Abstract
Retrotransposons such as LINE-1 and Alu comprise >25% of the human genome. While global hypomethylation of these elements has been widely reported in solid tumours, their epigenetic dysregulation is yet to be characterised in chronic lymphocytic leukemia (CLL), and there has been scant consideration of their evolutionary history that mediates sensitivity to hypomethylation. Here, we developed an approach for locus- and evolutionary subfamily-specific analysis of retrotransposons using the Illumina Infinium Human Methylation 450K microarray platform, which we applied to publicly-available datasets from CLL and other haematological malignancies. We identified 9,797 microarray probes mapping to 117 LINE-1 subfamilies and 13,130 mapping to 37 Alu subfamilies. Of these, 10,782 were differentially methylated (PFDR<0.05) in CLL patients (n=139) compared with healthy individuals (n=14), with enrichment at enhancers (P=0.002). Differential methylation was associated with evolutionary age of LINE-1 (r2=0.31, P=0.003) and Alu (r2=0.74, P=0.002) elements, with greater hypomethylation of older subfamilies (L1M, AluJ). Locus-specific hypomethylation was associated with differential expression of proximal genes, including DCLK2, HK1, ILRUN, TANK, TBCD, TNFRSF1B and TXNRD2, with higher expression of DCLK2 and TNFRSF1B associated with reduced patient survival. Hypomethylation at nine loci was highly frequent in CLL (>90% patients) but not observed in healthy individuals or other leukaemias, and was detectable in blood samples taken prior to CLL diagnosis in 9 of 82 individuals from the Melbourne Collaborative Cohort Study. Our results demonstrate differential methylation of retrotransposons in CLL by their evolutionary heritage that modulates expression of proximal genes.
Introduction
Retrotransposons, including long interspersed elements (LINE) and short interspersed elements (SINE), comprise more than 25% of the human genome. The most widely studied LINE and SINE are LINE-1 (L1) and Alu respectively, which can be further categorised into subfamilies according to sequence variants that inform upon their evolutionary heritage: L1 can be broadly categorised into L1M (mammalian-specific, oldest), L1P (primate-specific, intermediate) and L1H (human-specific, youngest) subfamilies; and Alu into AluJ (oldest), AluS (intermediate) and AluY (youngest).1,2 Retrotransposons are considered to be mobile within the human genome, but the ability to retrotranspose has been lost in the oldest L1 and Alu subfamilies due to sequence changes over time, while, among others, members of the L1PA, L1HS, AluS and AluY families have retained this ‘jumping’ ability.3 Of these, the AluY subfamily is the most active,4 while there are approximately 68 L1 sequences in the human genome that remain active in retrotransposition.5 De novo retrotransposition of an Alu element has been estimated to occur in 1 in every 21 births, and an L1 element in 1 in 212 births.6 Somatic retrotransposition is largely supressed through epigenetic mechanisms that silence expression of these elements, but hypomethylation of L1 and Alu has been reported in response to a range of environmental exposures, including benzene7 and tobacco,8 with differential sensitivity to environmental pollutants according to the evolutionary age of the subfamily.9
Deregulation of retrotransposon methylation is common in many cancers10 and is associated with patient prognosis. 11 It can serve to activate the expression of oncogenes, such as through locus-specific hypomethylation events and transcription from alternative transcriptional start sites located within retrotransposon elements.12 At the genome-wide level, it can also lead to chromosomal instability and the subsequent acquisition of genetic defects,13,14 with important consequences for patient prognosis. Hypomethylation of these elements is associated with their re-activation and can lead to retrotransposition events, which are frequent in colorectal, lung, prostate and breast cancers (47–93% of tumors)15 and are biased towards hypomethylated regions of the genome.16 Retrotransposition can initiate carcinogenesis in epithelial tissues17 and has been identified in adenomas, metaplasia and histologically-normal tissue surrounding tumors.18,19
The epigenome in chronic lymphocytic leukemia (CLL) is characterised by widespread hypomethylation at the point of diagnosis20 and continues to be reshaped during disease progression, with epigenetic changes associated with genetic evolution.21 However, to date there has been scant study of the role of retrotransposons in the development and progression of leukamia, with only pyrosequencing- based analysis of single L1 and Alu subfamilies performed.22,23 In this study, we used the Illumina Infinium HumanMethylation 450 BeadChip microarray platform (HM450K) to analyse locus- and subfamily-specific changes in the methylation of retrotransposons in CLL. We identified the HM450K probes mapping to retrotransposons and employed these to analyse the methylation of subfamilies by their evolutionary age. Sites identified as differentially methylated in CLL were analysed in other B-cell malignancies to assess their specificity to the disease, and in pre-diagnostic baseline samples from cohort study participants who went on to develop CLL to determine whether they are detectable prior to diagnosis.
Methods
Publicly-available HM450K and gene expression microarray datasets
Differentially methylated retrotransposon loci were identified in a discovery cohort of 139 CLL patients and CD19+ B-cells from 14 healthy individuals from the study of Kulis et al.,20 obtained through the International Cancer Genome Consortium (European Genome-phenome Archive accession number: EGAS00001000272; Online Supplementary Table S1). Validation was performed in a cohort of 24 CLL patients attending clinic in North-East England (Online Supplementary Table S2). Ethical approval for the validation study was granted by the North East - Newcastle & North Tyneside 1 Research Ethics Committee (REC reference number 17/NE/0361). All donors provided written informed consent.
Leading hits were analyzed in publicly-available datasets available through Gene Expression Omnibus (GEO). Details of studies and participants are summarized in the Online Supplementary Table S1. Data from healthy individuals was obtained from the studies of Hannum et al. (n=656; GSE40279),24 the European Prospective Investigation into Cancer and Nutrition (EPIC, n=329; GSE51057)25 and the Young Finns Study (n=184; GSE69270).26 DNA was extracted from whole blood (Hannum et al.)24 or buffy coat (EPIC and Young Finns Study) and therefore represent unfractionated leukocyte preparations. Examination in other haematological malignancies was performed using data from patients with acute lymphoblastic leukemia (ALL) (n=797; GSE47051),27 chronic myeloid leukemia (CML) (n=12; GSE106600),28 acute myeloid leukaemia (AML) (n=68; GSE62298),29 and lymphoma (n=31; GSE42372).30 Analysis of methylation by hematological cell type was achieved using data from the discovery cohort and the studies of Reinius et al. (n=6; GSE35069)31 and Lee et al. (n=4-6; GSE45461).32
Correlations between retrotransposon DNA methylation and the expression of proximal genes were examined within the discovery cohort.20 Comparison of gene expression in normal CD19+ B cells (n=32) and CLL (n=188) was performed using gene expression microarray data from GSE50006.
Pre-diagnostic samples from the Melbourne Collaborative Cohort Study
Leading hits unique to CLL patients were examined in prospective samples taken up to 18 years prior to diagnosis in order to reveal potential early epigenetic changes in disease development. HM450K data from 82 prospective CLL cases and 82 age-matched controls within the Melbourne Collaborative Cohort Study (MCCS) were used to examine DNA methylation changes prior to the diagnosis of CLL. The study protocol and performance of HM450K microarrays have been previously described.33
Identification of microarray probes mapping to retrotransposon subfamilies
HM450K probes mapping to CpG sites within retrotransposons were identified using the Data Integrator function of the UCSC Genome Browser34 with annotation from RepeatMasker35 for the human genome build GRCh37/hg19 (Online Supplementary Methods).
Statistical analyses
Analysis of methylation change by subfamily evolutionary age was performed by linear regression using estimates of time since sequence amplification (millions of years ago, MYA) from the studies of Kapitonov et al.36 and Khan et al.37 Differentially methylated loci in CLL were identified by t-test, with correction for false discovery rate (FDR) by the Benjamini-Hochberg method. All statistical analyses were performed in R (version 3.4.0) using the ggplot2, heatmap2, qqman and corrplot packages, and GraphPad Prism (version 7.0b).
Results
Distribution of HM450K probes mapping to retrotransposons
We identified 22,927 probes mapping to retrotransposons, 9,797 of which mapped to 117 L1 subfamilies and 13,130 to 37 Alu subfamilies. L1 elements were categorised into oldest (L1M, mammalian-wide), intermediate (L1P, primate-specific) and youngest (L1HS, human-specific and L1PA, primate-amplified) subfamilies. Alu elements were categorised into oldest (AluJ), intermediate (AluS) and youngest (AluY) subfamilies.1
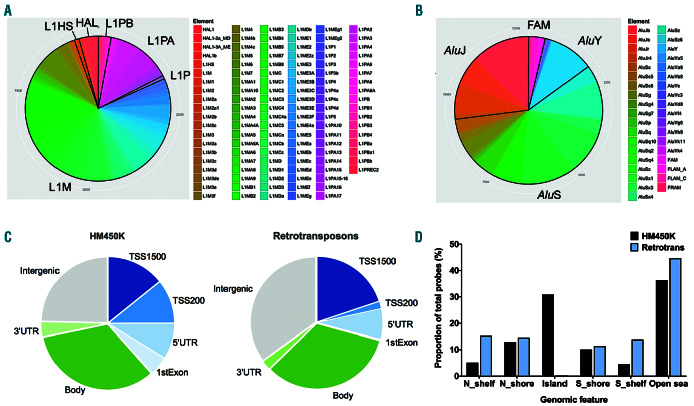
Figure 1.
Distribution of HM450K probes mapping to L1 and Alu elements. (A-B) The number of probes mapping to each subfamily are displayed. A total of 9,797 probes map to 117 L1 subfamilies, and 13,130 probes to 37 Alu subfamilies. Their categorisation into old (L1M, AluJ), intermediate (L1P, L1PB, AluS), young (L1HS, L1PA, AluY) and related (HAL, FAM) subfamilies are indicated. (C-D) Distribution of probes mapping to retrotransposons (‘Retrotransposons’; n=22,927) by genomic feature (C) and in relation to CpG islands (D), in comparison to all probes on the HM450K array (‘HM450K’; n=485,512). Annotation for genomic features and CpG islands was extracted from the Illumina annotation file. TSS1500 ≤ 1500 bp upstream of the transcription start site; TSS200 ≤ 200 bp upstream of the transcription start site: UTR: untranslated region; Body: gene body; N: north, upstream of a CpG island; S: south, downstream of a CpG island.
The distribution of subfamilies revealed a high proportion mapping to older L1 (L1M) and intermediate Alu (AluS) subfamilies (Figure 1A–B). A total of 7,581 probes map to L1M elements, 364 to L1P, 1,435 to L1PA, 62 to human-specific L1HS, and 355 to HAL1. For Alu, 3,563 probes map to AluJ elements, 7,626 to AluS, 1,468 to AluY, 73 to FAM, 310 to FLAM, and 90 to FRAM. A comprehensive list of probes by subfamily is provided in the Online Supplementary Table S3. The relative proportions of all Alu probes mapping to the AluJ (28%), AluS (60%) and AluY (12%) subfamilies did not significantly differ from their natural abundance in the human genome (26%, 62% and 13% respectively; P>0.70, Fisher’s exact test) as identified by RepeatMasker.35 Similarly, the proportion of L1 probes mapping to the L1M (80%), L1P (4%) and L1HS/L1PA (16%) subfamilies did not significantly differ from their frequency in the human genome (81%, 6% and 13% respectively; P>0.70). Therefore, the HM450K platform offers a representative means to interrogate retrotransposon methylation in the human genome.
In comparison to all >450,000 probes on the HM450K microarray, retrotransposon probes show relative enrichment for mapping to intergenic regions, and depletion of probes mapping to within 200 bases of transcriptional start sites (TSS200) and first exons (Figure 1C). Additionally, there is substantial depletion of probes mapping to CpG islands, with relative enrichment at north and south shelves (Figure 1D).
Retrotransposon elements are highly methylated in normal B cells
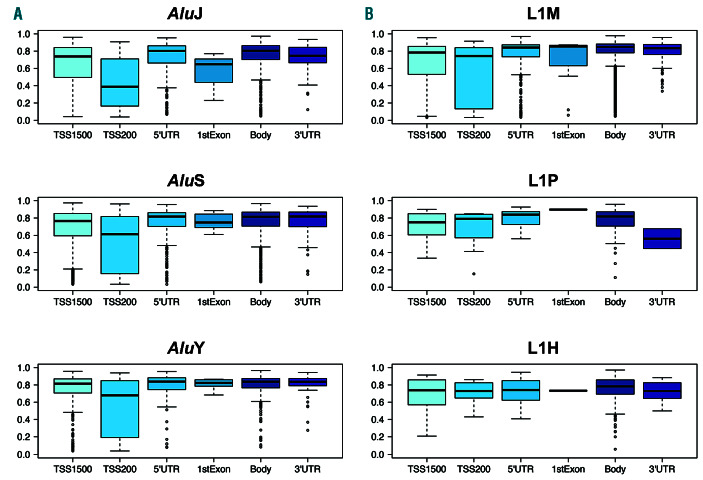
Retrotransposon elements predominantly showed high levels of methylation in normal CD19+ B cells, but displayed significant methylation variability by genomic feature location and by evolutionary age of the subfamily (Figure 2). Those CpG sites mapping to Alu sequences showed a highly significant trend of increasing methylation from the older to younger elements (P<2.2x10-16, ANOVA), with mean methylation levels (b ± standard deviation [SD]) of 0.72±0.21 for AluJ, 0.74±0.18 for AluS, and 0.78±0.17 for AluY. L1 elements did not display such a linear trend by evolutionary age, with the mean methylation of L1P elements (b: 0.73±0.19) lower than those of L1M (0.76±0.20) and L1H/PA (0.76±0.14) (P<0.008, ANOVA).
Analysis by genomic region revealed that those retrotransposons mapping to <200 bp upstream of TSS showed the greatest variability in methylation levels, while those mapping to 5’UTR and first exon regions displayed the least (Figure 2). Furthermore, at these regions L1M elements displayed greater variability than younger L1H/L1PA elements, while AluY similarly displayed less variability of methylation patterns in comparison with the older AluJ sequences.
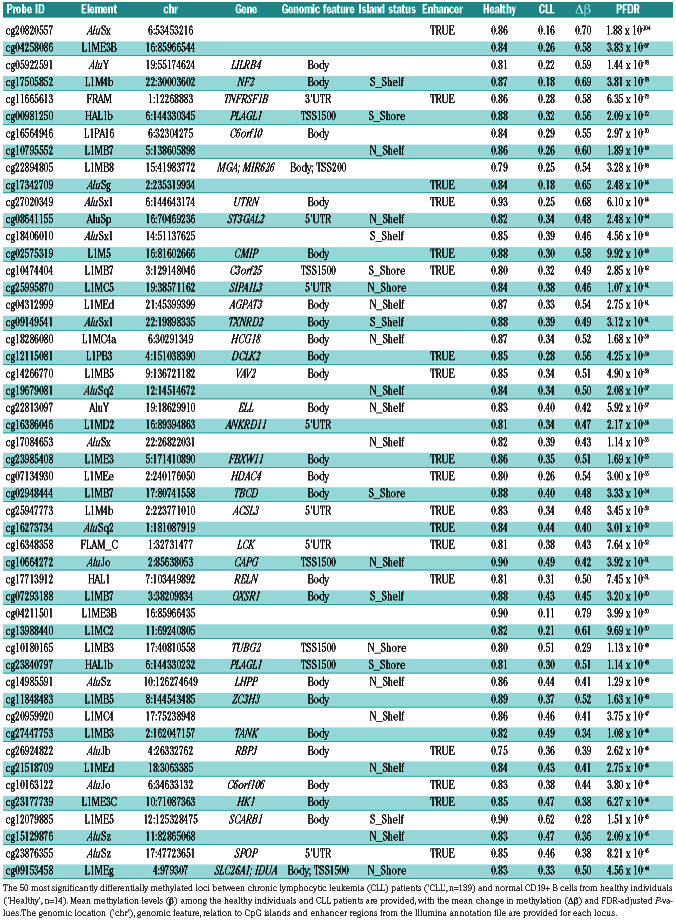
Table 1.
Leading differentially methylated retrotransposon loci in chronic lymphocytic leukemia.
Figure 2.
Methylation of L1 and Alu elements by genomic feature in normal CD19+ B cells. Methylation (b) of retrotransposon probes by Alu (A) and L1 (B) subfamilies and genomic feature, as reported by the Illumina annotation file. Retrotransposon probes were categorised as those mapping to within 1,500 or 200 bases of the transcriptional start site (TSS1500, TSS200), the 5’ untranslated region (5’UTR), first exon (1st exon), gene body (Body) and 3’ untranslated region (3’UTR). Lines indicate median values, boxes the interquartile range (IQR), whiskers the highest and lowest values within 1.5*IQR, and outliers are displayed as individual points.
Differential methylation of retrotransposon subfamilies by evolutionary age
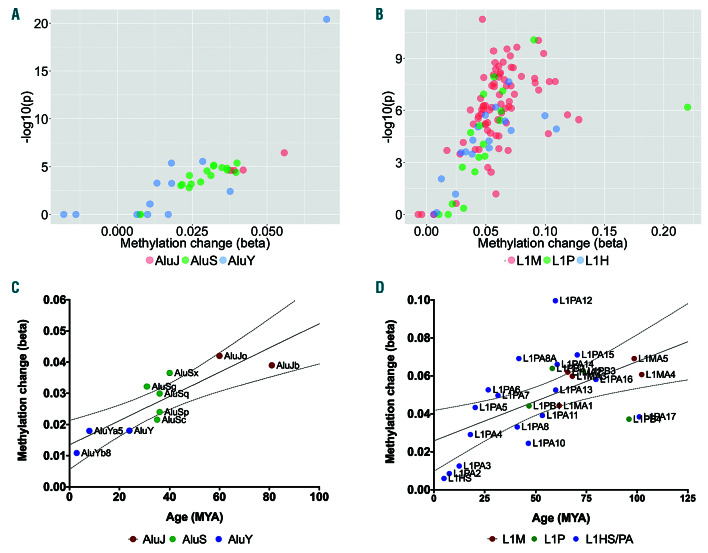
To identify changes in retrotransposon methylation in CLL, we analysed a publicly-available dataset of 139 CLL patients and normal CD19+ B-cells from 14 healthy individuals, 20 serving as the discovery cohort. Our analysis revealed that L1 and Alu elements are hypomethylated in CLL in a subfamily-specific manner. Greater changes in methylation were observed in the older AluJ subfamilies (mean Δb:-0.04) than in the intermediate age AluS (Δb:-0.03) and youngest AluY subfamilies (Δb:-0.02) (Figure 3A; PTrend<0.0001, ANOVA). L1 elements displayed a similar trend for greater hypomethylation among the oldest L1M subfamilies (mean Δb: -0.06) in comparison with the intermediate L1P (Δb: -0.05) and young L1HS & L1PA (Δb:-0.02) subfamilies (PTrend<0.0001, ANOVA), but displayed considerably greater variability by individual subfamily (Figure 3B).
Analysis of subfamilies by age of insertion into the human genome revealed a clear trend of greater hypomethylation of Alu elements in those integrated earliest (Figure 3C; r2=0.74, P=0.002). A similar but more subtle trend was also observed for hypomethylation of L1 subfamilies by time since integration (Figure 3D; r2=0.31, P=0.018).
Locus-specific hypomethylation of retrotransposons in CLL
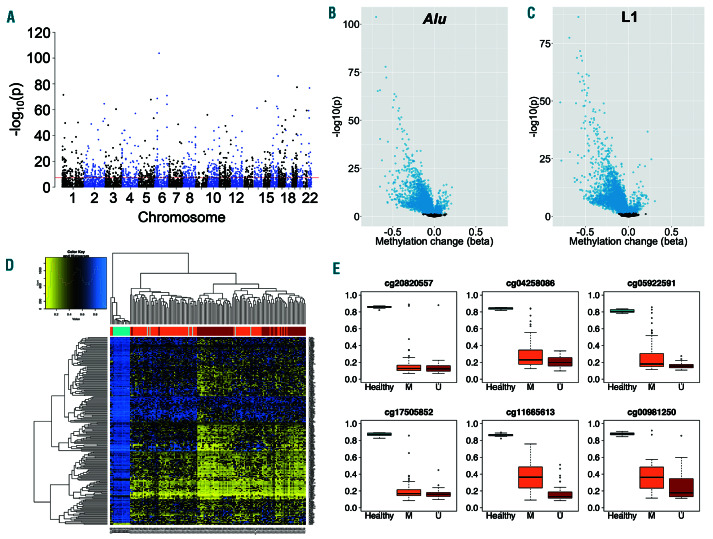
Epigenome-wide analysis of locus-specific retrotransposon methylation in the same 139 CLL patients and 14 healthy individuals revealed 10,782 loci distributed widely across the genome to be differentially methylated in CLL (PFDR<0.05; Figure 4A), with 5,896 mapping to Alu elements and 4,886 to L1 elements. Of these, 9,670 (89.7%) were hypomethylated and 1,112 (10.3%) were hypermethylated (Figure 4B-C), with differential methylation primarily occurring at sites that are highly methylated in normal B cells (>0.7 for 7,295 loci). The differentially methylated loci showed enrichment at enhancer regions (P=0.002, Fisher’s exact test), but no significant enrichment at CpG islands, shelves or shores (P=0.10-0.73). Unsupervised clustering identified distinct patterns of retrotransposon methylation in CLL in comparison with normal B cells, and an association with IGHV mutational status among CLL patients (Figure 4D).
The magnitude of hypomethylation was frequently large, with 43 of the 50 most significant loci displaying changes in methylation (Δb) of >0.40 (Table 1). At these 50 leading loci, methylation was uniformly high in normal CD19+ B cells from healthy individuals (range b: 0.71– 0.95), while hypomethylation was highly frequent in CLL and more common amongst patients with unmutated IGHV (Figure 4E).
We noted that single base changes (SNP) have been identified at the C or G position of target CpG sites for 43 of these 50 most significantly differentially methylated loci, but with extremely low minor allele frequencies (<0.01 for 41 of the 43 loci) in European populations (Online Supplementary Table S4).
Analysis in a validation cohort
To confirm our findings in the discovery cohort, we performed epigenome-wide analysis of retrotransposon methylation in a validation cohort of 24 CLL patients (Online Supplementary Table S2), in comparison to the same 14 healthy individuals from the discovery cohort. A total of 9,488 retrotransposon loci were differentially methylated in the validation CLL cohort (PFDR<0.05) with a very high level of correlation in the magnitude of methylation changes observed in the discovery and validation cohorts relative to normal CD19+ B cells (rho=0.76, P<2.2x10-16, Online Supplementary Figure S1). Of the 50 leading loci in the discovery cohort (Table 1), 41 were differentially methylated in the validation cohort (PFDR<0.0000001) with changes in methylation (Δb) of >0.40; the probes for the other nine loci were missing from the validation cohort dataset and could not be examined (Online Supplementary Table S5).
Figure 3.
Differential methylation of L1 and Alu subfamilies in chronic lymphocytic leukemia by evolutionary age. (A-B) Mean change in methylation (Δb) for probes mapping to each of the 37 Alu (A) and 117 L1 (B) subfamilies by the FDR-adjusted P-value. (C-D) Mean methylation change (Δb) of Alu (C) and L1 (D) subfamilies by time since integration into the genome (millions year ago, MYA). Mean and 95% Confidence Intervals (95% CI) are displayed.
DNA methylation of retrotransposon loci during B cell differentiation
Most epigenetic changes observed in CLL mirror those that occur during the process of B-cell development.21 In order to account for differential methylation that may be the product of B-cell maturation or proliferation, as opposed to being unique to CLL, we utilised publiclyavailable DNA methylation microarray data from CD19+ CD27– IgD+ naïve B cells (n=3) and CD19+ CD27+ IgA/G+ class-switched memory B cells (n=3) available from the study of Kulis et al.20 Of the 10,782 loci previously identified as differentially methylated in CLL, 8,898 (82.5%) did not display substantial changes in methylation (Δb>0.2) during B-cell maturation. Importantly only one of the leading 50 loci (Table 1), cg13988440, displayed such a change. Therefore, in contrast to many other studies of the CLL epigenome, our analysis has predominantly revealed changes in DNA methylation that are specific to the disease.
Effect of locus-specific hypomethylation upon proximal gene expression
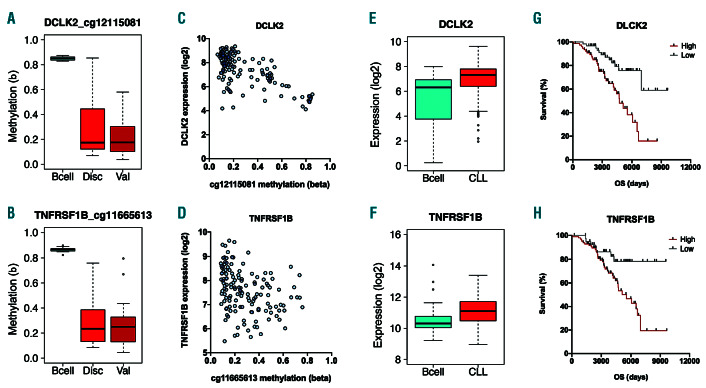
In order to explore the functional consequences of locus-specific retrotransposon hypomethylation events, we examined the effects of differential methylation at the leading 50 loci (Table 1) upon the expression of proximal genes using expression microarray data from the same 139 CLL patients within the discovery cohort. Thirty-seven of the leading 50 hits from the discovery cohort map to genes, with the other 13 mapping to intergenic regions that are >2 kb from the nearest transcriptional start site (Table 1). Of the 37 hits mapping to genes, differential methylation at 29 of the loci was confirmed in the validation cohort (Figure 5A-B; Online Supplementary Figure 1). Methylation at 7 of these 29 loci was correlated with expression of the proximal gene (Figure 5C–D; PFDR<0.05, Spearman rank correlation), illustrating the capacity for retrotransposon hypomethylation to modulate gene transcription. These seven genes were C6orf106 (ILRUN), DCLK2, HK1, TANK, TBCD, TNFRSF1B (TNFR2) and TXNRD2 (Online Supplementary Table S6). All showed negative correlations between methylation and expression (rho=-0.25 to -0.59), with the exception of TBCD which demonstrated a positive correlation (rho=0.25). Each of these seven genes displayed uniformly low levels of methylation at their promoter regions, thereby potentially facilitating retrotransposon-mediated regulation of gene expression (Online Supplementary Figure S2). In an independent cohort of 188 CLL patients and CD19+ B cells isolated from 32 healthy individuals, all seven genes were determined to be differentially expressed in CLL (Figure 5E–F; PFDR<0.05, Mann Whitney U test).
Figure 4.
Locus-specific differential methylation of retrotransposon elements in chronic lymphocytic leukemia. (A) Manhattan plot of retrotransposon loci showing differential methylation between chronic lymphocytic leukemia (CLL) patients (n=139) and normal CD19+ B cells from healthy individuals (n=14). The threshold (red line) indicates P<5x10-8. (B-C) Volcano plots for probes mapping to Alu (B) and L1 (C) elements. The change in methylation (Δb) and P-value (-log10(p)) are displayed, with significantly different loci (PFDR<0.05) highlighted in blue. (D) Heatmap displaying unsupervised clustering of the 200 most significantly differentially methylated loci. The colour scale indicates methylation level, from low (yellow) to high (blue). Healthy individuals (turquoise, n=14) and CLL patients with mutated (orange, n=75), unmutated (dark red, n=57) or unknown (grey, n=6) IGHV status are indicated. (E) Methylation of the six most significantly differentially methylated loci in healthy individuals (turquoise, n=14) and CLL patients with IGHV mutated (orange, n=75) and unmutated (dark red, n=57) disease. Lines indicate median values, boxes the interquartile range (IQR), whiskers the highest and lowest values within 1.5*IQR, and outliers are displayed as individual points.
In order to examine the clinical relevance of these genes, we examined their expression in the discovery cohort in relation to patient outcomes. High expression of DCLK2 and TNFRSF1B were associated with reduced patient survival (Figure 5G–H; PFDR<0.05, log-rank test), while high expression of HK1, ILRUN, TANK, and TBCD were associated with improved patient survival. In order to examine whether the associations with impaired patient prognosis were driven by IGHV status, analysis was then performed separately for IGHV mutated (n=75) and unmutated (n=57) patients, where status was known. Among IGHV mutated patients, significant associations with survival were retained for ILRUN and TANK expression (PFDR<0.05), while DCLK2 PFDR=0.0532) approached significance. HK1, TBCD and TNFRSF1B no longer achieved statistical significance in this restricted patient group (all PFDR<0.13). Among IGHV unmutated patients, significant associations were present for expression of TANK (PFDR=0.0210) and TNFRSF1B (PFDR=0.0005).
Stability of retrotransposon methylation across hematopoietic cell types
It is recognised that studies of DNA methylation in blood samples must take into account changes in the relative proportions of the cell types.38 We therefore examined our 10 leading loci (Table 1) in publicly-available HM450K datasets from isolated hematopoietic cell types to reveal differences in retrotransposon methylation by leukocyte cell type. We employed data obtained from samples of whole blood, peripheral blood mononuclear cells, CD16+ neutrophils, eosinophils, CD14+ monocytes, multipotent progenitors, pre-B cells, immature B cells, CD19+ B cells, CD4+ T-cells, CD8+ T cells, and CD56+ natural killer cells (each n=6). Nine of the loci (cg20820557, cg04258086, cg05922591, cg17505852, cg11665613, cg00981250, cg16564946, cg10795552, and cg17342709) were highly methylated across all cell types and displayed little variation between them (Figure 6A-B). In contrast, cg22894805 showed high methylation in lymphoid cells but low levels in myeloid cells (Figure 6C).
Figure 5.
Impact of retrotransposon methylation upon the expression of proximal genes. (A-B) Methylation at cg12115081 (A) and cg11665613 (B) in normal CD19+ B cells (‘Bcell’; n=14) and chronic lymphocytic leukemia (CLL) patients within the discovery cohort (‘Disc’; n=139) and validation cohort (‘Val’; n=24). (C-D) Correlation between retrotransposon methylation and proximal gene expression for cg12115081 and DCLK2 expression (C) and for cg11665613 with TNFRSF1B expression (D). (E-F) Expression of DCKL2 (E) and TNFRSF1B (F) in normal CD19+ B-cells (n=32) and CLL patients (n=188) in an independent cohort. (G-H) Kaplan-Meier plots for patient overall survival stratified by expression level (high/low) of DCLK2 (G) and TNFRSF1B (H) in the Discovery cohort.
Specificity of differential retrotransposon methylation to CLL
In order to determine whether the observed changes are specific to CLL or may also be present in other lymphoid and myeloid malignancies, we analysed the 10 most significantly altered loci (Table 1) in publicly-available HM450K datasets from healthy individuals and patients with other hematological malignancies. At each of these 10 loci we had observed mean methylation levels (b) of <0.32 in CLL patients in comparison to >0.79 in normal CD19+ B cells.
First, to assess the variability in methylation of these loci in the general population, we leveraged three largescale studies of blood samples from a total of 1,169 healthy individuals: the Hannum et al. study of human ageing (n=656);24 the European Prospective Investigation into Cancer and Nutrition (EPIC, n=329);25 and the Young Finns study (n=184).26 Our analysis confirmed the loci to be almost exclusively highly methylated in healthy individuals (Figure 6D–E). Eight of the loci (cg20820557, cg04258086, cg05922591, cg17505852, cg11665613, cg00981250, cg10795552, and cg17342709) displayed mean methylation levels (b) of 0.86–0.96 in each of the three cohorts of healthy individuals, with low variability across the populations (Online Supplementary Table S7). cg16564946 was also highly methylated but displayed more variation within the cohorts (mean b: 0.74±0.11 – 0.79±0.07). Methylation at cg22894805 measured in these unfractionated leukocyte samples was lower than previously observed in isolated CD19+ B-cells (Table 1 and Online Supplementary Table S7) due to variation in methylation by leukocyte cell type (Figure 6C), but was still different in each study compared to CLL patients (all P<0.0001; Mann Whitney U test).
In order to determine whether the identified loci may also be differentially methylated in other hematological malignancies, we examined datasets from patients with ALL (n=797),27 CML (n=12),28 AML (n=68),29 and lymphoma (diffuse large B-cell lymphoma and Burkitt’s lymphoma, n=31).30 Hypomethylation was predominantly confined to CLL at nine of the loci (cg20820557, cg04258086, cg05922591, cg17505852, cg11665613, cg00981250, cg16564946, cg10795552, and cg17342709), with mean methylation (b) levels of >0.74 in ALL, CML and AML (Figure 6D-E and Online Supplementary Table S7). Mean methylation levels were uniformly lower in lymphoma (b: 0.61–0.73), with up to 23% of patients having methylation levels of <0.40 at the loci. In contrast to the other nine loci, and in concordance with our analysis by hematopoietic cell type, cg22894805 had low methylation levels in myeloid malignancies (AML, CML) and higher levels in lymphoid malignancies (ALL, lymphoma) (Figure 6F).
Figure 6.
Differentially methylated loci in other haematological malignancies and leukocyte cell types. (A-C) Methylation (b) of three loci by blood cell type. Publiclyavailable datasets from whole blood (WB, n=6), peripheral blood mononuclear cells (PBMC, n=6), CD16+ neutrophils (Neutro, n=6), eosinophils (Eosin, n=6), CD14+ monocytes (Mono, n=6), multipotent progenitor cells (MPP, n=6), Pre-B I and Pre-B II cells (both n=6), immature B cells (B_imm, n=4), CD19+ B cells (n=14), CD4+ and CD8+ T cells (both n=6), and CD56+ natural killer cells (NK, n=6). Lines indicate median values, boxes the interquartile range (IQR), whiskers the highest and lowest values within 1.5*IQR, and outliers are displayed as individual points. (D-F) Methylation (β) of the same three loci in publicly-available datasets from three studies of healthy individuals (Hannum et al.24, n=656; EPIC, n=329; the Young Finns Study (YFS), n=184), CLL patients within the discovery cohort (n=139), and patients with ALL (n=797), AML (n=68), CML (n=12), and diffuse large B-cell and Burtkitt’s lymphomas (Lymph, n=31).
Together, our results indicate that the largest observed differences in retrotransposon methylation are highly specific to CLL and are not the product of changes in cell type proportions, including hypomethylation at cg11665613 that is associated with reactivation of TNFRSF1B expression.
Locus-specific hypomethylation of retrotransposons prior to diagnosis
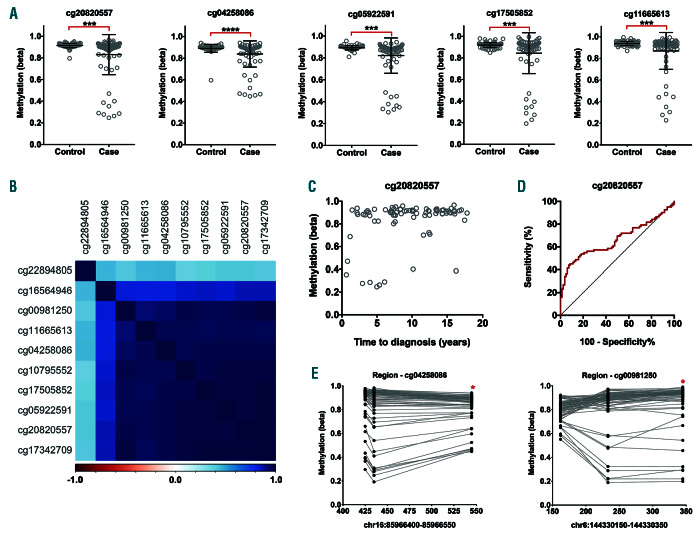
We sought to establish whether these epigenetic changes might constitute early events that are detectable prior to patient diagnosis. We analysed methylation of the 10 leading loci in blood samples taken from 82 individuals up to 18 years prior to their being diagnosed with CLL and 82 individually matched controls who remained free of disease at the same age at follow-up. For nine of the loci (cg20820557, cg04258086, cg05922591, cg17505852, cg11665613, cg00981250, cg16564946, cg10795552, and cg17342709) lower methylation levels were prospectively associated with the diagnosis of CLL (all P<0.003, Mann Whitney U test). Additionally, there was weak evidence of an association for cg22894805 (P=0.0501). Reduced methylation levels were, however, confined to 9 of the 82 CLL cases, with the remainder displaying normal, high levels of methylation at each locus (Figure 7A). Methylation across the 10 loci was very highly correlated (Figure 7B), with the same nine individuals displaying hypomethylation at each locus. There was weak evidence of a positive association between methylation and time to diagnosis (PFDR=0.06–0.12) at all loci except for cg10795552 (PFDR=0.51). Notably, hypomethylation was more commonly present among cases diagnosed <7 years after blood draw (7 of 24, 29%) than those diagnosed more than 7 years after (2 of 58, 3%) (Figure 7C). ROC curve analysis identified that a methylation cut-off of b=0.90 for cg2080557 provides 45% sensitivity and 91% specificity (area under curve [AUC]=0.66, P=0.0003) in predicting diagnosis with CLL (Figure 7D).
In order to determine whether hypomethylation is highly localised or more regional, we examined methylation at other CpG sites captured by the HM450K array that are within 200 bp of the target loci. Only two of these loci had regional CpG sites in close proximity that are captured by the array: two CpG sites within 119 bp of cg04258086 (cg04211501 and cg06951664); and two CpG sites up to 184 bp downstream of cg00981250 (cg23840797 and cg18593039). For both regions, the neighbouring CpG sites mapped to within the same retrotransposon element as the target (L1ME3B and HAL1 respectively). Methylation at the neighbouring CpG sites was observed to be highly similar to that of the target loci (Figure 7E), demonstrating that the observed epigenetic changes are not confined to a single CpG site.
Figure 7.
Hypomethylation of retrotransposon loci prior to chronic lymphocytic leukemia diagnosis. Analysis of 10 retrotransposon loci observed to be hypomethylated in chronic lymphocytic leukemia (CLL) patients, analysed by HM450K in prospective samples taken from future CLL patients within the Melbourne Collaborative Cohort Study. (A) Methylation (β) of five loci in 82 future CLL patients (Case) and 82 age-matched individuals who remained free of the disease (Control) (***P<0.005, ****P<0.0001). (B) Correlation in methylation between the ten loci among the 82 future CLL cases. The colour scale indicates negative (red) and positive (blue) correlations. (C) Correlation between methylation (β) of cg20820557 and time to diagnosis (years) in the 82 future CLL cases. (D) ROC curve analysis for cg20820557 in the prediction of CLL diagnosis among 82 future CLL cases and 82 age-matched healthy controls who remained free of the disease (area under curve [AUC]=0.66, P=0.0003). (E) Regional analysis of methylation (β) at HM450K probes mapping to within 200 bases of cg04258086 and cg00981250 (indicated by asterisks, *). Methylation at loci within the same patient are indicated by connecting lines.
Discussion
To the best of our knowledge, our study is the first to utilise the Illumina Infinium microarray platform to analyse locus- and subfamily-specific methylation of retrotransposons in human disease. The potential for this approach has previously been demonstrated through use of the HM27K and HM450K microarray platforms to analyse L1 and Alu elements categorised as young (L1H, AluY), intermediate (L1P, AluS) and old (L1M, AluJ) in healthy tissues39 and a lymphoblastoid cell line.40 We have built on this to identify, for the first time, differential methylation of LINE-1 and Alu elements by their evolutionary heritage in leukemia. Importantly, our analysis has identified novel epigenetic changes specific to CLL that are highly frequent among patients, that modulate the expression of proximal genes, and that can be detected prior to diagnosis in some individuals. Together, our results suggest that epigenetic dysregulation of retrotransposons may be implicated in the development and progression of CLL.
There remains significant ignorance regarding the genetic and epigenetic origins and evolution of CLL. It has recently been demonstrated that the genome and epigenome show co-evolution during disease progression.21,41 This co-evolution predominantly consists of hypomethylation away from promoters and transcriptional start sites at regions that are predominantly highly methylated in normal B cells21 which, hence, may describe general loss of epigenome maintenance as opposed to sitespecific selection for gene silencing or activation. Our study has revealed hypomethylation of retrotransposon elements in a subfamily-specific manner which, in accordance with the findings of Oakes et al.,21 primarily occurs away from CpG islands in regions that are highly methylated in normal B cells. Our findings may thus throw light on the underlying cause of this epigenetic evolution in CLL. Interestingly, we observed that this hypomethylation is significantly enriched at enhancer regions, which may implicate the dysregulation of retrotransposons in the transcriptional re-wiring of CLL cells.
To date there have only been two studies of retrotransposon methylation in CLL, both of which reported associations with genetic instability. Fabris et al.22 reported significant hypomethylation of L1 and Alu elements that was associated with 17p deletions, while Hoxha et al.23 reported associations between L1 and Alu methylation with telomere length. This is corroborated by recent findings that telomere length is associated with del(17p), del(11q) and mutations in ATM and SF3B1,42 together linking methylation of retrotransposons, telomere length and the acquisition of genetic abnormalities in CLL. This is further supported by the extensive evidence linking loss of retrotransposon methylation to their activation and genetic instability in solid tumours.13,14 However, the studies by Fabris et al.22 and Hoxha et al.23 used a pyrosequencingbased approach that was limited to the L1HS and AluSx subfamilies, on account of their relative abundance in the genome and subsequent suitability as surrogate markers of global DNA methylation patterns. Such an approach does not facilitate locus-specific analysis. While providing novel information about the disease, these two studies were unable to address the full complexity of retrotransposon DNA methylation in CLL. Indeed, we have demonstrated the utility and necessity of performing subfamilyand locus-specific analysis to fully appreciate their implication in malignancies. Comprehensive analysis of retrotransposons by high-throughput sequencing approaches is highly challenging due to the difficulty in mapping sequencing reads containing repetitive sequences to singular genomic regions, requiring tailored bioinformatics approaches such as RetroSeq.43 Hence, the approach that we have developed, to analyse retrotransposons using the widely-utilised HM450K platform, enables a comparatively simple and cost-effective means to perform this demanding work.
We have utilised this locus-specific analytical approach to reveal that hypomethylation of retrotransposons can modulate the expression of proximal genes in CLL. In particular, we identified hypomethylation of an L1PB3 sequence within the second intron of DCLK2 and a FRAM sequence within the 3’UTR of TNFRSF1B that are inversely associated with expression of the genes, both of which are overexpressed in CLL and for which higher expression levels are associated with reduced patient survival. TNFRSF1B encodes a member of the TNF-receptor superfamily that interacts with TRAF2 to promote cell proliferation and survival via stimulation of the non-canonical NF-kB pathway, and it is known to be overexpressed in CLL.44,45 Myeloid cell lines treated with the DNMT inhibitor 5-aza-2’-deoxycytidine show increased expression of TNFRSF1B,46 and our study has demonstrated that hypomethylation of the FRAM element within the 3’UTR is associated with increased expression of the gene. While many studies have focussed primarily on epigenetic changes at promoter and enhancer regions, DNA methylation at 3’UTR has been shown to be associated with differential gene expression and patient outcomes in solid tumours,47 although the underlying mechanisms have not been elucidated. Increased expression of the TNFRSF1B promotes cell survival and resistance to apoptosis,48 which may explain our observation of worse patient prognosis with high TNFRSF1B expression.
In contrast, the function of DCLK2 in CLL biology is less clear. This gene encodes a regulator of microtubule polymerisation, and while family members have been identified as potential prognostic markers in gastric and colorectal cancers,49,50 DCLK2 has not previously been reported as differentially methylated or expressed in CLL. Further work is required to establish the functional role of this gene in the disease. We also report that hypomethylation of an intronic AluSx1 element within TXNRD2 was associated with increased expression of the gene in CLL. While we observed no association with patient survival, a germline variant within 300 bp of this Alu element has been reported to be associated with shorter time to first treatment.51 The gene encodes a thioredoxin reductase that promotes malignant cell survival by providing protection against reactive oxygen species,52 and targeting these enzymes by use of chemotherapeutic agents such as auranofin may be effective in the treatment of CLL.53
In addition to the impact on the expression of proximal genes, epigenetic dysregulation of L1 and Alu elements may further be implicated in CLL biology through retrotransposition events. Recent work elsewhere has identified enrichment of transposable elements in the transcriptome of CLL patients, but without being able to identify the underlying cause.54 Our study has revealed widespread locus-specific epigenetic dysregulation of retrotransposon elements in the genome, which may serve to explain their increased expression in CLL, but the full implications of these observations remains to be elucidated. Hypomethylation of retrotransposon elements leads to their re-activation55 and potential retrotransposition into the genome. Somatic retrotransposition has been widely studied in solid tumours and is a highly frequent event in cancers of the lung, colon, breast and prostate.15 However, to date there have been no studies of this phenomenon in leukemias. Our findings, together with those observations elsewhere of reactivation of transposable elements54 and association of retrotransposon methylation with the acquisition of 17p deletions,22 indicate that the study of this phenomenon may help to identify novel genetic drivers of the disease. The most frequently mutated driver genes (NOTCH1, SF3B1, TP53, MYD88 and BIRC3) are mutated in only 2-12% of CLL patients,56 and large-scale studies have been unable to identify driver mutations in 25-40% of patients,57,58 underlining the need for further research in this area. While aberrant methylation was more frequently observed in the older and (presumed) inactive subfamilies such as L1M, hypomethylation of active AluY and L1HS elements was also identified, with two AluY sequences on chromosome 19 amongst the leading hits from our analysis.
We noted that rare genetic variants have been reported at the target CpG sites for many of the leading hits from this study. Genome-wide association studies have identified more than 20 genetic susceptibility loci for CLL,59 yet together these loci only account for 25% of heritable risk60 and therefore genetic predisposition to CLL remains to be fully explained. SNP are enriched within Alu elements,61 and such variants at CpG sites can lead to their elimination and thus loss of methylation and reactivation. However, the extremely low minor allele frequencies for the variants mapping to the 50 leading loci and our observation of normal methylation in most individuals prior to their CLL diagnosis suggest that these loci do not correspond to inherited predisposition to the disease. It nonetheless remains to be established whether the observed hypomethylation could originate through the acquisition of genetic changes during disease initiation, as the high frequency of methylated CpG sites within L1 and Alu sequences results in a high point mutation rate through the deamination of methylated-cytosine.
As our study has primarily involved analysis of publicly- available datasets, we have been unable to sequence regions of interest to examine regional changes in DNA methylation and detect genetic variants that may be implicated in the observed changes. Furthermore, we have not been able to examine whether these epigenetic changes lead to re-expression of the elements and somatic retrotransposition. However, our study has significant strengths. We have developed an approach that enables the locus-specific interrogation of L1 and Alu element methylation across the genome using the widely-utilized HM450K platform, thereby facilitating its ready application to studies elsewhere. We have also comprehensively examined the leading hits from our analysis, including examination in 1,169 healthy individuals to determine the frequency of hypomethylation in the general population, fractionated leukocytes to identify cell type-specific differences, and other hematological malignancies to determine the specificity of the observed changes to CLL.
In summary, we have identified locus- and subfamilyspecific hypomethylation of L1 and Alu elements that is highly frequent and specific to CLL, modulates the expression proximal genes, and was observable in a proportion of future CLL cases prior to their diagnosis. Further work is required to elucidate how these epigenetic changes may be implicated in leukaemogenesis and to investigate somatic retrotransposition as a potential driver of CLL.
Data availability
The datasets used in this study are publicly-available through the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home) and the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/). Accession numbers for all datasets used in this study can be found within Online Supplementary Table S1.
Supplementary Material
Acknowledgments
We would like to thank all patients involved in the study. We acknowledge the Edinburgh Clinical Research Facility for the performance of the DNA methylation microarrays from the validation cohort, and the Newcastle University Bioinformatics Support Unit for their assistance with the analysis of DNA methylation microarray data.
Funding Statement
Funding: This work was supported by funding from Bright Red (awarded to Timothy Barrow) and the JGW Patterson Foundation (grant number 30015.088.045/PA/IXS, awarded to Gordon Strathdee).
References
- 1.Mighell AJ, Markham AF, Robinson PA. Alu sequences. FEBS Lett. 1997;417(1):1-5. [DOI] [PubMed] [Google Scholar]
- 2.Price AL, Eskin E, Pevzner PA. Wholegenome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14(11):2245-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007; 23(4):183-191. [DOI] [PubMed] [Google Scholar]
- 4.Konkel MK, Walker JA, Hotard AB, et al. Sequence analysis and characterization of active human Alu subfamilies based on the 1000 Genomes Pilot Project. Genome Biol Evol. 2015;7(9):2608-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck CR, Collier P, Macfarlane C, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141(7):1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xing J, Zhang Y, Han K, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19(9):1516-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67(3):876-880. [DOI] [PubMed] [Google Scholar]
- 8.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180(5):462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byun HM, Motta V, Panni T, et al. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol. 2013;10:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao-Jie L, Hui-Ying X, Qi X, Jiang X, Shi-Jie M. LINE-1 in cancer: multifaceted functions and potential clinical implications. Genet Med. 2016;18(5):431-439. [DOI] [PubMed] [Google Scholar]
- 11.Benard A, van de Velde CJ, Lessard L, et al. Epigenetic status of LINE-1 predicts clinical outcome in early-stage rectal cancer. Br J Cancer. 2013;109(12):3073-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolff EM, Byun HM, Han HF, et al. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6(4):e1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daskalos A, Nikolaidis G, Xinarianos G, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer. 2009;124(1):81-87. [DOI] [PubMed] [Google Scholar]
- 14.Symer DE, Connelly C, Szak ST, et al. Human L1 retrotransposition is associated with genetic instability in vivo. Cell. 2002; 110(3):327-338. [DOI] [PubMed] [Google Scholar]
- 15.Tubio JMC, Li Y, Ju YS, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014; 345(6196):1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee E, Iskow R, Yang L, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016;26(6):745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ewing AD, Gacita A, Wood LD, et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 2015;25(10):1536-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doucet-O’Hare TT, Rodić N, Sharma R, et al. LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc Natl Acad Sci U S A. 2015; 112(35):E4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44(11):1236-1242. [DOI] [PubMed] [Google Scholar]
- 21.Oakes CC, Claus R, Gu L, et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2014;4(3):348-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabris S, Bollati V, Agnelli L, et al. Biological and clinical relevance of quantitative global methylation of repetitive DNA sequences in chronic lymphocytic leukemia. Epigenetics. 2011;6(2):188-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoxha M, Fabris S, Agnelli L, et al. Relevance of telomere/telomerase system impairment in early stage chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2014;53(7):612-621. [DOI] [PubMed] [Google Scholar]
- 24.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demetriou CA, Chen J, Polidoro S, et al. Methylome analysis and epigenetic changes associated with menarcheal age. PLoS One. 2013;8(11):e79391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kananen L, Marttila S, Nevalainen T, et al. Aging-associated DNA methylation changes in middle-aged individuals: the Young Finns study. BMC Genomics. 2016;17:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordlund J, Bäcklin CL, Wahlberg P, et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013; 14(9):r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maupetit-Mehouas S, Court F, Bourgne C, et al. DNA methylation profiling reveals a pathological signature that contributes to transcriptional defects of CD34. Mol Oncol. 2018;12(6):814-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira HJ, Heyn H, Vizoso M, et al. DNMT3A mutations mediate the epigenetic reactivation of the leukemogenic factor MEIS1 in acute myeloid leukemia. Oncogene. 2016;35(23):3079-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsunaga A, Hishima T, Tanaka N, et al. DNA methylation profiling can classify HIVassociated lymphomas. AIDS. 2014; 28(4):503-510. [DOI] [PubMed] [Google Scholar]
- 31.Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee ST, Xiao Y, Muench MO, et al. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res. 2012;40(22):11339-11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong Doo N, Makalic E, Joo JE, et al. Global measures of peripheral blood-derived DNA methylation as a risk factor in the development of mature B-cell neoplasms. Epigenomics. 2016;8(1):55-66. [DOI] [PubMed] [Google Scholar]
- 34.Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smit AFA, Hubley R, Green P. RepeatMasker Open-4.0. 2013 [Google Scholar]
- 36.Kapitonov V, Jurka J. The age of Alu subfamilies. J Mol Evol. 1996;42(1):59-65. [DOI] [PubMed] [Google Scholar]
- 37.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16(1):78-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price EM, Cotton AM, Peñaherrera MS, McFadden DE, Kobor MS, Robinson W. Different measures of “genome-wide” DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics. 2012;7(6):652-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Y, Joyce BT, Liu L, et al. Prediction of genome-wide DNA methylation in repetitive elements. Nucleic Acids Res. 2017; 45(15):8697-8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landau DA, Clement K, Ziller MJ, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26(6):813-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strefford JC, Kadalayil L, Forster J, et al. Telomere length predicts progression and overall survival in chronic lymphocytic leukemia: data from the UK LRF CLL4 trial. Leukemia. 2015;29(12):2411-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keane TM, Wong K, Adams DJ. RetroSeq: transposable element discovery from nextgeneration sequencing data. Bioinformatics. 2013;29(3):389-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hui D, Satkunam N, Al Kaptan M, Reiman T, Lai R. Pathway-specific apoptotic gene expression profiling in chronic lymphocytic leukemia and follicular lymphoma. Mod Pathol. 2006;19(9):1192-1202. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Coombes KR, Highsmith WE, Keating MJ, Abruzzo LV. Differences in gene expression between B-cell chronic lymphocytic leukemia and normal B cells: a metaanalysis of three microarray studies. Bioinformatics. 2004;20(17):3166-3178. [DOI] [PubMed] [Google Scholar]
- 46.Laurenzana A, Petruccelli LA, Pettersson F, et al. Inhibition of DNA methyltransferase activates tumor necrosis factor alphainduced monocytic differentiation in acute myeloid leukemia cells. Cancer Res. 2009;69(1):55-64. [DOI] [PubMed] [Google Scholar]
- 47.McGuire MH, Herbrich SM, Dasari SK, et al. Pan-cancer genomic analysis links 3’UTR DNA methylation with increased gene expression in T cells. EBioMedicine. 2019; 43:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11(9):372-377. [DOI] [PubMed] [Google Scholar]
- 49.Dai J, Li ZX, Zhang Y, et al. Whole genome messenger RNA profiling identifies a novel signature to predict gastric cancer survival. Clin Transl Gastroenterol. 2019; 10(1): e00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao T, Wang M, Xu L, Wen T, Liu J, An G. DCLK1 is up-regulated and associated with metastasis and prognosis in colorectal cancer. J Cancer Res Clin Oncol. 2016; 142(10):2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mosquera Orgueira A, Antelo Rodríguez B, Alonso Vence N, et al. The association of germline variants with chronic lymphocytic leukemia outcome suggests the implication of novel genes and pathways in clinical evolution. BMC Cancer. 2019;19(1):515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lincoln DT, Ali Emadi EM, Tonissen KF, Clarke FM. The thioredoxin-thioredoxin reductase system: over-expression in human cancer. Anticancer Res. 2003;23(3B):2425-2433. [PubMed] [Google Scholar]
- 53.Fiskus W, Saba N, Shen M, et al. Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Res. 2014;74(9):2520-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferreira PG, Jares P, Rico D, et al. Transcriptome characterization by RNA sequencing identifies a major molecular and clinical subdivision in chronic lymphocytic leukemia. Genome Res. 2014;24(2):212-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schulz WA, Steinhoff C, Florl AR. Methylation of endogenous human retroelements in health and disease. Curr Top Microbiol Immunol. 2006;310:211-250. [DOI] [PubMed] [Google Scholar]
- 56.Baliakas P, Hadzidimitriou A, Sutton LA, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29(2):329-336. [DOI] [PubMed] [Google Scholar]
- 57.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puente XS, Beà S, Valdés-Mas R, et al. Noncoding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015; 526 (7574):519-524. [DOI] [PubMed] [Google Scholar]
- 59.Berndt SI, Skibola CF, Joseph V, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet. 2013;45(8):868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Law PJ, Berndt SI, Speedy HE, et al. Genome-wide association analysis implicates dysregulation of immunity genes in chronic lymphocytic leukaemia. Nat Commun. 2017;8:14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng SK, Xue H. Alu-associated enhancement of single nucleotide polymorphisms in the human genome. Gene. 2006;368:110-116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this study are publicly-available through the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home) and the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/). Accession numbers for all datasets used in this study can be found within Online Supplementary Table S1.