Abstract
The quinolinate synthase of prokaryotes and photosynthetic eukaryotes, NadA, contains a [4Fe-4S] cluster with unknown function. We report crystal structures of Pyrococcus horikoshii NadA in complex with dihydroxyacetone phosphate (DHAP), iminoaspartate analogs, and quinolinate. DHAP adopts a nearly planar conformation and chelates the [4Fe-4S] cluster via its keto and hydroxyl groups. The active-site architecture suggests that the cluster acts as a Lewis acid in enediolate formation, similar to zinc in class II aldolases. The DHAP and putative iminoaspartate structures suggest a model for a condensed intermediate. The ensemble of structures suggests a two-state system, which may be exploited in early steps.
Graphical Abstract
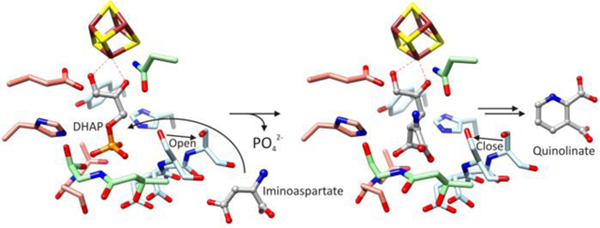
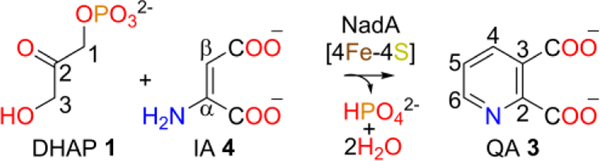
De novo biosynthesis of nicotinamide adenine dinucleotide (NAD) occurs via the intermediate quinolinate (QA) for which two major biosynthetic pathways are known.1,2 The pathway used by most bacteria, plants, and algae starts from L-aspartate and requires two enzymes, flavin adenine dinucleotide (FAD) dependent L-aspartate oxidase, NadB,3 and [4Fe-4S] cluster dependent QA synthase, NadA.4–6 In some organisms, NadB is replaced by NAD(P) dependent L-aspartate dehydrogenase.7 NadB catalyzes the oxidation of L-aspartate to iminoaspartate (IA) via reduction of FAD to FADH23,8,9 and structural studies suggest an enamine product.10 NadA catalyzes two condensations to form the pyridine ring of QA from IA and dihydroxyacetone phosphate (DHAP), eliminating phosphate and two waters (Scheme 1).3,11
Scheme 1.
NadA catalyzed reaction.
Labeling studies showed that the pyridine C4 originates from C1 of DHAP, indicating the mode of condensation.12 Two mechanisms have been proposed.1,3 Both start with DHAP and require a keto-aldo isomerization that precedes Schiff base formation between the imine nitrogen of IA and C3 of DHAP. Nasu et al. proposed that these steps occur after C1-Cβ bond formation and phosphate elimination (Fig. S1A),3 whereas Begley et al. proposed that ketose-aldose isomerization occurs at the outset, producing glyceraldehyde 3-phosphate (G3P) and allowing the Schiff base to form first (Fig. S1B).1 Biochemical studies suggest that DHAP rather than G3P condenses with IA.11,13 However, little is known about the mechanism and the role of the [4Fe-4S] cluster.
Crystal structures have been reported for Pyrococcus horikoshii NadA (PhNadA),14 Pyrocococcus furiosus NadA (PfNadA),15 and Thermotoga maritima NadA (TmNadA).16 The first structure of PhNadA showed a triangular monomer containing three domains and a bound malate identified the active site. However, the structure lacked a [4Fe-4S] cluster.14 The structure of PfNadA displayed an alternate domain arrangement and oligomeric state, but also lacked a [4Fe-4S] cluster.15 The structure of TmNadA, reported recently, revealed the location of the [4Fe-4S] cluster and identified a charge relay system.16 Finally, a second structure of PhNadA, also reported recently, showed a [4Fe-4S] cluster with bound QA.17
Here, we report high-resolution crystal structures of PhNadA with a bound [4Fe-4S] cluster and with bound DHAP, itaconate (IA analog), maleate, citraconate (enamine 4 analogs), and L-malate (L-aspartate analog). We also report a high-resolution crystal structure of PhNadA with bound QA. PhNadA was prepared with a [4Fe-4S] cluster by coexpression with the Escherichia coli suf operon using E. coli BL21 (DE3) as the host strain.18,19 Purification and crystallization were performed in an anaerobic chamber. Crystals were irradiated at beam lines NE-CAT 24-ID-E of the Advanced Photon Source (APS) and A1 and F1 of the Cornell High Energy Synchrotron Source (CHESS). The structure of PhNadA lacking a [4Fe-4S] cluster14 was used as a search model for molecular replacement. COOT,20 PHENIX,21 and CHIMERA22 were used for model building, refinement, and analysis. Difference electron density for the ligands is shown in Fig. S2 and data collection and refinement statistics are given in Tables S1-S3.
PhNadA contains three domains displaying pseudo three-fold symmetry that each contribute a cysteine residue for ligation to a centrally located [4Fe-4S] cluster (Fig. S3A-C). The overall structure resembles that of the enzymes IspH and Dph2.15 Each cysteine thiolate resides at the positive end of a 5-residue helix (Fig. S3D). IspH contains a similar cluster-binding motif except the helices are ~3 times longer.23 In contrast, Dph2 positions one of the thiolates near the positive end of a short helix; the other two reside in short turns.24
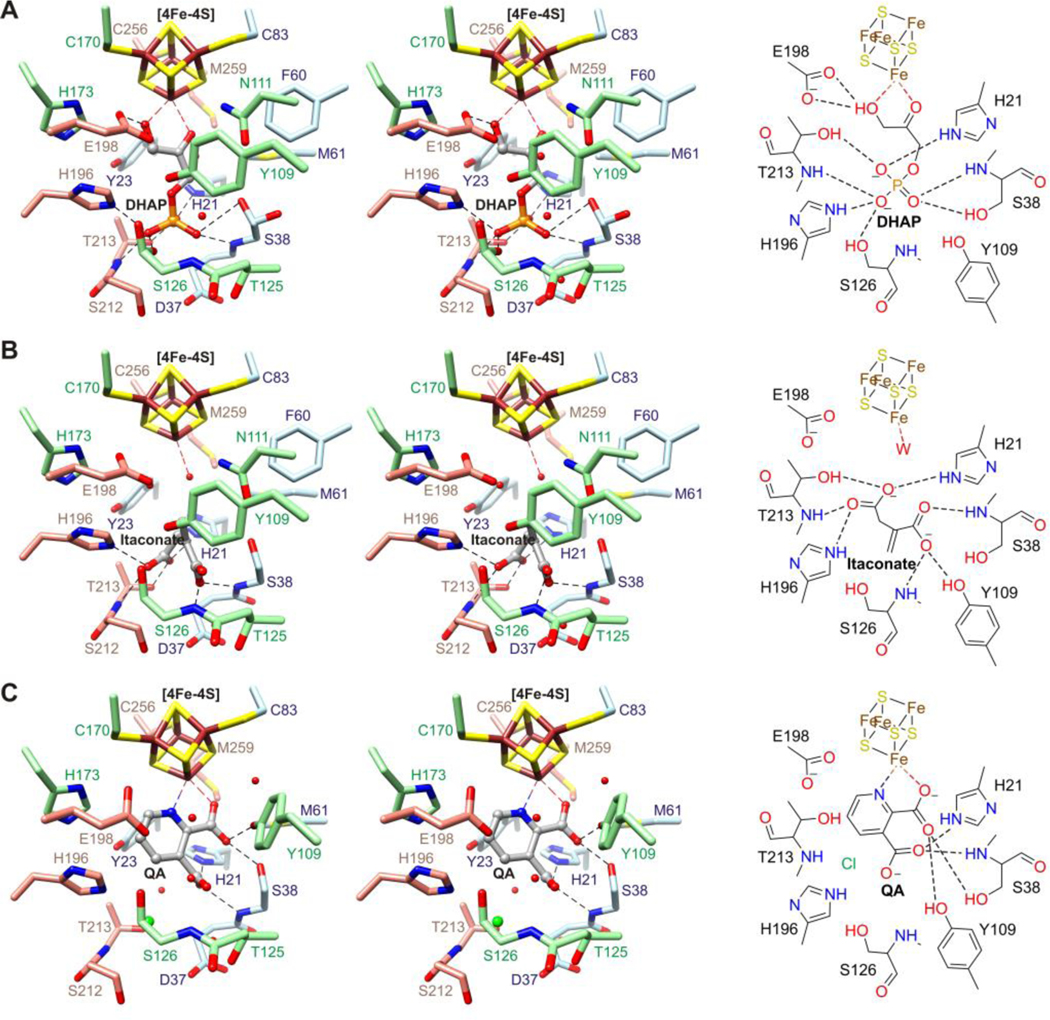
DHAP binds the unique (unligated) iron of the [4Fe-4S] cluster via its keto and hydroxyl groups and forms several hydrogen bonds with PhNadA via its phosphate (Fig. 1A). The conformation of DHAP is nearly planar having O2–C2–C3–O3, C1–C2–C3–O3, O1–C1–C2–C3, and O1–C1–C2–O2 torsional angles of −14.2°, −179.7°, −14.8°, and 179.6°, respectively. O3 is 2.6 Å from the carboxylate of Glu198 and sp3 geometry at C3 places the proR and proS protons near the hydroxyl of Tyr23 and the carboxylate of Glu198, respectively. O2 is 3.8 Å from the side chain NH2 of Asn111. The phosphate group is within hydrogen bonding distance of Nε2 of His21 and His196, the hydroxyl group of Ser38, Ser126, and Thr213, and the backbone NH of Ser38 and Thr213, and may also be stabilized by helix dipoles.
Figure 1.
Stereo diagrams (left) and schematic drawings (right) of the active site of PhNadA with bound (A) DHAP, (B) itaconate, and (C) QA. Black broken lines denote potential hydrogen bonds, red spheres denote waters, and the green sphere denotes chloride.
Itaconate is synclinal and occupies the DHAP phosphate site; two waters bind near the [4Fe-4S] cluster (Fig. 1B). The C1 carboxylate forms hydrogen bonds with the backbone NH of Ser38 and Ser126 and with the hydroxyl of Tyr109; the Cγ carboxylate forms hydrogen bonds with the hydroxyl and backbone NH of Thr213 and with Nε2 of His21 and His196. The methylene points toward the hydroxyl of Tyr109 and the carboxylate of Glu198.
QA chelates the unique iron of the [4Fe-4S] cluster via its pyridine nitrogen and the carboxylate at C2, which is also within hydrogen bonding distance of the hydroxyl of Ser38 and Tyr109 (Fig. 1C and Ref. 17). The carboxylate at C3 forms hydrogen bonds with Nε2 of His21 and with the backbone NH of Ser38. The binding mode is similar to that reported recently;17 however, in our structure chloride binds 3.2 Å from Nε2 of His196 and the backbone NH of Thr213, and Asn111 and Tyr109 are oriented differently.
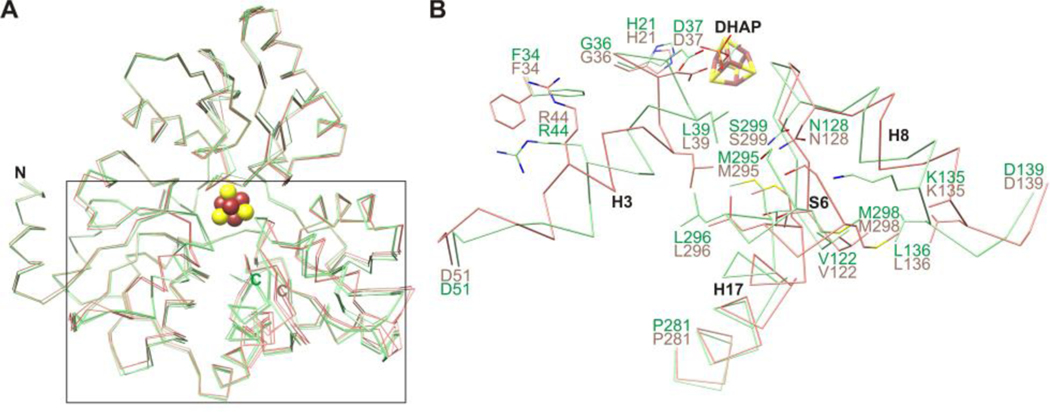
Maleate and citraconate bind similar to itaconate but adopt a more planar conformation consistent with a double bond between C2 and C3 (Fig. S4A and B). L-malate shows a unique binding mode in which the C4 carboxylate binds to the unique iron of the [4Fe-4S] cluster and no interactions are made with helix H3 (Fig. S4C). This leads to conformational changes that result in a more open active site cavity, similar to the holoenzyme structure. The ensemble of PhNadA structures suggests a two-state system (Fig. 2).
Figure 2.
(A) Superimposition of Cα traces showing open (holo and L-malate bound structures; salmon) and closed (DHAP, itaconate, maleate, and citraconate bound structures; light green) states of PhNadA. (B) Close-up of holo and DHAP bound structures indicating side chain clashes.
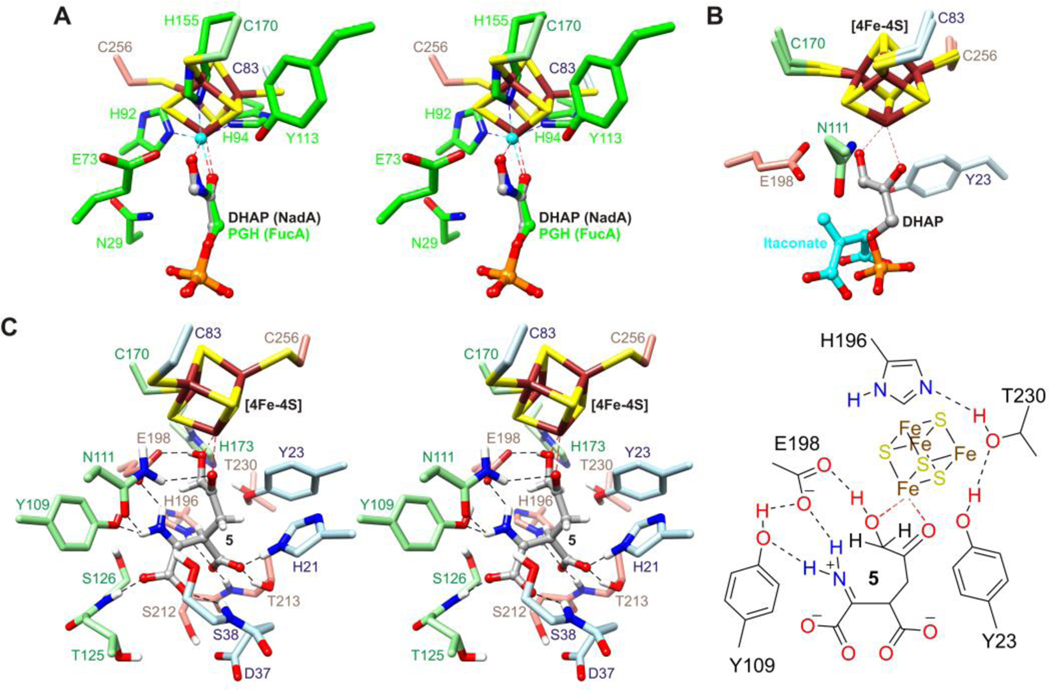
The structure of DHAP and the nearby architecture support an electrophilic role for the [4Fe-4S] cluster in enediolate formation with phosphate elimination disfavored prior to ternary complex formation. O3, C3, C2, O2, and C1 are nearly coplanar, consistent with an enediol(ate)-like structure, and the binding mode is strikingly similar to the mode of binding of phosphoglycolohydroxamate (PGH) to zinc in fuculose-1-phosphate aldolase 25 (FucA) (Fig. 3A). PGH is a cis enediolate analog used in studies of triose phosphate isomerase (TPI),26–28 methylglyoxal synthase (MGS),29 and class II aldolases,25,30 which form an enediolate from DHAP via polarization of the C2 carbonyl, proton abstraction from C3, and stabilization of the negative charge acquired by O2. TPI uses a histidine and lysine residue26,27,31–33 for electrophilic catalysis whereas class II aldolases use zinc;25,30,34 Fig. 3A suggests that a [4Fe-4S] cluster could play a similar role as zinc. The side chain NH2 of Asn111 in PhNadA is also well positioned to contribute to O2 oxyanion stabilization and assist DHAP binding.26,35,36 TPI and FucA utilize a glutamate residue as the catalytic base26,37 whereas MGS utilizes an aspartate residue.38 The side chains of Tyr23 and Glu198 in PhNadA, which are part of a predicted charge relay system and essential for activity,16,17 are well positioned to perform proton abstractions from C3 or the C3 hydroxyl. Biochemically, an enediol(ate) links intermediates 5 and 6 and may account for the TPI activity reported for NadA in the absence of IA (Fig. S5).13 The O1-C1 bond is “in-plane”, which stereoelectronically disfavors the elimination of phosphate from the enediolate.25,34,39 Phosphate elimination is likely suppressed further initially by the conformational changes that close the active site cavity in around the bound DHAP (Fig. 2), similar to the role of a mobile loop in TPI, the absence of which accelerates degradative loss of phosphate leading to methylglyoxal.40 The planarity of the O1-C1-C2-O2 torsion (~180°) raises the possibility of conjugation.
Figure 3.
(A) Stereo diagram of the superimposition of DHAP bound to the [4Fe-4S] cluster in PhNadA onto PGH bound to zinc (cyan) in FucA (green). (B) Superimposition of the [4Fe-4S] cluster with bound DHAP onto the [4Fe-4S] cluster in the itaconate bound structure of PhNadA. (C) Stereo diagram (left) and schematic drawing (right) of intermediate 5 based on the result in panel B, with C1 and Cβ joined, followed by energy minimization with positional restraints applied to the protein atoms.
The bound IA analogs clash severely with the DHAP phosphate (Fig. S6), requiring structural changes for ternary complex formation and phosphate elimination. The nature of these changes and the timing of phosphate elimination are unknown; however, the open state (Fig. 2) is likely required to provide the space needed for binding both substrates and to create an opening for ejecting phosphate. Helix H3 forms a portion of the phosphate-binding site (Fig. 1A) and its conformation depends on the liganded state (Fig. 2). Its conformation in the holo and L-malate bound structures, which is also a minor conformation in the itaconate bound structure, yields an open cavity from which phosphate can be eliminated from an orthogonal conformation (Fig. S7).
After phosphate is eliminated, the closed active site cavity neatly accommodates intermediate 5 and structurally similar intermediates (Fig. S1A). Itaconate, maleate, and citraconate adopt a similar conformation that is also adopted by malate in the absence of the [4Fe-4S] cluster.14 The conformation is stabilized by several hydrogen bonds and possibly by helix dipoles, suggesting that it is likely adopted by the IA moiety at some point along the reaction coordinate. Superimposition of the [4Fe-4S] cluster with bound DHAP onto the [4Fe-4S] cluster in the itaconate bound structure places the keto and hydroxyl groups by the two waters near the cluster and, without the phosphate, leads to geometry at the C1-Cβ bond consistent with intermediate 5: C1 and Cβ are 1.67 Å apart with C1-Cβ-Cα and C1-Cβ-Cγ angles of 108.6° and 133.0°, respectively (Fig. 3B). An energy-minimized model is given in Fig. 3C.
The pyridine nitrogen and the adjacent carboxylate in QA chelate the [4Fe-4S] cluster similar to the way the amino and carboxylate groups of S-adenosylmethionine bind a [4Fe-4S] cluster in radical SAM enzymes.17,41,42 From one point of view, the orientation of QA places C4 and hydroxylated C5 of intermediate 8 between the side chains of the essential residues Glu198 and Tyr23, suggesting a potential catalytic role (Fig. 1C). However, the reactions forming intermediate 8 and QA likely do not require enzyme catalysis.3 This suggests a simpler interpretation of QA formation from transitions between a two-state system starting from bound DHAP activated by the [4Fe-4S] cluster: a closed-to-open cavity transition allowing ternary complex formation, condensation, and phosphate elimination, and an open-to-closed cavity transition stabilizing a later intermediate associated with [4Fe-4S] cluster dependent keto-aldo isomerization.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Tadhg Begley and Dr. Angad Mehta for stimulating discussions, and Dr. Cynthia Kinsland for cloning PhNadA.
Funding Sources
This work is based upon research conducted at the APS on the NE-CAT beamlines, which are supported by award GM103403 from the NIH. Use of the APS is supported by the U.S. Department of Energy, under Contract No. DE-AC02-06CH11357. MacCHESS is supported by NIH grant GM103485 at the CHESS.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Materials and Methods, Tables S1-S3, and Figures S1-S8. (PDF)
The coordinates of PhNadA (5KTM), PhNadA/DHAP (5KTN), PhNadA/quinolinate (5KTO), PhNadA/itaconate (5KTP), PhNadA/maleate (5KTR), PhNadA/citraconate (5KTS), and PhNadA/L-malate (5KTT) have been deposited in the Protein Data Bank.
The authors declare no competing financial interests.
REFERENCES
- (1).Begley TP, Kinsland C, Mehl RA, Osterman A, and Dorrestein P. (2001), Vitam. Horm 61, 103–119. [DOI] [PubMed] [Google Scholar]
- (2).Gazzaniga F, Stebbins R, Chang SZ, McPeek MA, and Brenner C. (2009), Microbiol. Mol. Biol. Rev 73, 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Nasu S, Wicks FD, and Gholson RK (1982), J. Biol. Chem 257, 626–632. [PubMed] [Google Scholar]
- (4).Cicchillo RM, Tu L, Stromberg JA, Hoffart LM, Krebs C, and Booker SJ (2005), J. Am. Chem. Soc 127, 7310–7311. [DOI] [PubMed] [Google Scholar]
- (5).Gardner PR, and Fridovich I. (1991), Arch. Biochem. Biophys 284, 106–111. [DOI] [PubMed] [Google Scholar]
- (6).Ollagnier-de Choudens S, Loiseau L, Sanakis Y, Barras F, and Fontecave M. (2005), FEBS Lett. 579, 3737–3743. [DOI] [PubMed] [Google Scholar]
- (7).Yang Z, Savchenko A, Yakunin A, Zhang R, Edwards A, Arrowsmith C, and Tong L. (2003), J. Biol. Chem 278, 8804–8808. [DOI] [PubMed] [Google Scholar]
- (8).Seifert J, Kunz N, Flachmann R, Laufer A, Jany KD, and Gassen HG (1990), Biol. Chem. Hoppe Seyler 371, 239–248. [PubMed] [Google Scholar]
- (9).Tedeschi G, Negri A, Mortarino M, Ceciliani F, Simonic T, Faotto L, and Ronchi S. (1996), Eur. J. Biochem 239, 427–433. [DOI] [PubMed] [Google Scholar]
- (10).Bossi RT, Negri A, Tedeschi G, and Mattevi A. (2002), Biochemistry 41, 3018–3024. [DOI] [PubMed] [Google Scholar]
- (11).Suzuki N, Carlson J, Griffith G, and Gholson RK (1973), Biochim. Biophys. Acta 304, 309–315. [DOI] [PubMed] [Google Scholar]
- (12).Wicks FD, Sakakibara S, Gholson RK, and Scott TA (1977), Biochim. Biophys. Acta 500, 213–216. [DOI] [PubMed] [Google Scholar]
- (13).Reichmann D, Coute Y, and Ollagnier de Choudens S. (2015), Biochemistry 54, 6443–6446. [DOI] [PubMed] [Google Scholar]
- (14).Sakuraba H, Tsuge H, Yoneda K, Katunuma N, and Ohshima T. (2005), J. Biol. Chem 280, 26645–26648. [DOI] [PubMed] [Google Scholar]
- (15).Soriano EV, Zhang Y, Colabroy KL, Sanders JM, Settembre EC, Dorrestein PC, Begley TP, and Ealick SE (2013), Acta Crystallogr. D Biol. Crystallogr 69, 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cherrier MV, Chan A, Darnault C, Reichmann D, Amara P, Ollagnier de Choudens S, and Fontecilla-Camps JC (2014), J. Am. Chem. Soc 136, 5253–5256. [DOI] [PubMed] [Google Scholar]
- (17).Esakova OA, Silakov A, Grove TL, Saunders AH, McLaughlin MI, Yennawar NH, and Booker SJ (2016), J. Am. Chem. Soc 138, 7224–7227. [DOI] [PubMed] [Google Scholar]
- (18).Hanzelmann P, Hernandez HL, Menzel C, Garcia-Serres R, Huynh BH, Johnson MK, Mendel RR, and Schindelin H. (2004), J. Biol. Chem 279, 34721–34732. [DOI] [PubMed] [Google Scholar]
- (19).Mehta AP, Abdelwahed SH, Fenwick MK, Hazra AB, Taga ME, Zhang Y, Ealick SE, and Begley TP (2015), J. Am. Chem. Soc 137, 10444–10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010), Acta Crystallogr. D66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010), Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004), J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (23).Rekittke I, Wiesner J, Rohrich R, Demmer U, Warkentin E, Xu W, Troschke K, Hintz M, No JH, Duin EC, Oldfield E, Jomaa H, and Ermler U. (2008), J. Am. Chem. Soc 130, 17206–17207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhang Y, Zhu X, Torelli AT, Lee M, Dzikovski B, Koralewski RM, Wang E, Freed J, Krebs C, Ealick SE, and Lin H. (2010), Nature 465, 891–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dreyer MK, and Schulz GE (1996), J. Mol. Biol 259, 458–466. [DOI] [PubMed] [Google Scholar]
- (26).Davenport RC, Bash PA, Seaton BA, Karplus M, Petsko GA, and Ringe D. (1991), Biochemistry 30, 5821–5826. [DOI] [PubMed] [Google Scholar]
- (27).Komives EA, Chang LC, Lolis E, Tilton RF, Petsko GA, and Knowles JR (1991), Biochemistry 30, 3011–3019. [DOI] [PubMed] [Google Scholar]
- (28).Noble ME, Zeelen JP, and Wierenga RK (1993), Proteins 16, 311–326. [DOI] [PubMed] [Google Scholar]
- (29).Marks GT, Harris TK, Massiah MA, Mildvan AS, and Harrison DH (2001), Biochemistry 40, 6805–6818. [DOI] [PubMed] [Google Scholar]
- (30).Hall DR, Leonard GA, Reed CD, Watt CI, Berry A, and Hunter WN (1999), J. Mol. Biol 287, 383–394. [DOI] [PubMed] [Google Scholar]
- (31).Go MK, Koudelka A, Amyes TL, and Richard JP (2010), Biochemistry 49, 5377–5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Jogl G, Rozovsky S, McDermott AE, and Tong L. (2003), Proc. Natl. Acad. Sci. U. S. A 100, 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lodi PJ, and Knowles JR (1991), Biochemistry 30, 6948–6956. [DOI] [PubMed] [Google Scholar]
- (34).Fessner W-D, Schneider A, Held H, Sinerius G, Walter C, Hixon M, and Schloss JV (1996), Angew. Chem. Int. Ed. Engl 35, 2219–2221. [Google Scholar]
- (35).Kursula I, Partanen S, Lambeir AM, Antonov DM, Augustyns K, and Wierenga RK (2001), Eur. J. Biochem 268, 5189–5196. [DOI] [PubMed] [Google Scholar]
- (36).Plater AR, Zgiby SM, Thomson GJ, Qamar S, Wharton CW, and Berry A. (1999), J. Mol. Biol 285, 843–855. [DOI] [PubMed] [Google Scholar]
- (37).Joerger AC, Gosse C, Fessner WD, and Schulz GE (2000), Biochemistry 39, 6033–6041. [DOI] [PubMed] [Google Scholar]
- (38).Saadat D, and Harrison DH (1999), Structure 7, 309–317. [DOI] [PubMed] [Google Scholar]
- (39).Lolis E, and Petsko GA (1990), Biochemistry 29, 6619–6625. [DOI] [PubMed] [Google Scholar]
- (40).Pompliano DL, Peyman A, and Knowles JR (1990), Biochemistry 29, 3186–3194. [DOI] [PubMed] [Google Scholar]
- (41).Layer G, Moser J, Heinz DW, Jahn D, and Schubert WD (2003), EMBO J. 22, 6214–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Walsby CJ, Ortillo D, Broderick WE, Broderick JB, and Hoffman BM (2002), J. Am. Chem. Soc 124, 11270–11271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.