Abstract
Genome editing employs targeted nucleases as powerful tools to precisely alter the genome of target cells and regulate functional genes. Various strategies have been risen so far as the molecular scissors-mediated genome editing that includes zinc finger nuclease, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats—CRISPR-related protein 9. These tools allow researchers to understand the basics of manipulating the genome, create animal models to study human diseases, understand host–pathogen interactions and design disease targets. Targeted genome modification utilizing RNA-guided nucleases are of recent curiosity, as it is a fast and effective strategy that enables the researchers to manipulate the gene of interest, carry out functional studies, understand the molecular basis of the disease and design targeted therapies. CRISPR-Cas9, a bacterial defense system employed against viruses, consists of a single-strand RNA-guided Cas9 nuclease connected to the corresponding complementary target sequence. This powerful and versatile tool has gained tremendous attention among the researchers, owing to its ability to correct genetic disorders. To help illustrate the potential of this gene editor in unexplored corners of oncology, we describe the history of CRISPR-Cas9, its rapid progression in cancer research as well as future perspectives.
Keywords: CRISPR-Cas9, Genome editing, Glioblastoma multiforme, gRNA, Oncological research
Introduction
Genome/gene editing involves modification of genes in an organism using enzymes specific to the target gene sequence. This technique deals with grouping of advancements that enable researchers to change a living organism’s DNA [1]. These innovations permit hereditary material to be either included, expelled, or changed at specific areas in the genome [2]. A few ways to deal with genome altering include zinc finger nuclease, transcription activator-like effector nucleases, etc. A recent interest of most of the researchers is CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats and CRISPR associated protein-9), discovered in the year 2012 by Jennifer Doudna and Emmanuelle Charpentier, University of California Berkeley and University of Vienna [3]. The CRISPR-Cas9 framework has produced a great curiosity among researchers, since they are quicker, less expensive, increasingly precise, and more effective than other existing genome altering strategies [4].
As of now, based on the CRISPR database, CRISPR structures were found to be present in 202 (870 CRISPR sequences were found) out of 232 archaeal genomes analyzed and 3059 (8069 CRISPR sequences were found) out of 6782 bacterial genomes analyzed (https://crispr.i2bc.paris-saclay.fr/). Till date, CRISPR has provided a clear idea on how a gene product contributes to development as well as disease in an organism [5]. To demonstrate “what actually CRISPR is” and “the steps towards fulfilling the genome editor’s potential in cancer treatment” we include in this review, the emerging oncology-based applications of CRISPR, their role in cancer diagnosis and the future impact of this tool in medicine and diagnostics.
A Crispy History of CRISPR Technology
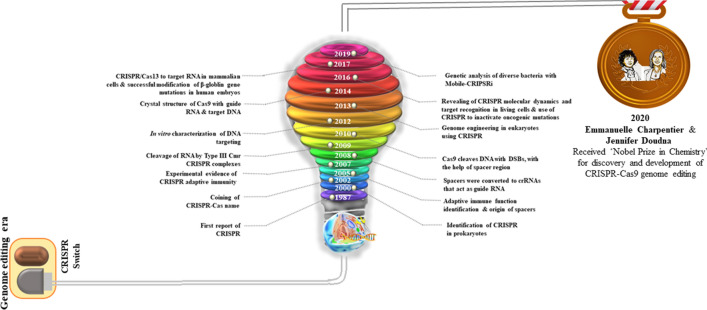
Initially, CRISPR was identified 30 years ago in Escherichia coli during the investigation of the gene responsible for isozyme conversion of alkaline phosphatase [6] (Fig. 1). At that point, it was not really conceivable to anticipate the function of these repeated sequences because of the absence of adequate DNA sequencing data. The genuine capacity of this sequence stayed mysterious until the mid-2000s. In 1993, CRISPRs were seen for the first time in Achaea, explicitly in Haloferax mediterranei [7], and, was further identified in an expanding number of bacterial and archaeal genomes. In the mid-2000s, the revelation of sequence similarity between the spacer areas of CRISPRs with the sequences of bacteriophages, archaea, and plasmids shed light on the exempting function of CRISPR as an immune system. Alongside, in 2002 certain genes that were proposed to encode DNA repair proteins for hyper thermophilic archaea were recognized to be closely associated with CRISPR and were assigned Cas (CRISPR-Associated) genes [8]. Relative genomic studies in this way recommended that CRISPR and Cas proteins work together, thereby comprising a putative acquired immunity framework to secure prokaryotic cells against attacking infections and plasmids, similar to the eukaryotic RNA interference (RNAi) framework [9].
Fig. 1.
Timeline of CRISPR-Cas9 technology
The capacity of the CRISPR-Cas framework as a prokaryotic acquired immunity framework was at last tentatively demonstrated in 2007, employing the lactic acid bacterium Streptococcus thermophilus [10]. Inclusion of the phage sequence into the spacer portion of the CRISPR of S. thermophilus made this strain impervious to the corresponding phage. Nevertheless, this bacterial resistance to the phage vanished when the protospacer sequence was deleted from the phage genome. Furthermore, the CRISPR-Cas sequence of S. thermophilus expressed in E. coli demonstrated heterologous protection against plasmid transformation and provided immunity against phage infections [11]. The CRISPR-Cas9 sequence of Streptococcus pyogenes was then employed to carry out genome editing in human nerve cells and kidney cells of mouse [12, 13]. Consequently, CRISPR-Cas tool was widely referred to as an acquired immunity system in prokaryotes.
Process of CRISPR-Cas Mediated Acquired Immune System
CRISPR-Cas mediated-acquired immunity basically involves three main stages namely, adaptation, expression and interference [10]. During the adaptation stage, the invading DNA from a virus/phage will be recognized by the Cas proteins, followed by fragmentation and incorporation of the fragments into the spacer region of CRISPR. Transcription of the modified CRISPR region occurs at the expression stage resulting in pre-CRISPR RNA (pre-crRNA), which is further processed into smaller units, producing crRNA through the action of Cas- endonucleases. At the interference stage, by exploiting the homology of the spacer arrangement present in crRNA, the foreign DNA is caught, and a complex with Cas protein having nuclease activity degrades the exogenous DNA (Fig. 2) [14].
Fig. 2.
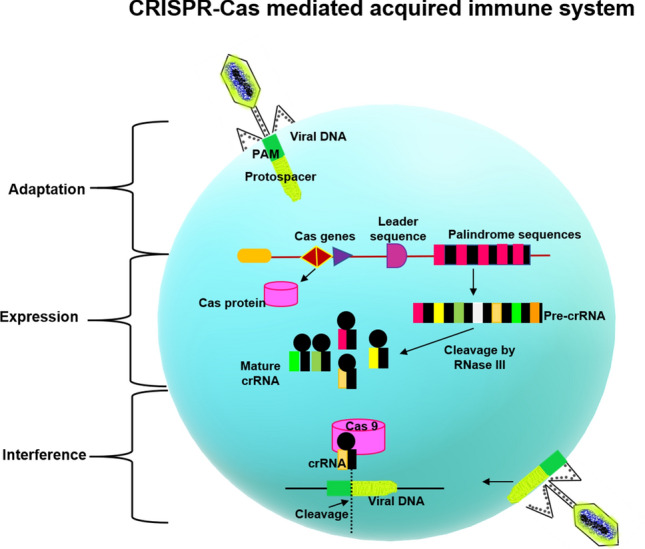
CRISPR-Cas mediated acquired immune system
Components of CRISPR-Cas9
The CRISPR-Cas9 framework comprises two key components that introduces a change into the target DNA: The protein called Cas9, goes about as 'molecular scissors', that can cut the two strands of DNA at a particular region in the genome and a small sequence of RNA called guide RNA (gRNA), a pre-designed RNA succession (around 20 bases complementary to the target DNA) situated inside a long RNA framework [15]. The gRNA is intended to discover and bind to a particular sequence in the DNA. The enzyme, Cas9 follows the gRNA to the similar area and makes a cut across the two strands of the DNA [15]. The only prerequisite for proper binding of Cas-gRNA complex is the Protospacer adjacent motif (PAM). The PAM motif is recognized by the PAM binding domain of the Cas9 protein, thereby creating a double stranded break on both the DNA strands [16]. At this stage, the cell identifies the damaged DNA and attempts to fix it, either by non-homology end joining (NHEJ) or homology-directed repair (HDR) [16]. Among these two methods, NHEJ often leads to insertion or deletion mutations during repair and is often considered to be an error-prone repair mechanism [17]. In contrast to this strategy, HDR, during the process of reconstruction of cleaved DNA, makes use of the donor DNA template to introduce mutations [18]. Researchers therefore utilize the HDR mechanism to bring about changes within one or more genes in the genome of a cell of interest for precise editing. To design sgRNA libraries and to investigate pooled CRISPR-Cas9 screens, several web-based and bioinformatics resources have been created. Some of the frequently employed tools are listed (Table 1).
Table 1.
Tools for designing sgRNA libraries
Unique Advantages of CRISPR in Cancer Therapy
It is well known that cancer is a complex disease and despites decades of research the statistics of cancer-related deaths is not spiking down. This unmet scientific need urges more effective and efficient therapeutic strategies. In addition to being a powerful research tool in cell and gene therapy, CRIPSR-Cas9 technology holds trump card in cancer treatment as well (Fig. 3). One possible attribute might be due to the regulation of CRIPSR-Cas9 on endogenous gene expression. As stated above, the gRNAs can recruit the inactive Cas9 protein to target specific DNA sites [19]. In the bound state, transcriptional inhibition or activation domains can be used to either inhibit or activate the expression of target genes in tumor cells [20]. Besides, in several cancers such as Ewing sarcoma, acute myeloblastic leukemia and tumors like Glioblastoma Multiforme (GBM), the epigenetic factors play a predominant role in cancer progression [21]. Hence by targeting the epigenetic machinery and its regulatory components, it would be plausible to deregulate cancer growth. This stratagem is also possible with CRISPR-Cas9, whereby the Cas9 protein can be tethered to either histone modifiers or other DNA methylation regulatory proteins to perform the epic ‘epigenome editing’ [22]. Nevertheless, CRISPR-Cas9 tools can also be employed to target the tumor makers in cancer cells directly, thereby offering an opportunity to eliminate tumor proliferation and invasion [23]. Also, in order to test the efficacy of drugs for optimizing the chemotherapy regimen in cancer treatment, in vitro and in vivo animal models that recapitulate the actual tumor microenvironment in patient setting is an obligatory requirement [24, 25]. Obviously, construction of such models would be expensive and time consuming. The advent of CRISPR-Cas9 forefronts has made this tedious job quite simpler and cheaper than the standard protocols. The best example to illustrate the essential role played by CRISPR-Cas9 technology in drug discovery process in oncology would be in ovarian cancer mouse model ID8, whereby CRIPSR-Cas9 mediated knockout of BRCA2 and TP53 genes, increased their sensitivity to the PARP inhibitor, rucaparib [26].
Fig. 3.
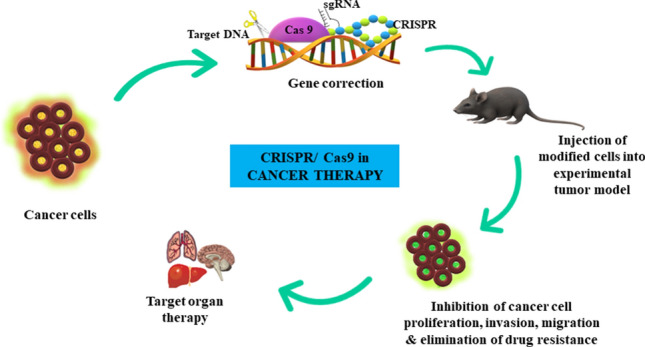
CRISPR/Cas9 in cancer therapy
As such, this tool offers many advantages over the traditional TALENs and ZFNs. These include easy prediction of off-target sites, precise targeting of multiple genome sites simultaneously and easy design of genomic targets. Besides, CRISPR has shown promising results in targeting epigenetic changes occurring during carcinogenesis, which was not possible by the previously discovered ZFNs and TALENs [27].
Applications of CRISPR in Cancer Research
CRISPR in Glioblastoma Multiforme (GBM)
GBM, a heterogenous primary brain tumor is often characterized by dismal prognosis emphasizing an unmet need for its treatment [28]. GBM is characterized by overexpression, dysregulation or activation of various membrane receptors such as epidermal growth factor receptor (EGFR), vascular endothelial growth factor (VEGF), mammalian target of rapamycin (mTOR), platelet derived growth factor receptor (PDGFR), protein kinase inhibitors, etc. [29]. Owing to the novel insights provided by CRISPR-Cas9 technology in various cancers, quest to determine its efficacy on alleviating GBM pathogenesis has now gained immense clinical importance. Huang et al. has recently employed CRISPR-Cas9 tool to knock out specifically EGFR exon 17 in glioma cells. Interestingly, this CRIPSR-mediated knockout increased the expression of UBX domain containing protein-1 (UBXN1), a suppressor of Nuclear Factor-ƙβ (NFƙβ) leading to the inhibition of glioma cell growth [30].
T-cell immune therapy often serves as an alternative to conventional treatment strategy, in the treatment of solid tumors [31]. Chimeric Antigen Receptor (CAR) is one of the most promising platforms in T-cell-based therapies [32]. However, till date CAR-T cells have not been that successful in GBM [33]. Surface expression of programmed cell death ligand 1 (PD-1) is one of the mechanisms, by which GBM cells hamper CAR-T cells. Hence, an attempt was made by Choi et al., employing CRISPR-Cas9 to knockout beta-2-microglobulin (B2M), PD1 and endogenous T-cell receptor (TRAC). The triple-gene edited version of CAR-T cells exhibited enhanced activity than their normal counterparts in a preclinical intracranial xenograft animal model [34]. It has been reported previously that in most of the GBM cases, the vascular basement protein called laminin-411(α4β1γ1) is over-expressed in grade IV gliomas, whereas their expression is comparatively lesser in the lower grades of gliomas [35]. Hence, laminin-411 is considered to be a progression related biomarker in glioma. Employing CRISPR-Cas9 tool, the α4 and β1 chains were knocked down in laminin-411 by Sun et al., in an intracranial mice model. The results showed a significant decrease in tumor growth and volume [35]. Besides, another study by MacLeod et al. showed that genome-wide CRISPR-Cas9 screening was able to assess the genes responsible for modulating the sensitivity of glioma stem cells to chemotherapy. Herein, it was found that the members of SOX family namely, SOCS3, DOT1L and USPS played a major role in GSCs growth and targeting them might improve GBM treatment chemo-resistance as well [36]. In a similar study, large-scale CRISPR-Cas9 loss of function screening of 4,574 genes responsible for membrane trafficking and invasion of GBM was performed. Among the genes studied, the strongest regulator of GBM invasion was found to be MAP4K4. Concomitant targeting of MAP4K4 employing sgRNAs in U138 human GBM resulted in decreased invasion and migration of the cells which insists that MAP4K4 would be a potential target candidate to limit GBM invasion [37]. The RecQ family of DNA helicases plays an important role in DNA replication and recombination [38]. Mutations in these genes result in pre-mature aging and cancer predisposition [39]. Previous studies on GBM pathogenesis have stated that RecQL4 is significantly upregulated both at mRNA and protein levels when compared with normal astrocytes [40]. CRISPR-Cas9 mediated knockout of RecQL4 in LN-18 and U87MG human glioma cells resulted in decreased proliferation and impaired cell viability thereby providing rationale for further elucidation of RecQL4 role in GBM pathogenesis [40]. Therefore, advancing CRISPR against GBM would possibly open up a new therapeutic avenue to combat glioma pathogenesis.
CRISPR in Lung Cancer
Lung cancer, with high morbidity and mortality rate, ranks first among all the malignancies in developed and developing countries such as India, Europe, China and the USA [41]. Various tumor suppressor genes associated with lung cancer include, tumor suppressor gene TP53 [42], Adenomatous polyposis coli (APC) [43], Bromodomain containing 7 (BRD7) [44], Cyclin Dependent Kinase Inhibitor 2A (CDKN2A) [45] and NM23 [46]. Proto-oncogenes constituting lung cancer are receptor tyrosine kinase (RTK) [47], MYC [48], JUN [49], FOS [50], RAF1 [51] and ERBB1&2 [52]. Mutations in tumor suppressor genes and overexpression of proto-oncogenes promoted the development of lung cancer [53]. The robust nature of CRIPSR-Cas9 in editing the genome of a cell was exploited in tumor gene therapy to knockout oncogenes which are mutated or overexpressed such as EGFR [54], NESTIN [55], Insulin-like growth factor 1 factor (IGF1R) [56] and remodeling and spacing factor 1 (RSF1) [57]. Knockout of the oncogenic mutant EGFR allele, using CRISPR-Cas9 repressed the proliferation and growth of lung cancer growth cell lines A549, H1975, and H1650 [58, 59]. In Kirsen Rat Sarcoma viral oncogene homolog (KRAS) mutant, non-small cell lung cancer cells (NSCLC), tenacious DNA damage and increased sensitivity to radio-therapy was observed after knockout of focal adhesion kinase (FAK) gene employing CRISPR-Cas9 tool [60]. Furthermore, it was reported by Liu et al., that knockout of NESTIN in A549 and H1299 lung cancer cells enhanced apoptosis, inhibiting the multiplication, colonization, and invasion of the tumor cells by suppressing their transition from epithelium to the mesenchymal area (EMT) [55]. Coming to its role on tumor suppressor genes, CRISPR-Cas9 gene editing technique is being widely employed to target, repair and restore the function of tumor suppressor genes thereby inhibiting tumorigenesis [61]. Among these, knockout of Keap1 in a KRAS- mouse model of lung adenocarcinoma utilizing CRISPR-Cas9 indicated overactivation of Nrf2 leading to glutaminolysis thereby inhibiting tumor progression [62]. Another study by Xu et al., affirmed that genetic knockout of the tumor-suppressor gene mitofusion 2 (MFN2) in the lung cancer cell line A549 promoted survival, cell viability, and cell growth, and invasion by upregulating the mTORC2/Akt signaling pathway [61]. Also, knockout of the tumor-silencer miR-1304 in human lung adenocarcinoma cells, A549 and NCI-H1975 employing CRISPR-Cas9 significantly increased heme oxygenase-1 (HO-1) expression, promoting the growth and survival of the cells [63].
So far, scarcely any investigations have analyzed tumor suppressor genes and proto-oncogenes in lung cancer utilizing the CRISPR-Cas9 tool, and this ought to be considered as a significant heading for future research. Targeted repair and restoration of the role of tumor-suppressors by utilizing CRISPR-Cas9 technology can possibly shed light on lung cancer treatment.
CRISPR in Breast Cancer
Breast cancer is the second most common type of cancer-related death in women, impacting 2.1 million women each year and 1.2 million cases diagnosed in 2018 [64]. Based on the expression of receptors such as Ki-67, estrogen receptor, ERBB2 and progesterone receptor, it can be sub-divided into triple negative breast cancer, luminal A, luminal B and HER2 enriched [65]. Among these, the estrogen positive breast cancers are very common accounting for 70% of breast cancer cases [66]. To improve prognosis and patient’s outcome, CRISPR-mediated genome editing was employed to elucidate the molecular markers as targets for breast cancer treatment. Treuren et al., used CRISPR-Cas9 to study the effect of Migration and Invasion Enhancer 1(MIEN 1) knockout in breast cancer cell lines namely MDA-MB-231 and its three metastatic variants MDA-MB-831, 1833 and 4175 [67]. However, it was reported that CRISPR-mediated gene deletion did not have any effect on the proliferation and survival of breast cancer cells. Mixed lineage kinase-3 (MLK3), a member of MAP3K pathway mediates signal transduction and activation of the ERK pathway [68]. Downregulation of MLK3 in highly metastatic TNBC-4T1 cells using CRISPR-Cas9 technology led to repression of cellular migration and invasion [69]. Liao et al., has reported that deletion of Ubr5, in a murine triple negative breast cancer model suppressed tumor growth and metastasis and promoted apoptosis in those cells [70]. Several functional proteins are also being investigated for their therapeutic potential against breast cancer. One such study focused on the microtubule associated serine/threonine like (MASTL) protein, which was over-expressed in estrogen receptor positive breast cancer, and contributed to poor prognosis and adverse clinical outcomes [71]. CRISPR-mediated knockout of MASTL in breast cancer cell lines reduced the proliferation of the cells and some therapeutic effects were observed in vivo as well [72]. The SRC family kinases, protein tyrosine phosphatase N23 (PTPN23) and FYN also plays a major role in breast cancer [73, 74]. The effect on CRISPR-Cas9 mediated knockout of both FYN and PTPN23 in vitro in Cal-51 (breast cancer cell line) cells and in vivo in an orthotopic xenograft mice model was performed. It was found that CRISPR-mediated knockout attenuated the growth of cancerous cell in both the models. Interestingly, the authors found that FYN was associated with chemoresistance, as it was over-expressed in the resistant cells when compared with their normal counterparts [74].
With regard to RNA-binding proteins, Zheng et al., employed CRISPR technology to screen PHD finger protein 5A (PHF5A), which is ought to be a key factor in breast cancer proliferation and migration [75]. Another similar report by Yang et al., denoted that PHF5A is a novel oncoprotein in lung adenocarcinoma [76]. Also, Hubert et al., has shown that suppression of PHF5A resulted in a significant decrease in GBM tumor growth in a xenograft animal model [77]. Thus, PHF5A would serve as potent therapeutic target, not only in breast cancer, but in other tumors like lung adenocarcinoma and GBM as well.
Recently, non-coding RNAs (ncRNAs) have gained interest in cancer research, and their dysregulation is associated with carcinogenesis [78]. One such study employing ncRNAs was carried out by Singh et al., in MCF-7 cell line and xenograft-induced animal model. It was observed that abrogation of BC-200, a ncRNA which regulates protein synthesis in estrogen receptor positive breast cancer cells, reduced cancer cell growth by over-expression of Bcl-2 [79]. Adding to this, abrogation of another ncRNA, miR-10b in MDA-MB-231 breast cancer cells, suppressed cancer cell migration as well [80]. These data provide insights on the efficacy of targeting ncRNAs for breast cancer treatment.
CRISPR in Liver Cancer
Liver cancer, which is more common in men, is the fourth main cause of cancer related death worldwide, with a 5-year survival rate of 19% [64]. Hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma are the two molecular subtypes of primary liver cancer [81]. Patients with liver cancer are characterized by poor prognosis, frequent recurrence and limited therapeutic options are available currently [82]. Hence, employing CRISPR technology to sort out potential targets would be a promising approach for liver cancer treatment. The oncogenic role of CXCR-4 a chemokine receptor, in promoting HCC proliferation, survival and migration via activation of P13K/Akt and Ras-MAPK is well-known [83]. CRISPR-mediated knockout of CXCR-4 in HepG2 cells resulted in suppression of cancer cell proliferation and migration. Also, CXCR-4 attenuation resulted in reduced tumor size in vivo [84]. Nuclear receptor coactivator (NCOA5) was also studied as a target for liver cancer in an in vitro study employing HCC cells. It was found that knockout of NCOA5 inhibited the migration of cells and suppressed the epithelial to mesenchymal transition (EMT) as well [85]. These aforementioned targets can be considered for elucidation of their potential in vivo or in clinical trials for liver cancer treatment.
CRISPR in Colorectal Cancer
Colorectal cancer (CRC) develops through a series selection of genetic changes. The initiating or gate-keeping mutation is primarily 2-hit mutations in APC, which occur in > 80% of the cases [86]. Subsequent activating mutations in KRAS occur in the early to intermediate adenoma stage, while loss-of-function mutations in SMAD4 and TP53 occur in the intermediate to late adenoma and late adenoma to adenocarcinoma stages, respectively [87, 88]. In addition to the commonly mutated APC, KRAS, mothers against decapentaplegic homolog 4 (SMAD4), and tumor protein 53 (TP53) genes, large-scale genome sequencing studies have identified numerous other genes that are infrequently mutated in CRC [89], revealing extensive inter and intra tumor genetic heterogeneity in cancer tissues. These complications in CRC raise the need for new technologies which can overcome problems of genetic factors. In the recent years, CRISPR gene editing has been employed to overcome the problem of genetic mutation that enhance chemoresistance and tumor progression in CRCs.
Takeda et al., has performed CRISPR-Cas9 screening and validated in organoids derived from intestinal tumors that developed in mice carrying a heterozygous loss-of-function mutation in APC and an activating mutation in KRAS. They have reported that co-occurrent mutations in receptors for activin and transforming growth factor-β (TGF-β) synergistically promote tumorigenesis, and shed light on the role of activin receptors in CRC which can be targeted for CRC treatment [90]. A study by Li et al. has reported the role of CD133 in colon cancer, established an in-vitro model by knocking out CD133 in colon cancer (LoVo) cell line using CRISPR-Cas9 gene editing system. The authors have suggested that knocking out of CD133 contributed to attenuate the abilities of colon cancer cells, including colony formation, cell proliferation, migration, and invasion. Surprisingly, CD133-knockout cells showed remarkable suppression of tumorigenicity of CRC, including cell proliferation and colony formation capacities, migration, and invasion. Therefore, modulation CD133 expression could be an effective treatment for the CD133 + group of patients [91]. Recently, Matano et al. has used the CRISPR-Cas9 genome-editing system to introduce multiple mutations into organoids derived from normal human intestinal epithelium. By modulating the culture conditions to mimic that of the intestinal niche, they selected isogenic organoids harboring mutations in the tumor suppressor genes APC, SMAD4 and TP53, and in the oncogenes KRAS and/or PIK3CA [92]. Their results suggest that multiple driver gene mutations contribute to niche-independent stem cell maintenance but not to metastatic progression of CRC tumors in mice. However, further studies will be required to elucidate the functional contribution of other molecular abrasions, such as copy number variations or epigenetic alterations and/ or aneuploidy in the malignant progression of human CRC in this system.
CRISPR in Pancreatic Cancer
Pancreatic ductal adenocarcinoma (PDAC) accounts for > 85% of all cases of pancreatic cancer. For all the stages combined, the first and fifth years relative survival rates are 28% and 7%, respectively [93]. Though there have been substantial advances in the development of novel treatment strategies such as targeted therapies directed against molecular alterations arising in cancer cells, PDAC still remains a lethal disease [94]. In this context, a study by Christina et al., revealed that, a novel microRNA factor, fork-head box protein A2 (FOXA2), acts as a tumor suppressor gene in pancreatic cancer (PC), affecting PC proliferation and invasiveness. CRISPR-Cas9 mediated knockout of FOXA2 resulted in enhanced tumor growth in a PC xenograft model, thereby highlighting the role of FOXA2 in alleviating PC pathogenesis [95]. Another study by Watanabe et al., clarified the prognostic impact of Lysine-specific demethylase 6A (KDM6A) deficiency on PDAC and the therapeutic potential of this subtype by knocking out KDM6A gene by CRISPR-Cas9. The study demonstrated that KDM6A deficiency is a characteristic of the malignant subtype of PDAC, which contributes to epigenetic inactivation of CDKN1A through the modulation of histone acetylation. Thus, KDM6A expression can be used as a prognostic biomarker in PDACs [96].
CRISPR in Ovarian Cancer
Ovarian cancer (OC) is the fifth most lethal cancer affecting women worldwide with a high incidence of 10–15 out of 100,000 females being affected worldwide [97]. Though most of the patients are diagnosed at an early stage, epithelial ovarian cancer (EOC) remains the most lethal gynaecologic cancer [98]. This is mainly attributed to the fact that nearly 70% of the patients relapse even after receiving optimal cytoreductive surgery (CRS) along with chemotherapy. This leads to a low 5-year survival rate among OC patients [99].
B-cell-specific Moloney murine leukemia virus integration site 1 (BMI1), a main constituent of Polycomb group proteins (PcG), is an important mediator of epigenetic modulation in vital cellular events and has been reported to be present in many human malignancies [100]. In a study by Zhao. Q et. al., the BMI1 gene was knocked out by CRISPR mediated gene editing to identify the molecular traits in EOC in both in-vitro (SKOV3) and in-vivo (BALB/c mice) models. They reported that the knockout of BMI1, responsible for the regulation of PI3K/ AKT pathway could be a novel prognostic biomarker for EOC [101].
Recent studies have used the CRISPR-Cas9 tool to elucidate synergistic gene knockouts, to overcome drug resistance in various cancers. A study in 2012 by Alan et. al., employed CombiGEM-CRISPR to create a library of 23,409 barcoded g-RNA combinations for EOC. Using these combinations, high-throughput pooled screening was performed to identify the gene pairs that inhibited proliferation of EOC. The study showed that the knockout of dual genes, using CRISPR-cas9 reduced the proliferation in OVCAR8 human PC cells. [102]. This dual knockout of genes can be used as a treatment strategy in targeted gene therapy.
Applications of CRISPR in Other Fields of Medicine
Even before elucidation of their use in genome editing, CRISPR loci was employed as genetic markers for genotyping, species identification, introducing transcriptional and epigenetic modifications [5]. For example, to trace out the microevolution of Yersinia pestis, the causative agent of plaque, CRISPR tool was used for genotyping [103]. Liu et al., reported that CRISPR tool was able to accurately differentiate the strains of the same serotype of Salmonella, which was not possible by the conventional serotyping techniques such as Amplified Fragment Length Polymorphism (AFLP), Multiple locus variable number tandem repeat analysis (MLVA), etc. [104]. This novel tool was employed to produce sequence-specific antimicrobial agents that can cleave genomes of pathogenic bacterial strains. For instance, a mouse skin colonization model was created by Bikard et al., so as to show that CRISPR-Cas treatment can be utilized to specifically inhibit Staphylococci. Undoubtedly, a significant decrease in bacterial growth was observed, highlighting their reliability as a valuable remedy for the rising tide of antibiotic resistant strains such as β-lactam and vancomycin resistant Staphylococci [105]. Nevertheless, Cas9 variants have emerged as toolboxes to induce specific single nucleotide changes within the genome. These variants rely on Cas9- nickases coupled to the enzyme, cytidine deaminase to bring about single nucleotide transitions such as G to A or C to T. One such work was reported by Komor et al., wherein the target base, cytidine was directly converted to uridine without any double stranded breaks in the DNA by fusing Cas9 protein with cytidine deaminase domain thereby acting as a key to open door for correction of variety of point mutations in human diseases [106].
CRISPR to Create Animal Models for Human Diseases
Disease modeling was one of the early uses of CRISPR-Cas9 in medical research. CRISPR-Cas9 can also be used to represent multi-genic disorders more easily than traditional transgenic systems [107]. Cancer, for instance, is a disorder resulting in various hereditary changes. Establishment of cancer models through the conventional transgenic procedures regularly is laborious and tedious. CRISPR-Cas9, in contrast, can legitimately induce somatic mutations in mice more efficiently than the transgenic systems [108, 109]. Duchenne muscular dystrophy (DMD), for instance, is brought about by mutation in the gene encoding for dystrophin. By focusing on two exons at the same time in the rat DMD gene with CRISPR-Cas9, Nakamura et al., over-ruled the expression of dystrophin in rats, thereby establishing a rat model of DMD [110]. Similar work was carried out by Xue et al., wherein CRISPR-Cas9 segments inserted into the mice through tail vein hydrodynamic infusion, effectively prompted both gain-of-function and loss-of-function (LOF) mutations in mouse liver cells which were responsible for hepatocarcinogenesis [108].
CRISPR Altered T-cells for Cancer Therapy
The most inspiring area in the utilizations of CRISPR-Cas9 is its potential for gene therapy. Only a while after its introduction into mammalian cells [12], CRISPR-Cas9 exhibited its potential in gene therapy by inducing mutations in HIV-1 virus so as to reduce its expression in human T-cells [111]. From that point forward, many efforts have been made to investigate the efficacy of CRISPR-Cas9 in battling infectious diseases and to induce protective mutations in the host cells as well [112].
At the point when it was revealed in 2012, researchers had incredible expectations that this gene editing tool could treat or fix hundreds to thousands of hereditary disorders. As a step towards it, from last year, specialists in the USA started testing its efficacy in individuals, a significant initial phase in deciding if the innovation can satisfy its medicinal potentiality. These clinical trials are trying to elucidate the safety and efficiency of CRISPR-Cas9 against blood disorders [113], cancer and patients with visual impairment [114]. Many such preliminaries are awaited upon to start soon.
Results from the specialists at the University of Pennsylvania, about the completed clinical trials, highlighted that “CRISPR treatment intended to enhance the efficacy of T-cells, the cancer-fighting immune cells was safe”. The outcomes are from three patients -two with myeloma and one with sarcoma—whose T-cells were removed and modified in the lab. CRISPR crippled genes in the T-cells, and equipped the cells with a “warhead”—a gene intended to guide the T-cells to tumor cells that have a particular protein on their surfaces [115]. Previously, genetically engineered cells called CAR T-cells, have been utilized in patients for quite a long time to fight cancer [116]; enhancing T-cells’ cancer fighting ability with the assistance of CRISPR is another interesting development. These findings were presented on December 7, 2019 at the American Society of Hematology meeting in Orlando, Fla. It was demonstrated that CRISPR-altered T-cells grabbed hold and was reproduced in the patients employed for the study. None of the three individuals had any side effects related with the altered cells [115]. This indeed is an uplifting news since the T-cell-based modifications to fight cancer, have caused high fevers, low pulse, seizures and adverse side effects. Though the treatment didn't have a significant impact on reducing the development of the patients' tumors, their safety and feasibility has been demonstrated through this study. Scientists in the mere future will be substantially more centered around the effectiveness in employing CRISPR-altered T-cells for therapy.
Adding on to the soup, similar trial on CRISPR-altered T-cells is under progress in China. Furthermore, CRISPR Therapeutics, a Cambridge, Mass-based organization, hopes to start three clinical trials in which altered T-cells target blood and kidney cancers [113].
CRISPR-Cas Systems in Diagnostics
CRISPR-Cas systems have been utilized by various researchers working on disease diagnostics as well. Pardee et al., combined CRISPR-Cas9 with nucleic acid sequence-based amplification (NASBA), an isothermal enhancement method, to precisely differentiate the closely related Zika infection strains in vitro and in a macaque model. The team attached a synthetic trigger sequence to NASBA-amplified viral RNA and utilized a sgRNA-Cas9 complex to create a strand break in the subsequent dsDNA. The presence or absence of a strain-specific PAM brought about either truncated or full-length DNA segments upon Cas9 cleavage. Full-length strands, enacted the trigger switch, which prompted a change in color on a paper-based sensor, enabling strain differentiation in vitro [117]. In another study, Muller et al., compiled CRISPR-Cas9 with optical DNA mapping to detect antibiotic resistance genes in bacteria. In this, guide RNA (gRNA)-Cas9 complex binds and cleaves nucleic acid sequences of plasmids containing genes conferring resistance and a fluorescent dye (YOYO-1) and netropsin freely binds to the DNA at AT-rich areas specifically, bringing about an emission intensity which is unique to every DNA fragment. Through this assay, researchers can recognize plasmids that confer an extended-spectrum β-lactamases (ESBLs), including cefotaxime 15 (CTX-M-15) and CTX-M-14 and carbapenemases including Klebsiella pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase 1 (NDM-1) [118]. Guk et al., coupled CRISPR-Cas9 with DNA fluorescent in situ hybridization (FISH) to identify methicillin-resistant Staphylococcus aureus (MRSA). This strategy uses a dead CRISPR associated protein (dCas9) framework wherein a sgRNA-dCas9 complex combined with a SYBR green I fluorescent test identifies the gene mecA of MRSA. While the complex detects the target DNA sequence, dCas9 does not induce DNA cleavage, subsequently making it reasonable for detection by FISH. This test can recognize MRSA at a concentration of 10 CFU/ml and can specifically distinguish S. aureus isolated with and without the mecA gene [119]. Further, CRISPR-Cas12 entered into genome editing era wherein, it was employed in a technique called DNA-endonuclease targeted CRISPR trans reporter (DETECTR) for detection of human papillomavirus from patients [120]. Another high-throughput technique combining CRISPR-Cas12 with a fluorescent-based point of care technology for accurate detection of African Swine Fever Virus (ASFV) has been developed. In this system, once the cas12a/crRNA detects and precisely binds to the target DNA, the cas12a/crRNA/DNA complex gets activated, degrading the fluorescent reporter [121]. Currently, CRIPSR-Cas13-based platforms, which integrates an isothermal- RPA (recombinase polymerase amplification) or a reverse transcription- RPA with the nuclease cas13a, termed as SHERLOCK (specific high-sensitivity enzyme reporter unlocking) has been developed. This cas13/crRNA complex, binds and targets the nucleic acids of pathogens along with the cleavage of a nontarget RNA-coupled fluorescence reporter, resulting in the emission of fluorescent signal which can be detected real-time. Employing SHERLOCK technique, Gootenberg has identified Escherichia coli, Pseudomonas aeruginosa, and has differentiated these strains from Mycobacterium tuberculosis, Staphylococcus aureus and Klebsiella pneumoniae [122]. The SHERLOCK technique along with heating unextracted diagnostic samples to obliterate nucleases (HUDSON) was employed by Myhrvold et al., which allows rapid detection of dengue virus, within 2 h from the body fluids of patients such as saliva, serum and whole blood [123]. COVID-19, which out-broke in November 2019, has become a pandemic and has spread globally. RT-PCR is the usual diagnostic method; however, inadequate access to reagents and increasing count of patients has put forth a vital need for easily affordable and highly sensitive techniques. One such protocol employing SHERLOCK to quickly diagnose SARS-CoV-2 was put forth by Feng Zhang et al. This technique involves isolation of RNA from patient sample followed by isothermal amplification, cas13 detection of target RNA sequence and visual reading of the reporter signal [124]. Besides, Broughton et al., has introduced a quick and accurate CRISPR-Cas12-based lateral flow assay to detect SARS-CoV-2 employing primers targeting the nucleoprotein and envelope of the virus [125]. Much more recently, a paper-based strip test known as Feluda, to detect and target SARS-CoV-2 within 30 min has been developed by CSIR-IGIB (Institute of Genomics and Integrative Biology) and Tata group. This Feluda test deploys CRISPR-Cas9-based forefront to detect the virus with about 98% specificity and 96% sensitivity. When compared with the qRT-PCR test and antigen-based tests, this CRISPR-Cas9-based paper-strips would be a “game changer” as these are of affordable cost (Rs. 500/kit), more rapid (results would return in an hour), highly specific and accurate, which would rule-out the false positive/negative results as well [126].
Challenges in Employing CRISPR/Cas Technology in cancer Treatment
As the saying “Every advantage has its own disadvantage” goes by, though CRISPR-based systems have laid a profound podium for cancer treatment, there are a lot of factors yet to be defined to increase its efficacy, especially if it is intended to treat cancer patients. First and foremost, the off-target effect of CRIPSR system makes it difficult for the researchers to target a specific genomic locus [127, 128]. This is because, CRISPR-based editing usually creates indels at undesired loci of the genome [125, 126]. Continuation of genetic modification at that point would obviously raise the risk of toxicity in adjacent normal cells and unwanted mutations. Hence, while designing CRISPR-based technology in oncology research, it is detrimental to identify and take control over the off-target events. In this regard, Kleinstiver et al., has formulated the ways by which Cas9 protein can be modified to enhance the specific recognition of target DNA and maintain specificity of CRISPR-based genome editing, which are under evaluation currently [129, 130]. Many such studies concerning ways to minimize the off-target effects are warranted in future. Besides, the editing efficiency is yet another hurdle which has to be sorted out before implementing CRISPR-based therapeutics for oncological therapy. The efficiency of NHEJ and HDR in repair of double stranded DNA break is one among the important determining factors for overall gene editing efficiency. The efficacy of NHEJ and HDR in DNA repair varies for different cell types. Broadly speaking, NHEJ is an error-prone mechanism, which incorporates indels at the site of cleavage; HDR, on the other side, replaces the target gene with a recombinant alternative sequence. However, HDR template requires a viral or non-viral vector for translocation into the nucleus [131]. Hence, strategies such as nanocarriers, to enhance the productivity of HDR template delivery into the nucleus along with co-delivery of CRISPR components would increase the specificity and efficiency of gene editing or gene correction in tumors. Though the off-target effect and efficient delivery is resolved, ‘how fit the edited cells are?’ is another unanswered question [132]. If the CRISPR- edited cells possess great adaptability and proliferation rate than the unedited counterparts, then these edited ones can reach the therapeutic threshold essential for fruitful treatment upshots. However, if the editing efficiency is low, or the edited cells are not able to adapt themselves like the unedited ones, this pitfall will have a huge effect on the expected therapeutic outcome. Hence, modification of the genome of the edited cells in vitro, followed by reinfusion of the cells back to the patients would help to overcome this obstacle at least partially. Last but not least, the immune response provoked by the Cas9 protein upon entry into patient’s body is another limiting factor. This is due to the presence of short peptides on the surface of Cas9 which acts as epitopes that can bind to the MHC molecules. In fact, previous literature has reported that the immunogenicity caused by CRISPR-Cas9 technique was the prime factor for destabilization of the host cell [133]. Hence, strategies to minimize the host response following delivery of CRISPR-Cas system has to be framed before carrying this system to patient setting.
Future Perspectives
CRISPR-based forefronts have turned out be crucial for advancements in science and cancer research. This is because of their wide implementation in basic research to understand the queries concerning how genes function and in the designing of therapies against diseases like cancer. In any case, there are yet a few difficulties related with this tool that should be addressed too. For instance, adeno-associated viral vectors (AVV) are commonly employed to deliver Cas9 components into the host cell. Though these vectors are serotype-associated target specific and show reduced carcinogenic risk, they have a maximum carrying capacity of 4.7 kB only, whereas, the size of Cas9 gene is 4–7 kB [134]. This huge size of Cas9 makes it hard to pack the protein in low immunogenic AVV utilized in vivo and in vitro gene delivery. Additionally, Cas9 from S. aureus and S. pyogenes has been appeared to cause irresistible and infectious ailments in clinical trials [135]. One potential technique to surpass this issue is to update Cas9 or utilize an alternate bacterial protein that can get away from the host immune reactions. Intellia Therapeutics has built up a lipid-nanoparticle-based CRISPR-Cas9 framework, to modify genes in rodents and non-human primates. With the birth of the "CRISPR babies" in November 2018 [136], it is the responsibility of the researchers to respond to the challenges of CRISPR in this era of genetic inequality and its long-term implications. Despite the fact that these difficulties endure, CRISPR-based advancements hold huge potential and are an incredible expansion to the genome altering tool compartment for the improvement of biotherapies and betterment of humanity.
Conclusion
When compared with the past genome altering techniques, this short RNA-guided Cas9 nuclease framework has a few points of interest namely, wide genome availability, capacity to target multiple sites in a single step, and simplicity in designing of the targets. Regardless of the challenges in utilization of the CRISPR-Cas9 framework for in vivo investigations, scientists have already started to work on this framework to build up animal models for various diseases and to figure out the function of genes. To address their contributions in oncological field, the CRISPR-Cas9 framework will probably offer a novel and less tedious methodology for gene therapy, regulation of gene expression, and new wave of drug targets. In the mere future, CRISPR will slash to identify life-threatening diseases and provide roadmap to develop vaccines, thereby “nearing the beginning of the end of cancers, infectious diseases and hereditary disorders”.
Acknowledgement
We would like to thank the Faculty of Central Inter-Disciplinary Research Facility, Sri Balaji Vidyapeeth for giving us constant support in drafting this review article.
Authors’ Contributions
All the authors contributed equally to the conception and design of this review article. TSA—conceptualized the idea for this review article and critically revised the work. PSD and KSS—performed the literature search and drafted the review article.
Funding
This review article did not receive any funding.
Data Availability
Not applicable.
Compliance with Ethical Standards
Conflict of interest
The authors have no conflict of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holman CM. A Fractured International Response to CRISPR-Enabled Gene Editing of Agricultural Products. Biotechnology Law Report. 2019;38(1):3–23. doi: 10.1089/blr.2019.29100.cmh. [DOI] [Google Scholar]
- 2.Howard HC, van El CG, Forzano F, Radojkovic D, Rial-Sebbag E, de Wert G, Cornel MC. One small edit for humans, one giant edit for humankind? Points and questions to consider for a responsible way forward for gene editing in humans. European Journal of Human Genetics. 2018;26(1):1–11. doi: 10.1038/s41431-017-0024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 4.Lino CA, Harper JC, Carney JP, Timlin JA. Delivering CRISPR: a review of the challenges and approaches. Drug Delivery. 2018;25(1):1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology. 1987;169(12):5429–5433. doi: 10.1128/JB.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mojica FJM, Juez G, Rodriguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Molecular Microbiology. 1993;9(3):613–621. doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- 8.Jansen R, van Embden JDA, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Molecular Microbiology. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 9.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biology Direct. 2006;1(1):7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Horvath P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 11.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Research. 2011;39(21):9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Zhang F. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Church GM. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrangou R, Marraffini LA. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Molecular cell. 2014;54(2):234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy. Nucleic Acids. 2015;4(11):e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta RM, Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. Journal of Clinical Investigation. 2014;124(10):4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodgers K, McVey M. Error-prone repair of DNA double-strand breaks. Journal of cellular physiology. 2016;231(1):15–24. doi: 10.1002/jcp.25053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang XD, Gao F, Liu MJ, Fan QL, Chen DK, Ma WT. Methods for Enhancing Clustered Regularly Interspaced Short Palindromic Repeats/Cas9-Mediated Homology-Directed Repair Efficiency. Frontiers in Genetics. 2019;10:551. doi: 10.3389/fgene.2019.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature methods. 2013;10(8):741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Huang B. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell. 2013;155(7):1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawlor ER, Thiele CJ. Epigenetic changes in pediatric solid tumors: promising new targets. Clinical cancer research. 2012;18(10):2768–2779. doi: 10.1158/1078-0432.CCR-11-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Gersbach CA. CRISPR–Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nature biotechnology. 2017;35(6):561–568. doi: 10.1038/nbt.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Felsher DW. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 24.Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Frontiers in Bioengineering and Biotechnology. 2016;4:12. doi: 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi R, Perkins G. Animal models for studying tumor microenvironment (TME) and resistance to lymphocytic infiltration. Cancer Biology & Therapy. 2018;19(9):745–754. doi: 10.1080/15384047.2018.1470722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walton J, Blagih J, Ennis D, Leung E, Dowson S, Farquharson M, McNeish IA. CRISPR/Cas9-mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer research. 2016;76(20):6118–6129. doi: 10.1158/0008-5472.CAN-16-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachdeva M, Sachdeva N, Pal M, Gupta N, Khan IA, Majumdar M, Tiwari A. CRISPR/Cas9: molecular tool for gene therapy to target genome and epigenome in the treatment of lung cancer. Cancer Gene Therapy. 2015;22(11):509–517. doi: 10.1038/cgt.2015.54. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh M, Shubham S, Mandal K, Trivedi V, Chauhan R, Naseera S. Survival and prognostic factors for glioblastoma multiforme: Retrospective single-institutional study. Indian Journal of Cancer. 2017;54(1):362. doi: 10.4103/ijc.IJC_157_17. [DOI] [PubMed] [Google Scholar]
- 29.Wayne A, Burgess A, Kaye AH, Morokoff A. Complexities of lysophospholipid signalling in glioblastoma. Journal of Clinical Neuroscience. 2014;21(6):893–898. doi: 10.1016/j.jocn.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Huang K, Yang C, Wang Q, Li Y, Fang C, Tan Y, Kang C. The CRISPR/Cas9 system targeting EGFR exon 17 abrogates NF-κB activation via epigenetic modulation of UBXN1 in EGFRwt/vIII glioma cells. Cancer Letters. 2017;388:269–280. doi: 10.1016/j.canlet.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Katz SG, Rabinovich PM. T Cell Reprogramming Against Cancer. Methods in molecular biology (Clifton, N.J.) 2020;2097:3–44. doi: 10.1007/978-1-0716-0203-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Rivière I. Clinical manufacturing of CAR T cells: foundation of a promising therapy. Molecular Therapy - Oncolytics. 2016;3:16015. doi: 10.1038/mto.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhavan D, Alizadeh D, Wang D, Weist MR, Shepphird JK, Brown CE. CAR T cells for brain tumors: Lessons learned and road ahead. Immunological Reviews. 2019;290(1):60–84. doi: 10.1111/imr.12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi BD, Yu X, Castano AP, Darr H, Henderson DB, Bouffard AA, Maus MV. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. Journal for Immunotherapy of Cancer. 2019;7(1):304. doi: 10.1186/s40425-019-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun T, Patil R, Galstyan A, Klymyshyn D, Ding H, Chesnokova A, Ljubimova JY. Blockade of a laminin-411 - Notch axis with CRISPR/Cas9 or a nanobioconjugate inhibits glioblastoma growth through tumor-microenvironment crosstalk. Cancer Research. 2019;79(6):1239–1251. doi: 10.1158/0008-5472.CAN-18-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacLeod G, Bozek DA, Rajakulendran N, Monteiro V, Ahmadi M, Steinhart Z, Angers S. Genome-Wide CRISPR-Cas9 Screens Expose Genetic Vulnerabilities and Mechanisms of Temozolomide Sensitivity in Glioblastoma Stem Cells. Cell Reports. 2019;27(3):971–986.e9. doi: 10.1016/j.celrep.2019.03.047. [DOI] [PubMed] [Google Scholar]
- 37.Prolo LM, Li A, Owen SF, Parker JJ, Foshay K, Nitta RT, Grant GA. Targeted genomic CRISPR-Cas9 screen identifies MAP4K4 as essential for glioblastoma invasion. Scientific Reports. 2019;9(1):14020. doi: 10.1038/s41598-019-50160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA Repair. Annual review of genetics. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brosh RM, Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Research. 2007;35(22):7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Król SK, Kaczmarczyk A, Wojtas B, Kaminska B. PO-017 Is RECQL4 a novel player in glioblastoma pathogenesis? ESMO Open. 2018 doi: 10.1136/esmoopen-2018-EACR25.62. [DOI] [Google Scholar]
- 41.Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, Jemal A. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States: Potentially Preventable Cancers in US. CA: A Cancer Journal for Clinicians. 2018;68(1):31–54. doi: 10.3322/caac.21440. [DOI] [PubMed] [Google Scholar]
- 42.Mogi A, Kuwano H. TP53 Mutations in Nonsmall Cell Lung Cancer. Journal of Biomedicine and Biotechnology. 2011;2011:1–9. doi: 10.1155/2011/583929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper, G. M. (2000). Tumor Suppressor Genes. The Cell: A Molecular Approach. 2nd edition. Retrieved from https://www.ncbi.nlm.nih.gov/books/NBK9894/
- 44.Gao Y, Wang B, Gao S. BRD7 Acts as a Tumor Suppressor Gene in Lung Adenocarcinoma. PLoS ONE. 2016;11(8):e0156701. doi: 10.1371/journal.pone.0156701. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Schuster K, Venkateswaran N, Rabellino A, Girard L, Pena-Llopis S, Scaglioni PP. Nullifying the CDKN2A/B Locus Promotes Mutant K-ras Lung Tumorigenesis. Molecular cancer research. 2014;12(6):912–923. doi: 10.1158/1541-7786.MCR-13-0620-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan M, Zhang W, Wang J, Al Yaghchi C, Ahmed J, Chard L, Wang Y. Efficiently Editing the Vaccinia Virus Genome by Using the CRISPR-Cas9 System. Journal of Virology. 2015;89(9):5176–5179. doi: 10.1128/JVI.00339-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oxnard GR, Binder A, Jänne PA. New Targetable Oncogenes in Non–Small-Cell Lung Cancer. Journal of Clinical Oncology. 2013;31(8):1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rapp UR, Korn C, Ceteci F, Karreman C, Luetkenhaus K, Serafin V, Potapenko T. Myc Is a Metastasis Gene for Non-Small-Cell Lung Cancer. PLoS ONE. 2009;4(6):e6029. doi: 10.1371/journal.pone.0006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu Y, Kinoshita I, Kikuchi J, Yamazaki K, Nishimura M, Birrer MJ, Dosaka-Akita H. Growth inhibition of non-small cell lung cancer cells by AP-1 blockade using a cJun dominant-negative mutant. British Journal of Cancer. 2008;98(5):915–922. doi: 10.1038/sj.bjc.6604267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elangovan IM, Vaz M, Tamatam CR, Potteti HR, Reddy NM, Reddy SP. FOSL1 Promotes Kras-induced Lung Cancer through Amphiregulin and Cell Survival Gene Regulation. American Journal of Respiratory Cell and Molecular Biology. 2018;58(5):625–635. doi: 10.1165/rcmb.2017-0164OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian H, Yin L, Ding K, Xia Y, Wang X, Wu J, He X. Raf1 is a prognostic factor for progression in patients with non-small cell lung cancer after radiotherapy. Oncology Reports. 2018;39:1966–1974. doi: 10.3892/or.2018.6277. [DOI] [PubMed] [Google Scholar]
- 52.Wang. ErbB Receptors and Cancer. Methods Mol Bio. 2017;1652:3–35. doi: 10.1007/978-1-4939-7219-7_1. [DOI] [PubMed] [Google Scholar]
- 53.Lee EYHP, Muller WJ. Oncogenes and Tumor Suppressor Genes. Cold Spring Harbor Perspectives in Biology. 2010;2(10):a003236. doi: 10.1101/cshperspect.a003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang H, Shrager JB. CRISPR/Cas-mediated genome editing to treat EGFR-mutant lung cancer a personalized molecular surgical therapy. EMBO Molecular Medicine. 2016;8(2):83–85. doi: 10.15252/emmm.201506006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu F, Zhang Y, Lu M, Wang C, Li Q, Gao Y, Meng X. Nestin servers as a promising prognostic biomarker in non-small cell lung cancer. American Journal of Translational Research. 2017;9(3):1392–1401. [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang C, Meng L, Yang B, Luo X. Application of CRISPR/Cas9 gene editing technique in the study of cancer treatment. Clinical Genetics. 2020;97(1):73–88. doi: 10.1111/cge.13589. [DOI] [PubMed] [Google Scholar]
- 57.Chen X, Sun X, Guan J, Gai J, Xing J, Fu L, Li Q. Rsf-1 Influences the Sensitivity of Non-Small Cell Lung Cancer to Paclitaxel by Regulating NF-κB Pathway and Its Downstream Proteins. Cellular Physiology and Biochemistry. 2017;44(6):2322–2336. doi: 10.1159/000486116. [DOI] [PubMed] [Google Scholar]
- 58.Koo T, Yoon AR, Cho HY, Bae S, Yun CO, Kim JS. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Research. 2017;45(13):7897–7908. doi: 10.1093/nar/gkx490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheung AHK, Chow C, Zhang J, Zhou Y, Huang T, Ng KC-K, To K-F. Specific targeting of point mutations in EGFR L858R-positive lung cancer by CRISPR/Cas9. Laboratory Investigation. 2018;98(7):968–976. doi: 10.1038/s41374-018-0056-1. [DOI] [PubMed] [Google Scholar]
- 60.Tang KJ, Constanzo JD, Venkateswaran N, Melegari M, Ilcheva M, Morales JC, Scaglioni PP. Focal Adhesion Kinase Regulates the DNA Damage Response and Its Inhibition Radiosensitizes Mutant KRAS Lung Cancer. Clinical Cancer Research. 2016;22(23):5851–5863. doi: 10.1158/1078-0432.CCR-15-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu K, Chen G, Li X, Wu X, Chang Z, Xu J, Dong L. MFN2 suppresses cancer progression through inhibition of mTORC2/Akt signaling. Scientific Reports. 2017;7(1):41718. doi: 10.1038/srep41718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Papagiannakopoulos T. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nature Medicine. 2017;23(11):1362–1368. doi: 10.1038/nm.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li C, Pu M, Li C, Gao M, Liu M, Yu C, Ren J. MicroRNA-1304 suppresses human non-small cell lung cancer cell growth in vitro by targeting heme oxygenase-1. Acta Pharmacologica Sinica. 2017;38(1):110–119. doi: 10.1038/aps.2016.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018 GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA a cancer journal for clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 65.Yersal O, Barutca S. Biological subtypes of breast cancer: Prognostic and therapeutic implications. World Journal of Clinical Oncology. 2014;5(3):412–424. doi: 10.5306/wjco.v5.i3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dai X, Xiang L, Li T, Bai Z. Cancer Hallmarks, Biomarkers and Breast Cancer Molecular Subtypes. Journal of Cancer. 2016;7(10):1281–1294. doi: 10.7150/jca.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Treuren T, Vishwanatha JK. CRISPR deletion of MIEN1 in breast cancer cells. PLoS ONE. 2018 doi: 10.1371/journal.pone.0204976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brancho D, Ventura JJ, Jaeschke A, Doran B, Flavell RA, Davis RJ. Role of MLK3 in the Regulation of Mitogen-Activated Protein Kinase Signaling Cascades. Molecular and Cellular Biology. 2005;25(9):3670–3681. doi: 10.1128/MCB.25.9.3670-3681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rattanasinchai C, Llewellyn BJ, Conrad SE, Gallo KA. MLK3 regulates FRA-1 and MMPs to drive invasion and transendothelial migration in triple-negative breast cancer cells. Oncogenesis. 2017;6(6):e345–e345. doi: 10.1038/oncsis.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao L, Song M, Li X, Tang L, Zhang T, Zhang L, Ma X. E3 Ubiquitin Ligase UBR5 Drives the Growth and Metastasis of Triple-Negative Breast Cancer. Cancer Research. 2017;77(8):2090–2101. doi: 10.1158/0008-5472.CAN-16-2409. [DOI] [PubMed] [Google Scholar]
- 71.Zhuge BZ, Du BR, Meng L, Zhang YQ. MASTL is a potential poor prognostic indicator in ER+ breast cancer. European Review for Medical and Pharmacological Sciences. 2017;21:2413–2420. [PubMed] [Google Scholar]
- 72.Alvarez Fernandez M, Sanz-Flores M, Sanz-Castillo B, Salazar-Roa M, Partida D, Zapatero-Solana E, Malumbres M. Therapeutic relevance of the PP2A-B55 inhibitory kinase MASTL/Greatwall in breast cancer. Cell Death & Differentiation. 2017;25(5):828–840. doi: 10.1038/s41418-017-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hochgrafe F, Zhang L, O’Toole SA, Browne BC, Pinese M, Porta Cubas A, Daly RJ. Tyrosine phosphorylation profiling reveals the signaling network characteristics of Basal breast cancer cells. Cancer Research. 2010;70(22):9391–9401. doi: 10.1158/0008-5472.CAN-10-0911. [DOI] [PubMed] [Google Scholar]
- 74.Zhang S, Fan G, Hao Y, Hammell M, Wilkinson JE, Tonks NK. Suppression of protein tyrosine phosphatase N23 predisposes to breast tumorigenesis via activation of FYN kinase. Genes & Development. 2017;31(19):1939–1957. doi: 10.1101/gad.304261.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng YZ, Xue M-Z, Shen H-J, Li X-G, Ma D, Gong Y, Shao Z-M. PHF5A Epigenetically Inhibits Apoptosis to Promote Breast Cancer Progression. Cancer Research. 2018;78(12):3190–3206. doi: 10.1158/0008-5472.CAN-17-3514. [DOI] [PubMed] [Google Scholar]
- 76.Yang Y, Zhu J, Zhang T, Liu J, Li Y, Zhu Y, Wu Q. PHD-finger domain protein 5A functions as a novel oncoprotein in lung adenocarcinoma. Journal of Experimental & Clinical Cancer Research. 2018;37(1):65. doi: 10.1186/s13046-018-0736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, Paddison PJ. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes & Development. 2013;27(9):1032–1045. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Brien A, Zhou T, Tan C, Alpini G, Glaser S. Role of Non-Coding RNAs in the Progression of Liver Cancer: Evidence from Experimental Models. Cancers. 2019;11(11):1652. doi: 10.3390/cancers11111652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh R, Gupta SC, Peng W-X, Zhou N, Pochampally R, Atfi A, Mo Y-Y. Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death & Disease. 2016;7(6):e2262. doi: 10.1038/cddis.2016.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El Fatimy R, Subramanian S, Uhlmann EJ, Krichevsky AM. Genome Editing Reveals Glioblastoma Addiction to MicroRNA-10b. Molecular Therapy. 2017;25(2):368–378. doi: 10.1016/j.ymthe.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chaisaingmongkol J, Budhu A, Dang H, Rabibhadana S, Pupacdi B, Kwon SM, Wang XW. Common Molecular Subtypes among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell. 2017;32(1):57–70. doi: 10.1016/j.ccell.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bruix J, Han KH, Gores G, Llovet JM, Mazzaferro V. Liver cancer: Approaching a personalized care. Journal of hepatology. 2015;62(1):S144–S156. doi: 10.1016/j.jhep.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chatterjee S, Azad BB, Nimmagadda S. The Intricate Role of CXCR4 in Cancer. Advances in cancer research. 2014;124:31–82. doi: 10.1016/B978-0-12-411638-2.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X, Zhang W, Ding Y, Guo X, Yuan Y, Li D. CRISPR/Cas9-mediated genome engineering of CXCR4 decreases the malignancy of hepatocellular carcinoma cells in vitro and in vivo. Oncology Reports. 2017;37(6):3565–3571. doi: 10.3892/or.2017.5601. [DOI] [PubMed] [Google Scholar]
- 85.He J, Zhang W, Li A, Chen F, Luo R. Knockout of NCOA5 impairs proliferation and migration of hepatocellular carcinoma cells by suppressing epithelial-to-mesenchymal transition. Biochemical and Biophysical Research Communications. 2018;500(2):177–183. doi: 10.1016/j.bbrc.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 86.Testa U, Pelosi E, Castelli G. Colorectal Cancer: Genetic Abnormalities, Tumor Progression, Tumor Heterogeneity, Clonal Evolution and Tumor-Initiating Cells. Medical Sciences. 2018;6(2):31. doi: 10.3390/medsci6020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakayama M, Oshima M. Mutant p53 in colon cancer. Journal of Molecular Cell Biology. 2019;11(4):267–276. doi: 10.1093/jmcb/mjy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen HT, Duong H-Q. The molecular characteristics of colorectal cancer: Implications for diagnosis and therapy (Review) Oncology Letters. 2018;16(1):9–18. doi: 10.3892/ol.2018.8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamagishi H, Kuroda H, Imai Y, Hiraishi H. Molecular pathogenesis of sporadic colorectal cancers. Chinese Journal of Cancer. 2016;35:4. doi: 10.1186/s40880-015-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takeda H, Kataoka S, Nakayama M, Ali MAE, Oshima H, Yamamoto D, Oshima M. CRISPR-Cas9–mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proceedings of the National Academy of Sciences. 2019;116(31):15635–15644. doi: 10.1073/pnas.1904714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li W, Cho M-Y, Lee S, Jang M, Park J, Park R. CRISPR-Cas9 mediated CD133 knockout inhibits colon cancer invasion through reduced epithelial-mesenchymal transition. PLoS ONE. 2019;14(8):e0220860. doi: 10.1371/journal.pone.0220860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Sato T. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nature Medicine. 2015;21(3):256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 93.Rawla P, Sunkara T, Gaduputi V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World Journal of Oncology. 2019;10(1):10–27. doi: 10.14740/wjon1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Regel I, Mayerle J, Ujjwal Mukund M. Current Strategies and Future Perspectives for Precision Medicine in Pancreatic Cancer. Cancers. 2020;12(4):1024. doi: 10.3390/cancers12041024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vorvis C, Hatziapostolou M, Mahurkar-Joshi S, Koutsioumpa M, Williams J, Donahue TR, Iliopoulos D. Transcriptomic and CRISPR/Cas9 technologies reveal FOXA2 as a tumor suppressor gene in pancreatic cancer. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2016;310(11):G1124–G1137. doi: 10.1152/ajpgi.00035.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Watanabe S, Shimada S, Akiyama Y, Ishikawa Y, Ogura T, Ogawa K, Tanaka S. Loss of KDM6A characterizes a poor prognostic subtype of human pancreatic cancer and potentiates HDAC inhibitor lethality. International Journal of Cancer. 2019;145(1):192–205. doi: 10.1002/ijc.32072. [DOI] [PubMed] [Google Scholar]
- 97.Ovarian cancer statistics. (2018, August 22). World Cancer Research Fund. Retrieved from https://www.wcrf.org/dietandcancer/cancer-trends/ovarian-cancer-statistics
- 98.Lisio MA, Fu L, Goyeneche A, Gao Z, Telleria C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. International Journal of Molecular Sciences. 2019;20(4):952. doi: 10.3390/ijms20040952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanda ET, Budroni M, Cesaraccio R, Palmieri G, Palomba G, Capobianco G, Cossu A. Epidemiology of ovarian cancer in North Sardinia, Italy, during the period 1992–2010. European Journal of Gynaecological Oncology. 2015;36(1):69–72. [PubMed] [Google Scholar]
- 100.Siddique HR, Saleem M. Role of BMI1, a Stem Cell Factor, in Cancer Recurrence and Chemoresistance: Preclinical and Clinical Evidences. Stem Cells. 2012;30(3):372–378. doi: 10.1002/stem.1035. [DOI] [PubMed] [Google Scholar]
- 101.Zhao Q, Qian Q, Cao D, Yang J, Gui T, Shen K. Role of BMI1 in epithelial ovarian cancer: investigated via the CRISPR/Cas9 system and RNA sequencing. Journal of Ovarian Research. 2018;11(1):31. doi: 10.1186/s13048-018-0406-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wong ASL, Choi GCG, Cui CH, Pregernig G, Milani P, Adam M, Lu TK. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proceedings of the National Academy of Sciences. 2016;113(9):2544–2549. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cui Y, Li Y, Gorgé O, Platonov ME, Yan Y, Guo Z, Yang R. Insight into Microevolution of Yersinia pestis by Clustered Regularly Interspaced Short Palindromic Repeats. PLoS ONE. 2008;3(7):e2652. doi: 10.1371/journal.pone.0002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. Novel Virulence Gene and Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) Multilocus Sequence Typing Scheme for Subtyping of the Major Serovars of Salmonella enterica subsp. enterica. Applied and Environmental Microbiology. 2011;77(6):1946–1956. doi: 10.1128/AEM.02625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nature Biotechnology. 2014;32(11):1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Im W, Moon J, Kim M. Applications of CRISPR/Cas9 for Gene Editing in Hereditary Movement Disorders. Journal of Movement Disorders. 2016;9(3):136–143. doi: 10.14802/jmd.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Jacks T. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514(7522):380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ventura A. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakamura K, Fujii W, Tsuboi M, Tanihata J, Teramoto N, Takeuchi S, Nishihara M. Generation of muscular dystrophy model rats with a CRISPR/Cas system. Scientific Reports. 2014;4:5635. doi: 10.1038/srep05635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Scientific Reports. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kang H, Minder P, Park MA, Mesquitta WT, Torbett BE, Slukvin II. CCR5 Disruption in Induced Pluripotent Stem Cells Using CRISPR/Cas9 Provides Selective Resistance of Immune Cells to CCR5-tropic HIV-1 Virus. Molecular Therapy. Nucleic Acids. 2015;4:e268. doi: 10.1038/mtna.2015.42. [DOI] [PubMed] [Google Scholar]
- 113.CRISPR Clinical Trials: Will CRISPR Cure These Diseases? (2020, February 17). Retrieved from https://www.synthego.com/blog/crispr-cure-diseases
- 114.Ramirez, V. B. (2019, July 28). First Human CRISPR Trial in the US Aims to Cure Inherited Blindness. Singularity Hub. Retrieved from https://singularityhub.com/2019/07/28/first-human-crispr-trial-in-the-us-aims-to-cure-inherited-blindness/
- 115.Positive results in first-in-U.S. trial of CRISPR-edited immune cells. (2020, August 27). Penn Today. Retrieved from https://penntoday.upenn.edu/news/positive-results-first-us-trial-crispr-edited-immune-cells
- 116.Zhao L, Cao YJ. Engineered T Cell Therapy for Cancer in the Clinic. Frontiers in Immunology. 2019;10:2250. doi: 10.3389/fimmu.2019.02250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, Collins JJ. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell. 2016;165(5):1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 118.Müller V, Rajer F, Frykholm K, Nyberg LK, Quaderi S, Fritzsche J, Westerlund F. Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Scientific Reports. 2016;6(1):37938. doi: 10.1038/srep37938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guk K, Keem JO, Hwang SG, Kim H, Kang T, Lim E-K, Jung J. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosensors and Bioelectronics. 2017;95:67–71. doi: 10.1016/j.bios.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 120.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.He Q, Yu D, Bao M, Korensky G, Chen J, Shin M, Du K. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence-based point-of-care system. Biosensors and Bioelectronics. 2020;154:112068. doi: 10.1016/j.bios.2020.112068. [DOI] [PubMed] [Google Scholar]
- 122.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Sabeti PC. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang, F., Abudayyeh, O. O., & Gootenberg, J. S. (2020). A protocol for detection of COVID-19 using CRISPR diagnostics. A protocol for detection of COVID-19 using CRISPR diagnostics, 8, v.20200321.
- 125.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Chiu CY. CRISPR–Cas12-based detection of SARS-CoV-2. Nature Biotechnology. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.India’s new paper Covid-19 test could be a ‘game changer.’ (2020, October 4). BBC News. Retrieved from https://www.bbc.com/news/world-asia-india-54338864
- 127.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nature reviews. Genetics. 2016;17(5):300–312. doi: 10.1038/nrg.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kleinstiver BP, Prew MS, Tsai SQ, Topkar V, Nguyen NT, Zheng Z, Joung JK. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen N, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 variants with undetectable genome-wide off-targets. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yin H, Song CQ, Dorkin JR, Zhu LJ, Li Y, Wu Q, Anderson DG. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nature biotechnology. 2016;34(3):328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cox DBT, Platt RJ, Zhang F. Therapeutic Genome Editing: Prospects and Challenges. Nature medicine. 2015;21(2):121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sun W, Ji W, Hall JM, Hu Q, Wang C, Beisel CL, Gu Z. Efficient Delivery of CRISPR-Cas9 for Genome Editing via Self-Assembled DNA Nanoclews. Angewandte Chemie (International ed. in English) 2015;54(41):12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Naso MF, Tomkowicz B, Perry WL, Strohl WR. Adeno-Associated Virus (AAV) as a Vector for Gene Therapy. Biodrugs. 2017;31(4):317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Crudele JM, Chamberlain JS. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nature Communications. 2018;9:3497. doi: 10.1038/s41467-018-05843-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.New Details About the Infamous “CRISPR Babies” Experiment Have Just Been Revealed. (2020, February 20). Retrieved from https://www.sciencealert.com/china-s-failed-experiment-proves-we-re-not-ready-for-human-gene-editing
- 137.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nature Methods. 2014;11(2):122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 139.Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature Methods. 2015;12(10):982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.MacPherson CR, Scherf A. Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nature Biotechnology. 2015;33(8):805–806. doi: 10.1038/nbt.3291. [DOI] [PubMed] [Google Scholar]
- 141.Perez AR, Pritykin Y, Vidigal JA, Chhangawala S, Zamparo L, Leslie CS, Ventura A. GuideScan software for improved single and paired CRISPR guide RNA design. Nature biotechnology. 2017;35(4):347–349. doi: 10.1038/nbt.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mendoza BJ, Trinh CT. Enhanced guide-RNA design and targeting analysis for precise CRISPR genome editing of single and consortia of industrially relevant and non-model organisms. Bioinformatics. 2018;34(1):16–23. doi: 10.1093/bioinformatics/btx564. [DOI] [PubMed] [Google Scholar]
- 143.Heigwer F, Zhan T, Breinig M, Winter J, Brügemann D, Leible S, Boutros M. CRISPR library designer (CLD): software for multispecies design of single guide RNA libraries. Genome Biology. 2016;17(1):55. doi: 10.1186/s13059-016-0915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Singh R, Kuscu C, Quinlan A, Qi Y, Adli M. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Research. 2015;43(18):e118. doi: 10.1093/nar/gkv575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nature methods. 2015;12(9):823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chari R, Yeo NC, Chavez A, Church GM. sgRNA Scorer 2.0–a species independent model to predict CRISPR/Cas9 activity. ACS synthetic biology. 2017;6(5):902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu H, Xiao T, Chen C-H, Li W, Meyer CA, Wu Q, Liu XS. Sequence determinants of improved CRISPR sgRNA design. Genome Research. 2015;25(8):1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wong N, Liu W, Wang X. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biology. 2015;16(1):218. doi: 10.1186/s13059-015-0784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Research. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Research. 2014;42(W1):W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bae S, Park J, Kim J-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stemmer M, Thumberger T, del Keyer M, S., Wittbrodt, J., & Mateo, J. L. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE. 2015;10(4):e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31(7):1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Prykhozhij SV, Rajan V, Gaston D, Berman JN. CRISPR MultiTargeter: A Web Tool to Find Common and Unique CRISPR Single Guide RNA Targets in a Set of Similar Sequences. PLoS ONE. 2015;10(3):e0119372. doi: 10.1371/journal.pone.0119372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianné J, Renaud J-B, Concordet J-P. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biology. 2016;17(1):148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Root DE. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu LJ, Holmes BR, Aronin N, Brodsky MH. CRISPRseek: A Bioconductor Package to Identify Target-Specific Guide RNAs for CRISPR-Cas9 Genome-Editing Systems. PLoS ONE. 2014;9(9):e108424. doi: 10.1371/journal.pone.0108424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.