Abstract
Pancreatic cancer (PC) is one of the most malignant gastrointestinal tumors and the 5‐year survival is only 9%. The expression of miRNAs in serum has been proved to be related to tumorigenesis and development of cancers. The miRNA targets and gene targets were predicted in microRNA.org, miRDB, TargetScan, and RNAInter. The expression data of STK31 (Serine/Threonine Kinase 31) and miRNAs generated from PC samples was from TCGA and the relationship of expression of STK31 and miR‐543 was confirmed in PC samples from our center. Double luciferase reporter gene assay was used to demonstrate the direct binding between miR‐543 and STK31. The effect of expression level of miRNAs on survival time was assessed by Kaplan–Meier curves. The Go Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of miR‐543‐related genes were performed. The results showed that miR‐543 had a statistically significant correlation with the expression of STK31 and contained the direct binding site with STK31. The expression level of miR‐543 may affect the survival of PC. The results of GO and KEGG pathway analysis showed that miR‐543 might play a key role in Insulin signaling pathway. MiR‐543 could be combined with STK31 and affect the expression of STK31. The expression of miR‐543 could also predict the survival of patients with PC, which suggested that miR‐543 might play an important role in PC. The GO and KEGG pathway analysis also displayed that miR‐543 was involved in several other pathways of pancreas.
Keywords: miR‐543, Pancreatic cancer, STK31, Survival
miR‐543 has the sequence of STK31 binding site and its expression was negatively correlated with STK31 expression in PC. miR‐543 was also correlated with 1548 genes expression and the enrichment study was found that they enriched in Insulin signaling pathway, ErbB signaling pathwayn and T cell receptor signaling pathway. Our results indicate that miR‐543 may play an important role and is a promising therapeutic target in PC.

1. INTRODUCTION
Pancreatic cancer (PC) is one of the most malignant gastrointestinal tumors worldwide, with more than 56,770 estimated newly diagnosed cases per year in United States in 2019. 1 As the most frequent cause of cancer deaths, PC is an extremely lethal disease with 45,750 cancer deaths annually. For all stages combined, PC has the worst prognosis with the lowest 5‐year survival (9%). 1 Although it is well known that the serum carbohydrate antigen (CA) 19‐9, CA125, and carcinoembryonic antigen (CEA) are the most commonly molecular markers used to assist diagnosis and predict prognosis, the accuracy of the prediction is not ideal and our understanding of the mechanism is limited. 2 , 3 , 4 , 5 , 6 Therefore, there is a great need to identify novel biomarkers to improve the prediction of prognosis of PC and explain its possible molecular mechanism.
MicroRNAs (miRNAs) are a family of small noncoding RNAs (21–23 nucleotides), which play important roles in regulating gene expression by binding imperfectly to the 3´‐untranslated region of target mRNAs. 7 , 8 After the first report about the role of miR‐15 and miR‐16 in chronic lymphocytic leukemia (CLL) in 2002, 9 lots of studies have drawn the conclusion that the expression of miRNAs in plasma/serum is associated with many cancers, including PC. 9 , 10 , 11 , 12 Therefore, miRNAs may be potentially stable noninvasive biomarkers in PC. However, these miRNAs, which could definitely affect some gene expression in tissues, seem more important to the prediction of diagnosis and prognosis in cancers. Previously, study has suggested that STK31 plays an important role in PC and we recently found that miR‐543 was related to the expression of STK31. It have been found that miR‐543 expression could affect tumor proliferation, invasion, and migration in several cancers. 13 , 14 , 15 Thus, we hypothesize that miR‐543 is associated with the development of PC by interacting with STK31 and it is supposed to be a promising marker for prediction of prognosis of PC.
Here, we identify the relationship between miR‐543 and STK31 and confirm the value in aiding to predicting clinical prognosis of PC. And we also attempt to explore the potential function of miR‐543 in PC including the genetic networks of its downstream.
2. METHODS
2.1. Public data sets
TCGA public databases for pancreatic cancer (PAAD data set, released on 1 June 2015 https://tcga‐data.nci.nih.gov/tcga/tcgaHome2.jsp) 16 was used. It contained multiomics information including the clinical, gene, and miRNA expression data of PC, which could be used to evaluate the expression pattern of miRNAs and genes. In TCGA, 170 PC samples had gene expression, miRNA expression, and clinical information. RNA‐Seq by Expectation‐Maximization (RSEM) and Reads Per Million (RPM) was used to present the expression of genes and miRNAs from RNA‐Seq, respectively.
2.2. Study population
Fifty frozen tumor tissues from PC patients stored in Pancreas Biobank from the First Affiliated Hospital of Nanjing Medical University, who underwent initial surgical resection between December 2016 and April 2017 in the pancreas center of the First Affiliated Hospital of Nanjing Medical University, were tested. There was no treatment before surgery for these patients and there was a documented survival of at least 3 months from the date of surgery, which mean that we excluded those people who died from surgical complications. All patients agreed to participate in the study and were achieved informed consent. This study was approved by the ethical board of the institute of the First Affiliated Hospital of Nanjing Medical University.
2.3. RNA/MIRNA extraction and quantitative RT‐PCR
After tumor tissue collection, they were stored in liquid nitrogen before used. RNA and miRNA isolation were performed as described previously. 17 Briefly, total RNA was extracted from tissues using TRIzol Reagent (Invitrogen) to denature the tissues and purified using a RNeasy Mini Kit (Qiagen). MiRNA was extracted using TRIzol LS Reagent and collected using a miRNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The STK31 and miR‐543 expression levels in the tissue samples were normalized using ACTIN and U6, respectively. The reverse transcriptase reactions were performed with a TaqMan RNA RT Kit (ABI) for STK31 and TaqMan miRNA RT Kit and stem‐loop RT primers (TIANGEN) for miR‐543 using the StepOnePlus Real‐Time System (Thermo). To confirm the relationship between STK31 and miR‐543, SYBR Green qRT PCR was performed. The primers were listed: STK31‐F: TGTCTGGTGAGGTACATTGACT, STK31‐R: GCTCCCCAAAAAGGTTGTGC; ACTIN‐F: AGCGAGCATCCCCCAAAGTT, ACTIN‐R: GGGCACGAAGGCTCATCATT; miR‐543‐F: ACATTCGCGGTGCACTTCTT miR‐543‐R: CAGTGCGTGTCGTGGAGT and U6‐F: CTCGCTTCGGCAGCACA, U6‐F: AACGCTTCACGAATTTGCGT. All reactions were carried out in three times and it included the no‐template controls. The CT values were determined using the fixed threshold settings. To calculate the relative expression levels of the miR‐543 and STK31, we used a standard PCR procedure to analyze the data. The 2−△CT were calculated to represent the expression levels and △CT the following formulas: △CTSTK31 = CTSTK31−CTACTIN for the gene STK31 and △CTmiR‐543 = CTmiR‐543−CTU6 for miR‐543 expression.
2.4. MIRNA target prediction
microRNA.org (http://www.microrna.org), miRDB (http://www.mirdb.org/), TargetScan (http://www.targetscan.org/vert_72/), and RNAInter (http://www.rna‐society.org/raid/home.html) were used to predict the targets of miRNAs. microRNA.org is a comprehensive resource of microRNA target predictions and expression profiles and target predictions are based on miRanda algorithm which incorporates current biological knowledge on target rules and on the use of an up‐to‐date compendium of mammalian microRNAs. 18 miRDB is an online database for miRNA target prediction and functional annotations which uses common features associated with miRNA binding and target downregulation to identify and predict miRNA targets with machine learning methods. 19 , 20 TargetScan predicts biological targets of miRNAs by searching for the presence of conserved 8mer, 7mer, and 6mer sites which match the seed region of each miRNA. 21 , 22 STK31 was performed as targeted gene being regulated and miR‐543 as miRNA playing the regulating function. RNAInter is a comprehensive resource for RNA interactome data obtained from the literature and other databases, containing over 41 million RNA‐associated interactions of RCI, RDI, RHI, RPI, and RRI 23 .
2.5. Dual‐luciferase reporter assays
A dual‐luciferase reporter assay was used to assess the direct binding site between STK31 and miR‐543. To construct the STK31 promoter luciferase vector, sequences (730 bp) include the wild‐type 3′‐untranslated region (UTR) of STK31 and ACTIN were synthesized by GeneBay (Nanjing) and subcloned into the BsiWI‐XhoI restrictive sites of the pEZX‐FR02 Vector. 293T cells were co‐transfected with the pEZX‐FR02 reporter vectors and miR‐543 mimic using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.). After 48 h incubation at 37°C with 5% of CO2, luciferase activity was evaluated. Firefly luciferase activity was normalized to Renilla luciferase (Promega Corporation) gene activity with a dual‐luciferase reporter assay (Promega Corporation). miR‐543 mimics (5′‐AAACAUUCGCGGUGCACUUCUU‐3′) and its negative control (NC mimics; 5′‐UUCUCCGAACGUGUCACGUTT‐3′) were synthesized by Guangzhou RiboBio Co., Ltd. This reporter assay was performed three times.
2.6. Statistical analysis
Spearman correlation analysis was performed to establish the relationship between all miRNAs and STK31 expression to find these potentially functional miRNAs both in the data from our center and TCGA database. Data of functional experiments were assayed by comparing mean ±SD using a unpaired Student's t‐test for independent two groups and one‐way ANOVA followed by Tukey's test for independent multiple groups and were assumed for *p < 0.05, **p < 0.001, ***p < 0.0001. Kaplan–Meier curves were performed to evaluate the prognostic value of miRNAs and STK31 in PC, and log‐rank testing was used to state the statistical significance. When we focused on the miR‐543, the relationship between all genes (20,532 genes, including STK31) with miR‐543 was analyzed also by spearman correlation test (p < 1 × 10−6). The Gene Ontology (GO) knowledgebase (http://www.geneontology.org/) is the largest source to provide structured and well‐established vocabulary for annotating proteins from cellular component (CC), biological process (BP), and molecular function (MF) aspects 24 , 25 . The Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp/kegg/) is a database resource for understanding high‐level functions and utilities of the biological system and predicting the potential pathways for genes. 26 , 27 The R package “clusterProfiler (v3.12.0)” 28 was used to analyze the GO term and KEGG pathway for these miR‐543‐related genes and dotplot was used to present the identified GO term and KEGG pathway. The significant threshold of these enrichment analysis was FDR <0.05. All of the statistical analyses were performed with R software (version 3.6.3) and p < 0.05 was considered as statistical significance for all the tests.
3. RESULTS
3.1. Prediction of STK31‐associated miRNAs
In the databases of microRNA.org, TargetScan, RNAInter, and miRDB, 6 miRNAs, 58 miRNAs, 46 miRNAs, and 16 miRNAs were predicted to correlate with STK31 in Homo sapiens, Mus musculus, or Rattus norvegicus. Since the prediction principles of these databases are different, 21 miRNAs, which we selected, appeared in at least two databases (RNAInter, TargetScan, and miRDB: miR‐8084, miR‐7243, miR‐3140, miR‐561, miR‐4772, miR‐589, miR‐1912; RNAInter and miRDB: miR‐196c; RNAInter and TargetScan: miR‐153, miR‐3120; RNAInter and microRNA.org: miR‐490, miR‐300, miR‐381, miR‐543, miR‐495, miR‐186; TargetScan and miRDB: miR‐223, miR‐4777, miR‐3145, miR‐335, miR‐6417.) and these miRNAs were studied by expression correlations with STK31 in TCGA (Figure 1).
FIGURE 1.
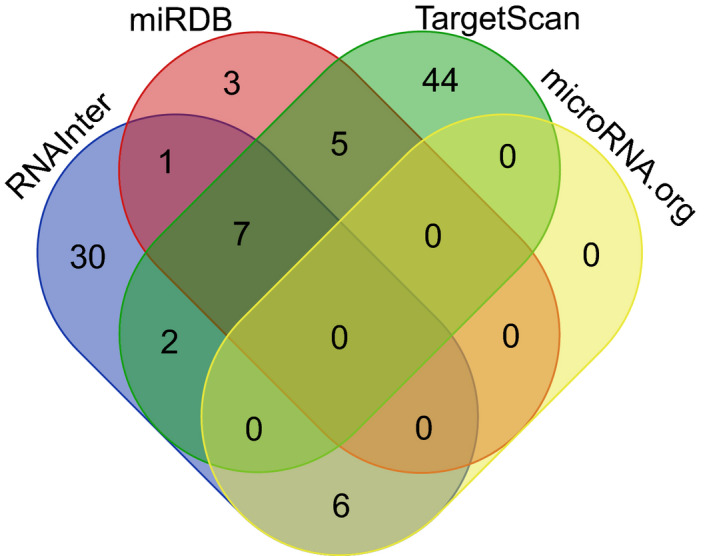
Venn diagram for predicted STK31‐associated miRNA in four database
3.2. Expression correlations between STK31 and MIRNAS in TCGA
From TCGA database, the expression level of STK31 and all these 15 of 21 miRNAs predicted by Public Database was acquired. The other six miRNAs (miR‐8084, miR‐7243, miR‐4772, miR‐196c, miR‐4777, and miR‐6417) were not contained in TCGA miRNA database. All these eight miRNAs, whose median reads per million (RPM) was greater than 1, were then selected to analyze the expression correlation with the expression of STK31 (Figure 2). Three miRNAs (miR‐543, miR‐495, and miR‐381) were significantly correlative with STK31 (R = −0.20, spearman p = 0.0076; R = −0.17, p = 0.0207; R = −0.29, p < 0.0001, respectively).
FIGURE 2.
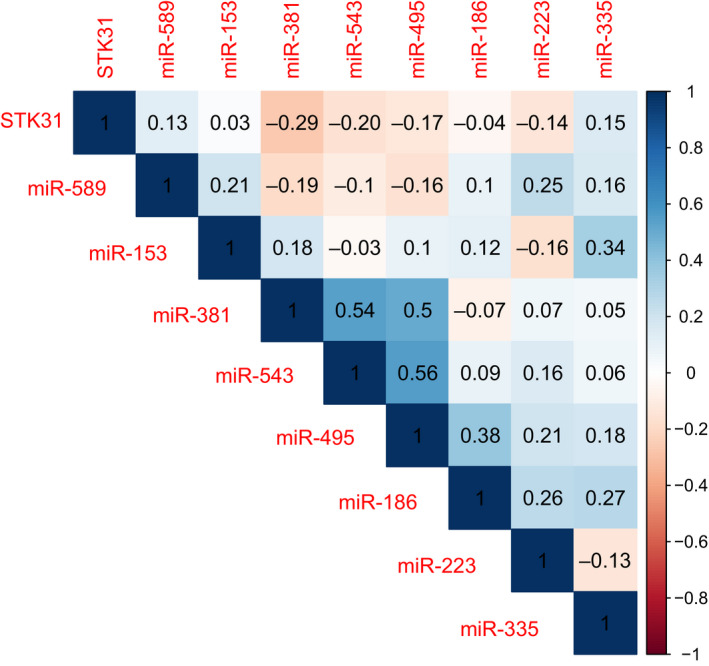
The expression correlation between the expression of STK31 and 10 miRNAs in TCGA
3.3. Relationship between MIR‐543 and prognosis of PC
After the expression correlation analysis, the prognostic correlation analysis was performed to assess the relationship between their expression and prognosis. The results showed that the high expression of miR‐543 suggested the lower overall survival (MST 485 days in high expression of SKT31 and 666 days in low, log‐rank p = 0.046, Figure 3A). But, different expression levels of miR‐543 could not predict the disease‐free survival (log‐rank p = 0.270, Figure 3B). In contrast to miR‐543, high expression of STK31 suggested the higher overall survival (log‐rank p = 0.030, Figure 3C) but not in disease‐free survival (log‐rank p = 0.093, Figure 3D).
FIGURE 3.
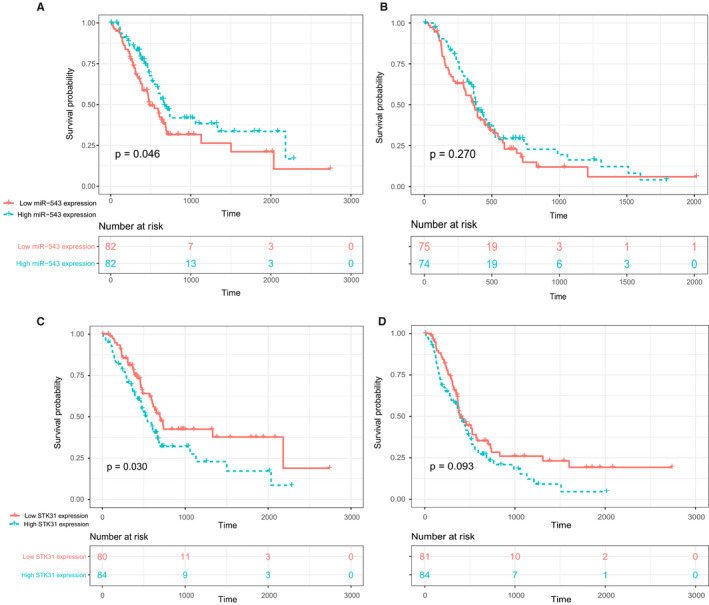
Overall survival (A) and disease‐free survival (B) of PC in low‐ and high‐miR‐543 expression groups and OS (C) and disease‐free survival (D) of PC in low‐ and high‐STK31 expression groups in TCGA data set
3.4. Validate the expression correlations between STK31 and MIR‐543
In addition to the analysis in TCGA database, we also confirmed the relationship between the expression of miR‐543 and gene STK31 in 50 pancreatic cancer tissues from our Pancreas Biobank. The basic characteristics of these patients are shown in Table 1. The results were consistent with the TCGA database and they confirmed that miR‐543 expression was negatively correlated with STK31 expression (Spearman R = −0.28, p = 0.0437, Figure 4A,B). With dual‐luciferase reporter assays, we found that the miR‐543 mimics co‐transfected with pEZX‐FR02‐STK31 significantly decreased the promoter activity of the reporter gene, leading to lower STK31 expression (Figure 4C).
TABLE 1.
Clinical characteristics of patients with PC from our Pancreas Biobank
| Characteristic | |
|---|---|
| Age (y, Mean ± SD) | 61.7 ± 10.8 |
| Gender N(%) | |
| Male | 28 (56) |
| Female | 22 (44) |
| CA19‐9 (U/ml, IQR) | 173(53–524) |
| CEA (U/ml, IQR) | 3.1(1.7–5.3) |
| T stage N(%) | |
| T1 | 5 (10) |
| T2 | 33 (66) |
| T3 | 4 (8) |
| T4 | 5 (10) |
| Unknown | 3 (6) |
| N stage N(%) | |
| N0 | 25 (50) |
| N1/N2 | 25 (50) |
| M stage N(%) | |
| M0 | 50 (100) |
| M1 | 0 (0) |
| TNM stage N(%) | |
| I | 17 (34) |
| II | 19 (38) |
| III | 11 (22) |
| Unknown | 3 (6) |
| Grade N(%) | |
| I/I–II/II | 11 (22) |
| II–III/III | 37 (74) |
| Unknown | 2 (4) |
FIGURE 4.
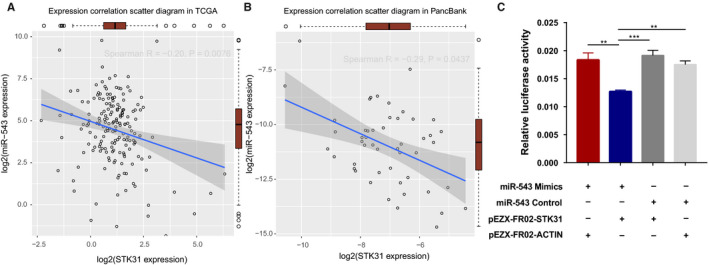
The relationship between STK31 and miR‐543. A, Expression correlation scatter diagram in TCGA and the red box present the expression of STK31 and miR‐543. B, Expression correlation scatter diagram in our Pancreas Biobank and the red box present the expression of STK31 and miR‐543. C, Reporter gene assays with constructs containing the STK31 promoter in 293T cell lines. miR‐543 control or mimics was co‐transfected into cells. Data shown are means ± SD
3.5. The functional and pathway enrichment analysis of MIR‐543‐related genes
We then predicted the targets of miR‐543 in miRDB and TargetScanHuman. In all, 1208 and 758 genes were identified as potential target genes of miR‐543. After removing the represented genes, 1548 genes were left (Figure 5A, Table S1). Based on the GO term and KEGG analysis, a total of 19 GO_BP terms, 27 GO_CC terms, 19 GO_MF terms, and 13 KEGG pathways were enriched for the miR‐543‐related genes. The GO terms included “dendrite development” (BP GO:0016358; FDR, 0.0002841753), “ubiquitin‐like protein transferase activity” (MF GO:0019787; FDR, 0.002539626), and “protein serine/threonine phosphatase activity” (MF GO:0004722; FDR, 0.011598712) (Figure 5B). In addition, significant pathways included “Insulin signaling pathway” (FDR, 0.00919), “ErbB signaling pathway” (FDR, 0.00919) and “T cell receptor signaling pathway” (FDR, 0.012167) (Figure 5C).
FIGURE 5.
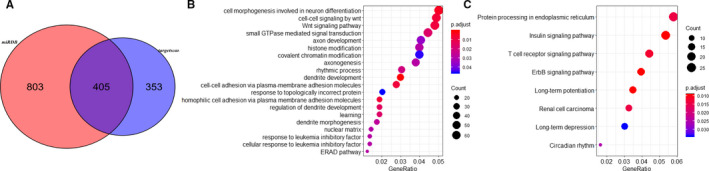
The functional and pathway enrichment analysis of miR‐543‐related genes. A, Venn Diagram for the predicted the targets of miR‐543 in miRDB and TargetScanHuman. B, GO term enrichment of miR‐543‐related genes. C, KEGG enrichment of miR‐543‐related genes
4. DISCUSSION
A great deal of evidence suggests that miRNAs are stable diagnostic and prognostic biomarkers for cancers. Previously, several studies have been related to miR‐543. Dysregulation of miR‐543 expression could influence tumor proliferation, invasion, and even migration in colorectal cancer, 14 hepatocellular carcinoma, clear cell renal cell carcinoma 13 cervical cancer, 29 and gastric cancer. 30 But the results of these studies are not completely consistent in different cancers. Sun et al. demonstrated that they identified miR‐543 as tumor promoter and plays a vital role in CRC metastasis by direct targeting PTEN directly. 14 Yu et al. also demonstrated that miR‐543 was a tumor promoter and dramatically overexpressed in hepatocellular carcinoma (HCC). The overexpression of miR‐543 could promote the ability of proliferation and invasion in HCC cell line by targeting PAQR3. 15 Moreover, miR‐543 promoted cell proliferation by targeting SIRT1 31 and cell migration and invasion by targeting sPOP 30 in gastric cancer. But Li and his team came to the conclusion that miR‐543 was decreased in endometrial cancer and suppressed endometrial cancer oncogenicity via targeting FAK and TWIST1 32 and XU et al. reported that miR‐543 might function as a tumor suppressor in patients with glioma. 33 The different function of miR‐543 in different types of tumor suggested that miR‐543 might play different roles in different cancers. Up to now, there is no report about miR‐543 in PC has been published. Therefore, we investigated the possible role of miR‐543 in PC.
Based on the previous study of STK31, 34 we further studied its related miRNAs in current study and found a novel potentially important miRNAs, miR‐543, in PC. First, we searched in predictions tools for STK31‐related miRNAs. After we identified STK31‐related miRNAs, we analyzed the relationship between these miRNAs and the survival time of PC in TCGA database. The results showed that miR‐543 was significantly related with the prognosis of PC. In addition, we confirmed the expression correlationship between miR‐543 and STK31 in PC tissue specimen. At last, we predicted the miR‐543‐related genes and uncovered that miR‐543 might play roles in pathways of insulin signaling, ErbB signaling pathway, and T cell receptor signaling in PC.
Previous studies have strongly suggested that miR‐543 can regulate cell invasion and impedes apoptosis by activation of the Wnt/β‐catenin pathway though Smad7 35 and promotes cell proliferation and metastasis through the Wnt/β‐catenin pathway by targeting Dickkopf 1 in cancers. 36 These results are consistent with our findings in bioinformatic analysis in PC. We also found that cell–cell signaling by Wnt and Wnt signaling pathway was enriched in miR‐543‐related genes. Moreover, researches have confirmed that miR‐543 is involved in regulating cell invasion and metastasis in renal cell carcinoma. And in this study, cell–cell adhesion via plasma‐membrane adhesion molecules in GO_BP and renal cell carcinoma in KEGG were also enriched. Neamah WH et al. 37 studied the aryl hydrocarbon receptor (AhR) and found that miR‐543 could target these anti‐inflammatory, which was consistent with our findings of the relationship between miR‐543 and T cell receptor signaling pathway.
In previous studies, we have displayed that STK31 expression was significantly higher in patients with poorer prognosis, suggesting that STK31 had potential clinical value. 34 And we also found that STK31 was reactivated by demethylation. As one of the common mechanisms regulating gene expression, we predicted that the expression of STK31 was regulated by miR‐543 and had the sequence of miR‐543 binding. We had confirmed their expression correlationship in PC tissues by qPCR. STK31 is one of miR‐543‐regulated genes and miR‐543 may play an important role in PC. And we also confirmed the direct binding between miR‐543 and STK31 by dual‐luciferase reporter assays. Despite the exciting probability of therapeutic targets in PC, there are also some challenges existed. The major one is that miRNA‐based therapeutics can cause unexpected side effects because miRNAs have more than one downstream‐regulated gene and it is difficult to ensure tumor‐specific delivery and retention of miRNAs. Therefore, off‐target effects are likely to be happened. 38 To overcome this shortcoming, we need to carry out adequate animal and clinical trials. Another limitation is that miRNAs inhibitors are not stable in vivo. 38 MiRNA introduced into mice via the tail vein will be cleared in 30 mins from the circulatory system 39 . We need to improve existing methods to make inhibitors more stable. Once we could solve these problems, miRNA must be a very meaningful therapeutic targets.
In conclusion, this study showed that miR‐543 can directly binding with STK31 and its expression was negatively correlated with STK31 expression in PC. In addition to the relationship with STK31, miR‐543 was also correlated with 1548 genes expression and the enrichment study was found that they enriched in several KEGG pathway, such as Insulin signaling pathway, ErbB signaling pathway, and T cell receptor signaling pathway, which provided comprehensive insight into the potential molecular mechanisms in PC. Taken together, our results indicate that miR‐543 may play an important role and is a promising therapeutic target in PC.
CONFLICTS OF INTEREST
All authors have reviewed the final version of the manuscript and approved it for publication. The authors have no conflicts of interest to declare.
AUTHOR'S CONTRIBUTIONS
Weizhong Yuan & Hao Gao: They contributed equally to this work. They were mainly responsible for acquisition of data, analysis and interpretation of data, and drafting the article. Guangfu Wang: He provided assistance for data acquisition and statistical analysis. Yi Miao & Kuirong Jiang: They were responsible for revising it critically for important intellectual content. Kai Zhang & Junli Wu: They contributed to the conception and design of the study and coordinate the work of all parties. Kai Zhang was also responsible for the verification of data and analytical methods; Junli Wu also contributed to the final approval of the version to be submitted.
Supporting information
Table S1
ACKNOWLEDGMENTS
This work was supported by grants from Youth Program of National Natural Science Foundation of China (81703301); the National Natural Science Foundation of China (81672449); The Project of Invigorating Health Care through Science, Technology and Education, Jiangsu Provincial Medical Outstanding Talent (JCRCA2016009); the Innovation Capability Development Project of Jiangsu Province (BM2015004).
Yuan W, Gao H, Wang G, et al. Higher miR-543 levels correlate with lower STK31 expression and longer pancreatic cancer survival. Cancer Med. 2020;9:9632–9640. 10.1002/cam4.3559
Weizhong Yuan and Hao Gao contributed equally to this work.
Contributor Information
Kai Zhang, Email: zhangkai@njmu.edu.cn.
Junli Wu, Email: junliwu1973@hotmail.com.
DATA AVAILABILITY STATEMENT
Some or all data generated or used during the study are available from the corresponding author by request.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. [DOI] [PubMed] [Google Scholar]
- 2. Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet. 2016;388(10039):73–85. [DOI] [PubMed] [Google Scholar]
- 3. Chen YE, Gao S‐G, Chen J‐M, et al. Serum CA242, CA199, CA125, CEA, and TSGF are biomarkers for the efficacy and prognosis of cryoablation in pancreatic cancer patients. Cell Biochem Biophys. 2015;71(3):1287–1291. [DOI] [PubMed] [Google Scholar]
- 4. Shi Q, Feng KE, Xia L, et al. Combined use of serum miR‐499a‐5p and CA199 increases the diagnostic sensitivity of pancreatic cancer. Clin Lab. 2019;65(11). 10.7754/Clin.Lab.2019.190416. [DOI] [PubMed] [Google Scholar]
- 5. Lei XF, et al. Application values of detection of serum CA199, CA242 and CA50 in the diagnosis of pancreatic cancer. J Biol Regul Homeost Agents. 2017;31(2):383–388. [PubMed] [Google Scholar]
- 6. Scara S, Bottoni P, Scatena R. CA 19–9: biochemical and clinical aspects. Adv Exp Med Biol. 2015;867:247–260. [DOI] [PubMed] [Google Scholar]
- 7. Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353(17):1768–1771. [DOI] [PubMed] [Google Scholar]
- 8. Zhang B, Pan X, Cobb GP, et al. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. [DOI] [PubMed] [Google Scholar]
- 9. Calin GA, et al. Frequent deletions and down‐regulation of micro‐ RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xue J, et al. Circulating microRNAs as promising diagnostic biomarkers for pancreatic cancer: a systematic review. Onco Targets Ther. 2019;12:6665–6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Michael IP, Saghafinia S, Hanahan D. A set of microRNAs coordinately controls tumorigenesis, invasion, and metastasis. Proc Natl Acad Sci U S A. 2019;116(48):24184–24195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zou X, Wei J, Huang Z, et al. Identification of a six‐miRNA panel in serum benefiting pancreatic cancer diagnosis. Cancer Med. 2019;8(6):2810–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang F, et al. MicroRNA‐543 promotes the proliferation and invasion of clear cell renal cell carcinoma cells by targeting Kruppel‐like factor 6. Biomed Pharmacother. 2018;97:616–623. [DOI] [PubMed] [Google Scholar]
- 14. Sun J, Zhou J, Dong M, et al. Dysregulation of MicroRNA‐543 expression in colorectal cancer promotes tumor migration and invasion. Mol Carcinog. 2017;56(1):250–257. [DOI] [PubMed] [Google Scholar]
- 15. Yu L, et al. MicroRNA‐543 acts as an oncogene by targeting PAQR3 in hepatocellular carcinoma. Am J Cancer Res. 2014;4(6):897–906. [PMC free article] [PubMed] [Google Scholar]
- 16. Weinstein JN, Collisson EA, Mills GB, et al. The cancer genome Atlas Pan‐Cancer analysis project. Nat Genet. 2013;45(10):1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fu Y, Liu X, Chen Q, et al. Downregulated miR‐98‐5p promotes PDAC proliferation and metastasis by reversely regulating MAP4K4. J Exp Clin Cancer Res. 2018;37(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Betel D, Wilson M, Gabow A, et al. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36(Database issue):D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agarwal V, Bell GW, Nam J‐W, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 e05005–e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia DM, Baek D, Shin C, et al. Weak seed‐pairing stability and high target‐site abundance decrease the proficiency of lsy‐6 and other microRNAs. Nat Struct Mol Biol. 2011;18(10):1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin Y, Liu T, Cui T, et al. RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res. 2020;48(D1):D189–D197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Resource TGO. 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanehisa M, Sato Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29(1):28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu G, Wang L‐G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu X, Gan L, Zhang J. miR‐543 inhibites cervical cancer growth and metastasis by targeting TRPM7. Chem Biol Interact. 2019;302:83–92. [DOI] [PubMed] [Google Scholar]
- 30. Xu J, et al. miRNA‐543 promotes cell migration and invasion by targeting SPOP in gastric cancer. Onco Targets Ther. 2018;11:5075–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li J, Dong G, Wang BO, et al. miR‐543 promotes gastric cancer cell proliferation by targeting SIRT1. Biochem Biophys Res Commun. 2016;469(1):15–21. [DOI] [PubMed] [Google Scholar]
- 32. Bing LI, Hong C, Li‐Xin S, et al. MicroRNA‐543 suppresses endometrial cancer oncogenicity via targeting FAK and TWIST1 expression. Arch Gynecol Obstet. 2014;290(3):533–541. [DOI] [PubMed] [Google Scholar]
- 33. Xu L, Yu JU, Wang Z, et al. miR‐543 functions as a tumor suppressor in glioma in vitro and in vivo. Oncol Rep. 2017;38(2):725–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang K, et al. The clinical value, regulatory mechanisms, and gene network of the cancer‐testis gene STK31 in pancreatic cancer. Oncotarget. 2017;8(21):35154–35164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen DW, et al. MicroRNA‐543 promotes cell invasion and impedes apoptosis in pituitary adenoma via activating the Wnt/beta‐catenin pathway by negative regulation of Smad7. Biosci Biotechnol Biochem. 2019;83(6):1035–1044. [DOI] [PubMed] [Google Scholar]
- 36. Chen ZY, et al. MiR‐543 promotes cell proliferation and metastasis of renal cell carcinoma by targeting Dickkopf 1 through the Wnt/beta‐catenin signaling pathway. J Cancer. 2018;9(20):3660–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neamah WH, Singh NP, Alghetaa H, et al. AhR activation leads to massive mobilization of myeloid‐derived suppressor cells with immunosuppressive activity through regulation of CXCR2 and MicroRNA miR‐150‐5p and miR‐543‐3p that target anti‐inflammatory genes. J Immunol. 2019;203(7):1830–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kong YW, Ferland‐McCollough D, Jackson TJ, et al. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–e258. [DOI] [PubMed] [Google Scholar]
- 39. Trang P, Wiggins JF, Daige CL, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19(6):1116–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Data Availability Statement
Some or all data generated or used during the study are available from the corresponding author by request.


