Abstract
A substantial amount of research is being conducted on zonation markers to identify hepatic injuries and disorders based on the structural and functional zonation of the liver. In contrast to metabolic zonation, hepatocyte ploidy reflects the capability of liver regenerative turnover. Nonetheless, many knowledge gaps remain in the understanding of the links between liver disorders and altered zonation and ploidy, partially owing to the lack of sufficient zonation markers. Under this setting, we recapitulated the currently known and prospective markers used to identify normal and altered liver zonation in different disorders. Furthermore, we discussed new findings from studies that have used advanced methodologies to identify potential markers with greater accuracy. We also elaborated on the perspectives and future applications of zonation research in the early detection of various liver diseases.
1. Introduction
The concept of liver zonation can be traced back to the 1960s when it was first recognized in mice by Neporent and Glicksman [1]. Since that time, zonation has been studied in liver diseases [2]. With a uniform anatomical structure, the liver consists of hexagonal lobules in the form of a honeycomb. At each angle of the lobules, the portal veins, hepatic arterioles, and bile ducts build up the “portal triads,” whereas central veins are located in the center of the lobules (Figure 1). Hepatocytes are arranged in a spongy manner along the porta-central axis, and their biochemical and physiological functions vary. Six to eight periportal hepatocytes (zone 1) are located in the vicinity of afferent periportal zones, while two to three pericentral hepatocytes (zone 3) are adjacent to the efferent pericentral zones. Zone 2, with six to ten hepatocytes, is located in the middle lobules with unclear boundaries between zones 1 and 3 [3]. All these structural characteristics are termed liver zonation [4].
Figure 1.
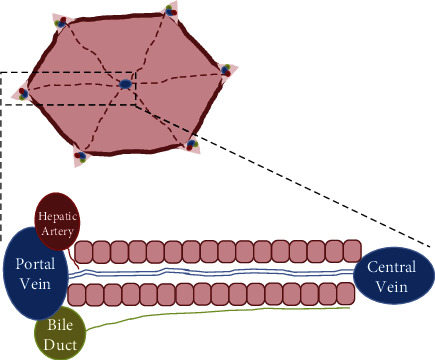
Diagram of the zonal areas in liver lobules. The liver consists of a regular arrangement of hexagonal lobules. At each angle of the lobules, the portal veins, hepatic arterioles, and bile ducts constitute the “portal triads”, whereas central veins are in the center of the lobules.
Liver spatial heterogeneity is manifested by metabolic zonation, as characterized by the varying expression profiles of metabolic genes from the periportal to the pericentral zones, as well as polyploidy of the hepatocytes [5]. In contrast to the distribution of metabolic zonation, hepatocyte polyploidy reflects the capability of liver regenerative turnover [6]. The diploid Axin2+ hepatocytes were documented to be more likely the progenitors to fuel polyploid hepatocytes in normal physiology and responses to various stresses [6]. However, other reports also argued that after acute injury, hepatocytes proliferated and regenerated the liver in all regions, in which limited contribution was observed from Axin2+ hepatocytes [7]. Moreover, in chronic liver diseases, hepatocyte proliferation was impaired, and instead, the cholangiocytes, located in bile ducts, played a vital role in substituting hepatocytes [8]. In the severely damaged liver, the cholangiocytes were thought to be one of the sources of hepatic progenitor cells, which bore the ability to differentiate into hepatocytes [9, 10]. Previous findings suggested that hepatocytes with different ploidy could be used to distinguish the different zones of the liver. In rodent liver, hepatocytes are diploid with two haploid chromosome sets (2c) at birth, while in adults, most hepatocytes are polyploid, tetraploid (4c), octoploid (8c), or even contain more haploid chromosome sets (e.g., 16c). Nonetheless, the spatial distribution of diploidy and overall polyploidy is still warranted to be studied. Some studies demonstrated that hepatocytes around pericentral zones are diploid, differing from polyploid hepatocytes around the periportal zones [6]. However, there are still some conflicting reports on this aspect, as multinucleated hepatocytes are found next to the pericentral zones [11]. In addition, some studies have argued that 4c and 8c polyploid hepatocytes accumulate in the midlobule zone, rather than in the periportal and pericentral zones (Figure 2) [12]. These inconsistent findings challenge the feasibility of ploidy as a zonation reference.
Figure 2.
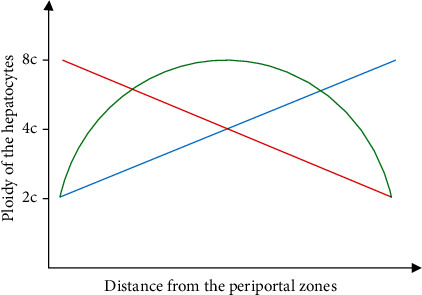
Differential distribution profiles of the hepatocyte ploidy. Although a large number of studies have been performed to shed light on the distribution of hepatocyte ploidy, there are still remarkable controversies to date. Some studies demonstrate that the hepatocytes around the pericentral regions are diploid, different from the polyploid hepatocytes around the periportal regions (red line). However, other reports suggest that multinucleated hepatocytes are adjacent to the pericentral regions (blue line). In the meantime, it has been argued that 4c and 8c polyploid hepatocytes localize in the midzone, rather than in the periportal and pericentral regions (green line).
Genes and biomarkers are being studied as zonation markers to identify liver disorders. Furthermore, more prospective zonation markers are being discovered using new technologies. Nonetheless, more efforts are warranted to improve the poor accuracy and low sensitivity of the currently known liver zonation markers in diagnosing liver injuries and disorders. In the current review, we recapitulated the known and potential markers of liver zonation and assessed their applications and limitations in identifying liver injuries and disorders. Some newly discovered molecules are also discussed as prospective zonation markers to identify liver disorders.
2. Current Markers Used to Identify Liver Zonation
2.1. The Basis of Functional Zonation of the Liver
As an essential metabolic organ, the liver exerts functional zonation to efficiently conduct metabolic processes [13]. The portal veins supply the liver with nutrient-rich blood (75% of the total blood supply to the liver), while hepatic arterioles provide the liver with oxygen-rich blood (25% of the total blood supply to the liver). The blood enters the sinusoids and is concentrated in the pericentral zones. The liver cell plate, which is composed of 15–25 hepatocytes, localizes from the periportal to the pericentral zones, and these hepatocytes can be divided into three different zones based on their metabolic activity (zones 1–3). The function of these hepatocytes relies on their location within the liver plate (Figure 3) [3]. Moreover, pericentral regions are more specialized in antioxidative stress and detoxification activities, carried out by glutamine synthetase (GS) and cytochrome P450 (CYP450) enzymes, as well as glycolysis, lipogenesis, and bile acid synthesis. Periportal regions undertake several energy-consuming processes, including cholesterol (CHO) synthesis, urea synthesis, gluconeogenesis, and fatty acid oxidation, and exhibit the highest level of albumin expression (Figure 3) (Table 1) [13, 14].
Figure 3.
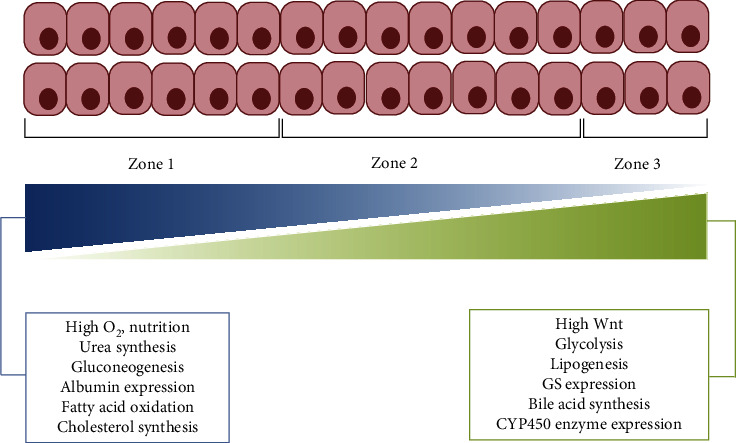
A schematic displays the gradient patterns of basal metabolites across the three zones. The liver cell plate, composed of 15–25 hepatocytes, lies between the periportal and pericentral zones. Six to eight periportal hepatocytes (zone 1) enclose the periportal zone, while two to three pericentral hepatocytes (zone 3) are located near the pericentral zone. The boundary of zone 2, consisting of six to ten hepatocytes, in the middle lobules is still not well-defined. From zone 1 to zone 3, the concentration of O2, the synthesis of urea and cholesterol, the gluconeogenesis, the expression of albumin, and the oxidation of fatty acid decrease, whereas Wnt signaling, the expression of glutamine synthetase (GS), and the activity of cytochrome P450 (CYPs) enzyme, the glycolysis, the lipogenesis, and the synthesis of bile acid otherwise increase.
Table 1.
Current biochemical indicators used as markers to identify liver zonation.
| Zone | Biological processes | Biochemical indicators | Detection methods | References |
|---|---|---|---|---|
| Pericentral zones | Glutamine synthetase expression | Glutamine synthetase | Glutamine synthetase assay | [13] |
| Cytochrome P450 (CYPs) enzyme expression | CYP450 | CYP450 ELISA | ||
| Bile acid synthesis | Bile acids | Total bile acid assay | ||
| Lipogenesis | Acetyl-CoA carboxylase | Acetyl-CoA carboxylase assay and western blot | [23] | |
| ATP citrate lyase | ATP citrate lyase assay | |||
| Glycolysis | Glycolytic enzymes | Protein levels of hexokinase II and lactate dehydrogenase-A | [43] | |
| Lactate | Lactic acid assay | |||
| Lactate dehydrogenase | Lactate dehydrogenase assay | |||
|
| ||||
| Periportal zones | Cholesterol synthesis | Cholesterol | Total cholesterol assay | [13] |
| Urea synthesis | Urea | Urea assay | ||
| Albumin expression | Albumin | Albumin assay | [14] | |
| Fatty acid oxidation | Carnitine palmitoyltransferase-1 | Carnitine palmitoyltransferase-1 assay | [23] | |
| Gluconeogenesis | Gluconeogenic genes and protein expression | qRT-PCR and western blot | ||
| Glucose | Blood glucose monitor | |||
2.2. Markers for Normal Functional Zonation of the Liver
Although CYtP450 enzymes are expressed in all hepatocytes, most xenobiotic metabolism is performed by approximately 50% of the hepatocytes located in the pericentral regions [15]. Moreover, GS, which is expressed in the pericentral regions, catalyzes the condensation of ammonia by converting glutamate to glutamine [16]. Therefore, the expression of CYPs and GS may be a potential marker for the functional zonation of the pericentral regions.
Wnt/β-catenin, a master regulator of hepatocyte proliferation and liver metabolic zonation, is activated in the pericentral zones. Adenomatous polyposis bacillus, a negative regulator of Wnt signaling, is absent in the periportal regions and is recognized as a zonation-keeper of the liver [3]. Progenitor cells are widely identified by their preferential expression of progenitor cell markers, such as Axin2 and Tbx3. The expression of Axin2, a transcriptional target gene downstream of Wnt/β-catenin signaling, offers a reliable readout of the response of hepatocytes to Wnt signaling. Moreover, the Wnt protein acts as a transient signal that maintains the pluripotency of stem cells in the liver, as Tbx3, another target gene downstream of Wnt signaling, is identified as a stem cell marker expressed by pericentral hepatocytes [6]. Thus, Axin2 and Tbx3 could be used as functional zonation markers for the pericentral regions in order to maintain liver topology (Table 2).
Table 2.
Current genes used as markers to identify liver zonation.
| Zone | Gene symbol | Biological function | Detection methods | Sources | References |
|---|---|---|---|---|---|
| Pericentral zones | Cyp1a | CYPs serve to detoxify xenobiotics prior to being drained into the pericentral zone. | Frozen section + immunostaining | Rat anti-CYP1A2 antibody | [15] [44] |
| Cyp3a | Rabbit anti-CYP3A1 antibody, Biotrend, Germany | ||||
| Cyp2c | Rat anti-CYP2C6 antibody | ||||
| Cyp2e1 | Frozen/FFPE section + immunostaining | Rabbit anti-CYP2E1 antibody, Sigma-Aldrich, USA | |||
| Gs | GS catalyzes the condensation of ammonia by converting glutamate to glutamine. | Frozen section + immunostaining | Mouse anti-GS antibody, BD Bioscience, Germany RNAscope probes: NM 008131, region 103-973 Anti-GS (clone 2B12) antibody, Sigma-Aldrich, USA |
[16] | |
| In situ hybridization | |||||
| Western blot | |||||
| qRT-PCR | |||||
| Axin2 | A universal transcriptional target of Wnt/β-catenin signaling is Axin2, whose expression offers a reliable readout of hepatocytes responding to Wnt. | In situ hybridization | RNAscope probes: NM 015732, region 330-1287 Chicken anti-GFP antibody, Abcam ab13970, UK |
[6] | |
| Frozen section + immunostaining (GFP-tag model) | |||||
| Tbx3 | Tbx3 is another target gene of Wnt signaling, which has been identified as a progenitor marker of hepatocytes. | In situ hybridization | RNAscope probes: NM 198052 | ||
|
| |||||
| Periportal zones | Arg1 | Arginase converts L-arginine to L-ornithine and urea, affecting hepatic nitric oxide synthase (NOS) activities by decreasing L-arginine bioavailability. | Frozen section + immunostaining Western blot qRT-PCR | Rabbit anti-arginase-1 antibody, Sigma-Aldrich, USA Anti-arginase-1 antibody, BD Transduction Laboratories, CA |
[17] |
| Cps1 | CPS is an ATP-dependent enzyme and converts ammonia and carbon dioxide (CO2) to carbamoyl phosphate. | Frozen section + immunostaining qRT-PCR | Rabbit anti-CPS1 antibody, Abcam, UK | ||
| Mfsd2α | Mfsd2a, previously recognized as a protein that maintains blood-brain barrier function, is a periportal zonation marker. | Frozen section + immunostaining (RFP-tag model) | Rabbit anti-RFP antibody, Rockland 600-401-379, USA | [18] | |
Note: Cyp: cytochrome P450; Gs: glutamine synthetase; Tbx3: T-box transcription factor 3; Arg1: Arginase1; Cps1: carbamoyl phosphate synthase 1; Mfsd2a: major facilitator superfamily domain-containing 2a; FFPE: formalin-fixed paraffin-embedded.
In the periportal regions, hepatocytes express a high level of arginase and carbamoyl phosphate synthetase-1 (CPS1) to carry out metabolic functions. Arginase converts L-arginine to L-ornithine and urea, and thus affects the activity of hepatic nitric oxide synthase by reducing L-arginine bioavailability. CPS1, an ATP-dependent enzyme, converts ammonia and carbon dioxide to carbamoyl phosphate [17]. Thus, both arginase and CPS1 could serve as functional zonation markers of the periportal regions. Major facilitator superfamily domain containing 2a (Mfsd2a), which was previously recognized as a protein that maintains the blood-brain barrier, could also be a functional zonation marker of the periportal zones (Table 2) [18].
3. Disordered Liver Zonation Points to Abnormal Metabolism and Potential Risks of Morbidity
The great potential of liver zonation resides in its potential to aid the diagnosis of abnormal liver metabolism and risks for various diseases. In 1996, Lorraine et al. investigated zonal metabolic enzymes and found that marked metabolic zonation of normal lobules was retained in fibrotic hepatic lobules in humans; however, this zonation pattern was lost in cirrhotic lobules [19]. Although diverse liver disorders and injuries may alter zonation, reports on diagnosis using zonation markers are still limited to only a few liver diseases and injuries, including nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), and hepatocellular carcinoma (HCC), as discussed below.
3.1. Hepatic Metabolic Diseases
The liver is the main organ for the metabolism of fats, carbohydrates, urea, bilirubin, and vitamins [20]. Thus, biochemical indicators are closely involved in the metabolic processes in the liver and are frequently used as markers to identify liver zonation. With an increasing number of cases of hepatic metabolic diseases that cause severe damage to liver functions and even systemic impairments, fundamental studies on markers are necessary to determine the hepatic metabolic zonation profiles in both healthy and diseased conditions. For example, canonical Wnt/β-catenin and hypoxia-inducible factor signaling pathways were found to be connected with altered liver zonation together with the pathogenesis of hepatic metabolic diseases [21, 22]. Lipogenesis mainly occurs in pericentral hepatocytes, while fatty acid oxidation occurs in periportal hepatocytes [23]. Furthermore, fatty acid uptake is greater in periportal regions, and the expression of fatty acid-binding protein tends to decline from the periportal to pericentral zones. Together, these findings suggest that the genes and biomarkers involved in fatty acid metabolism could be prospective zonation markers for identifying hepatic metabolic diseases [24].
NAFLD, which is present in 25% of the world's population, starts with the uncontrolled accumulation of fat in hepatocytes, causing inflammation and fibrosis in the liver [25]. In NAFLD patients, hepatic steatosis is obvious in pericentral hepatocytes [14]. A high-fat diet was found to induce the deposition of triglycerides (TGs) in areas surrounding the periportal zones in NAFLD animal models, suggesting that TGs might be a marker to identify NAFLD (Table 3) [26]. Different types of high-fat diets (e.g., solid vs. liquid) might result in divergent deposition patterns of TGs, which is indicative of different zonation patterns [27]. NASH frequently causes hepatic fibrosis by inducing hepatocyte injury and inflammatory cell infiltration. Although the level of serum bile acids is elevated, and the lysophosphatidylcholine level is reduced, in NASH patients, the liver zonation changes in NASH patients, and their contribution to NASH remain largely unknown [28]. The enrichment of arachidonic acid (AA) in pericentral hepatocytes leads to enhanced oxidative stress and consequent damage to pericentral regions in mice with NASH [29]. Additionally, the spatial distribution of the remodeling enzyme, lysophosphatidylcholine acetyltransferase 2, plays a role in changing the zonal location of AA-containing lipids [29]. Thus, AA-containing lipids might be used as markers for predicting the risk of zonal NASH. However, there is still no reported zonation marker to determine altered glycolysis and gluconeogenesis under hepatic metabolic diseases.
Table 3.
Newly-defined molecules as prospective markers to identify liver zonation.
| Zone | Potential markers | Biological processes/function | Detection methods | References |
|---|---|---|---|---|
| Pericentral zone | ICAM1 | This gene encodes cell surface glycoproteins mainly expressed on endothelial cells. | Immunostaining (protein) Diffusion pseudotime (DPT) analysis and self-organizing maps (SOMs) (gene) |
[40] [45] |
| GLUL | The protein encoded by this gene plays a role in acid-base homeostasis, ammonia and glutamate detoxification, and cell proliferation. | Immunostaining (protein) DPT and SOMs (gene) |
||
| ENG | This gene encodes a transmembrane protein, which is a glycoprotein of the vascular endothelium. | Immunostaining (protein) DPT and SOMs (gene) |
||
| miR-93 | miR93 inhibits the expression of transcription factor 7-like 1 (Tcf7l1). | qRT-PCR | [34] | |
|
| ||||
| Periportal zone | ALB | The protein encoded by this gene plays a role in the regulation of blood plasma colloid osmotic pressure and acts as a carrier protein. | DPT and SOMs (gene) | [40] [45] |
| BTNL9 | The expression of the protein encoded by this gene is found in several types of cancers. | DPT and SOMs (gene) | [40] | |
| ANPEP | The protein encoded by this gene is expressed in plasma membranes. | Immunostaining (protein) DPT and SOMs (gene) |
||
| PCK1 | This gene is the main control point for the regulation of gluconeogenesis. | Immunostaining (protein) DPT and SOMs (gene) |
[40] [45] [46] |
|
| MTHFS | The protein encoded by this gene is an enzyme that catalyzes the conversion of 5-formyltetrahydrofolate to 5,10-methenyltetrahydrofolate. | Immunostaining (protein) DPT and SOMs (gene) |
[40] [46] |
|
| GATM | This gene encodes a mitochondrial enzyme that is involved in creatine biosynthesis. | Immunostaining (protein) DPT and SOMs (gene) |
||
| Liver fatty acid-binding protein (L-FABP) | This protein controls fatty acid uptake. | Immunostaining | [24] | |
| Triglyceride (TG) (high-fat diet (HFD)-induced nonalcoholic fatty liver disease (NAFLD) mouse model) | TG is the composition of lipids. | TG assay (liver and plasma) | [26] | |
| miR-99a and miR-100 | They inhibit the expression of frizzled class receptor 8 (Fzd8). | qRT-PCR | [34] | |
Note: ICAM1: intercellular adhesion molecule 1; GLUL: glutamate-ammonia ligase; ENG: endoglin; ALB: albumin; BTNL9: butyrophilin 9; ANPEP: alanyl aminopeptidase; PCK1: phosphoenolpyruvate carboxykinase 1; MTHFS: methenyltetrahydrofolate synthetase; GATM: glycine amidinotransferase.
3.2. Hepatocellular Carcinoma (HCC)
HCC is a highly prevalent cancer and is the third leading cause of death among cancers [21]. Unfortunately, limited therapeutic choices are available for patients with advanced HCC, yielding an urgent demand for the early detection and diagnosis of this disease [30]. Alpha-fetoprotein (AFP) is a known marker for HCC, and its levels are elevated up to 70% in the sera of HCC patients. Thus far, AFP has been widely used as an adjuvant diagnostic index. Nonetheless, the pathophysiological knowledge of AFP expression in different liver zones remains elusive [30]. Similarly, neuropilin 1, a transmembrane glycoprotein with increased expression in HCC, is also a novel HCC marker [31]. Although considerable progress has been made in identifying biomarkers for HCC, little insight has been gained on their likely zonated expression in HCC patients. Intriguingly, HCC manifests a high predilection of invasion towards portal veins, leading to an extremely poor prognosis [32]. Portal veins are located near the bile ducts. To this end, HCC patients with a bile duct thrombus display differential progression of HCC, including a higher rate of lymphovascular invasion, macrovascular invasion, and poor differentiation [33].
The Wnt signaling pathway, which is more active in pericentral liver regions, is closely implicated in HCC. Wnt receptors (e.g., frizzled class receptor 7 (Fzd7) and Fzd8) are pericentrally zonated. In contrast, Wnt inhibitors (e.g., catenin beta interacting protein 1 and transcription factor 7-like 1 (Tcf7l1)) are periportally zonated [34]. Moreover, microRNAs (miRNAs), which are also differentially expressed among zones in the liver, contribute to the zonated activity of Wnt signaling. For example, the pericentrally upregulated expression of miR-93 inhibits Tcf7l1 expression, leading to increased Wnt signaling activity in the pericentral zones. In contrast, the periportally downregulated expression of miR-99a and miR-100 leads to a loss in their capability to inhibit the expression of Fzd8, and thus enhances Wnt activity in the periportal zones [34]. Under this setting, the expression patterns of these miRNAs might be used as zonation markers to detect early HCC.
3.3. Liver Injuries following Toxin Exposure
Exposure to chemicals and even particulate matter can give rise to liver impairments. Along with the increasing understanding of injury-related zonation, some markers have been suggested to predict the risk of liver injury. For example, the accumulation of copper and iron is graded by location (pericentral, midzonal, and periportal zones) [35]. From the pericentral to the periportal zones, the concentration of iron gradually decreases, while the concentration of copper increases [36]. Moreover, the zonated accumulation of some metals, particularly copper and iron, between the periportal and pericentral zones, is altered, and even reversed, in rats upon exposure to 3,3′,4,4′,5-pentachlorobiphenyl [36], suggesting that copper and iron zonation might be potential indicators for detecting the risk of liver injury induced by chemicals.
Other toxic xenobiotics, such as CCl4, an established chemical to induce animal models of hepatic fibrosis, are metabolically activated by CYPs, such as CYP1A, CYP2C, CYP2E1, and CYP3A, and cause hepatocyte necrosis in a pericentral pattern [15]. Acetaminophen intoxication has also been shown to cause necrosis of pericentral hepatocytes [34]. Therefore, CYPs might be used as a potential indicator of pathological changes in the pericentral regions [15]. Moreover, miRNAs are stable in circulation and could be potential biomarkers to indicate the risk of hepatic injuries. A retrospective study revealed that most pericentral miRNAs were enriched in plasma, but not in the periportal zones in acetaminophen-treated mice [34]. Additionally, L-arginine treatment induced a clear zonation of liver injury, as characterized by congested sites around pericentral zones and invasion of inflammatory cells around periportal zones [37]. In addition to metabolic functions, the localization of toxins is also crucial in determining their zonated toxicity. For instance, graphene oxide changes the expression levels of some proteins, such as E-cadherin, due to its preferential accumulation in the periportal zones [38]. Therefore, different toxins may elicit distinct effects on liver zonation, which makes it difficult to obtain universal zonation markers to detect liver injuries for all toxins. Therefore, both universal and specific markers are required.
4. Continued Efforts in Search of New Zonation Markers
Although some biomarkers and genes have been reported as markers for identifying liver zonation in both normal and pathological states, issues in the early detection of liver disorders have not been addressed. To overcome the technical challenges, some emerging strategies and techniques, such as single-cell RNA sequencing (scRNA-seq), are being employed to shed light on the heterogeneity of hepatocytes and disease-related cellular reprogramming [14]. In combination with single-molecule fluorescence in situ hybridization, scRNA-seq can generate a high-resolution visual map of RNA expression in spatial and global dimensions of the liver, making it feasible to explore more accurate zonation markers in the future [39].
With the aid of scRNA-seq technology, which helped determine hepatic transcriptomes at the single-cell level, hepatocytes and liver sinusoidal endothelial cells (LSECs) were identified to reveal marked features of zonation in mouse and human livers [40, 41]. The zonation profiles for hepatocytes are shown below (Table 3). By analyzing hepatocyte-characterized zonation, some genes, such as ALB and PCK1, are located in periportal zones, while GLUL is expressed in pericentral zones. By analyzing LSEC-characterized zonation, 806 genes (out of a total of 1198 genes) were found to have zonal expression profiles, including ICAM1 and ENG (central region), and BTNL9, ANPEP, MTHFS, and GATM (periportal region). Lymphatic vessel endothelial hyaluronan receptor 1 and CD14, which have been validated at the protein level, are recognized as markers to distinguish between central and midzonal LSECs [42]. Thus, zonal protein profiles of these genes might be further developed as prospective markers to distinguish the different zones of the liver (Table 3) [40].
5. Concluding Remarks and Perspectives
Liver zonation is important for the physiology of the liver, but disordered zonation is also closely associated with diverse liver diseases. Liver zonation is currently defined through zonation markers, such as biochemical indicators and genes. However, these zonation markers are subject to change under various pathological conditions, and the changes in these markers, in turn, could be used to recognize liver disorders and injuries, especially at the early stage. Despite recent encouraging progress, more selective zonation markers with higher accuracy are needed for early diagnosis.
In general, more work is required in the study of zonation markers. First, specific zonation markers have not yet been identified to represent certain liver disorders, and extensive work is needed to find accurate zonation markers for the early detection of liver disorders. Second, new markers, or marker toolboxes, should be identified with the aid of advanced biotechnologies, such as scRNA-seq. With these new markers, more information would be obtained to elucidate liver metabolism under normal physiological and diseased conditions. Third, given biomarkers for liver regeneration and progenitor cells, a new opportunity to decipher the mechanism(s) underlying the formation of liver zonation would be possible. Therefore, additional work to determine the molecular bases for self-renewal regulation is warranted. Finally, current methods to identify liver zonation mainly rely on immunohistochemical staining, the polymerase chain reaction, and other sophisticated technologies. However, these methods involve difficult and expensive experimental procedures. Thus, simplified readouts should be developed for the detection of altered liver zonation, such as serum biochemical markers, which will be insightful in detecting changes in liver zonation through conventional examinations.
Contributor Information
Sijin Liu, Email: sjliu@rcees.ac.cn.
Shuguang Chen, Email: cshuguang6@sina.com.
Data Availability
No data were used to support the study.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Qiuyuan Yang and Shuping Zhang equally contributed to this work.
References
- 1.Neporent L. M., Glicksman A. S. Liver glycogen zonation in the mouse following fructose and glucose injection. The American Journal of Anatomy. 1961;109(1):15–23. doi: 10.1002/aja.1001090103. [DOI] [PubMed] [Google Scholar]
- 2.Demura H. The adrenal glands in liver disease. The Tohoku Journal of Experimental Medicine. 1962;78(3):274–292. doi: 10.1620/tjem.78.274. [DOI] [PubMed] [Google Scholar]
- 3.Torre C., Perret C., Colnot S. Molecular determinants of liver zonation. Progress in Molecular Biology and Translational Science. 2010;97:127–150. doi: 10.1016/B978-0-12-385233-5.00005-2. [DOI] [PubMed] [Google Scholar]
- 4.Gebhardt R., Matz-Soja M. Liver zonation: novel aspects of its regulation and its impact on homeostasis. World Journal of Gastroenterology. 2014;20(26):8491–8504. doi: 10.3748/wjg.v20.i26.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katsuda T., Hosaka K., Matsuzaki J., et al. Transcriptomic dissection of hepatocyte heterogeneity: linking ploidy, zonation, and stem/progenitor cell characteristics. Cellular and Molecular Gastroenterology and Hepatology. 2020;9(1):161–183. doi: 10.1016/j.jcmgh.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang B., Zhao L., Fish M., Logan C. Y., Nusse R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature. 2015;524(7564):180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun T., Pikiolek M., Orsini V., et al. AXIN2+ pericentral hepatocytes have limited contributions to liver homeostasis and regeneration. Cell Stem Cell. 2020;26(1):97–107.e6. doi: 10.1016/j.stem.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Raven A., Lu W.-Y., Man T. Y., et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547(7663):350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irie T., Asahina K., Shimizu-Saito K., Teramoto K., Arii S., Teraoka H. Hepatic progenitor cells in the mouse extrahepatic bile duct after a bile duct ligation. Stem Cells and Development. 2007;16(6):979–988. doi: 10.1089/scd.2007.0037. [DOI] [PubMed] [Google Scholar]
- 10.Russell J. O., Lu W.-Y., Okabe H., et al. Hepatocyte-specific β-catenin deletion during severe liver injury provokes cholangiocytes to differentiate into hepatocytes. Hepatology. 2019;69(2):742–759. doi: 10.1002/hep.30270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeCluyse E. L., Witek R. P., Andersen M. E., Powers M. J. Organotypic liver culture models: meeting current challenges in toxicity testing. Critical Reviews in Toxicology. 2012;42(6):501–548. doi: 10.3109/10408444.2012.682115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanami S., Ben-Moshe S., Elkayam A., Mayo A., Bahar Halpern K., Itzkovitz S. Dynamic zonation of liver polyploidy. Cell and Tissue Research. 2017;368(2):405–410. doi: 10.1007/s00441-016-2427-5. [DOI] [PubMed] [Google Scholar]
- 13.Kietzmann T. Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biology. 2017;11:622–630. doi: 10.1016/j.redox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong X., Kuang H., Liu T., Lin J. D. A single-cell perspective of the mammalian liver in health and disease. Hepatology. 2020;71(4):1467–1473. doi: 10.1002/hep.31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghallab A., Myllys M., Holland C. H., et al. Influence of liver fibrosis on lobular zonation. Cell. 2019;8(12):p. 1556. doi: 10.3390/cells8121556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hakvoort T. B. M., He Y., Kulik W., et al. Pivotal role of glutamine synthetase in ammonia detoxification. Hepatology. 2016;65(1):281–293. doi: 10.1002/hep.28852. [DOI] [PubMed] [Google Scholar]
- 17.Malleske D. T., Rogers L. K., Velluci S. M., et al. Hyperoxia increases hepatic arginase expression and ornithine production in mice. Toxicology and Applied Pharmacology. 2006;215(1):109–117. doi: 10.1016/j.taap.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Pu W., Zhang H., Huang X., et al. Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nature Communications. 2016;7(1):13369–13369. doi: 10.1038/ncomms13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Racine-Samson L., Scoazec J., D'Errico A., et al. The metabolic organization of the adult human liver: a comparative study of normal, fibrotic, and cirrhotic liver tissue. Hepatology. 1996;24(1):104–113. doi: 10.1002/hep.510240118. [DOI] [PubMed] [Google Scholar]
- 20.Zabaleta N., Hommel M., Salas D., Gonzalez-Aseguinolaza G. Genetic-based approaches to inherited metabolic liver diseases. Human Gene Therapy. 2019;30(10):1190–1203. doi: 10.1089/hum.2019.140. [DOI] [PubMed] [Google Scholar]
- 21.Kietzmann T. Liver zonation in health and disease: hypoxia and hypoxia-inducible transcription factors as concert masters. International Journal of Molecular Sciences. 2019;20(9):p. 2347. doi: 10.3390/ijms20092347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perugorria M. J., Olaizola P., Labiano I., et al. Wnt-β-catenin signalling in liver development, health and disease. Nature Reviews. Gastroenterology & Hepatology. 2019;16(2):121–136. doi: 10.1038/s41575-018-0075-9. [DOI] [PubMed] [Google Scholar]
- 23.Hijmans B. S., Grefhorst A., Oosterveer M. H., Groen A. K. Zonation of glucose and fatty acid metabolism in the liver: mechanism and metabolic consequences. Biochimie. 2014;96:121–129. doi: 10.1016/j.biochi.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Bass N. M., Barker M. E., Manning J. A., Jones A. L., Ockner R. K. Acinar heterogeneity of fatty acid binding protein expression in the livers of male, female and clofibrate-treated rats. Hepatology. 1989;9(1):12–21. doi: 10.1002/hep.1840090104. [DOI] [PubMed] [Google Scholar]
- 25.De Chiara F., Ureta Checcllo C., Ramón Azcón J. High protein diet and metabolic plasticity in non-alcoholic fatty liver disease: myths and truths. Nutrients. 2019;11(12):p. 2985. doi: 10.3390/nu11122985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaemers I. C., Stallen J. M., Kunne C., et al. Lipotoxicity and steatohepatitis in an overfed mouse model for non-alcoholic fatty liver disease. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2011;1812(4):447–458. doi: 10.1016/j.bbadis.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Buettner R., Parhofer K. G., Woenckhaus M., et al. Defining high-fat-diet rat models: metabolic and molecular effects of different fat types. Journal of Molecular Endocrinology. 2006;36(3):485–501. doi: 10.1677/jme.1.01909. [DOI] [PubMed] [Google Scholar]
- 28.Soto-Gutierrez A., Gough A., Vernetti L. A., Taylor D. L., Monga S. P. Pre-clinical and clinical investigations of metabolic zonation in liver diseases: the potential of microphysiology systems. Experimental Biology and Medicine (Maywood, N.J.) 2017;242(16):1605–1616. doi: 10.1177/1535370217707731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall Z., Bond N. J., Ashmore T., et al. Lipid zonation and phospholipid remodeling in nonalcoholic fatty liver disease. Hepatology. 2017;65(4):1165–1180. doi: 10.1002/hep.28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Stefano F., Chacon E., Turcios L., Marti F., Gedaly R. Novel biomarkers in hepatocellular carcinoma. Digestive and Liver Disease. 2018;50(11):1115–1123. doi: 10.1016/j.dld.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Lin J., Zhang Y., Wu J., et al. Neuropilin 1 (NRP1) is a novel tumor marker in hepatocellular carcinoma. Clinica Chimica Acta. 2018;485:158–165. doi: 10.1016/j.cca.2018.06.046. [DOI] [PubMed] [Google Scholar]
- 32.Kim J. M., Kwon C. H. D., Joh J.-W., et al. C-reactive protein may be a prognostic factor in hepatocellular carcinoma with malignant portal vein invasion. World Journal of Surgical Oncology. 2013;11(1):92–92. doi: 10.1186/1477-7819-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navadgi S., Chang C.-C., Bartlett A., McCall J., Pandanaboyana S. Systematic review and meta-analysis of outcomes after liver resection in patients with hepatocellular carcinoma (HCC) with and without bile duct thrombus. HPB : the official journal of the International Hepato Pancreato Biliary Association. 2016;18(4):312–316. doi: 10.1016/j.hpb.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Moshe S., Shapira Y., Moor A. E., et al. Spatial sorting enables comprehensive characterization of liver zonation. Nature Metabolism. 2019;1(9):899–911. doi: 10.1038/s42255-019-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whittemore J. C., Newkirk K. M., Reel D. M., Reed A. Hepatic copper and iron accumulation and histologic findings in 104 feline liver biopsies. Journal of Veterinary Diagnostic Investigation. 2012;24(4):656–661. doi: 10.1177/1040638712445765. [DOI] [PubMed] [Google Scholar]
- 36.Klaren W. D., Vine D., Vogt S., Robertson L. W. Spatial distribution of metals within the liver acinus and their perturbation by PCB126. Environmental Science and Pollution Research International. 2018;25(17):16427–16433. doi: 10.1007/s11356-017-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu N., Shen Y., Liu F., et al. Morphological and immunobiochemical analysis of the liver in L-arginine induced experimental chronic pancreatitis. Pancreatology. 2017;17(2):247–254. doi: 10.1016/j.pan.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y., Feng W., Liu R., Xia T., Liu S. Graphene oxide causes disordered zonation due to differential intralobular localization in the liver. ACS Nano. 2020;14(1):877–890. doi: 10.1021/acsnano.9b08127. [DOI] [PubMed] [Google Scholar]
- 39.Ben-Moshe S., Itzkovitz S. Spatial heterogeneity in the mammalian liver. Nature Reviews Gastroenterology & Hepatology. 2019;16(7):395–410. doi: 10.1038/s41575-019-0134-x. [DOI] [PubMed] [Google Scholar]
- 40.Aizarani N., Saviano A., Sagar M. L., et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572(7768):199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong X., Kuang H., Ansari S., et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Molecular Cell. 2019;75(3):644–660.e5. doi: 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strauss O., Phillips A., Ruggiero K., Bartlett A., Dunbar P. R. Immunofluorescence identifies distinct subsets of endothelial cells in the human liver. Scientific Reports. 2017;7(1) doi: 10.1038/srep44356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Rosa V., Monti M., Terlizzi C., Fonti R., Del Vecchio S., Iommelli F. Coordinate modulation of glycolytic enzymes and OXPHOS by imatinib in BCR-ABL driven chronic myelogenous leukemia cells. International Journal of Molecular Sciences. 2019;20(13):p. 3134. doi: 10.3390/ijms20133134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morrison V. M., Burnett A. K., Forrester L. M., Wolf C. R., Craft J. A. The contribution of specific cytochromes P-450 in the metabolism of 7,12-dimethylbenz[a]anthracene in rat and human liver microsomal membranes. Chemico-Biological Interactions. 1991;79(2):179–196. doi: 10.1016/0009-2797(91)90081-H. [DOI] [PubMed] [Google Scholar]
- 45.Halpern K. B., Shenhav R., Matcovitch-Natan O., et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542(7641):352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlen M., Fagerberg L., Hallstrom B. M., et al. Tissue-based map of the human proteome. Science. 2015;347(6220, article 1260419) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used to support the study.


