The dual anticytokine and antiviral actions of baricitinib reduce SARS-CoV-2 infectivity in organoids and morbidity in people.
Abstract
Using AI, we identified baricitinib as having antiviral and anticytokine efficacy. We now show a 71% (95% CI 0.15 to 0.58) mortality benefit in 83 patients with moderate-severe SARS-CoV-2 pneumonia with few drug-induced adverse events, including a large elderly cohort (median age, 81 years). An additional 48 cases with mild-moderate pneumonia recovered uneventfully. Using organotypic 3D cultures of primary human liver cells, we demonstrate that interferon-α2 increases ACE2 expression and SARS-CoV-2 infectivity in parenchymal cells by greater than fivefold. RNA-seq reveals gene response signatures associated with platelet activation, fully inhibited by baricitinib. Using viral load quantifications and superresolution microscopy, we found that baricitinib exerts activity rapidly through the inhibition of host proteins (numb-associated kinases), uniquely among antivirals. This reveals mechanistic actions of a Janus kinase-1/2 inhibitor targeting viral entry, replication, and the cytokine storm and is associated with beneficial outcomes including in severely ill elderly patients, data that incentivize further randomized controlled trials.
INTRODUCTION
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has become the biggest public health challenge of this century with more than 1 million fatalities and 34 million cases of coronavirus disease 2019 (COVID-19) across all countries in the first 10 months since it emerged (1–4). It is essential that more effective treatments are found before a vaccine is developed and made widely available (5, 6). We previously reported that use of artificial intelligence (AI) based on a knowledge graph of more than 1 billion relationship edges enabled the rapid identification of the once daily orally administered drug, baricitinib, approved as a treatment for adult rheumatoid arthritis (RA), as a potential therapeutic (7–11). Baricitinib was predicted to have a dual mode of action, reducing viral infectivity through the inhibition of numb-associated kinases (NAKs) and, thus, viral endocytosis and its better known well-described anti-inflammatory mechanism through blockade of Janus kinase 1/2 (JAK1/2) (12–14).
In severe cases, the so-called “cytokine storm” can result in profound lung damage and the development of acute respiratory distress syndrome (ARDS), the leading cause of death in COVID-19 (15, 16). SARS-CoV-2 infection also results in damage to many other organs including the kidneys, brain, and vasculature via mechanisms including endothelial cell disruption and intussusceptive angiogenesis (17, 18). Plasma levels of the proinflammatory cytokine interleukin-6 (IL-6) signaling predominantly through JAK/STAT (signal transducer and activator of transcription) have been reported to be a prognostic indicator of mortality (19, 20), and using samples from a randomized phase 2b trial in RA (4), we demonstrated that baricitinib reduces IL-6 levels in a dose-dependent manner (21), the first time this has been shown in patients. Therapeutics capable of clearing the virus and reducing the cytokine-mediated inflammation may be beneficial. Purported antiviral agents have, at best, a small impact on disease remission in hospitalized patients and, in some cases, show no differences in viral loads compared to control arms (22). Dexamethasone, known to modulate inflammation-mediated lung injury, resulted in lower mortality among those who were receiving either invasive mechanical ventilation or oxygen alone at randomization (23). In many cases, patients are unable to withstand the exuberant inflammation associated with ARDS. Furthermore, high viral loads cause disseminated end-organ compromise through vascular endotheliitis, thrombosis, and other associated effects (17, 24).
In this situation, an assessment of the efficacy of baricitinib would include demonstration of reduced viral infectivity and associated end-organ damage and control of the excessive cytokine-mediated inflammation. Here, we explore the clinical effect of baricitinib therapy patients and matched controls from two European cohorts. Since COVID-19 is associated with multisystem organ damage, we also investigated whether the cytokine-mediated inflammation could induce the expression of the SARS-CoV-2 receptor angiotensin-converting enzyme 2 (ACE2) (25, 26) in extrapulmonary systems. We show that type-1 interferons (IFNs), specifically IFN-α2, whose levels are increased in patients with severe COVID-19 (27), increase ACE2 expression in three-dimensional (3D) cultures of primary human liver cells, resulting in increased viral load, and that this induction is fully inhibited by clinically relevant concentrations of baricitinib.
RESULTS
Clinical results
A total of 601 patients were enrolled in the University of Pisa (n = 179) and Albacete Hospital (n = 422) cohorts, between mid-March and mid-April 2020, when these regions were the global epicenters of the pandemic, under severe capacity constraints. Of these, 37 patients were treated with baricitinib in the Pisa cohort and 46 were treated in the Albacete cohort, all of European descent; eighty-three controls were included using propensity score–matching systems, both in the Italian and in the Spanish cohorts, as per the CONSORT diagram (Fig. 1).
Fig. 1. CONSORT flow diagram for University of Pisa and Albacete Hospital cohorts.
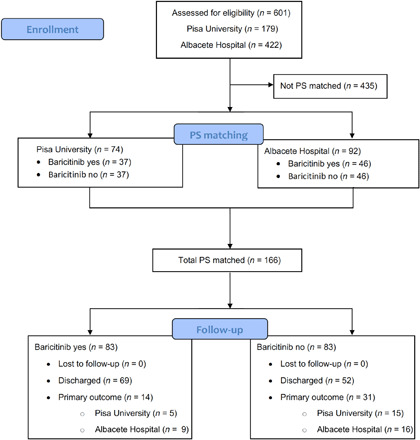
PS, propensity score.
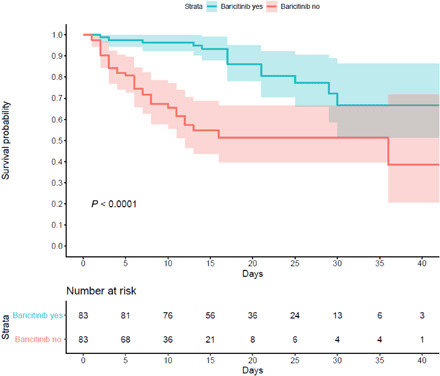
Tables 1 and 2 present baseline data of participants from Pisa and Albacete, respectively, showing clinical characteristics immediately following admission, and Table 3 presents propensity score–matched population data from both sites, comprising the group taking baricitinib and the control group. Male sex was predominant, and the Albacete patients were older than those from Pisa (80.9 versus 66.0 years) in treated groups. Most individuals received concomitant “antiviral therapy” with hydroxychloroquine and lopinavir/ritonavir, antibiotics, corticosteroids, and low–molecular weight heparin (LMWH). In the merged matched population, the primary composite end point of death or invasive mechanical ventilation occurred in 14 (16.9%) patients in the baricitinib-treated group compared to 29 (34.9%) in the control group (P < 0.001) (Table 3). In the multivariate Cox regression analysis adjusted for all the covariates included in the matching of the two cohorts, baricitinib was independently associated as a protective variable with the primary outcome [hazard ratio (HR) 0.29; 95% confidence interval (CI) 0.15 to 0.58; P < 0.001] (Table 4). Figure 2 presents the Kaplan-Meier curves for the primary outcome in the treated and control groups in the merged cohorts. Baricitinib’s favorable effect appeared “early” and was maintained until termination of the follow-up (P < 0.0001). Further analyses including only participants who received five or more baricitinib doses yielded similar results as those presented. Analyses of the propensity score–matched cohorts from the University of Pisa (fig. S3A) and Albacete Hospital (fig. S3B) are shown independently. Similar curves and statistics can be observed, compared with the final merged cohort (Fig. 2).
Table 1. Characteristics of patients receiving or not receiving baricitinib in the unmatched and matched study population from the University of Pisa (Italy).
All data are medians with the interquartile range or number of participants (%). COPD, chronic obstructive pulmonary disease; PaO2/FiO2, ratio of arterial oxygen partial pressure to fractional inspired oxygen; ALT, alanine aminotransferase; ARB, angiotensin-receptor blocker; AST, aspartate aminotransferase; SOFA, sequential organ failure assessment; ULN, upper limit of normal.
| Baricitinib group (n = 37) | Control group (n = 142) | PS matched control group (n = 37) | |
| Age | 66.0 (48.0–84.0)** | 76.5 (62.5–83)** | 65 (40–90) |
| Male sex | 27 (73) | 95 (66.9) | 26 (70.3) |
| Interval between symptom onset and admission |
6 (3.5–9) | 7 (3–8) | 7 (4–7.5) |
| Coexisting conditions | |||
| Hypertension | 16 (43.2) | 79 (55.9) | 18 (48.6) |
| Cardiovascular disease | 9 (24.3) | 56 (39.4) | 6 (16.2) |
| Solid cancer | 6 (16.2) | 22 (15.5) | 9 (24.3) |
| Diabetes | 7 (18.9) | 32 (22.5) | 8 (21.6) |
| COPD | 1 (2.7)* | 27 (19.0)* | 0 (0.0) |
| Chronic kidney failure | 2 (5.4) | 16 (11.3) | 1 (2.7) |
| Charlson comorbidity index | 2 (0–4) | 2 (1–5) | 1 (0–6) |
| Medications at baseline | |||
| ACE inhibitor or ARB | 9 (24.3) | 43 (30.4) | 9 (24.3) |
| Direct oral anticoagulant or warfarin | 5 (13.5) | 24 (16.9) | 1 (2.7) |
| SOFA score | 3 (1–5) | 3 (2–4) | 3 (2–4) |
| Baseline PaO2/FiO2 | 242 (143–341) | 254 (200–298) | 252 (169–335) |
| Noninvasive mechanical ventilation | 17 (45.9)*** | 19 (13.4)*** | 13 (35.1) |
| Baseline laboratory tests | |||
| C-reactive protein (mg/dl) | 5.7 (0.0–18.0) | 8.3 (3.7–16.1) | 11.2 (0.0–25.4) |
| Lymphocyte count | 1010 (400–1620) | 830 (580–1160) | 740 (145–1335) |
| ALT (U/liter) | 39 (13–65) | 25 (16–45) | 28 (0–58) |
| AST (U/liter) | 43 (12–74) | 33 (24–50) | 32 (10–54) |
| ALT > 3× ULN | 1 (2.7%) | 8 (5.6%) | 1 (2.7%) |
| AST > 3× ULN | 1 (2.7%) | 11 (7.7%) | 2 (5.4%) |
| Total bilirubin (mg/dl) | 0.51 (0.31–0.71) | 0.49 (0.35–0.77) | 0.48 (0.08–0.88) |
| Concomitant treatment | |||
| Hydroxychloroquine | 34 (91.9)* | 102 (71.8)* | 34 (91.9) |
| Other antibiotics | 33 (89.2) | 109 (76.8) | 34 (91.9) |
| Proteases inhibitors | 30 (81.1)* | 89 (62.7)* | 29 (78.4) |
| LMWH (enoxaparin) | 36 (97.3)*** | 98 (69)*** | 36 (97.3) |
| Steroids | 27 (73.0)** | 65 (45.8)** | 28 (75.7) |
| Primary outcome | 5 (13.5)*** | 66 (46.5)*** | 13 (35.1)* |
| Invasive mechanical ventilation | 4 (10.8) | 19 (13.4) | 9 (24.3) |
| Died without intubation | 1 (2.7) | 47 (33.1) | 4 (5.4) |
*P < 0.05
**P < 0.01
***P < 0.001
Table 2. Characteristics of patients receiving or not receiving baricitinib in the unmatched and matched study population from the Albacete Hospital (Spain).
IMV, invasive mechanical ventilation. All data are means (SD) or number of participants (%).
| Baricitinib group (n = 46) | Control group (n = 376) | PS matched control group (n = 46) | |
| Age | 80.9 (5.8) | 82.7 (6.3) | 80.6 (6.3) |
| Male sex | 30 (65.2) | 201 (53.5) | 30 (65.2) |
| Interval between symptom onset and admission |
7.4 (5.2) | 7.3 (4.9) | 7.3 (5.1) |
| Coexisting conditions | |||
| Hypertension | 34 (73.9) | 296 (78.7) | 35 (76.1) |
| Cardiovascular disease | 18 (39.1) | 167 (44.4) | 15 (32.6) |
| Solid cancer | 2 (4.3) | 20 (5.3) | 1 (2.2) |
| Diabetes | 21 (45.7) | 139 (37.0) | 14 (30.4) |
| COPD | 11 (23.9) | 84 (22.3) | 12 (26.1) |
| Chronic kidney failure | 5 (10.9) | 64 (17.0) | 6 (13.0) |
| Charlson comorbidity index | 2.9 (2.3) | 2.0 (1.9) | 3.2 (2.8) |
| Medications at baseline | |||
| ACE inhibitor or ARB | 24 (52.2) | 196 (52.3) | 26 (56.5) |
| Direct oral anticoagulant or warfarin | 8 (17.4) | 65 (17.3) | 4 (8.7) |
| Antiaggregants | 14 (30.4) | 121 (32.2) | 15 (32.6) |
| Statins | 23 (50.0) | 158 (42.0) | 21 (45.7) |
| Insulin | 9 (19.6) | 42 (11.2) | 4 (4.3) |
| Oral hypoglycemic agents | 15 (32.6) | 109 (29.0) | 13 (28.3) |
| Antidepressants | 10 (21.7) | 123 (32.7) | 11 (23.9) |
| Inhaled therapy for COPD | 12 (26.1) | 95 (25.3) | 15 (32.6) |
| Baseline PaO2/FiO2 | 284 (109) | 280 (107) | 282 (96) |
| Baseline laboratory tests | |||
| D-dimer (μg/liter) | 6944 (18,052) | 6182 (26,894) | 5443 (16,872) |
| Lactate dehydrogenase (U/liter) | 387 (136) | 372 (288) | 370 (166) |
| C-reactive protein (mg/liter) | 147.2 (98.6) | 137.6 (118.0) | 141.8 (145.8) |
| Ferritin (ng/ml) | 1357 (1094)** | 878 (975)** | 1039 (927) |
| Leucocyte count (per μl) | 9414 (4790) | 8986 (4711) | 7690 (3675) |
| Lymphocyte count (per μl) | 987 (905) | 967 (777) | 934 (517) |
| Hemoglobin (g/dl) | 13.7 (2.1) | 13.2 (2.1) | 13.7 (3.2) |
| Fibrinogen (mg/dl) | 395 (78) | 378 (175) | 375 (70) |
| Creatinine (mg/dl) | 1.2 (0.5) | 1.3 (0.91) | 1.1 (0.5) |
| AST (U/liter) | 35.5 (23.4) | 41.4 (40.3) | 40 (46) |
| Gamma-glutamyltransferase (U/liter) | 63.3 (49.5) | 54.2 (74.3) | 65.5 (119.2) |
| ALT (U/liter) | 33.8 (25.3) | 30.8 (57.3) | 31.0 (25.5) |
| ALT (U/liter) after treatment, at discharge | 47.5 (45.8) | – | – |
| ALT > 2× ULN after treatment, at discharge | 5 (9.1) | – | – |
| ALT > 3× ULN after treatment, at discharge | 3 (5.5) | – | – |
| Chest x-ray | |||
| Interstitial pattern | 46 (100) | 345 (93.0) | 44 (95.7) |
| Opacities | 38 (82.6)* | 233 (63.7)* | 32 (69.6) |
| Severity score | 3.4 (2.1)** | 2.2 (2.2)** | 2.4 (2.1) |
| Concomitant treatment | |||
| Hydroxychloroquine | 45 (97.8)* | 321 (85.4)* | 46 (100) |
| Antibiotics | 46 (100) | 365 (97.1) | 45 (97.8) |
| Lopinavir/ritonavir | 39 (84.8) | 288 (76.8) | 42 (91.3) |
| LMWH (enoxaparin) | 46 (100)** | 322 (85.6)** | 46 (100) |
| Glucocorticoids | 44 (95.7)*** | 266 (70.7)*** | 42 (91.3) |
| Anakinra | 18 (39.1)*** | 29 (7.7)*** | 10 (21.7) |
| Primary outcome (mortality or IMV) | 9 (19.6)** | 157 (41.8)** | 16 (34.8) |
*P < 0.05
**P < 0.01
***P < 0.001
Table 3. Common characteristics of patients receiving or not receiving baricitinib in the propensity score–matched populations from the University of Pisa and the Albacete Hospital.
PS, propensity score matching. All data are means (SD) or number of participants (%).
|
Baricitinib group (n = 83) |
PS control group (n = 83) |
|
| Age | 74.0 (12.5) | 74.1 (13.6) |
| Male sex | 43 (51.8) | 42 (50.6) |
| Coexisting conditions | ||
| Hypertension | 50 (60.2) | 53 (63.9) |
| Cardiovascular disease | 27 (32.5) | 21 (25.3) |
| Solid cancer | 8 (9.6) | 10 (12.0) |
| Diabetes | 28 (33.7) | 22 (26.5) |
| COPD | 12 (14.5) | 12 (14.5) |
| Chronic kidney failure | 7 (8.4) | 7 (8.4) |
| Charlson comorbidity index | 2.7 (2.3) | 2.8 (2.7) |
| Medications at baseline | ||
| ACE inhibitor or ARB | 33 (39.8) | 35 (42.2) |
| Baseline PaO2/FiO2 | 268 (101) | 266 (86) |
| Baseline laboratory tests | ||
| C-reactive protein (mg/liter) | 86.0 (100.6) | 82.2 (123.9) |
| Lymphocyte count (per μl) | 1052 (831) | 40 (46) |
| ALT (U/liter) | 38.7 (26.5) | 34.6(29.9) |
| Concomitant treatment | ||
| Hydroxychloroquine | 79 (95.2) | 80 (96.4) |
| Antibiotics | 79 (95.2) | 79 (95.2) |
| Lopinavir/ritonavir | 69 (83.1) | 71 (85.5) |
| LMWH | 82 (98.8) | 82 (98.8) |
| Glucocorticoids | 71(85.5) | 70 (84.3) |
| Primary outcome | 14 (16.9)* | 29 (34.9)* |
| Time to outcome | 19.9 (9.1)** | 13.1 (9.7)** |
*P < 0.01
**P < 0.001
Table 4. Multivariate Cox regression analyses for the primary outcome in the propensity score–matched populations from the University of Pisa and the Albacete Hospital.
Selection bias was addressed by propensity score analysis. Briefly, this is a two-phase technique used to estimate a treatment effect in comparative groups selected by nonrandom means. In the first phase of a propensity score analysis, variables that influence selection to group assignment are used to model the probability of receiving treatment (or of being in the reference group, in this case, the baricitinib group). The resulting probability is the propensity score. In the second phase, the propensity score is used to adjust for preexisting group differences in the analysis of the relevant outcomes. There are several ways to use propensity scores such as stratification variables, matching patients on the basis of their propensity score, or their use as a weighting or adjustment variable during multivariate analysis. In the current study, each baricitinib patient was matched to a control patient on the basis of comparable propensity scores. Assuming that all relevant covariates are included in the propensity score model, the group effect observed in a propensity score analysis represents an unbiased estimate of the true treatment effect.
| HR (95% CI) | P | |
| Baricitinib | 0.29 (0.15–0.58) | 0.0001 |
| Age | 1.01 (0.98–1.04) | 0.470 |
| Male sex | 1.13 (0.54–2.34) | 0.750 |
| Hypertension | 1.31 (0.52–3.32) | 0.572 |
| Diabetes | 0.51 (0.23–1.17) | 0.113 |
| Chronic obstructive lung disease |
0.51 (0.17–1.54) | 0.230 |
| Cardiovascular disease | 1.41 (0.68–2.92) | 0.351 |
| Chronic kidney disease | 1.45 (0.51–4.15) | 0.491 |
| Solid cancer | 1.18 (0.49–2.87) | 0.709 |
| Charlson comorbidity index |
1.03 (0.90–1.17) | 0.680 |
| Baseline PaO2/FiO2 | 1.00 (1.00–1.00) | 0.823 |
| Lymphocyte count (per μl) |
1.00 (1.00–1.00) | 0.657 |
| ALT | 1.01 (1.00–1.03) | 0.026 |
| Hydroxychloroquine | 2.77 (0.28–27.41) | 0.384 |
| Lopinavir/ritonavir | 1.18 (0.38–3.61) | 0.776 |
| Glucocorticoids | 1.79 (0.60–5.34) | 0.299 |
| LMWH | 0.10 (0.01–1.33) | 0.081 |
| Antibiotics | 2.34 (0.29–18.90) | 0.427 |
Fig. 2. Kaplan-Meier analysis of the propensity score–matched cohorts from Pisa University and Albacete Hospital cohorts.
Safety issues with baricitinib
A major role of observational studies is to provide relevant information regarding toxicity. Seven (19%) individuals from Pisa reported transaminitis [2× to 5× upper limit of normal (ULN)] within 72 hours of commencing baricitinib, leading to cessation of the drug in four of these patients. In three patients and in our previously published small series (21), we continued to treat despite this and the liver function abnormalities self-resolved. One patient developed a severe lymphocytopenia (<500 cells/mm3) during treatment, leading to drug discontinuation. A total of five (14%) individuals from Pisa developed infectious complications during baricitinib treatment as follows: one bacteremia due to Enterococcus faecalis arising from the urinary tract, two episodes of central venous catheter–related bacteremia due to coagulase-negative staphylococci, one urinary tract infection by New Delhi metallo-beta-lactamase (NDM)–producing Klebsiella pneumoniae, and one severe facial herpes simplex infection. In the unmatched control group, a total of 21 (14.8%) patients developed an infection: 8 bloodstream infections, 8 urinary tract infections, and 5 with pneumonia. In the propensity score–matched control group, six (16.2%) episodes of infection occurred: one bacteremia due to Enterobacter aerogenes arising from the urinary tract, one bacteremia by NDM-producing K. pneumoniae, three urinary tract infections (due to Escherichia coli, E. faecalis, and K. pneumoniae), and one pneumonia by Pseudomonas aeruginosa.
Apart from transaminitis, adverse events were difficult to ascribe to baricitinib because of rapidly evolving clinical/capacity constraints. We did not observe any signs of coagulopathy or thrombosis caused by baricitinib in any of our patients, although this has been described as a potential toxicity associated with longer-term use in RA (12–14). However, most were anticoagulated with LMWH.
In the Spanish cohort, a transaminitis of 2× ULN was observed after baricitinib treatment in eight (17%) patients and at 3× ULN in four (9%) patients; the drug was not stopped in any individual based on this, and abnormalities normalized despite the continuation of treatment. Other adverse events probably or possibly related to baricitinib were observed in nine (20%) patients: two oral candidiasis, one bacteremia for Enterococcus faecium, one bacterial pneumonia with negative cultures, three new atrial fibrillation (all had previous heart disease), one hypertensive episode, one episode of heart failure in the presence of known heart disease, one case of urinary obstruction, one episode of diarrhea, and one gastrointestinal bleed in an individual with gastric malignancy. In the unmatched control group from Albacete (n = 376), we observed 17 (4.5%) urinary tract infections and one herpes zoster reactivation. Other complications were gastrointestinal (nausea, vomiting, or diarrhea) in 30.7%, hepatic in 4.5%, renal in 21.1%, neurologic in 9.8%, cutaneous in 4%, and arrhythmias in 5.1%.
IFNs sensitize 3D primary human liver cultures to SARS-CoV-2 infections by inducing ACE2
Organotypic 3D cultures of primary human liver cells are susceptible to infection with SARS-CoV-2 as previously reported (21), with virus signals being enriched in proximity to ACE2 proteins (Fig. 3A). Thus, we evaluated the effect(s) of cytokines on ACE2 expression. To evaluate cytokine effects on human liver cells, we focused on IFNs (IFN-α2, IFN-β, and IFN-γ), interleukins (IL-1β, IL-6, IL-10, and IL-18), and tumor necrosis factor–α (TNFα), serum levels of which are increased in patients with moderate-to-severe disease (15, 16, 28). We detected greater than fivefold induction of ACE2 expression after 16 hours of exposure with IFN-α2 and IFN-β, whereas IFN-γ, TNFα, and the other interleukins tested did not induce ACE2 in liver microtissues (Fig. 3B). As IFN-α2 serum levels in patients exceed those of IFN-β, we focused our subsequent analyses on IFN-α2. Coexposure of liver spheroids to IFN-α2 in combination with other cytokines did not significantly amplify IFN-mediated ACE2 induction (Fig. 3C). On the basis of these data, we thus hypothesized that increased levels of IFN-α2 in patients with severe SARS-CoV-2 infection can stimulate ACE2 expression in parenchymal cells and further increase virulence.
Fig. 3. Baricitinib inhibits cytokine-mediated increased infectivity of SARS-CoV-2 in organotypic primary human liver culture.
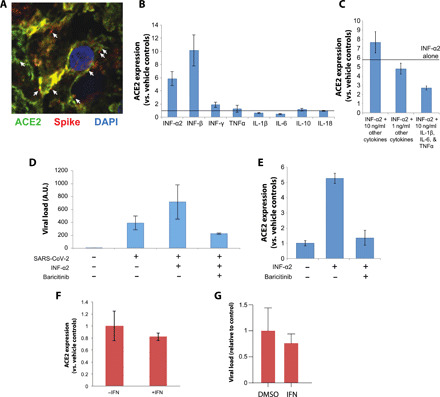
(A) Immunofluorescence confocal imaging of a liver spheroid 48 hours after infection with SARS-CoV-2. Viral spike protein is shown in red, ACE2 in green, and DAPI (4′,6-diamidino-2-phenylindole) in blue. Arrows indicate examples of where spike protein and ACE2 signals are in close proximity. (B) Liver spheroids were treated with different cytokines (10 ng/ml), and the fold increase in ACE2 transcript levels are shown relative to controls (indicated by the solid line). Note that IFN-α2 and IFN-β significantly induce ACE2 levels. N = 2 technical replicates. (C) Combinatorial cytokine exposure does not result in increased ACE2 induction compared to IFN-α2 alone. “Other cytokines” corresponds to IFN-β, IFN-γ, TNFα, IL-1β, IL-6, IL-10, and IL-18. (D) IFN-α2 increases viral load in hepatocyte spheroids, and this effect is fully inhibited by baricitinib. N = 2 to 3 biological replicates. (E) IFN-α2–mediated induction of ACE2 is fully prevented by baricitinib. N = 3 biological replicates. All cytokine concentrations were 10 ng/ml unless stated otherwise. (F) ACE2 in lung organoids is not induced even by very high concentrations of IFN-α2 (50 ng/ml). N = 3 biological replicates. (G) By contrast, IFN-α2 slightly reduces viral load in lung organoids. N = 3 biological replicates. Error bars indicate SEM. A.U., arbitrary units; DMSO, dimethyl sulfoxide.
Next, we evaluated the effects of IFN-α2–mediated induction of ACE2 on viral load and found that IFN-α2 increased viral copy numbers in 3D organotypic liver cultures by approximately twofold (Fig. 3D). Exposure to therapeutically relevant concentrations of baricitinib (800 nM) fully abolished ACE2 induction by IFN-α2 and efficiently blocked the increased infectivity in cytokine-treated 3D liver microtissues even beyond the levels observed in non–cytokine-exposed samples (Fig. 3, D and E), in agreement with our previous report (21). In contrast, in lung organoids, IFN-α2 did not induce ACE2 even at higher concentrations and had opposite, i.e., mild antiviral, effects (50 ng/ml compared to 10 ng/ml for liver spheroids; Fig. 3, F and G), suggesting that IFN effects differ between pulmonary organs and liver. Furthermore, baricitinib did not reduce viral levels in monkey kidney epithelial cells (Vero cells) and human adenocarcinomic alveolar basal epithelial cells (A549 cells) (fig. S4). These results demonstrate that infection and replication mechanisms in primary cells and cell lines are distinctly different and incentivize the use of organotypic culture systems for studies of infectious disease biology.
Baricitinib efficiently blocks IFN signaling
To gain a detailed understanding of the molecular effects of baricitinib, we sequenced primary liver spheroids using RNA sequencing (RNA-seq), resulting in the first comprehensive map of IFN response genes in human hepatocytes. In infected and noninfected liver spheroids, IFN-α2 significantly induced a total of 407 and 696 genes, respectively [log2 fold change (FC) > |1| and false discovery rate (FDR) < 0.05; Fig. 4A]. Genes that were differentially expressed irrespective of infection (n = 271) were strongly enriched in viral defense pathways, antigen presentation, and endosomal trafficking (Fig. 4B). IFN strongly regulated genes associated with platelet activation (29), but this effect was blunted by viral infection, demonstrating intricate pathway-specific fine-tuning of host gene expression by SARS-CoV-2.
Fig. 4. Baricitinib reverses IFN-mediated gene expression signature alterations.
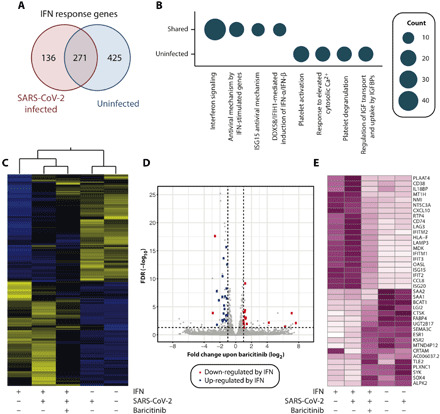
(A) Venn diagram depicting the overlap of differentially expressed genes upon IFN-α2 (10 ng/ml) in SARS-CoV-2–infected and uninfected liver spheroids. (B) Circle plots illustrating significantly deregulated genes falling into specific Reactome terminologies in infected and noninfected samples (FDR < 0.05). Circle diameter is indicative for the number of genes per category. (C) Heatmap representation of IFN-responsive genes (n = 832). z scores of normalized TPMs (transcripts per million mapped reads) are plotted (purple, high; white, low). (D) Volcano plot showing the differentially expressed genes in all infected samples upon treatment with baricitinib. Blue and red dots indicate genes that are significantly up- and down-regulated upon IFN treatment. Note that with the exception of SAA1/2, baricitinib results in inverse changes to gene expression compared to IFN, thus ameliorating IFN-induced gene expression alterations. (E) Heatmap visualization of genes for which significant effects on gene expression were detected for baricitinib and IFN.
Baricitinib reversed IFN-induced gene expression changes (Fig. 4C). Genes that were highly induced upon IFN treatment were significantly down-regulated by baricitinib, including the IFN-stimulated genes (ISG) 15 and 20, chemokines (CCL8 and CXCL10), major histocompatibility complex components (CD74, LAG3, and LAMP3), and several members of the IFN-induced protein (IFIT) family. In contrast, baricitinib reverses the IFN-mediated inhibition of immunoreceptor signaling (SYK), metabolic remodelers (BCAT1 and KSR2), and transcriptional regulators (SOX4 and TLE2; Fig. 4, D and E). The only genes that escaped baricitinib action were serum amyloid A1 (SAA1) and its paralog SAA2, whose expression levels were decreased upon IFN treatment and even further reduced upon baricitinib.
Baricitinib binds to host NAKs
Ligand binding studies as carried out by Sorrell et al. (30) documented that all three of these closely related enzymes are effectively inhibited by baricitinib [BIKE Kd (dissociation constant) = 39.8 nM; AAK1 Kd = 17.2 nM; GAK Kd = 134.4 nM] (figs. S1 and S2). While the most relevant NAK in the inhibition of viral entry is unclear, BIKE (BMP-2-inducible protein kinase) was not expressed in liver cells, suggesting that AAK1 (AP2-associated protein kinase 1) and GAK (cyclin G-associated kinase) are sufficient to mediate viral entry. To disentangle the effects of baricitinib on replication and viral entry, we evaluated viral loads 4 hours after infection using baricitinib concentrations close to the relevant Kd values (100 nM). dSTORM superresolution microscopy revealed anti-nucleocapsid protein immunoreactivity clusters throughout the infected samples (Fig. 5A). In contrast, virus signals were almost absent in baricitinib-treated samples (Fig. 5B). The density of anti-nucleocapsid staining in baricitinib-treated spheroids was almost absent and in baricitinib-treated samples (P < 0.001) and reduced to levels found in noninfected controls, demonstrating that baricitinib efficiently blocked viral entry at nanomolar concentrations (Fig. 5C). These results were corroborated by quantitative polymerase chain reaction (qPCR) analyses, which showed significant reductions in intracellular viral loads, irrespective of IFN-mediated ACE2 induction (Fig. 5D). Combined, these data corroborate a dual-effect model for baricitinib in which the compound (i) inhibits the induction of IFN response genes, such as ACE2, resulting in reduced infectivity during the cytokine storm, and (ii) blocks viral entry by inhibition of NAKs (Fig. 5E).
Fig. 5. Baricitinib blocks viral entry of SARS-CoV-2.
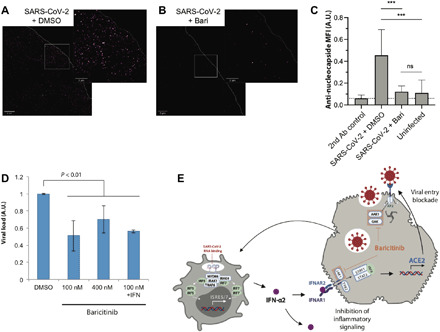
Superresolution dSTORM microscopy of short-term (4 hours) infected liver spheroids stained for nucleocapsid treated with vehicle control (A) or baricitinib (100 nM) (B). (C) Relative mean fluorescence intensities (MFIs) for regions with dimensions of 20 μm by 20 μm of infected and treated organoids and secondary antibody–only controls; five regions per 3D tissue culture. Bars are means ± SD; ***P < 0.001 two-tailed Student’s t test. Ab, antibody; ns, not significant. (D) qPCR analysis of viral load in organotypic primary human liver culture following short-term (4 hours) infections corroborates inhibition of viral entry. (E) Suggested mechanism of dual baricitinib antiviral action on viral entry and inflammatory signaling. Baricitinib inhibits on viral entry by inhibition of the NAKs AAK1 and GAK. In addition, baricitinib blocks inflammatory JAK/STAT signaling, resulting in reduced expression of the IFN target gene and SARS-CoV-2 receptor ACE2. A plasmacytoid dendritic cell is shown on the left, and a hepatocyte is shown on the right.
Model building was also used to assess the potential impact of selected point mutations that occur with differing frequencies naturally, on baricitinib binding to BIKE (Ala58➔Asp, Ala58➔Val, Arg134➔Gln, Arg134➔Leu, Glu131➔Gln, Glu131➔Lys, and Ala135➔Thr), and modeled baricitinib binding to both AAK1 (Ala72➔Thr and Gly132➔Val) and GAK (Glu124➔Val, Gly128➔Glu, and Gln129➔Arg). None of these point mutations appear likely to destabilize the structure of the kinase catalytic domain (i.e., disrupt packing in the hydrophobic core). In addition, none of these point mutations appear likely to disrupt baricitinib binding (table S1). All of the hydrogen bonds and van der Waals interactions observed between the drug and wild-type BIKE appear to be preserved for each point mutant (fig. S2A). The same holds true for the predicted hydrogen bonds and van der Waals interactions between the drug and mutant forms of AAK1 (fig. S2B) and the drug and mutant forms of GAK (fig. S2C), respectively.
DISCUSSION
Effective and well-tolerated treatments for patients infected with the novel coronavirus have proved elusive. Here, we link antiviral and anticytokine activities of baricitinib in hospitalized patients with COVID-19 pneumonia and in spheroid models of SARS-CoV-2 infection. During the pandemic’s exponential phase, in investigator-led studies in both Italy and Spain, the treatment of hospitalized patients with COVID-19 using the JAK/STAT pathway inhibitor baricitinib has been associated with clinical improvement, findings that require assessment in randomized trials (31). The inclusion of a large cohort with a median age of 81 years merits particular attention here.
Baricitinib was generally well tolerated with a reduction in inflammation and substantially improved outcomes. In the matched populations, 16.9% of the baricitinib-treated patients died and/or progressed to invasive mechanical ventilation, compared to 34.9% in the standard-of-care group (P < 0.001; Fig. 2). Note that baricitinib’s effects were evident from the first treatment days and maintained over follow-up (Fig. 2). Although most recommended treatment regimens support the use of 4 mg for 14 days, lower doses and shorter durations may be beneficial for certain populations including elderly patients described here. It is also remarkable from our data that in the propensity score–matched populations from both sites, most of the participants received similar treatment regimens with hydroxychloroquine, lopinavir/ritonavir, LMWH, and glucocorticoids. In Cox proportional hazard analyses, these treatments did not affect survival, while baricitinib was beneficial in univariate, multivariate, and time-related analyses (Table 4).
The main toxicities observed were hepatic, infectious, gastrointestinal, and cardiovascular, although in most cases, a definite causative relationship with baricitinib use could not be ascertained. An elevation in plasma transaminases occurred in some patients, and as previously observed, continuation of baricitinib was possible with resolution of liver function abnormalities. A recent case report in an 87-year-old severely unwell patient from Foggia, Italy showed that its use was associated with rapid clinical improvement in stark contrast to her infected husband and son who did not receive baricitinib and died (32). While these reports are inconclusive, in aggregate, these data strongly support the continued investigation of baricitinib for COVID-19 in ongoing randomized control trials, including ACTT-II (NCT04401579), where baricitinib is given in combination with remdesivir, TACTIC-R (NCT04390464), and others including NCT04373044. Multivariate analyses that we performed showed no differences in corticosteroid type (prednisolone, dexamethasone, hydrocortisone, or methylprednisolone), duration, dose, and outcome.
Age appears the most relevant risk factor for adverse outcomes related to COVID-19 (16). In patients with COVID-19 pneumonia, immune dysfunction is present, even in mild cases (25), and immune responses in older adults are slower, less coordinated, and less efficient, so-called immunosenescence (33). Until now, there have been no clinical trials or post-authorization studies that have demonstrated efficacy or safety of commonly used drugs in much older adults hospitalized with COVID-19, an especially vulnerable and susceptible population, and age is often an exclusion criterion in trials, as is frailty.
One concern in early disease is amelioration of the antiviral activity of type-1 IFNs, which signal through JAK/STAT pathways. Our study using viral infection assays in liver spheroids confirms that ACE2 is an ISG, supporting previous data (34). This induction should increase viral infectivity rather than reduce it. This was observed in human hepatocytes, which normally express low levels of this receptor, in contrast to endothelial cells where ACE2 is abundantly expressed and contributes to the local control of perfusion. This, in turn, suggests that extrapulmonary effects of COVID-19 may be mediated by type-1 IFN–mediated increases in ACE2 expression, both on endothelial and parenchymal cells, resulting in endotheliitis (24) and liver injury in up to 60% of severely ill patients (35). While deficiency in type-1 IFN immunity is associated with life-threatening COVID-19 pneumonia (36), induction of IFNs and ISGs is detected in some critically ill patients (37). In aggregate with our findings in liver spheroids, these data suggest that type I IFNs play an important bivalent role in COVID pathobiology that requires tight regulation and lead us to hypothesize that JAK/STAT inhibitors could be beneficial early in the course of the disease by reducing IFN-I–induced ACE2 expression. Notably, we found important qualitative differences between the response of liver spheroids, where IFNs induced ACE2 and increased infectivity, and lung organoids, where ACE2 and viral load were not affected by IFN signaling. Vascular endothelial cells express high levels of ACE2 (38); however, while endothelial cells are highly responsive to IFN signaling (39), whether ACE2 constitutes an IFN response gene remains to be elucidated. Combined, these data suggest that the effects of baricitinib might differ between organ systems, and we could speculate that the anti-inflammatory effects might be most beneficial in those tissues in which ACE2 is an IFN response gene, including liver. It will be interesting to see whether further systematic analyses of the effect of IFNs on ACE2 expression and infectivity across human tissues will confirm these hypotheses.
To further validate our data in early patients and strengthen the safety database, we also present results for an additional 48 patients with mild-moderate pneumonia treated at two hospitals in Italy (Table 5 and table S1); a transient transaminitis was observed in two individuals, and all patients recovered uneventfully, with the most common drug-induced adverse event in 131 baricitinib-treated individuals presented here.
Table 5. Baseline demographic, clinical, and laboratory characteristics of patients with COVID-19 treated with either baricitinib or with standard COVID-19 therapy and results at 2 weeks from the Hospital of Prato.
Standard univariate statistical tests were performed to compare baricitinib-treated patients to age- and sex-matched controls. These consisted of the Mann-Whitney U test for pairwise comparisons, the Wilcoxon test for paired data, and Fisher’s exact test for categorical variables. Kaplan-Meier product-limit estimation and the log-rank test were used to perform a survival analysis between groups. A dose of 4 mg daily of baricitinib was given for 14 days. SpO2, peripheral capillary oxygen saturation; SBP, systolic blood pressure; DBP, diastolic blood pressure; WBC, white blood cells; MEWS, modified early warning score; CVD, cardiovascular disease; NA, not applicable; IQR, interquartile range; ICU, intensive care unit.
|
Features at baseline (all patients received hydroxychloroquine and lopinavir/ritonavir) |
Baricitinib group | Control group* | P value |
| Patient number, N (%) | 23 (100) | 18 (100) | |
| Male/female, N (%) | 20/3 (87/13) | 14/4 (78/22) | 0.679 |
| Age years, median (IQR) | 62.5 (57.75–72.25) | 64.1 (55.7–70.1) | 0.776 |
| Days interval from symptoms onset and therapy starting |
6 (4–6.25) | 5.5 (4–5.25) | 0.924 |
| Cough, N (%) | 17 (73.9) | 15 (83.3) | 0.709 |
| Dyspnea, N (%) | 20 (86.9) | 14 (77.8) | 0.679 |
| Sputum production, N (%) | 7(30.4) | 9 (50) | 0.334 |
| Headache, N (%) | 8 (34.8) | 7 (38.9) | 0.757 |
| Diarrhea, N (%) | 5 (21.7) | 5 (27.8) | 0.524 |
| Ageusia/anosmia, N (%) | 9 (39.1) | 8 (44.4) | 0.860 |
| Hypertension, N (%) | 5 (21.7) | 6 (33.3) | 0.489 |
| Diabetes, N (%) | 6 (26) | 4 (22.2) | 1.000 |
| COPD, N (%) | 5 (21.7) | 4 (22.2) | 1.000 |
| CVD, N (%) | 4 (17.4) | 2 /11.1) | 0.679 |
| Malignancy, N (%) | 1 (4.3) | 1 (5.5) | 1.000 |
| Fever (°C) | 38 (37.5–38.6) | 37.9 (37.6–38.9) | 0.912 |
| Respiratory rate (N/min) | 18 (16.5–23.2) | 21 (18–24) | 0.524 |
| SpO2 (%) | 94 (90–95.5) | 92 (91–93) | 0.357 |
| PaO2/FiO2, median (IQR) | 293 (199–296) | 271.4 (264–283) | 0.356 |
| Pulse rate, median (IQR) | 84 (72.3–89.1) | 88 (86–94.5) | 0.129 |
| SBP mm/Hg, median (IQR) | 110 (100–130) | 105 (98–115.6) | 0.789 |
| DBP mm/Hg, median (IQR) | 70 (60–84) | 65.5 (60–68.5) | 0.589 |
| WBC (×109/liter), median (IQR) | 7.6 (5.7–10.4) | 7.9 (7.1–8.6) | 0.757 |
| Neutrophils (×109/liter), median (IQR) | 6,3 (4.2–7.8) | 7.1 (6.4–8.1) | 0.224 |
| Lymphocytes (×109/liter), median (IQR) | 0.6 (0.5–1.1) | 0.72 (0.6–0.8) | 0.524 |
| Hemoglobin (g/liter), median (IQR) | 116 (102–133.2) | 127 (108–136) | 0.565 |
| Platelets (×109/liter), median (IQR) | 207 (174–232) | 368 (340–415) | 0.002 |
| ALT (IU/liter), median (IQR)† | 27.6 (22.7–53.1) | 44 (36–50) | 0.176 |
| AST (IU/liter), median (IQR) | 31 (25.2–47.3) | 44 (34.7–48) | 0.235 |
| ALT (IU/liter) > upper normal limit N (%) | 8 (34.7) | 9 (50) | 0.358 |
| ALT (IU/liter) > upper normal limit, median (IQR) |
50 (45.5–62.7) | 55 (45–68) | 0.707 |
| AST (IU/liter) > upper normal limit N/% | 10 | 11 | 0.350 |
| AST (IU/liter) > upper normal limit, median (IQR) |
51.5 (44.5–76.5) | 67 (55–80) | 0.302 |
| Creatinine (mg/dl), median (IQR) | 1.0 (0.9–1.3) | 1.1 (0.9–1.2) | 0.789 |
| CRP (mg/dl), median (IQR) | 9.12 (5.9–16.5) | 4.3 (1.5–5.2) | 0.001 |
| Procalcitonin (ng/ml), median (IQR) | 0.5 (0.3–1.0) | 1.1 (0.8–2.2) | 0.589 |
| IL-6 (pg/ml)‡, median (IQR) | 29.2 (7.1–39.4) | 24.2 (5.2–27.6) | 0.189 |
| MEWS, median (IQR) | 2 (1–3.1) | 3 (3–4) | 0.544 |
| Results at 2 weeks after therapy | |||
| ICU admission N (%) | 0 | 5 (33) | 0.011 |
| Discharged, N (%) | 18 (78.2) | 1 (5.5) | <0.0001 |
| SpO2, median (IQR) | 97 (94.8–98.1) | 92.4 (85.5–93.2) | <0.0001 |
| PaO2/FiO2 value, median (IQR) | 428.7 (306.1–457) | 277.8 (144–345) | 0.002 |
| Lymphocytes (×109/liter), median (IQR) | 1.3 (1.2–1.9) | 0.8 (0.6–0.9) | 0.019 |
| CRP (mg/dl), median (IQR) | 0.87 (0.58–2.9) | 5.2 (2.1–12.3) | <0.0001 |
| IL-6 (pg/ml)‡, median (IQR) | 6.1 (3.2–7.4) | NA | NA |
| ALT (IU/liter), median (IQR) | 37 (24.1–57.4 | NA | NA |
| AST (IU/liter), median (IQR) | 55.4 (28–64.3) | NA | NA |
*Standard therapy group: Patients with COVID-19 under standard respiratory therapy commenced antiretrovirals (Kaletra) and hydroxychloroquine before starting the therapy with baricitinib.
†Normal ALT and AST values: 10 to 40 IU/liter.
‡IL-6 normal value: <7 pg/ml.
The susceptibility of liver spheroids to SARS-CoV-2 infection was, at first, unexpected to us but ties in with the transaminitis observed. Note that SARS-CoV-2 can productively infect human gut enterocytes, as shown using human small intestinal organoids (40). We have analyzed ACE2 expression in available proteomics and RNA-seq data and detected low-level ACE2 transcripts or protein with average FPKM (fragments per kilobase of transcript per million mapped reads) data across three donors of 0.95 (41). These levels are consistent with previous data indicating low but detectable levels of ACE2 in liver cells (42). We have also interrogated the The Cancer Genome Atlas with the UALCAN database and found low levels of ACE2 transcripts in peritumoral liver cells in hepatocellular carcinoma data.
The demonstration that baricitinib inhibited viral infectivity in vitro, as well as having an overall anti-inflammatory effect in vivo, confirms predictions arising from the use of AI and the comprehensive biomedical knowledge graph (7–9). It also demonstrates that compiling such an AI-enriched database, with its associated algorithms, enables the identification of relationships, in this case, the ability of a single approved drug to both inhibit viral infectivity and ameliorate the exuberant inflammatory consequences of viral infection. It also enables network effects to be identified quickly, which is of enormous importance when trying to identify therapeutics in the midst of a global pandemic (10).
We did not observe any thrombotic or vascular events in our cohort, a previously raised possible concern with the use of baricitinib and also an increasing concern in general with COVID-19 infection (14, 43, 44), although most patients (86%) here received LMWH. The short half-life of baricitinib (t1/2 = 12.5 hours) versus the anti–IL-6 antibodies such as tocilizumab (t = 13 days), oral once/daily administration, and lack of drug-drug interactions (it is excreted largely unchanged), we believe, lend itself to use during a short-term viral infection and also the possibility of utility in low- and middle-income countries. A glomerular filtration rate (GFR) of <30 ml/min is a limitation for the higher 4-mg dose, but a 2-mg dose can be used: We highlight from the Spanish cohort that specifically shows that baricitinib may be safe and effective with the lower 2-mg dose and fewer treatment days. Randomized trials are needed in this elderly population, and note, for example, that many other studies (e.g., for remdesivir) exclude elderly individuals or those with a GFR of <50 ml/min. Other features such as its potential to cross the blood-brain barrier and help ameliorate neurologic sequelae may also be beneficial in selected patients, as we have described (45, 46). Trials of other JAK inhibitors such as ruxolitinib are ongoing, and the same principles may apply (47), although our knowledge graph specifically called out baricitinib among these related molecules (9). We noted that the brain:plasma ratio for baricitinib measured 5× that for ruxolitinib (45). Furthermore, the computational docking results (figs. S3 and S4) suggest that any one or two or all three of the NAKs could be the baricitinib target(s) relevant to our clinical findings, although BIKE is unexpressed on liver spheroids so the target is likely to be AAK1 and/or GAK. We consider it unlikely that baricitinib is acting on an alternative protein target(s) that may not even be a protein kinase, but we cannot rule out this possibility. For example, TMPRSS2, a membrane-bound serine protease that primes the spike protein (48), is also well known to be important for viral entry via ACE2. IL-6 signaling (49) is related to its expression, and baricitinib, which reduces IL-6 levels in a dose-dependent manner, may affect its expression. Should additional clinical studies confirm the benefits of baricitinib in managing advanced SARS-CoV-2 infections, efforts aimed at identification of the precise molecular target(s) would be useful here.
Notwithstanding our findings supporting the hypothesis that baricitinib is clinically active and safe in this setting, there are several limitations in our study that need to be acknowledged. This was not a randomized trial comparing baricitinib to a placebo control group. Therefore, known and unknown confounding variables could have compromised the results. The propensity score–matching procedure eliminated much of the group imbalance in known confounding variables, but some imbalance remained in important parameters such as existing comorbidities. In addition, the sample size from the final baricitinib cohort was small, limiting overall statistical power. Multiple statistical tests were performed without an adjustment for multiplicity, which increases the risk of a type I statistical error. Given these limitations, we eagerly await the completion of the baricitinib randomized trials that are currently ongoing.
In summary, we have confirmed dual actions of baricitinib, demonstrating its ability to inhibit viral entry into primary human hepatocyte spheroids and the reduction in inflammatory markers in patients with COVID-19 as per our suggested model (Fig. 5C). In addition, we show that baricitinib prevents the type-1 IFN–mediated increase in expression of ACE2, the receptor for SARS-CoV-2.
METHODS
Patients and statistics
Two observational studies from Italy and Spain were conducted, approved by the Internal Review Boards (IRBs) of each institution. Entry criteria for each institution included patients with radiologically defined COVID-19 pneumonia and laboratory-confirmed infection, as diagnosed by a positive SARS-CoV-2 RT-PCR (reverse transcription PCR) test by nasopharyngeal swab. All patients enrolled had a blood oxygen saturation (SaO2) < 94% at baseline but did not require mechanical ventilation. The Pisa, Italy cohort included all consecutive cases with SARS-CoV-2 pneumonia and PaO2/FiO2 (P/F) ratio of <300 mmHg at admission diagnosed between the period of 7 and 31 March 2020 and had moderate-to-severe or severe disease. The COVID-AGE study from Albacete, Spain (NCT04362943) was conducted at Complejo Hospitalario Universitario of Albacete, dedicated to older adults; thus, baricitinib doses were lower than in Italy. The Albacete cohort included patients ≥70 years with SARS-CoV-2 pneumonia, diagnosed between 9 March and 20 April 2020, not requiring mechanical ventilation, and again having moderate-to-severe or severe disease.
Written informed consent was obtained from each patient in Italy and verbal informed consent in Spain as approved by the local IRB. Baricitinib was administered at a dose of 4 mg/day for 14 days in conjunction with standard of care in Italy and at lower doses of 2 or 4 mg/day for 3 to 11 days in the Spanish cohort because of age-related factors. Any patient with confirmed COVID-19 infection who received at least three doses of baricitinib was included in the intent-to-treat analysis. Standard clinical and laboratory data were collected including patients’ demographics, comorbidities, oxygen support, adverse events, laboratory values, concomitant therapies, and clinical outcomes. Exclusion criteria in all patients included a history of active or latent tuberculosis infection (QuantiFERON Plus-test positivity, QIAGEN, Germany), pregnancy, and/or lactation. The primary outcome was death from any cause or intensive care unit admission needing invasive mechanical ventilation during hospitalization.
For both cohorts, propensity score matching was used to create the control selection sample using patients admitted in the same period of time in both hospitals, not treated with baricitinib. Cases and controls were matched for relevant predictors of respiratory failure and death, as well as baseline treatments. The following variables, all of them clinically relevant for mortality and respiratory failure, were included in the propensity score estimation: age, sex, chronic obstructive pulmonary disease, arterial hypertension, cardiovascular disease, diabetes mellitus, chronic kidney failure, Charlson comorbidity index, baseline P/F ratio, lymphocyte count, therapy with steroids, LMWH, and “antiviral” therapy with hydroxychloroquine or protease inhibitors (lopinavir/ritonavir). Propensity score matching was conducted with the statistical package MatchIt (v4.0.2), using the recommendations from Ho et al. (50) to improve parametric models and preprocessing of nonparametric data. After the propensity score was determined for each patient, those treated with baricitinib were matched to 1:1 to a control patient using a greedy matching procedure with replacement, targeting the average treatment effect on the treated replacement. Continuous variables were tested using Wilcoxon-Mann-Whitney rank sum test; categorical variables were tested using Pearson’s chi-square or Fisher’s exact test. For every test performed, the P value was always above 0.05, and therefore, we can say that the groups were homogeneous regarding the control variables. Validity of the balance in potential confounding variables was assessed visually with normality plots using the “R” function [“geom:smooth()” using formula “y ~ x”], also using the Wilcoxon rank sum test with continuity correction analyses for continuous variables, and with Pearson’s chi-square tests for categorical ones. Although the matching figures were not perfect, no statistical differences were found for any variables included in the propensity score in the merged Pisa and Albacete cohorts. Mortality reduction was analyzed with Cox proportional hazard models adjusted for all the variables included in the propensity score matching, to further control for the small deviations in the matching procedure, plus the presence of active cancer, alanine aminotransferase, and antibiotic use. Safety data are presented in a descriptive manner. Last, individual and merged data from Pisa and Albacete were used to analyze the primary outcome, survival, or mechanical ventilation, in baricitinib and control groups using Kaplan-Meier analysis, including 95% CIs. The global significance was determined using a Peto-Peto analysis that prioritizes the first part of the curves but with increased robustness compared to Taron-Ware analysis. This decision was taken after the observation that the main differences between baricitinib and control patients were present from the beginning of the treatment. Statistical significance was established at P < 0.05 using a two-sided test statistic. All analyses were done using the statistical package R.
Human spheroid and organoid culture, cytokine exposures, and SARS-CoV-2 infections
SARS-CoV-2 (GenBank accession number MT093571) was isolated from a nasopharyngeal sample of a patient in Sweden on Vero E6 cells. Organotypic 3D primary human liver cultures with physiological transcriptomic, proteomic, and metabolomic profiles were cultured as previously described (51–53). In short, cryopreserved primary human hepatocyte (BioIVT, USA) were thawed, and the isolated cell suspensions were seeded into 96-well ultralow attachment plates (Corning) with 1500 cells per well. In short, cells were seeded in 100 μl of Williams E medium supplemented with 2 mM l-glutamine, penicillin (100 U/ml), streptomycin (100 μg/ml), insulin (10 μg/ml), transferrin (5.5 μg/ml), sodium selenite (6.7 ng/ml), 100 nM dexamethasone, and 10% fetal bovine serum. Spontaneous self-aggregation of the hepatocytes occurred, and after few days, a single spheroid could be observed per well. Once spheroids were sufficiently compact (after day 5), cell aggregates were preexposed to baricitinib and/or cytokines.
Formed liver spheroids and pulmonary organoids were exposed for 1 day with reconstituted human IFN-α2 (BioLegend, no. 592706), IFN-β (R&D Systems, no. 8499-IF-010), IFN-γ (BioLegend, no. 570206), IL-1β (BioLegend, no. 579406), IL-6 (R&D Systems, no. 206-IL-050), IL-10 (R&D Systems, no. 217-IL-010), IL-18 (BioLegend, no. 592104), and TNFα (R&D Systems, no. 210-TA-100) either alone or in combination with baricitinib before viral infections. Cytokine concentrations were 10 ng/ml unless stated otherwise. Following these preexposures, cells were infected with SARS-CoV-2 at a multiplicity of infection of 0.1 in triplicate for 48 hours. After 48 hours, spheroids were washed with phosphate-buffered saline (PBS), pooled (32 wells per condition), and lysed using TRIzol (Thermo Fisher Scientific). RNA was extracted using Direct-zol mini kit (Zymo Research), and relative levels of ACE2 and viral RNA were determined by qRT-PCR as described (54).
Confocal microscopy
Primary human hepatocyte spheroids were fixed in 4% paraformaldehyde for at least 4 hours and washed with PBS. For cryoprotection, spheroids were incubated with 30% sucrose in PBS at 4°C overnight or until the microtissues sank. Subsequently, they were washed with PBS and transferred into micromolds for OCT (optimal cutting temperature) cryo-mount embedding. OCT-embedded spheroids were frozen in an isopropanol dry ice bath and were sectioned at 8-μm thickness on a CryoStar NX70 cryostat. Sections were washed twice with PBS for 10 min and blocked with PBTA buffer [5% bovine serum albumin (BSA), 0.25% Triton X-100, and 0.01% NaN3 in PBS] for 2 hours at room temperature. Subsequently, the blocked sections were incubated overnight at 4°C with the monoclonal primary antibody anti-1A9 (diluted in PBTA to a final concentration of 5 μg/ml). Samples were washed 3 × 15 min with PBS at room temperature before incubation with the secondary antibody (donkey anti-mouse diluted in PBTA at 1:500) for 2 hours at room temperature. Unbound secondary antibody was washed out three times with PBS (15 min each) at room temperature, and the slides were mounted with 4′,6-diamidino-2-phenylindole Gold Antifade.
Superresolution microscopy
Sections, prepared as above, were thawed and rehydrated using PBS then blocked for 30 min using a blocking buffer containing 10% goat serum, 2% BSA, and 0.3% Triton X-100. Primary antibody against nucleocapsid was diluted at 1:500 in 10% goat serum and 2% BSA and incubated overnight at 4°C. Sections were then washed with PBS 3 × 10 min. Primary antibodies were detected with Alexa Fluor 647–conjugated secondary goat anti-mouse F(ab′)2 (Abcam, ab98758) diluted to 500 ng/ml in 10% goat serum and 2% BSA for 1 hour at room temperature. Last, sections were washed 5 × 10 min and mounted using an ONI B-Cubed imaging buffer. Superresolution images were acquired on the Nanoimager S Mark II from ONI (Oxford Nanoimaging) equipped with 405-nm/150-mW, 473-nm/1-W, 560-nm/1-W, and 640-nm/1-W lasers and dual emission channels split at 640 nm. To achieve single-molecule blinking, samples were irradiated with the 640-nm laser, and then, 10,000 frames were acquired in an appropriate focal plane at 30 Hz. Single-molecule data were filtered on the basis of photon count, precision, and sigma value in NimOS. All samples were filtered using the same parameters.
RNA sequencing
RNA was isolated from 3D liver tissue samples and were ribosomal RNA (rRNA) depleted, followed by strand-specific RNA library generation (Takara SMART-Seq Stranded kit). RNA libraries were sequenced paired end (75 + 75 cycles) on an Illumina NextSeq 500 device. On average, 20 million reads per sample were obtained, low-quality reads were removed, and ends were trimmed using Trimmomatic (version 0.36). rRNA contamination was determined by mapping to a customized human rRNA reference genome (HISAT2 version 2.1.0). Approximately, 8 to 15% rRNA contamination was found in the samples, which is below the manufacturer’s expected range between 15 and 35%. Nonaligned reads were further mapped to the human reference genome (downloaded from University of California, Santa Cruz, hg38). After mapping to the human genome, generated .bam files were processed using samtools (version 1.10). Bedgraph files were generated using Homer (version 4.11). Last, count tables were generated using the tool Subread (version 2.0.0).
Differential gene expression analysis was performed using both DESeq2 (version 1.26) and edgeR (version 3.28). For both methods, genes with zero counts were filtered out, and more than eight counts in at least 2 of the 12 samples were considered further. In DESeq2, independent filtering and Cook’s distance were set to false. In edgeR, the glmLRT method was used. Only genes with log2 FC > |1| and FDR < 0.05 were considered. When intersecting the number of differentially expressed genes identified using the two methods, we found an overlap of >90% and continued the analysis by using the data obtained from DESeq2. Pathway analysis was performed using the R package ReactomePA (version 1.30.0).
Computational structure analysis
3D atomic-level structures of NAKs BIKE [Protein Data Bank (PDB) ID 4w9x], AAK1 (PDB ID 4wsq), and GAK (PDB ID 4y8d) were compared at the level of amino acid sequence and 3D structure (figs. S1 and S2). Superposition computational docking using the experimental structure of the BIKE-baricitinib protein:drug complex (PDB ID 4w9x) was used to investigate the modes of binding of baricitinib to both AAK1 and GAK (fig. S2). Figures were generated using UCSF Chimera (55).
Supplementary Material
Acknowledgments
We are grateful to the patients who contributed data. Funding: V.M.L. acknowledges support by the Swedish Research Council (grant agreement numbers 2016-01153, 2016-01154, and 2019-01837), the Strategic Research Programmes in Diabetes (SFO Diabetes) and Stem Cells and Regenerative Medicine (SFO StratRegen), and the EU/ EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant 875510). D.G. is partially funded by Ricerca Corrente Linea 1 and 3. J.S. and S.O. wish to thank the Imperial BRC, ECMC, the NIHR, BSAC, and AAC. The RCSB Protein Data Bank is supported by grants to S.K.B. from the NSF (DBI-1832184), the NIH (R01GM133198), and the Department of Energy (DE-SC0019749). P.A. acknowledges support by CIBERFES, Instituto de Salud Carlos III, Ministerio de Economía y Competitividad, España, and Ayuda cofinanciada por el Fondo Europeo de Desarrollo Regional (FEDER) Una Manera de hacer Europa (grant number CB16/10/00408). This study was supported by the Imperial BRC and ECMC, the NIHR, and AAC. Role of the funding sources: none declared. Author contributions: Study concept: J.S., F.Me., A.M., P.A., and V.M.L. Clinical data collection: G.S.N., M.F., G.T., L.G., A.V., F.Mo., L.R.R., F.F., A.A.C., S.D.M., L.C., P.M.S.-J., D.C.B., L.S.M., M.P., M.M.R., F.A.P., R.S.S.-T., R.G.-M., F.Me., and P.A. Experimental analyses of liver cultures: S.Y., J.X.S., and V.M.L. In vitro viral assays: V.M., J.X.S., S.Y., and A.M. Statistical analyses: G.D. and P.A. Superresolution microscopy: J.H.F., Z.F.N., and A.G.M. RNA-seq: C.S. and C.K. Manuscript writing: J.S., S.Y., P.A., and V.M.L. P.R., S.O., Y.-J.T., W.L., H.Z., and J.M.P. contributed to data analysis. F.L., L.G.L., N.C., A.P., L.N., D.M., D.G., F.C., and A.F. contributed to patient cohort data. S.K.B. and S.D. contributed to modeling studies. All authors contributed to data collection, helped revise and write the final manuscript, and approved the final manuscript. Competing interests: The conflicts of J.S. can be found at https://nature.com/onc/editors (it includes a lecture fee from Eli Lilly for discovering the role of baricitinib here). P.R. is an employee of BenevolentAI and received an honoraria for a lecture from Eli Lilly for lecturing with J.S. M.F. received grants and speaker honoraria from MSD, Angelini, Shionogi, and Nordic Pharma. L.G. is a cofounder of Quipu, a spin-off company of the University of Pisa and National Research Center of Pisa, Italy, and received a research grant from Pfizer and speaker honoraria from Boehringer Ingelheim, Corman, Sanofi-Aventis, and Sevier. F.M. has participated in advisory boards and/or received speaker honoraria from Angelini, Correvio, MSD, Pfizer, Astellas, Gilead, BMS, Jansenn, ViiV, BioMerieux, Biotest, Becton-Dickinson, Pfizer, and Shionogi. D.G. has received a research grant from Eli Lilly and has served on the Scientific Advisory Board for Eli Lilly and Company. S.D. has no competing interest. S.K.B. received a speaker honorarium from Incyte Pharmaceuticals. V.M.L. is the co-founder, the CEO, and a shareholder of HepaPredict AB. In addition, V.M.L. discloses consultancy work for EnginZyme AB. G.S.N. has received a speaker honorarium from Eli Lilly. The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/sciadv.abe4724/DC1
REFERENCES AND NOTES
- 1.Fauci A. S., Lane H. C., Redfield R. R., Covid-19—Navigating the uncharted. N. Engl. J. Med. 382, 1268–1269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paules C. I., Marston H. D., Fauci A. S., Coronavirus infections—More than just the common cold. JAMA 323, 707–708 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Wu Z., McGoogan J. M., Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA 323, 1239–1242 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Tanaka Y., Emoto K., Cai Z., Aoki T., Schlichting D., Rooney T., Macias W., Efficacy and safety of baricitinib in Japanese patients with active rheumatoid arthritis receiving background methotrexate therapy: A 12-week, double-blind, randomized placebo-controlled study. J. Rheumatol. 43, 504–511 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Baden L. R., Rubin E. J., Covid-19 - The search for effective therapy. N. Engl. J. Med. 382, 1851–1852 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullard A., COVID-19 vaccine development pipeline gears up. Lancet 395, 1751–1752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Rawling M., Savory E., Stebbing J., Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 395, e30–e31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson P. J., Corbellino M., Stebbing J., Baricitinib for COVID-19: A suitable treatment?—Authors’ reply. Lancet Infect. Dis. 20, 1013–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stebbing J., Krishnan V., de Bono S., Ottaviani S., Casalini G., Richardson P. J., Monteil V., Lauschke V. M., Mirazimi A., Youhanna S., Tan Y.-J., Baldanti F., Sarasini A., Terres J. A. R., Nickoloff B. J., Higgs R. E., Rocha G., Byers N. L., Schlichting D. E., Nirula A., Cardoso A., Corbellino M.; Sacco Baricitinib Study Group , Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol. Med. 12, e12697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz M. B., Vera V., Sinclair D. A., Can artificial intelligence identify effective COVID-19 therapies? EMBO Mol. Med. 12, e12817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Titanji B. K., Farley M. M., Mehta A., Connor-Schuler R., Moanna A., Cribbs S. K., O’Shea J., De Silva K., Chan B., Edwards A., Gavegnano C., Schinazi R. F., Marconi V. C., Use of baricitinib in patients with moderate and severe COVID-19. Clin. Infect. Dis. 2020, ciaa879 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridman J. S., Scherle P. A., Collins R., Burn T. C., Li Y., Li J., Covington M. B., Thomas B., Collier P., Favata M. F., Wen X., Shi J., Gee R. M., Haley P. J., Shepard S., Rodgers J. D., Yeleswaram S., Hollis G., Newton R. C., Metcalf B., Friedman S. M., Vaddi K., Selective inhibition of JAK1 and JAK2 is efficacious in rodent models of arthritis: preclinical characterization of INCB028050. J. Immunol. 184, 5298–5307 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Genovese M. C., Kremer J., Zamani O., Ludivico C., Krogulec M., Xie L., Beattie S. D., Koch A. E., Cardillo T. E., Rooney T. P., Macias W. L., de Bono S., Schlichting D. E., Smolen J. S., Baricitinib in patients with refractory rheumatoid arthritis. N. Engl. J. Med. 374, 1243–1252 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Taylor P. C., Keystone E. C., van der Heijde D., Weinblatt M. E., Del Carmen Morales L., Gonzaga J. R., Yakushin S., Ishii T., Emoto K., Beattie S., Arora V., Gaich C., Rooney T., Schlichting D., Macias W. L., de Bono S., Tanaka Y., Baricitinib versus placebo or adalimumab in rheumatoid arthritis. N. Engl. J. Med. 376, 652–662 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., Guan L., Wei Y., Li H., Wu X., Xu J., Tu S., Zhang Y., Chen H., Cao B., Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 395, 1054–1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ackermann M., Verleden S. E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., Li W. W., Li V. W., Mentzer S. J., Jonigk D., Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 383, 120–128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross O., Moerer O., Weber M., Huber T. B., Scheithauer S., COVID-19-associated nephritis: Early warning for disease severity and complications? Lancet 395, e87–e88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F., Li L., Xu M. D., Wu J., Luo D., Zhu Y. S., Li B. X., Song X. Y., Zhou X., Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J. Clin. Virol. 127, 104370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan Q., Yang K., Wang W., Jiang L., Song J., Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 46, 846–848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P., COVID-19: Combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 20, 400–402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F. G., Horby P. W., Cao B., Wang C., Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395, 1569–1578 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.RECOVERY Collaborative Group, Horby P., Lim W. S., Emberson J. R., Mafham M., Bell J. L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., Prudon B., Green C., Felton T., Chadwick D., Rege K., Fegan C., Chappell L. C., Faust S. N., Jaki T., Jeffery K., Montgomery A., Rowan K., Juszczak E., Baillie J. K., Haynes R., Landray M. J., Dexamethasone in hospitalized patients with Covid-19—Preliminary report. N. Engl. J. Med. , (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varga Z., Flammer A. J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A. S., Mehra M. R., Schuepbach R. A., Ruschitzka F., Moch H., Endothelial cell infection and endotheliitis in COVID-19. Lancet 395, 1417–1418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., Choe H., Farzan M., Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F., Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilk A. J., Rustagi A., Zhao N. Q., Roque J., Martínez-Colón G. J., Kechnie J. L. M., Ivison G. T., Ranganath T., Vergara R., Hollis T., Simpson L. J., Grant P., Subramanian A., Rogers A. J., Blish C. A., A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070–1076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang K.-J., Su I.-J., Theron M., Wu Y.-C., Lai S.-K., Liu C.-C., Lei H.-Y., An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 75, 185–194 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Cheng X., Tang Y., Qiu X., Wang Z., Fu G., Wu J., Kang H., Wang J., Wang H., Chen F., Xiao X., Billiar T. R., Lu B., The role of type 1 interferons in coagulation induced by gram-negative bacteria. Blood 135, 1087–1100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorrell F. J., Szklarz M., Abdul Azeez K. R., Elkins J. M., Knapp S., Family-wide structural analysis of human numb-associated protein kinases. Structure 24, 401–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ottaviani S., Stebbing J., What is the best drug to treat COVID-19? The need for randomized controlled trial. Med. (N.Y.), 10.1016/j.medj.2020.04.002 (2020). [DOI] [PMC free article] [PubMed]
- 32.Lo Caputo S., Corso G., Clerici M., Santantonio T. A., Baricitinib: A chance to treat COVID-19? J. Med. Virol. 2020, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolich-Zugich J., Knox K. S., Rios C. T., Natt B., Bhattacharya D., Fain M. J., SARS-CoV-2 and COVID-19 in older adults: What we may expect regarding pathogenesis, immune responses, and outcomes. GeroSci. 42, 505–514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziegler C. G. K., Allon S. J., Nyquist S. K., Mbano I. M., Miao V. N., Tzouanas C. N., Cao Y., Yousif A. S., Bals J., Hauser B. M., Feldman J., Muus C., Wadsworth M. H. II, Kazer S. W., Hughes T. K., Doran B., Gatter G. J., Vukovic M., Taliaferro F., Mead B. E., Guo Z., Wang J. P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J. M. S., Taylor C. J., Lin B., Waghray A., Mitsialis V., Dwyer D. F., Buchheit K. M., Boyce J. A., Barrett N. A., Laidlaw T. M., Carroll S. L., Colonna L., Tkachev V., Peterson C. W., Yu A., Zheng H. B., Gideon H. P., Winchell C. G., Lin P. L., Bingle C. D., Snapper S. B., Kropski J. A., Theis F. J., Schiller H. B., Zaragosi L.-E., Barbry P., Leslie A., Kiem H.-P., Flynn J. A. L., Fortune S. M., Berger B., Finberg R. W., Kean L. S., Garber M., Schmidt A. G., Lingwood D., Shalek A. K., Ordovas-Montanes J.; HCA Lung Biological Network , SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181, 1016–1035.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C., Shi L., Wang F.-S., Liver injury in COVID-19: Management and challenges. Lancet 5, 428–430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., Bastard P., Liu Z., le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I. K. D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A. A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W. M., Razooky B. S., Hoffmann H. H., Michailidis E., Moens L., Han J. E., Lorenzo L., Bizien L., Meade P., Neehus A. L., Ugurbil A. C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M. F., Mansouri D., al-Saud H., al-Mulla F., Almourfi F., al-Muhsen S. Z., Alsohime F., al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L. R., D’Angio’ M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A. J., Tompkins M. F., Alba C., Vandernoot I., Goffard J. C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P. E., Keles S., Çölkesen F., Ozcelik T., Yasar K. K., Senoglu S., Karabela Ş. N., Rodríguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C.; COVID-STORM Clinicians; COVID Clinicians; Imagine COVID Group; French COVID Cohort Study Group; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; NIAID-USUHS/TAGC COVID Immunity Group, Snow A. L., Dalgard C. L., Milner J. D., Vinh D. C., Mogensen T. H., Marr N., Spaan A. N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M. J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R. P., Zhang S. Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C. M., Abel L., Notarangelo L. D., Cobat A., Su H. C., Casanova J. L., Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, eabd4570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J., Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 27, 883–890.e2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamming I., Timens W., Bulthuis M. L. C., Lely A. T., Navis G. J., van Goor H., Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 203, 631–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia H., Thelwell C., Dilger P., Bird C., Daniels S., Wadhwa M., Endothelial cell functions impaired by interferon in vitro: Insights into the molecular mechanism of thrombotic microangiopathy associated with interferon therapy. Thromb. Res. 163, 105–116 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Lamers M. M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T. I., Ravelli R. B. G., van Schayck J. P., Mykytyn A. Z., Duimel H. Q., van Donselaar E., Riesebosch S., Kuijpers H. J. H., Schipper D., van de Wetering W. J., de Graaf M., Koopmans M., Cuppen E., Peters P. J., Haagmans B. L., Clevers H., SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilarnau N., Vorrink S. U., Ingelman-Sundberg M., Lauschke V. M., A 3D cell culture model identifies Wnt/β-catenin mediated inhibition of p53 as a critical step during human hepatocyte regeneration. Adv. Sci. 7, 2000248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paizis G., Tikellis C., Cooper M. E., Schembri J. M., Lew R. A., Smith A. I., Shaw T., Warner F. J., Zuilli A., Burrell L. M., Angus P. W., Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut 54, 1790–1796 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levi M., Thachil J., Iba T., Levy J. H., Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 7, e438–e440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott I. C., Hider S. L., Scott D. L., Thromboembolism with Janus kinase (JAK) inhibitors for rheumatoid arthritis: How real is the risk? Drug Saf. 41, 645–653 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Richardson P. J., Ottaviani S., Prelle A., Stebbing J., Casalini G., Corbellino M., CNS penetration of potential anti-COVID-19 drugs. J. Neurol. 267, 1880–1882 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gavegnano C., Haile W. B., Hurwitz S., Tao S., Jiang Y., Schinazi R. F., Tyor W. R., Baricitinib reverses HIV-associated neurocognitive disorders in a SCID mouse model and reservoir seeding in vitro. J. Neuroinflammation 16, 182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong J., Tang J., Ye C., Dong L., The immunology of COVID-19: Is immune modulation an option for treatment? Lancet Rheumatol. , (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 81, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Handle F., Puhr M., Schaefer G., Lorito N., Hoefer J., Gruber M., Guggenberger F., Santer F. R., Marques R. B., van Weerden W. M., Claessens F., Erb H. H. H., Culig Z., The STAT3 inhibitor galiellalactone reduces IL6-mediated AR activity in benign and malignant prostate models. Mol. Cancer Ther. 17, 2722–2731 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Ho D. E., Imai K., King G., Stuart E. A., Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Polit. Anal. 15, 199–236 (2017). [Google Scholar]
- 51.Bell C. C., Hendriks D. F. G., Moro S. M. L., Ellis E., Walsh J., Renblom A., Puigvert L. F., Dankers A. C. A., Jacobs F., Snoeys J., Sison-Young R. L., Jenkins R. E., Nordling Å., Mkrtchian S., Park B. K., Kitteringham N. R., Goldring C. E. P., Lauschke V. M., Ingelman-Sundberg M., Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 6, 25187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell C. C., Lauschke V. M., Vorrink S. U., Palmgren H., Duffin R., Andersson T. B., Ingelman-Sundberg M., Transcriptional, functional, and mechanistic comparisons of stem cell-derived hepatocytes, HepaRG cells, and three-dimensional human hepatocyte spheroids as predictive in vitro systems for drug-induced liver injury. Drug Metab. Dispos. 45, 419–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vorrink S. U., Ullah S., Schmidt S., Nandania J., Velagapudi V., Beck O., Ingelman-Sundberg M., Lauschke V. M., Endogenous and xenobiotic metabolic stability of primary human hepatocytes in long-term 3D spheroid cultures revealed by a combination of targeted and untargeted metabolomics. FASEB J. 31, 2696–2708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R. A., Stahl M., Leopoldi A., Garreta E., del Pozo C. H., Prosper F., Romero J. P., Wirnsberger G., Zhang H., Slutsky A. S., Conder R., Montserrat N., Mirazimi A., Penninger J. M., Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 181, 905–913.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/sciadv.abe4724/DC1


