Abstract
In both mouse and human, Notch1 activation is the main initial driver to induce T-cell development in hematopoietic progenitor cells. The initiation of this developmental process coincides with Notch1-dependent repression of differentiation towards other hematopoietic lineages. Although well described in mice, the role of the individual Notch1 target genes during these hematopoietic developmental choices is still unclear in human, particularly for HES4 since no orthologous gene is present in the mouse. Here, we investigated the functional capacity of the Notch1 target genes HES1 and HES4 to modulate human Notch1-dependent hematopoietic lineage decisions and their requirement during early T-cell development. We show that both genes are upregulated in a Notch-dependent manner during early T-cell development and that HES1 acts as a repressor of differentiation by maintaining a quiescent stem cell signature in CD34+ hematopoietic progenitor cells. While HES4 can also inhibit natural killer and myeloid cell development like HES1, it acts differently on the T- versus B-cell lineage choice. Surprisingly, HES4 is incapable of repressing B-cell development, the most sensitive hematopoietic lineage with respect to Notch-mediated repression. In contrast to HES1, HES4 promotes initiation of early T-cell development, but ectopic expression of HES4, or HES1 and HES4 combined, is insufficient to induce T-lineage differentiation. Importantly, knockdown of HES1 or HES4 significantly reduces human T-cell development. Overall, we show that the Notch1 target genes HES1 and HES4 have non-redundant roles during early human T-cell development which may relate to differences in mediating Notch-dependent human hematopoietic lineage decisions.
Introduction
Multipotent hematopoietic stem cells (HSC) reside in the bone marrow after birth and give rise to all the different blood cell types.1 This process is orchestrated by integrating cell-intrinsic mechanisms and external stimuli which results in the activation of molecular networks that drive differentiation towards specific hematopoietic lineages.2,3 Such events are also important during the early stages of intrathymic T-cell development. Following colonization of thymus-seeding progenitors from the bone marrow, Delta-like-ligand-4 (DLL4), which is expressed by thymic epithelial cells, triggers the Notch1 receptor on these multipotent precursors. Activation of Notch1 induces T-cell differentiation through upregulation of T-lineage genes and this also prevents differentiation towards other hematopoietic lineages.4-6 In mice, the regulatory network that drives early T-cell development has been well-studied. Activation of TCF1 contributes to the induction of T-lineage genes,7 while Gata3 upregulation is important to impair B-lineage potential.8 Subsequently, residual natural killer (NK) cell potential is repressed by BCL11B that completes T-cell commitment.9 During these steps, Notch activation remains essential to guide the cells along the T-lineage differentiation pathway and for proliferation and survival. In human, however, the regulatory network that supports these processes is less clear. Due to species-specific gene expression patterns and functions, extrapolation of murine data is often not possible. One important difference between human and mouse is the need for Notch signaling during T-cell development. In both, strong Notch1 activation is necessary to induce T-cell specification at the expense of B- and myeloid lineage potential.10 In human, however, Notch signaling then needs to be reduced for efficient differentiation towards committed thymocytes,11 while in mouse high Notch activity is maintained.12 Thereafter, Notch also differentially modulates the differentiation towards T-cell receptor (TCR)-and TCR-T-cells in mouse versus human.13-15
Activation of Notch1 results in the expression of many different target genes whose individual roles in controlling hematopoietic lineage decisions are still unclear. These include some members of the HES gene family that encode bHLH transcription factors that function as homoand heterodimers and repress their target genes through recruitment of co-repressors.16 HES1 is a well-known target and is essential for the inhibition of myeloid development during early T-lineage differentiation in mouse through repression of C/EBP-17 It also plays a prominent role in Notch-driven T-cell acute lymphoblastic leukemia (T-ALL).18 Importantly, the human genome also encodes HES4, a HES1 paralogue that is absent in the mouse19 (Online Supplementary Figure S1), but present in Xenopus and Zebrafish. It has been documented that, depending on the species, biological setting and tissue, HES4 can have overlapping or antagonistic roles compared to HES1.20-24 Although HES4 displays clear Notch-dependent regulation in Notch-driven cancers,25 it is unclear if HES4 can mediate non-malignant Notch-driven hematopoietic lineage decisions.
Given that the individual roles of the various Notch target genes are still unclear, we studied the function of HES1 and HES4 during the early Notch-dependent hematopoietic lineage decisions in human. We show that they have both different and similar impacts on the differentiation of human hematopoietic progenitor cells (HPC) towards various blood cell types when overexpressed, and that both are essential during early human T-cell development.
Methods
Isolation of CD34+ hematopoietic precursor cells
Umbilical cord blood (CB) was obtained with informed consent and used with approval of the Medical Ethical Commission of Ghent University Hospital (Belgium). Mononuclear cells were isolated through Lymphoprep density-gradient (ELITech) and CD34+ HPC were purified using CD34-magnetic beads (Miltenyi Biotec).
Other methods are available in the Online Supplementary Material and Methods.
Results
HES4, like HES1, displays Notch-dependent expression and regulation during human T-lineage differentiation
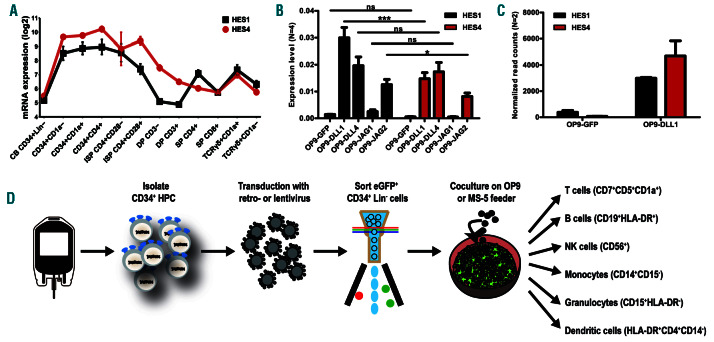
HES4 was shown to be Notch dependent in leukemic T cells,25 but whether HES4 displays Notch-dependent expression during non-malignant T-cell development is unclear. Gene expression analysis showed that both HES1 and HES4 are expressed during similar Notch-dependent stages4,14 of human T-cell development (Figure 1A). This Notch-dependent HES4 expression was functionally validated through exposure of CB CD34+Lin– HPC onto OP9 stromal cells that express different Notch ligands. HES1 and HES4 expression in HPC was only induced in conditions that permit T-lineage differentiation (OP9-DLL1, OP9-DLL4 and OP9-Jagged2 (JAG2)), but not in B-lineage permissive conditions (OP9-GFP and OP9-JAG1; Figure 1B).26 Similarly, short 48 hour (h) co-culture experiments onto OP9-GFP versus OP9-DLL1 stromal cells confirmed the Notch-dependent expression of both HES genes in intrathymic CD34+ cells, cells that have already experienced in vivo Notch activation (Figure 1C). Publicly available data revealed that, in contrast to HES1, HES4 is not expressed in HSC in the bone marrow (Online Supplementary Figure S2A). However, HES4 expression is detected in monocytes while lymphocytes and dendritic cells (DC) express lower levels. Protein staining confirmed the higher HES4 levels in monocytes compared to lymphocytes (Online Supplementary Figure S2B). These findings illustrate that both HES1 and HES4 are Notch-dependently induced during human T-cell development. Given that no information is available on the role of HES4 in human hematopoiesis, we studied this using viral gene perturbation approaches in human CD34+ HPC that are differentiated towards various hematopoietic lineages in wellestablished in vitro co-culture systems (Figure 1D).
HES4 lacks the potential to repress B-cell development
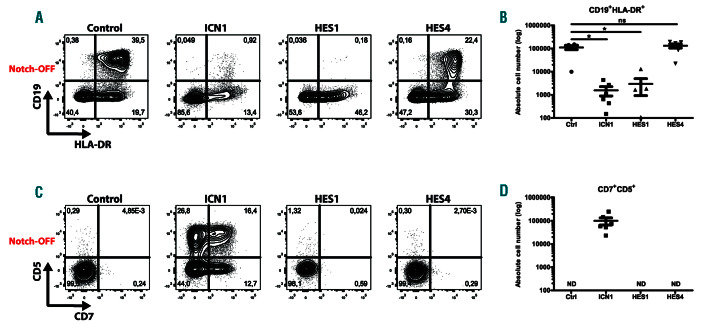
DLL4-dependent Notch1 activation is essential to induce T-lineage differentiation at the expense of B-cell development.6,13,27 Since both HES1 and HES4 are induced following Notch1 activation, we investigated if these genes have the potential to repress B-cell development through enforced expression in CB HPC. This resulted in 50-fold higher expression levels of each gene compared to ex vivo isolated CD34+ thymocytes that reflect physiological Notch activation (Online Supplementary Figure S3A), and protein staining suggests that slightly higher levels of HES4 were obtained compared to HES1 (Online Supplementary Figure S3B). HES1- and HES4-transduced HPC were cultured on MS-5 stromal cells in B-lineage differentiation conditions and compared to HPC transduced with the constitutive active form of Notch1 (ICN1) that mimics Notch activation and represses B-cell development. 13,27 Whereas control transduced HPC efficiently differentiate into CD19+HLA-DR+ B-cells, enforced ICN1 expression inhibited B-cell development (Figure 2A-B). Remarkably, while HES1 also inhibited B-lineage differentiation, continuous HES4 expression did not impact B-cell development (Figure 2A-B), and combined overexpression of HES1 and HES4 showed a dominant repression of HES1 (Online Supplementary Figure S4A-B). Consistently, CD7+CD5+ T-cell precursors were generated in cultures where ICN1 was overexpressed, but not within control transduced conditions (Figure 2C-D). In contrast, enforced expression of either HES1 or HES4 (Figure 2C-D), or HES1 and HES4 combined (Online Supplementary Figure S4C-D), was insufficient to induce the development of CD7+CD5+ T-lineage progenitors. These findings show that HES1 and HES4 have distinct capacities to mediate the Notch1dependent T- versus B-cell lineage decision and that both HES proteins, individually or combined, are insufficient to impose T-cell fate.
Figure 1.
HES4, like HES1, is expressed Notch dependently during human T-cell differentiation. (A) Line plots show log2 normalized probe intensities for HES1 (black) and HES4 (red) gene expression levels during normal T-cell development. Data shows the average expression of two independent experiments from two different donors. Error bars indicate the standard error of the mean (SEM). (B) Quantitative RT-PCR of HES1 (black bars) and HES4 (red bars) in CB CD34+Lin- precursors cultured for 3 days on OP9-GFP, OP9-DLL1, OP9-DLL4, OP9-JAG1 and OP9-JAG2. Data shows the average expression of four independent experiments, relative to the mean of GAPDH and YHWAZ mRNA levels. Error bars indicate SEM. ***P<0.001; *P<0.05 (two-way ANOVA); ns: not significant. (C) DESeq2-normalized read counts of HES1 (black bars) and HES4 (red bars) in thymic CD34+ cells cultured for 2 days on OP9-GFP and OP9-DLL1. Graphs show the average of two independent experiments and error bars indicate SEM. (D) Schematic overview illustrating the experimental workflow and the differentiation markers used to identify the different hematopoietic lineages.
HES4, but not HES1, promotes induction of T-lineage differentiation
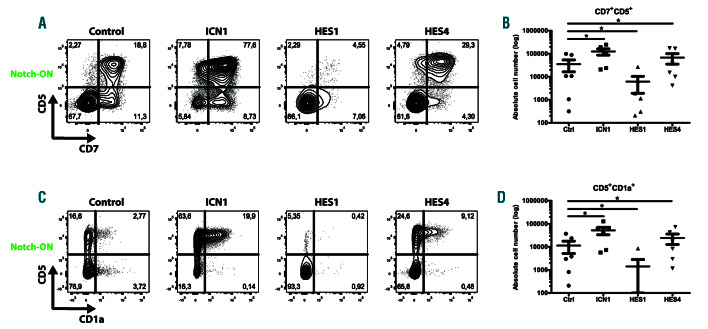
To further study the impact of HES1 and HES4 on early T-cell development, we performed perturbation experiments in OP9-DLL4 co-cultures that support induction of T-lineage differentiation in human HPC as illustrated through the emergence of CD7+CD5+ T-cell precursors in control transduced cells (Figure 3A). We analyzed these cultures after 2 weeks to focus on the efficiency and kinetics of induction of T-lineage specification and commitment. Ectopic expression of ICN1 promoted the generation of early CD7+CD5+ and CD5+CD1a+ T-cell precursors from HPC (Figure 3A, C) and yielded higher numbers of these cells (Figure 3B, D). Enforced expression of HES1, however, resulted in a significant reduction in the frequency and absolute number of CD7+CD5+ specified and CD5+CD1a+ committed T-cells (Figure 3A-D). In contrast, high levels of HES4 improved the differentiation of HPC towards the T-cell lineage, although less efficient compared to ICN1 overexpression (Figure 3A-D). This positive role of HES4 on early T-lineage differentiation was counteracted by HES1 when both were overexpressed (Online Supplementary Figure S5A-B). Because previous work from our lab demonstrated that JAG2, DLL1 and DLL4, but not JAG1, have the potential to initiate T-cell development in human HPC,26 we investigated if enforced HES4 expression could alter these Notch ligand requirements to induce T-lineage differentiation. Whereas overexpression of ICN1 in HPC was sufficient to induce T-cell development in OP9-GFP, OP9-JAG1 and OP9-DLL4 co-cultures, HES4- transduced cells, like the control, could only generate CD7+CD5+ T-cell progenitors on OP9-DLL4 stromal cells (Online Supplementary Figure S6A-D). Overall, these findings illustrate that, while continuous HES1 expression inhibits T-lineage specification and commitment, HES4 can enhance induction of T-cell development in the presence of Notch signaling when present at higher levels compared to endogenous HES1 expression. However, HES4 is not sufficient to alter the Notch ligand requirements to induce T-lineage differentiation in HPC.
NK-cell development is severely hampered by HES1 and HES4
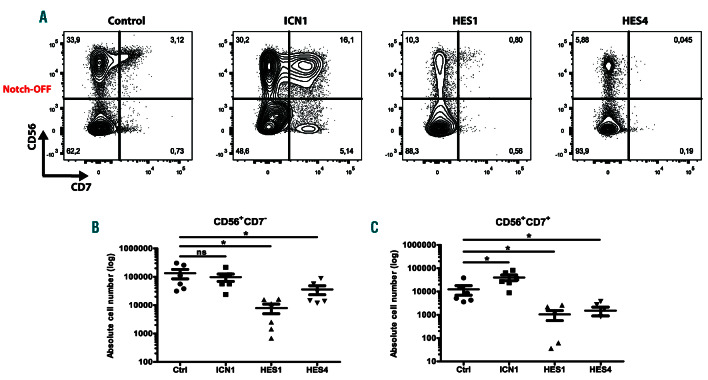
In human, Notch1 activation is permissive for NK-lineage differentiation,28 and our laboratory previously illustrated that this is mediated by the Notch1 target gene DTX1.11 Here, we studied the impact of ectopic HES1 and HES4 expression on NK-cell development through co-culture of control, ICN1-, HES1- and HES4-transduced HPC on MS-5 stromal cells with cytokines supporting NK-cell differentiation. As documented previously,28 enforced ICN1 expression enhanced NK-cell development and resulted in a significant increase in CD56+ NK-cells that express CD7, a Notch-dependent marker during early T-cell development (Figure 4A, C). In contrast, both HES1 and HES4 reduced NK-cell development, and this was the case for both CD56+CD7– and CD56+CD7+ subsets (Figure 4A-C). To investigate if HES4 overexpression can improve the generation of CD56+CD7+ NK-cells in the presence of Notch signaling, analogous to the Notch signaling requirement for HES4 to enhance differentiation towards CD7+ T-lineage precursors, we cultured control and HES4-transduced HPC on MS5-DLL4 stromal cells in the presence of NK-lineage supporting cytokines. However, also in these conditions, continuous HES4 expression impaired the development of CD56+CD7+ NK-cells (Online Supplementary Figure S7A-B). These findings illustrate that both HES1 and HES4 can repress NK-cell development.
Figure 2.
HES4 is not sufficient to inhibit B-cell development. Flow cytometric analysis (A, C) and absolute cell numbers (B, D) of control, ICN1-, HES1- and HES4- transduced CD34+Lin– CB progenitors cultured on the MS-5 feeder (Notch-OFF) for 3 weeks in the presence of the B-lineage supporting cytokines IL-7 and SCF, showing the development of CD19+HLA-DR+ B-lineage cells (A-B) and CD7+CD5+ T-lineage cells (C-D). Data shows the average of six independent experiments and error bars indicate SEM. *P<0.05 (non-parametric paired Wilcoxon test); ns: not significant; ND: not detectable. Flow cytometry plots shown are representative for six independent experiments.
HES1 and HES4 inhibit the development of myeloid cells
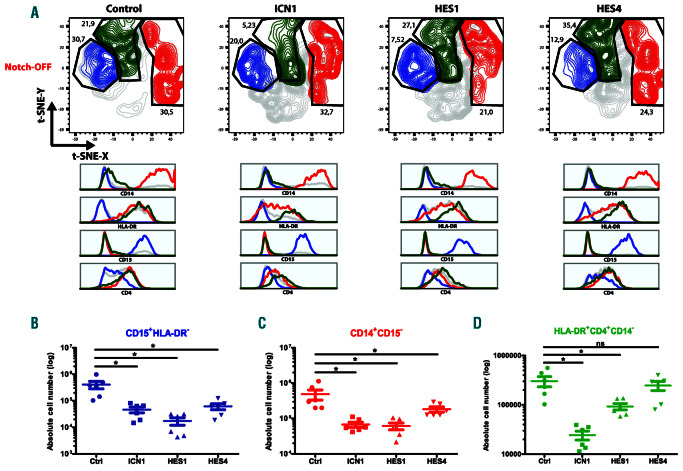
Notch1-mediated repression of myeloid lineage differentiation is critical following thymic colonization of HPC4,6 and genetic experiments in mice revealed an important role for HES1 in this process.17 To investigate if HES1 and HES4 can mediate repression of human myeloid development downstream of Notch1, we cultured control, ICN1-, HES1- and HES4-transduced HPC in conditions that support myeloid differentiation. Consistent with earlier work, enforced expression of ICN1 reduced myeloid development.13,27,29 While this was mainly reflected by a decrease in the absolute cell numbers for CD15+HLA-DR– granulocytes and CD14+CD15– monocytes, for HLADR+ CD4+CD14–DC both the frequency and absolute cell numbers were decreased (Figure 5A-D). In case of continuous HES1 and HES4 expression, however, we observed a reduction in both the percentage and absolute cell number of granulocytes (Figure 5A-B) and monocytes (Figure 5A, C). In contrast, while the absolute numbers of DC was decreased by HES1 overexpression, this was not the case upon ectopic expression of HES4. Furthermore, the frequencies of DC were increased in these cultures (Figure 5A, D). Thus, both HES1 and HES4 have the potential to repress monocyte and granulocyte differentiation downstream of Notch1, whereas DC development is less impaired, consistent with the intrathymic development of these antigen-presenting cells.
HES1, but not HES4, maintains a quiescent stem cell signature in CD34+ cells
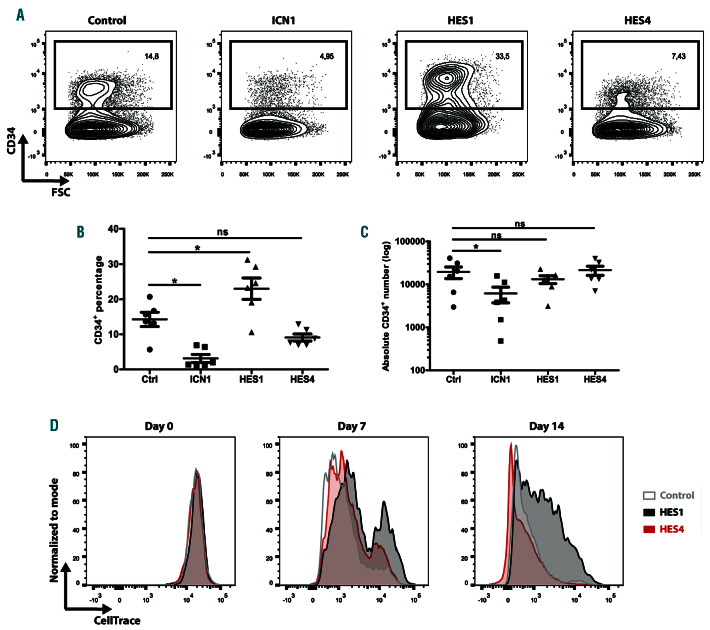
Enforced expression of HES1 can maintain a stem cell signature in multiple cell types.16,30,31 To compare the effect of HES1 and HES4 on HSC maintenance, we tracked the frequency and number of CD34+ HPC, following ICN1, HES1 or HES4 transduction, in OP9-DLL4 T-lineage differentiation conditions. In agreement with previous studies,13,27 ectopic expression of ICN1 induced T-cell development and resulted in a significant decrease in the percentage and absolute number of CD34+ HPC (Figure 6A-C). Although conditions with elevated HES1 levels did not yield higher numbers of CD34+ cells (Figure 6C), we observed a significant and more than 2-fold increase in their frequency (Figure 6A-B). In contrast, enforced expression of HES4 did not affect the fraction of CD34+ HPC (Figure 6A-C). In addition, since earlier work illustrated that overexpression of HES1 results in the generation of quiescent HSC,18 we evaluated cellular proliferation. While HPC that express higher levels of HES1 displayed reduced proliferation, HES4-transduced cells proliferated similarly as the control (Figure 6D). These findings suggest that HES1, but not HES4, can induce maintenance of a quiescent stem cell state by repressing differentiation and proliferation of CD34+ HPC.
RNA sequencing reveals HES1- and HES4-induced molecular pathways during early human hematopoiesis
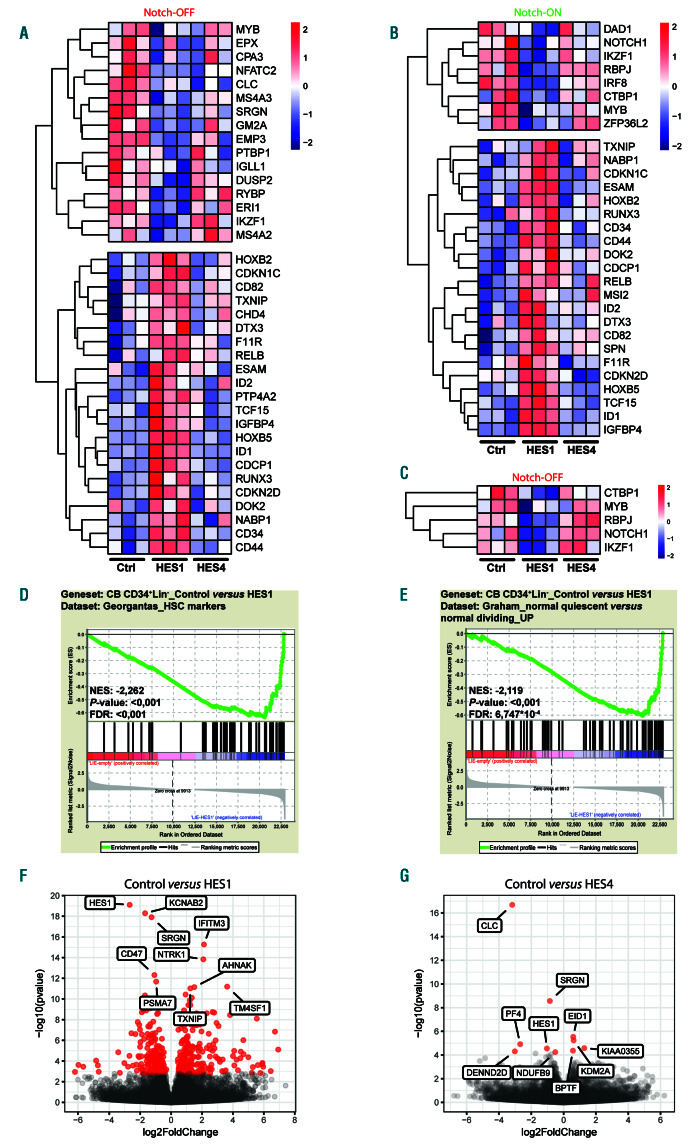
To reveal specific HES1- and HES4-induced molecular changes, we performed RNA sequencing in control, HES1- and HES4-transduced CD34+Lin– HPC that were cultured in the absence or presence of Notch signaling on OP9-GFP or OP9-DLL1, respectively. Enforced expression of HES1 in the absence of Notch1 activation (OP9-GFP, Notch-OFF, Figure 7A) resulted in a significant downregulation of genes associated with the differentiation of B-cells (MYB,32 GM2A,33 DUSP233 and IGLL134), NK-cells (ERI1,35 EMP336 and SRGN37) and myeloid cells (EPX,38 CLC,39 SRGN,37 MS4A240 and MS4A341) (Figure 7A). CEBPA expression was not detected in these experiments. However, given the essential role of HES1 in repression of CEBPA-dependent myeloid differentiation in mouse,17 we evaluated the impact of HES1 and HES4 on CEBPA expression in the HL-60 leukemic cell line and, consistently, HES1 was very potent in repressing CEBPA expression, while HES4 was less efficient (Online Supplementary Figure S8A). In OP9- DLL1-cultured CD34+ HPC (Notch-ON, Figure 7B), we observed significant repression of Notch signaling and Tlineage associated genes by HES1, including NOTCH1, RBPJ, MYB42 and DAD143 (Figure 7B). Surprisingly, on OP9-GFP stromal cells where no or little Notch1 triggering occurs, we also observed downregulation of these genes upon HES1 overexpression (Figure 7C). Also IRF8 expression, which is induced by Notch signaling for DC deve-lopment,44 was repressed by HES1 (Figure 7B). On both OP9 stromal co-cultures, expression of IKZF1, which is essential for differentiation of lymphoid-primed multipotent progenitors into common lymphoid progenitors (CLP),45 was downregulated by high levels of HES1 (Figure 7A-B). In contrast, HES1 overexpression showed significant upregulation of genes associated with stem cell maintenance, such as CD34, CD44, HOXB246, HOXB546, ESAM47 and ID148, and genes involved in cell cycle arrest, including IGFBP4, CDKN2D and CDKN1C, which was independent of Notch signaling (Figure 7A-B). These findings were confirmed by gene set enrichment analysis (GSEA) that revealed significant enrichment of HSC markers and genes involved in CD34+ quiescence in the HES1-transduced condition (Figure 7D-E). Genes associated with the maintenance of multipotent HSC at the expense of both CLP and granulocyte-monocyte progenitor (GMP) differentiation were also enriched upon HES1 overexpression (Online Supplementary Figure S8BC). In contrast, HES4 overexpression did not result in large gene expression changes. Among the significantly downregulated genes, we found CLC and SRGN (Figure 7A, G), which were previously associated with respectively myeloid and NK-cell development.37,39 Moreover, upon HES4 overexpression, we observed a slight, but non-significant upregulation of NOTCH1, RBPJ and IKZF1 in the absence of Notch signaling (Figure 7C), while this was not the case when the cells were cultured on the OP9-DLL1 feeder (Figure 7B). Furthermore, high levels of HES1 or HES4 resulted in a significant downregulation of HES1 itself (Figure 7F-G). Comparison of the promotor sequences of the putative HES1 and HES4 target genes did not reveal a difference in the relative abundance of E-box, N-box and C-site motifs between HES1 and HES4 targets (Online Supplementary Figure S9A). However, using HOMER motif analysis, we observed a difference in the top enriched transcription factor binding sites within HES1 and HES4 target gene promoters (Online Supplementary Figure S9B-C). Thus, using RNA sequencing, we identified potential downstream targets of HES1 and HES4 that may modulate Notch-driven lineage decisions during human hematopoiesis and this may involve co-regulation by specific transcription factors.
Figure 3.
HES4, but not HES1, promotes induction of T-lineage differentiation. Flow cytometry analysis (A, C) and absolute cell numbers (B, D) of control, ICN1-, HES1- and HES4-transduced CD34+ Lin- CB precursors cultured on the OP9-DLL4 feeder (Notch-ON) for 2 weeks in the presence of the T-lineage specific cytokines IL-7, SCF and FLT3L, showing the development of CD7+ CD5+ (A-B) and CD5+ CD1a+ (C-D) T-lineage cells. Data shows the average of six independent experiments and error bars indicate the standard error of the mean (SEM). *P<0.05 (non-parametric paired Wilcoxon test). Flow cytometry plots shown are representative for six independent experiments.
HES1 and HES4 are both essential for inducing human T-cell development in HPC
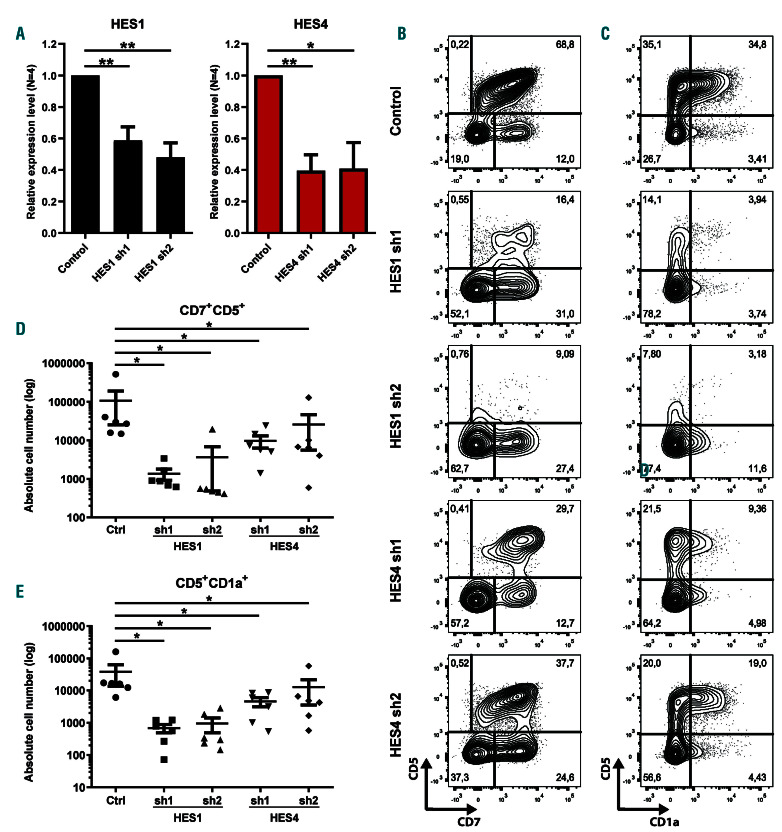
To investigate if HES1 and HES4 are required downstream of Notch during early human T-lineage differentiation, we performed HES1 and HES4 short hairpin RNA (shRNA)-mediated knockdown experiments in HPC that were cultured on OP9-DLL4 in T-lineage supporting conditions. For both genes, we used two independent shRNA which induced 40-60% knockdown (Figure 8A). Compared to the control, both the frequency and absolute numbers of CD7+CD5+ T-lineage specified and CD5+CD1a+ committed T-cell progenitors were significantly reduced upon knockdown of either HES1 or HES4 (Figure 8B-E). However, we did not observe a diversion to the B- or myeloid lineage in these cultures (data not shown), probably as a result of insufficient knockdown. Nevertheless, these findings illustrate that both HES1 and HES4 have essential and non-redundant roles during early human T-cell development.
Figure 4.
HES1 and HES4 repress NK cell development. (A) Flow cytometric analysis of control, ICN1-, HES1- and HES4-transduced CB CD34+Lin– precursors cultured on the MS-5 feeder (Notch-OFF) for 3 weeks in the presence of the NK-lineage specific cytokines IL-7, SCF, FLT3L and IL-15, showing the development of CD56+CD7- and CD56+CD7+ NK cells. Plots shown are representative for six independent experiments. (B-C) Absolute numbers of CD56+CD7– (B) and CD56+CD7+ (C) NK cells generated in corresponding cultures shown in (A). Data shows the average of six independent experiments. Error bars indicate the standard error of the mean (SEM). *P<0.05 (non-parametric paired Wilcoxon test); ns: not significant.
Discussion
It is well established that Notch1 activation in thymusseeding precursors allows progenitor cells to differentiate towards the T-cell lineage, while development towards other hematopoietic cell types is repressed.4-6,13 However, the individual roles of various Notch1 target genes during these developmental choices are still unknown. Therefore, we investigated the potential of HES1 and HES4 to mediate human Notch1-dependent hematopoietic lineage decisions. We illustrate that both genes are essential downstream of Notch1 to induce human T-cell development and that they have both similar and different potentials to repress the differentiation towards alternative, non T-cell lineages.
In murine hematopoiesis, HES1 is a repressor of intrathymic myeloid differentiation during the Notchdependent induction of T-cell development.17 HES1 is also required for sustaining leukemic growth in Notch-driven T-ALL, both in mouse and human.18,49 Previous overexpression studies in human have shown that HES1 inhibits B- and NK-lineage differentiation, but failed to reveal a block in monocyte development.27 In agreement with previous work in mice,17 we show here that HES1 can repress myeloid development in human and that this is mediated through repression of several genes with known roles in monocyte, granulocyte and dendritic cell development, thereby providing novel molecular insights in this process. The discrepancy in monocyte differentiation compared to previous work27 may relate to differences in expression levels following transduction since we used a codon-optimized construct to facilitate comparison with HES4. This could also explain the more efficient block in B-cell development in our results.
Our novel RNA sequencing data reveal mechanisms through which HES1 specifically represses T-, B- and NKassociated genes, thereby validating the developmental impacts in our co-cultures. In light of the requirement for HES1 downstream of Notch1 during T-cell development, the repression of T-lineage differentiation by HES1 is striking. While this may result from the high expression levels in our perturbation settings, this may also reflect stage-specific requirements for HES1 in this process that are recurrent and may involve HES1 oscillation as observed during embryonic development.16 Although it is unclear if the latter occurs in developing thymocytes, Notch activation is required at different stages of early T-cell development and recurrent Notch activation events therein seem essential to balance between proliferation and differentiation.4,50 While HES1 is required to repress myeloid development in early T-cell precursors, it could later be essential to maintain thymocytes in a quiescent state, which is necessary during TCR rearrangements.4 Although less evident in human, this fits the high expression level of Hes1 in DN3a thymocytes that are active TCR-rearranging cells.51 A role for HES1 as a regulator of cell quiescence also concurs with the observation that continuous expression in HPC maintains cells in a CD34+ precursor state, consistent with previous studies.18,27 Here, we show that this is correlated with the upregulation of genes involved in stem cell maintenance, quiescence and cell cycle arrest. However, while HES1 is dispensable in mice for HSC maintenance,18 our data suggests that HES1 represses differentiation in human by maintaining a quiescent stem cell signature.
Figure 5.
Myeloid differentiation is hampered by HES1 and HES4. (A) Flow cytometry analysis of control, ICN1-, HES1- and HES4-transduced CB CD34+Lin- progenitors cultured on the MS-5 feeder (Notch-OFF) for 1 week in the presence of the myeloid-lineage specific cytokines SCF, FLT3L, TPO, GM-CSF and G-CSF, showing the development of CD15+HLA-DR– granulocytes (blue), CD14+CD15– monocytes (red) and HLA-DR+CD4+CD14– dendritic cells (green). t-SNE plots shown are representative of six independent experiments. (B-D) Absolute numbers of CD15+HLA-DR- granulocytes (B), CD14+CD15– monocytes (C) and HLA-DR+CD4+CD14– dendritic cells (D) generated in corresponding cultures shown in (A). Data shows the average of six independent experiments and error bars indicate the standard error of the mean (SEM). *P<0.05 (non-parametric paired Wilcoxon test); ns: not significant.
For the first time, we study the role of HES4 as a downstream modulator of Notch during non-malignant human hematopoiesis. In agreement with previous studies that compared the function of HES1 and HES4 in brain and bone development,20,21 our data suggests that both HES proteins have similar, and also different roles during human Notch-dependent hematopoietic lineage decisions. Compared to HES1, which is a strong repressor with a clear impact on HPC differentiation, enforced HES4 expression has a similar impact on myeloid and NK-cell development, but not on B- and T-cell differentiation. We believe that this lower repressive capacity of HES4 compared to HES1 is not due to lower expression levels following transduction since we obtained a similar mRNA increase for both HES genes, and protein staining even suggested higher amounts of HES4 protein compared to HES1. These different repressive capacities may, however, reflect a more stringent role for HES genes in the repression of myeloid fate compared to B-lineage differentiation, which is in agreement with a genetic study in mice that showed that intrathymic Notch signaling is more important for the repression of myeloid development than for preventing B-lineage diversion.52 One exception to the robust repression of myeloid development by HES genes concerns their impact on DC development. Although HES1 overexpression did result in a significant reduction in the absolute number of DC, their frequency was increased compared to control transduced cells. Strikingly, HES4 was not sufficient to robustly repress DC development and this higher tolerance towards HES genes is in agreement with the intrathymic and Notch-dependent development of DC.53 Interestingly, in contrast to HES1, HES4 did promote T-cell development in the presence of Notch1 signaling. This places HES4 in line with other Notch targets, such as IL7R and TCF7, that have clear stimulating roles on T-cell development following Notch1 activation.11,54 However, HES4 by itself, or in combination with HES1, is insufficient to induce T-cell development,26 indicating that other downstream targets, or combinations thereof, are important for driving this process. Nevertheless, the clear distinct roles of HES1 and HES4 on B- and T-cell development may be important to control the initial Notch-dependent B- versus T-cell lineage decision in which HES1 represses the B-cell lineage while HES4 permits further T-cell development. Following this initial bifurcation, both HES1 and HES4 may then be required to repress NK-lineage differentiation while initially still permitting DC development. As such, HES1 and HES4 may have unique roles during these early stages of human T-cell differentiation, which are not-redundant as illustrated through our loss-of-function studies. Unfortunately, these knockdown studies did not result in an increased development of other, non T-cell lineages, but we believe this may result from the incomplete loss of HES1 and HES4 expression in these experiments.
In contrast to HES1, HES4 overexpression did not result in large gene expression changes. This might reflect the immature state of the transduced HPC since gene expression profiling was performed 48 h after transduction in an effort to discover direct regulatory events. However, the lack of HES4 expression in HSC suggests a minimal role for this protein at this immature stage, and the precise role for HES4 in gene regulation may therefore only be revealed at later differentiation stages, such as following Notch1 activation in immature thymocytes. Other genome-wide approaches, such as ChIP-seq in HES4-expressing thymocytes, will be required to identify other HES4 target genes. Remarkably, while we did not observe clear differences in the relative abundance of specific HES binding motifs (E-box, N-box and C-site) in the promotors of the predicted HES1 and HES4 target genes, both HES proteins may differentially regulate downstream target gene expression through interactions with other transcription factors that are distinct for HES1 and HES4. Such mechanism could explain their different impact on hematopoietic lineage decisions. Intriguingly, we identified HES1 as a downstream target of both HES1 and HES4, thereby confirming the autoregulation of HES116 and suggesting a novel feedback loop between HES4 and HES1 that may also contribute to a possible transient repression of HES1, thereby allowing early thymocytes to escape from the repressive effects of HES1 to support their further differentiation. Such a direct repressive role of HES4 on HES1 expression seems essential given that protein interactions through the formation of HES1-HES4 heterodimers seemingly fail to counteract the strong repressive activity of HES1 homodimers. We anticipate that single cell (sc)RNA sequencing will provide more detailed insights into the actual sequence of HES1 and HES4 expression during early human T-cell development.
Figure 6.
HES1, but not HES4, induces stem cell maintenance and quiescence in CD34+ cells. (A) Flow cytometric analysis of control, ICN1-, HES1- and HES4-transduced CB CD34+Lin– precursors cultured on the OP9-DLL4 feeder for 2 weeks in the presence of the T-lineage specific cytokines IL-7, SCF and FLT3L, showing the maintenance of CD34+ HPC. Plots shown are representative of six independent experiments. (B-C) Frequency (B) and absolute numbers (C) of CD34+ cells maintained in corresponding cultures shown in (A). Data shows the average of six independent experiments. Error bars indicate the standard error of the mean (SEM). *P<0.05 (non-parametric paired Wilcoxon test); ns: not significant. (D) Flow cytometry analysis of control (grey), HES1 (black)- and HES4 (red)-transduced CB CD34+Lin– HPC cultured on the OP9-DLL4 feeder for 2 weeks in the presence of the T-lineage supporting cytokines IL-7, SCF and FLT3L, showing proliferation as measured by the dilution of the CellTrace Violet dye at indicated time points.
Figure 7.
Gene expression analysis of HES1 and HES4 during human hematopoiesis. (A) Heat map representation of differentially expressed genes in control versus HES1-, or HES4-transduced CB CD34+Lin– cells cultured for 1 day on OP9-GFP (Notch-OFF), highlighting genes involved in the differentiation towards the B-, natural killer (NK)- and myeloid lineage (top) and HSC maintenance and quiescence (bottom). (B) Heat map representation of differentially expressed genes in control versus HES1-, or HES4-transduced CB CD34+Lin– progenitors cultured for 1 day on OP9-DLL1 (Notch-ON), highlighting genes involved in the differentiation towards the T-lineage (top) and HSC maintenance and quiescence (bottom). (C) Heat map representation of differentially expressed genes in control versus HES1-, or HES4-transduced CB CD34+Lin– cells cultured for 1 day on OP9-GFP (Notch-OFF), highlighting genes associated with Notch signaling and T-cell development. (D-E) GSEA shows a significant enrichment of HSC markers (D) and genes involved in quiescence (E) in the HES1-transduced condition compared to control. (F-G) Volcano plot representation of control versus HES1 (F)-, or HES4 (G)-transduced CB CD34+Lin– precursors cultured for 1 day on OP9-GFP, highlighting the expression of the top 10 differentially expressed genes. Red dots indicate significance (padj<0.05; n=3).
Figure 8.
Non-redundant roles for HES1 and HES4 during early human T-cell development. (A) Quantitative RT-PCR of HES1 (black bars) and HES4 (red bars) in CB CD34+Lin– cells transduced with a control or two different HES1 or two different HES4 single hairpin RNA (shRNA), respectively, and cultured for 3 days on OP9-DLL4 to induce Notch activation. Data shows the average expression of four independent experiments, relative to the mean of ACTB and GAPDH mRNA levels, and normalized to expression in the control shRNA condition. Error bars indicate the standard error of the mean (SEM). **P<0.01; *P<0.05 (paired Student’s t-test). (B-C) Flow cytometry analysis of control, HES1 and HES4 shRNA-transduced CD34+Lin– hematopoietic progenitor cells (HPC) cultured on the OP9-DLL4 stromal cells for 2 weeks in the presence of the T-lineage supporting cytokines IL-7, SCF and FLT3L, showing the development of CD7+CD5+ (B) and CD5+CD1a+ (C) T-cell precursors. Contour plots shown are representative for six independent experiments. (D-E) Absolute numbers of CD7+CD5+ T-lineage specified (D) and CD5+CD1a+ T-lineage committed (E) progenitors generated in corresponding cultures shown in (B) and (C), respectively. Data shows the average of six independent experiments and error bars indicate the standard error of the mean (SEM). *P<0.05 (non-parametric paired Wilcoxon test).
Given that HES4 is not present in the mouse genome, this work thus improves our understanding of the mechanisms through which Notch activation controls human blood cell development. This is an important issue since we have previously documented that Notch activation has different impacts on T-cell development in human versus mouse.4,14,55 Our work shows that both HES1 and HES4 are important to support the Notch1-dependent induction of human Tcell development and that this may be regulated through both different and similar repressive mechanism on other hematopoietic lineages. These findings thus underscore the distinct regulatory functions of each member of the complex network of Notch1 downstream target genes.
Supplementary Material
Acknowledgments
The authors thank Juan-Carlos Zuniga-Pflucker (University of Toronto) for OP9 stromal cells, Dr. Francois and Dr. Van Nooten (Department of Cardiac Surgery; University Hospital Ghent) for thymus tissue, Dr. Conny Matthys (Navelstrengbloedbank; University Hospital Ghent) for cord blood samples and Karin Weening (University of Ghent) for assistance with vector cloning.
Funding Statement
Funding: This work was supported by grants of the Fund for Scientific Research Flanders (FWO), the Concerted Research Action of Ghent University (GOA), the Foundation against Cancer (STK) and the Interuniversity Attraction Poles Program (IUAP) from the Belgian Science Policy.
References
- 1.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120-136. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Emerson SG. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene. 2002;21(21):3295-3313. [DOI] [PubMed] [Google Scholar]
- 3.Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taghon T, Waegemans E, Van de Walle I. Notch signaling during human T cell development. Curr Top Microbiol Immunol. 2012;360:75-97. [DOI] [PubMed] [Google Scholar]
- 5.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8(1):9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Smedt M, Hoebeke I, Reynvoet K, Leclercq G, Plum J. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or Tcell lineage in thymus microenvironment. Blood. 2005;106(10):3498-3506. [DOI] [PubMed] [Google Scholar]
- 7.Weber BN, Chi AW, Chavez A, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011; 476(7358):63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Ojeda ME, Klein Wolterink RG, Lemaitre F, et al. GATA-3 promotes T-cell specification by repressing B-cell potential in pro-T cells in mice. Blood. 2013; 121(10):1749-1759. [DOI] [PubMed] [Google Scholar]
- 9.Hosokawa H, Romero-Wolf M, Yui MA, et al. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat Immunol. 2018;19(12): 1427-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waegemans E, Van de Walle I, De Medts J, et al. Notch3 activation is sufficient but not required for inducing human T-lineage specification. J Immunol. 2014;193(12):5997-6004. [DOI] [PubMed] [Google Scholar]
- 11.Van de Walle I, Dolens AC, Durinck K, et al. GATA3 induces human T-cell commitment by restraining Notch activity and repressing NK-cell fate. Nat Commun. 2016;7:11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch U, Fiorini E, Benedito R, et al. Deltalike 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205(11): 2515-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Smedt M, Reynvoet K, Kerre T, et al. Active form of Notch imposes T cell fate in human progenitor cells. J Immunol. 2002; 169(6):3021-3029. [DOI] [PubMed] [Google Scholar]
- 14.Van de Walle I, De Smet G, De Smedt M, et al. An early decrease in Notch activation is required for human TCR-alphabeta lineage differentiation at the expense of TCR-gammadelta T cells. Blood. 2009;113(13):2988-2998. [DOI] [PubMed] [Google Scholar]
- 15.Van de Walle I, Waegemans E, De Medts J, et al. Specific Notch receptor-ligand interactions control human TCR-alphabeta/gammadelta development by inducing differential Notch signal strength. J Exp Med. 2013; 210(4):683-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development. 2007;134(7):1243-1251. [DOI] [PubMed] [Google Scholar]
- 17.De Obaldia ME, Bell JJ, Wang X, et al. T cell development requires constraint of the myeloid regulator C/EBP-alpha by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14(12):1277-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wendorff AA, Koch U, Wunderlich FT, et al. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity. 2010;33(5):671-684. [DOI] [PubMed] [Google Scholar]
- 19.Zhou M, Yan J, Ma Z, et al. Comparative and evolutionary analysis of the HES/HEY gene family reveal exon/intron loss and teleost specific duplication events. PLoS One. 2012;7(7):e40649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai G, Cheung I, Shulha HP, et al. Epigenetic dysregulation of hairy and enhancer of split 4 (HES4) is associated with striatal degeneration in postmortem Huntington brains. Hum Mol Genet. 2015;24(5):1441-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McManus M, Kleinerman E, Yang Y, et al. Hes4: A potential prognostic biomarker for newly diagnosed patients with high-grade osteosarcoma. Pediat Blood Cancer. 2017; 64(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El Yakoubi W, Borday C, Hamdache J, et al. Hes4 controls proliferative properties of neural stem cells during retinal ontogenesis. Stem Cells. 2012;30(12):2784-2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murato Y, Hashimoto C. Xhairy2 functions in Xenopus lens development by regulating p27(xic1) expression. Dev Dyn. 2009; 238(9):2179-2192. [DOI] [PubMed] [Google Scholar]
- 24.Cakouros D, Isenmann S, Hemming SE, et al. Novel basic helix-loop-helix transcription factor hes4 antagonizes the function of twist-1 to regulate lineage commitment of bone marrow stromal/stem cells. Stem Cells Dev 2015;24(11):1297-1308. [DOI] [PubMed] [Google Scholar]
- 25.Stoeck A, Lejnine S, Truong A, et al. Discovery of biomarkers predictive of GSI response in triple-negative breast cancer and adenoid cystic carcinoma. Cancer Discov. 2014;4(10):1154-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van de Walle I, De Smet G, Gartner M, et al. Jagged2 acts as a Delta-like Notch ligand during early hematopoietic cell fate decisions. Blood. 2011;117(17):4449-4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoebeke I, De Smedt M, Van de Walle I, et al. Overexpression of HES-1 is not sufficient to impose T-cell differentiation on human hematopoietic stem cells. Blood. 2006;107(7):2879-2881. [DOI] [PubMed] [Google Scholar]
- 28.De Smedt M, Taghon T, Van de Walle I, De Smet G, Leclercq G, Plum J. Notch signaling induces cytoplasmic CD3 epsilon expression in human differentiating NK cells. Blood. 2007;110(7):2696-2703. [DOI] [PubMed] [Google Scholar]
- 29.Klinakis A, Lobry C, Abdel-Wahab O, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473(7346):230-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahmann I, Brohl D, Zyrianova T, et al. Oscillations of MyoD and Hes1 proteins regulate the maintenance of activated muscle stem cells. Genes Dev. 2019;33(9-10):524-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sueda R, Imayoshi I, Harima Y, Kageyama R. High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 2019;33(9-10): 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23(3):275-286. [DOI] [PubMed] [Google Scholar]
- 33.Young K, Borikar S, Bell R, Kuffler L, Philip V, Trowbridge JJ. Progressive alterations in multipotent hematopoietic progenitors underlie lymphoid cell loss in aging. J Exp Med. 2016;213(11):2259-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansson R, Zandi S, Anderson K, et al. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112(4):1048-1055. [DOI] [PubMed] [Google Scholar]
- 35.Thomas MF, Abdul-Wajid S, Panduro M, et al. Eri1 regulates microRNA homeostasis and mouse lymphocyte development and antiviral function. Blood. 2012;120(1):130-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crinier A, Milpied P, Escaliere B, et al. Highdimensional single-cell analysis identifies organ-specific signatures and conserved NK cell subsets in humans and mice. Immunity. 2018;49(5):971-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolset SO, Pejler G. Serglycin: a structural and functional chameleon with wide impact on immune cells. J Immunol. 2011;187(10): 4927-4933. [DOI] [PubMed] [Google Scholar]
- 38.Gorgens A, Radtke S, Mollmann M, et al. Revision of the human hematopoietic tree: granulocyte subtypes derive from distinct hematopoietic lineages. Cell Rep. 2013;3(5): 1539-1552. [DOI] [PubMed] [Google Scholar]
- 39.Grootens J, Ungerstedt JS, Nilsson G, Dahlin JS. Deciphering the differentiation trajectory from hematopoietic stem cells to mast cells. Blood Adv. 2018;2(17):2273-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng S, Papalexi E, Butler A, Stephenson W, Satija R. Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol Syst Biol. 2018; 14(3):e8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishibashi T, Yokota T, Satoh Y, et al. Identification of MS4A3 as a reliable marker for early myeloid differentiation in human hematopoiesis. Biochem Biophys Res Commun. 2018;495(3):2338-2343. [DOI] [PubMed] [Google Scholar]
- 42.Allen RD, 3rd, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13(9):1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong NA, Kabra NH, Hsieh SN, Cado D, Winoto A. In vivo overexpression of Dad1, the defender against apoptotic death-1, enhances T cell proliferation but does not protect against apoptosis. J Immunol. 1999; 163(4):1888-1893. [PubMed] [Google Scholar]
- 44.Lee J, Zhou YJ, Ma W, et al. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat Immunol. 2017;18(8):877-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dege C, Hagman J. Mi-2/NuRD chromatin remodeling complexes regulate B and Tlymphocyte development and function. Immunol Rev. 2014;261(1):126-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ali MAE, Fuse K, Tadokoro Y, et al. Functional dissection of hematopoietic stem cell populations with a stemness-monitoring system based on NS-GFP transgene expression. Sci Rep. 2017;7(1):11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yokota T, Oritani K, Butz S, et al. The endothelial antigen ESAM marks primitive hematopoietic progenitors throughout life in mice. Blood. 2009;113(13):2914-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perry SS, Zhao Y, Nie L, Cochrane SW, Huang Z, Sun XH. Id1, but not Id3, directs long-term repopulating hematopoietic stemcell maintenance. Blood. 2007;110(7):2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Espinosa L, Cathelin S, D'Altri T, et al. The Notch/Hes1 pathway sustains NF-kappaB activation through CYLD repression in T cell leukemia. Cancer Cell. 2010;18(3):268-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taghon T, Van de Walle I, De Smet G, et al. Notch signaling is required for proliferation but not for differentiation at a well-defined beta-selection checkpoint during human Tcell development. Blood. 2009;113(14):3254-3263. [DOI] [PubMed] [Google Scholar]
- 51.Wong GW, Knowles GC, Mak TW, Ferrando AA, Zuniga-Pflucker JC. HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRbeta-selected mouse thymocytes. Blood. 2012;120(7):1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feyerabend TB, Terszowski G, Tietz A, et al. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 2009;30(1):67-79. [DOI] [PubMed] [Google Scholar]
- 53.Martin-Gayo E, Gonzalez-Garcia S, Garcia-Leon MJ, et al. Spatially restricted JAG1- Notch signaling in human thymus provides suitable DC developmental niches. J Exp Med. 2017;214(11):3361-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez-Garcia S, Garcia-Peydro M, Alcain J, Toribio ML. Notch1 and IL-7 receptor signalling in early T-cell development and leukaemia. Curr Top Microbiol Immunol. 2012;360:47-73. [DOI] [PubMed] [Google Scholar]
- 55.Taghon T, Rothenberg EV. Molecular mechanisms that control mouse and human TCRalphabeta and TCR-gammadelta T cell development. Semin Immunopathol. 2008; 30(4):383-398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.