Abstract
Saliva has been proposed as an alternative to upper airway swabs when testing for severe acute respiratory syndrome coronavirus 2. Although some studies have suggested higher viral loads and clinical sensitivity when testing saliva, studies have been relatively small and have given rise to contradictory results. To better understand the relative performance characteristics of saliva and upper airway samples, I performed a rapid systematic review (registered on PROSPERO as CRD42020205035), focusing on studies that included at least 20 subjects who provided diagnostic saliva and upper airway samples on the same day, which were tested by nucleic acid amplification methods and for which a confusion matrix could be constructed for based on a composite reference standard. Nineteen studies comprising 21 cohorts that met predetermined acceptance criteria were identified following a search of PubMed, medRxiv, and bioRxiv. Seven of these cohorts were incorporated into a meta-analysis using a random effects model, which suggests that nasopharyngeal swabs are somewhat more sensitive than saliva samples for the diagnosis of early disease in ambulatory patients, such as in drive-through centers or community health centers. Nevertheless, the difference is modest, and the reduced need for personal protective equipment for saliva sampling may justify the difference. Conclusions are limited by the significant heterogeneity of disease prevalence in the study populations and variation in the approaches to saliva sample collection.
Rapid identification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral infection is important for treatment of symptomatic individuals, for disrupting transmission by asymptomatic carriers, and for understanding the dynamics of infection in communities.1 Although the use of flocked swabs to obtain nasopharyngeal (NP) specimens for testing has constituted the gold standard for upper respiratory system (URS) sampling, uses of specimens obtained by swabbing the oropharynx, midturbinate, and anterior nares (AN; alias nasal) have all served as alternatives, both as a result of supply shortages and because these sites can be self-sampled, reducing clinician exposure and the need for personal protective equipment. Saliva, which can also be obtained without clinician assistance, has been proposed as a safe, easy, and comfortable way to obtain samples for coronavirus disease 2019 (COVID-19) testing,2 but published studies have been based on heterogeneous study populations and have given conflicting results.3, 4, 5, 6, 7, 8 In this article, we set out to answer the question.
“When nucleic acid amplification tests for SARS-CoV-2 are employed for initial diagnosis, what is the relative sensitivity for detection of virus when saliva samples are used rather than nasopharyngeal, oropharyngeal, midturbinate, or anterior nares swabs?”
To answer this question, I conducted a rapid systematic review based on Preferred Reporting Items for Systematic Reviews and Meta-Analyses principles,9 using analysis that relies on a composite reference standard (CRS)10 based on both swabs and saliva samples; this approach is not biased against either sample type.
Materials and Methods
The study is registered on PROSPERO (https://www.crd.york.ac.uk/prospero; CRD42020205035), but a complete protocol has not been published. PubMed, medRxiv, and bioRxiv were searched over the interval from January 1, 2020, to August 17, 2020. Preliminary searches showed that a combination of the terms COVID-19 and saliva was able to capture all or nearly all relevant articles in which synonymous terms, such as SARS-CoV-2, novel coronavirus, or oral fluid, appeared. For this reason, a simple search string for saliva COVID-19 sensitivity was used with all three databases to identify articles for further screening. Several additional articles were identified after initial peer review. Following the initial identification of articles, the titles and abstracts were screened to eliminate articles not meeting the prespecified inclusion criteria. Articles remaining after this process were rescreened, particularly because many of the articles reviewed were in the form of research letters that did not have an abstract. Ultimately, 19 articles that met inclusion criteria were available for analysis, as shown in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses flow diagram11 (Supplemental Figure S1).
To be included in this systematic review, studies were required to include a minimum of 20 individual subjects; each subject must have had both a saliva specimen and at least one of several URS swab specimens [nasopharyngeal swabs (NPSs), oropharyngeal swabs, midturbinate swabs, or AN (or nasal) swabs] obtained on the same day. Articles that reported on tongue and cheek swabbing, without a specific effort to soak up saliva, were not included. Each of these specimens must have undergone analysis for SARS-CoV-2 sequences using either an isothermal amplification or an RT-PCR–based method. Results must have been reported in a manner that allowed construction of a confusion matrix, based on a single sample-pair per patient, including the saliva- and upper airway–based test. If an article reported patients who were tested multiple times, and a confusion matrix could not be constructed that reflected results of the first NPS/saliva pair, that study was excluded. Studies in which discrepant analysis was used to resolve diagnostic conflicts between the two sites were not to be included unless data could be analyzed independently of the discrepant analysis. In the event that multiple time points were included in one of the included studies, only the first time point was to be used in my analysis. If confusion matrices could only be constructed from data involving multiple time points from the same patients, the study was excluded. No attempt was made to obtain data from the investigators involved in these published studies.
When articles that were identified on bioRxiv or medRxiv were compared with those on PubMed, an additional six duplicate articles were eliminated. Four articles that were not identified by the search strategy were added.1 The potential for bias associated with each study was evaluated using the QUADAS212 , 13 instrument. The risk of spectrum bias was assessed from the perspective of testing as an initial diagnostic method for ambulatory patients; the bias assessment does not constitute a judgement on the quality of the study, which may have been performed to demonstrate assay validity, assessment of recovery, or other purposes different than that for which I evaluated potential bias. Seven articles with a low risk of bias that were deemed appropriate to include in a meta-analysis were analyzed using a diagnostic effects model (der Simion–Laird),14 as implemented by OpenMetaAnalyst15 software program.
A predetermined data extraction form included study author, type of study, inclusion and exclusion criteria, setting, sample types, swab types, transport medium, manufacturer or description of nucleic acid amplification assays, as well as space to record study results in the form of confusion matrices. After initial compilation and tabulation of data, a second review was undertaken to determine whether saliva samples involved coughing or in other ways included sputum in the sample; this step was not part of the original protocol.
Because the choice of any particular sample type as a gold standard provides a biased estimate of relative sensitivity, which compared with all other sample types, a CRS10 was computed for each study on the basis of all sample types included in the study, when possible. For one study in which results were not presented in a manner that made this possible, a CRS was computed individually for comparisons of each upper airway sample with the saliva sample. Equivocal results and assay failures were not used in the calculation of sensitivity or in the construction of the CRS for each study. Confidence limits for sensitivity were computed using the Newcombe efficient score method,16 as implemented in the Vassarstats Clinical Calculator 1 (http://vassarstats.net, last accessed January 12, 2021) (Table 1 ).3, 4, 5, 6, 7, 8 , 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Criteria for performing a formal meta-analysis were as follows: i) studies used the same amplification technology (such as RT-PCR); ii) studies used the same upper airway sample site (AN, midturbinate, and NP could be included together, but not admixed with studies based on oropharynx samples); iii) studies enrolled a similar patient mix (eg, symptomatic, asymptomatic, or hospitalized) and similar clinical environment (drive-through/community health center or hospital).
Table 1.
Study Characteristics and Sampling Method Sensitivity
| Authors | Patient characteristics | Setting | Potential for spectrum bias∗ | n | Positive, % | Sensitivity, % (95% CI) |
|||
|---|---|---|---|---|---|---|---|---|---|
| Saliva | NP swab | OP swab | Nasal self-swab | ||||||
|
Rose3 |
Not stated | Drive-through testing center | Low | 100 | 9 | 89 (51–99) | 100 (63–100) | ||
|
Jamal17 |
Symptomatic inpatients with COVID-19 | Hospital | High | 18 | 94 | 88 (66–98) | 94 (69–100) | ||
|
Landry4 |
Symptomatic outpatients | Drive-through testing center | Low | 124 | 28 | 86 (69–95) | 94 (79–99) | ||
|
Pasomsub5 |
Symptomatic, asymptomatic travelers and contacts |
Hospital respiratory infection clinic | Low | 200 | 11 | 86 (62–96) | 90 (68–98) | ||
|
Dogan18 |
Hospitalized, possible COVID-19, moderate to severe disease |
Hospital | High | 161 | 37 | 58 (45–71) | 92 (81–97) | 93 (83–98) | |
|
Yokota19 |
Asymptomatic | Airport | Low | 1763 | 0.2 | 80 (30–99) | 100 (46–100) | ||
|
Yokota19 |
Asymptomatic | Contact tracing | Low | 161 | 29 | 94 (81–98) | 87 (74–95) | ||
|
Byrne20 |
Symptomatic patients | Hospital | High | 110 | 13 | 86 (56–97) | |||
|
Griesemer6 |
Symptomatic patients and asymptomatic contacts in a known hot spot |
Drive-through testing center | Low | 227 | 41 | 87 (78–93) | 98 (92–100) | 92 (84–97) | |
|
Griesemer6 |
Symptomatic and asymptomatic | Medical center testing tent | Low | 236 | 5.2 | 50 (22–78) | 100 (70–100) | 42 (16–71) | |
|
Miller21 |
Symptomatic or asymptomatic with previous positive PCR | Physician offices | High | 91 | 40 | 97 (84–100) | 94 (80–99) | ||
|
Hanson7† |
Symptomatic | Drive-through testing center | Low | 354 | 24 | 94 (86–98) | 93 (85–97) | 86 (77–93) | |
|
L'Helgouach22 |
Asymptomatic health care workers |
Hospital | High | 92 | 4.3 | 100 (40–100) | 0 (0–60) | ||
|
Iwasaki23 |
Symptomatic, some previously diagnosed with COVID-19 |
Hospital | High | 76 | 13 | 90 (54–99) | 90 (54–99) | ||
|
Becker8 |
Probably symptomatic‡ | Community testing environment | Low | 77 | 20 | 60 (33–83) | 100 (93–100) | ||
|
SoRelle24§ |
Symptomatic | Not stated | Uncertain | 83 | 47 | 82 (66–92) | 100 (89–100) | ||
|
Kojima25 |
Symptomatic and previously tested at drive-through center, and close contacts exposed, previously tested |
Patient homes | High | 45 | 64 | 90 (72–97)¶ 66 (46–81)‖ |
79 (60–91) | 86 (67–95) | |
|
McCormick-Baw26 |
Symptomatic, some hospitalized | Hospital emergency department | High | 156 | 32 | 96 (87–99) | 98 (90–100) | ||
|
Rao27 |
Confirmed positives | Quarantine center | High | 217 | 74 | 93 (87–96) | 55 (47–63) | ||
|
Wong28∗∗ |
Confirmed positive hospitalized patients | Hospital | High | 44 | 95 | 95 (83–99) | 95 (83–99) | ||
|
Güçlü29 |
Mix of NP swab positive and negative inpatients and outpatients | Hospital | High | 64 | 44 | 96 (80–100) | 86 (66–95) | ||
COVID-19, coronavirus disease 2019; ID, identifier; MT, midturbinate; NP, nasopharyngeal; OP, oropharynx.
Potential for spectrum bias was evaluated in terms of the enrolled cohort. Although a group of 200 consecutively enrolled hospital patients would not be considered as experiencing selection bias, it would be viewed as having a high potential for spectrum bias (with regards to this study) because all patients were sufficiently ill as to require hospitalization. Similarly, a group of patients selected on the basis of RT-PCR Ct values would be considered biased (no matter what those values were).
Data are not presented in a way that allows generation of a composite reference that includes all three specimen types. Sensitivity values of saliva samples and nasal samples are each computed from separate composite references that include saliva/NP and nasal/NP, respectively.
Although the article did not explicitly identify these patients as symptomatic, at the time the work was done symptomatic patients were the focus of most community testing.
NP samples were tested using RT-PCR, whereas the saliva samples were tested using ID NOW. Thus, index test bias (from the perspective of this systematic review) was intentionally built into the design of this study.
Supervised saliva collection.
Unsupervised saliva collection.
Data reflect information from the first test of 44 confirmed positive patients shown in Figure 4 of the article.28
Results
A total of 149 articles were considered for inclusion; of these, 19 studies comprising 21 cohorts met inclusion criteria (Supplemental Figure S1). A brief summary of the studies included in this review may be found in Table 1, and a brief discussion of each articles (including the results used in this review) is presented in the Supplemental Appendix S1.3, 4, 5, 6, 7, 8 , 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Twelve of the included cohorts involved ≥100 patients. Two of the studies separately presented data for two cohorts; one study presented data for a single cohort in which two approaches were used to collect saliva specimens.25
The risk of spectrum bias associated with the study population, or method of recruitment, was rated as either high or unclear for 12 studies. This was the most common concern raised in the quality assessment. Studies with a high or unclear risk of bias were characterized by failure to present patient symptom status (one study), inclusion of subjects who had previously tested positive or been hospitalized for SARS-CoV-2 (eight studies), focus on health care workers (one study), or possibly allowing provider discretion, rather than sequential or random consent, for enrollment (two studies). Although the reviewed studies did not explicitly state that the interpretation of tests based on saliva samples was conducted without knowledge of upper airway results (or vice versa), the manner in which RT-PCR testing for SARS-CoV-2 is conducted in clinical laboratories makes it highly unlikely that the specimen flow or the interpretation of RT-PCR tests would experience significant bias. One study intentionally built in a bias associated with testing saliva specimens using the ID NOW assay (Abbott Scarborough, Inc., Scarborough, ME), while using RT-PCR for testing NP swabs.24 Only seven studies reported limits of detection for their RT-PCR assays.
In one study, a midturbinate swab served as the sole upper airway sample; and in one study, an NP/oropharynx swab was obtained. In all the remainder, an NP swab sample was evaluated. Four studies also evaluated AN swabs, and one evaluated oropharynx swabs. The prevalence of SARS-CoV-2 infection within the cohorts, as measured using the CRS, varied from 0.02% to 95%. In 15 of the 21 cohorts studied, NP swabs demonstrated a sensitivity of ≥90% by comparison with the composite reference standard. In three of the four studies that also evaluated AN swabs, NP swabs appeared to be somewhat superior. When compared with saliva samples, anterior nasal swabs performed better in one study, equally well in a second, and less well in a third study associated with a low risk of spectrum bias, with widely overlapping CIs in all cases.
In 2 of the 21 cohorts represented in Table 1, saliva and upper airway samples demonstrated identical sensitivity patient groups, whereas in 7 cohorts, saliva demonstrated greater sensitivity. In one cohort, supervised saliva collection was more sensitive than URS swab, and unsupervised saliva collection was less sensitive.25 In the remaining cohorts, saliva samples demonstrated lower sensitivity than URS swabs in comparison with the CRS. However, most studies demonstrated wide overlap in the CIs for saliva and URS specimens. The studies in which overlap was not observed included a study of hospitalized patients who were moderately to severely ill18 and a study of patients in a quarantine center who were tested 8 to 10 days after initial confirmation of SARS-CoV-2 infection.27 One of these studies reported saliva to be more sensitive, whereas the other reported NP swabs to be more sensitive. Both studies were rated as having a high risk of spectrum bias. The third study, which was thought to have a low risk of spectrum bias, found saliva to be less sensitive than NP swab.8 More important, five of the seven cohorts in which saliva provided greater sensitivity were thought to be associated with a high risk of spectrum bias with respect to initial diagnostic testing of a community population.21 , 22 , 25 , 27 , 29 Of 10 cohorts that included patients who had previously tested positive for SARS-CoV-2, or included hospitalized patients, 4 showed higher sensitivity for saliva specimens,21 , 25 , 27 , 29 2 showed sensitivity identical to that for NP swabs,23 , 28 and 4 showed higher sensitivity for NPSs or midturbinate swabs.17 , 18 , 20 , 26 A single study of asymptomatic health care workers identified four positives from saliva samples and none using NP swabs.22 In contrast, seven of nine cohorts believed to have a low risk of spectrum bias with respect to diagnosis demonstrated higher sensitivity for NP swabs than for saliva specimens.3, 4, 5, 6 , 8 , 19
Three studies suggested similar or greater sensitivity for saliva sampling than for nasal self-swabs, but CIs are wide and one study is associated with a significant risk of spectrum bias.6 , 7 , 25
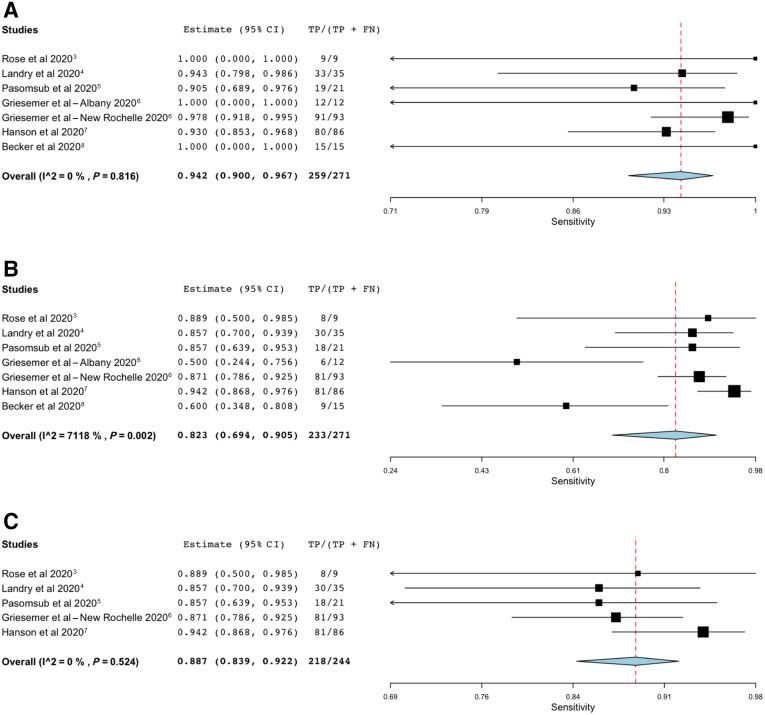
Seven cohorts associated with either drive-through centers or community health centers, and believed to have a low risk of spectrum bias (Supplemental Appendix S1), were included in the initial meta-analysis.3, 4, 5, 6, 7, 8 Studies based on hospitalized patients were excluded from the meta-analysis, because they tend not to be representative of those in whom initial diagnostic testing is performed. Results of this meta-analysis are shown as forest plots in Figure 1 .
Figure 1.
A: OpenMetaAnalyst forest plot for sensitivity of nasopharyngeal swab samples, measured against the composite reference standard. B: OpenMetaAnalyst forest plot for sensitivity of saliva samples, measured against the composite reference standard. The value for I2 indicates substantial heterogeneity, which appears to result from inclusion of the study by Becker et al,8 and the Albany cohort from Griesemer et al.6C: OpenMetaAnalyst forest plot for sensitivity of saliva samples, measured against the composite reference standard, when the study by Becker et al8 and the Albany cohort from Griesemer et al6 are not included. This post-hoc analysis was not part of the prespecified analysis plan. FN, false negative; TP, true positive.
Figure 1 presents computer output for the meta-analyses. Sensitivity of NP swabs, compared with the reference standard, was estimated at 94% (Figure 1A), with lower and upper bounds of 90% and 97%, respectively. There was no statistical suggestion of heterogeneity [τ2 = 0.000; Q (df = 6) = 2.946; P = 0.816; I2 = 0]. In contrast (Figure 1B), the sensitivity of saliva sampling was estimated at 82%, with lower and upper bounds of 69% and 91%, respectively. There was a strong suggestion of heterogeneity [τ2 = 0.631; Q (df = 6) = 20.822; P = 0.002; I2 = 71.184], in spite of the fact that tests of heterogeneity have relatively low power for analyses that include small numbers of studies.30 This suggested that it would be appropriate to conduct an unplanned exploratory analysis. After inspection of the forest plot (Figure 1B), the Becker et al8 study and the Albany cohort6 were removed and the saliva meta-analysis was repeated (Figure 1C). This procedure gave an estimate of 89% for the sensitivity of saliva samples, with lower and upper bounds of 84% and 92%, respectively. Once these studies were removed from the meta-analysis, there was no suggestion of heterogeneity [τ2 = 0.000; Q (df = 6) = 3.209; P = 0.524; I2 = 0). In each analysis, the sensitivity estimate for saliva samples remains lower than that of NP swabs. The upper and lower bounds for the NP swab sensitivity estimate do not include the estimate for saliva samples. Also, the bounds for the saliva sample estimate do not include the NPS estimate.
It is difficult to ascertain why the study by Becker et al8 is an outlier. This was one of two studies in this review to use an RNA stabilizer with saliva (Supplemental Table S1). The other study to do this found a higher sensitivity for saliva samples than NP swabs when saliva sample collection was supervised, but a lower sensitivity when saliva sample collection was unsupervised.25 The precise approach used for saliva collection was different between these two studies, but hints at a possibility that mixing of stabilizer with saliva may not have been optimal; it seems unreasonable to suppose that use of RNA stabilizer is intrinsically unfavorable to subsequent PCR analysis. There is no evidence to suggest that use of viral transport medium, phosphate-buffered saline, or Tris-EDTA during saliva collection or transport significantly affects subsequent assay sensitivity. Similarly, it is difficult to understand why the Albany cohort was an outlier.6
Discussion
Nasopharyngeal swabs have been the gold standard for diagnosis of SARS-CoV-2 infection. The studies examined give us no reason to question that belief. Only three of the studies considered herein show a diagnostic sensitivity of <90% for NP swabs; one of these, a study of hospital workers, showed no positive results from NP swabbing. My meta-analysis, based on cohorts undergoing initial diagnostic testing, gives an estimate of 94% (range, 91% to 97%) sensitivity for NP swabs; estimating the sensitivity by simply adding the positives for seven cohorts included in the meta-analysis gives an estimate of 95% (range, 92% to 98%) sensitivity for NP swabs. These estimates are similar to that obtained by repeated testing,31 and provide a fairly high degree of confidence regarding NP swab sensitivity for initial diagnosis of SARS CoV-2 infection when assessed using the composite reference standard.
The meta-analysis suggests that, when employed for initial diagnosis of SARS-CoV-2 infection in ambulatory patients, saliva samples are somewhat less likely than NP swab samples to give a positive nucleic acid amplification test result when compared with the CRS. Nevertheless, the performance of saliva-based testing is generally good, and the reduction of sensitivity may be offset by considerations of patient comfort (which might promote more frequent surveillance), as well as reduction in requirements for swabs and personal protective equipment.
Three cohorts included in this review dealt with testing of asymptomatic individuals.19 , 22 Data from two of the three cohorts suggest superiority for saliva, although only one, a contact-tracing cohort, includes more than five positive subjects. Another study by Wyllie et al,2 which did not meet the formal inclusion criteria for this systematic review, arrived at the same conclusion. The low numbers of positive specimens identified in these studies limits confidence in any conclusion of superiority for saliva, but supports the conclusion that saliva sampling is sufficiently sensitive for screening, as does a study that appeared after the inclusion period for this systematic review.32
The focus of this systematic review was initial diagnosis of SARS-CoV-2 infection in ambulatory subjects, and our conclusion regarding the relative sensitivity of saliva versus NP swabs should not be assumed to apply to patients who are hospitalized or who present later in the course of disease; several of the studies included in this review suggest that saliva sampling may be more sensitive later in disease, particularly when efforts are made to ensure that sputum is included in the specimen.27 , 28 None of the studies included in this meta-analysis involved attempts to collect lower-airway secretions together with saliva, and the heterogeneity of collection techniques used in studies reported herein precludes any conclusion regarding the optimal method for collecting saliva.
This analysis has several limitations. The use of a CRS, which defines the false-positive rate as zero for all assays, introduces a downward bias in the estimates of sensitivity.33 This bias varies somewhat based on study size and precise cell frequencies, but is expected to result in a similar degree of bias (about 1% to 1.5%) for estimates of both saliva sample performance and upper airway sample performance; the conclusions regarding relative sensitivity are not changed. The use of an abbreviated search strategy may have missed articles that otherwise met inclusion criteria. The inclusion criteria excluded small studies (<20 subjects) that might nevertheless have provided additional information regarding relative performance. The design and the review were the product of a single individual, which may have introduced bias into both decisions regarding study design and inclusion of articles in the meta-analysis. Finally, the studies included in the review demonstrate considerable heterogeneity in assay design, patient population, and disease prevalence. Thus, this systematic review and meta-analysis may provide more insight into the range of potential results expected for laboratories introducing saliva-based testing than they do for the specific performance of any detailed sampling and assay strategy.
Despite these weaknesses, this systematic review has several offsetting strengths. It focuses on sample sets that have been taken at the same time, thus differing from other systematic reviews,34, 35, 36, 37, 38 as well as a release from the Norwegian Institute of Public Health (https://www.fhi.no/globalassets/dokumenterfiler/rapporter/2020/saliva-sample-for-testing-sars-cov-2-infection-memo-2020.pdf, last accessed December 14, 2020). The total number of samples included in the meta-analysis is substantially greater than that of any individual study, based on simultaneous sample comparison, and allows for meta-analysis–based sensitivity estimates. This likely yields a more accurate assessment of performance in community-based initial testing. Although there is no true gold standard for diagnosis of COVID-19 infection, the use of a CRS, a relatively unbiased approach to generation of a reference standard, is more likely to provide a better sense of the performance difference associated with saliva sampling versus upper airway sampling than alternative approaches based on intrinsically biased comparisons.
The sensitivity difference between tests based on saliva samples and those based on NPSs is modest and, for broad community testing, justified by both patient comfort and the reduced need for personal protective equipment.
Acknowledgments
I thank Yuan-Po Tu (The Everett Clinic), Ted Schutzbank (Meridian Biosciences), Robert Becker (Food and Drug Administration), Karen Heichman (Bill and Melinda Gates Foundation), and those posting on the Association for Molecular Pathology discussion group, CHAMP, for valuable discussions that led to this work.
Footnotes
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.12.008.
Supplemental Data
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRIMSA) flow diagram (adapted from http://www.equator-network.org/reporting-guidelines/prisma, last accessed December 21, 2020) outlining the search and selection process applied during the literature search and review.
References
- 1.Tu Y.-P., O’Leary T.J. Testing for severe acute respiratory syndrome–coronavirus 2: challenges in getting good specimens, choosing the right test, and interpreting the results. Crit Care Med. 2020;48:1680–1689. doi: 10.1097/CCM.0000000000004594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. 2020;383:1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rose D., Ranoa E., Holland R.L., Alnaji F.G., Green K.J., Wang L., Brooke C.B., Burke M.D., Fan T.M., Hergenrother P.J. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. BioRxiv. 2020 [Epub] doi: 10.1011/2020.06.18.159434. [Google Scholar]
- 4.Landry M.L., Criscuolo J., Peaper D.R. Challenges in use of saliva for detection of SARS CoV-2 RNA in symptomatic outpatients. J Clin Virol. 2020;130:104567. doi: 10.1016/j.jcv.2020.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Sungkanuparph S., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.05.001. [Epub ahead of print] doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griesemer S., Van Slyke G., Ehrbar D., Strle K., Yildirim T., Centurioni D., Walsh A., Chang A., Waxman M., St George K. Evaluation of specimen types and saliva stabilization solutions for SARS-CoV-2 testing. medRxiv. 2020 doi: 10.1128/JCM.01418-20. [Epub] doi: 10.1101/2020.06.16.20133041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson K.E., Barker A.P., Hillyard D.R., Gilmore N., Barrett J.W., Orlandi R.R., Shakir S.M. Self-collected anterior nasal and saliva specimens versus healthcare worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01824-20. e01824-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker D., Sandoval E., Amin A., Hoff P.D.E., Diets A., Leonetti N., Lim Y.W., Elliott C., Laurent L., Grzymski J., Lu J.T., De Hoff P., Diets A., Leonetti N., Lim Y.W., Elliott C., Laurent L., Grzymski J., Lu J.T., Hoff P.D.E., Diets A., Leonetti N., Lim Y.W., Elliott C., Laurent L., Grzymski J., Lu J.T. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. medRxiv. 2020 [Epub] doi: 10.1101/2020.05.11.20092338. [Google Scholar]
- 9.McGrath T.A., Alabousi M., Skidmore B., Korevaar D.A., Bossuyt P.M.M., Moher D., Thombs B., McInnes M.D.F. Recommendations for reporting of systematic reviews and meta-analyses of diagnostic test accuracy: a systematic review. Syst Rev. 2017;6:194. doi: 10.1186/s13643-017-0590-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baughman A.L., Bisgard K.M., Cortese M.M., Thompson W.W., Sanden G.N., Strebel P.M. Utility of composite reference standards and latent class analysis in evaluating the clinical accuracy of diagnostic tests for pertussis. Clin Vaccin Immunol. 2008;15:106–114. doi: 10.1128/CVI.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moher D., Liberati A., Tetzlaff J., Altman D.G. The PRISMA Group: Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:194. [PMC free article] [PubMed] [Google Scholar]
- 12.Whiting P.F., Rutjes A.W.S., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M.G., Sterne J.A.C., Bossuyt P.M.M. Quadas-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 13.Schueler S., Schuetz G.M., Dewey M. The revised QUADAS-2 tool. Ann Intern Med. 2012;156:323. doi: 10.7326/0003-4819-156-4-201202210-00018. author reply 323-324. [DOI] [PubMed] [Google Scholar]
- 14.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 15.Wallace B.C., Dahabreh I.J., Trikalinos T.A., Lau J., Trow P., Schmid C.H. Closing the gap between methodologists and end-users: R as a computational back-end. J Stat Softw. 2012;49:1–15. [Google Scholar]
- 16.Newcombe R.G. Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17:857–872. doi: 10.1002/(sici)1097-0258(19980430)17:8<857::aid-sim777>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 17.L’Helgouach N., Champigneux P., Santos-Schneider F., Molina L., Espeut J., Alali M., Baptiste J., Cardeur L., Dubuc B., Foulongne V., Galtier F., Makinson A., Marin G., Picot M.-C., Prieux-Lejeune A., Quenot M., Checa-Robles F.J., Salvetat N., Vetter D., Reynes J., Molina F. EasyCOV: LAMP based rapid detection of SARS-CoV-2 in saliva. medRxiv. 2020 [Epub] doi: 10.1101/2020.05.30.20117291. [Google Scholar]
- 18.Miller M., Jansen M., Bisignano A., Mahoney S., Wechsbergrg C., Albanese N., Castillo L., Farinas P., Lazarin G.A., Jaremko M. Validation of a self-administrable, saliva-based RT-qPCR test detecting SARS-CoV-2. medRxiv. 2020 [Epub] doi: 10.1101/2020.06.05.20122721. [Google Scholar]
- 19.Güçlü E., Koroglu M., Yürümez Y., Toptan H., Kose E., Güneysu F., Karabay O. Comparison of saliva and oro-nasopharyngeal swab sample in the molecular diagnosis of COVID-19. Rev Assoc Med Bras. 2020;66:1116–1121. doi: 10.1590/1806-9282.66.8.1116. [DOI] [PubMed] [Google Scholar]
- 20.Wong S.C.Y., Tse H., Siu H.K., Kwong T.S., Chu M.Y., Yau F.Y.S., Cheung I.Y.Y., Tse C.W.S., Poon K.C., Cheung K.C., Wu T.C., Chan J.W.M., Cheuk W., Lung D.C. Posterior oropharyngeal saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:2939–2946. doi: 10.1093/cid/ciaa797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasaki S., Fujisawa S., Nakakubo S., Kamada K., Yamashita Y., Fukumoto T., Sato K., Oguri S., Taki K., Senjo H., Sugita J., Hayasaka K., Konno S., Nishida M., Teshima T. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect. 2020;81:e145–e147. doi: 10.1016/j.jinf.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byrne R.L., Kay G.A., Kontogianni K., Brown L., Collins A.M., Cuevas L.E., Ferreira D., Fraser A.J., Garrod G., Hill H., Menzies S., Mitsi E., Owen S.I., Williams C.T., Hyder-Wright A., Adams E.R., Cubas-Atienzar A.I. Saliva offers a sensitive, specific and non-invasive alternative to upper respiratory swabs for SARS-CoV-2 diagnosis. Emerg Infect Dis. 2020;26:2769–2770. doi: 10.3201/eid2611.203283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamal A.J., Mozafarihashjin M., Coomes E., Powis J., Li A.X., Paterson A. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa848. [Epub ahead of print] doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick-Baw C., Morgan K., Gaffney D., Cazares Y., Jaworski K., Byrd A., Molberg K., Cavuoti D. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.01109-20. e01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokota I., Shane P.Y., Okada K., Unoki Y., Yang Y., Inao T., Sakamaki K., Iwasaki S., Hayasaka K., Sugita J., Nishida M., Fujisawa S., Teshima T. Mass screening of asymptomatic persons for SARS-CoV-2 using saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1388. [Epub ahead of print] doi: 10.1093/cid/ciaa1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojima N., Turner F., Slepnev V., Bacelar A., Deming L., Kodeboyina S., Klausner J.D. Self-collected oral fluid and nasal swab specimens demonstrate comparable sensitivity to clinician-collected nasopharyngeal swab specimens for the detection of SARS-CoV-2. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1589. [Epub ahead of print] doi: 10.1093/cid/ciaa1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SoRelle J.A., Mahimainathan L., McCormick-Baw C., Cavuoti D., Lee F., Thomas A., Sarode R., Clark A.E., Muthukumar A. Saliva for use with a point of care assay for the rapid diagnosis of COVID-19. Clin Chim Acta. 2020;510:685–686. doi: 10.1016/j.cca.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dogan O.A., Kose B., Agaoglu N.B., Yildiz J., Alkurt G., Kendir Demirkol Y., Irvem A., Dinler-Doganay G., Doganay L. Does sampling saliva increase detection of SARS-CoV-2 by RT-PCR? comparing saliva with oro-nasopharyngeal swabs. J Virol Methods. 2021;290:114049. doi: 10.1016/j.jviromet.2020.114049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao M., Rashid F.A., Sabri F.S.A.H., Jamil N.N., Zain R., Hashim R., Amran F., Kok H.T., Samad M.A.A., Ahmad N. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS-CoV-2. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1156. [Epub ahead of print] doi:/ 10.1093/cid/ciaa1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins J.P.T., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. Br Med J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long D.R., Gombar S., Hogan C.A., Greninger A.L., Shah V.O., Bryson-Cahn C., Stevens B., Rustagi A., Jerome K.R., Kong C.S., Zehnder J., Shah N.H., Weiss N.S., Pinsky B.A., Sunshine J. Occurrence and timing of subsequent SARS-CoV-2 RT-PCR positivity among initially negative patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa722. [Epub ahead of print] doi: 10.1093/cid/ciaa722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Babady N.E., McMillen T., Jani K., Viale A., Robilotti E.V., Aslam A., Diver M., Sokoli D., Mason G., Shah M.K., Korenstein D., Kamboj M. Performance of severe acute respiratory syndrome coronavirus 2 real-time RT-PCR tests on oral rinses and saliva samples. J Mol Diagn. 2021;23:3–9. doi: 10.1016/j.jmoldx.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takwoingi Y., Guo B., Riley R.D., Deeks J.J. Performance of methods for meta-analysis of diagnostic test accuracy with few studies or sparse data. Stat Methods Med Res. 2017;26:1896–1911. doi: 10.1177/0962280215592269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khurshid Z., Zohaib S., Joshi C., Moin S.F., Zafar M.S., Speicher D.J. Saliva as a non-invasive sample for the detection of SARS-CoV-2: a systematic review. medRxiv. 2020 [Epub] doi: 10.1101/2020.05.09.20096354. [Google Scholar]
- 35.Czumbel L.M., Kiss S., Farkas N., Mandel I., Hegyi A., Nagy Á., Lohinai Z., Szakács Z., Hegyi P., Steward M.C., Varga G. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front Med. 2020;7:465. doi: 10.3389/fmed.2020.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peeters E., Kaur Dhillon Ajit Singh S., Vandesompele J., Mestdagh P., Hutse V., Arbyn M. Rapid systematic review of the sensitivity of SARS-CoV-2 molecular testing on saliva compared to nasopharyngeal swabs. medRxiv. 2020 [Epub] doi: 10.1101/2020.08.05.20168716. [Google Scholar]
- 37.Subsoontorn P., Lohitnavy M., Kongkaew C. The diagnostic accuracy of nucleic acid point-of-care tests for human coronavirus: a systematic review and meta-analysis. Sci Rep. 2020;10:22349. doi: 10.1038/s41598-020-79237-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarrom D., Elston L., Washington J., Prettyjohns M., Cann K., Myles S., Groves P. The effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review. BMJ Evid Based Med. 2020 doi: 10.1136/bmjebm-2020-111511. [Epub ahead of print] doi: 10.1136/bmjebm-2020-111511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRIMSA) flow diagram (adapted from http://www.equator-network.org/reporting-guidelines/prisma, last accessed December 21, 2020) outlining the search and selection process applied during the literature search and review.