Abstract
Alzheimer’s disease (AD) is a chronic neurodegenerative disease characterized by intracellular formation of neurofibrillary tangles and extracellular deposition of β-amyloid protein (Aβ) in the extracellular matrix. The pathogenesis of AD has not yet been fully elucidated and little is known about global alterations in the brain proteome that are related to AD. To identify and quantify such AD-related changes in the brain, we employed a tandem mass tags (TMT) approach coupled to high-resolution mass spectrometry. We compared the proteomes of frontal cortex from AD patients with corresponding age-matched brain samples. LC-MS/MS analysis carried out on an Orbitrap Fusion Lumos Tribrid mass spectrometer led to identification of 8,066 proteins. Of these, 432 proteins were observed to be significantly altered (>1.5 fold) in their expression in AD brains. Proteins whose abundance was previously known to be altered in AD were identified including secreted phosphoprotein 1 (SPP1), somatostatin (SST), SPARC related modular calcium binding 1 (SMOC1), dual specificity phosphatase 26 (DUSP26) and neuronal pentraxin 2 (NPTX2). In addition, we identified several novel candidates whose association with AD has not been previously described. Of the novel molecules, we validated chromogranin A (CHGA), inner membrane mitochondrial protein (IMMT) and RAS like proto-oncogene A (RALA) in an additional set of 20 independent brain samples using targeted parallel reaction monitoring (PRM) mass spectrometry assays. The differentially expressed proteins discovered in our study, once validated in larger cohorts, should help discern the pathogenesis of AD.
Keywords: AD, TMT, Proteome, Neurodegeneration
Introduction
Alzheimer’s disease (AD) is a chronic neurodegenerative disorder that manifests typically with memory loss and progressively affects other domains resulting in functional impairment. Patients with AD generally progress through preclinical, mild cognitive impairment (MCI) and dementia stages (Albert et al. 2011). The major pathological hallmarks of the disease are accumulation of extracellular plaques and formation of intracellular neurofibrillary tangles, which lead to synaptic dysfunction and eventually neuronal death (Glenner & Wong 1984; Lee et al. 1991). Existing therapies do not prevent progression of the disease and a number of disease-modifying drugs have failed in recent trials. Development of novel therapies for treating AD is difficult in the light of our limited understanding of its pathogenesis. Thus, further elucidation of molecular mechanisms and cellular signaling pathways that are responsible for neuronal cell death is critical for discovering novel targets for developing treatment strategies for AD.
Several studies have employed animal models of AD to detect disease-related changes in the brain (Martin et al. 2008; Robinson et al. 2011; Yang et al. 2011; Shevchenko et al. 2012). One limitation of such animal studies is that they generally fail to recapitulate the pathology of Alzheimer’s disease in humans. Nevertheless, animal models represent different aspects of the disease which develop over time and can thus be studied experimentally (Sasaguri et al. 2017), which is not possible with human post mortem brain samples that reflect late disease. Identification of disease-related changes in the brain using proteomics analysis requires well-characterized samples. Baltimore Longitudinal Study of Aging (BLSA) is perhaps the most comprehensive and largest cohort used for research into various aspects of aging. The BLSA, supported by the National Institutes of Aging, has been in existence since 1958. The sample description and enrollment criteria of the BLSA have been described previously (Stone & Norris 1966).
Several transcriptome-based studies quantified the transcript from the postmortem human tissue (Psych et al. 2015; Kang et al. 2011). Transcriptome analysis of the distant region of the healthy and AD post-mortem tissues has also been reported (Twine et al. 2011). Despite remarkable finding in these studies, other studies have reported disagreement in predicting protein abundance based on transcripts particularly in the brain samples (Carlyle et al. 2017; Seyfried et al. 2017). A number of mass spectrometry-based proteomics studies have been used for identification of AD biomarkers (Diner et al. 2017; Edgar et al. 1999; Andreev et al. 2012; Hashimoto et al. 2012). Edgar et al. used hippocampal tissue and 2D electrophoresis for the proteome analysis of AD and controls (Edgar et al. 1999). Another study employed label-free quantitation of brain samples from AD and corresponding control aged brains (Andreev et al. 2012). Proteome analysis of hippocampal neurons isolated from postmortem brain using laser capture microdissection were also carried out in AD and controls (Hashimoto et al. 2012). Quantitative iTRAQ based proteome profiling were also carried out in hippocampus, parietal cortex and cerebellum (Manavalan et al. 2013). Chang et al. performed SWATH analysis on synaptosomes isolated from hippocampus specimens in the Alzheimer’s disease cases and subjects with normal (Chang et al. 2015). Seyfried et al. carried out proteomic analysis of various stages of AD from the cortex (Seyfried et al. 2017). However, with few exceptions, the majority of these studies identified a limited number of proteins. Thus, there remains a need of a more detailed analysis of the brain proteome for the understanding of the AD pathogenesis.
In this study, we employed an Orbitrap Fusion Lumos Tribrid mass spectrometer, which is a latest generation mass spectrometer, and coupled it with TMT-based multiplexed proteomic analysis to identify brain protein expression profile in the control and AD samples. Our study identified several known as well novel molecules involved in AD pathogenesis. A subset of the identified molecules was validated using parallel reaction monitoring (PRM) assays and ummunofluorescence.
Material and Methods
Brain samples
The brain tissues used for analysis were collected from the autopsy cohort of the Baltimore Longitudinal Study of Aging (BLSA) (Troncoso et al. 1998). Human postmortem tissues were acquired under Johns Hopkins proper Institutional Review Board (IRB) protocols with consent from family (NA_00034084; The Baltimore Longitudinal Study of Aging Autopsy Program: A Prospective Study of the Effects of Aging on Cognition and Brain Pathology). The study was not pre-registered. The relevant metadata including, disease status, neuropathological criteria, age, sex, post-mortem interval and APOE genotype are provided in Table 1. We used medial frontal gyrus samples for proteomic analysis for discovery (4 controls and 5 AD) and validation (8 controls and 12 AD) studies. The mean age was 88±6.8 yrs for controls and 87±5.7 yrs for AD. Postmortem neuropathological evaluation of amyloid plaque distribution carried out according to the Consortium to Establish a Registry for Alzheimer’s disease (CERAD) (Mirra et al. 1991). No randomization methods were used during allocation of subjects to different experimental groups in the study. We used 100 mg of brain tissues for the protein extraction and then a part of the homogenized material (100 ug) was used for further analysis. The tissue were homogenized in the presence of liquid nitrogen to a fine powder using a pulverizer and subsequently lysed in 4% SDS by sonication. After the tissue lysis the protein amount was estimated using BCA (Bicinchoninic Acid) protein assay.
Table 1.
A list of previously reported AD proteins identified in this study
| Sample | Control/AD | CERAD | BRAAK | AGE | SEX | ApoE |
|---|---|---|---|---|---|---|
| 1 | Control | 1 | 2 | 80 | M | 3/3 |
| 2 | Control | 0 | 4 | 99 | M | 3/3 |
| 3 | Control | 0 | 3 | 82 | M | 2/3 |
| 4 | Control | 0 | 3 | 95 | M | 2/3 |
| 5 | Control | 0 | 2 | 84 | M | 2/3 |
| 6 | Control | 0 | 4 | 86 | M | 2/3 |
| 7 | Control | 0 | 2 | 91 | M | 2/3 |
| 8 | Control | 0 | 3 | 86 | M | 2/3 |
| 9 | AD | 1 | 3 | 94 | M | 3/4 |
| 10 | AD | 3 | 4 | 83 | F | 3/3 |
| 11 | AD | 3 | 5 | 81 | M | 3/3 |
| 12 | AD | 3 | 6 | 82 | M | 2/3 |
| 13 | AD | 3 | 6 | 83 | F | 3/4 |
| 14 | AD | 3 | 6 | 94 | F | 2/3 |
| 15 | AD | 3 | 4 | 92 | M | 3/3 |
| 16 | AD | 3 | 5 | 88 | M | 3/3 |
| 17 | AD | 3 | 4 | 98 | M | 3/3 |
| 18 | AD | 3 | 6 | 86 | M | 4/4 |
| 19 | AD | 3 | 6 | 92 | M | 3/4 |
| 20 | AD | 3 | 6 | 88 | M | 3/3 |
In-solution trypsin digestion of proteins and TMT labelling
100 μg of protein from each brain sample was reduced and alkylated with 5 mM DTT for 20 min at 60°C and 10 mM iodoacetamide for 20 min at room temperature, respectively (Sathe et al. 2018a). The proteins were precipitated using ice-cold acetone in 1:5 protein to acetone ratio. The precipitated proteins were dissolved in the 100 μl of 2 M urea in 50 mM TEABC. Protein digestion was carried out using lys-C (1:100) (Wako, Catalog # 25–02543) for 4 h followed by trypsin (1:20) (Promega, Catalog # V511A) at 37°C for 12–16 h. Resulting peptides were cleaned using Sep-Pak C18 material (Waters, Catalog # WAT023501). For C18 clean-up, the peptides were acidified in 1% formic acid followed by centrifugation at 10,000 rpm for 10 minutes for removal of undigested proteins. For peptide clean-up, we used solvent A (0.1% formic acid) and solvent B (40% acetonitrile in 0.1% formic acid) as the mobile phase. The peptides were passed through the Sep-Pak column, washed with solvent A and eluted from the column using solvent B. The eluted peptides were dried using vacufuge and then dissolved in 100 mM of TEABC. Peptides from brain samples were labeled using TMT as per the manufacturer’s instruction (Catalog # 90110, Thermo Fisher Scientific). Peptides derived from control brain samples were labeled with TMT tags as 126, 127N, 128N and 128C and patient samples with TMT tag as 129N, 129C, 130N, 130C and 131. The labeling reaction was quenched using 5% hydroxylamine followed by pooling of samples.
Basic-pH RPLC fractionation
The labeled peptides were fractionated using basic pH reversed phase liquid chromatography into 96 fractions as described previously (Selvan et al. 2014). Briefly, the mixture of TMT labeled peptides was dissolved in 1 ml of bRPLC solvent A (7 mM TEABC, pH 8.5) and fractionated by bRPLC chromatography on a XBridge C18, 5 μm 250 × 4.6 mm column (Waters, Milford, MA) using an increasing gradient of bRPLC solvent B (7mM TEABC, pH 8.5, 90% acetonitrile) on an Agilent 1260 HPLC system. The temperature of the column was not controlled and the flow rate of the mobile phase was0.3 ml/min. The eluted peptide absorbance was measured at 280 nm. The total run time was 90 min and the collected volume was 27 ml. A total of 96 fractions were generated which were then concatenated into 24 fractions and vacuum dried.
LC-MS/MS analysis
These dried peptides were reconstituted in 20 μl of 0.1% formic acid for the each fraction and 9 μl injected in duplicate. These fractions were analyzed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, Bremen, Germany) connected with Easy-nLC 1200 nanoflow liquid chromatography system (Thermo Scientific, Bremen, Germany). Dried peptides for each fraction were reconstituted in 0.1% formic acid and loaded onto an enrichment column (100 μm × 2 cm, Nanoviper) at a flow rate of 3 μl/min. Peptides were resolved on an analytical column (75 μm × 50 cm, RSLC C18) at a flow rate of 300 nl/min using a gradient of 5–30 % solvent B (0.1 % formic acid in 95 % acetonitrile) for 80 min. The acquisition time was set to 90 min. The data were acquired in a data-dependent mode. The precursor MS scans (from m/z 350–1500) were acquired in the Orbitrap at a resolution of 120,000 (at 200 m/z). The parameters used for the MS1 was AGC target 2 × 105 and ion filling time set 50 ms. The most abundant ions with charge state ≥2 were isolated in 3 sec cycle and fragmented using HCD fragmentation with 38% normalized collision energy and detected at a mass resolution of 30,000 (at 200 m/z). The parameters for MS/MS was used as AGC target was set as 5 × 104 and ion filling time set 100 ms dynamic exclusion was set for 30 s with a 10-ppm mass window.
Data analysis
The MS/MS database searches were carried out using SEQUEST search algorithm through Proteome Discoverer platform (version 2.1, Thermo Scientific) against Human RefSeq protein database (Version 73, containing protein entries with common contaminants). The workflow included spectrum selector, SEQUEST search nodes, and percolator. Trypsin was specified as the protease and a maximum of two missed cleavages were allowed. The search parameters involved carbamidomethylation at cysteine, TMT 10-plex (+229.163) modification at N-termini of peptides and lysine were set as fixed modification while oxidation of methionine was set as a variable modification. MS and MS/MS mass tolerances were set to 10 ppm and 0.1 Da, respectively. False discovery rate of 1% was set at the PSM level as well as protein level. The mass spectrometry data generated from this study have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via PRIDE partner repository with the dataset identifier PXD010560.
Bioinformatics analysis
To identify the functional role of the altered proteins, we obtained molecular function and localization of the proteins from Human Protein Reference Database (HPRD) (Peri et al. 2003). The involvement of these proteins in biological processes was also obtained from HPRD. Pathway analysis was performed using Funrich (Version 3.1.3) (https://www.FunRich.org) (Pathan et al., 2015) (Kandasamy et al. 2010). Principal component analysis (PCA) of all quantified proteins in AD and control samples and Welch’s t-test statistic of all differentially regulated proteins were calculated using Perseus (Version 1.6) (Tyanova et al., 2016). The Perseus output was used for generating a volcano plot. The heat map was generated using Perseus.
PRM analysis
The peptides were analyzed on Q Exactive HF mass spectrometer (Thermo Scientific, Bremen, Germany) interfaced with RSLC nanoflow liquid chromatography system (Thermo Scientific, San Jose, CA). Peptide digests were reconstituted in 0.1% formic acid and loaded onto a trap column (100 μm × 2 cm, Nanoviper) at a flow rate of 3μl/min. PRM assay were developed for the targets similar to earlier study (Sathe et al. 2018b; Sathe et al. 2020). The AD and control samples were randomized using simple randomization without any blinding. For simple randomization of the samples, we used sample identifiers and order them randomly in MS Excel using the RAND function to generate an equal number of pseudo-random numbers. Sorting by the column of random numbers will produce a randomized running order for the experiment. For targeted analysis, 2 μg of brain peptides from each sample were resolved on an analytical column (75 μm × 50 cm, RSLC C18). A linear gradient of 4% to 35% buffer B over 75 min was used for the analysis. Peptides from selected target proteins were monitored using a targeted inclusion list. The parameters used for the PRM analysis were set as follows: Orbitrap resolution of 35,000, AGC target value of 5 × 105, injection time of 100 ms, isolation window of 2 m/z, HCD normalized collision energy of 27 and starting mass of m/z 110. PRM data were analyzed using the Skyline software package. Acquired raw files were loaded to Skyline. Peak integration was verified manually and the results were exported for further statistical analysis.
Immunofluorescence analysis of the altered molecules in brain
Antibodies against neuronal pentraxin 2 (NPTX2-Proteintech Cat# 10889–1-AP, RRID: AB_2153875), inner membrane mitochondrial protein (IMMT - LifeSpan Cat# LS-B248–100, RRID: AB_10965300), SPARC related modular calcium binding 1(SMOC1-LsBio - Cat# LS-B12376–200), dual specificity phosphatase 26 (DUSP26-LS-A106538–100) and somatostatin (LifeSpan Cat# LS-B7046–50, RRID: AB_11180164) was procured from the LsBio Seattle WA. Sections were deparaffinized in xylene and rehydrated in 100% and 95% ethanol. After washing with tap water, antigen retrieval was performed in 1x HistoVT One (pH7.0) (Nacalai: 06380–05) at 95°C. Then, all samples were blocked in PBS (Phosphate Buffered Saline) with 5% normal goat serum and 0.2% Triton x-100 for 1h at RT. Primary antibodies (Table 2) were incubated in blocking buffer for 16h at 4°C. On the following day, samples were washed in PBS for 5min × 3, then Alexa fluor secondary antibodies (abcam; selected according to color and host species of primary antibodies) and DAPI (1μg/mL final; Roche: 10 236 276 001) in PBS with 0.5% Tween 20 were applied and incubated for 1h at RT. Samples were washed in PBS once for 5min. For quenching lipofuscin autofluorescence, we used TrueBlackTM Lipofuscin Autofluorescence Quencher (Biotium: 23007) diluted 1/40 in 70% ethanol, applied to the samples and incubated for 50s at RT. To facilitate the TrueBlack reaction, samples were constantly swirled by hand during the incubation. Then, samples were washed in PBS for 5min × 3 and cover slipped using ProLongTM Gold Antifade reagent (Invitrogen: P36930). Samples were kept at 4°C at least 1 day before imaging. Immunofluorescent images were taken on a Zeiss LSM 700 confocal microscope in the Microscope Facility of the Johns Hopkins School of Medicine.
Table 2.
A summary of the antibodies used for immunofluroscence
| Antibody | Company | Catalog # | Host | Dilution | Target |
|---|---|---|---|---|---|
| SMOC1 | LSBIO | LS-B12376 | Rabbit | 1/100 | SPARC related modular calcium binding 1 |
| SST | LSBIO | LS-B7046 | Rabbit | 1/100 | Somatostatin |
| DUSP26 | Atlas Antibodies |
HPA018221 | Rabbit | 1/100 | Dual specificity protein phosphatase 26 |
| IMMT | Atlas Antibodies | HPA036164 | Rabbit | 1/100 | Inner Membrane Mitochondrial Protein |
| NPTX2 | Proteintech | NBP131254 | Rabbit | 1/100 | Neuronal Pentraxin 2 |
| SYP | Abcam | ab8049 | Mouse | 1/500 | Synaptophysin (presynaptic protein) |
| MTCO1 | Abcam | Ab14705 | Mouse | 1/100 | Mitochondrially Encoded Cytochrome C Oxidase I |
| ALDH1L1 | Millipore | MABN495 | Mouse | 1/1000 | Astrocyte |
| MAP2 | Abcam | ab92434 | Chicken | 1/1000 | Neuron |
Statistical analysis
TMT data analysis was performed with the Perseus software package and calculation of the p-value were generated. (Tyanova et al. 2016). Graph Pad Prism version 5.04 (GraphPad Software Inc., San Diego, CA, USA) was used for the statistical analysis of PRM data. The distribution type was analyzed using D’Agostino & Pearson omnibus normality test. For data sets with normal distribution pattern, students’t test was used to examine group comparisons in the validation studies described below. A p-value of 0.05 or less was considered significant.
Results
Demographics and clinical features of the subjects
For the discovery and PRM-based targeted proteomic analyses, we used 20 brain samples. The important characteristics of the samples are shown in Table 1, Supplementary table 1. These samples include 8 controls and 12 AD patients. There were no significant differences in the age between the control and AD samples. The median age of the individuals at the time of death used for this analysis was 83 years for AD patients and 82 years for controls..
Quantitative proteomic analysis of LC-MS/MS data
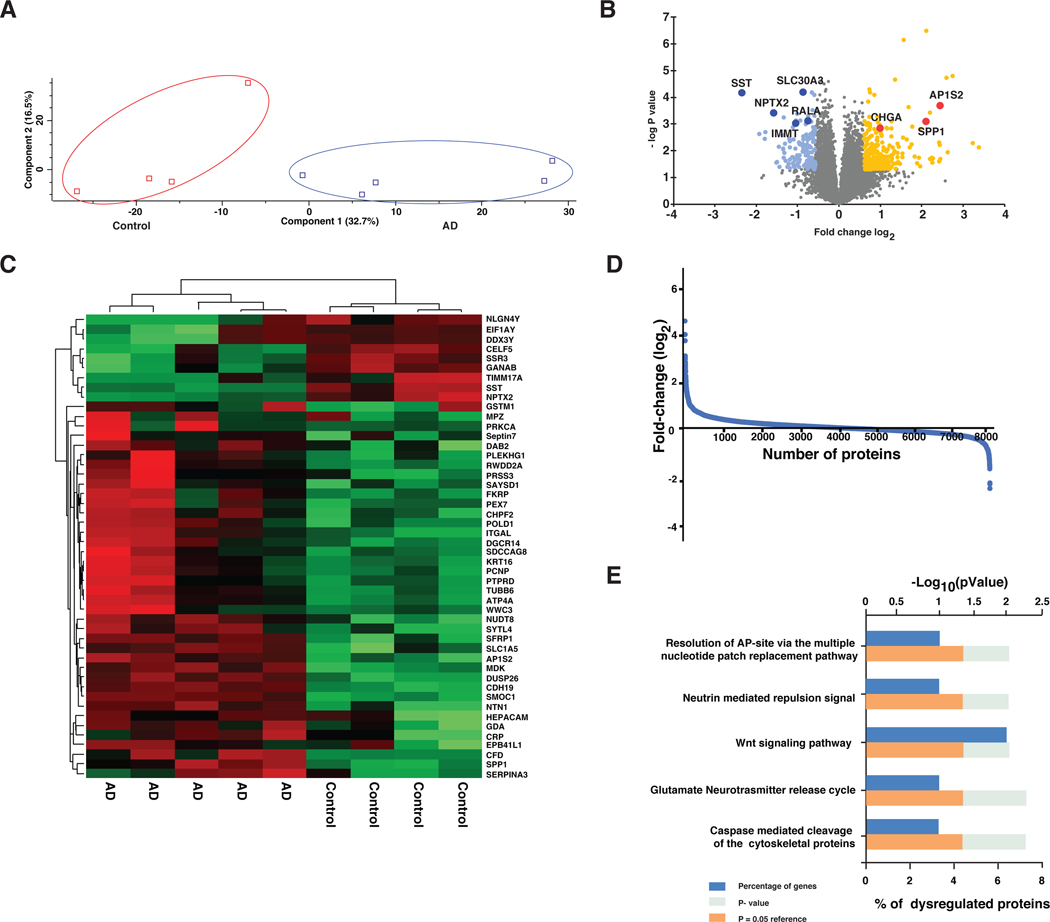
To identify proteomics changes in the brain of patients with Alzheimer’s disease, we carried out a differential proteomic analysis of medial frontal gyrus obtained from postmortem tissues of 5 AD patients and 4 cognitively normal individuals using a TMT-based quantitative proteomics approach. The workflow for the experiment is summarized in Figure 1. The fractions were analyzed by LC-MS/MS individually which led to 98,033 peptides from 8,066 proteins. Of these 8,066 proteins 7,073 proteins were detected with ≥2 unique peptides. A list of proteins identified in this study is provided in Supplementary table 2. The identified proteins were quantified based on the TMT ratios of peptides derived from these proteins. To determine whether the proteomic analysis could separate the two groups, a principal component analysis was carried out using Perseus. PCA analysis revealed a clear separation of the AD and control groups (Figure 2 A). The distribution of the fold change of the identified proteins were shown in the Figure 2 D. From the identified 8,066 proteins, 432 proteins were significantly altered proteins - 247 proteins were upregulated >1.5-fold and 131 proteins were downregulated <1.5-fold in AD brain tissues. A Volcano plot representing the fold change distribution of the differentially expressed proteins in AD brain s is provided in the Figure 2B. A heat map depicting selected proteins identified in this study has been provided in Figure 2C. The list of differentially expressed proteins in AD patients is provided in Supplementary table 3.
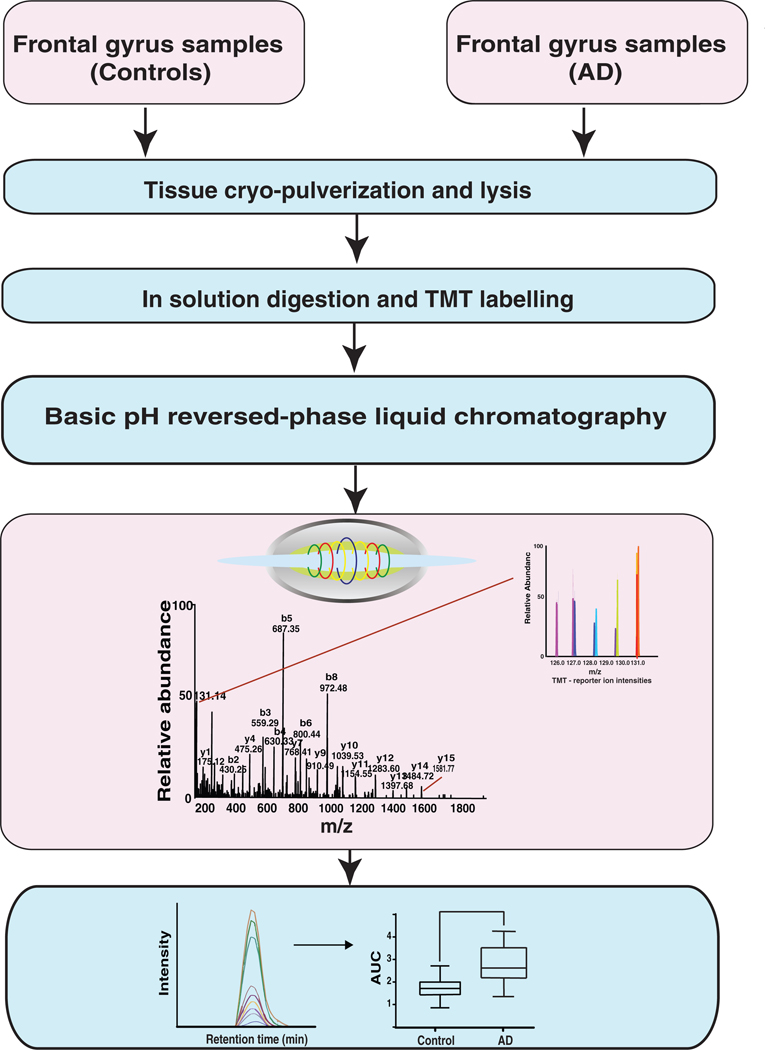
Figure 1.
A schematic of the workflow used to study the proteomic changes in the brain of AD patients. The brain samples were homogenized in liquid nitrogen and lysed in 2% SDS buffer. Brain proteomes from cognitively normal individuals were compared with that of AD patients using a TMT-based quantitative proteomics approach in the discovery step. Validation was carried out using parallel reaction monitoring (PRM) assays for a subset of molecules that were found to be altered in abundance in AD.
Figure 2.
Summary of TMT-based discovery experiments (A) PCA plot for the brain proteome data (B) Volcano plot for the proteome data for identification significantly altered proteins in AD brain (C) Heat map of the significantly altered proteins in AD brain (D) Distribution of fold-change values (log2 ratios) (AD/Controls) for proteins identified in the study (E) Enriched altered signaling pathways in AD brain
Bioinformatics analysis of altered proteins in AD brain
For a detailed biological understanding of altered proteins in AD brain, we carried out a bioinformatics analysis of the altered proteins. We classified the proteins based on their subcellular localization, biological processes and molecular functions. This annotation of the proteins is based on manually curated Human Protein Reference Database (Peri et al. 2003). The distribution of proteins was visualized based on their subcellular localization (Supplementary figure 1A), biological processes (Supplementary figure 1B) and molecular function (Supplementary figure 1C). In this analysis, we found that the majority of altered proteins were localized to the cytoplasm (17%) and extracellular space (15%) followed by plasma membrane (13%) and nucleus (12%). We observed that the majority of the altered proteins were involved in signal transduction (21%) followed by metabolism (18%) and cell growth and maintenance (10%). FunRich was used to carry out biological pathway analysis of proteins dysregulated in AD, which revealed enrichment of several pathways including glutamate neurotransmitter release cycle, Wnt and neutrin signaling pathways and caspase mediated cleavage of cytoskeletal proteins. A graphical representation of the altered pathways is shown in figure 2E.
Alterations of synaptic proteins in AD
In Alzheimer’s disease, there is a gradual decline in the cognition. Loss of synapses and the association of synaptic proteins with dementia has been described previously (DeKosky & Scheff 1990). The correlation between cognitive impairment and synaptic loss is better compared to hallmarks of AD - tau and amyloid-β pathologies - has also been described (Blennow et al. 1996). To identify the role of synaptic proteins in AD pathogenesis, we compared our proteomics dataset with a published study that investigated synaptic proteins in AD (Bereczki et al. 2018). Bereczki et al. detected 851 synaptic proteins in their data. Our study detected 735 synaptic proteins out of the 851 of which 45 proteins were significantly differentially expressed in the AD brains compared to controls. Twenty-four of these proteins were upregulated and 21 were downregulated in AD.
Targeted analysis of proteins
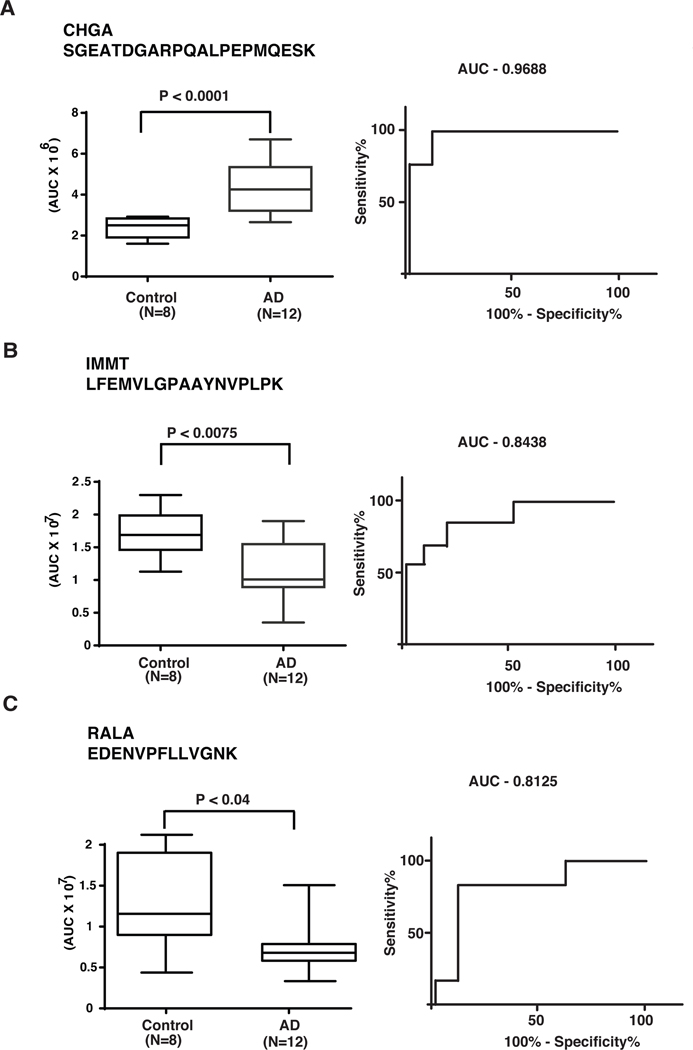
From our proteomics analysis, we identified a number of candidates with >1.5-fold alteration in AD brains. Of these proteins, we selected chromogranin A (CHGA), RAS like proto-oncogene A (RALA) and inner membrane mitochondrial protein (IMMT) for validation on additional control and AD brain samples using targeted parallel reaction monitoring assays. The PRM assays were carried out using an independent set of 8 controls and 12 AD brain samples. We monitored the peptides with retention time scheduling. PRM data on each sample were acquired in triplicates. We were able to confirm significant higher levels of CHGA Figure 4A and lower level of IMMT Figure 4B and RALA Figure 4C in brains of AD affected patients using PRM.
Figure 4.
PRM-based validation of candidate proteins by parallel reaction monitoring. The length of the box is thus the interquartile range of the sample. The other dimension of the box does not represent anything in particular. A line is drawn across the box at the sample median. Whiskers sprout from the two ends of the box until they reach the sample maximum and minimum. The crossbar at the far end of each whisker is optional and its length signifies nothing.: (A) Chromogranin A (CHGA); quantification shown in box plot with significant change with P value P < 0.0001 and ROC curve (B) Inner Membrane Mitochondrial Protein (IMMT); quantification shown in box plot with significant change with P value P < 0.0075 and ROC curve (C) Ras-related protein Ral-A (RalA); quantification shown in box plot with significant change with P value P < 0.04 and ROC curve
Immunofluorescence analysis of selected proteins
A subset of significantly altered proteins was used for the immunofluorescence analysis to detect its localization in cell type (neuron vs astrocyte) and cell localization (cytoplasm/nucleus/organelle). These includes secreted modular calcium-binding protein 1 (SMOC1), dual Specificity Phosphatase 26 (DUSP26), inner membrane mitochondrial protein (IMMT), somatostatin (SST) and neuronal pentraxin-2 (NPTX2). The immunofluorescence analysis were carried out on the control brain samples. We have observed SMOC1 signals scatter apart from nuclei (Supplementary figure 2A). Similarly we have also observed SST signal in the neuron and DUSP26 showing strong cytoplasmic signal in the neuron (Supplementary figure 2B and C). IMMT shows neuronal mitochondrial localization and NPTX2 signal is placed next to presynaptic marker (SY38) and absent in red signals, indicating majority of NPTX2 is in postsynapse.
Discussion
Proteomic profiling of AD brains
In this study, we employed an Orbitrap Fusion Lumos Tribrid mass spectrometer - a latest generation mass spectrometer - coupled with advanced sample preparation methods to increase the depth of the proteome. Isobaric labeling methods permit multiplexing and provide relative quantitation of proteins across samples without changing their analytical properties. In this study, we carried out a TMT-based quantitative proteomic analysis of the human brain tissues to investigate proteomic changes in Alzheimer’s individuals as compared to control individuals without any evidence of Alzheimer’s disease. From this analysis, we identified 8,066 proteins (7,073 proteins with ≥2 unique peptides). To evaluate the ability of the proteomic data to differentiate AD samples from controls, we used principal component analysis. A distinct separation of brains from Alzheimer disease and controls was observed, with PC1 32.7% and PC2 16.5% (Fig. 2A). To identify statistically significantly altered proteins in AD, we plotted p-values and fold-changes from AD and control samples using a volcano plot. This analysis led identification of 432 proteins that were differentially regulated (>1.5 fold-change) in patients with AD as compared to age-matched controls. The distribution of the fold-changes of the identified proteins suggests that the majority of proteins (95%) were unchanged in the AD brain compared to controls. To understand biological pathways involved in the AD pathogenesis, these 432 significantly altered proteins were used for enrichment analysis. Among the enriched networks, we identified several molecules involved in the neutrin mediated repulsion signal, glutamate neurotrasmitter release cycle and Wnt signaling pathway. The differentially expressed proteins identified in our analysis include a number of previously reported molecules in addition to novel proteins. Finally, multiplexed parallel reaction monitoring (PRM) assays were developed to quantitate a subset of these differentially altered proteins in the AD brain samples.
Known markers identified in the dataset
We carried out analysis of control and AD brain samples to identify differentially expressed proteins in AD brain. From this analysis, we identified 432 differentially expressed proteins. These differentially altered proteins included several known AD markers, such as SPP1, NPTX2, SST and SLC30A3 whose expression correlated with AD. This validates our approach of discovering altered proteins in AD brain. A partial list of proteins previously reported to be AD markers is provided in Table 3. Apart from known altered proteins, we also detected several new molecules which were not studied earlier in the AD context. A partial list of these novel proteins is provided in Table 4.
Table 3.
A list of previously reported AD proteins identified in this study
| Gene Symbol | Protein | Fold-change (This study) | P-Value | Number of samples used (N) |
|---|---|---|---|---|
| SPP1 | Secreted phosphoprotein 1 | 4.6 | 0.0008 | 9 |
| CHGA | Chromogranin A | 2.0 | 0.001 | 9 |
| SLC30A3 | Solute carrier family 30 member 3 | 0.5 | 0.00006 | 9 |
| NPTX2 | Neuronal pentraxin 2 | 0.3 | 0.0003 | 9 |
| SST | Somatostatin | 0.1 | 0.00007 | 9 |
Table 4.
A list of novel differentially expressed proteins in AD brains
| Gene Symbol | Protein | Fold-change (This study) | P-Value | Number of samples used (N) |
|---|---|---|---|---|
| ATP4A | ATPase H+/K+ transporting subunit | 13.8 | 0.005 | 9 |
| MDK | Midkine | 6.0 | 0.00001 | 9 |
| SMOC1 | SPARC related modular calcium binding 1 | 4.3 | 0.0000003 | 9 |
| IMMT | Inner membrane mitochondrial protein | 0.4 | 0.001 | 9 |
| RALA | RAS like proto-oncogene A | 0.5 | 0.0007 | 9 |
Osteopontin (SPP1) is marker of neuroinflammation. It is a cytokine expressed by cytotoxic T cells and it activates and recruit’s macrophages at the site. Its expression in pyramidal neurons is increased in AD patients (Wung et al. 2007). It is also observed to be upregulated in the AD transgenic mouse models (Wirths et al. 2010). High levels of osteopontin have been observed in the CSF of familial AD individuals carrying known genetic mutations (Ringman et al. 2012). Another study also reported increased level of osteopontin in the cerebrospinal fluid of patients with Alzheimer’s disease and its levels also correlate with a decline in cognition (Comi et al. 2010). It is observed that the OPN plays a major role in the Aβ clearance by prominent monocyte-macrophage recruitment into AD brains and by promoting macrophage polarization headed for an anti-inflammatory, highly phagocytic phenotype (Rentsendorj et al. 2018). Our study validates this upregulation of osteopontin in the brain of AD patients. We observed 4.6-fold upregulation of osteopontin in brains of AD patients as compared to controls. The MS/MS spectra along with quantitation were shown in figure 3A.
Figure 3.
Representative MS/MS spectra of peptides identified along with quantitation provided in the box plot: The length of the box is thus the interquartile range of the sample. The other dimension of the box does not represent anything in particular. A line is drawn across the box at the sample median. Whiskers sprout from the two ends of the box until they reach the sample maximum and minimum. The crossbar at the far end of each whisker is optional and its length signifies nothing. A) Osteopontin (SPP1) along with its reported ion quantification shown in box plot with significant change with P value P < 0.0001 (B) Somatostatin (SST) along with its reported ion quantification shown in box plot with significant change with P value P < 0.0013 (C) Chromogranin A (CHGA) along with its reported ion quantification shown in box plot with significant change with P value P < 0.0045; and, D) Inner Membrane Mitochondrial Protein (IMMT) along with its reported ion quantification shown in box plot with significant change with P value P < 0.0009.
Somatostatin (SST) is a neuropeptide and it has been reported that its concentration is lower in the CSF and brain of AD patients (Bissette & Myers 1992; Saiz-Sanchez et al. 2010). It is also observed that lowering concentration of somatostatin is proportional to the amyloid accumulation, which is central event in a cascade leading to AD. Insulin-degrading-enzyme (IDE) is one of the proteases known to degrade Aβ, and thus an exciting target for AD therapy. Somatostatin plays an important role in controlling IDE activity (Tundo et al. 2012). Similar to the role of IDE in degradation of Aβ, there is another mechanism by which somatostatin degrades Aβ. During aging, the expression of the neprilysin decreases and it is known that neprilysin reduces the accumulation of both soluble and fibrillar Aβ (Iwata et al. 2002). It is also known that somatostatin increases the activity of neprilysin and helps in degradation of Aβ. Overall, these findings clearly indicate that aging reduces somatostatin expression, which leads to accumulation of Aβ suggesting that somatostatin receptors can be potential pharmacological targets for the treatment of Alzheimer disease (Saito et al. 2005). There is a reported trend toward increased DNA methylation of the SST promoter with increasing age (Grosser et al. 2014). It was observed that somatostatin was significantly decreased in AD and its level also correlated with dementia scores (Strittmatter et al. 1997). We also observed that somatostatin was downregulated 10-fold in the brain of AD patients. We have also observed SST localized in the neuron. The MS/MS spectra along with quantitation were shown in figure 3B.
Solute carrier family 30 member 3 (SLC30A3) is involved in intracellular zinc homeostasis. Altered expression of SLC30A3 has been defined in AD patients. It is observed that SCLA30A expression is reduced with increasing age as well as in the AD patients (Olesen et al. 2016). Another study shows the role of zinc in depression in people with dementia and highlights zinc metabolism as a therapeutic target (Whitfield et al. 2015). The zinc transporter-3 knock-out mice leads to cognitive loss and show a phenocopy for the synaptic and memory deficits of Alzheimer’s disease (Adlard et al. 2010). In our analysis, we observed a 2- fold downregulation of SLC30A3 in AD brains. NPTX2 is a member of a family of neuronal pentraxins that include neuronal pentraxin 1 (NPTX1) and neuronal pentraxin receptor (NPTXR). NPTX2 neuronal pentraxin is a synaptic protein involved in excitatory synapse formation. NPTX2 clusters alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)-type glutamate receptors at the synapses, which results in non-apoptotic cell death of dopaminergic nerve cells (Chang et al. 2010). NPTX2 also enables excitatory synapse formation, learning and memory by clearance of extracellular debris to anchor α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) channels (Elbaz et al. 2015). NPTX2 levels in AD patients correlates with cognition. Its expression is also reduced in AD brains. CSF levels of NPTX2 are also found to be lower in AD patients as compared to controls (Xiao et al. 2017). Downregulation of CSF and brain NPTX2 directly correlates with cognitive status and hippocampal volume. GluA4 expression was reduced and correlated with NPTX2 expression supporting the idea that GluA4 expression is dependent on NPTX2 expression in AD. NPTX2 is required for homeostatic scaling of pyramidal neuron excitatory synapses on PV interneurons (Chang et al. 2010). The deletion of NPTX2 results in a selective reduction of pyramidal neuron drive of PV interneurons in developing mouse cortex (Gu et al. 2013). GluA4 is biomarker of PV-interneuron function that is dependent on NPTX2 expression. This shows failure of adaptive control of pyramidal neuron-PV circuits, which could be another pathophysiological mechanism contributing to cognitive failure in AD. Our proteomic analysis also corroborates the earlier published data. In our analysis, we observed 3.3-fold downregulation of NPTX2 in the AD brains.
We have also identified higher abundance of the SPARC related modular calcium binding 1 (SMOC1) in the frontal cortex of the AD brain compared to age matched controls. SMOC1 is also known for its amyloid plaque physiology. It co-localize with Aβ plaques in the brain of AD human and mouse model (Bai et al. 2020). We have also identified the higher abundance of the SMOC1 in the AD brain in our mass spectrometry data. Another protein dual specificity phosphatase 26 (DUSP26) is a tyrosine phosphatase and its increased expression in the hippocampus of AD patients is already reported. DUSP26 selectively regulate the production of Aβ42 in neuronal cells under hypoxic stress. Targeting DUSP26 can be used as therapeutic targets to halt AD progression (Jung et al. 2016). In our mass spectrometry analysis we identified 6.7 fold up-regulation of DUSP26 in the AD brain and it showed strong signal in the neurons.
Targeted analysis of a subset of proteins
Targeted proteomic analysis has become routine practice in biomarker research. These targeted mass spectrometry-based assays have several advantages including multiplexing, reproducibility, accuracy and that fact that they do not require specific antibodies (Henderson et al. 2017; Shi et al. 2016; Rifai et al. 2006). PRM assays have been successfully used for biomarker analysis for many diseases including lung adenocarcinoma, ovarian cancer and also Alzheimer’s disease. From our discovery analysis, we selected a subset of proteins that were altered in AD samples - including Chromogranin, MICOS complex subunit MIC60 and Ras-related protein Ral-A. Chromogranin is a secretory protein that belongs to the chromogranin/secretogranin family (Taupenot et al. 2003), which is a precursor for various bioactive peptides including vasostatin I and II, chromacin, pancreastatin, parastatin (Lawrence et al. 2011). In an earlier study, higher levels of chromogranin were found in AD brains (Weiler et al. 1990). Chromogranin activates microglia which release neurotoxins. These neurotoxins initiate apoptotic death of cortical neurons through apoptotic mediators (Ciesielski-Treska et al. 2001). In our study, we found 2-fold higher levels of chromogranin in AD brains as compared to the controls. Based on this, we validated these results on control and AD brain samples using parallel reaction monitoring. We again detected significantly higher levels (2-fold) of CHGA in AD patients. A unique peptide for CHGA - SGEATDGARPQALPEPMQESK - was monitored and confirmed at higher abundance in AD brain compared to controls with a significant P value p < 0.0002 (Figure 4A). The MS/MS spectrum for the same peptide is shown in Figure 3C.
MICOS complex subunit MIC60 is a part of MICOS complex. MIC60 is also known as inner membrane mitochondrial protein (IMMT). MICOS is a large protein complex of the inner mitochondrial membrane which play important roles in cristae junction and membrane architecture maintenance (Alkhaja et al. 2012; Ott et al. 2015). MICOS downregulation leads to a decrease in cell proliferation, increase in apoptosis and abnormal mitochondrial function (John et al. 2005). Mitochondrial dysfunction has been reported earlier in Alzheimer’s disease. Altered brain metabolism in AD reflects impaired mitochondrial function (Mosconi et al. 2010; Toledo et al. 2017). Another evidence of mitochondrial dysfunction in AD is the generation of reactive oxygen species (ROS) (Bonda et al. 2014; Wang et al. 2014). ROS is generated because of defects in the ETC resulting in DNA, lipid and protein damage. In our analysis, we observed a 2-fold down regulation of IMMT in AD brains as compared to the controls. We also validated these results using parallel reaction monitoring. The unique peptide (LFEMVLGPAAYNVPLPK) for IMMT was monitored and showed a lower abundance in AD compared to controls with a significant P value p < 0.0075 as shown in Figure 4B. The MS/MS spectrum for the same peptide is shown in figure 3D.
Ras-related protein Ral-A (RalA) is a protein encoded by the RALA gene located on chromosome 7 (Rousseau-Merck et al. 1988). RALA is multifunctional GTPase involved in multiple cellular processes including cell migration, proliferation, oncogenic transformation and membrane trafficking. It also acts as a GTP sensor and regulates integrin mediated exocytosis (Balasubramanian et al. 2010). Although the role of the RALA is not clear in AD pathology, association of other Small GTPases has been previously described in AD (Qu et al. 2019; Besga et al. 2017). In our discovery analysis, we detected a decreased abundance of RALA in the brain of AD patients. We also independently validated these results using additional samples by parallel reaction monitoring. We monitored the peptide - EDENVPFLLVGNK - by parallel reaction monitoring and observed significantly lower (P < 0.0008) abundance in AD brains as shown in Figure 4C.
Limitations of the study
The study has several limitations, chief among them being the small sample sizes for discovery and validation. This is unfortunately due to a lack of well curated postmortem brain samples and the overwhelming majority of similar studies, with few exceptions are limited by this issue (Ping et al. 2018; Portelius et al. 2017). Another limitation is that the BLSA is a unique cohort of highly educated participants (the mean education is approximately 17 years) so findings may not generalize to the population. This study also limited its sampling to the medial frontal gyrus. While tau pathology and neuronal degeneration begin in the entorhinal cortex and the hippocampus, amyloid deposition, which is known to precede tau pathology, starts in the default mode network comprising of the precuneus, medial orbitofrontal, and posterior cingulate cortices (Palmqvist et al. 2017). In a follow-up study, we plan to analyze biomarker changes in the medial temporal lobes. This study also compared changes between cognitively normal and subjects with AD, which could potentially bias discovery of alterations that are downstream of more proximate events. In the future, we plan to look for similar changes in asymptomatic and prodromal AD subjects. Despite these limitations, we have also identified known altered AD proteins in our study along with novel molecules.
Conclusions
We used well characterized AD and control brain samples from the Baltimore Longitudinal Study of Aging to detect differentially expressed proteins in brains of AD individuals. We detected 8,066 proteins out of which 432 show differential expression. We confirmed differential expression of CHGA, IMMT and RALA in an independent set of brain samples of AD and control samples using parallel reaction monitoring. The novel proteins identified in this study can be further studied to investigate their functional role in AD pathogenesis.
Supplementary Material
Acknowledgments
The authors thank members of the Pandey laboratory for valuable suggestions.
Funding
This study was supported, in part, by NIA grant U19AG03365 (MA), NINDS grant P50NS38377 (AP), an NIH shared instrumentation grant (S10OD021844). This work was supported by the DBT/Wellcome Trust India Alliance Margdarshi Fellowship (grant number IA/M/15/1/502023) awarded to AP and philanthropic support from the Thomas Lantry Foundation to AM
Abbreviations Used
- AD
Alzheimer’s disease
- AGC
automatic gain control
- BCA
Bicinchoninic Acid
- BLSA
Baltimore Longitudinal Study of Aging
- CERAD
Consortium to Establish a Registry for Alzheimer’s disease
- CSF
Cerebrospinal fluid
- DTT
dithiothreitol
- GO
gene ontology
- HCD
high energy collision dissociation
- HPRD
human protein reference database
- HPLC
high pressure liquid chromatography
- IAA
iodoacetamide
- IHC
immunohistochemistry
- LC
liquid chromatography
- MCI
mild cognitive impairment
- MS
mass spectrometry
- PPM
part per million
- PCA
Principal component analysis
- PRM
parallel reaction monitoring
- PTM
Post-translational modifications
- bRPLC
basic pH reversed phase chromatography
- SDS
sodium dodecyl sulfate
- TEABC
triethylammonium bicarbonate
- TMT
tandem mass tags
--Human subjects --
Involves human subjects:
If yes: Informed consent & ethics approval achieved:
=> if yes, please ensure that the info “Informed consent was achieved for all subjects, and the experiments were approved by the local ethics committee.” is included in the Methods.
ARRIVE guidelines have been followed:
Yes
=> if it is a Review or Editorial, skip complete sentence => if No, include a statement in the “Conflict of interest disclosure” section: “ARRIVE guidelines were not followed for the following reason:
“
(edit phrasing to form a complete sentence as necessary).
=> if Yes, insert in the “Conflict of interest disclosure” section:
“All experiments were conducted in compliance with the ARRIVE guidelines.” unless it is a Review or Editorial
Footnotes
Consent for publication
All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
References
- Adlard PA, Parncutt JM, Finkelstein DI and Bush AI (2010) Cognitive loss in zinc transporter-3 knock-out mice: a phenocopy for the synaptic and memory deficits of Alzheimer’s disease? J Neurosci 30, 1631–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert MS, DeKosky ST, Dickson D. et al. (2011) The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 7, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhaja AK, Jans DC, Nikolov M. et al. (2012) MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell 23, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev VP, Petyuk VA, Brewer HM et al. (2012) Label-free quantitative LC-MS proteomics of Alzheimer’s disease and normally aged human brains. J Proteome Res 11, 3053–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Wang X, Li Y. et al. (2020) Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer’s Disease Progression. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian N, Meier JA, Scott DW, Norambuena A, White MA and Schwartz MA (2010) RalA-exocyst complex regulates integrin-dependent membrane raft exocytosis and growth signaling. Curr Biol 20, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereczki E, Branca RM, Francis PT et al. (2018) Synaptic markers of cognitive decline in neurodegenerative diseases: a proteomic approach. Brain 141, 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besga A, Chyzhyk D, Gonzalez-Ortega I, Echeveste J, Grana-Lecuona M, Grana M and Gonzalez-Pinto A (2017) White Matter Tract Integrity in Alzheimer’s Disease vs. Late Onset Bipolar Disorder and Its Correlation with Systemic Inflammation and Oxidative Stress Biomarkers. Front Aging Neurosci 9, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissette G and Myers B (1992) Somatostatin in Alzheimer’s disease and depression. Life Sci 51, 1389–1410. [DOI] [PubMed] [Google Scholar]
- Blennow K, Bogdanovic N, Alafuzoff I, Ekman R and Davidsson P (1996) Synaptic pathology in Alzheimer’s disease: relation to severity of dementia, but not to senile plaques, neurofibrillary tangles, or the ApoE4 allele. J Neural Transm (Vienna) 103, 603–618. [DOI] [PubMed] [Google Scholar]
- Bonda DJ, Wang X, Lee HG, Smith MA, Perry G and Zhu X (2014) Neuronal failure in Alzheimer’s disease: a view through the oxidative stress looking-glass. Neurosci Bull 30, 243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlyle BC, Kitchen RR, Kanyo JE et al. (2017) A multiregional proteomic survey of the postnatal human brain. Nat Neurosci 20, 1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MC, Park JM, Pelkey KA, Grabenstatter HL, Xu D, Linden DJ, Sutula TP, McBain CJ and Worley PF (2010) Narp regulates homeostatic scaling of excitatory synapses on parvalbumin-expressing interneurons. Nat Neurosci 13, 1090–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang RY, Etheridge N, Nouwens AS and Dodd PR (2015) SWATH analysis of the synaptic proteome in Alzheimer’s disease. Neurochem Int 87, 1–12. [DOI] [PubMed] [Google Scholar]
- Ciesielski-Treska J, Ulrich G, Chasserot-Golaz S, Zwiller J, Revel MO, Aunis D and Bader MF (2001) Mechanisms underlying neuronal death induced by chromogranin A-activated microglia. J Biol Chem 276, 13113–13120. [DOI] [PubMed] [Google Scholar]
- Comi C, Carecchio M, Chiocchetti A. et al. (2010) Osteopontin is increased in the cerebrospinal fluid of patients with Alzheimer’s disease and its levels correlate with cognitive decline. J Alzheimers Dis 19, 1143–1148. [DOI] [PubMed] [Google Scholar]
- DeKosky ST and Scheff SW (1990) Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity. Ann Neurol 27, 457–464. [DOI] [PubMed] [Google Scholar]
- Diner I, Nguyen T and Seyfried NT (2017) Enrichment of Detergent-insoluble Protein Aggregates from Human Postmortem Brain. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar PF, Schonberger SJ, Dean B, Faull RL, Kydd R and Cooper GJ (1999) A comparative proteome analysis of hippocampal tissue from schizophrenic and Alzheimer’s disease individuals. Mol Psychiatry 4, 173–178. [DOI] [PubMed] [Google Scholar]
- Elbaz I, Lerer-Goldshtein T, Okamoto H and Appelbaum L (2015) Reduced synaptic density and deficient locomotor response in neuronal activity-regulated pentraxin 2a mutant zebrafish. FASEB J 29, 1220–1234. [DOI] [PubMed] [Google Scholar]
- Glenner GG and Wong CW (1984) Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun 120, 885–890. [DOI] [PubMed] [Google Scholar]
- Grosser C, Neumann L, Horsthemke B, Zeschnigk M and van de Nes J (2014) Methylation analysis of SST and SSTR4 promoters in the neocortex of Alzheimer’s disease patients. Neurosci Lett 566, 241–246. [DOI] [PubMed] [Google Scholar]
- Gu Y, Huang S, Chang MC, Worley P, Kirkwood A and Quinlan EM (2013) Obligatory role for the immediate early gene NARP in critical period plasticity. Neuron 79, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Bogdanovic N, Nakagawa H, Volkmann I, Aoki M, Winblad B, Sakai J and Tjernberg LO (2012) Analysis of microdissected neurons by 18O mass spectrometry reveals altered protein expression in Alzheimer’s disease. J Cell Mol Med 16, 1686–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CM, Bollinger JG, Becker JO, Wallace JM, Laha TJ, MacCoss MJ and Hoofnagle AN (2017) Quantification by nano liquid chromatography parallel reaction monitoring mass spectrometry of human apolipoprotein A-I, apolipoprotein B, and hemoglobin A1c in dried blood spots. Proteomics Clin Appl 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata N, Takaki Y, Fukami S, Tsubuki S and Saido TC (2002) Region-specific reduction of A beta-degrading endopeptidase, neprilysin, in mouse hippocampus upon aging. J Neurosci Res 70, 493–500. [DOI] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ and Zha J (2005) The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell 16, 1543–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Nah J, Han J, Choi SG, Kim H, Park J, Pyo HK and Jung YK (2016) Dual-specificity phosphatase 26 (DUSP26) stimulates Abeta42 generation by promoting amyloid precursor protein axonal transport during hypoxia. J Neurochem 137, 770–781. [DOI] [PubMed] [Google Scholar]
- Kandasamy K, Mohan SS, Raju R. et al. (2010) NetPath: a public resource of curated signal transduction pathways. Genome Biol 11, R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F. et al. (2011) Spatio-temporal transcriptome of the human brain. Nature 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence B, Gustafsson BI, Kidd M, Pavel M, Svejda B and Modlin IM (2011) The clinical relevance of chromogranin A as a biomarker for gastroenteropancreatic neuroendocrine tumors. Endocrinol Metab Clin North Am 40, 111–134, viii. [DOI] [PubMed] [Google Scholar]
- Lee VM, Balin BJ, Otvos L Jr. and Trojanowski JQ (1991) A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science 251, 675–678. [DOI] [PubMed] [Google Scholar]
- Manavalan A, Mishra M, Feng L, Sze SK, Akatsu H and Heese K (2013) Brain site-specific proteome changes in aging-related dementia. Exp Mol Med 45, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B, Brenneman R, Becker KG, Gucek M, Cole RN and Maudsley S (2008) iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer’s mice: understanding the interface between physiology and disease. PLoS One 3, e2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirra SS, Heyman A, McKeel D. et al. (1991) The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41, 479–486. [DOI] [PubMed] [Google Scholar]
- Mosconi L, Berti V, Glodzik L, Pupi A, De Santi S and de Leon MJ (2010) Pre-clinical detection of Alzheimer’s disease using FDG-PET, with or without amyloid imaging. J Alzheimers Dis 20, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen RH, Hyde TM, Kleinman JE, Smidt K, Rungby J and Larsen A (2016) Obesity and age-related alterations in the gene expression of zinc-transporter proteins in the human brain. Transl Psychiatry 6, e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott C, Dorsch E, Fraunholz M, Straub S and Kozjak-Pavlovic V (2015) Detailed analysis of the human mitochondrial contact site complex indicate a hierarchy of subunits. PLoS One 10, e0120213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist S, Scholl M, Strandberg O. et al. (2017) Earliest accumulation of beta-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nat Commun 8, 1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R. et al. (2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res 13, 2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping L, Duong DM, Yin L, Gearing M, Lah JJ, Levey AI and Seyfried NT (2018) Global quantitative analysis of the human brain proteome in Alzheimer’s and Parkinson’s Disease. Sci Data 5, 180036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelius E, Brinkmalm G, Pannee J, Zetterberg H, Blennow K, Dahlen R, Brinkmalm A and Gobom J (2017) Proteomic studies of cerebrospinal fluid biomarkers of Alzheimer’s disease: an update. Expert Rev Proteomics 14, 1007–1020. [DOI] [PubMed] [Google Scholar]
- Psych EC, Akbarian S, Liu C. et al. (2015) The PsychENCODE project. Nat Neurosci 18, 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Pan C, He SM, Lang B, Gao GD, Wang XL and Wang Y (2019) The Ras Superfamily of Small GTPases in Non-neoplastic Cerebral Diseases. Front Mol Neurosci 12, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsendorj A, Sheyn J, Fuchs DT et al. (2018) A novel role for osteopontin in macrophage-mediated amyloid-beta clearance in Alzheimer’s models. Brain Behav Immun 67, 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifai N, Gillette MA and Carr SA (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol 24, 971–983. [DOI] [PubMed] [Google Scholar]
- Ringman JM, Schulman H, Becker C. et al. (2012) Proteomic changes in cerebrospinal fluid of presymptomatic and affected persons carrying familial Alzheimer disease mutations. Arch Neurol 69, 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RA, Lange MB, Sultana R. et al. (2011) Differential expression and redox proteomics analyses of an Alzheimer disease transgenic mouse model: effects of the amyloid-beta peptide of amyloid precursor protein. Neuroscience 177, 207–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Merck MF, Bernheim A, Chardin P, Miglierina R, Tavitian A and Berger R (1988) The ras-related ral gene maps to chromosome 7p15–22. Hum Genet 79, 132–136. [DOI] [PubMed] [Google Scholar]
- Saito T, Iwata N, Tsubuki S, Takaki Y, Takano J, Huang SM, Suemoto T, Higuchi M and Saido TC (2005) Somatostatin regulates brain amyloid beta peptide Abeta42 through modulation of proteolytic degradation. Nat Med 11, 434–439. [DOI] [PubMed] [Google Scholar]
- Saiz-Sanchez D, Ubeda-Banon I, de la Rosa-Prieto C, Argandona-Palacios L, Garcia-Munozguren S, Insausti R and Martinez-Marcos A (2010) Somatostatin, tau, and beta-amyloid within the anterior olfactory nucleus in Alzheimer disease. Exp Neurol 223, 347–350. [DOI] [PubMed] [Google Scholar]
- Sasaguri H, Nilsson P, Hashimoto S. et al. (2017) APP mouse models for Alzheimer’s disease preclinical studies. EMBO J 36, 2473–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe G, Mangalaparthi KK, Jain A, Darrow J, Troncoso J, Albert M, Moghekar A and Pandey A (2020) Multiplexed Phosphoproteomic Study of Brain in Patients with Alzheimer’s Disease and Age-Matched Cognitively Healthy Controls. OMICS 24, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe G, Na CH, Renuse S, Madugundu A, Albert M, Moghekar A and Pandey A (2018a) Phosphotyrosine profiling of human cerebrospinal fluid. Clin Proteomics 15, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe G, Na CH, Renuse S, Madugundu AK, Albert M, Moghekar A and Pandey A (2018. b) Quantitative Proteomic Profiling of Cerebrospinal Fluid to Identify Candidate Biomarkers for Alzheimer’s Disease. Proteomics Clin Appl, e1800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvan LD, Renuse S, Kaviyil JE et al. (2014) Phosphoproteome of Cryptococcus neoformans. J Proteomics 97, 287–295. [DOI] [PubMed] [Google Scholar]
- Seyfried NT, Dammer EB, Swarup V. et al. (2017) A Multi-network Approach Identifies Protein-Specific Co-expression in Asymptomatic and Symptomatic Alzheimer’s Disease. Cell Syst 4, 60–72 e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko G, Wetterhall M, Bergquist J, Hoglund K, Andersson LI and Kultima K (2012) Longitudinal characterization of the brain proteomes for the tg2576 amyloid mouse model using shotgun based mass spectrometry. J Proteome Res 11, 6159–6174. [DOI] [PubMed] [Google Scholar]
- Shi T, Song E, Nie S, Rodland KD, Liu T, Qian WJ and Smith RD (2016) Advances in targeted proteomics and applications to biomedical research. Proteomics 16, 2160–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JL and Norris AH (1966) Activities and attitudes of participants in the Baltimore longitudinal study. J Gerontol 21, 575–580. [DOI] [PubMed] [Google Scholar]
- Strittmatter M, Cramer H, Reuner C, Strubel D, Hamann G and Schimrigk K (1997) Molecular forms of somatostatin-like immunoreactivity in the cerebrospinal fluid of patients with senile dementia of the Alzheimer type. Biol Psychiatry 41, 1124–1130. [DOI] [PubMed] [Google Scholar]
- Taupenot L, Harper KL and O’Connor DT (2003) The chromogranin-secretogranin family. N Engl J Med 348, 1134–1149. [DOI] [PubMed] [Google Scholar]
- Toledo JB, Arnold M, Kastenmuller G. et al. (2017) Metabolic network failures in Alzheimer’s disease: A biochemical road map. Alzheimers Dement 13, 965–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troncoso JC, Cataldo AM, Nixon RA et al. (1998) Neuropathology of preclinical and clinical late-onset Alzheimer’s disease. Ann Neurol 43, 673–676. [DOI] [PubMed] [Google Scholar]
- Tundo G, Ciaccio C, Sbardella D, Boraso M, Viviani B, Coletta M and Marini S (2012) Somatostatin modulates insulin-degrading-enzyme metabolism: implications for the regulation of microglia activity in AD. PLoS One 7, e34376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twine NA, Janitz K, Wilkins MR and Janitz M (2011) Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by Alzheimer’s disease. PLoS One 6, e16266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M and Cox J (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang W, Li L, Perry G, Lee HG and Zhu X (2014) Oxidative stress and mitochondrial dysfunction in Alzheimer’s disease. Biochim Biophys Acta 1842, 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler R, Lassmann H, Fischer P, Jellinger K and Winkler H (1990) A high ratio of chromogranin A to synaptin/synaptophysin is a common feature of brains in Alzheimer and Pick disease. FEBS Lett 263, 337–339. [DOI] [PubMed] [Google Scholar]
- Whitfield DR, Vallortigara J, Alghamdi A, Hortobagyi T, Ballard C, Thomas AJ, O’Brien JT, Aarsland D and Francis PT (2015) Depression and synaptic zinc regulation in Alzheimer disease, dementia with lewy bodies, and Parkinson disease dementia. Am J Geriatr Psychiatry 23, 141–148. [DOI] [PubMed] [Google Scholar]
- Wirths O, Breyhan H, Marcello A, Cotel MC, Bruck W and Bayer TA (2010) Inflammatory changes are tightly associated with neurodegeneration in the brain and spinal cord of the APP/PS1KI mouse model of Alzheimer’s disease. Neurobiol Aging 31, 747–757. [DOI] [PubMed] [Google Scholar]
- Wung JK, Perry G, Kowalski A. et al. (2007) Increased expression of the remodeling- and tumorigenic-associated factor osteopontin in pyramidal neurons of the Alzheimer’s disease brain. Curr Alzheimer Res 4, 67–72. [DOI] [PubMed] [Google Scholar]
- Xiao MF, Xu D, Craig MT et al. (2017) NPTX2 and cognitive dysfunction in Alzheimer’s Disease. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Qiao H and Tian X (2011) Proteomic analysis of cerebral synaptosomes isolated from rat model of alzheimer’s disease. Indian J Exp Biol 49, 118–124. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.