Abstract
The continuing emergence and development of pathogenic microorganisms that are resistant to antibiotics constitute an increasing global concern, and the effort in new antimicrobials discovery will remain relevant until a lasting solution is found. A new bacterial strain, designated JFL21, was isolated from seafood and identified as B. amyloliquefaciens. The antimicrobial substance produced by B. amyloliquefaciens JFL21 showed low toxicity to most probiotics but exhibited strong antimicrobial activities against multidrug-resistant foodborne pathogens. The partially purified antimicrobial substance, Anti-JFL21, was characterized to be a multiple lipopeptides mixture comprising the families of surfactin, fengycin, and iturin. Compared with commercially available polymyxin B and Nisin, Anti-JFL21 not only could exhibit a wider and stronger antibacterial activity toward Gram-positive pathogens but also inhibit the growth of a majority of fungal pathogens. After further separation through gel filtration chromatography (GFC), the family of surfactin, fengycin, and iturin were obtained, respectively. The results of the antimicrobial test pointed out that only fengycin family presented marked antimicrobial properties against the indicators of L. monocytogenes, A. hydrophila, and C. gloeosporioides, which demonstrated that fengycins might play a major role in the antibacterial and antifungal activity of Anti-JFL21. Additionally, the current study also showed that the fengycins produced by B. amyloliquefaciens JFL21 not only maintained stable anti-Listeria activity over a broad pH and temperature range, but also remained active after treatment with ultraviolet sterilization, chemical reagents, and proteolytic enzymes. Therefore, the results of this study suggest the new strain and its antimicrobials are potentially useful in food preservation for the biological control of the multidrug-resistant foodborne pathogens.
Keywords: fengycin, Bacillus amyloliquefaciens, antimicrobial activity, multidrug-resistant foodborne pathogens, probiotics, lipopeptides
Introduction
Foodborne diseases have been a serious global public health issue and most of them are caused by foodborne pathogens such as Salmonella, Shigella, Vibrio, Escherichia coli O157, Yersinia enterocolitica, Listeria monocytogenes, Staphylococcus aureus, and Aspergillus flavus, etc. (World Health Organization, 2015; European Food Safety Authority, 2018; Tack et al., 2019). According to the data reported by the World Health Organization (WHO), foodborne diseases have caused approximate 600 million illness and 420, 000 deaths in the world and at least $100 billion costs in low- and middle-income countries every year, and the actual number of cases is also likely to be underestimated due to the high failure rate of reporting foodborne diseases worldwide (World Health Organization, 2015, 2019). Although antibiotics are essential to treat infections caused by foodborne pathogens, their overuse and misuse has been linked to the emergence and spread of multiple resistant strains during the past decades and has led to public awareness (Garedew et al., 2015; Kuch et al., 2018; U.S. Centers for Disease Control and Prevention, 2019). According to the government data of U.S. Centers for Disease Control and Prevention (CDC), antibiotic-resistant infections from foodborne germs (bacteria and fungi) still cause more than 2.8 million illnesses and 35,000 deaths in the United States each year despite attempts to combat the problem (U.S. Centers for Disease Control and Prevention, 2019). Thus, there is an urgent need for alternatives to antibiotics to fight against the foodborne pathogens.
In recent years, many studies have shown that the cyclic lipopeptides (CLPs) produced by Bacillus spp. have potent antimicrobial activity against antibiotic-resistant strains and can be generally divided into three main families: surfactin, fengycin, and iturin (Chi et al., 2015; Jemil et al., 2017; Perez et al., 2017; Piewngam et al., 2018). Due to their specific amphiphilic structure, CLPs primarily destroy target organisms by directly disrupting the integrity of the plasma membrane or cell wall in a detergent-like manner, and thus display a lower propensity to develop resistance than do conventional antibiotics (Banat et al., 2010; Mandal et al., 2013; Singh and Abraham, 2014; Patel et al., 2015; Ndlovu et al., 2017). Besides, CLPs are biodegradable, biocompatible, eco-friendly, relatively non-toxic, and resistant to extreme conditions of temperatures, pH, and salinity (Pradhan et al., 2013; Ben Ayed et al., 2017). Because of these attractive characteristics, naturally produced antimicrobial CLPs have received increasing attention as promising new antibiotic candidates for food, pharmaceutical, and biomedical applications.
Considering the increasingly prominent problems of antibiotic contamination and food safety, this research aimed to search for candidate Bacillus strains that could produce a potent antimicrobial agent to combat various foodborne pathogens with multi-drug resistant profiles. Besides, FITR, HPLC, and MALDI-TOF MS analysis were exploited to identify the structural characteristics of the partially purified antimicrobial substances Anti-JFL21. Moreover, the different lipopeptide families in Anti-JFL21 was further separated, and the effective antimicrobial family was elucidated. what’s more, the fengycins stability after the treatment of heating, pH change, ultraviolet sterilization, enzymes, and chemical reagents was determined to evaluate the possible incorporation in the food production chain.
Materials and Methods
Microorganisms and Cultivation Conditions
In this study, the three Bacillus isolates named JFL21, LQG17, and LQG36, with excellent antimicrobial properties were, respectively, isolated from the gut of hairtail, fermented soybean, and pickle purchased from Guangzhou farmers market. Bacillus subtilis 168, one of the model organisms of the Bacillus genus, was purchased from the Bacillus Genetic Stock Center (BGSC). The thirty-three indicator strains used for evaluation of antibacterial and antifungal activity were selected for their importance as probiotics or pathogens in food and seafood products. The source and culture conditions of the thirty-three indicator strains are listed in Table 1. The bacterial or fungal cultures were preserved in 25% glycerol at −80°C.
TABLE 1.
Drug resistance of representative bacterial pathogens, pathogenic fungi, and probiotics in food and seafood products.a
| Indicator microorganisms | Sourceb | Mediac | T(°C) | Antibiotic sensitivityd |
|||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||||
| Lactic acid bacteria (Probiotics) | |||||||||||||||
| Lactobacillus plantarum | Yogurt | MRS | 37 | +++ | ++ | − | − | +++ | − | +++ | − | +++ | − | − | − |
| Pediococcus pentosaceus | Tilapia gut | MRS | 37 | ++ | ++ | − | − | +++ | − | +++ | − | ++ | − | − | − |
| Lactobacillus casei | Pickle | MRS | 37 | +++ | − | − | +++ | − | − | − | − | ++ | − | − | − |
| Lactococcus lactis | Pleurotus eryngii | MRS | 37 | +++ | ++ | +++ | ++ | +++ | ++ | +++ | − | ++ | − | − | − |
| Leuconostoc mesenteroides | Shrimp gut | MRS | 37 | +++ | +++ | +++ | +++ | +++ | ++ | +++ | ++ | +++ | − | − | − |
| Gram-positive pathogen | |||||||||||||||
| Listeria monocytogenes | ATCC 19111 | BHI | 37 | +++ | − | +++ | − | ++ | − | ++ | ++ | +++ | − | − | − |
| Staphylococcus aureus | ATCC 12600 | TSA | 37 | +++ | ++ | +++ | ++ | +++ | +++ | ++ | ++ | +++ | − | − | − |
| Staphylococcus epidermidis | ATCC 14990 | BHI | 37 | − | ++ | ++ | − | − | + | + | + | +++ | + | − | − |
| Staphylococcus warneri | ATCC 27836 | TSA | 37 | − | − | +++ | + | − | ++ | − | − | ++ | − | − | − |
| Staphylococcus haemolyticus | ATCC 29970 | TSA | 37 | +++ | − | +++ | +++ | − | ++ | − | − | +++ | − | − | − |
| Bacillus cereus | ATCC 14579 | TSA | 37 | + | ++ | + | + | ++ | ++ | + | +++ | +++ | − | − | − |
| Gram-negative pathogen | |||||||||||||||
| Vibrio parahaemolyticus | ATCC 17802 | BHI | 37 | ++ | − | − | − | − | + | − | + | ++ | − | − | − |
| Vibrio harveyi | ATCC 33843 | TSA | 37 | +++ | − | ++ | − | − | − | + | +++ | +++ | − | − | − |
| Vibrio vulnificus | ATCC 27562 | BHI | 37 | +++ | − | − | − | − | + | +++ | + | + | − | − | − |
| Vibrio campbellii | ATCC 33863 | TSA | 37 | − | ++ | +++ | − | − | + | − | − | ++ | − | − | − |
| Pseudomonas aeruginosa | ATCC 10145 | TSA | 37 | − | − | +++ | − | − | − | − | − | +++ | − | − | − |
| Aeromonas hydrophila | ATCC 7966 | TSA | 37 | − | +++ | +++ | ++ | − | ++ | + | − | ++ | − | − | − |
| Indicator microorganisms | Sourceb | Mediac | T(°C) |
Antibiotic sensitivityd |
|||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | ||||
| Escherichia coli O157:H7 | ATCC 35150 | TSA | 37 | +++ | + | ++ | − | − | ++ | + | − | ++ | − | − | − |
| Salmonella choleraesuis | ATCC 10708 | TSA | 37 | + | +++ | ++ | − | − | + | − | + | + | − | − | − |
| Salmonella typhimurium | CMCC(B) 50115 | TSA | 37 | +++ | − | +++ | − | − | ++ | − | + | + | − | − | − |
| Shigella flexneri | ATCC 29903 | TSA | 37 | +++ | +++ | +++ | − | − | + | ++ | − | + | − | − | − |
| Yersinia enterocolitica | ATCC 9610 | TSA | 37 | − | − | − | − | − | ++ | + | ++ | +++ | − | − | − |
| Proteus mirabilis | ATCC 29906 | TSA | 37 | +++ | + | − | − | − | + | − | ++ | − | − | − | − |
| Cronobacter sakazakii | ATCC 51329 | TSA | 37 | +++ | ++ | + | − | − | ++ | + | + | ++ | + | − | − |
| Klebsiella pneumoniae | ATCC 13883 | TSA | 37 | − | +++ | +++ | ++ | ++ | ++ | ++ | − | ++ | − | − | − |
| Enterobacter aerogenes | CMCC(B) 45103 | TSA | 37 | − | − | +++ | − | − | + | − | − | ++ | − | − | − |
| Pathogenic fungi | |||||||||||||||
| Rhizopus oryzae | GIM 3.126 | PDA | 28 | − | − | − | − | − | − | − | − | − | − | − | − |
| Aspergillus niger | Grape | PDA | 28 | − | − | − | − | − | − | − | − | − | − | − | ++ |
| Aspergillus flavus | GIM 3.18 | PDA | 28 | − | − | − | − | − | − | − | − | − | − | − | − |
| Aspergillus fumigatus | GIM 3.19 | PDA | 28 | − | − | − | − | − | − | − | − | − | − | +++ | − |
| Peronophythora litchii | Litchi | PDA | 28 | − | − | − | − | − | − | − | − | − | − | − | ++ |
| Colletotrichum gloeosporioides | Mango | PDA | 28 | − | − | − | − | − | − | − | − | − | − | − | − |
| Penicillium polonicum | Orange | PDA | 28 | − | − | − | − | − | − | − | − | − | − | +++ | − |
aThe experimental concentration of each antibiotic was 100 ug/ml. 1, Ampicillin; 2, chloramphenicol; 3, Tetracycline; 4, Chlorotetracycline; 5, Erythromycin; 6, kanamycin; 7, Azithromycin; 8, Apramycin; 9, Gentamicin; 10, Spectinomycin; 11, Benomyl; 12, Actidione. bATCC, American Type Culture Collection; CMCC, National Center for Medical Culture Collections; Rhizopus oryzae, Aspergillus flavus, and Aspergillus fumigatus were purchased from Guangdong Culture Collection Center; The other indicator bacteria were isolated from different food samples and stored in our laboratory. cMRS, de Man, Rogosa and Sharpe medium; TSA, Trypticase Soya Agar; BHI, Brain Heart Infusion; PDA, Potato Dextrose Agar. dThe inhibition zone (IZ) were measured based on the agar disk diffusion assay. −: Sensitivity, IZ ≤ 9; +: Slight resistance, 9 ≤ IZ < 15 mm; ++: Moderate resistance, 15 ≤ IZ < 21 mm; +++: High resistance, IZ ≥ 21 mm.
Measurement of Drug Resistance of Probiotics and Pathogens
Drug resistance of probiotics and pathogens were determined according to the agar disk diffusion assay (Xu et al., 2014; Perez et al., 2017). Briefly, Sterile Oxford cups (10 mm × 6 mm × 8 mm, height × inner diameter × outer diameter) were placed on the assay medium seeded with the fresh culture suspension of different indicator strains (about 108 colony forming units/mL for bacterial cells and 106 spores/mL for fungal strains). Each cup was added with 100 μL of commonly used antibiotics (100 μg/mL) (Table 1). Besides, the same amount of sterile water, anhydrous ethanol, DMSO, and 0.01 mol/L HCl that applied to prepare antibiotics were used as the negative control to subtract the inhibitory activity of the solvents. After incubation 24 h at 37°C for bacteria and 7 days at 28°C for fungal strains, the inhibition zones were measured and recorded as a mean diameter (mm). All tests were conducted in triplicate.
Identification of Bacterial Strain and Phylogenetic Analysis
Genomic DNA from the Bacillus spp. was extracted using the HiPure Bacterial DNA Kit (Magen, China) according to the manufacturer’s protocol. The 16S rRNA gene fragment from each of the isolates was amplified using universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) according to the method described by Dimkić et al. (2017). The amplified product was purified with the QIAquick PCR purification kit (Qiagen, Germany) and sent for sequencing to Tianyi Huiyuan Bioscience & Technology Inc. (Guangzhou, China). The sequences obtained were deposited in GenBank under the accession numbers MT159453 for Bacillus sp. JFL21, MT159454 for Bacillus sp. LQG17, and MT159455 for Bacillus sp. LQG36. The homology comparison of the obtained sequences with previously sequenced genes in the GenBank database was performed, using the National Center for Biotechnology Information’s Blast search program (Bethesda, United States). The most closely related sequences of strain types were aligned using Clustal W software, and phylogenetic trees were constructed in MEGA version 7 using the Neighbor-joining method with 1,000 bootstrap repetitions (Dimkić et al., 2017; Perez et al., 2017).
Production of Antimicrobial Compounds
An inoculum of the glycerol stock of each of the Bacillus strains was streaked onto a Luria-Bertani (LB) plate which was incubated for 24 h at 37°C. A single colony was then inoculated into LB broth medium at 30°C and 200 r/min for 16 h (approximately 5 × 107 CFU/mL) to prepare seed cultures. A 1% (v/v) of inoculum was transferred into a 1 L shake flask containing 400 ml of Landy medium which was composed of the following: glucose 20 g/L, yeast extract 1 g/L, L-glutamic acid 5 g/L, KCl 0.5 g/L, MgSO4 0.5 g/L, KH2PO4 1 g/L, L-phenylalanine 2 mg/L, MnSO4 5 mg/L, FeSO4 0.15 mg/L, CuSO4 0.16 mg/L. The initial pH was adjusted to 7.0 and submerged fermentation was carried out in the rotary shaker at 30°C and 200 r/min for 48 h to produce the antimicrobial substance. After fermentation, the culture was centrifuged at 10,000 × g for 15 min at 4°C to remove microbial cells. The cell-free supernatants (CFS) was further sterilized by filtration through a cellulose filter with a pore size of 0.45 μm. Thereafter the filtered CFS (100 μL) from different Bacillus strains was tested for antimicrobial activity against thirty-three indicator strains (Table 1) using the agar disk diffusion assay mentioned above. The same amount of uninoculated Landy medium was used as negative control.
Extraction and Partial Purification of the Antimicrobial Compounds
The partial purification of the antimicrobial substance was carried out by a combination of acid precipitation and methanol extraction method (Sharma et al., 2015; Ndlovu et al., 2017). Briefly, the filtered CFS was adjusted to pH 2.0 by the addition of 6N HCl and allowed to precipitate at 4°C overnight. The acid precipitate was then harvested by centrifugation at 10,000 × g for 15 min at 4°C, and the pellet was washed twice with the 100 mL pH 2.0 distilled water (prepared through a MilliQ system from Millipore, Billerica, United States). This pellet was then resuspended in distilled water by raising the pH to 7.0, lyophilized, and solvent-extracted with methanol. The methanol-soluble fraction was dried using a rotary vacuum evaporator at 45°C. After the removal of methanol, a minimum quantity of methanol was added to dissolve the antimicrobial compound, which was again lyophilized and weighed for quantification. The crude extract from the CFS of Bacillus sp. JFL21 and Bacillus sp. LQG17 was designated as Anti-JFL21 and Anti-LQG17, respectively.
Determination of the Antimicrobial Activity of Anti-JFL21 and Anti-LQG17
Anti-JFL21 and Anti-LQG17 were prepared with methanol at the final concentration of 1 mg/mL and then tested for antimicrobial activity against the thirty-three indicator strains. Besides, the remaining supernatant after the extraction of Anti-JFL21 or Anti-LQG17 were separately readjusted to pH 7, and the antimicrobial experiment was also carried out to verify whether the antimicrobial substances were sufficiently extracted. Antimicrobial activity was measured by implementing the agar disk diffusion assay mentioned above. Meanwhile, the antimicrobial spectrum of Anti-JFL21 and Anti-LQG17 against bacteria and fungi was compared to those of Nisin (1 mg/mL, Sigma) and polymyxin B (1 mg/mL, Sigma). The same amount of sterile water, methanol, and 0.02 mol/L HCl that applied to prepare samples were used as negative controls.
Fourier Transformation Infra-Red (FTIR) Analysis
Fourier transformation infra-red spectroscopy analysis was performed to identify the structural groups of the purified biosurfactant using a Bruker Vertex 70v FTIR spectrometer (Bruker, Germany). In this experiment, 1 mg of Anti-JFL21 was ground with 100 mg of KBr in a pestle and pressed with load for 30 s to obtain translucent pellets. The FTIR spectra were collected at a frequency range from 4000 to 500 cm–1 (Sharma et al., 2015).
Reversed-Phase HPLC Analysis
The powder of Anti-JFL21 and Anti-LQG17 was dissolved in an aqueous solution of 40% methanol at 1 mg/mL and filtered using a 0.22 μm pore filter (Millipore, United States). A 20 μL aliquot was injected into an Inertsil ODS-SP C18 column (4.6 mm ID × 25 cm L, 5 μm particle diameter) in the HPLC system (Shimadzu, Japan) to separate and identify the isoforms, according to published methods with some modifications (Yang et al., 2015). In brief, the equal volumes of 1 mg/mL standard surfactin, fengycin and iturin (purity of 90% or greater, Sigma-Aldrich, United States) were fully mixed and used to confirm the RP-HPLC fraction groups of three lipopeptide families. The mobile phases consisted of water (A) and acetonitrile (B), and all of them contained 0.1% trifluoroacetic acid (TFA). The detailed gradient strategy for the acetonitrile-water mobile phase system was as follows: 0–3 min, 45% acetonitrile to 50% acetonitrile; 3–8 min, 50% acetonitrile to 80% acetonitrile; 8–25 min, 80% acetonitrile to 100% acetonitrile; 25–30 min, 100% acetonitrile. The total flow rate of the mobile phases was kept at 0.8 mL/min, and the products were monitored by absorbance at 215 nm. All HPLC solvents were prepared fresh daily and filtered under vacuum before use.
Matrix-Assisted Laser Desorption Ionization – Time of Flight Mass Spectrometry (MALDI-TOF MS) Analysis
Anti-JFL21 was further subjected to MALDI-TOF MS analysis and the methanol solvent was used as a negative control. The experiments were conducted using an ultrafleXtremeTM MALDI-TOF MS instrument (Bruker Daltonics, Germany) equipped with a 337 nm pulsed nitrogen laser. All tested samples (2 μL) were mixed with an equal volume of the matrix (a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile with 0.1% TFA), as described previously (Kim et al., 2010; Piewngam et al., 2018). The sample was spotted onto the gold-coated plate and air-dried. Then the target plate was loaded into an ultrafleXtremeTM MALDI-TOF MS instrument (Bruker Daltonics, Germany). The mass spectrum was analyzed in the range of 500–3500 Da. MALDI-TOF MS/MS coupled with LIFT mode in the same spectrometer was used to analyze the fragment ions of the selected precursor ions for further characterization of the amino acid sequence. Other identification score criteria used were those recommended by the manufacturer.
Isolation and Purification of Different Lipopeptides Family
To clarify which lipopeptide classes were responsible for antibacterial and antifungal activity, the family of iturin, fengycin, and surfactin were separated through Sephadex LH-20 gel filtration chromatography (GFC) (Kim et al., 2010; Zhang et al., 2013). L. monocytogenes, A. hydrophila, and C. gloeosporioides were used as indicator strains for quantifying antimicrobial effects since they were found to be the most sensitive indicator among Gram-positive pathogens, Gram-negative pathogens, and fungal pathogens, respectively. Briefly, the dried Anti-JFL21 was dissolved in 40% methanol (v/v) and prepared to a final concentration of 15 mg/mL, and filtered through a 0.2 μm membrane. To separate and purify the three lipopeptide family of Anti-JFL21, the filtrate (2 mL) of Anti-JFL21 was applied to a Sephadex LH-20 column (16 mm ID × 100 cm L) and fractionated by size-exclusion chromatography by using 40% methanol as eluent. A total of 100 fractions was collected (3 mL/tube) using a fraction collector (model DBS 100; Shanghai Hu Xi Analysis Instrument Factory Co. Ltd., Shanghai, China), and the flow rate of the eluent was kept at 0.5 mL/min. Then, each fraction was further detected using a UV spectrophotometer at 215 nm and analyzed by RP-HPLC, and tested for the antimicrobial activity.
Determination of the Minimal Inhibitory Concentration of the Fengycins From Different Sources
After separation through GFC, the isolated active fractions containing only fengycin were pooled and dried with a vacuum freeze dryer. To further evaluate and compare the antimicrobial efficacy of fengycins isolated from Anti-JFL21 with the commercial fengycins standard (Sigma-Aldrich, United States), the minimal inhibitory concentration (MIC) was measured according to the agar disk diffusion assay (Sharma et al., 2015). L. monocytogenes, A. hydrophila, and C. gloeosporioides were used as indicator microorganisms, which were typically representative of Gram-positive pathogen, Gram-negative pathogen, and fungal pathogen, respectively. In brief, fengycins and its commercial standard was dissolved in methanol at a final concentration of 0.8 mg/mL and diluted by two-fold dilution to a variety of concentrations. Samples of different concentrations were then used for measuring of the antimicrobial activity against indicator strains and the MIC value was defined as the lowest concentration of samples that inhibits the visible growth of a tested strain after a chosen incubation period. The same amount of methanol was used as negative controls and the experiments were repeated independently three times.
Biochemical Characterization of Fengycins
The powder of fengycins isolated from Anti-JFL21 was then dissolved in sterile phosphate-buffered saline (PBS) at pH 7.0 at a final concentration of 1 mg/mL and used for investigating the effect of temperature, pH, ultraviolet sterilization. The effect of temperature, pH, ultraviolet sterilization, enzymes, and chemical reagents on the antimicrobial activity of anti-JFL21 was evaluated by the agar disk diffusion assay using L. monocytogenes as an indicator strain (Sabaté and Audisio, 2013; Chalasani et al., 2015; Lee et al., 2016). The untreated sample was used as positive controls and its activity was defined as 100%. Negative controls were also performed without fengycins to subtract the inhibitory activity of the enzymes and chemical reagents.
In brief, the aliquots of fengycins were separately incubated under ultraviolet sterilization or in a water bath at different temperatures (37, 60, 80, and 100°C) for 2 h, and the residual antimicrobial activity was determined after cooling the treated samples to room temperature. Besides, the effect of pH on fengycins activity was examined over a pH range of 1.0–13.0 by adding 1N NaOH or HCl as appropriate. After incubation in the various pH for 2 h at 25°C, the samples were neutralized to pH 7.0 and its residual activity was measured. To evaluate stability to different enzymes, fengycins were incubated at 37°C for 2 h with 1 mg/mL (final concentration) of the following enzymes (Sigma, United States): cellulase, α-amylase, proteinase K, papain, bromelain, trypsin, and pepsin. The enzymes after incubation were inactivated by heating at 80°C for 10 min and then residual antimicrobial activities were measured. Also, the effects of various chemical reagents (EDTA, SDS, ZnSO4, and MnCl2) on fengycins activity were assayed after preincubating the fengycins with 1 mM (final concentration) of these reagents at 25°C and 100 r/min for 2 h.
Statistical Analysis
The diameters of the zones of inhibition against various indicator strains were expressed as mean values ± SD. All the assays were repeated at least in three separate experiments. The student’s t-test was utilized to determine the statistically significant difference in the residual anti-Listeria activity after the various treatments. The P-values of less than 0.05 (p < 0.05) were considered significant.
Results
Measurement of Drug Resistance of the Probiotics and Pathogens
To investigate the drug resistance of probiotics and pathogens used in this study, the inhibitory effects of the twelve commonly used antibiotics against thirty-three indicator strains were examined. The negative controls (sterile water, anhydrous ethanol, DMSO, and 0.01 mol/L HCl) did not show any antimicrobial ability against all the indicator strains (data not shown), which suggest that the solvents itself did not affect the antimicrobial activity of the antibiotics against the tested indicator strains. As presented in Table 1, all the pathogenic microorganisms used in this study were resistant to several different antibiotics, indicating all of them were multidrug-resistant. In particular, all pathogens except for C. sakazakii and K. pneumoniae were resistant to at least six antibiotics. Furthermore, conventional antibiotics usually did not have selective antimicrobial effects, and they inhibit the growth of probiotics as well as pathogens. Thus, it is important to develop alternative antibiotics that are less toxic to most probiotics but can inhibit or kill pathogens extensively and potently.
Characterization of Bacillus Strains Isolated From Fermented Food and Seafood Products
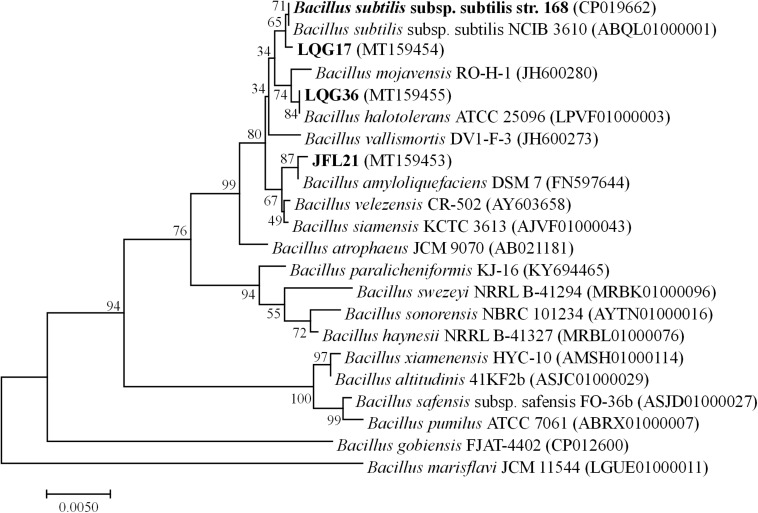
A phylogenetic tree based on the 16S rDNA sequences of the four Bacillus spp. was constructed and shown in Figure 1. As can be seen, the strain Bacillus sp. JFL21 was assigned in a node with 99.86% of support with B. amyloliquefaciens DSM 7, while the strain Bacillus sp. LGQ17 was clustered together with the members of other B. subtilis groups supported by a sequence similarity of 99.93%. Moreover, Bacillus sp. LGQ36 was designated as B. halotolerans ATCC 25096 because their 16S rDNA gene sequences were 100% identical.
FIGURE 1.
Phylogenetic tree based on 16SrDNA gene sequences from four Bacillus strains using the Neighbor-joining method (1000 bootstrap replicates).
Comparison of the Antimicrobial Activities of Metabolites From Different Bacillus spp.
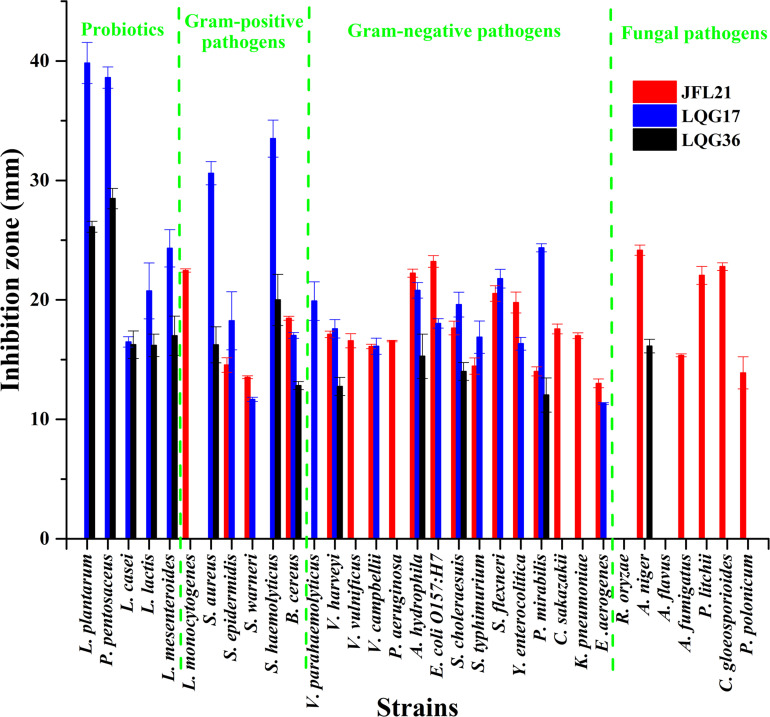
As presented in Figures 2, 3, the CFS produced by B. subtilis LGQ17 and B. halotolerans LGQ36 had an obvious inhibitory effect on the majority of indicators except for pathogenic fungi, while the CFS produced by B. amyloliquefaciens JFL21 exhibited a marked antimicrobial activity toward the majority of indicators except for probiotics. Among them, the CFS produced by B. amyloliquefaciens JFL21 exhibited broader inhibitory spectrum toward pathogens but showed no inhibitory effect on probiotics, and thus has higher application value. However, the CFS produced by B. subtilis 168 failed to show any antibacterial or antifungal effect against all indicators, which may be due to its poor capability to produce antimicrobial substances. Based on the antimicrobial spectrum and inhibitory efficacy, the antimicrobial substance produced by B. amyloliquefaciens JFL21 and B. subtilis LGQ17 were selected for the additional extraction and characterization.
FIGURE 2.
Antimicrobial spectrum of cell-free supernatants from different Bacillus strains. JFL21, LQG17, and LQG36 were referred to as the cell-free supernatants produced by Bacillus amyloliquefaciens JFL21, Bacillus natto LQG17, and Bacillus halotolerans LQG36, respectively.
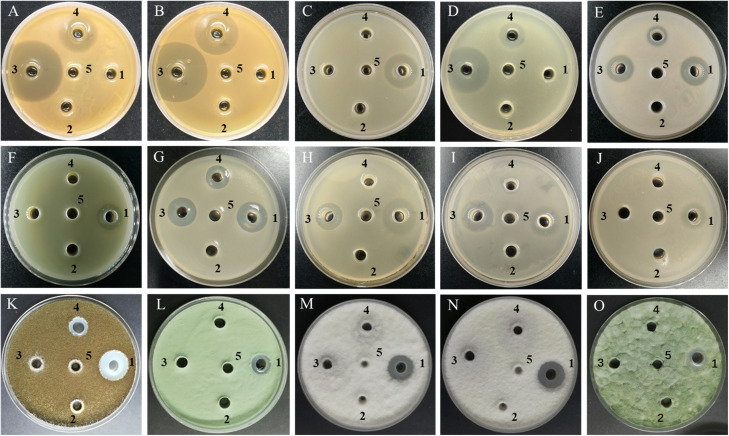
FIGURE 3.
Antimicrobial activity of cell-free supernatants from different Bacillus strains against some representative indicator microorganisms. (A) L. plantarum; (B) L. pentosus; (C) L. monocytogenes; (D) S. aureus; (E) B. cereus; (F) P. aeruginosa; (G) A. hydrophila; (H) E. coli O157:H7; (I) S. typhimurium; (J) C. sakazakii; (K) A. niger; (L) A. fumigatus; (M) P. litchii; (N) C. gloeosporioides; (O) P. polonicum; 1–4, the cell-free supernatants produced by Bacillus amyloliquefaciens JFL21, Bacillus subtilis 168, Bacillus subtilis LQG17, and Bacillus halotolerans LQG36, respectively; 5, the uninoculated Landy medium.
Inhibitory Spectrum of the Bioactive Substance Produced From B. amyloliquefaciens JFL21 and B. subtilis LGQ17
As showed in Table 2, Anti-JFL21 exhibited significant antibacterial and antifungal activity against most indicators while Anti-LQG17 showed a weak antibacterial effect against a few indicators. To further check whether the antimicrobial substances were adequately extracted, the remaining supernatant after the extraction of Anti-JFL21 or Anti-LQG17 were also tested for the antimicrobial activity. The result revealed that the remaining supernatant after the extraction of Anti-LQG17 (CFS-LQG17) still has an obvious antibacterial effect on many indicators, which indicated Anti-LQG17 maybe not the primary antimicrobial substances produced by B. subtilis LGQ17 and the main active ingredients need to be clarified by subsequent experiments. However, the remaining supernatant after the extraction of Anti-JFL21 (CFS- JFL21) did not inhibit the growth of every indicator (data not shown), confirming the antimicrobial substances were sufficiently extracted from the CFS of B. amyloliquefaciens JFL21. Additionally, the antimicrobial activity of Anti-JFL21 was also compared with those of commercially available nisin and polymyxin B, which were represented as bacteriocin and lipopeptide, respectively (Table 2). As can be observed, Anti-JFL21 not only exhibited unique advantages in inhibiting the growth of a broad range of fungal pathogens but also showed a wider and stronger antibacterial activity toward Gram-positive pathogens when compared with polymyxin B and Nisin. What’s more, although Anti-JFL21 also inhibited the growth of some probiotics, the inhibitory activity against probiotics was significantly lower when compared to pathogens. These results suggest that Anti-JFL21 could be expected to a promising alternative source of biological preservatives in the food industry for controlling various multidrug-resistant foodborne pathogens and extending the shelf-life of food products.
TABLE 2.
Inhibition spectrum of Anti-JFL21 and Anti-LQG17a.
| Indicator strains | Inhibition zone (mm) |
||||
| Anti-JFL21 (1 mg/mL) | Anti-LQG17 (1 mg/mL) | CFS-LQG17 (100 ul)b | Polymyxin B (1 mg/mL) | Nisin (1 mg/mL) | |
| Probiotics | |||||
| L. plantarum | 14.36 ± 1.22 | 12.46 ± 0.89 | 29.24 ± 0.87 | − | 15.07 ± 0.35 |
| P. pentosaceus | − | − | 26.12 ± 0.56 | − | 14.13 ± 0.62 |
| L. casei | 13.22 ± 0.67 | − | − | − | 21.76 ± 0.96 |
| L. lactis | − | − | 15.33 ± 2.25 | − | 17.34 ± 0.74 |
| L. mesenteroides | − | − | 20.59 ± 1.37 | − | 19.51 ± 1.69 |
| G+ pathogen | |||||
| L. monocytogenes | 22.96 ± 0.74 | − | − | − | − |
| S. aureus | 14.51 ± 1.48 | − | 25.22 ± 1.14 | − | − |
| S. epidermidis | 13.22 ± 0.46 | − | 14.12 ± 0.57 | 12.17 ± 0.31 | 17.15 ± 0.54 |
| S. warneri | 12.98 ± 0.51 | − | 13.02 ± 0.79 | 14.22 ± 1.04 | − |
| S. haemolyticus | − | − | 18.02 ± 0.66 | − | − |
| B. cereus | 19.80 ± 0.25 | 12.99 ± 0.49 | 14.88 ± 0.42 | − | − |
| G– pathogen | |||||
| V. parahaemolyticus | − | − | 21.12 ± 0.85 | 15.34 ± 1.63 | − |
| V. harveyi | 15.65 ± 1.20 | − | − | 22.45 ± 0.19 | − |
| V. vulnificus | − | − | − | − | − |
| V. campbellii | 14.55 ± 1.67 | − | − | 16.42 ± 0.47 | − |
| P. aeruginosa | 20.25 ± 0.57 | − | − | − | − |
| A. hydrophila | 22.72 ± 0.34 | 11.98 ± 0.73 | 20.96 ± 0.08 | 21.13 ± 0.87 | − |
| E. coli O157:H7 | 21.45 ± 0.96 | − | 13.08 ± 1.45 | 15.36 ± 0.95 | − |
| S. choleraesuis | 19.65 ± 0.42 | − | 12.66 ± 0.75 | 15.74 ± 0.32 | − |
| S. typhimurium | 17.94 ± 1.03 | − | − | 15.89 ± 0.45 | − |
| S. flexneri | 15.73 ± 2.86 | − | 20.02 ± 0.17 | 15.93 ± 0.22 | − |
| Y. enterocolitica | 16.85 ± 2.51 | − | − | 17.96 ± 2.01 | − |
| P. mirabilis | 20.71 ± 0.11 | 10.02 ± 1.15 | 17.46 ± 0.32 | − | − |
| C. sakazakii | 20.57 ± 0.92 | − | − | 14.86 ± 1.15 | − |
| K. pneumoniae | 17.34 ± 0.34 | − | − | 17.03 ± 0.84 | − |
| E. aerogenes | 16.58 ± 1.54 | − | − | 14.65 ± 0.73 | − |
| Pathogenic fungi | |||||
| R. oryzae | − | − | − | − | − |
| A. niger | 21.76 ± 0.15 | − | − | − | − |
| A. flavus | 18.28 ± 0.75 | − | − | − | − |
| A. fumigatus | 20.13 ± 0.68 | − | − | − | − |
| P. litchii | 16.55 ± 0.91 | − | − | − | − |
| C. gloeosporioides | 22.24 ± 0.49 | − | − | − | − |
| P. polonicum | 19.21 ± 1.13 | − | − | − | − |
a−: no inhibition zone; bCFS-LQG17: the remaining supernatant after the extraction of Anti-LQG17.
Structure Analysis of Crude Lipopeptides by FTIR
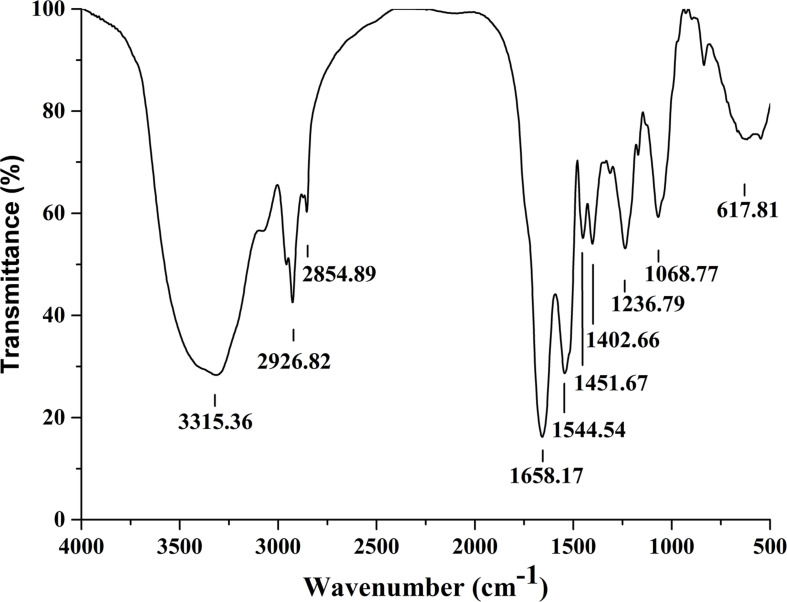
Fourier transformation infra-red spectral peaks of Anti-JFL21 were exhibited in Figure 4. The broader absorption peak of 3315.36 cm–1 indicates the presence of –OH or –NH groups. Peaks at 2926.82 cm–1, 2854.89 cm–1, 1451.67 cm–1, and 1402.66 cm–1 confirm the –C–H stretching (−CH3, −CH2) of the aliphatic chain of the lipid. A similar stretching for –C–H of lipid was also found by Pradhan et al. (2013) and Al-Wahaibi et al. (2014). The presence of amide bond (617.81 cm–1), N–H bending of secondary amides (1544.54 cm–1), and the carbonyl group (C = O) of amide (1658.17 cm–1) confirms the peptide fraction in the sample. C–O bending of esters was characterized by the peaks at 1236.79 cm–1 and 1068.77 cm–1. The same bending and stretching were reported for peptide groups in the previous research (Pradhan et al., 2013; Sharma et al., 2015; Varjani and Upasani, 2016). Thus, the nature of the antimicrobial substance produced by B. amyloliquefaciens JFL21 was speculated as cyclic lipopeptide compounds.
FIGURE 4.
Fourier transformation infra-red spectra analysis of the lipopeptides mixture Anti-JFL21 obtained from Bacillus amyloliquefaciens JFL21.
Lipopeptides Quantification by RP-HPLC Analysis
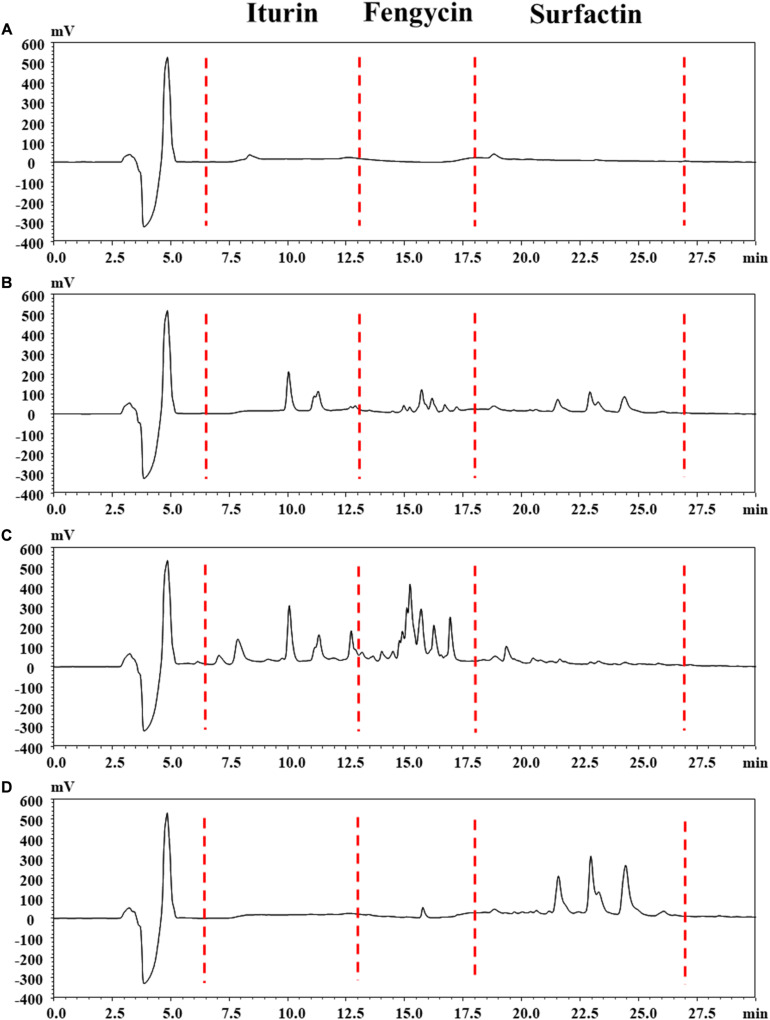
The reversed-phase HPLC (RP-HPLC) analysis was carried out to characterize the structural properties of Anti-JFL21 and Anti-LGQ17, and the results were presented in Figure 5. The peaks at retention time 4.86, 8.37, and 18.81 min maybe methanol solvent or impurity peak (Figure 5A). The retention times of iturin, fengycin, and surfactin were identified in the ranges of 6.5–13, 13–18, and 18–27 min, respectively, according to our previous results (Figure 5B). For the lipopeptides extract Anti-JFL21, three peaks clusters were observed on the RP-HPLC profile which exhibited similar retention times as the three standard groups (Figure 5C). The RP-HPLC profiles of Anti-JFL21 showed the major peaks which corresponded to iturin and fengycin families, while traces of surfactin families were also detected but the content was relatively low. Furthermore, the peaks of the iturin and fengycin families in Anti-JFL21 were relatively more than those of the three standards, which may be due to the more analogs or homologs of three lipopeptide families in Anti-JFL21. Meanwhile, RP-HPLC analysis also revealed that Anti-LGQ17 was mainly composed of four surfactin analogs (Figure 5D). These results suggested that different Bacillus species could synthesize different CLPs family, and the antimicrobial properties of these CLPs exhibited significant differences.
FIGURE 5.
Reversed phase HPLC chromatogram analysis of the lipopeptides mixture Anti-JFL21 and Anti-LQG17. (A) methyl alcohol; (B) complexes with final concentrations of 0.33 mg/mL of commercial standard iturin, fenycin, and surfactin; (C) 1 mg/mL Anti-JFL21 obtained from Bacillus amyloliquefaciens JFL21; (D) 1 mg/mL Anti-LQG17 obtained from Bacillus natto LQG17.
Identification of the Active Compounds by MALDI-TOF MS
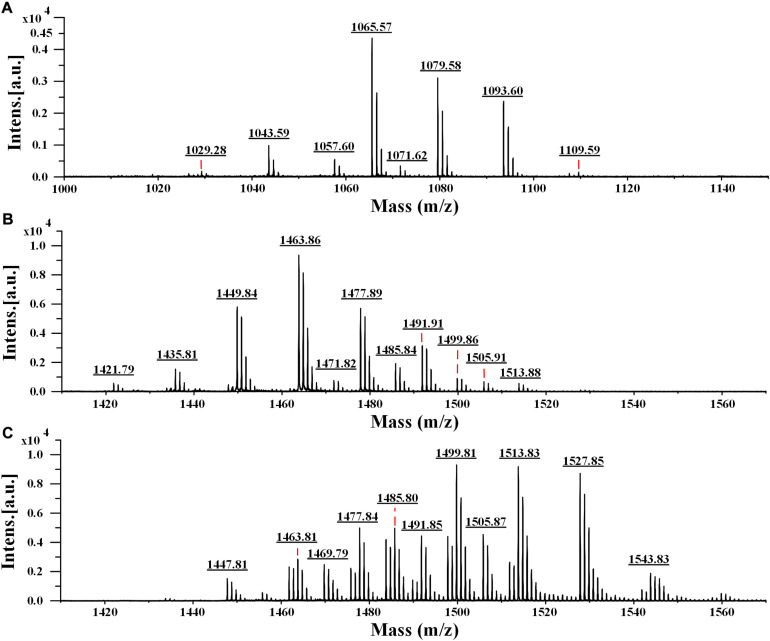
To determine the accurate molecular mass, Anti-JFL21 was further subjected to MALDI-TOF MS analysis. Anti-LGQ17 was not selected for further analysis since it did not show significant antimicrobial activity. As shown in Figure 5, the MALDI-TOF MS analysis result of Anti-JFL21 showed two well-resolved groups of peaks at m/z values range of 1029.28–1109.59 Da (Figure 6A) and 1421.79–1513.88 Da (Figure 6B), which could be, respectively, attributed to the isomers of iturin and fengycin, as previously described (Torres et al., 2015; Yang et al., 2015; Sarwar et al., 2018). As can be observed in Figure 6A, the protonated molecular ion ([M + H]+) peaks of the iturin A homologs were detected at m/z 1029.28, 1043.59, 1057.60, and 1071.62 Da, and the peaks with the differences of 14 Da may correspond to the molecular weight of one CH2 group. Besides, peaks in the m/z of 1065.57, 1079.58, and 1093.60 Da may be assigned as the sodium adducts ([M + Na]+) of C14, C15, and C16 homologs of iturin A, and m/z = 1109.59 Da could be attributed to the potassium adducts ([M + K]+) for C16 iturin. However, the absence of a typical peak of surfactin (e.g., m/z 1008, 1022, 1034, 1046, and 1058) may be due to its low relative content in the sample, which is consistent with the results of RP-HPLC analysis. On the other hand, several fengycin homologs were detected as [M + H]+ at m/z 1421.79, 1435.81, 1449.84, 1463.86, 1477.89, and 1491.91 Da (Figure 6B), which may correspond to the different fatty acid length. Moreover, the [M + Na]+ and [M + K]+ adducts of fengycin isoforms were also observed at m/z 1471.82, 1485.84, 1499.86, and 1513.88 Da (Figure 6B). Compared with the groups of fengycin peaks of Anti-JFL21, the commercial fengycin standard was also identified with many of the same ions of [M + H]+, [M + Na]+, and [M + K]+, such as the 1463.81, 1477.84, 1485.80, 1491.85, 1499.81, and 1505.87 Da (Figure 6C). In addition, the difference between the ion of 1447.81 Da detected in the commercial fengycin standard and the 1449.84 Da identified in Anti-JFL21 only 2 Da, which may be due to the presence of a double bond in fatty acids chain (Pathak et al., 2012; Sa et al., 2018). The major m/z peaks of Anti-JFL21 and the fengycin standard were summarized in the Table 3.
FIGURE 6.
MALDI-TOF MS analysis of the lipopeptides mixture Anti-JFL21 and the commercial fengycin standard. (A) the peaks of Anti-JFL21 in the range of m/z = 1000–1150 Da; (B) the peaks of Anti-JFL21 in the range of m/z = 1410–1570 Da; (C) the peaks of the commercial fengycin standard in the range of m/z = 1410–1570 Da.
TABLE 3.
Possible assignments of major m/z peaks of Anti-JFL21 and the commercial fengycin standarda.
| Samples | Detected precursor ions (m/z) |
Two fingerprint ions (m/z) | Possible assignmentb | Amino acid at position 6 and 10 | ||
| [M + H]+ | [M + Na]+ | [M + K]+ | ||||
| Anti-JFL21 | 1029.28 | − | − | / | C13 iturin A | / |
| 1043.59 | 1065.57 | − | / | C14 iturin A | / | |
| 1057.60 | 1079.58 | − | / | C15 iturin A | / | |
| 1071.62 | 1093.60 | 1109.59 | / | C16 iturin A | / | |
| 1421.79 | − | − | 952.25, 1066.42 | C14 fengycin A2 | Ala6, Val10 | |
| 1435.81 | − | − | 952.35, 1066.44 966.37, 1080.48 |
C15 fengycin A2 C14 fengycin A1 |
Ala6, Val10 Ala6, Ile10 | |
| 1449.84 | 1471.82 | − | 952.30, 1066.41 966.31, 1080.40 980.34, 1094.45 |
C16 fengycin A2 C15 fengycin A1 C14 fengycin B2 |
Ala6, Val10 Ala6, Ile10 Val6, Val10 | |
| 1463.86 | 1485.84 | − | 952.32, 1066.44 966.35, 1080.46 |
C17 fengycin A2 C16 fengycin A1 |
Ala6, Val10 Ala6, Ile10 | |
| 1477.89 | 1499.86 | − | 952.43, 1066.50 966.37, 1080.48 980.39, 1094.51 994.39, 1108.52 |
C18 fengycin A2 C17 fengycin A1 C16 fengycin B2 C15 fengycin B1 |
Ala6, Val10 Ala6, Ile10 Val6, Val10 Val6, Ile10 | |
| 1491.91 | 1513.88 | − | 952.26, 1066.18 966.19, 1080.16 980.20, 1094.20 994.35, 1108.50 |
C19 fengycin A2 C18 fengycin A1 C17 fengycin B2 C16 fengycin B1 |
Ala6, Val10 Ala6, Ile10 Val6, Val10 Val6, Ile10 | |
| 1505.91 | − | − | 980.39, 1094.47 994.42, 1108.53 |
C18 fengycin B2 C17 fengycin B1 |
Val6, Val10 Val6, Ile10 | |
| fengycin standard | 1447.81 | 1469.79 | − | 966.45, 1080.55 994.47, 1108.55 |
*C15 fengycin A1 *C13 fengycin B1 |
Ala6, Ile10 Val6, Ile10 |
| 1463.81 | 1485.80 | − | 966.37, 1080.46 994.38, 1108.48 |
C16 fengycin A1 C14 fengycin B1 |
Ala6, Ile10 Val6, Ile10 | |
| 1477.84 | 1499.81 | − | 966.39, 1080.47 994.40, 1108.49 |
C17 fengycin A1 C15 fengycin B1 |
Ala6, Ile10 Val6, Ile10 | |
| 1491.85 | 1513.83 | − | 966.34, 1080.45 994.33, 1108.44 |
C18 fengycin A1 C16 fengycin B1 |
Ala6, Ile10 Val6, Ile10 | |
| 1505.87 | 1527.85 | 1543.83 | 994.34, 1108.46 | C17 fengycin B1 | Val6, Ile10 | |
a−, no detected; /, not tested or studied; *, indicates one double bond in the fatty acid chain. bPossible assignments based on Pathak et al. (2012), Torres et al. (2015); Yang et al. (2015), and Sarwar et al. (2018).
Characterization of Fengycin Isoforms by MALDI-TOF MS/MS
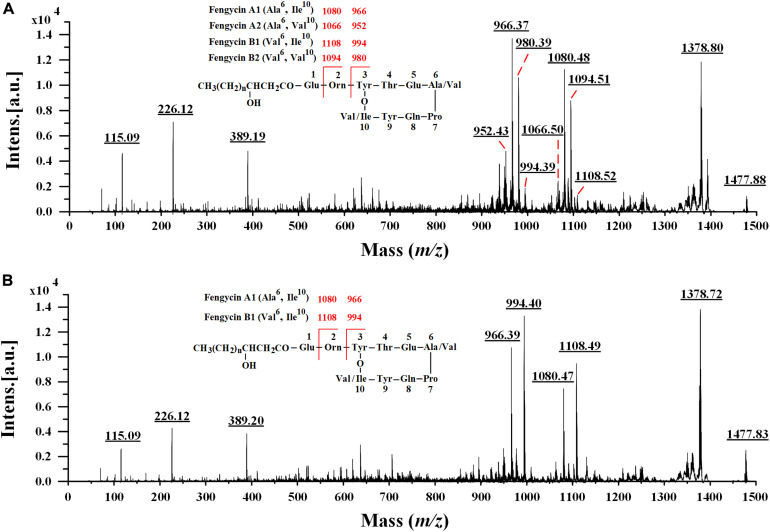
In the earlier literature, fengycins have been mainly divided into fengycin A and fengycin B, which are characterized by the existence of Ala and Val residues at position 6 of the peptide ring, respectively (Vanittanakom et al., 1986; Wang et al., 2004). Recently, a new variant of fengycin, termed as fengycin A2 or fengycin B2 with the presence of Val instead of Ile at position 10 was also observed (Pathak et al., 2012; Cochrane and Vederas, 2016). Since many fengycin isoforms may have the same nominal mass and molecular formula, it is difficult for MALDI-TOF MS to definitely discriminate different fengycin isomers. To obtain more precise amino acid sequence information, the fengycin protonated parent ions ([M + H]+) of Anti-JFL21 together with the commercial fengycin standard were selected for further MALDI-TOF MS/MS analysis. The representative MS/MS spectra of the precursor ion of 1477 Da from Anti-JFL21 compared to the fengycin standard was displayed in Figure 7.
FIGURE 7.
Comparison of MALDI-TOF MS/MS spectra of the lipopeptides mixture Anti-JFL21 and the commercial fengycin standard. MS/MS spectra of the precursor ion of 1477.88 Da from Anti-JFL21 (A) compared to the precursor ion of 1477.84 Da from the commercial fengycin standard (B).
In the MS/MS spectrum of the precursor ion of 1477.88 Da (Figure 7A), it can be quickly identified as C18 fengycin A2 (Ala6, Val10), C17 fengycin A1 (Ala6, Ile10), C15 fengycin B1 (Val6, Ile10) and C16 fengycin B2 (Val6, Val10) based on the presence of four typical paired product ions of 952 and 1066 Da, 966 and 1080 Da, 980 and 1094 Da, and 994 and 1108 Da, respectively (Wang et al., 2004; Pathak et al., 2012). Similarly, two paired product ions of 966 and 1080 Da, and 994 and 1108 Da were appeared in MS/MS spectrum of the parent ion at m/z 1477.84 Da (Figure 7B), which enable it to be assigned corresponding to C17 fengycin A1 and C15 fengycin B1. Unlike Anti-JFL21, the fengycin standard were failed to detect the two paired specific fingerprint ions of 952 and 1066 Da, and 980 and 1094 Da (Figure 7B and Table 3), which indicated the fengycin standard was only composed of fengycin A1 and fengycin B1. Besides, the product ion of 1378 Da may be resulted from the loss of Val while a series of fragment ions at m/z 389 Da, 226 Da, and 115 Da may specifically represented the sequence of Pro-Gln-Tyr, Pro-Gln, and Pro-H2O, respectively (Yu et al., 2012; Yang et al., 2015). In addition, other fengycin precursor peaks, fingerprint ions, and structural assignments were summarized and showed in Table 3.
Isolation and Purification of Lipopeptides
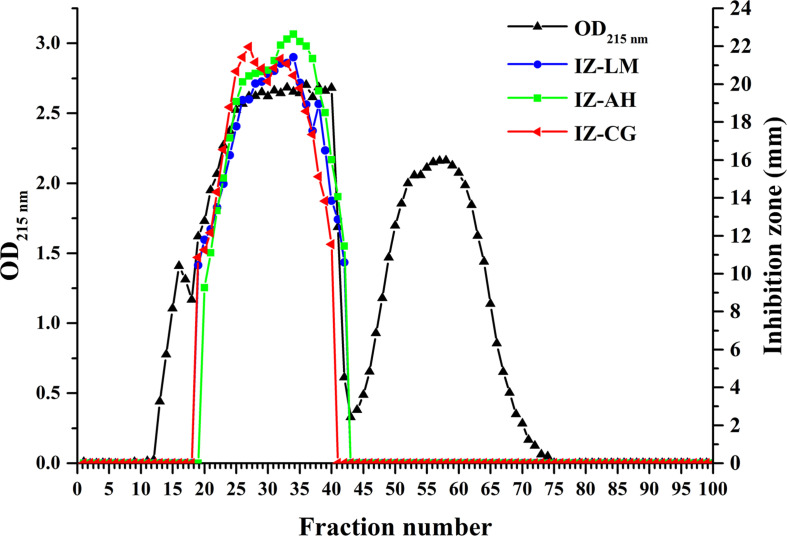
To assess the antimicrobial contribution of individual lipopeptide classes, Anti-JFL21 was subjected to further isolation and purification by GFC. As shown in Figure 8, the spectrophotometer analysis at 215 nm indicated the possible presence of lipopeptides in the fractions from 13 to 66, which was also confirmed by RP-HPLC (data not shown). Besides, RP-HPLC results revealed that the fractions in the range of 13–18, 19–42, and 43–72 were mainly identified as single surfactin, fengycin, and iturin families, respectively (Supplementary Figure 1). The fractions containing fengycins exhibited the primary antimicrobial activity against L. monocytogenes, A. hydrophila, and C. gloeosporioides, whereas the rest of the fractions comprising only surfactins or iturins did not present any antagonistic effect on the three indicator strains (Figure 8 and Supplementary Figure 2). These results demonstrated that fengycins play a key role in antimicrobial activity.
FIGURE 8.
Gel filtration chromatography and antimicrobial activity of the different fractions separated from the lipopeptides mixture Anti-JFL21 produced by Bacillus amyloliquefaciens JFL21. IZ-LM, IZ-AH, and IZ-CG were referred to as the inhibition zone of the different fractions separated from Anti-JFL21 against L. monocytogenes, A. hydrophila, and C. gloeosporioides, respectively.
Comparison of Minimal Inhibitory Concentration of the Fengycins Isolated From Anti-JLF21 and the Commercial Fengycins Standard
To compare the antimicrobial efficacy of the fengycins isolated from Anti-JFL21 with the commercial fengycins standard, their MIC was determined and compared using L. monocytogenes, A. hydrophila, and C. gloeosporioides as the indicator strains (Table 4). The result showed that the fengycins isolated from Anti-JFL21 could display the antimicrobial effect toward L. monocytogenes, A. hydrophila, and C. gloeosporioides at a low MIC range of 25–50 ug/mL. Besides, the commercial fengycins standard was found to active against C. gloeosporioides at low MIC concentration (25 ug/mL) but fail to inhibit the growth of L. monocytogenes and A. hydrophila even at the highest tested concentration (800 μg/ml). These results suggest that the fengycins isolated from Anti-JFL21 are more efficient in inhibiting pathogenic bacteria as compared to the commercial fengycins standard, which may be contributed to the greater variety of structural analogs, especially in the variation at position 10 (Val/Ile) (Table 3).
TABLE 4.
Minimum inhibitory concentration of different fengycin sources against some representative indicator strains.
| Indicator strains | MIC (ug/ml) |
|
| Fengycins isolated from Anti-JFL21 | The commercial Fengycins standard | |
| L. monocytogenes | 25 | >800 |
| A. hydrophila | 50 | >800 |
| C. gloeosporioides | 25 | 25 |
Biochemical Characterization of Fengycins
To evaluate the possible application in the food industry, the antimicrobial stability of fengycins produced by B. amyloliquefaciens JFL21 under various conditions was determined. L. monocytogenes, an important foodborne pathogen was used as an indicator strain. As shown in Table 5, the fengycins did not show loss of its inhibitory activity against L. monocytogenes after the treatment for 2 h under the ultraviolet sterilization, over a pH range of 5.0 to 9.0, or at temperatures below 80°C. Although there was a very significant reduction in the activity after incubation for 2 h at 100°C, pH 1.0, and pH 13.0, the residual activity remained 90, 85, and 72%, respectively. Thus, our results indicate that the fengycins were resistant to the various harsh conditions and the sterilization process such as commercial pasteurization and ultraviolet sterilization, and thus can be used as a natural preservative in the food industry.
TABLE 5.
Effect of enzymes, chemical reagents, and physical factors on fengycins anti-Listeria activitya.
| Factors | Treatment for 2 h | Relative activity (%)a |
| Control | Untreated | 100 |
| pH | 1 | 85 ± 3.8** |
| 3 | 93 ± 2.6* | |
| 5 | 99 ± 1.1 | |
| 7 | 102 ± 1.6 | |
| 9 | 101 ± 0.7 | |
| 11 | 88 ± 3.2** | |
| 13 | 72 ± 5.9** | |
| Heat | 37°C | 99 ± 1.9 |
| 60°C | 100 ± 0.5 | |
| 80°C | 97 ± 1.5 | |
| 100°C | 90 ± 4.3* | |
| Light | Ultraviolet | 101 ± 0.3 |
| Enzymes (1 mg/mL) | Cellulase | 104 ± 2.6 |
| α-Amylase | 99 ± 1.4 | |
| Proteinase K | 98 ± 0.5 | |
| Papain | 100 ± 2.7 | |
| Bromelain | 101 ± 1.0 | |
| Trypsin | 100 ± 0.2 | |
| Pepsin | 98 ± 5.3 | |
| Chemical reagents (1 mM) | EDTA | 102 ± 2.4 |
| SDS | 97 ± 1.8 | |
| MnCl2 | 100 ± 1.1 | |
| ZnSO4 | 92 ± 6.4 |
aThe relative antimicrobial activity was measured by using L. monocytogenes as an indicator strain. Values are given as mean ± SD from triplicate determinations (n = 3). The value represents the mean ± SE of triple determinations. *p < 0.05, **p < 0.01.
Discussion
The increased emergence and evolution of multiple drug-resistant foodborne pathogens in recent decades has become a worldwide vexing problem of public concern. In this work, antibiotics susceptibility test results indicated that all foodborne pathogens used in the current study were multidrug-resistant according to the recent drug-resistance definition (Table 1; Magiorakos et al., 2012). Likewise, Lee et al. (2017) disclosed that 27 (82%) of 33 L. monocytogenes isolates collected from ready-to-eat seafood and food processing environments were resistant to at least four antibiotics. Besides, a high proportion of multidrug-resistant isolates from food was also commonly reported in other foodborne pathogens such as Y. enterocolitica (66/70, 94.3%) (Ye et al., 2015), Salmonella (30/42, 71.4%) (Tirziu et al., 2015), and S. aureus (62/93, 66.67%) (Ma et al., 2018). Thus, the development of new antimicrobial agents or other alternative strategies is required promptly.
Bacillus is an important genus producing a wide array of bioactive secondary metabolites, including lipopeptides, polyketides, lantibiotics, siderophores, lytic enzymes, and peptides (Cawoy et al., 2015; Dimkić et al., 2017). Moreover, the majority of Bacillus spp. such as B. subtilis, B. amyloliquefaciens, and B. licheniformis have been generally recognized as safe microorganisms for application in the food industry and considered to be important biological control agents (Torres et al., 2015; Lastochkina et al., 2019). In this study, three Bacillus isolates from various food sources, namely B. amyloliquefaciens JFL21, B. subtilis LQG17, and B. halotolerans LQG36 were obtained and their antimicrobial properties were compared. Results revealed that the CFS produced by these Bacillus strains exhibited different inhibitory spectrum or inhibitory efficacy against the thirty-three indicators (Figures 2, 3), which suggested that different Bacillus species or isolates may be capable of producing different kinds or amounts of bioactive metabolites. Hence, screening and characterization of the Bacillus spp. with remarkable biological properties still receives a great scientific interest due to the application potential of these strains and their active metabolites (Li et al., 2015).
In recent years, CLPs have received considerable attention because of their efficient biosurfactant and antimicrobial properties (Perez et al., 2017; Sarwar et al., 2018). In this work, the lipopeptides mixture Anti-JFL21 not only exhibited strong antimicrobial activities against the majority of multidrug-resistant foodborne pathogens but also showed relatively low toxicity to most probiotics (Figures 2, 3). This may be since all probiotics used in this study belongs to lactic acid bacteria (LAB), and Anti-JFL21 have a relatively weak inhibitory effect on LAB. The similar phenomenon was also observed by Lee et al. (2016), who reported that the lipopeptides mixture containing four surfactin and four bacillomycin D analogs, inhibited the growth of many Gram-positive pathogens (e.g., B. cereus and L. monocytogenes) and fungal pathogen (e.g., A. nidulans and F. moniliforme), but it did not inhibit LAB such as L. plantarum and L. lactis. Moreover, Todorov et al. (2018) also found that L. plantarum has a natural high resistance to a glycopeptide antibiotic when compared to L. monocytogenes, which was attributed to the presence of D-Ala-D-Lactate in its peptidoglycan rather than in the D-Ala-D-Ala dipeptide. Thus, it is speculated that the weak inhibitory effect of lipopeptides Anti-JFL21 on LAB may be due to the difference of cell wall composition between LAB and other indicator bacteria. However, the specific mechanism of the relatively weak bacteriostatic effect of CLPs on LAB still needs to be further explored.
Historically, the elucidation of specific effective antimicrobial components is crucial to the accuracy of the drug application or antiseptic addition. In this research, the family of surfactin, fengycin, and iturin were successfully separated by GFC, and the results of the antimicrobial test demonstrated that fengycins might play a major role in the antibacterial and antifungal activity of Anti-JFL21 (Figure 8 and Supplementary Figures 1,2). To our knowledge, this study is the first report that fengycin is a primary active compound of B. amyloliquefaciens against a broad range of foodborne pathogens including Gram-positive pathogens, Gram-negative pathogens, and fungal pathogens. However, unlike some previous studies, which displayed that surfactin and iturin presented important antibacterial and antifungal activity, respectively (Loiseau et al., 2015; Narendra Kumar et al., 2017; Sudarmono et al., 2019). Surfactins produced by B. subtilis LQG17 showed a weak antibacterial effect against few bacterial indicators, and surfactins and iturins produced by B. amyloliquefaciens JFL21 were not able to inhibit either bacteria or fungi. This may be due to the production and structural diversity of each lipopeptide family are influenced by the various Bacillus species or isolates (Sabaté and Audisio, 2013; Fan et al., 2017; Sudarmono et al., 2019), and the effective isoforms of surfactin and iturin synthesized by B. subtilis LQG17 or B. amyloliquefaciens JFL21 was few or not enough to reach the antimicrobial concentration. This phenomenon was also found by Sabaté and Audisio (2013), who reported that the surfactin produced by B. subtilis C4 inhibited the L. monocytogenes with a concentration of 0.125 mg/mL while 1 mg/mL of the surfactin produced by B. subtilis M1 was necessary to inhibit the pathogen.
Indeed, it is well known that different isoforms and homologs of the lipopeptide family could exhibit different properties and activities, which depend in particular on the chain length (Ben Ayed et al., 2014). In this work, RP-HPLC and MALDI-TOF MS analysis also revealed that each lipopeptide family produced by B. amyloliquefaciens JFL21 was composed of different isoforms and fengycins exhibited variation in the length of the β-hydroxy-fatty acid from 14 to 19 carbons units (Figure 6B and Table 3). Our results are consistent with previous works reported by Jemil et al. (2017) and Perez et al. (2017) that showed the co-production of different homologous compounds of each lipopeptide family. However, the quantity of fengycin isoforms may be different in the various Bacillus species or isolates. For example, B. subtilis N6–34 could produce multiple fengycin homologs with chain lengths from C15 to C17 whereas B. amyloliquefaciens S13-3 only synthesize C16 fengycin variant (Yamamoto et al., 2015; Sa et al., 2018).
Except for the difference of the fatty acid chain length, fengycin isoforms also vary in the lactone rings of amino acid composition. According the previous literature, the variations at position 6 (Ala/Val), and variations at position 10 (Val/Ile) were mostly observed (Pathak et al., 2012; Cochrane and Vederas, 2016). Since the precursor ions mainly cleaved at the Glu–Orn and Orn–Tyr bonds, resulting in the specific octapeptide ring ions at m/z 952 and 1066 Da, 966 and 1080 Da, 980 and 1094 Da, and 994 and 1108 Da, respectively (Wang et al., 2004; Pathak et al., 2012; Yang et al., 2015; Sa et al., 2018). Moreover, the extra stability of the charged octapeptide ring system suppressed further fragmentation along the peptide backbone, and thus these specific product ions have been used as diagnostics for identifying fengycin variants with the amino acids replacement within the macrocyclic moiety (Pathak et al., 2012). Based on these specific product ions, the fengycin identified in the lipopeptides mixture Anti-JFL21 was readily assigned as four fenycin variants (fengycin A1, A2, B1, and B2) with chain lengths from C13 to C19, whereas the commercial fengycin standard was determined as two fenycin variants (fengycin A1 and B1) with lengths from C13 to C18. In addition, the MALDI-TOF MS/MS analysis revealed that a precursor ion of fengycin could be simultaneously identified as multiple fengycin variants, which indicated that it is a mixture of structural analogs and the further component purification are still required to obtain the mono variant or isoform.
Previously, fengycin was mainly characterized by strong antifungal activities against a wide range of plant pathogens, especially filamentous fungi (Guo et al., 2014; Fan et al., 2017; Zihalirwa Kulimushi et al., 2017). Nevertheless, it is particularly noteworthy that the fengycin produced by B. amyloliquefaciens JFL21 not only exhibited a remarkable antifungal activity comparable to the commercial fengycin standard but also showed stronger antibacterial efficacy against Gram-positive and Gram-negative foodborne pathogen as compared to the commercial fengycin standard (Table 4). This may be due to the commercial fengycin standard only contained two fengycin variants with the variations at position 6 (Ala/Val), whereas the fengycin identified in the present study was composed of four fengycin variants with the variations not only at position 6 (Ala/Val) but also at position 10 (Val/Ile) (Table 3). In addition, the various content combinations of homologs or isoforms may also lead to the different antimicrobial properties (Sabaté and Audisio, 2013; Kaur et al., 2017; Sa et al., 2018). However, to better clarify the structure-function relationships, the separation, characterization, and antimicrobial test of specific fengycin variants or isoforms are still highly necessary in the future study.
In general, an antimicrobial substance in a food item is by itself not likely to ensure complete safety, and therefore often used in combination with either other antimicrobial compounds, chemical reagents, or technologies such as ultraviolet sterilization, heat, and various pH pressure (Zacharof and Lovitt, 2012; Lee et al., 2016). The current study shows that the fengycins produced by B. amyloliquefaciens JFL21 not only maintained anti-Listeria activity over a broad pH and temperature range, but it was also active after treatment with ultraviolet sterilization, chemical reagents, and proteolytic enzymes (Table 5). The similar properties of highly thermostable and resistant to many proteolytic enzymes and a broad range of pH were also found in other Bacillus-derived antimicrobial CLPs such as surfactin (Sabaté and Audisio, 2013) and a lipopeptide mixture (Ben Ayed et al., 2014).
To date, the active mechanism of fengycins is less well known compared with iturins and surfactins. The possible mechanism for the antifungal activity of fengycins is that they interact with sterol and phospholipid molecules in membranes and thus alter the structure and permeability of the fungal cell membrane (Farzand et al., 2019; Hanif et al., 2019). However, the mechanism of interaction may vary and depend on the structural features of fengycins and pathogen species (Kaur et al., 2017; Sarwar et al., 2018). Recently, Piewngam et al. (2018) firstly revealed that β-OH-C17 fengycin B could potentially be used as quorum-sensing blockers to combat antibiotic-resistant S. aureus, which laid the foundation for probiotic Bacillus to comprehensively eradicate intestinal S. aureus colonization. Consequently, extensive studies are still required for fully understanding and characterizing the exact antimicrobial role of fengycins toward individual foodborne pathogen.
Conclusion
In this study, three Bacillus strains with excellent antimicrobial properties named JFL21, LQG17, and LQG36, were isolated from food or seafood sources and identified closely related to the species of B. amyloliquefaciens, B. subtilis, and B. halotolerans, respectively. Besides, the model organism B. subtilis 168 was also exploited to compare the antimicrobial properties with the three Bacillus strains. The results showed that the capacity to produce antimicrobial substances varied greatly among different Bacillus species or isolates. Among them, the antimicrobial substances produced by B. amyloliquefaciens JFL21 was low toxicity to most probiotics but exhibited strong and extensive antimicrobial activities against multidrug-resistant foodborne pathogens, and thus have more application potential. Subsequently, the FITR, HPLC, and MALDI-TOF MS analysis revealed that the partially purified antimicrobial compounds, Anti-JFL21, was composed of multiple lipopeptides of the surfactin, fengycin, and iturin families. After the separation of different lipopeptide classes, the result of this work demonstrated that fengycins not only play a critical role in antimicrobial activity but also remain highly stable against a variety of enzymes, chemical reagents, and extreme conditions. To the best of our knowledge, this study is the first report that fengycin is a primary active compound of B. amyloliquefaciens against a broad range of foodborne pathogenic microorganisms including Gram-positive pathogens, Gram-negative pathogens, and fungal pathogens.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
L-ZL, L-QG, and J-FL conceived and designed this experiment. L-ZL performed data analysis and was a major contributor in drafting the work. Q-WZ and TW helped revise the manuscript critically. L-ZL and Q-WZ carried out the antimicrobial experiment. L-ZL and Z-QZ performed the purification, characterization, and comparison of the antimicrobial metabolites from different Bacillus. HZ and C-FZ performed the isolation and purification of lipopeptides. TW and Q-YX performed the biochemical characterization of fengycin. All authors reviewed and approved the submitted manuscript to be published.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This study was funded by the National Natural Science Foundation of China (Grant number 31901693), the Key-Area Research and Development Program of Guangdong Province (Grant numbers 2018B020205003, 2018B020205001, and 2020B020226006), and Guangdong Basic and Applied Basic Research Foundation (Grant number 2020A1515010756).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.579621/full#supplementary-material
Reversed-phase HPLC analysis of the different fractions separated from the lipopeptides mixture Anti-JFL21. (A) fraction 16; (B) fraction 30; (C) fraction 55.
Antimicrobial activity of different lipopeptide components separated from the lipopeptides mixture Anti-JFL21 by Sephadex LH-20 gel column chromatography. (A–C) L. monocytogenes; (D–F) A. hydrophila; (G–I) C. gloeosporioides; CK, 1 mg/mL Anti-JFL21; 13–18, the fractions from 11 to 18; 25–30, the fractions from 25 to 30; 55–60, the fractions from 55 to 60.
References
- Al-Wahaibi Y., Joshi S., Al-Bahry S., Elshafie A., Al-Bemani A., Shibulal B. (2014). Biosurfactant production by Bacillus subtilis B30 and its application in enhancing oil recovery. Colloids Surf. B Biointerfaces 114 324–333. 10.1016/j.colsurfb.2013.09.022 [DOI] [PubMed] [Google Scholar]
- Banat I. M., Franzetti A., Gandolfi I., Bestetti G., Martinotti M. G., Fracchia L., et al. (2010). Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 87 427–444. 10.1007/s00253-010-2589-0 [DOI] [PubMed] [Google Scholar]
- Ben Ayed H., Hmidet N., Béchet M., Chollet M., Chataigné G., Leclère V., et al. (2014). Identification and biochemical characteristics of lipopeptides from Bacillus mojavensis A21. Process Biochem. 49 1699–1707. 10.1016/j.procbio.2014.07.001 [DOI] [Google Scholar]
- Ben Ayed H., Hmidet N., Béchet M., Jacques P., Nasri M. (2017). Identification and natural functions of cyclic lipopeptides from Bacillus amyloliquefaciens An6. Eng. Life Sci. 17 536–544. 10.1002/elsc.201600050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawoy H., Debois D., Franzil L., De Pauw E., Thonart P., Ongena M. (2015). Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Microb. Biotechnol. 8 281–295. 10.1111/1751-7915.12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani A. G., Gunaseelan D., Sushma N., Ramkrishna S., Utpal R. (2015). An antimicrobial metabolite from Bacillus sp.: significant activity against pathogenic bacteria including multidrug-resistant clinical strains. Front. Microbiol. 6:1335. 10.3389/fmicb.2015.01335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Z., Rong Y. J., Li Y., Tang M. J., Chi Z. M. (2015). Biosurfactins production by Bacillus amyloliquefaciens R3 and their antibacterial activity against multi-drug resistant pathogenic E. coli. Bioprocess Biosyst. Eng. 38 853–861. 10.1007/s00449-014-1328-9 [DOI] [PubMed] [Google Scholar]
- Cochrane S. A., Vederas J. C. (2016). Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med. Res. Rev. 36 4–31. 10.1002/med.21321 [DOI] [PubMed] [Google Scholar]
- Dimkić I., Stanković S., Nišavić M., Petković M., Ristivojević P., Fira D., et al. (2017). The profile and antimicrobial activity of Bacillus lipopeptide extracts of five potential biocontrol strains. Front. Microbiol. 8:925. 10.3389/fmicb.2017.00925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Food Safety Authority (2018). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 16 1–262. 10.2903/j.efsa.2018.5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H. Y., Ru J. J., Zhang Y. Y., Wang Q., Li Y. (2017). Fengycin produced by Bacillus subtilis 9407 plays a major role in the biocontrol of apple ring rot disease. Microbiol. Res. 199 89–97. 10.1016/j.micres.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Farzand A., Moosa A., Zubair M., Khan R. A., Massawe C. V., Tahir A. H., et al. (2019). Suppression of Sclerotinia sclerotiorum by the induction of systemic resistance and regulation of antioxidant pathways in tomato using fengycin produced by Bacillus amyloliquefaciens FZB42. Biomolecules 9:613. 10.3390/biom9100613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garedew L., Hagos Z., Addis Z., Tesfaye R., Zegeye B. (2015). Prevalence and antimicrobial susceptibility patterns of Salmonella isolates in association with hygienic status from butcher shops in Gondar town, Ethiopia. Antimicrob. Resist. Infect. Control 4:21. 10.1186/s13756-015-0062-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q. G., Dong W. X., Li S. Z., Lu X. Y., Wang P. P., Zhang X. Y., et al. (2014). Fengycin produced by Bacillus subtilis NCD-2 plays a major role in biocontrol of cotton seedling damping-off disease. Microbiol. Res. 169 533–540. 10.1016/j.micres.2013.12.001 [DOI] [PubMed] [Google Scholar]
- Hanif A., Zhang F., Li P. P., Li C. C., Xu Y. J., Zubair M., et al. (2019). Fengycin produced by Bacillus amyloliquefaciens FZB42 inhibits Fusarium graminearum growth and mycotoxins biosynthesis. Toxins 11:295. 10.3390/toxins11050295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemil N., Manresa A., Rabanal F., Ben Ayed H., Hmidet N., Nasri M. (2017). Structural characterization and identification of cyclic lipopeptides produced by Bacillus methylotrophicus DCS1 strain. J. Chromatogr. B 1060 374–386. 10.1016/j.jchromb.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Kaur P. K., Joshi N., Singh I. P., Saini H. S. (2017). Identification of cyclic lipopeptides produced by Bacillus vallismortis R2 and their antifungal activity against Alternaria alternata. J. Appl. Microbiol. 122 139–152. 10.1111/jam.13303 [DOI] [PubMed] [Google Scholar]
- Kim P. I., Ryu J., Kim Y. H., Chi Y. T. (2010). Production of biosurfactant lipopeptides Iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J. Microbiol. Biotechnol. 20 138–145. 10.4014/jmb.0905.05007 [DOI] [PubMed] [Google Scholar]
- Kuch A., Goc A., Belkiewicz K., Filipello V., Ronkiewicz P., Gołębiewska A., et al. (2018). Molecular diversity and antimicrobial susceptibility of Listeria monocytogenes isolates from invasive infections in Poland (1997–2013). Sci. Rep. 8:14562. 10.1038/s41598-018-32574-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastochkina O., Seifikalhor M., Aliniaeifard S., Baymiev A., Pusenkova L., Garipova S., et al. (2019). Bacillus spp.: efficient biotic strategy to control postharvest diseases of fruits and vegetables. Plants 8:97. 10.3390/plants8040097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. Y., Ha J. H., Lee M. K., Cho Y. S. (2017). Antimicrobial susceptibility and serotyping of Listeria monocytogenes isolated from ready-to-eat seafood and food processing environments in Korea. Food Sci. Biotechnol. 26 287–291. 10.1007/s10068-017-0038-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Lee J., Nam Y. D., Lee J. S., Seo M. J., Yi S. H. (2016). Characterization of antimicrobial lipopeptides produced by Bacillus sp. LM7 isolated from chungkookjang, a Korean traditional fermented soybean food. Int. J. Food Microbiol. 221 12–18. 10.1016/j.ijfoodmicro.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Li X., Yang H., Zhang D. L., Li X., Yu H. M., Shen Z. Y. (2015). Overexpression of specific proton motive force-dependent transporters facilitate the export of surfactin in Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 42:93. 10.1007/s10295-014-1527-z [DOI] [PubMed] [Google Scholar]
- Loiseau C., Schlusselhuber M., Bigot R., Bertaux J., Berjeaud J. M., Verdon J. (2015). Surfactin from Bacillus subtilis displays an unexpected anti-Legionella activity. Appl. Microbiol. Biotechnol. 99 5083–5093. 10.1007/s00253-014-6317-z [DOI] [PubMed] [Google Scholar]
- Ma Y., Zhao Y. Y., Tang J. N., Tang C., Chen J., Liu J. (2018). Antimicrobial susceptibility and presence of resistance & enterotoxins/enterotoxin-likes genes in Staphylococcus aureus from food. CyTA J. Food 16 76–84. 10.1080/19476337.2017.1340341 [DOI] [Google Scholar]
- Magiorakos A. P., Srinivasan A., Carey R. B., Carmeli Y., Falagas M. E., Giske C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- Mandal S. M., Barbosa A. E. A. D., Franco O. L. (2013). Lipopeptides in microbial infection control: scope and reality for industry. Biotechnol. Adv. 31 338–345. 10.1016/j.biotechadv.2013.01.004 [DOI] [PubMed] [Google Scholar]
- Narendra Kumar P., Swapna T. H., Khan M. Y., Reddy G., Hameeda B. (2017). Statistical optimization of antifungal iturin A production from Bacillus amyloliquefaciens RHNK22 using agro-industrial wastes. Saudi J. Biol. Sci. 24 1722–1740. 10.1016/j.sjbs.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndlovu T., Rautenbach M., Vosloo J. A., Khan S., Khan W. (2017). Characterisation and antimicrobial activity of biosurfactant extracts produced by Bacillus amyloliquefaciens and Pseudomonas aeruginosa isolated from a wastewater treatment plant. AMB Express 7:108. 10.1186/s13568-017-0363-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Ahmed S., Eswari J. S. (2015). Therapeutic cyclic lipopeptides mining from microbes: latest strides and hurdles. World J. Microbiol. Biotechnol. 31 1177–1193. 10.1007/s11274-015-1880-8 [DOI] [PubMed] [Google Scholar]
- Pathak K. V., Keharia H., Gupta K., Thakur S. S., Balaram P. (2012). Lipopeptides from the banyan endophyte, Bacillus subtilis K1: mass spectrometric characterization of a library of fengycins. J. Am. Soc. Mass Spectrom 23:1716. 10.1007/s13361-012-0437-4 [DOI] [PubMed] [Google Scholar]
- Perez K. J., Viana J. D. S., Lopes F. C., Pereira J. Q., Dos Santos D. M., Oliveira J. S., et al. (2017). Bacillus spp. Isolated from puba as a source of biosurfactants and antimicrobial lipopeptides. Front. Microbiol. 8:61. 10.3389/fmicb.2017.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piewngam P., Zheng Y., Nguyen T. H., Dickey S. W., Joo H.-S., Villaruz A. E., et al. (2018). Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562 532–537. 10.1038/s41586-018-0616-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A. K., Pradhan N., Mall G., Panda H. T., Sukla L. B., Panda P. K., et al. (2013). Application of lipopeptide biosurfactant isolated from a halophile: Bacillus tequilensis CH for inhibition of biofilm. Appl. Biochem. Biotechnol. 171 1362–1375. 10.1007/s12010-013-0428-3 [DOI] [PubMed] [Google Scholar]
- Sa R. B., An X., Sui J. K., Wang X. H., Ji C., Wang C. Q., et al. (2018). Purification and structural characterization of fengycin homologues produced by Bacillus subtilis from poplar wood bark. Aust. Plant Pathol. 47 259–268. 10.1007/s13313-018-0552-1 [DOI] [Google Scholar]
- Sabaté D. C., Audisio M. C. (2013). Inhibitory activity of surfactin, produced by different Bacillus subtilis subsp. subtilis strains, against Listeria monocytogenes sensitive and bacteriocin-resistant strains. Microbiol. Res. 168 125–129. 10.1016/j.micres.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Sarwar A., Hassan M. N., Imran M., Iqbal M., Majeed S., Brader G., et al. (2018). Biocontrol activity of surfactin A purified from Bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol. Res. 209 1–13. 10.1016/j.micres.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Sharma D., Ansari M. J., Gupta S., Al Ghamdi A., Pruthi P., Pruthi V. (2015). Structural characterization and antimicrobial activity of a biosurfactant obtained from Bacillus pumilus DSVP18 grown on potato peels. Jundishapur J. Microbiol. 8:e21257. 10.5812/jjm.21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Abraham J. (2014). Ribosomally synthesized peptides from natural sources. J. Antibiot. 67 277–289. 10.1038/ja.2013.138 [DOI] [PubMed] [Google Scholar]
- Sudarmono P., Wibisana A., Listriyani L. W., Sungkar S. (2019). Characterization and synergistic antimicrobial evaluation of lipopeptides from Bacillus amyloliquefaciens isolated from oil-contaminated soil. Int. J. Microbiol. 2019:3704198. 10.1155/2019/3704198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack D. M., Marder E. P., Griffin P. M., Cieslak P. R., Dunn J., Hurd S., et al. (2019). Preliminary incidence and trends of infections with pathogens transmitted commonly through food - foodborne diseases active surveillance network, 10 U.S. sites, 2015-2018. MMWR Morb. Mortal. Wkly. Rep. 68 369–373. 10.15585/mmwr.mm6816a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirziu E., Lazar R., Sala C., Nichita I., Morar A., Seres M., et al. (2015). Salmonella in raw chicken meat from the Romanian seaside: frequency of isolation and antibiotic resistance. J. Food Prot. 78 1003–1006. 10.4315/0362-028x.jfp-14-460 [DOI] [PubMed] [Google Scholar]
- Todorov S. D., De Paula O. L., Camargo A. C., Lopes D. A., Nero L. A. (2018). Combined effect of bacteriocin produced by Lactobacillus plantarum ST8SH and vancomycin, propolis or EDTA for controlling biofilm development by Listeria monocytogenes. Rev. Argent. Microbiol. 50 48–55. 10.1016/j.ram.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Torres M. J., Petroselli G., Daz M., Erra-Balsells R., Audisio M. C. (2015). Bacillus subtilis subsp. subtilis CBMDC3f with antimicrobial activity against Gram-positive foodborne pathogenic bacteria: UV-MALDI-TOF MS analysis of its bioactive compounds. World J. Microbiol. Biotechnol. 31 929–940. 10.1007/s11274-015-1847-9 [DOI] [PubMed] [Google Scholar]
- U.S. Centers for Disease Control and Prevention (2019). Antibiotic Resistance Threats in the United States, 2019. Available online at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed March 27, 2020). [Google Scholar]
- Vanittanakom N., Loeffler W., Koch U., Jung G. (1986). Fengycin–a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot. 39 888–901. 10.7164/antibiotics.39.888 [DOI] [PubMed] [Google Scholar]
- Varjani S. J., Upasani V. N. (2016). Carbon spectrum utilization by an indigenous strain of Pseudomonas aeruginosa NCIM 5514: production, characterization and surface active properties of biosurfactant. Bioresour. Technol. 221 510–516. 10.1016/j.biortech.2016.09.080 [DOI] [PubMed] [Google Scholar]
- Wang J. Q., Liu J. Z., Wang X. L., Yao J., Yu Z. S. (2004). Application of electrospray ionization mass spectrometry in rapid typing of fengycin homologues produced by Bacillus subtilis. Lett. Appl. Microbiol. 39 98–102. 10.1111/j.1472-765X.2004.01547.x [DOI] [PubMed] [Google Scholar]
- World Health Organization (2015). WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007-2015. Available online at: http://apps.who.int/iris/bitstream/handle/10665/199350/9789241565165_eng.pdf (accessed April 25, 2019). [Google Scholar]
- World Health Organization (2019). International Food Safety Conference. Available online at: https://www.who.int/news-room/events/international-food-safety-conference (accessed March 27, 2020). [Google Scholar]
- Xu H.-M., Rong Y.-J., Zhao M.-X., Song B., Chi Z.-M. (2014). Antibacterial activity of the lipopetides produced by Bacillus amyloliquefaciens M1 against multidrug-resistant Vibrio spp. isolated from diseased marine animals. Appl. Microbiol. Biotechnol. 98 127–136. 10.1007/s00253-013-5291-1 [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Shiraishi S., Suzuki S. (2015). Are cyclic lipopeptides produced by Bacillus amyloliquefaciens S13-3 responsible for the plant defence response in strawberry against Colletotrichum gloeosporioides? Lett. Appl. Microbiol. 60 379–386. 10.1111/lam.12382 [DOI] [PubMed] [Google Scholar]
- Yang H., Li X., Li X., Yu H., Shen Z. (2015). Identification of lipopeptide isoforms by MALDI-TOF-MS/MS based on the simultaneous purification of iturin, fengycin, and surfactin by RP-HPLC. Anal. Bioanal. Chem. 407 2529–2542. 10.1007/s00216-015-8486-8 [DOI] [PubMed] [Google Scholar]
- Ye Q. H., Wu Q. P., Hu H. J., Zhang J. M., Huang H. X. (2015). Prevalence, antimicrobial resistance and genetic diversity of Yersinia enterocolitica isolated from retail frozen foods in China. FEMS Microbiol. Lett. 362:fnv197. 10.1093/femsle/fnv197 [DOI] [PubMed] [Google Scholar]
- Yu S. M., Oh B. T., Lee Y. H. (2012). Biocontrol of green and blue molds in postharvest satsuma mandarin using Bacillus amyloliquefaciens JBC36. Biocontrol Sci. Technol. 22 1181–1197. 10.1080/09583157.2012.719150 [DOI] [Google Scholar]
- Zacharof M. P., Lovitt R. W. (2012). Investigation of shelf life of potency and activity of the Lactobacilli produced bacteriocins through their exposure to various physicochemical stress factors. Probiotics Antimicrob. Proteins 4 187–197. 10.1007/s12602-012-9102-2 [DOI] [PubMed] [Google Scholar]
- Zhang B., Dong C. J., Shang Q. M., Cong Y., Kong W. J., Li P. L. (2013). Purification and partial characterization of Bacillomycin L produced by Bacillus amyloliquefaciens K103 from lemon. Appl. Biochem. Biotechnol. 171 2262–2272. 10.1007/s12010-013-0424-7 [DOI] [PubMed] [Google Scholar]
- Zihalirwa Kulimushi P., Argüelles Arias A., Franzil L., Steels S., Ongena M. (2017). Stimulation of fengycin-type antifungal lipopeptides in Bacillus amyloliquefaciens in the presence of the maize fungal pathogen Rhizomucor variabilis. Front. Microbiol. 8:850. 10.3389/fmicb.2017.00850 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reversed-phase HPLC analysis of the different fractions separated from the lipopeptides mixture Anti-JFL21. (A) fraction 16; (B) fraction 30; (C) fraction 55.
Antimicrobial activity of different lipopeptide components separated from the lipopeptides mixture Anti-JFL21 by Sephadex LH-20 gel column chromatography. (A–C) L. monocytogenes; (D–F) A. hydrophila; (G–I) C. gloeosporioides; CK, 1 mg/mL Anti-JFL21; 13–18, the fractions from 11 to 18; 25–30, the fractions from 25 to 30; 55–60, the fractions from 55 to 60.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.