Abstract
Transmission of genetic material through high quality gametes to progeny requires accurate homologous chromosome recombination and segregation during meiosis. A failure to accomplish these processes can have major consequences in reproductive health, including infertility and development disorders in offspring. Sirtuins, a family of NAD+-dependent protein deacetylases and ADP-ribosyltransferases, play key roles in genome maintenance, metabolism and aging. In recent years, Sirtuins have emerged as regulators of several reproductive processes and interventions aiming to target Sirtuin activity are of great interest in the reproductive biology field. Sirtuins are pivotal to protect germ cells against oxidative stress, a major determinant influencing ovarian aging and the quality of gametes. Sirtuins also safeguard the integrity of the genome through epigenetic programs required for regulating gene repression, DNA repair and chromosome segregation, among others. Although these functions are relatively well characterized in many somatic tissues, how they contribute to reproductive functions is not well understood. This review summarizes our current knowledge on the role of Sirtuins in female reproductive systems and discusses the underlying molecular pathways. In addition, we highlight the importance of Sirtuins as anti-aging factors in the ovary and summarize current preclinical efforts to identify treatments to extend female reproductive longevity.
Introduction
Sexual reproduction depends on the generation of eggs capable of giving rise to viable embryos upon fertilization. Oocyte development initiates in the fetus when germ cell progenitors colonize the embryonic gonadal ridges, proliferate, and enter meiosis (Bolcun-Filas & Handel, 2018). During fetal development, oocytes undergo meiotic recombination, which involves the pairing and synapsis of maternal and paternal chromosomes and the exchange of genetic material between them. Oocytes that successfully complete this process then arrest the cell cycle in prophase of meiosis I (MI), and form the pool of primordial follicles, also known as the ovarian reserve. Because women are born with a finite number of gametes, the ovarian reserve reflects a woman’s reproductive life span. Before sexual maturity, some primordial follicles initiate growth and progress through the primary, secondary and antral stages and eventually die by atresia. At pubertal onset, few antral follicles complete growth and, upon ovulation, oocytes contained in these follicles resume meiosis to become mature eggs released into the reproductive tract. After meiotic resumption, oocytes first segregate homologous chromosomes to complete MI and then they arrest at metaphase of meiosis II (MII). Fertilization of the MII-arrested egg by a sperm triggers sister chromatid separation to complete MII. Women with diminished ovarian reserve have a shorted reproductive lifetime and reduced fertility (Huhtaniemi et al., 2018). Similarly, chromosome segregation errors during female meiosis are a major source of infertility and birth defects (Webster & Schuh, 2017). Therefore, identifying the mechanisms required for high-quality gamete production is therefore of vital importance in order to develop preventive, diagnostic and curative strategies.
The Sir2 family of proteins, also called Sirtuins, are conserved throughout evolution (Vaquero, 2009). The mammalian genome encodes seven Sirtuin genes, named SIRT1-SIRT7. Sirtuin expression has been detected in oocytes of different mammalian species, including human (Zhao et al., 2016), mouse (Kawamura et al., 2010), and pigs (Ma, Zhang, Zhang, Han, & Rui, 2015). However, the specific requirements of mammalian sirtuins during female meiosis are not fully understood. In somatic cells, SIRT1, SIRT6 and SIRT7 localize and function in the nucleus while SIRT4 and SIRT5 function in mitochondria. SIRT3 is predominantly mitochondrial but also translocates to the nucleus when cells are under stress. SIRT2 is mostly found in the cytoplasm but it also shuttles into the nucleus during the G2/M cell-cycle transition. Sirtuins are sensors of cellular stress, promoting cellular adaptation under these conditions by regulating mitochondrial and nuclear function. Sirtuins deacylate and ADP-ribosylate a wide range of substrates in the presence of NAD+, an essential cofactor for Sirtuin activity (Johnson & Imai, 2018). Sirtuins are activated through different mechanisms including an increase in the NAD+/NADH ratio or through phosphorylation by stress response kinases such as AMP-activated protein kinase (AMPK) or c-Jun N-terminal kinase 1(JNK1)(Vaquero & Reinberg, 2009). Nuclear Sirtuins control fundamental aspects of genome biology through epigenetic modifications and the regulation of key transcription factors and other chromatin factors, whereas mitochondrial Sirtuins regulate major metabolic and ROS detoxification enzymes.
At the physiological level, Sirtuins are central in controlling aging and longevity pathways. SIRT3, SIRT6 and SIRT7 are both associated with human and rodent longevity (Bellizzi et al., 2005; Donlon et al., 2018; Tian et al., 2019) and mice overexpressing SIRT6 throughout the body or SIRT1 specifically in the brain both correlate with increased lifespan (Kanfi et al., 2012; Satoh et al., 2013). Consistently, deficiencies of Sirt1, Sirt6 and Sirt7 in mice are associated with premature aging syndromes in mouse knockout models (Cheng et al., 2003; Mostoslavsky et al., 2006; Vazquez et al., 2016), and deficiencies of Sirt2, Sirt3 and Sirt4 result in increased cancer risk (Haigis, Deng, Finley, Kim, & Gius, 2012; Jeong, Hwang, & Seong, 2016; H. S. Kim et al., 2011; Serrano et al., 2013). Because ovarian function ceases at menopause (~50 years of age), the ovary is the most rapidly aging organ in women. Furthermore, because oocytes must sustain function over decades, this remarkable longevity increases their chances of accumulating genomic damage, a potent driver of cellular senescence and organismal aging (Lopez-Otin, Blasco, Partridge, Serrano, & Kroemer, 2013). Here, we review the literature focused on Sirtuin functions in mouse oocyte development, health and longevity and we highlight important areas that remain unsolved.
Nuclear Sirtuins
SIRT1
In somatic cells, SIRT1 participates in several cellular processes including regulation of heterochromatin and gene silencing (Vaquero et al., 2007; Vaquero et al., 2004), telomere maintenance (Palacios et al., 2010) and the repair of DNA damage (Bosch-Presegue & Vaquero, 2014; Dobbin et al., 2013). SIRT1 is a key regulator of reproductive function at multiple levels and the most comprehensively evaluated Sirtuin to date. Homozygous deletion of Sirt1 in mice causes developmental defects and partial embryonic lethality. Both male and female mice that survive to adulthood are sterile (McBurney et al., 2003). Sirt1−/− females have normal follicle development, but they have defective estrous cycles and fail to ovulate (McBurney et al., 2003). Because administration of stimulatory gonadotropins to Sirt1−/− females restores ovulation, defects in the hypothalamic-pituitary-gonadal axis are likely the main cause of the primary infertility phenotype.
In addition to regulating the hormone axis, SIRT1 also regulates oocyte production and quality (Table 1). When prophase I-arrested oocytes are treated with high doses of the SIRT1 selective inhibitor Ex527, approximately 50% fail to resume meiosis and those that progress to metaphase of MII have an increased percentage of abnormal spindles and chromosome misalignment (Di Emidio et al., 2014). Consistently, embryos derived from Sirt1−/− eggs after in vitro fertilization (IVF) have reduced developmental potential, are less efficient in reaching the 2-cell stage and give rise to reduced number of live pups (Coussens, Maresh, Yanagimachi, Maeda, & Allsopp, 2008).
Table 1:
Summary of phenotypes associated with Sirtuin deficiency in oocytes
| SIRTUIN | Mouse knockout models | Knockdown/Inhibition models |
|---|---|---|
| SIRT1 | • Infertile • Impaired hypothalamic-pituitary-gonadal signaling |
• Arrest at prophase I (∼50%) Chromosome and spindle abnormalities at MII (∼25%) |
| SIRT2 | • Normal oocyte spindle assembly | • Reduced polar body extrusion rates (∼50%) • Poor and aberrant K-MT attachments in MI (50%) • Chromosome and spindle abnormalities at MII (∼35%) • Aneuploidy at MII (∼23%) • Increased H4K16ac and acetylated α-tubulin at MII |
| SIRT3 | • Fertile • Normal oocyte meiotic maturation • Impaired in vitro embryo development after IVF • Increased ROS levels |
• Normal oocyte meiotic maturation • Increased ROS levels |
| SIRT4 | • N/A | • Normal oocyte meiotic maturation |
| SIRT5 | • N/A | • N/A |
| SIRT6 | • N/A | • Reduced polar body extrusion rates (∼40%) • Chromosome and spindle abnormalities at MI (∼27%) • Aberrant K-MT attachments at MI (50%) • Aneuploidy at MII (∼30%) • Increased H4K16ac in prophase I and MII |
| SIRT7 | • Age dependent subfertility • Defects in chromosome synapsis during meiotic recombination • Reduced primordial follicle pool • Normal oocyte meiotic maturation • Aneuploidy at MII (∼50%) |
• Reduced NEBD and polar body extrusion rates (~50%) • Chromosome and spindle abnormalities at MI (∼50%) • Enlarged polar body, symmetrical divisions and diminished actin cap at MII • Increased ROS levels and diminished ATP production • Impaired in vitro embryo development after IVF • Increased γH2Ax and apoptosis in early embryos |
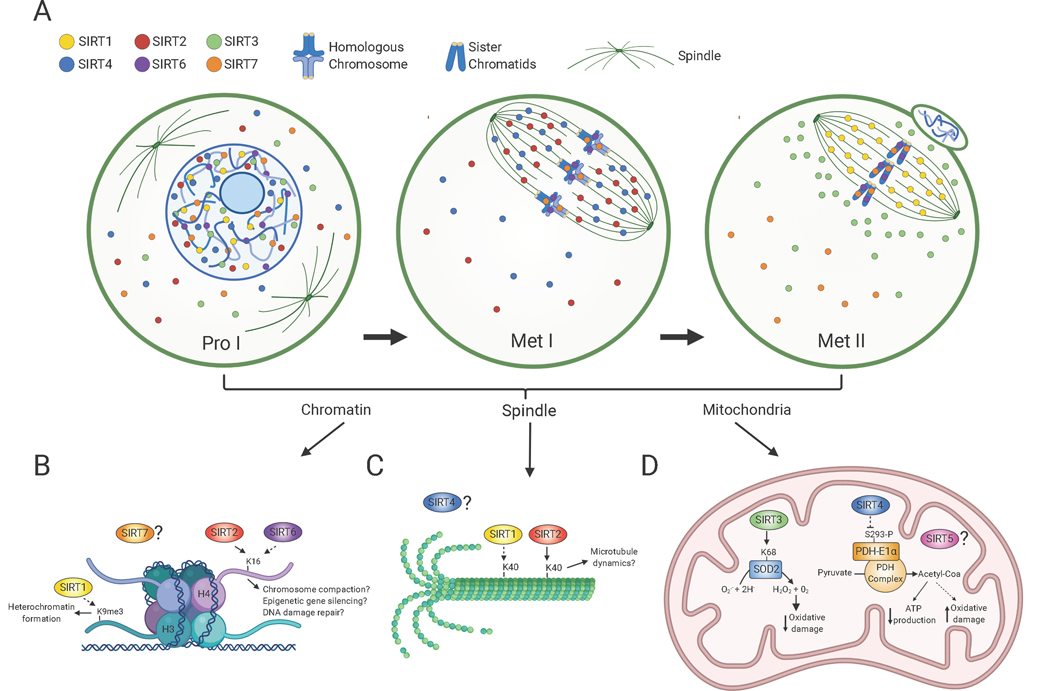
In somatic cells, SIRT1 promotes heterochromatin formation through SUV39H1 methyltransferase regulation and the spreading of its substrate histone H3K9 trimethylation (H3K9me3). In prophase I and metaphase II-arrested mouse oocytes and eggs, SIRT1 activation correlates with increased levels of H3K9me3 suggesting the existence of a similar SIRT1-SUV39H1 interplay during female meiosis (Nevoral et al., 2019). Because SIRT1 is predominantly nuclear in prophase I-arrested oocytes but then localizes to the spindle at metaphase II (Figure 1), SIRT1-dependent H3K9me3 regulation most likely happens during prophase I. H3K9me3 is enriched at pericentromeric regions where it plays important roles in sister chromatid cohesion and chromosome segregation (Guenatri, Bailly, Maison, & Almouzni, 2004). H3K9me3 is also crucial for gene repression and facultative heterochromatin formation and DNA repair (Ayrapetov, Gursoy-Yuzugullu, Xu, Xu, & Price, 2014; Becker, Nicetto, & Zaret, 2016). Further investigation should elucidate how SIRT1-mediated regulation of this epigenetic mark contributes to meiotic progression.
Figure 1:
Sirtuin distribution in oocytes during meiotic divisions and its associated molecular functions. A) Scheme denotes Sirtuin localization in oocytes at prophase I (Pro I) and at Metaphase I and II (Met I, Met II). SIRT1 is mostly nuclear in prophase I-arrested oocytes and on the spindle in Met II-arrested eggs. SIRT2 is found throughout the ooplasm and nucleus in prophase I and relocalizes to the spindle in Met I. SIRT6 and SIRT7 are chromatin-bound at all developmental stages, but SIRT7 is also present to a lesser extent in the ooplasm. SIRT3 and SIRT4 are predominantly nuclear in prophase I. SIRT3 redistributes around the spindle in Met II eggs and SIRT4 reallocates to the spindle in Met I. The cellular distribution of SIRT5 in oocytes is currently unknown. B-D) Molecular functions of Sirtuins during meiotic maturation in chromatin (B), the spindle (C) and mitochondria (D). B) SIRT1 promotes H3K9me3 deposition and heterochromatin formation and SIRT2 and SIRT6 regulates H4K16ac levels with potential implications in chromosome compaction, gene silencing and DNA damage repair signaling. C) SIRT1 and SIRT2 both regulate acetylation of α-tubulin suggesting a potential role in spindle dynamics. D) SIRT3 stimulates SOD2 activity and ROS balance and SIRT4 limits Pyruvate Dehydrogenase (PDH) complex activity leading to reduced ATP production. Solid and dashed arrows denote potential direct and indirect roles, respectively. Created with BioRender.com.
The localization of SIRT1 to spindle microtubules in MII-arrested eggs suggests a role of SIRT1 in spindle regulation(Nevoral et al., 2019) (Figure 1B). Activation of SIRT1 with BML-278 results in diminished α-tubulin acetylation at lysine 40 (K40) residue and, conversely, SIRT1 inhibition with Sirtinol increases α-tubulin acetylation levels. Acetylation of α-tubulin is associated microtubule stability (Janke & Montagnac, 2017) but whether SIRT1 regulates spindle dynamics remains to be investigated.
Interestingly, there are sexually dimorphic phenotypes when mice harbor a point mutation in the conserved histidine (H355Y, SirT1y/y) critical for SIRT1 enzymatic activity. Males expressing this catalytically inactive mutant SIRT1 are sterile but females are fertile (Seifert et al., 2012). Therefore, the catalytic activity of SIRT1 is required to sustain fertility in male mice, but not in females. Whether SIRT1 activity is required later as females age, if SIRT1 functions in a non-catalytic fashion, or if there are compensation from other Sirtuins, are still open questions.
SIRT2
SIRT2 is an important regulator of mitosis at multiple levels. SIRT2 directly deacetylates histone H4 acetylated at K16 (H4K16ac) during the G2/M transition thereby inducing chromatin condensation (Vaquero et al., 2006). SIRT2 also interacts and activates the methyl-transferase PR-Set7 resulting in spreading of H4K20me1, another epigenetic mark required for chromosome compaction and mitotic progression (Serrano et al., 2013). Furthermore, SIRT2 positively regulates the anaphase-promoting complex via coactivators CDH1 and CDC20 (H. S. Kim et al., 2011). SIRT2 acts as α-tubulin deacetylase with important implications in microtubule dynamics (North, Marshall, Borra, Denu, & Verdin, 2003). Surprisingly, despite the vast body of literature describing the roles of SIRT2 during somatic cell division, there are few studies describing SIRT2 in gamete development (D. Qiu et al., 2018; L. Zhang et al., 2014). In mouse oocytes, SIRT2 depletion via siRNA results in either meiotic arrest at metaphase I or in the production of aneuploid metaphase II eggs. The arrested oocytes presented altered spindle morphologies, chromosome misalignment and aberrant and poor kinetochore-microtubule (K-MT) attachments (D. Qiu et al., 2018; L. Zhang et al., 2014).
At the molecular level, SIRT2 appears to regulate several pathways required for oocyte maturation including chromatin structure and spindle function. SIRT2 depletion in oocytes leads to increased levels of H4K16ac and acetylated tubulin (L. Zhang et al., 2014). SIRT2 localization is dynamic during oocyte meiotic maturation: it is first dispersed throughout the oocyte during prophase I and then moves onto the spindle during metaphase (Figure 1A). Therefore, it is possible that SIRT2-dependent epigenetic regulation via histone deacetylation occurs shortly after meiotic resumption and later its deacetylase activity targets the spindle microtubules. Consistent with this model, H4K16ac is deacetylated concurrently with other histone marks during the prophase I-to-metaphase I transition (J. M. Kim, Liu, Tazaki, Nagata, & Aoki, 2003; L. Zhang et al., 2014). SIRT2 preferentially deacetylates H4K16ac in mitotic cells (Vaquero et al., 2006), strongly indicating that the hyperacetylation of H4K16ac in SIRT2-depleted oocytes is due to the lack of the SIRT2 histone deacetylase activity (Figure 1B). Regulation of H4K16ac is pivotal for several nuclear processes, including higher order chromatin structure (Shogren-Knaak et al., 2006), however if SIRT2 knockdown oocytes have compromised chromatin compaction is not known.
SIRT2 knock-down in oocytes leads to an increase in aberrant K-MT attachments and reduced inter-kinetochore tension. Because centromeres are devoid of H4K16ac (Horikoshi et al., 2013), SIRT2 may also be pivotal for kinetochore structure and function (D. Qiu et al., 2018; L. Zhang et al., 2014). SIRT2 also associates with the meiotic spindle and its knock-down in oocytes leads to increased α-tubulin K40 acetylation. As mentioned, SIRT2 is an α-tubulin deacetylase in somatic cells (North et al., 2003), suggesting that SIRT2 regulates spindle acetylation levels directly (Figure 1C). Importantly, tubulin acetylation is associated with stable microtubules (I. Yu, Garnham, & Roll-Mecak, 2015), raising the possibility that SIRT2-dependent spindle deacetylation may impact the dynamics of spindle assembly and maintenance. Future experiments will be required to elucidate the biological significance of SIRT2-spindle interactions.
Despite the importance of SIRT2 as a meiotic regulator in in vitro depletion and inhibition studies, Sirt2−/− mice are born at normal Mendelian ratios and have normal embryonic development (H. S. Kim et al., 2011; Serrano et al., 2013). In addition, oocytes from whole body Sirt2 knockout females have normal spindle assembly (Bertoldo et al., 2020)(Table 1). Further experiments addressing the fertility of Sirt2−/− mice and whether other Sirtuins could compensate for the lack of SIRT2 protein will help clarify the in vivo function of SIRT2 in gametogenesis.
SIRT6
In somatic cells, SIRT6 plays important roles in genome maintenance by transcriptional regulation (Kawahara et al., 2009), repairing DNA damage (Toiber et al., 2013) and regulating chromosome segregation (Tasselli et al., 2016; Tasselli, Zheng, & Chua, 2017). SIRT6 harbors both deacylase and ADP-ribosyltransferase enzymatic activities. SIRT6 is an emerging regulator of female reproduction function (Han et al., 2015). Upon in vitro maturation of mouse oocytes, Sirt6 siRNA-mediated depletion correlates with reduced meiotic maturation and an increased production of aneuploid MII eggs. At metaphase I, SIRT6-knockdown oocytes have enlarged spindles, chromosome misalignment and abnormal K-MT attachments. In addition, embryos derived via IVF from these MII eggs are less efficient in developing to the blastocyst stage (Ge et al., 2019). Consistent with its documented chromatin-based role in somatic cells (Tasselli et al., 2017), SIRT6 is a chromatin-bound protein at every phase of oocyte maturation assessed (Figure 1A). In somatic cells, SIRT6 regulates several epigenetic marks including histone H3K56ac, H3K9ac and H3K18ac. However, in SIRT6-depleted oocytes, H3K9ac and H3K56ac are unchanged, while H4K16ac is significantly increased (Han et al., 2015)(Figure 1B). It is possible that SIRT6-dependent regulation of H3K9ac and H3K56ac occurs focally at specific genomic sites, or that other Sirtuin members could compensate for the lack of SIRT6 activity. However, these results could also indicate that SIRT6 epigenetic targets may differ in a cell-type specific manner. SIRT6 is a critical regulator of mitosis through its epigenetic-based control of H3K18ac at pericentromeric chromatin (Tasselli et al., 2016). Because epigenetic changes at centromeric chromatin are thought to be critical for the establishment of amphitelic K-MT attachments, it will be important to determine whether SIRT6-dependent H3K18ac regulation plays a similar role in meiotic cells.
SIRT7
In somatic cells, SIRT7 participates in several processes including regulating transcription (Blank & Grummt, 2017),and DNA double-strand break repair (Vazquez, Thackray, & Serrano, 2017). In transcription, SIRT7 fulfills both activation and repression functions. As a transcriptional repressor, SIRT7 deacetylates histone H3K18ac and H3K36ac at specific promoters; as a transcriptional activator, SIRT7 deacetylates and modulates the activity of specific transcription factors and different components of the basal transcriptional machinery. SIRT7 is also recruited to DNA damage sites where it promotes epigenetic changes and facilitates the recruitment of DNA repair proteins. Early studies of SIRT7 found high protein levels of this sirtuin in mouse testis (Ford et al., 2006). However, the role of SIRT7 in male reproduction still remains unexplored.
In female mice, SIRT7 is important for establishment of the ovarian reserve and reproductive longevity (Vazquez, Blengini, Hernandez, Serrano, & Schindler, 2019). Mice lacking SIRT7 have reduced numbers of primordial follicles resulting in early onset infertility. Mechanistically, SIRT7 appears to be required for synapsis of homologous chromosomes, an essential step in repairing double-strand DNA breaks during meiotic prophase. Defects in the early events of the DNA-damage response have been linked with chromosome synapsis defects (Pittman et al., 1998; Romanienko & Camerini-Otero, 2000). However, the recruitment of single stranded DNA binding Replication Protein A (RPA1) and the strand exchange protein Disruption of meiotic control 1 (DMC1), proteins required for chromosome homology search after double strand break formation, are both normal in Sirt7−/− oocytes. These data suggest that SIRT7 plays a direct role in the synapsis process. In somatic cells, SIRT7 is a histone H3K18 deacetylase (Barber et al., 2012). Interestingly, the chromatin regions that fail to synapse correlate with higher levels of H3K18 acetylation suggesting that this epigenetic marks regions of chromosome asynapsis. Curiously, H3K18ac also occurred at the rare asynaptic sites in wild-type oocytes. Therefore, deacetylation of H3K18ac does not depend upon SIRT7 activity. Further examination is therefore required to determine the mechanism by which SIRT7 promotes chromosome synapsis. It is possible that SIRT7 targets its other histones substrates, including succinylated H3K122(Li et al., 2016) and acetylated H3K36 (W. W. Wang et al., 2019). Further experiments will be required to define whether these histone modifications are involved during the meiotic process and its relationship with SIRT7 function.
Recent evidence have shown that SIRT7 silences Long interspersed elements-1 (LINE-1) retrotransposons in somatic cells by positioning them in the nuclear periphery (Vazquez, Thackray, et al., 2019), a highly repressive environment. Despite their importance in genetic evolution, retrotransposon expression and movement induce DNA damage and mutations, and their expression has been linked to aging and age-related pathologies. Importantly, LINE-1 retrotransposition during meiotic prophase drives the natural process of oocyte fetal loss (Tharp, Malki, & Bortvin, 2020). Whether SIRT7 regulates the establishment of the ovarian reserve through LINE-1 regulation has not yet been explored.
The role of SIRT7 during oocyte meiotic maturation is less clear (Table 1). Knockdown of SIRT7 using siRNA results in many abnormalities: aneuploidy at MII and embryos with reduced developmental potential (Gao et al., 2018). SIRT7-depleted oocytes mature slower, with delays in nuclear envelope break down (NEBD) and in the extrusion of the first polar body. In addition, MI oocytes have abnormal spindles and disorganized chromosomes, and SIRT7-depleted eggs also display enlarged polar bodies and symmetrical divisions associated with a diminished cortical actin cytoskeletal, suggesting a role in regulating polarity. In somatic cells, SIRT7 regulates the expression of nuclear encoded mitochondrial genes (Ryu et al., 2014), and Sirt7−/− mice have impaired oxidative respiration and mitochondrial dysfunction. Similarly, in SIRT7-depleted oocytes, the production of reactive oxygens and total ATP levels are reduced suggesting that mitochondrial dysfunction also occurs in germ cells (Gao et al., 2018). Importantly, pretreatment of SIRT7-depleted oocytes with the antioxidant NAC prevented the formation of the aforementioned phenotypes, highlighting the importance of SIRT7 in redox homeostasis during meiotic maturation and early development.
Although Sirt7−/− females have more aneuploid eggs, spindle formation, chromosome alignment and polar body extrusion appeared normal. Why SIRT7-RNAi oocytes are more severe in phenotype compared to oocytes from SIRT7 knockout mice is still an open question (Table 1). One possibility is phenotypic penetrance differences in knockout oocytes. Many, but not all, oocytes undergo apoptosis and are removed from the germline before sexual maturity is reached. Therefore, the most severely affected oocytes cannot be examined, and only the healthiest survive for maturation studies. Alternatively, lack of SIRT7 in mice may allow for developmental compensation. In this regard, SIRT6, the closest family member to SIRT7, or another member of the HDAC superfamily (Seto & Yoshida, 2014), could compensate during meiotic maturation in Sirt7−/− mice. Further experiments will be required to address this question and to further untangle the molecular mechanisms modulated by SIRT7.
Mitochondrial Sirtuins
SIRT3
SIRT3 is the major deacetylase in somatic cell mitochondria, where it plays critical roles in metabolism and redox homeostasis. To promote ROS balance, SIRT3 directly deacetylates and activates the mitochondrial enzyme superoxide dismutase (SOD2), required for the partitioning of superoxide radical (O2-) to oxygen (O2) and hydrogen peroxide (H2O2) (Bause & Haigis, 2013). SIRT3 also regulates all complexes of the electron transport chain to ensure ATP production and minimize ROS generation.
SIRT3 protein is detected in human oocytes, granulosa and theca cells of developing follicles. SIRT3 protein is also present in the corpus luteum, the remaining tissue after ovulation, but it is absent in corpus albicans, the scar that is left after the corpus luteum involutes (Fu et al., 2014; Pacella-Ince, Zander-Fox, & Lan, 2014). In addition, SIRT3 mRNA levels increase as oocytes mature and its expression peaks in zygotes (Zhao et al., 2016). Although this expression pattern would indicate a critical role SIRT3 in follicle growth and gamete development, Sirt3−/− mice are fertile (Iljas & Homer, 2020) and produce eggs that can be fertilized in vitro (Kawamura et al., 2010). In addition, Sirt3−/− females have normal ovarian reserves, and follicle growth and oocyte maturation occurs efficiently (Iljas & Homer, 2020). Sirt3 knockout and knockdown oocytes, however, have increased levels of ROS (Iljas & Homer, 2020; Kawamura et al., 2010; L. Zhang et al., 2015; Zhao et al., 2016). Given the known deleterious effects of oxidative damage on meiotic progression (X. Zhang, Wu, Lu, Guo, & Ma, 2006), it is surprising that SIRT3-deficient oocytes undergo normal in vitro maturation (Iljas & Homer, 2020). Whether Sirt3-knockout and knockdown eggs have an increased occurrence of aneuploidy is not known.
Studies of Sirt3−/− egg fertilization and embryo development reveal requirements in cell culture-related stress conditions. When Sirt3−/− eggs are fertilized and cultured in vitro they have increased levels of oxidative damage and are less efficient in reaching the blastocyst stage (Kawamura et al., 2010). However, when Sirt3−/− eggs are fertilized in vitro, but then transferred into pseudopregnant mothers to complete development in vivo, embryos implant and develop to full term highlighting that SIRT3 function is more critical under stressful conditions such as in vitro culture. Curiously, in vivo-fertilized 1-cell zygotes flushed from Sirt3−/− females can develop efficiently to blastocyst stage in vitro (Iljas & Homer, 2020), suggesting that if reducing one stressful in vitro parameter (i.e. fertilization or embryo culture), can rescue the defective stress response.
Retrospective analysis of data from patients affected by infertility shows that in vivo maturation of oocytes through controlled ovarian stimulation results in higher pregnancy rates than in vitro maturation techniques (Zhao et al., 2016). In this study, in vitro-matured oocytes had reduced SIRT3 mRNA levels compared to those in vivo matured. Those oocytes in vitro matured had compromised mitochondrial biogenesis and diminished developmental potential. Importantly, SIRT3 reintroduction into human oocytes by microinjection followed by in vitro maturation and fertilization, improved blastocyst formation highlighting again the beneficial role of SIRT3 for in vitro culture conditions.
Similar to its function in somatic cells, SIRT3 also regulates SOD2 acetylation in oocytes (L. Zhang et al., 2015). SIRT3-knockdown oocytes have increased levels of acetylated SOD2 at residue K68 (Figure 1D). In somatic cells, SIRT3 deacetylates several SOD2 lysines (Chen et al., 2011; X. Qiu, Brown, Hirschey, Verdin, & Chen, 2010; Tao et al., 2010) but whether this also occurs in oocytes is unknown. Curiously, upon expression in mouse oocytes, an K68 acetylated mimetic of SOD2 (K68Q), but not other SOD2 lysine mutants, increases cellular oxidative damage suggesting that this is the primary, or sole, site required for SOD2 activation and ROS detoxification in oocytes. Oocytes from obese female mice have high ROS levels and reduced SIRT3 protein levels (L. Zhang et al., 2015). Oocytes from obese females adapt to high nutrient environments by promoting mitochondria biogenesis and by increasing oxidative phosphorylation, which, in turn, results in oxidative stress and poor reproductive outcomes (Igosheva et al., 2010). Importantly, SIRT3 reintroduction via microinjection in mouse oocytes prevents oxidative damage through SOD2 activity regulation, highlighting a potential benefit of SIRT3 activation in protecting oocytes from the stress-induced effects of obesity. In contrast, females completely lacking Sirt3 maintained on a high fat diet have similar reproductive competencies as WT animals, including similar ovarian reserve, oocyte maturation and fertility rates (Iljas & Homer, 2020). Despite having no obvious reproductive abnormalities, Sirt3−/− oocytes from obese females have defects in ROS balance and mitochondrial dysfunction, suggesting the existence of compensatory mechanism to cope with this stress. Because Sirt3−/− oocytes have increased SIRT1 protein levels, it is possible that SIRT1 is the compensatory sirtuin under these circumstances.
SIRT4
In somatic cells, SIRT4 plays important roles in mitochondrial function and cellular metabolism. SIRT4 is a weak deacetylase but exhibits ADP ribosyltransferase, deacylase and lipoamidase activities (Min, Gao, & Yu, 2018). As a lipoamidase, SIRT4 limits the activity of the pyruvate dehydrogenase (PDH) complex via enzymatic hydrolysis of the lipoamide cofactor (Mathias et al., 2014). The activity of the PDH complex is also inhibited by phosphorylation of the pyruvate dehydrogenase E1α subunit (PDH-E1α). The role of SIRT4 in reproduction is mostly unexplored but potentially important. In oocytes, SIRT4 overexpression, but not its depletion, perturbs meiotic maturation (Zeng et al., 2018). Oocytes where SIRT4 is overexpressed have abnormal spindle morphology, chromosome misalignment and reduced polar body extrusion rates. In addition, these oocytes have mitochondrial dysfunction characterized by altered mitochondrial distribution, reduced mitochondrial transmembrane potential, reduced ATP levels and increased ROS production (Figure 1D). How SIRT4 expression in oocytes is linked to these phenotypes is not understood. In somatic cells, SIRT4 localizes in mitochondria where it plays key roles in cellular metabolism (van de Ven, Santos, & Haigis, 2017). In contrast, in oocytes endogenous SIRT4 resides in the cytoplasm with some accumulation in the nucleus and it redistributes from chromatin to the spindle upon meiotic resumption (Figure 1A) (Zeng et al., 2018). In prophase I-arrested oocytes, mitochondria have a homogenous distribution(Dalton & Carroll, 2013), strongly suggesting that SIRT4 may also localize to mitochondria in germ cells. The nuclear and spindle localization, suggests a function involving genome and spindle regulation. PDH-E1α phosphorylation in SIRT4-overexpressing oocytes is increased at serine 293 (Zeng et al., 2018). In addition, SIRT4 overexpression together with a mutant PDH-E1α protein, in which S293 has been mutated to alanine to preclude phosphorylation, partially restores ATP levels and reduces oxidative damage. These results suggest that SIRT4-dependent PDH regulation also occurs in oocytes. Further studies will be required to understand how this functional relationship prevents oxidative damage during meiotic maturation. SIRT4 limits PDH activity (Mathias et al., 2014) and, therefore, SIRT4 activity should reduce cellular respiration and probably ROS production. Further experiments will be required to elucidate the molecular pathways controlled by SIRT4 and its relationship with oocyte maturation.
SIRT5
SIRT5 is primarily localized in mitochondria where it regulates mitochondrial metabolism including amino acid degradation and the urea cycle, the tricarboxylic acid cycle and fatty acid metabolism (Kumar & Lombard, 2018). Similar to SIRT4, SIRT5 deacetylase activity is weak or absent. SIRT5 deacylase activity, however, is strong towards large acyl moieties including succinyl, malonyl, or glutaryl groups. Sirt5−/− mice are born at submendelian ratios in a C57BL/6 genetic background (J. Yu et al., 2013), but whether this reduction is due to fertility defects has not been studied. SIRT5 mRNA is present in human and mouse oocytes (Kawamura et al., 2010; Zhao et al., 2016). SIRT5 expression is higher in human MII eggs compared to prophase I-arrested oocytes. Despite this evidence suggests a potential role for SIRT5 during oocyte maturation, the specific functions of this sirtuins in female meiosis need to be determined.
Reproductive aging
Both female and male reproductive competencies decline with age (Chamani & Keefe, 2019; Kaufman, Lapauw, Mahmoud, T’Sjoen, & Huhtaniemi, 2019). Aging in males is associated with a moderate decline in fertility. In contrast, in females the reproductive system, and in particular the ovary, ages rapidly resulting in significant reduction in fecundity after the age of 35 years followed by a precipitous decline until menopause. Factors determining the rate of ovarian aging include the depletion of the ovarian reserve as a consequence of primordial follicle recruitment and accumulation of DNA damage in quiescent follicles due to lifetime exposure to genotoxic agents. Therefore, ovarian aging is characterized by a reduction in number and quality of oocytes, which ultimately results in compromised fertility. Sirtuins have anti-aging properties in many tissues and significant evidence suggests that the ovary may also benefit from them (Figure 2).
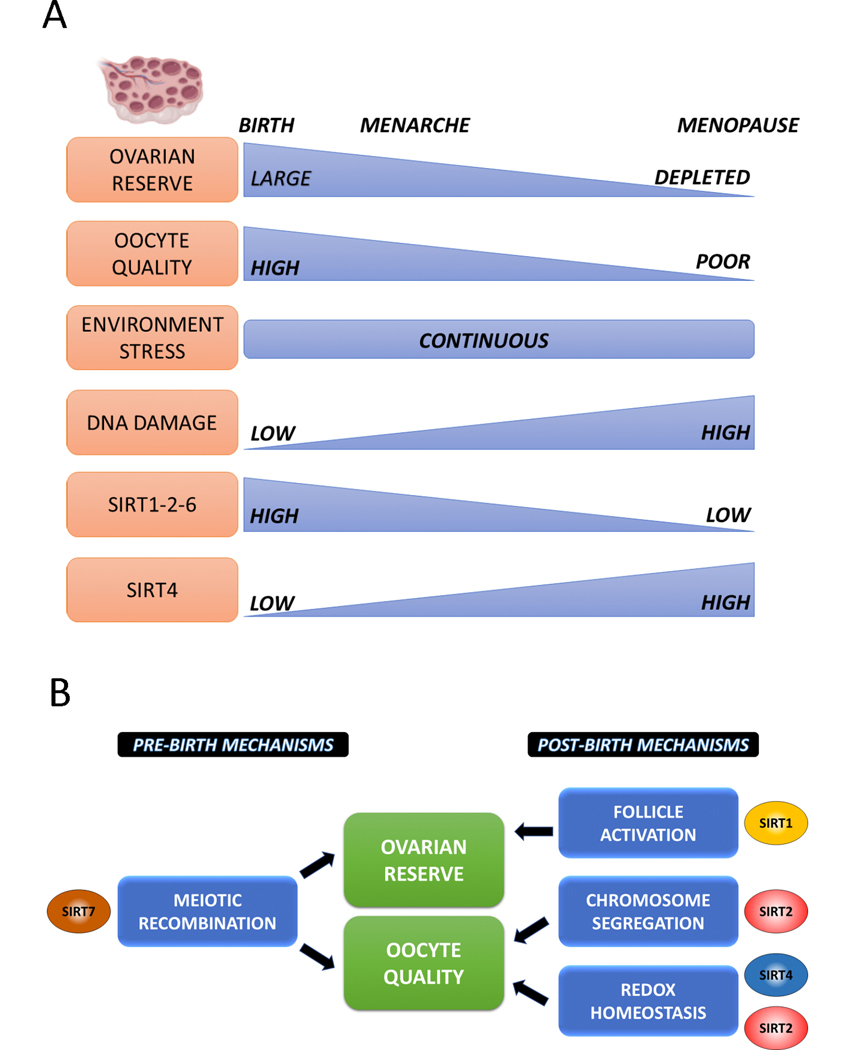
Figure 2.
Sirtuins in reproductive aging. (A) Summary of events underlying ovarian aging and its relationship with Sirtuin protein levels. (B) Sirtuin dependent mechanisms related to its anti-aging properties in the ovary.
A decline in the cellular response to oxidative stress is thought to exacerbate the age-dependent accumulation of genome damage in oocytes (S. Wang et al., 2020). Because Sirtuins are important for the elimination of ROS in mouse oocytes, Sirtuin downregulation in oocytes from older females may lead to oxidative damage accumulation and compromised oocyte quality. Consistent with this model, SIRT1, SIRT2 and SIRT6 protein levels decline in oocytes from aged animals (Di Emidio et al., 2014; Ge et al., 2019; L. Zhang et al., 2014). However, only SIRT2 reduction has been associated with increased ROS levels in aged oocytes. Importantly, 14-months old female transgenic mice overexpressing SIRT2 (Bertoldo et al., 2020) have improved fertility associated with improved oocyte quality, as assessed by normal spindle formation, chromosome alignment and reduced aneuploidy rates. These transgenic females have reduced ROS levels in their gametes. How SIRT2 downregulation in aged oocytes is linked to increased ROS production is not known. One possible mechanism involves 6-phosphate dehydrogenase (G6PD) regulation. G6PD, which is activated and deacetylated by SIRT2 in somatic cells (Y. P. Wang et al., 2014), catalyzes the first reaction of pentose phosphate pathway resulting in the generation NADPH, an important cofactor in many cellular reactions including the reduction of glutathione, a key player to combat oxidative stress (Adeoye, Olawumi, Opeyemi, & Christiania, 2018). G6PD acetylation levels are reduced in SIRT2-overexpressing oocytes but whether it improves ROS balance has not been formally demonstrated. Transgenic mice overexpressing SIRT1 via a Zp3-driven Cre in growing oocytes manifest age-dependent changes in fecundity including an initial reproductive delay and increased reproductive longevity (Long et al., 2019). At the histological level, SIRT1 overexpression correlates with increased numbers of primordial follicles and reduced atretic ones. In these mice, SIRT1 overexpression initiates in growing oocytes highlighting and suggesting an important role of SIRT1 in follicle development. In contrast to SIRT1, SIRT2 and SIRT6 levels that decrease with age, SIRT4 protein levels increase and its depletion alleviates many phenotypes associated with oocyte aging such as improved spindle morphology and chromosome alignment and better mitochondrial performance (Zeng et al., 2018).
The axis involving SIRT2 and Budding uninhibited by benzimidazole‐related 1 protein (BUBR1) may also mediate the anti-aging effects of SIRT2 in oocytes (D. Qiu et al., 2018). Levels of BUBR1, a protein involved in the spindle assembly checkpoint, decline in testis and ovary during the normal aging process and BUBR1 knockout mice develop a premature aging phenotype (Baker et al., 2004). SIRT2 interacts with and deacetylates BUBR1 and a BUBR1 mutant protein mimicking constitutive deacetylation at the specific SIRT2 lysine targets can rejuvenate aged oocytes (D. Qiu et al., 2018). Further investigation is required to provide additional insights into the underlying SIRT2 dependent molecular mechanisms during reproductive aging.
Defects in telomere length maintenance are hallmarks of genome instability and aging (Lopez-Otin et al., 2013). Short telomeres stimulate the DNA damage response resulting in the activation of senescence and apoptosis pathways (Johnson & Imai, 2018). In this sense, oocytes and embryos derived from old female mice have reduced telomere length (Ge et al., 2019; Rocca, Foresta, & Ferlin, 2019). In somatic cells, SIRT6 regulates telomeric chromatin structure through histone H3K9 deacetylation, which in turn favors efficient telomere replication in S-phase (Michishita et al., 2008). Similar to somatic cells, SIRT6-knockdown mouse oocytes and embryos have dysfunctional telomeres. SIRT6 protein levels decline in aged oocytes and SIRT6 reintroduction followed by IVF results in embryos with elongated telomeres, reduced DNA damage at telomeric sites and overall better survival.
Sirtuins require NAD+ to perform their enzymatic reactions and oocytes from old mice have reduced levels of NAD+ (Bertoldo et al., 2020), suggesting that Sirtuin activity may be compromised in germ cells during aging. The major pathway for NAD+ biosynthesis in mammals uses nicotinamide as a precursor, which is first converted to nicotinamide mononucleotide (NMN) by the enzyme nicotinamide phosphoribosyltransferase and then it is further transformed to NAD+ by the enzyme Nicotinamide mononucleotide adenylyltransferase (NMAT)(Johnson & Imai, 2018). Importantly, restoration of NAD+ levels with supplements of NMN improves the yield of oocytes and ovulated eggs and the quality of in vitro matured oocytes, establishing a causal link between NAD+ abundance and oocyte senescence. Similarly, transgenic mice overexpressing NMNAT1, which localizes to the nucleus, but not NMAT3, found in the mitochondria, increases oocyte production suggesting that nuclear NAD+ consuming enzymes are important mediators of this phenotype. Remarkably, NMN supplementation in aged female mice improves fertility outcomes, as reflected by shorter time for the first litter and increased litter sizes. Further examination should delineate the molecular pathways activated upon NAD+ levels restoration via NMN supplementation.
Calorie restriction (CR), an intervention that reduces the caloric intake without compromising the ingestion of essential nutrients, partially preserves the ovarian reserve (Garcia et al., 2019; Li et al., 2015; Luo et al., 2012; Nelson, Gosden, & Felicio, 1985; Selesniemi, Lee, Muhlhauser, & Tilly, 2011) and improves oocyte quality (Selesniemi et al., 2011). In rodents, CR limits follicle recruitment therefore delaying ovarian reserve depletion. Indeed, under these circumstances, ovaries have increased primordial follicle numbers and reduced atretic ones. It is well documented that CR increases NAD+ levels resulting in Sirtuin activation (Vaquero & Reinberg, 2009). CR increases SIRT1 and SIRT6 protein levels in the ovary but what the downstream effectors are or whether other Sirtuins respond to this intervention are not known (Luo et al., 2012). CR reduces mTOR signaling as measured by reduced phosphorylation of p70-S6K (Zhou et al., 2014). mTOR is also a major energy sensor that becomes activated with high nutrients to promote anabolism and cell growth (Saxton & Sabatini, 2017). SIRT1 is a negative regulator of mTOR signaling by interacting with TSC2 (Ghosh, McBurney, & Robbins, 2010), a component of the mTOR inhibitory complex, in MEFs and HeLA cells but whether this functional interplay may also exist in the ovary is not known. CR also increases FOXA3 expression, a transcription factor which is thought to be important to maintain follicles in a quiescent state. SIRT1 deacetylates and activates FOXA3 to prevent cellular senescence in the lung, but it remains to be seen whether the SIRT1/FOXO3a axis is important for ovarian reserve preservation during CR.
Consistent with Sirtuins being central mediators of the cellular response to CR, Sirtuin-activating compounds recapitulate the effects of CR in the ovary. Resveratrol, a polyphenol found in the skin of red grapes and red wine, activates SIRT1 and improves fertility of aged female mice (Liu et al., 2013). Long term exposure to resveratrol helps extend the reproductive lifespan. Resveratrol increases the quantity of primordial and primary oocytes and the number of eggs at ovulation. In addition, egg quality is enhanced as measured by parameters such as spindle morphology and chromosome alignment. Resveratrol administration also increases telomere length in oocytes which is consistent with previous observation that SIRT1 overexpression contributes to telomere maintenance in somatic cell (Palacios et al., 2010).
Conclusions
Sirtuins are critical regulators of reproductive health at multiple levels, including endocrine signaling and germ cell development. In females, Sirtuins have been mostly studied in cellular and mouse models of Sirtuin removal or depletion, albeit with different outcomes (Table 1). These studies have unveiled critical roles for Sirtuins during oocyte maturation. Sirtuins protect oocytes from oxidative stress, a major problem encountered during in vitro maturation techniques (Agarwal, Aponte-Mellado, Premkumar, Shaman, & Gupta, 2012; Soto-Heras & Paramio, 2020). Sirtuins also promote epigenetic changes during oocyte maturation but what are the readers of these epigenetic marks and the downstream biological functions remain mostly unknown.
Because the age of child-bearing is increasing in the Western world and because of the desire for women to have their own biological children, Sirtuins are attractive targets for sustaining and prolonging quality reproductive health (Figure 2). Sirtuin activation have the potential to improve the generation of high-quality gametes, with important implications in assisted reproductive technologies. Sirtuins are also critical regulator of the ovarian reserve (Luo et al., 2012; Vazquez, Blengini, et al., 2019), a major determinant of the reproductive lifespan. While SIRT1 preserves the ovarian reserve by limiting follicle activation, SIRT7 regulates embryonic meiotic recombination and therefore the initial establishment of primordial follicle pool. From a translational point of view, this knowledge is critical as it predicts that interventions aiming to maximize the ovarian reserve should target sirtuins activity during gestation, possibly as maternal supplements, and later on in life to limit follicle recruitment and ovarian reserve depletion. Importantly, diminished ovarian reserve has been associated with other disease risks indicating that Sirtuin activity on the female reproductive system may benefit overall women’s health (Pal, Bevilacqua, Zeitlian, Shu, & Santoro, 2008; Quinn & Cedars, 2018).
Our current knowledge about the role of Sirtuins in oogenesis has increased considerably in the last decade. However, there are still several open questions that should be addressed to provide a full perspective of the contribution of sirtuins in oocyte physiology and female fertility: First, despite the key role of Sirtuins in the DNA damage response in somatic cells (Vazquez et al., 2017), we still do not know whether Sirtuins regulate DNA repair pathways in meiosis. In this sense, a recent link between SIRT7 and chromosome synapsis during meiotic recombination (Vazquez, Blengini, et al., 2019) supports this possibility. Second, whether Sirtuins regulate chromosome segregation or spindle function during meiotic divisions is still poorly understood. Sirtuins regulate epigenetic marks known to be enriched atpericentromeric and centromeric chromatin (Tasselli et al., 2016; Vaquero et al., 2007) but how this may impact the recruitment of kinetochore proteins and further chromosome segregation is not known. Moreover, although we have some evidence suggesting a role of Sirtuins in meiotic spindle regulation, particularly SIRT1, SIRT2 and SIRT4 (figure 1A–1C), no clear mechanism has been described so far. Third, another important issue is whether Sirtuins regulate gene silencing in prophase I-arrested oocytes. The well-established role of nuclear Sirtuins as gene silencers, suggests that they may be involved in genome compaction and global transcription repression that occurs in prophase I-arrested oocytes prior to meiotic resumption. Finally, a remaining question is whether Sirtuins limit attrition of fetal oocytes. In somatic cells, nuclear Sirtuins are important to silence DNA repetitive elements, including LINE-1 retrotransposons (Van Meter et al., 2014; Vazquez, Thackray, et al., 2019), which are known to drive oocyte attrition during oocyte development.
Future research should address not only these important issues but also define the specific contribution of the Sirtuin family members in female and male reproduction. Understanding these questions, will undoubtedly expand our basic knowledge in germ cells and mammalian genome dynamics and will provide significant advances in our understanding of the molecular pathways related with human infertility.
Data sharing
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Acknowledgements
We apologize to the many colleagues whose work could not be cited due to space limitations. We thank the members of the Schindler and Vaquero labs for constructive discussions, in particular Leela Biswas for assisting with the oocyte cartoon. We also thank the HGINJ and IJC for institutional support. The Schindler lab is supported by grants from the National Institutes of Health (R01-HD091331 and R01-GM112801). The Chromatin Biology group is funded through Ministry of Economy and Competitiveness (SAF2017-88975R). BNV is supported by a Beatriu de Pinós Fellowship of the Catalonian Government Agency AGAUR (BP-2016-00250).
Footnotes
Conflict of interest
The authors have no conflicts of interest to declare.
References
- Adeoye O, Olawumi J, Opeyemi A, & Christiania O (2018). Review on the role of glutathione on oxidative stress and infertility. JBRA Assist Reprod, 22(1), 61–66. doi: 10.5935/1518-0557.20180003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A, Aponte-Mellado A, Premkumar BJ, Shaman A, & Gupta S (2012). The effects of oxidative stress on female reproduction: a review. Reprod Biol Endocrinol, 10, 49. doi: 10.1186/1477-7827-10-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, & Price BD (2014). DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci U S A, 111(25), 9169–9174. doi: 10.1073/pnas.1403565111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, . . . van Deursen JM (2004). BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet, 36(7), 744–749. doi: 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, . . . Chua KF (2012). SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature, 487(7405), 114–118. doi: 10.1038/nature11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause AS, & Haigis MC (2013). SIRT3 regulation of mitochondrial oxidative stress. Exp Gerontol, 48(7), 634–639. doi: 10.1016/j.exger.2012.08.007 [DOI] [PubMed] [Google Scholar]
- Becker JS, Nicetto D, & Zaret KS (2016). H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes. Trends Genet, 32(1), 29–41. doi: 10.1016/j.tig.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, . . . De Benedictis G (2005). A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics, 85(2), 258–263. doi: 10.1016/j.ygeno.2004.11.003 [DOI] [PubMed] [Google Scholar]
- Bertoldo MJ, Listijono DR, Ho WJ, Riepsamen AH, Goss DM, Richani D, . . . Wu LE (2020). NAD(+) Repletion Rescues Female Fertility during Reproductive Aging. Cell Rep, 30(6), 1670–1681 e1677. doi: 10.1016/j.celrep.2020.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank MF, & Grummt I (2017). The seven faces of SIRT7. Transcription, 8(2), 67–74. doi: 10.1080/21541264.2016.1276658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, & Handel MA (2018). Meiosis: the chromosomal foundation of reproduction. Biol Reprod, 99(1), 112–126. doi: 10.1093/biolre/ioy021 [DOI] [PubMed] [Google Scholar]
- Bosch-Presegue L, & Vaquero A (2014). Sirtuins in stress response: guardians of the genome. Oncogene, 33(29), 3764–3775. doi: 10.1038/onc.2013.344 [DOI] [PubMed] [Google Scholar]
- Chamani IJ, & Keefe DL (2019). Epigenetics and Female Reproductive Aging. Front Endocrinol (Lausanne), 10, 473. doi: 10.3389/fendo.2019.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, & Xiong Y (2011). Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep, 12(6), 534–541. doi: 10.1038/embor.2011.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, . . . Chua KF (2003). Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A, 100(19), 10794–10799. doi: 10.1073/pnas.1934713100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens M, Maresh JG, Yanagimachi R, Maeda G, & Allsopp R (2008). Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One, 3(2), e1571. doi: 10.1371/journal.pone.0001571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton CM, & Carroll J (2013). Biased inheritance of mitochondria during asymmetric cell division in the mouse oocyte. J Cell Sci, 126(Pt 13), 2955–2964. doi: 10.1242/jcs.128744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Emidio G, Falone S, Vitti M, D’Alessandro AM, Vento M, Di Pietro C, . . . Tatone C (2014). SIRT1 signalling protects mouse oocytes against oxidative stress and is deregulated during aging. Hum Reprod, 29(9), 2006–2017. doi: 10.1093/humrep/deu160 [DOI] [PubMed] [Google Scholar]
- Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, . . . Tsai LH (2013). SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci, 16(8), 1008–1015. doi: 10.1038/nn.3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlon TA, Morris BJ, Chen R, Masaki KH, Allsopp RC, Willcox DC, . . . Willcox BJ (2018). Analysis of Polymorphisms in 59 Potential Candidate Genes for Association With Human Longevity. J Gerontol A Biol Sci Med Sci, 73(11), 1459–1464. doi: 10.1093/gerona/glx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, & Guarente L (2006). Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev, 20(9), 1075–1080. doi: 10.1101/gad.1399706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Wada-Hiraike O, Hirano M, Kawamura Y, Sakurabashi A, Shirane A, . . . Fujii T (2014). SIRT3 positively regulates the expression of folliculogenesis- and luteinization-related genes and progesterone secretion by manipulating oxidative stress in human luteinized granulosa cells. Endocrinology, 155(8), 3079–3087. doi: 10.1210/en.2014-1025 [DOI] [PubMed] [Google Scholar]
- Gao M, Li X, He Y, Han L, Qiu D, Ling L, . . . Gu L (2018). SIRT7 functions in redox homeostasis and cytoskeletal organization during oocyte maturation. FASEB J, fj201800078RR. doi: 10.1096/fj.201800078RR [DOI] [PubMed] [Google Scholar]
- Garcia DN, Saccon TD, Pradiee J, Rincon JAA, Andrade KRS, Rovani MT, . . . Schneider A (2019). Effect of caloric restriction and rapamycin on ovarian aging in mice. Geroscience, 41(4), 395–408. doi: 10.1007/s11357-019-00087-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Li C, Li C, Huang Z, Zeng J, Han L, & Wang Q (2019). SIRT6 participates in the quality control of aged oocytes via modulating telomere function. Aging (Albany NY), 11(7), 1965–1976. doi: 10.18632/aging.101885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh HS, McBurney M, & Robbins PD (2010). SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One, 5(2), e9199. doi: 10.1371/journal.pone.0009199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, & Almouzni G (2004). Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol, 166(4), 493–505. doi: 10.1083/jcb.200403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Deng CX, Finley LW, Kim HS, & Gius D (2012). SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res, 72(10), 2468–2472. doi: 10.1158/0008-5472.CAN-11-3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Ge J, Zhang L, Ma R, Hou X, Li B, . . . Wang Q (2015). Sirt6 depletion causes spindle defects and chromosome misalignment during meiosis of mouse oocyte. Sci Rep, 5, 15366. doi: 10.1038/srep15366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikoshi N, Kumar P, Sharma GG, Chen M, Hunt CR, Westover K, . . . Pandita TK (2013). Genome-wide distribution of histone H4 Lysine 16 acetylation sites and their relationship to gene expression. Genome Integr, 4(1), 3. doi: 10.1186/2041-9414-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhtaniemi I, Hovatta O, La Marca A, Livera G, Monniaux D, Persani L, . . . Misrahi M (2018). Advances in the Molecular Pathophysiology, Genetics, and Treatment of Primary Ovarian Insufficiency. Trends Endocrinol Metab, 29(6), 400–419. doi: 10.1016/j.tem.2018.03.010 [DOI] [PubMed] [Google Scholar]
- Igosheva N, Abramov AY, Poston L, Eckert JJ, Fleming TP, Duchen MR, & McConnell J (2010). Maternal diet-induced obesity alters mitochondrial activity and redox status in mouse oocytes and zygotes. PLoS One, 5(4), e10074. doi: 10.1371/journal.pone.0010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iljas JD, & Homer HA (2020). Sirt3 is dispensable for oocyte quality and female fertility in lean and obese mice. FASEB J, 34(5), 6641–6653. doi: 10.1096/fj.202000153R [DOI] [PubMed] [Google Scholar]
- Janke C, & Montagnac G (2017). Causes and Consequences of Microtubule Acetylation. Curr Biol, 27(23), R1287–R1292. doi: 10.1016/j.cub.2017.10.044 [DOI] [PubMed] [Google Scholar]
- Jeong SM, Hwang S, & Seong RH (2016). SIRT4 regulates cancer cell survival and growth after stress. Biochem Biophys Res Commun, 470(2), 251–256. doi: 10.1016/j.bbrc.2016.01.078 [DOI] [PubMed] [Google Scholar]
- Johnson S, & Imai SI (2018). NAD (+) biosynthesis, aging, and disease. F1000Res, 7, 132. doi: 10.12688/f1000research.12120.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, . . . Cohen HY (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature, 483(7388), 218–221. doi: 10.1038/nature10815 [DOI] [PubMed] [Google Scholar]
- Kaufman JM, Lapauw B, Mahmoud A, T’Sjoen G, & Huhtaniemi IT (2019). Aging and the Male Reproductive System. Endocr Rev, 40(4), 906–972. doi: 10.1210/er.2018-00178 [DOI] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, . . . Chua KF (2009). SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell, 136(1), 62–74. doi: 10.1016/j.cell.2008.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Uchijima Y, Horike N, Tonami K, Nishiyama K, Amano T, . . . Kurihara H (2010). Sirt3 protects in vitro-fertilized mouse preimplantation embryos against oxidative stress-induced p53-mediated developmental arrest. J Clin Invest, 120(8), 2817–2828. doi: 10.1172/JCI42020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, . . . Deng CX (2011). SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell, 20(4), 487–499. doi: 10.1016/j.ccr.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Liu H, Tazaki M, Nagata M, & Aoki F (2003). Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol, 162(1), 37–46. doi: 10.1083/jcb.200303047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, & Lombard DB (2018). Functions of the sirtuin deacylase SIRT5 in normal physiology and pathobiology. Crit Rev Biochem Mol Biol, 53(3), 311–334. doi: 10.1080/10409238.2018.1458071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Fu YC, Xu JJ, Lin XH, Chen XC, Zhang XM, & Luo LL (2015). Caloric restriction promotes the reserve of follicle pool in adult female rats by inhibiting the activation of mammalian target of rapamycin signaling. Reprod Sci, 22(1), 60–67. doi: 10.1177/1933719114542016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, . . . Yu W (2016). SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun, 7, 12235. doi: 10.1038/ncomms12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zheng P, & Dean J (2010). Maternal control of early mouse development. Development, 137(6), 859–870. doi: 10.1242/dev.039487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Yin Y, Ye X, Zeng M, Zhao Q, Keefe DL, & Liu L (2013). Resveratrol protects against age-associated infertility in mice. Hum Reprod, 28(3), 707–717. doi: 10.1093/humrep/des437 [DOI] [PubMed] [Google Scholar]
- Long GY, Yang JY, Xu JJ, Ni YH, Zhou XL, Ma JY, . . . Luo LL (2019). SIRT1 knock-in mice preserve ovarian reserve resembling caloric restriction. Gene, 686, 194–202. doi: 10.1016/j.gene.2018.10.040 [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, & Kroemer G (2013). The hallmarks of aging. Cell, 153(6), 1194–1217. doi: 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LL, Chen XC, Fu YC, Xu JJ, Li L, Lin XH, . . . Zhang XM (2012). The effects of caloric restriction and a high-fat diet on ovarian lifespan and the expression of SIRT1 and SIRT6 proteins in rats. Aging Clin Exp Res, 24(2), 125–133. doi: 10.3275/7660 [DOI] [PubMed] [Google Scholar]
- Ma R, Zhang Y, Zhang L, Han J, & Rui R (2015). Sirt1 protects pig oocyte against in vitro aging. Anim Sci J, 86(9), 826–832. doi: 10.1111/asj.12360 [DOI] [PubMed] [Google Scholar]
- Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, . . . Cristea IM (2014). Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell, 159(7), 1615–1625. doi: 10.1016/j.cell.2014.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, . . . Lemieux M (2003). The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol, 23(1), 38–54. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/12482959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, . . . Chua KF (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature, 452(7186), 492–496. doi: 10.1038/nature06736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Z, Gao J, & Yu Y (2018). The Roles of Mitochondrial SIRT4 in Cellular Metabolism. Front Endocrinol (Lausanne), 9, 783. doi: 10.3389/fendo.2018.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, . . . Alt FW (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell, 124(2), 315–329. doi: 10.1016/j.cell.2005.11.044 [DOI] [PubMed] [Google Scholar]
- Nelson JF, Gosden RG, & Felicio LS (1985). Effect of dietary restriction on estrous cyclicity and follicular reserves in aging C57BL/6J mice. Biol Reprod, 32(3), 515–522. doi: 10.1095/biolreprod32.3.515 [DOI] [PubMed] [Google Scholar]
- Nevoral J, Landsmann L, Stiavnicka M, Hosek P, Moravec J, Prokesova S, . . . Kralickova M (2019). Epigenetic and non-epigenetic mode of SIRT1 action during oocyte meiosis progression. J Anim Sci Biotechnol, 10, 67. doi: 10.1186/s40104-019-0372-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, & Verdin E (2003). The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell, 11(2), 437–444. doi: 10.1016/s1097-2765(03)00038-8 [DOI] [PubMed] [Google Scholar]
- Pacella-Ince L, Zander-Fox DL, & Lan M (2014). Mitochondrial SIRT3 and its target glutamate dehydrogenase are altered in follicular cells of women with reduced ovarian reserve or advanced maternal age. Hum Reprod, 29(7), 1490–1499. doi: 10.1093/humrep/deu071 [DOI] [PubMed] [Google Scholar]
- Pal L, Bevilacqua K, Zeitlian G, Shu J, & Santoro N (2008). Implications of diminished ovarian reserve (DOR) extend well beyond reproductive concerns. Menopause, 15(6), 1086–1094. doi: 10.1097/gme.0b013e3181728467 [DOI] [PubMed] [Google Scholar]
- Palacios JA, Herranz D, De Bonis ML, Velasco S, Serrano M, & Blasco MA (2010). SIRT1 contributes to telomere maintenance and augments global homologous recombination. J Cell Biol, 191(7), 1299–1313. doi: 10.1083/jcb.201005160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, . . . Schimenti JC (1998). Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell, 1(5), 697–705. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9660953 [DOI] [PubMed] [Google Scholar]
- Qiu D, Hou X, Han L, Li X, Ge J, & Wang Q (2018). Sirt2-BubR1 acetylation pathway mediates the effects of advanced maternal age on oocyte quality. Aging Cell, 17(1). doi: 10.1111/acel.12698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, & Chen D (2010). Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab, 12(6), 662–667. doi: 10.1016/j.cmet.2010.11.015 [DOI] [PubMed] [Google Scholar]
- Quinn MM, & Cedars MI (2018). Cardiovascular health and ovarian aging. Fertil Steril, 110(5), 790–793. doi: 10.1016/j.fertnstert.2018.07.1152 [DOI] [PubMed] [Google Scholar]
- Rocca MS, Foresta C, & Ferlin A (2019). Telomere length: lights and shadows on their role in human reproduction. Biol Reprod, 100(2), 305–317. doi: 10.1093/biolre/ioy208 [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, & Camerini-Otero RD (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell, 6(5), 975–987. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/11106738 [DOI] [PubMed] [Google Scholar]
- Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, . . . Auwerx J (2014). A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab, 20(5), 856–869. doi: 10.1016/j.cmet.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, . . . Imai S (2013). Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab, 18(3), 416–430. doi: 10.1016/j.cmet.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, & Sabatini DM (2017). mTOR Signaling in Growth, Metabolism, and Disease. Cell, 169(2), 361–371. doi: 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- Seifert EL, Caron AZ, Morin K, Coulombe J, He XH, Jardine K, . . . McBurney MW (2012). SirT1 catalytic activity is required for male fertility and metabolic homeostasis in mice. FASEB J, 26(2), 555–566. doi: 10.1096/fj.11-193979 [DOI] [PubMed] [Google Scholar]
- Selesniemi K, Lee HJ, Muhlhauser A, & Tilly JL (2011). Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. Proc Natl Acad Sci U S A, 108(30), 12319–12324. doi: 10.1073/pnas.1018793108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L, Martinez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, . . . Vaquero A (2013). The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev, 27(6), 639–653. doi: 10.1101/gad.211342.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, & Yoshida M (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol, 6(4), a018713. doi: 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, & Peterson CL (2006). Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science, 311(5762), 844–847. doi: 10.1126/science.1124000 [DOI] [PubMed] [Google Scholar]
- Soto-Heras S, & Paramio MT (2020). Impact of oxidative stress on oocyte competence for in vitro embryo production programs. Res Vet Sci, 132, 342–350. doi: 10.1016/j.rvsc.2020.07.013 [DOI] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, . . . Gius D (2010). Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell, 40(6), 893–904. doi: 10.1016/j.molcel.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, . . . Chua KF (2016). SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol, 23(5), 434–440. doi: 10.1038/nsmb.3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasselli L, Zheng W, & Chua KF (2017). SIRT6: Novel Mechanisms and Links to Aging and Disease. Trends Endocrinol Metab, 28(3), 168–185. doi: 10.1016/j.tem.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharp ME, Malki S, & Bortvin A (2020). Maximizing the ovarian reserve in mice by evading LINE-1 genotoxicity. Nat Commun, 11(1), 330. doi: 10.1038/s41467-019-14055-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Firsanov D, Zhang Z, Cheng Y, Luo L, Tombline G, . . . Gorbunova V (2019). SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species. Cell, 177(3), 622–638 e622. doi: 10.1016/j.cell.2019.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, . . . Mostoslavsky R (2013). SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell, 51(4), 454–468. doi: 10.1016/j.molcel.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Ven RAH, Santos D, & Haigis MC (2017). Mitochondrial Sirtuins and Molecular Mechanisms of Aging. Trends Mol Med, 23(4), 320–331. doi: 10.1016/j.molmed.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, & Gorbunova V (2014). SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun, 5, 5011. doi: 10.1038/ncomms6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A (2009). The conserved role of sirtuins in chromatin regulation. Int J Dev Biol, 53(2–3), 303–322. doi: 10.1387/ijdb.082675av [DOI] [PubMed] [Google Scholar]
- Vaquero A, & Reinberg D (2009). Calorie restriction and the exercise of chromatin. Genes Dev, 23(16), 1849–1869. doi: 10.1101/gad.1807009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, & Reinberg D (2007). SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature, 450(7168), 440–444. doi: 10.1038/nature06268 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, & Reinberg D (2004). Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell, 16(1), 93–105. doi: 10.1016/j.molcel.2004.08.031 [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, . . . Reinberg D (2006). SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev, 20(10), 1256–1261. doi: 10.1101/gad.1412706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez BN, Blengini CS, Hernandez Y, Serrano L, & Schindler K (2019). SIRT7 promotes chromosome synapsis during prophase I of female meiosis. Chromosoma. doi: 10.1007/s00412-019-00713-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez BN, Thackray JK, & Serrano L (2017). Sirtuins and DNA damage repair: SIRT7 comes to play. Nucleus, 8(2), 107–115. doi: 10.1080/19491034.2016.1264552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez BN, Thackray JK, Simonet NG, Chahar S, Kane-Goldsmith N, Newkirk SJ, . . . Serrano L (2019). SIRT7 mediates L1 elements transcriptional repression and their association with the nuclear lamina. Nucleic Acids Res, 47(15), 7870–7885. doi: 10.1093/nar/gkz519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez BN, Thackray JK, Simonet NG, Kane-Goldsmith N, Martinez-Redondo P, Nguyen T, . . . Serrano L (2016). SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J, 35(14), 1488–1503. doi: 10.15252/embj.201593499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng Y, Li J, Yu Y, Zhang W, Song M, . . . Liu GH (2020). Single-Cell Transcriptomic Atlas of Primate Ovarian Aging. Cell, 180(3), 585–600 e519. doi: 10.1016/j.cell.2020.01.009 [DOI] [PubMed] [Google Scholar]
- Wang WW, Angulo-Ibanez M, Lyu J, Kurra Y, Tong Z, Wu B, . . . Liu WR (2019). A Click Chemistry Approach Reveals the Chromatin-Dependent Histone H3K36 Deacylase Nature of SIRT7. J Am Chem Soc. doi: 10.1021/jacs.8b12083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Zhou LS, Zhao YZ, Wang SW, Chen LL, Liu LX, . . . Ye D (2014). Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J, 33(12), 1304–1320. doi: 10.1002/embj.201387224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster A, & Schuh M (2017). Mechanisms of Aneuploidy in Human Eggs. Trends Cell Biol, 27(1), 55–68. doi: 10.1016/j.tcb.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Yu I, Garnham CP, & Roll-Mecak A (2015). Writing and Reading the Tubulin Code. J Biol Chem, 290(28), 17163–17172. doi: 10.1074/jbc.R115.637447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS, . . . Auwerx J (2013). Metabolic characterization of a Sirt5 deficient mouse model. Sci Rep, 3, 2806. doi: 10.1038/srep02806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Jiang M, Wu X, Diao F, Qiu D, Hou X, . . . Wang Q (2018). SIRT4 is essential for metabolic control and meiotic structure during mouse oocyte maturation. Aging Cell, e12789. doi: 10.1111/acel.12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Han L, Ma R, Hou X, Yu Y, Sun S, . . . Wang Q (2015). Sirt3 prevents maternal obesity-associated oxidative stress and meiotic defects in mouse oocytes. Cell Cycle, 14(18), 2959–2968. doi: 10.1080/15384101.2015.1026517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hou X, Ma R, Moley K, Schedl T, & Wang Q (2014). Sirt2 functions in spindle organization and chromosome alignment in mouse oocyte meiosis. FASEB J, 28(3), 1435–1445. doi: 10.1096/fj.13-244111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu XQ, Lu S, Guo YL, & Ma X (2006). Deficit of mitochondria-derived ATP during oxidative stress impairs mouse MII oocyte spindles. Cell Res, 16(10), 841–850. doi: 10.1038/sj.cr.7310095 [DOI] [PubMed] [Google Scholar]
- Zhao HC, Ding T, Ren Y, Li TJ, Li R, Fan Y, . . . Qiao J (2016). Role of Sirt3 in mitochondrial biogenesis and developmental competence of human in vitro matured oocytes. Hum Reprod, 31(3), 607–622. doi: 10.1093/humrep/dev345 [DOI] [PubMed] [Google Scholar]
- Zhou XL, Xu JJ, Ni YH, Chen XC, Zhang HX, Zhang XM, . . . Fu YC (2014). SIRT1 activator (SRT1720) improves the follicle reserve and prolongs the ovarian lifespan of diet-induced obesity in female mice via activating SIRT1 and suppressing mTOR signaling. J Ovarian Res, 7, 97. doi: 10.1186/s13048-014-0097-z [DOI] [PMC free article] [PubMed] [Google Scholar]