Abstract
Autism spectrum disorder (ASD) is associated with several oropharyngeal abnormalities, including buccal sensory sensitivity, taste and texture aversions, speech apraxia, and salivary transcriptome alterations. Furthermore, the oropharynx represents the sole entry point to the gastrointestinal (GI) tract. GI disturbances and alterations in the GI microbiome are established features of ASD, and may impact behavior through the “microbial-gut-brain axis.” Most studies of the ASD microbiome have used fecal samples. Here, we identified changes in the salivary microbiome of children aged 2–6 years across three developmental profiles: ASD (n = 180), nonautistic developmental delay (DD; n = 60), and typically developing (TD; n = 106) children. After RNA extraction and shotgun sequencing, actively transcribing taxa were quantified and tested for differences between groups and within ASD endophenotypes. A total of 12 taxa were altered between the developmental groups and 28 taxa were identified that distinguished ASD patients with and without GI disturbance, providing further evidence for the role of the gut-brain axis in ASD. Group classification accuracy was visualized with receiver operating characteristic curves and validated using a 50/50 hold-out procedure. Five microbial ratios distinguished ASD from TD participants (79.5% accuracy), three distinguished ASD from DD (76.5%), and three distinguished ASD children with/without GI disturbance (85.7%). Taxonomic pathways were assessed using the Kyoto Encyclopedia of Genes and Genomes microbial database and compared with one-way analysis of variance, revealing significant differences within energy metabolism and lysine degradation. Together, these results indicate that GI microbiome disruption in ASD extends to the oropharynx, and suggests oral microbiome profiling as a potential tool to evaluate ASD status.
Keywords: microbiome, autism spectrum disorder, developmental delay, oropharynx, saliva, gastrointestinal disturbance
Lay Summary:
Previous research suggests that the bacteria living in the human gut may influence autistic behavior. This study examined genetic activity of microbes living in the mouth of over 300 children. The microbes with differences in children with autism were involved in energy processing and showed potential for identifying autism status.
Introduction
The microbiome of the gastrointestinal (GI) tract is essential for mammalian physiology, aiding digestion, synthesis, and absorption of important nutritional components such as amino acids, folate, and B vitamins [Preidis & Versalovic, 2009]. Accumulating evidence suggests that the GI microbiome also influences host behavior and neurodevelopment through the “microbial-gut-brain axis” [Cryan & O’Mahony, 2011]. This axis represents an evolving concept of microbial-mediated cross talk between the central nervous system (CNS) and GI tract that occurs through several different modalities, including direct neural activation, immune modulation, and hormonal, peptidergic, and epigenetic signaling [de Theije et al., 2014].
Although the exact mechanisms remain enigmatic, it is also now increasingly clear that alterations in the GI microbiome and gut-brain axis occur in a range of neuropsychiatric and neurodevelopmental disorders, including autism spectrum disorder (ASD) [Mayer, Padua, & Tillisch, 2014]. In fact, a disproportionate number of ASD patients suffer from GI comorbidities, including constipation, chronic diarrhea, abdominal pain, and gastroesophageal reflux [Horvath & Perman, 2002]. Although nearly 50% of ASD risk is attributable to genetic variations (such as nucleotide polymorphisms and copy number variants) [Sandin et al., 2017], it is possible that gene–environment interactions could act through the gut-brain axis (under the influence of the GI microbiome) and significantly modulate ASD risk [Finegold et al., 2002]. Microbial influence on serotonin levels provides a striking example of this [O’Mahony, Clarke, Borre, Dinan, & Cryan, 2015]. Polymorphisms in the serotonin transporter gene contribute to the risk of ASD [Yirmiya et al., 2001]. However, the majority of serotonin synthesis occurs in intestinal enterochromaffin cells, through the conversion of tryptophan into serotonin via tryptophan hydroxylase [Reigstad et al., 2015]. The GI microbiome enhances serotonin synthesis via the effects of short-chain fatty acids on enterochromaffin cells [Yano et al., 2015]. Once synthesized, most serotonin acts within the gut to promote intestinal motility, although some of it passes into the peripheral circulation and can potentially impact the CNS, particularly during early brain development. Thus, disturbance of the gut microbiome could alter serotonin signaling, acting in concert with a child’s genetic background.
Building on this idea of gene–environment interactions, there is accumulating evidence for disrupted gut-brain signaling in ASD [Frye, Rose, Slattery, & MacFabe, 2015]. For example, a recent study of 13 children with regressive-onset autism and GI comorbidities identified increased levels of fecal Clostridium and nonspore-forming anaerobes compared to those seen in typically developing (TD) controls [Finegold et al., 2002]. Disturbances in fecal Clostridium abundance have also been reported in two additional ASD microbiome studies [Parracho, Bingham, Gibson, & McCartney, 2005; Song, Liu, & Finegold, 2004]. Other investigations have noted alterations in the Bacteroides/Firmicutes ratio in children with ASD, though the directionality of those changes conflict [Finegold et al., 2010; Tomova et al., 2015; Williams et al., 2011]. Additional reports also implicate Lactobacillus, Prevotella, Sutterella, Desulfovibrio, and Veillonellaceae alterations in patients with ASD [Adams, Johansen, Powell, Quig, & Rubin, 2011; Vuong & Hsiao, 2017; Wang et al., 2013]. The lack of consensus between studies is challenging, but may be explained, in part, by the relatively small sample sizes used to explore a highly heterogeneous disorder. The small size of these investigations has also prevented subdivision of ASD participants into phenotypic subtypes. A larger scale approach could provide valuable insights into the relationship of the microbiome to autistic behavior, GI pathology, and immune function.
The potential role of the microbiome in ASD also gains strong support from several animal studies that have modulated social behaviors through dysbiosis, and ameliorated those symptoms with restoration of gut microbes [Buffington et al., 2016; Kumar & Sharma, 2016]. Parallel findings have even been reported in humans with ASD. For example, studies of antibiotic therapy with vancomycin or cycloserine [Urbano et al., 2014] have been able to temporarily mitigate some of the behavioral symptoms in ASD patients [Sandler et al., 2000]. A recent study of fecal microbiota transfer therapy in 18 children with ASD also demonstrated improvements in bacterial diversity alongside improvements in parent-reported GI and ASD symptoms [Kang et al., 2017]. These effects persisted for 8 weeks after intervention.
It is worth noting that nearly all studies of the ASD microbiome have focused on the lower GI tract. However, the oropharynx, which serves as the sole entry point to the GI tract, also represents a site of ASD pathology [Jaber, 2011]. Children with ASD suffer from increased rates of motor (speech; Tierney et al., 2015) and sensory (food texture; Cermak, Curtin, & Bandini, 2010) pathology in the mouth and we have previously described epitranscriptomic changes in the saliva of children with ASD [Hicks, Ignacio, Gentile, & Middleton, 2016]. This led us to posit that perturbations in the oral microbiome might also occur in children with ASD.
Here, we interrogate the human oral microbiome using high-throughput shotgun metatranscriptome data from the oropharynx of 180 children with ASD, 106 TD controls, and 60 children with nonautistic developmental delay (DD). We hypothesized that organisms identified by previous studies with altered abundance in the lower GI tract of ASD individuals would demonstrate changes in transcriptional activity in the oropharynx. Furthermore, we posited that specific microbiome communities would: (a) differentiate ASD endophenotypes and (b) correlate with expression of mRNAs related to neuro-hormone signaling and metabolic regulation.
Methods
This cross-sectional, observational, case control study was approved by the Independent Review Boards at the Penn State College of Medicine and the State University of New York Upstate Medical University. Written informed consent was obtained from the parents of all children who participated in the study.
Participants
Children ages 2–6 years (n = 346) were enrolled in the study. Participants were divided into three groups (ASD, n = 180; TD, n = 106; DD, n = 60) based on developmental status. ASD was defined by a clinician consensus diagnosis, using criteria specified in the Diagnostic and Statistical Manual of the American Psychiatric Association, Fifth Edition (DSM-5). TD participants included children with negative ASD screening on the Modified Checklist for Autism in Toddlers—Revised, and children who met typical developmental milestones on standardized physician assessment (e.g., survey of well-being in young children; parents’ evaluation of developmental status). The DD group included children with an ICD-10 diagnosis of DD (e.g., expressive speech delay, intellectual disability, behavioral concern) who did not meet DSM-5 criteria for ASD on clinician assessment. Children with feeding tube dependence, active tooth decay, fever, upper respiratory infection, or current use of oral antibiotics were excluded from all groups. Children with a family history of ASD in a first degree relative or a chronic medical condition requiring routine care by a pediatric specialist were excluded from the TD group. Phenotypic subgroup analysis examined ASD children with: (a) attention deficit hyperactivity disorder (ADHD; n = 43) or (b) GI disturbance (n = 39) relative to ASD children without the given comorbidity. ADHD was identified through parental survey, which asked “Does your child have a diagnosis of ADHD/ADD?” Positive answers were confirmed by ICD-10 diagnosis (F90.0), or stimulant medication (e.g., methylphenidate-based prescription) on chart review when possible. GI disturbance was defined as constipation (K59.0), gastroesophageal reflux (K21), chronic abdominal pain (R10), food sensitivities (T78.1), or recurrent diarrhea (K59.1, R19.7) reported by parental survey and confirmed through chart review of associated ICD-10 diagnosis codes.
Data Collection
Parents of all participants were administered a child medical/demographic survey and the Vineland Adaptive Behavior Scale—Second Edition (VABS-II) at the time of enrollment. Most of the ASD (n = 138) and DD participants (n = 21) were administered the Autism Diagnostic Observation Schedule—Second Edition (ADOS-2), or previous assessment scores were documented through chart review when available. ADOS-2 administration was performed by a trained certified health professional. The participant characteristics that were collected included: (a) demographic information (age, sex, ethnicity, body mass index); (b) oral/GI factors (time of collection, time of last meal, time of last tooth brushing, probiotic use, history of GI disturbance, medical/food allergies, dietary restrictions); and (c) medical history (birth age, birth delivery route, birth weight, asthma status, vaccination status). These factors were selected based on potential relevance to the profile of the oral microbiome.
Sample Collection and RNA Analysis
Saliva was collected from each participant at the time of enrollment. Following an oral water rinse, an ORAcollect swab (DNA Genotek, Ottawa, Canada) was used to obtain saliva from the sublingual and parotid regions of the mouth in a nonfasting state, at least 15 min after the most recent consumption of food or drink. Swabs were stored at −20°C prior to processing at the State University of New York Upstate Molecular Analysis Core Facility. Salivary RNA was extracted using a standard Trizol technique and the RNeasy mini column (Qiagen, Valencia, CA). Yield and quality of RNA was checked with an Agilent Bioanalyzer prior to library construction and quantification with next generation sequencing. Multiplexed samples were processed on a NextSeq 500 Instrument (Illumina, San Diego, CA) at a targeted depth of 10 million single end 50 base reads per sample. After adapter trimming and quality control analysis, RNA reads were aligned to the Human Microbiome Database using k-SLAM software. Sequence alignment with the k-mer method was used for comprehensive taxonomic classification and identification of microbial genes, as previously described [Ainsworth, Sternberg, Raczy, & Butcher, 2017]. Only taxa with raw read counts of 10 or more in at least 20% of samples were interrogated for differential abundance. Individual RNA transcripts were not subjected to analysis. Instead, we interrogated the pathways and ontologies represented by the community of microbial transcripts through cross-referencing the Kyoto Encyclopedia of Genes and Genomes (KEGG) microbial database using MicrobiomAnalyst software. This database consists of 82 KEGG Ontology (KO) Pathway sets, 11 KEGG Metabolism sets, and 20 Clusters of Orthologous Groups Function sets. Mapping was limited to those transcripts present at raw read counts of five or more in at least 10% of samples. Both taxonomic and pathway level data were analyzed for differences between groups following quantile normalization, using MetaboAnalyst software [Dhariwal et al., 2017] to perform nonparametric comparisons of the observed abundance counts between groups. These data sets will be made publicly available on the NCBI Sequence Read Archive following acceptance for publication.
Statistical Analysis
Differences in medical, demographic, and neuropsychological characteristics between ASD and TD or DD groups were assessed with a two-tailed Student’s t test, with significant differences defined by an uncorrected P < 0.05. Taxa with the greatest abundance (present in the largest concentrations) and prevalence (present in the largest number of samples) were reported at the species and phylum levels. The Shannon alpha diversity index and Bray–Curtiss index of beta diversity (homogeneity of group dispersions method) were calculated from the taxonomic profiles and compared across the three groups. Differential taxon expression across all participants was visualized with a multivariate partial least squares discriminant analysis (PLS-DA) and variable importance in projection was determined for each taxon. Individual taxa differences among the three groups were investigated with nonparametric Kruskal–Wallis testing, followed by posthoc between group comparisons (ASD:TD or ASD:DD) with a Mann–Whitney U test. Differences in KEGG pathway transcripts between diagnostic groups were evaluated using a one-way analysis of variance with a false discovery rate (FDR) correction for multiple testing set at (P < 0.05). Post hoc testing was performed between all three groups using a Tukey’s Honestly Significantly Difference test.
Taxon associations with a predefined set of ASD endophenotypes were assessed as follows: (a) taxon differences between ASD participants with/without GI disturbance; or with/without ADHD were examined with a nonparametric Mann–Whitney U test. (b) ASD participants were divided into three adaptive behavior groups (0-, 1-, or 2- SD below the mean value of 100) for Communication, Socialization, and Activities of Daily Living subscales of the VABS-II. A three group comparison was chosen to differentiate ASD participants with “minimal,” “moderate,” and “severe” impairment within each subdomain, in light of previous reports that the GI microbiome differed among children with varying autism severity [Finegold et al., 2010]. Between-groups taxonomic differences were assessed with nonparametric Kruskal–Wallis testing for the three VABS-II categories. (c) Relationships between autistic behavior measures on the ADOS-2 (Social Affect, Restrictive/Repetitive Behavior, and Comparison Score) and oral taxon activity were assessed with Pearson’s correlations. Factors with Benjamini–Hochberg FDR correction <0.05 were reported for each phenotype-taxonomic comparison.
Relationships between oral microbe activity, metabolomic pathways (KEGG IDs), and clinical characteristics were assessed with Pearson’s correlation (for continuous variables) or Spearman’s rank test (for dichotomous variables). Diagnostic accuracies of taxon levels in the oral microbiome were assessed with a multivariate logistic regression analysis, comparing: (a) ASD:TD; (b) ASD:DD; and (c) GI disturbance phenotypes across diagnoses. Classification accuracy was visualized with a receiver operating characteristic (ROC) curve, using the first 50% of samples from each group (chosen at random) and a 10-fold cross-validation procedure. The remaining 50% of samples were used to validate the predictive model for each comparison. Area under the curve (AUC) was calculated and 95% confidence intervals (CI) were reported.
Results
Participant Characteristics
The ASD group (n = 180) had a mean age of 53 (±16) months, was 85% males, and was 59% Caucasian (Table 1). TD participants (n = 106) were, on average, 10 months younger (43 ± 16 months), were 60% males, and were 63% Caucasian. The DD group (n = 60) had an average age of 50 (±13) months, was 70% males, and was 67% Caucasian. There was no difference in average collection time between ASD (12:29 p.m. ± 2:48), TD (12:21 p.m. ± 2:43), and DD (12:43 p.m. ± 2:43) subjects. There were also no differences between groups in time since last meal, or time since last tooth brushing. Only 3% of ASD and DD children were taking a probiotic, compared with 0% of TD children. ASD participants had higher rates of GI disturbance (22%) than the TD group (3%), but not the DD group (20%). More ASD participants had a food or medicine allergy (21%) than TD (9%) and DD (8%) participants, but they had similar rates of dietary restrictions. There was no difference between groups in birth weight, though children in the ASD group had higher rates of cesarean section (19%) than TD (9%) and DD (8%) participants. There were no differences between groups in rates of asthma or vaccination. The ASD group had lower mean VABS-II scores on the Socialization (73 ± 13) and Activities of Daily Living (75 ± 14) domains relative to TD and DD groups. Average scores on the VABS-II Communication scale in the ASD group (72 ± 18) differed from TD participants (103 ± 15), but not DD participants (76 ± 17). ASD subjects had higher ADOS-2 scores on the Social Affect domain (13 ± 5) and the Restrictive and Repetitive Behavior domain (3 ± 1) relative to DD participants (6 ± 4; and 2 ± 1, respectively). Their ADOS-2 comparison scores (7 ± 2) were also higher than DD participants (4 ± 2).
Table 1.
Participant Characteristics
| Clinical characteristics | ASD (n = 180) | TD (n = 106) | DD (n = 60) |
|---|---|---|---|
| Demographics | |||
| Age, mean (SD), years | 53 (16) | 43 (16)* | 50 (13) |
| Male (%), No. | 154 (86) | 64 (60)* | 43 (70)* |
| Caucasian (%), No. | 107 (59) | 67 (63) | 40 (67) |
| Body mass index (SD), kg/m2 | 16.5 (2.8) | 16.4 (2.0) | 17.0 (3.1) |
| Oral/GI factors | |||
| Time of collection (SD) | 12:29 (2:48) | 12:21 (2:43) | 12:43 (2:38) |
| Time since last meal (SD), hr | 3 (3) | 3 (3) | 2 (2) |
| Time of last tooth brush (SD), hr | 8 (5) | 5 (4) | 5 (3) |
| Food/medical allergies (%), No. | 38 (21) | 9 (9)* | 5 (8)* |
| Dietary restrictions (%), No. | 25 (14) | 8 (8) | 11 (18) |
| Probiotic use (%), No. | 5 (3) | 0 (0) | 2 (3) |
| GI disturbance (%), No. | 39 (22) | 3 (3)* | 12 (20) |
| Medical characteristics | |||
| Cesarean section (%), No. | 35 (19) | 9 (9)* | 5 (8)* |
| Birth weight (SD), kg | 3.3 (0.9) | 3.2 (0.7) | 3.2 (1.2) |
| Asthma (%), No. | 18 (10) | 8 (8) | 10 (17) |
| Fully vaccinated (%), No. | 169 (94) | 97 (92) | 58 (97) |
| Neuropsychiatric characteristics | |||
| ADHD (%), No. | 43 (23) | 10 (9)* | 17 (24) |
| Vineland communication (SD) | 72 (18) | 103 (15)* | 76 (17) |
| Vineland socialization (SD) | 73 (13) | 107 (17)* | 80 (19)* |
| Vineland ADL (SD) | 75 (14) | 104 (18)* | 81 (18)* |
| ADOS social affect (SD) | 13 (5) | - | 6 (4)* |
| ADOS RRB (SD) | 3 (1) | - | 2 (1)* |
| ADOS comparison (SD) | 7 (2) | - | 4 (2) |
Note. Characteristics with significant (P < 0.05) between-group differences on Student’s two-tailed t test are denoted with asterisks. Vineland domain standard scores are shown (where a score of 100 is average). ADOS subdomain and comparison scores are shown. Abbreviations: ADHD, attention-deficit hyperactivity disorder; ADL, activities of daily living; ADOS, Autism Diagnostic Observation Schedule; ASD, autism spectrum disorder; DD, developmental delay; GI, gastrointestinal; RRB, restricted and repetitive behavior; TD, typically developing.
Microbial Diversity Profiles
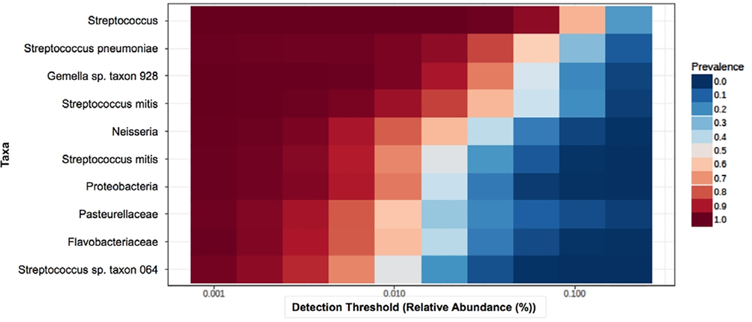
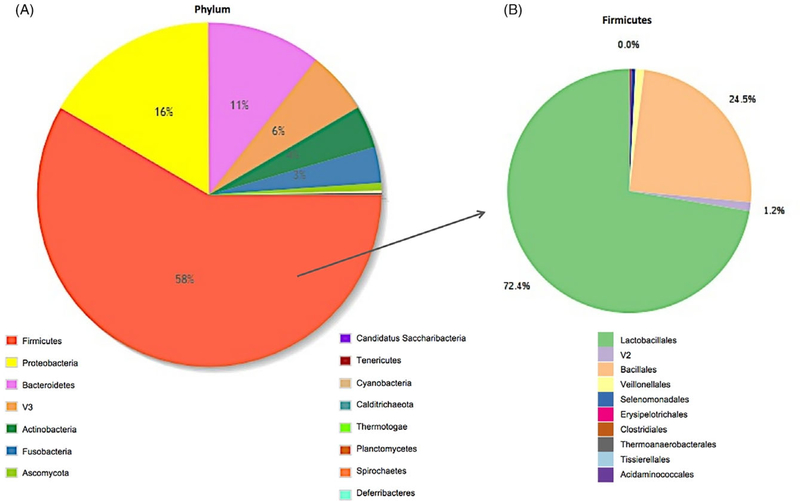
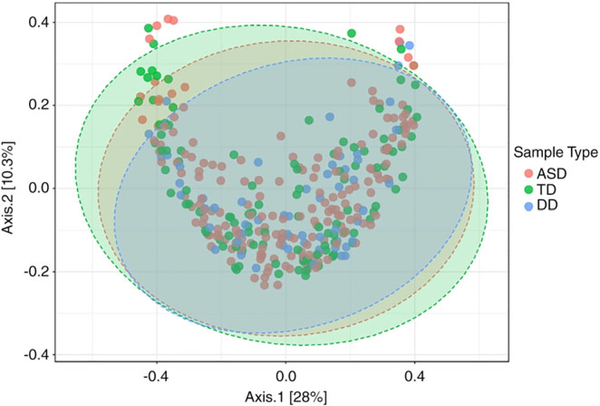
Among all samples, there was an average of 790,031 taxonomic reads per sample. The mean read count did not differ between ASD (785,766), TD (823,480), and DD (738,335) groups. Taxonomic reads were filtered to include only the taxa with counts of ≥10 in ≥20% of samples. Of the 753 taxa meeting these criteria, 41 were present in all samples. The core, oral microbiome (defined as taxa present in >70% of samples with relative abundance >0.5%) included 10 taxa (Fig. 1): Streptococcus (3.9 × 107 total raw reads), Streptococcus pneumoniae (2.0 × 107), Gemella sp. oral taxon 928 (3.5 × 107), Streptococcus mitis (2.0×107), Neisseria (9.2 × 106), S. mitis B6 (7.3 × 106), Proteobacteria (5.1 × 106), Pasteurellacae (6.0 × 106), Flavobacteriaceae (5.6 × 106), and Streptococcus sp. oral taxon 064 (3.6 × 106). The most abundant oral phyla among all samples was Firmicutes (58% of reads), followed by Proteobacteria (16%) and Bacteroides (11%; Fig. 2A). The most prominent taxonomic orders within the Firmicutes phylum were Lactobacillales (72% of reads) and Bacillales (25%; Fig. 2B). There was no difference in Shannon alpha diversity between ASD, TD, and DD groups at the species (P = 0.60; F = 1.01; Fig. S1A, Supporting Information), or phylum levels (p = 0.48; F = 0.73; Fig. S1B, Supporting Information). Bray–Curtis beta diversity, measured with a homogeneity of group dispersions technique, demonstrated significant differences (P = 0.04, F = 3.25) between ASD, TD, and DD groups (Fig. 3). The greatest between-sample diversity was present in the TD group. The DD group displayed the least distribution relative to ASD and TD groups.
Figure 1.
The core oral microbiome. The 10 oral taxa with the highest transcriptional activity across all participants (n = 346) are shown. Relative abundance (x-axis) for all 10 taxa exceeded 0.5% of the oral microbiome, and each taxa was present in counts of 10 or more in at least 70% of samples (prevalence, shown in red-blue scale).
Figure 2.
The core oral phyla. Abundance of oral transcripts at the phylum level across all participants (n = 346) are shown as percentage of the total (A). Firmicutes (58%) was the most abundant phylum, followed by Proteobacteria (16%) and Bacteroides (11%). Among the Firmicutes phylum (B) Lactobacillales was most abundant (72.4%) order, followed by Bacillales (24.5%).
Figure 3.
Bray–Curtis beta diversity. Microbial diversity between participants was calculated using a homogeneity of group dispersions technique for autism spectrum disorder (ASD) (red; n = 180), typically developing (TD) (green; n = 106), and developmental delay (DD) (blue; n = 60) groups. There was a significant difference (P = 0.04, F = 3.25) between groups, with the greatest between-samples diversity in the TD group. This two-dimensional plot accounts for 38.3% of the variance among participants. Confidence intervals of 95% are shown by the colored ovals.
Microbial Differences
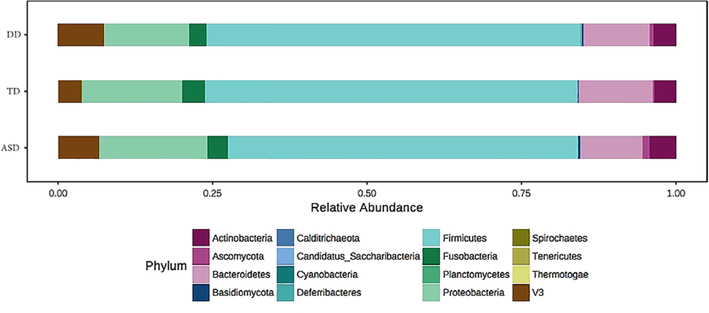
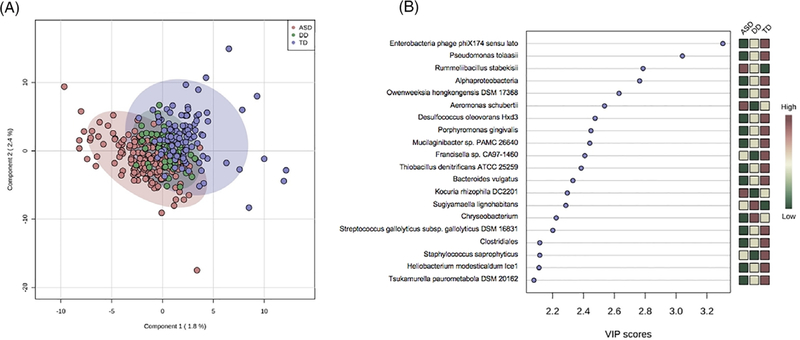
Differences between ASD, TD, and DD groups were explored at the phylum and species levels with a Kruskal–Wallis test. There were 12 taxa with significant differences (FDR < 0.05) between ASD, TD, and DD groups. There were six taxa with differential expression (FDR ≤ 0.05) between ASD and TD groups on Mann Whitney U test (Table 2). Two taxa were elevated in children with ASD (Limnohabitans sp. 63ED37–2, FDR = 0.01; Planctomycetales, FDR = 0.04) and four were decreased (Ramlibacter tataouinensis TTB310, FDR = 0.001; Mucilaginibacter sp. PAMC 26640, FDR = 0.001; Bacteroides vulgatus, FDR = 0.05; Gemmata sp. SH-PL17, FDR = 0.05). Three taxa showed significant differences (FDR ≤ 0.05) between ASD and DD children. Two taxa were elevated in children with ASD (Brucella, FDR = 0.05; Enterococcus faecalis OG1RF, FDR = 0.05) and one was decreased (Flavobacteriumsp.PK15,FDR=0.05). Phylum differences were observed between the three diagnostic groups (Fig. 4) for Planctomycetes (χ2 = 31.0, FDR = 3.2E-06), Cyanobacteria (χ2 = 14.8, FDR = 0.005), and Calditrichaeota (χ2 = 9.6, FDR = 0.04). These differences resulted largely from ASD/TD variation (Table S1, Supporting Information). Only Planctomycetes differed between ASD and both TD (fold change (FC) = 1.28, FDR = 0.001) and DD groups (FC = 0.03; FDR = 0.02). Notably, no changes were observed in the Firmicutes:Bacteroides ratio of the ASD group, though Bacteroides displayed nominally lower expression in ASD vs. TD participants (FC = 0.89, FDR = 0.051). A PLS-DA was used to visualize differences in taxonomic profiles at the species level between ASD, TD, and DD groups in two dimensions. A model accounting for 4% of the variance between groups resulted in partial separation of ASD, TD, and DD participants (Fig. 5A). The 20 taxa most critical for the differential group projection are shown (Fig. 5B). Of these 20 taxa, 14 demonstrate relative reductions in ASD samples and three are increased in saliva of ASD participants relative to TD and DD groups.
Table 2.
Taxon Differences Between ASD/TD and ASD/DD Groups at the Species Level
| Taxon | FC | FDR |
|---|---|---|
| ASD vs. TD | ||
| Mucilaginibacter sp. PAMC 26640 | 0.17 | 0.001 |
| R. tataouinensis TTB310 | 0.85 | 0.001 |
| Limnohabitans sp. 63ED37-2 | 1.05 | 0.01 |
| Planctomycetales | 1.21 | 0.04 |
| B. vulgatus | 0.43 | 0.05 |
| Gemmata sp. SH-PL17 | 0.86 | 0.05 |
| Cyanobacteria | 2.38 | 0.06 |
| Bacteroides ovatus | 0.23 | 0.07 |
| Thiobacillus denitrificans ATCC 25259 | 0.53 | 0.10 |
| Porphyromonas gingivalis TDC60 | 1.24 | 0.10 |
| ASD vs. DD | ||
| Brucella | 2.79 | 0.05 |
| Flavobacterium sp. PK15 | 0.41 | 0.05 |
| E. faecalis OG1RF | 2.27 | 0.05 |
| C. minutus PCC 6605 | 0.62 | 0.11 |
| Comamonas testosterone TK102 | 0.69 | 0.11 |
| Pseudomonadaceae | 0.77 | 0.11 |
| Cellulomonas fimi ATCC 484 | 1.63 | 0.11 |
| Flavobacterium psychrophilum | 0.62 | 0.11 |
| Flavobacterium crassostreae | 0.74 | 0.11 |
| M. luteus NCTC 2665 | 1.34 | 0.11 |
Note. The 10 species with the largest differences among autism spectrum disorder (ASD), typically developing (TD), and nonautistic developmental delay (DD) groups on Mann–Whitney U test are shown. Fold changes (FC) among ASD/TD and ASD/DD groups are listed.
Figure 4.
Oral phyla abundance across autism spectrum disorder (ASD), typically developing (TD), and developmental delay (DD) children. The relative abundance of 16 oral phyla is shown for children with autism spectrum disorder (ASD; n = 180), typically developing (TD; n = 106), and nonautistic DD (n = 60). Nonparametric Kruskal–Wallis testing revealed significant differences false discovery rate (FDR < 0.05) among the three groups for Planctomycetes (χ2 = 31.0, FDR = 3.2E-06), Cyanobacteria (χ2 = 14.8, FDR = 0.005), and Calditrichaeota (χ2 = 9.6, FDR = 0.04).
Figure 5.
Oral taxonomic profiles distinguish autism spectrum disorder (ASD) children from typically developing (TD) and developmental delay (DD) peers. A PLS-DA was used to visualize differences in taxonomic profiles at the species level between ASD, TD, and DD groups in two dimensions (A). A model accounting for 4% of the variance between groups resulted in partial separation of ASD participants (red) from TD (blue) and DD (green) peers. The 20 taxa most critical for group projection are shown, based on variable importance in projection score (B). The majority of these taxa (14) are reduced (green boxes) in ASD samples relative to TD and DD groups. Three taxa are elevated in ASD participants (red boxes) and three demonstrated intermediate expression patterns (yellow boxes).
Microbiome Variations Among ASD Phenotypes
Variations among microbiome elements at the phylum and taxon level were explored among common ASD phenotypes (Table 3). Differential expression among ASD subjects with/without ADHD, and with/without GI disturbance was investigated with a non-parametric Mann–Whitney approach. There were no taxa or phyla with differential expression among ASD children with and without ADHD. There were no phyla and 28 taxa with significant differences (FDR < 0.05) between ASD patients with and without GI disturbance (Table S2, Supporting Information). Three of these taxa were down-regulated in ASD children with GI disturbance and 25 were upregulated. None of the 28 taxa overlapped with those identified in ASD:TD and ASD:DD comparisons. Domain standard scores for adaptive behaviors (Communication, Socialization, and Activities of Daily Living) were characterized as 0-, 1-, or 2SD below the mean value (100) and phyla/taxon differences across ASD participants were identified with a Kruskal–Wallis test. There were one phylum (Calditrichaeota) and five taxa with differences across ASD Communication groups (Acinetobacter, Micrococcus luteus, Moraxella, Porphyromonas, and Pasteurellaceae bacterium). There were no differences across Socialization, or Activities of Daily Living phenotypes at the phyla or taxon level. Relationships of microbiome elements with Restrictive/Repetitive Behavior, Socialization, and Comparison Scores on ADOS-2 were interrogated using a Pearson correlation. At the phylum level, actinobacteria levels were significantly correlated (R > 0.20, FDR < 0.05) with ADOS-2 Social Affect (R = 0.24; FDR = 0.036). There were no phyla correlated with ADOS Restrictive/Repetitive Behavior Scores. At the taxon level, there were four elements correlated with ADOS-2 Restrictive/Repetitive Behavior Scores (Moraxella bovoculi, S. mitis, Riemerella anatipestifer, and Chryseobacterium sp. IHB B 17019). There were no taxa correlated with ADOS-2 Socialization or Comparison Scores. None of the taxa or phyla associated with ASD endophenotypes overlapped with those identified in ASD:TD and ASD:DD comparisons. Thus, the microbes with differential activity in the ASD group may contribute to the appearance of autistic traits at a critical threshold, but do not display a dose–response relationship with the abundance of autistic traits.
Table 3.
Differential Phyla and Species Profiles Among ASD Phenotypes
| ASD phenotypes | Phyla (#) | Species (#) |
|---|---|---|
| Medical traits | ||
| With (n = 39) and without (n = 141) GI disturbance | 0 | 28 see Table S2, Supporting Information for list |
| With (n = 70) and without (n = 110) ADHD | 0 | 0 |
| Adaptive behavior (binned into groups of 0, 1, or 2 SD below mean) | ||
| Communication (n = 38, 43, 64) | 1 | 5 |
| Calditrichaeota, χ2 = 9.7, FDR = 0.039 | Acinetobacter, χ2 = 23.0, FDR = 0.0019 | |
| M. luteus, χ2 = 21.5, FDR = 0.0033 | ||
| Moraxella, χ2 = 18.2, FDR = 0.014 | ||
| Porphyromonas, χ2 = 15.3, FDR = 0.049 | ||
| Pasteurellaceae, χ2 = 15.1, FDR = 0.049 | ||
| Socialization (n = 25, 61, 59) | 0 | 0 |
| Activities of daily living (n = 35, 64, 46) | 0 | 0 |
| Autistic characteristics | ||
| Social affect score | 1 Actinobacteria, R = 0.24, FDR = 0.036 | 0 |
| Restrictive/repetitive behavior score | 0 | 4 |
| M. bovoculi, R = 0.40, FDR = 0.0003 | ||
| S. mitis, R = 0.35, FDR = 0.005 | ||
| R. anatipestifer, R = 0.34, FDR = 0.005 | ||
| Chryseobacterium sp. IHB B 17019, R = 0.31, FDR = 0.030 | ||
| ADOS score comparison | 0 | 0 |
Note. Comparison of phyla and species level data among ASD phenotypes was completed for medical traits, adaptive behavior, and autistic characteristics. ASD subjects with/without gastrointestinal disturbance; and with/without ADHD were compared by Mann Whitney U test. The number of phyla/species differences is listed for each comparison. Adaptive behaviors (measured by Vineland Adaptive Behavior Scale —Second Edition) were defined as 0-, 1-, or 2- SD below the mean score (100) and microbial differences were compared across groups with a nonparametric Kruskal–Wallis test. Autistic traits were quantified with the Autism Diagnostic Observation Schedule—Second Edition, and associations with oral microbiome transcriptional activity were determined with Pearson Correlation analysis. Here, a positive R value denotes a direct relationship between microbial transcription activity and abundance of autistic traits. Individual phylum/species differences are denoted, with the exception of species differences among GI phenotypes, which can be found in Table S2, Supporting Information. Abbreviations: ADHD, attention-deficit hyperactivity disorder; ADOS, Autism Diagnostic Observation Schedule; ASD, autism spectrum disorder.
Relationship of Oral Microbiome Elements and Clinical Characteristics
There were no significant (R ≥ 0.20; FDR < 0.05) relationships between clinical characteristics and individual phylum levels on Spearman (dichotomous variables) or Pearson correlation analysis (continuous variables). Individual taxa showed relationships with age, body mass index, time of collection, time since last meal, and time since last tooth brushing (Table 4). None of the taxa associated with clinical features overlapped with taxa identified as “altered” in ASD patients, or among ASD endophenotypes. The largest number of taxon associations (21) was found with time of saliva collection, and 15 of these taxa were from the Streptococcus genus. The strongest correlation was found between time since last toothbrush and Candida dubliniensis CD36 (R = 0.43; FDR = 0.048). Notably, dietary restrictions, food/medicine allergies, probiotic use, and vaccination status showed no correlations with oral taxonomic concentrations.
Table 4.
Relationship Between Oral Taxa and Clinical Characteristics
| Clinical characteristic | Taxon | R | FDR |
|---|---|---|---|
| Age | -S. lithotrophicus ES1 | −0.23 | 4.5E-4 |
| -Klebsiella pneumoniae | 0.21 | 0.012 | |
| -Snodgrassella alvi B2 | −0.21 | 0.018 | |
| -R. anatipestifer Yb2 | −0.20 | 0.022 | |
| Sex | - | - | - |
| Ethnicity | - | - | - |
| Body mass index | -S. pneumoniae ST556 | 0.34 | 4.5E-8 |
| -S. pneumoniae Taiwan 19F-14 | 0.29 | 1.0E-5 | |
| -S. pneumoniae PCS8235 | 0.23 | 2.0E-5 | |
| -Streptococcus infantarius subsp. CJ18 | 0.21 | 6.4E-5 | |
| Collection time | -S. pneumoniae SPNA45 | 0.26 | 0.0014 |
| -Streptococcus pseudopneumoniae IS7493 | 0.26 | 0.0014 | |
| -S. pneumoniae OXC141 | 0.25 | 0.0023 | |
| -Streptococcus oralis | 0.24 | 0.0035 | |
| -S. alvi wkB2 | −0.24 | 0.0037 | |
| -S. pneumoniae TIGR4 | 0.24 | 0.0037 | |
| -S. pneumoniae 70585 | 0.23 | 0.0057 | |
| -S. pneumoniae PCS8235 | 0.23 | 0.0071 | |
| -S. pneumoniae Hungary19A-6 | 0.22 | 0.0077 | |
| -S. pneumoniae R6 | 0.22 | 0.0081 | |
| -Haemophilus influenzae 86-028NP | 0.22 | 0.0082 | |
| -Porphyromonas | −0.22 | 0.0083 | |
| -S. pneumoniae gamPNI0373 | 0.22 | 0.0095 | |
| -Streptococcus sp. oral taxon 064 | 0.21 | 0.0095 | |
| -S. pneumoniae Taiwan19F-14 | 0.21 | 0.0095 | |
| -Bacteroides caecimuris | −0.21 | 0.013 | |
| -S. pneumoniae ATCC 700669 | 0.21 | 0.014 | |
| -S. pneumoniae D39 | 0.20 | 0.016 | |
| -Tannerella forsythia KS16 | −0.20 | 0.016 | |
| -S. lithotrophicus ES-1 | −0.20 | 0.017 | |
| -S. pneumoniae | 0.20 | 0.018 | |
| Time since last meal | -Planococcus maritimus | 0.28 | 0.0061 |
| -Brevibacillus laterosporus LMG 15441 | 0.26 | 0.016 | |
| -Actinomyces meyeri | 0.24 | 0.027 | |
| Last tooth brush | -C. dubliniensis CD36 | 0.43 | 0.048 |
| Food/medical allergies | - | - | - |
| Dietary restrictions | - | - | - |
| Probiotic use | - | - | - |
| Cesarean section | - | - | - |
| Birth weight | - | - | - |
| Asthma | - | - | - |
| Vaccination status | - | - | - |
Note. Relationships between species level data and clinical/demographic characteristics are shown. Pearson analysis was employed for continuous variables and Spearman Rank analysis was used for dichotomous variables.
Classification Accuracy
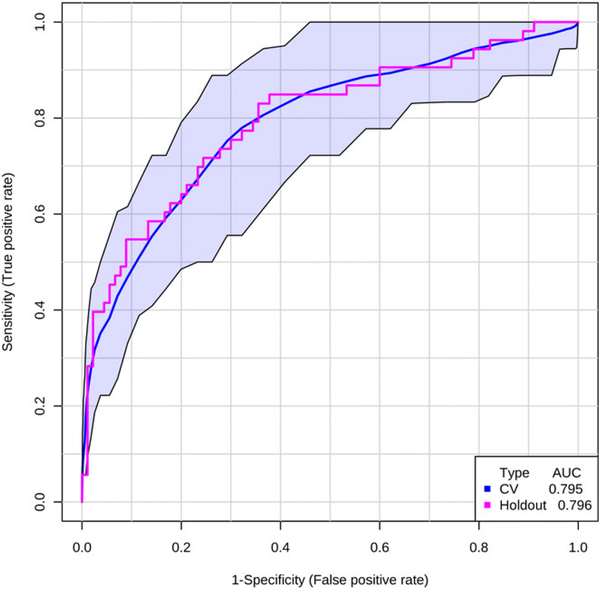
The utility of individual taxa to identify ASD status and GI phenotype was explored with a multivariate logistic regression analysis and classification accuracy was visualized by ROC curve analysis. For each comparison, 50% of the participants in each group were used to identify ratios between taxa with predictive accuracy, which were then tested in the remaining 50% of naïve “hold-out” samples. Five ratios, involving eight taxa (Mucilaginibacter/R. tataouinensis, Sphingomonadales/Planctomycetales, Alphaproteobacteria/Cyanobacteria, Alphaproteobacteria/Limnohabitans, and R. tataouinensis/Thiobacillus denitrificans) correctly identified 66/90 ASD participants and 38/53 TD participants in the training set, demonstrating an AUC of 0.795 (95% CI: 0.711–0.872). This panel of taxa demonstrated nearly identical performance in the hold-out set (Fig. 6A), identifying 73/90 ASD children and 33/53 TD children (AUC = 0.796). Three ratios, involving five taxa (Chamaesiphon minutus/Lactococcus lactis, Pseudomonadaceae/Lactococcus lactis, Flavobacterium sp. PK15/Burkholderiales) correctly identified 64/90 ASD children and 20/30 DD children in the training set, demonstrating an AUC of 0.770 (95% CI: 0.643–0.867). These three ratios performed similarly in the hold-out set of naïve samples, identifying 82/90 ASD children and 21/30 DD children (AUC = 0.765; Fig. 6B). Taxon levels also demonstrated utility for differentiating ASD children with GI disturbance from ASD children without GI disturbance. Three ratios, involving five taxa (Neisseria meningitidis M04–240196/Sideroxydans lithotrophicus ES-1, Neurospora crassa OR 74A/Acidipropionibacterium acidiproprionici, Enterobacterales/Neurospora crassa OR 74A), correctly identified 17/19 ASD children with GI disturbance and 51/70 ASD children without GI disturbance in the training set (AUC = 0.839; 95% CI: 0.759–0.958). In the hold-out set, this panel of taxa identified 7/20 ASD children with GI disturbance and 67/71 children without GI disturbance (AUC = 0.857; Fig. 6C).
Figure 6.
Transcriptional activity of oral taxa differentiates autism spectrum disorder (ASD) participants. The ability of taxonomic RNA profiles to identify ASD status was explored with multivariate logistic regression analyses and visualized on receiver operator characteristic curve. The first 50% of subjects in each comparison were used to build cross-validation (CV) curves (blue), that were tested in the remaining 50% of naïve holdout samples (pink). Five ratios, involving eight taxa differentiated ASD and typically developing (TD) children with an area under the curve (AUC) of 0.795 (95% Confidence interval (CI): 0.711–0.872) on CV and 0.796 on holdout testing (A). Three ratios, involving five taxa differentiated ASD and developmental delay (DD) children with an AUC of 0.770 (95% CI: 0.643–0.867) on CV and 0.765 on holdout testing (B). Finally, three ratios, involving five taxa identified ASD children with gastrointestinal (GI) disturbance relative to ASD peers without GI disturbance in both CV (AUC = 0.839; 95% CI: 0.759–0.958) and holdout models (AUC = 0.857) (C).
Metabolomic Pathway Profiling
The functional properties of microbial RNA transcripts measured in the oropharynx were investigated through alignment to the KEGG microbial database. KEGG pathways were filtered to include those with five or more alignments in at least 10% of the samples and quantile normalized. This resulted in 113 total KEGG pathway sets. Among the 113 pathways, seven demonstrated differential abundance (FDR < 0.05) between ASD, TD, and DD groups (Table 5). KEGG pathways with differential representation included Microbial Energy Metabolism, Translation Ribosome Structure and Biogenesis, Pyrimidine Metabolism, Lysine Degradation, Nucleotide Metabolism, Carbon Metabolism, and Nucleotide Transport and Metabolism (Fig. S2, Supporting Information). A Pearson analysis was used to identify phylogenetic groups most highly related to these metabolomic pathways (Table 5). Six KEGG IDs displayed significant (R > 0.4, FDR < 0.05) associations with activity of three phyla. Notable relationships were observed between K00415 (ubiquinol cytochrome C reductase; UBCR2) and both Ascomycota (R = 0.45, FDR = 1.6E-16) and Cyanobacteria (R = 0.46, FDR = 2.8E-17). UBCR2 is involved in oxidative phosphorylation, is disrupted in patients with mitochondrial respiratory chain deficiencies, and is implicated in Alzheimer’s, Parkinson’s, and Huntington’s disease. Ascomycota activity was also associated with K14221 (tRNA-Asp; R = 0.53; FDR = 4.5E-24) and K14226 (tRNA-His; R = 0.58; FDR = 1.5E-30), the latter of which is implicated in myoclonic epilepsy. Additional phyla associated with disrupted metabolomic pathways were Spirochaetes (K04069, pyruvate formate lyase activating enzyme, R = 0.42, FDR = 2.2E-14; and K04043, DNAk, R = 0.44, FDR = 9.7E-16) and Cyanobacteria (K01979, ssUrRNA, R = 0.47, FDR = 2.1E-18).
Table 5.
Metabolic Pathways Differentially Regulated in the Oral Microbiome of ASD, TD, and DD Children
| KEGG pathway | χ2 | FDR | Mann–Whitney | KO | Phyla | R | FDR |
|---|---|---|---|---|---|---|---|
| Energy metabolism | 24.8 | 0.00047 | ASD > TD ASD > DD | K04043 | Spirochaetes | 0.44 | 9.7E-16 |
| K00415 | Ascomycota | 0.45 | 1.6E-16 | ||||
| K00415 | Cyanobacteria | 0.46 | 2.8E-17 | ||||
| Translation ribosomal structure and biogenesis | 18.2 | 0.0062 | ASD > TD ASD > DD | K01979 | Cyanobacteria | 0.47 | 2.1E-18 |
| Pyrimidine metabolism | 15.8 | 0.013 | ASD < TD ASD < DD | ||||
| Lysine degradation | 15.3 | 0.013 | ASD > TD | K04069 | Spirochaetes | 0.42 | 2.2E-14 |
| Nucleotide metabolism | 14.5 | 0.016 | ASD < TD ASD < DD | ||||
| Carbon metabolism | 12.6 | 0.030 | ASD > TD ASD > DD | ||||
| Nucleotide transport and metabolism | 12.6 | 0.030 | ASD < TD ASD < DD | K14226 | Ascomycota | 0.58 | 1.5E-30 |
| K14221 | Ascomycota | 0.53 | 4.5E-24 | ||||
Note. Nonparametric Kruskal–Wallis testing was used to interrogate 113 KEGG pathways for differences in representation among the oral microbiome in children with ASD, TD, or nonautistic DD. KEGG pathways with FDR < 0.05 are shown. A Mann–Whitney test was used as a post hoc contrast between the groups. Individual KO pathways significantly (R > 0.40) associated with individual phylogenies across samples are displayed. Abbreviations: ASD, autism spectrum disorder; DD, developmental delay; FDR, false detection rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; KO, KEGG Orthology; TD, typical development.
Discussion
To the best of our knowledge, this study comprises the largest investigation of the microbiome in children with ASD, and the first to utilize oropharyngeal samples. It identifies distinct oral microtranscriptomic activity in ASD children relative to both TD peers and nonautistic peers with DD. These taxonomic patterns show some overlap with previous reports of the gut microbiome, but also identify novel changes in the oropharynx.
Like the gut, the oropharynx is a site of significant pathology in ASD [Cermak et al., 2010; Tierney et al., 2015]. Children with ASD experience increased rates of motor (speech apraxia) and sensory (food texture and taste) dysfunction. In addition, the oropharynx represents the sole point of entry to the GI tract and a major site of host-environment interaction. Sensory and motor innervation of the oropharynx by five cranial nerves (V, VII, IX, X, and XII) provides major linkages between the oropharynx and CNS and a plausible exchange pathway for the gut-brain axis (which also exerts major influences via cranial nerve X) [Bercik et al., 2011]. Thus, it is not surprising that particular microtranscriptome profiles are enhanced in ASD children with GI disturbance. Notably, we found that several of these “alterations” are also associated with specific autistic features. For example, M. luteus levels are decreased in both ASD children with GI disturbance and ASD children with adaptive communication scores more than two standard deviations below the mean. Similarly, levels of R. anatipestifer and Actinobacteria demonstrated correlations with measures of restricted/repetitive behavior, and social affect, respectively, and were “altered” in children with GI disturbance. Such trends are particularly striking when considering that ASD phenotypes unrelated to the GI tract (ADHD) showed no differences in microbiome profiles at the phylum or species levels.
In the context of recent studies highlighting the genetic contributions to ASD [Sandin et al., 2017], it is unlikely that microbial shifts represent the sole driver of autistic behavior. However, alterations in the microbiome have been linked with atypical social, communicative, and repetitive behavior in animal models [Buffington et al., 2016; Kumar & Sharma, 2016]. One mechanism for this link may be metabolomic disruptions [De Angelis et al., 2013]. Here we show that the microbial RNA profiles disrupted in children with ASD (relative to DD and TD peers) differentially target metabolic pathways in the oropharynx. It is well established that microbial activity in the GI tract plays an important role in the metabolism of compounds essential to host nutrition [LeBlanc et al., 2013; Preidis & Versalovic, 2009]. Here, we identify upregulation of microbial RNAs related to Lysine Degradation in the oropharynx of children with ASD. Lysine is a ketogenic amino acid whose degradation results in glutamate production. Glutamate is a key neurotransmitter involved in learning and memory. Increased levels have been reported in plasma [Aldred, Moore, Fitzgerald, & Waring, 2003; Shinohe et al., 2006] and the CNS of patients with ASD [Bejjani et al., 2012; Brown, Singel, Hepburn, & Rojas, 2013]. Interestingly, we also found evidence of increased “Energy Metabolism” and “Carbon Metabolism” transcripts in the oral microbiota of ASD children relative to TD and DD children. The KEGG Energy Metabolism entry includes a set of subcategories (oxidative phosphorylation, photosynthesis, carbon fixation, methane metabolism, nitrogen metabolism, and sulfur metabolism). Of these, further inspection strongly suggests that the increase in “Energy Metabolism” in ASD children is driven by increased expression of bacterial transcripts involved in Oxidative phosphorylation (1.6-fold) and Methane metabolism (1.2-fold). Indeed, oxidative phosphorylation by QCRC2 (a pathway implicated in CNS pathology such as Alzheimer’s and Parkinson’s disease) was strongly associated with cyanobacteria activity; and cyanobacteria activity was elevated in ASD participants relative to TD peers.
A second mechanism by which host–microbial interactions may lead to altered social behavior is through toxicological effects [Vuong & Hsiao, 2017]. For example, here we report alterations in oral Cyanobacteria in children with ASD at the phylum (Fig. 4), and species (Table 2) level, and show that levels of Cyanobacteria may be used to differentiate children with autism from TD peers. Cyanobacteria are water-borne pathogens that produce cyanotoxins and can lead to serious illness (e.g., GI disturbance, hay fever, pruritus). The cyanobacteria neurotoxin β-N-methylamino-L-alanine has been proposed to contribute to neurodegenerative conditions such as Parkinson’s and Alzheimer’s diseases. In addition, Son et al. [2015] have previously reported disruptions in cyanobacteria levels in the fecal microbiome of children with ASD relative to neurotypical siblings.
Clinical Implications
The microtranscriptome profiles found in the oropharynx of children with ASD may provide an objective tool for screening, diagnosing, or classifying patients. We show here that the levels of eight oral taxa may distinguish children with ASD from TD peers, while a panel of five taxa classifies ASD and DD subjects, both with nearly 80% accuracy. Previously, we have demonstrated that microRNA levels in saliva may differentiate children with ASD from healthy controls [Hicks & Middleton, 2016]. It is intriguing to consider that some perturbations in salivary microRNA may be driven by host interactions with the microbiome. Given the role of microRNA as an intercellular signaling molecule and its importance in normal brain development, microbial-microRNA cross talk may be one mechanism by which the gut-brain-axis functions. This interaction deserves further study.
Large-scale individual profiling of the microbiome also highlights a potential avenue for therapeutic targets. Several previous studies have demonstrated changes in autism symptoms or traits with antimicrobial or probiotic interventions [Kang et al., 2017; Sandler et al., 2000; Urbano et al., 2014]. These studies successfully reset the gut microbiota using untargeted approaches. Given the heterogeneity of taxonomic features that become evident when large numbers of ASD children are studied alongside specific measures of behavioral features, it seems that a more individualized approach could improve treatment success. For instance, based on these findings, probiotics targeted at the restoration of Micrococcus species in children with autism, GI disturbance, and communication difficulties may provide individualized benefit. Alternatively, antibiotics selected to specifically target Riemerella species in ASD children with GI difficulties and repetitive behaviors might be of clinical utility. Perhaps the greatest benefit to the oral microbiome approach is it allows easily repeated microbiome collections on-demand, over time, so that one can track changes in these microbial communities in response to targeted therapy.
Limitations
It is impossible to control for every variable that could conceivably influence the oral microbiome across ASD, TD, and DD groups. The present study included a rigorous collection of relevant factors (Table 1) so that the results could be interpreted with full transparency. It is worth noting that the only oral/GI factors that differed between ASD, TD, and DD groups were GI disturbance rates, and food/medical allergy rates, and the latter was not associated with expression patterns of any oral taxa. One oral taxon (R. anatipestifer Yb2) was associated with GI disturbance (Table S2, Supporting Information) and weakly associated with age (Table 4). A second oral taxon (S. lithotrophicus ES1) with utility for detecting GI disturbance among ASD subjects was also associated with age and collection time. Thus, it is possible that several microbial factors identified in the present study are confounded by changes in the GI tract over time. Longitudinal analyses of the oral microbiome among developing children would be useful in elucidating these relationships.
A second factor to consider when comparing results of the present study to previous literature is the use of high-throughput metatranscriptomic sequencing, rather than a 16S rRNA approach. In the present study, the resulting values provide a direct measure of transcriptional activity within the microbiome from different species and taxa, rather than focusing on microbial abundance. This approach allows for a functional interrogation of RNA properties through KO databases, but also makes comparisons to previous literature somewhat difficult. Thus, patterns of microbial disturbance previously reported in the fecal microbiome may be missed with this approach if abundance did not translate directly to transcriptional activity (i.e., the bacteria in those studies were not actively transcribing gene products).
The characterization of ASD subgroups as it relates to oral microbe transcriptional activity should be interpreted with caution. In this study, ASD group assignment was based on DSM-5 criteria that were interpreted by multiple providers across several medical sites, and phenotypic subgrouping was based on VABS-II and ADOS-2 evaluation for a subset of participants. The largest previous microbiome study in ASD characterized 59 ASD patients with the child behavior checklist [Son et al., 2015], while the current study characterizes only 138 of its 180 ASD participants with the ADOS-2. Furthermore, the ADOS-2 was administered by clinicians rather than research-reliable administrators. The ADOS-2 scores reported here are subdomain and total scores, which are not typically used quantitatively due to their psychometric properties. Finally, designation of ASD participants into ADHD or GI subgroups is based on parental report and medical record validation, not standardized scales such as the child behavior checklist or the GI severity index. Such scales would provide meaningful, quantifiable data for subgroup analyses and should be considered in future studies of the ASD microbiome.
Conclusions
There is mounting evidence that the GI microbiome is disrupted in children with ASD [Finegold et al., 2010; Mulle, Sharp, & Cubells, 2013]. The present study shows that this disruption may extend to the oropharynx, influencing the transcriptional activity of the microbial community. Such shifts appear to be associated with ASD comorbidities (such as GI disturbance), as well as social and repetitive behaviors. The mechanism for this relationship may result from alterations of microbial metabolism, or through pathogenic microbial–host relationships, but will certainly require further study. Oral taxonomic and functional profiling may provide utility as objective markers of ASD phenotypes.
Supplementary Material
Table S2. Taxon Differences between ASD patients with and without GI disturbance.
Figure S1. Alpha diversity dose not differ between ASD, TD, and DD participants. Estimates of alpha diversity among children with autism spectrum disorder (ASD; red), typical development (TD; green) and non-autistic developmental delay (DD; blue) did not differ at the species (A) or phylum (B) levels. Box plots demonstrate mean diversity for each group with 95% confidence interval. Individual participants are represented by block dots.
Table S1. Phylum differences between ASD, DD, and DD participants.
Figure S2. Metabolism pathways with differential expression among ASD, TD, and DD groups Among the 113 KEGG pathways represented by microbial RNA expression, seven demonstrated significant differences (FDR < 0.05) among children with autism spectrum disorder (ASD; red), typical development (TD; green), and non-autistic developmental delay (DD; blue). Relative RNA representation for the seven KEGG pathways is shown.
Acknowledgments
The authors would like to thank R.C., J.B., and C.D.G. for assistance with study design. The authors would also like to thank J.R. and M.B. for assistance with participant identification. The authors acknowledge A.T., M.C, F.G., C.G., L.P., T.W., A.S., S.B., R.P., E.C., J.V., and N.V. for assistance with participant recruitment and sample collection. Finally, the authors thank D.W. and X.Z. for their guidance with statistical analysis. Funding was provided by Quadrant Biosciences Inc. (Research agreement with SDH), NIH STTR (R41 MH111347-0), and Kirson-Kolodner-Fedder Charitable Fund.
Conflict of Interest
Drs. Hicks and Middleton are coinventors of preliminary patents for salivary RNA biomarkers in disorders of the CNS that are assigned to the SUNY Upstate and Penn State Research Foundations and licensed to Quadrant Biosciences, Inc. Dr. Hicks is also a consultant for Motion Intelligence, Inc. These conflicts of interest are currently managed by the Penn State College of Medicine and SUNY Upstate Medical University. Mr. Uhlig, Mr. Williams, and Ms. Wagner are each affiliated with (employed by) Quadrant Biosciences Inc., which has exclusive rights to the IP resulting from this research. The remaining authors have no conflicts to disclose.
Footnotes
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Contributor Information
Steven D. Hicks, Penn State College of Medicine, Division of Academic General Pediatrics, Department of Pediatrics, Hershey, PA.
Richard Uhlig, Quadrant Biosciences, Inc., Syracuse, NY.
Parisa Afshari, State University of New York Upstate Medical University, Departments of Neuroscience and Physiology, Syracuse, NY.
Jeremy Williams, Quadrant Biosciences, Inc., Syracuse, NY.
Maria Chroneos, Penn State College of Medicine, Division of Academic General Pediatrics, Department of Pediatrics, Hershey, PA.
Cheryl Tierney-Aves, Penn State College of Medicine, Division of Rehabilitation and Development, Department of Pediatrics, Hershey, PA.
Kayla Wagner, State University of New York Upstate Medical University, Departments of Neuroscience and Physiology, Syracuse, NY.
Frank A. Middleton, State University of New York Upstate Medical University, Departments of Neuroscience and Physiology, Syracuse, NY State University of New York Upstate Medical University, Department of Pediatrics, Syracuse, NY.
References
- Adams JB, Johansen LJ, Powell LD, Quig D, & Rubin RA (2011). Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterology, 11(1), 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth D, Sternberg MJ, Raczy C, & Butcher SA (2017). k-SLAM: Accurate and ultra-fast taxonomic classification and gene identification for large metagenomic data sets. Nucleic Acids Research, 45(4), 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldred S, Moore KM, Fitzgerald M, & Waring RH (2003). Plasma amino acid levels in children with autism and their families. Journal of Autism and Developmental Disorders, 33(1), 93–97. [DOI] [PubMed] [Google Scholar]
- Bejjani A, O’Neill J, Kim JA, Frew AJ, Yee VW, Ly R, … Levitt JG (2012). Elevated glutamatergic compounds in pregenual anterior cingulate in pediatric autism spectrum disorder demonstrated by 1H MRS and 1H MRSI. PLoS One, 7(7), e38786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Park AJ, Sinclair D, Khoshdel A, Lu J, Huang X, … Verdu EF (2011). The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut–brain communication. Neurogastroenterology and Motility, 23(12), 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Singel D, Hepburn S, & Rojas DC (2013). Increased glutamate concentration in the auditory cortex of persons with autism and first-degree relatives: A 1H-MRS study. Autism Research, 6(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffington SA, Di Prisco GV, Auchtung TA, Ajami NJ, Petrosino JF, & Costa-Mattioli M (2016). Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell, 165(7), 1762–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak SA, Curtin C, & Bandini LG (2010). Food selectivity and sensory sensitivity in children with autism spectrum disorders. Journal of the American Dietetic Association, 110(2), 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, & O’Mahony SM (2011). The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterology and Motility, 23(3), 187–192. [DOI] [PubMed] [Google Scholar]
- De Angelis M, Piccolo M, Vannini L, Siragusa S, De Giacomo A, Serrazzanetti DI, et al. (2013). Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One, 8(10), e76993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Theije CG, Wopereis H, Ramadan M, van Eijndthoven T, Lambert J, Knol J, et al. (2014). Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain, Behavior, and Immunity, 37, 197–206. [DOI] [PubMed] [Google Scholar]
- Dhariwal A, Chong J, Habib S, King IL, Agellon LB, & Xia J (2017). MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Research, 45, W180–W188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finegold SM, Dowd SE, Gontcharova V, Liu C, Henley KE, Wolcott RD, … Green JA III. (2010). Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe, 16(4), 444–453. [DOI] [PubMed] [Google Scholar]
- Finegold SM, Molitoris D, Song Y, Liu C, Vaisanen ML, Bolte E, … Kaul A (2002). Gastrointestinal microflora studies in late-onset autism. Clinical Infectious Diseases, 35, S6–S16. [DOI] [PubMed] [Google Scholar]
- Frye RE, Rose S, Slattery J, & MacFabe DF (2015). Gastrointestinal dysfunction in autism spectrum disorder: The role of the mitochondria and the enteric microbiome. Microbial Ecology in Health and Disease, 26(1), 27458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SD, Ignacio C, Gentile K, & Middleton FA (2016). Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatrics, 16(1), 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SD, & Middleton FA (2016). A comparative review of microRNA expression patterns in autism spectrum disorder. Frontiers in Psychiatry, 7(1), 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath K, & Perman JA (2002). Autism and gastrointestinal symptoms. Current Gastroenterology Reports, 4(3), 251–258. [DOI] [PubMed] [Google Scholar]
- Jaber MA (2011). Dental caries experience, oral health status and treatment needs of dental patients with autism. Journal of Applied Oral Science, 19(3), 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, … Krajmalnik-Brown R (2017). Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome, 5(1), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H, & Sharma B (2016). Minocycline ameliorates prenatal valproic acid induced autistic behaviour, biochemistry and blood brain barrier impairments in rats. Brain Research, 1630, 83–97. [DOI] [PubMed] [Google Scholar]
- LeBlanc JG, Milani C, de Giori GS, Sesma F, Van Sinderen D, & Ventura M (2013). Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Current Opinion in Biotechnology, 24(2), 160–168. [DOI] [PubMed] [Google Scholar]
- Mayer EA, Padua D, & Tillisch K (2014). Altered brain-gut axis in autism: Comorbidity or causative mechanisms? Bioessays, 36(10), 933–939. [DOI] [PubMed] [Google Scholar]
- Mulle JG, Sharp WG, & Cubells JF (2013). The gut microbiome: A new frontier in autism research. Current Psychiatry Reports, 15(2), 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony SM, Clarke G, Borre YE, Dinan TG, & Cryan JF (2015). Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behavioural Brain Research, 277, 32–48. [DOI] [PubMed] [Google Scholar]
- Parracho HM, Bingham MO, Gibson GR, & McCartney AL (2005). Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. Journal of Medical Microbiology, 54(10), 987–991. [DOI] [PubMed] [Google Scholar]
- Preidis GA, & Versalovic J (2009). Targeting the human microbiome with antibiotics, probiotics, and prebiotics: Gastroenterology enters the metagenomics era. Gastroenterology, 136(6), 2015–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reigstad CS, Salmonson CE, Rainey JF, Szurszewski JH, Linden DR, Sonnenburg JL, et al. (2015). Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. The FASEB Journal, 29(4), 1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, & Reichenberg A (2017). The heritability of autism spectrum disorder. JAMA, 318(12), 1182–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler RH, Finegold SM, Bolte ER, Buchanan CP, Maxwell AP, Väisänen ML, … Wexler HM (2000). Short-term benefit from oral vancomycin treatment of regressive-onset autism. Journal of Child Neurology, 15(7), 429–435. [DOI] [PubMed] [Google Scholar]
- Shinohe A, Hashimoto K, Nakamura K, Tsujii M, Iwata Y, Tsuchiya KJ, … Mori N (2006). Increased serum levels of glutamate in adult patients with autism. Progress in Neuro-Psychopharmacology & Biological Psychiatry, 30(8), 1472–1477. [DOI] [PubMed] [Google Scholar]
- Son JS, Zheng LJ, Rowehl LM, Tian X, Zhang Y, Zhu W, … Li E (2015). Comparison of fecal microbiota in children with autism spectrum disorders and neurotypical siblings in the Simons simplex collection. PLoS One, 10(10), e0137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Liu C, & Finegold SM (2004). Real-time PCR quantitation of clostridia in feces of autistic children. Applied and Environmental Microbiology, 70(11), 6459–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney C, Mayes S, Lohs SR, Black A, Gisin E, & Veglia M (2015). How valid is the checklist for autism spectrum disorder when a child has apraxia of speech? Journal of Developmental and Behavioral Pediatrics, 36(8), 569–574. [DOI] [PubMed] [Google Scholar]
- Tomova A, Husarova V, Lakatosova S, Bakos J, Vlkova B, Babinska K, & Ostatnikova D (2015). Gastrointestinal microbiota in children with autism in Slovakia. Physiology & Behavior, 138, 179–187. [DOI] [PubMed] [Google Scholar]
- Urbano M, Okwara L, Manser P, Hartmann K, Herndon A, & Deutsch SI (2014). A trial of D-cycloserine to treat stereotypies in older adolescents and young adults with autism spectrum disorder. Clinical Neuropharmacology, 37(3), 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong HE, & Hsiao EY (2017). Emerging roles for the gut microbiome in autism spectrum disorder. Biological Psychiatry, 81(5), 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, & Conlon MA (2013). Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Molecular Autism, 4(1), 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BL, Hornig M, Buie T, Bauman ML, Paik MC, Wick I, … Lipkin WI (2011). Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One, 6(9), e24585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, … Hsiao EY (2015). Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell, 161(2), 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yirmiya N, Pilowsky T, Nemanov L, Arbelle S, Feinsilver T, Fried I, & Ebstein RP (2001). Evidence for an association with the serotonin transporter promoter region polymorphism and autism. American Journal of Medical Genetics, 105(4), 381–386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Taxon Differences between ASD patients with and without GI disturbance.
Figure S1. Alpha diversity dose not differ between ASD, TD, and DD participants. Estimates of alpha diversity among children with autism spectrum disorder (ASD; red), typical development (TD; green) and non-autistic developmental delay (DD; blue) did not differ at the species (A) or phylum (B) levels. Box plots demonstrate mean diversity for each group with 95% confidence interval. Individual participants are represented by block dots.
Table S1. Phylum differences between ASD, DD, and DD participants.
Figure S2. Metabolism pathways with differential expression among ASD, TD, and DD groups Among the 113 KEGG pathways represented by microbial RNA expression, seven demonstrated significant differences (FDR < 0.05) among children with autism spectrum disorder (ASD; red), typical development (TD; green), and non-autistic developmental delay (DD; blue). Relative RNA representation for the seven KEGG pathways is shown.