JAK2, CALR and MPL driver mutations activate JAK/STAT signalling in BCR-ABL1 translocation negative classic myeloproliferative neoplasms (MPN). Detection of these mutations in patients is included in major criteria for establishing diagnoses of polycythemia vera (PV), essential thrombocythemia (ET) and primary myelofibrosis (PMF).1,2 JAK2 Val617Phe and exon 12 mutations are detected in approximately 98% of PV, while JAK2 Val617Phe, CALR exon 9 frameshift mutations and MPL exon 10 mutations collectively are detectable in 83% of ET and 92% of PMF.2 Mutations in these genes are typically mutually exclusive, however, JAK2 Val617Phe/JAK2 exon 12,3,4 JAK2/CALR,5,6 JAK2/MPL4,6,7 and very rarely CALR/MPL8 comutations have been reported. Further, the presence of multiple independent JAK2 Val617Phe mutations has been demonstrated in the majority of ET patients in one case series, and is one of several lines of evidence that suggest a predisposition in some patients to the development of JAK2 (and other MPN driver) mutations, the nature of which has not been elucidated but may include, genetic predisposition, a preceding clonal mutation or a permissive microenvironment.9
Herein we describe two unusual cases of patients with an MPN, each harboring two driver mutations (JAK2/CALR and JAK2/MPL) and the use of single cell DNA sequencing analysis to further investigate the clonal architecture of these neoplasms. In both cases we demonstrate that the driver mutations represent independent clones and give insight into the potential clonal complexity of this rare phenomenon.
The first patient studied was a 70-year-old male with a long (approximately 30-year) history of PV, which transformed to myelofibrosis (MF) 14 years prior to analysis. Approximately 10 years after fibrotic transformation (i.e., 4 years prior to analysis) the patient commenced treatment with azacitidine and ruxolitinib for leukemic transformation. At this timepoint, trisomy of 1q was observed in his peripheral blood by G-banded karyotyping and confirmed by fluorescence in situ hybridization (FISH) analysis of 1q21 (CKS1B) in 62% of analyzed cells. His blast count normalized and he remains stable on azacitidine and ruxolitinib with post-polycythemic MF without blast excess.
Next generation sequencing (NGS)-based gene panel analysis of 26 genes involved in myeloid malignancy (previously described10) was performed on a stored bone marrow aspirate DNA sample from 14 years prior at the point of myelofibrotic transformation (the earliest timepoint available). This testing detected a JAK2 p.(Val617Phe) (NM_004972.3:c.1849G>T) at a variant allele frequency (VAF) of 62% as well as a CALR p.(Lys385Asnfs*47) (NM_004343.3:c.1154_1155ins TTGTC) mutation at a VAF of 17%. Testing of a peripheral blood sample stored at the time of leukemic transformation again demonstrated the JAK2 and CALR mutations but in addition a TP53 p.(His178Pro) (NM_000546.5:c.533A>C) mutation was detected at a VAF of approximately 5%.
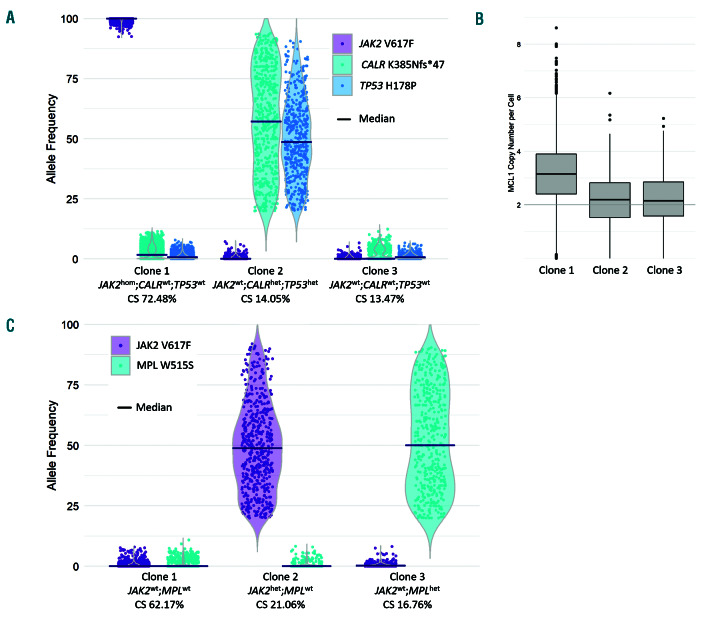
We sought to understand the clonal relationship of the JAK2 and CALR mutations in this patient’s disease by performing single cell mutation analysis of mononuclear cells collected from a recent peripheral blood sample using a custom Tapestri gene panel to sequence 10,564 single cells (Mission Bio) (see Online Supplementary Materials and Methods). The previously identified JAK2, CALR and TP53 mutations were genotyped in 96%, 52% and 94% of all filtered cells, and detected in 64%, 26% and 29% of genotyped cells with an aggregate single cell VAF (by read count) of 62%, 16% and 16%, respectively (Online Supplementary Table S1). Considering only the 38% of cells with genotypes determined for all three mutations, subclone analysis identified the presence of three major subclones (Figure 1A and Online Supplementary Table S2). The JAK2 mutation was detected as a homozygote in 72.48% of the cells comprising major clones (i.e., clone size [CS] 72.48%) in the absence of CALR and TP53 mutations. Heterozygous CALR and TP53 mutations were identified together within the second clone (CS 14.05%) and a wild-type (i.e., JAK2/CALR/TP53 mutation negative) clone was also detected (CS 13.47%). Amplicon-based copy number analysis of the single cell data also detected the trisomy of 1q (observed as copy number gain of panel gene MCL1 located at 1q21.3) and specifically identified this amplification within the JAK2 mutant clone (mean copy number 3.20) (Figure 1B). These data are consistent with the existence of two distinct clones, both with additional lesions suggestive of advancing forms of MPN.11
The second patient studied was a 74-year-old male who presented with persistent thrombocytosis (platelets 560-590x109/L) in association with symptomatic splenomegaly. A JAK2 p.(Val617Phe) (c.1849G>T) mutation was detected in his peripheral blood sample by an allele-specific assay and a subsequent bone marrow biopsy showed a mildly hypercellular bone marrow with fibrosis (grade MF-1 to MF-2/3), mild granulocytic hyperplasia, increased megakaryopoiesis and megakaryocytic atypia, which in conjunction with the JAK2 mutation was considered most consistent with PMF. He was commenced on ruxolitinib and responded clinically with decreased abdominal pain.
A 26 gene myeloid NGS panel performed on his bone marrow aspirate (as above) detected a MPL p.(Trp515Ser) (NM_005373.2:c.1544G>C) mutation at a VAF of 19% and an established pathogenic SRSF2 p.(Pro95Arg) (NM_003016.4:c.284C>G) mutation at a VAF of 53% in addition to the previously known JAK2 mutation at a VAF of 15%. Given that spliceosome mutations, including those in SRSF2, occur exclusively in a heterozygote state in myeloid malignancies due to their dependency on the wild-type allele,12,13 this finding is consistent with the entire hematopoietic compartment sampled being comprised of a single heterozygous SRSF2 mutant clone with subclonal JAK2 and MPL mutations. In order to further investigate the clonal relationship between the JAK2 and MPL mutations within this SRSF2 mutant parental clone, single cell analysis was performed on 8,181 total nucleated cells from a peripheral blood sample (see Online Supplementary Materials and Methods). The previously identified JAK2 and MPL mutations were genotyped in 91% and 44% of all cells, and detected in 21% and 23% of genotyped cells with an aggregate single cell VAF (by read count) of 11% and 13%, respectively (Online Supplementary Table S1). Mutually exclusive heterozygous JAK2 (CS 21.06%) and MPL (CS 16.76%) subclones were detected in the 36% of cells with genotypes determined for both mutations. A JAK2/MPL mutation negative population was also detected (CS 62.17%) (Online Supplementary Table S2). This analysis demonstrates the independent origin of the JAK2 and MPL mutations within the context of an SRSF2 mutant parental clone in this patient (Figure 1C).
Figure 1.
Single cell clone analysis of two myeloproliferative neoplasm patients. (A and C) Major clone analysis of patients 1 and 2, respectively, demonstrating allele frequency distribution of single cells within each of the major clones identified by the presence of JAK2 Val617Phe, CALR Lys385Asnfs*47 and TP53 His178Pro mutations in patient 1, and JAK2 Val617Phe and MPL Trp515Ser mutations in patient 2. Zygosity (wild-type [wt], heterozygous [het], homozygous [hom]) is inferred from the median allele frequency (indicated). Variant data were filtered and major clones identified as indicated in the supplementary methods. (B) MCL1 copy number in patient 1 is shown for the corresponding clones, indicating a copy number gain of chromosome 1q21.3 within the JAK2hom;CALRwt;TP53wt clone.
The analysis of both of these rare and illustrative patients at the single cell level provides interesting and important insights into the clonal architecture of both comutated MPN and MPN in general. Firstly, we have unambiguously established the existence of these driver mutations in different subclones. Molecular characterization of MPN at a subclonal level has previously been performed using colony assays grown on medium from single cells in the presence of cytokines and growth factor to generate sufficient material for further characterization. Whilst this technique has given important insights to date,3,4 it is technically limited by the number of cells that can be assessed, the potential for mixed colonies as well as the preferential growth of particular clones with exogenous stimulation. Although different biases in the form of cell doublets and allelic drop-out may exist within single cell sequencing technologies, direct sequencing of >8,000 individual primary cells conclusively demonstrates the occurrence of JAK2/CALR/MPL driver mutations in these patients in distinct clones within each patient.
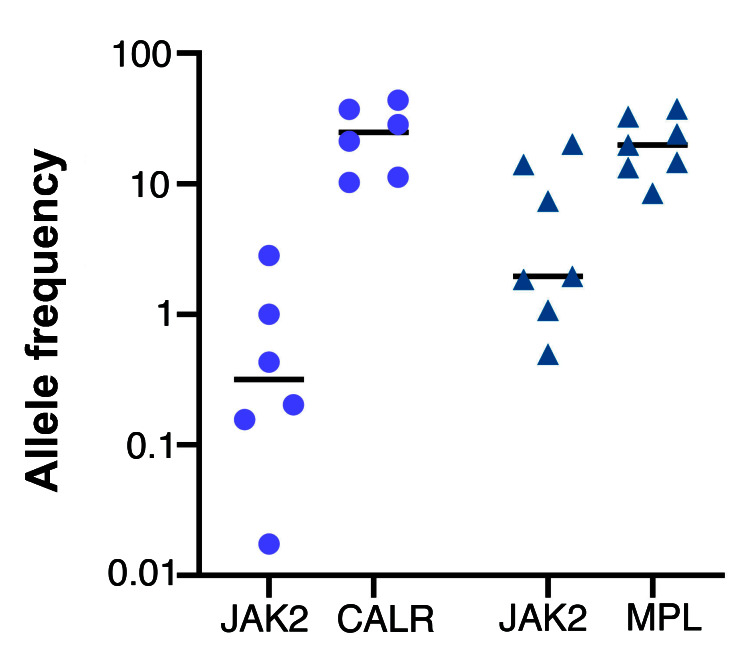
The majority of comutated cases of MPN described in the literature to date have been noted to have JAK2 mutations present at low VAF (typically <4%)4,6 compared to the other driver mutation (CALR/MPL). We reviewed the data from 2,664 consecutive cases of clinical NGS MPN sequencing performed at our own institution (Peter MacCallum Cancer Center, Melbourne, Australia) over 5 years (2014-2019) and identified 16 comutated cases (six JAK2/CALR, seven JAK2/MPL, three double-mutated JAK2) among 1,082 JAK2 (n=740), CALR (n=282) or MPL (n=73) mutated patients and noted similar findings (Figure 2). The biological relevance of the low VAF JAK2 mutation in these types of cases is unclear and the possibility that it may represent a “bystander” mutation outside the true disease compartment cannot be excluded. Indeed it is noted that the JAK2 Val617Phe is one of the most common mutations observed in age related clonal hematopoiesis.14 In contrast, single cell analysis of our cases suggest that they truly represent biclonally driven MPN given (i) the high fraction of clones containing each driver mutation (ii) the acquisition of further genomic abnormalities (1q21 gain and TP53 mutation) providing evidence of clonal evolution within both compartments. Unfortunately, no historical sample was present in case 1 from the polycythemic phase of illness to understand the time course of the acquisition of JAK2/CALR mutations. We note however, the extreme rarity of CALR mutations in PV.15
The second case demonstrates that biclonal MPN driver mutations can arise from a common progenitor cell, a feature that has previously been observed in the context of single driver mutations.16 This possibility is not excluded for the first case given the relatively focussed mutation analysis of 26 genes only. The finding of biclonality in these two cases does not exclude the possibility that co-mutation of driver genes may occur within a single clone in other cases, especially considering the background incidence of JAK2 mutations in age related clonal hematopoiesis.
Figure 2.
Allele frequencies in six JAK2/CALR and seven JAK2/MPL comutated patients detected among a series of 2,664 consecutive diagnostic samples assessed for myeloproliferative neoplasms. The median allele frequency for JAK2 is significantly lower than for either comutated CALR (JAK2 vs. CALR, median 0.32% [range: 0.02-2.83%] versus 24.90% [range: 10.30-43.98%]) or comutated MPL (JAK2 vs. MPL, median 1.96% [range 0.50%- 20.12%] versus 19.88% [range: 8.51-37.61%]).
In summary we have described two cases of biclonal MPN characterized by single cell sequencing confirming the presence of driver mutations in different clones. These data provide insights into the potential molecular complexity of MPN including intralineage clonal evolution. Moreover, these data highlight the importance of routine assessment of all three canonical driver mutations (i.e., JAK2/CALR/MPL) in patients with MPN in order to accurately characterize disease biology.
Supplementary Material
Acknowledgments
The authors would like to thank the Snowdome Foundation and the Wilson Center for Lymphoma Genomics for their funding support.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Lyon, France: International Agency for Research on Cancer, 2017. [Google Scholar]
- 2.Tefferi A, Pardanani A. Myeloproliferative Neoplasms: a contemporary review. JAMA Oncol. 2015;1(1):97-105. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Kralovics R, De Libero G, Theocharides A, Gisslinger H, Skoda RC. Clonal heterogeneity in polycythemia vera patients with JAK2 exon12 and JAK2-V617F mutations. Blood. 2008;111(7):3863-3866. [DOI] [PubMed] [Google Scholar]
- 4.Beer PA, Jones AV, Bench AJ, et al. Clonal diversity in the myeloproliferative neoplasms: independent origins of genetically distinct clones. Br J Haematol. 2009;144(6):904-908. [DOI] [PubMed] [Google Scholar]
- 5.Nomani L, Bodo J, Zhao X, Durkin L, Loghavi S, Hsi ED. CAL2 immunohistochemical staining accurately identifies CALR mutations in myeloproliferative neoplasms. Am J Clin Pathol. 2016; 146(4):431-438. [DOI] [PubMed] [Google Scholar]
- 6.Usseglio F, Beaufils N, Calleja A, Raynaud S, Gabert J. Detection of CALR and MPL mutations in low allelic burden JAK2 V617F essential thrombocythemia. J Mol Diagn. 2017;19(1):92-98. [DOI] [PubMed] [Google Scholar]
- 7.Lasho TL, Pardanani A, McClure RF, et al. Concurrent MPL515 and JAK2V617F mutations in myelofibrosis: chronology of clonal emergence and changes in mutant allele burden over time. Br J Haematol. 2006;135(5):683-687. [DOI] [PubMed] [Google Scholar]
- 8.Bernal M, Jimenez P, Puerta J, Ruiz-Cabello F, Jurado M. Co-mutated CALR and MPL driver genes in a patient with myeloproliferative neoplasm. Ann Hematol. 2017;96(8):1399-1401. [DOI] [PubMed] [Google Scholar]
- 9.Lambert JR, Everington T, Linch DC, Gale RE. In essential thrombocythemia, multiple JAK2-V617F clones are present in most mutantpositive patients: a new disease paradigm. Blood. 2009;114(14):3018-3023. [DOI] [PubMed] [Google Scholar]
- 10.Yannakou CK, Jones K, McBean M, et al. ASXL1 c.1934dup;p.Gly646Trpfs*12-a true somatic alteration requiring a new approach. Blood Cancer J. 2017;7(12):656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcellino BK, Hoffman R, Tripodi J, et al. Advanced forms of MPNs are accompanied by chromosomal abnormalities that lead to dysregulation of TP53. Blood Adv. 2018;2(24):3581-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim E, Ilagan JO, Liang Y, et al. SRSF2 Mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SC, Dvinge H, Kim E, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22(6):672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauveau A, Nibourel O, Tondeur S, et al. Absence of CALR mutations in JAK2-negative polycythemia. Haematologica. 2017; 102(1):e15-e16.27758825 [Google Scholar]
- 16.Ortmann CA, Kent DG, Nangalia J, et al. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372(7):601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.