Abstract
Water-soluble phenolic acids are major bioactive compounds in the medicinal plant species Salvia miltiorrhiza. Phenolic acid biosynthesis is induced by methyl jasmonate (MeJA) in this important Chinese herb. Here, we investigated the mechanism underlying this induction by analyzing a transcriptome library of S. miltiorrhiza in response to MeJA. Global transcriptome analysis identified the MeJA-responsive R2R3-MYB transcription factor-encoding gene SmMYB1. Overexpressing SmMYB1 significantly promoted phenolic acid accumulation and upregulated the expression of genes encoding key enzymes in the phenolic acid biosynthesis pathway, including cytochrome P450-dependent monooxygenase (CYP98A14). Dual-luciferase (dual-LUC) assays and/or an electrophoretic mobility shift assays (EMSAs) indicated that SmMYB1 activated the expression of CYP98A14, as well as the expression of genes encoding anthocyanin biosynthesis pathway enzymes, including chalcone isomerase (CHI) and anthocyanidin synthase (ANS). In addition, SmMYB1 was shown to interact with SmMYC2 to additively promote CYP98A14 expression compared to the action of SmMYB1 alone. Taken together, these results demonstrate that SmMYB1 is an activator that improves the accumulation of phenolic acids and anthocyanins in S. miltiorrhiza. These findings lay the foundation for in-depth studies of the molecular mechanism underlying MeJA-mediated phenolic acid biosynthesis and for the metabolic engineering of bioactive ingredients in S. miltiorrhiza.
Subject terms: Secondary metabolism, Transcription factors
Introduction
Salvia miltiorrhiza Bunge (Lamiaceae family) is a valuable traditional Chinese medicinal plant species whose common name is Danshen1,2. Dried roots and rhizomes of Danshen have historically been used to treat cardiovascular and cerebrovascular diseases, and they are reported to have pharmacological activities3–5. Several Danshen products, including Fufang Danshen dripping pills, are used widely in clinical practice6,7. The bioactive ingredients in S. miltiorrhiza include water-soluble phenolic acids such as salvianolic acid A (Sal A), salvianolic acid B (Sal B), caffeic acid (CA), and rosmarinic acid (RA)8–12. In commercial Danshen decoctions, Sal B is the major marker component used for quality control according to the official Chinese Pharmacopoeia13,14. These compounds have attracted increased amounts of attention in recent years.
Given the widespread use of Danshen, it is important to improve the production of phenolic compounds in S. miltiorrhiza to meet clinical demands. Phenolic acid biosynthesis in S. miltiorrhiza involves two parallel pathways: the general phenylpropanoid pathway and the tyrosine-derived pathway13,15. In the phenylpropanoid pathway, l-phenylalanine is metabolized to 4-coumaroyl-CoA via phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), and 4-coumarate-CoA ligase (4CL)16,17. In the tyrosine-derived pathway, 3,4-dihydroxyphenyllactic acid (DHPL) is generated from l-tyrosine via tyrosine aminotransferase (TAT) and 4-hydroxyphenylpyruvate reductase (HPPR), as well as additional unresolved catalytic steps, to form 3,4-dihydroxyphenyllactic acid7. These two products are subsequently catalyzed by rosmarinic acid synthase (RAS) and a cytochrome P450-dependent monooxygenase (CYP98A14) to form RA15. Other detailed steps in these pathways have not yet been characterized, and the enzymes used for the biosynthesis of water-soluble phenolic acid medicinal substances such as Sal B have not yet been identified7,18,19. 4-Coumaroyl-CoA in the phenylpropanoid pathway of phenolic acid biosynthesis is also the primary precursor for anthocyanin biosynthesis (Fig. 1). Important catalytic enzymes in the anthocyanin biosynthesis pathway include chalcone synthase (CHS), chalcone isomerase (CHI), flavone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), and anthocyanidin synthase (ANS)13,20,21.
Fig. 1. Phenolic acid biosynthesis pathway and branched anthocyanin biosynthesis pathway.
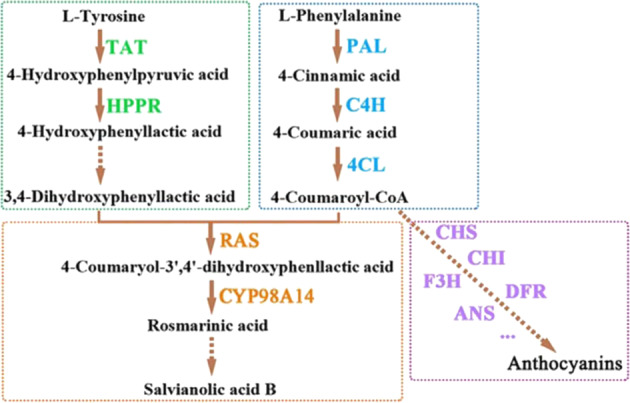
The green dotted frame indicates the tyrosine-derived pathway, the blue dotted frame indicates the phenylpropanoid pathway, the orange dotted frame indicates the downstream biosynthesis pathway of phenolic acids, and the purple dotted frame indicates the anthocyanin biosynthesis pathway
Metabolic engineering is an efficient approach for improving the production of active ingredients in medicinal plants. This process requires a deep understanding of the biosynthesis pathways of active ingredients, including both biosynthesis-related and regulatory genes22–25. Although great progress has been made in the isolation and functional identification of biosynthesis-related genes involved in medicinal compounds, little is known about the transcriptional regulation of phenolic acid biosynthesis in S. miltiorrhiza. In addition, phenolic acid biosynthesis is induced by MeJA treatment18,19, but the underlying regulatory mechanism is not clear. To determine how MeJA regulates phenolic acid biosynthesis, we previously performed a transcriptome analysis of hairy roots of S. miltiorrhiza under MeJA treatment and demonstrated that MYB transcription factor (TF) genes are differentially expressed in response to MeJA19, which implied that MYB TFs may be involved in regulating the biosynthesis of phenolic acids in S. miltiorrhiza.
MYB TFs are classified into four subfamilies based on the number of repeats of the conserved MYB domain (involved in DNA binding) in their N-terminal regions: 1R-MYBs, R2R3-MYBs, R1R2R3-MYBs, and 4R-MYBs. R2R3-MYBs (two-repeat MYBs) are the most common type of MYB TF in plants26–28 and have many roles in regulating plant development, growth, defense, secondary metabolism, and other biological functions28. For example, the R2R3-MYB TF EsMYBA1 can activate the promoters of dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS) genes to promote anthocyanin accumulation in the medicinal plant species Epimedium sagittatum29. The apple MYB TF MdMYB3 has been reported to regulate anthocyanin biosynthesis and flower development30. Moreover, in S. miltiorrhiza, overexpressing SmMYB36 in hairy roots significantly decreased the expression of most genes of the phenylpropanoid pathway (such as PAL1 and C4H1) and the tyrosine pathway (such as TAT1 and HPPR1), thereby inhibiting phenolic acid accumulation31. SmMYB39 negatively regulates the transcription and enzyme activities of C4H and TAT, thus decreasing rosmarinic acid contents, as demonstrated in transgenic hairy roots overexpressing SmMYB3932. However, very few R2R3-MYB TFs that act as positive regulators in promoting phenolic acid accumulation have been identified, and the molecular mechanism regulating phenolic acid biosynthesis induced by MeJA in S. miltiorrhiza remains unknown.
Since MYB TFs have important roles in plant secondary metabolism and have a positive effect on MeJA-inducible phenolic acid biosynthesis, it is important to investigate the roles of MYB TFs in regulating phenolic acid biosynthesis in S. miltiorrhiza. In the current study, we identified the MeJA-responsive R2R3-MYB transcription factor-encoding gene SmMYB1 from an S. miltiorrhiza MeJA-induced transcriptome library and demonstrated that SmMYB1 activated CYP98A14, CHI, and ANS expression and separately promoted the accumulation of both phenolic acids and anthocyanins. Moreover, SmMYB1 was shown to interact with SmMYC2 to form an MYB1/MYC2 complex that additively promotes the expression of CYP98A14 compared to the action of SmMYB1 alone. Our findings thus provide insights into the molecular mechanisms underlying MeJA-induced phenolic acid and anthocyanin biosynthesis in S. miltiorrhiza.
Materials and methods
Plant materials and elicitor preparation
S. miltiorrhiza plants were grown in a greenhouse at Zhejiang Chinese Medical University or cultivated on Murashige and Skoog (MS) media at 25 °C under a 16 h light/8 h dark photoperiod4,33. Nicotiana benthamiana seeds were sown in pots under the same conditions as those of S. miltiorrhiza4,34. Different tissues from a single one-year-old S. miltiorrhiza plant, including root, stem, leaf, petiole, calyx, and petal tissues, were collected for RNA isolation. The elicitors methyl jasmonate (MeJA), abscisic acid (ABA), gibberellic acid 3 (GA3), yeast extract (YE), salicylic acid (SA), and ethylene (Eth) were prepared as described by Zhang et al.34. Authentic tanshinone and phenolic acid standards (Aladdin, China) were prepared according to the methods of Shi et al.4.
Isolation and characterization of SmMYB1
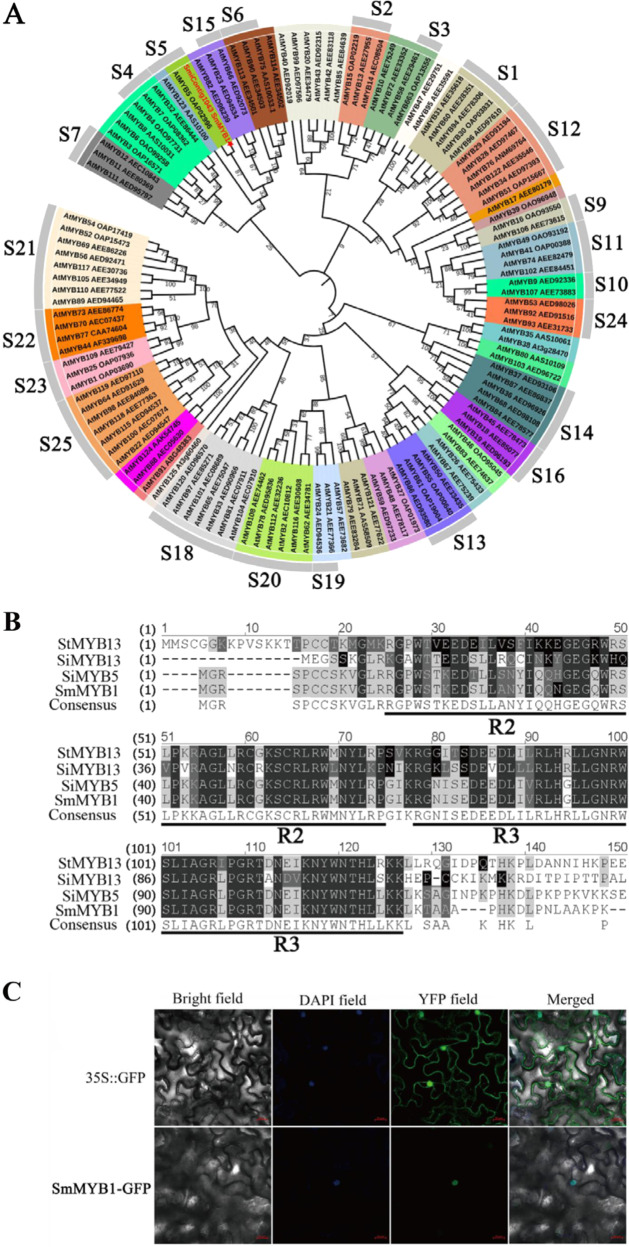
The open reading frame (ORF) of SmMYB1 was cloned as described by Zhou et al.35. The primer pair used for gene cloning is listed in Supplementary Table S1. SmMYB1 was subjected to BLAST-Protein (BLASTP) analysis via the nonredundant (NR) protein sequence database (www.ncbi.nlm.nih.gov). Scutellaria taiwanensis StMYB13 (accession number AKA59794.1), Scutellaria indica SiMYB13 (AKA59777.1), and Sesamum indicum SiMYB5 (XP_011070362.1), which are highly homologous to SmMYB1 (MN400427), were subjected to amino acid sequence alignment using Vector NTI software (Invitrogen, USA). Sequence alignment and phylogenetic analysis were performed using ClustalX36. A phylogenetic tree was constructed based on amino acid sequences by the neighbor-joining method using MEGA 6.0 software, and the reliability of each node in the tree was evaluated using the bootstrap method, with 1000 replicates37.
Quantitative real-time PCR (qRT-PCR)
The qRT-PCR analysis was performed as described by Shi et al.38. Different tissues (root, stem, leaf, petiole, calyx, and petal tissues) were collected from a single one-year-old plant and from hairy roots treated with MeJA, abscisic acid (ABA), gibberellin (GA3), yeast extract (YE), salicylic acid (SA) and ethylene (Eth), after which the tissue samples were frozen in liquid nitrogen. Total RNA was extracted as described previously34. A qRT-PCR assay was performed using a GoTaq-qPCR Master Mix kit (Promega, China) on an Applied Biosystems StepOne Real-time PCR System (USA). The internal control was the S. miltiorrhiza Actin gene. All the primers used for qRT-PCR are listed in Supplementary Table S1. Gene expression levels were quantified using the comparative Ct method4,5.
Subcellular localization of SmMYB1
The full-length ORF of SmMYB1 (without the stop codon) was fused to a green fluorescent protein (GFP) gene under the control of the CaMV 35S promoter within a pMON530 vector to generate a SmMYB-GFP fusion protein as described previously4,5 (Supplementary Fig. S1). The constructs were subsequently transferred into Agrobacterium tumefaciens GV3101, which were then infiltrated into five-week-old N. benthamiana leaves for transient transformation, as described previously5. pMON530-GFP was used as a control. To visualize the nuclei, 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) solution (10 μg/mL) was injected into N. benthamiana leaves with a syringe 3 h prior to microscopy observations. YFP and DAPI signals were excited at 488 and 405 nm, respectively. At 48 h after infiltration, the GFP signals were visualized using a confocal spectral microscope imaging system (Leica TCS SP5).
Generating S. miltiorrhiza hairy roots
The complete ORF of SmMYB1 was cloned into the SpeI and BstEII restriction enzyme sites of pCAMBIA2300+ under the control of the CaMV 35S promoter and NOS terminator to generate a pCAMBIA2300+–SmMYB1 construct (Supplementary Fig. S2). The DNA sequence encoding SRDX (LDLDLELRLGFA) was inserted into the ORF of SmMYB1 before the stop codon (TAA)26. This sequence was then subcloned into the SpeI and BstEII sites of a pCAMBIA2300+ vector to generate a pCAMBIA2300+-SmMYB1-SRDX construct (Supplementary Fig. S2). All the constructs were introduced into Agrobacterium strain C58C1, which were then transformed into S. miltiorrhiza to produce transgenic lines with hairy roots, as described previously5,38.
Measuring phenolic acid and anthocyanin contents
Hairy roots grown in shaker flasks were harvested and dried in an oven. The dried roots were ground to a powder, after which 200 mg samples were extracted with 16 mL of methanol/dichloromethane (3:1, v/v), sonicated for 1 h, and centrifuged at room temperature. Phenolic acid was extracted from the samples for HPLC analysis, as described previously5,12,14. The phenolic acids Sal A, Sal B, CA, and RA were extracted from the selected transgenic lines and quantified by comparison with standard curves and the corresponding retention times. Total phenolic acids were designated as TPA. Anthocyanin contents were determined by spectrophotometry by measuring the absorbance at 530 nm and 657 nm, with solutions without extracts used as blank controls39.
Purification of recombinant proteins
cDNA from SmMYB1 was amplified by PCR and cloned into a pETMALC-H vector (Supplementary Fig. S3), which was then transformed into Escherichia coli BL21 cells. All the primers used are listed in Supplementary Table S1. SmMYB1 proteins were expressed in E. coli via induction with 0.5 mM isopropyl b-d-1-thiogalactopyranoside (IPTG), followed by incubation at 37 °C for 3 h and purification using amylose resin (Shanghai Sangon Biotech Co., Ltd.).
Electrophoretic mobility shift assays (EMSAs)
EMSAs were performed using a Light Shift Chemiluminescent EMSA Kit (Thermo, USA) according to the manufacturer’s instructions, with minor modifications. Briefly, ~2 mg of the purified fusion protein was inputted into the binding reaction12, which was performed at 25 °C for 15 min in a thermal cycler (Bio-Rad). Biotin-labeled probes were synthesized by Shanghai Sangon Biotech Co., Ltd.; the sequences of the probes are listed in Supplementary Table S1.
Dual-luciferase (dual-LUC) assays
Dual-LUC assays were performed as reported previously14. The primer pairs used to clone the promoters of genes involved in phenolic acid biosynthesis are listed in Supplementary Table S1. The promoter regions of candidate genes (~2000 bp) were inserted into A pGreen 0800-LUC vector driving a firefly LUC reporter gene, and the Renilla luciferase (REN) gene was controlled by the 35S promoter (Fig. S4). The recombinant vectors were subsequently transferred into A. tumefaciens (GV3101) together with a helper plasmid (pSoup-P19) encoding a cosuppressor. N. benthamiana leaves were then transiently transformed with the constructs by infiltrating the Agrobacterium mixture into the abaxial sides of the leaves. The infected area was harvested, extracts were evaluated using a Dual-Luciferase Reporter Assay System (Promega, Madison, USA), and, after 48 h, the fluorescence values of LUC and REN were detected with a luminometer. The LUC/REN ratio indicated the ability of SmMYB1 to bind to and activate gene promoters.
Yeast two-hybrid (Y2H) assays
A Y2H assay was conducted using the Matchmaker Gold Yeast Two-Hybrid System according to the manufacturer’s instructions (Takara, Japan). The complete ORFs of SmMYB1 and SmMYC2 were amplified by PCR and cloned into the pGADT7 and pGKBT7 vectors, respectively, using the primers listed in Supplementary Table S1. The recombinant vectors were subsequently transformed into yeast strain AH109, and the autoactivation of pGKBT7-SmMYB1 was tested.
The bait and prey constructs were cotransformed into AH109 yeast cells and grown on double dropout (DDO) media for three days. The transformed colonies were then plated onto selection media to assess the possible interactions between proteins (Clontech, USA).
Bimolecular fluorescence complementation (BiFC) assays
To conduct BiFC assays, the ORF of SmMYB1 fused to cYFP and the ORF of SmMYC2 fused to nYFP were cloned into pXY104 and pXY106 vectors, respectively (Supplementary Fig. S5). The resulting constructs were introduced into A. tumefaciens strain GV3101, which were then infiltrated into 6-week-old N. benthamiana leaves using a 1 mL needleless syringe, as described previously40. The infected leaves were examined 48 h after infiltration. YFP fluorescent signals were captured using a confocal spectral microscopy imaging system (Leica TCS, SP5).
Statistical analysis
All experiments performed in this study, including those involving PCR identification, qRT-PCR, HPLC, dual-LUC assays, and EMSAs, were repeated three times. Gene expression levels, phenolic acid contents, and anthocyanin contents were expressed as the means ± standard deviations (SDs). SPSS 11.5 software (SPSS) was used to analyze statistical significance by single-sample t-tests and one-way analysis of variance.
Results
Isolation and characterization of SmMYB1
We used a previously established MeJA-treated Danshen hairy root transcriptome database (0 h, untreated control; treated for 1 and 6 h with MeJA) (available in the Sequence Read Archive [SRA] of the National Center for Biotechnology Information [NCBI] public database under accession number GSE100970) to identify R2R3-MYB TFs related to phenolic acid biosynthesis in S. miltiorrhiza19. The expression levels of forty-two R2R3-MYB TF-encoding genes in the transcriptome database were upregulated in response to MeJA treatment (RPKM fold change > 2, Supplementary Fig. S6). Among these, we selected the MYB TF-encoding gene SmMYB1 (SmiContig1043, GenBank accession number MN400427), whose expression profile was similar to that of the phenolic acid biosynthesis pathway genes TAT1, PAL1, HPPR1, RAS1, and CYP98A14, for functional analysis (Fig. S6).
The full-length cDNA sequence of SmMYB1 contained an ORF of 645 bp. We constructed a phylogenetic tree comprising SmMYB1 and 125 R2R3-MYB TFs in A. thaliana (Fig. 2A). BLAST-Protein (BLASTP) analysis showed that SmMYB1 shared the highest homology (85.6%) with Sesamum indicum MYB5 (SiMYB5); both proteins contained two conserved R2 and R3 repeats at the N-terminus, as revealed by amino acid sequence alignment (Fig. 2B). SmMYB1 shares the closest evolutionary relationships with members of the S5 subfamily (AtMYB5 and AtMYB123), S15 subfamily (AtMYB23 and AtMYB66), S6 subfamily (AtMYB75, AtMYB90, AtMYB113, and AtMYB114), S4 subfamily (AtMYB4, AtMYB7, and AtMYB32), and S7 subfamily (AtMYB11, AtMYB12, and AtMYB111).
Fig. 2. Characterization and subcellular localization of SmMYB1.
A Phylogenetic tree comprising SmMYB1 in S. miltiorrhiza and 125 R2R3-MYB TFs in A. thaliana generated by the neighbor-joining method using MEGA 6.05 software. The numbers on the nodes indicate the bootstrap values after 1000 replicates. B Multiple amino acid sequence alignment of R2R2-MYB TFs. The underlines represent conserved domains. C Subcellular localization of SmMYB1 in N. benthamiana leaf epidermal cells. DAPI was used as a positive control
Expression analysis of SmMYB1
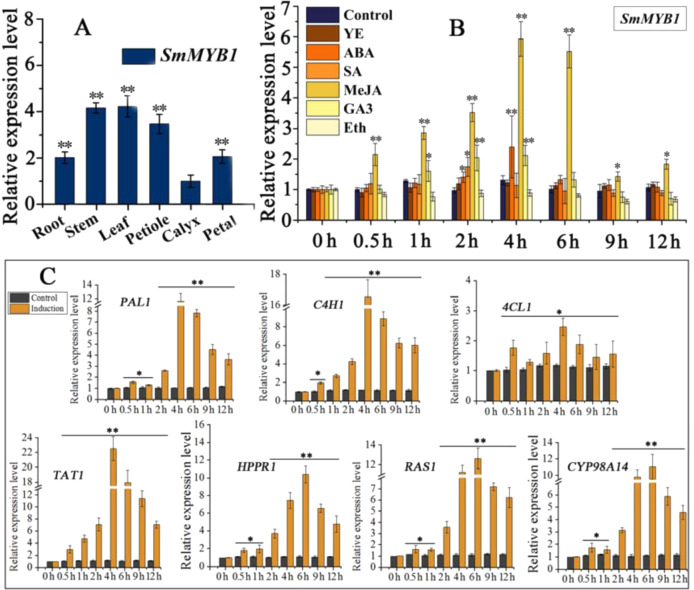
SmMYB1 is expressed in the nucleus, as revealed by transient expression analysis of epidermal cells from young N. benthamiana leaves (Fig. 2C). We examined the expression profiles of SmMYB1 in different tissues of S. miltiorrhiza by qRT-PCR. SmMYB1 was expressed ubiquitously in all tissues examined, including root, stem, leaf, petiole, calyx, and petal tissues, with the highest expression in the leaf tissue and the lowest in the calyx tissue (Fig. 3A). We also examined the expression of SmMYB1 in S. miltiorrhiza hairy roots after treatment with YE, ABA, SA, MeJA, GA3, and Eth. SmMYB1 expression was significantly upregulated in response to MeJA, with expression peaking after 4 h of MeJA treatment to levels 4.5-fold higher than those of the control (Fig. 3B). Seven genes for phenolic acid biosynthesis, including PAL1, TAT1, RAS1, and CYP98A14, showed various degrees of upregulation after MeJA treatment (Fig. 3C). These results indicate that SmMYB1 is induced in response to MeJA treatment and is expressed at the highest levels in S. miltiorrhiza leaves.
Fig. 3. Expression profiles of SmMYB1 and genes involved in the phenolic acid biosynthesis pathway in response to treatment with MeJA.
A Expression patterns of SmMYB1 in different tissues. The fold changes of the relative gene expression level in all the other tissues are normalized to the expression level in the calyx tissue. B Effect of elicitors (yeast extract, YE; abscisic acid, ABA; salicylic acid, SA; methyl jasmonate, MeJA; gibberellic acid, GA3; ethylene, Eth) on SmMYB1 expression. C Expression profiles of genes in the phenolic acid biosynthesis pathway after treatment with MeJA. The fold changes in the relative gene expression level were all normalized to the control expression without induction at the 0 h time point in B and C. The error bars indicate the standard errors of the means. The asterisks indicate significant differences between the calyx versus the other tissues in A, between the treatment versus the control in B, and according to C t-tests at two different significance levels (**P < 0.01; *P < 0.05). The experiment was replicated three times
SmMYB1 promotes phenolic acid accumulation and activates the CYP98A14 promoter
To investigate the role of SmMYB1 in phenolic acid biosynthesis, we selected three SmMYB1-overexpressing (SmMYB1-OE) hairy root lines and three SRDX repression lines (SmMYB1-SRDX) for further analysis (Supplementary Fig. S7). SmMYB1 was expressed at significantly higher levels in all of these lines than in the control (Supplementary Fig. S8).
We performed HPLC to measure phenolic acid yields from the transgenic hairy root lines. The average phenolic acid yield in the SmMYB1-OE lines was 121.64 mg/g dry weight (DW) compared to only 60.84 mg/g DW in the control. By contrast, the average phenolic acid yield in the SmMYB1-SRDX lines was 34.91 mg/g DW (Fig. 4A). In addition, the expression of the phenolic acid biosynthesis-related genes PAL1, C4H1, 4CL1, TAT1, HPPR1, RAS1, and CYP98A14 was upregulated in the SmMYB1-overexpression lines but strongly downregulated in the SmMYB1-SRDX lines compared to the control. The expression of TAT1 and CYP98A14 was most significantly affected in these lines (Fig. 4B). Taken together, these results indicate that SmMYB1 positively regulates phenolic acid levels in S. miltiorrhiza.
Fig. 4. Expression of key genes and phenolic acid contents in transgenic SmMYB1 hairy roots.
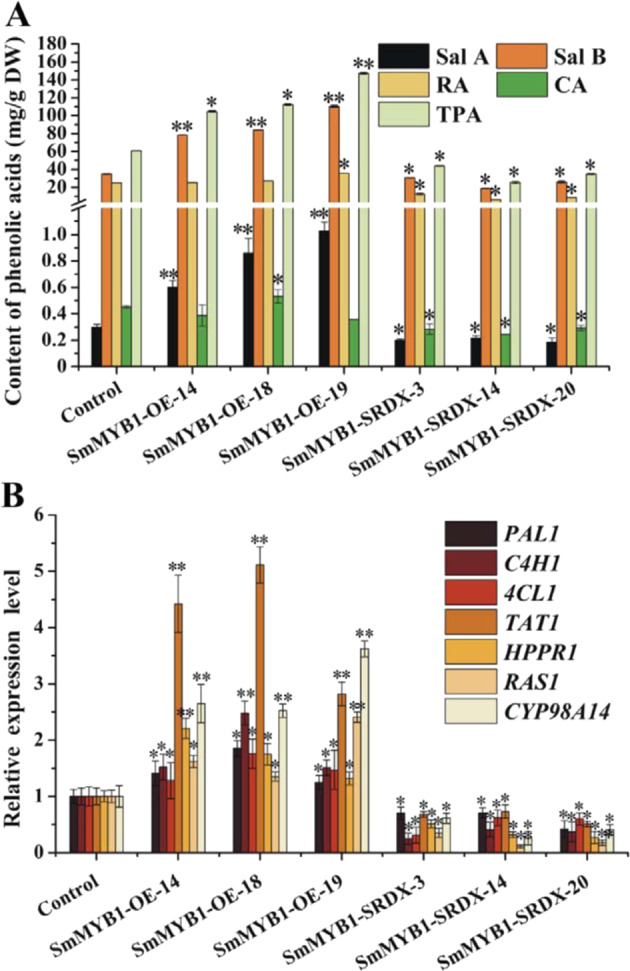
A Phenolic acid (salvianolic acid A, Sal A; salvianolic acid B, Sal B; rosmarinic acid, RA; caffeic acid, CA; total phenolic acids, TPA) contents in SmMYB1-OE and SmMYB1-SRDX lines, as measured by HPLC. B Expression of genes involved in phenolic acid biosynthesis in SmMYB1-OE (overexpression) and SmMYB1-SRDX (repression) lines. The fold changes of the relative gene expression level in transgenic hairy roots were normalized to those in the control. The error bars indicate the standard errors of the means. The asterisks on the bars indicate significant differences compared with the control according to t-tests at two different significance levels (**P < 0.01; *P < 0.05). The experiment was replicated three times
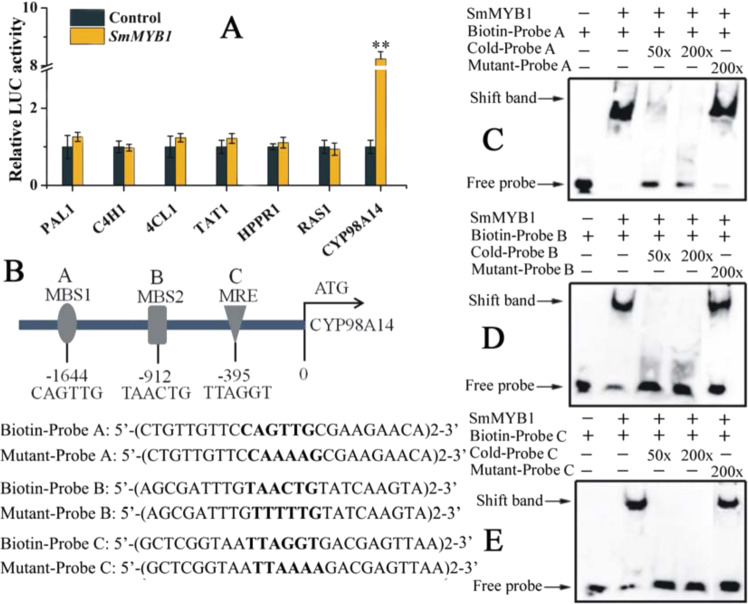
The promoters of all seven of these phenolic acid biosynthesis pathway genes contain MYB-binding elements (Supplementary Fig. S9). We, therefore, performed dual-LUC assays to determine whether SmMYB1 activates the promoters of these genes in vivo. Among these genes, a significantly higher LUC/REN ratio was detected only when pGreen 0800::CYP98A14 constructs WERE USED, with the ratio increasing more than 7-fold in the presence of SmMYB1 compared to that of the control (Fig. 5A). We searched for cis-elements that bind to MYB proteins within the promoter region of CYP98A14 using the PlantCARE program (http://bioinformatics.psb.ugent.be/beg/tools/plantcare). Three MYB-binding cis-elements were found: the CAGTTG (MBS1), TAACTG (MBS2), and TTAGGT (MRE) motifs (Fig. 5B). To further verify the binding of SmMYB1 to the MYB-binding cis-elements in the CYP98A14 promoter, we performed EMSAs, the results of which confirmed that SmMYB1 binds to the promoter region of CYP98A14 (Fig. 5C–E). These findings indicate that SmMYB1 activates CYP98A14 expression both in vivo and in vitro, possibly through direct binding of the cis-element in the promoter of CYP98A14.
Fig. 5. Validation of the targeted genes bound by SmMYB1.
A Dual-LUC assay results. Shown is the effector construct that contained SmMYB1 driven by the CaMV 35S promoter. The promoters of candidate genes (PAL1, C4H1, 4CL1, TAT1, HPPR1, RAS1, and CYP98A14) fused to LUC were used as reporter constructs. An effector vector not carrying SmMYB1 served as a negative control. Fold changes in the relative LUC activity were normalized to the control activity. The error bars indicate the standard errors of the means. The asterisks indicate significant differences according to t-tests at two different significance levels (**P < 0.01; *P < 0.05). The experiment was replicated three times. B MYB-binding elements in CYP98A14 and the probe sequences for EMSA analysis. C–E EMSA of SmMYB1. The Cold-Probe sequences were the same as the labeled probes but without biotin labels
SmMYB1 regulates anthocyanin accumulation by affecting CHI and ANS expression
Phenylalanine is a common precursor for phenolic acid and anthocyanin biosynthesis (Fig. 1). Therefore, we measured anthocyanin accumulation in the transgenic hairy root lines. Anthocyanin levels were significantly higher in the SmMYB1-OE lines than in the control and drastically lower in the SmMYB1-SRDX lines (Fig. 6A, B). Furthermore, the expression of anthocyanin biosynthesis pathway genes CHS, CHI, F3H, ANS, and DFR was significantly upregulated in the SmMYB1-OE lines compared with that in the control, whereas that of CHS, CHI, ANS, and DFR were significantly downregulated in the SmMYB1-SRDX lines (Fig. 6C).
Fig. 6. Role of SmMYB1 in promoting anthocyanin accumulation in S. miltiorrhiza transgenic hairy root lines.
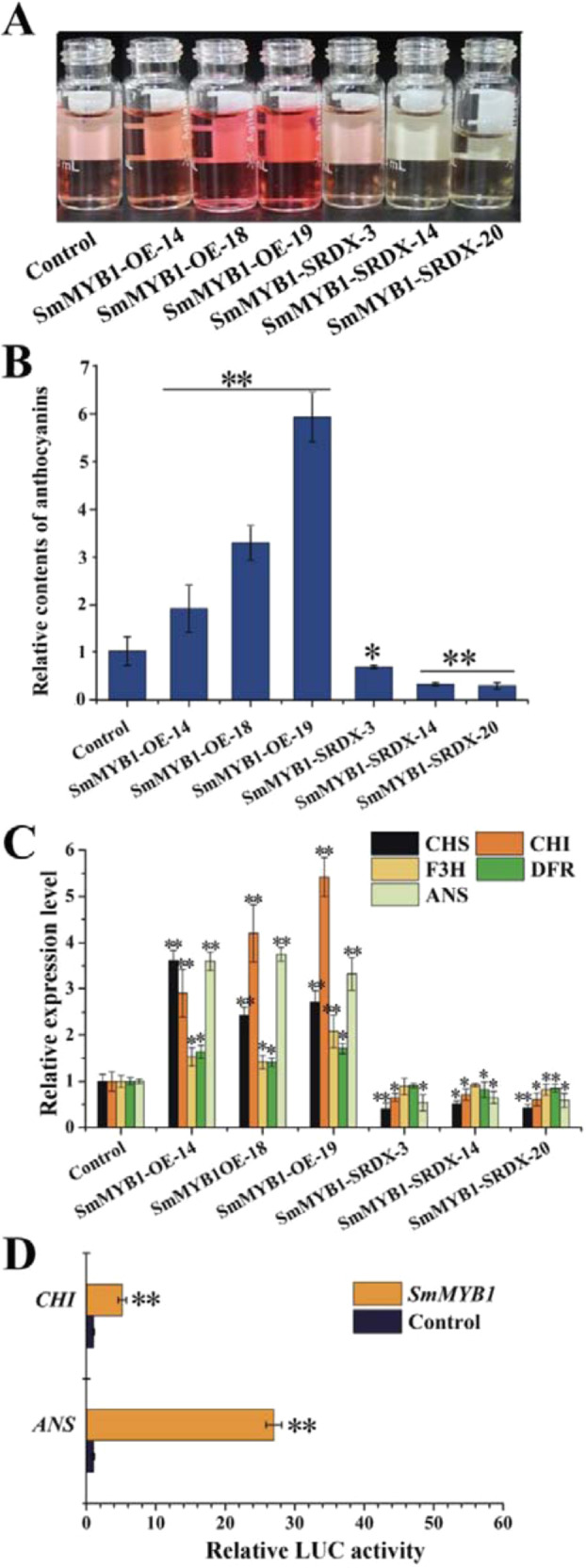
A, B Anthocyanin contents and the appearance of extracts from transgenic hairy root lines. Fold changes of the relative anthocyanin contents in transgenic hairy roots were normalized to the contents in the control. Control: empty vector; SmMYB-OE: SmMYB overexpression; SmMYB-SRDX: SmMYB-SRDX repression. A Expression profiles of genes involved in anthocyanin biosynthesis. Fold changes of the relative gene expression level in transgenic hairy roots were normalized to the level in the control. B Activation of CHI and ANS expression, as revealed by dual-LUC assays. The effector construct contained SmMYB1 driven by the CaMV 35S promoter. The promoters of candidate genes (CHI, ANS) fused to LUC were used as reporter constructs. An effector vector not carrying SmMYB1 served as a negative control. The error bars indicate the standard errors of the means. The asterisks indicate significant differences according to t-tests at two different significance levels (**P < 0.01; *P < 0.05). The experiment was replicated three times
Since overexpressing SmMYB1 significantly increased the expression of CHI and ANS, we searched for MYB-binding elements in the promoters of these genes. We then performed a dual-LUC assay to determine whether SmMYB1 activates the transcription of CHI and ANS in vivo. The LUC/REN ratio using CHI and ANS increased 5.16-fold and 26.98-fold, respectively, in the presence of SmMYB1 compared to that of the control (Fig. 6D). These findings indicate that SmMYB1 increases CHI and ANS expression, thereby promoting anthocyanin biosynthesis. Furthermore, overexpressing SmMYB1 not only increased phenolic acid accumulation but also increased anthocyanin biosynthesis in the S. miltiorrhiza hairy root lines.
Validation of the protein-protein interaction between SmMYB1 and SmMYC2
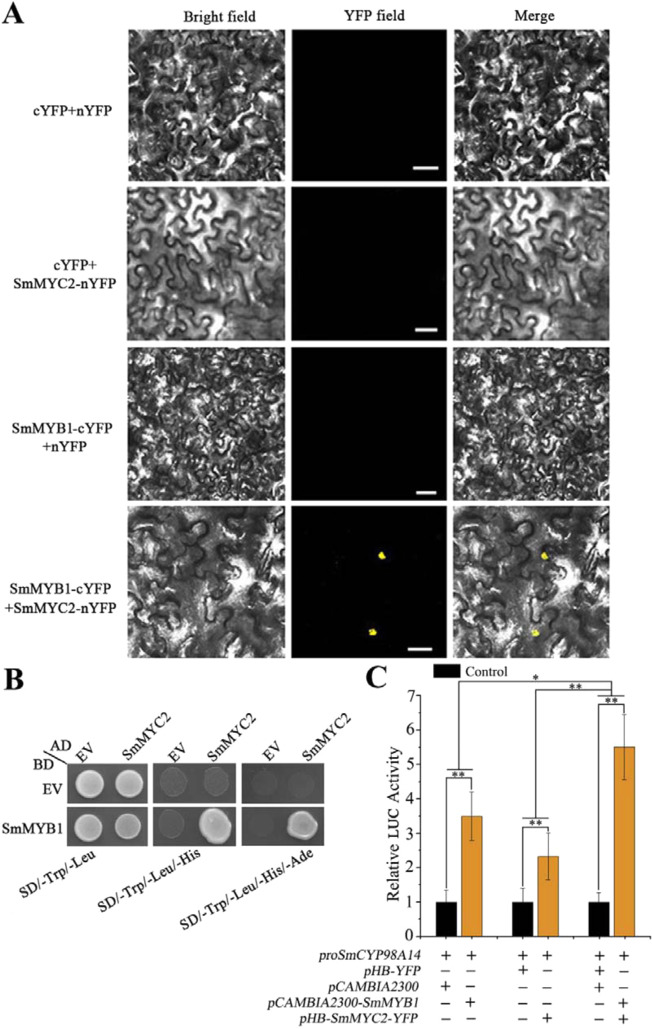
MYC2 is a central transcriptional regulator of jasmonate signaling41. Analysis of a previously generated transcriptome dataset19 indicated that SmMYC2 is strongly induced by MeJA. Therefore, we performed BiFC and Y2H assays to investigate the interaction between SmMYB1 and SmMYC2 (GenBank accession number MN400428). In the BiFC assay, the polypeptides cYFP and nYFP were able to bind together only when fusion constructs expressing SmMYB1-cYFP and nYFP-SmMYC2 were paired together. Specifically, yellow fluorescence was observed only when N. benthamiana leaves expressing the resulting full-length YFP were exposed to blue light, whereas leaves harboring the control constructs did not fluoresce under blue light (Fig. 7A). In the Y2H assay, yeast cells harboring the recombinant plasmids SmMYC2-AD and SmMYB1-BD grew normally on SD media lacking the nutrients l-tryptophan, l-leucine, l-histidine HCl monohydrate, and adenine hemisulfate (SD/−Trp/−Leu/−His/−Ade) (Fig. 7B).
Fig. 7. SmMYB1 interacts with SmMYC2 to promote CYP98A14 expression.
A BiFC assay; scale bars, 50 µm. B Y2H assay. AD: pGADT7 vector; BD: pGBKT7 vector. C Coexpressing SmMYB1 and SmMYC2 enhanced the activation of the reporter construct, as revealed by dual-LUC assays. The effector constructs contained SmMYB1 or SmMYC2 driven by the CaMV 35S promoter. proCYP98A14::LUC was used as the reporter construct. Effector vectors not carrying SmMYB1 or SmMYC2 were used as negative controls. Fold changes of the relative LUC activity were normalized to the activity of the control. The asterisks indicate significant differences according to t-tests at two different significance levels (**P < 0.01; *P < 0.05). The experiment was replicated three times
SmMYB1/SmMYC2 additively activates the CYP98A14 promoter
Overexpressing SmMYC2 increased phenolic acid production in S. miltiorrhiza by activating the expression of CYP98A1442,43. Thus, the finding that SmMYC2 has the same target as SmMYB1 prompted us to examine whether SmMYC2 and SmMYB1 together would activate CYP98A14 expression to a greater extent than either protein would separately. We used dual-LUC assays to assess the promoter activity of CYP98A14 with the effectors SmMYB1 and SmMYC2 delivered individually or in combination into N. benthamiana leaf cells. As expected, the activation of the CYP98A14 promoter was significantly enhanced by coexpressing SmMYC2 and SmMYB1 compared to that as a result of either protein alone (Fig. 7C). Taken together, these results suggest that SmMYB1 increases the transcriptional activation activity of CYP98A14 by possibly interacting with SmMYC2 to additively promote phenolic acid biosynthesis.
Discussion
SmMYB1 might serve as an additional link between MeJA induction and phenolic acid biosynthesis
Both biotic and abiotic elicitors, such as YE, Ag+, Co2+, α-amino isobutyric acid, β-aminobutyric acid (BABA), MeJA, GA3, Eth, SA, and wounding, increase the accumulation of secondary metabolites in plants, and in turn, the following processes have been shown to be affected: the control of tanshinone and phenolic acid biosynthesis in S. miltiorrhiza43–46; anthocyanin, lignin and suberin biosynthesis in Arabidopsis47–49; stilbene biosynthesis in Vitis vinifera50; and proanthocyanidin and flavonol biosynthesis in apple51. MeJA can be used to simulate natural wounding. In Arabidopsis, cell wall damage induces lignin biosynthesis, which is regulated by a reactive oxygen species- and jasmonic acid-dependent process47. Whether phenolic acid accumulation can be induced by wounding through SmMYB1 under jasmonic acid-dependent processes in S. miltiorrhiza needs to be further studied.
MYB TFs play key roles in regulating secondary metabolite biosynthesis in plants. Although various phenolic acid biosynthesis-related genes have been isolated and characterized, we need to identify a few R2R3-MYB TFs that function as positive regulators in promoting phenolic acid accumulation; in this regard, it is valuable to elucidate the molecular mechanism through which phenolic acid biosynthesis in S. miltiorrhiza is regulated. In the current study, the R2R3-MYB TF-encoding gene SmMYB1 was chosen for further analysis based on transcriptional data mining, because SmMYB1 (SmiContig1043) was strongly responsive to MeJA induction at the two MeJA induction points and exhibited an expression profile similar to that of biosynthesis pathway genes (TAT1, PAL1, HPPR1, RAS1, and CYP98A14) in our transcriptome dataset. In addition, SmMYB1 exhibited expression patterns similar to the pattern exhibited by key phenolic acid biosynthesis pathway genes, such as RAS1 and CYP98A14 (Fig. S6).
S. miltiorrhiza contains 110 R2R3-MYB family members, as revealed by genomic analysis28. Based on the conserved motifs in their amino acid sequences, these R2R3-MYBs could be further divided into 37 subgroups in S. miltiorrhiza28,31. Predictions made by comparing R2R3-MYBs in S. miltiorrhiza with those in other plant species suggested that S3, S4, S5, S6, S7, S13, and S21 subgroup members are involved in phenolic acid biosynthesis, while S4 and S20 subgroup members regulate terpenoid biosynthesis28,31. SmMYB1 is closely evolutionarily related to subgroup five proteins (Fig. 2A), suggesting that it might regulate phenolic acid biosynthesis32,52. Here, we isolated and identified the novel R2R3-MYB TF SmMYB1 from S. miltiorrhiza. The average total phenolic acid content was 2-fold greater in the SmMYB1-OE hairy root lines than in the control but only 57% of the control amount in the SmMYB1-SRDX lines (Fig. 4A). These results indicate that SmMYB1 positively regulates MeJA-mediated phenolic acid biosynthesis in S. miltiorrhiza.
In fact, except for SmMYB1, another MYB TF (whose unigene ID is SmiContig1859), which shows the greatest upregulation under MeJA induction in our transcriptome data set, was identified recently, designated as SmMYB2. SmMYB2 cannot interact with SmMYC2 like SmMYB1 but significantly increases the accumulation of salvianolic acid by activating the same target CYP98A14 gene as SmMYB1 does in overexpression transgenic hairy roots of S. miltiorrhiza45. In addition to SmMYB2, SmMYB1 is obviously another important R2R3-MYB member identified by us that serves as a link between MeJA induction and phenolic acid biosynthesis in S. miltiorrhiza.
SmMYB1 positively regulates phenolic acid biosynthesis in S. miltiorrhiza by activating CYP98A14
In the current study, overexpressing SmMYB1 not only upregulated the expression of its target gene CYP98A14 but also upregulated the expression of six genes in the phenolic acid biosynthesis pathway. Overexpressing SmERF1L1 promotes the expression of DXS2, DXR, HMGS, CPS1, and KSL1, but only DXR is a target of SmERF1L112. In SmERF128-overexpressing transgenic hairy roots, the expression of CYP76AH1, KSL1, CPS1, HDS, MDS, and DXR was activated, but only CYP76AH1, KSL1, and CPS1 were confirmed to be targets of SmERF12853. Based on these findings, it appears that the ectopic expression of certain TF-encoding genes upregulates the expression of not only their target genes but also other genes in the same biosynthesis pathway.
Although several MYB TFs, such as MYB36 and MYB39, have been functionally identified in S. miltiorrhiza31,32, direct evidence for the regulation of their target genes in vivo and in vitro is lacking. In the current study, we isolated and functionally identified the R2R3-MYB TF SmMYB1, which upregulated the expression of genes (including PAL1, C4H1, 4CL1, TAT1, HPPR1, CYP98A14, and RAS1) in the phenolic acid biosynthesis pathway in SmMYB1-overexpressing lines and consequently increased their phenolic acid contents. We verified that SmMYB1 activated CYP98A14 expression via dual-LUC assays and EMSA (Fig. 5A–E). Whether SmMYB1 can directly activate CYP98A14 expression needs to be further validated by ChIP-qPCR experiments.
SmMYB1 positively regulates anthocyanin accumulation in S. miltiorrhiza
We also investigated anthocyanin accumulation and the expression of anthocyanin biosynthesis-related genes in S. miltiorrhiza. Interestingly, overexpressing SmMYB1 upregulated the expression of anthocyanin biosynthesis-related genes such as CHI and ANS, consequently increasing anthocyanin contents. Dual-LUC assays further confirmed that SmMYB1 promotes anthocyanin biosynthesis, most likely by activating CHI and ANS. However, CHS, CHI, F3H, and FLS are the targets of the R2R3-MYB TFs MYB11, MYB12, and MYB111 in A. thaliana54. VvMYB14 and VvMYB15 specifically activate the promoter of the STS gene and regulate stilbene biosynthesis in grapevine50, and MsMYB22 activates flavonol pathways by binding to the FLS gene promoter in apple51. ODO1, a member of the R2R3-type MYB family, regulates the biosynthesis of fragrance-related volatiles by activating the promoter of the 5-enol-pyruvylshikimate-3-phosphate synthase (EPSPS) gene in the shikimate biosynthesis pathway in petunia flowers55. AtMYB12 was shown to target the ENO and DAHPS genes in the shikimate pathway and PAL1, CHS, and F3H in the phenylpropanoid pathway in A. thaliana, which not only increases the supply of carbon from primary metabolism but also fuels the shikimate and phenylalanine biosynthesis pathways to supply more aromatic amino acids for secondary metabolism54. Therefore, different R2R3-MYB TFs function in different regulatory pathways in different plant species.
The R2R3-MYB TF SmMYB39 negatively regulates phenolic acid accumulation in Danshen32, while overexpressing SmMYB111 and SmMYB2 promotes phenolic acid accumulation in S. miltiorrhiza hairy root lines23,42,45. Because only a few R2R3-MYB TFs with confirmed roles in promoting phenolic acid accumulation have been identified, the molecular mechanism through which phenolic acid biosynthesis is regulated in S. miltiorrhiza remains unknown. Taken together, our results provide new insights into the regulatory mechanism utilized by the R2R3-MYB TFs SmMYC2 and SmMYB1, which not only positively regulate phenolic acid biosynthesis but also promote anthocyanin accumulation in S. miltiorrhiza hairy root lines.
SmMYC2 interacts with SmMYB1 to additively activate the expression of CYP98A14
MYC2 is a central transcriptional regulator of jasmonate signaling41. SmMYC2 activates the expression of CYP98A14 to promote phenolic acid accumulation in S. miltiorrhiza42,43,56. To date, the molecular mechanism through which SmMYC2 coregulates phenolic acid biosynthesis together with MYB proteins in S. miltiorrhiza remains unknown. Previous studies demonstrated that various R2R3-MYB TFs do not act alone but instead interact with other proteins, such as bHLH and WD40 TFs, to form transcriptional complexes that regulate flavonoid biosynthesis51,54. In the present study, we demonstrated that SmMYC2 interacts with SmMYB1 to additively activate the expression of CYP98A14. The APETALA2/ethylene-responsive factor (AP2/ERF) TFs SmERF1L1 and SmERF128 positively regulate diterpenoid tanshinone biosynthesis by activating the expression of genes involved in the tanshinone biosynthesis pathway in S. miltiorrhiza12,53. It would be interesting to determine whether AP2/ERF TFs interact or cooperate with MYB TFs or MYC2 to promote phenolic acid or anthocyanin accumulation in S. miltiorrhiza.
A proposed model for the role of SmMYB1 in phenolic acid biosynthesis
In S. miltiorrhiza, overexpression of SmMYC2 increases the production of phenolic acids in transgenic hairy roots42. Based on the present findings and those of previous studies in A. thaliana and other plant species57–59, we propose a model describing the role of SmMYB1 in modulating phenolic acid biosynthesis in S. miltiorrhiza. According to this model, SmMYB1 is induced by MeJA signals and activates the expression of CYP98A14 by binding to cis-elements (MBS1, MBS2, MRE) in its promoter, thereby promoting phenolic acid accumulation. As a positive regulator of MeJA signaling, SmMYC2 possibly interacts with SmMYB1 to additively activate the expression of CYP98A14, which jointly promotes phenolic acid accumulation in S. miltiorrhiza (Fig. 8).
Fig. 8. Proposed model for the role of SmMYB1 in modulating phenolic acid biosynthesis in S. miltiorrhiza.
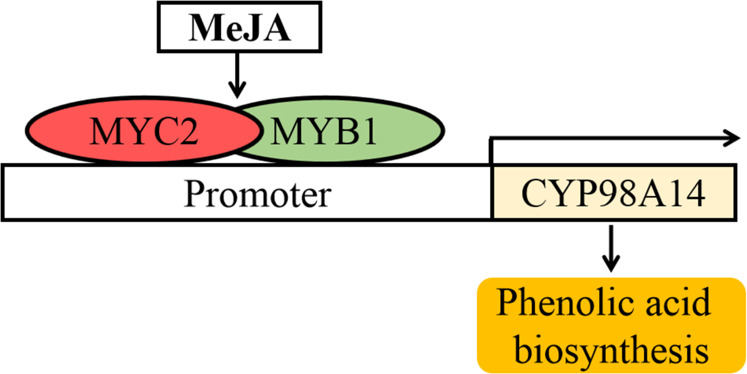
As TFs in response to MeJA, MYC2 and MYB1 can interact to additively promote CYP98A14 expression, thereby promoting the accumulation of phenolic acids
Supplementary information
Acknowledgements
This work was supported by the National Key R&G Program of China (2018YFC1706200), the National Natural Science Fund (81522049, 31571735, 31270007), the “Dawn” Program of the Shanghai Education Commission (16SG38), the Shanghai Science and Technology Committee Project (17JC1404300, 15430502700), the Zhejiang Provincial Ten Thousands Program for Leading Talents of Science and Technology Innovation (2018R52050), the Zhejiang Natural Science Fund (LY20H280008), and the Zhejiang Provincial Program for the Cultivation of High-level Innovative Health talents, Pre-Research Projects of Zhejiang Chinese Medical University (2018ZG30). We appreciate the great experimental support from the Public Platform of Medical Research Center, Academy of Chinese Medical Science, Zhejiang Chinese Medical University.
Author contributions
G.K. and W.Z. conceived and designed the study; W.Z., M.S., C.D., F.H., S.L., and Y.W. performed the research; W.Z., M.S., C.D., and F.H. analyzed the data; and W.Z., C.D., and G.K. wrote the paper.
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
These authors contributed equally: Wei Zhou, Min Shi, Changping Deng.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-00443-5).
References
- 1.Xu H, et al. Metabolic regulation and genetic engineering of pharmaceutical component tanshinone biosynthesis in Salvia miltiorrhiza. J. Med. Plants Res. 2010;4:2591–2597. doi: 10.5897/JMPR09.636. [DOI] [Google Scholar]
- 2.Shi, M., Liao, P., Nile, S., Georgiev, M. & Kai, G. Biotechnological exploration of transformed root culture for value-added products. Trends Biotechnol. 10.1016/j.tibtech.2020.06.012 (2020) [DOI] [PubMed]
- 3.Zhou L, Zuo Z, Chow M. Danshen: An overview of its chemistry pharmacology pharmacokinetics and clinical use. J. Clin. Pharmacol. 2005;45:1345–1359. doi: 10.1177/0091270005282630. [DOI] [PubMed] [Google Scholar]
- 4.Shi M, et al. Enhanced diterpene tanshinone accumulation and bioactivity of transgenic Salvia miltiorrhiza hairy roots by pathway engineering. J. Agric. Food Chem. 2016;64:2523–2530. doi: 10.1021/acs.jafc.5b04697. [DOI] [PubMed] [Google Scholar]
- 5.Zhou W, et al. Molecular cloning and characterization of two 1-deoxy-d-xylulose-5-phosphate synthase genes involved in tanshionone biosynthesis in Salvia miltiorrhiza. Mol. Breed. 2016;36:124. doi: 10.1007/s11032-016-0550-3. [DOI] [Google Scholar]
- 6.Ren J, Fu L, Nile SH, Zhang J, Kai G. Salvia miltiorrhiza in treating cardiovascular diseases: a review on its pharmacological and clinical applications. Front. Pharmacol. 2019;10:75. doi: 10.3389/fphar.2019.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi M, Huang F, Deng C, Wang Y, Kai G. Bioactivities biosynthesis and biotechnological production of phenolic acids in Salvia miltiorrhiza. Crit. Rev. Food Sci. 2019;10:1–12. doi: 10.1080/10408398.2018.1474170. [DOI] [PubMed] [Google Scholar]
- 8.Liao P, et al. Molecular cloning characterization and expression analysis of a new gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Salvia miltiorrhiza. Acta Physiol. Plant. 2009;31:565–572. doi: 10.1007/s11738-008-0266-z. [DOI] [Google Scholar]
- 9.Deng C, et al. ABA-responsive transcription factor bZIP1 is involved in modulating biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza. J. Exp. Bot. 2020;71:5948–5962. doi: 10.1093/jxb/eraa295. [DOI] [PubMed] [Google Scholar]
- 10.Kai G, et al. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 2011;13:319–327. doi: 10.1016/j.ymben.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Guo J, et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. PNAS. 2013;110:12108–12113. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Q, et al. The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 2019;274:368–375. doi: 10.1016/j.foodchem.2018.08.119. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Pathway engineering for phenolic acid accumulations in Salvia miltiorrhiza by combinational genetic manipulation. Metab. Eng. 2014;21:71–80. doi: 10.1016/j.ymben.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Sun M, et al. The biosynthesis of phenolic acids is positively regulated by the JA-responsive transcription factor ERF115 in Salvia miltiorrhiza. J. Exp. Bot. 2019;70:243–254. doi: 10.1093/jxb/ery349. [DOI] [PubMed] [Google Scholar]
- 15.Di P, et al. ¹³C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 2013;8:1537–1548. doi: 10.1021/cb3006962. [DOI] [PubMed] [Google Scholar]
- 16.Zhao S, Hu Z, Liu D, Leung F. Two divergent members of 4-coumarate: coenzyme A ligase from Salvia miltiorrhiza Bunge: cDNA cloning and functional study. J. Integr. Plant Biol. 2006;48:1355–1364. doi: 10.1111/j.1744-7909.2006.00302.x. [DOI] [Google Scholar]
- 17.Huang B, et al. Characterization and expression profiling of tyrosine aminotransferase gene from Salvia miltiorrhiza (Dan-shen) in rosmarinic acid biosynthesis pathway. Mol. Biol. Rep. 2008;35:601–612. doi: 10.1007/s11033-007-9130-2. [DOI] [PubMed] [Google Scholar]
- 18.Xiao Y, et al. Methyl jasmonate dramatically enhances the accumulation of phenolic acids in Salvia miltiorrhiza hairy root cultures. Physiol. Plant. 2009;137:1–9. doi: 10.1111/j.1399-3054.2009.01257.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhou W, et al. Comprehensive transcriptome profiling of Salvia miltiorrhiza for discovery of genes associated with the biosynthesis of tanshinone and salvianolic acids. Sci. Rep. 2017;7:10554. doi: 10.1038/s41598-017-10215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkel-Shirley B. Flavonoid biosynthesis: a colorful model for genetics biochemistry cell biology and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porth I, Hamberger B, White R, Ritland K. Defense mechanisms against herbivory in Picea: sequence evolution and expression regulation of gene family members in the phenylpropanoid pathway. BMC Genomics. 2011;12:608. doi: 10.1186/1471-2164-12-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng C, et al. Tanshinone production could be increased by the expression of SmWRKY2 in Salvia miltiorrhiza hairy roots. Plant Sci. 2019;284:1–8. doi: 10.1016/j.plantsci.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Li S, et al. SmMYB111 is a key factor to phenolic acid biosynthesis and interacts with both SmTTG1 and SmbHLH51 in Salvia miltiorrhiza. J. Agric. Food Chem. 2018;66:8069–8078. doi: 10.1021/acs.jafc.8b02548. [DOI] [PubMed] [Google Scholar]
- 24.Cao W, et al. Transcription factor SmWRKY1 positively promotes the biosynthesis of tanshinones in Salvia miltiorrhiza. Front. Plant Sci. 2018;9:554. doi: 10.3389/fpls.2018.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Patra B, Pattanaik S, Wang Y, Yuan L. GATA and phytochrome interacting factor transcription factors regulate light-induced vindoline biosynthesis in Catharanthus roseus. Plant Physiol. 2019;180:1336–1350. doi: 10.1104/pp.19.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34:733–739. doi: 10.1046/j.1365-313X.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 27.Dubos C, et al. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Li C, Lu S. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genomics. 2014;15:277. doi: 10.1186/1471-2164-15-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, et al. A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway. PLos ONE. 2013;8:e70778. doi: 10.1371/journal.pone.0070778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vimolmangkang S, Han Y, Wei G, Korban S. An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013;13:176. doi: 10.1186/1471-2229-13-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding K, et al. SmMYB36 a novel R2R3-MYB transcription factor enhances tanshinone accumulation and decreases phenolic acid content in Salvia miltiorrhiza hairy roots. Sci. Rep. 2017;7:5104. doi: 10.1038/s41598-017-04909-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, et al. Cloning and characterization of a putative R2R3 MYB transcriptional repressor of the rosmarinic acid biosynthetic pathway from Salvia miltiorrhiza. PLos ONE. 2013;8:e73259. doi: 10.1371/journal.pone.0073259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kai G, et al. Characterization expression profiling and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotechnol. Bioproc. E. 2010;15:236–245. doi: 10.1007/s12257-009-0123-y. [DOI] [Google Scholar]
- 34.Zhang L, et al. Molecular cloning and expression analysis of a new putative gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Salvia miltiorrhiza. Acta Physiol. Plant. 2011;33:953–961. doi: 10.1007/s11738-010-0627-2. [DOI] [Google Scholar]
- 35.Zhou W, et al. Mapping of Ppd-B1 a major candidate gene for late heading on wild emmer chromosome arm 2BS and assessment of its interactions with early heading QTLs on 3AL. PLoS ONE. 2016;11:e0147377. doi: 10.1371/journal.pone.0147377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi M, et al. Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins. Sci. Rep. 2016;6:20919. doi: 10.1038/srep20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W, Gong Y, Huang C, Gao F. Molecular cloning and function analysis of flavonoid 3’-hydroxylase gene in the purple-fleshed sweet potato (Ipomoea batatas) Mol. Biol. Rep. 2012;39:295–302. doi: 10.1007/s11033-011-0738-x. [DOI] [PubMed] [Google Scholar]
- 40.Lv Z, et al. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and botrytis cinerea in Artemisia annua. Plant Cell Physiol. 2016;57:1961–1971. doi: 10.1093/pcp/pcw118. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki-Sekimoto Y, Saito H, Masuda S, Shirasu K, Ohta H. Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signal. Behav. 2014;9:e27639. doi: 10.4161/psb.27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang N, et al. Overexpression of SmMYC2 increases the production of phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 2017;18:1804. doi: 10.3389/fpls.2017.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, et al. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 2016;6:22852. doi: 10.1038/srep22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge X, Wu J. Tanshinone production and isoprenoid pathways in Salvia miltiorrhiza hairy roots induced by Ag+ and yeast elicitor. Plant Sci. 2005;168:487–491. doi: 10.1016/j.plantsci.2004.09.012. [DOI] [Google Scholar]
- 45.Deng C, et al. SmMYB2 promotes salvianolic acid biosynthesis in the medicinal herb Salvia miltiorrhiza. J. Integr. Plant Biol. 2020;62:1688–1702. doi: 10.1111/jipb.12943. [DOI] [PubMed] [Google Scholar]
- 46.Hao X, et al. Effects of methyl jasmonate and salicylic acid on the tanshinone production and biosynthetic genes expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl Biochem. 2015;62:24–31. doi: 10.1002/bab.1236. [DOI] [PubMed] [Google Scholar]
- 47.Denness L, et al. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 2011;156:1364–1374. doi: 10.1104/pp.111.175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stracke R, et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gou M, et al. The MYB107 transcription factor positively regulates biosynthesis. Plant Physiol. 2017;173:1045–1058. doi: 10.1104/pp.16.01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Höll J, et al. The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell. 2013;25:4135–4149. doi: 10.1105/tpc.113.117127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang N, et al. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana) Plant J. 2017;90:276–292. doi: 10.1111/tpj.13487. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, et al. Overexpression of SmMYB9b enhances tanshinone concentration in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2017;36:1297–1309. doi: 10.1007/s00299-017-2154-8. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Ji A, Xu Z, Luo H, Song J. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol. Biol. 2019;100:83–93. doi: 10.1007/s11103-019-00845-7. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015;6:8635. doi: 10.1038/ncomms9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verdonk J, et al. ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell. 2005;17:1612–1624. doi: 10.1105/tpc.104.028837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du T, et al. SmbHLH37 Functions antagonistically with SmMYC2 in regulating jasmonate-mediated biosynthesis of phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 2018;9:1720. doi: 10.3389/fpls.2018.01720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chini A, Boter M, Solano R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009;276:4682–4692. doi: 10.1111/j.1742-4658.2009.07194.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhao S, Zhang J, Tan R, Yang L, Zheng X. Enhancing diterpenoid concentration in Salvia miltiorrhiza hairy roots through pathway engineering with maize C1 transcription factor. J. Exp. Bot. 2015;66:7211–7266. doi: 10.1093/jxb/erv418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.