL-12 treatment and the addition of IFN-γ blocking antibody abrogated this effect (Figure 6B, right). Parallel EBOV infections of CD40−/− pmacs demonstrated that agonistic anti-CD40 mAb did not restrict EBOV infection, but IL-12 treatment remained effective at blocking virus replication and the presence of infectious virus in the supernatant (Figure 6C). Further, the IL-12 inhibition of EBOV infection was reversed by the presence of blocking IFN-γ antibody.
Figure 6. IL-12 stimulates IFN-γ that is required for CD40 restriction of virus replication.
A) Enriched pmacs were obtained from Ifnar−/− and Ifnar/CD40−/− mice. Cells were stimulated with CD40 agonist, IL-12 or IFN-γ and IRF-1 gene expression was quantified by qRT-PCR. B-C) Peritoneal cells from male WT (B) or CD40−/− (C) mice were incubated with agonistic CD40 mAb or IL-12 for 24 hours. Some cells were also treated with blocking anti-IFN-γ antibody as noted during antibody or cytokine treatment. After incubation, non-adherent cells were removed and enriched pmacs were infected with EBOV (Mayinga)(MOI = 0.0015) under BSL-4 conditions. At 24 hours of infection, cells were fixed, stained with anti-VP40 antibody and Hoescht dye, and infected cells were quantified by microscopy. In parallel, 24-hour supernatants from infected cells were titered on Vero cells and quantified by the same microscopic method. D-E) Enriched pmacs from male C57BL/6 Ifngr−/− mice were stimulated with the indicated stimuli. Twenty-four hours after stimulation, RNA was isolated from cells and IL-12p35 and IRF-1 mRNA was quantified by qRT-PCR (D) or infected with WT EBOV (MOI 0.0015) or rVSV/EBOV GP (MOI=5) and viral RNA was quantified 24 hours following infection by qRT-PCR (E). All experiments were performed three times. For all experiments, * indicates p<0.05.
In additional studies to investigate the requirement for IFN-γ, resident pmacs were isolated and enriched from IFN-γ receptor-deficient mice (Ifngr−/−) that do not respond to IFN-γ. While IL-12p35 production was not disrupted in these cells, stimulation of IFN-γ-dependent IRF-1 expression did not occur following CD40 ligation or the addition of IL-12 or IFN-γ (Figure 6D). We also found that none of these stimuli inhibited rVSV/EBOV GP or EBOV infection in the IFNγR−/− pmacs (Figure 6E). These in vitro experiments indicate that CD40 signaling in macrophages leads to IL-12 dependent IFN-γ production.
CD40 is required for protection against acute virus infection in vivo
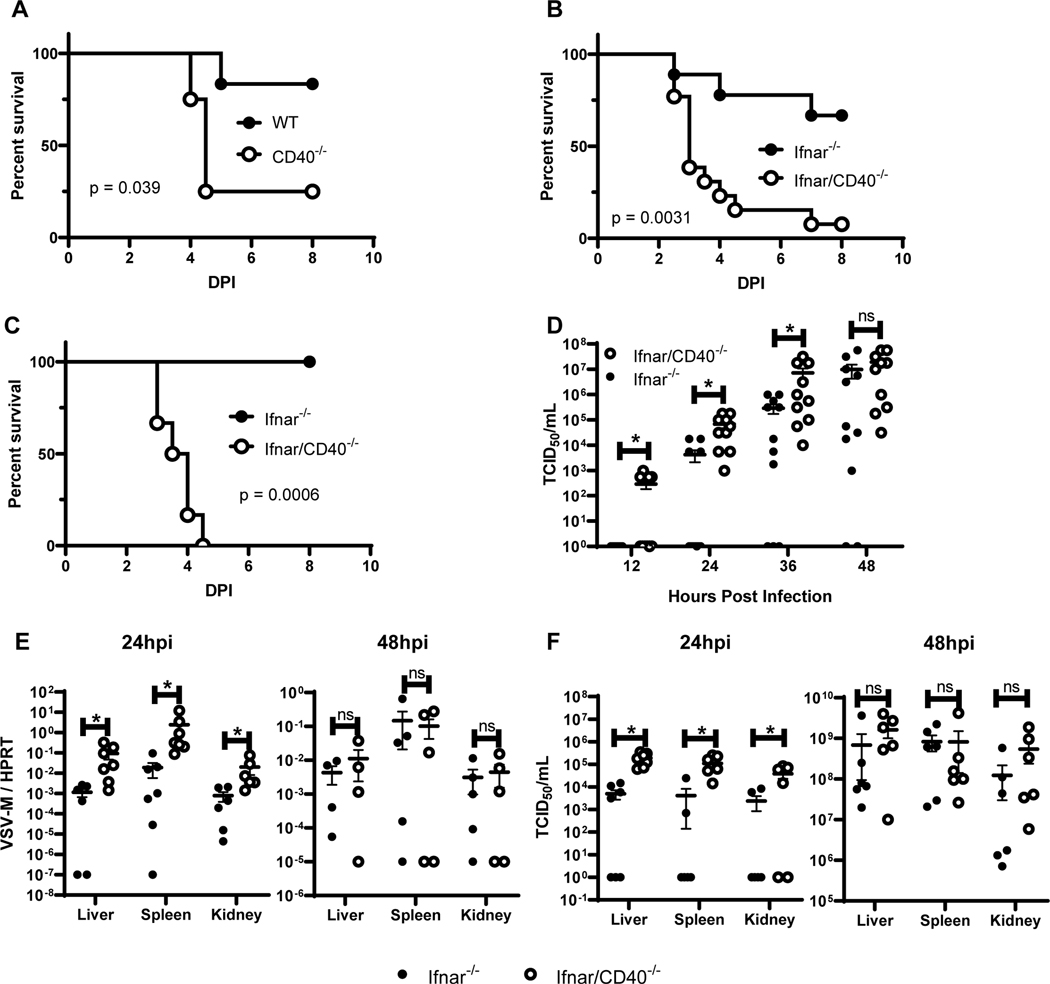
To investigate the role of CD40 signaling during acute viral challenge, we performed in vivo studies with rVSV/EBOV GP. We first compared WT and CD40−/− mice treated with the monoclonal antibody MAR-1 to block type I IFN/IFNAR interactions acutely. This allowed us to assess the effect of our recombinant virus in mice with typical immune system development by only transiently blocking type I IFN responses. We found that CD40−/− mice were significantly more sensitive to an otherwise low-lethal dose of the virus (Figure 7A).
Figure 7. CD40 restricts rVSV/EBOV GP infection within the first 12 hours of infection and exacerbates pathogenesis.
A) Survival curves of female WT (n=6) and CD40−/− (n=4) mice treated with 1 μg anti-IFNAR (MAR-1) antibody one day prior to challenge with a sublethal dose of rVSV/EBOV GP. Mice were monitored daily. B) Survival curve of female Ifnar−/− (n=9) and Ifnar/CD40−/− (n=13) mice that were challenged with a dose of rVSV/EBOV GP which was sublethal to Ifnar−/− mice by i.p. injection. Mice were monitored daily (DPI=days post infection). C) Survival curve of female Ifnar−/− (n=6) and Ifnar/CD40−/−(n=6) mice infected with a dose of rVSV/EBOV GP that was sublethal to Ifnar−/− mice by retro-orbital injection. Mice were monitored daily. D) Female Ifnar−/− (n=8) and Ifnar/CD40−/− (n=8) mice were infected i.p. with a dose of rVSV/EBOV GP that is sublethal to Ifnar−/− mice. Serum was collected at 12-hour intervals and viremia was assessed by end point dilutions on Vero cells. E/F) Female Ifnar−/− (n=12) and Ifnar/CD40−/− (n=12) mice were infected i.p. with a dose of rVSV/EBOV GP that is sublethal to Ifnar−/− mice. At 24 or 48 hours following infection, mice were anesthetized, perfused through the heart with PBS, and euthanized prior to organ harvest. RNA was isolated and qRT-PCR analysis of viral mRNA was performed (E). In parallel organs were homogenized and titers were assessed on Vero cells (n=4 organs/group) (F). All experiments were performed three times. For all experiments, * indicates p<0.05.
We then repeated these in vivo challenges with Ifnar−/− mice. Ifnar−/− and Ifnar/CD40−/− mice were infected i.p. with a low dose of rVSV/EBOV GP that was sublethal for Ifnar−/− mice. Mice lacking CD40 exhibited significantly increased mortality (Figure 7B). Ifnar/CD40−/− mice also succumbed to a dose of virus that was non-lethal to Ifnar−/− mice when virus was administered by retro-orbital infection, providing evidence that CD40 signaling also controlled virus infection when virus was administered by alternative routes (Figure 7C).
Timing of viremia was also explored in Ifnar−/− and Ifnar/CD40−/− mice. Serum was obtained at 12-hour intervals following i.p. rVSV/EBOV GP administration and the infectious virus present was determined by end point dilution of serum samples on Vero cells (TCID50) (Figure 7D). Ifnar/CD40−/− mice exhibited viremia as rapidly as 12 hours after infection, and viremia was significantly higher than in Ifnar−/− mice through the first 36 hours. Of note, the CD40-deficient animals attained maximal viral titers earlier than the CD40-sufficient animals, suggestive of an accelerated disease process. We also examined virus replication in the visceral organs: liver, spleen and kidney, of Ifnar−/− and Ifnar/CD40−/− mice at 24 and 48 hours following a dose of rVSV/EBOV GP that was sublethal to Ifnar−/− mice. Viral loads were elevated in all organs of CD40-deficient animals at 24 hours compared to organs from CD40-sufficient mice, but not significantly different by 48 hours, consistent with the observed kinetics of viremia (Figure 7E). Titers from these organs reflected similar changes in viral replication (Figure 7F). In total, our findings demonstrate that CD40 serves to restrict virus replication at early times during infection. Loss of CD40 expression mice rendered more sensitive to rVSV/EBOV GP infection.
Discussion:
Here we show that CD40 signaling in peritoneal cells protects against acute challenge by a wide range of RNA viruses at 12–24 hours of infection. Our findings support that CD40 signaling on pmacs is responsible for the protection as enriched pmacs were protected against EBOV (Mayinga) or rVSV/EBOV GP after CD40 signaling. Macrophages are thought to be critical target cells for EBOV infection at early times during infection (36, 62, 63), so our model system has allowed us to study the in vivo and ex vivo effects of CD40 ligation on a highly relevant cell type. While the importance for CD40 ligation and signaling is well appreciated in the development and maintenance of adaptive immune responses, a critical role for CD40 in early innate immune control of an acute viral infection was unexpected. During in vivo rVSV/EBOV GP infection, viremia was exacerbated within the first 12–36 hours of infection when CD40 was absent, providing compelling evidence that CD40 contributes to early host defenses against acute viral infection. Mortality in Ifnar/CD40−/− mice occurred on average at 3.3 days of infection, a time that precedes the development of adaptive immune responses (9, 48, 49). Thus, we postulated that CD40 orchestrates important early innate immune responses that restrict virus replication and effectively control viral pathogenesis, supporting a potentially novel and unanticipated role for CD40 signaling during acute virus infection.
While our rVSV/EBOV GP studies primarily used pmacs and mice on an Ifnar−/− background, this genetic background did not appear to alter CD40 signaling responses, as several key experiments in WT peritoneal cells or pmacs, with or without acute IFN blockade, provided similar results. Both rVSV/EBOV GP and WT EBOV infection in wild-type pmacs was controlled by either addition of IL-12 or an agonistic anti-CD40 mAb, just as these proteins restricted infection in Ifnar−/− pmacs. Further, our in vivo studies using WT or CD40−/− mice treated with the IFNAR-specific mAb, MAR-1, demonstrated that CD40−/− mice were as sensitive to rVSV/EBOV GP as Ifnar/CD40−/− mice, rapidly succumbing to an infection that was sublethal to CD40-competent mice. Finally, while virus pathogenesis was not observed in WT mice due to limited spread of our recombinant virus, studies with WT mice demonstrated that CD40 ligation reduced infection in the peritoneal cavity, whereas CD154 blockade and T cell depletions enhanced infection. In total these experiments provide strong evidence that the in vivo and ex vivo effects of CD40 ligation are similar in WT and Ifnar−/− mice.
We also defined the predominant signaling pathway required for protection following CD40 ligation. CD40/TRAF6 interactions elicited IL-12 expression and IL-12 synthesis led to IFN-γ production and signaling to control both our BSL2 model virus and EBOV (Mayinga) in enriched pmacs. This CD40-dependent signaling pathway is well established to enhance adaptive immunity, but has not been previously appreciated to play a critical role in innate immunity against acute virus infection of macrophages. While IL-12 is reported to protect against a variety of infectious pathogens (9, 40, 64, 65), to our knowledge this is the first report that IL-12 mediates protection against filoviruses. Signaling events through this pathway are of interest in the filovirus field as we have previously shown that IFN-γ treatment early during infection protects mice from lethal EBOV challenge by restricting virus replication (40). The current studies extend our prior work by identifying a mechanism by which IFN-γ production is elicited during the course of acute infection. Further, our new findings highlight IL-12 as another potential post-exposure therapeutic agent against early EBOV infection. Given the interest in predictive biomarkers during EBOV infection (66), these studies highlight receptors and cytokines that at very early times during infection may significantly influence the course of disease.
These studies identified that CD154 expressed on lymphocytes within the peritoneum of the naïve mouse interacts with macrophage CD40, leading to early control of virus replication. Our data support that peritoneal lymphocytes stimulate resident macrophages via the CD154/CD40 axis to limit acute rVSV/EBOV GP infection over the first 24 hours of infection. Thus, within the peritoneum, these cell-cell interactions mediated by CD40/CD154 ligation restrict virus replication days prior to the induction of virus-specific adaptive immunity.
Intriguingly, CD40/CD154 signaling can synergize with other innate immune signals, such as TLR signaling, to enhance cytokine production (67). As the EBOV glycoprotein binds to TLR4 and stimulates TLR4 signaling in macrophages (68, 69), it is possible that virus glycoprotein interactions with TLR4 serve as a second signal to synergize with CD40 signaling. Future studies are needed to more specifically examine the role of TLR4 and potential additional ‘second signals’ in CD40-mediated innate immune protection against EBOV infection of macrophages.
Experiments investigating the increase in replication and spread of virus in CD40-deficient animals raise a series of questions. It is of interest that, while the relative levels of virus in organs from Ifnar−/− and Ifnar/CD40−/− mice differ early in infection, the change in virus load between 24 and 48 hours in organs is not comparable to the increases in serum viral titer seen in that same time frame. While the most likely explanation is that these differences are attributable to the technique utilized, as organs from each time point are necessarily from different animals, it is also possible that organ titers display different kinetics than serum titers. Along these same lines, we note that by 48 hours of infection virus load in organs and viremia were no longer restricted by the expression of CD40, with equivalent levels of viremia evident in both Ifnar−/− and Ifnar/CD40−/− mice. Yet, most CD40-sufficient mice survived this challenge, whereas the Ifnar/CD40−/− mice did not. An explanation consistent with our observations is that the lack of CD40 signaling that results in earlier virus replication causes overall dysregulated cytokine responses, exacerbating disease. Infection of macrophages and DCs is thought to drive the pathogenic immune response in EBOV infection, and we are targeting macrophages by delivering the virus i.p., Thus, it is possible that control of virus replication and associated cytokine responses in macrophages play crucial roles in determining outcomes. Consistent with this, it is well appreciated that dysregulated cytokine production exacerbates EBOV disease in humans (70–72). Thus, the ability of the CD40-sufficient mice to survive, despite significant viremia by day 2 of infection, remains an intriguing area of future investigation.
Our experiments with apilimod which provided evidence of the ability of IL-12 to control rVSV/EBOV GP yielded somewhat unexpected results as PIKfyve inhibition by apilimod directly reduces EBOV (and rVSV/EBOV GP) entry into Vero cells, Huh7 cells, HEK 293T cells and purified human monocyte-derived macrophages due to inhibition of vesicular trafficking (73, 74). In our studies, it appears that the effect of apilimod on IL-12 production outweighed the inhibitory effect of apilimod on virus entry. This is consistent with work by Dyall et al (57) which found that apilimod does not protect mice from lethal EBOV challenge. Together, these data support that when CD154 is present, apilimod-mediated suppression of IL-12 signaling offsets the direct inhibitory effects of apilimod on virus entry. This result further emphasizes the critical importance of adequate production of IL-12 by pmacs in protection from viral pathogenesis.
Filoviruses infect a broad range of cells over the course of infection (62, 63) and many of these cells, such as endothelial and epithelial cell populations also express CD40. The ability of CD40 signaling to control virus replication in these other cells needs to be examined. While CD40 regulates differentiation of some epithelial cells such as keratinocytes (75, 76), examination of the impact of CD40 signaling on innate immune responses in these cells is in its infancy. Interestingly, many of these CD40-expressing populations are also capable of producing IL-12 (77, 78) and thus might play a critical role in the innate immune response to EBOV in different tissues. Given that macrophages are the predominant cell infected early in EBOV infection, we would expect that CD40 expression on additional cell types might contribute to inhibition of virus at later times during infection as the virus disseminates.
Cell population(s) responsible for IFN-γ production within the peritoneal cavity remains to be explored. T cells, NK cells, and additional innate lymphoid cell populations are capable of robust IFN-γ production, all of which are present in the peritoneal cavity (Figure S (79). While we looked at the effect of T cell and NK cell depletion in this study, we don’t differentiate between the dual roles of CD154 presentation and response to IL-12 and subsequent IFN-γ production. Recent work from Weizman et. al. suggests that type 1 innate lymphoid cells (ILC1s) are capable of robust interferon gamma production and facilitate control of mouse cytomegalovirus when delivered i.p. (80), and others have shown that ILC1s produce IFN-γ downstream of CD40 signaling in the gut (81). These studies suggest that ILC1s may be a source of IFN-γ in the peritoneal cavity. Intriguingly, blocking IFN-γ availability with an anti-IFN-γ mAb increases virus infection in both CD40-sufficient and CD40-deficient pmacs, suggesting that additional pathways which are independent of CD40 signaling also stimulate IFN-γ-mediated control of rVSV/EBOV GP infection.
Of broader interest, we show that ex vivo CD40-dependent protection is not unique for filoviruses, but extends to other globally relevant viral pathogens that target macrophages, such as alphaviruses, rhabdoviruses and orthomyxoviruses. Studies on the in vivo restriction by CD40 signaling against other RNA viruses are warranted. These studies lay the groundwork for the investigation of CD40 signaling in an entirely new context and provide insight into potential therapeutic targets common to several RNA viruses.
Supplementary Material
Acknowledgements
We would like to thank Dr. Hillel Haim and members of the Maury, Bishop, Varga and Butler laboratories for their thoughtful comments on the manuscript.
Funding sources: WJM: NIH R21 AI139902, R21 AI144215 and University of Iowa Department of Microbiology and Immunology Developmental Grant Award; KR: NIH T32 GM007337 and T32 GM067795; GAB: NIH AI119163 and VA Merit Review I01 BX001702; NSB: NIH R01AI127481 and R01AI125446; JMW: NIH AI114776; SMV: NIH RO1AI124093
This material is based upon work supported in part by facilities and equipment provided by the Dept. of Veterans Affairs, VHA, Office of Research and Development. We acknowledge the staff at the University of Iowa College of Medicine and Holden Comprehensive Cancer Center Radiation and Free Radical Research (RFRR) Core for radiation services. The RFRR core facility is supported by funding from NIH P30 CA086862.
Some of the data presented herein were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine / Holden Comprehensive Cancer Center core research facility at the University of Iowa. The facility is funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran’s Administration Medical Center.
Abbreviations:
- ABSL2 –
Animal Biosafety level 2
- Ab -
Antibody
- APCs –
Antigen presenting cells
- DCs –
Dendritic cells
- EBOV –
Ebola virus
- GP –
Glycoprotein
- GFP –
Green fluorescent protein
- IgG –
Immunoglobulin G
- IAV -
Influenza A virus
- IFN -
Interferon
- IFNAR –
Interferon alpha/beta receptor
- Ifnar -
Interferon alpha/beta receptor gene
- IFN-γ –
Interferon gamma
- IL -
Interleukin
- i.p. –
Intraperitoneal
- i.v. -
Intravenous
- mAb –
Monoclonal antibody
- pmacs –
Peritoneal macrophages
- PBMCs -
Peripheral blood mononuclear cells
- qRT-PCR –
Quantitative reverse transcription polymerase chain reaction
- rVSV/EBOV GP –
Recombinant VSV expressing EBOV glycoprotein
- rVSV/G –
Recombinant VSV expressing it native glycoprotein
- RRV -
Ross river virus
- TCID –
Tissue culture infectious dose
- VSV -
Vesicular stomatitis virus
- VLP -
Virus like particle
Footnotes
Competing Interests
All authors declare no competing interests.
References:
- 1.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. 2009. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev 229:152–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hollenbaugh D, Grosmaire LS, Kullas CD, Chalupny NJ, Braesch-Andersen S, Noelle RJ, Stamenkovic I, Ledbetter JA, Aruffo A. 1992. The human T cell antigen gp39, a member of the TNF gene family, is a ligand for the CD40 receptor: expression of a soluble form of gp39 with B cell co-stimulatory activity. EMBO J 11:4313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alderson MR, Armitage RJ, Tough TW, Strockbine L, Fanslow WC, Spriggs MK. 1993. CD40 expression by human monocytes: regulation by cytokines and activation of monocytes by the ligand for CD40. J Exp Med 178:669–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner DH Jr., Stout RD, Suttles J. 1994. Role of the CD40-CD40 ligand interaction in CD4+ T cell contact-dependent activation of monocyte interleukin-1 synthesis. Eur J Immunol 24:3148–54. [DOI] [PubMed] [Google Scholar]
- 5.Demangel C, Palendira U, Feng CG, Heath AW, Bean AG, Britton WJ. 2001. Stimulation of dendritic cells via CD40 enhances immune responses to Mycobacterium tuberculosis infection. Infect Immun 69:2456–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazouz N, Ooms A, Moulin V, Van Meirvenne S, Uyttenhove C, Degiovanni G. 2002. CD40 triggering increases the efficiency of dendritic cells for antitumoral immunization. Cancer Immun 2:2. [PubMed] [Google Scholar]
- 7.Van den Eertwegh AJ, Noelle RJ, Roy M, Shepherd DM, Aruffo A, Ledbetter JA, Boersma WJ, Claassen E. 1993. In vivo CD40-gp39 interactions are essential for thymus-dependent humoral immunity. I. In vivo expression of CD40 ligand, cytokines, and antibody production delineates sites of cognate T-B cell interactions. J Exp Med 178:1555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei XF, Ohkawara Y, Stampfli MR, Mastruzzo C, Marr RA, Snider D, Xing Z, Jordana M. 1998. Disruption of antigen-induced inflammatory responses in CD40 ligand knockout mice. J Clin Invest 101:1342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell KA, Ovendale PJ, Kennedy MK, Fanslow WC, Reed SG, Maliszewski CR. 1996. CD40 ligand is required for protective cell-mediated immunity to Leishmania major. Immunity 4:283–9. [DOI] [PubMed] [Google Scholar]
- 10.Bishop GA, Moore CR, Xie P, Stunz LL, Kraus ZJ. 2007. TRAF proteins in CD40 signaling. Adv Exp Med Biol 597:131–51. [DOI] [PubMed] [Google Scholar]
- 11.Kelsall BL, Stuber E, Neurath M, Strober W. 1996. Interleukin-12 production by dendritic cells. The role of CD40-CD40L interactions in Th1 T-cell responses. Ann N Y Acad Sci 795:116–26. [DOI] [PubMed] [Google Scholar]
- 12.Yoshimoto T, Nagase H, Ishida T, Inoue J, Nariuchi H. 1997. Induction of interleukin-12 p40 transcript by CD40 ligation via activation of nuclear factor-kappaB. Eur J Immunol 27:3461–70. [DOI] [PubMed] [Google Scholar]
- 13.Seder RA, Gazzinelli R, Sher A, Paul WE. 1993. Interleukin 12 acts directly on CD4+ T cells to enhance priming for interferon gamma production and diminishes interleukin 4 inhibition of such priming. Proc Natl Acad Sci U S A 90:10188–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimoto T, Takeda K, Tanaka T, Ohkusu K, Kashiwamura S, Okamura H, Akira S, Nakanishi K. 1998. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-gamma production. J Immunol 161:3400–7. [PubMed] [Google Scholar]
- 15.Subauste CS. 2009. CD40 and the immune response to parasitic infections. Semin Immunol 21:273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okwor I, Jia P, Uzonna JE. 2015. Interaction of Macrophage Antigen 1 and CD40 Ligand Leads to IL-12 Production and Resistance in CD40-Deficient Mice Infected with Leishmania major. J Immunol 195:3218–26. [DOI] [PubMed] [Google Scholar]
- 17.de Oliveira FA, Barreto AS, Bomfim LG, Leite TR, Dos Santos PL, de Almeida RP, da Silva AM, Duthie MS, Reed SG, de Moura TR, Ribeiro de Jesus A. 2015. Soluble CD40 Ligand in Sera of Subjects Exposed to Leishmania infantum Infection Reduces the Parasite Load in Macrophages. PLoS One 10:e0141265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nunes MP, Cysne-Finkelstein L, Monteiro BC, de Souza DM, Gomes NA, DosReis GA. 2005. CD40 signaling induces reciprocal outcomes in Leishmania-infected macrophages; roles of host genotype and cytokine milieu. Microbes Infect 7:78–85. [DOI] [PubMed] [Google Scholar]
- 19.Marovich MA, McDowell MA, Thomas EK, Nutman TB. 2000. IL-12p70 production by Leishmania major-harboring human dendritic cells is a CD40/CD40 ligand-dependent process. J Immunol 164:5858–65. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi T, Rao SP, Meylan PR, Kornbluth RS, Catanzaro A. 1999. Role of CD40 ligand in Mycobacterium avium infection. Infect Immun 67:3558–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gold JA, Parsey M, Hoshino Y, Hoshino S, Nolan A, Yee H, Tse DB, Weiden MD. 2003. CD40 contributes to lethality in acute sepsis: in vivo role for CD40 in innate immunity. Infect Immun 71:3521–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nolan A, Weiden MD, Hoshino Y, Gold JA. 2004. Cd40 but not CD154 knockout mice have reduced inflammatory response in polymicrobial sepsis: a potential role for Escherichia coli heat shock protein 70 in CD40-mediated inflammation in vivo. Shock 22:538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. 1994. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity 1:167–78. [DOI] [PubMed] [Google Scholar]
- 24.Ebihara H, Theriault S, Neumann G, Alimonti JB, Geisbert JB, Hensley LE, Groseth A, Jones SM, Geisbert TW, Kawaoka Y, Feldmann H. 2007. In vitro and in vivo characterization of recombinant Ebola viruses expressing enhanced green fluorescent protein. J Infect Dis 196 Suppl 2:S313–22. [DOI] [PubMed] [Google Scholar]
- 25.Reed LJ MH. 1938. A simple method of estimating fifty percent endpoints. American Journal of Epidemiology 27:493–497. [Google Scholar]
- 26.Alfson KJ, Avena LE, Beadles MW, Staples H, Nunneley JW, Ticer A, Dick EJ Jr., Owston MA, Reed C, Patterson JL, Carrion R Jr., Griffiths A. 2015. Particle-to-PFU ratio of Ebola virus influences disease course and survival in cynomolgus macaques. J Virol 89:6773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shtanko O, Sakurai Y, Reyes AN, Noel R, Cintrat JC, Gillet D, Barbier J, Davey RA. 2018. Retro-2 and its dihydroquinazolinone derivatives inhibit filovirus infection. Antiviral Res 149:154–163. [DOI] [PubMed] [Google Scholar]
- 28.Nolan T, Hands RE, Bustin SA. 2006. Quantification of mRNA using real-time RT-PCR. Nat Protoc 1:1559–82. [DOI] [PubMed] [Google Scholar]
- 29.Shtanko O, Reyes AN, Jackson WT, Davey RA. 2018. Autophagy-Associated Proteins Control Ebola Virus Internalization Into Host Cells. J Infect Dis 218:S346–S354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlesinger LS, Horwitz MA. 1990. Phagocytosis of leprosy bacilli is mediated by complement receptors CR1 and CR3 on human monocytes and complement component C3 in serum. J Clin Invest 85:1304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlesinger LS. 1993. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J Immunol 150:2920–30. [PubMed] [Google Scholar]
- 32.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A 107:11531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bishop GA, Warren WD, Berton MT. 1995. Signaling via major histocompatibility complex class II molecules and antigen receptors enhances the B cell response to gp39/CD40 ligand. Eur J Immunol 25:1230–8. [DOI] [PubMed] [Google Scholar]
- 34.Zarzycka B, Seijkens T, Nabuurs SB, Ritschel T, Grommes J, Soehnlein O, Schrijver R, van Tiel CM, Hackeng TM, Weber C, Giehler F, Kieser A, Lutgens E, Vriend G, Nicolaes GA. 2015. Discovery of small molecule CD40-TRAF6 inhibitors. J Chem Inf Model 55:294–307. [DOI] [PubMed] [Google Scholar]
- 35.Glik A, Mazar J, Rogachev B, Zlotnik M, Douvdevani A. 2005. CD40 ligand expression correlates with resolution of peritonitis and mononuclear cell recruitment. Perit Dial Int 25:240–7. [PubMed] [Google Scholar]
- 36.Bray M, Geisbert TW. 2005. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol 37:1560–6. [DOI] [PubMed] [Google Scholar]
- 37.Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. 1999. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis 179 Suppl 1:S248–58. [DOI] [PubMed] [Google Scholar]
- 38.Davis KJ, Anderson AO, Geisbert TW, Steele KE, Geisbert JB, Vogel P, Connolly BM, Huggins JW, Jahrling PB, Jaax NK. 1997. Pathology of experimental Ebola virus infection in African green monkeys. Involvement of fibroblastic reticular cells. Arch Pathol Lab Med 121:805–19. [PubMed] [Google Scholar]
- 39.Mateo M, Carbonnelle C, Reynard O, Kolesnikova L, Nemirov K, Page A, Volchkova VA, Volchkov VE. 2011. VP24 is a molecular determinant of Ebola virus virulence in guinea pigs. J Infect Dis 204 Suppl 3:S1011–20. [DOI] [PubMed] [Google Scholar]
- 40.Rhein BA, Powers LS, Rogers K, Anantpadma M, Singh BK, Sakurai Y, Bair T, Miller-Hunt C, Sinn P, Davey RA, Monick MM, Maury W. 2015. Interferon-gamma Inhibits Ebola Virus Infection. PLoS Pathog 11:e1005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers KJ, Brunton B, Mallinger L, Bohan D, Sevcik KM, Chen J, Ruggio N, Maury W. 2019. IL-4/IL-13 polarization of macrophages enhances Ebola virus glycoprotein-dependent infection. PLoS Negl Trop Dis 13:e0007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brunton B, Rogers K, Phillips EK, Brouillette RB, Bouls R, Butler NS, Maury W. 2019. TIM-1 serves as a receptor for Ebola virus in vivo, enhancing viremia and pathogenesis. PLoS Negl Trop Dis 13:e0006983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrell SA, Maheshwari RK, Mohanty SB, Friedman RM. 1982. Effect of human interferon on vesicular stomatitis virus released from bovine embryonic kidney cells. Am J Vet Res 43:565–8. [PubMed] [Google Scholar]
- 44.Agrawal H, Jacob N, Carreras E, Bajana S, Putterman C, Turner S, Neas B, Mathian A, Koss MN, Stohl W, Kovats S, Jacob CO. 2009. Deficiency of type I IFN receptor in lupus-prone New Zealand mixed 2328 mice decreases dendritic cell numbers and activation and protects from disease. J Immunol 183:6021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohamadzadeh M, Coberley SS, Olinger GG, Kalina WV, Ruthel G, Fuller CL, Swenson DL, Pratt WD, Kuhns DB, Schmaljohn AL. 2006. Activation of triggering receptor expressed on myeloid cells-1 on human neutrophils by marburg and ebola viruses. J Virol 80:7235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lesley R, Kelly LM, Xu Y, Cyster JG. 2006. Naive CD4 T cells constitutively express CD40L and augment autoreactive B cell survival. Proc Natl Acad Sci U S A 103:10717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Butler NS, Moebius J, Pewe LL, Traore B, Doumbo OK, Tygrett LT, Waldschmidt TJ, Crompton PD, Harty JT. 2011. Therapeutic blockade of PD-L1 and LAG-3 rapidly clears established blood-stage Plasmodium infection. Nat Immunol 13:188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landay ME. 1982. Antibody response following subcutaneous and intraperitoneal injection of Bacteroides fragilis in rabbits. Eur J Clin Microbiol 1:248–51. [DOI] [PubMed] [Google Scholar]
- 49.Hu Z, Molloy MJ, Usherwood EJ. 2016. CD4(+) T-cell dependence of primary CD8(+) T-cell response against vaccinia virus depends upon route of infection and viral dose. Cell Mol Immunol 13:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukundan L, Bishop GA, Head KZ, Zhang L, Wahl LM, Suttles J. 2005. TNF receptor-associated factor 6 is an essential mediator of CD40-activated proinflammatory pathways in monocytes and macrophages. J Immunol 174:1081–90. [DOI] [PubMed] [Google Scholar]
- 51.Kennedy MK, Picha KS, Fanslow WC, Grabstein KH, Alderson MR, Clifford KN, Chin WA, Mohler KM. 1996. CD40/CD40 ligand interactions are required for T cell-dependent production of interleukin-12 by mouse macrophages. Eur J Immunol 26:370–8. [DOI] [PubMed] [Google Scholar]
- 52.Tomura M, Yu WG, Ahn HJ, Yamashita M, Yang YF, Ono S, Hamaoka T, Kawano T, Taniguchi M, Koezuka Y, Fujiwara H. 1999. A novel function of Valpha14+CD4+NKT cells: stimulation of IL-12 production by antigen-presenting cells in the innate immune system. J Immunol 163:93–101. [PubMed] [Google Scholar]
- 53.Bi Z, Quandt P, Komatsu T, Barna M, Reiss CS. 1995. IL-12 promotes enhanced recovery from vesicular stomatitis virus infection of the central nervous system. J Immunol 155:5684–9. [PubMed] [Google Scholar]
- 54.Komatsu T, Srivastava N, Revzin M, Ireland DD, Chesler D, Reiss CS. 1999. Mechanisms of cytokine-mediated inhibition of viral replication. Virology 259:334–41. [DOI] [PubMed] [Google Scholar]
- 55.Moller JR, Rager-Zisman B, Quan PC, Schattner A, Panush D, Rose JK, Bloom BR. 1985. Natural killer cell recognition of target cells expressing different antigens of vesicular stomatitis virus. Proc Natl Acad Sci U S A 82:2456–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai X, Xu Y, Cheung AK, Tomlinson RC, Alcazar-Roman A, Murphy L, Billich A, Zhang B, Feng Y, Klumpp M, Rondeau JM, Fazal AN, Wilson CJ, Myer V, Joberty G, Bouwmeester T, Labow MA, Finan PM, Porter JA, Ploegh HL, Baird D, De Camilli P, Tallarico JA, Huang Q. 2013. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem Biol 20:912–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dyall J, Nelson EA, DeWald LE, Guha R, Hart BJ, Zhou H, Postnikova E, Logue J, Vargas WM, Gross R, Michelotti J, Deiuliis N, Bennett RS, Crozier I, Holbrook MR, Morris PJ, Klumpp-Thomas C, McKnight C, Mierzwa T, Shinn P, Glass PJ, Johansen LM, Jahrling PB, Hensley LE, Olinger GG Jr., Thomas C, White JM. 2018. Identification of Combinations of Approved Drugs With Synergistic Activity Against Ebola Virus in Cell Cultures. J Infect Dis doi: 10.1093/infdis/jiy304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wada Y, Lu R, Zhou D, Chu J, Przewloka T, Zhang S, Li L, Wu Y, Qin J, Balasubramanyam V, Barsoum J, Ono M. 2007. Selective abrogation of Th1 response by STA-5326, a potent IL-12/IL-23 inhibitor. Blood 109:1156–64. [DOI] [PubMed] [Google Scholar]
- 59.Sedegah M, Finkelman F, Hoffman SL. 1994. Interleukin 12 induction of interferon gamma-dependent protection against malaria. Proc Natl Acad Sci U S A 91:10700–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schijns VE, Haagmans BL, Horzinek MC. 1995. IL-12 stimulates an antiviral type 1 cytokine response but lacks adjuvant activity in IFN-gamma-receptor-deficient mice. J Immunol 155:2525–32. [PubMed] [Google Scholar]
- 61.Chace JH, Hooker NA, Mildenstein KL, Krieg AM, Cowdery JS. 1997. Bacterial DNA-induced NK cell IFN-gamma production is dependent on macrophage secretion of IL-12. Clin Immunol Immunopathol 84:185–93. [DOI] [PubMed] [Google Scholar]
- 62.Connolly BM, Steele KE, Davis KJ, Geisbert TW, Kell WM, Jaax NK, Jahrling PB. 1999. Pathogenesis of experimental Ebola virus infection in guinea pigs. J Infect Dis 179 Suppl 1:S203–17. [DOI] [PubMed] [Google Scholar]
- 63.Geisbert TW, Hensley LE, Larsen T, Young HA, Reed DS, Geisbert JB, Scott DP, Kagan E, Jahrling PB, Davis KJ. 2003. Pathogenesis of Ebola hemorrhagic fever in cynomolgus macaques: evidence that dendritic cells are early and sustained targets of infection. Am J Pathol 163:2347–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamanaka M, Yu P, Yasui T, Yoshida K, Kawabe T, Horii T, Kishimoto T, Kikutani H. 1996. Protective role of CD40 in Leishmania major infection at two distinct phases of cell-mediated immunity. Immunity 4:275–81. [DOI] [PubMed] [Google Scholar]
- 65.Soong L, Xu JC, Grewal IS, Kima P, Sun J, Longley BJ Jr., Ruddle NH, McMahon-Pratt D, Flavell RA. 1996. Disruption of CD40-CD40 ligand interactions results in an enhanced susceptibility to Leishmania amazonensis infection. Immunity 4:263–73. [DOI] [PubMed] [Google Scholar]
- 66.Eisfeld AJ, Halfmann PJ, Wendler JP, Kyle JE, Burnum-Johnson KE, Peralta Z, Maemura T, Walters KB, Watanabe T, Fukuyama S, Yamashita M, Jacobs JM, Kim YM, Casey CP, Stratton KG, Webb-Robertson BM, Gritsenko MA, Monroe ME, Weitz KK, Shukla AK, Tian M, Neumann G, Reed JL, van Bakel H, Metz TO, Smith RD, Waters KM, N’Jai A, Sahr F, Kawaoka Y. 2017. Multi-platform ‘Omics Analysis of Human Ebola Virus Disease Pathogenesis. Cell Host Microbe 22:817–829 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vanden Bush TJ, Bishop GA. 2008. TLR7 and CD40 cooperate in IL-6 production via enhanced JNK and AP-1 activation. Eur J Immunol 38:400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okumura A, Pitha PM, Yoshimura A, Harty RN. 2010. Interaction between Ebola virus glycoprotein and host toll-like receptor 4 leads to induction of proinflammatory cytokines and SOCS1. J Virol 84:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lai CY, Strange DP, Wong TAS, Lehrer AT, Verma S. 2017. Ebola Virus Glycoprotein Induces an Innate Immune Response In vivo via TLR4. Front Microbiol 8:1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. 2010. Human fatal zaire ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, Speranza E, Munoz-Fontela C, Haldenby S, Rickett NY, Garcia-Dorival I, Fang Y, Hall Y, Zekeng EG, Ludtke A, Xia D, Kerber R, Krumkamp R, Duraffour S, Sissoko D, Kenny J, Rockliffe N, Williamson ED, Laws TR, N’Faly M, Matthews DA, Gunther S, Cossins AR, Sprecher A, Connor JH, Carroll MW, Hiscox JA. 2017. Transcriptomic signatures differentiate survival from fatal outcomes in humans infected with Ebola virus. Genome Biol 18:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kash JC, Walters KA, Kindrachuk J, Baxter D, Scherler K, Janosko KB, Adams RD, Herbert AS, James RM, Stonier SW, Memoli MJ, Dye JM, Davey RT Jr., Chertow DS, Taubenberger JK. 2017. Longitudinal peripheral blood transcriptional analysis of a patient with severe Ebola virus disease. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nelson EA, Dyall J, Hoenen T, Barnes AB, Zhou H, Liang JY, Michelotti J, Dewey WH, DeWald LE, Bennett RS, Morris PJ, Guha R, Klumpp-Thomas C, McKnight C, Chen YC, Xu X, Wang A, Hughes E, Martin S, Thomas C, Jahrling PB, Hensley LE, Olinger GG Jr., White JM. 2017. The phosphatidylinositol-3-phosphate 5-kinase inhibitor apilimod blocks filoviral entry and infection. PLoS Negl Trop Dis 11:e0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiu S, Leung A, Bo Y, Kozak RA, Anand SP, Warkentin C, Salambanga FDR, Cui J, Kobinger G, Kobasa D, Cote M. 2018. Ebola virus requires phosphatidylinositol (3,5) bisphosphate production for efficient viral entry. Virology 513:17–28. [DOI] [PubMed] [Google Scholar]
- 75.Peguet-Navarro J, Dalbiez-Gauthier C, Moulon C, Berthier O, Reano A, Gaucherand M, Banchereau J, Rousset F, Schmitt D. 1997. CD40 ligation of human keratinocytes inhibits their proliferation and induces their differentiation. J Immunol 158:144–52. [PubMed] [Google Scholar]
- 76.Denfeld RW, Hollenbaugh D, Fehrenbach A, Weiss JM, von Leoprechting A, Mai B, Voith U, Schopf E, Aruffo A, Simon JC. 1996. CD40 is functionally expressed on human keratinocytes. Eur J Immunol 26:2329–34. [DOI] [PubMed] [Google Scholar]
- 77.Aragane Y, Riemann H, Bhardwaj RS, Schwarz A, Sawada Y, Yamada H, Luger TA, Kubin M, Trinchieri G, Schwarz T. 1994. IL-12 is expressed and released by human keratinocytes and epidermoid carcinoma cell lines. J Immunol 153:5366–72. [PubMed] [Google Scholar]
- 78.Walter MJ, Kajiwara N, Karanja P, Castro M, Holtzman MJ. 2001. Interleukin 12 p40 production by barrier epithelial cells during airway inflammation. J Exp Med 193:339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ebbo M, Crinier A, Vely F, Vivier E. 2017. Innate lymphoid cells: major players in inflammatory diseases. Nat Rev Immunol 17:665–678. [DOI] [PubMed] [Google Scholar]
- 80.Weizman OE, Adams NM, Schuster IS, Krishna C, Pritykin Y, Lau C, Degli-Esposti MA, Leslie CS, Sun JC, O’Sullivan TE. 2017. ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell 171:795–808 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brasseit J, Kwong Chung CKC, Noti M, Zysset D, Hoheisel-Dickgreber N, Genitsch V, Corazza N, Mueller C. 2018. Divergent Roles of Interferon-gamma and Innate Lymphoid Cells in Innate and Adaptive Immune Cell-Mediated Intestinal Inflammation. Front Immunol 9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.