Abstract
Multiple myeloma (MM) is a hematologic malignancy strongly characterized by genomic instability, which promotes disease progression and drug resistance. Since we previously demonstrated that LIG3-dependent repair is involved in the genomic instability, drug resistance and survival of MM cells, we here investigated the biological relevance of PARP1, a driver component of the alternative nonhomologous end joining pathway, in MM. We found a significant correlation between higher PARP1 mRNA expression and poor prognosis of MM patients. PARP1 knockdown or its pharmacological inhibition by olaparib impaired MM cell viability in vitro and was effective against in vivo xenografts of human MM. Anti-proliferative effects induced by PARP1 inhibition were correlated with an increase of DNA double-strand breaks, activation of the DNA damage response and apoptosis. Importantly, by comparing a gene expression signature of PARP-inhibitor sensitivity to our plasma cell dyscrasia gene expression profiling, we identified a subset of MM patients who could benefit from PARP inhibitors. In particular, gene set enrichment analysis suggested that high MYC expression correlates with sensitivity to PARP inhibitors in MM. Indeed, we identified MYC as a promoter of PARP1-mediated repair in MM and, consistently, we demonstrated that cytotoxic effects induced by PARP inhibition are mostly detectable in MYC-proficient MM cells. Taken together, our findings indicate that MYC-driven MM cells are addicted to PARP1 alternative non-homologous end joining repair, which therefore represents a druggable target in this still incurable disease.
Introduction
Genomic instability represents a key hallmark of cancer since it progressively promotes the acquisition of features that lead to tumorigenesis. Indeed, almost all cancers are characterized by the tendency to accumulate genetic aberrations, such as gene copy-number variations, translocations and mutations, which finally confer growth and survival advantages.1
Genomic instability is strongly fostered by alterations of DNA repair pathways, which compromise “genomic guardian” mechanisms involved in the prevention of neoplastic transformation. At the same time, the DNA damage response machinery represents a specific Achilles heel of tumors, since it could be therapeutically exploited by designing anticancer strategies targeting cancer DNA repair vulnerabilities with a synthetic lethality approach.2 The discovery that homologous recombination- deficient tumor cells are specifically sensitive to inhibition of poly(ADP ribose) polymerase 1 (PARP1)3-5 provides an example of how to therapeutically exploit these vulnerabilities and led to the rapid development of several other DNA damage response inhibitors such as ATM, ATR, CHK1 and CHK2, DNA-PK and WEE1 inhibitors.6 However, it remains unclear why a substantial number of patients who lack homologous recombination mutations still benefit from PARP inhibition,7 a condition defined as “PARPness”.8
Double-strand breaks in DNA are mainly repaired by classical non-homologous end joining (NHEJ)9 and homologous recombination.10 Recently, a third repair pathway, named alternative NHEJ11 has been demonstrated to function as an error-prone, back-up pathway when the two major mechanisms are defective, contributing to the pathogenesis of several tumors.12,15
Multiple myeloma (MM) is a hematologic malignancy characterized by the growth of malignant plasma cells harboring numerous karyotypic aberrations.16 Indeed, the presence of specific cytogenetic abnormalities defines subgroups of MM patients with different prognoses requiring risk-adapted treatment. Although new therapeutics have prolonged the survival of MM patients, a cure for this disease is still an unmet need, and the identification of novel and actionable molecular drivers might provide innovative therapeutic approaches. It is known that genomic instability is a hallmark of MM but, to date, specific Achilles heels have not been identified.
PARP1 is the best-characterized member of the PARP family and plays a crucial role in the alternative NHEJ pathway.17,18 Indeed, it senses DNA damage via its DNA binding domain,19 and subsequently synthesizes poly (ADP-ribose) polymers which are added to itself and other acceptor proteins, thus recruiting other DNA repair proteins, including DNA ligase 3 (LIG3).
We have provided evidence that upregulation of LIG3- mediated DNA repair plays a pivotal role in genomic instability and survival of MM cells.20 Starting from these data, we here report that PARP1 is crucial for survival of MYC-addicted MM cells, and we provide the rational framework for the use of PARP inhibitors as a therapeutic strategy in this still incurable disease.
Methods
A more detailed description of the methods used is provided in the Online Supplementary Methods.
Cell lines and primary tumor specimens
Cell lines were obtained from the American Type Culture Collection or kindly provided by sources indicated in the Online Supplementary Methods.
Analysis of cell viability and apoptosis
Cell viability was analyzed by Cell Titer-Glo assay (Promega, Madison, WI, USA); apoptosis was evaluated by flow cytometric analysis (Attune NxT Flow cytometer, Thermo Fisher Scientific, IL, USA) following annexin V-7-aminoactinomycin D staining (BD Pharmingen).
Cell cycle analysis
The cell cycle was analyzed by propidium iodide flow cytometry (BD Pharmingen), according to the manufacturer’s instructions, using an Attune NxT Flow cytometer (Thermo Fisher Scientific).
Western blot analysis
Whole cell protein extracts were prepared from MM cells and from peripheral blood mononuclear cells in NP40 Cell Lysis Buffer (Novex®) containing a cocktail of protease inhibitors (Sigma, Steinheim, Germany). Cell lysates were loaded and separated by polyacrylamide gel electrophoresis. Proteins were transferred onto membranes by a Trans-Blot® TurboTM Transfer Starter System for 7 min. After protein transfer, the membranes were blotted with antibodies listed in the Online Supplementary Table and visualized with a C-DiGit® Blot Scanner (LI-COR) using ECL Western Blotting Detection Reagents (Thermo Fisher Scientific). Images were captured using Image Studio® software (LI-COR, version 5.0).
Immunofluorescence
Cells were harvested, centrifuged onto glass slides (Cytospin 4, Thermo Scientific), and fixed in 4% paraformaldehyde in phosphate- buffered saline (PBS), pH 7.4, for 12 min at 22°C, followed by three 5 min washes in PBS. Cells were permeabilized (0.1% Triton X-100 in PBS, 15 min), washed in PBS (3 times, for 5 min each), and incubated for 1 h at 22°C with blocking buffer (1.5% bovine serum albumin in PBS). They were reacted for more than 12 h at 4°C with primary antibodies listed in the Online Supplementary Table, washed in PBS (3 times, for 5 min each), and incubated for 1 h at 22°C in the dark, with appropriate secondary antibodies. Cells were washed three times in PBS and mounted under coverslips with Vectashield with DAPI (Vector Laboratories). Images were acquired with an SP2 Leica Zeiss confocal laser-scanning microscope with a 63× oil objective.
Animals and in vivo models of human multiple myeloma
Male CB-17 severe combined immunodeficient (SCID) mice (6- to 8-weeks old; Harlan Laboratories, Inc., IN, USA) were housed and monitored in our Animal Research Facility. Experimental procedures and protocols had been approved by the Magna Graecia University Institutional Review Board and were conducted according to protocols approved by the National Directorate of Veterinary Services (Italy). Mice were subcutaneously inoculated with 5x106 H929 or ABZB cells and treatment started when palpable tumors became detectable (100-200 mm3). Animals were treated daily with olaparib (Selleckchem) (100 mg/kg) or vehicle (10% v/v dimethylsulfoxide in 10% w/v Kleptose (hydroxypropyl-bcyclodextrin) in purified de-ionized water) via oral gavage. Tumor sizes were measured as described elsewhere,21 and the investigator was blinded to group allocation.
Statistical analysis
Each experiment was performed at least three times and values are reported as means ± standard deviations. Comparisons between groups were made with the Student t-test, while statistical significance of differences among multiple groups was determined by GraphPad software (www.graphpad.com). Graphs were obtained using Graphpad Prism version 6.0. P-values of less than 0.05 were accepted as statistically significant.
Results
High PARP1 expression occurs in multiple myeloma and predicts for poor prognosis
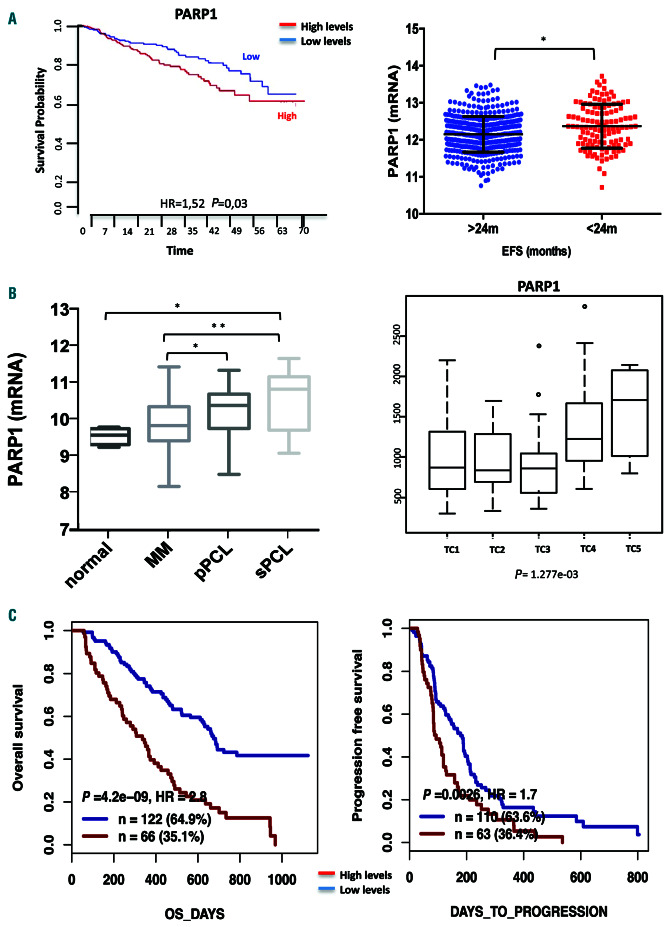
To understand the role of PARP1 in the pathophysiology of MM, the prognostic relevance of its mRNA expression was investigated. Analysis of public MM datasets revealed that higher PARP1 mRNA expression was significantly correlated with shorter overall survival and event-free survival (Figure 1A), thus highlighting its pivotal contribution to disease pathogenesis. Indeed, PARP1 mRNA expression increased during disease progression and in high-risk MM subgroups harboring t(4;14) and t(14;16) translocations (translocations/cyclins classes TC4 and TC5, respectively) (Figure 1B, Online Supplementary Figure S1A).22 Of note, no significant influence of copy number on PARP1 gene expression was found (Online Supplementary Figure S1B), suggesting that high expression of PARP1 in MM patients could be derived from deregulation of transcriptional or post-transcriptional mechanisms that normally regulate PARP1 expression. Notably PARP1 mRNA was the most expressed among other PARP family members. Furthermore, mRNA expression of other PARP family members did not retain any prognostic relevance, except for PARP2 (Online Supplementary Figure S1C, D).
Figure 1.
High PARP1 levels are associated with poor prognosis in multiple myeloma patients. (A) Data obtained interrogating the GSE24080 dataset using PRECOG software (https://precog.stanford.edu/index.php). Prognostic relevance of PARP1 mRNA (NM_208644_at) expression, on overall survival and event-free survival of patients with multiple myeloma (MM). (B) PARP1 mRNA expression analysis from proprietary datasets. Left panel: PARP1 levels in healthy donors as compared to those in cases of MM and primary and secondary plasma cell leukemia. Right panel: PARP1 expression in MM patients according to translocations/cyclins classification. (C) Analysis of the GSE9782 dataset using GenomicScape software (www.genomicscape.com). Prognostic relevance of PARP1 levels on overall and progression-free survival of MM patients treated with bortezomib. *P<0.05; **P<0.01. EFS: event-free survival; pPCL: primary plasma cell leukemia; sPCL: secondary plasma cell leukemia; TC: translocations/cyclins; HR: hazard ratio; OS: overall survival.
Moreover analysis of the GSE9782 dataset revealed that higher levels of PARP1 mRNA predicted poor overall and progression-free survival for patients who received bortezomib- based therapy (Figure 1C). Overall, these findings strongly suggest the involvement of PARP1 in the genomic instability of MM, which promotes disease progression and drug resistance.
PARP1 is a therapeutic target in multiple myeloma
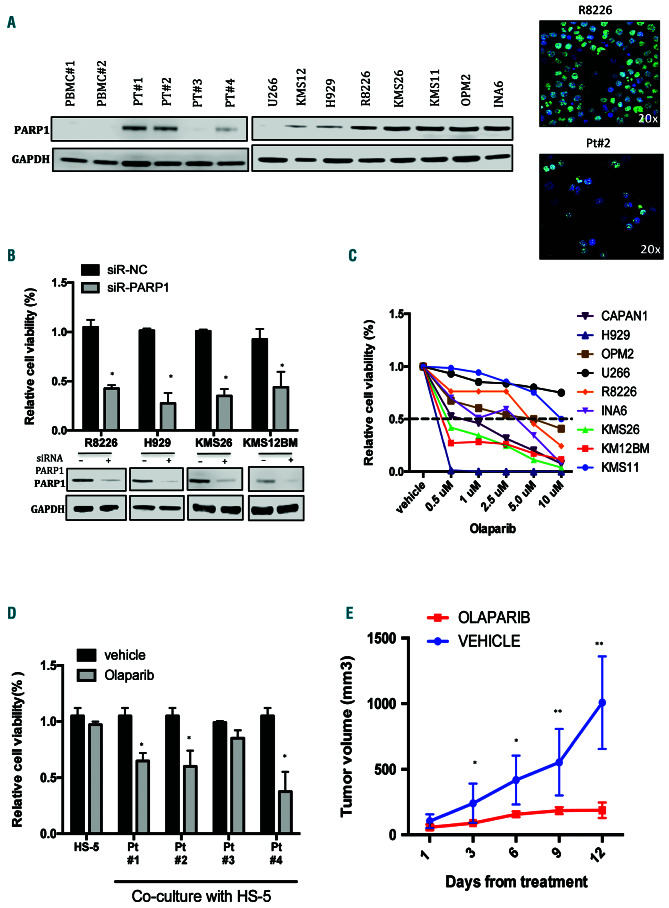
Based on clinical findings, which suggested a relevant role of PARP1 in the pathogenesis of MM, protein levels were analyzed in a panel of MM cell lines, primary cells from MM patients and peripheral blood mononuclear cells from healthy donors. As shown in Figure 2A, there was upregulation of PARP1 protein expression with a nuclear distribution in MM cells, as compared to the level of expression in peripheral blood mononuclear cells from healthy donors. In particular PARP1 protein was undetectable in only one of four primary MM cells evaluated and in the U266 cell line.
To investigate its biological relevance in MM cells, knockdown experiments were performed to evaluate whether PARP1 was required for MM cell viability. Importantly, PARP1 downregulation significantly reduced viability and increased apoptosis of MM cell lines (Figure 2B, Online Supplementary Figure S2A).
The effects induced by a clinically available PARP1/2 inhibitor, olaparib, on MM cell survival were investigated. Importantly, MM cells were highly sensitive to olaparib with an median inhibitory concentration (IC50) <10 mM observed in seven of eight tested cell lines (Figure 2C). In particular, the IC50 was 0.5 mM in NCI-H929, KMS-12BM, KMS26 and INA-6 cell lines, as in the highly sensitive BRCA2-defective CAPAN1 cells, a pancreatic adenocarcinoma cell line that was used as a positive control cell line in this experiment.23 Moreover, olaparib induced an increase of apoptotic cell death and a reduction of clonogenic growth in a dose-dependent manner (Online Supplementary Figure SB, C). Importantly, olaparib impaired the viability of primary MM plasma cells co-cultured with stromal cells (Figure 2C), thus overcoming the protective role of the bone marrow microenvironment. To confirm the translational relevance of our in vitro findings, the in vivo anti-MM activity of olaparib was evaluated and was shown to result in significant inhibition of tumor growth (Figure 2E).
Finally, the basal expression of the other olaparib target, PARP2, was evaluated. Notably, its protein expression was much lower than that of PARP1 (Online Supplementary Figure S2D), thus suggesting a minor role of PARP2 in driving olaparib activity in MM cell lines.
Overall, these findings strongly suggest that PARP1 could be a promising therapeutic target in MM.
PARP1 inhibition triggers the DNA damage response in multiple myeloma cells
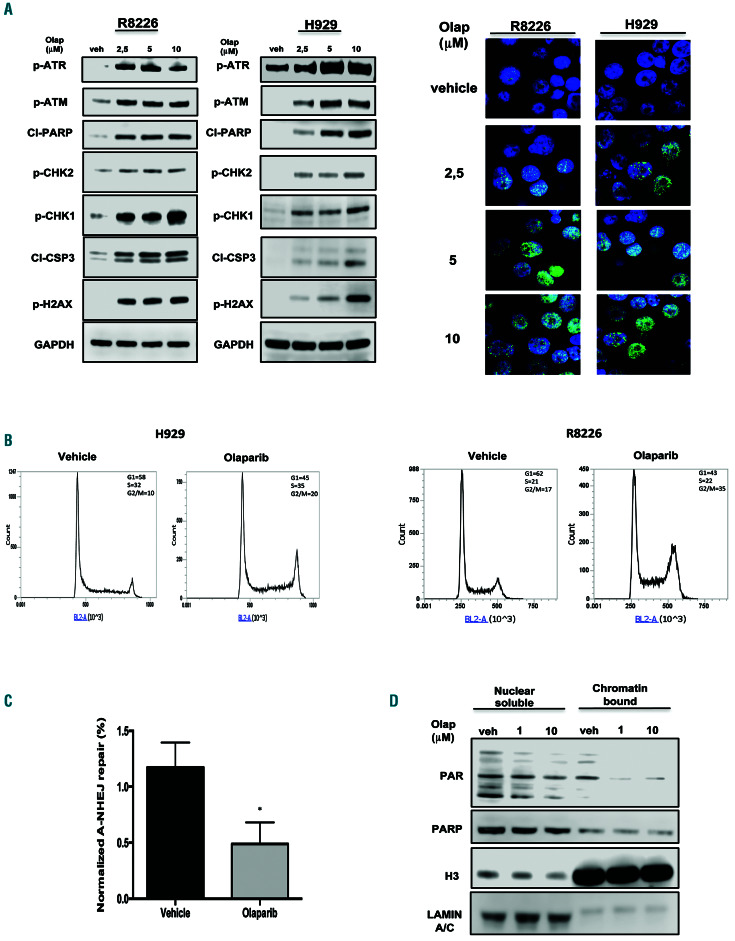
Since PARP1 plays a critical role in the repair of doublestrand breaks, the effect of PARP1 inhibition on the DNA damage response was investigated. Importantly, olaparib treatment induced a relevant increase of unrepaired DNA damage, along with a significant activation of the DNA damage response and apoptosis signaling, as demonstrated by increased phosphorylation of ATM, ATR, CHK1, CHK2, and H2AX, occurring together with PARP1 and caspase-3 cleavage (Figure 3A). Similar results were also obtained after PARP1 knockdown in MM cell lines or after olaparib treatment in primary MM cells (Online Supplementary Figure S3A, B).
Cell cycle analysis revealed that there was also G2-arrest after olaparib treatment (Figure 3B), an effect that was abrogated by caffeine24 (Online Supplementary Figure S3C), thus suggesting that checkpoint activation induced by olaparib treatment depends on ATM/ATR signaling.
To investigate molecular mechanisms underpinning the increase of the DNA double-strand breaks observed after olaparib treatment, functional experiments were performed. To this aim, the activity of alternative NHEJ repair was evaluated given the pivotal role exerted by PARP1 in this DNA repair pathway. Indeed, alternative NHEJ repair was clearly reduced in MM cells treated with olaparib as compared to that in cells treated with vehicle alone (Figure 3C, Online Supplementary Figure S3D).
To investigate whether PARP trapping onto damaged DNA is responsible for the cytotoxic activity of PARP inhibitors, R8226 cell lysates were fractionated into nuclear-soluble and chromatin-bound fractions. Notably, although PARylation was reduced by treatment, olaparib alone did not significantly increase PARP1 chromatin binding as compared to the nuclear-soluble fraction (Figure 3D), suggesting that the effects of olaparib in MM are correlated to inhibition of double-strand break repair rather than to PARP-trapping activity.
MYC drives sensitivity to PARP inhibition
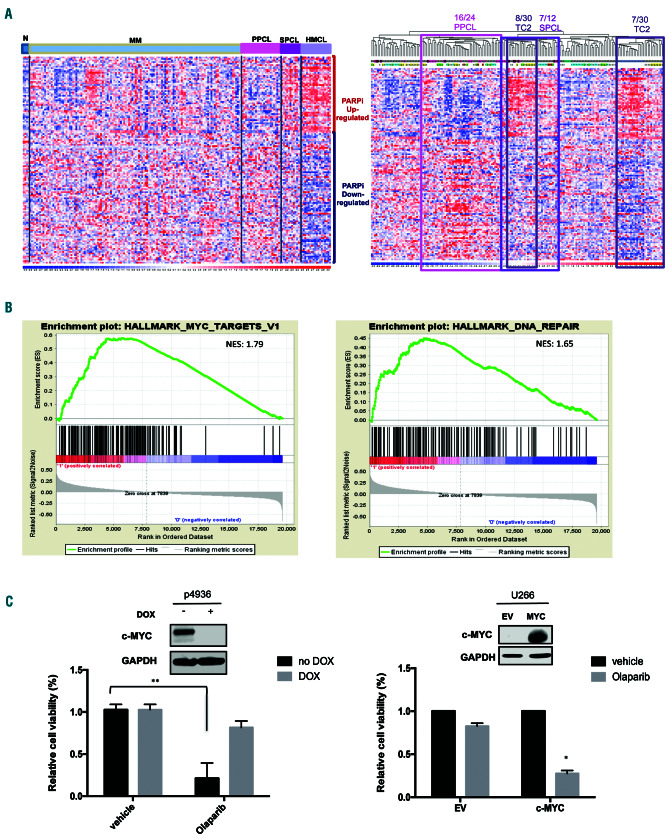
To identify predictive biomarkers for PARP inhibition in MM, a published PARP inhibition response gene expression signature25 was evaluated in a proprietary dataset of plasma cell dyscrasias, previously analyzed by global gene expression profiling using microarray technology.26 A large proportion of secondary plasma cell leukemia (PCL) samples and human myeloma cell lines showed a PARP inhibitor-positive expression pattern, MM samples showed heterogeneous expression levels, whereas primary PCL cases mostly showed an opposite expression trend. Notably, among MM TC classes, half the TC2 cases were grouped together with the majority of secondary PCL and all human myeloma cell line PARP inhibitor-positive clusters (Figure 4A). Therefore, in order to identify concordantly modulated sets of genes that were potentially associated with the PARP inhibitor signature in MM, PARP inhibitor-positive and -negative TC2 cases were compared by gene set enrichment analysis.27 Interestingly, groups of genes regulated by MYC and involved in DNA repair resulted among the most significantly upregulated in PARP inhibitor-positive versus PARP inhibitor-negative cases (Figure 4B, Online Supplementary Figure S4A). Similar results were also obtained comparing PARP inhibtor--positive and -negative PCL cases. Accordingly, the highest median levels of expression of MYC transcripts were reached in secondary PCL and human myeloma cell lines across the groups of plasma cell dyscrasias and in MM class TC2 (Online Supplementary Figure S4B).
Figure 2.
Addiction of multiple myeloma cells to PARP1. (A) Left panel: immunoblot of PARP1 performed on peripheral blood mononuclear cells collected from healthy donors, multiple myeloma (MM) cell lines and patients (Pts). GAPDH was used as a loading control. Right panel: immunofluorescence analysis showing subcellular distribution of PARP1 in R8226 cells and a patient’s (Pt#2) MM cells (magnification 20x): PARP1 (green); DAPI (blue) was used for nuclear staining. (B) Indicated cell lines were transfected with scramble control or PARP1-siRNA. Top: a Cell Titer-Glo assay was performed 4 days after transfection. Bottom: immunoblot analysis of PARP1. GAPDH was used as a loading control. Analysis was performed 48 h after transfection. (C) Indicated MM cells were treated with increasing dose of olaparib. Left panel: a Cell Titer-Glo assay was performed 7 days after treatment. Results are expressed as a percentage of vehicle-treated cells. (D) Cell viability of CD138+ cells from four different MM patients co-cultured with HS-5 stromal cells and treated with olaparib 5 uM or vehicle. The assay was performed 4 days after treatment. (E) In vivo growth of H929 subcutaneous xenograft daily treated with vehicle or olaparib (100 mg/kg) via oral gavage. Averaged tumor volume of each group ± standard deviation is shown. Data are representative of at least three independent experiments. *P<0.05; **P<0.01.
Figure 3.
Olaparib activates the DNA damage response in multiple myeloma cells. (A) R8226 and H929 cells were treated with olaparib. Left panel: Immunoblot analysis was performed 24 h after treatment. Right panel: immunofluorescence evaluation by γ-H2AX foci. Representative images of unrepaired double-strand breaks are shown. DAPI (blue) was used for nuclear staining. (B) Cell cycle analysis performed 24 h after treating H929 and R8226 cells with olaparib (2.5 mM). (C) Alternative non-homologous end junction repair was evaluated by EJ2-GFP assay on R8226 cells 48 h af ter treatment with olaparib (2.5 mM) treatment. (D) Western blot analysis of nuclear-soluble and chromatin-bound fractions prepared from R8226 cells. Cells were treated for 30 min with vehicle or olaparib as indicated. Histone H3 and LAMIN A/C were used as positive markers for chromatin and nuclear-soluble fractions, respectively. Data are representative of at least three independent experiments. *P<0.05; **P<0.01. Olap: olaparib; A-NHEJ: alternative non-homologous end junction; PAR: poly (ADP)-ribose; PARP: poly (ADP)-ribose polymerase; H3: histone H3.
Figure 4.
MYC drives sensitivity to PARP inhibition. (A) Hierarchical agglomerative clustering of samples of plasma cell dyscrasias on PARP inhibitor gene expression signature. Types of samples (normal [N], multiple myeloma [MM], primary plasma cell leukemia [PPCL], secondary plasma cell leukemia [SPCL], human myeloma cell lines [HMCL]) and MM translocations/cyclins classes (TC1, TC2, TC3, TC4, TC5) are depicted in different colors. The color scale bar represents the relative gene expression changes normalized by the standard deviation. PPCL, SPCL and MM-TC2 significant clusters are highlighted: 16/24 PPCL, P=4x10-4; 7/12 SPCL P=8.4x10-3; 8/30 TC2, P=6.5x10-3; 7/30 TC2, P=9.4x10-3. (B) Enrichment plots of selected significant (nominal P<0.05) gene sets by gene set enrichment analysis on PARP inhibitor-positive versus PARP inhibitor-negative MM-TC2 cases. Normalized enrichment scores (NES) are indicated. (C) Lef t panel: cell viability analysis in P493-6 cells after 7 days of exposure to vehicle or olaparib (5 mM), in the presence of doxycycline (DOX: 500 ng/mL) (low MYC) or in its absence (high MYC). Right panel: cell viability analysis in U266 empty vector (EV) and U266 MYC-positive cells after 7 days of exposure to vehicle or olaparib (5 mM). Data are representative of at least three independent experiments. *P<0.05; **P<0.01.
To validate the bioinformatics results, basal levels of MYC protein were evaluated in olaparib-treated MM cell lines. Notably, olaparib-insensitive U266 cells did not express MYC protein (Online Supplementary Figure S4C).28 Next, olaparib was tested on P936 B cell-like lymphoma cells, a cellular model in which MYC levels can be turned on or off by adding doxycycline. Notably, olaparib treatment effectively induced cell death only in Myc-on conditions. Conversely, as formal proof of our hypothesis, overexpression of MYC in U266 cells restored cell death upon PARP1 knockdown or PARP1 inhibitor treatment (Figure 4C, Online Supplementary Figure S4D). Moreover, the lack of correlation between PARP inhibitor sensitivity and the evidence of homologous recombination deficiency (as evaluated by RAD51 protein levels), or the presence of MM characteristic cytogenetic abnormalities (13q deletion, 1q gain, hyperdyploidy) or somatic mutations (BRAF, NRAS, KRAS, DIS3) (Online Supplementary Figure 4E), further support the hypothesis that PARP-inhibition triggers MYC-dependent synthetic lethality in MM.
Figure 5.
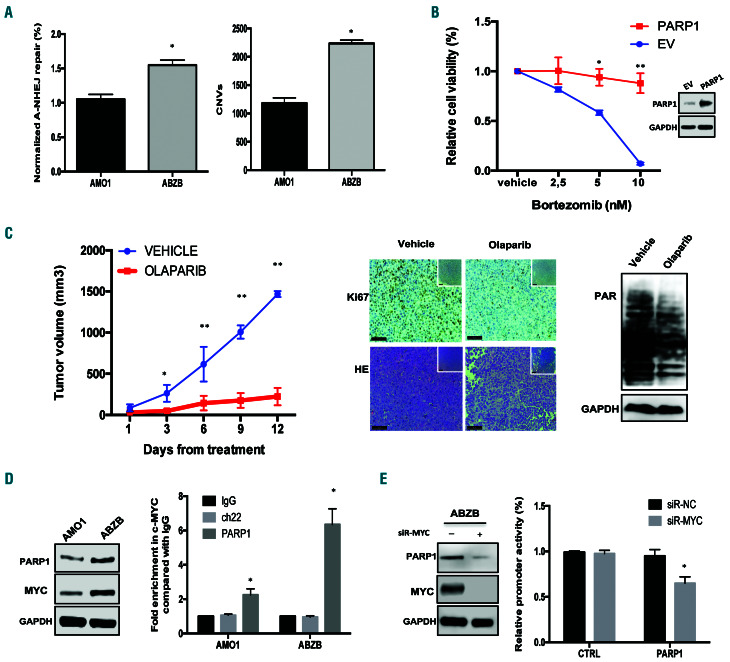
Bortezomib-resistant cells are highly sensitive to PARP inhibitors. (A) Left: alternative non-homologous end junction repair (A-NHEJ) was evaluated by an EJ2-GFP assay on AMO1 and ABZB cells. Right: Af fymetrix CytoScan HD Array analysis of copy-number variations, using genomic DNA from AMO1 and ABZB cells. (B) A Cell Titer-Glo Assay was performed on AMO1 cells transfected with PARP1 ORF or control ORF (EV) and then treated for 24 h with increasing doses of bortezomib. PARP1 overexpression was confirmed by western blot analysis. (C) In vivo growth of luciferase gene-marked ABZB xenografts treated daily with vehicle or olaparib (100 mg/kg) via oral gavage. Left: averaged tumor volume of each group ± standard deviation is shown. *P<0.05; **P<0.01. Right: PAR levels and Ki67 and HE expression were evaluated respectively by western blot and immunohistochemical analysis (20x, 40x insets) from a representative ABZB xenograf t per group. (D) Left: Immunoblot of PARP1 and MYC performed on AMO1 and ABZB cells. GAPDH was used as a loading control. Right: quantitative polymerase chain reaction for PARP1 promoter performed after chromatin immunoprecipitation with MYC antibody in AMO1 and ABZB cells compared with negative (Ch22) and IgG controls. (E) Lef t: ABZB cells were transfected with siRNA -normal control (NC) or siRNA-MYC. Immunoblot of PARP1 and MYC was performed 48 h after transfection. Right: promoter activity of transfected PARP1 and negative CTRL promoter constructs in ABZB cells co-transfected with either siRNA-NC or siRNA-MYC. Data are representative of at least three independent experiments. *P<0.05; **P<0.01.
Bortezomib-resistant cells are highly sensitive to PARP inhibition
Based on clinical data, which suggest a potential role in bortezomib resistance (Figure 1C), the effects of PARP1- inhibition in this context were also investigated. Notably, AMO1 bortezomib-resistant (ABZB) cells showed greater alternative NHEJ repair and copy-number variations as compared to their sensitive counterpart AMO1 cells (Figure 5A), thus suggesting an association between an increase of genomic instability and acquisition of bortezomib resistance. Interestingly, enforced expression of PARP1 in AMO1 cells significantly antagonized the effect induced by bortezomib on cell viability (Figure 5B).
Importantly, olaparib showed significant growth inhibitory activity against ABZB cells in vitro and in vivo (Figure 5C, Online Supplementary Figure S5A, B). Since higher levels of PARP1 and MYC proteins were observed in ABZB cells than in AMO1 cells (Figure 5D), we next investigated a potential role of MYC in PARP1 transcription.
Notably, analysis of a MM patients’ dataset (GSE24080), showed a significant positive correlation between PARP1 and MYC expression (Online Supplementary Figure S5C). Indeed, bioinformatic screening (cistrome.org) revealed significant enrichment of MYC binding consensus sequences to PARP1 promoter (Online Supplementary Figure S5D), which was next validated by chromatin immunoprecipitation analysis (Figure 5E). Short interfering RNA-mediated knockdown of MYC induced downregulation of PARP1 protein and decreased promoter activity (Figure 5E), indicating that MYC binds to the PARP1 promoter, modeling its transcription.
Discussion
The DNA damage response is an attractive area of investigation because of the opportunity to selectively kill cancer cells addicted to compensatory DNA repair pathways by synthetic lethality.29 There is experimental evidence that alternative NHEJ is an error-prone DNA repair pathway30-35 and master driver of genomic instability in cancer.36
In this study, we investigated the involvement of PARP1, a pivotal component of error-prone alternative NHEJ repair, in the pathogenesis of MM genomic instability. 37 The alternative NHEJ repair pathway is involved in important processes of B-lineage differentiation such as VD-J recombination and class switch recombination,38 critical steps in which errors in the rearrangement of germline DNA could generate translocation of protooncogenes, such as MYC,39 into the immunoglobulin loci, thus potentially contributing to myelomagenesis.
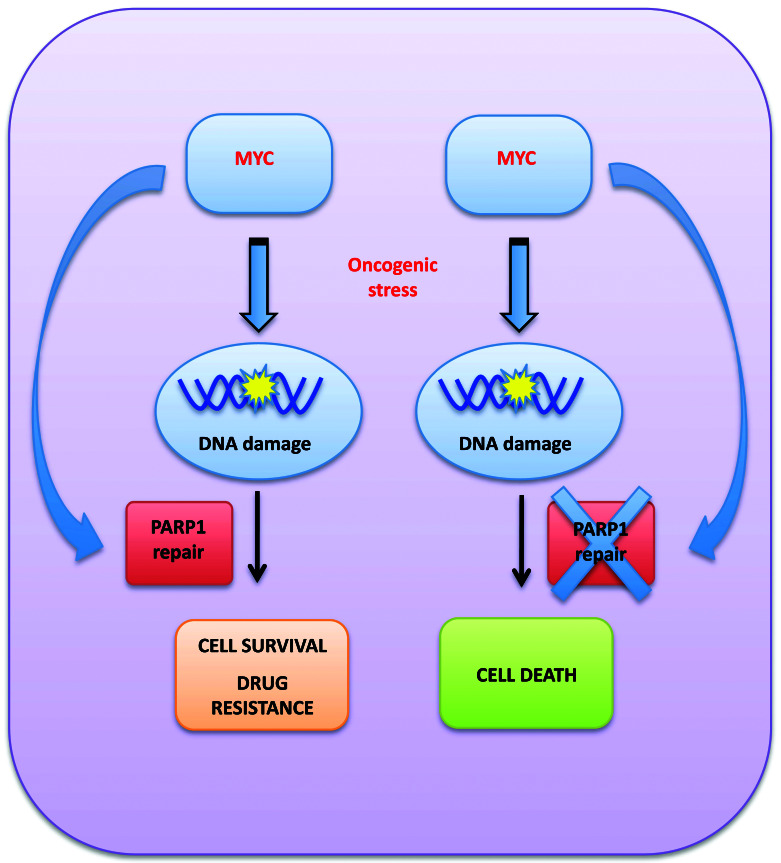
Figure 6.
Cartoon describing the addiction of MYC-driven multiple myeloma cells to PARP1.
Consistently, we found that high PARP1 mRNA expression is significantly correlated with poor event-free and overall survival in MM patients, and increases during disease progression and in high-risk cases. Moreover, PARP1 mRNA is the most expressed among PARP family members, thus suggesting a critical role of PARP1 in MM pathogenesis. In fact, we found that PARP1 knockdown induces MM cell death.
With the aim of translating these findings into the design of a new therapeutic strategy, we provide evidence that olaparib, an available PARP inhibitor, induces anti- MM activity at clinical doses,40 even against bortezomibresistant MM cells. Growth inhibitory effects induced by olaparib were correlated to inhibition of alternative NHEJ41 which led to an increase of unrepaired DNA damage and apoptotic cell death.
To investigate mechanisms leading to PARPi sensitivity in MM, we used a PARP inhibition sensitivity gene expression signature.25 Interestingly our analysis highlighted a pivotal role of MYC, a driver transcription factor hyperactivated in the majority of MM patients.42,43 Indeed, the signature was enriched in patients with TC2 MM and in subgroups with secondary PCL, which display the highest MYC expression.44 Consistently, we found that PARP inhibition induces cytotoxic effects against MM, which mostly occur in MYC-proficient cells.
Our findings are consistent with recent reports showing that MYC-dependent Burkitt lymphoma45 and neuroblastoma46 are sensitive to PARP inhibition. Indeed, MYC drives genomic instability by deregulation of the DNA damage response, thus increasing the dependency of cancer cells on low-fidelity DNA repair pathways.47-49 Moreover, MYC is a strong inducer of replication stress, which requires activation of the DNA damage response to repair DNA damage and to sustain cell survival.50,51
Based on these findings, we hypothesize that MYC-driven MM cells switch their DNA repair machinery to errorprone PARP-mediated alternative NHEJ. This event allows them to overcome DNA damage overload from oncogenic stress, promoting cell survival and, at the same time, the acquisition of new genetic changes leading to disease progression (Figure 6). Indeed, it could be hypothesized that MYC induces alternative NHEJ repair to balance the downregulation of homologous recombination induced by bortezomib,52,53 contributing to the development of drug resistance and, at the same time, making MM cells more dependent on PARP1-mediated DNA repair to survive. Consistently, we also report here that bortezomibresistant MM cells are extremely sensitive to PARP inhibition. This finding is of translational relevance, since it could support the design of a maintenance therapy with PARP inhibitors as a strategy to prevent disease progression and acquisition of drug resistance.
In conclusion, our study demonstrates addiction of MYC-driven MM cells to PARP1, which could be exploited as a new opportunity for synthetic lethality in the clinical scenario of precision oncology. Moreover, our findings highlight the role of MYC as a driver of PARP1-mediated repair, identifying a novel mechanism of genome stability and survival regulation in MM and a potential biomarker of PARPness in this still incurable disease.
Supplementary Material
Acknowledgments
This work was supported by the Italian Association for Cancer Research (AIRC) with “Special Program for Molecular Clinical Oncology–5 per mille”, 2010/15 and its Extension Program” n. 9980, 2016/18 (PI: PT); and also by “Innovative Immunotherapeutic Treatments of Human Cancer” Multi Unit Regional n. 16695 (cofinanced by AIRC and the CARICAL Foundation), 2015/18 (PI: PT). We thank Dr. Ivana Criniti for her study coordination support and editorial assistance.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-674. [DOI] [PubMed] [Google Scholar]
- 2.Murata S, Zhang C, Finch N, et al. Predictors and modulators of synthetic lethality: an update on PARP inhibitors and personalized medicine. Biomed Res Int. 2016;2016: 2346585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim G, Ison G, McKee AE, et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCAmutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin Cancer Res. 2015;21(19):4257-4261. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramaniam S, Beaver JA, Horton S, et al. FDA approval summary: rucaparib for the treatment of patients with deleterious BRCA mutation-associated advanced ovarian cancer. Clin Cancer Res. 2017;23(23): 7165-7170. [DOI] [PubMed] [Google Scholar]
- 5.[No authors listed]. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(17):1700. [DOI] [PubMed] [Google Scholar]
- 6.Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what's next after PARP inhibitors? Nat Rev Clin Oncol. 2018;15(9):564-576. [DOI] [PubMed] [Google Scholar]
- 7.Pilie PG, Gay CM, Byers LA, O'Connor MJ, Yap TA. PARP inhibitors: extending benefit beyond BRCA-mutant cancers. Clin Cancer Res. 2019;25(13):3759-3771. [DOI] [PubMed] [Google Scholar]
- 8.Pilie PG, Tang C, Mills GB, Yap TA. State-ofthe- art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16(2):81-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pannunzio NR, Watanabe G, Lieber MR. Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J Biol Chem. 2018;293(27):10512-10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229-257. [DOI] [PubMed] [Google Scholar]
- 11.Chang HHY, Pannunzio NR, Adachi N, Lieber MR. Non-homologous DNA end joining and alternative pathways to doublestrand break repair. Nat Rev Mol Cell Biol. 2017;18(8):495-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan J, Li L, Small D, Rassool F. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood. 2010;116(24): 5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman EA, Lu F, Bashllari D, et al. Alternative NHEJ pathway components are therapeutic targets in high-risk neuroblastoma. Mol Cancer Res. 2015;13(3):470-482. [DOI] [PubMed] [Google Scholar]
- 14.Tobin LA, Robert C, Nagaria P, et al. Targeting abnormal DNA repair in therapyresistant breast cancers. Mol Cancer Res. 2012;10(1):96-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceccaldi R, Liu JC, Amunugama R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518(7538):258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neri P, Bahlis NJ. Genomic instability in multiple myeloma: mechanisms and therapeutic implications. Expert Opin Biol Ther. 2013;13(Suppl 1):S69-82. [DOI] [PubMed] [Google Scholar]
- 17.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiology ical outcomes, and clinical targets. Mol Cell. 2010;39(1):8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paddock MN, Bauman AT, Higdon R, et al. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA Repair (Amst). 2011;10(3):338-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang G, Liu C, Chen SH, et al. Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 2018;46(7):3446-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caracciolo D, Di Martino MT, Amodio N, et al. miR-22 suppresses DNA ligase III addiction in multiple myeloma. Leukemia. 2019;33(2):487-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leone E, Morelli E, Di Martino MT, et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin Cancer Res. 2013;19(8):2096-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCabe N, Lord CJ, Tutt AN, et al. BRCA2- deficient CAPAN-1 cells are extremely sensitive to the inhibition of poly (ADP-ribose) polymerase: an issue of potency. Cancer Biol Ther. 2005;4(9):934-936. [DOI] [PubMed] [Google Scholar]
- 24.Sarkaria JN, Busby EC, Tibbetts RS, et al. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59(17):4375-4382. [PubMed] [Google Scholar]
- 25.McGrail DJ, Lin CC, Garnett J, et al. Improved prediction of PARP inhibitor response and identification of synergizing agents through use of a novel gene expression signature generation algorithm. NPJ Syst Biol Appl. 2017;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lionetti M, Barbieri M, Todoerti K, et al. Molecular spectrum of BRAF, NRAS and KRAS gene mutations in plasma cell dyscrasias: implication for MEK-ERK pathway activation. Oncotarget. 2015;6(27): 24205-24217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge- based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dib A, Gabrea A, Glebov OK, Bergsagel PL, Kuehl WM. Characterization of MYC translocations in multiple myeloma cell lines. J Natl Cancer Inst Monogr. 2008;(39):25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278(5340):1064-1068. [DOI] [PubMed] [Google Scholar]
- 30.Iliakis G, Murmann T, Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat Res Genet Toxicol Environ Mutagen. 2015;793: 166-175. [DOI] [PubMed] [Google Scholar]
- 31.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36(11):3847-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood RD, Doublie S. DNA polymerase theta (POLQ), double-strand break repair, and cancer. DNA Repair (Amst). 2016;44:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhuang J, Jiang G, Willers H, Xia F. Exonuclease function of human Mre11 promotes deletional nonhomologous end joining. J Biol Chem. 2009;284(44):30565-30573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor RM, Whitehouse CJ, Caldecott KW. The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res. 2000;28(18):3558-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simsek D, Brunet E, Wong SY, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7(6):e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caracciolo DM M., Tagliaferri P., Tassone P. Alternative non-homologous end joining repair: a master regulator of genomic instability in cancer[Review]. Prec Cancer Med. 2019;2:8. [Google Scholar]
- 37.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12(5):335-348. [DOI] [PubMed] [Google Scholar]
- 38.Boboila C, Jankovic M, Yan CT, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci U S A. 2010;107(7):3034-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikulasova A, Ashby C, Tytarenko RG, et al. Microhomology-mediated end joining drives complex rearrangements and over expression of MYC and PVT1 in multiple myeloma. Haematologica. 2020:105(4): 1055-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361(2):123-134. [DOI] [PubMed] [Google Scholar]
- 41.Howard SM, Yanez DA, Stark JM. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015;11(1):e1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chesi M, Robbiani DF, Sebag M, et al. AIDdependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13(2):167-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuehl WM, Bergsagel PL. MYC addiction: a potential therapeutic target in MM. Blood. 2012;120(12):2351-2352. [DOI] [PubMed] [Google Scholar]
- 44.Weinhold N, Kirn D, Seckinger A, et al. Concomitant gain of 1q21 and MYC translocation define a poor prognostic subgroup of hyperdiploid multiple myeloma. Haematologica. 2016;101(3):e116-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maifrede S, Martin K, Podszywalow- Bartnicka P, et al. IGH/MYC translocation associates with BRCA2 deficiency and synthetic lethality to PARP1 inhibitors. Mol Cancer Res. 2017;15(8):967-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colicchia V, Petroni M, Guarguaglini G, et al. PARP inhibitors enhance replication stress and cause mitotic catastrophe in MYCN-dependent neuroblastoma. Oncogene. 2017;36(33):4682-4691. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Owonikoko TK, Sun SY, et al. c-Myc suppression of DNA double-strand break repair. Neoplasia. 2012;14(12):1190-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muvarak N, Kelley S, Robert C, et al. c-MYC generates repair errors via increased transcription of alternative-NHEJ factors, LIG3 and PARP1, in tyrosine kinase-activated leukemias. Mol Cancer Res. 2015;13(4):699-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W, Liu B, Wu W, et al. Targeting the MYCN-PARP-DNA damage response pathway in neuroendocrine prostate cancer. Clin Cancer Res. 2018;24(3):696-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cottini F, Hideshima T, Suzuki R, et al. Synthetic lethal approaches exploiting DNA damage in aggressive myeloma. Cancer Discov. 2015;5(9):972-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kotsantis P, Petermann E, Boulton SJ. Mechanisms of oncogene-induced replication stress: jigsaw falling into place. Cancer Discov. 2018;8(5):537-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neri P, Ren L, Gratton K, et al. Bortezomibinduced "BRCAness" sensitizes multiple myeloma cells to PARP inhibitors. Blood. 2011;118(24):6368-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pawlyn C, Loehr A, Ashby C, et al. Loss of heterozygosity as a marker of homologous repair deficiency in multiple myeloma: a role for PARP inhibition? Leukemia. 2018;32(7): 1561-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.