Abstract
B cells are an integral part of the adaptive immune system and regulate innate immunity. Derived from hematopoietic stem cells, B cells mature through a series of cell fate decisions. Complex transcriptional circuits form and dissipate dynamically during these lineage restrictions. Genomic aberrations of involved transcription factors underlie various B-cell disorders. Acquired somatic aberrations are associated with cancer, whereas germline variations predispose to both malignant and nonmalignant diseases. We review the opposing role of transcription factors during B-cell development in health and disease. We focus on early B-cell leukemia and discuss novel causative gene–environment cooperation and their implications for precision medicine. Childhood leukemia is frequently initiated during fetal hematopoiesis. Clinical silent preleukemic clones are detectable in cord blood of a large number of healthy newborns. These predisposing alterations cooperate with environmental factors to trigger leukemia onset. Understanding of the underlying principles is a prerequisite for the development of measures to prevent leukemia in children.
Introduction
B cells are white blood cells of the lymphocyte subtype. They play an essential role in humoral immunity of the adaptive immune system by secretion of antibodies (1). An antibody response is elicited against a specific antigen when unique B-cell receptors (BCR) expressed on the cell surface of B cells recognize and bind the antigen (1). In addition, B cells produce cytokines, present antigens, and function as regulators of the second branch of the immune system, innate immunity (2). All mature blood cells, including B cells, are generated by hematopoietic stem cells (HSC) and differentiate through the serial action of transcription factors (TF) that determine their cell fate at specific decision-making stages during development (1). B cells arise in the bone marrow of mammals, and their development is tightly regulated. Disturbances of B-cell development can cause diseases ranging from benign lymphoproliferation to malignant leukemia and lymphoma (1). Therefore, a key to understanding these diseases is to unravel physiologic B-cell development. Recent studies have revealed a growing number of genetic alterations affecting B-cell TFs that directly cause or predispose individuals to B-cell acute lymphoblastic leukemia (B-ALL) development (3, 4). These novel insights have revised the way we view early B-cell development. Here, we review these recent findings and how they advance our understanding of early B-cell development and associated disease.
Transcriptional and Epigenetic Regulation Determines Normal Early B-cell Development
Early B-cell Differentiation
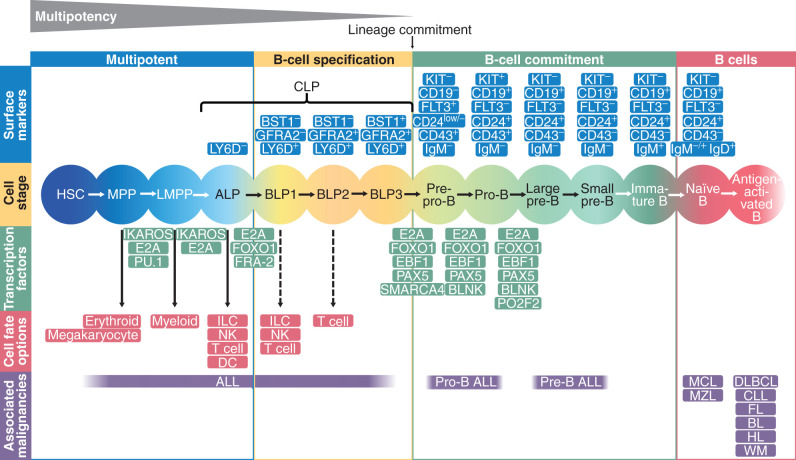
Pluripotent stem cells give rise to multiple cell types during development. The differentiation process is tightly regulated by lineage-specific TFs and epigenetic modification resulting in stepwise lineage commitment, differentiation, and lineage-specific gene expression (5). The B-cell differentiation process can be subdivided into distinct stages based on the expression of cell-surface markers and the differentiation potential of the cells (Fig. 1). As a first step, HSCs lose their self-renewal capacity. They begin to express the surface marker tyrosine kinase receptor Flt3 and transition to multipotent progenitors (MPP; ref. 6). MPPs that have lymphoid and myeloid potential, but retain only limited potential to differentiate along the erythroid and megakaryocyte lineage, are referred to as lymphoid-primed MPPs (LMPP; ref. 7). LMPPs are the precursors to the common lymphoid progenitors (CLP) that give rise to B cells, natural killer (NK) cells, dendritic cells (DC), innate lymphoid cells (ILC), and T cells and retain only a low myeloid potential (8). CLPs are subdivided into Ly6D (lymphocyte antigen 6 complex, locus D)-negative, all-lymphoid progenitors (ALP) and B-cell–biased lymphoid progenitors (BLP) that express Ly6D on the cell surface (9). ALPs retain the potential to differentiate into T cells, B cells, NK cells, and DCs, whereas BLPs are restricted to the B lineage. It has recently been shown in mice that BLPs can be further separated into three subsets (BLP1–3) according to the expression of the cell-surface proteins GFRA2 (GDNF Family Receptor Alpha 2) and BST1 (bone marrow stroma cell antigen 1; ref. 10). BLP1 (Ly6D+GFRA2−BST1−) and BLP2 (Ly6D+GFRA2+BST1−) retain the potential to differentiate into T cells and/or NK cells, whereas BLP3 (Ly6D+GFRA2+BST1+) loses this potential. BLP3s finally give rise to fully committed pro-B cells that can be identified by expression of CD19.
Figure 1.
Transcriptional regulation determines normal B-cell development. A global network of regulatory circuits and epigenetic factors drives differentiation of HSCs through B-cell lymphopoiesis. The first stages of B-cell development are marked by multipotent cell types that can also develop in very different hematopoietic cell types. Potential other developmental paths are given in red boxes. The most important TFs that drive the differentiation from one progenitor to another are presented in green between the developmental stages. Surface markers are given in blue and hematopoietic malignancies associated with the cell stage in purple. The multipotent cells are succeeded by BLPs that are already primed toward becoming B cells but still have other lymphoid options. The BLP stages are characterized by the expression of the surface proteins BST1 and GFRA2. After full lineage commitment, BLP3 develop into pro-B cells. The TFs E2A, EBF1, and PAX5, in combination with several epigenetic factors, play central roles in the B-cell fate decision. ALL, acute lymphoblastic leukemia; BL, Burkitt lymphoma; CLL, chronic lymphocytic leukemia; DLBCL, diffuse large B-cell lymphoma; FL, follicular center cell lymphoma; HL, Hodgkin lymphoma; MCL, mantle cell lymphoma; MZL, marginal zone B-cell lymphoma; WM, Waldenström macroglobulinemia.
Networks of Transcription and Epigenetic Factors Drive Early B-cell Differentiation
The contribution of specific TFs to the B-cell differentiation process has been previously reviewed thoroughly (11, 12). Briefly, current knowledge on the regulation of B-cell development is mainly based on studies in mice, and it is not clear whether the same statements also hold true for human B-cell development. Specification to the B-lymphoid lineage is initiated at the MPP to LMPP differentiation stage and regulated by three main TFs: PU.1 [purine-rich (PU) sequence binding factor], Ikaros (Ikzf1), and E2A (Tcf3). Likewise, there is strong cooperation between E2A, EBF1, and PAX5 in the commitment of cells to the B-cell lineage. In the absence of PAX5, B-cell development is arrested at the pro–B-cell stage (13) and commitment to the B-cell lineage is lacking (Fig. 1). These cells are capable of differentiating into multiple hematopoietic lineages, including T cells, NK cells, and myeloid cells (14, 15). Similar capacities for multilineage differentiation were reported in E2A-deficient progenitors and EBF1-deficient progenitors (15, 16). These progenitors express myeloid, ILC, and T-cell lineage genes and retain the potential to differentiate into these lineages. In contrast, ectopic expression of E2A or EBF1 restricts their alternative differentiation potential and promotes B-cell fate specification (16).
More precise, time-resolved analysis of TF expression patterns underlying the B-cell lineage commitment were recently shown using inducible systems for EBF1 expression and E2A inhibition (17, 18). Miyai and colleagues overexpressed an Id3–ER (estrogen receptor) fusion protein, whose nuclear translocation is induced by 4-hydroxytamoxifen (4-OHT) in hematopoietic stem and progenitor cells (17). In the presence of 4-OHT, E2A activity is repressed and B-cell development arrested at the MPP stage. These progenitor cells were named induced leukocyte stem (iLS) cells, because they retained the potential to differentiate into T, B, or myeloid lineage. B-cell differentiation of iLS cells was induced upon withdrawal of 4-OHT and regaining of E2A activity. Time-resolved analysis showed that the TF program was separated into three waves. Strikingly, TFs not specific to the B lineage, such as EGR1, NR4A2, and KLF4, were rapidly induced before the late onset of master regulators (including EBF1 and PAX5) in the third wave of commitment. Supporting this idea, Fra-2, a member of the activator protein 1 (AP-1) family belonging to the dimeric basic region-leucine zipper TFs, has been shown to be a critical regulator of FOXO1 in early B-cell differentiation (19). Many epigenetic regulators, such as SMARCA4, UHRF1, DNMT1, and EZH2 are also implicated in establishment of the B-cell fate. A similar hierarchy of transcriptional and epigenetic events was demonstrated in B-cell programming using EBF1 induction in developmentally arrested Ebf1−/− pre–pro-B cells (20) and in the differentiation of the Th cell 17 lineage (Th17; ref. 21), suggesting a general mode for differentiation of immune cells.
Chromatin Dynamics During B-cell Differentiation
Recent studies have indicated that the three-dimensional chromatin organization changes dramatically during B-cell development (20). TFs such as Pax5 are critical for the establishment of the B-cell lineage–specific genome structure. In uncommitted MPP cells, the transcriptionally inactive EBF1 locus is located at the nuclear lamina. After differentiating to the pro–B-cell stage, EBF1 relocates from the lamina to the inside of the nucleus to establish interactions associated with B lineage–specific transcriptional programs (20). These findings indicate that TFs fulfill a dual role in regulating lineage-specific gene expression programs and in establishing the three-dimensional genomic architecture during B-cell differentiation. However, it is still unclear how these processes are organized. Fundamental regulators of chromatin structure including CTCF (CCCTC-binding factor) and cohesin complexes may cooperate with TFs to regulate genome organization (22). Brahma-related gene-1 (Brg-1), a chromatin remodeler, is recruited to chromatin in B-cell progenitors and is critical for B-cell differentiation (23). Further studies are necessary to define regulating factors, control regions in the genome, and cell-type–specific genome structures.
Genetic Dysregulation of TFs Involved in Early B-cell Development Causes Various Benign and Malignant Blood Disorders
Genetic alterations of TFs involved in early B-cell development have been identified in a wide variety of blood disorders in humans (24) and add new, unexpected insights about B-cell development. Germline variants need to be compatible with embryogenesis and life in general. In some cases, de novo mutations are more severe or earlier in onset compared with transmitted ones. They can predispose to benign as well as malignant disorders. In contrast, acquired somatic variants have a more drastic effect on protein function and are restricted to malignant disease (ref. 24; Table 1).
Table 1.
Genetic alterations of hematopoietic TFs predisposing to B-ALL development
| Risk | Gene | Mutation type | Consequence | Tumor type | ||
|---|---|---|---|---|---|---|
| Low-penetrance susceptibility | CEBPE | Intronic SNP | Dysregulation | B-ALL | ||
| GATA3 | Intronic SNP | Dysregulation | Ph+ | |||
| IKZF1 | Intronic SNP, 3′UTR SNP | Dysregulation | B-ALL | |||
| ERG | ||||||
| ARID5B | Intronic SNP | Dysregulation | Hyperdiploid | |||
| IKZF3 | ||||||
| High-penetrance susceptibility | ETV6 | Missense | Loss of function | |||
| IKZF1 | Missense | Loss of function | ||||
| PAX5 | Missense | Lower transcriptional activity | ||||
| High-penetrance somatic variants | ETV6 | Gene fusion | Transcriptional dysregulation | ETV6–RUNX1+ pre–B-ALL | ||
| CNA and/or missense | Loss of function | |||||
| TCF3 | Gene fusion | Transcriptional dysregulation | TCF3–PBX1+ pre–B-ALL | |||
| ZNF384 | Gene fusion | Transcriptional dysregulation | ||||
| MEF2D | Gene fusion | Transcriptional dysregulation | ||||
| CEBPE | CNA and/or missense | Loss of function | ||||
| GATA3 | CNA and/or missense | Loss of function | ||||
| IKZF1 | CNA and/or missense | Loss of function | ||||
| ERG | CNA and/or missense | Loss of function | ||||
| PAX5 | Gene fusions | Loss of function | ||||
| IKZF3 | CNA and/or missense | Loss of function | ||||
| EBF1 | CNA and/or missense | Loss of function | ||||
| BTG1 | CNA and/or missense | Loss of function |
Abbreviations: CNA, copy-number alteration; Ph, Philadelphia chromosome; UTR, unstranslated region.
Germline Variation
Inherited or de novo germline variants of a growing number of hematopoietic TFs (including IKZF1, E2A/TCF3, PAX5, and ETV6) have been associated with benign blood disorders and familial B-ALL or lymphoma (25,26,27,28,29,30).
IKZF1 is a key regulator of both lymphoid and myeloid differentiation and implicated in proliferation restriction. Transmitted germline IKZF1 mutations were recently linked to common variable immunodeficiency syndrome (CVID; ref. 31). CVID is a frequent but genetically heterogeneous primary immunodeficiency (incidence of 1:50,000–1:25,000) clinically characterized by recurrent infections, due to markedly decreased numbers of isotype-switched mature B cells and corresponding low levels of serum IgG-type antibodies (and commonly also IgM and/or IgA). The CVID subtype caused by IKZF1 mutations presented with B-cell immune deficiency, autoimmunity, and susceptibility to B-ALL. IKZF1 comprises an N-terminal DNA-binding domain (DBD) and a C-terminal dimerization domain. Several isoforms have been described. IKZF1 mainly functions as a transcriptional repressor and binds to DNA as a homo- or heterodimer associating with its own isoforms or other family members [IKZF2 (HELIOS), IKZF3 (AIOLOS), or IKZF4 (EOS)] at pericentromeric heterochromatin regions. The identified CVID-associated mutations included mostly loss-of-function deletions and missense mutations affecting the DBD. They acted by haploinsufficiency. In addition, de novo germline mutations of IKZF1 DBD were reported that were autosomal dominant and acted in a dominant-negative manner. They were associated with early-onset combined immunodeficiency presenting with severe defects of both the innate and adaptive immune system (32). Besides low numbers of B cells and associated dysgammaglobulinemia, these variants caused multi-lineage abnormalities, including myeloid cells and lymphoid cells. Familial acute lymphoblastic leukemia (ALL) was observed in carriers with both de novo as well as transmitted loss-of-function IKZF1 variants, and it is currently assumed that almost 1% of “sporadic” B-ALL cases might be due to underlying germline IKZF1 mutations (26). These B-ALL–associated IKZF1 germline variants are not restricted to specific functional domains; many of these variants have no effect on TF activity but strongly influence stem cell–like features and cell– and cell–stroma interaction, and decrease drug responsiveness (26). Taken together, these studies identified IKZF1 as an immune deficiency and leukemia predisposition gene.
B-cell development is impaired at the early LMPP stage in mice deficient in the TF E2A/TCF3. In humans, a recurrent heterozygous dominant-negative de novo mutation in E2A/TCF3 (E555K) was recently identified in patients presenting with profound reduction of CD19+ B cells and agammaglobulinemia (33). B cells lacked a functional BCR, and differentiation was blocked at the common lymphoid precursor to pro–B-cell stage. However, some developmental progression along the B lineage takes place even in the complete absence of E2A/TCF3, as evidenced by a case with a homozygous nonsense E2A/TCF3 mutation and with severe hypogammaglobulinemia combined with B-ALL that was recently described (34).
Pax5 is an essential regulator of B-cell development and absolutely required to exit the pro–B-cell stage. A rare PAX5 germline variant (p.Gly183Ser) in the DBD associated with lower, but not lacking, transcriptional activity, was identified in three kindreds with susceptibility to B-ALL (25, 29). Leukemic cells displayed loss of hererozygosity by structural variations on chromosome 9p and retention of only the mutant variant. Consistently, sporadic ALL cases with combined 9p loss and somatic PAX5 variants affecting Gly183 were also observed (29). The lack of more frequent or more functionally disabling germline PAX5 mutations might be due to its functions in brain development and spermatogenesis.
The TFs ETV6 and RUNX1 are involved in early hematopoiesis of other blood cell lineages (e.g., megakaryocytic and erythroid development; ref. 35), but recent findings suggest broader roles in early hematopoiesis, impacting the development of multiple lineages including the B-cell lineage. Rare germline autosomal dominant loss-of-function mutations were recently identified in ETV6, which cause thrombocytopenia and red cell macrocytosis but also predispose to B-ALL (27, 28, 30, 36). The majority of familial mutations cluster within the ETS domain, but a mutation in the linker region (P214L) has been identified recurrently (37). These variants act in a dominant-negative fashion due to homo- and hetero-oligomerization of mutant ETV6 with other ETS family members and transcriptional repressors. They impair transcriptional activity and nuclear localization. In close to 1% of 4,405 unselected sporadic ALL cases, likely damaging germline risk variants were identified in ETV6 (27). It has recently been suggested that ETV6 may directly regulate PAX5 expression through the recruitment of SIN3A and HDAC3 to the PAX5 locus (37). Thus, mutant ETV6 may contribute to a block in B-cell differentiation, lineage infidelity, and leukemogenesis.
Germline mutations in the RUNX1 transcription factor are known to cause familial platelet disorder with associated myeloid malignancy (FPDMM). Affected family members usually present with moderate thrombocytopenia. Some cases developed mainly myeloid but also lymphoid malignancies. Although both dominant-negative and haplo-insufficient mutations are associated with platelet disorders, dominant-negative RUNX1 mutations affect hematopoiesis in a broader fashion and may increase the risk of leukemia (37).
In general, coding variants in B-cell TFs can lead to dramatic changes in transcriptional activities (mostly deleterious) and confer a very significant increase in ALL risk. For example, comparing the frequency of ETV6 variants in ALL cases with that in general population, we estimate that pathogenic variants in this gene carry an approximately 23-fold increase in relative risk (27). There is an extreme paucity of data on genome-wide assessment of rare ALL risk variants, and it is highly probable that many other ALL risk genes are yet to be discovered in the TF gene family. Besides rare variants linked to leukemia predisposition, common variants associated with disease susceptibility have also been uncovered (24). Genome-wide association studies (GWAS) of ALL susceptibility have identified at least 11 risk loci for this cancer, many of which reside within or in close proximity to TF genes (e.g., ARID5B, IKZF1, CEBPE, GATA3), including B-cell TFs as well (24). These common polymorphisms are almost always intronic, although they overlap with putative regulatory DNA elements and potentially influence gene transcription in cis (24). The effects of these common ALL risk variants are modest, with an up to 2-fold increase in RR. Even cumulatively, these variants explain only a very small fraction of absolute risk of ALL (38). Intriguingly, most genes identified from ALL GWAS are rarely affected by somatic alterations (with the exception of IKZF1). Interestingly, several of these TF genes are known to regulate myeloid or T lineage development (CEBPE and GATA3), raising the question of whether ALL risk variants promote expression of these TFs in the wrong lineages and consequently disrupt proper differentiation.
Somatic Variations
In B-ALL, somatic alterations involving TF genes can be largely divided into two types: chromosomal rearrangements resulting in fusion TFs or focal copy-number alterations and sequence mutations that directly affect TF activity. Interestingly, TFs involved in fusions are frequently not affected by concomitant copy-number alterations or mutations, suggesting that the gene fusion itself gives rise to novel functions important for leukemogenesis (3).
More than half of the gene fusion events in B-ALL involve one or more TF genes (3, 39). Some of those are among the first recognized genomic features of this cancer (e.g., ETV6–RUNX1 or TCF3–PBX1) and impact risk stratification, patient treatment, and outcome. In these two chimeric TF proteins, the DBD of ETV6 and TCF3 is replaced by that of RUNX1 and PBX1 (40, 41), respectively, thus causing global transcriptional deregulation. Although TCF3 and PBX1 are both directly involved in lymphoid development (42), ETV6 and RUNX1 are more involved in early hematopoiesis of other blood cell lineages, and the pathogenesis remains incompletely understood. ETV6 deletion is frequently observed in cases with ETV6-RUNX1, but ETV6 deletion alone rarely occurs in ALL, again suggesting that loss of endogenous TF activity is not the main pathogenic mechanism of these fusion genes. Recent genomic profiling studies have identified a plethora of novel fusion genes involving other hematopoietic TFs, for example, ZNF384 (39, 43), MEF2D (44), and PAX5. Whereas ZNF384 (30) and MEF2D (45) are not affected by copy-number alterations or mutations, PAX5 deletion is very common in B-ALL (∼30% of cases; ref. 46) and can be concomitant with PAX5 fusions (3). Both result in loss of PAX5 TF activity. A number of other hematopoietic TF genes are often targeted by copy-number alterations and/or mutations [e.g., EBF1 (46), IKZF1 (47), BTG1 (46)]. Gene fusions represent initiating events during early leukemogenesis, while small genomic aberrations often occur as late secondary events to potentiate and promote leukemogenic effects. There have been extensive studies on how TF gene fusions or mutations alter hematopoiesis, and the prevailing theory is that these genomic defects directly disrupt B-lymphoid cell development and create differentiation blockade (11, 48). The expansion of the immature progenitor cell pool increases the chances of acquiring oncogenic mutations and subsequent leukemic transformation. However, many TFs have complex functions (above and beyond transcription regulation during hematopoiesis), and it would be an oversimplification to assume that leukemia mutations in TF only affect B-cell differentiation.
Integrating germline and somatic genomic features of ALL offers a unique opportunity to identify interactions between leukemia and host genomes. For example, ARID5B risk variants are highly enriched in ALL with a hyperdiploid karyotype (24), whereas germline ETV6 variants and ETV6–RUNX1 fusion genes are mutually exclusive in ALL (27). Deregulation of TF genes probably drives preleukemic cells down a specific oncogenic pathway defined by characteristic somatic events. This type of integrated analysis is likely to shed new light on the roles of TF genes in ALL pathogenesis and lymphoid cell biology in general.
TFs Determine Molecular Subtypes and Prognostic Risk Groups of Childhood B-ALL
The genomic landscape of childhood B-ALL has been studied extensively (3, 49). Today, >90% of childhood ALL cases can be classified into specific genetic subgroups linked to distinct prognostic characteristics and treatment responses that have been extensively reviewed elsewhere (49). In general, B-ALLs are characterized by a very low mutational burden. However, in more than a third of pediatric patients with ALL (35%–50%), genetic alterations of B-cell TFs constitute the primary oncogenic event and determine the biological and clinical characteristics of the disease (49). Interchromosomal translocations generate fusion genes encoding chimeric TFs. Depending on the specific fusion gene present, prognostic risk groups can be determined, including low-risk (t12;21 coding for ETV6–RUNX1), intermediate-risk (t1;19 coding for TCF3–PBX1), and high-risk groups [KMT2A (MLL) translocations (11q23) and t17;19 coding for TCF3–HLF; ref. 49]. Some of the translocations can be acquired already in utero (50). Among them, MLL translocations are commonly strongly oncogenic and lead to poor prognostic infant ALL, usually without associated secondary mutations (51). However, ETV6–RUNX1 and TCF3–PBX1 mainly block B-cell differentiation and lead to expansion of pre–B-cell clones. These TF fusion genes are not sufficient to generate overt leukemia but depend on cooperating oncogenic secondary lesions to cause leukemia (50). These cooperating secondary aberrations frequently also affect B-cell TFs and are remarkably restricted to and recurrent for specific primary lesions (52). For instance, ETV6–RUNX1 is most frequently combined with loss of the second allele of ETV6, PAX5 deletion or downregulation, and mutations or loss of expression of the transcription cofactors BTG1 and TBL1XR1 (Transducin Beta Like 1 X-Linked Receptor 1). TCF3–PBX1 and TCF3–HLF are associated with PAX5 deletion/downregulation and TCF3 mutations (53). TCF3–HLF is further associated with deletions of BTG1 and VPREB1, although the number of studied cases is still low due to the rareness of this group (less than 1% of B-ALL cases). In general, PAX5 and IKZF1 deletions are common in ALL cases (15%) and are increased in high-risk ALL (up to 28% and 70%, respectively; refs. 3, 47). Importantly, lineage-specific targeted treatment may lead to occurrence of relapse due to cell adaptation. Recently, it was shown that 65% (13 of 20) of B-ALL cases relapsed after treatment with chimeric antigen receptor (CAR)-T cells targeting CD19. This was due to leukemic cells evolving to become CD19 negative (54). Therefore, therapies not only need to be adapted to the specific B-ALL developmental lineage but also need to take into account cooperative driver mutations to be successful (55).
Impaired Cell Fate during B-ALL Development
“Classic” bifurcating tree maps for the formation of the different blood cell types from HSCs depict strict compartments and a single route for the generation of differentiated cells, including B cells. However, recent work presents blood formation as the result of a continuous lineage priming (56), suggesting that individual hematopoietic precursors have a multitude of options as opposed to the classical sequential restriction-binary switch model (57). This new model would imply that the structure of the hematopoietic system is much less rigid than previously thought, and that the system could be more versatile. Previously identified precursor cells in fact seem to correspond to an amalgam of cells with plural differentiation potential (58). This is well exemplified by B-cell development as a lineage decision-making process where the ordered expression of TFs orchestrates B-cell lineage priming (Fig. 1). However, this process is much more plastic than described in the classic model. Plasticity of B-cell development was revealed by enforced expression of C/EBPα and C/EBPβ in B-cell precursors, which led to a reprogramming into macrophages (59). Recently, this plasticity has been further demonstrated by transient expression of the transcription factor Hoxb5 in precursor B cells, which was sufficient for stable conversion of B cells into T cells in vivo (60). This cell fate conversion occurs, in part, through the repression of TFs ensuring B-cell lineage priming (Ebf1, Pax5, Bcl11a, Foxp1, and Foxo1). Repression of Pax5 and Ebf1, for instance, is crucial for B-cell to T-cell conversion (15). This B-cell to T-cell conversion proves that a cell can adopt an alternative fate after having committed to another cell lineage, supporting the pair-wise model of hematopoiesis proposed by Brown and Ceredig (58). This pair-wise model does not assign a single path from HSCs to each of the various terminally differentiated blood cell types. Rather, the pair-wise model of hematopoiesis suggests a rainbow of pair-wise developmental options, gradually biased from the HSCs toward producing a specific blood cell type (Fig. 1).
A similar scenario also occurs in leukemogenesis, where leukemia cells largely belong to just one cell lineage, though many different leukemia types arise in a single stem/progenitor cell, which can give rise to many types of cells. ETV6–RUNX1 and BCR–ABLp190 are two of the most frequent drivers of B-ALL (49). Thus, at the start of B-ALL, a new but extraneous (malignant) fate must be imposed on the leukemia cell of origin to develop B-ALL (Fig. 1). The specific link between the ETV6–RUNX1 and BCR–ABLp190 oncogenes and human B-ALL development can be explained by two different interpretations. The classical explanation postulates that the fusion genes (ETV6–RUNX1 or BCR–ABLp190) are created in a committed/differentiated target B cell (61). Under this view, the phenotype of the leukemic B cell is conferred by the B-cell phenotype of the target cell. The second interpretation to explain this specific association between ETV6–RUNX1 and BCR–ABLp190 oncogenes and B-ALL is that these B-ALL–associated oncogenes are capable of imposing a leukemic B-cell fate onto a non-B target cell (62). Thus, the establishment of a B-cell tumor identity would require the first oncogenic hit to enforce an aberrant lineage program (62). It would be challenging, however, to verify this in human B-ALL, because the leukemias have evolved and undergone many mutations at the time of diagnosis (63). To prove that ETV6–RUNX1 and BCR–ABLp190 are able to impose a leukemic B-cell fate in non-B target cells, an experimental design limiting the expression of these oncogenes to non-B target cells would be needed, because under no other circumstances would it be possible to rule out a later role for ETV6–RUNX1 or BCR–ABLp190 once the leukemic B-cell phenotype is established. When the expression of either ETV6–RUNX1 or BCR–ABLp190 is restricted to hematopoietic stem/precursor cells in mice, the animals indeed develop exclusively B-ALL, which resembles the human disease (64, 65). These mouse models were designed to initiate ETV6–RUNX1 or BCR–ABLp190 expression in the HSC/progenitor population but to turn it down in committed B cells. The fact that only B-ALL emerges under these particular circumstances indicates that ETV6–RUNX1 and BCR–ABLp190 are able to impose a specific malignant B-cell fate. These findings link B-cell leukemogenesis with the aberrant B-cell lineage programming of early progenitors and show that oncogenes such as ETV6–RUNX1 and BCR–ABLp190 are able to define tumor cell identity during leukemogenesis. A similar scenario, where the induction of a new tumoral identity by the tumor genetic alteration occurs at the stem cell level, has also been described in other types of hematopoietic neoplasias and solid tumors (62). Thus, the oncogene-mediated restriction of the spectrum of options available to HSC to just one pathway/fate is central to the initiation of leukemia. It explains the association between specific oncogenes and the final phenotype of the hematopoietic neoplasia it triggers. Altogether, this evidence supports the idea that both normal B-cell development and B-cell leukemogenesis are cell lineage decision-making processes, with the leukemia-initiating events being “drivers” of leukemic B-cell lineage commitment.
Leukemic B-cell Priming and the Loss of B-cell Lineage–Specific Genes
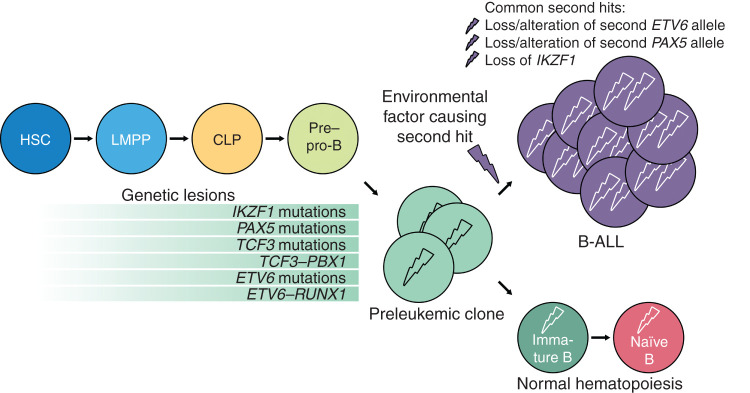
B-ALL is initiated by a first (pre)leukemic insult in a cell with the biological potential, intrinsic or acquired, to cause leukemia (Fig. 2). The first hit restricts the leukemia-initiating cells to a single-cell lineage. However, a single oncogenic insult is (with few exceptions) insufficient for B-ALL development, as shown in studies of twins with concordant childhood ALL and identical preleukemic translocations in their blood cells (66). Additional hits are necessary to convert the leukemia-initiating cell into a leukemic stem cell (LSC). Because the first hit imposes the leukemic B-cell lineage identity, what is the role of the second hit, mainly PAX5 and IKZF1 deletions, in this scenario? It has recently been shown that these lineage-specific genes are prominent DNA damage hotspots during leukemic transformation of B-cell precursors (67). The B-cell TF downregulation would not have an instructive role in the genesis of B-ALL, just a permissive one, preventing cells with the first oncogenic hit from being successfully terminally reprogrammed into leukemic B cells. This finding contrasts with Pax5 function in normal B cells, where deletion does not restrict precursor B cells in their lineage fate (68) and reprograms B cells into functional T lymphocytes (ref. 69; Fig. 2). In this regard, it has been proposed that the metabolic gatekeeper function of B-cell TFs may allow silent preleukemic clones to remain in a latent state (70). Thus, PAX5 and IKZF1 would limit the amount of cellular ATP to levels that are insufficient for malignant transformation of precursor B cells (65, 70). However, the development of B-ALL in mice where the expression of the first hit is restricted to HSCs indicates that the role of the B-cell TF loss during the leukemic B-cell priming might rely on a different function. The recent discovery of Ikaros acting as a guardian preventing autoimmunity by promoting BCR anergy and restraining TLR signalling is of immediate relevance to the persistence of preleukemic clones (71).
Figure 2.
Scheme of leukemic hematopoiesis. Cells develop from HSCs to pre–pro-B cells and acquire genetic lesions during these stages. It is not clear at what stage each of the mutations actually occurs. Common mutations of hematopoietic TFs are shown on light green background. These mutations lead to the formation of preleukemic clones. Without a second mutagenic event, normal hematopoiesis will be sustained. In case of a secondary oncogenic event, probably triggered by an environmental factor, B-ALL arises.
In p53-deficient cancers, the p53-mediated DNA damage response, which usually limits the reprogramming capacity of the first hit to ensure cell genomic integrity, is lost (62). B-cell TFs might have a similar role in constraining the malignant reprogramming function of the first hit. In this regard, P53 and PAX5 alterations seem to be mutually exclusive in human B-ALL development (63), and it has recently been shown that the reduction of Pax5 activity drastically accelerates the appearance of precursor B-ALL in mice where BCR–ABLp190 expression is restricted to hematopoietic stem/precursor cells (65). These results align with the fact that preleukemic clones carrying BCR–ABLp190 oncogenic lesions are frequently found in neonatal cord blood (72). However, they often remain silent, because the majority of these carriers do not develop B-ALL, supporting the theory that the first hit (BCR–ABLp190 gene) creates a preleukemic clone that remains clinically silent until secondary mutational events give rise to a full blown leukemia. Overall, these findings suggest that PAX5 downregulation does not have an instructive role in the genesis of B-ALL, just a permissive one, preventing cells with the first hit (BCR–ABLp190 gene) from being successfully terminally reprogrammed into leukemic B cells. Thus, reestablishing Pax5 function might be a therapeutic strategy for the eradication of leukemic cells and for blocking disease progression. As predicted, it has been shown that restoring endogenous Pax5 expression in leukemic B cells can trigger disease remission in mice (73). It is remarkable that the presence of Pax5 mediates B-cell commitment in normal development and that its absence is required to establish B-cell identity in ALL development (Fig. 2). However, although these results suggest that Pax5 downregulation plays a role in facilitating the restriction of cell lineage options to a leukemic B-cell lineage fate (lineage infidelity), such an activity has yet to be directly demonstrated. It would not be surprising if other important B-cell TFs (e.g., E2A/TCF3) contribute to the B-ALL development through a similar mechanism.
Gene–Environment Cooperations Are Novel Determinants in B-ALL Development
The ETV6–RUNX1 fusion gene is frequently found in neonatal cord blood, but only a few ETV6–RUNX1 carriers actually develop B-ALL (74). Similarly, pathogenic germline variants involving key lymphoid TFs, like PAX5 and IKZF1, predispose to B-ALL development (49). These acquired and germline alterations confer a low risk of developing B-ALL and represent the first oncogenic hit in the process of B-cell leukemogenesis. This first hit creates a preleukemic clone, but it needs secondary postnatal genetic alterations (“second hits”) to establish an irreversibly transformed state leading to the appearance of B-ALL. However, the mechanisms of leukemogenesis in individuals carrying a genetic predisposition remain uncertain. Identifying the factors causing the irreversibly transformed state has been particularly difficult because of the inherent challenge of detecting these stages in healthy children. In this regard, preclinical models of both ETV6–RUNX1-associated and PAX5-associated leukemia predisposition have been instrumental in uncovering a “gene–environment cooperation” as a novel determinant in the genesis of B-ALL (64, 75). This “gene–environment cooperation” refers to the increased likelihood of B-ALL development as a result of an increased sensitivity to specific environmental exposures in the presence of a genetic predisposition (Fig. 2). The cooperating oncogenic mutations can be triggered, for example, by environmental infectious exposure. Only together do both steps (genetic predisposition plus infection exposure) lead to overt leukemia in a proportion of predisposed mice, mimicking human B-ALL incidence associated with the same genetic alterations (64, 75). However, the second cooperating oncogenic mutation seems to be unique to each genetic predisposition. Consistent with this, wild-type mice (lacking genetic predisposition) never present with B-ALL when exposed to identical environmental infectious exposure (64, 75). Although these findings show that infectious exposure plays a role in enhancing B-ALL susceptibility of preleukemic carriers, this could be due to either direct induction of cooperating oncogenic mutations or by causing epigenetic reprogramming that, in turn, influences the specific second hit that cooperates with each predisposing alteration. However, such a mechanism has yet to be demonstrated. To this end, our capacity to model these early leukemia predisposition alterations triggering B-ALL initiation in vivo by infection exposure has opened new opportunities to study early B-ALL development and will help to unlock the mechanisms involved in infection-driven leukemogenesis. In addition, it will facilitate the discovery of how other relevant environmental factors might promote leukemogenesis in predisposed individuals. It will be exciting to see if elucidation of the gene–environment interaction in B-ALL development will lead to strategies for the prevention of B-ALL in children at risk.
Translational Implications of the Gene–Environment Cooperation
Prevention of cancer onset is one of the biggest scientific and clinical challenges in oncology. Although genomic profiling tests for the identification of children at risk are available (74), due to the lack of adapted therapeutic strategies, the identification of preleukemic clones or B-ALL–associated germline variants in children has no clinical consequence at present. Early detection of children at risk would require novel means of differentiating “true” children at risk that require intervention and the majority of predisposed children who will never develop B-ALL. The earliest possible identification of “true” children at risk will likely facilitate the prevention of progression to clinically relevant disease. Thus, the final challenge is to understand how these genetic variants contribute to B-ALL development, to develop preventive measures to reduce the incidence of the disease. To this end, preclinical models of B-ALL–associated germline variants in children will be essential tools for testing therapeutic options (e.g., vaccinations) to prevent the occurrence of this disease (64, 65, 75).
Conclusion
Early B-cell development and leukemia are both cell lineage–deciding processes where developmental options become restricted. The study of normal B-cell development has guided the current understanding of the molecular basis of B-cell malignancies. The genomic and molecular characterization of B-cell malignancies has contributed to our understanding of the molecular mechanisms that underlie normal B-cell development. The role reversal of TFs, like Pax5, in normal and leukemic B-cell development is remarkable. Although the molecular nature of B-ALL has now been defined, from a therapeutic perspective, it would be important to know whether the correlation between increased B-cell TF deletions and high-risk ALL is related to their function in promoting malignant B-cell lineage identity reprogramming. Although the exact nature of the “gene–environment cooperation” remains unknown, we have now the ability to model genetic B-ALL predisposition. This unlocks new opportunities for studying how B-ALL emerges. If we can understand how the “gene–environment interaction” is regulated, then we might learn how to intervene before a preleukemic condition evolves into leukemia. This knowledge would advance medicine and would have conceptual implications for other types of cancer associated with genetic predisposition. We expect that further understanding of both normal B cell and B-ALL development will ultimately generate the answers to these remaining clinical questions and will lead to the development of new therapeutic approaches to prevent disease leukemia development in children.
Disclosure of Potential Conflicts of Interest
D. Hein reports grants from the German Federal Office for Radiation Protection during the conduct of the study. C. Vicente-Dueñas reports grants from Instituto de Salud Carlos III–FEDER (CPII19/00024) and Instituto de Salud Carlos III - FEDER (PI17/00167) during the conduct of the study. No potential conflicts of interest were disclosed by the other authors.
Acknowledgments
The authors thank all the scientists who have contributed to this exciting field and apologize to those colleagues they were unable to cite. The authors thank all members of their groups for useful suggestions and for their critical reading of the manuscript. J.J. Yang has been supported by NIH grants P50GM115279 and P30CA21765. T. Ikawa has been supported by grants from the Japan Society for the Promotion of Science (16K15506 and 26293229) and Takeda Science Foundation. A. Borkhardt has been supported by the Katharina Hardt Stiftung. A. Borkhardt and U. Fischer have been supported by the German Federal Office for Radiation Protection (BfS)-Germany (FKZ: 3618S32275). U. Fischer has been supported by the German Carreras Foundation (DJCLS 21R/2019) and the Düsseldorf School of Oncology at the Heinrich-Heine University Düsseldorf. Research in I. Sánchez-García's group is partially supported by FEDER and by SAF2015-64420-R MINECO/FEDER, UE, RTI2018-093314-B-I00 MCIU/AEI/FEDER, UE, by Junta de Castilla y León (UIC-017, CSI001U16, and CSI234P18). The I. Sánchez-Garcia lab is a member of the EuroSyStem and the DECIDE Network funded by the European Union under the FP7 program. A. Borkhardt and I. Sánchez-Garcia have been supported by the German Carreras Foundation (DJCLS 07R/2019) and the Fundacion Unoentrecienmil (CUNINA project). A. Borkhardt, U. Fischer, C. Vicente-Dueñas, and I. Sánchez-García have been supported by the German Federal Office for Radiation Protection (BfS)-Germany (FKZ: 3618S32274). C. Vicente-Dueñas's group is partially supported by FEDER, “Miguel Servet” Grant (CPII19/00024 - AES 2017-2020) from the Instituto de Salud Carlos III (Ministerio de Economía y Competitividad), “Fondo de Investigaciones Sanitarias/Instituto de Salud Carlos III” (PI17/00167).
Footnotes
Blood Cancer Discov 2020;1:224–33
References
- 1.Cooper MD.The early history of B cells. Nat Rev Immunol 2015;15:191–7. [DOI] [PubMed] [Google Scholar]
- 2.Pulendran B, Ahmed R.Translating innate immunity into immunological memory: implications for vaccine development. Cell 2006;124:849–63. [DOI] [PubMed] [Google Scholar]
- 3.Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet 2019;51:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijayakrishnan J, Qian M, Studd JB, Yang W, Kinnersley B, Law PJ, et al. Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat Commun 2019;10:5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki K, Miyazaki M, Murre C.The establishment of B versus T cell identity. Trends Immunol 2014;35:205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, et al. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 2001;15:659–69. [DOI] [PubMed] [Google Scholar]
- 7.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 2005;121:295–306. [DOI] [PubMed] [Google Scholar]
- 8.Kondo M, Weissman IL, Akashi K.Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 1997;91:661–72. [DOI] [PubMed] [Google Scholar]
- 9.Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev 2009;23:2376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen CT, Ahsberg J, Sommarin MNE, Strid T, Somasundaram R, Okuyama K, et al. Dissection of progenitor compartments resolves developmental trajectories in B-lymphopoiesis. J Exp Med 2018;215:1947–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang SH, Carotta S, Nutt SL.Transcriptional control of pre-B cell development and leukemia prevention. Curr Top Microbiol Immunol 2014;381:189–213. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K.Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol 2006;7:382–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin H, Grosschedl R.Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 1995;376:263–7. [DOI] [PubMed] [Google Scholar]
- 14.Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol 2008;9:203–15. [DOI] [PubMed] [Google Scholar]
- 15.Nechanitzky R, Akbas D, Scherer S, Gyory I, Hoyler T, Ramamoorthy S, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol 2013;14:867–75. [DOI] [PubMed] [Google Scholar]
- 16.Ikawa T, Kawamoto H, Wright LY, Murre C.Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity 2004;20:349–60. [DOI] [PubMed] [Google Scholar]
- 17.Miyai T, Takano J, Endo TA, Kawakami E, Agata Y, Motomura Y, et al. Three-step transcriptional priming that drives the commitment of multipotent progenitors toward B cells. Genes Dev 2018;32:112–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R, Cauchy P, Ramamoorthy S, Boller S, Chavez L, Grosschedl R.Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev 2018;32:96–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johanson TM, Lun ATL, Coughlan HD, Tan T, Smyth GK, Nutt SL, et al. Transcription-factor-mediated supervision of global genome architecture maintains B cell identity. Nat Immunol 2018;19:1257–64. [DOI] [PubMed] [Google Scholar]
- 20.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 2010;11:635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature 2013;496:461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 2017;544:503–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bossen C, Murre CS, Chang AN, Mansson R, Rodewald HR, Murre C.The chromatin remodeler Brg1 activates enhancer repertoires to establish B cell identity and modulate cell growth. Nat Immunol 2015;16:775–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gocho Y, Yang JJ.Genetic defects in hematopoietic transcription factors and predisposition to acute lymphoblastic leukemia. Blood 2019;134:793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auer F, Ruschendorf F, Gombert M, Husemann P, Ginzel S, Izraeli S, et al. Inherited susceptibility to pre B-ALL caused by germline transmission of PAX5 c.547G>A. Leukemia 2014;28:1136–8. [DOI] [PubMed] [Google Scholar]
- 26.Churchman ML, Qian M, Te Kronnie G, Zhang R, Yang W, Zhang H, et al. Germline genetic IKZF1 variation and predisposition to childhood acute lymphoblastic leukemia. Cancer Cell 2018;33:937–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moriyama T, Metzger ML, Wu G, Nishii R, Qian M, Devidas M, et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: a systematic genetic study. Lancet Oncol 2015;16:1659–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noetzli L, Lo RW, Lee-Sherick AB, Callaghan M, Noris P, Savoia A, et al. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat Genet 2015;47:535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet 2013;45:1226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topka S, Vijai J, Walsh MF, Jacobs L, Maria A, Villano D, et al. Germline ETV6 mutations confer susceptibility to acute lymphoblastic leukemia and thrombocytopenia. PLoS Genet 2015;11:e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, et al. Loss of B cells in patients with heterozygous mutations in IKAROS. N Engl J Med 2016;374:1032–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boutboul D, Kuehn HS, Van de Wyngaert Z, Niemela JE, Callebaut I, Stoddard J, et al. Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J Clin Invest 2018;128:3071–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boisson B, Wang YD, Bosompem A, Ma CS, Lim A, Kochetkov T, et al. A recurrent dominant negative E47 mutation causes agammaglobulinemia and BCR(-) B cells. J Clin Invest 2013;123:4781–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Ali M, Yang J, Chan KW, Ben-Mustapha I, Mekki N, Benabdesselem C, et al. Homozygous transcription factor 3 gene (TCF3) mutation is associated with severe hypogammaglobulinemia and B-cell acute lymphoblastic leukemia. J Allergy Clin Immunol 2017;140:1191–4. [DOI] [PubMed] [Google Scholar]
- 35.Hock H, Meade E, Medeiros S, Schindler JW, Valk PJ, Fujiwara Y, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev 2004;18:2336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang MY, Churpek JE, Keel SB, Walsh T, Lee MK, Loeb KR, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet 2015;47:180–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hock H, Shimamura A.ETV6 in hematopoiesis and leukemia predisposition. Semin Hematol 2017;54:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst 2013;105:733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian M, Zhang H, Kham SK, Liu S, Jiang C, Zhao X, et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res 2017;27:185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monica K, LeBrun DP, Dedera DA, Brown R, Cleary ML.Transformation properties of the E2a-Pbx1 chimeric oncoprotein: fusion with E2a is essential, but the Pbx1 homeodomain is dispensable. Mol Cell Biol 1994;14:8304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zelent A, Greaves M, Enver T.Role of the TEL-AML1 fusion gene in the molecular pathogenesis of childhood acute lymphoblastic leukaemia. Oncogene 2004;23:4275–83. [DOI] [PubMed] [Google Scholar]
- 42.de Pooter RF, Kee BL.E proteins and the regulation of early lymphocyte development. Immunol Rev 2010;238:93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature 2018;562:373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun 2016;7:13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosoya T, Maillard I, Engel JD.From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol Rev 2010;238:110–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007;446:758–64. [DOI] [PubMed] [Google Scholar]
- 47.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009;360:470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Y, Yoshida T, Georgopoulos K.Transcriptional circuits in B cell transformation. Curr Opin Hematol 2017;24:345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pui CH, Nichols KE, Yang JJ.Somatic and germline genomics in paediatric acute lymphoblastic leukaemia. Nat Rev Clin Oncol 2019;16:227–40. [DOI] [PubMed] [Google Scholar]
- 50.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 1999;354:1499–503. [DOI] [PubMed] [Google Scholar]
- 51.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 2002;30:41–7. [DOI] [PubMed] [Google Scholar]
- 52.Papaemmanuil E, Rapado I, Li Y, Potter NE, Wedge DC, Tubio J, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet 2014;46:116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duque-Afonso J, Feng J, Scherer F, Lin CH, Wong SH, Wang Z, et al. Comparative genomics reveals multistep pathogenesis of E2A-PBX1 acute lymphoblastic leukemia. J Clin Invest 2015;125:3667–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gardner R, Wu D, Cherian S, Fang M, Hanafi LA, Finney O, et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood 2016;127:2406–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pui CH, Pei D, Raimondi SC, Coustan-Smith E, Jeha S, Cheng C, et al. Clinical impact of minimal residual disease in children with different subtypes of acute lymphoblastic leukemia treated with response-adapted therapy. Leukemia 2017;31:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 2017;19:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akashi K, Traver D, Miyamoto T, Weissman IL.A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000;404:193–7. [DOI] [PubMed] [Google Scholar]
- 58.Ceredig R, Rolink AG, Brown G.Models of haematopoiesis: seeing the wood for the trees. Nat Rev Immunol 2009;9:293–300. [DOI] [PubMed] [Google Scholar]
- 59.Xie H, Ye M, Feng R, Graf T.Stepwise reprogramming of B cells into macrophages. Cell 2004;117:663–76. [DOI] [PubMed] [Google Scholar]
- 60.Zhang M, Dong Y, Hu F, Yang D, Zhao Q, Lv C, et al. Transcription factor Hoxb5 reprograms B cells into functional T lymphocytes. Nat Immunol 2018;19:279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chessells JM, Hardisty RM, Rapson NT, Greaves MF.Acute lymphoblastic leukaemia in children: classification and prognosis. Lancet 1977;2:1307–9. [DOI] [PubMed] [Google Scholar]
- 62.Vicente-Duenas C, Hauer J, Cobaleda C, Borkhardt A, Sanchez-Garcia I.Epigenetic priming in cancer initiation. Trends Cancer 2018;4:408–17. [DOI] [PubMed] [Google Scholar]
- 63.Ma X, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018;555:371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez-Hernandez G, Hauer J, Martin-Lorenzo A, Schafer D, Bartenhagen C, Garcia-Ramirez I, et al. Infection exposure promotes ETV6-RUNX1 precursor B-cell leukemia via impaired H3K4 demethylases. Cancer Res 2017;77:4365–77. [DOI] [PubMed] [Google Scholar]
- 65.Martin-Lorenzo A, Auer F, Chan LN, Garcia-Ramirez I, Gonzalez-Herrero I, Rodriguez-Hernandez G, et al. Loss of Pax5 exploits Sca1-BCR-ABL(p190) susceptibility to confer the metabolic shift essential for pB-ALL. Cancer Res 2018;78:2669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maia AT, van der Velden VH, Harrison CJ, Szczepanski T, Williams MD, Griffiths MJ, et al. Prenatal origin of hyperdiploid acute lymphoblastic leukemia in identical twins. Leukemia 2003;17:2202–6. [DOI] [PubMed] [Google Scholar]
- 67.Boulianne B, Robinson ME, May PC, Castellano L, Blighe K, Thomas J, et al. Lineage-specific genes are prominent DNA damage hotspots during leukemic transformation of B cell precursors. Cell Rep 2017;18:1687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nutt SL, Heavey B, Rolink AG, Busslinger M.Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 1999;401:556–62. [DOI] [PubMed] [Google Scholar]
- 69.Rolink AG, Nutt SL, Melchers F, Busslinger M.Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature 1999;401:603–6. [DOI] [PubMed] [Google Scholar]
- 70.Chan LN, Chen Z, Braas D, Lee JW, Xiao G, Geng H, et al. Metabolic gatekeeper function of B-lymphoid transcription factors. Nature 2017;542:479–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwickert TA, Tagoh H, Schindler K, Fischer M, Jaritz M, Busslinger M.Ikaros prevents autoimmunity by controlling anergy and Toll-like receptor signaling in B cells. Nat Immunol 2019;20:1517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cazzaniga G, van Delft FW, Lo Nigro L, Ford AM, Score J, Iacobucci I, et al. Developmental origins and impact of BCR-ABL1 fusion and IKZF1 deletions in monozygotic twins with Ph+ acute lymphoblastic leukemia. Blood 2011;118:5559–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu GJ, Cimmino L, Jude JG, Hu Y, Witkowski MT, McKenzie MD, et al. Pax5 loss imposes a reversible differentiation block in B-progenitor acute lymphoblastic leukemia. Genes Dev 2014;28:1337–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schafer D, Olsen M, Lahnemann D, Stanulla M, Slany R, Schmiegelow K, et al. Five percent of healthy newborns have an ETV6-RUNX1 fusion as revealed by DNA-based GIPFEL screening. Blood 2018;131:821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin-Lorenzo A, Hauer J, Vicente-Duenas C, Auer F, Gonzalez-Herrero I, Garcia-Ramirez I, et al. Infection exposure is a causal factor in B-cell precursor acute lymphoblastic leukemia as a result of Pax5-inherited susceptibility. Cancer Discov 2015;5:1328–43. [DOI] [PubMed] [Google Scholar]