Abstract
Bone marrow failure (BMF) related to hypoplasia of hematopoietic elements in the bone marrow is a heterogeneous clinical entity with a broad differential diagnosis including both inherited and acquired causes. Accurate diagnostic categorization is critical to optimal patient care and detection of genomic variants in these patients may provide this important diagnostic and prognostic information. We performed real-time, accredited (ISO15189) comprehensive genomic characterization including targeted sequencing and whole exome sequencing in 115 patients with BMF syndromes (median age 24 years, range: 3 months - 81 years). In patients with clinical diagnoses of inherited BMF syndromes, acquired BMF syndromes or clinically unclassifiable BMF we detected variants in 52% (12 of 23), 53% (25 of 47) and 56% (25 of 45) respectively. Genomic characterization resulted in a change of diagnosis in 30 of 115 (26%) including the identification of germline causes for 3 of 47 and 16 of 45 cases with pre-test diagnoses of acquired and clinically unclassifiable BMF respectively. The observed clinical impact of accurate diagnostic categorization included choice to perform allogeneic stem cell transplantation, disease-specific targeted treatments, identification of at-risk family members and influence of sibling allogeneic stem cell donor choice. Multiple novel pathogenic variants and copy number changes were identified in our cohort including in TERT, FANCA, RPS7 and SAMD9. Whole exome sequence analysis facilitated the identification of variants in two genes not typically associated with a primary clinical manifestation of BMF but also demonstrated reduced sensitivity for detecting low level acquired variants. In conclusion, genomic characterization can improve diagnostic categorization of patients presenting with hypoplastic BMF syndromes and should be routinely performed in this group of patients.
Introduction
Bone marrow failure (BMF) related to hypoplasia of hematopoietic elements in the bone marrow is a heterogeneous clinical entity with a broad differential diagnosis. BMF is most commonly an acquired disorder related to an exogenous marrow insult such as drugs (e.g., cytotoxic chemotherapy), toxins or immune destruction of hemopoietic progenitors (acquired aplastic anemia [aAA]). Less commonly, BMF may be the result of an inherited/genetic cause such as Fanconi anemia (FA), Shwachman-Diamond syndrome (SDS), Diamond-Blackfan anemia (DBA) and other rare genetic disorders. Moreover, myelodysplastic syndrome (MDS) can present in the minority of cases (approximately 10%) with marrow hypoplasia (hMDS) rather than the typical hypercellular form with the notable exception that most children and adolescents with MDS will be categorized as refractory cytopenia of childhood (RCC) with the majority of these cases being hypocellular.1
An accurate diagnosis of the precise etiology and subtype of BMF is critical to providing appropriate clinical care. However, the clinical and morphological features of the bone marrow biopsy of the various BMF entities (both acquired and genetic) are non-specific with significant overlap limiting the ability to differentiate the underlying type of BMF. Whilst investigations such as telomere length assessment and chromosomal fragility testing can be helpful in suggesting an underlying diagnosis (of dyskeratosis congenita [DKC] and FA respectively), these techniques have significant technical limitations.2,3 In addition, they do not provide the clinical utility of knowing a specific causative genetic variant that can provide extra information regarding risk stratification, prediction of inheritance patterns and screening of asymptomatic/pre-symptomatic family members.
Genomic evaluation using next generation sequencing (NGS) is a powerful technique to identify variants in a large number of genes in order to resolve the diagnostic and therapeutic challenges faced by physicians treating BMF. Indeed, literature to date suggests a significant contribution to diagnostic rate and reclassification using NGS in patients with inherited/genetic BMF syndromes (IBMFS).4 Moreover, in the acquired setting, detection of variants in patients with aAA may inform of the risk of transformation to hematological malignancy and response to immunosuppression5,6 as well as aiding the diagnostic categorization between aAA and hMDS.4 In some diagnostic laboratories, detection of germline variants is now being performed by whole exome sequencing (WES) as a single approach applicable to a broad range of phenotypes, whereas targeted panel testing is more typically used in the acquired/hematological malignancy setting due to a requirement for greater sensitivity to detect subclonal variants.
Here we describe the results of the Melbourne Genomic Health Alliance Bone Marrow Failure Flagship - a multi-institutional study of 115 patients which aimed to prospectively assess the diagnostic impact of comprehensive genomic evaluation including both WES and targeted NGS panel testing in patients presenting with BMF characterized by bone marrow hypoplasia.
Methods
Recruitment and ethics
Eligible patients were recruited from four participating institutions in Victoria, Australia. Inclusion criteria were (i) age ≥3 months, (ii) a clinicopathological diagnosis (based on current guidelines)1,8 of either aAA, IBMFS, hMDS or a BMF syndrome characterized by marrow hypoplasia/aplasia but not able to be definitively categorized. All patients received pre-test counselling and assessment. Standardized pre-test counselling was provided by the treating hematologist or participants were referred to the study hematologist for the purpose of the study. Participants received written information in the form of the Patient Information Consent Form (PICF). Genetic counsellors and clinical geneticists were available at each of the study sites for referral if required. Research was conducted after Institutional Review Board ethics approval (HREC/13/MH/326 and HREC/17/PMCC/163) and all research was conducted in accordance with the Declaration of Helsinki.
Genomic characterization
All patients underwent baseline testing with (i) WES and (ii) targeted panel sequencing with a hybridization-based NGS BMF panel (a subset of the Peter MacCallum Cancer Center PanHaem panel).9 Sequence variants alone were analyzed from WES and both sequence variants as well as genome-wide copy number changes were assessed from the targeted BMF panel. Genes analyzed by either WES or targeted panel sequencing are shown in Figure 1 and listed in the Online Supplementary Table S1. Details of bioinformatic processing of sequence data are provided in the Online Supplementary Appendix. Briefly, both aligned WES and targeted panel data underwent variant calling using a typical germline variant caller (GATK HaplotypeCaller10) and GATK recommended best practices.11,12 In addition the targeted panel aligned sequence data was processed through a dedicated bioinformatics pipeline which included variant calling with GATK4/Mutect2 (https://software.broadinstitute.org/gatk/) in order to improve detection of low level acquired variants. Copy number variation (CNV) was estimated by comparing read counts from targeted panel data to a reference pool comprising normal samples with similar technical artefacts and visualized using CNspector.13
Depending on the clinical context and genomic findings, patients underwent further characterization with RNA-studies (including whole transcriptome RNA sequencing), Sanger sequencing and/or droplet digital PCR (ddPCR) (see the Online Supplementary Appendix). Sequencing was performed on peripheral blood, bone marrow, buccal epithelial cells or cultured skin fibroblasts depending on the clinical context.
All variants were described according to Human Genome Variation Society (HGVS) nomenclature and germline variants classified according to the American College of Medical Genetics guidelines for the interpretation of sequence variants.14 Accredited (International Organization for Standardization [ISO] 15189) diagnostic reports were issued to referrers for all patients in real-time. In addition, genomic results were reviewed in centralized multidisciplinary case conferences attended by the treating clinicians, molecular hematopathologists, medical scientists, clinical geneticists and genetic counsellors.
Results
Cohort description
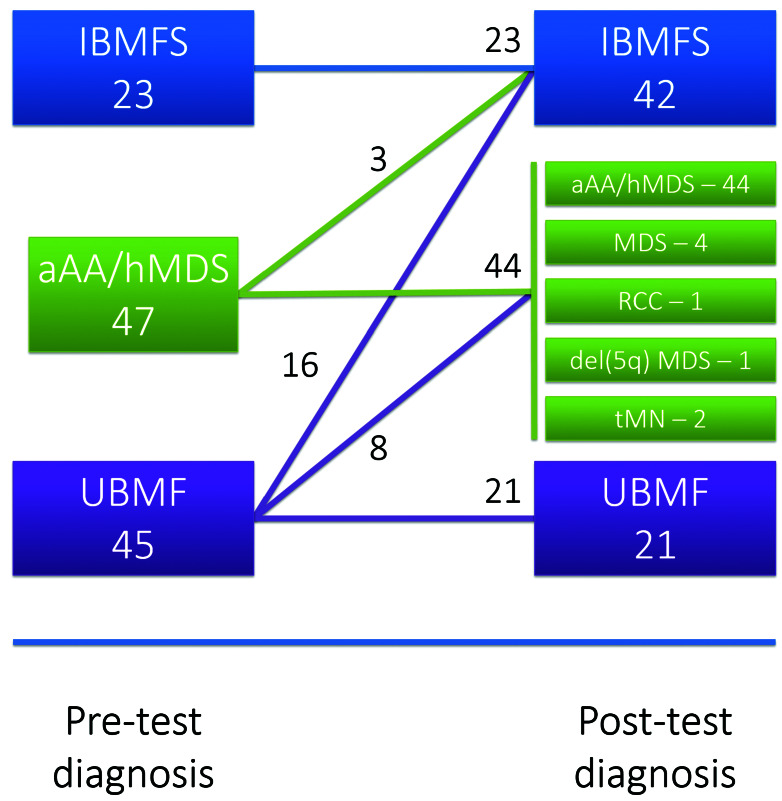
115 patients were recruited from May 2017 to August 2018. The median age of the overall cohort was 24 years (range: 3 months - 81 years) with a male to female (M:F) ratio of 1:1.4. Upon enrolment, the referring clinical team were asked to assign a BMF subtype based on the available clinicopathological features to capture the “pre-test diagnosis” for each patient, noting that these had been assigned on varied degrees of evidence. Any genomic evaluation completed before enrolment was considered part of “standard-of-care” investigations and were performed according to individual treating teams/institutional guidelines. For analysis purposes, patients were divided into three different pre-test diagnosis cohorts (i) patients with a pre-diagnosis of an IBMFS (n=23; median age 10 [range: 2-36 years]; M:F ratio 1:1.88), (ii) patients with a pre-test diagnosis of aAA or hMDS (n=47; median age 32 [range: 7-80 years]; M:F ratio 1:1.24) and (iii) patients with a pretest diagnosis of clinically unclassifiable BMF (UBMF) (n=45; median age 25 [range: 1-81 years]; M:F ratio 1:1.37) (Figure 2). If the referring clinical team could not definitively assign a pre-test diagnosis of IBMFS (including subtype) or aAA/hMDS, the patient was assigned to the UBMF category. After comprehensive genomic characterization was completed, multidisciplinary re-review of all clinical and pathological features incorporating newly acquired genomic data was performed and a “post-test diagnosis” was assigned. Clinical management of patients after assignment of post-test diagnosis was captured in regard to (i) proceeding to allogeneic stem cell transplant, (ii) screening of relatives for identified causative variants and (iii) disease-specific interventions.
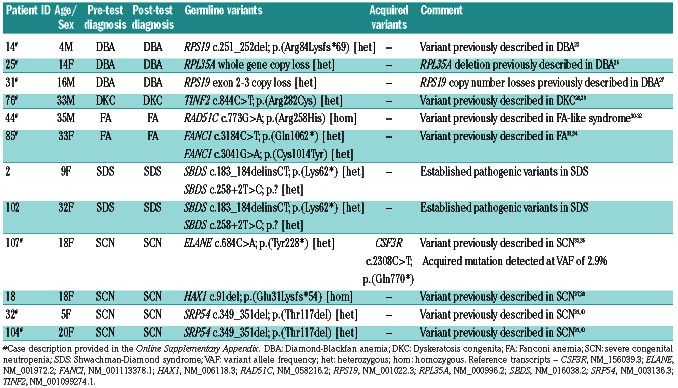
Table 1.
Causative genomic variants in patients with the clinical diagnosis of an inherited bone marrow failure syndrome.
Genomic characterization identifies causative variants in the majority of patients with a pre-test diagnosis of an IBMFS
In patients with a pre-test diagnosis of an IBMFS, genomic characterization confirmed an IBMFS by detecting causative germline variants in 52.2% (12 of 23). The genomic variants detected in this cohort are listed in Table 1. In all cases where a causative germline variant was identified, the post-test diagnosis was consistent with the pre-test diagnosis. One acquired CSF3R variant was detected in this group in a patient with severe congenital neutropenia (SCN) and a germline ELANE variant who had life-long treatment with G-CSF. A causative germline variant was not identified in patients with a pretest diagnosis of SCN in 71.4% (10 of 14) of cases despite genomic characterization.
Despite no change in the pre-test diagnosis in this group, the observed clinical impacts of rendering a precise genomic diagnosis to this group of patients with established clinical diagnoses included one patient (#76) where the identification of a specific variant in TINF2 permitted screening of his pre-symptomatic children for DKC. In addition, this group included an adult patient (#44) with the observation of a homozygous variant in RAD51C (Arg258His) resulting in a Fanconi-like syndrome (associated with positive chromosomal fragility studies) with subsequent implications for breast and ovarian cancer risk screening in siblings. Further clinicopathological details for cases are presented in the Online Supplementary Appendix.
Both acquired and inherited variants were detected in patients with a pre-test diagnosis of aAA/hMDS
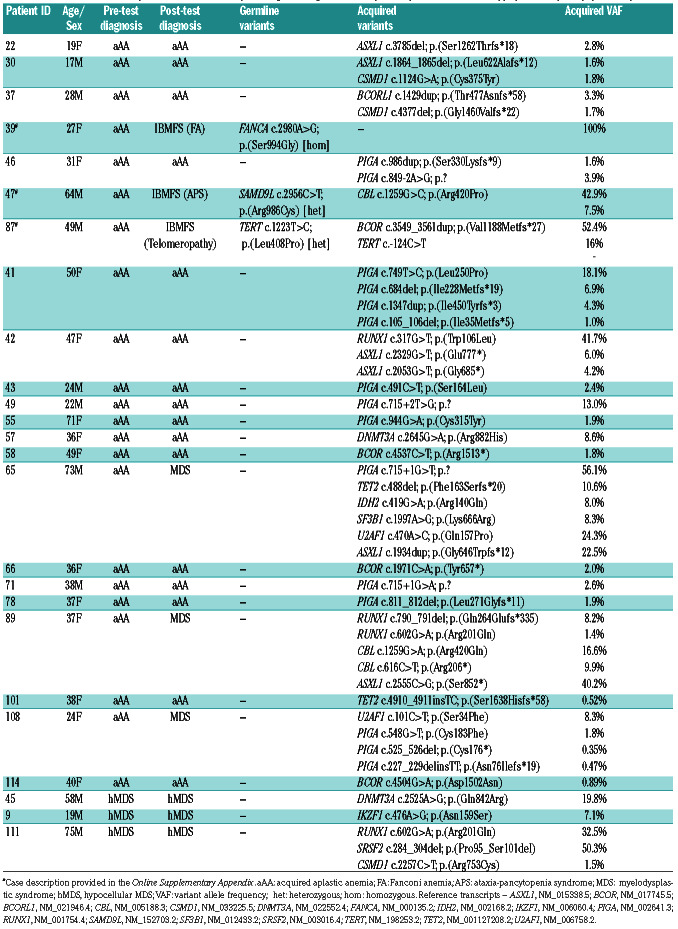
In patients with a pre-test diagnosis of aAA/hMDS, acquired variants were detected in 53.2% of patients (25 of 47) (see Table 2). Forty-one of 47 patients had a pretest diagnosis of aAA (22 of 41 [53.7%] with variants) and 6 of 47 had a pre-test diagnosis of hMDS (3 of 6 [50%] with variants). Variant allele frequency (VAF) of acquired variants in this group ranged from 0.35%- 56.1%. Acquired variants were detected in PIGA (n=15), ASXL1 (n=6), BCOR (n=4), RUNX1 (n=4), CBL (n=3), CSMD1 (n=3), DNMT3A (n=2), TET2 (n=2), U2AF1 (n=2), SRSF2 (n=1), SF3B1 (n=1), TERT (n=1), IKZF1 (n=1), IDH2 (n=1) and BCORL1 (n=1). Three patients in this group had multiple subclonal PIGA variants detectable, a phenomenon previously described in patients with paroxysmal nocturnal hemoglobinuria (PNH).15 No patients in this group had a clinical diagnosis of overt PNH. Over the limited follow-up available, 3 of 25 patients with acquired variants progressed with an aggressive hematological malignancy (acute myeloid leukemia or high-grade MDS) during the study period compared to 0 of 22 patients with no acquired variant detected (P=0.2368, Fisher’s exact test).
Table 2.
Variants detected in patients with a clinicopathological diagnosis of acquired aplastic anemia or hypoplastic myelodysplastic syndrome.
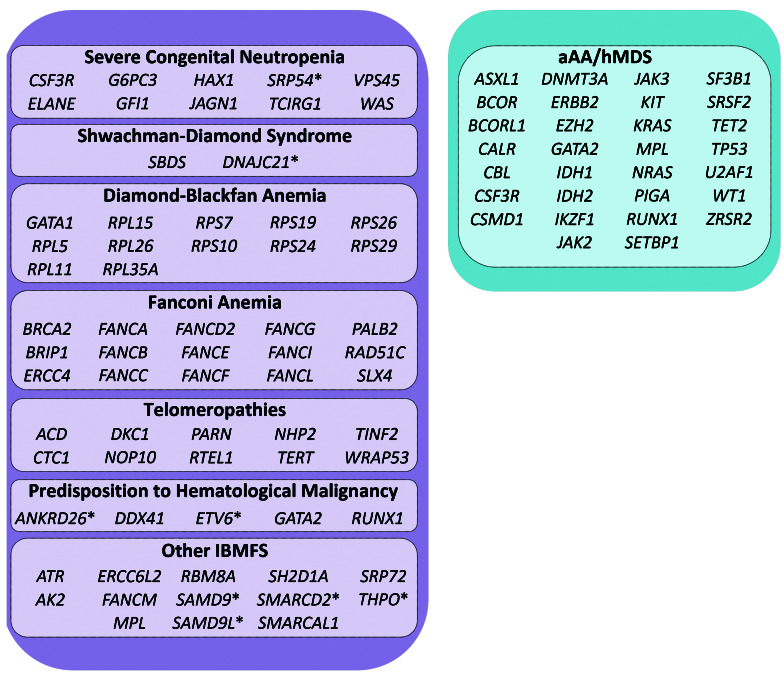
Figure 1.
Genes involved in inherited and acquired bone marrow failure syndromes. Genes causative of an inherited bone marrow failure syndrome (IBMFS), and genes associated with a risk of transformation to hematological malignancy or response to immunosuppression in patients with acquired aplastic anemia (aAA) and hypoplastic myelodysplastic syndrome (hMDS) were assessed in this study. Four genes, CSF3R, GATA2, MPL and RUNX1, were assessed in both germline and acquired settings. Genes targeted by whole exome sequencing (WES) only (i.e., not included on targeted panel) are denoted with an asterisk. Genes KMT2D and PSTPIP1 are not represented in this figure; variants detected in these two genes were considered ‘off-target’ findings.
Six of 47 (12.8%) patients in this group had a change of diagnostic categorization as a result of genomic characterization. Three of these six patients had a pre-test diagnosis of aAA and were re-categorized as having a myelodysplastic syndrome (transformed from underlying aAA) after genomic characterization. In addition to the genomic testing results themselves, the re-categorization of these patients was also the result of a re-assessment of the patient’s disease that was prompted by the availability of genomic testing (including repeat of investigations such as morphological assessment of bone marrow biopsy). The other three patients had a change in pre-test diagnosis from aAA/hMDS to an IBMFS. These cases include a 64-year-old male (#47) who presented with pancytopenia and was found to have bone marrow hypoplasia and morphological dysplasia as well as an acquired pathogenic CBL variant. A pathogenic SAMD9L variant was identified during germline testing. Detailed clinical review revealed a family history of early onset ataxia in a number of family members and subsequently the patient was diagnosed with ataxia-pancytopenia syndrome – one presentation of the SAMD9L group of inherited disorders16 (see the Online Supplementary Appendix). In addition, a 49-year-old male (#87) presented with pancytopenia and severe bone marrow hypoplasia and was diagnosed and treated for aAA. Genomic testing identified a germline variant in TERT (Leu408Pro). Identification of the TERT variant prompted telomere length testing and the patient was found to have telomere lengths <1st percentile. In addition, an acquired TERT promoter variant was detected in this patient adding further evidence to the pathogenicity of the germline variant. The patient had no other clinical features of DKC (see the Online Supplementary Appendix). Finally, a 27-year-old female (#39) was diagnosed with aAA after presenting with pancytopenia. She was commenced on eltrombopag but had no improvement in her blood counts. Genomic characterization revealed a homozygous FANCA variant (Ser994Gly) that resulted in exon skipping (demonstrated by RNA sequencing) with further testing demonstrating equivocal chromosomal fragility but failure of monoubiquitination in FANCD2 consistent with a diagnosis of FA (see the Online Supplementary Appendix).
Genomic characterization clarifies diagnostic categorization in patients with clinically unclassifiable BMF
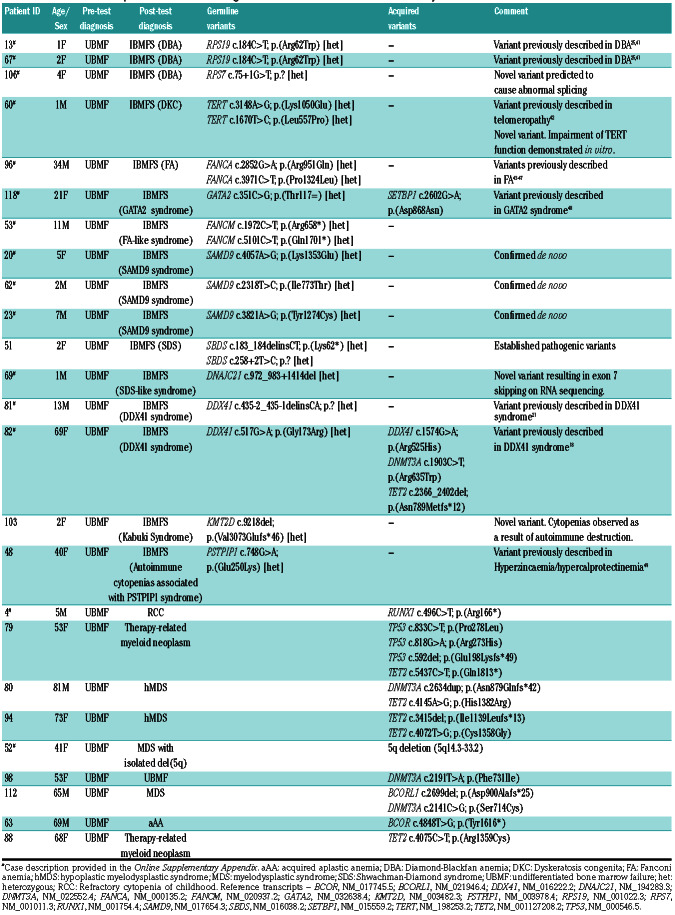
In patients with a pre-test diagnosis of UBMF, genomic characterization identified a causative germline or acquired variant in 25 of 45 (55.6%). In 24 of 25 patients this resulted in a change in the diagnostic categorization from a pre-test diagnosis of UBMF to a specific post-test diagnosis. Therefore genomic characterization resulted in diagnostic categorization in 24 of 45 (53.3%) of patients in this diverse group of patients. Variants detected in this cohort are listed in Table 3. Causative germline variants were detected in 16 patients and acquired variants were detected in nine patients. The diverse inherited and acquired diagnoses included FA, SDS, DKC, DBA, aAA, hMDS, therapy-related myeloid neoplasm (tMN) and MDS with isolated del(5q).
Table 3.
Variants detected in patients with a clinical diagnosis of undifferentiated bone marrow failure syndromes.
Figure 2.
Pre- and post-test diagnosis classification of patients. Numbers of patients classified into each diagnostic group, (i) inherited bone marrow failure syndrome (IBMFS), (ii) acquired aplastic anemia (aAA) or hypoplastic MDS (hMDS) or (iii) clinically unclassifiable bone marrow failure (UBMF) as evaluated prior to genomic testing (pre-test diagnosis) and again following genomic testing (posttest diagnosis) with transitions indicated by the connecting lines (in addition three patients changed diagnosis within the aAA/hMDS category from aAA to MDS). RCC: refractory cytopenia of childhood; tMN: therapy related myeloid neoplasm.
Three patients in this group received a post-test diagnosis of hMDS or MDS after genomic characterization. This was the result of re-interpretation of the clinical and pathological features in light of supportive molecular evidence for these diagnoses. Notably, one patient retained a post-test diagnosis of UBMF despite having a DNMT3A mutation detected due to inadequate pathological and clinical features to support a diagnosis of aAA, hMDS, MDS or tMN.
Five patients in this group presented with hypoplastic BMF and a monosomy 7 detectable by either conventional karyotyping, fluorescence in situ hybridization or copy number analysis by targeted sequencing. Three of these (#20, #23, #62) had SAMD9 variants that were all confirmed to be de novo through testing of parental samples. Multiple somatic reversion events were noted in these patients including acquired sequence variants as well as progressive resolution of monosomy 7 over time (see the Online Supplementary Appendix). One patient (#118) with monosomy 7 had a synonymous GATA2 variant (Thr117=) which has previously been associated with GATA2 haploinsufficiency syndrome.17
Two patients in this group had pathogenic DDX41 variants identified. One of these patients was a 69-year-old female (#82) who presented with severe neutropenia and was found to have a severely hypocellular bone marrow. Genomic characterization revealed a DDX41missense variant (Gly173Arg) which has been described previously in five patients presenting with a similar clinical phenotype.18 In addition, targeted panel sequencing in this patient identified a hotspot somatic DDX41 Arg525His variant present at approximately 3% VAF.
Finally, this group included a 5-year-old male (#4) with a pre-test diagnosis of UBMF with onset just after birth. Targeted panel sequencing identified a RUNX1 nonsense variant (Arg166*) at low VAF (4.8%). This variant was quantitatively monitored over time using ddPCR and was noted to be acquired, steadily increasing and reaching 11.8% over a 12-month period. After multidisciplinary review it was decided to proceed to an allogeneic bone marrow transplant in this patient.
Contribution to diagnostic categorization from targeted panel sequencing versus WES
All germline variants in genes common to the targeted panel and whole exome were detected. In eight patients, variants were detected in the whole exome in genes not covered in the targeted panel. These included variants in SAMD9, SAMD9L, DNAJC21, SRP54, KMT2D and PSTPIP1. These genes can be divided into two groups: (i) genes where the primary phenotype of pathogenic variants is an IBMFS (SAMD9, SAMD9L, DNAJC21 and SRP54) and (ii) genes where BMF is either an infrequent or not the primary clinical manifestation (KMT2D and PSTPIP1). Mutations in genes in this latter category were detected through an extended analysis of the WES data given the absence of other causative mutations found in these two patients and the remaining strong suspicion of a genetic etiology.
In total, 65 acquired variants were detected across the entire cohort. All of these variants were detected by targeted panel sequencing with a bioinformatic pipeline optimized for acquired variant detection. Only 16 of 65 (24.6%) of these variants were detected by WES using a standard germline bioinformatics pipeline. Of the 49 acquired variants not called by the WES pipeline, 24 were visible after manual inspection of aligned reads in whole exome data based on variant detection in the targeted panel data. The mean depth of targeted panel sequencing was 795x (range: 576-1,025) with 98.9% of bases sequenced to ≥100x coverage, compared to a mean depth of 198x (range: 117-260) and 77.5% ≥100x coverage for WES. The VAF of the variants that were not detected by WES (and associated pipeline) were all less than 15%.
Discussion
We have described a cohort of 115 patients presenting with a diverse range of hypocellular BMF that have undergone prospective comprehensive genomic evaluation for germline and acquired causes. In our cohort we were able to identify pathogenic germline or acquired variants in 54% (62 of 115) of the entire cohort including in 52% (12 of 23) of patients with a clinical diagnosis of IBMFS, 53% (25 of 47) of patients with a clinical diagnosis of aAA/hMDS and 56% (25 of 45) of patients with clinically unclassifiable BMF. Overall, genomic characterization resulted in a change in diagnostic categorization in 30 of 115 (26.1%) patients. The observed clinical impacts in our patient cohort after comprehensive genomic characterization included the choice to perform allogeneic stem cell transplantation, disease-specific treatments (such as corticosteroid for DBA and lenalidomide for MDS with del(5q)), identification of at-risk family members, enhanced screening of patients for secondary malignancy and influence of sibling allogeneic stem cell donor choice. We have also described multiple novel genomic findings including novel pathogenic variants in TERT (Leu408Pro and Leu557Pro), RPS7 (c.75+1G>T) and SAMD9 (Lys1353Glu, Tyr1274Cys and Ile773Thr) and have confirmed recent novel findings of other groups including (i) the association of biallelic FANCM variants with chemotherapy toxicity in the absence of an overt FA phenotype,19 (ii) the association of DDX41 Gly173Arg with a hypocellular MDS phenotype18 and (iii) the presence of acquired reversion variants in SAMD9/SAMD9L.20
The diverse spectrum of inherited and acquired diseases identified in our cohort highlights the diagnostic utility of comprehensive genomic characterization for patients with a clinical presentation of BMF. Despite the range of underlying diagnoses that may manifest as hypoplastic BMF there may be minimal or no specific features on conventional (non-genomic) diagnostic testing to indicate the specific underlying diagnosis. In addition, clinical features such as abnormal digits or oral leukoplakia cannot be relied upon to suggest an underlying genetic cause as they may be absent or unrecognized due to lack of physician familiarity with these rare diseases. Moreover, as more literature and larger cohorts emerge with genetic annotation, the clinical phenotypes associated with specific genes are expanding. For example, the expansion of the phenotype of pathogenic variants in DDX41 to include a hypocellular MDS-like phenotype in the case of Gly173Arg18 rather than the initial description of presentation in later life with acute myeloid leukemia after minimal antecedent overt hematological abnormality.21 Indeed, in our cohort we observed a pathogenic DDX41 variant in a 13-year-old male (#81) with isolated severe stable thrombocytopenia and no evidence of hematological malignancy.
A significant proportion of patients (47.8%) with a clinical diagnosis of IBMFS did not have causative variants detected. The vast majority (10 of 11) of these were in patients with SCN. This may be explained by the significant baseline rate of “variant-negative” cases in this condition described in the literature to date22 as well as the potential for other causes of neutropenia. Given these patients had no pathogenic variants detectable despite comprehensive genomic analysis (including WES), one possible future strategy for identifying causative variants may be to evaluate the non-coding regions of existing pathogenic genes (e.g., ELANE, HAX1) for deep intronic variants resulting in splicing abnormalities that are missed by methods focused on coding regions.
In the acquired BMF setting, we noted six patients who were re-categorized as either hMDS, MDS, tMN or refractory cytopenia of childhood (RCC) after genomic characterization. Whilst there are no absolutely specific molecular abnormalities that distinguish these disorders from aAA complicated by clonal hematopoiesis or age-related clonal hematopoiesis, the presence of the genomic abnormalities detected were sufficient for the treating team to re-evaluate the clinical and morphological features of these six cases and assign post-diagnoses of hMDS/MDS/RCC/tMN. This reflects the way in which this type of genomic data is increasingly integrated into diagnostic decision making in a real-world context, especially in areas where morphological interpretation is challenging and inter-observer agreement is poor.23,24 Another practical aspect regarding diagnostic workup is the limited availability of specific non-genomic tests (e.g., telomere length analysis) in the accredited setting which may have contributed to a variable rate of performance of these tests in the cohort.
In our cohort 39% (45 of 115) of patients were categorized as clinically UBMF before genomic testing. The relatively large number of patients in this category may be the result of numerous factors including a referral bias for patients to this study with atypical clinicopathological features in order to access novel genomic technologies. In addition, patients could also be included at an early timepoint in their disease course and it is possible that repeated history and physical examination over time in patients (including assessing response to therapy) would have enabled more definitive categorization before genomic testing. Moreover, there is an increasing recognition of the possibility of occult IBMFS in patients diagnosed as acquired BMF (e.g., aAA), indeed in our cohort three patients harbored occult IBMFS. This recognition, in turn, may lead to a desire by treating clinical teams to more comprehensively exclude IBMFS rather than relying on a clinical diagnosis of a purely acquired BMFS - hence their inclusion in a UBMF category. Reliable clinical and laboratory features to identify those with a low likelihood of IBMFS (e.g., the presence of a PNH clone, severity/time course/persistence of neutropenia in SCN) would be of significant value in resource-limited settings in order to avoid expensive and time consuming genetic testing in this potentially expanding subgroup. The approach to genomic characterization in our cohort included both targeted BMF panel sequencing and WES for all patients. Our choice of targeted panel sequencing in addition to WES was based on the requirement for enhanced sensitivity due to the clinical context of BMF mandating assessment for both germline and acquired genomic lesions. In addition to the detection of acquired variants in aAA/hMDS (which may be present at VAF less than 5%), clinically relevant reversion variants in inherited conditions such as SAMD9/SAMD9L can also exist which require sensitive testing methodologies. As expected (given the greater depth of sequencing able to be cost-effectively achieved), targeted panel sequencing was clearly superior at detecting low-level acquired variants when compared to WES. Using a bioinformatics pipeline optimized for acquired variant detection (GATK4/Mutect2) we were able to detect acquired variants down to a VAF of <0.5%. Despite their very low level, detection of these variants can be highly instructive for diagnostic categorization by demonstration of clonality (to distinguish from reactive conditions) as well as providing a baseline genomic profile for future monitoring of clonal evolution. Conversely, one potential benefit of WES for investigation of germline variants is in the context of phenotypic uncertainty in order to identify pathogenic variants across a broad gene list. In our cohort, two patients had pathogenic variants detected in genes which are not typically associated with BMF (KMT2D and PSTPIP1). The other genes originally not present in the targeted panel due to their recent description (SAMD9, SAMD9L, DNAJC21) have been added to a future iteration of the targeted panel. However the need for panel re-design and subsequent costly laboratory validation highlights a recognized drawback of targeted panel sequencing. Nevertheless, given the relative specificity of the clinical presentation and the requirement for sensitive detection of acquired variants, targeted panel sequencing is a methodological choice highly suited for this clinical context, compared to which, WES offers minimal additional clinical utility. Copy number analysis was possible using either of these sequencing strategies and facilitated the identification of pathogenic copy number losses (RPL35A, RPS19) as well as clinically relevant acquired monosomies. RNA sequencing was valuable for determining the effect of putative splice variants on mRNA transcripts (FANCA, DNAJC21).
In conclusion, we have described the impact of genomic characterization on diagnostic categorization in a cohort of patients with hypoplastic BMF. In addition to multiple novel genomic findings, this personalized genomic approach is of significant utility to rapidly providing accurate diagnostic categorization to inform clinical decisionmaking in this diverse and challenging group of diseases. The incorporation of a multidisciplinary review further strengthens the impact and clinical value of this approach.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the support of Maddie Riewoldt’s Vision, all participants of the multidisciplinary meetings and the patients and families. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government. Current work in GL’s laboratory is supported by the National Health and Medical Research Council (NHMRC, 1086020), Australian Research Council (ARC, DP170102235) and Maddie Riewoldt’s Vision.
Funding Statement
Funding: The study was funded by the State Government of Victoria (Department of Health and Human Services) and the 10 member organizations of the Melbourne Genomics Health Alliance.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th edn ed. Lyon, France: International Agency for Research on Cancer, 2017. [Google Scholar]
- 2.Oostra AB, Nieuwint AW, Joenje H, de Winter JP. Diagnosis of fanconi anemia: chromosomal breakage analysis. Anemia. 2012;2012:238731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai TP, Wright WE, Shay JW. Comparison of telomere length measurement methods. Philos Trans R Soc Lond B Biol Sci. 2018; 373(1741). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghemlas I, Li H, Zlateska B, et al. Improving diagnostic precision, care and syndrome definitions using comprehensive next-generation sequencing for the inherited bone marrow failure syndromes. J Med Genet. 2015; 52(9):575-584. [DOI] [PubMed] [Google Scholar]
- 5.Kulasekararaj AG, Jiang J, Smith AE, et al. Somatic mutations identify a subgroup of aplastic anemia patients who progress to myelodysplastic syndrome. Blood. 2014; 124(17):2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshizato T, Dumitriu B, Hosokawa K, et al. Somatic mutations and clonal hematopoiesis in aplastic anemia. N Engl J Med. 2015;373(1):35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bono E, McLornan D, Travaglino E, et al. Clinical, histopathological and molecular characterization of hypoplastic myelodysplastic syndrome. Leukemia. 2019; 33(10):2495-2505. [DOI] [PubMed] [Google Scholar]
- 8.Killick SB, Bown N, Cavenagh J, et al. Guidelines for the diagnosis and management of adult aplastic anaemia. Br J Haematol. 2016;172(2):187-207. [DOI] [PubMed] [Google Scholar]
- 9.Ryland GL, Jones K, Chin M, et al. Novel genomic findings in multiple myeloma identified through routine diagnostic sequencing. J Clin Pathol. 2018;71(10):895-899. [DOI] [PubMed] [Google Scholar]
- 10.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markham JF, Yerneni S, Ryland GL, et al. CNspector: a web-based tool for visualisation and clinical diagnosis of copy number variation from next generation sequencing. Sci Rep. 2019;9(1):6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen W, Clemente MJ, Hosono N, et al. Deep sequencing reveals stepwise mutation acquisition in paroxysmal nocturnal hemoglobinuria. J Clin Invest. 2014;124(10):4529-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen DH, Below JE, Shimamura A, et al. Ataxia-Pancytopenia Syndrome Is Caused by Missense Mutations in SAMD9L. Am J Hum Genet. 2016;98(6):1146-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wehr C, Grotius K, Casadei S, et al. A novel disease-causing synonymous exonic mutation in GATA2 affecting RNA splicing. Blood. 2018;132(11):1211-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebert M, Passet M, Raimbault A, et al. Clinical and molecular characteristics of DDX41-mutated patients in a large cohort of sporadic MDS/AML. Blood. 2018; 132(Suppl 1):S797. [Google Scholar]
- 19.Catucci I, Osorio A, Arver B, et al. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet Med. 2018;20(4):452-457. [DOI] [PubMed] [Google Scholar]
- 20.Tesi B, Davidsson J, Voss M, et al. Gain-offunction SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood. 2017;129(16):2266-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewinsohn M, Brown AL, Weinel LM, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127(8):1017-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boztug K, Klein C. Genetic etiologies of severe congenital neutropenia. Curr Opin Pediatr. 2011;23(1):21-26. [DOI] [PubMed] [Google Scholar]
- 23.Sasada K, Yamamoto N, Masuda H, et al. Inter-observer variance and the need for standardization in the morphological classification of myelodysplastic syndrome. Leuk Res. 2018;69:54-59. [DOI] [PubMed] [Google Scholar]
- 24.Parmentier S, Schetelig J, Lorenz K, et al. Assessment of dysplastic hematopoiesis: lessons from healthy bone marrow donors. Haematologica. 2012;97(5):723-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willig TN, Draptchinskaia N, Dianzani I, et al. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: wide variations in phenotypic expression. Blood. 1999; 94(12):4294-4306. [PubMed] [Google Scholar]
- 26.Farrar JE, Vlachos A, Atsidaftos E, et al. Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011; 118(26):6943-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulirsch JC, Verboon JM, Kazerounian S, et al. The genetic landscape of Diamond- Blackfan anemia. Am J Hum Genet. 2018;103(6):930-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vulliamy T, Beswick R, Kirwan MJ, Hossain U, Walne AJ, Dokal I. Telomere length measurement can distinguish pathogenic from non-pathogenic variants in the shelterin component, TIN2. Clin Genet. 2012;81(1):76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112(9):3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia–like disorder. Nat Genet. 2010;42(5):406-409. [DOI] [PubMed] [Google Scholar]
- 31.Somyajit K, Saxena S, Babu S, Mishra A, Nagaraju G. Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 2015;43(20):9835-9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somyajit K, Subramanya S, Nagaraju G. Distinct roles of FANCO/RAD51C protein in DNA damage signaling and repair: implications for Fanconi anemia and breast cancer susceptibility. J Biol Chem. 2012; 287(5):3366-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savage SA, Ballew BJ, Giri N, et al. Novel FANCI mutations in Fanconi anemia with VACTERL association. Am J Med Genet A. 2016;170A(2):386- 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mantere T, Haanpaa M, Hanenberg H, et al. Finnish Fanconi anemia mutations and hereditary predisposition to breast and prostate cancer. Clin Genet. 2015;88(1):68-73. [DOI] [PubMed] [Google Scholar]
- 35.Dale DC, Person RE, Bolyard AA, et al. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96(7):2317-2322. [PubMed] [Google Scholar]
- 36.Makaryan V, Zeidler C, Bolyard AA, et al. The diversity of mutations and clinical outcomes for ELANE-associated neutropenia. Curr Opin Hematol. 2015;22(1):3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith BN, Ancliff PJ, Pizzey A, Khwaja A, Linch DC, Gale RE. Homozygous HAX1 mutations in severe congenital neutropenia patients with sporadic disease: a novel mutation in two unrelated British kindreds. Br J Haematol. 2009;144(5):762-770. [DOI] [PubMed] [Google Scholar]
- 38.Carlsson G, Elinder G, Malmgren H, et al. Compound heterozygous HAX1 mutations in a Swedish patient with severe congenital neutropenia and no neurodevelopmental abnormalities. Pediatr Blood Cancer. 2009; 53(6):1143-1146. [DOI] [PubMed] [Google Scholar]
- 39.Bellanne-Chantelot C, Schmaltz-Panneau B, Marty C, et al. Mutations in the SRP54 gene cause severe congenital neutropenia as well as Shwachman-Diamond-like syndrome. Blood. 2018;132(12):1318-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carapito R, Konantz M, Paillard C, et al. Mutations in signal recognition particle SRP54 cause syndromic neutropenia with Shwachman-Diamond-like features. J Clin Invest. 2017;127(11):4090-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169-175. [DOI] [PubMed] [Google Scholar]
- 42.Cronkhite JT, Xing C, Raghu G, et al. Telomere shortening in familial and sporadic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178(7):729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ameziane N, Errami A, Leveille F, et al. Genetic subtyping of Fanconi anemia by comprehensive mutation screening. Hum Mutat. 2008;29(1):159-166. [DOI] [PubMed] [Google Scholar]
- 44.Gille JJ, Floor K, Kerkhoven L, Ameziane N, Joenje H, de Winter JP. Diagnosis of Fanconi anemia: mutation analysis by multiplex ligation- dependent probe amplification and PCR-based sanger sequencing. Anemia. 2012;2012:603253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bottega R, Nicchia E, Cappelli E, et al. Hypomorphic FANCA mutations correlate with mild mitochondrial and clinical phenotype in Fanconi anemia. Haematologica. 2018;103(3):417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan NV, Tipping AJ, Joenje H, Mathew CG. High frequency of large intragenic deletions in the Fanconi anemia group A gene. Am J Hum Genet. 1999;65(5):1330-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Z, Geng J, Yao RE, et al. Molecular defects identified by whole exome sequencing in a child with Fanconi anemia. Gene. 2013;530(2):295-300. [DOI] [PubMed] [Google Scholar]
- 48.Wehr C, Grotius K, Casadei S, et al. A novel disease-causing synonymous exonic mutation in GATA2 affecting RNA splicing. Blood. 2018;132(11):1211-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holzinger D, Fassl SK, de Jager W, et al. Single amino acid charge switch defines clinically distinct proline-serine-threonine phosphatase- interacting protein 1 (PSTPIP1)- associated inflammatory diseases. J Allergy Clin Immunol. 2015;136(5):1337-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.