Abstract
The genetics underlying Cyto-Nuclear Incompatibility (CNI) was studied in Pelargonium interspecific hybrids. We created hybrids of 12 closely related crop wild relatives (CWR) with the ornamental P. × hortorum. Ten of the resulting 12 (F1) interspecific hybrids segregate for chlorosis suggesting biparental plastid inheritance. The segregation ratios of the interspecific F2 populations show nuclear interactions of one, two, or three nuclear genes regulating plastid function dependent on the parents. We further validated that biparental inheritance of plastids is common in section Ciconium, using diagnostic PCR primers. Our results pave the way for using the diverse species from section Ciconium, each with its own set of characteristics, as novel sources of desired breeding traits for P. × hortorum cultivars.
Keywords: Pelargonium, cyto-nuclear incompatibility, interspecific hybridization, biparental inheritance, plastid
Introduction
Several closely related species from Pelargonium sect. Ciconium have been used for producing hybrids that are sold world-wide, commonly known as “garden geraniums,” that are some of the most popular and iconic ornamentals. However, there are genetic barriers to establishing crosses and making new hybrids, including Cyto-Nuclear Incompatibility (CNI) that can cause cytoplasmic male sterility (CMS), dwarf growth (DG) and chlorosis in hybrid offspring (Greiner et al., 2015; Postel and Touzet, 2020). Nearly all angiosperms have uniparental maternal organelle inheritance. Unusually, Pelargonium × hortorum as well as the species P. zonale display biparental inheritance of their organelles (Baur, 1909; Tilney-Bassett et al., 1992; Weihe et al., 2009). Inheritance of organelles in plants with biparental transmission was found to be non-Mendelian in these studies, even though the expression of organelles is managed by the nuclear genome (Barkan and Small, 2014; Börner et al., 2015; Zhang and Lu, 2019). Phenotypic effects of plastid types in otherwise equal nuclear genomic backgrounds were recently demonstrated in Arabidopsis (Flood et al., 2020), but other such studies are so far rare.
Pelargonium species are an attractive model system to study CNI as different organellar effects can be evaluated in offspring with equal nuclear-genomic backgrounds, using established crossing techniques. There is a long history of observations of CNI in Pelargonium starting in the twentieth century when the foundations were laid for some of the cultivars we have today (e.g., Sweet 1820, 1822). Subsequently, more detailed studies of CNI, especially plastid-induced, were carried out in Pelargonium (Baur, 1909; Tilney-Bassett, 1973, 1974, 1975), which, based on segregation ratios, ultimately found support for a two-gene model of complementary nuclear genomic alleles that control the inheritance of organelles in Pelargonium (Tilney-Bassett, 1976, 1984, 1988; Tilney-Bassett and Birky, 1981; Tilney-Bassett and Abdel-Wahab, 1982; Tilney-Bassett et al., 1989b).
To further advance our knowledge of CNI in Pelargonium, we have performed a section-wide survey of most of the crop wild relatives (CWR) of P. × hortorum and its supposed ancestors P. inquinans and P. zonale (James et al., 2004) to investigate the inheritance of organelles in general and plastids in particular. There are currently 17 species recognized in Pelargonium section Ciconium (van der Walt and Vorster, 1988; Röschenbleck et al., 2014) which are all considered the CWR of P × hortorum. Phylogenetic relationships among these species have recently been reconstructed based on 76 plastome exon sequences (van de Kerke et al., 2019 and references therein). We further investigated if chlorosis in the hybrid offspring can be correlated with a particular plastid type (e.g., the combined plastid proteome, metabolome, and transcriptome inherited from one parent). Given the ubiquitous occurrence of chlorosis in crosses between species of P. sect. Ciconium and in other sections in the genus (Sweet 1820–1822, Horn, 1994; Breman pers. obs.), we expect that biparental inheritance of organelles is more common than is currently reported in the published literature.
Finally, based on segregation ratios over one of the crossing series, we deduced the underlying model of interacting genes which can explain the occurrence of chlorotic phenotypes in these crosses, and hence CNI. We did this by disentangling the effects of each possible plastome type on chlorosis in the F1 species hybrids.
Materials and Methods
We established novel interspecific crossings between twelve related diploid species of P. section Ciconium and P. × hortorum (species and acronyms mentioned in Table 1). We verified the hybrid status of the offspring using phenotyping, especially by evaluating leaf morphology, as well as flower color and shape (for an example see Figure 1 for all others see Supplementary Figure 4). In addition, hybrid status and ploidy level of obtained F1 hybrids were verified by flow-cytometry using P. × hortorum as internal reference. Flow-cytometry was performed by Iribov bv (Heerhugowaard, Netherlands) on freshly collected leaf material using a Partec CA-II flowcytometer according to De Laat et al. (1987). Nuclei were stained with a High-Resolution Kit (Partec).
TABLE 1.
Plant materials used in this study.
| Species | Herbarium voucher | Species acronym used in the text | Institutea |
| P. acetosum | 1243 | ACET | NHM |
| P. acraeum | 1975 | ACRA | STEU |
| P. alchemilloides | 1885 | ALCH | STEU |
| P. articulatum | S1026 | ARTI | SYN |
| P. barklyi | S1027 | BARK | SYN |
| P. frutetorum | S1087 | FRUT | SYN |
| P. inquinans | 0682 | INQU | STEU |
| P. multibracteatum | 2902 | MULT | STEU |
| P. peltatum | 1890 | PELT | STEU |
| P. quinquelobatum | S1044 | QUIN | SYN |
| P. ranunculophyllum | A3651 | RANU | MSUN(*) |
| P. tongaense | 3074 | TONG | STEU |
| P. zonale | 1896 | ZONA | STEU |
| P. elongatum | 0854 | ELON | STEU |
| P. aridum | S1088 | ARID | SYN |
| P. × hortorum “Pinto white” (PW) | PEZ-BD8517 | HORT | SYN |
| P. × hortorum “Tango White” (TW) | NA | HORT | NA |
Herbarium voucher information. aSTEU, Stellenbosch University, RSA; AL, Albers/MSUN, Münster Germany; SYN, Syngenta collection number; NHM, Natural History Museum London UK.
FIGURE 1.
Example of the phenotype of an F1 hybrid generated from the HORT × ARID cross. (A) Shows the leaf and flower phenotypes of P. × hortorum; (B) shows the leaf and flower phenotypes of P. aridum; (C) shows the leaf phenotype of F1 HORT × ARID; (D) shows the flower phenotype of F1 HORT × ARID.
Interspecific F1 Hybrids
The F1 hybrids generated in this study were produced from the diploid HORT cultivar “Pinto White” (PW) crossed with the species outlined above (listed in Table 1 and Figure 2). In addition, we attempted one interspecific cross at the tetraploid level using HORT “Tango White” (TW) and P. articulatum (ARTI). For all crosses, plants were moved to a pre-cleaned greenhouse and manually pollinated by using dedicated small paint brushes, made of animal hair, at 1-day intervals from the moment of flowering, dependent on the species. When seed development did not take place or was impaired, embryo rescue (Table 2) was performed as follows: at 2–3 weeks after pollination, embryos were collected, dissected and put on tissue culture in dedicated cabinets using an approach similar to Kamlah et al. (2019).
FIGURE 2.
Schematic overview of the experimental setup for the F1 crosses put in a phylogenetic context. The tree displayed tree is based on van de Kerke et al. (2019). The position of P. ranunculophyllum is inferred from our plastome assembly data (Breman et al. in prep), combined with those of van de Kerke et al. (2019) and Breman et al. (in prep). On the top row are all accessions used, represented by their respective floral and leaf phenotypes and structured by their presumed phylogenetic distance to “HORT” (van de Kerke et al., 2019); “Generations established” indicates results obtained with “F1” indicating only F1 plants were obtained, no F2 could be generated; “F2” meaning the plants were fertile and could produce F2 offspring; red asterisks denote crosses that failed for reasons explained in the text. The bottom row indicates the number of plants used across the seasons.
TABLE 2.
F1 offspring overview.
| Hybrid | Origins of plastid | Phenotype | # offspring obtained |
| HORT × ZONA | Maternal | Chlorotic | 144 |
| HORT × ZONA | Paternal | Mostly Green | |
| HORT × ZONA | Biparental | Variegated | |
| HORT × ACET | Paternal | Green Vir | 7 |
| HORT × ACET | Maternal | Lethal | |
| HORT × FRUT | NP | Green | 72 |
| HORT × INQU | NP | Green | 2 |
| HORT × ACRA | Paternal | Green | 24 |
| HORT × QUIN | Maternal | Lethal | 12 |
| HORT × QUIN | Paternal | Chlorotic Vir | |
| HORT × QUIN | Biparental | Variegated | |
| HORT × MULT | Paternal | Chlorotic Vir | 21 |
| HORT × ALCH | Paternal | Chlorotic Vir | 8 |
| HORT × TONG | Paternal | Chlorotic | 36 |
| HORT × ARID | Paternal | Chlorotic | 10 |
| HORT × ARID | Maternal | Lethal | |
| HORT × PELT | * | – | – |
| HORT × RANU | * | – | – |
| HORT × BARK | Paternal | Lethal | 2 |
| HORT4 × ARTI | Either | Green | – |
| HORT4 × ARTI | Eihter | Chlorotic | |
| HORT4 × ARTI | Either | Lethal |
Vir indicates plants were virescent. *Crosses failed either because (paternal) plants would not flower or no fruit was ever observed. HORT4 refers to a tetraploid cultivar from HORT called “Tango White” All other HORT refer to the diploid “Pinto White” cultivar.
Interspecific F2 Populations
In order to evaluate the nuclear background of CNI, we created F2 progeny of particular F1 individuals (Table 3). We selected F1 plants which we assume to contain either one, or both parental cytotypes based on overall leaf coloration. We hypothesized that green and chlorotic plants contained one parental type (at that point unknown which one) and that variegated plants contained both (biparental). We selected from these a number of individuals for subsequent self-pollination to generate the F2 populations: six plants in total representing three phenotypes encountered in the HORT × ZONA F1 which includes 2 green (denoted as: HORT × ZONAG’), 2 variegated (denoted as: “HORT × ZONAV”) and 2 chlorotic (denoted as: “HORT × ZONAC”) plants (see Figures 3, 5). In addition, we included one cross (three green plants, the only surviving phenotype) involving P. acetosum (ACET). We also selected plants from a crossing involving P. frutetorum (FRUT) and P. inquinans (INQU) as positive controls for the evaluation. This is because Pinto White contains a plastid that is considered to have originated from the P. inquinans ancestor (James et al., 2004) and the plastid of P. frutetorum is indistinguishable from that of PW and P. inquinans (Breman et al., in prep). Therefore, we expected these crosses not to display chlorosis in the F2.
TABLE 3.
Genotypes detected in F1 and F2 offspring, using diagnostic PCR, for the HORT × ZONA cross.
| Pedigree | Plant/cross | Phenotype | Origin of plastid |
| F0 | Hortorum | G | Wild-type M |
| Zonale | G | Wild-type P | |
| F1 | 8542 | G | P |
| 8542 | C | M | |
| 8552 | G | P | |
| 8552 | V | M | |
| 8570 | C | M | |
| 8570 | G | P | |
| 8570 | C | P | |
| 8570 | V | M | |
| PEZ-BD8542 | 8618 | C | G-P |
| 8618 | G | G-P | |
| 8618 | C | NA | |
| 8619 | G | G-P | |
| 8619 | C | G-P | |
| 8620 | C | G-M | |
| 8620 | C | G-M | |
| 8620 | C | G-M | |
| 8620 | C | G-M | |
| 8627 | C | G-P | |
| 8627 | G | G-P | |
| 8628 | C | G-M | |
| 8628 | V | biparental | |
| 8628 | G | G-P | |
| 8628 | C | G-M | |
| 8629 | C | G-M | |
| PEZ-BD85552 | 8621 | C | NA |
| 8621 | G | G-P | |
| 8623 | C | G-M | |
| 8623 | G | G-M | |
| 8630 | C | G-P | |
| 8630 | G | G-P | |
| 8631 | C | G-M | |
| 8631 | G | G-M | |
| 8632 | Lethal | G-M | |
| 8632 | C | G-M | |
| PEZ-BD8570 | 8624 | C | G-M |
| 8625 | C | G-M | |
| 8625 | C | G-M | |
| 8625 | G | G-M | |
| 8626 | G | G-P | |
| 8626 | C | G-P | |
| 8634 | Lethal | G-M | |
| 8634 | G | G-P | |
| 8634 | C | G-M |
Structured by cross. (G)-P and (G)-M denotes (grand) paternally-and maternally inherited plastids.
FIGURE 3.
Experimental setup for obtaining F2 crosses of F1 parents with different phenotypes.
FIGURE 5.
Tracing plastids throughout the pedigrees. Correlation between phenotype and genotype, subsequent segregation in F2 offspring is also indicated. (A,B) TWO HORT × ZONA crosses; (C) the HORT × ACET cross; (D) the HORT × FRUT cross. Text in subscript denotes the plastid type found in the parental population.
We also evaluated a subset of plants for evaluation of segregation for CNI phenotype patterns that are expressed during the pre-seedling phase. We selected three F1 parents of HORT × ACETG, four HORT × ZONAG and one parent each of HORT × FRUTG and HORT × ACRAG. Because fruit-set was low this season for HORT × ZONA we pooled these to enable Chi2 testing. We feel pooling was justified, because these plants share parentage, and have the same phenotype and plastid.
Plant Rearing
Plants were grown in a greenhouse from seeds and leaf material was collected from the first primary leaves for DNA extraction. See Table 1 for the full list of plant material used with Herbarium accession numbers and see Supplementary Figures 6A–K for representative phenotypes of each F1 plant.
DNA Extraction, Primer Design, and Genotyping
Genomic DNA was extracted from leaf material using a modified CTAB protocol (Bakker et al., 1998) followed by RNAse treatment. We designed specific primers for plastome-typing parents and F1 offspring We used the Long Single Copy region (LSC) of assembled plastomes (Breman et al., in prep) for Pelargonium section Ciconium species. LSC has been shown to contain numerous indels (Chumley et al., 2006; Guisinger et al., 2008, 2011; Weng et al., 2017; Breman et al., in prep) which can be used to create genotype-specific primer sites. Visual inspection of sequence alignments, combined with parsimony analysis and using the “Apomorphy list” command in PAUP∗4b10 for windows (Swofford, 2002), was performed to find suitable primer sites and to check for unique autapomorphies therein. We specifically scanned for regions with a unique indel or multiple unique substitutions, allowing for genotype-specific primers. Amplicon sizes were designed to be < 500 bp, allowing for shorter PCR thermo-profiles. Candidate primer pairs were evaluated using Oligocalc (Kibbe, 2007)1 checking for differences between melting temperatures (ΔTm), self-priming and hairpin formation. Primers were accepted when ΔTm between forward and reverse primers was < 3°C and with only one hairpin and/or one self-priming was predicted. Further, we required a primer site to have a minimum Illumina read coverage of 20. A GC content of 40–50% was preferred, but this was not always possible. A GC content of 40–50% is considered best for ensuring stable binding during annealing and increase the primer pairs efficiency. Finally, we submitted the primers to a BLAST search (set for analyzing short sequences) to compare to all available Pelargonium sequences to verify target-specificity. Occasionally a single primer would have a significant hit to Pelargonium species outside section Ciconium, but this never occurred for both primers of a pair.
Primers were tested in vitro, using a panel of 16 section Ciconium species representing the range of parental plastid variation we would encounter in our offspring. Primer candidates were evaluated using the target accession and an annealing temperature gradient ranging from 49 to 60°C. Primers that amplified were subsequently tested against the panel of accessions at the highest possible temperature for which it showed amplification of the target. For PCR profiles and reaction conditions see Supplementary Figure 3.
Phenotyping of F1 and F2 Plants
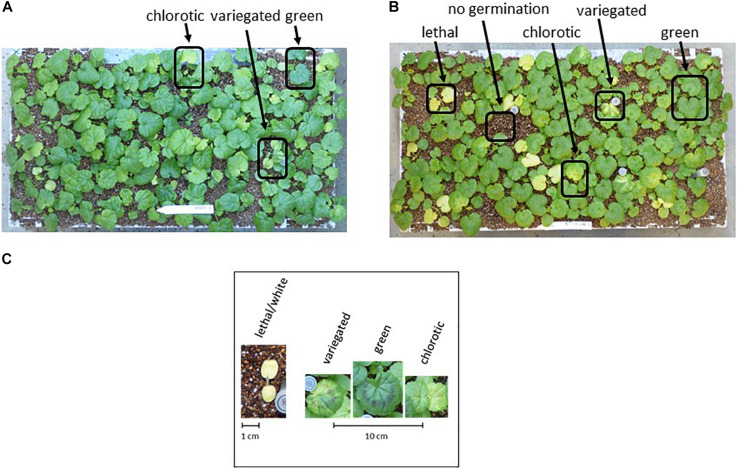
Leaf color phenotyping was performed at the seedling stage (Figures 4A,B). In order to consistently compare phenotypes across populations per cross, we took photos of seedlings at 2-week intervals during the seedling stage until the development of the first two primary leaves (Figure 4). We used the following four leaf-phenotyping categories based on a visual assessment of the phenotypes: (1) “Green”: leaf phenotype comparable to parents; (2) “Chlorotic,” plants are lighter green than either parent or even yellow; (3) “White,” plants germinate, but die within 2 weeks. Seeds that failed to germinate are added to this category; (4) “Variegated,” plants display more than one chlorotic phenotype in the same individual, presumably due to heteroplasmy (see Figures 4B,C, 5).
FIGURE 4.
Chlorosis phenotypes. (A) Segregation for chlorosis among progeny in the F2 HORT × FRUT. (B) Segregation for chlorosis in F1 HORT × ZONA. Examples of “green,” “chlorotic” and “variegated” plants are indicated, as well as “lethal” phenotypes and seeds that failed to germinate. (C) Scaled close-up of examples of phenotypes.
Ratios of the four phenotypes for each tested F2 population were compared and fitted to a one-, two- and three-gene model of inheritance of nuclear genomic alleles (calculated using the spreadsheet from Montoliu, 2012). We assumed four phenotypes and combined these according to five different scenarios, each representing assumptions on expected phenotypic ratios and their expression. The first scenario tested considers four phenotypes (i.e., “not affected,” “mildly,” “severely,” and “lethal”). The second and third scenarios consider there to be three phenotypes (“not affected,” “affected,” and “lethally affected”). Finally, the fourth and fifth scenarios consider only two phenotypes (“affected” vs. “not affected”). We then evaluated these five different scenarios by binning individuals differently. E.g., under scenarios two and three only green plants are considered to be unaffected but the lethal category consisted either of only the white or the white and severely affected plants (Table 4A). Thereby we further assumed different parental genotypes and their expected phenotypic ratios leading to eight testable phenotypic ratios representing models of one, two, or three loci involved (Table 4B).
TABLE 4A.
Crosses matching genetic models of inheritance.
| One gene model |
Two gene model |
Three gene model |
Observed ratios | |||||||
| Scenarios | Crosses meeting criteria | a | b | c | d | e | f | g | h | |
| Scenario 1 | F2_hort_x_zonaV | M*** | – | – | – | – | M*** | – | M*** | ∼4:5:1:3 |
| Scenario 2 | F2_hort_x_zonaG | – | – | M*** | – | – | – | – | – | ∼7:7:1 |
| F2_hort_x_zonaV | M*** | M*** | – | – | – | M*** | – | M*** | ∼1:2:1 | |
| F2_hort_x_frut | – | M*** | – | – | – | M*** | – | M*** | ∼1:3:0 | |
| Scenario 3 | None | – | – | – | – | – | – | – | – | – |
| Scenario 4 | None | – | – | – | – | – | – | – | – | – |
| Scenario 5 | F2_hort_x_zonaG | – | – | M*** | – | – | – | – | – | ∼1:1!50:50 |
| F2_hort_x_zonaV | M*** | M*** | – | – | – | M*** | – | M*** | ∼1:2!75:25 | |
| F2_hort_x_frut | – | M*** | – | – | – | M*** | – | M*** | ∼1:3!75:25 | |
| Fruit/seed phase | ||||||||||
| Scenario 1 | None | – | – | – | – | – | – | – | – | – |
| Scenario 2 | None | – | – | – | – | – | – | – | – | – |
| Scenario 3 | None | – | – | – | – | – | – | – | – | – |
| Scenario 4 | F2_hort_x_frut | – | – | M*** | – | – | – | – | – | 1:1!50:50 |
| F2_hort_x_zonaG | —- | M*** | – | – | – | M*** | – | M*** | ∼1:3!25:75 | |
| F2_hort_x_acet | – | M*** | – | – | M*** | – | M*** | ∼1:3!25:75 | ||
| Scenario 5 | F2_hort_x_frut | – | M*** | – | – | M*** | – | – | ∼1:4 | |
| F2_hort_x_acra | – | – | M*** | – | – | – | – | – | ∼1:1.5 | |
| F2_hort_x_zonaG | – | M*** | – | – | – | M*** | – | – | ∼1:1.3 | |
M: mendelian model applies, ***P < 0.001 under the χ2-test! indicates ratios matching particular model. For all observed ratios and counts of phenotypes under each scenario please see Supplementary Material 5.
TABLE 4B.
Possible parental genotype and expected phenotypic ratios.
| Lettercode | Genetic model and expected ratios |
| a | F1xF1 = AaxAa 25:50:25 |
| b | F1xF1 = AaxAa 25:75 |
| c | F1xF1 = AaxAA 50:50 |
| d | F1xF1 = AaBbxAaBb 6.25:18.75:18.75:56.25 |
| e | F1xF1 = AABbxAaBB 25:25:25:25 |
| f | F1xF1 = AABbxAaBB 25:75 |
| g | F1xF1 = AaBbCcxAaBbCC 6.25:18.75:56.25:18.75 |
| h | F1xF1 = AaBbCcxAaBbCC 25:75 |
For evaluating seed phenotypes, we used a similar approach, distinguishing four phenotypes: (1) “normal,” not affected by CNI, 2); “bleached,” seed contains endosperm that is still filled, but the seed is bleached; (3) “watery,” in this case the endosperm is bleached and not properly filled; (4) “lethal,” seeds with this phenotype displayed early aborted or undeveloped embryos. For examples see Figure 6.
FIGURE 6.
Seed phenotypes displaying signs of Cyto-nuclear incompatibility (see text for further explanation). (A) Normal seed phenotype; (B) bleached seed phenotype; (C) seed containing no or watery endosperm; (D) no embryo development.
As for leaf phenotypes, for seed phenotyping we evaluated five scenarios as well. We assumed four (“normal,” “bleached,” “empty,” “lethal,” scenario 1), three (“not affected,” “affected,” and “lethally affected,” scenarios 2 and 3) and two (“affected” vs. “not affected,” scenarios 4 and 5) phenotypes.
Genotyping F1 Plants and F2 Populations
We plastome-typed F1 plants using our diagnostic primers described above (Table 3). In those cases where the F1 population segregated for chlorosis, we tested accessions representing each phenotype. We then typed F2 plants from each population, and plastome types were then associated with the measured leaf phenotypes to establish the correlation, and thus effect, of each plastid type in the segregating offspring.
Results
Crossing Results
Interspecific F1 Hybrids
From thousands of pollination attempts we created a total of 314 F1 hybrid plants from crossing our species panel to the ornamental cultivar P. × hortorum PW (see Table 2). Twelve interspecific crosses were successful in producing F1 plants (Table 2). For three crosses embryo rescue (ER) was needed In order to produce scorable progeny, whereas three attempted crossings failed. Attempts to cross HORT with P. elongatum (ELON) failed, but this was expected given the difference in basic chromosome numbers between the two accessions (HORT × = 9, P. elongatum × = 4 (Gibby and Westfold, 1986; Gibby et al., 1990). The other two failed due to a lack of flowering HORT with P. peltatum (HORT × PELT) or poor greenhouse conditions (such as too high humidity or temperature) for the paternal source HORT with P. ranuncluophyllum (HORT × RANU). Except for HORT × ZONA (Baur, 1909 and many others since), HORT × ACET and HORT × QUIN (Hondo et al., 2014, 2015), these crosses are novel and were never reported in literature before. Remarkably, in 10 cases the F1 offspring displayed segregation for leaf color phenotype (e.g., chlorosis). When segregation did occur, it ranged from varying levels of chlorosis to nearly green for some crosses to spanning the full range of possible phenotypes from lethal white plantlets to nearly fully green plants (Figure 4).
Establishment of the F2 Populations
For all F1 crosses we were able to obtain an F2 generation (Figure 3) with varying degrees of success, e.g., the green F1 “HORT × ZONA” cross used to produce the F2 yielded significantly more offspring as well as a slightly higher germination success than the variegated or chlorotic parents did (Table 5). The seed phenotypes for F2 crosses which were used in this study are shown in Figure 6.
TABLE 5.
F2 material obtained using selection of plants from Table 1.
| Cross | G | V | C | Lethal* | # seeds | Sum | Germination % | Marker(s) found | Ratio |
| F2_hort_x_zona C | 8 | 1 | 51 | 34 | 125 | 94 | 0.78 | 5 FRIN, 1 ZONA | ∼1:6:4 |
| F2_hort_x_zonaG | 139 | 3 | 138 | 20 | 332 | 300 | 0.84 | 1 FRIN, 5 ZONA | ∼7:7:1 |
| F2_hort_x_zonaV | 52 | 4 | 79 | 48 | 230 | 183 | 0.76 | 2 FRIN, 1 ZONA, 2 FRIN, and ZONA | ∼1:2:1 |
| F2_hort_x_acet C | 33 | 2 | 49 | 28 | 116 | 112 | 0.95 | ACET | 1 |
| F2_hort_x_frutG | 24 | 3 | 63 | 0 | 90 | 90 | 1.00 | FRIN | 1 |
| F2_hort_x_inquG | 144 | 0 | 0 | 0 | 144 | 144 | 1.00 | FRIN | 1 |
Chlorosis phenotypes of F1 parents and plant counts are given. “G” denotes a green plant, “C” a chlorotic one and “V” a variegated plant. For the description of the chlorosis categories see the text. ∗Lethal plants are counted without the seeds that failed to germinate.
Primer Design and PCR Verification
We designed 11 primer-pairs targeting single accessions (e.g., genotype-specific primers) or a group of accessions (Table 6). All primer-pairs performed as expected, except BART, which amplified ARTI but not BARK. We therefore used this primer only for detecting ARTI. For gel photo documentation accompanying the primer pair evaluations we refer to Supplementary Figures 2, 3. All primers worked across a range of template DNA concentrations (0.1 ng/μl up to > 5 ng/μl). A 1/10th dilution of the extracts generally increased PCR performance.
TABLE 6.
Primer pair details.
| Primer pair name | Target(s) | Sequence 5′–>3′ | Plastome region |
| FRIN | P. inquinans, frutetorum and × hortorum | AAAGGCCAGATTGGGCGGC | F: IGS and R; 5′ of rna polymerase beta subunit 2 exon |
| GACGAATTCGGTCCGATTCAACAC | |||
| ZONA | P. zonale | GAATTGTAATGCGGAGCTGC | F and R: IGS |
| AAGAAAGAGGATATAGCCGGAC | |||
| ACET | P. acetosum | GAATCCCCACCTACACTACAC | F and R: MATK exon, 3′end |
| CCTTGACTAAAGCGCAATTTTG | |||
| ACRA | P. acraeum | GACCCTATCTCTCTGTATTC | F and R: IGS |
| TTTGGTCTCCGAAAAGAAAAGG | |||
| ALRA | P. alchemilloides and ranunculophyllum | GGATCTTATCTATTCTCTATTC | F and R: IGS just downstream of trnK-UUU small exon |
| CGATCTAGATCTAATTGTAC | |||
| MUQU | P. multibracteatum and quinquelobatum | GGTTTCGCGTCAATTGC | F and R in flanking IGS’s of atpH, atpH exon is entirely covered by fragment |
| CTGAATTTAGCTATGATTTCG | |||
| ARID | P. aridum | CTGAACTGAACTCAAATGGA | F and R: in IGS, fragment contains trnH-IS and trnI-LE |
| ATTGCGAGGATCCTACTTTG | |||
| BARK | p. barklyi | GAAAGATCTATTCGAGTCGAG | F: in IGS, R: in intron between tnrL-UAA exons |
| GGGGCCTCATTACATTAATC | |||
| PELT | P. peltatum | CTCAAAAGAAGGGTAGAAGGG | F and R: in IGS’s surrounding trnS-GGA |
| CCCTGTCTGCTCTTTCCAA | |||
| TONG | P. tongaense | GATCTCAAAGCAAAGAGAGC | F: IGS, R: in ndhJ exon |
| CTTGGCTAGTGTATACCATTTG | |||
| BART | P. articulatum and P. barklyi | GAATCCAAAAGAAATGAAATG | F and R: IGS between atpB and rbcL |
| AAAAGGAATAGGTTTTGTAG |
Plastome regions were identified using genbank ID: DQ897681.1 (P. × hortorum, Chumley et al., 2006). F refers to forward, R refers to reverse.
Phenotyping and Genotyping the F1 and F2 Population for HORT × ZONA and HORT × ACET
For a full overview of the tests for all scenarios under all eight genetic models (Tables 4A,B) we refer to Supplementary Figure 5. We discuss here those crosses that demonstrated Mendelian patterns of segregation as well as the models under which this applies. We found that the F1 plants segregate for chlorosis, with no obvious Mendelian patterns of segregation (Table 4A and Supplementary Figure 5), but that they are otherwise phenotypically homozygous, i.e., non-segregating. When genotyping the F1 plants, we found that green individuals contained the P. zonale type plastid (ZONA), whereas chlorotic individuals contained that of P. frutetorum/P. inquinans (FRIN) (Tables 2, 3 plastids of “maternal origin”). A small minority (< 5%) of the plants displayed (partial) variegation and this percentage reduced, for most, as the plant aged with most settling into a single phenotype. From these we detected either the FRIN or the ZONA plastids, but as we recovered both from the F2 offspring (see below) they must actually have contained both. We have evaluated plastid types in all phenotypes of F2 offspring (structured per F1 cross, Figure 5) for the HORT × ZONA cross series. We found FRIN and ZONA plastid types in the F2 (Table 3) and, in general, F2 offspring always contained the same plastid as was detected in the F1 plant (for example, see Figure 5), except for the variegated plants. In the F1 HORT × ZONA variegated plants we found only one of the plastids, either FRIN or ZONA, but in the F2 we detected both, even once in one variegated individual (Figure 5A). We analyzed the bleached and green tissue from this plant and found that white tissue predominantly contained the FRIN type and green contained the ZONA type (Figure 5A).
When pooling the green and light green plants and treating these as one (scenarios 4 and 5) phenotype, subsequent testing for Mendelian patterns of segregation did not yield a clear pattern (Supplementary Figure 5), as was the case for three phenotypic categories. When we categorized the phenotype ratios as “affected” or “not affected,” we saw that they matched those expected under either a one- or two-gene model for all crosses assuming lethal interactions are also possible between alleles. The populations where the ratios conformed to the one gene model are F2 HORT × ZONAV and F2 HORT × ZONAC. The segregation ratios in this “affected vs. not-affected” analysis pointed to one lethal combination of alleles and two combinations that yield viable or affected plants. When pooling light green and yellow plants and subsequently testing for Mendelian patterns of segregation, a pattern emerges for the F2 HORT × ZONAV and the F2 HORT × ZONAG populations (scenario 2). In contrast, when analyzing the observations for the F2 HORT × ZONAC plants there did not appear to be a pattern. The patterns for the F2 HORT × ZONAV and the F2 HORT × ZONAG populations did point to a genetic difference in the F1 population (and therefore also in the F0 populations). With the green populations following the one gene model whereby the F1 was Aa × AA.
The ratios for the plants phenotyped for F2 seeds and their corresponding possible underlying genetic models are listed in Table 4A. We deduced that there were likely one (in HORT × FRUT and in HORT × ZONA) and two loci (in HORT × ACET) interacting in this phase of plant development. Given that the phenotypic ratios under scenarios 1–4 did are similar to, but not exactly what would expected when of one, two, or three genes interact. We suspect that more complex interactions, possibly involving more than two or even three genes, played are role or that the loci involved are linked in some cases with aggravating or moderating effects of linked loci. This appears especially to be the case for HORT × ACRA where ratios under scenarios 1–4 are: ∼2:1:2:20 (3 loci); ∼2:1:2 (2 loci); ∼1:1:10 (3 loci), and ∼1:12 (3 loci), respectively (see Supplementary Figure 5 for more details).
Positive Controls
Our positive controls HORT × INQU and HORT × FRUT yielded 100% green plants in the F2. In the F2 this was maintained for HORT × INQU for both plant and seed phenotypes, but surprisingly, the F2 of HORT × FRUT displayed segregation for chlorosis and seed phenotypes (Figures 4A and 6) indicative of the one gene model of segregation with a heterozygous parent with possible lethal combinations expressed in the pre-seedling phase as well (Table 4A and Figure 6).
Genotyping Phylogenetically More Distant F1 Hybrids
We recovered two plastid types in the offspring of F1 of HORT × QUIN (Table 2). We found segregation for chlorosis and detected both the FRIN type as well as the MUQU type plastids in the offspring. None of these plants were fully green. In the F1 HORT with P. aridum (HORT × ARID) we found segregation for chlorosis, with the majority of offspring lethal and one plant surviving a full season. For F1 HORT × ARID We detected FRIN and ARID plastids in the offspring. In the F1 HORT × ALCH, F1 HORT × TONG, F1 HORT × ACRA, F1 HORT × MULT and HORT × BARK, we detected only the paternal plastids (Supplementary Figure 2). This is similar to the F1 HORT × ACET cross in that we detected only one type in the offspring suggesting lethal interactions with the FRIN type plastid. In the F1 HORT × ARTI cross we find segregation for chlorosis and no correlation between phenotype and genotype, we detected both the FRIN and ARTI type plastids. For an overview of all the results (see Table 2).
Discussion
We show that biparental inheritance occurs throughout the section and that hybridization is relatively easy, both observations have important implications for interpreting current concepts of Pelargonium section Ciconium evolution. This study further demonstrates that using multiple interspecific crosses can be used to gain insight into the genetics underlying organelle management and expression, potentially uncovering drivers of speciation. Our studies expand on the two-interacting gene model found to regulate plastid inheritance in Ciconium which was inferred 50 years ago by Tilney-Bassett et al. (1989b, 1992). While a limited number of crosses between P. × hortorum and section Ciconium have been previously reported (e.g., Hondo et al., 2014, 2015), we have greatly expanded on this by covering nearly all of the CWR in the section including those that are phylogenetically more distantly related.
Biparental Inheritance of Plastids and Evolutionary Implications
We have found maternal (P. frutetorum/inquinans; FRIN) and paternal (other Ciconium plastid types) inheritance in nearly all our offspring indicating that the ability to inherit and express more than one plastid is the rule rather than the exception in Pelargonium section Ciconium. Even though it was demonstrated before on a limited scale (Baur, 1909; Tilney-Bassett et al., 1992; Weihe et al., 2009), it was never demonstrated to be so ubiquitous. This has important implications for the study of Ciconium speciation as bi-parental inheritance may provide an escape from the acquisition of deleterious plastid mutations (Mullers ratchet), because there is the possibility for an additional plastome types to occur in the individual plant. Also, it may allow to occupy new niches quicker and perhaps even allow populations that have become separate in space and time to reconnect (Greiner et al., 2011; Apitz et al., 2013; Greiner and Bock, 2013; Greiner et al., 2015; Barnard-Kubow et al., 2016, 2017; Sobanski et al., 2019).
Plastid Effects
We have found evidence that in our crosses the FRIN plastid caused bleaching in the HORT × ZONA crosses and that it was possibly lethal for the HORT × ACET cross given the absence of any offspring containing FRIN. The observation that ZONA plastids caused less chlorosis than FRIN in these types of crosses is not new in itself and this study confirms what was already hinted at by Tilney-Bassett and Almouslem (1989a) and more recently confirmed by Weihe et al., 2009 who observed that the “inquinans plastid” caused bleaching. The F1 HORT × ZONA plants were, in some cases viable when containing the FRIN plastid allowing us to evaluate the effects of both plastid types in subsequent generations. As to which part of the plastome is the root cause we can only speculate, but a number of genes have been demonstrated to be under selection in the Geraniaceae plastomes (Shikanai et al., 2001; Blazier et al., 2016a, b; Ruhlman et al., 2017; Weng et al., 2017; Ruhlman and Jansen, 2018). More surprising was the find that the F2 HORT × FRUT showed a segregation for chlorosis, even though the F1 did not. This hints at a slight incompatibility between the FRIN type plastid and either the HORT or FRUT parent. This is surprising given that we cannot distinguish the plastids. Therefore, given the segregation ratios (Table 4A), one nuclear gene, either originating from HORT or FRUT, must be slightly divergent and must be responsible for this effect. Given that this segregation was not the case for The HORT × INQU F2 population and no segregation occurs when selfing HORT, we deduce that one of the alleles originating from FRUT was responsible.
F2 Segregation Pointing to Two or Three Epistatically Interacting Genes
We demonstrate, in a second generation series of plants that, irrespective of plastid type, there was segregation for chlorosis. Chlorotic phenotypes of the F2 did not appear to show Mendelian inheritance patterns under a one or two allele model in all cases. However, nuclear alleles must be involved because the plastid backgrounds are the same for each plant (Stubbe, 1958, 1959, 1989; Amoatey and Tilney-Basset, 1994; Barr and Fishman, 2010; Li et al., 2013). For the F2 HORT × ZONAV population both the one gene model and the two gene model did seem to be equally good at explaining the results. The observed numbers conformed well to the F1 HORT × ZONAV population being heterozygous. As outlined above, ratios for the three phenotypic categories do not shed much light on the underlying genetics, but when we categorize the phenotype ratios in a binary way, “affected or not affected,” we see the ratios for all crosses matching or approaching ratios for phenotypes that resemble the situation where one combination is lethal and two combinations of alleles are not. For the HORT × ZONAC population the ratio is more akin (10:1 under the two phenotypes scenario 5, Supplementary Figure 5A) to the ratio’s expected (9:1 under the two phenotypes scenario 4, Supplementary Figure 5A) under a two gene interaction model whereby heterozygous combinations are lethal and the homozygous combinations of at least one allele are not. The ratios for the HORT × ACET cross hint at a possible trihybrid segregation, whereby two alleles interact in a lethal way, because of the following reasoning: If segregation was perfect we would expect the following phenotypic ratio’s under the three gene model; 27:9:9:9:3:3:3:1 but we observe 25:9:1:5 under the four phenotypes scenario 1 (Supplementary Figure 5A). For this pattern to occur we would have to assume there are two alleles that interact in a lethal way, causing the deviation from the expected ratio’s, but also that there is a third allele which in turn moderates some of these effects or may cause extra lethality.
The ratios of CNI phenotypes observed in the seeds points to a similar type of interactions further explaining why we observe sometimes skewed segregation ratios. In the case of the HORT × ACET cross we observe mendelian segregation of under gene models b, f, h (25:75 phenotype ratios under the one, two, and three gene models) with the majority of the individuals being lethal. When we view the ratios of all phenotypes for HORT × ACET and HORT × ACRA (10:3:2:1 and 20:2:1:2, respectively, Supplementary Figure 5B) these, similarly as for the seedlings evaluated, reminiscent of ratios for the two gene model whereby heterozygous combinations are lethal and the homozygous combinations of at least one allele are not. Thus, combining the observations of both seed and seedling phase of plant development, would yield for the HORT × ACET cross a series of at least five loci involved in development and expression of organelles. For the HORT × FRUT at least two loci would be required to explain the observations, one acting in each stage of development we studied.
Model of > 3 Interacting Nuclear Genes
The observation that the HORT × ACET cross needs two and a three gene model to explain the observed patterns may indicate that those crosses which consist of combination that are phylogenetically further removed from HORT may be subject to the effects of more than three genes. As mentioned above CNI plays a role in embryo and fruit development as well. This in turn could point to a more complex model of genetic interactions involving more loci than we thus far proposed. The machinery for synthesis and management of organelles consists of numerous PPR genes that each act during a different step of these processes (Barkan and Small, 2014; Börner et al., 2015; Zhang and Lu, 2019). These can perhaps be viewed as a genetic “block chain” whereby no mismatch of combinations is allowed in order to result in a viable, green and self-sustaining plant. In our interspecific crosses there were ample opportunities for these mismatches to occur. While we have no hard evidence for this we do see from the numbers of plants recovered from our crossing attempts decreases with increased phylogenetic distance. In other words for the plants from the crosses of e.g., HORT × ARID we obtained one plant only using the same effort as was invested in the other crosses. This one plant may represent the rare, fortunate gene combination that allows the individual to survive under ideal conditions, while all other combinations are lethal. Given that phylogenetically close crosses (HORT × FRUT, HORT × ZONA, HORT × ACET) require the one, two, or three gene model with the assumption of lethality to explain the phenotypic ratios for both the seedling and seed phase we evaluated, we may just be viewing the tip of the iceberg for the phylogenetically more distant crosses. Generally, genes thought to be involved in chloroplast management and expression are Whirly genes (Maréchal et al., 2009; Isemer et al., 2012; Krupinska et al., 2014, 2019), involved in importing proteins into chloroplasts (Krausea et al., 2005; Chateigner-Boutin et al., 2008; Mackenzie and Kundariya, 2019), and PPR genes, acting at the level of RNA editing (Takenaka et al., 2013; Wang et al., 2016a, b; Rojas et al., 2019; Small et al., 2020). These genes are good candidates to study in Pelargonium and a closer study of the proteins they encode for as well as the type of RNA editing taking place, may explain both biparental inheritance as well as early stage processes of speciation.
Data Quality
Our approach to phenotyping contains a number of potential sources of error possibly obscuring more nuanced phenotypic differences. We evaluated the seedlings at two points in time to correct for differences in development phase and possible environmental effects on the stability of the phenotypes. Differences in ambient temperature at each point can, potentially severely, affect the expression of chlorosis (pers. observations all authors). Furthermore, the interpretation of the photos, while allowing for reviewing the phenotyping afterward is subject to interpretation. Defining a plant as “affected” or not is sometimes context dependent. In the initial germination phase seedlings were germinated under controlled conditions and all at the same time to insure that we were comparing plants in equal phases of development. Great care was taken to make sure the photos of each set were taken at the same day to reduce chance of observing changed phenotypes when environmental conditions change. A further reduction of errors in interpretation can, in the future, be achieved by germinating seeds under even more controlled conditions and using automated imaging software, for interpretation of chlorotic phenotypes (see for an example of this approach Flood et al., 2020).
Seed phenotypes in Pelargonium related to CNI have not been studied before. We have chosen very clear-cut categories and in doing so may have underestimated the actual level of CNI. Nevertheless, our phenotypes are reminiscent of what is regularly encountered in relation to mutated organelle expressing PPR genes in Arabidopsis thaliana (Chi et al., 2008; Du et al., 2017; Zhang et al., 2017). Finally, in some cases we found a discrepancy in plastid types detected, between parents and the offspring of the variegated plants. Probably, variegated plants are able to manage and express both plastids and subsequently one type is outcompeted but not completely removed. This competition was demonstrated in Oenothera and occurs at a cellular level (Sobanski et al., 2019).
Crossings
In our study we have obtained at least one individual F1 hybrid plants for the majority of interspecific crosses attempted (except for P. ranunculophyllum). Most were obtainable from seed showing high compatibility of the genomes and plastids. We attribute the two unsuccessful crosses to suboptimal greenhouse climate conditions as we observed that for a pollination to be successful abiotic factors such as climate and humidity are important (reviewed by Lohani et al., 2020). The chance to obtain a (viable) F1 plant further roughly correlates with previously published plastome based phylogenetic distances (Figure 1).
Our approach in this study is reminiscent of the study recently published by Flood et al. (2020) who used cybrids to study the effects of different plastids types in equal nuclear genomic backgrounds. We have used F1 generation crosses which, though different from the cybrids in the sense that the nuclear genome is hybrid, is still uniform and allows us to study the effects of different types of organelles. Our approach is different that this study focuses more on an evolutionary, rather than at the population level as was the case in Flood et al. (2020).
Conclusion and Future Applications
The insight from this study further open up possibilities for breeding of currently available Pelargonium cultivars with their crop wild relatives. Now we could conceivably start breeding in plastids that, for instance, perform better in warmer/colder/dryer climates allowing for the adjustment of cultivars to different climates (Deng et al., 2004; Cortés and Blair, 2018; Westerbergh et al., 2018) and other abiotic factors (Mezghani et al., 2019; Wang et al., 2019; Ribera et al., 2020; Singh et al., 2020). Especially, photosynthesis would be an interesting trait to focus on as differences between the species are, likely, more dramatic than those observed between the different populations of A. thaliana which has been the focus so far when studying the effects of plastid types and photosynthetic efficiency (Flood et al., 2011; Cruz et al., 2016; Flood et al., 2020). The fact that different types of plastids have a different effect in a similar nuclear background means that breeding efforts that wish to incorporate crop wild relatives to increase genetic diversity or introduce new traits should consider the organellar background of the material as well. Knowing the effects can aid in making more informed decisions as to which species to attempt a cross with and which not. This then can lead to more focused and mores successful breeding attempts.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
FCB, FTB, RCS, and MES conceived the study. FCB carried out the analysis. FCB and FTB wrote the manuscript. FCB, SP, and JWK did the laboratory work PCR. MS-S did laboratory work embryo rescue. All authors read the draft and gave feedback.
Conflict of Interest
RCS and MS-S were employed by the Syngenta Seeds BV, Netherlands. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Frank Becker for his assistance with testing primers, Toni Lockers for her work on the countless pollinations, Dennis Reus Harald Gorter for the greenhouse management and Ewa Schoutsen and Anja Kolk for performing embryo rescue. Finally we thank the two reviewers who’s comments improved this manuscript.
Funding. This research was funded by the Dutch Foundations for applied scientific research (TTW). Grant number: 14531 “Pelargonium genomics for overcoming cytonuclear incompatibility and bridging species barriers” of the Green Genetics program.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.614871/full#supplementary-material
Diagnostic PCR targeted on HORT × ZONA and HORT × ACET.
Gel images other HORT × Ciconium F1 crosses displaying plastid types as identified using PCR.
PCR mastermix and thermo profile.
Overview of representative hybrid plants of F1 plants obtained in this study.
Full overview of all scenarios and gene models tested.
(A–K) Representative phenotypes for all F1 plants created in the course of this study.
References
- Amoatey H. M., Tilney-Basset R. A. E. (1994). A test of the complementary gene model for the control of biparental plastid inheritance in zonal Pelargoniums. Heredity 72 69–77. 10.1038/hdy.1994.8 [DOI] [Google Scholar]
- Apitz J., Weihe A., Pohlheim F., Börner T. (2013). Biparental inheritance of organelles in Pelargonium: evidence for intergenomic recombination of mitochondrial DNA. Planta 237 509–515. 10.1007/s00425-012-1768-x [DOI] [PubMed] [Google Scholar]
- Bakker F. T., Hellbrügge D., Culjam A., Gibby M. (1998). Phylogenetic relationships within Pelargonium sect. Peristera (Geraniaceae) inferred from nrDNA and cpDNA sequence comparisons. Syst. Evol. 211 273–287. 10.1007/bf00985364 [DOI] [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65 415–442. [DOI] [PubMed] [Google Scholar]
- Barnard-Kubow K. B., McCoy M. A., Galloway L. F. (2017). Biparental chloroplast inheritance leads to rescue from cytonuclear incompatibility. New Phytol. 213 1466–1476. 10.1111/nph.14222 [DOI] [PubMed] [Google Scholar]
- Barnard-Kubow K. B., So N., Galloway L. F. (2016). Cytonuclear incompatibility contributes to the early stages of speciation. Evolution 70 2752–2766. 10.1111/evo.13075 [DOI] [PubMed] [Google Scholar]
- Barr C. M., Fishman L. (2010). The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics 184 455–465. 10.1534/genetics.109.108175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur E. (1909). Das Wesen und die Earblichkeitsverhältnisse der ‘arietates albomarginatae hort’ von Pelargonium zonale. Z. Indukt. Abstammungs-Vererbungsl 1 330–351. 10.1007/bf01990603 [DOI] [Google Scholar]
- Blazier J. C., Jansen R. K., Mower J. P., Govindu M., Zhang J., Weng M.-L., et al. (2016a). Variable presence of the inverted repeat and plastome stability in Erodium. Ann. Bot. 117 1209–1220. 10.1093/aob/mcw065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier J. C., Ruhlman T. A., Wenig M.-L., Rehman S. K., Sabir J. S. M., Jansen R. K. (2016b). Divergence of RNA polymerase a subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci. Rep. 6:24595. 10.1038/srep24595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner T., Aleynikova A. Y., Zubo Y. O., Kusnetsov V. V. (2015). Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta 1847 761–769. 10.1016/j.bbabio.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin A. L., Ramos-Vega M., Guevara-Garcia A., Andres C., de la Luz Gutiérrez-Nava M., Cantero A., et al. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56 590–602. 10.1111/j.1365-313X.2008.03634.x [DOI] [PubMed] [Google Scholar]
- Chi W., Ma J., Zhang D., Guo J., Chen F., Lu C., et al. (2008). The Pentratricopeptide Repeat Protein DELAYED GREENING1 Is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147 573–584. 10.1104/pp.108.116194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumley T. W., Palmer J. D., Mower J. P., Fourcade M. H., Calie P. J., Boore J. L., et al. (2006). The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23 2175–2190. 10.1093/molbev/msl089 [DOI] [PubMed] [Google Scholar]
- Cortés A. J., Blair M. W. (2018). Genotyping by sequencing and genome–environment associations in wild common bean predict widespread divergent adaptation to drought. Front. Plant Sci. 9:128. 10.3389/fpls.2018.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J. A., Savage L. J., Zegarac R., Hall C. C., Satoh-Cruz M., Davis G. A., et al. (2016). Dynamic environmental photosynthetic imaging reveals emergent phenotypes. Cell Syst. 2 365–377. 10.1016/j.cels.2016.06.001 [DOI] [PubMed] [Google Scholar]
- De Laat A., Godhe W., Vogelzang M. (1987). Determination of ploidy of single plants and populations by flow cytometry. Plant Breed. 99 303–307. 10.1111/j.1439-0523.1987.tb01186.x [DOI] [Google Scholar]
- Deng J., Hondo K., Kakihara F., Masahira K. (2004). Seasonal variation of flowering characteristics and high-temperature stress tolerance in Geraniums. J. High Technol. Agric. 16 173–182. 10.2525/jshita.16.173 21291192 [DOI] [Google Scholar]
- Du L., Zhang J., Qu S., Zhao Y., Su B., Lv X., et al. (2017). The pentatricopeptide repeat protein pigment-defective mutant2 is Involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis. Plant Cell Physiol. 58 747–759. 10.1093/pcp/pcx004 [DOI] [PubMed] [Google Scholar]
- Flood P. J., Harbinson J., Aarts M. G. M. (2011). Natural genetic variation in plant photosynthesis. Trends Plant Sci. 16 327–335. 10.1016/j.tplants.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Flood P. J., Theeuwen T. P. J. M., Schneeberger K., Keizer P., Kruijer W., Severing E., et al. (2020). Reciprocal cybrids reveal how organellar genomes affect plant phenotypes. Nat. Plants 6 13–21. 10.1038/s41477-019-0575-9 [DOI] [PubMed] [Google Scholar]
- Gibby M., Albers F., Prinsloo B. (1990). Karyological studies in Pelargonium sectt. Ciconium, Dibrachya, and Jenkinsonia (Geraniaceae). Plant Syst. Evol. 170 151–159. 10.1007/BF00937700 [DOI] [Google Scholar]
- Gibby M., Westfold J. (1986). A cytological study of Pelargonium sect. Eumorpha. Syst. Evol. 153 205–222. 10.1007/BF00983688 [DOI] [Google Scholar]
- Greiner S., Rauwolf U., Meurer J., Hermann R. G. (2011). The role of plastids in plant speciation. Mol. Ecol. 20 671–691. 10.1111/j.1365-294X.2010.04984.x [DOI] [PubMed] [Google Scholar]
- Greiner S., Bock R. (2013). Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. Bioessays 35 354–365. 10.1002/bies.201200137 [DOI] [PubMed] [Google Scholar]
- Greiner S., Sobanski J., Bock R. (2015). Why are most organelle genomes transmitted maternally? Bioessays 37 80–94. 10.1002/bies.201400110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger M. M., Kuehl J. V., Boore J. L., Jansen R. K. (2008). Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. U. S. A. 105 18424–18429. 10.1073/pnas.0806759105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger M. M., Kuehl J. V., Boore J. L., Jansen R. K. (2011). Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28 583–600. 10.1093/molbev/msq229 [DOI] [PubMed] [Google Scholar]
- Hondo K., Iwasaki C., Kakihara F. (2014). Characteristics of progeny obtained from the cross between Geraniums (Pelargonium x hortorum) and wild species in section Ciconium, Including heat tolerance and odor of leaves. Environ. Control Biol. 52 37–43. 10.2525/ecb.52.37 21291192 [DOI] [Google Scholar]
- Hondo K., Sukhumpinij P., Kakihara F. (2015). Flower color and pigments in yellow-flowered hybrid progeny raised from the interspecific cross Pelargonium quinquelobatum × white-flowered geraniums. Sci. Hortic. 195 145–153. 10.1016/j.scienta.2015.09.014 [DOI] [Google Scholar]
- Horn W. (1994). Interspecific crossability and inheritance in Pelargonium. Plant Breed. 113 3–17. 10.1111/j.1439-0523.1994.tb00696.x [DOI] [Google Scholar]
- Isemer R., Mulisch M., Schäfer A., Kirchner S., Koop H.-U., Krupinska K. (2012). Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 586 85–88. 10.1016/j.febslet.2011.11.029 [DOI] [PubMed] [Google Scholar]
- James C. M., Gibby M., Barrett J. A. (2004). Molecular studies in Pelargonium (Geraniaceae). A taxonomic appraisal of section Ciconium and the origin of the “Zonal” and “Ivy-leaved” cultivars. Plant Syst. Evol. 243 131–146. 10.1007/s00606-003-0074-2 [DOI] [Google Scholar]
- Kamlah R., Pinker I., Plaschil S., Olbricht K. (2019). Hybridization between Pelargonium acetosum L’Hér. and Pelargonium× peltatum. J. Appl. Bot. Food Qual. 92 49–56. 10.5073/JABFQ.2019.092.007 [DOI] [Google Scholar]
- Kibbe, (2007). OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35 W43–W46. 10.1093/nar/gkm234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausea K., Kilbienski I., Mulisch M., Rödiger A., Schäfer A., Krupinska K. (2005). DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 579 3707–3712. 10.1016/j.febslet.2005.05.059 [DOI] [PubMed] [Google Scholar]
- Krupinska K., Braun S., Nia M. S., Schäfer A., Hensel G., Bilger W. (2019). The nucleoid-associated protein WHIRLY1 is required for the coordinate assembly of plastid and nucleus-encoded proteins during chloroplast development. Planta 249 1337–1347. 10.1007/s00425-018-03085-z [DOI] [PubMed] [Google Scholar]
- Krupinska K., Oetke S., Desel C., Mulisch M., Schäfer A., Hollmann J., et al. (2014). WHIRLY1 is a major organizer of chloroplast nucleoids. Front. Plant Sci. 5:432. 10.3389/fpls.2014.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Qi X., Li X., Li L., Zhong C., Huang H. (2013). Maternal inheritance of mitochondrial genomes and complex inheritance of chloroplast genomes in Actinidia Lind.: evidences from interspecific crosses. Mol. Genet. Genomics. 288 101–110. 10.1007/s00438-012-0732-6 [DOI] [PubMed] [Google Scholar]
- Lohani N., Singh M. B., Bhalla P. L. (2020). High temperature susceptibility of sexual reproduction in crop plants. J. Exp. Bot. 71 555–568. 10.1093/jxb/erz426 [DOI] [PubMed] [Google Scholar]
- Mackenzie S. A., Kundariya H. (2019). Organellar protein multi-functionality and phenotypic plasticity in plants. Phil. Trans. R. Soc. B 375:20190182. 10.1098/rstb.2019.0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A., Parent J.-S., Véronneau-Lafortune F., Joyeux A., Lang B. F., Brisson B. (2009). Whirly proteins maintain plastid genome stability in Arabidopsis. PNAS 106 14693–14698. 10.1073/pnas.0901710106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezghani N., Khoury C.-K., Carver D., Achicanoy H. A., Simon P., Flores F. M., et al. (2019). Distributions and conservation status of carrot wild relatives in Tunisia: a case study in the western Mediterranean basin. Crop Sci. 59 2317–2328. 10.2135/cropsci2019.05.0333 [DOI] [Google Scholar]
- Montoliu L. (2012). Mendel: a simple excel workbook to compare the observed and expected distributions of genotypes/phenotypes in transgenic and knockout mouse crosses involving up to three unlinked loci by means of a χ2 test. Transgenic Res. 21 677–681. 10.1007/s11248-011-9544-4 [DOI] [PubMed] [Google Scholar]
- Postel Z., Touzet P. (2020). Cytonuclear genetic incompatibilities in plant speciation. Plants 9:487. 10.3390/plants9040487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribera A., Bai Y., Wolters A.-M. A., van Treuren R., Kik C. (2020). A review on the genetic resources, domestication and breeding history of spinach (Spinacia oleracea L.). Euphytica 216:48 10.1007/s10681-020-02585-y [DOI] [Google Scholar]
- Rojas M., Yu Q., Williams-Carrier R., Maliga P., Barkan A. (2019). Engineered PPR proteins as inducible switches to activate the expression of chloroplast transgenes. Nat. Plants 5 505–511. 10.1038/s41477-019-0412-1 [DOI] [PubMed] [Google Scholar]
- Röschenbleck J., Albers F., Müller K., Weinl S., Kudla J. (2014). Phylogenetics, character evolution and a subgeneric revision of the genus Pelargonium (Geraniaceae). Phytotaxa 159 31–76. 10.11646/phytotaxa.159.2.1 24943631 [DOI] [Google Scholar]
- Ruhlman T. A., Jansen R. K. (2018). “Aberration or analogy? The atypical plastomes of Geraniaceae,” in Advances in Botanical Research 85: Plastid Genome Evolution, eds Chaw S.-M., Jansen R. K. (Amsterdam, NL: Elsevier; ), 223–262. 10.1016/bs.abr.2017.11.017 [DOI] [Google Scholar]
- Ruhlman T. A., Zhang Z., Blazier J. C., Sabir J. M. S., Jansen R. K., et al. (2017). Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am. J. Bot. 104 559–572. 10.3732/ajb.1600453 [DOI] [PubMed] [Google Scholar]
- Shikanai T., Shimizu K., Ueda K., Nishimura Y., Kuroiwa T., Hashimoto T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in Tobacco. Plant Cell Physiol. 42 264–273. 10.1093/pcp/pce031 [DOI] [PubMed] [Google Scholar]
- Singh M., Kumar S., Basandrai A. K., Basandrai D., Malhotra N., Saxena D. R., et al. (2020). Evaluation and identification of wild lentil accessions for enhancing genetic gains of cultivated varieties. PLoS One 15:e0229554. 10.1371/journal.pone.0229554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I. D., Schallenberg-Rüdinger M., Takenaka M., Mireau H., Ostersetzer-Biran O. (2020). Plant organellar RNA editing: what 30 years of research has revealed. Plant J. 101 1040–1056. 10.1111/tpj.14578 [DOI] [PubMed] [Google Scholar]
- Sobanski J., Giavalisco P., Fischer A., Kreiner J. M., Walther D., Schöttler M. A., et al. (2019). Chloroplast competition is controlled by lipid biosynthesis in evening primroses. PNAS 116 5665–5674. 10.1073/pnas.1811661116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbe W. (1958). Dreifarbenpanschierung bei Oenothera: II. Wechselwirkungen zwischen Geweben mit zweierblich verschiedenen Plastidensorten. Z. für Vererbungslehre 89 189–203. 10.1007/bf00890107 [DOI] [Google Scholar]
- Stubbe W. (1959). Genetische analyse des zusammenwirkens von genom und plastom bei Oenothera. Z. für Vererbungslehre 90 288–298. 10.1007/bf00888761 [DOI] [Google Scholar]
- Stubbe W. (1989). Oenothera—An ideal system for studying the interaction of genome and plastome. Plant Mol. Biol. Rep. 7 245–257. 10.1007/BF02668633 [DOI] [Google Scholar]
- Swofford D. L. (2002). PAUP∗: Phylogenetic analysis using parsimony (∗and other methods), version 4. 10.1007/bf02668633 [DOI] [Google Scholar]
- Tilney-Bassett R. A. E. (1973). The control of plastid inheritance in Pelargonium II. Heredity 30 1–13. 10.1038/hdy.1973.1 [DOI] [Google Scholar]
- Tilney-Bassett R. A. E. (1974). The control of plastid inheritance in Pelargonium III. Heredity 33 353–360. 10.1038/hdy.1974.102 [DOI] [Google Scholar]
- Tilney-Bassett R. A. E. (1975). “Genetics of variegated plants,” in Genetics and biogenesis of mitochondria and chloroplasts. eds Birky C. W., Perlman P. S, Byers T. J. (Columbus: Ohio State University Press; ), 268–308. [Google Scholar]
- Tilney-Bassett R. A. E. (1976). The control of plastid inheritance in Pelargonium IV. Heredity 37 95–107. 10.1038/hdy.1976.68 [DOI] [Google Scholar]
- Tilney-Bassett R. A. E. (1984). “The genetic evidence for nuclear control of chloroplast biogenesis in higher plants,” in Chloroplast biogenesis. Society for Experimental Biology Seminar Series 21, ed. Ellis R. J. (Cambridge: Cambridge University Press; ), 13–50. [Google Scholar]
- Tilney-Bassett R. A. E. (1988). “Inheritance of plastids in Pelargonium,” in The Division and Segregation of Organelles. Society for Experimental Biology Seminar Series 35, eds Boffey S. A., Lloyd D. (Cambridge: Cambridge University Press; ), 115–129. [Google Scholar]
- Tilney-Bassett R. A. E., Abdel-Wahab O. A. L. (1982). Irregular segregation at the Pr locus controlling plastid inheritance in Pelargonium: gametophytic lethal or incompatibility system? Theor. Appl. Genet. 62 185–191. 10.1007/BF00293357 [DOI] [PubMed] [Google Scholar]
- Tilney-Bassett R. A. E., Almouslem A. B. (1989a). Variation in plastid inheritance between Pelargonium cultivars and their hybrids. Heredity 63 145–153. 10.1038/hdy.1989.86 [DOI] [Google Scholar]
- Tilney-Bassett R. A. E., Almouslem A. B., Amoate H. M. (1992). Complementary genes control biparental plastid inheritance in Pelargonium. Theor. Appl. Genet. 85 317–324. 10.1007/BF00222876 [DOI] [PubMed] [Google Scholar]
- Tilney-Bassett R. A. E., Almouslem A. B., Amoatey H. M. (1989b). “The complementary gene model for biparental plastid inheritance,” in Physiology, Biochemistry, and Genetics of Non Green Plastids, eds Boyer C. D., Shannon J. C., Hardison R. C. (Rockville, MD: The American Society of Plant Physiologists; ), 265–266. [Google Scholar]
- Tilney-Bassett R. A. E., Birky C. W., Jr. (1981). The mechanism of the mixed inheritance of chloroplast genes in Pelargonium: evidence from gene frequency distributions among the progeny of crosses. Theor. Appl. Genet. 60 43–53. 10.1007/BF00275177 [DOI] [PubMed] [Google Scholar]
- van de Kerke S. J., Shrestha B., Ruhlman T. A., Weng M.-L., Jansen R. K., Jones C. S., et al. (2019). Plastome based phylogenetics and younger crown node age in Pelargonium. Mol. Phylogenet. Evol. 137 33–43. 10.1016/j.ympev.2019.03.021 [DOI] [PubMed] [Google Scholar]
- van der Walt J. J. A., Vorster P. J. (1988). Pelargoniums of Southern Africa. Volumes I-III. Annals of Kirstenbosch botanic gardens. Pretoria: National Botanic Gardens of South Africa. [Google Scholar]
- Wang D., Liu H., Zhai G., Wang L., Shao J., Tao Y. (2016b). OspTAC2 encodes a pentatricopeptide repeat protein and regulates rice chloroplast development. J.Genet. Genomics 43 601–608. 10.1016/j.jgg.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Wang J., Li Y., Li C., Yan C., Zhao X., Yuan C., et al. (2019). Twelve complete chloroplast genomes of wild peanuts: great genetic resources and a better understanding of Arachis phylogeny. BMC Plant Biol. 19:504. 10.1186/s12870-019-2121-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wu Y., Messing J. (2016a). Genome-wide analysis of pentatricopeptide-repeat proteins of an aquatic plant. Planta 244 893–899. 10.1007/s00425-016-2555-x [DOI] [PubMed] [Google Scholar]
- Weihe A., Apitz J., Salinas A., Pohlheim F., Börner T. (2009). Biparental inheritance of plastidial and mitochondrial DNA and hybrid variegation in Pelargonium. Mol. Genet. Genomics 282 587–593. 10.1007/s00438-009-0488-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M.-L., Ruhlman T. A., Jansen R. K. (2017). Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 214 842–851. 10.1111/nph.14375 [DOI] [PubMed] [Google Scholar]
- Westerbergh A., Lerceteau-Köhler E., Sameri M., Bedada G., Lundquist P.-O. (2018). Towards the development of perennial barley for cold temperate climates—evaluation of wild barley relatives as genetic resources. Sustainability 10:1969 10.3390/su10061969 [DOI] [Google Scholar]
- Zhang J., Xiao J., Li Y., Su B., Xu H., Shan X., et al. (2017). PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J. Exp. Bot. 68 5615–5627. 10.1093/jxb/erx360 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lu C. (2019). The enigmatic roles of PPR-SMR proteins in plants. Adv. Sci. 6:1900361. 10.1002/advs.201900361 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diagnostic PCR targeted on HORT × ZONA and HORT × ACET.
Gel images other HORT × Ciconium F1 crosses displaying plastid types as identified using PCR.
PCR mastermix and thermo profile.
Overview of representative hybrid plants of F1 plants obtained in this study.
Full overview of all scenarios and gene models tested.
(A–K) Representative phenotypes for all F1 plants created in the course of this study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.