Metabolomic profiling identifies small peptides that may provide early diagnosis of thermal stress and bleaching in coral.
Abstract
Understanding the response of the coral holobiont to environmental change is crucial to inform conservation efforts. The most pressing problem is “coral bleaching,” usually precipitated by prolonged thermal stress. We used untargeted, polar metabolite profiling to investigate the physiological response of the coral species Montipora capitata and Pocillopora acuta to heat stress. Our goal was to identify diagnostic markers present early in the bleaching response. From the untargeted UHPLC-MS data, a variety of co-regulated dipeptides were found that have the highest differential accumulation in both species. The structures of four dipeptides were determined and showed differential accumulation in symbiotic and aposymbiotic (alga-free) populations of the sea anemone Aiptasia (Exaiptasia pallida), suggesting the deep evolutionary origins of these dipeptides and their involvement in symbiosis. These and other metabolites may be used as diagnostic markers for thermal stress in wild coral.
INTRODUCTION
The exchange of metabolites between organisms and their environment gave rise to the complexity of life on Earth. Examples include bacterial communities surrounding deep-ocean vents (1), the plant rhizosphere (2), the human microbiome (3), and the coral holobiont (4). The exchange of metabolites between stony corals (Scleractinia), their dinoflagellate algal photosymbionts (Symbiodiniaceae), and associated microbes is the foundation of coral reef ecosystems (5–9), which cover ca. 255,000 km2 of the planet surface (10). Under ambient conditions, the algal cells provide 90 to 95% of host energetic requirements in the form of lipids, carbohydrates, amino acids, and O2 (11). In return, nitrogen and other inorganic compounds from the coral are recycled by the algae to fuel cell metabolism (12, 13). Environmental stress, often caused by climate change, disrupts this symbiotic relationship, resulting in coral bleaching (5). With mass bleaching and mortality on the rise worldwide, these climatic changes have resulted in catastrophic reef losses (5, 7). During bleaching, coral susceptibility to disease increases (14), while growth and reproduction rates decrease (15, 16). The holobiont is the fundamental unit of selection with the host cnidarian playing an important role in recovery and resilience (17). Despite the importance of coral reefs and the services they provide, we know little about the metabolomic signatures of coral bleaching (18). Such understanding will allow the identification of bleaching biomarkers for diagnosis and provide insights into pathways that trigger, or prevent, the breakdown of the symbiosis. Untargeted, polar metabolite profiling was applied to holobionts of Montipora capitata and Pocillopora acuta (Fig. 1A) to connect biochemistry to physiology and ecology (19, 20) by identifying metabolic features associated with the short-term thermal stress response. These species were chosen because they display two widely different responses to thermal stress. Previous work has shown that Hawaiian M. capitata can meet 100% of its daily metabolic energy requirements through heterotrophic feeding during periods of bleaching (17), whereas P. acuta from the same reef system typically experiences higher mortality rates during bleaching because of lower thermal tolerance and a lack of effective feeding (21).
Fig. 1. Analysis of Hawaiian stony corals.
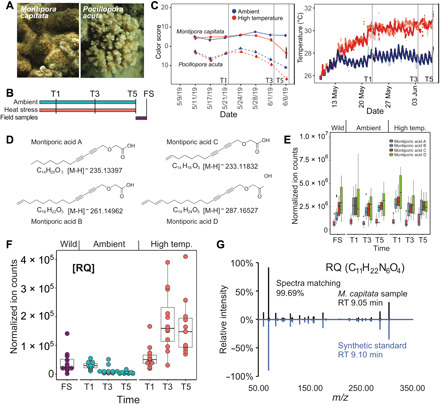
(A) Images of M. capitata and P. acuta from Kāne‘ohe Bay, O‘ahu. Photo credit: D. Bhattacharya, Rutgers University. (B) Experimental design. FS refers to field samples. These are the wild coral individuals that were examined at the end of the thermal stress experiment. (C) Color scores for the (right image) ambient- and high-temperature treated coral species M. capitata and P. acuta at the Hawaiʻi Institute of Marine Biology. Ambient-temperature tanks are shown in variations of red, and high-temperature tanks are shown in variations of blue. The color scores represent a proxy for algal symbiont density in coral holobionts with low values indicating bleaching phenotype. The sharp score decrease for P. acuta under ambient tank conditions is explained by the unexpected warming event that occurred in Kāneʻohe Bay, Oʻahu from which the culture water was drawn. Vertical gray lines indicate sampling points T1 (22 May 2019), T3 (3 June 2019), and T5 (7 June 2019). (D) Structures of MAs identified in the coral holobiont. (E) Accumulation of total MAs in M. capitata over the duration of the tank experiments and from wild populations. (F) Metabolite arginine-glutamine (RQ) in M. capitata changes over time during thermal stress (T1 to T5). (G) The metabolite C11H22N6O4 that showed accumulation under heat stress matches the synthetic standard of RQ dipeptide in retention time and MS2 spectra.
Coral nubbins from four colonies were subjected to heat stress (2.7° to 3.2°C above ambient temperature) for 5 weeks (T1 to T5) (see Materials and Methods) (Fig. 1B). We sampled corals for polar metabolomic analysis at 11, 23, and 27 days after temperature shift (T1, T3, and T5, respectively). The T2 and T4 nubbins were collected 16 and 25 days after thermal stress was initiated in case data needed to be added between T1, T3, and T5. After the end of the temperature stress experiments, nubbins were sampled from wild corals (from the same colonies used in the tank experiments) to serve as a nontreatment control. Because metabolites are shared among all members of the holobiont, we targeted the metaorganism rather than attempting to analyze its individual components. During the experiments, Kāneʻohe Bay was at the beginning of an unexpected warming event of ca. 2°C. This led to thermal stress in the ambient condition tanks because they drew water from the bay without any tank temperature control. The impact of this natural fluctuation was recorded in photographic bleaching scores that measure color as a proxy for algal abundance in the coral nubbins (Fig. 1C) (22). M. capitata showed little visible effect from the warming event, whereas P. acuta in the ambient tanks was affected in a similar fashion as the high-temperature treatment (Fig. 1C). For this reason, analysis of the M. capitata data was prioritized, with P. acuta used to verify the presence of metabolites in a second coral species and to study general trends in metabolites that are candidates for markers of bleaching progression.
RESULTS
Results of polar metabolomic analysis
Untargeted hydrophilic interaction liquid chromatography (HILIC)–mass spectrometry (LC-MS) revealed thousands of features from all the coral samples, many of which showed a notable difference in accumulation under the tested ambient- and high-temperature treatments (fig. S1A). These metabolic features include known central metabolites such as amino acids, nucleotides, and sugar phosphates. In addition, high-resolution LC–tandem MS (LC-MS2) allowed the detection, quantitation, and structural elucidation of many secondary metabolites. Examples of which are montiporic acids (MAs) (fig. S1B), which were first discovered in coral eggs of Montipora digitata from Australia (23). These compounds are disubstituted acetylenes with a carboxyl group linked to two alkyne (carbon-carbon triple bond) groups, followed by an unbranched alkane tail. The four known MAs (MA-A to MA-D; Fig. 1D) have antimicrobial activity, are cytotoxic against leukemia cells, and reduce the photosynthetic competency of coral symbionts (23, 24). MAs are highly abundant in M. capitata samples from both ambient and heat-stressed conditions as well as in the wild samples (Fig. 1E and figs. S1C and S2A). This could be because MAs have been found in high abundance in coral eggs (24) and M. capitata was preparing to spawn. We found evidence of MAs in P. acuta. However, the ion counts of MAs in P. acuta was >1000-fold less than in M. capitata (figs. S1C and S2B), suggesting that the biosynthetic activity of MAs in different coral species can vary substantially, likely reflecting their different biological functions. It is unclear whether MAs contribute to stress resistance in M. capitata by controlling symbiont density before warming events. Nonetheless, their high abundance in this species, which exceeds the total concentration of free amino acids (fig. S2A), suggests that coral animals divert copious resources to the production of secondary metabolites.
Although some metabolites, such as glucose, display a depletion in heat-stressed nubbins over the time course, we were interested in other known and previously unidentified metabolomic features in the holobiont that change under high-temperature treatment and may diagnose thermal stress before visible bleaching (18). By comparing the polar metabolite profiles of the ambient and heat-stressed time courses, many features that have significantly increased intensities in the heat-stressed corals were observed (25). One feature, mass/charge ratio (m/z) 303.177 at a retention time of 9.05 min, was detected under positive ionization mode in M. capitata and showed the largest relative increase of ion counts throughout the duration of the time course under heat stress (Fig. 1F). This feature also showed a time-dependent increase under high temperature in P. acuta (fig. S3A). On the basis of the accurate mass and isotopic fine structure, this molecule was assigned the chemical formula of C11H22N6O4. To gain insights into the structure of this metabolite, the MS2 spectra was collected using parallel reaction monitoring and the high-quality MS2 pseudospectra were generated by calculating the correlation between MS1 and MS2 spectra. Through systematic comparisons of the accurate mass, retention time, and the MS2 spectra for chemically synthesized pure standards of various dipeptides and tripeptides (table S1), C11H22N6O4 was positively identified to be arginine-glutamine (RQ; Fig. 1G).
Accumulation of the RQ dipeptide under heat stress could result from increased proteolysis and/or insufficient peptide clearance. Among the metabolites that show significantly different levels between ambient and heat-stressed conditions, the majority match the accurate mass of dipeptides (Fig. 2, A to C). These putative dipeptides are significantly enriched under the heat-stressed condition at both T3 (P = 4.8 × 10−13, Fisher’s exact test) and T5 (P = 5.6 × 10−14) time points. To confirm the chemical identities of these putative dipeptides, chemical standards were synthesized and MS2 spectra from the coral samples and the standards were collected. The structures of three additional metabolites were determined: C11H22N4O4 ([M + H]+ m/z 275.1714 [lysine-glutamine, KQ]; Fig. 2, D and G), C11H23N5O3 ([M + H]+ m/z 274.1874 [arginine-valine, RV]; Fig. 2, E and H), and C9H19N5O3 ([M + H]+ m/z 246.1561 [arginine-alanine, RA]; Fig. 2, F and I). Note that some features not identified as dipeptides also increased in intensity with thermal stress; however, the majority are not statistically significant (Fig. 2C). In comparison, the stress-sensitive and bleached P. acuta showed many metabolites with decreased ion counts after heat stress, possibly reflecting a dampened metabolism. However, the RQ and KQ dipeptides showed significant increases in this coral at T3 and T5 (fig. S3A). The similarities and clear dipeptide production differences between these two coral species suggest that their production may not be explained solely by proteolysis due to metabolic shutdown before cell death. However, an accumulation of proteogenic dipeptides during a time course stress experiment is linked to autophagy in Arabidopsis thaliana (26). Dipeptides also serve a diversity of other functions such as small-molecule regulators [e.g., H+ buffers (27), antioxidants (28), and glucose regulators (29)]. RQ has been shown to ameliorate the impacts of oxygen imbalance in retinopathy of prematurity in murine models (30) and may have beneficial effects on the reversal of oxygen-induced lung damage (31). Some known primary metabolites have incrementally increased intensities during prolonged thermal stress in M. capitata. Under high temperature, methionine, methionine sulfoxide, and cytidine 5′-diphosphate (CDP)–choline are all significantly increased at T5 (Fig. 3, A to D). The elevated level of methionine is of particular interest because of its involvement in scavenging reactive oxygen species (ROS), as well as initiating and maintaining epigenetic modifications (32, 33). Methionine and methionine sulfoxide are both up-regulated in response to prolonged heat stress, whereas S-adenosyl-l-methionine (SAM) shows a downward trend (Fig. 3C).
Fig. 2. Dipeptide production by stony corals under thermal stress.
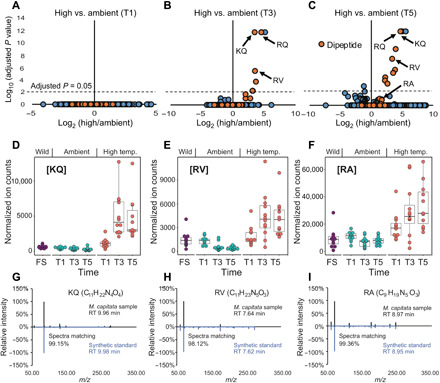
(A to C) Volcano plots of M. capitata metabolites generated from positive ionization mode data when comparing ambient- and high-temperature treatments. Putative dipeptides are shown with the filled orange circles. RA at T3 had an adjusted P = 1 and is not marked on the plot. (D to F) Accumulation of dipeptides KQ, RV, and RA under heat stress. The P values are all <0.005 when comparing ambient T5 and heat-stressed T5 using Student’s t test. (G to I) The metabolites that showed accumulation under heat stress matched synthetic standards of KQ, RV, and RA dipeptides in retention time and MS2 spectra.
Fig. 3. Production of known metabolites by M. capitata under thermal stress.
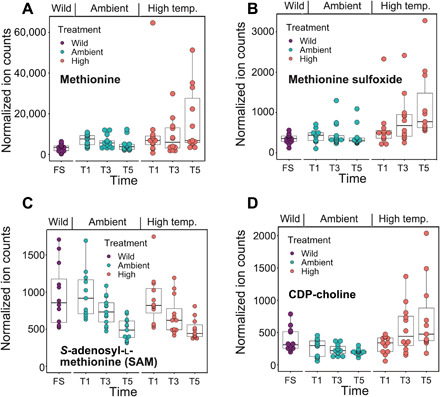
(A) Accumulation of methionine, (B) accumulation of methionine sulfoxide, and (C) reduction of SAM. Differences between the T5 ambient- and high-temperature groups are significant, as determined by a two-tailed Student’s t test with P ≤ 0.05 for the analyses shown in (A) and (B). SAM showed a downward trend during the treatment but was not significantly different between the ambient- and high-temperature groups.
For the dipeptides discussed above, nubbins from each colony were also analyzed separately to visualize colony-to-colony and within-colony variation. The accumulation of dipeptides under heat stress is a robust result among all four colonies; however, different colonies and different nubbins from the same colony show varying degrees of this response (fig. S4). Colony 248, for instance, shows relatively lower variation between nubbins when analyzing all four dipeptides, compared to high variation between nubbins in colonies 289 and 291. In addition, colony 291 has some of the highest level of accumulation of each dipeptide across all time points in the ambient- and high-temperature samples (fig. S4). These results may indicate that each colony has varying rates of turnover for each metabolite, possibly because of differing adaptation to thermal stress. Additional work is needed to elucidate the basis of metabolite variation within and between colonies.
Network analysis
To gain additional insights into dipeptide function in corals, we generated co-occurrence networks that included the verified and predicted dipeptides with known metabolic features. To address the hypothesis that dipeptide production is co-regulated and clusters under thermal stress, Louvain communities (34) in networks were detected and an average dipeptide by community (ADPC) score was computed for each treatment (table S2). This analysis shows that the ADPC score for dipeptides becomes smaller from T1 to T5, indicative of greater clustering (Fig. 4A). This pattern is apparent in the networks of M. capitata at T5, whereby large clusters of dipeptides occur in the thermal stress networks (fig. S5). Here, the focus was on two subnetworks in the T5 thermal stress treatment that offer key insights. The first shows that MA-A and the highly abundant MA-D (Fig. 1E) link two key stress responses, carbohydrate metabolism and protective osmolyte production (Fig. 4B). The production of organic osmolytes such as betaine, sorbitol (and its precursor, glucose), sucrose, and trimethylamine N-oxide are all associated with the stress response (35). These are all co-regulated and increase in abundance (see metabolite intensity in Fig. 4B) in the coral holobiont, with the algal symbiont as the likely source of these compounds (36). Organic acids are also important intermediates in energy production and as sensing molecules (37). On the left side of this subnetwork are metabolites involved in carbohydrate metabolism, including pyruvate, which can be fed into the tricarboxylic acid (TCA) cycle to produce adenosine 5′-triphosphates. A second subnetwork (Fig. 4C) shows the co-regulation of many dipeptides, including RQ and other characterized compounds (KQ, RV, and RA) and putative compound retention time (RT). In addition to the functions described above as a potential response to oxygen stress, dipeptides and free amino acids (right side of the subnetwork) are important, readily assimilated sources of nitrogen for the algal symbiont, and can be used to generate energy via the TCA cycle or metabolized in the purine pathway (36). The putative methylation-related metabolites methionine and methionine sulfoxide discussed above are in this subnetwork. A key metabolite involved in phospholipid metabolism, cell signaling, and glutamate transport and an intermediate in betaine synthesis (35), CDP-choline also shows steady accumulation in M. capitata from T1 to T5 (Fig. 3D). In support of our findings, a recent study that focused on metabolites predictive of coral bleaching sensitivity identified betaine as a key marker of resilience in M. capitata (38).
Fig. 4. Network analysis of M. capitata metabolites.
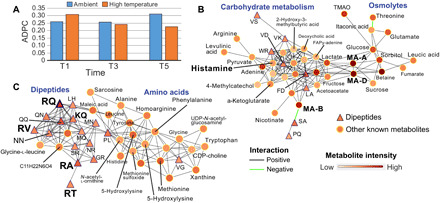
(A) ADPC scores for ambient and thermal treatments at T1, T3, and T5. (B) Subnetwork at thermal stress T5 showing the relationship between carbohydrate metabolism and osmolyte accumulation. Dipeptides are shown as triangles (under both positive and negative ionization modes), and other metabolites are shown as circles with annotations, when available. Metabolite intensity and type of correlation are shown in the legend. (C) Subnetwork at thermal stress T5 showing the relationship between dipeptide and amino acid accumulation.
Comparison to Aiptasia
The observation that many dipeptides were up-regulated in M. capitata as bleaching progressed raised the question whether we would observe differences in dipeptides based on the presence of the symbionts. To address this issue and because it is difficult to obtain aposymbiotic corals, we generated ultra-high–performance LC-MS (UHPLC-MS) data from the sea anemone model Aiptasia (Exaiptasia pallida). Often used as a model for stony corals, the anemone Aiptasia is a member of a sister lineage to stony corals and it also harbors dinoflagellate symbionts, but it does not biomineralize (8). Comparison of data gathered from animals in a symbiotic or a stable aposymbiotic state (i.e., alga-free for >1 year; see Materials and Methods) showed a significant difference in the ion count intensities of multiple known and unknown compounds present in corals (fig. S3B), including RQ and other dipeptides (fig. S6). Because the experimental design differs between the coral and Aiptasia work, no direct link between dipeptides and increasing thermal stress in the latter can be concluded. These results, however, indicate that RQ can be produced in Aiptasia without the presence of the algal symbiont, suggesting the host and/or other associated microbes as a potential source for the metabolite. The increased production of RQ in symbiotic Aiptasia and in thermally stressed M. capitata supports its hypothesized role as a response to the presence of algal symbionts.
DISCUSSION
A controlled time course experiment was used to create a polar metabolite–based understanding of the coral response to thermal stress in M. capitata and P. acuta, focusing on metabolites exhibiting increased production that could serve as biomarkers for thermal stress. These results provide the platform to address differences in resilience to thermal stress between coral colonies that are located in close proximity in reefs or are generated using controlled breeding experiments. Elucidating how metabolites associated with DNA and gene modification events, such as methylation, can alter expression patterns and effect resilience is of importance to conservation efforts.
Finding a high abundance of MAs and their central position in the metabolite networks suggests that coral animals divert a large part of their metabolic resources to the production of secondary metabolites. These polyacetylenic carboxylic acids are more abundant than many metabolites in central metabolic pathways such as glycolysis and the TCA cycle and were, unexpectedly, also detected in P. acuta albeit at a much lower level. Equally unexpected was the potentially critical role of dipeptides, such as RQ, in coral stress biology. We postulate that RQ may be a host response to oxygen stress resulting from redox imbalance in the coral or the algal symbionts caused by the high-temperature treatment. With regard to known metabolites such as methionine and methionine sulfoxide, their increase under thermal stress may be due to the role of methionine in scavenging ROS, with ROS generated from the symbiotic breakdown driving methionine oxidation that increases methionine sulfoxide levels. Furthermore, apoptotic and necrotic pathways triggered by thermal stress could release additional methionine from cellular pools. The reduction of SAM availability may be due to competing pathways for methionine between its role as a cofactor for epigenetic modifiers through SAM and in the biosynthesis of high dimethylsulfoniopropionate (DMSP) production in stressed corals (39). DMSP/dimethyl sulfoxide production may be linked to the selection of coral-associated bacterial communities, raising the possibility that corals restructure their microbiome to be as beneficial as possible during times of stress. SAM also plays a critical role as a methyl donor for epigenetic modifier enzymes such as DNA methyltransferases and histone methyltransferases (40) that provide regulation of gene expression and expression variability. Therefore, the limited availability of SAM could further exacerbate energetic demands due to metabolic depression and the cost of bleaching stress response because epigenetic regulation of the expression of essential genes in stress response and repair would be further hampered. By generating additional structures of previously unidentified stress-associated coral metabolites and their elevated levels in different species and populations, we hope, in the future, to understand the intersection of the (epi)genetic architecture of these metaorganisms and coral resilience in the field.
MATERIALS AND METHODS
Cultivation of coral nubbins
From the waters of Kāne‘ohe Bay, HI, four colonies of each coral species M. capitata and P. acuta were identified and collected under SAP 2019-60. Each of the four colonies for each species was fragmented into 30 pieces at the Hawaiʻi Institute of Marine Biology, located on Moku o Loʻe in Kāne‘ohe Bay, HI, and hot-glued to labeled plugs. The 30 glued nubbins of each genotype were then randomly distributed among six tanks (~32 liters; 48.3 cm by 38.1 cm by 17.8 cm; L × W × H), leaving five replicates per genotype in each tank for a total of 40 coral nubbins per tank and 120 coral nubbins per species. Tanks were placed in a flow-through system that had a steady supply of water directly from Kāne‘ohe Bay with an average flow rate of (173.8 ± 73 liters hour−1, mean ± SD). Each tank was fitted with a submersible pump (Hydor 200 gph), a HOBO Water Temp Pro temperature logger (operation range, −40° to 70°C; resolution, 0.02°C at 25°; accuracy, ±0.21°C from 0° to 50°C; Onset Computer Corp.), an Apex temperature probe (Neptune Systems), and two heaters (Aqueon 300-W Heater set to 31°C and DaToo 300-W Glass Heater set to 34°C). The temperature in the tanks was controlled by powering off and on the heaters based on set points in the Apex aquarium controller (Neptune Systems). Light was set for a 12-hour light/12-hour dark cycle using Arctic T247 lights (Ocean Revive).
Assessment of coral bleaching
Each sample was photographed using a digital camera with a red/blue/green color standard. Red/blue/green values that were extracted in ImageJ (41) from the coral were standardized to the color standards by dividing the experimental value observed in the coral against the corresponding actual recorded value from the color standards (22). Using the normalized intensity values from each color channel, a bleaching score was quantified as PC1 from principle components analysis of these data. As stress is prolonged and bleaching becomes more pronounced, the red/blue/green color readings from the coral will equalize around the same number because white is an equal expression of all colors. All nubbins available at each time points were used for color assessment.
Experimental design
Once fragmented, the coral nubbins were allowed an acclimation period of 5 days at ambient temperature (26.84 ± 0.50, mean ± SD) before initiating the temperature ramping. Tanks were randomly assigned to treatment groups, with tanks 1, 4, and 6 in the ambient treatment and tanks 2, 3, and 5 in the high-temperature treatment. For each tank, the species were cocultured, with M. capitata and P. acuta nubbins alternating in each row of plugs. The high-temperature treatment tanks were set to increase by ~0.4°C every 2 days for a total of 9 days (13 May to 22 May 2019) until they were between 30.5° and 31.0°C (Fig. 1C). Treatments of 30.33 ± 0.35 (mean ± SD) for high and 27.67 ± 0.34 (mean ± SD) for ambient were held for the remainder of the experiment, which lasted 16 days (Fig. 1C). High tanks were, on average, 2.66°C above ambient when the ramp was completed. Temperature readings from the HOBO loggers were confirmed using spot checks with a handheld digital certified thermometer (Control Company accuracy, ± 0.05°C; resolution, 0.001°C) about two to three times daily. Light measurements were taken to quantify photosynthetically active radiation (photosynthetic photon flux density in μmol photons m−2 s−1) using an underwater cosine light sensor and meter (MQ-510 Full Spectrum Underwater Quantum Meter, Apogee) and did not differ substantially between tanks (ambient, 345 ± 25.2 μmol photons m−2 s−1 and high, 340 ± 29.8 μmol photons m−2 s−1; means ± SD). The pH on the total scale was measured with a glass probe (METTLER TOLEDO InLab Expert Pro pH probe no. 51343101; accuracy, ±0.2 mV and resolution, 0.1 mV) and handheld meter (Thermo Fisher Scientific Orion Star A series A325) based on a tris standard (Dickson laboratory University of California San Diego) and showed no significant differences between the treatments (ambient, 7.91 ± 0.07 and high, 7.91 ± 0.04; means ± SD; t = 0.30332, df = 78.732, P = 0.7624).
Coral sampling
Sampling of the nubbins began after the onset of color score divergence between the treatment groups (Fig. 1C). Three sampling points were selected on 22 May 2019 (T1), 3 June 2019 (T3), and 7 June 2019 (T5) on the basis of reaching maximum treatment temperature (Fig. 1C), where the coral color score began to diverge by treatment and where the coral color score differences were maximized between treatments within species within the experimental time frame. Coral nubbins were selected for sampling randomly using a random number generator, and color scores were recorded for each nubbin after sampling. Corals were sampled at ~14:30 at each time point by removing them from their treatment, only touching the plastic bases and inserting them into sterile Whirl-Paks that were immediately submerged in liquid nitrogen and transferred to −80°C until metabolite extraction.
Metabolite extraction from coral nubbins
Metabolites were extracted using a protocol optimized for water-soluble polar metabolite analysis on LC-MS. The extraction buffer was a solution of 40:40:20 (methanol:acetonitrile:water) (v/v/v) + 0.1 M formic acid. The extraction buffer was stored at −20°C before usage. Immediately preceding the metabolite extraction, 1 ml of extraction buffer was added to a 2-ml glass Dounce homogenizer that had chilled on ice. Pieces of the −80°C preserved nubbins were then clipped, weighed, and added to the cold extraction buffer in the Dounce and left to incubate for 5 min. The pestle of the Dounce was then used to homogenize the coral tissue until there was a visible accumulation of coral skeleton at the bottom of the Dounce and the homogenate was visibly pigmented. An additional 500-μl aliquot of cold 40:40:20 + 0.1M formic acid extraction buffer was then used to rinse down the sides of the Dounce and pestle. The total 1.5-ml volume was then strained through a sterile 100-μm cell strainer into a 50-ml receptacle. There was a visible amount of skeleton collected in the strainer. The rest of the homogenate was then transferred to a 1.5-ml Eppendorf tube, vortexed for 10 s, and then centrifuged for 10 min at 16,000g at 4°C. After centrifugation, there was a pellet at the bottom of the tube. A final 500-μl aliquot of the homogenate was then pipetted to a second clean Eppendorf tube, to which 44 μl of 15% NH4HCO3 was added to neutralize the acid in the buffer. This was the final extract and was ready to be loaded to instrument vials for analysis.
Cultivation of Aiptasia
The animals were held at 27°C under a 12-hour light/12-hour dark cycle at 25 μmol photons m−2 s−1. All individuals were from the clonal population CC7, which naturally contains the algal symbionts of the Symbiodiniaceae clade A species Symbiodinium linuchae (42). CC7 animals were made aposymbiotic as previously described (43). This resulted in the strain CC7-Apo. CC7-Apo animals subsequently exposed to algae of the clonal axenic strain SSB01 (Symbiodiniaceae clade B species Breviolum minutum) (44) and cultured under standard conditions. After these anemone strains were created, each strain was continued and propagated though asexual reproduction in long-term culture for >1 year before experimentation. By using long-term, stable anemone cultures, we intended to minimize the effects of the stress on the animals required to generate the CC7-Apo and CC7-SSB01 anemones that were used for metabolomic analysis. Aiptasia were aliquoted individually to the 1.5-ml Eppendorf tubes in 500 μl of artificial seawater, flash-frozen, and stored at −80°C until processing at Rutgers University.
Metabolite extraction from Aiptasia
Metabolites were extracted using a protocol optimized for water-soluble polar metabolite analysis on LCMS. The extraction buffer used was a solution of 40:40:20 (methanol:acetonitrile:water) (v/v/v) + 0.1 M formic acid and was stored at −20°C before usage. CC7-Apo and CC7-SSB01 animals were removed from the artificial seawater storage, and intragroup animals were pooled to produce six replicates, each having an input weight of 25 mg in a 1.5-ml Eppendorf tube. Replicates were kept on dry ice before extraction. A total of 500 μl of extraction buffer was added; then, samples were vortexed for 10 s and transferred to crushed ice to incubate for 10 min. Samples were then centrifuged for 10 min, and the supernatant was transferred to a 1.5-ml Eppendorf tube; the procedure was repeated for a second round of extraction. A total of 1 ml of supernatant was then centrifuged, and a final 500-μl aliquot of the homogenate was pipetted to a clean Eppendorf tube, to which 44 μl of 15% NH4HCO3 was added to neutralize the acid in the buffer. This was the final extract and was ready to be loaded to instrument vials for analysis.
UHPLC conditions
The HILIC separation was performed on a Vanquish Horizon UHPLC system (Thermo Fisher Scientific, Waltham, MA) with XBridge BEH Amide column (150 mm by 2.1 mm, 2.5-μm particle size; Waters, Milford, MA) using a gradient of solvent A [95%:5% H2O:acetonitrile with 20 mM acetic acid and 40 mM ammonium hydroxide (pH 9.4)] and solvent B [20%:80% H2O:acetonitrile with 20 mM acetic acid and 40 mM ammonium hydroxide (pH 9.4)]. The gradient was 0 min, 100% B; 3 min, 100% B; 3.2 min, 90% B; 6.2 min, 90% B; 6.5 min, 80% B; 10.5 min, 80% B; 10.7 min, 70% B; 13.5 min, 70% B; 13.7 min, 45% B; 16 min, 45% B; 16.5 min, 100% B; and 22 min, 100% B. The flow rate was 300 μl/min. The injection volume was 5 μl, and the column temperature was 25°C. The autosampler temperature was set to 4°C, and the injection volume was 5 μl.
Full-scan MS
The full-scan MS analysis was performed on a Thermo Fisher Scientific Q Exactive Plus with a HESI source, which was set to a spray voltage of −2.7 kV under negative mode and 3.5 kV under positive mode. The sheath, auxiliary, and sweep gas flow rates were 40, 10, and 2 (arbitrary unit), respectively. The capillary temperature was set to 300°C, and aux gas heater was 360°C. The S-lens radio frequency (RF) level was 45. The m/z scan range was set to 72 to 1000 m/z under both positive and negative ionization mode. The automatic gain control (AGC) target was set to 3e6, and the maximum injection time (IT) was 200 ms. The resolution was set to 70,000.
Parallel reaction monitoring MS
The MS2 spectra generation was performed on a Thermo Fisher Scientific Q Exactive Plus with a HESI source, which was set to a spray voltage of −2.7 kV under negative mode and 3.5 kV under positive mode. The sheath, auxiliary, and sweep gas flow rates were 40, 10, and 2 (arbitrary unit), respectively. The capillary temperature was set to 300°C, and aux gas heater was 360°C. The S-lens RF level was 45. The m/z scan ranges were specified for the four dipeptides and monitored for the full 22-min run time. The AGC target was set to 2 × 105, and the maximum IT was 100 ms. The resolution was set to 17,500. The isolation window was set to 2.0 m/z. Collision energy was set to a stepwise 30, 50, and 80 normalized collision energy (NCE). Our results suggest that a single MS2 spectrum may contain irrelevant m/z signals and the pseudospectrum, generated by correlating multiple MS2 spectra to the MS1-extracted ion chromatogram, is an effective approach to “clean” the MS2 spectrum. The pseudospectra were generated using a modified version of COVINA (45).
Synthesis of standards
The dipeptide and tripeptide standards were synthesized and purified to 95% purity by GenScript USA (Piscataway, NJ). All standards were shipped at 25°C and stored at −20°C until they were prepared for analysis on the mass spectrometer. Parallel reaction monitoring was used to generate the MS2 spectra for comparison with samples. A total of 12 pmol of each standard was used in the analysis.
Data analysis
The MS1 data were processed using Maven (46). The compound annotation was based on accurate mass and retention time match to the metabolite standards from the in-house library. The feature detection for untargeted metabolomics was done using Compound Discoverer (Thermo Fisher Scientific, version 3.1). Before the groupwise comparison, the signal intensities were normalized to the sample weight. Significance was determined by the groupwise signal intensity comparison of each metabolite at each time point using a Student’s t test that assumed unequal variance. P values were then adjusted using the Benjamini-Hochberg correction (false discovery rate, <0.05). Of those features with adjusted P < 0.05, the Fisher’s exact test was then applied to determine whether the putative dipeptides were significantly enriched in those features.
Data quality
Before running the samples, the LC-MS system was evaluated for performance readiness by running commercially available standard mixtures and in-house standard mixtures to assess the mass accuracy, signal intensities, and the consistency of retention time. All known metabolites in the mixture were detected within 5 parts per million mass accuracy. Process blanks matching the composition of the extraction solvent were used in every sample batch to assess background signals and ensure that there was no carryover from one run to the next. In addition, the sample queue was randomized with respect to species and treatment to eliminate the potential for batch effects.
Network analysis
The R package DGCA (47) was used to determine the correlation between pairs of metabolites respectively under ambient and stressed conditions for M. capitata. The pairwise correlation was calculated with the function matCorr using the Pearson method. The functions matCorSig and adjustPVals were used to calculate and adjust (with the Benjamini-Hochberg method) the correlation P values, respectively. Only pairs with an adjusted P ≤ 0.05 were retained to construct the co-occurrence networks. To gain insights into dipeptide function, Louvain communities (34) in networks were detected and a dipeptide score was computed using the following formula to determine the ADPC score (data file S2) in which n = nodes, dp = dipeptides, and okm = other known metabolites
Supplementary Material
Acknowledgments
Funding: This work was supported by a seed grant awarded to D.B. from the office of the Vice Chancellor for Research and Innovation at Rutgers University and by NSF grants NSF-OCE 1756616 and NSF-OCE 1756623 to D.B. and H.M.P. D.B. was supported by a NIFA-USDA Hatch grant (NJ01170). The metabolomics analysis performed by the Rutgers Cancer Institute of New Jersey Metabolomics Shared Resource was supported, in part, by funding from NCI-CCSG P30CA072720-5923. Author contributions: D.B. and X.S. conceived the project. A.W. and E.N.C. performed the bioinformatic analyses. J.S.P. performed the network analysis. H.M.P. helped design the experiments and provided coral tissues for analysis. H.M.P. and D.C. conducted the coral experiment. P.A.C. provided Aiptasia tissues. All authors provided feedback on the manuscript and experimental design. A.W., E.N.C., X.S., and D.B. wrote the article with help from all the authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The untargeted and targeted metabolomic data from M. capitata, P. acuta, and Aiptasia are available in data files S1 and S2, respectively. Raw experimental data and analytical code are available at https://github.com/hputnam/Coral_Hospital/releases/tag/v1 and will be archived at BCO-DMO upon acceptance for publication. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/1/eabd4210/DC1
REFERENCES AND NOTES
- 1.Huber J. A., Mark Welch D. B., Morrison H. G., Huse S. M., Neal P. R., Butterfield D. A., Sogin M. L., Microbial population structures in the deep marine biosphere. Science 318, 97–100 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Pascale A., Proietti S., Pantelides I. S., Stringlis I. A., Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 10, 1741 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maruvada P., Leone V., Kaplan L. M., Chang E. B., The human microbiome and obesity: Moving beyond associations. Cell Host Microbe 22, 589–599 (2017). [DOI] [PubMed] [Google Scholar]
- 4.J. E. N. Veron, The biogeography & evolution of the Scleractinia, in Corals in Space & Time (Cornell Univ. Press, 1995). [Google Scholar]
- 5.National Academies of Sciences, Engineering, and Medicine, in A Research Review of Interventions to Increase the Persistence and Resilience of Coral Reefs (National Academy of Sciences, 2019). [Google Scholar]
- 6.Muscatine L., Porter J. W., Reef corals: Mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460 (1977). [Google Scholar]
- 7.Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., Harvell C. D., Sale P. F., Edwards A. J., Caldeira K., Knowlton N., Eakin C. M., Iglesias-Prieto R., Muthiga N., Bradbury R. H., Dubi A., Hatziolos M. E., Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Tivey T. R., Parkinson J. E., Weis V. M., Host and symbiont cell cycle coordination is mediated by symbiotic state, nutrition, and partner identity in a model cnidarian-dinoflagellate symbiosis. MBio 11, e02626-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourne D. G., Morrow K. M., Webster N. S., Insights into the coral microbiome: Underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Spalding M., Grenfell A., New estimates of global and regional coral reef areas. Coral Reefs 16, 225–230 (1997). [Google Scholar]
- 11.Falkowski P. G., Dubinsky Z., Muscatine L., Porter J. W., Light and the bioenergetics of a symbiotic coral. Bioscience 34, 705–709 (1984). [Google Scholar]
- 12.Yonge C. M., The significance of the relationship between corals and zooxanthellæ. Nature 128, 309–311 (1931). [Google Scholar]
- 13.Davy S. K., Allemand D., Weis V. M., Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ainsworth T. D., Fine M., Roff G., Hoegh-Guldberg O., Bacteria are not the primary cause of bleaching in the Mediterranean coral Oculina patagonica. ISME J. 2, 67–73 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Baird A. H., Marshall P. A., Mortality, growth and reproduction in scleractinian corals following bleaching on the Great Barrier Reef. MEPS 237, 133–141 (2002). [Google Scholar]
- 16.Ainsworth T. D., Krause L., Bridge T., Torda G., Raina J. B., Zakrzewski M., Gates R. D., Padilla-Gamiño J. L., Spalding H. L., Smith C., Woolsey E. S., Bourne D. G., Bongaerts P., Hoegh-Guldberg O., Leggat W., The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 9, 2261–2274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grottoli A. G., Rodrigues L. J., Palardy J. E., Heterotrophic plasticity and resilience in bleached corals. Nature 440, 1186–1189 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Hillyer K. E., Dias D. A., Lutz A., Wilkinson S. P., Roessner U., Davy S. K., Metabolite profiling of symbiont and host during thermal stress and bleaching in the coral Acropora aspera. Coral Reefs 36, 105–118 (2017). [Google Scholar]
- 19.Nagler M., Nägele T., Gilli C., Fragner L., Korte A., Platzer A., Farlow A., Nordborg M., Weckwerth W., Eco-metabolomics and metabolic modeling: Making the leap from model systems in the lab to native populations in the field. Front. Plant Sci. 9, 1556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters K., Worrich A., Weinhold A., Alka O., Balcke G., Birkemeyer C., Bruelheide H., Calf O. W., Dietz S., Dührkop K., Gaquerel E., Heinig U., Kücklich M., Macel M., Müller C., Poeschl Y., Pohnert G., Ristok C., Rodríguez V. M., Ruttkies C., Schuman M., Schweiger R., Shahaf N., Steinbeck C., Tortosa M., Treutler H., Ueberschaar N., Velasco P., Weiß B. M., Widdig A., Neumann S., van Dam N. M., Current challenges in plant eco-metabolomics. Int. J. Mol. Sci. 19, E1385 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahr K. D., Tran T., Jury C. P., Toonen R. J., Abundance, size, and survival of recruits of the reef coral Pocillopora acuta under ocean warming and acidification. PLOS ONE 15, e0228168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edmunds P. J., Gates R. D., Gleason D. F., The tissue composition of Montastraea franksi during a natural bleaching event in the Florida Keys. Coral Reefs 22, 54–62 (2003). [Google Scholar]
- 23.Fusetani N., Toyoda T., Asai N., Matsunaga S., Maruyama T., Montiporic acids A and B, cytotoxic and antimicrobial polyacetylene carboxylic acids from eggs of the scleractinian coral Montipora digitata. J. Nat. Prod. 59, 796–797 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Hagedorn M., Farrell A., Carter V., Zuchowicz N., Johnston E., Padilla-Gamiño J., Gunasekera S., Paul V., Effects of toxic compounds in Montipora capitata on exogenous and endogenous zooxanthellae performance and fertilization success. PLOS ONE 10, e0118364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matthews J. L., Crowder C. M., Oakley C. A., Lutz A., Roessner U., Meyer E., Grossman A. R., Weis V. M., Davy S. K., Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc. Natl. Acad. Sci. U.S.A. 114, 13194–13199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thirumalaikumar V. P., Wagner M., Balazadeh S., Skirycz A., Autophagy is responsible for the accumulation of proteogenic dipeptides in response to heat stress in Arabidopsis thaliana. FEBS J., 10.1111/febs.15336 , (2020). [DOI] [PubMed]
- 27.Thwaites D. T., Kennedy D. J., Raldua D., Anderson C. M., Mendoza M. E., Bladen C. L., Simmons N. L., H/dipeptide absorption across the human intestinal epithelium is controlled indirectly via a functional Na/H exchanger. Gastroenterology 122, 1322–1333 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z., Zhao Y., Wang X., Lin R., Zhang Y., Ma H., Guo Y., Xu L., Zhao B., The novel dipeptide Tyr-Ala (TA) significantly enhances the lifespan and healthspan of Caenorhabditis elegans. Food Funct. 7, 1975–1984 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Vahdatpour T., Nokhodchi A., Zakeri-Milani P., Mesgari-Abbasi M., Ahmadi-Asl N., Valizadeh H., Leucine-glycine and carnosine dipeptides prevent diabetes induced by multiple low-doses of streptozotocin in an experimental model of adult mice. J. Diabetes Investig. 10, 1177–1188 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw L. C., Li Calzi S., Li N., Moldovan L., Sengupta-Caballero N., Quigley J. L., Ivan M., Jun B., Bazan N. G., Boulton M. E., Busik J., Neu J., Grant M. B., Enteral Arg-Gln dipeptide administration increases retinal docosahexaenoic acid and neuroprotectin d1 in a murine model of retinopathy of prematurity. Invest. Ophthalmol. Vis. Sci. 59, 858–869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma L., Li N., Liu X., Shaw L., Li Calzi S., Grant M. B., Neu J., Arginyl-glutamine dipeptide or docosahexaenoic acid attenuate hyperoxia-induced lung injury in neonatal mice. Nutrition 28, 1186–1191 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Luo S., Levine R. L., Methionine in proteins defends against oxidative stress. FASEB J. 23, 464–472 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moskovitz J., Berlett B. S., Poston J. M., Stadtman E. R., The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc. Natl. Acad. Sci. U.S.A. 94, 9585–9589 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blondel V. D., Guillaume J.-L., Lambiotte R., Lefebvre E., Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp. 10, P10008 (2008). [Google Scholar]
- 35.Burg M. B., Ferraris J. D., Intracellular organic osmolytes: Function and regulation. J. Biol. Chem. 283, 7309–7313 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillyer K. E., Tumanov S., Villas-Bôas S., Davy S. K., Metabolite profiling of symbiont and host during thermal stress and bleaching in a model cnidarian-dinoflagellate symbiosis. J. Exp. Biol. 219, 516–527 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Krüger A., Grüning N. M., Wamelink M. M., Kerick M., Kirpy A., Parkhomchuk D., Bluemlein K., Schweiger M. R., Soldatov A., Lehrach H., Jakobs C., Ralser M., The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid. Redox Signal. 15, 311–324 (2011). [DOI] [PubMed] [Google Scholar]
- 38.T. N. Roach, J. Dilworth, H. Christian Martin, A. D. Jones, R. Quinn, C. Drury, Metabolomic signatures of coral bleaching history. bioRxiv 2020.05.10.087072 [Preprint]. 2020. 10.1101/2020.05.10.087072. [DOI] [PubMed]
- 39.Raina J. B., Tapiolas D. M., Forêt S., Lutz A., Abrego D., Ceh J., Seneca F. O., Clode P. L., Bourne D. G., Willis B. L., Motti C. A., DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502, 677–680 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Wang Z., Long H., Chang C., Zhao M., Lu Q., Crosstalk between metabolism and epigenetic modifications in autoimmune diseases: A comprehensive overview. Cell. Mol. Life Sci. 75, 3353–3369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sunagawa S., Wilson E. C., Thaler M., Smith M. L., Caruso C., Pringle J. R., Weis V. W., Medina M., Schwarz J. A., Generation and analysis of transcriptomic resources for a model system on the rise: The sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10, 258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiang T., Hambleton E. A., DeNofrio J. C., Pringle J. R., Grossman A. R., Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. J. Phycol. 49, 447–458 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Baumgarten S., Simakov O., Esherick L. Y., Liew Y. J., Lehnert E. M., Michell C. T., Li Y., Hambleton E. A., Guse A., Oates M. E., Gough J., Weis V. M., Aranda M., Pringle J. R., Voolstra C. R., The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 11893–11898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su X., Chiles E., Maimouni S., Wondisford F. E., Zong W., Song C., In-source CID ramping and covariant ion analysis of hydrophilic interaction chromatography metabolomics. Anal. Chem. 92, 4829–4937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melamud E., Vastag L., Rabinowitz J. D., Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82, 9818–9826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenzie A. T., Katsyv I., Song W. M., Wang M., Zhang B., DGCA: A comprehensive R package for differential gene correlation analysis. BMC Syst. Biol. 10, 106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/1/eabd4210/DC1


