Abstract
Purpose
2D creatine CEST (2D-CrCEST), with a slice thickness of 10–20 mm and temporal resolution (τRes) of ~30s, has previously been shown to capture the creatine recovery (CR) kinetics in healthy controls and in patients with abnormal creatine kinase (CK) kinetics following the mild plantar flexion exercise (PFE). Since the distribution of disease burden may vary across the muscle length for many musculoskeletal disorders, there is a need to increase coverage in slice-encoding direction. Here, we demonstrate the feasibility of 3D-CrCEST with τRes of ~30s and also propose an improved voxel-wise B1+-calibration approach for CrCEST.
Methods
The current 7T study with enrollment of five volunteers, involved collecting the baseline CrCEST imaging for the first 2 minutes, followed by 2 minutes of PFE and then 8 minutes of post-exercise CrCEST imaging to detect the temporal evolution of creatine concentration post-exercise.
Results
Very good repeatability of 3D-CrCEST findings for activated muscle groups on intra-day and inter-day basis was established, with coefficient of variance of creatine recovery constants (τCr) being 7–15.7%, 7.5%, 5.8%, for lateral gastrocnemius, medial gastrocnemius and peroneus longus, respectively. We also established a good intra- and inter-day scan repeatability for 3D-CrCEST and also showed good correspondence between τCr-measurements using 2D-CrCEST and 3D-CrCEST acquisitions.
Conclusion
In this study, for the first time, we have demonstrated the feasibility and the repeatability of 3D-CrCEST method in calf muscle with improved B1+-correction to measure Cr recovery kinetics within a large 3D volume of calf muscle.
Keywords: Creatine, Plantar flexion exercise, Creatine Kinase, Phosphocreatine, Creatine recovery kinetics, Creatine CEST, CrCEST
INTRODUCTION
Cellular phosphocreatine (PCr) pool serves as a rapid mobilizable energy reservoir in tissues with high fluctuating energy demands, such as skeletal muscle, heart and brain. Among all tissues of high energy demand, the skeletal muscle retains the highest reservoir of PCr (20–35 mM) (1). During the period of higher than regular energy needs, such as mild/intense exercise or neuronal function, creatine kinase (CK) isoenzymes catalyzes the conversion of PCr and adenosine diphosphate (ADP) to creatine (Cr) and adenosine triphosphate (ATP) in a reversible reaction (PCr + ADP + H+ ⇋ Cr + ATP). At the onset of higher energy demand by skeletal muscle, PCr is broken down to provide ATP needed for muscle contraction and ATP further hydrolyses to ADP and inorganic phosphate. Upon the cessation of exercise activity, ATP supplied by mitochondria as a result of glycolysis makes the conversion of Cr to PCr a highly favored reaction. This rate of conversion is determined by multiple factors, such as mechanisms intrinsic to skeletal muscle contractile function, the acidity, the restoration of energy balance and oxygen transport through blood circulation. The rate of PCr (1/τPCr) or Cr (1/τCr) recovery after exercise, governed by the CK reaction, is strongly coupled to net mitochondrial oxidative phosphorylation (OXPHOS) or ATP synthesis, with longer τPCr (or τCr) for a particular muscle group being suggestive of lower OXPHOS capacity (2). Further, the abnormality in CK kinetics has been implicated in systemic disorders, such as primary mitochondrial disorders (2), diabetes mellitus (3), Alzheimer’s disease (4), Parkinson’s disease (5) and cardiovascular disease (6), as well as other musculoskeletal diseases and disorders, such as exercise-induced or sports-related muscle injury (7), and other muscular pathologies including Duchenne (8,9) and Becker (8,10) dystrophies and distal myopathies (11).
As a noninvasive imaging biomarker, phosphorous magnetic resonance spectroscopy (31PMRS) has traditionally been used to the measure the metabolic response of exercised skeletal muscle in humans and has contributed immensely (12) to the vital understanding of muscle energetics. However, 31PMRS suffers from poor spatial resolution as well as low sensitivity. On the other hand, proton magnetic resonance spectroscopy (1HMRS) cannot be used to assess the CK pathways as it can only measure total creatine without any differentiation between PCr and Cr (13). It was previously shown (14) that amine protons on creatine undergoes chemical exchange at ~1.8 ppm off-resonance downfield from the water peak and the resultant chemical exchange saturation transfer (CEST) effect is linearly proportional to the Cr concentration in the physiological pH range. Though multiple factors can affect CrCEST-weighted (CrCESTW) image, the temporal decay of CrCEST can primarily be attributed to creatine decay itself (refer: discussion section). Further, CrCEST was also shown to have three orders of magnitude higher sensitivity than 31PMRS (13). Subsequently, 2D creatine CEST (2D-CrCEST) was developed and optimized at 3 Tesla (15) and 7 Tesla (13) MRI scanners. With sufficient in-plane resolution and temporal resolution (τRes) of ~30s, 2D-CrCEST made it possible to visualize and quantify OXPHOS capacity on the inter-muscular basis. However, it suffers from the lack of spatial coverage in slice-encoding direction, which may be very valuable in the case of many musculoskeletal diseases and disorders with different disease distributions along the muscular length. Being a single slice method, it also precludes any correction for motion occurred during the acquisition. Hence, there is a need to extend the CrCEST method to 3D acquisition for increasing the coverage in slice-encoding direction, as well as for improving the signal to noise ratio (SNR) of time-series of CrCEST maps. However, the need to retain the temporal resolution of ~30–35s, sufficient to follow the time course of creatine recovery (CR) kinetics in vivo, made the implementation of 3D-CrCEST challenging.
In this manuscript, we have demonstrated the feasibility of performing 3D-CrCEST MRI to measure OXPHOS capacity, with sufficient spatial and temporal resolution following plantar flexion exercises (PFE). The repeatability of 3D-CrCEST findings following PFE has also been investigated on five healthy volunteers. Additionally, we have also compared the CR kinetics of 3D-CrCEST and 2D-CrCEST under the similar exercise conditions on two healthy volunteers.
METHODS
Five healthy human volunteers (4 males, 1 female; aged 24–55 years old) participated in the approved University of Pennsylvania Institutional Review Board study protocol. Written informed consents were obtained from all volunteers before the scans. Imaging experiments were performed on a 7T MRI scanner (MAGNETOM Terra, Siemens Healthcare, Erlangen, Germany) using a single-channel transmit/28-channel receive phased array 1H knee coil (Quality Electrodynamics, Mayfield, USA). All pulse sequences were custom prototypes developed in-house.
The CrCEST study involved two minutes of baseline CrCEST acquisition, followed by two minutes of mild PFE and then eight minutes of post-exercise CrCEST acquisition. Water saturation shift referencing (WASSR) images (16) and relative B1+ maps (17) were also acquired to correct for the effect of B0- and B1+-inhomogeneity on the CEST-weighted (CESTW) images. With post-exercise calibration data starting around ~11 minutes post-exercise and lasting for additional ~4–5 minutes, the total post-exercise acquisition time was ~15–16 minutes. To assess the intra-day scan repeatability, all five volunteers also underwent at least two sets of 3D-CrCEST experiment within same session following separate PFEs. Between both intra-day sessions, participants were pulled out of the scanner and rested for 10 minutes before repositioning. A minimum of 20 minutes separation was maintained between successive PFEs to ensure full creatine recovery. To assess the inter-day scan repeatability, two volunteers underwent 3D-CrCEST acquisition following the PFE on at least two different days. Experiments and simulations were also carried out to compare the impact of the implemented acquisition modules on the accuracy of τCr-measurements for 2D-CrCEST and 3D-CrCEST.
PFE
An MR-compatible pneumatical ergometer (Trispect, Ergospect, Innsbruck, Austria) was used in the manual mode with the applied air pressure at 8 pound-per-square-inch (psi) and the push pedal frequency at 30 beats per minute (BPM). We kept the plantar flexion angle (PFA) of Φ = 40° and also modified our exercise setup to minimize ankle rotation while pushing the paddle.
CrCEST imaging
The CrCEST imaging involved a magnetization preparation module (supplementary Information Fig. S1) consisting of a 5×100ms long Hanning-windowed saturation pulse train (100ms pulse duration, 99% duty cycle, B1rms of 2.9μT), followed by a single shot TurboFLASH (tfl) read out with centric phase-encoding order and tfl-TR = 3.5ms, tfl-TE = 1.47ms, BW=710 Hz/pixel, with shot TR of 5s. Other imaging parameters were: voxel size = 1.4×1.4×5mm3; FOV = 160×160×40 mm3; FOV phase = 100%; averages = 1; number of slices = 8. Slices at both ends of imaging volumes were discarded due to fold-over artifacts. Raw CEST images were acquired at varying saturation frequency offsets (FOs) from ±1.2 to ±2.4 ppm (relative to water resonance set as 0 ppm) with a step-size of ±0.6 ppm, with following order: +1.2, −1.2., +1.8, −1.8, +2.4, −2.4 ppm (refer to appendix A.1 for the justification of chosen FO range). k-space strategies to maintain the temporal resolution of ~30s during CrCEST time-series: The spiral acquisition with elliptical scanning in transverse orientation was performed with centric phase-encoding order. CrCEST imaging was further accelerated using GRAPPA (GeneRalized Auto-calibrating Partial Parallel Acquisition (18)) with acceleration factors: along PE = 2, along 3D = 1, Reference lines = 24. A turbo-factor of 536 was needed to enable a single-shot readout of a matrix size 112×112×8.
WASSR imaging
WASSR images (16), corresponding to FOs from ±0 to ±0.9 ppm with a step-size of ±0.15 ppm and scan time of 80s, were acquired with a 2×100ms long saturation pulse train (100ms pulse duration, 99% duty cycle, B1rms of 0.29μT).
B1+ imaging
Data for generating a B1+ map were obtained using a flip crush pulse sequence (19) with three flip angles (20°, 40° and 80°), with the detailed implementation described in appendix A.3.
To suppress the fat signal, a chemical-shift-selective fat saturation pulse was applied immediately prior to image readout (Supplementary Information Fig. S1). Two additional dummy shots were performed for CEST, WASSR, B1+ map sequences to ensure Mz-value stay the same before every shot (appendix A.4).
Reference Voltage
A constant voltage of 320V was used for CEST, WASSR and B1+- acquisitions, rather than optimizing it on subject-wise basis. Since gastrocnemius muscle is the most utilized muscle groups, that choice of reference voltage ensured the relative B1+ of ~0.8–1.1 in those regions.
Data Processing
Data processing was performed using in-house Matlab (version R2019a) scripts, as described in (20). First, CEST asymmetry ratio was computed from CESTW images at frequency offset of (Δω = +1.8 ppm) using the following equation:
where MSat (± Δω) are the equilibrium magnetization achieved with train of saturation pulses applied at ± Δω relative to water peak.
B0 map generated using WASSR imaging (16) was used to perform B0-correction of CESTW images (appendix A.2). Afterwards the effect of B1+-inhomogeneity was corrected using calibration data (appendix B).
Voxel-wise B1+-Calibration
B1+-calibration curves were generated on the voxel-wise basis using the polynomial of 2nd degree and B0-corrected CrCEST values were corrected on voxel-wise basis (appendix B). In comparison, older works (13,15,21) on CrCEST employed a linear B1+-correction (Eqn. C.3. of appendix C).
Residuals of Fitting
The residual of fitting was calculated on multi voxel basis as: ; where ycalib and yfit are column vectors representing multi voxel calibration data and corresponding fits. The averaging, denoted by 〈…〉, was performed over the mask consisting of the entire calf muscle.
ΔCrCEST time-series
For the better visual assessment of time-series of CrCEST maps, we also calculated the difference between CrCEST maps at intermediate time points (t) and at last time point (tend): ΔCrCEST(t) = CrCEST(t)- CrCEST(tend).
Such subtraction suppresses the effect of baseline across all frames, making temporal trend more conspicuous.
Curve fitting for CrCEST
Muscle-group-specific (MGS) masks were constructed (appendix D) and post-exercise CrCEST time-series curves, averaged over MGS masks on slice-wise basis, were calculated. Expression for the mono-exponential decay was fitted to underlying post-exercise CrCEST time-series curve: , where A01 represents post-exercise baseline contribution and where τCr is the decay time constant for CrCEST contrast.
Statistical Analysis
We used a linear mixed model to regress τCr-measurements in selected muscle group on a random intercept for each participant. We then computed the intraclass correlation coefficient (ICC) to quantify reliability of measurements within participants.
SNR comparison between 3D vs. 2D sequence for M0 image
As CESTW image intensity depended on B0-, B1+-inhomogeneity, we calculated SNR for M0 image instead. M0 image, for our case, is equivalent to CESTWimage with saturation pulse applied at 100 ppm offset. At-rest 2D M0 images with 2 repetitions were acquired for different values of tfl-FA=5°, 10°, slice thickness =5, 10 mm and parallel acquisition = yes/no, whereas at-rest 3D M0 images were only acquired with tfl-FA = 5° and parallel acquisitions with other sequence parameters as described before.
For the purpose of measuring SNR, a selected healthy volunteer was scanned twice with the same sequence and the difference image was calculated. Finally, SNR was calculated as: √2x (mean signal/standard deviation of the difference image). The reported SNR corresponds to LG of the volunteer.
Effective SNR (eSNR) comparison of at-rest CrCEST maps for 2D and 3D acquisitions
Using at-rest CrCEST maps extracted using data acquired with two repetitions, the difference map was calculated. The eSNR of at-rest CrCEST was calculated as: .
At-rest CrCEST data with 2 repetitions were acquired for 2D acquisition with four different combinations of parameter values (tfl-FA=5°, 10°, slice-thickness = 5, 10 mm and parallel acquisition = yes/no) and for 3D acquisition with two different combinations of parameters values (tfl-FA = 5°, slice-thickness = 5, 10 mm and parallel acquisitions).
RESULTS
CrCEST-values, for single-shot acquisition, were found to be appreciably reduced compared to corresponding values with four-shot acquisition (CrCEST% difference = 0.64–0.87) (appendix A.6). The experimental and simulated results will be presented later on, to show that the choice of the acquisition parameters, used in our experiments, will have no appreciable impact on τCr-measurements.
Proposed B1+-calibration approach
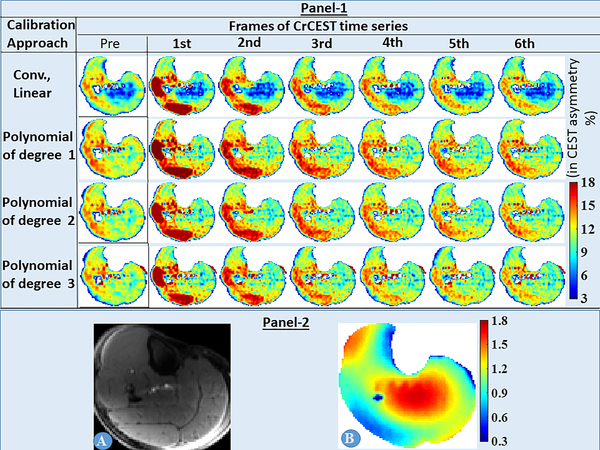
As evident from Fig. 1, the conventional linear B1+-calibration approach led to artificially suppressed CrCEST asymmetry values in the center of calf muscle, where relative B1+-values were higher (~1.3–1.8). However, this artifact was eliminated for the proposed calibration approach using polynomials of degree n (=1, 2, 3). The polynomial of degree 2, with highest drop in fitting residual for an additional degree of freedom (DOF) (Tab. 2), was chosen as the optimal candidate for B1+-calibration.
Fig. 1.
Panel-1: The effect of various B1+-calibration approaches on time-series of CrCEST map is shown for the middle slice (5th out of 8 slices) of volunteer #1. Panel-2: Reference image (A) and B1+ map (B) for the same slice are also shown.
The conventional linear B1+-calibration approach led to artificially suppressed CrCEST asymmetry values in the center portions of CrCEST maps, where relative B1+-values were higher (~1.3–1.8). However, for the proposed calibration approach using polynomials of degree n (=1,2,3), this artifact was eliminated.
Tab. 2:
Residuals of fit calculated for different candidates being utilized for the B1+-calibration of CrCEST maps, along with respective degree of freedom and the drop in residuals for each additional unit of DOF.
It should be noted that the polynomial of degree 2 results in highest drop in residual for an additional unit of DOF and hence, was chosen at the optimal candidate.
| B1 calibration model | Residual of fit (Res) | Degree of freedom (DOF) | Drop in “Res” for each “additional” unit of DOF |
|---|---|---|---|
| Old Calibration approach i.e. linear calibration with no offset | 0.1346 | 1 | x |
| Linear with offset | 0.1121 | 2 | 0.0225 |
| Polynomial of degree 2 | 0.0527 | 3 | 0.0594 |
| Polynomial of degree 3 | 0.0431 | 4 | 0.0096 |
Temporal trends are more conspicuous in Supplementary Information Fig. S3, panel 1, depicting the effect of different B1+-calibration approaches on ΔCrCEST time-series. Evidently, the conventional linear B1+-calibration approach led to higher post-exercise elevation and lower τCr values. Also, the temporal trend of ΔCrCEST time-series showed similar temporal trends of decay for all three candidates, i.e. polynomials of degrees 1, 2, 3.
Post-exercise CrCEST time-series from 3D acquisition
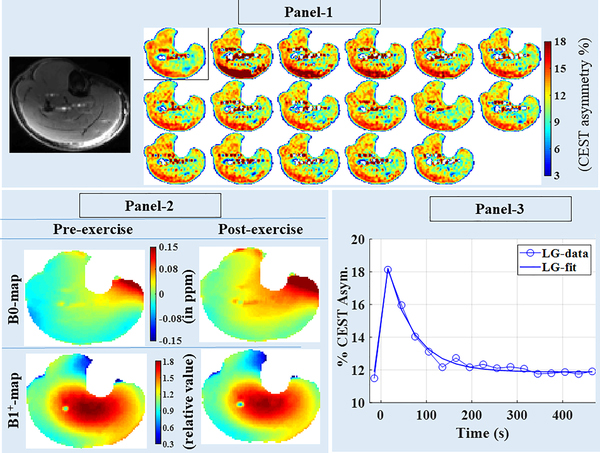
Fig. 2, panel-1 shows post-exercise CrCEST time-series for volunteer 2, corresponding to the middle slice (5th out of 8 slices). Only one out of four pre-exercise repetitions and selected time frames of post-exercise CrCEST time-series are being presented. Lateral gastrocnemius (LG) was the major contributor to the exercise load during PFE for this volunteer.
Fig. 2:
Panel-1: Time-series of CrEST maps showing exercise-induced changes in skeletal muscle, using images from 5th slice of volunteer #2. First CrCEST frame correspond to pre-exercise level, whereas the remaining sixteen frames correspond to sixteen time points post-exercise with 30s temporal resolution. Panel-2: B1+ maps and B0 maps calculated using data acquired immediately after pre-exercise CrCEST and post-exercise CrCEST are being presented. Whereas the B1+ map did not change appreciably post-exercise, B0 maps were appreciably different with most of changes attributed to post-exercise shift in leg position (also refer to Supplementary Information Fig. S2). Panel-3: Time-series plot of CrCEST averaged over the entire lateral gastrocnemius and corresponding fit is shown.
Unlike B1+ map, B0 maps were appreciably different between pre- and post-exercise sessions (Fig. 2, Panel-2). The central frequency drift in a 10-minute window was estimated to be ~0.02 ppm (~6Hz) (appendix A.5), which was not appreciable. Hence, post-exercise B0 map shift was found to be mostly due to shift in the leg position post-exercise and WASSR map acquired post-exercise was sufficient to correct for this B0 map shift.
Muscle-specific τCr measurements using 3D-CrCEST
LG was the common muscle group activated across all healthy volunteers. In addition, some volunteers also utilized medial gastrocnemius (MG) and peroneus longus (PL).
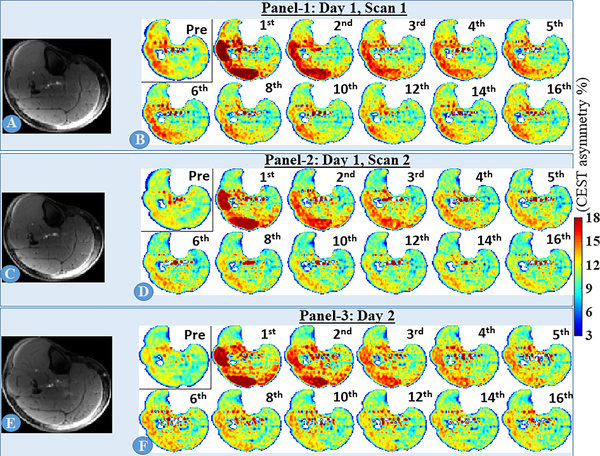
The time-series of CrCEST maps corresponding to the 5th slice of volunteer 1, after smoothing with 3×3×3 kernel, is shown in Fig. 3, 1st panel. Post-exercise, LG and PL muscles showed elevated level of CrCEST contrast, which returned to baseline after a few time frames. CrCEST time-series from two other sessions (inter- and intra-day) are also shown (Fig. 3, 2nd and 3rd panels). For a better visual assessment, corresponding ΔCrCEST time-series is shown in Supplementary Information Fig. S4. Inter- and intra-day scans showed very good correspondences even by visual inspection and corresponding coefficient of variances (COVs) of τCr were 10.3% and 5.8% for LG and PL, respectively (Tab. 3).
Fig. 3:
inter- and intra-day CrCEST time series for volunteer #1, corresponding to the middle slice (5th out of 8 slices). A, C, D are matching anatomical (reference) slices for three inter-/intra-day scans and B, D and F are respective time-series for CrCEST maps. Only one out of four pre-exercise repetitions and selected time frames of post-exercise CrCEST time series are presented, with time frame number written on right top corner of each frame. Both lateral gastrocnemius and peroneus longus were utilized during PFE.
Tab. 3:
Creatine recovery constants measured across various volunteers, including intra-day and inter-day scans. A minimum of 20 minutes separation was maintained between successive PFEs to ensure full creatine recovery.
The slice with the thickest portion of LG was chosen as the slice of interest. When comparing between two sessions, similar slices were manually chosen based on thickness of LG and appearance of fibula within those slices.
| Volunteer | Age (year), gender (m/f)* | Sessions | Creatine rate constants over multiple repetitions (in seconds) | Coefficient of variance (in %) | ||||
|---|---|---|---|---|---|---|---|---|
| LG | MG | PL | LG | MG | PL | |||
| #1 | 41, m | Day 1, session 1 | 70.57 | x | 58.57 | 10.27 | x | 5.80 |
| Day 1, session 2 | 59.18 | x | 54.76 | |||||
| Day 2, session 1 | 71.60 | x | 61.50 | |||||
| #2 | 41, m | Day 1, session 1 | 63.27 | x | x | 8.37 | x | x |
| Day 1, session 2 | 69.38 | x | x | |||||
| Day 2, session 1 | 58.74 | x | x | |||||
| #3 | 24, m | Day 1, session 1 | 55.75 | x | x | 15.70 | x | x |
| Day 1, session 2 | 69.67 | x | x | |||||
| # 4 | 55, f | Day 1, session 1 | 26.50 | 36.46 | x | 7.00 | 7.55 | x |
| Day 1, session 2 | 24 | 40.57 | x | |||||
| # 5 | 42,m | Day 1, session 1 | 61.24 | x | x | 11.07 | x | x |
| Day 1, session 2 | 52.35 | x | x | |||||
“m” for male; “f” for female
Tab. 3 summarizes the inter-muscular variability in 3D-CrCEST based τCr-measurements activated muscle groups (LG, MG, PL) across repeated scans from all healthy volunteers. Except for the volunteer #4, LG was the major contributor to PFE workload across all volunteers showing highest post-exercise CrCEST asymmetry elevation. For volunteer #4, MG was the major contributor to PFE workload and LG was a minor contributor. On the muscle group basis, COVs of τCr-measurements were 7–15.7%, 7.5%, 5.8%, for LG, MG and PL, respectively. Further, if we only consider the single largest contributor to PFE workload in our study, i.e. MG for volunteer #4 and LG for all other volunteers, the COVs were 7–15.7% (averaged COV = 10.5%) for τCr.
The linear mixed model had the resulting ICC of 0.87 for τCr of LG muscle group, a muscle group utilized by all volunteers during PFE. As an exploratory analysis, we also fitted a similar model using τCr values in the single largest contributor muscle group to PFE for each volunteer. The single largest contributor, defined as the muscle group with largest post-exercise CrCEST elevation, was MG for volunteer #4 and LG for all other volunteers. The resulting ICC of this model is 0.719. Both of these ICC values indicate a high level of repeatability in τCr-measurements.
Supplementary Information Fig. S5 shows time-series of ΔCrCEST maps from volunteer #5 showing exercise-induced changes in skeletal muscle, using images from 2nd to 7th slices. Considering that the volunteer is healthy, no substantial difference is expected along the slice-encoding direction. As our processing does not assume or exploit any spatial correlation across the slice, the gradual and consistent changes captured across slices underscores the ability of 3D-CrCEST to capture variations in post-exercise creatine along muscle length faithfully. Supplementary Information Tab. S1 shows the stability of CrCEST asymmetry values from LG, PL across slices for volunteer 1.
Tab. 4 presents the detailed comparison between 2D vs. 3D acquisitions with different sequence parameter combinations (tfl-FA=5°,10°; slice thickness = 5,10 mm; parallel acquisition or PAT = ‘Yes’, ‘No’), represented by respective sequence parameter combination codes (SPCCs), for both M0 images and at-rest CrCEST maps.
Tab. 4:
2D and 3D acquisitions of M0 images and at-rest CrCEST maps are compared for signal-to-noise ratio (SNR) and effective SNR (eSNR), respectively.
Please notice that CrCEST maps are derived values and hence, we have used the term effective SNR. We have also arbitrarily defined the so-called sequence parameter combination codes (SPCCs), to enable easy references to combinations of sequence parameters from the text in the result section.
| Acquisition types | Sequence Parameter combination codes (SPCCs) | Acquisition details | SNR/ eSNR | |
|---|---|---|---|---|
| M0-image | At-rest “reconstructed” CrCEST-map | |||
| 2D Acquisition; No parallel acquisition |
1 | Thickness = 10 mm; tfl-FA = 10° |
585 | 33.95 |
| 2 | Thickness = 5mm; tfl-FA = 10° |
315 | 18.79 | |
| 3 | Thickness = 5mm; tfl-FA = 5° |
288 | 16.55 | |
| 2D Acquisition; Parallel acquisition |
4 | Thickness = 5mm; tfl-FA = 5° |
228 | 11.70 |
| 3D Acquisition; Parallel acquisition |
5 | Thickness = 10mm; tfl-FA = 5° |
513 | 20.47 |
| 6 | Thickness = 5mm; tfl-FA = 5° |
280 | 10.95 | |
SNR comparison between 3D vs. 2D sequence for M0 image
Among 2D vs. 3D acquisitions of M0 images, the SNR was highest (≈585) for 2D acquisition with tfl-FA=10°, slice thickness = 10mm and no parallel acquisition (PAT) (SPCC:1 for Tab. 4). The highest SNR for 3D acquisition was ~510, achieved for tfl-FA=5°, slice thickness = 10mm and PAT = ‘yes’ (SPCC:5). The lower SNR for 3D acquisition can be mostly attributed to the reduced tfl-FA and the implementation of PAT. Reducing the thickness by half to 5mm reduced SNR by ~50% for either of the acquisitions (SPCC:1 vs. SPCC:2; SPCC:5 vs. SPCC:6). Even with 2D acquisitions, SNRs of M0 images were evidently reduced with the reduction in tfl-FA (SPCC:2 vs. SPCC:3) and with the implementation of PAT (SPCC:3 vs. SPCC:4).
eSNR comparison of at-rest CrCEST maps for 2D and 3D acquisitions
Similar to trend seen with M0 images, eSNR of at-rest CrCEST maps was highest (≈34) for 2D acquisition with TFL-FA=10°, slice thickness = 10 mm and no PAT (SPCC:1 of Tab. 4). In case of 2D acquisition, eSNR of at-rest CrCEST map decreased with both the reduction in TFL-FA (SPCC:2 vs. SPCC:3 of Tab. 4) and the implementation of PAT (SPCC:3 vs. SPCC:4 of Tab. 4), resulting in eSNR of 11.7 for 2D acquisition with TFL-FA=5° and parallel acquisition (PAT) (SPCC:4 of Tab. 4). However, eSNR was lowest for 3D acquisition with TFL-FA=5° and parallel acquisition (PAT), which could be attributed to drop in CrCEST asymmetry resulting from longer readout, lower TFL-FA and PAT = ‘Yes’ (Tab. 4). For thicker slice of 10 mm, the eSNR for at-rest 3D-CrCEST went up to 20.5 (SPCC:5 of Tab. 4).
Comparison of post-exercise CrCEST time-series for 2D and 3D acquisitions
The saturation modules were kept identical for 2D and 3D-CrCEST, including fat-saturation module. So, only differences on the acquisition side can affect the accuracy of τCr-measurement between 2D and 3D-CrCEST. Accordingly, simulations were also carried out to compare the impact of the implemented acquisition modules of 2D and 3D acquisitions on the accuracy of τCr-measurements (refer Appendix E). Simulations showed that our choices of acquisition parameters would alter the individual CrCEST maps; however, after subtracting the baseline results, those choices would have no appreciable impact on either the temporal trend of measured CrCEST or on τCr-measurements.
For a better visual comparison, time-series of ΔCrCEST maps were calculated for experimentally acquired 2D and 3D acquisitions. For a fair comparison, the thickness of 3D acquisition was increased to 10mm (identical to thickness of 2D acquisition) for this analysis. On the other hand, tfl-FA of 5° with parallel acquisition was used for 3D acquisition, whereas 2D acquisition involved tfl-FA of 10° without parallel acquisition. Supplementary Information Fig. S6 depicts ΔCrCEST time-series, extracted using both acquisitions, for two volunteers (#1, #2). For both volunteers, the temporal trends of ΔCrCEST time-series are visually similar between 2D and 3D acquisition, though the temporal trend for 2D ΔCrCEST time-series appears to be smoother and less noisy. The better SNR for 2D acquisition can be attributed to utilization of the shorter readout length, higher tfl-FA and the absence of PAT in case of 2D acquisition. Volunteer #1 utilized LG and PL during exercise in both acquisitions, while only LG was activated for volunteer #2. Corresponding τCr values were also consistent between both 2D and 3D acquisition (Tab. 5), with the muscle-group-specific COV of τCr between both acquisitions <13.6%.
Tab. 5:
Comparison of Cr recovery constants measured on volunteers 1 and 2, utilizing 2D-CrCEST and 3D-CrCEST in the same session. A minimum of 20 minutes separation was maintained between successive PFEs to ensure full creatine recovery.
| Volunteer | Creatine rate constant (in seconds) measured using 2D CrCEST | Creatine rate constant (in seconds) measured using 3D CrCEST | Coefficient of variance (in %) |
|---|---|---|---|
| # 1 | LG: 74.56 Per: 55.30 |
LG: 71.60 Per: 61.50 |
LG: 2.86 Per: 7.51 |
| # 2 | LG: 71.17 | LG: 58.74 | LG: 13.53 |
DISCUSSION
In this study, we have demonstrated for the first time the feasibility of the 3D-CrCEST method to measure CR kinetics within a large 3D volume of exercised calf muscle with a very good repeatability. Though healthy volunteers in this study mostly utilized LG, MG and PL, other muscle groups are known to be utilized by patient population with significantly reduced exercise capacity and sometime even healthy volunteers can utilize soleus. We have also observed a good agreement between Cr recovery rates measured using either 3D-CrCEST or 2D-CrCEST.
With the CrCEST experiment requiring six frequency offsets and TR of 5s needed to allow the sufficient magnetization recovery following the acquisition of CESTW images at particular frequency offset, the temporal resolution of 30 seconds was achieved. This temporal resolution was sufficient to follow CR kinetics even in healthy volunteers, where τCr-values are expected to be lower (~40–70s). In patients with compromised CR kinetics, the τCr-values would be even longer. So, we would expect better methodological accuracies and comparable or better repeatability in diseased population. Since T1 of musculoskeletal tissues are 23–55% higher than corresponding values at 3T (22) and that allows for longer readout, this 3D application is better suited for ultra-high field of 7T.
Multiple factors can affect the spatial distribution of CrCESTW images and hence, CrCEST maps as well. The amine group is also present on other species of CK reaction, such as PCr, ATP and ADP. However, they do not contribute to CrCEST contrast appreciably, as their exchange rates with water are orders of magnitude smaller than the exchange rate of amine group on Cr (13). Other proton exchanging species such as glycogen, lactate, etc. may also be contributing; however, their contribution is not expected to be impacted by PFE and hence, would not affect the post-exercise temporal trend of CrCEST data. The simulation (appendix E) also showed that the choice of acquisition parameters may alter the baseline CrCEST as well. As such, it’s debatable to what extent the baseline CrCEST captures the true creatine concentration at baseline. However, as shown in the simulation (appendix E), these factors would not have appreciable impacts on capturing the temporal trend, which is the central theme of our work.
3D-CrCEST provides a unique opportunity to assess the variability of CR kinetics profile across the muscle length on intramuscular and intermuscular basis, with the temporal resolution (30s) and total scan time identical to that of 2D-CrCEST. However, compared to 2D acquisition, a smaller tfl-FA, the implementation of parallel acquisition and longer readout needed for 3D acquisition resulted in the compromise on SNR. Experimental results showed that a longer readout time required for 3D-CrCEST acquisition led to decreased CrCEST contrast relative to the baseline, which is consistent with simulations for 3D acquisition vs. 2D acquisition (Appendix E, Supplementary Information Fig. S8.F). Given the result from the simulations showing the similarity of τCr-measurements between 2D and 3D-CrCEST, it was justified to enroll only two participants to show the similarity of τCr measurement between both types of acquisitions experimentally. Though 2D-CrCEST, with single slice coverage, is sufficient to assess the CR kinetics compromise in skeletal muscle disorders attributed to “systemic” mitochondrial energy deficiency, 3D-CrCEST would still provide a more comprehensive assessment of such systemic disorders particularly in those patients with significant or complete fatty replacement of specific muscle groups. Additionally, 3D-CrCEST is uniquely placed to investigate and study medically complex skeletal muscle disorders with varying disease distributions across the muscle (lengthwise, intramuscular basis), including muscle fibrosis resulting from muscular dystrophies, myopathies, peripheral artery disease (PAD) and severe injuries. Further, though it has been widely believed that some amount of exercise, such as strengthening exercise, flexibility training, aerobic exercise, would have beneficial effects on patients with musculoskeletal diseases, such as Duchenne and Becker dystrophies, Limb-girdle muscular dystrophy etc., there is an unmet need for an imaging biomarker to assess the effect of such exercise on CR kinetics. 3D-CrCEST may serve as a very valuable tool in such context.
Also, a drop in in vivo pH would lead to a significant drop in percentage of CrCEST asymmetry, as well as an appreciable reduction (approximately 20–30% drop from pH = 7.0 to pH = 6.5) in the forward reaction of creatine enzyme kinetics (23). So, it is crucial to optimize the exercise workload so as to avoid any acidification due to lactate generation. Additional work is needed to understand the effect of exercise-induced pH changes on the CrCEST-based CR kinetics in vivo and as such, the current method should not be used in patients with significant acidosis.
Though currently we are limited to coverage of six slices (discarding two slices at edges), larger coverage may be possible in future with further technological developments with respect to sequence development, parallel transmission and coil design. One can also increase the slice thickness to increase the coverage. For studies aiming at assessing CR kinetics profile along the muscle length on both inter-muscular and intra-muscular basis, 3D-CrCEST would also allow for the motion correction, if the calf position of the patient is shifted between different time points of post-exercise acquisition. A motion correction strategy exploiting low-rank approximation of Z-spectrum has recently been published by Wech and Koester (24). Though our 3D-CrCEST data does not have high resolution on Z-spectrum, an approach similar to theirs exploiting low rank-approximation in the tempo-spatial domain can be utilized as well.
The τCr-values reported here are similar to PCr recovery constants reported in few 31PMRS studies (25,26), though they are higher than values reported in other 31PMRS-based PCr recovery studies (27–31). Since these differences cannot be attributed to inter-subject variations alone, significant components of these variations can be attributed to factors, such as different exercise loads, partial-volume effect and the usage of ergometers of different designs. Most of PCr studies utilize sensitivity profile of radiofrequency (RF) coil for signal localization. Due to the partial-volume effect, the signal may come from exercising as well non-exercising muscle groups or combinations of two different exercised muscle groups with different recovery constants. Different ergometers may load differently, and different exercise loads can also lead to appreciable changes in recovery rate constant values. Even the leg positioning and knee angle can influence the distribution of the exercise load (32) and thus, can affect underlying CR kinetics. Since mostly gastrocnemius (GA), PL and soleus get utilized in PFE, only these muscle groups can be probed using CrCEST. With straight leg positioning, it’s mostly GA sometimes assisted by PL and soleus accounting for most of the exercise load. To probe the OXPHOS capacity of soleus, the PFE with bended knee experiment can be performed to make soleus the major contributor to the exercise.
We would also like to point out that the reported study is technical in nature, aimed at demonstrating the feasibility of 3D-CrCEST sequence with temporal resolution and total scan time identical to 2D-CrCEST. Given the small sample size, no valid clinical inference should be made based on this study. Although we expect the performance of 3D-CrCEST to be similar to that of 2D-CrCEST that were validated against the 31PMRS, we did not directly validate 3D-CrCEST with 31PMRS. Additional limitations include higher COV of τCr-values and temporal resolution of 30s being used. COVs of τCr-values, measured using 3D-CrCEST, were higher than values usually reported for non-dynamic MRI experiments. However, this is in line with other 31PMRS studies (33–35), where high (COV) (~20–30%) with respect to half-life of PCr recovery were reported and a large component of those variabilities were attributed to physiological variations (35). Further, if we only consider the single largest contributor to PFE workload in our study, i.e. MG for volunteer #4 and LG for all other volunteers, the COVs were 7–15.7% (averaged COV = 10.5%) for τCr-values. While non-dynamic MRI, such as glutamate weighted CEST (36), suffers from methodological variations only with origins in acquisition and processing, CrCEST and 31PMRS-based PCr recovery experiments of exercised muscle suffer from two additional sources of variabilities, namely biological/physiological variations and variabilities across various sessions and volunteers originating from PFE paradigm itself. Physiological variations are very difficult to control, and therefore, we did not attempt to manipulate this aspect of the experiment. Healthy volunteers enrolled in our study utilized GA muscle as the major contributor. GA is the lone bi-articular leg muscle crossing both the knee and ankle. It has been shown that the positioning of both joints affect plantar flexion torque of GA (37). The study by Niess et al. (32) further confirmed the influence of knee angle on the distribution of work-load, leading to the change in the metabolic profile measured using 31PMRS and recommended the PFE with straight leg with fixed knee to be the optimal paradigm. Our PFE paradigm included straight leg positioning as well, with position of coil fixing the knee in place. We kept the plantar flexion angle (PFA) of Φ = 40°, so that all participants can consistently push to maximum angle. Additionally, we modified the ergometer setup to restrict the ankle rotation “θ” so as to avoid inversion and eversion of plantar flexion surface while pushing the ergometer paddle. On the methodological side, the coarse temporal resolution of 30s may also have contributed to increased variability of quantification, though it was found to be sufficient to follow CR kinetics even in healthy volunteers with faster expected τCr-values. Nonetheless, since we expect τCr-values to be significantly increased in patients with compromised CR kinetics (2), our current temporal resolution would be sufficient for that purpose.
Further, the low temporal resolution combined with the lower SNR (<20) of time-series of 3D-CrCEST maps and the physiological variation on the local scale did not allow for the fitting of the recovery rate on voxel-wise basis, rather we did muscle-group-specific slice-wise fit. However, it is possible to consider multiple regions of interest (ROIs) within the same muscle group, which could be chosen even along the muscle length, and get the recovery constants reliably for respective ROIs. In future, we would be developing the acquisition strategies aimed at reducing the temporal resolution and would also be working on developing denoising methods to make voxel-wise fit feasible.
CONCLUSIONS
In summary, this work has demonstrated for the first time the feasibility of in vivo 3D-CrCEST imaging on ultra-high-field scanners (7T) to measure creatine recovery kinetics in healthy human calf muscle following PFE.
Supplementary Material
Supplementary Information Fig. S1. Pulse sequence diagram showing the implementation of CrCEST and WASSR sequences. The basic sequence structures are same for both CrCEST and WASSR, with only differences being range of offset frequency values being scanned, saturation powers being applied (2.9 μT for CrCEST vs. 0.29 μT for WASSR), and saturation duration (500 ms for CrCEST vs. 200 ms for WASSR).
The pulse sequence consisted of saturation pulse train followed by spoiler gradients. Following that, a chemical-shift-selective fat saturation pulse was applied to suppress the fat signal, followed by another set of spoiler gradients and then image readout using TurboFLASH readout was implemented.
Supplementary Information Fig. S2. To assess the effect of the field drift, series of WASSR maps from the calf muscle at-rest were acquired for 10 minutes 40 seconds (temporal resolution = 70s; number of repetitions = 9; 2 dummy shots worth 10s). ΔB0 map, calculated as the difference between 9th (last) and 1st WASSR maps, is shown here. B0-shift was mostly less than 0.02 ppm (~6Hz). The center of frequency drift measured at the end of the scan, measured at the scanner, was ~6Hz.
Supplementary Information Fig. S3: Panel-1: The effect of various B1+-calibration approaches on ΔCrCEST time-series is shown for the middle slice (5th out of 8 slices). Panel-2: Reference image (A) and B1+ map (B) for the same slice are also shown.
This figure contains information similar to Fig. 1, except ΔCrCEST time-series is shown here instead of CrCEST time-series as temporal trends are more conspicuous in former. Evidently, the conventional linear B1+-calibration approach led to higher post-exercise elevations and lower creatine decay constants. ΔCrCEST time-series showed similar temporal trends for all other three candidates, i.e. polynomials of degrees 1,2,3.
Supplementary Information Fig. S4: Inter- and intra-day ΔCrCEST time-series for volunteer #1, corresponding to the middle slice (5th out of 8 slices). A, C, D are matching anatomical (reference) slices for three inter-/intra-day scans and B, D and F are respective time-series for CrCEST maps. Only one out of four pre-exercise repetitions and selected time frames of post-exercise CrCEST time-series are being presented, with frame number written on right top corner of each frame. Both lateral gastrocnemius and peroneus longus were utilized during PFE.
This figure contains information similar to Fig. 3, except ΔCrCEST time-series is shown here instead of CrCEST time-series, as the temporal trends are more conspicuous in former.
Supplementary Information Fig. S5: Time-series of ΔCrEST maps showing exercise-induced changes in skeletal muscle, using images from 2nd −7th slices of volunteer #5. Only one out of four pre-exercise CrCEST maps and selected time frames (first six out of sixteen) of post-exercise CrCEST maps are shown.
The temporal decay trends for LG and PL were similar across all slices (#2–7). The extent of participation of MG to PFE exercise load, as judged by post-exercise CrCEST elevation, varied across slices with earlier slices showing more involvement of LG.
Supplementary Information Fig. S6: Time-series of ΔCrEST maps showing exercise-induced changes in skeletal muscle of two volunteers for both 2D and 3D acquisitions. Only one out of four pre-exercise CrCEST maps and selected time frames (first six out of sixteen) of post-exercise CrCEST maps are shown.
For a fair comparison, the thickness of 3D acquisition was increased to 10 mm (comparable to thickness of 2D comparison) for this analysis. On the other hand, parallel acquisition with tfl-FA of 5o was used for 3D acquisition, whereas 2D acquisition involved tfl-FA of 10o without parallel acquisition.
Supplementary Information Fig. S7: Typical muscle-group-specific masks chosen for two volunteers (#1, #4). Masks were constructed based on the reference image and CrCEST map/ ΔCrCEST map immediately post-exercise, as described in appendix D.
Supplementary Information Fig. S8: The evolution of transverse magnetization (Mxy) immediately after excitation pulses for 2D and 3D acquisitions are shown in (A) and (D). For longer readout duration, Mxy tends to reach to a steady state. The point spread function (PSF) computed along the phase encoding direction for both the 2D and 3D GRE sequences with centric phase encoding order are displayed in (B) and (E). The blurring effects of these PSFs, typically characterized by the broadness of the peak at half the maximum magnitude, are all less than 2 pixels.
The correlation between Mz(0) and PSF(0) for 2D and 3D acquisitions are plotted in (C) and (F). The linear correlations fit well for both type of acquisition. The 3D acquisition with longer readout reveal smaller slope, indicating declined resolving power due to longer readout.
Supplementary Information Tab. S1: CrCEST asymmetry values for the activated muscle groups of volunteer #1, namely LG and PL, at rest and at the 1st time point of post-exercise acquired during day-1, session-1. LG and PL were the active muscle groups for this volunteer. The reported standard deviation corresponds to voxel-wise CrCEST distribution across a particular muscle group located within the slice.
ACKNOWLEGEMENTS
“Research reported in this publication was supported by the National Institute of Biomedical imaging and Bioengineering of the National Institutes of Health under Award Number P41EB015893 and National Institute On Aging of the National Institutes of Health under Award Number R56AG062665.”
APPENDICES
Appendix A: Justification for various sequence related choices
A.1. Justification for the range of B0-sampling
In case of the 2D implementation, a smaller range of B0-inhomogeneity across the slice was expected (|ΔB0| <0.1 ppm) and accordingly, the offset frequency step size of ±0.3 ppm (>2x|ΔB0|) was chosen (13). A larger FO range was needed to allow for larger B0-inhomogeneity range expected (|ΔB0| <0.15–0.2 ppm) in case of 3D acquisition. Some extra room (>2x|ΔB0|) was allowed to make the protocol more robust for cases where B0-inhomogeneity can be larger, such as in case of suboptimal shimming or shimming over the calf muscle of many patients with metallic transplants.
A.2. Interpolation of CrCEST data needed for B0-correction
We restricted CrCEST acquisition to only three frequency-offset pairs to achieve the temporal resolution sufficient for capturing underlying recovery kinetics. Nonetheless, given the flatness of CrCEST asymmetry curve around 1.8 ppm (Fig. 1d of (15)), this limited sampling of the asymmetry curve was found to be sufficient for the B0-interpolation. To perform the interpolation needed for B0-correction, we fitted the polynomial of 2nd order to raw CESTW images acquired at previously mentioned FOs.
A.3. B1+-Imaging
Data for generating a B1+ map were obtained using a flip crush pulse sequence (19) with three flip angles (20°, 40° and 80°). All other imaging parameters were identical to CEST imaging and it took approximately 25s to acquire a B1+ imaging data set including two dummy shots preceding the sequence. Data corresponding to 40° and 80° were used to generate B1+ map for imaging volume with lower B1+ (relative B1+ ≤1.0); whereas 20° and 40° data were used for imaging volume with higher B1+ (relative B1+>1.0).
A.4. T1 recovery and Dummy Shots
The CrCEST sequence consisted of a saturation module, followed by an acquisition module and a T1 recovery delay. With T1 of muscle being approximately 1500ms at 7T (22), shot TR = 5s ( 3.3xT1) was insufficient for the full recovery of magnetization before every shot. By adding two dummy shots and with proper choice of the minimum time delay dictated by the Bloch simulation, Mz-value was ensured to stay the same before every shot. TR of 5s was found to be sufficient for this purpose. However, since it’s vital not to unnecessary waste too much time post-exercise, we synchronized the end portion of the exercise with the start of post-exercise CrCEST acquisition as follows: About ~115s into the exercise, we started with CrCEST acquisition preceded by two dummy shot. To avoid any motion artifacts contaminating the first frame of post-exercise CrCEST, participants were asked to stop the exercise at the start of 2nd dummy shots and that allowed 4–5s delay before the start of data acquisition.
A.5. Assessing the center frequency and B0 map drifts during ~10.5 minutes acquisition window
To assess the effect of the field drift, series of WASSR maps from the at-rest calf muscle were acquired for 10 minutes 40 seconds (temporal resolution = 70s; number of repetitions = 9; 2 dummy shots worth 10s). ΔB0 map, calculated as the difference between 9th WASSR map and first WASSR map, is shown in Supplementary Information Fig. S2. B0-shift was mostly less than 0.02 ppm (~6Hz). On the scanner measurement of the center of frequency drift at the end of the scan was also ~6Hz. Multiple such scans on three different days generated similar results.
A.6. Effect of readout lengths on CrCEST
With T1 of muscle ≈ 1500ms (22), the readout time of 1822ms (≈turbo-factor of 536×3.4ms) following magnetization preparation corresponds to T1 recovery of 56%. Region of interest (ROI)-averaged at-rest CrCEST asymmetry values in LG, MG, soleus and PL muscles were compared for different number of shots and resultant different readout times (Tab. 1). CrCEST-values for single shot acquisition were found to be appreciably reduced compared to corresponding values with four-shot acquisition (CrCEST% difference = 0.64–0.87). However, the decrease in CrCEST values due to the usage of single-shot acquisition would affect successive time points of post-exercise decay curves in a similar manner and would not affect the creatine decay constants appreciably.
Tab. 1:
Pre-exercise CEST asymmetry values in four major muscle groups, namely, LG, MG, soleus, PL, of a single volunteer, for varying number (1, 2, 4) of shots. Reported values correspond to CrCEST value averaged over ROI (CrCESTROI) consisting of respective muscle groups, selected on the middle (4th out of 8) slice of the imaging volume. Mean and standard deviation correspond to CrCESTROI calculated over three repeated acquisitions.
| # Shots | CEST Asymmetry % | |||
|---|---|---|---|---|
| LG | MG | Soleus | PL | |
| 4 | 13.06±0.19 | 10.84±0.26 | 12.29±0.19 | 12.12±0.21 |
| 2 | 12.72±0.27 | 10.63±0.29 | 12.06±0.21 | 11.95±0.16 |
| 1 | 12.19±0.22 | 10.11±0.28 | 11.61±0.24 | 11.48±0.17 |
Appendix B: Voxel-wise B1+-calibration
To generate the CEST B1+-calibration curves, CEST data were acquired at eight different values of = 1.45, 2.03, 2.25, 2.47, 2.68, 2.9, 3.19, 3.40 micro-tesla (μT). CEST contrast vs. B1,rms curves were generated from the calibration data on the voxel-wise basis using the polynomial of nth degree (n=1,2,3):
| (B.1.) |
where is the root mean square of calculated B1+-value. The nominal value of B1,rms (i.e. ) is 2.9 μT.
Optimal choice for the calibration
We evaluated the fit to calibration data for n = 1,2,3 and corresponding residual values were calculated. Polynomial of higher degree is guaranteed to lower the residual of fitting; hence, the metric for choosing the appropriate model was the drop in residual values for each additional DOF. Based on this metric, polynomial of degree 2 was the optimal choice.
To suppress the effect of low SNR (~10–30) on calibration, the fitting was performed using a sliding window of size 3×3×3 centered on the voxels of interest.
B1+-correction using the B1+-calibration curve
The regular CESTW image, acquired at , is equivalent to
| (B.2.) |
The B0, B1 corrected CEST asymmetry value, equivalent to CESTW acquired at , can be given by:
| (B.3.) |
Subtracting (B.2.) from (B.3) yields:
| (B.4.) |
Hence, the measured CEST value at any voxel, with underlying B1 value (B1rel) can be corrected using:
| (B.5) |
Where, .
Appendix C: Linear voxel-wise B1+-calibration
In the linear B1+-calibration model, CEST contrast vs. B1rms curves were generated from the calibration data on the voxel-wise basis using the following equation (equivalent to the polynomial of degree n=1s):
| (C.1.) |
where .
The B0,B1+-corrected CEST asymmetry value can be given by:
| (C.2.) |
The old B1+-calibration approach, as described in the method section, is equivalent to the linear calibration model (or polynomial of degree 1) with p0 = 0.
Setting p0 = 0 and taking the ratio of (C.1.) and (C.2.)
| (C.3.) |
The old calibration approach, with only one DOF and no offset, only needs B1,rel (or B1+ map) and no additional calibration data is needed.
Appendix D. Mask selection for activated muscle groups
In our experiment, one or more muscle groups from among LG, MG and PL were the major contributor to PFE. So, our analysis focused on the major contributing muscle group/s from these three groups only. Masks were constructed based on the reference image and CrCEST map/ΔCrCEST map immediately post-exercise.
LG
For LG muscle, the mask consisted of the entire muscle falling within that slice. Masks depicted in the 1st and 2nd row of the Supplementary Information Fig. S7 shows the LG mask chosen for volunteers #1, #4.
PL
When constructing mask for PL, it was crucial to avoid voxels with ghosting artifacts originating from pulsation of arteries as that would contaminate the analysis. Mask depicted in the 1st row of the Supplementary Information Fig. S7 shows the PL mask chosen for volunteer #1.
MG
Only a part of MG showed appreciable participation with average post-exercise elevation in CrCEST asymmetry of approximately 5% or larger, and only those parts were used to construct corresponding ROIs. Entire width of the muscle group was used along the radial direction. Mask depicted in the 2nd row of the Supplementary Information Fig. S7 shows MG mask chosen for volunteer #4.
Appendix E. Effect of 2D vs. 3D acquisitions on the Creatine rate constant measured via CrCEST
The saturation parameters were kept identical for 2D and 3D-CrCEST acquisitions, including fat-saturation module. So, only differences on the acquisition side between 2D and 3D-CrCEST can affect the accuracy of reconstruction. Accordingly, we investigated the effect of alteration in point spread function (PSF) of the implemented acquisition modules on the accuracy of creatine rate constants measurement.
The signal at any spatial and temporal coordinates can be given by:
| (E.1.) |
where is the true spatial distribution and PSF is the point spread function of the implemented acquisition module. The symbol “⊗” indicates convolution operation. With GRE sequences, PSF depends on the initial magnetization, which is time dependent in our current case, . So we are including time dependence for .
We performed simulations to learn the effect of PSF on image contrasts and hence, on the creatine time constants measurements. Simulated results (presented at the end of this appendix E) showed that blurring effect was negligible, with less than 2-voxel wide (Eqn. (E.2.)), and that left amplitude-loss effect (ALE) as the only possible contributor to differences in reconstructions.
| (E.2.) |
In Eqn. (E.2), we have substituted , where is a delta function.
In the absence of significant blurring, ALE was considered as a local scaling (Eqn. (E.3), below) function on the true spatial distribution of magnetization (=aM+b, where a and b are two constants governed by both the parameters (θ, TR, N) of the GRE sequence and local T1 and T2*), then
| (E.3.) |
Using the following functional form for , the signal can be written as
Using the notation for the baseline contribution, previous expression can be written as,
For 3D acquisition:
| (E.4.) |
For 2D acquisition:
| (E.5.) |
Therefore, after the baseline is subtracted, the temporal trend of image intensity of the GRE sequence, as primarily decided by its PSF(0,t) when the blurring effect is little, is linearly correlated with the temporal trend of [Cr(r,t)]. And this relation is not affected by the 2D or 3D acquisition.
Simulated Result Showing effect of PSF on the Reconstruction
For a spoiled-gradient-echo (GRE) sequence, Mz(i) is the longitudinal magnetization before the i+1th RF excitation pulse, which follows the Bloch equation during the TR interval after the ith RF pulse:
| (E.6) |
where M0 is the equilibrium magnetization, θ is the flip angle of each RF pulse, and T1 is the longitudinal relaxation time.
Taking six evenly-sampled initial longitudinal magnetizations before the first RF excitation pulse, Mz(0) = [1, 0.8, 0.6, 0.4, 0.2, 0], their respective longitudinal magnetizations during the successive RF pulse trains were calculated for both 2D and 3D GRE sequences applied in this work (2D: TFL-TR = 3.1 ms, θ = 10°, turbo-factor = 120; 3D: TFL-TR = 3.5 ms, θ = 5°, turbo-factor = 536). As shown in Figure S8 (2D: A; 3D: D), these transient states vary depending on the initial Mz(0), but all approach to the steady state determined only by the sequence parameters (θ, TR).
Note that the transverse magnetizations during the echo train are only proportional to Mz as: Mxy(i)=Mz(i)sin(θ)exp(−TE/T2*). Using the simulated Mz as the modulation transfer functions (MTF) in k-space with centric phase-encoding order, the corresponding point spread function, as the Fourier transform pair in the spatial domain along the phase-encoding direction, were computed for both the 2D and 3D GRE sequences, and displayed in Supplementary Information Fig. S8 (2D: acquisition B; 3D acquisition: E). The blurring effects of these PSFs, typically characterized by the broadness of the peak at half the maximum magnitude, are all less than 2 pixels. The amplitude-loss effect can be described by the peak magnitude of each PSF in the spatial domain, PSF(0), which is equal to the area of the MTF in k-space. Note that the PSFs here were calculated based on Cartesian sampling for both 2D and 3D k-space trajectories. Although the 3D GRE acquisition in this study adopted a spiral acquisition, its PSF would be only slightly different from its Cartesian counterpart in terms of both blurring and amplitude-loss effects (38).
The correlation between Mz(0) and PSF(0) is plotted in Supplementary Information Fig. S8 (2D acquisition: C; 3D acquisition: F) with a linear function as Mz(0) = a PSF (0) + b, where a and b are two constants governed by the parameters (θ, TR, N) of 2D and 3D sequences, respectively.
REFERENCES
- 1.Fitch CD. Significance of abnormalities of creatine metabolism Rowland P, editor. Amsterdam: Exerpta Medica; 1977. [Google Scholar]
- 2.DeBrosse C, Nanga RPR, Wilson N, D’Aquilla K, Elliott M, Hariharan H, Yan F, Wade K, Nguyen S, Worsley D, Parris-Skeete C, McCormick E, Xiao R, Cunningham ZZ, Fishbein L, Nathanson KL, Lynch DR, Stallings VA, Yudkoff M, Falk MJ, Reddy R, McCormack SE. Muscle oxidative phosphorylation quantitation using creatine chemical exchange saturation transfer (CrCEST) MRI in mitochondrial disorders. JCI Insight 2016; 1(18):e88207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popovich BK, Boheler KR, Dillmann WH. Diabetes decreases creatine kinase enzyme activity and mRNA level in the rat heart. Am J Physiol 1989; 257(4 Pt 1):E573–577. [DOI] [PubMed] [Google Scholar]
- 4.David S, Shoemaker M, Haley BE. Abnormal properties of creatine kinase in Alzheimer’s disease brain: correlation of reduced enzyme activity and active site photolabeling with aberrant cytosol-membrane partitioning. Brain Res Mol Brain Res 1998; 54(2):276–287. [DOI] [PubMed] [Google Scholar]
- 5.Takubo H, Shimoda-Matsubayashi S, Mizuno Y. Serum creatine kinase is elevated in patients with Parkinsons disease: a case controlled. Parkinsonism Relat D 2003; 9:S43–S46. [DOI] [PubMed] [Google Scholar]
- 6.Ooi DS, Isotalo PA, Veinot JP. Correlation of antemortem serum creatine kinase, creatine kinase-MB, troponin I, and troponin T with cardiac pathology. Clin Chem 2000; 46(3):338–344. [PubMed] [Google Scholar]
- 7.Armstrong RB, Warren GL, Warren JA. Mechanisms of exercise-induced muscle fibre injury. Sports Med 1991; 12(3):184–207. [DOI] [PubMed] [Google Scholar]
- 8.Zatz M, Rapaport D, Vainzof M, Passosbueno MR, Bortolini ER, Pavanello RDM, Peres CA. Serum Creatine-Kinase (Ck) and Pyruvate-Kinase (Pk) Activities in Duchenne (Dmd) as Compared with Becker (Bmd) Muscular-Dystrophy. Journal of the Neurological Sciences 1991; 102(2):190–196. [DOI] [PubMed] [Google Scholar]
- 9.Ozawa E, Hagiwara Y, Yoshida M. Creatine kinase, cell membrane and Duchenne muscular dystrophy. Mol Cell Biochem 1999; 190(1–2):143–151. [PubMed] [Google Scholar]
- 10.Aston JP, Kingston HM, Ramasamy I, Walters EG, Stansbie D. Plasma pyruvate kinase and creatine kinase activity in Becker muscular dystrophy. J Neurol Sci 1984; 65(3):307–314. [DOI] [PubMed] [Google Scholar]
- 11.Mastaglia FL, Laing NG. Distal myopathies: clinical and molecular diagnosis and classification. J Neurol Neurosurg Psychiatry 1999; 67(6):703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boesch C Musculoskeletal spectroscopy. J Magn Reson Imaging 2007; 25(2):321–338. [DOI] [PubMed] [Google Scholar]
- 13.Kogan F, Haris M, Singh A, Cai K, Debrosse C, Nanga RP, Hariharan H, Reddy R. Method for high-resolution imaging of creatine in vivo using chemical exchange saturation transfer. Magn Reson Med 2014; 71(1):164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haris M, Nanga RP, Singh A, Cai K, Kogan F, Hariharan H, Reddy R. Exchange rates of creatine kinase metabolites: feasibility of imaging creatine by chemical exchange saturation transfer MRI. NMR Biomed 2012; 25(11):1305–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kogan F, Haris M, Debrosse C, Singh A, Nanga RP, Cai K, Hariharan H, Reddy R. In vivo chemical exchange saturation transfer imaging of creatine (CrCEST) in skeletal muscle at 3T. J Magn Reson Imaging 2014; 40(3):596–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PC. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med 2009; 61(6):1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Cai K, Haris M, Hariharan H, Reddy R. On B1 inhomogeneity correction of in vivo human brain glutamate chemical exchange saturation transfer contrast at 7T. Magn Reson Med 2013; 69(3):818–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002; 47(6):1202–1210. [DOI] [PubMed] [Google Scholar]
- 19.Stollberger R, Wach P. Imaging of the active B1 field in vivo. Magn Reson Med 1996; 35(2):246–251. [DOI] [PubMed] [Google Scholar]
- 20.Cai K, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nat Med 2012; 18(2):302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haris M, Singh A, Cai K, Kogan F, McGarvey J, Debrosse C, Zsido GA, Witschey WR, Koomalsingh K, Pilla JJ, Chirinos JA, Ferrari VA, Gorman JH, Hariharan H, Gorman RC, Reddy R. A technique for in vivo mapping of myocardial creatine kinase metabolism. Nat Med 2014; 20(2):209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan CD, Saranathan M, Bangerter NK, Hargreaves BA, Gold GE. Musculoskeletal MRI at 3.0 T and 7.0 T: a comparison of relaxation times and image contrast. Eur J Radiol 2013; 82(5):734–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goudemant JF, Vanderelst L, Dupont B, Vanhaverbeke Y, Muller RN. Ph and Temperature Effects on Kinetics of Creatine-Kinase in Aqueous-Solution and in Isovolumic Perfused Heart - a P-31 Nuclear Magnetization-Transfer Study. Nmr in Biomedicine 1994; 7(3):101–110. [DOI] [PubMed] [Google Scholar]
- 24.Wech T, Kostler H. Robust motion correction in CEST imaging exploiting low-rank approximation of the z-spectrum. Magn Reson Med 2018; 80(5):1979–1988. [DOI] [PubMed] [Google Scholar]
- 25.Khushu S, Rana P, Sekhri T, Sripathy G, Tripathi RP. Bio-energetic impairment in human calf muscle in thyroid disorders: a 31P MRS study. Magn Reson Imaging 2010; 28(5):683–689. [DOI] [PubMed] [Google Scholar]
- 26.Meyerspeer M, Robinson S, Nabuurs CI, Scheenen T, Schoisengeier A, Unger E, Kemp GJ, Moser E. Comparing localized and nonlocalized dynamic 31P magnetic resonance spectroscopy in exercising muscle at 7 T. Magn Reson Med 2012; 68(6):1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Layec G, Haseler LJ, Hoff J, Hart CR, Liu X, Le Fur Y, Jeong EK, Richardson RS. Short-term training alters the control of mitochondrial respiration rate before maximal oxidative ATP synthesis. Acta Physiol 2013; 208(4):376–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mancini DM, Walter G, Reichek N, Lenkinski R, Mccully KK, Mullen JL, Wilson JR. Contribution of Skeletal-Muscle Atrophy to Exercise Intolerance and Altered Muscle Metabolism in Heart-Failure. Circulation 1992; 85(4):1364–1373. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida T, Abe D, Fukuoka Y. Phosphocreatine resynthesis during recovery in different muscles of the exercising leg by P-31-MRS. Scand J Med Sci Spor 2013; 23(5):E313–E319. [DOI] [PubMed] [Google Scholar]
- 30.Larson-Meyer DE, Newcomer BR, Hunter GR, Joanisse DR, Weinsier RL, Bamman MM. Relation between in vivo and in vitro measurements of skeletal muscle oxidative metabolism. Muscle Nerve 2001; 24(12):1665–1676. [DOI] [PubMed] [Google Scholar]
- 31.Valkovic L, Chmelik M, Just Kukurova I, Jakubova M, Kipfelsberger MC, Krumpolec P, Tusek Jelenc M, Bogner W, Meyerspeer M, Ukropec J, Frollo I, Ukropcova B, Trattnig S, Krssak M. Depth-resolved surface coil MRS (DRESS)-localized dynamic (31) P-MRS of the exercising human gastrocnemius muscle at 7 T. NMR Biomed 2014; 27(11):1346–1352. [DOI] [PubMed] [Google Scholar]
- 32.Niess F, Fiedler GB, Schmid AI, Laistler E, Frass-Kriegl R, Wolzt M, Moser E, Meyerspeer M. Dynamic multivoxel-localized P-31 MRS during plantar flexion exercise with variable knee angle. Nmr in Biomedicine 2018; 31(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Layec G, Bringard A, Le Fur Y, Vilmen C, Micallef JP, Perrey S, Cozzone PJ, Bendahan D. Reproducibility assessment of metabolic variables characterizing muscle energetics in vivo: A 31P-MRS study. Magn Reson Med 2009; 62(4):840–854. [DOI] [PubMed] [Google Scholar]
- 34.McCully KK, Turner TN, Langley J, Zhao Q. The reproducibility of measurements of intramuscular magnesium concentrations and muscle oxidative capacity using 31P MRS. Dyn Med 2009; 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards LM, Tyler DJ, Kemp GJ, Dwyer RM, Johnson A, Holloway CJ, Nevill AM, Clarke K. The reproducibility of 31-phosphorus MRS measures of muscle energetics at 3 Tesla in trained men. PLoS One 2012; 7(6):e37237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanga RPR, DeBrosse C, Kumar D, Roalf D, McGeehan B, D’Aquilla K, Borthakur A, Hariharan H, Reddy D, Elliott M, Detre JA, Epperson CN, Reddy R. Reproducibility of 2D GluCEST in healthy human volunteers at 7 T. Magn Reson Med 2018; 80(5):2033–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landin D, Thompson M, Reid M. Knee and Ankle Joint Angles Influence the Plantarflexion Torque of the Gastrocnemius. J Clin Med Res 2015; 7(8):602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin Q Point spread functions of the T-2 decay in k-space trajectories with long echo train. Magnetic Resonance Imaging 2012; 30(8):1134–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information Fig. S1. Pulse sequence diagram showing the implementation of CrCEST and WASSR sequences. The basic sequence structures are same for both CrCEST and WASSR, with only differences being range of offset frequency values being scanned, saturation powers being applied (2.9 μT for CrCEST vs. 0.29 μT for WASSR), and saturation duration (500 ms for CrCEST vs. 200 ms for WASSR).
The pulse sequence consisted of saturation pulse train followed by spoiler gradients. Following that, a chemical-shift-selective fat saturation pulse was applied to suppress the fat signal, followed by another set of spoiler gradients and then image readout using TurboFLASH readout was implemented.
Supplementary Information Fig. S2. To assess the effect of the field drift, series of WASSR maps from the calf muscle at-rest were acquired for 10 minutes 40 seconds (temporal resolution = 70s; number of repetitions = 9; 2 dummy shots worth 10s). ΔB0 map, calculated as the difference between 9th (last) and 1st WASSR maps, is shown here. B0-shift was mostly less than 0.02 ppm (~6Hz). The center of frequency drift measured at the end of the scan, measured at the scanner, was ~6Hz.
Supplementary Information Fig. S3: Panel-1: The effect of various B1+-calibration approaches on ΔCrCEST time-series is shown for the middle slice (5th out of 8 slices). Panel-2: Reference image (A) and B1+ map (B) for the same slice are also shown.
This figure contains information similar to Fig. 1, except ΔCrCEST time-series is shown here instead of CrCEST time-series as temporal trends are more conspicuous in former. Evidently, the conventional linear B1+-calibration approach led to higher post-exercise elevations and lower creatine decay constants. ΔCrCEST time-series showed similar temporal trends for all other three candidates, i.e. polynomials of degrees 1,2,3.
Supplementary Information Fig. S4: Inter- and intra-day ΔCrCEST time-series for volunteer #1, corresponding to the middle slice (5th out of 8 slices). A, C, D are matching anatomical (reference) slices for three inter-/intra-day scans and B, D and F are respective time-series for CrCEST maps. Only one out of four pre-exercise repetitions and selected time frames of post-exercise CrCEST time-series are being presented, with frame number written on right top corner of each frame. Both lateral gastrocnemius and peroneus longus were utilized during PFE.
This figure contains information similar to Fig. 3, except ΔCrCEST time-series is shown here instead of CrCEST time-series, as the temporal trends are more conspicuous in former.
Supplementary Information Fig. S5: Time-series of ΔCrEST maps showing exercise-induced changes in skeletal muscle, using images from 2nd −7th slices of volunteer #5. Only one out of four pre-exercise CrCEST maps and selected time frames (first six out of sixteen) of post-exercise CrCEST maps are shown.
The temporal decay trends for LG and PL were similar across all slices (#2–7). The extent of participation of MG to PFE exercise load, as judged by post-exercise CrCEST elevation, varied across slices with earlier slices showing more involvement of LG.
Supplementary Information Fig. S6: Time-series of ΔCrEST maps showing exercise-induced changes in skeletal muscle of two volunteers for both 2D and 3D acquisitions. Only one out of four pre-exercise CrCEST maps and selected time frames (first six out of sixteen) of post-exercise CrCEST maps are shown.
For a fair comparison, the thickness of 3D acquisition was increased to 10 mm (comparable to thickness of 2D comparison) for this analysis. On the other hand, parallel acquisition with tfl-FA of 5o was used for 3D acquisition, whereas 2D acquisition involved tfl-FA of 10o without parallel acquisition.
Supplementary Information Fig. S7: Typical muscle-group-specific masks chosen for two volunteers (#1, #4). Masks were constructed based on the reference image and CrCEST map/ ΔCrCEST map immediately post-exercise, as described in appendix D.
Supplementary Information Fig. S8: The evolution of transverse magnetization (Mxy) immediately after excitation pulses for 2D and 3D acquisitions are shown in (A) and (D). For longer readout duration, Mxy tends to reach to a steady state. The point spread function (PSF) computed along the phase encoding direction for both the 2D and 3D GRE sequences with centric phase encoding order are displayed in (B) and (E). The blurring effects of these PSFs, typically characterized by the broadness of the peak at half the maximum magnitude, are all less than 2 pixels.
The correlation between Mz(0) and PSF(0) for 2D and 3D acquisitions are plotted in (C) and (F). The linear correlations fit well for both type of acquisition. The 3D acquisition with longer readout reveal smaller slope, indicating declined resolving power due to longer readout.
Supplementary Information Tab. S1: CrCEST asymmetry values for the activated muscle groups of volunteer #1, namely LG and PL, at rest and at the 1st time point of post-exercise acquired during day-1, session-1. LG and PL were the active muscle groups for this volunteer. The reported standard deviation corresponds to voxel-wise CrCEST distribution across a particular muscle group located within the slice.