Abstract
The isobaric carrier approach, which combines small isobarically labeled samples with a larger isobarically labeled carrier sample, finds diverse applications in ultrasensitive mass spectrometry analysis of very small samples, such as single cells. To enhance the growing use of isobaric carriers, we characterized the trade-offs of using isobaric carriers in controlled experiments with complex human proteomes. The data indicate that isobaric carriers directly enhance peptide sequence identification without simultaneously increasing the number of protein copies sampled from small samples. The results also indicate strategies for optimizing the amount of isobaric carrier and analytical parameters, such as ion accumulation time, for different priorities such as improved quantification or an increased number of identified proteins. Balancing these trade-offs enables adapting isobaric carrier experiments to different applications, such as quantifying proteins from limited biopsies or organoids, building single-cell atlases, or modeling protein networks in single cells. In all cases, the reliability of protein quantification should be estimated and incorporated in all subsequent analyses. We expect that these guidelines will aid in explicit incorporation of the characterized trade-offs in experimental designs and transparent error propagation in data analysis.
Keywords: single-cell proteomics, isobaric carrier, quantification accuracy, benchmarking, data reliability, optimizing mass spectrometry analysis
Graphical Abstract
INTRODUCTION
Mass spectrometry (MS) has become the most powerful method for analyzing proteins in bulk samples composed of many cells.1,2 However, MS analysis of smaller samples, such as single cells, is more challenging because the ions analyzed by the MS detectors may be insufficient for accurate quantification and sequence identification.3–8 To mitigate these challenges, we introduced the isobaric carrier concept as a part of Single Cell ProtEomics by MS (SCoPE-MS),9,10 and the concept has been used by multiple laboratories as recently reviewed.11 The isobaric carrier approach employs tandem mass tags to label small samples of interest (e.g., proteomes of single cells) and a carrier sample (e.g., the proteome of 100 cells) and then combines all labeled samples to be analyzed together by tandem mass spectrometry (MS/MS), as illustrated in Figure 1.
Figure 1.
Schematic diagram of the isobaric carrier concept. The peptides from small samples and from a larger (carrier) sample are labeled with isobaric tags, mixed, and analyzed by MS/MS. Some sets, as the standards listed in Table 1, may have more than one isobaric carrier sample. This approach increases the intensity of precursor ions (1), provides RIs for quantification (2), and facilitates sequence identifications based on the peptide fragments pooled across all samples (3).
The isobaric carrier concept has been applied to single-cell protein analysis, detection of mutant proteoforms, phosphorylation, and protein synthesis.11 This growing use of the concept motivated us to benchmark its benefits and limitations in controlled experiments and to extend the previously suggested approaches for optimizing ultrasensitive MS experiments. 12 We explored the role of isobaric carriers in (i) facilitating the detection of precursor ions in MS1 survey scans and (ii) facilitating sequence identification by providing peptide fragment ions to MS2 spectra. These benefits must be balanced with possible adverse effects on quantification. Specifically, large levels of isobaric carriers may enable identifying peptides whose single-cell copies are insufficiently sampled by the MS detector to support accurate quantification. 11
A fundamental and general challenge to single-cell analysis is sampling sufficient copies from each molecule type to support its reliable quantification. This remains, for example, a significant bottleneck for advanced single-cell RNA-sequencing approaches.13,14 Nonetheless, some applications such as building cell-type and cell-state atlases can benefit from analyzing a large number of genes and single cells even if only a few copies are sampled from most messenger RNAs.15 Other applications, such as building biophysical models, demand accurate quantification and require sampling a larger number of copies per gene.4,6,16 MS approaches for single-cell analysis have already demonstrated the ability to sample 20-fold more copies per gene compared to established single-cell RNA-seq methods.17
Sampling many copies per gene is challenging and often comes at the cost of decreased throughput for both single-cell RNA-seq14,15 and MS analysis.4,17 Thus, experimental designs should take into account the costs and benefits of each analytical strategy and choose whether to emphasize throughput or quantitative accuracy. This optimization has already been discussed,4,12,17 but the trade-offs of varying the number of cells in the isobaric carriers remain incompletely characterized. Here, we sought to empirically characterize these trade-offs and use the data to provide recommendations for experimental designs.
MATERIALS AND METHODS
Sample Preparation
All analyses used samples corresponding to 100× standards prepared in bulk and diluted 100-fold to model the single-cell SCoPE2 set. These standards were prepared as previously described.17 Specifically, U937 and Jurkat cells were collected from exponentially growing cultures, washed twice with cold phosphate-buffered saline, and counted under a hemocytometer to estimate cell density. The cells were then lysed in high-performance liquid chromatography (LC)-grade water according to the mPOP protocol:18 a 15 min freeze step at −80 °C, followed by a 10 min heating step at 90 °C. Following lysis, samples were digested at 37 °C for 3 h with 10 ng/μL of Promega Trypsin Gold in 100 mM triethylammonium bicarbonate buffer (TEAB). The bulk digested material was then serially diluted and labeled to generate the 16-plex design schemes shown in Table 1. These standards were prepared as 100× bulk samples with a total volume of 100 μL, and 1% (1 μL) of each bulk sample was then injected for MS analysis to simulate a single SCoPE2 experiment with two carrier channels of the indicated cell number and six single-cell channels, Table 1. No biological replicates were prepared of these bulk samples.
Table 1.
Standards with Variable Amounts of Isobaric Carriersa
| carrier size | 126 | 127N | 127C | 128N | 128C | 129N | 129C | 130N | 130C | 131N | 131C | 132N | 132C | 133N | 133C | 134N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100-cell | 50J | 50U | 1J | 1U | 1J | 1U | 1J | 1U | 1J | 1U | ||||||
| 200-cell | 100J | 100U | 1J | 1U | 1J | 1U | 1J | 1U | 1J | 1U | ||||||
| 300-cell | 150J | 150U | 1J | 1U | 1J | 1U | 1J | 1U | 1J | 1U | ||||||
| 400-cell | 200J | 200U | 1J | 1U | 1J | 1U | 1J | 1U | 1J | 1U | ||||||
| 600-cell | 300J | 300U | 1J | 1U | 1J | 1U | 1J | 1U | 1J | 1U | ||||||
| 800-cell | 400J | 400U | 1J | 1U | 1J | 1U | 1J | 1U | 1J | 1U |
“We prepared a series of diluted standards approximating SCoPE2 sets with different amounts of cells in their isobaric carriers. U stands for U937 (a cell line of monocytes), and J stands for Jurkat cells (a cell line of T-cells). The number in front of the letter indicates the number of cell equivalents in each sample injected for MS analysis.
MS Analysis
The standards were analyzed with the MS methods used for SCoPE2 samples.17 Specifically, all samples were separated on IonOpticks Odyssey (ODY-25075C18A) analytical columns with a 20 cm length and a 75 μm inner diameter. It was run using a Dionex RSLC3000 nano LC. All samples were analyzed using a Thermo Scientific Q-Exactive mass spectrometer. RAW files were searched using MaxQuant 1.6.7.0.19 The human SwissProt FASTA database (20,375 entries, downloaded 8/22/2020) was used for searching data from U-937 and Jurkat cells. Trypsin was specified as the digest enzyme, and a maximum of two missed cleavages was allowed for peptides between 5 and 26 amino acids long. Methionine oxidation (+15.99491 Da) and protein N-terminal acetylation (+42.01056 Da) were specified as variable modifications. TMTpro (+304.207145) was specified as a fixed modification on lysine and peptide N-terminus. RAW files were also searched using SpectroMine 2.1.200828.47784.20 The human SwissProt FASTA database (20,375 entries, downloaded 8/22/2020) was used for searching data from U-937 and Jurkat cells. Trypsin was specified as the digest enzyme, and a maximum of two missed cleavages was allowed for peptides between 7 and 52 amino acids long. Methionine oxidation (+15.99491 Da) and protein N-terminal acetylation (+42.01056 Da) were specified as variable modifications. TMTpro (+304.207145) was specified as a fixed modification on lysine and peptide N-terminus. MaxQuant results from Cong et al. were downloaded from the publication’s Supporting Information.21
Data Availability
All data sets associated with this article have been deposited at massIVE with ID: MSV000086004 and at scope2.slavovlab.-net/mass-spec/Isobaric-carrier-optimization.
Data Analysis and Visualization
Data analysis followed a previously reported approach, Data-driven Optimization of MS (DO-MS), for evaluating and optimizing MS experiments.12 Specifically, Figures 2 and 4 were generated by plotting variables reported by MaxQuant. The Pearson correlation values displayed in Figure 5a were computed from the subset of peptides observed in all samples. For each sample, the correlation was computed between the vector of single-cell Jurkat/U937 reporter ion (RI) ratios (the mean taken over the three replicates of each cell type before computing each ratio) and the corresponding vector of ratios estimated from the isobaric carrier ratios (Jurkat/U937 RI ratios). Error bars were calculated by repeated sampling with replacement of a subset of the peptides. The analysis for Figure 5b was performed the same way but subsetting first for those peptides with a single-cell RI intensity >2000. To control for the shape of RI distributions in each sample, we sampled subsets as follows: the range of RI values >2000 was divided into 10 bins of a width of 0.24 on a log10-scale starting at log10(2000), and then, one peptide was sampled from each bin. The correlations were computed using the sampled subsets of peptides with RI intensities having similar distributions.
Figure 2.
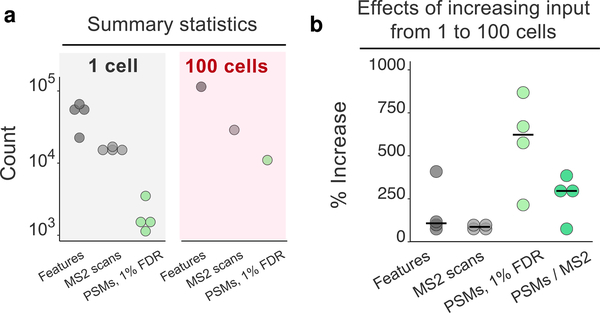
Increasing input from 1 to 100 cells primarily benefits the identification rate of MS2 spectra. Replicates of 1-cell and 100-cell HeLa samples were analyzed by label-free proteomics by Cong et al.21 (a) Number of peptide-like features (unique isotopic envelopes resolved with respect to m/z and retention time with a charge ≥+2), the number of MS2 spectra recorded, and the number of peptide-spectral matches (PSMs) determined by MaxQuant at a 1% false-discovery rate (FDR). (b) Percent increase in each metric from 1 to 100 cell inputs. The percent increase in PSMs and identification rate (PSMs/MS2: the number of PSMs at a 1% FDR divided by the number of MS2 scans obtained) is greater than the percent increase in peptide-like features or MS2 scans obtained. The black bars denote medians.
Figure 4.
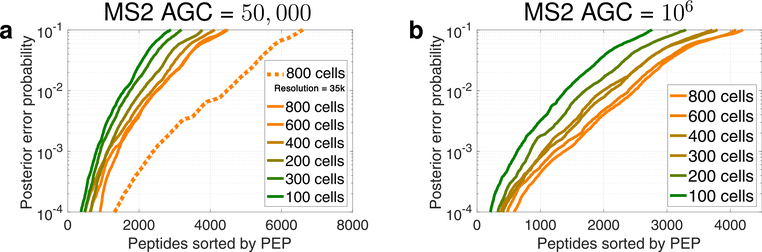
Number of identified peptides. Increasing the number of cells in the isobaric carrier results in a larger number of confidently identified peptides at any level of confidence as quantified by the posterior error probability (PEP). This trend is observed both with the low MS2 AGC target (50,000) shown in (a) and with the high MS2 AGC target (1,000,000) shown in (b). The MS2 scans of the run displayed with a dotted curve were performed at 35k resolution to demonstrate the potential advantage of low AGC for scanning and identifying more peptides. The MS2 scans of all other runs were performed at 70k resolution. The PEPs are estimated by MaxQuant using only the mass spectra23 and do not include additional features, such as retention time.3 These plots are standard components of the DO-MS reports,12 and the full reports are included in the Supporting Information and at scope2.slavovlab.net.
Figure 5.
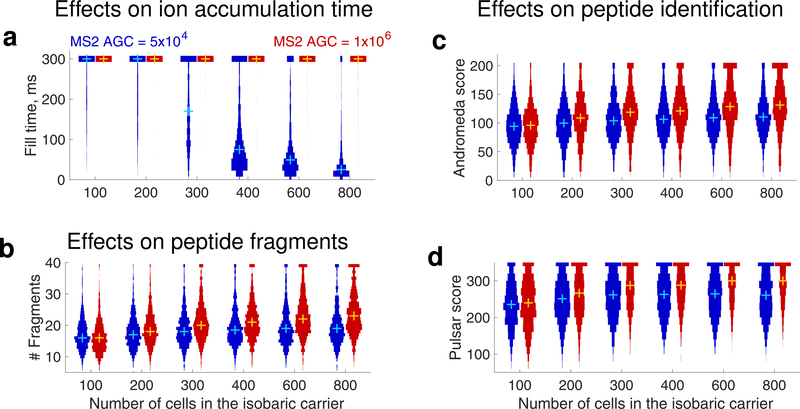
Effects of increasing the size of the isobaric carrier on peptide accumulation and sequence identification. (a) Distributions of MS2 accumulation times for all peptides identified across all displayed experiments with standards listed in Table 1. (b) Number of peptide fragments detected per PSM for all peptides that are identified in each experiment. For visualization purposes, peptides with more than 39 fragments were set to have 39 fragments. The confidence of sequence identification for peptides identified across all experiments is shown as distributions of scores computed either by Andromeda (c) or by Pulsar (d). These scores quantify the confidence of PSMs;20,25 For visualization purposes, Andromeda scores exceeding 200 were set to 200 and Pulsar scores exeeding 350 were set to 350. The distributions shown here can be generated by DO-MS (DO-MS is software freely available at do-ms.slavovlab.net) and can be used to evaluate the regime of analysis for any particular set of experiments.12 To enable controlled comparisons, the distributions show data only for the subset of peptides identified across all levels of isobaric carriers.12,24 The plus marks denote medians.
RESULTS AND DISCUSSION
Peptide Sequence Identification Is a Major Bottleneck
To understand the major benefits of using the isobaric carrier, we began our analysis by considering the major challenges and bottlenecks for ultrasensitive MS analysis without using the isobaric carrier. Specifically, we aimed to evaluate the effects of the isobaric carrier on three stages of MS/MS data acquisition and analysis shown in Figure 1: (1) detecting peptide-like features (precursor ions) during MS1 survey scans, (2) quantifying peptides from the small samples based on the RIs, and (3) identifying peptide sequences from the fragment ions.
We evaluated the efficiency of each of these three steps for 1-cell and 100-cell samples analyzed by Cong et al.21 The data shown in Figure 2a demonstrate that even for single cells, the mass spectrometer detected tens of thousands of peptide-like features and conducted MS2 scans on over 10,000 of these features. These numbers are comparable to the corresponding numbers for 100-cell samples and indicate that detecting precursor ions is not a limiting factor; indeed, the number of detected ions exceeds the number of MS2 scans that can be performed with the method setting.
Thus, while the isobaric carrier can enhance the detection of precursor ions (stage 1 in Figure 1), such an enhancement is unlikely to significantly increase the number of MS2 scans conducted because feature detection is not a limiting factor, as shown in Figure 2; what is limiting is the speed of the mass spectrometer and the time for performing MS2 scans of the large number of identified peptide-like features. If needed, the feature detection can be further enhanced by increasing the accumulation times for survey scans, as can be afforded by narrower isolation windows, for example, as implemented by BoxCar data acquisition.22
Despite the large number of MS2 scans, the rate of assigning confident peptide sequences is relatively low for the 1-cell samples and increases by over 250% for the 100-cell sample, Figure 2b. This increase is likely due to increased diversity and abundance of observed peptide fragment ions for the 100-cell sample. Thus, obtaining enough peptide fragments for confident sequence identification is a major bottleneck in the analysis of small samples, such as individual cells. The peptide fragments provided by the isobaric carrier (as illustrated in Figure 1) can help overcome this bottleneck.
Theoretical Expectations
While the isobaric carrier can bolster peptide sequence identification, it does not increase the number of ion copies sampled from the small-sample peptides. Therefore, the isobaric carrier may support confident peptide identification even when the peptide copies sampled from the small samples are insufficient to support reliable quantification.
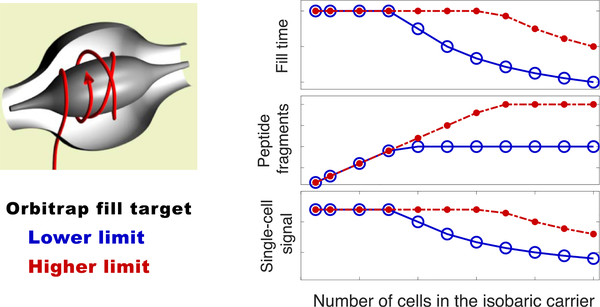
Before empirically examining the effects of increased amounts of isobaric carriers, we consider the theoretically expected effects as shown in Figure 3. Without an isobaric carrier, the rate of accumulating ions from the small samples is low and thus the ion accumulation is likely to use the maximum allowed accumulation time before reaching the operator-defined automatic gain control (AGC) target. As the amount of the isobaric carrier increases, the rate of ion accumulation increases as well, and accumulated ions begin to reach the AGC target before the maximum fill time. Thus, the fill time begins to decrease, resulting in decreased sampling of ions from the small samples. The shorter fill times can decrease the cycle times and thus increase the number of analyzed peptides, which can be sent for MS2 scans and reliably identified thanks to the fragment ions originating from the isobaric carrier and the small samples.
Figure 3.
Theoretically expected effects of increased isobaric carrier. Increasing the number of cells in the isobaric carrier increases the rate of accumulating peptide fragments for MS2 analysis. When the target AGC is reached, accumulation of ions stops, which may increase the speed of the analysis at the expense of decreased sampling of peptides from the small samples.
Increasing the AGC target increases the size of the isobaric carrier at which accumulation times (and ion sampling from the small samples) begin to decrease, Figure 3. Thus, higher AGC targets may increase the copy number of ions sampled from the small samples at the expense of more time needed for MS2 analysis of each peptide. This theoretical example illustrates a clear trade-off that we explore next empirically in controlled experiments.
Effects of Isobaric Carriers on Peptide Identification
To empirically benchmark the effects of the isobaric carrier, we created bulk standards that, when diluted 100-fold, model SCoPE2 standards with two carrier channels and six single-cell channels, as shown in Table 1. The sets have isobaric carriers with sizes ranging from 100-cell to 800-cell equivalents, Table 1. Standards with larger carriers result in identifying more peptides (four), including additional less abundant peptides not identified in the standards with a smaller carrier. The larger carriers might support much higher peptide identification rates with different parameters (faster ion accumulation and MS2 scans), but we kept these parameters constant across all experiments to allow for controlled comparisons. Thus, the 256 ms MS2 transient times required for 70k resolution do not allow taking advantage of the short accumulation times for large-size carriers and low AGC targets, about 40 ms for the 800-cell carrier, as shown in Figure 5a; see DO-MS reports. To demonstrate the potential for identifying more peptides, we reduced the MS2 transient times (by reducing the resolution to 35,000), which increased the number of MS2 scans and peptide identifications at the low AGC target, as shown by the dotted curve in Figure 4a.
If this compositional difference is not taken into account, the distributions of peptide quantities cannot be meaningfully compared; comparisons between distributions containing different subsets of peptides may lead to substantial biases. This phenomenon is well known as missing not at random or nonignorable missingness24 and can cause erroneous interpretations of proteomics data.12 To avoid such problems, we controlled for different peptide compositions by focusing the analysis only on the subset of peptides identified across all samples.
As theoretically expected for Figure 3, increasing the carrier amount results in reaching the low AGC target before the maximum fill times, Figure 5a: for carrier channels exceeding 300 cells, we observe that the low AGC target may be reached before the maximum fill time. However, the high AGC target is not reached by most peptides within 300 ms even with an 800-cell isobaric carrier, Figure 5a.
As the amount of isobaric carrier increases, some peptides begin to reach the low AGC target (50,000) at about the 300-cell carrier. For larger carriers, the accumulated peptide fragments and the confidence of peptide identification remain constant, Figure 5b–d, while the small-sample signal decreases proportionately to the decreased fill times, Figure 6a. The high AGC target results in different trends: the accumulated peptide fragments and the confidence of peptide identification increase with the carrier size, Figure 5b–d, while the small-sample signal remains constant, Figure 6a. This effect of an increased MS2 AGC target is consistent with previous observations,26 and previously used analytical parameters for SCoPE-MS and SCoPE2 analysis have corresponded to the high AGC target regime.10,17 In this regime, the isobaric carrier does not limit MS2 accumulation times and ions are accumulated for the maximum time allowed, as shown in Figure 5a.
Figure 6.
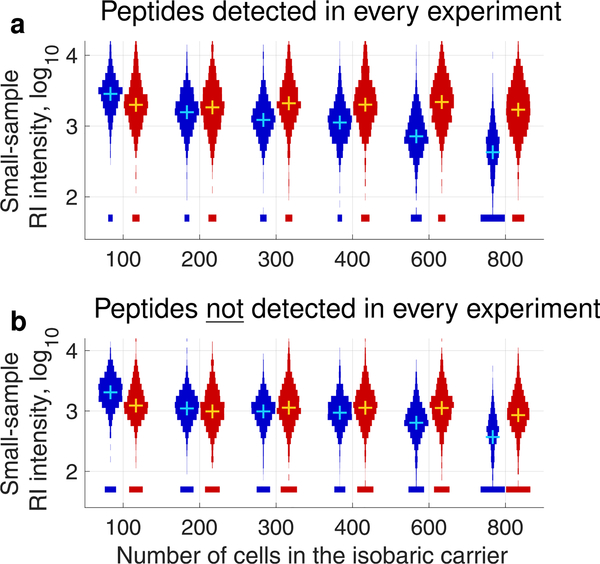
Effects of increasing the size of the isobaric carrier on the RI intensities in the small samples. Both (a) and (b) show distributions of RI intensities from the small samples of the standards listed in Table 1. As shown in Figure 5, the blue distributions correspond to MS2 AGC = 50,000, and the red distributions correspond to MS2 AGC = 1,000,000. (a) Only the RI intensities for peptides identified across all experiments are shown to allow for a well-controlled comparison.12,24 (b) Only the RI intensities for peptides not identified across all experiments are shown to evaluate whether some of these peptides have a high enough RI intensity to be quantifiable. The means and medians of these distributions cannot be meaningfully compared because of nonignorable missing data.24
In addition to comparing the distributions of small-sample RI intensities for the peptides detected across all carrier levels (Figure 6a), we also compared the small-sample RI intensities for the peptides detected only in some samples, Figure 6b. While this comparison is not well controlled, it allows to evaluate whether the additional peptides detected with larger isobaric carriers are sampled with sufficient copy numbers to allow quantification in the small samples. These additional peptides are likely less abundant and sampled with fewer copies. Indeed, the distributions of small-sample RI intensities tend to decrease with the increase of isobaric carrier for both MS2 AGC targets, Figure 6b. However, many of these additional peptides have high enough RI intensities to have the potential to support quantification, especially with the higher AGC MS2 target, Figure 6b.
Effects of Isobaric Carriers on Peptide Quantification
The number of ion copies sampled per peptide is an important determinant of quantification accuracy.4,17 Thus, the decreased ion copy number sampling at the low MS2 AGC target and the high carrier are likely to adversely affect quantification accuracy. However, the magnitude of this effect is unclear because other factors contribute significantly to the accuracy of quantification, such as the efficiency of sample preparation and the unintentional coisolation of multiple precursor ions for MS2 analysis. Indeed, the same ion sampling efficiency results in more accurate protein quantification in standards diluted to single-cell levels than in single-cell SCoPE2 sets.17 This fact has underscored the importance of sample preparation recently reviewed by Kelly7 and demands the use of direct benchmarks for quantification accuracy.
To benchmark relative quantification in the single-cell channels, we used the quantification derived from the two carrier channels present in each set, Table 1: we correlated the peptide fold changes estimated in the small samples to the corresponding peptide fold changes estimated from the isobaric carrier channels, Figure 7a. The results indicate that for the high AGC target (106), the correlations remain relatively constant as the size of the carrier increases. In stark contrast, the correlations decline for the low AGC target, Figure 7a, in parallel to the decline of RI intensities shown in Figure 6a. Similarly, we correlated the peptide fold changes estimated in the small samples to each other in Figure S1. The same trend is observed as when correlating to fold changes estimated from the isobaric carrier channel. These results indicate that sampling fewer copies from single cells is not simply a function of a large isobaric carrier and hardware limitations; rather, it is a reflection of analytical parameters that favor the speed of analysis (e.g., lower AGC) and the number of identified peptides over sampling many ion copies. While this regime results in less quantitative data, the increased throughput might offer worthwhile advantages for some applications, such as building single-cell atlases.15
Figure 7.
Effects of increasing the size of the isobaric carrier for two target levels of peptide quantification. (a) Correlations between the protein fold changes (between monocytes and T-cells) estimated from the small samples and the carrier samples. All peptides quantified across all samples at a 1% FDR are used for this analysis. For the lower AGC series, there were 966 unique peptides and 382 unique proteins quantified in every experiment. For the higher AGC series, there were 982 unique peptides and 349 unique proteins quantified in every experiment. (b) Correlations between fold changes as in (a) but only for peptides whose RI intensities in the small samples are larger than 2000. In both (a) and (b), error bars represent the 90% confidence intervals computed from resampling subsets of the data. For the lower AGC series, there were 725 unique peptides and 298 unique proteins quantified in every experiment. For the higher AGC series, there were unique 657 peptides and 240 unique proteins quantified in every experiment.
To further test the interpretation that the quantitative accuracy is lower because of decreased sampling of ion copies, we evaluated the quantitative accuracy for the subset of peptides having RI intensities above 2000. The high correlations between fold change vectors (ρ > 0.9) indicate good accuracy for all carrier sizes and both AGC targets, Figure 7b. These results affirm that in our experiments, quantitative accuracy depends on sampling enough ion copies from the small samples. Furthermore, the results emphasize that the sampling efficiency and quantitative accuracy are peptide- and protein-specific; a single experiment contains both well-quantified and poorly quantified proteins. Therefore, we see a great benefit in estimating the reliability of quantification for each protein and using this reliability for further quantitative analysis of the data, for example, for correcting estimates of the fraction of explained variance.27
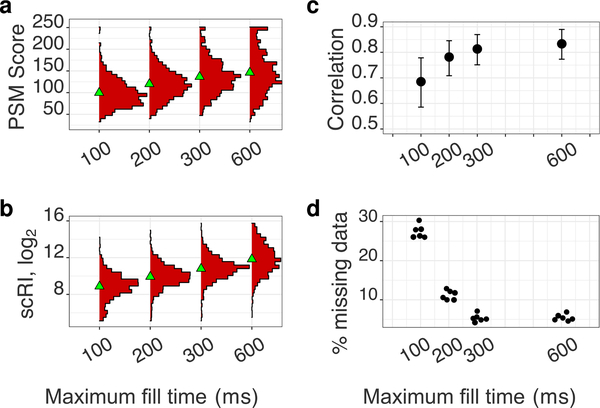
The data presented so far illustrate the effects of parameters that indirectly alter the MS2 accumulation time. To further demonstrate these effects, we directly limited MS2 accumulation times to 100, 200, 300, and 600 ms. The results demonstrate that longer accumulation times result in higher confidence of peptide identification (Figure 8a), more sample ions from the small samples (Figure 8b), improved quantification (Figure 8c), and decreased missing data (Figure 8d). Importantly, the trends in Figure 8c,d demonstrate that the improvements in the quantification of the small samples (lysates diluted to the single-cell level) saturate at about 300 ms. This corresponds to the default accumulation time used in this and previous SCoPE2 analysis.17 Different parameters (and less abundant proteins) may benefit from longer accumulation times, and we recommend using this analysis (available via DO-MS12) to establish optimal accumulation times.
Figure 8.
Effects of increasing the maximum MS2 fill time from 100 to 600 ms for a standard with a 100-cell isobaric carrier. All peptides quantified across all samples at a 1% FDR are used for this analysis. (a) The confidence of sequence identification for peptides is shown as distributions of scores computed by Andromeda.25 (b) Distributions of RI intensities from the small samples (scRI) of a standard with a 100-cell isobaric carrier across all maximum fill times. (c) Correlations between the protein fold changes (between monocytes and HEK-293) estimated from the small samples and the isobaric carrier samples. (d) Fraction of missing RI intensities from the small samples as a function of maximum fill time.
A fundamental challenge to analyzing very small samples, such as individual mammalian cells, is sampling sufficient copies from each molecule to support its reliable quantification. While single-cell RNA-seq methods have generated much useful data without overcoming this challenge,14,15 some applications require accurate quantification that can be achieved only by sampling enough ion copies.
The data in Figure 6 indicate that the sampling challenge is not created by the isobaric carrier approach but may be exacerbated by it in two ways. First, the isobaric carrier approach enables identifying the sequence of peptides that may be insufficiently sampled to be reliably quantified in the small samples. Second, poor experimental design (such as an insufficient ion accumulation time or a very large carrier amount and a low limit on total ion accumulation) may reduce the copy number of sampled ions and thus undermine quantification. The data presented here indicate that both pitfalls can be overcome by estimating the sampling efficiency for each peptide and then using for further quantification peptides with enough sampled copies to support reliable quantification, Figure 7b. We suggest that experimental designs optimize the isobaric-carrier amount and the ion accumulation times to reflect the relative priorities of throughput (the number of proteins and cells analyzed per unit time) and quantification accuracy (the copy number of ions sampled). The results should emphasize sampling efficiency and reliability estimates for each quantified protein rather than merely the number of identified peptides and proteins.
The data presented here illustrate a fundamental trade-off between throughput (the number of cells and proteins analyzed) and the number of copies sampled per peptide from the small samples. The cornerstone of this trade-off is the fact that the isobaric carrier does not amplify (boost) the RI intensities of peptides from the small samples (e.g., single cells). Therefore, delivering a sufficient number of ion copies from small samples is essential for accurate quantification, as previously suggested4,11,17 and demonstrated here in Figure 7. Thus, two modalities of ultrasensitive MS analysis emerge: analysis aiming to maximize quantitative accuracy must increase the delivery of analytes to the MS detector;11 analysis aiming to maximize throughput must reduce the time spent per analyte. To balance these competing priorities, we offer the following guidelines for LC–MS/MS experiments employing the isobaric carrier concept.
Guidelines
The size of the isobaric carrier should reflect the project priorities. The data presented here reinforce previous suggestions that isobaric carriers that are about 200-fold larger than the small samples provide most of the needed increase in peptide fragments to enhance sequence identification without adverse effects on quantification.9,17 Larger carrier sizes can further benefit peptide sequence identification (Figures 4 and 5) but will result in identifying additional less abundant peptides that are sampled with fewer copy numbers in the small samples (Figure 6). If more than one carrier is used, optimization depends on the sum of all cells in all carriers.
Evaluate whether the isobaric carrier affects peptide sampling. We recommend estimating whether the carrier levels and the AGC target result in reduced accumulation times and sampling of proteins from the small sample. This can be visualized by plotting the distributions of accumulation times and RI intensities as illustrated in Figures 5 and 6. These distributions can be automatically generated by DO-MS.12
Estimate the reliability of quantification for each protein. Estimate the sampling error on a per-protein basis as previously demonstrated,17 benchmark protein quantification as shown in Figure 7, or estimate the reliability of quantification based on the consistency of the quantification of peptides originating from the same protein as demonstrated by Franks et al.27 If the reliability of quantification is limited by counting noise, improved sampling can increase reliability. However, if the reliability is limited by other factors, such as sample preparation, improved copy-number sampling may have limited benefits.
Incorporate estimates of reliability in all subsequent analyses. Data points should be weighted based on their reliability, with weights proportional to the reliability. Use error propagation methods to reflect the noise in the final results. For example, correlations between noisy variables can be divided by the corresponding reliability to estimate the fraction of explained variance independent from the noise.27
Coisolation in the Context of the Isobaric Carrier
As with any approach using tandem mass tags, quantification of samples employing isobaric carriers can be severely undermined by coisolating ions. Therefore, methods employing isobaric carriers should minimize coisolation by using narrow isolation windows to sample ions for MS2 scans, by aiming to sample them when the abundance of the ions is the highest (the apex of its elution peak), and by employing high-performance chromatography that affords sharp elution peaks with minimal overlaps. These approaches have significantly decreased coisolation and improved quantification in SCoPE2 analysis when compared to SCoPE-MS.17
The degree of coisolation can vary across a set of isobarically labeled samples analyzed by the same LC–MS/MS run. This variation is due to the fact that the coisolating analytes can vary in abundance across different samples. The degree of this variation depends on the samples analyzed together and is larger for samples that differ more, such as an isobaric carrier sample.
CONCLUSIONS
Our data and analysis suggest that a principal benefit of the isobaric carrier is enhanced peptide sequence identification. Increasing the amount of isobaric carriers may allow faster peptide analysis and identification rate, but the associated decrease in accumulation times decreases the copy numbers of ions sampled from the small samples. Thus, the amounts of the isobaric carrier must reflect the balance of peptide sampling and the depth of quantification that are best suited for the analysis. Then, the quantification errors should be estimated and reflected in any subsequent analysis.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Gray Huffman, Edward Emmott, and Prof. Barry Karger for constructive comments. This work was funded by a New Innovator Award from the NIGMS from the National Institutes of Health to N.S. under award number DP2GM123497, an Allen Distinguished Investigator award through the Paul G. Allen Frontiers Group to N.S., a CZI Seed Networks grant from the Chan Zuckerberg Initiative to N.S., and a Merck Exploratory Science Center Fellowship, Merck Sharpe & Dohme Corp., to N.S..
Footnotes
The authors declare no competing financial interest.
Data analysis code: scope2.slavovlab.net.
GitHub repository: github.com/SlavovLab/isobaric-carrier.
Supplemental website: scope2.slavovlab.net/mass-spec/Isobaric-carrier-optimization.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00675.
Correlations between protein fold changes estimated from the single-cell channels of standards with different numbers of cells in the isobaric carrier (PDF)
DO-MS reports for all experiments (ZIP)
Contributor Information
Harrison Specht, Department of Bioengineering and Barnett Institute, Northeastern University, Boston, Massachusetts 02115, United States.
Nikolai Slavov, Department of Bioengineering, Barnett Institute, and Department of Biology, Northeastern University, Boston, Massachusetts 02115, United States.
REFERENCES
- (1).Cravatt BF; Simon GM; Yates JR III The biological impact of mass-spectrometry-based proteomics. Nature 2007, 450, 991. [DOI] [PubMed] [Google Scholar]
- (2).Zhang Y; Fonslow BR; Shan B; Baek M-C; Yates JR III Protein analysis by shotgun/bottom-up proteomics. Chem. Rev 2013, 113, 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chen AT; Franks A; Slavov N DART-ID increases single-cell proteome coverage. PLoS Comput. Biol 2019, 15, No. e1007082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Specht H; Slavov N Transformative opportunities for single-cell proteomics. J. Proteome Res 2018, 17, 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Marx V A dream of single-cell proteomics. Nat. Methods 2019, 16, 809–812. [DOI] [PubMed] [Google Scholar]
- (6).Slavov N Unpicking the proteome in single cells. Science 2020, 367, 512–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kelly RT Single-Cell Proteomics: Progress and Prospects. Mol. Cell. Proteomics 2020, 19, 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Orsburn B The Single Cell Proteomics Revolution; Bioanalysis Zone, 2020. [Google Scholar]
- (9).Budnik B; Levy E; Harmange G; Slavov N Massspectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. bioRxiv 2018, 102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Budnik B; Levy E; Harmange G; Slavov N SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018, 19, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Slavov N Single-cell protein analysis by mass spectrometry. Curr. Opin. Chem. Biol 2021, 60, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Huffman RG; Chen A; Specht H; Slavov N DO-MS: Data-Driven Optimization of Mass Spectrometry Methods. J. Proteome Res 2019, 18, 2493–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Klein AM; Mazutis L; Akartuna I; Tallapragada N; Veres A; Li V; Peshkin L; Weitz DA; Kirschner MW Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ziegenhain C; Vieth B; Parekh S; Reinius B; Guillaumet-Adkins A; Smets M; Leonhardt H; Heyn H; Hellmann I; Enard W Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 2017, 65, 631–643. [DOI] [PubMed] [Google Scholar]
- (15).Mereu E; et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol 2020, 38, 747–755. [DOI] [PubMed] [Google Scholar]
- (16).Munsky B; Neuert G; van Oudenaarden A Using gene expression noise to understand gene regulation. Science 2012, 336, 183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Specht H; Emmott E; Petelski AA; Gray Huffman R; Perlman DH; Serra M; Kharchenko P; Koller A; Slavov N Single-cell mass-spectrometry quantifies the emergence of macrophage heterogeneity. bioRxiv 2019, 665307. [Google Scholar]
- (18).Specht H; Harmange G; Perlman DH; Emmott E; Niziolek Z; Budnik B; Slavov N Automated sample preparation for high-throughput single-cell proteomics. bioRxiv 2018, 399774. [Google Scholar]
- (19).Sinitcyn P; Tiwary S; Rudolph J; Gutenbrunner P; Wichmann C; Yılmaz Ş; Hamzeiy H; Salinas F; Cox J MaxQuant goes Linux. Nat. Methods 2018, 15, 401. [DOI] [PubMed] [Google Scholar]
- (20).Muntel J; Gandhi T; Verbeke L; Bernhardt OM; Treiber T; Bruderer R; Reiter L Surpassing 10,000 identified and quantified proteins in a single run by optimizing current LC-MS instrumentation and data analysis strategy. Mol. Omics 2019, 15, 348–360. [DOI] [PubMed] [Google Scholar]
- (21).Cong Y; Liang Y; Motamedchaboki K; Huguet R; Truong T; Zhao R; Shen Y; Lopez-Ferrer D; Zhu Y; Kelly RT Improved Single-Cell Proteome Coverage Using Narrow-Bore Packed NanoLC Columns and Ultrasensitive Mass Spectrometry. Anal. Chem 2020, 92, 2665–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Meier F; Geyer PE; Virreira Winter S; Cox J; Mann M BoxCar acquisition method enables single-shot proteomics at a depth of 10,000 proteins in 100 minutes. Nat. Methods 2018, 15, 440–448. [DOI] [PubMed] [Google Scholar]
- (23).Cox J; Mann M MaxQuant enables high peptide identification rates, individualized ppb range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 2008, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- (24).Little RJ; Rubin DB Statistical Analysis with Missing Data; John Wiley & Sons, 2019; Vol. 793. [Google Scholar]
- (25).Cox J; Neuhauser N; Michalski A; Scheltema RA; Olsen JV; Mann M Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res 2011, 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- (26).Tsai C-F; Zhao R; Williams SM; Moore RJ; Schultz K; Chrisler WB; Pasa-Tolic L; Rodland KD; Smith RD; Shi T; et al. An improved Boosting to Amplify Signal with Isobaric Labeling (iBASIL) strategy for precise quantitative single-cell proteomics. Mol. Cell. Proteomics 2020, 19, 828–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Franks A; Airoldi E; Slavov N Post-transcriptional regulation across human tissues. PLoS Comput. Biol 2017, 13, No. e1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets associated with this article have been deposited at massIVE with ID: MSV000086004 and at scope2.slavovlab.-net/mass-spec/Isobaric-carrier-optimization.