Abstract
Protein S-acylation (commonly known as palmitoylation) is a widespread reversible lipid modification, which plays critical roles in regulating protein localization, activity, stability, and complex formation. The deregulation of protein S-acylation contributes to many diseases such as cancer and neurodegenerative disorders. The past decade has witnessed substantial progress in proteomic analysis of protein S-acylation, which significantly advanced our understanding of S-acylation biology. In this review, we summarized the techniques for the enrichment of S-acylated proteins or peptides, critically reviewed proteomic studies of protein S-acylation at eight different levels, and proposed major challenges for the S-acylproteomics field. In summary, proteome-scale analysis of protein S-acylation comes of age and will play increasingly important roles in discovering new disease mechanisms, biomarkers, and therapeutic targets.
Keywords: ABE, APT, Click Chemistry, MLCC, Palmitoylation, PAT, Proteomics, S-acylation, S-palmitoylation
Graphical Abstract
1. Introduction
In eukaryotic cells, many proteins are subjected to different lipid modifications such as protein S-acylation, myristoylation, prenylation, farnesylation, and geranylgeranylation.1 Among these, protein S-acylation is a reversible lipid modification, where long-chain fatty acids are covalently attached to specific cysteine residues of proteins via labile thioester bonds.2 It is also one of the most common post-translational modifications — in human cells, proteins encoded by about 4,000 genes are subjected to protein S-acylation (swisspalm.org/releases). S-acylated proteins can be modified by different types of long-chain fatty acids, such as palmitate (C16:0), stearate (C18:0), myristate (C14:0), palmitoleate (C16:1), oleate (C18:1), arachidonate (C20:4), and eicosapentaenoate (C20:5).3-7 Because palmitate is the predominant fatty acid, protein S-acylation is more commonly (albeit less accurately) called protein S-palmitoylation or simply palmitoylation.
Protein S-acylation was first reported in 1979.8 After a quiescent decade, the following three decades have witnessed a steady growth of S-acylation publications, suggesting an increasing appreciation of the functional importance of protein S-acylation. Studies have demonstrated that reversible protein S-acylation dynamically regulates the localization, activity, stability, trafficking, and complex formation of proteins.9 In cells, the reversibility of protein S-acylation provides an important mechanism for mediating rapid trafficking of membrane proteins between subcellular organelles, as well as for regulating protein segregation or clustering in membrane compartments like lipid rafts.10 In addition, S-acylation may cross-talk with other post-translational modifications (PTMs), such as S-nitrosylation, ubiquitination, and phosphorylation, thereby forming dynamic regulatory programs.11 Hence, protein S-acylation plays critical roles in a wide range of biological processes such as cell signaling, metabolism, immune response, and host-pathogen interactions.10 Its aberration contributes to many diseases including cancer and neurodegenerative disorders.11,12
In eukaryotes, protein S-acylation can be spontaneous, but it is believed to be predominantly catalyzed by a group of highly conserved Asp-His-His-Cys (DHHC) motif-containing palmitoyl S-acyltransferases (PATs) encoded by ZDHHC genes.13,14 Different species have varied numbers of DHHC-PATs: 5-7 in yeasts, 24 in Arabidopsis, and 23-24 in mammals.13,15 In human cells, 23 PATs are encoded by genes ZDHHC1-24 excluding ZDHHC10. All these PATs are integral membrane proteins with a range of cellular localizations and tissue expression patterns.16 Most PATs are localized in the endoplasmic reticulum (ER) and Golgi apparatus, and at least three PATs (DHHC-5/20/21) are primarily localized in the plasma membrane.16 Protein S-acylation is reversible, and de-S-acylation is catalyzed by enzymes such as acyl-protein thioesterases (APTs), palmitoyl-protein thioesterases (PPTs), and α/β hydrolase fold domain (ABHD) proteins.17 These enzymes are localized in varied subcellular localizations, such as cytosol (APT1/2 and ABHD17A/B/C), lysosomes (PPT1/2), and mitochondria (ABHD10).18-22 For convenience, all these de-S-acylation enzymes are hereinafter referred to as APTs.
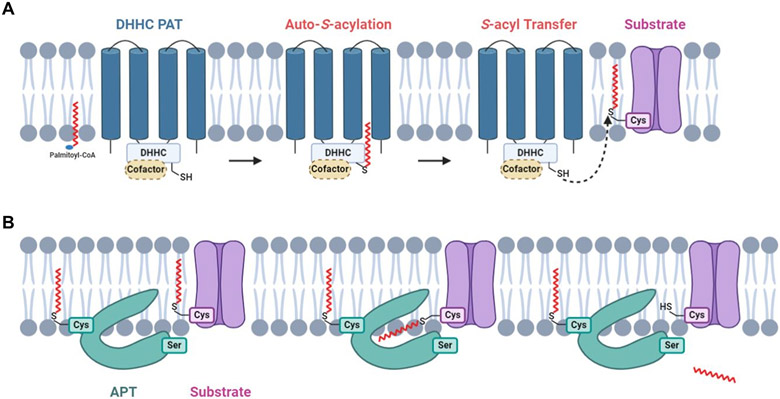
Although it is still incompletely understood,23 studies suggested that DHHC-PATs usually catalyze S-acylation in two steps: a DHHC-PAT is autoacylated to form a transient acyl-intermediate and then the fatty acyl group is transferred from the DHHC motif to its substrate protein in close proximity (Fig. 1A).24 Most DHHC-PATs are able to catalyze S-acylation without the help of a protein cofactor. However, some DHHC-PATs require cofactors for proper functioning. These enzymes and cofactors form protein complexes such as Erf2-Erf4, DHHC5-GOLGA7, DHHC6-SELENOK (Selenoprotein K), DHHC9-GCP16 (Golgi complex-associated protein of 16 kDa).25-28 The cofactors were shown to stabilize cognate DHHC-PATs or acyl-DHHC intermediates.26,29,30 In protein de-S-acylation, APTs can be S-acylated and tethered to membrane, where they remove the S-acyl groups of their substrate proteins (Fig. 1B).31
Figure 1. The biochemistry of protein S-acylation and de-S-acylation.
(A) DHHC-PATs are integral membrane proteins with active sites oriented toward the cytosol. A DHHC-PAT is first autoacylated on the DHHC cysteine residue, followed by a transfer of the S-acyl group to the acceptor cysteine residue of a substrate protein. Certain DHHC-PATs require a cofactor for proper functioning. (B) De-S-acylation enzymes can remove S-acyl groups from substrate S-acylproteins. A subpopulation of de-S-acylation enzymes are S-acylated and tethered to membrane, where their hydrophobic pockets capture S-acyl chains of substrate S-acylproteins, allowing enzymatic de-S-acylation.
Traditionally, protein S-acylation was analyzed using [3H]-palmitate labeling, followed by immunoprecipitation and days to weeks of film exposure.32 However, this method only allows the analysis of S-acylation on the basis of individual proteins, and it is hazardous, time-consuming, and sometimes insufficiently sensitive. To facilitate the detection and quantification of S-acylated proteins, various methods for the enrichment of S-acylated proteins/peptides have been developed since 200433. These methods were coupled with mass spectrometry-based proteomics methods for global identification and quantification of S-acylated proteins and S-acylation sites. This review aims to summarize the progress in these aspects and provide our view of major challenges in the S-acylproteomics field. For reviews on the enzymology of protein S-acylation and its functions in pathophysiology, readers are directed to references14,34,35.
2. Methods for the Enrichment of S-acylated Proteins
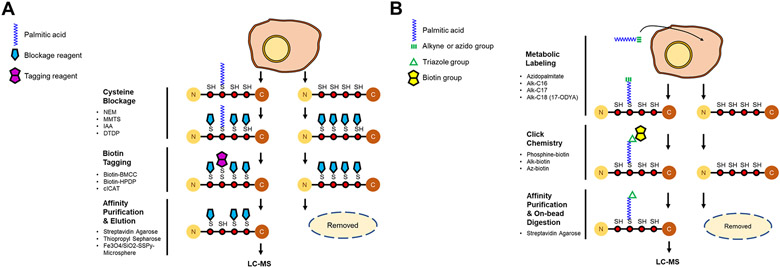
For proteome-scale analysis of protein S-acylation, a key step is to enrich S-acylated proteins with high specificity. Since 2004, various methods have been developed to enrich S-acylated proteins. These methods can be broadly classified into two categories: one is termed acyl-biotinyl exchange (ABE) and the other metabolic labeling with a palmitic acid analog followed by click chemistry (MLCC) (Fig. 2).
Figure 2. Schematic of the methods for the enrichment of S-acylated proteins.
(A) In acyl-biotinyl exchange (ABE), free thiols on cysteines are blocked by an alkylation reagent (shown as a blue pentagon). S-acyl groups are specifically cleaved off by neutral hydroxylamine, and the newly exposed free thiols are labeled by a biotin-tag (shown as a purple double pentagon). The biotin-tagged (formerly S-acylated) proteins are captured by streptavidin affinity purification, eluted by a reducing agent, and analyzed by LC-MS/MS. (B) In metabolic labeling with a palmitic acid analog followed by click chemistry (MLCC), an alkyne- or azide (shown as a green triple bond)-functionalized palmitate analog is incorporated into S-acylation sites by the endogenous S-acylation machinery. After cell lysis, the palmitate analog-labeled (i.e., S-acylated) proteins are conjugated with a biotin analog (shown as a yellow double pentagon) by click chemistry (a triazole is shown as a green triangle), selectively enriched by streptavidin affinity purification, and digested by trypsin on beads.
2.1. ABE-related methods
ABE is a cysteine-centric method (Fig. 2A). In ABE, free cysteine residues are blocked by an alkylating reagent, and then S-acylated cysteine residues are converted into free cysteine residues by neutral hydroxylamine, which specifically cleaves thioester bonds. The newly formed free cysteine residues (i.e., formerly S-acylated cysteine residues) are conjugated with a biotin analog (e.g., biotin-HPDP) through cysteine alkylation. Consequently, S-acylated proteins are converted into biotinylated proteins, so they can be enriched by streptavidin affinity purification.
The first ABE method was reported by the Green group in 2004.33 Nevertheless, the method was only coupled with streptavidin blotting to quantify a single protein α7/5HT3A. The first ABE-based S-acylproteomics study was reported by the Davis lab in 2006.36 In this study, free cysteine residues were blocked by N-ethylmaleimide (NEM). Following hydroxylamine treatment, newly formed free thiols were reacted with biotin-HPDP, and the in vitro biotinylated proteins (i.e., formerly S-acylated proteins) were enriched by streptavidin affinity purification. Subsequently, various ABE-related S-acylproteomic methods were developed to enable site-specific analysis, simplify the procedure, and further improve the specificity (Fig. 2A, Table S1).
To enable site-specific S-acylproteomic analysis, at least six different methods were developed. In 2008, Zhang et al. reported a method called palmitoyl-cysteine isolation capture and analysis (PICA).37 In PICA, free cysteine residues were blocked with methyl methanethiosulfonate (MMTS), followed by the cleavage of thioester bonds with neutral hydroxylamine. Subsequently, newly formed thiols were labeled with cleavable isotope-coded affinity tag (cICAT). After protein digestion, cICAT-labeled tryptic peptides were purified on an avidin affinity column and identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). In 2010, we published a different approach named palmitoyl protein identification and site characterization (PalmPISC).38 In PalmPISC, disulfide bonds were reduced by tris(2-carboxyethyl) phosphine (TCEP), and all free thiols were alkylated with NEM. Subsequently, S-acyl groups were cleaved off by neutral hydroxylamine, and the newly formed thiols were conjugated with biotin-HPDP. After protein digestion, in vitro biotinylated peptides (i.e., formerly S-acylated peptides) were enriched by streptavidin affinity purification and analyzed by LC-MS/MS for peptide identification and site localization. In 2011, Forrester et al. developed a method termed acyl resin-assisted capture (acyl-RAC).39 In this method, free cysteine residues were blocked by MMTS, followed by hydroxylamine treatment to cleave off S-acyl groups. Subsequently, newly formed free thiols were captured with thiol-reactive thiopropyl Sepharose resin. Captured proteins were subjected to onresin digestion, followed by washing to remove non-S-acylated peptides. Afterward, the remaining S-acylated peptides were released by dithiothreitol and analyzed by LC-MS/MS. In 2015, Gould et al. developed a method similar to acyl-RAC, where phenylmercury resin was used to substitute thiopropyl Sepharose resin.40 In 2017, Collins reported yet another method called site-specific ABE (ssABE).41 Akin to our PalmPISC method, ssABE used TCEP to reduce disulfide bonds, iodoacetamide to alkylate free cysteine residues, neutral hydroxylamine to remove S-acyl groups, biotin-HPDP to label newly formed cysteine residues, trypsin to digest proteins, and streptavidin agarose to enrich formerly S-acylated peptides. In 2019, Zareba-Koziol published a method named PANIMoni, standing for palmitoylation and nitrosylation interplay monitoring.42 The PANIMoni method essentially extends our PalmPISC method to analyze both S-acylation and S-nitrosylation. In common, the six methods rely on in vitro chemical reactions and protein digestion to capture originally S-acylated peptides, followed by LC-MS/MS analysis for peptide identification and site localization.
ABE enrichment is time-consuming and labor-intensive, requiring multiple rounds of protein precipitation and solubilization to remove excess chemical reagents, so they will not interfere with subsequent reactions. To simplify the procedure, multiple improvements have been made. Hurst et al. showed that NEM can be scavenged by 2,3-dimethyl-1,3-butadiene in an aqueous Diels-Alder 4+2 cyclo-addition reaction. Thus, the multiple precipitation steps to remove excess NEM can be eliminated.43 The above-mentioned ssABE approach used Amicon Ultra spin filter columns with a 30 kD molecular weight cutoff for buffer exchange.41 The ssABE procedure is still time-consuming, as it requires at least 10 lengthy centrifugal spins. However, it eliminates multiple precipitation-solubilization cycles and thus is less labor-intensive. The aforementioned acyl-RAC method substitute the two-step biotinylation and streptavidin affinity purification with one-step thiopropyl Sepharose reaction.39 Zhang et al. generated a novel magnetic microsphere modified with 2,2’-dithiodipyridine (Fe3O4/SiO2-SSPy microsphere), which can directly react with free (formerly S-acylated) cysteine residues exposed by neutral hydroxylamine, akin to the thiopropyl Sepharose in the acyl-RAC method.44
Nevertheless, the ABE methods invariably suffer from high background resulting from the co-isolation of non-S-acylated forms.45 Quantitative proteomic analysis showed that about two-thirds of proteins enriched by ABE were background proteins.45 The high background may severely mask the detection of low-abundance S-acylated proteins and compress the signal-to-noise ratios of many S-acylated proteins. To address this issue, we developed a new method named low-background ABE (LB-ABE).45 We found that certain non-S-acylated cysteine residues could not be completely blocked by NEM, even after longer incubation time. The residual free cysteine residues can react with biotin-HPDP, resulting in their in vitro biotinylation. Consequently, a fraction of non-S-acylated proteins, especially those containing multiple non-S-acylated cysteine residues, can be co-isolated with S-acylated proteins by streptavidin affinity purification. By using 2,2′-dithiodipyridine (DTDP), a thiol-reactive reagent similar to HPDP, residual free cysteines can be further blocked, preventing their biotinylation by biotin-HPDP and enrichment by affinity purification. Using the LB-ABE method, the level of co-isolated background proteins was reduced by >70%.45 Notably, by coupling the LB-ABE method with LC-MS/MS, we identified 2,895 highconfidence candidate S-acylated proteins from human prostate cancer LNCaP cells.45
2.2. MLCC-related methods
MLCC is a palmitate-centric method (Fig. 2B). In MLCC, an azide- or alkyne-functionalized palmitate analog is metabolically incorporated into S-acylated proteins during cell culture. After cell lysis, palmitate analog-labeled proteins are conjugated with a biotin analog by click chemistry. The in vitro biotinylated (formerly S-acylated) proteins can be isolated by streptavidin affinity purification.
The first MLCC method was reported by the Ploegh group in 2007.46 However, similar to the first ABE method, the MLCC method was only coupled with streptavidin blotting to detect fatty-acylated proteins.46 The first MLCC-based S-acylproteomic analysis was published by the Berthiaume lab a year later.47 In this study, ω-azido-palmitate was metabolically incorporated into mitochondrial proteins, and the labeled proteins were conjugated with phosphine-biotin by the Staudinger ligation method, followed by MS analysis. Subsequently, many MLCC-derived methods were developed (Fig. 2B, Table S2). Major differences include 1) different palmitate analogs such as azido-palmitate, alkynyl palmitate (Alk-C16), and alkynyl stearate (Alk-C18, also known as 17-octadecylnoic acid or 17-ODYA), 2) different affinity tags such as phosphine-biotin and biotin-azide, and 3) different conjugation methods such as click chemistry (azide-alkyne) and Staudinger ligation (azide-phosphine). For a comprehensive review of MLCC, see reference48.
2.3. ABE and MLCC are highly complementary
ABE and MLCC are complementary methods with different strengths and weaknesses.49 Firstly, ABE can be used to analyze protein S-acylation in all types of biological samples including clinical tissues and biofluids, for which MLCC is not very suitable because it requires metabolic labeling. Secondly, both ABE and MLCC can be applied to in vitro cultured cells. Nevertheless, MLCC generally requires the optimization of the concentration of palmitate analogs and the metabolic labeling time, in order to achieve optimal sensitivity and minimize cell death. In addition, MLCC is not very suitable for cells with high lipogenic activity (e.g., many cancer cell lines), where S-acylated proteins are predominantly labeled by native palmitate. In comparison, ABE can be uniformly applied to any cell type. Thirdly, ABE has the potential of uncovering the entire S-acylated proteome. In comparison, MLCC is biased toward S-acylproteins with rapid turnover.50
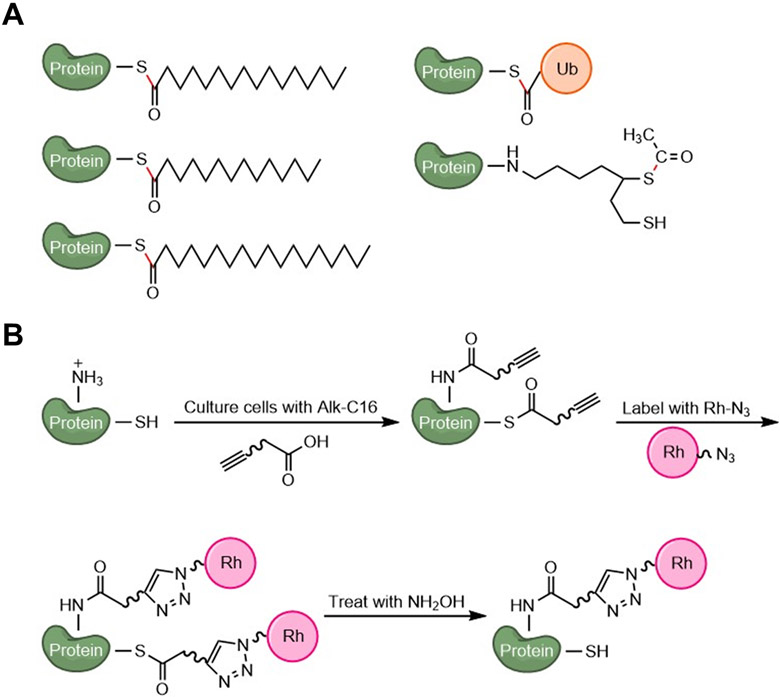
Fourthly, ABE enables global site-specific analysis of S-acylation. However, it cannot distinguish different thioester-linked modifications, which are removed by neutral hydroxylamine during sample processing. In contrast, MLCC has not yet been successfully applied to comprehensively analyze S-acylation sites. This is largely because MLCC methods introduce a bulky (400-900 Da) group that is very hydrophobic. Thus, tagged S-acylated peptides can easily get lost during sample preparation, transfer, and storage. They are also hard to separate on Ci8 reverse-phase columns.49 Fifthly, MLCC allows studying S-acylation dynamics, for which ABE is not very suitable due to the removal of S-acyl groups. Sixthly, MLCC might allow for live-cell imaging of S-acylation,51 which can hardly be achieved by ABE. Lastly, ABE cannot distinguish different S-acyl species or different thioester-linked modifications (Fig. 3A), while MLCC can detect N- or O-acylation in addition to S-acylation (Fig. 3B). For example, it was shown that TNFα can be long-chain fatty acylated on K19 and K20 residues, so it can be detected or enriched by MLCC52. To distinguish S-acylation from other fatty acylation, hydroxylamine treatment needs to performed (Fig. 3B). Collectively, ABE and MLCC are highly complementary and best used in combination.50,53
Figure 3. Illustration of post-translational modifications that can be detected by ABE or MLCC.
(A) Representative thioester-linked PTMs that cannot be distinguished by ABE. Thioester bonds are shown in red. (B) Representative fatty acylation (S-acylation and N-acylation) that can be detected by MLCC. Hydroxylamine treatment enables the distinction between S-acylation and N-acylation.
3. Proteomic Analysis of Protein S-acylation
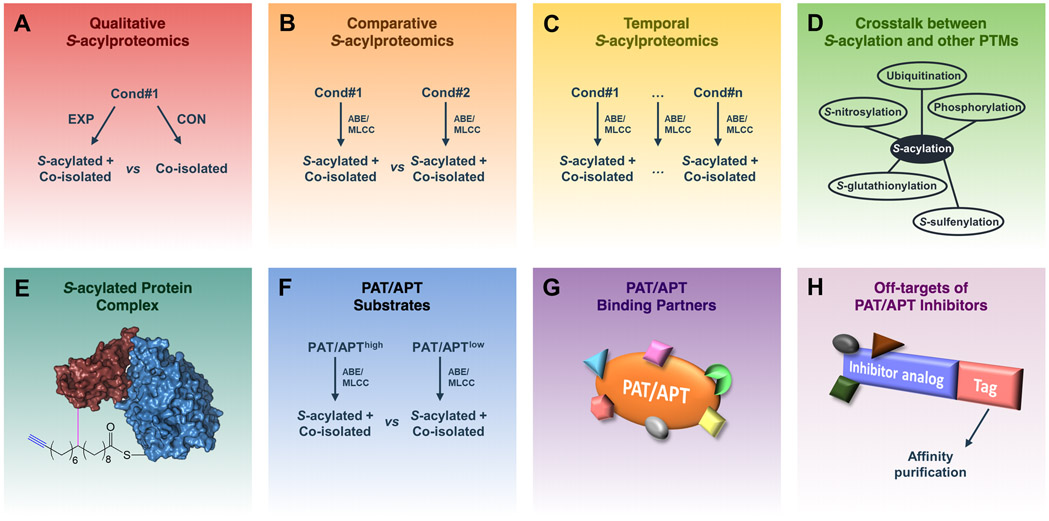
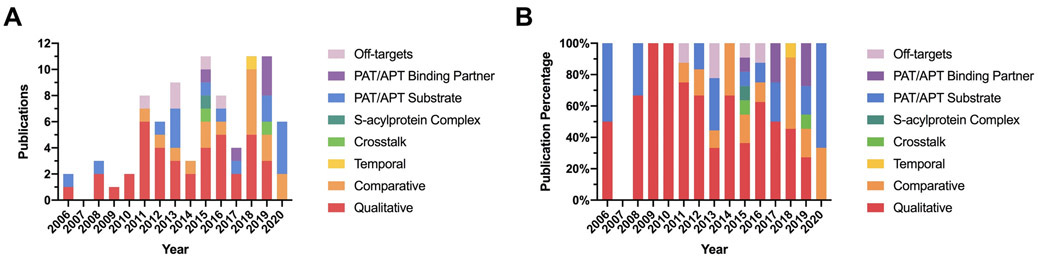
Due to a lack of S-acylprotein-specific antibodies, S-acylproteomics studies were exclusively conducted by ABE/MLCC enrichment followed by MS analysis. These studies can be broadly classified into eight types: 1) qualitative S-acylproteomics for the identification of S-acylproteins and S-acylation sites, 2) comparative S-acylproteomics to compare two different S-acylproteomes, 3) temporal S-acylproteomics to study S-acylation dynamics, 4) global analysis of the crosstalk between S-acylation and other protein modifications, 5) proteomic identification of S-acylated protein complexes, 6) global identification of PAT/APT substrates, 7) global identification of PAT/APT-binding partners, and 8) proteomic identification of off-targets of PAT/APT inhibitors (Fig. 4). As shown in Figure 5, early S-acylproteomics studies were dominated by qualitative S-acylproteomics analysis, whereas recent years have witnessed more functional S-acylproteomics studies.
Figure 4. Illustration of different types of proteomics applications in S-acylation studies.
(A) Qualitative S-acylproteomics focuses on profiling S-acylproteins and S-acylation sites as well as distinguishing them from co-isolated non-S-acylated forms. (B) Comparative S-acylproteomics compares the differences between S-acylproteomes under different conditions. (C) Temporal S-acylproteomics investigates the dynamic changes of protein S-acylation at different time points. (D) Proteomic analysis of the cross-talk between S-acylation and other PTMs. (E) Proteomic analysis of S-acylated protein complexes identifies proteins associated with the S-acylated form of a target protein. (F) Proteomic analysis of PAT/APT substrates by identifying proteins whose S-acylation levels are regulated by a specific PAT/APT. (G) Proteomic analysis of PAT/APT-binding partners. (H) Proteomic identification of off-targets of a PAT/APT inhibitor.
Figure 5. Stacked histogram of the numbers of publications from 2006 to 2020, which were grouped according to the study types.
(A) Stacked histogram in absolute numbers. (B) Stacked histogram in percentages.
3.1. Qualitative S-acylproteomics
Currently, neither ABE nor MLCC can enrich S-acylated proteins to a purity of 100%. In addition, so far, no techniques for proteome-scale identification of intact S-acylated peptides have been established. Thus, for proteomic identification of candidate S-acylproteins and S-acylation sites, it is essential to include an appropriate control to exclude co-isolated non-S-acylated proteins from the list of total identified proteins. To distinguish putative S-acylproteins from co-isolated contaminant proteins, at least a semi-quantitative proteomic comparison needs to be conducted, followed by statistical analysis.
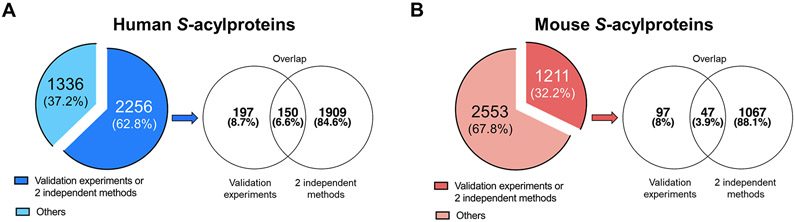
In the past 14 years, 40 S-acylproteomics papers were published to catalog S-acylated proteins in different species, especially human (14 papers) and mouse (13 papers), with gradually increasing numbers of identified S-acylprotein candidates (Table S3). However, it should be noted that many of these studies did not perform statistical analysis or apply stringent significance cutoffs to filter out co-isolated non-S-acylated proteins. Therefore, caution needs to be excised as regards the validity of some claimed candidate S-acylproteins. Nevertheless, with the development of more advanced S-acylproteomics techniques, hundreds to thousands of putative S-acylproteins were reported in single studies even when stringent cutoffs were applied. For instance, using LB-ABE and label-free proteomics, our lab identified nearly 3,000 putative S-acylproteins from cultured human LNCaP cells, with stringent cutoffs of FDR<1% and enrichment fold >2.45 Of note, according to the SwissPalm database (v3), which compiled nearly 800 S-acylation studies published before 06/28/2019, ~3,600 human and ~3,800 mouse genes may encode S-acylproteins (Fig. 6). This suggests that protein S-acylation is a pervasive PTM and that the entire S-acylproteome can be mapped in single studies in the near future.
Figure 6. Categorization of known human and mouse S-acylated proteins.
(A) Distribution of known human S-acylated proteins. (B) Distribution of known mouse S-acylated proteins. The numbers were retrieved from the SwissPalm database (v3).
An S-acylprotein may contain one to multiple S-acylated cysteine residues. However, no consensus sites for S-acylation have yet been established, although studies have shown that S-acylation is commonly observed on cysteine residues proximal to the N/C-termini of proteins, within cysteine-rich motifs, or adjacent to the cytosolic side of transmembrane domains.54 A more recent study suggested that membrane protein S-acylation is determined by the accessibility of PATs to cysteines on membrane-embedded proteins, rather than a preferred substrate sequence motif.55 Although computational algorithms have been developed to predict S-acylation sites,56,57 their reliability is still uncertain. Thus, experimental identification of S-acylation sites needs to be conducted. So far, six site-specific S-acylproteomics studies have been performed to identify novel S-acylation sites (Table S3). As mentioned above, MLCC is not very suitable for site-specific analysis because of technical challenges associated with high hydrophobicity. Unsurprisingly, all these site-specific studies employed ABE-related methods to enrich originally S-acylated peptides. However, only three of them included a control group and filtered out background non-S-acylation sites from all identified sites.38,41,58 In one study, we applied PalmPISC to analyze the lipid raft and non-raft fractions of human DU145 cells, and identified 127 putative S-acylation sites with high confidence (p<0.05, >2.4-fold enrichment) and additional 39 S-acylation sites with medium confidence (p<0.095, >2-fold enrichment). In another study, Fang et al. applied acyl-RAC to analyze human SW480 cells and identified 151 putative S-acylation sites (>2-fold enrichment).58 In yet another study, Collins et al. applied ssABE to analyze mouse forebrain and identified 906 putative S-acylation sites (FDR<0.05, >3-fold enrichment). Given that >3,500 human/mouse genes encode S-acylproteins and that many S-acylproteins may contain two or more S-acylation sites, it is expected that an S-acylproteome may contain >5,000 S-acylation sites. Thus, more powerful techniques still await to be developed to cover the complete S-acylproteome at the site-specific level.
3.2. Comparative S-acylproteomics
Since 2011, 16 comparative S-acylproteomics studies were conducted to identify S-acylproteins that were differentially expressed between two conditions (Table S4). Both ABE and MLCC methods have been successfully applied in these studies. For quantitative comparison, the most commonly used technique is label-free quantitation (LFQ; in 6 studies), followed by duplex stable-isotope labeling by amino acids in cell culture (SILAC2plex; in 4 studies). Interestingly, the comparative S-acylproteomics studies were almost exclusively on human or mouse samples (8 studies per species).
Among these comparative S-acylproteomics studies, the most comprehensive ones were conducted by us and collaborators, using LB-ABE coupled with label-free proteomics.59,60 In one study, we compared the S-acylproteomes of whole cell lysates, membrane fractions, small extracellular vesicles (EVs), and large EVs isolated from human PC3 cells.59 A total of 2,408 putative S-acylproteins were identified, among which 141 S-acylproteins are highly abundant in all the four groups, whereas 388 S-acylproteins are of varied abundance. Of note, this study reported the first S-acylprotein signature of EVs and demonstrated that small and large EVs harbor proteins associated with different biological processes and subcellular origin. In the other study, we compared the S-acylproteomes of 1) cisplatin-sensitive T24S versus cisplatin-resistant T24R bladder cancer cells, 2) T24S cells treated with versus without 2-bromopalmitate (2BP), a general S-acylation inhibitor, and 3) T24R cells treated with versus without 2BP.60 Overall, the study identified 3,695 putative S-acylproteins, of which 200-500 were differentially expressed in each comparison. Interestingly, the well-known immune checkpoint protein PD-L1 was found to be highly S-acylated in T24R cells, compared with T24S cells. In addition, pharmacological inhibition of fatty acid synthase suppressed PD-L1 S-acylation and abundance. Thus, the results highlighted the role of protein S-acylation in mediating bladder cancer chemoresistance. Taken together, the two studies demonstrated the effectiveness of our LB-ABE-based S-acylproteomics method in comprehensive comparison of S-acylproteomes.
3.3. Temporal S-acylproteomics
So far, only one S-acylproteomics study was performed to exam the temporal changes of S-acylproteins (Table S5).50 In this study, Won and Martin coupled MLCC with 6-plex tandem mass tags (TMT6plex) to analyze the changes of the S-acylproteomes of human 293T and HAP1 cells. These cells were treated with or without hexadecylfluorophosphonate (HDFP), a generic serine lipase inhibitor, for 45, 90, 180, 360, and 480 min. Unsupervised hierarchical clustering analysis revealed four clusters with different kinetic features. Notably, the most rapidly cycling S-palmitoylated cluster includes well characterized signaling proteins, such as NRAS, MTDH, and GNAS. As TMT reagents become available for 16plex and 27plex analyses61,62, we envision that more dynamic S-acylproteomics studies will be conducted in the near future to better understand the roles of dynamic S-acylation in cell signaling and pathogenesis.
3.4. Proteomic analysis of the crosstalk between S-acylation and other PTMs
Two proteomic studies were conducted to analyze the crosstalk between protein S-acylation and other cysteine modifications (Table S6).40,42 In one study, Gould et al. comprehensively identified cysteine residues that were modified by S-acylation, S-glutathionylation, S-nitrosylation, and S-sulfenylation from mouse liver.40 A total of 2,596 sites in 1,319 mouse liver proteins were mapped. The study suggested that the four cysteine modifications were largely mutually exclusive — they target discrete protein populations and cysteine residues, with minimal complementation between the modifications. However, it should be noted that no control samples were included to distinguish modified peptides from co-enriched unmodified peptides that contain cysteine residues. Therefore, it remains unclear how many of the identified sites were truly modified. In the other study, Zeraba-Koziol et al. developed a method called PANIMoni to study the crosstalk of S-acylation and S-nitrosylation of postsynaptic density proteins in mouse models of chronic stress.42 A total of 813 S-acylated and 620 S-nitrosylated cysteine sites were identified from 465 and 360 proteins, respectively. Somewhat contradictory to the above-mentioned conclusion by Gould et al., this study showed that the two modifications may crosstalk in the regulation of 122 proteins, including receptors, scaffolding proteins, regulatory proteins, and cytoskeletal proteins. Taken together, the studies suggested that, although the crosstalk between S-acylation and other cysteine modifications is not widespread, it does occur on certain functionally important proteins.
In addition, many studies have shown that, at least for some proteins, S-acylation prevents protein ubiquitination and degradation. Our and others’ S-acylproteomic studies also suggested that many kinases and phosphatases can be S-acylated. Thus, it would be interesting to determine whether the crosstalk between S-acylation and ubiquitination or phosphorylation is widespread, as well as the biological functions of such cross-talks. To address the questions, more sensitive methods need to be developed, due to the generally low stoichiometry of these modifications.
3.5. Proteomic analysis of S-acylated protein complexes
Studies have shown that protein S-acylation regulates the formation of protein complexes.63 Thus, to understand the functions of protein S-acylation, it is critical to distinguish the binding partners of S-acylated proteins from those of unmodified forms. However, it is much more challenging to analyze membrane-associated S-acylated protein complexes than cytosolic protein complexes, because the former is often destroyed during cell lysis and hard to reconstitute in vitro. To address this issue, Peng and Hang developed a bifunctional fatty acid chemical reporter, x-alk-16, for characterizing S-palmitoylated protein complexes in living cells (Table S7).64 This reporter is equipped with an alkyne for biorthogonal detection of S-palmitoylated proteins, as well as a diazirine for in-cell photo-crosslinking of S-palmitoylated protein complexes. In this study, to characterize the S-acylated IFITM3 protein complex, HA-tagged IFITM3 was overexpressed in HEK293T cells, which were labeled with or without x-alk-16 and then irradiated with or without UV light at 365 nm. After cell lysis, IFITM3 protein complexes were enriched by anti-HA immunoprecipitation. Label-free proteomics comparison of photo-crosslinked and non-photo-crosslinked groups led to the identification of 12 putative IFITM3-interacting proteins, among which CANX and BCAP31 were further confirmed. Although promising, it seems that the x-alk-16 compound has not been commercialized, hindering its adoption by other research groups.
3.6. S-acylproteomic identification of PAT/APT substrates
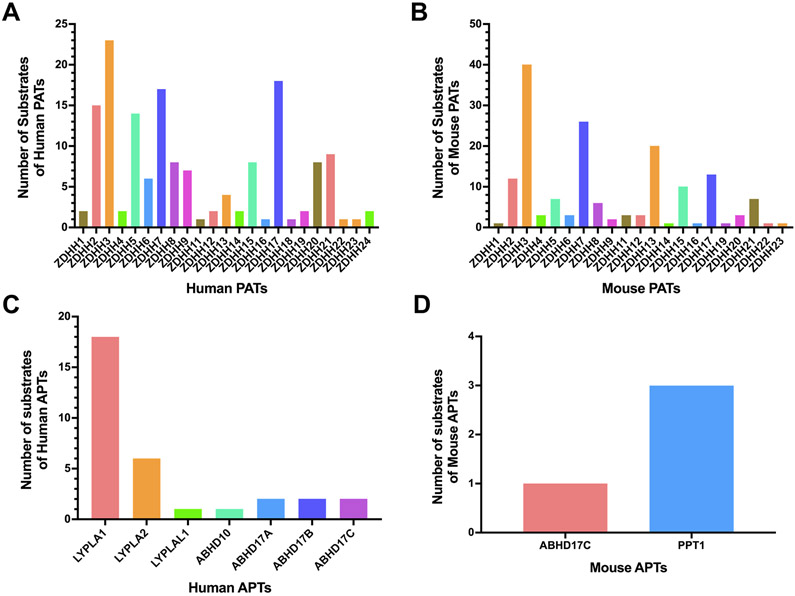
To understand the biological functions of the PATs/APTs in physiology and diseases, it is essential to reconstruct the PAT/APT-substrate networks. So far, most PAT/APT-substrate pairs have been established on the basis of individual proteins, predominantly via site mutagenesis. Our survey of the SwissPalm database and literature suggested that 0-40 protein substrates have been mapped to each PAT/APT (Fig. 7, Table S8-11). Interestingly, the identified PAT substrates are strongly biased towards DHHC2/3/5/7/17 in humans and DHHC3/7/13/17 in mice (Fig. 7A-B). This is probably because these PATs are appealing to many groups due to their functional importance in major diseases such as Huntington’s disease (DHHC13/17)65,66, type I diabetes (DHHC17)67, and cancer (DHHC2/3/5/7/13/17)35. In comparison, much less is known about the substrates for each APT, with the exception of APT1 (Fig. 7C-D).
Figure 7. Histograms of known substrates of human and mouse PATs/APTs.
(A) Histogram of known protein substrates of human PATs. (B) Histogram of known protein substrates of mouse PATs. (C) Histogram of known protein substrates of human APTs. (D) Histogram of known protein substrates of mouse APTs.
To facilitate the discovery of novel PAT/APT-substrate pairs, quantitative or semi-quantitative S-acylproteomics methods have been applied to compare the differences of S-acylproteomes in wild-type and PAT/APT-deficient cells or tissues. Thus far, a total of 15 S-acylproteomic studies have been reported to identify the substrates of human DHHC2, DHHC3, and APT1, mouse DHHC5/7/13/17 and APT1, fly DHHC8, plant TIP1, fungal AkrA, as well as yeast Ark1/2, Erf2, Swf1, and Pfa3/4/5 (Table S12). Interestingly, 12 of these studies employed ABE-related methods, probably due to their wider applicability than MLCC. Up to 362 candidate substrate proteins were identified for each study, based on the significant changes of S-acylprotein abundance.
However, it should be noted that the genetic manipulation of a PAT/APT may cause significant changes in the abundance of proteins, leading to the false discovery of candidate substrates.68 Thus, quantitative proteomics analysis needs to be conducted in parallel with the quantitative S-acylproteomics analysis, in order to minimize false discoveries. Nevertheless, only two out of the 15 studies also conducted quantitative proteomics analysis.69,70 In one study, Li et al. applied MLCC to enrich S-acylated proteins from neural stem cells derived from control and Zdhhc5-deficient mice, and used SILAC to quantify the S-acylproteome difference.69 Among ~300 candidate S-acylproteins, about 20 were >2-fold decreased in Zdhhc5-deficient cells, compared with control cells. However, a proteomic analysis of unenriched tryptic digests of the paired samples showed that the S-acylprotein level changes actually reflected differences in overall membrane protein abundance. Therefore, it remains unclear which of the identified candidate DHHC5 substrates are authentic substrates or false discoveries. In the other study, Shen et al. applied acyl-RAC to enrich S-acylated peptides from livers of wild-type and Zdhhc13-deficient mice.70 A total of 1,905 S-acylation sites corresponding to 883 proteins were identified. Using LFQ, a number of differentially abundant S-acylated peptides were identified, but the number was not specified in the paper. In addition, TMT10plex-based quantitative proteomics comparison of the paired samples was conducted, so as to remove the effects of changes in protein expression. Consequently, 400 S-acylation sites corresponding to 254 proteins were accepted as candidate DHHC13 substrate sites. Among these, Cys104 of MCAT and Cys618 of CTNND1 were validated as DHHC13 substrate sites. The remaining 13 studies did not conduct quantitative proteomics analysis in parallel. Nevertheless, a small number of candidate substrates were successfully validated by orthogonal methods in some studies. Taken together, quantitative S-acylproteomics analysis alone provides useful information for identifying potential enzyme substrates, yet caution needs to be exercised to avoid data over-interpretation.
In short, integrated quantitative S-acylproteomics and proteomics analyses have a great potential of uncovering PAT/APT-substrate networks on a large scale. Given that a mammalian genome encodes >30 PATs/APTs and >3,500 S-acylated proteins, only the tip of the iceberg of the entire network has been mapped so far. A collective effort in the S-acylation field is required to navigate the uncharted territory, which will provide great insights into the S-acylation biology and reveal novel disease biomarkers and therapeutic targets.
3.7. Proteomic identification of PAT/APT-binding partners
Accumulating evidence has suggested that, akin to the MAPK signaling cascade, a PAT may modulate the S-acylation levels of indirect substrates via activating an S-acylation cascade. For instance, DHHC16 can S-acylate DHHC6 and thus regulates its turnover, localization, and activity.71 Hence, to reconstruct the PAT/APT substrate network, it is important to distinguish direct from indirect substrates of each enzyme. However, this cannot be achieved by the above-mentioned integrative S-acylproteomics and proteomics analysis. To identify direct substrates, one strategy is to overlap all candidate substrates with binding partners of each PAT/APT. In addition, a PAT/APT may play non-enzymatic functions, e.g., serving as a scaffold protein. Thus, the identification of PAT/APT-binding partners is equally important for understanding the non-enzymatic functions.
So far, five proteomic studies were conducted to identify binding partners of human/mouse PATs/APTs (Table S13). Two studies focused on discovering DHHC5-associated proteins.29,72 In one study, Wang et al. applied BioID to identify proteins proximal to DHHC5 in mouse 3T3-L1 cells, resulting in the identification of 10 DHHC5-associated proteins including CD36.72 In the other study, Woodley et al. used affinity purification-mass spectrometry (AP-MS) to identify DHHC5-binding partners in human HeLa cells, leading to the identification of 304 and 39 candidate interacting proteins in plasma membrane and endosome, respectively.29 Surprisingly, the two studies did not identify any common DHHC5-binding partners. Two additional studies analyzed PPT1-binding partners.73,74 In one study, Scifo et al. applied AP-MS to analyze the human PPT1 interactome in human SH-SY5Y cells, resulted in the identification of 22 candidate PPT1-interacting proteins.73 In the other study, Sapir et al. employed AP-MS to investigate the PPT1 interactome in the mouse brain, leading to the identification of 64 candidate PPT1-interacting proteins.74 Nevertheless, only one protein (VCP) was identified in both studies. Taken together, the limited number of studies suggested that PATs/APTs might form different complexes in different species or cell types. It is of great interest to uncover the common core complex as well as species- and cell-type-specific complexes for each PAT/APT.
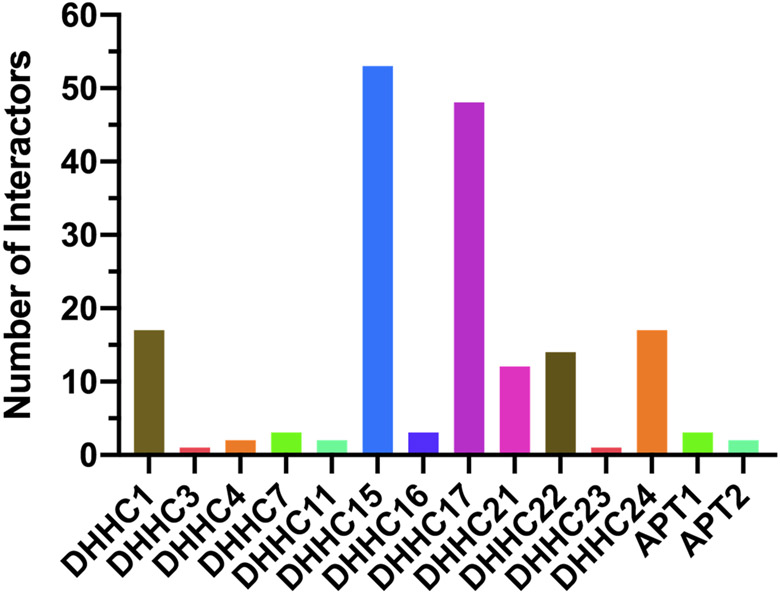
More recently, Luck et al. published so-far the largest Human Reference Interactome (HuRI) map, reporting 52,569 interactions between 8,274 human proteins.75 According to the latest HuRI online database, 53 interacting proteins were discovered for DHHC15, followed by smaller numbers of interacting proteins for 11 other PATs and two APTs (Figure 8). Nevertheless, DHHC5 or PPT1 interactomes have not yet been covered in the current version. We expect that more comprehensive PAT/APT interactomes will be reported in the near future.
Figure 8. Histogram of protein interactors of human PATs/APTs.
The numbers were retrieved from the HuRI database (http://www.interactome-atlas.org/).
3.8. Proteomic identification of off-targets of PAT/APT inhibitors
Due to the important roles of protein S-acylation in physiology and disease, multiple PAT/APT inhibitors have been developed to investigate the functions of S-acylation. To assess inhibitor selectivity and predict possible adverse events in vivo, it is important to profile the off-targets for each inhibitor. Table S14 shows that five such studies have been reported.
Despite many efforts, no inhibitors for individual PATs have yet been successfully developed. Nonetheless, protein S-acylation can be broadly inhibited by lipid-based inhibitors such as 2-bromopalmitate (2BP) and nonlipid-based inhibitors such as cerulenin and tunicamycin. To identify the off-targets of 2BP, two approaches akin to MLCC were developed. In one approach, an ω-azido analog of 2BP (2BPN3) was synthesized for metabolic labeling, click conjugation, and streptavidin enrichment of potential 2BP targets.76 Surprisingly, about 450 putative targets were highly enriched (≥5-fold relative to the control), of which only five are DHHC-PATs. In the other approach, two terminal alkyne analogs of 2BP, 16C-BYA and 18C-BYS, were synthesized to label and enrich 2BP targets, leading to the identification of >1,200 proteins including three DHHC-PATs. Similarly, cerulenin was found to target −140 proteins including DHHC20.77 Collectively, the studies suggested that, although 2BP and cerulenin do target certain DHHC-PATs, both have a large number of off-targets. Thus, more potent and specific PAT inhibitors are awaited.
For APTs, several small-molecule inhibitors have been successfully developed, such as the APT1/2 inhibitor Palmostatin B/M, APT1-specific inhibitor ML348, and APT2-specific inhibitor ML349.78-81 To determine the off-targets of Palmostatin B/M, Rusch el al. designed and synthesized two palmostatin B/M-based activity-based proteome profiling (ABPP) probes, and applied them to probe targets in an SDS-PAGE gel.82 Differentially abundant gel bands were excised and proteins were identified by LC-MS/MS. A total of 59 proteins including APT1/2 and PPT1 were identified. To determine ML349 targets, Won et al. synthesized biotinylated-ML349 and applied it to enrich ML349-interacting proteins. In addition, o-ML349-biotin and unconjugated azide-PEG3-biotin, both of which have very low inhibitory activity against APT2, were used as negative controls. Label-free proteomics analysis identified about 10 candidate ML349 targets including APT2. Taken together, the studies suggest that APT inhibitors are relatively target-specific but still have some off-targets.
4. Important Challenges for S-acylproteomics
4.1. Global analysis of native and intact S-acylated peptides
Currently, the S-acylproteomics field is still plagued by a lack of effective methods for global analysis of native and intact S-acylated peptides, which would provide definitive evidence of protein S-acylation and reveal the identities of fatty S-acyl species. Of note, neither ABE nor MLCC can address this issue, because the former removes fatty S-acyl groups whereas the latter utilizes unnatural fatty acids. The challenges are at least two folds. Firstly, no methods have been developed to efficiently separate native S-acylated peptides from all other non-S-acylated peptides. Although a pan anti-palmitoylation antibody was reported58, it remains unclear whether it can efficiently enrich S-palmitoylated peptides. Secondly, due to their high hydrophobicity, intact S-acylated peptides tend to be lost during sample processing and are difficult to separate by conventional C18 reversed-phase columns. Therefore, ground-breaking technologies need to be developed to address both challenges.
4.2. Proteomic analysis of S-acylation site stoichiometry
Our and other labs have shown that proteins may be S-acylated at varied levels. For example, our recent study showed that many signaling proteins have low (~1%) S-acylation stoichiometry, whereas caveolin-1 is almost 100% S-acylated.45 Techniques such as PEG switch83 and acyl-PEG exchange84 enabled the determination of S-acylation site stoichiometry, but they are antibodybased and only allow the analysis of individual proteins. Innovative approaches need to be developed to enable the proteome-scale analysis of S-acylation site stoichiometry.
4.3. Single cell S-acylproteomics
S-acylated proteins and complexes may be present at different levels in different cell states and types. Thus, it is critical to understand the dynamic changes of protein S-acylation at the single-cell resolution. Due to the low abundance of most S-acylated proteins, current S-acylproteomics analysis requires micrograms to milligrams of protein lysates. To achieve single cell S-acylproteomics, miniatured and streamlined sample processing methods need to be developed to minimize sample loss, and more sensitive proteomic methods have to be established to detect and quantify a tiny amount of S-acylated proteins or peptides.
5. Summary
The past 14 years have witnessed significant progress in the S-acylproteomics field, where the techniques are becoming mature to cover the near-complete S-acylproteome.45 The S-acylproteomics field has switched the gear from qualitative S-acylproteomics to functional S-acylproteomics, which provided novel insights into the roles of S-acylation in physiology and disease. By coupling with novel PAT/APT probes and inhibitors,85-87 we envision that S-acylproteomics will play an increasingly important role in understanding disease mechanisms and discovering novel biomarkers and therapeutic targets. In addition, we expect that innovative S-acylproteomics technologies will be developed to address key challenges in the field and revolutionize the study of protein S-acylation.
Supplementary Material
Supporting Table S1. A list of ABE-related methods for the enrichment of S-acylated proteins.
Supporting Table S2. A list of MLCC-related methods for the enrichment of S-acylated proteins.
Supporting Table S3. A list of quantitative S-acylproteomics studies to identify S-acylated proteins and/or S-acylation sites.
Supporting Table S4. A list of comparative S-acylproteomics studies.
Supporting Table S5. A list of temporal S-acylproteomics studies.
Supporting Table S6. A list of proteomic studies of S-acylation cross-talk with other post-translational modifications.
Supporting Table S7. A list of proteomic studies of S-acylprotein complexes.
Supporting Table S8. A list of human PAT-substrate pairs in the SwissPalm database (v3).
Supporting Table S9. A list of mouse PAT-substrate pairs in the SwissPalm database (v3).
Supporting Table S10. A list of human APT-substrate pairs in the SwissPalm database (v3).
Supporting Table S11. A list of mouse APT-substrate pairs in the SwissPalm database (v3).
Supporting Table S12. A list of proteomic studies of PAT/APT substrates.
Supporting Table S13. A list of proteomic studies of PAT/APT-binding partners.
Supporting Table S14. A list of proteomic studies of the off-targets of PAT/APT inhibitors.
ACKNOWLEDGMENT
We acknowledge financial support from the National Cancer Institute (1R01CA218526 to W.Y.). Because of space limitations, it was impossible to include a very comprehensive list of references for all the work discussed. We apologize to the authors whose important contributions were not described or cited.
Funding Sources
This work was supported by NCI 1R01CA218526 (W.Y.).
ABBREVIATIONS
- 17-ODYA
17-octadecylnoic acid
- 2BP
2-bromopalmitate
- 2BPN3
ω-azido analog of 2BP
- ABE
acyl-biotinyl exchange
- ABHD
α/β hydrolase fold domain
- ABPP
activity-based proteome profiling
- acyl-RAC
acyl resin-assisted capture
- AP-MS
affinity purification mass spectrometry
- APT
acylprotein thioesterase
- biotin-HPDP
N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide
- cICAT
cleavable isotope-coded affinity tag
- DHHC
Asp-His-His-Cys
- DTDP
2,2′-dithiodipyridine
- ER
endoplasmic reticulum
- EV
extracellular vesicles
- FDR
false discovery rate
- HDFP
hexadecylfluorophosphonate
- HuRI
human reference interactome map
- LB-ABE
low-background acyl-biotinyl exchange
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- LFQ
label-free quantitation
- MLCC
metabolic labeling with a palmitic acid analog followed by click chemistry
- MMTS
methyl methanethiosulfonate
- NEM
N-ethylmaleimide
- PalmPISC
palmitoyl-protein identification and site characterization
- PANIMoni
palmitoylation and nitrosylation interplay monitoring
- PAT
palmitoyl S-acyltransferase
- PICA
palmitoyl-cysteine isolation capture and analysis
- PPT
palmitoyl protein thioesterase
- SILAC2plex
duplex stable-isotope labeling by amino acids in cell culture
- ssABE
site-specific acyl-biotinyl exchange
- TCEP
tris(2-carboxyethyl) phosphine
- TMT6plex
6-plex tandem mass tags
REFERENCES
- (1).Jiang H; Zhang X; Chen X; Aramsangtienchai P; Tong Z; Lin H Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies. Chem. Rev 2018, 118 (3), 919–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Linder ME; Deschenes RJ Palmitoylation: Policing Protein Stability and Traffic. Nat. Rev. Mol. Cell Biol 2007, 8 (1), 74–84. [DOI] [PubMed] [Google Scholar]
- (3).Muszbek L; Laposata M Covalent Modification of Platelet Proteins by Palmitate. Blood 1989, 74 (4), 1339–1347. [PubMed] [Google Scholar]
- (4).Muszbek L; Laposata M Myristoylation of Proteins in Platelets Occurs Predominantly through Thioester Linkages. J. Biol. Chem 1993, 268 (11), 8251–8255. [PubMed] [Google Scholar]
- (5).Muszbek L; Laposata M Covalent Modification of Proteins by Arachidonate and Eicosapentaenoate in Platelets. J. Biol. Chem 1993, 268 (24), 18243–18248. [PubMed] [Google Scholar]
- (6).Muszbek L; Haramura G; Cluette-Brown JE; Van Cott EM; Laposata M The Pool of Fatty Acids Covalently Bound to Platelet Proteins by Thioester Linkages Can Be Altered by Exogenously Supplied Fatty Acids. Lipids 1999, 34 Suppl, S331–7. [DOI] [PubMed] [Google Scholar]
- (7).Liang X; Nazarian A; Erdjument-Bromage H; Bornmann W; Tempst P; Resh MD Heterogeneous Fatty Acylation of Src Family Kinases with Polyunsaturated Fatty Acids Regulates Raft Localization and Signal Transduction. J. Biol. Chem 2001, 276 (33), 30987–30994. [DOI] [PubMed] [Google Scholar]
- (8).Schmidt MF; Schlesinger MJ Fatty Acid Binding to Vesicular Stomatitis Virus Glycoprotein: A New Type of Post-Translational Modification of the Viral Glycoprotein. Cell 1979, 17 (4), 813–819. [DOI] [PubMed] [Google Scholar]
- (9).Conibear E; Davis NG Palmitoylation and Depalmitoylation Dynamics at a Glance. J. Cell Sci 2010, 123 (Pt 23), 4007–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jin J; Zhi X; Wang X; Meng D Protein Palmitoylation and Its Pathophysiological Relevance. J. Cell. Physiol 2020, jcp.30122. [DOI] [PubMed] [Google Scholar]
- (11).Blanc M; David F; Abrami L; Migliozzi D; Armand F; Bürgi J; van der Goot FG SwissPalm: Protein Palmitoylation Database. F1000Research 2015, 4, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sanders SS; Martin DDO; Butland SL; Lavallée-Adam M; Calzolari D; Kay C; Yates JR; Hayden MR Curation of the Mammalian Palmitoylome Indicates a Pivotal Role for Palmitoylation in Diseases and Disorders of the Nervous System and Cancers. PLoS Comput. Biol 2015, 11 (8), e1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mitchell DA; Vasudevan A; Linder ME; Deschenes RJ Protein Palmitoylation by a Family of DHHC Protein S-Acyltransferases. J. Lipid Res 2006, 47 (6), 1118–1127. [DOI] [PubMed] [Google Scholar]
- (14).De I; Sadhukhan S Emerging Roles of DHHC-Mediated Protein S-Palmitoylation in Physiological and Pathophysiological Context. Eur. J. Cell Biol 2018, 97 (5), 319–338. [DOI] [PubMed] [Google Scholar]
- (15).Linder ME; Jennings BC Mechanism and Function of DHHC S-Acyltransferases. Biochem. Soc. Trans 2013, 41 (1), 29–34. [DOI] [PubMed] [Google Scholar]
- (16).Ohno Y; Kihara A; Sano T; Igarashi Y Intracellular Localization and Tissue-Specific Distribution of Human and Yeast DHHC Cysteine-Rich Domain-Containing Proteins. Biochim. Biophys. Acta 2006, 1761 (4), 474–483. [DOI] [PubMed] [Google Scholar]
- (17).Won SJ; Cheung See Kit M; Martin BR Protein Depalmitoylases. Crit. Rev. Biochem. Mol. Biol 2018, 53 (1), 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Vesa J; Hellsten E; Verkruyse LA; Camp LA; Rapola J; Santavuori P; Hofmann SL; Peltonen L Mutations in the Palmitoyl Protein Thioesterase Gene Causing Infantile Neuronal Ceroid Lipofuscinosis. Nature 1995, 376 (6541), 584–587. [DOI] [PubMed] [Google Scholar]
- (19).Duncan JA; Gilman AG A Cytoplasmic Acyl-Protein Thioesterase That Removes Palmitate from G Protein Alpha Subunits and P21(RAS). J. Biol. Chem 1998, 273 (25), 15830–15837. [DOI] [PubMed] [Google Scholar]
- (20).Lin DT; Conibear E ABHD17 Proteins Are Novel Protein Depalmitoylases That Regulate N-Ras Palmitate Turnover and Subcellular Localization. Elife 2015, 4, e11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Yokoi N; Fukata Y; Sekiya A; Murakami T; Kobayashi K; Fukata M Identification of PSD-95 Depalmitoylating Enzymes. J. Neurosci 2016, 36 (24), 6431–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Cao Y; Qiu T; Kathayat RS; Azizi SA; Thorne AK; Ahn D; Fukata Y; Fukata M; Rice PA; Dickinson BC ABHD10 Is an S-Depalmitoylase Affecting Redox Homeostasis through Peroxiredoxin-5. Nat. Chem. Biol 2019, 15 (12), 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Stix R; Lee CJ; Faraldo-Gómez JD; Banerjee A Structure and Mechanism of DHHC Protein Acyltransferases. J. Mol. Biol 2020, 432 (18), 4983–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Jennings BC; Linder ME DHHC Protein S-Acyltransferases Use Similar Ping-Pong Kinetic Mechanisms but Display Different Acyl-CoA Specificities. J. Biol. Chem 2012, 287 (10), 7236–7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lobo S; Greentree WK; Linder ME; Deschenes RJ Identification of a Ras Palmitoyltransferase in Saccharomyces Cerevisiae. J. Biol. Chem 2002, 277 (43), 41268–41273. [DOI] [PubMed] [Google Scholar]
- (26).Ko PJ; Woodrow C; Dubreuil MM; Martin BR; Skouta R; Bassik MC; Dixon SJ A ZDHHC5-GOLGA7 Protein Acyltransferase Complex Promotes Nonapoptotic Cell Death. Cell Chem. Biol 2019, 26 (12), 1716–1724.e9. [DOI] [PubMed] [Google Scholar]
- (27).Fredericks GJ; Hoffmann FKW; Rose AH; Osterheld HJ; Hess FM; Mercier F; Hoffmann PR Stable Expression and Function of the Inositol 1,4,5-Triphosphate Receptor Requires Palmitoylation by a DHHC6/Selenoprotein K Complex. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (46), 16478–16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Swarthout JT; Lobo S; Farh L; Croke MR; Greentree WK; Deschenes RJ; Linder ME DHHC9 and GCP16 Constitute a Human Protein Fatty Acyltransferase with Specificity for H- and N-Ras. J. Biol. Chem 2005, 280 (35), 31141–31148. [DOI] [PubMed] [Google Scholar]
- (29).Woodley KT; Collins MO S-Acylated Golga7b Stabilises DHHC5 at the Plasma Membrane to Regulate Cell Adhesion. EMBO Rep. 2019, 20 (10), e47472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Fredericks G; Hoffmann F; Hondal R; Rozovsky S; Urschitz J; Hoffmann P Selenoprotein K Increases Efficiency of DHHC6 Catalyzed Protein Palmitoylation by Stabilizing the Acyl-DHHC6 Intermediate. Antioxidants 2017, 7 (1), 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kong E; Peng S; Chandra G; Sarkar C; Zhang Z; Bagh MB; Mukherjee AB Dynamic Palmitoylation Links Cytosol-Membrane Shuttling of Acyl-Protein Thioesterase-1 and Acyl-Protein Thioesterase-2 with That of Proto-Oncogene H-Ras Product and Growth-Associated Protein-43. J. Biol. Chem 2013, 288 (13), 9112–9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Schlesinger MJ; Magee AI; Schmidt MF Fatty Acid Acylation of Proteins in Cultured Cells. J. Biol. Chem 1980, 255 (21), 10021–10024. [PubMed] [Google Scholar]
- (33).Drisdel RC; Green WN Labeling and Quantifying Sites of Protein Palmitoylation. Biotechniques 2004, 36 (2), 276–285. [DOI] [PubMed] [Google Scholar]
- (34).Zaballa ME; van der Goot FG The Molecular Era of Protein S-Acylation: Spotlight on Structure, Mechanisms, and Dynamics. Crit. Rev. Biochem. Mol. Biol 2018, 53 (4), 420–451. [DOI] [PubMed] [Google Scholar]
- (35).Ko P; Dixon SJ Protein Palmitoylation and Cancer. EMBO Rep. 2018, 19 (10), e46666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Roth AF; Wan J; Bailey AO; Sun B; Kuchar JA; Green WN; Phinney BS; Yates JR; Davis NG Global Analysis of Protein Palmitoylation in Yeast. Cell 2006, 125 (5), 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zhang J; Planey SL; Ceballos C; Stevens SM Jr; Keay SK; Zacharias DA Identification of CKAP4/P63 as a Major Substrate of the Palmitoyl Acyltransferase DHHC2, a Putative Tumor Suppressor, Using a Novel Proteomics Method. Mol. Cell. Proteomics 2008, 7 (7), 1378–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Yang W; Di Vizio D; Kirchner M; Steen H; Freeman MR Proteome Scale Characterization of Human S-Acylated Proteins in Lipid Raft-Enriched and Non-Raft Membranes. Mol. Cell. Proteomics 2010, 9 (1), 54–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Forrester MT; Hess DT; Thompson JW; Hultman R; Moseley MA; Stamler JS; Casey PJ Site-Specific Analysis of Protein S-Acylation by Resin-Assisted Capture. J. Lipid Res 2011, 52 (2), 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gould NS; Evans P; Martinez-Acedo P; Marino SM; Gladyshev VN; Carroll KS; Ischiropoulos H Site-Specific Proteomic Mapping Identifies Selectively Modified Regulatory Cysteine Residues in Functionally Distinct Protein Networks. Chem. Biol 2015, 22 (7), 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Collins MO; Woodley KT; Choudhary JS Global, Site-Specific Analysis of Neuronal Protein S-Acylation. Sci. Rep 2017, 7 (1), 4681–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zareba-Koziol M; Bartkowiak-Kaczmarek A; Figiel I; Krzystyniak A; Wojtowicz T; Bijata M; Wlodarczyk J Stress-Induced Changes in the S-Palmitoylation and S-Nitrosylation of Synaptic Proteins. Mol. Cell. Proteomics 2019, 18 (10), 1916–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hurst CH; Turnbull D; Plain F; Fuller W; Hemsley PA Maleimide Scavenging Enhances Determination of Protein S-Palmitoylation State in Acyl-Exchange Methods. Biotechniques 2017, 62 (2), 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhang X; Zhang Y; Fang C; Zhang L; Yang P; Wang C; Lu H Ultradeep Palmitoylomics Enabled by Dithiodipyridine-Functionalized Magnetic Nanoparticles. Anal. Chem 2018, 90 (10), 6161–6168. [DOI] [PubMed] [Google Scholar]
- (45).Zhou B; Wang Y; Yan Y; Mariscal J; Di Vizio D; Freeman MR; Yang W Low-Background Acyl-Biotinyl Exchange Largely Eliminates the Coisolation of Non-S-Acylated Proteins and Enables Deep S-Acylproteomic Analysis. Anal. Chem 2019, 91 (15), 9858–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Hang HC; Geutjes EJ; Grotenbreg G; Pollington AM; Bijlmakers MJ; Ploegh HL Chemical Probes for the Rapid Detection of Fatty-Acylated Proteins in Mammalian Cells. J. Am. Chem. Soc 2007, 129 (10), 2744–2745. [DOI] [PubMed] [Google Scholar]
- (47).Kostiuk MA; Corvi MM; Keller BO; Plummer G; Prescher JA; Hangauer MJ; Bertozzi CR; Rajaiah G; Falck JR; Berthiaume LG Identification of Palmitoylated Mitochondrial Proteins Using a Bio-Orthogonal Azido-Palmitate Analogue. FASEB J. 2008, 22 (3), 721–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gao X; Hannoush RN A Decade of Click Chemistry in Protein Palmitoylation: Impact on Discovery and New Biology. Cell Chem. Biol 2018, 25 (3), 236–246. [DOI] [PubMed] [Google Scholar]
- (49).Zhou B; An M; Freeman MR; Yang W Technologies and Challenges in Proteomic Analysis of Protein S-Acylation. J. Proteomics Bioinform 2014, 7 (9), 256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Won SJ; Martin BR Temporal Profiling Establishes a Dynamic S-Palmitoylation Cycle. ACS Chem. Biol 2018, 13 (6), 1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yoon HI; Yhee JY; Na JH; Lee S; Lee H; Kang SW; Chang H; Ryu JH; Lee S; Kwon IC; et al. Bioorthogonal Copper Free Click Chemistry for Labeling and Tracking of Chondrocytes In Vivo. Bioconjug. Chem 2016, 27 (4), 927–936. [DOI] [PubMed] [Google Scholar]
- (52).Jiang H; Khan S; Wang Y; Charron G; He B; Sebastian C; Du J; Kim R; Ge E; Mostoslavsky R; et al. SIRT6 Regulates TNF-α Secretion through Hydrolysis of Long-Chain Fatty Acyl Lysine. Nature 2013, 496 (7443), 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Jones ML; Collins MO; Goulding D; Choudhary JS; Rayner JC Analysis of Protein Palmitoylation Reveals a Pervasive Role in Plasmodium Development and Pathogenesis. Cell Host Microbe 2012, 12 (2), 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Smotrys JE; Linder ME Palmitoylation of Intracellular Signaling Proteins: Regulation and Function. Annu. Rev. Biochem 2004, 73 (1), 559–587. [DOI] [PubMed] [Google Scholar]
- (55).Rodenburg RNP; Snijder J; van de Waterbeemd M; Schouten A; Granneman J; Heck AJR; Gros P Stochastic Palmitoylation of Accessible Cysteines in Membrane Proteins Revealed by Native Mass Spectrometry. Nat. Commun 2017, 8 (1), 1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Zhou F; Xue Y; Yao X; Xu Y CSS-Palm: Palmitoylation Site Prediction with a Clustering and Scoring Strategy (CSS). Bioinformatics 2006, 22 (7), 894–896. [DOI] [PubMed] [Google Scholar]
- (57).Ning W; Jiang P; Guo Y; Wang C; Tan X; Zhang W; Peng D; Xue Y GPS-Palm: A Deep Learning-Based Graphic Presentation System for the Prediction of S-Palmitoylation Sites in Proteins. Brief. Bioinform 2020, 1–12. [DOI] [PubMed] [Google Scholar]
- (58).Fang C; Zhang X; Zhang L; Gao X; Yang P; Lu H Identification of Palmitoylated Transitional Endoplasmic Reticulum ATPase by Proteomic Technique and Pan Antipalmitoylation Antibody. J. Proteome Res 2016, 15 (3), 956–962. [DOI] [PubMed] [Google Scholar]
- (59).Mariscal J; Vagner T; Kim M; Zhou B; Chin A; Zandian M; Freeman MR; You S; Zijlstra A; Yang W; et al. Comprehensive Palmitoyl-Proteomic Analysis Identifies Distinct Protein Signatures for Large and Small Cancer-Derived Extracellular Vesicles. J. Extracell. vesicles 2020, 9 (1), 1764192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Shahid M; Kim M; Jin P; Zhou B; Wang Y; Yang W; You S; Kim J S-Palmitoylation as a Functional Regulator of Proteins Associated with Cisplatin Resistance in Bladder Cancer. Int. J. Biol. Sci 2020, 16 (14), 2490–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Thompson A; Wölmer N; Koncarevic S; Selzer S; Böhm G; Legner H; Schmid P; Kienle S; Penning P; Höhle C; et al. TMTpro: Design, Synthesis, and Initial Evaluation of a Proline-Based Isobaric 16-Plex Tandem Mass Tag Reagent Set. Anal. Chem 2019, 91 (24), 15941–15950. [DOI] [PubMed] [Google Scholar]
- (62).Wang Z; Yu K; Tan H; Wu Z; Cho J-H; Han X; Sun H; Beach TG; Peng J 27-Plex Tandem Mass Tag Mass Spectrometry for Profiling Brain Proteome in Alzheimer’s Disease. Anal. Chem 2020, 92 (10), 7162–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Haag SM; Gulen MF; Reymond L; Gibelin A; Abrami L; Decout A; Heymann M; Van Der Goot FG ; Turcatti G; Behrendt R; et al. Targeting STING with Covalent Small-Molecule Inhibitors. Nature 2018, 559 (7713), 269–273. [DOI] [PubMed] [Google Scholar]
- (64).Peng T; Hang HC Bifunctional Fatty Acid Chemical Reporter for Analyzing S-Palmitoylated Membrane Protein-Protein Interactions in Mammalian Cells. J. Am. Chem. Soc 2015, 137 (2), 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Sutton LM; Sanders SS; Butland SL; Singaraja RR; Franciosi S; Southwell AL; Doty CN; Schmidt ME; Mui KK; Kovalik V; et al. Hip14l-Deficient Mice Develop Neuropathological and Behavioural Features of Huntington Disease. Hum. Mol. Genet 2013, 22 (3), 452–465. [DOI] [PubMed] [Google Scholar]
- (66).Milnerwood AJ; Parsons MP; Young FB; Singaraja RR; Franciosi S; Volta M; Bergeron S; Hayden MR; Raymond LA Memory and Synaptic Deficits in Hip14/DHHC17 Knockout Mice. Proc. Natl. Acad. Sci. U. S. A 2013, 110 (50), 20296–20301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Berchtold LA; Storling ZM; Ortis F; Lage K; Bang-Berthelsen C; Bergholdt R; Hald J; Brorsson CA; Eizirik DL; Pociot F; et al. Huntingtin-Interacting Protein 14 Is a Type 1 Diabetes Candidate Protein Regulating Insulin Secretion and Beta-Cell Apoptosis. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (37), E681–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Wan J; Savas JN; Roth AF; Sanders SS; Singaraja RR; Hayden MR; Yates JR 3rd; Davis NG Tracking Brain Palmitoylation Change: Predominance of Glial Change in a Mouse Model of Huntington’s Disease. Chem. Biol 2013, 20 (11), 1421–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Li Y; Martin BR; Cravatt BF; Hofmann SL DHHC5 Protein Palmitoylates Flotillin-2 and Is Rapidly Degraded on Induction of Neuronal Differentiation in Cultured Cells. J. Biol. Chem 2012, 287 (1), 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Shen LF; Chen YJ; Liu KM; Haddad ANS; Song IW; Roan HY; Chen LY; Yen JJY; Chen YJ; Wu JY; et al. Role of S-Palmitoylation by ZDHHC13 in Mitochondrial Function and Metabolism in Liver. Sci. Rep 2017, 7 (1), 2182–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Abrami L; Dallavilla T; Sandoz PA; Demir M; Kunz B; Savoglidis G; Hatzimanikatis V; van der Goot FG Identification and Dynamics of the Human ZDHHC16-ZDHHC6 Palmitoylation Cascade. Elife 2017, 6, 10.7554/eLife.27826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Wang J; Hao JW; Wang X; Guo H; Sun HH; Lai XY; Liu LY; Zhu M; Wang HY; Li YF; et al. DHHC4 and DHHC5 Facilitate Fatty Acid Uptake by Palmitoylating and Targeting CD36 to the Plasma Membrane. Cell Rep. 2019, 26 (1), 209–221.e5. [DOI] [PubMed] [Google Scholar]
- (73).Scifo E; Szwajda A; Soliymani R; Pezzini F; Bianchi M; Dapkunas A; Debski J; Uusi-Rauva K; Dadlez M; Gingras AC; et al. Proteomic Analysis of the Palmitoyl Protein Thioesterase 1 Interactome in SH-SY5Y Human Neuroblastoma Cells. J. Proteomics 2015, 123, 42–53. [DOI] [PubMed] [Google Scholar]
- (74).Sapir T; Segal M; Grigoryan G; Hansson KM; James P; Segal M; Reiner O The Interactome of Palmitoyl-Protein Thioesterase 1 (PPT1) Affects Neuronal Morphology and Function. Front. Cell. Neurosci 2019, 13, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Luck K; Kim DK; Lambourne L; Spirohn K; Begg BE; Bian W; Brignall R; Cafarelli T; Campos-Laborie FJ; Charloteaux B; et al. A Reference Map of the Human Binary Protein Interactome. Nature 2020, 580 (7803), 402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Davda D; El Azzouny MA; Tom CT; Hernandez JL; Majmudar JD; Kennedy RT; Martin BR Profiling Targets of the Irreversible Palmitoylation Inhibitor 2-Bromopalmitate. ACS Chem. Biol 2013, 8 (9), 1912–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Zheng B; Zhu S; Wu X Clickable Analogue of Cerulenin as Chemical Probe to Explore Protein Palmitoylation. ACS Chem. Biol 2015, 10 (1), 115–121. [DOI] [PubMed] [Google Scholar]
- (78).Dekker FJ; Rocks O; Vartak N; Menninger S; Hedberg C; Balamurugan R; Wetzel S ; Renner S; Gerauer M; Scholermann B; et al. Small-Molecule Inhibition of APT1 Affects Ras Localization and Signaling. Nat. Chem. Biol 2010, 6 (6), 449–456. [DOI] [PubMed] [Google Scholar]
- (79).Hedberg C; Dekker FJ; Rusch M; Renner S; Wetzel S; Vartak N; Gerding-Reimers C; Bon RS; Bastiaens PIH; Waldmann H Development of Highly Potent Inhibitors of the Ras-Targeting Human Acyl Protein Thioesterases Based on Substrate Similarity Design. Angew. Chem. Int. Ed. Engl 2011, 50 (42), 9832–9837. [DOI] [PubMed] [Google Scholar]
- (80).Adibekian A; Martin BR; Chang JW; Hsu KL; Tsuboi K; Bachovchin DA; Speers AE; Brown SJ; Spicer T; Fernandez-Vega V; et al. Confirming Target Engagement for Reversible Inhibitors in Vivo by Kinetically Tuned Activity-Based Probes. J. Am. Chem. Soc 2012, 134 (25), 10345–10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Won SJ; Davda D; Labby KJ; Hwang SY; Pricer R; Majmudar JD; Armacost KA; Rodriguez LA; Rodriguez CL; Chong FS; et al. Molecular Mechanism for Isoform-Selective Inhibition of Acyl Protein Thioesterases 1 and 2 (APT1 and APT2). ACS Chem. Biol 2016, 11 (12), 3374–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Rusch M; Zimmermann TJ; Bürger M; Dekker FJ; Görmer K; Triola G; Brockmeyer A; Janning P; Böttcher T; Sieber SA; et al. Identification of Acyl Protein Thioesterases 1 and 2 as the Cellular Targets of the Ras-Signaling Modulators Palmostatin B and M. Angew. Chem. Int. Ed. Engl 2011, 50 (42), 9838–9842. [DOI] [PubMed] [Google Scholar]
- (83).Howie J; Reilly L; Fraser NJ; Vlachaki Walker JM; Wypijewski KJ; Ashford ML ; Calaghan SC; McClafferty H; Tian L; Shipston MJ; et al. Substrate Recognition by the Cell Surface Palmitoyl Transferase DHHC5. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (49), 17534–17539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Percher A; Ramakrishnan S; Thinon E; Yuan X; Yount JS; Hang HC Mass-Tag Labeling Reveals Site-Specific and Endogenous Levels of Protein S-Fatty Acylation. Proc. Natl. Acad. Sci. U. S. A 2016, 113 (16), 4302–4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Azizi SA; Kathayat RS; Dickinson BC Activity-Based Sensing of S-Depalmitoylases: Chemical Technologies and Biological Discovery. Acc. Chem. Res 2019, 52 (11), 3029–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Hernandez JL; Majmudar JD; Martin BR Profiling and Inhibiting Reversible Palmitoylation. Curr. Opin. Chem. Biol 2013, 17 (1), 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Hamel LD; Lenhart BJ; Mitchell DA; Santos RG; Giulianotti MA; Deschenes RJ Identification of Protein Palmitoylation Inhibitors from a Scaffold Ranking Library. Comb. Chem. High Throughput Screen 2016, 19 (4), 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Table S1. A list of ABE-related methods for the enrichment of S-acylated proteins.
Supporting Table S2. A list of MLCC-related methods for the enrichment of S-acylated proteins.
Supporting Table S3. A list of quantitative S-acylproteomics studies to identify S-acylated proteins and/or S-acylation sites.
Supporting Table S4. A list of comparative S-acylproteomics studies.
Supporting Table S5. A list of temporal S-acylproteomics studies.
Supporting Table S6. A list of proteomic studies of S-acylation cross-talk with other post-translational modifications.
Supporting Table S7. A list of proteomic studies of S-acylprotein complexes.
Supporting Table S8. A list of human PAT-substrate pairs in the SwissPalm database (v3).
Supporting Table S9. A list of mouse PAT-substrate pairs in the SwissPalm database (v3).
Supporting Table S10. A list of human APT-substrate pairs in the SwissPalm database (v3).
Supporting Table S11. A list of mouse APT-substrate pairs in the SwissPalm database (v3).
Supporting Table S12. A list of proteomic studies of PAT/APT substrates.
Supporting Table S13. A list of proteomic studies of PAT/APT-binding partners.
Supporting Table S14. A list of proteomic studies of the off-targets of PAT/APT inhibitors.