Abstract
Spurred by availability of automatic segmentation software, in vivo MRI investigations of human hippocampal subfield volumes have proliferated in the recent years. However, a majority of these studies apply automatic segmentation to MRI scans with approximately 1 × 1 × 1 mm3 resolution, a resolution at which the internal structure of the hippocampus can rarely be visualized. Many of these studies have reported contradictory and often neurobiologically surprising results pertaining to the involvement of hippocampal subfields in normal brain function, aging, and disease. In this commentary, we first outline our concerns regarding the utility and validity of subfield segmentation on 1 × 1 × 1 mm3 MRI for volumetric studies, regardless of how images are segmented (i.e., manually or automatically). This image resolution is generally insufficient for visualizing the internal structure of the hippocampus, particularly the stratum radiatum lacunosum moleculare, which is crucial for valid and reliable subfield segmentation. Second, we discuss the fact that automatic methods that are employed most frequently to obtain hippocampal subfield volumes from 1 × 1 × 1 mm3 MRI have not been validated against manual segmentation on such images. For these reasons, we caution against using volumetric measurements of hippocampal subfields obtained from 1 × 1 × 1 mm3 images.
Keywords: 1 mm3, FreeSurfer, hippocampal subfields, MRI, volumetry
In this commentary, we outline our concerns regarding the utility and validity of subfield segmentation on 1 × 1 × 1 mm3 MRI for volumetric studies. This image resolution is generally insufficient for visualizing the internal structure of the hippocampus, particularly the stratum radiatum lacunosum moleculare, which is crucial for valid and reliable subfield segmentation. Second, automatic methods that are employed most frequently to obtain hippocampal subfield volumes from 1 × 1 × 1 mm3 MRI have not been validated against manual segmentation on such images.
1. INTRODUCTION
The subfields of the human hippocampus (as demarcated in Figure 1) have distinct cytoarchitecture, neurochemistry, and function, and each plays a distinct role in multiple cognitive processes, including episodic memory and spatial navigation (Bakker, Kirwan, Miller, & Stark, 2008; Brown, Hasselmo, & Stern, 2014; Carr et al., 2017; Daugherty, Bender, Raz, & Ofen, 2016; Duncan, Tompary, & Davachi, 2014; Kyle, Smuda, Hassan, & Ekstrom, 2015; Yassa & Stark, 2011). Moreover, hippocampal subfields are thought to be selectively vulnerable to several diseases and pathological conditions, such as hypoxia/ischemia, Alzheimer's disease (AD), temporal lobe epilepsy and depression (Braak & Braak, 1991; Goubran et al., 2016; Schmidt‐Kastner & Freund, 1991; Small, Schobel, Buxton, Witter, & Barnes, 2011). Thus, valid and reliable volumetry of hippocampal subfields estimated from MRI may yield useful biomarkers for studying disease mechanisms or for early diagnosis and clinical trials. The interest in in vivo hippocampal subfields research grew out of extensive investigations in animal models and postmortem human studies, which proliferated with the advent of freely available software for automating hippocampal subfield segmentation. In most respects, the development of automatic hippocampal subfield segmentation is a worthy development that strengthens the translational connection between animal and human studies, and between ex vivo and in vivo research. It broadens the community contributing to the study of cognitive and clinical neuroscience and allows for the enhancement of the knowledge base by enabling re‐analysis of archival data. These positive developments, however, come with limitations that need to be critically examined.
FIGURE 1.
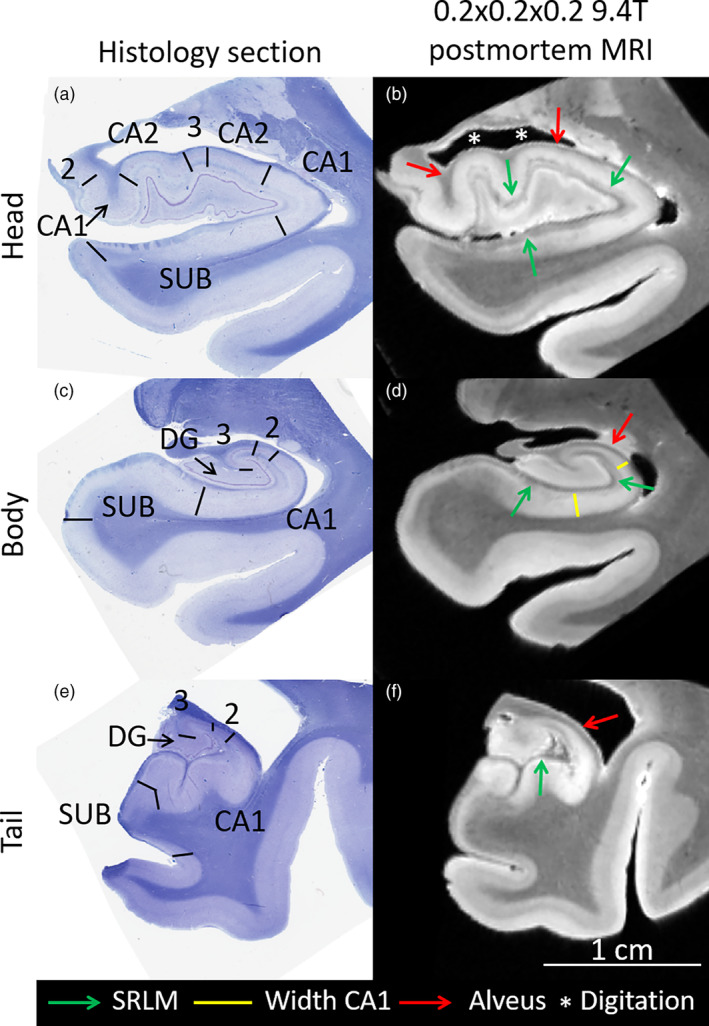
Coronal histology sections (Kluver Barrera stained) and ex vivo 0.2 × 0.2 × 0.2 mm3 MRI in the same subject of the hippocampal head (a,b), body (c,d), and tail (e,g). The black lines in the histology sections demarcate the cytoarchitectonic border between the subfields, traced by an expert analyzing digital histology sections at full 0.5 × 0.5 μm2 resolution. The green arrows point to the stratum radiatum lacunosum moleculare (SRLM) layer, which appears hypointense in the MRI. Note that the width of the cornu ammonis and subiculum are determined by the location of the SLRM, a critical landmark for segmentation of the subfields (yellow lines). Moreover, on the ex vivo MRI, the alveus (outer hypointense band, red arrows) can be seen, which is helpful for demarcating the outer border of the hippocampus and specifically its digitations (white asterisks). CA, cornu ammonis, SUB, subiculum (includes pre‐ and parasubiculum), DG, dentate gyrus
TABLE 1.
Differences in hippocampal subfield volumes in older adults, patients with MCI and patients with AD using automatic FreeSurfer 6.0 segmentation of T1‐weighted MRI and manual segmentation of proton density (PD)‐weighted MRI. Bold font indicates significance after Bonferroni correction (0.05/24)
| Older adults vs. MCI | Older adults vs. AD | MCI vs. AD | ||||
|---|---|---|---|---|---|---|
| FS6‐T1 | Effect size (95% CI) | p‐value | Effect size (95% CI) | p‐value | Effect size (95% CI) | p‐value |
| CA1 | 1.19 (0.28; 2.10) | .02 | 2.35 (1.49; 3.21) | 0.000000001 | 0.97 (0.07; 2.00) | .01 |
| CA2/3/4/DG a | 1.59 (0.65; 2.53) | .001 | 2.08 (1.16; 2.91) | 0.00000007 | 0.45 (0.54; 1.45) | .25 |
| SUB b | 1.11 (0.21; 2.01) | .03 | 2.78 (1.86; 3.70) | 0.00000000003 | 1.56 (0.44; 2.67) | 0.0006 |
| Whole hippocampus | 1.56 (0.62; 2.50) | .002 | 2.70 (1.79; 3.61) c | 0.00000000004 | 0.93 (0.10; 1.96) | .01 |
| Man‐PD | ||||||
| CA1 | 1.47 (0.54; 2.40) | 0.0001 | 1.62 (0.83; 2.39) | 0.00000008 | 0.04 (0.94; 1.03) | .69 |
| CA2/3/4/DG | 0.19 (−0.67; 1.05) | .93 | 1.03 (0.32; 1.75) | 0.023 | 0.65 (0.36; 1.65) | .21 |
| SUB | 1.00 (0.11; 1.89) | .06 | 1.81 (1.02; 2.60) | 0.000002 | 0.63 (0.38; 1.64) | .09 |
| Whole hippocampus | 1.08 (0.18; 1.98) | .02 | 1.98 (1.17; 2.80) | 0.0000003 | 0.45 (0.46; 1.54) | .11 |
Abbreviations: AD, Alzheimer's disease; CA, cornu ammonis; CI, confidence interval; DG, dentate gyrus; Man, manual; MCI, Mild Cognitive Impairment; PD, proton density; SUB, subiculum.
This label includes CA2, CA3, CA4 and GC‐DG.
This label includes subiculum, presubiculum and parasubiculum.
cThe slightly higher reported effect sizes are likely due to a previously reported bias in FreeSurfer 6.0 where larger hippocampal volumes are over‐segmented to a larger extent than smaller hippocampal volumes (Schmidt et al., 2018), which could potentially explain the larger effect size in the current study.
Although all available automatic segmentation methods were developed using high resolution images, over 200 peer‐reviewed publications (see Appendix 1) have applied these automatic methods to ~1 × 1 × 1 mm3 T1‐weighted images (the actual resolution of these images varies but is close to 1 mm3 isotropic; for simplicity, we will refer to this resolution as ~1 mm3 isotropic hereafter). Most of these studies investigated hippocampal subfield volumes, and of these, many reported biologically implausible results. For instance, reports of smaller volumes of the cornu ammonis (CA) 4, dentate gyrus (DG), or the granular cell layer of DG in early AD patients compared to controls (Broadhouse et al., 2019; Mak et al., 2017; Marizzoni et al., 2018; Zhao et al., 2019) are at odds with the classic pathological findings showing that these subfields do not accumulate AD pathology (neurofibrillary tangles) until later stages of the disease (Braak & Braak, 1991). Moreover, many of the hippocampal subfield volumetric findings reported in ~1 mm3 isotropic MRI studies have not been replicated. For example, a recent review of findings on hippocampal subfield volumes in schizophrenia and bipolar disorder (Haukvik, Tamnes, Söderman, & Agartz, 2018), reported discrepant results in most studies included in the review.
In this commentary, we therefore express our concerns regarding hippocampal subfield volumetry estimated from ~1 mm3 isotropic MRI. We note that these concerns are not limited to T1‐weighted MRI but rather apply to all sequences with a similar resolution. First, we point out that ~1 mm3 isotropic resolution is insufficient for visualizing inner structures of the hippocampus, regardless of how the images are subsequently segmented (i.e., manually or automatically). Second, we review the validation of automatic hippocampal subfield segmentation on ~1 mm3 isotropic MRI and, in most instances, the lack thereof. Finally, we express our concerns about alternative, indirect validation approaches of automatic hippocampal subfield segmentation on ~1 mm3 isotropic MRI.
2. IMAGE RESOLUTION AND VISUALIZATION OF INNER STRUCTURES OF THE HIPPOCAMPUS
A very important consideration for choosing an imaging protocol is how well the resulting image allows for visualization of the inner structure of the hippocampus, which is critically important for valid and reliable segmentation. Although not all subfield boundaries are apparent on in vivo MRI, one important landmark, the stratum radiatum lacunosum moleculare (SRLM), can be visualized with appropriate tissue contrast and image resolution. The SRLM is a layer of CA and subiculum (Note that the subicular portion of the SRLM lacks the stratum radiatum) (Figures 1 and 2) that spans the entire extent of the hippocampus and is critical for determining a large portion of the borders between these subfields and the DG (Duvernoy et al., 2005; Insausti & Amaral, 2012). It appears as a thin hypointense layer in T2‐weighted (or, for example, proton density weighted) MRI, and to a lesser extent, as a hyperintense layer on T1‐weighted MRI. Because the SRLM denotes the boundary between the DG and the subiculum and CA regions, visualizing this structure enables the characterization of selective regional atrophy or thinning in these subfields (Figures 1 and 2). In the hippocampal head, identifying SRLM is necessary for visualizing the hippocampal digitations, which show substantial individual differences in depth and number, and are related to hippocampal volume and CA1 neuronal count (Adler et al., 2018; Simic, Kostovic, Winblad, & Bogdanovic, 1997; Figures 1 and 2).
FIGURE 2.
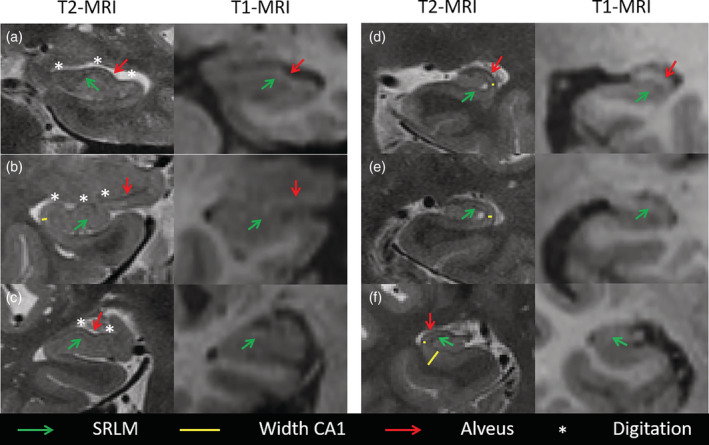
Comparison of 0.4 × 0.4 × 2.6 mm3 T2‐ and 1 × 1 × 1 mm3 T1‐weighted MRI in the same subjects demonstrating better visualization of the inner structure of the hippocampus on 0.4 × 0.4 × 2.6 mm3 T2‐weighted MRI; a and d‐f are cognitively normal (CN) older adults and b‐c are patients with Mild Cognitive Impairment (MCI). The green arrows point out the hypointense band stratum radiatum lacunosum moleculare (SRLM) which can be observed on the high‐resolution T2‐weighted MRI scans but not on the 1 mm3 isotropic T1‐weighted MRI scans. The visualization of SRLM allows for accurate depiction of the width of cornu ammonis (CA) as indicated by the yellow lines in T2‐weighted MRI but not on T1‐weighted MRI. Similarly, the high resolution and the depiction of SRLM and alveus (outer hypointense band, red arrows) in the T2‐weighted MRI, but not T1‐weighted MRI, allow for clear visualization of the digitations (white asterisks) in the hippocampal head, which is crucial for accurate segmentation. Note that the alveus is visible as a hyperintense band on T1‐weighted MRI but is less sharp than on the T2‐weighted MRI
Because the SLRM is a thin structure, ~1 mm3 isotropic scans are insufficient for visualizing this important landmark. A recent postmortem study indicated that the hypointense band in 0.2 × 0.2 × 0.2 mm3 T2‐weighted MRI corresponding to SRLM is ~0.87 mm thick (range 0.77–1.05 mm) in nondemented older adults and 0.65 mm (range 0.52–0.88 mm) in patients with AD (Adler et al., 2018). To the best of our knowledge, no information is available on other populations or age groups at the time of this writing. Thus, visualizing SRLM in vivo requires MRI with high in‐plane resolution. Consistent visualization of this landmark is crucial for separating the DG from the other subfields, even if SRLM is not segmented and measured as a separate structure. Given that the SRLM is thinner than a single 1 mm voxel, and thus can rarely be visualized on ~1 mm3 isotropic MRI scans, it is unlikely that these scans can yield hippocampal subfield volume estimate by either manual or automatic segmentation. This difficulty is illustrated in Figure 2, which shows within‐subject comparisons of high‐resolution T2‐weighted MRI and 1 mm3 isotropic T1‐weighted MRI scans, and in Figure 3, which demonstrates a qualitative within‐subject comparison of an automatic segmentation method applied to both T1‐weighted scans and high resolution T2‐weighted scans. Finally, Figure S1 shows an automatic segmentation of a 1 mm3 isotropic T1‐weighted MRI scan compared to manual segmentation of a high resolution proton density weighted image.
FIGURE 3.
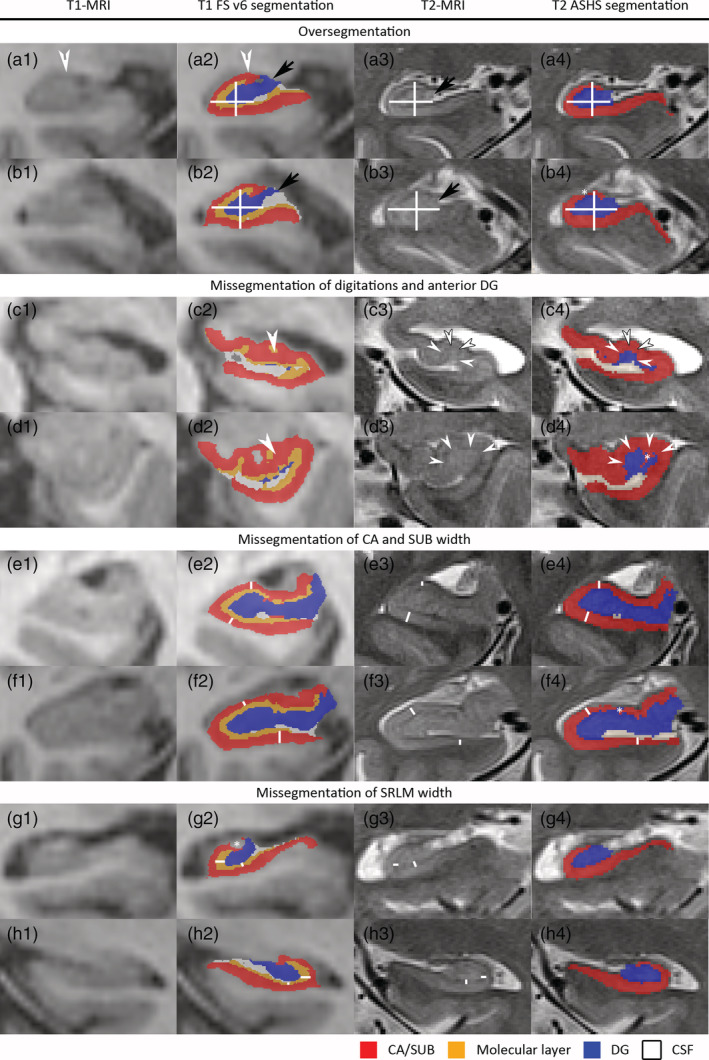
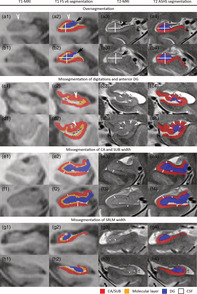
Qualitative within‐subject comparison of automatic segmentation of hippocampal subfields on T1‐ and T2‐weighted MRI. To make a fair comparison, we only selected automatic methods. Specifically, we selected the two methods that are most commonly used for T1‐ and T2‐weighted MRI respectively. We selected FreeSurfer (FS) version 6.0 (Iglesias et al., 2015) for T1‐weighted MRI and Automatic Segmentation for Hippocampal Subfields (ASHS) (Yushkevich et al., 2015) for T2‐weighted MRI. MRI scans from two patients with Mild Cognitive Impairment and two cognitively normal older adults were selected. The T1‐weighted MRI was registered to the T2‐weighted MRI using rigid registration, as done by the ASHS pipeline (Xie et al., 2019). Specifically, the T2‐weighted MRI was up‐sampled to isotropic resolution and rigid registration in the “Greedy” tool (https://github.com/pyushkevich/greedy) was performed using the mutual information image similarity metric with multi‐resolution optimization (×4, ×2, and ×1 subsampling). The T2‐weighted MRI image was further resampled to 0.2 × 0.2 × 0.2 mm3 resolution and the T1‐weighted MRI and its segmentation were resliced into the 0.2 × 0.2 × 0.2 mm3 up‐sampled T2‐weighted MRI space, thus preserving as much of the detail in the segmentation as possible. The quality of image registration was verified in ITK‐SNAP. Moreover, grids are displayed so the registration quality can be checked. To facilitate the comparison of the two different methods, we collapsed the labels for the different subiculum and cornu ammonis (CA) regions into one label (red) and the dentate gyrus (DG) and granular cell layer into another label (blue). We kept the label for the molecular layer (orange) generated by FS (note that this label is often referred to as stratum radiatum lacunosum moleculare or SRLM). For ASHS, a general agreement can be observed between the features in the T1‐weighted MRI and the segmentation boundaries; however, there is less agreement for FS. Both the FS and ASHS segmentations contained errors, which are inherent to automatic segmentations. In this figure, we display four typical FS segmentation errors, but also point out errors in the ASHS segmentations (b4, d4, f4). Given the size of the FS segmentation errors, it is unlikely that partial volume effects on T2‐weighted MRI and small registration errors can fully explain the errors observed on T1‐weighted MRI. While the observed FS errors may seem small, hippocampal subfields are very thin and small errors can therefore have a large impact on the measurements of these granular regions. Figures a1‐a4 and b1–b4 exemplify over‐segmentation of the hippocampus, in general, where the outer boundary of the segmentation does not match that of the hippocampus on MRI. The black arrows (a2–3, b2–3) indicate the over‐segmentation of the DG, such that cerebrospinal fluid and the fimbria is included in the DG label, even though there is a separate fimbria label in FS, which is not displayed in these images. The white arrows (a1–2) indicate the over‐segmentation of the CA (likely CA2 and CA3), including cerebrospinal fluid and alveus in the CA label, even though there is a separate label for the alveus which is not displayed in these images. Over‐segmentation is further exemplified by the white crosses showing the width and height of the hippocampus, which is fairly well‐matched by the ASHS segmentation but not by the FS segmentation. An asterisk indicates a small segmentation error for the ASHS segmentation. The second block shows the mis‐segmentation of the anterior DG and the digitations. The SRLM or dark band is indicated by white arrows in c3 and d3; this indicates the border of the DG (blue) with surrounding subfields. As shown in c2 and d2, the FS segmentation does not match the anatomy as hardly any DG is present and the yellow label does not match the SRLM (hypointense on the T2‐weighted MRI). This means that DG is erroneously included in the molecular and CA labels. ASHS, on the other hand, shows a more accurate segmentation of the DG, although an error (*) can be observed in d4. The third block shows the mis‐segmentation of CA and the subiculum. The width of CA and the subiculum is indicated by white lines in several places on the T2‐weighted MRI (e3 and f3) and is also marked in approximately the same location in the ASHS and FS segmentation. As can be observed in e2 and f2, the width of CA/subiculum is not correctly picked up by FS. FS also shows inconsistencies in the width measurement, such that this region is sometimes thinner and sometimes thicker than the actual width. Although the ASHS segmentation (e4 and f4) is not perfectly matched to the MRI, it is fairly close and more consistent with the actual width. Note that if the width of the CA/subiculum is not correctly estimated, the volume measurements will also be off. The fourth block shows the mis‐segmentation of the SRLM width. The SRLM width is indicated by white lines on the T2‐weighted MRI and approximately the same location in the FS segmentation. While the orange label of the molecular layer in the FS segmentation sometimes approximates the actual SRLM width, in other places it is much thicker or thinner. As the thickness of SRLM is not correctly estimated, the volume measurements will also be inaccurate. Moreover, note a small FS segmentation error in g2
3. REVIEW OF VALIDATION OF SUBFIELD SEGMENTATION ON ~1 MM3 ISOTROPIC IMAGES
The preferred validation, or “the gold standard,” of automatic segmentation would be a comparison with annotated histology sections. This would require matched in vivo and ex vivo imaging data with hippocampal subfield boundaries traced on ex vivo histology, all within the same subject. Such data sets are exceedingly rare (e.g., only 15, sometimes partial, specimens were available in Goubran et al., 2016 and two in Wisse et al., 2016). It is therefore common to validate automatic segmentation against expert manual segmentation—the “bronze standard.” Such validation typically compares the accuracy of the automatic segmentation relative to reliable manual segmentation and then compares the resulting value(s) to the inter‐rater and intra‐rater reliability of manual segmentation. When errors made by an algorithm relative to reliable manual segmentation are statistically equivalent to the disagreements made between different expert raters, it can be argued that the algorithm is an acceptable stand‐in for manual segmentation. In other words, if the reliability of an automated algorithm compared with a manual rater is similar to the reliability of two manual raters, the algorithm can be considered as an acceptable stand‐in for manual segmentation. See Box 1 for more information on the terminology used here and in the sections below.
Box 1: Terminology.
Accuracy: Accuracy is the agreement of a measurement relative to an external standard.
Reliability: Reliability is the internal consistency of a measure, which can be evaluated by the agreement between repeated measures (e.g., between raters or multiple assessments). This is similar to precision.
Construct validity or validity: Construct validity is the degree to which a test measures what it purports to be measuring. In the case of hippocampal subfield segmentation, construct validity refers to the degree to which the segmentation protocol correctly reflects known hippocampal subfield anatomy.
Convergent validity: Convergent validity is a form of construct validity. It refers to the degree to which two measures of constructs that theoretically should be related, are in fact related.
Face validity: Face validity is the degree to which a procedure appears effective in terms of its stated aims. In application to hippocampal subfield segmentation, it may correspond to the qualitative similarities between a segmentation on in vivo MRI as compared to an atlas image with histological labeling.
Prior: A prior in the context of FreeSurfer subfield segmentation is prior knowledge of hippocampal subfield anatomy obtained by combining anatomical annotations of ex vivo MRI scans in the FreeSurfer 6.0 atlas. It can be thought of as the “average” hippocampal subfield anatomy along with statistical information describing how individual anatomies deviate from the average. This information is captured relative to the overall shape of the hippocampus. This statistical representation of anatomy is coupled with information on MRI appearance of hippocampal subfields from the ex vivo atlas (e.g., SRLM voxels have lighter appearance than CA1 voxels). When segmenting a new MRI scan, FreeSurfer uses information both from this prior model and from the MRI scan being segmented to determine how to deform the atlas to match intensity features in the new MRI. However, when intensity features in the new MRI are not informative (e.g., when the intensity values within the hippocampus are largely homogeneous), it is unlikely that these features influence the deformation of the atlas, and so the final segmentation is likely to be driven by atlas information (i.e., the prior) alone.
We next discuss several different automatic segmentation methods. In the first section, we discuss the most commonly used method for automatic segmentation of hippocampal subfields, implemented in FreeSurfer software (Iglesias et al., 2015; Van Leemput et al., 2009). This method has not been validated against manual segmentation as applied to ~1mm3 isotropic T1‐weighted images. In the second section, we discuss the validation of two other automatic methods for hippocampal subfield segmentation.
3.1. Hippocampal subfield segmentation in the different FreeSurfer versions
The original van Leemput et al. method implemented in FreeSurfer 5.3 was evaluated against manual segmentation in high‐resolution MRI scans (0.38 × 0.38 × 0.38 mm3) (Van Leemput et al., 2009), but only in a few slices of the hippocampal body and not against manual segmentations of the same protocol in lower‐resolution ~1 mm3 isotropic data, to which it is commonly applied. Although FreeSurfer 5.3 has been deprecated, as stated on the software website, (https://surfer.nmr.mgh.harvard.edu/fswiki/HippocampalSubfieldSegmentation) and subfield segmentation performed with this version has been criticized for low construct validity (Box 1) of the segmentation protocol (de Flores et al., 2015; Wisse, Biessels, & Geerlings, 2014), FreeSurfer 5.3 is nonetheless still being used by multiple research groups to segment hippocampal subfields (Duan et al., 2020; Izzo, Andreassen, Westlye, & van der Meer, 2020; Takaishi et al., 2020).
The hippocampal subfield segmentation module in FreeSurfer 6.0 (Iglesias et al., 2015) uses an ex vivo atlas derived from 15 ex vivo MRI scans of the hippocampus and accompanying detailed annotations of 13 hippocampal subfields, which improved the face validity of this version over version 5.3. This, together with the large number of experiments performed in the Iglesias et al. paper (2015), may lead readers to conclude that FreeSurfer 6.0 has been completely validated. However, the segmentations provided by FreeSurfer 6.0 on in vivo MRI scans have not been validated against histological annotations in the same subjects (gold standard), nor have these segmentations been validated against manual segmentation on the same MRI scans using the FreeSurfer 6.0 13‐label protocol (bronze standard). As such, the validity of these algorithms when applied to ~1 mm3 isotropic T1‐weighted scans remains unknown.
In the absence of extensive validation, researchers should not assume that automatic methods produce anatomically accurate segmentations on images that lack sufficient anatomical detail that would allow manual segmentation. While it is theoretically possible that an automatic algorithm might detect and exploit some anatomical features in ~1 mm3 isotropic MRI scans that a trained expert cannot, a more likely explanation is that automatic methods fill in missing information (e.g., SRLM) by using anatomical priors (Box 1). The creators of Freesurfer's automatic tool acknowledged this on their website: “When segmenting 1 mm scans, the position of the internal boundaries between the hippocampal substructures largely relies on prior knowledge acquired from our ex vivo training data and summarized in our statistical atlas.” (Iglesias, 2019). The resulting interpolation of subfields based on priors and visible features, such as the overall hippocampus boundary, is likely to ignore individual variation in hippocampal subfield anatomy, such as variation in the ratio of DG and CA thickness or variation in the number and location of digitations in the hippocampal head (see Figure 3).
The authors of this commentary therefore express concerns that automatic subfield volumetric measurements generated by FreeSurfer using ~1 mm3 isotropic images—currently the most widely used method for subfield segmentation—likely do not capture variation in subfield volumes per se, but rather act as a proxy of total volume. They are, therefore, likely unable to capture specific variability in anatomical features, for example, selective thinning in some regions or patterns of digitations in the hippocampal head (see, for example, findings by Elman et al. in support of this statement (Elman et al., 2018). The limitations of automatic subfield segmentation on ~1 mm3 isotropic T1‐weighted images have been pointed out previously (de Flores, La Joie, Landeau, et al., 2015; Wisse et al., 2014), and similar limitations would, of course, apply to volume estimates from manual segmentation of images of the same resolution. Moreover, the creators of the FreeSurfer 6.0 subfield segmentation algorithm (Iglesias et al., 2015) stated in their paper that the use of subfield segmentation based on ~1 mm3 isotropic T1‐weighted images was better suited “as seed and target regions in functional and diffusion MRI studies,” and cautioned against the interpretation of subfield volumes in quantitative analyses (Iglesias et al., 2015). These concerns also hold for the subfield segmentation algorithm recently introduced in FreeSurfer version 7.0 as well as future versions that provide similar automatic subfield volumetric measurements on ~1 mm3 isotropic images.
The findings presented in Box 2 support our concerns regarding subfield segmentation on ~1 mm3 isotropic images for the estimation of volumes. In short, we compared FreeSurfer 6.0 segmentation of 1 mm3 isotropic T1‐weighted images to manual segmentation of high‐resolution proton density‐weighted images with respect to the ability to capture subfield volume associations with MCI and AD, similar to a previous comparison of Freesurfer 5.3 with manual segmentations (de Flores, La Joie, Landeau, et al., 2015). The hippocampal subfield volumes generated by FreeSurfer 6.0 failed to reveal significant AD‐related differences in the expected subfield, CA1, which is the first region in the hippocampus to accrue neurofibrillary tangle pathology (Braak & Braak, 1991). In contrast, smaller CA1 volume in β‐amyloid positive MCI patients compared to controls was found in the manually segmented proton density dataset in the same subjects, in line with multiple in vivo MRI studies (reviewed by de Flores, La Joie, & Chetelat, 2015). Although these types of comparisons cannot replace the comparison of automated or manual segmentations against histology annotations in the same subject, we believe that a segmentation method—manual or automated—that is insensitive to clinically relevant changes in diseases will have limited utility.
Box 2: FreeSurfer 6.0 comparison with manual segmentation in controls, MCI and AD patients, as in De Flores et al. (de Flores, La Joie, Landeau, et al., 2015).
In this box, we aim to compare hippocampal subfield volumes between older adults, patients with Mild Cognitive Impairment (MCI) and Alzheimer's disease (AD). Similar to De Flores et al. (de Flores, La Joie, Landeau, et al., 2015), we aim to compare the performance of automatic segmentations implemented in FreeSurfer (FS) 6.0 (Iglesias et al., 2015) on 1 × 1 × 1 mm3 T1‐weighted MRI and manual segmentations on high‐resolution 0.4 × 0.4 × 2 mm3 (2 mm gap) proton density weighted images (which have similar contrast as T2‐weighted images). Twenty‐eight older adults (mean age: 70.3 ± 6.5 years; 46.4% men; education: 12.4 ± 4.1 years), 9 β‐amyloid positive (A+, cut off based on 3 standard deviations from a young control group) patients with MCI (mean age: 70.0 ± 5.5 years; 42.9% men; education: 10.7 ± 4.8 years) and 13 A+ patients with AD (mean age: 65.2 ± 10.3 years; 30.8% men; education: 10.8 ± 4.3 years) were included. Note that a smaller number of subjects was included than in de Flores, La Joie, and Chetelat (2015) because the FreeSurfer 6.0 pipeline failed in some subjects and only A+ patients were included. All manual segmentations were performed by author RLJ. The intraclass correlation coefficient was 0.94 for cornu ammonis (CA) 1, 0.89 for subiculum and 0.94 for CA2/3/dentate gyrus (La Joie et al., 2010). All subfield volumes were corrected for intracranial volume and analyses of covariance were performed, including age, gender, and education as covariates. Based on Braak staging of neurofibrillary tangle pathology (Braak & Braak, 1991), which is closely related to neurodegeneration (Bobinski et al., 1997; Fukutani et al., 1995), the earliest and strongest volume loss is expected in CA1, then the subiculum, while the other CA regions and the dentate gyrus are expected to be affected in later stages.
The Table 1 demonstrates that segmentations generated by FS 6.0 do not reflect the expected pattern of atrophy, with only CA2/3/4/dentate gyrus surviving Bonferroni correction in the comparison between A+ MCI and older adults, and with all regions showing a similar effect when comparing older adults with A+ patients with AD. Splitting up the subfields in the different labels provided by FS 6.0 does not change the results (see Table S1). Conversely, using manual segmentation, CA1 was found as the most atrophied region in A+ MCI patients as compared to older adults, whereas, as expected, both CA1 and subiculum were significantly smaller compared to A+ AD. CA2/3/4/dentate gyrus showed the weakest effect size when comparing older adults to both A+ MCI and A+ AD patients. Note that, based on a comparison of 95% CI, the effect sizes are equivalent, except in CA2/3/4/dentate gyrus comparing A+ MCI patients and older adults. See Figure S1 for a comparison of the manual and automatic segmentation.
These results indicate that FreeSurfer 6.0 hippocampal subfield volumes measured on standard T1‐weighted images do not capture the expected pattern of atrophy over the course of AD, while an appropriate manual segmentation performed on high resolution images in the same subjects does. Note that this comparison does not allow any conclusions about the application of FreeSurfer 6.0 package to high resolution T2‐weighted images.
3.2. Validation of other automatic methods against manual segmentation
In this section, we discuss two methods that developed an approach to compare automatic segmentations of hippocampal subfields against manual segmentations on ~1 mm3 isotropic images. Two studies (Caldairou et al., 2016; Pipitone et al., 2014) down‐sampled manual subfield segmentations obtained on high resolution (Caldairou: 0.6 × 0.6 × 0.6 mm3; Pipitone: 0.3 × 0.3 × 0.3 mm3) T1 and T2‐weighted images to ~1 mm3 isotropic resolution (Caldairou: 1x1x1 mm3; Pipitone: 0.9x0.9x0.9 mm3) T1‐weighted MRI scans from the same subjects. The down‐sampled data from the same subjects served to evaluate the performance of the automatic segmentation algorithms. Although the Dice Similarity Coefficient (DSC) values presented by Caldairou et al. (2016) exceeded 0.80, they were considerably lower, that is, between 0.41–0.65, in the Pipitone et al. (2014) paper. These lower DSC values may be due to the higher complexity of the segmentation protocol (Pipitone et al., 2014; Winterburn et al., 2013), the smaller number of atlases, and the wider age range of subjects included in the atlas set. The explanation for the higher DSC values in the Caldairou et al. (2016) paper could be two‐fold. One possibility is that there is enough subtle signal information to infer the location of DG/CA/subiculum from 1 mm3 T1‐weighted MRI (these are larger, and geometrically less complex labels than in the protocol (Winterburn et al., 2013) used in Pipitone et al. (2014) paper), which would mean that it is acceptable to use their method in ~1 mm3 isotropic MRI. The other possibility is that the segmentation is driven by a shape prior, given that the authors discuss using a strong prior, and the location of the subfields is sufficiently predictable from the shape prior, for example, because this is a young population, to get the high DSC values. Additionally, the high DSC values may be partially explained by the fact that the included subfield or subfield groups were relatively large in size, a factor that positively affects DSC values. Indeed, reported correlation coefficients were considerably lower in the Caldairou et al. study: 0.28–0.64. We would like to stress that this evaluation speaks to the consistency of this specific method in this cohort but does not generalize to provide evidence that all automatic methods can accurately measure subfields in ~1 mm3 isotropic images, nor that the same method will perform adequately in other populations. In fact, the relatively low correlation coefficients in the Caldairou et al. (2016) and the relatively low DSC values in the Pipitone et al. (2014) warrant further caution toward obtaining hippocampal subfield volumes from 1 mm3 isotropic MRI scans. Finally, applying one of these methods to ~1 mm3 isotropic MRI scans from other populations will not allow for careful assessment of the quality of the segmentations given the limited detail available in these images.
4. CONCERNS REGARDING ANOTHER VALIDATION APPROACH FOR SUBFIELD SEGMENTATION ON ~1 MM3 ISOTROPIC MRI
Finally, we caution against validating FreeSurfer 6.0 ~ 1 mm3 isotropic T1‐weighted MRI (FS‐T1) segmentations against Freesurfer 6.0 applied on a combination of T1‐weighted MRI and high‐resolution T2‐weighted MRI (FS‐T1T2) as a standard. This comparison offers little information on validity, as FS‐T1T2 has not yet been validated against manual segmentation or histology. Moreover, the validation of FS‐T1 against FS‐T1T2 is inherently biased, given that segmentation of both FS‐T1 and FS‐T1T2 is based on some combination of intensity features and shape priors. To illustrate this point, suppose that in both cases, 100% of segmentation information came from shape priors (Box 1). One would then observe a perfect correlation between FS‐T1 and FS‐T1T2. Measuring the correlation between FS‐T1 and FS‐T1T2 therefore has the unfortunate side effect of measuring the strength of the priors. A high correlation could either be due to T1 and T1 + T2 providing similar information for segmentation, or due to a strong reliance on a prior (e.g., that the hippocampus has a certain internal structure, that each structure has a typical shape and volume, and that by warping the outer surface, the inner structures are similarly warped). The inability to disambiguate the two factors driving the metric make it flawed for evaluating FS‐T1 anatomical validity. Relating to this point, the use of the FreeSurfer subfield package has in the past been justified by high test–retest reliability, but test–retest reliability does not speak to construct validity. In contrast, high test–retest reliability likely shows the strength of the prior. Note that these concerns also hold for FreeSurfer version 7.0 and any similar future versions.
5. ALTERNATIVE METHODS TO OBTAIN GRANULAR STRUCTURAL MEASURES OF THE HIPPOCAMPUS USING ~1 MM3 ISOTROPIC MRI
We are unfortunately not able to provide alternatives for obtaining hippocampal subfield volumetric measures from ~1 mm3 isotropic MRI, given the above stated limitations of these kind of images. However, alternative approaches exist for obtaining more granular measures than those available via whole hippocampal volume using ~1 mm3 isotropic MRI. For example, researchers have divided the hippocampus along its long axis (head, body and tail; e.g., Bernasconi et al., 2003; Chen, Chuah, Sim, & Chee, 2010; Daugherty, Yu, Flinn, & Ofen, 2015; Malykhin, Carter, Seres, & Coupland, 2010) and have also used surface deformation‐based methods (e.g., Apostolova et al., 2012; Wang et al., 2006). While these methods cannot make inferences about the inner subfields, they can nonetheless provide more granular and highly relevant measures of hippocampal structure that may be more sensitive to functional and clinical correlates than whole hippocampal volume.
6. CONCLUSION
The interest in MRI‐based hippocampal subfield research has significantly increased in recent years due to availability of public‐domain datasets, such as the ADNI (Weiner et al., 2017) and human connectome datasets (Van Essen et al., 2013), as well as by publicly available automatic tools, such as FreeSurfer (Iglesias et al., 2015; Van Leemput et al., 2009), ASHS (Yushkevich et al., 2015), and MAGeT (Pipitone et al., 2014; Winterburn et al., 2013). Although these developments enable large‐scale subfield analyses, it is important to remain cautious regarding the increasing application of automatic segmentation methods to inappropriate data sets (e.g., ~1 mm3 isotropic T1‐weighted MRI scans) for several reasons. First, the resolution of these images is insufficient for visualizing the inner structures of the hippocampus, particularly the SRLM, that are crucial for either manual or automatic subfield segmentation. Second, automatic subfield segmentation on ~1 mm3 isotropic images has not been validated against manual segmentation for some methods, including FreeSurfer (Iglesias et al., 2015; Van Leemput et al., 2009), the most commonly used approach. We are therefore concerned that subfield volumetric data from ~1 mm3 isotropic MRI scans are not capturing subfield volumes as intended, but rather represent a proxy of total volume and are not able to capture specific variability in anatomical features. It should be noted that there are some methods that have validated automatic hippocampal subfield segmentations against manual segmentations down‐sampled to ~1 mm3 isotropic MRI scans (Caldairou et al., 2016; Pipitone et al., 2014). However, the results require careful scrutiny before considering the application of such methods to a particular data set.
Although our concerns are partly based on reasoning and formal comparisons of images acquired with different weighting (T1 vs. T2) and resolution, we believe the arguments outlined in this commentary are strong enough to warrant caution against hippocampal subfield segmentation on ~1 mm3 isotropic MRI scans, a caution supported by other research groups (e.g., Elman et al., 2018; Giuliano et al., 2017; Iglesias et al., 2015). We recommend that future studies further compare hippocampal subfield segmentation on ~1 mm3 isotropic MRI scans with higher resolution MRI scans. We believe that such studies will provide additional data highlighting the need for caution when attempting to segment hippocampal subfields on ~1 mm3 isotropic images and replicate some of the previous comparison papers (de Flores, La Joie, Landeau, et al., 2015; Mueller et al., 2018).
CONFLICT OF INTERESTS
None of the authors has any disclosures.
Supporting information
Appendix S1. Supporting Information.
Appendix 1.
ACKNOWLEDGMENTS
The authors thank David Wolk for sharing screenshots of MRI scans included in the current paper. The study was supported by Fondation Plan Alzheimer (Alzheimer Plan 2008‐2012) (GC), Programme Hospitalier de Recherche Clinique (PHRCN 2011‐A01493‐38 and PHRCN 2012 12‐006‐0347) (GC), Agence Nationale de la Recherche (LONGVIE 2007) (GC), Region Basse‐Normandie; Association France Alzheimer et maladies apparentees AAP 2013 (GC), the Fondation Philippe Chatrier (RDF), a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC; RGPIN‐2017‐06178) (RKO), a grant from the Canadian Institutes of Health (PJT‐162292) (RKO) and new investigator grant from the Alzheimer Society of Canada (RKO), National Institute of Health grants R01 AG055121 (LW), R01 EB020062 (LW), R01 AG034613 (CELS), R01 AG056014 (PY), R01 AG011230 (NR) and the donors of Alzheimer's Disease Research, a program of the BrightFocus Foundation (LEMW).
Wisse LEM, Chételat G, Daugherty AM, et al. Hippocampal subfield volumetry from structural isotropic 1 mm3 MRI scans: A note of caution. Hum Brain Mapp. 2021;42:539–550. 10.1002/hbm.25234
Rosanna K. Olsen and Valerie A. Carr shared the last authorship.
Funding information Agence Nationale de la Recherche, Grant/Award Number: LONGVIE 2007; Alzheimer Society of Canada, Grant/Award Number: New investigator grant; Alzheimer's Disease Research, a program of the BrightFocus Foundation; Association France Alzheimer et maladies apparentees AAP 2013; Canadian Institutes of Health Research, Grant/Award Number: PJT‐162292; Discovery Grant from the Natural Sciences and Engineering Research Council of Canada, Grant/Award Number: RGPIN‐2017‐06178; Fondation Plan Alzheimer, Grant/Award Number: Alzheimer Plan 2008‐2012; National Institutes of Health, Grant/Award Numbers: AG011230, AG034613, AG055121, AG056014, EB020062; Programme Hospitalier de Recherche Clinique, Grant/Award Numbers: PHRCN 2011‐A01493‐38, PHRCN 2012 12‐006‐0347; the Fondation Philippe Chatrier
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Adler, D. H. , Wisse, L. E. , Ittyerah, R. , Pluta, J. B. , Ding, S. , Xie, L. , … Schuck, T. (2018). Characterizing the human hippocampus in aging and Alzheimer's disease using a computational atlas derived from ex vivo MRI and histology. Proceedings of the National Academy of Sciences, 115, 4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova, L. G. , Green, A. E. , Babakchanian, S. , Hwang, K. S. , Chou, Y. Y. , Toga, A. W. , & Thompson, P. M. (2012). Hippocampal atrophy and ventricular enlargement in normal aging, mild cognitive impairment (MCI), and Alzheimer disease. Alzheimer Disease and Associated Disorders, 26, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, A. , Kirwan, C. B. , Miller, M. , & Stark, C. E. (2008). Pattern separation in the human hippocampal CA3 and dentate gyrus. Science, 319, 1640–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi, N. , Bernasconi, A. , Caramanos, Z. , Antel, S. B. , Andermann, F. , & Arnold, D. L. (2003). Mesial temporal damage in temporal lobe epilepsy: A volumetric MRI study of the hippocampus, amygdala and parahippocampal region. Brain, 126, 462–469. [DOI] [PubMed] [Google Scholar]
- Bobinski, M. , Wegiel, J. , Tarnawski, M. , Bobinski, M. , Reisberg, B. , de Leon, M. J. , … Wisniewski, H. M. (1997). Relationships between regional neuronal loss and neurofibrillary changes in the hippocampal formation and duration and severity of Alzheimer disease. Journal of Neuropathology and Experimental Neurology, 56, 414–420. [DOI] [PubMed] [Google Scholar]
- Braak, H. , & Braak, E. (1991). Neuropathological stageing of Alzheimer‐related changes. Acta Neuropathologica, 82, 239–259. [DOI] [PubMed] [Google Scholar]
- Broadhouse, K. M. , Mowszowski, L. , Duffy, S. , Leung, I. , Cross, N. , Valenzuela, M. J. , & Naismith, S. L. (2019). Memory performance correlates of hippocampal subfield volume in mild cognitive impairment subtype. Frontiers in Behavioral Neuroscience, 13, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, T. I. , Hasselmo, M. E. , & Stern, C. E. (2014). A high‐resolution study of hippocampal and medial temporal lobe correlates of spatial context and prospective overlapping route memory. Hippocampus, 24, 819–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldairou B, Bernhardt BC, Kulaga‐Yoskovitz J, Kim H, Bernasconi N, Bernasconi A (2016): A surface patch‐based segmentation method for hippocampal subfields. In International Conference on Medical Image Computing and Computer‐Assisted Intervention, 379‐387. Cham: Springer.
- Carr, V. A. , Bernstein, J. D. , Favila, S. E. , Rutt, B. K. , Kerchner, G. A. , & Wagner, A. D. (2017). Individual differences in associative memory among older adults explained by hippocampal subfield structure and function. Proceedings of the National Academy of Sciences of the United States of America, 114, 12075–12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. H. , Chuah, L. Y. , Sim, S. K. , & Chee, M. W. (2010). Hippocampal region‐specific contributions to memory performance in normal elderly. Brain and Cognition, 72, 400–407. [DOI] [PubMed] [Google Scholar]
- Daugherty, A. M. , Bender, A. R. , Raz, N. , & Ofen, N. (2016). Age differences in hippocampal subfield volumes from childhood to late adulthood. Hippocampus, 26, 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty, A. M. , Yu, Q. , Flinn, R. , & Ofen, N. (2015). A reliable and valid method for manual demarcation of hippocampal head, body, and tail. International Journal of Developmental Neuroscience, 41, 115–122. [DOI] [PubMed] [Google Scholar]
- de Flores, R. , La Joie, R. , & Chetelat, G. (2015). Structural imaging of hippocampal subfields in healthy aging and Alzheimer's disease. Neuroscience, 309, 29–50. [DOI] [PubMed] [Google Scholar]
- de Flores, R. , La Joie, R. , Landeau, B. , Perrotin, A. , Mezenge, F. , de La Sayette, V. , … Chetelat, G. (2015). Effects of age and Alzheimer's disease on hippocampal subfields: Comparison between manual and FreeSurfer volumetry. Human Brain Mapping, 36, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y. , Lin, Y. , Rosen, D. , Du, J. , He, L. , & Wang, Y. (2020). Identifying morphological patterns of hippocampal atrophy in patients with mesial temporal lobe epilepsy and Alzheimer disease. Frontiers in Neurology, 11, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, K. , Tompary, A. , & Davachi, L. (2014). Associative encoding and retrieval are predicted by functional connectivity in distinct hippocampal area CA1 pathways. The Journal of Neuroscience, 34, 11188–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy, H. M. , Cattin, E. , Naidich, T. , Fatterpekar, G. M. , Raybaud, C. , Risold, P. Y. , … Scarabino, T. (2005). The human hippocampus. Germany: Springer Verlag Berlin Heidelberg. [Google Scholar]
- Elman, J. A. , Panizzon, M. S. , Gillespie, N. A. , Hagler, D. J., Jr. , Fennema‐Notestine, C. , Eyler, L. T. , … Franz, C. E. (2018). Genetic architecture of hippocampal subfields on standard resolution MRI: How the parts relate to the whole. Human Brain Mapping, 40(5), 1528–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutani, Y. , Kobayashi, K. , Nakamura, I. , Watanabe, K. , Isaki, K. , & Cairns, N. J. (1995). Neurons, intracellular and extracellular neurofibrillary tangles in subdivisions of the hippocampal cortex in normal ageing and Alzheimer's disease. Neuroscience Letters, 200, 57–60. [DOI] [PubMed] [Google Scholar]
- Giuliano, A. , Donatelli, G. , Cosottini, M. , Tosetti, M. , Retico, A. , & Fantacci, M. E. (2017). Hippocampal subfields at ultra high field MRI: An overview of segmentation and measurement methods. Hippocampus, 27, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubran, M. , Bernhardt, B. C. , Cantor‐Rivera, D. , Lau, J. C. , Blinston, C. , Hammond, R. R. , … Khan, A. R. (2016). In vivo MRI signatures of hippocampal subfield pathology in intractable epilepsy. Human Brain Mapping, 37, 1103–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haukvik, U. K. , Tamnes, C. K. , Söderman, E. , & Agartz, I. (2018). Neuroimaging hippocampal subfields in schizophrenia and bipolar disorder: A systematic review and meta‐analysis. Journal of Psychiatric Research, 104, 217–226. [DOI] [PubMed] [Google Scholar]
- Iglesias, J. E . (2019). Segmentation of hippocampal subfields Retrieved from https://surfer.nmr.mgh.harvard.edu/fswiki/HippocampalSubfields.
- Iglesias, J. E. , Augustinack, J. C. , Nguyen, K. , Player, C. M. , Player, A. , Wright, M. , … Alzheimer's Disease Neuroimaging Initiative . (2015). A computational atlas of the hippocampal formation using ex vivo, ultra‐high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage, 115, 117–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insausti, R. , & Amaral, D. G. (2012). Hippocampal formation In Mai J. K. & Paxinos G. (Eds.), The human nervous system. San Diego: Elsevier Academic Press. [Google Scholar]
- Izzo, J. , Andreassen, O. A. , Westlye, L. T. , & van der Meer, D. (2020). The association between hippocampal subfield volumes in mild cognitive impairment and conversion to Alzheimer's disease. Brain Research, 1728, 146591. [DOI] [PubMed] [Google Scholar]
- Kyle, C. T. , Smuda, D. N. , Hassan, A. S. , & Ekstrom, A. D. (2015). Roles of human hippocampal subfields in retrieval of spatial and temporal context. Behavioural Brain Research, 278, 549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Joie, R. , Fouquet, M. , Mezenge, F. , Landeau, B. , Villain, N. , Mevel, K. , … Chetelat, G. (2010). Differential effect of age on hippocampal subfields assessed using a new high‐resolution 3T MR sequence. NeuroImage, 53, 506–514. [DOI] [PubMed] [Google Scholar]
- Mak, E. , Gabel, S. , Su, L. , Williams, G. B. , Arnold, R. , Passamonti, L. , … Rowe, J. B. (2017). Multi‐modal MRI investigation of volumetric and microstructural changes in the hippocampus and its subfields in mild cognitive impairment, Alzheimer's disease, and dementia with Lewy bodies. International Psychogeriatrics, 29, 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malykhin, N. V. , Carter, R. , Seres, P. , & Coupland, N. J. (2010). Structural changes in the hippocampus in major depressive disorder: Contributions of disease and treatment. Journal of Psychiatry & Neuroscience, 35, 337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marizzoni, M. , Ferrari, C. , Jovicich, J. , Albani, D. , Babiloni, C. , Cavaliere, L. , … Hoffmann, K. (2018). Predicting and tracking short term disease progression in amnestic mild cognitive impairment patients with prodromal Alzheimer's disease: Structural brain biomarkers. Journal of Alzheimer's Disease, 69(1), 3–14. [DOI] [PubMed] [Google Scholar]
- Mueller, S. G. , Yushkevich, P. A. , Das, S. , Wang, L. , Van Leemput, K. , Iglesias, J. E. , … Paz, K. (2018). Systematic comparison of different techniques to measure hippocampal subfield volumes in ADNI2. NeuroImage: Clinical, 17, 1006–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipitone, J. , Park, M. T. , Winterburn, J. , Lett, T. A. , Lerch, J. P. , Pruessner, J. C. , … Alzheimer's Disease Neuroimaging Initiative . (2014). Multi‐atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. NeuroImage, 101, 494–512. [DOI] [PubMed] [Google Scholar]
- Schmidt, M. F. , Storrs, J. M. , Freeman, K. B. , Jack, C. R., Jr. , Turner, S. T. , Griswold, M. E. , & Mosley, T. H., Jr. (2018). A comparison of manual tracing and FreeSurfer for estimating hippocampal volume over the adult lifespan. Human Brain Mapping, 39, 2500–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt‐Kastner, R. , & Freund, T. F. (1991). Selective vulnerability of the hippocampus in brain ischemia. Neuroscience, 40, 599–636. [DOI] [PubMed] [Google Scholar]
- Simic, G. , Kostovic, I. , Winblad, B. , & Bogdanovic, N. (1997). Volume and number of neurons of the human hippocampal formation in normal aging and Alzheimer's disease. The Journal of Comparative Neurology, 379, 482–494. [DOI] [PubMed] [Google Scholar]
- Small, S. A. , Schobel, S. A. , Buxton, R. B. , Witter, M. P. , & Barnes, C. A. (2011). A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nature Reviews. Neuroscience, 12, 585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi, M. , Asami, T. , Yoshida, H. , Nakamura, R. , Yoshimi, A. , & Hirayasu, Y. (2020). Smaller volume of right hippocampal CA2/3 in patients with panic disorder. Brain Imaging and Behavior, 1–7. [DOI] [PubMed] [Google Scholar]
- Van Essen, D. C. , Smith, S. M. , Barch, D. M. , Behrens, T. E. , Yacoub, E. , Ugurbil, K. , & WU‐Minn HCP Consortium . (2013). The WU‐Minn human connectome project: An overview. NeuroImage, 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput, K. , Bakkour, A. , Benner, T. , Wiggins, G. , Wald, L. L. , Augustinack, J. , … Fischl, B. (2009). Automated segmentation of hippocampal subfields from ultra‐high resolution in vivo MRI. Hippocampus, 19, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Miller, J. P. , Gado, M. H. , McKeel, D. W. , Rothermich, M. , Miller, M. I. , … Csernansky, J. G. (2006). Abnormalities of hippocampal surface structure in very mild dementia of the Alzheimer type. NeuroImage, 30, 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner, M. W. , Veitch, D. P. , Aisen, P. S. , Beckett, L. A. , Cairns, N. J. , Green, R. C. , … Alzheimer's Disease Neuroimaging Initiative . (2017). The Alzheimer's Disease Neuroimaging Initiative 3: Continued innovation for clinical trial improvement. Alzheimers Dement, 13, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterburn, J. L. , Pruessner, J. C. , Chavez, S. , Schira, M. M. , Lobaugh, N. J. , Voineskos, A. N. , & Chakravarty, M. M. (2013). A novel in vivo atlas of human hippocampal subfields using high‐resolution 3 T magnetic resonance imaging. NeuroImage, 74, 254–265. [DOI] [PubMed] [Google Scholar]
- Wisse, L. , Adler, D. H. , Ittyerah, R. , Pluta, J. B. , Robinson, J. L. , Schuck, T. , … Elliott, M. A. (2016). Comparison of in vivo and ex vivo MRI of the human hippocampal formation in the same subjects. Cerebral Cortex, 27(11), 5185–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. , Biessels, G. J. , & Geerlings, M. I. (2014). A critical appraisal of the hippocampal subfield segmentation package in FreeSurfer. Frontiers in Aging Neuroscience, 6(), 261–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, L. , Wisse, L. E. , Pluta, J. , de Flores, R. , Piskin, V. , Manjón, J. V. , … Wolk, D. A. (2019). Automated segmentation of medial temporal lobe subregions on in vivo T1‐weighted MRI in early stages of Alzheimer's disease. Human Brain Mapping, 40, 3431–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa, M. A. , & Stark, C. E. (2011). Pattern separation in the hippocampus. Trends in Neurosciences, 34, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Pluta, J. B. , Wang, H. , Xie, L. , Ding, S. L. , Gertje, E. C. , … Wolk, D. A. (2015). Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Human Brain Mapping, 36, 258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W. , Wang, X. , Yin, C. , He, M. , Li, S. , & Han, Y. (2019). Trajectories of the hippocampal subfields atrophy in the Alzheimer's disease: A structural imaging study. Frontiers in Neuroinformatics, 13, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.
Appendix 1.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


