Fig. 5.
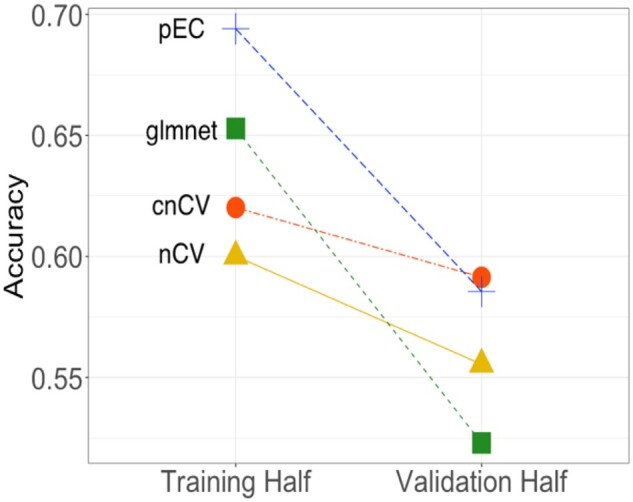
RNA-seq accuracy comparisons. RNA-seq dataset from a MDD study (Mostafavi et al., 2014) is split into a Training Half and a Validation Half. The final training model for each method is tested on the independent validation half to assess the degree of overfitting. Standard nCV (triangle) uses 3592 genes in its model, cnCV (circle) uses 399 genes in its model and pEC (plus) uses 3415 genes in its model. These methods use ReliefF feature selection and random forest classification. Glmnet (square) selects 56 genes by penalized logistic regression feature selection. The cnCV validation accuracy is highest and has the lowest overfitting
