Significance
Bacterial phenotypes require the concerted expression of multiple genes, usually coordinated by a transcriptional regulator. Although the functions of many genes in sequenced bacterial genomes can be inferred, the regulatory networks that coordinate their expression are only known in a few model systems. Using a bioinformatic and experimental approach, we solve the DNA-specificity code of extracytoplasmic function sigma factors (ECF σs), a major class of bacterial regulators. We develop and use a high-stringency pipeline to predict the genes regulated by 67% of ECF σs in >10,000 species, providing a comprehensive look at the role of a broadly distributed family of gene regulatory proteins. This conceptual and computational framework is potentially applicable to other bacterial regulators.
Keywords: transcriptional regulation, sigma factors, bioinformatics, phylogenetic footprinting
Abstract
Bacterial genomes are being sequenced at an exponentially increasing rate, but our inability to decipher their transcriptional wiring limits our ability to derive new biology from these sequences. De novo determination of regulatory interactions requires accurate prediction of regulators’ DNA binding and precise determination of biologically significant binding sites. Here we address these challenges by solving the DNA-specificity code of extracytoplasmic function sigma factors (ECF σs), a major family of bacterial regulators, and determining their putative regulons. We generated an aligned collection of ECF σs and their promoters by leveraging the autoregulatory nature of ECF σs as a means of promoter discovery and analyzed it to identify and characterize the conserved amino acid–nucleotide interactions that determine promoter specificity. This enabled de novo prediction of ECF σ specificity, which we combined with a statistically rigorous phylogenetic footprinting pipeline based on precomputed orthologs to predict the direct targets of ∼67% of ECF σs. This global survey indicated that some ECF σs are conserved global regulators controlling many genes throughout the genome, which are important under many conditions, while others are local regulators, controlling a few closely linked genes in response to specific stimuli in select species. This analysis reveals important organizing principles of bacterial gene regulation and presents a conceptual and computational framework for deciphering gene regulatory networks.
The genomes of medically, industrially, and environmentally important bacteria are being sequenced at a rapidly increasing rate. However, the utility of these sequences is limited by our inability to decipher their transcriptional wiring. Because adaptation to new environments or conditions is driven both by changes in the regulation of existing genes and by acquisition of novel functions, deciphering gene regulation from genome sequences is a key aspect of understanding cellular lifestyles, the evolution of pathogenesis, intrinsic antibiotic resistance, and biofilm growth.
In bacteria, a major point of transcriptional regulation is the use of sigma factors (σs) to direct RNA polymerase (RNAP) to specific promoters (1). A vast majority of σs belong to the σ70 family, which consists of four phylogenetically and structurally related groups (2). The Group 1 housekeeping σs are highly conserved, universally essential, and responsible for recognizing thousands of promoters in each bacterial genome. The Group 2 to 4 alternative σs are active under specific growth or environmental conditions and effect specialized transcriptional programs by directing RNAP to a smaller set of distinct promoters. Group 4 σs, also called extracytoplasmic function (ECF) σs, are the most abundant and diverse group, often representing 5 to 10% of the regulatory repertoire (3, 4). ECF σs regulate genes involved in differentiation, metal homeostasis, outer membrane integrity, oxygen response, and other processes (5–8). Despite their importance, the vast majority of ECF σs remain uncharacterized, and no computational method exists for determining the set of genes regulated by an ECF σ (its regulon).
ECF σs are small (∼200 amino acids) and contain two well-conserved globular domains, σ4 and σ2, that interact respectively with the -35 and -10 regions of the core promoter (1). ECF σs are less proficient at promoter melting than other σs, resulting in a requirement for near consensus promoters, and often regulate their own promoter (3, 4). Together, these attributes make ECF σs amenable to computational approaches. Previous work identified the autoregulatory promoters of some ECF σs by separating σs into groups and searching their upstream regions for overrepresented bipartite motifs (3, 4, 9). However, the promoter motifs discovered were not suitable for determining individual ECF σ regulons. First, the groups were broad, often containing multiple ECF σs from the same genome. Since multiple ECF σs in a single bacterium are unlikely to regulate identical regulons, these motifs are likely an ensemble of multiple distinct promoters. Second, this method cannot determine motifs of nonautoregulatory ECF σs. Finally, identifying the true regulon requires a method that can discriminate against the large fraction of false positive sites predicted by most methods (10–12).
Here we overcome these hurdles by computationally and experimentally analyzing the protein determinants of promoter recognition to elucidate how amino acid identities at key positions determine the promoter specificity of ECF σs. This protein-DNA code enables prediction and rational reengineering of the promoter specificity of all ECF σs. We then developed a statistically rigorous phylogenetic footprinting pipeline and used it to predict ECF σ regulons. We identified regulons, which encompassed a broad range of functions, for ∼67% of ECF σs. ECF σs can generally be classified into global and local regulators. Whereas local ECF σs have small regulons and a sparse distribution within clades, global ECF σs regulate many promoters and are found in more members of a bacterial clade. Our analysis reveals important organizing principles of bacterial regulatory network evolution and provides a truly global survey of gene regulatory interactions in bacteria.
Results
ECF σs Interact with DNA in a Conserved Fashion.
Predicting and rationally engineering the specificity of a DNA binding protein requires conserved interactions between specific positions in the protein and the DNA. To determine whether ECF σs fulfill this requirement, we constructed a large database of ECF σs, determined their putative autoregulatory promoters, and performed a mutual information (MI) analysis to determine whether specific amino acid positions covaried with specific nucleotide positions across evolutionary time.
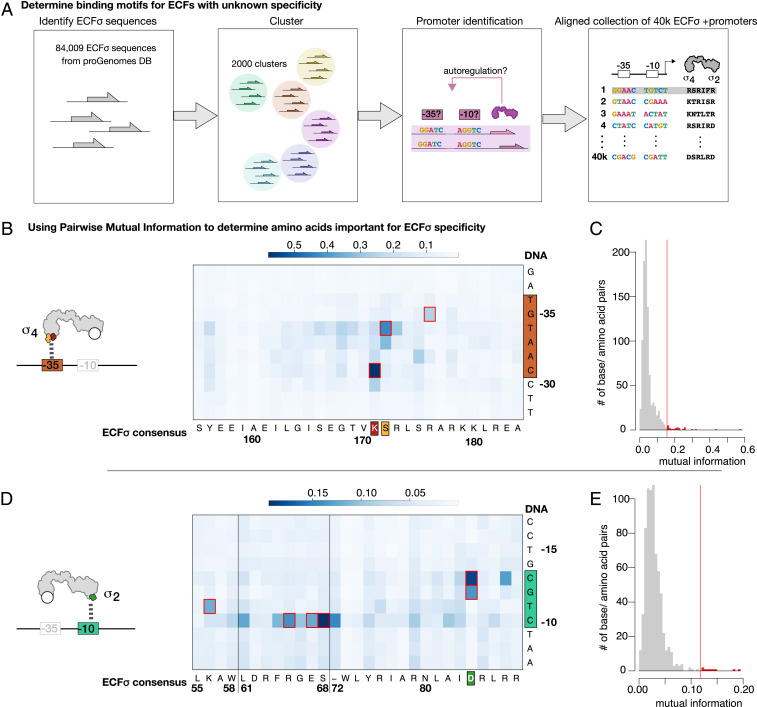
We first identified 84,009 nonduplicate ECF σs from 10,503 genomes in the proGenomes database (13). proGenomes implements eggNOG 4.5 (14), a hierarchical gene orthology framework, to provide consistent functional and taxonomic annotations of >25,000 bacterial genomes. We determined the putative autoregulatory promoter motifs of these ECF σs by clustering them based on their protein sequences (Methods) and searching for overrepresented motifs in the upstream sequences of each cluster. A schematic of this process is depicted in Fig. 1A. To maximize our chances of identifying motifs, we applied two distinct clustering methods [K-mer distance and eggNOG orthology groups (14); Dataset S1], separately searched both the 150 bp and the 300 bp upstream of the ECF σ translation start site, and considered sequences upstream of the ECF σ operon (if applicable), and divergently transcribed genes (if applicable). We identified 91,552 potential promoters belonging to 41,665 unique ECF σs. Motifs were aligned, and putative promoter sequence(s) were associated with the respective ECF σ sequences, generating a single, weighted collection of matched and aligned promoters and ECF σ sequences (Dataset S2).
Fig. 1.
MI analysis reveals conserved ECF σ-promoter interaction. (A) Schematic depicting the process for generating an aligned collection of ECF σs and their putative promoters. ECF σs in the proGenomes database are identified and clustered based on their protein sequence. The upstream DNA sequences associated with each cluster are then mined for autoregulatory motifs, which are aligned, resulting in an aligned collection of ECF σ sequences and their aligned respective putative autoregulatory motifs. (B–E) MI maps (B and D) and distributions (C and E) for the interaction between σ4 domain and -35 promoter motif (B and C) and for the interaction between σ2 and -10 promoter motif (D and E). The red lines in the distributions of MI values represent 7 SDs (as quantified using median absolute distance) above the median. High MI values were not due to phylogenetic artifacts and were significantly higher than random (SI Appendix, Fig. S3). Positions boxed in red are previously identified base-specific interactions.
We next performed a MI analysis on this collection to identify interacting amino acid–nucleotide pairs (Methods). This approach assumes that mutations in a specificity-determining amino acid are compensated for by mutations in the cognate promoter position (and vice versa), leading to high covariation (and therefore MI). Only a few amino acid – nucleotide pairs had high MI (Fig. 1 B–E), and these were consistent with specificity determining interactions observed in the crystal structures of three divergent ECF σs bound to their cognate promoters: Escherichia coli RpoE (15, 16) and Mycobacterium tuberculosis SigL (17) and SigH (18, 19) (Fig. 1 B and D, red boxes), suggesting that the mechanism of ECF σ–promoter interaction is conserved in a majority of ECF σs despite their extensive sequence divergence. This implies that our collection of matching ECF σs and putative promoters can serve as a dictionary in which the promoter specificity conferred by any amino acid at key DNA recognizing positions can be determined.
The Identity of Key Amino Acid Positions Determines Promoter Specificity.
Our MI analysis implies that the identity of the amino acid at key positions should have a predictable effect on the promoter specificity of an ECF σ, enabling both prediction of natural ECF σ promoters and the ability to design ECF σs with arbitrary promoter specificity. We tested this proposition by experimentally characterizing the effect of all possible substitutions at each of three amino acid–nucleotide pairs chosen on the basis of their high MI and presence in ECF σ structures (interactions equivalent to E. coli RpoE Arg171/-31, Ser172/-34, and Asn84/-13 and Asn84/-12). For each pair, we constructed all 19 possible amino acid substitutions and tested them against all four possible promoter variants using a previously described multiplasmid system (9) in which a heterologously expressed ECF σ drives in vivo GFP expression from a test promoter in E. coli (depicted at the top of Fig. 2). We used a different ECF σ for the mutational analysis of each position to highlight the general nature of this dictionary. To simplify nomenclature, we will refer to amino acid positions in ECF σs by the equivalent amino acid in E. coli RpoE, e.g., Arg171RpoE describes the position aligned with E. coli RpoE Arg171.
Fig. 2.
Comparison of experimental and predicted effects of single amino acid mutations on promoter specificity. Using a previously described (9) E. coli multiplasmid system where heterologous ECF σs expressed from one plasmid are used to drive GFP expression from the putative ECF σ promoter in a second plasmid (Top), we tested the activity of ECF σs variants containing all possible amino acid variants at the position equivalent to E. coli RpoE Ser172, Arg171, and Asn84 (Middle). Single amino acid changes drastically altered promoter specificity (Bottom) to match the promoter specificity of natural ECF σs with those amino acid identities (Bottom, gray shading).
The Arg171RpoE mutants were constructed in ECF32_1122 (from Erwinia amylovora). As expected, mutations at this position drastically altered -31 specificity (Fig. 2 and Dataset S3). Importantly, the experimentally determined -31 nucleotide preference of most active ECF σ mutants agreed strongly with the -31 promoter nucleotides of naturally occurring ECF σs with that same amino acid in the Arg171RpoE position. Similarly, mutations at the Ser172RpoE position, constructed in ECF14_1324 [from Streptomyces coelicolor A3 (2)], affected -34 specificity (Fig. 2, Methods, and Dataset S3). Changes in specificity were well correlated to the specificity of naturally occurring ECF σs with the same Ser172RpoE identity in our dataset (R2 = 0.37; P < 0.02). Finally, mutations at the Asn84RpoE position, constructed in ECF11_987 (from Vibrio parahaemolyticus), affected both -12 and -13 nucleotide specificity (Fig. 2, Dataset S3, and Methods), and changes were strongly correlated to those exhibited by natural ECF σs with that same amino acid at Asn84RpoE (Fig. 2; R2 = 0.49; P < 10−16).
Taken together, these comprehensive mutagenesis studies, performed on three distinct ECF σs, from phylogenetically diverse clades demonstrate the stringent yet diverse promoter specificity of ECF σs. All three ECF σs tested were exquisitely sensitive to both single nucleotide changes to their cognate promoters and single amino acid changes in both the σ2 and σ4 domains. Importantly, the changes in specificity caused by the amino acid mutations correlated strongly with the promoter preferences of natural ECF σs containing those amino acids, which enables the de novo prediction of ECF σ promoter specificity based on our aligned collection of ECF σs and promoters.
Modeling Studies Confirm the Importance of Specificity Determinants.
Although most high MI interactions identified amino acid–nucleotide pairs previously implicated in promoter recognition, some highlighted uncharacterized specificity determining interactions. To better understand how these positions may affect promoter specificity, we modeled these noncanonical interactions on existing ECF σ structures, similar to previous studies (20).
First, MI analysis uncovered covariation between the amino acid at the Phe175RpoE position and the -32 nucleotide. In our collection, many natural ECF σs with an arginine at Phe175RpoE had a nontemplate strand T at the -32 promoter position. Modeling these changes onto the E. coli RpoE DNA-bound σ4 structure (15), we found that a phenylalanine to arginine substitution at Phe175RpoE would lead to the loss of three nonpolar interactions between the phenylalanine side chain and the methyl, C6, and ribose C2′ atoms of the -32 template strand T. The compensatory nontemplate strand -32T mutation places the -32 template strand adenine N7 (a hydrogen bond acceptor) near the guanidine group of arginine at Phe175RpoE likely forming an H bond that could compensate for the loss of the nonpolar interactions (SI Appendix, Fig. S1A). This result strongly suggests that ECF σs with an arginine at Phe175RpoE likely require -32T to maintain activity, consistent with our predictions.
Second, our analysis identified a correlation between acidic residues at Lys56RpoE and a -11 A. Previous studies (16) implicated Lys56RpoE primarily in interactions with the -12 nucleotide and suggested that the -11T was predominantly recognized by Ile77RpoE, Ala60RpoE, and Asn80RpoE. To better understand the role of acidic residues at Lys56RpoE, we modeled an acidic substitution at the Lys56RpoE position onto a preexisting structure (16). Basic amino acids at Lys56RpoE would form a hydrogen bond with O4 of -11T, explaining their affinity for -11T. By contrast, the -11A nucleotide would not be able to form a hydrogen bond with a basic residue at Lys56RpoE but could do so with acidic residues at this position (SI Appendix, Fig. S1B), explaining the observed correlation.
Taken together, our modeling results strongly support the hypothesis that the sparse and modular correlations observed in the MI analysis are causal drivers of ECF specificity and highlight the importance of previously unappreciated interactions for the promoter specificity of ECF σs.
De Novo Prediction of ECF Promoter Specificity.
That only a few specific amino acids in an ECF σ make sequence-specific DNA contacts suggests that de novo prediction of promoter specificity based on the primary sequence of a σ may be attainable. Such an approach is suitable for inferring the promoter motifs not only of autoregulatory ECF σs but also of nonautoregulatory and recently diverged ECF σs, which cannot be accurately predicted through clustering-based approaches.
To predict the promoter specificity of ECF σs, we first selected one or two amino acid positions most correlated to the identity of each nucleotide position in the promoter based on the results of our MI analysis and published structures (SI Appendix, Table S1). We build the putative -35 and -10 PWMs of an ECF σ by considering nucleotide position separately. At each promoter position we evaluate the DNA specificity of natural ECF σs that have the same amino acid(s) at the specificity determining positions as our ECF σ of interest. By concatenating these individual sequence preferences, we determine PWMs representing the -35 and -10 sequence specificity of a given ECF σ (Fig. 3A). We determine how efficiently a given ECF σ will activate a given promoter by scoring every possible position in the promoter sequence using the -35 and -10 PWMs and a 16-, 17-, or 18-bp spacer as previously described (21).
Fig. 3.
ECF σ specificity can be accurately predicted based on the identity of key residues. (A) Schematic depicting the prediction of ECF σ motifs based on the identity of amino acids at specific positions. For a given ECF σ, its promoter specificity is determined in a piecewise fashion. For each position in the promoter, the identity of the interacting amino acid(s) is determined. Then, the nucleotide preference of natural ECF σs containing the same amino acid is assigned to the novel ECF σ. (B) Comparison between the experimentally determined (exp.; Top) and predicted (pred.; Bottom) activity of 86 ECF σs on 26 promoters (exp. data from ref. 9). (C) Correlation between actual (log-fold activation) and predicted (maximum PWM promoter score) activity of the 86 ECF σs on 26 promoters depicted in B. The solid blue line represents the median log-fold induction of all ECF σs on all promoters, and the dashed blue lines represent 3 SDs (as qualified by the median absolute distance) from the median.
We assessed the predictive power of this model by testing our ability to predict 2,236 ECF σs–promoter interactions (86 representative ECF σs × 26 promoters) for which in vivo experimental data are available (9) (Fig. 3B). The predicted activity of these 86 ECF σs was strongly correlated to their experimentally measured activity (R = 0.44; P < 10−16; Fig. 3C and Dataset S4). This correlation was comparable to that observed when scoring the experimentally determined PWM of E. coli RpoE on its native promoters (21) (in vitro R = 0.45; in vivo R = 0.60), suggesting that the promoter specificity of diverse ECF σs can be accurately predicted from their primary sequence.
Phylogenetic Footprinting of ECF σ Regulons.
Having established an accurate de novo method for predicting the -35 and -10 promoter specificity of ECF σs, we next sought to determine the putative regulons of all 84,009 ECF σs in our dataset. Previous studies on σ70 and E. coli RpoE highlighted that even when the -35 and -10 PWMs have been experimentally determined, stringently identifying functional promoters is difficult. For example, using optimized -35 and -10 PWMs to scan the E. coli genome recovered 86% of known σ70 promoters and 88% of known RpoE promoters but exhibited an FPR of ∼80% (12) and 92% (11), respectively. Such a high FPR would obfuscate the true ECF σ regulons and preclude biological interpretation of their function.
To overcome these high FPR rates and precisely determine which genes may be regulated by a particular ECF σ, we developed a phylogenetic footprinting approach (Fig. 4A). Phylogenetic footprinting determines true ECF σ binding sites by assessing their conservation in related species. Because natural selection over the course of evolution preferentially maintains functional DNA elements, such as bona fide ECF σ binding sites, this method can identify the conserved regulon of an ECF σ of interest within a set of genomes with a much lower FPR than other methods. Our pipeline uses precomputed orthologous groups (OGs) from eggNOG 4.5 (14) as implemented in the proGenomes+ database (13) to rapidly and robustly identify orthologous genes between organisms.
Fig. 4.
Phylogenetic footprinting identifies real ECF σ promoters. (A) Schematic of phylogenetic footprinting pipeline and its use for ECF σ regulon determination. The score of each OG in each species is the score of the best match of the predicted PWMs in the 300 bp upstream of the gene(s) assigned to the OG in that genome. These scores are normalized within species (which also normalized for GC%) and summed across species for each OG. OGs with high scores in many species are highly conserved targets. To determine the significance of scores, PWMs are shuffled and rescored multiple times to assess how often high scores occur by chance in the specific set of genomes queried. This corrects for genomic diversity (or lack thereof) and the number of genomes queried. (B) Distribution of 5′UTR lengths in predicted ECF σ promoters in all clades (black), Actinobacteria (orange), and Firmicutes (blue). The distribution of 5′UTR lengths is consistent with known bacterial promoters, exhibiting a peak (gray) at 20 to 40 bp, likely representing the minimal ribosome binding site. Leaderless transcripts (5′UTR length ∼ 0) are found in Actinobacteria but not in Firmicutes, as expected for these classes. (C) UP-element subsites are phased upstream of predicted ECF σ promoters and preferentially found at the distal subsite, consistent with previous work on E. coli RpoE which suggested that the distal site is more effective at activating transcription for this ECF σ (22).
Briefly, the 300 bp upstream of each gene in each genome is scored with the predicted PWMs of the ECF σ ortholog from that genome. Next, per gene scores are summed for all genes in an OG. OGs with high scores are likely to be regulated by an ECF σ of interest in the genomes of interest. To determine the significance threshold for OG scores, we repeat the pipeline 100+ times using randomized PWMs. This generates a background distribution that represents the conservation of arbitrary sequences between the genomes being queried (Methods and Fig. 4A). OGs with scores substantially higher than those generated by randomization are likely regulated by the ECF σs of interest.
The statistical power of phylogenetic footprinting (as assessed by the randomized distribution of OG scores, above) depends on the number and diversity of the genomes to which it is applied: nonfunctional sites may be conserved between closely related species or randomly in a small number of genomes. Conversely, regulons in distantly related species may not be conserved. Therefore, to maximize the size of the conserved regulon, we reclustered the ECF σs in our dataset within taxonomic families and orders (Dataset S5) prior to applying this method (Dataset S1). We found at least one significantly (FDR < 0.05) regulated eggNOG orthology group (effectively, at least one gene) for a majority (67%) of ECF σs. Dataset S6 consists of >600,000 ECF σ–gene interactions occurring at >250,000 specific promoters—a global survey of ECF σ function.
The Properties of Predicted ECF σ Promoters Are Similar to Experimentally Characterized Promoters.
To assess the accuracy of these predictions, we explored whether these putative ECF σ promoters exhibited properties of bacterial promoters. Because our phylogenetic footprinting approach considers only predicted -35 and -10 specificities, the presence of additional promoter characteristics (e.g., 5′UTR length, initiating nucleotide, and UP-element) would be associated with these predicted ECF σ promoters only if real promoters were being preferentially identified.
We first explored 5′UTR length. Previous transcriptome studies (23) identified a conserved distribution of 5′UTR lengths across bacterial phyla. The distribution of 5′UTR lengths of our predicted ECF σ promoters was similar to this distribution (Fig. 4B), exhibiting a peak around ∼20 to 40 bp, which corresponds to the minimal ribosome binding footprint (24). As expected, the distribution of predicted 5′UTR lengths was similar in all bacterial phyla, with the exception of extremely short 5′UTRs corresponding to leaderless mRNAs, whose prevalence is known to vary across species (23) (Fig. 4B and SI Appendix, Table S2).
We next explored the region downstream of the -10 element. Many bacterial promoters contain a pyrimidine at the −1 position of the nontemplate strand and a purine at the +1 position, allowing favorable base-stacking interactions between the −1 template strand purine and the +1 nontemplate strand purine (25). Consistent with this, we found that a pyrimidine–purine pair was present at one of the two likely transcription start sites (TSS; equivalent to the −1 or +1 position counted from the flipped out -10 base) in 67.3% of putative ECF σ promoters. Pyrimidine–purine pairs were enriched relative to their purine–pyrimidine counterparts at the two likely TSS positions but not before or after (SI Appendix, Fig. S2). For the 67.3% of promoters with pyrimidine–purine pairs, we assumed that the purine base was the initiating nucleotide (+1). Although both adenine and guanine are commonly used as initiating nucleotides in all phyla, the predicted promoters of leaderless mRNA promoters were highly enriched for adenine initiating nucleotides (SI Appendix, Table S2), as expected considering the importance of the AUG codon in recruiting ribosomes to these mRNAs (23).
Finally, we searched for UP elements, which consist of phased A/T tracts (26) with extremely narrow minor grooves upstream of the -35 element that enhance transcription by interacting with the C-terminal domain of one or both alpha subunits of RNAP (αCTD). UP-element subsites are located on the same face of the DNA-helix as RNAP, immediately upstream of the -35 element (proximal UP-element subsite), one DNA helix turn upstream (distal UP-element subsite), or two DNA helix turns upstream (far distal UP-element subsite). To determine whether our predicted ECF σ promoters had UP elements, which would indicate that promoters were correctly predicted, we predicted DNA shape upstream of all 251,594 predicted ECF σ promoters using DNAshapeR (27) and defined putative UP-element subsites as two adjacent nucleotides with a minor groove width <3.5A. We found that ∼25% of putative ECF σ promoters contained at least one UP-element subsite. UP-element subsites were predominantly located at the proximal, distal, and far-distal subsites (Fig. 4C), and the frequency of UP elements was consistent with their reported prevalence by bacterial phyla (28) (SI Appendix, Table S3).
These data demonstrate that our predicted ECF σ promoters share many characteristics with experimentally determined promoters, including 5′UTR length, pyrimidine–purine pairs near the start site, and UP elements upstream of the -35, and suggest that our phylogenetic footprinting approach correctly identifies biologically relevant and active bacterial promoters. The strength of these signals indicates that our pipeline achieves a low false positive rate when identifying promoters, a prerequisite for biological interpretation.
ECF σ Regulons Are Accurately Predicted.
Since our putative ECF σ promoters share many properties with active bacterial promoters, we next assessed whether the specific regulon predictions were correct. Although no regulon information exists for the overwhelming majority of ECF σs, ∼50% of ECF σs have been reported to autoregulate (3, 9) (Fig. 1). Taking advantage of this positive control, we assessed the fraction of ECF σs predicted to autoregulate. We found that 52.9% of ECF σs with significant regulons (P < 0.05) included themselves in their regulon, a percentage which did not vary substantially when the p-value threshold for significance was raised (55.3% autoregulatory at P < 0.01). As expected, ECF σs for which autoregulatory promoters were previously identified (Fig. 1) were more likely to autoregulate than ECF σs for which we did not previously identify autoregulatory promoters (70% for ECF σs with previously identified autoregulatory promoters, 33% for other ECF σs), highlighting the accurate and precise regulons determined by combining our DNA-specificity predictions and phylogenetic footprinting pipeline.
To more specifically probe the accuracy of regulon predictions, we assessed coexpression of predicted ECF σ regulons in 13 diverse bacterial species using publicly available gene expression data (29). We reasoned that the expression of genes in true ECF σ regulons, likely including the ECF σ itself, should be correlated across experimental conditions, as quantified by comparing the correlation between the expression of the predicted ECF σ regulon and the same number of randomly selected genes. We found that 41% of the predicted ECF σ regulons examined (37 out of 90; Dataset S7) were significantly more correlated than expected by random chance, suggesting coregulation. The lack of significant correlations for the remaining ECF σs may be due to incorrect regulon predictions, an insufficiently diverse set of gene expression data (i.e., no conditions that activated the σ), or small but correctly predicted regulons that fail to reach the threshold of statistical significance. Consistent with the idea that small regulons may be difficult to validate using this method, significantly correlated regulons had more genes on average (15.7) than regulons without significant correlations (7.3; P < 0.01). Taken together these data strongly suggest that our model of ECF σ promoter specificity combined with our phylogenetic footprinting approach to regulon determination correctly identifies the promoter regions and members of ECF σ regulons.
ECF σs Function as Both Global and Local Regulators.
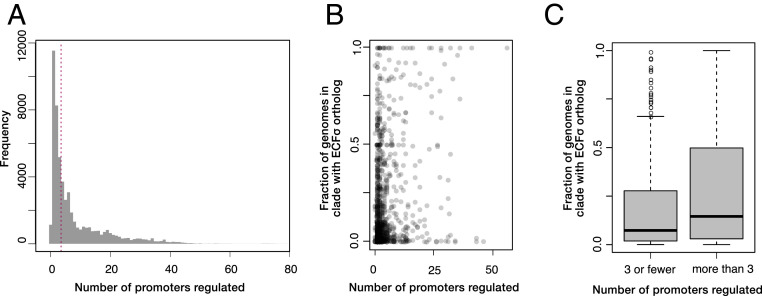
Studies of genome-wide gene regulatory networks in eukaryotes (30), archaea (31), and bacteria (32) have identified hierarchical network structures consisting of global and local transcriptional regulators. These differ in the number of regulated genes, their conservation, and the diversity of conditions in which they exert their effects. That some ECF σs may regulate small regulons was suggested in the original manuscript identifying the ECF σ family (33), but most of the well-characterized ECF σs such as RpoE in γ-proteobacteria and SigR/SigH in Actinobacteria are global regulators that control the expression of many genes in response to diverse conditions (11, 34–36). To determine whether most ECF σs function as global regulators, we first quantified the number of promoters they regulated and then examined whether they possessed the properties of global and local regulators. We found that ECF σs had regulons varying from 1 to 85 promoters (Fig. 5A). As expected, ECF σs known to have large regulons also had large average predicted regulons (e.g., RpoE, 17 promoters; SigR/SigH, 41 promoters; Dataset S8). However, nearly half of ECF σs in our dataset (47.8%) were predicted to regulate three or fewer promoters. We eliminated the two most obvious technical explanations for this observation, small ECF σs clusters or low-information motif predictions, by assessing the correlation between these factors and predicted regulon size. Although both motif information content and cluster size were significantly correlated to regulon size (all P < 10−16), together these parameters only explained a small amount of the variability in regulon size (R2 < 0.1), suggesting that much of the variation in regulon size is biologically meaningful. We therefore sought to determine if the ECF σs predicted to have large regulons are bona fide global regulators.
Fig. 5.
ECF σ regulon size and distribution within species. (A) Number of genes in ECF σ regulons. Red line separates ECF σs with three and four promoters. (B) Scatterplot of ECF σs eggNOG orthology groups showing the median number of regulated promoters and the fraction of genomes within a clade that have the ECF σ. (C) Boxplot of fraction of genomes with each ECF σ group separated by ECF σs with large or small regulons.
A key feature of global regulators is their importance under diverse conditions. To determine whether ECF σs with large (>3 promoters) predicted regulons are important in more conditions than ECF σs with small (≤3 promoters), we used a previously published dataset (37) in which 5,647 genome wide transposon fitness experiments were performed on 37 diverse bacterial species. A total of 136 ECF σs from our dataset, 77 of which had regulon predictions, were represented in these experiments, allowing us to determine if predicted regulon size correlated to the number of conditions in which each ECF σ affects cellular fitness. We found that the number of regulated promoters was significantly (P < 0.0001) correlated to the fraction of conditions in which the ECF σ was important for cellular fitness (t < −3). On average, ECF σs predicted to regulate large regulons had significant phenotypes in 4.2% of tested conditions, whereas ECF σs with small regulons had significant phenotypes in 0.4% of conditions (t test P < 0.0001). Several RpoE and RpoE2 homologs in Shewanella species were predicted to regulate large regulons but exhibited few significant phenotypes. Since these ECF σs have been reported to be important under a wide range of growth and stress conditions (38), the lack of significant phenotypes is likely due to experimental issues (such as compensatory mutations or lack of relevant conditions). Notably, the regulons of these ECF σs appear to be correctly predicted (Dataset S6) (38).
Another important characteristic of global regulators is their conservation within bacterial clades. To determine whether ECF σs with large predicted regulons are more conserved than those with small predicted regulons, we grouped ECF σs by their eggNOG orthology groups and assessed the fraction of genomes within the relevant bacterial clade encoding a member of each orthology group (Fig. 5B). ECF σs with large regulons were present on average in 28% of genomes, while those with small regulons were found on average in 16.9% of genomes within a clade (Fig. 5C). Due to the varying diversity within clades and within eggNOG orthology groups, our measure of penetrance within clades likely underestimates the ultimate conservation of ECF σs. Despite this, the statistically significant (t test P < 10−5) difference in distribution suggests that ECF σs with large regulons, such as RpoE in the γ-proteobacteria and SigR/SigH in the Actinobacteria, are conserved in a more species within those clades, while ECF σs with smaller regulons tend to have a patchier distribution.
One explanation for the patchier distribution of ECF σs with small regulons is that they were acquired through horizontal gene transfer (HGT). To determine whether ECF σs are commonly horizontally acquired, we applied tetranucleotide profiling (39) to all genomes in our dataset to identify regions of atypical sequence content associated with HGT events and assessed whether ECF σs were present in these regions. Regions of atypical sequence content, which canonically include rRNAs and tRNAs, were efficiently identified (69% of rRNAs and 46% of tRNAs), but ECF σs were not more likely to be horizontally acquired than other genes in most bacterial phyla (Dataset S9). However, ECF σs in both Bacteroidetes and Cytophagia (both members of the Fibrobacteres, Chlorobi, and Bacteroidetes superphylum) were more frequently horizontally acquired (21 and 18%, respectively) than the already high HGT rate of all genes in these phyla (16 and 13%, respectively). Many ECF σs in Bacteroidetes have been implicated in the regulation of carbohydrate utilization pathways, raising the possibility that these ECF σs are being transferred as part of carbohydrate utilization loci. Across all clades, ECF σs with small (≤3 promoters) regulons were more likely to be horizontally acquired than ECF σs with large (>3 promoters) regulons (9.7 vs. 7.9%, t test P < 10 to 12), suggesting that these ECF σs may be transferred together with their regulon. Consistent with this idea, we found that many of the genes regulated by ECF σs with small regulons were located within 2 kb of the ECF σ (25.0%, compared with 7.8% for ECF σs with >3 regulated promoters).
Taken together, these data suggest that ECF σs function both as local regulators, controlling the expression of a few promoters often located nearby in the genome, likely in response to specific stimuli and as global regulators, controlling the expression of many promoters and functioning to maintain cellular homeostasis under diverse conditions.
The Role of ECF σs in Bacterial Clades.
In addition to providing a database of putative ECF σs regulon predictions across clades to guide future discovery, our analysis also revealed specific regulatory interactions of biological interest which confirm and extend previous analyses. The full dataset (Dataset S6) provides additional intriguing connections.
A major finding of our study was that almost half of the ECF σs function as local regulators. This regulatory function is exemplified by two broadly distributed groups of σs: ECF41 and ECF42 groups. Previous work had noted the conserved synteny of the genes surrounding these ECF σs and had experimentally validated the small regulon of these ECF σs in a model organism (40, 41). Our analyses support these findings and expand them to numerous bacterial clades, highlighting the local nature of these regulators. Our recapitulation of these results also highlights the high stringency of our ECF σ regulon prediction.
The Actinobacteria are an expansive, diverse phylum of high-GC gram-positive organisms canonically associated with soil ecosystems but also relevant in human health and disease: Streptomyces produce antibiotics, Bifidobacteria are microbiome constituents, and Mycobacteria are important pathogens. Despite their varied niches, all Actinobacteria contain a homologous ECF σ, known as SigR in Streptomyces species and SigH in Mycobacterium species, which responds to redox and translation stress. We identified a core regulon consisting of 6 genes (iscA, trxB, clpC, SCO3296, rbpA, and sigH/R) which were significantly regulated in all six orders and an additional 18 genes predicted to be regulated in four to five orders (Dataset S8). Many of these conserved regulon members encode classical oxidative stress response proteins, such as thioredoxins, members of the Clp complex, and Fe-S cluster repair proteins. The regulon of this ECF σ in the Corynebacteriales (which includes Mycobacterium) contains six unique genes. Intriguingly, three of these (Rv3054c, Rv3463c, and Rv1334) were among the most strongly activated genes in a recent overexpression study (42). Both Rv3054c and Rv3463c (the two most SigH activated proteins in M. tuberculosis) encode reductases, and Rv1334 has been shown to be essential for survival in macrophages (43). Although the functions of these genes remain to be determined, their integration into the SigH regulon of Corynebacteriales, but not of the other orders, suggests a role for these proteins in responding to redox and translation stresses unique to the Corynebacteriales, potentially signaling a role for these proteins in infection.
SigX, an important ECF σ in Pseudomonas species (44), illustrates another aspect of regulon evolution. This ECF σ is important for growth, biofilm formation, and virulence. It is predicted to regulate 28 genes, many of which are involved in fatty acid synthesis and other membrane functions, consistent with previous work (44, 45). SigX is also predicted to regulate nearby genes cmpX, a conserved transmembrane protein, and oprF, the major Pseudomonas outer membrane porin. Despite its importance in Pseudomonas, SigX is found only in a few other closely related γ-proteobacterial clades (Paraglaciecola, Glaciecola, and Colwellia). Surprisingly, a SigX homolog is found in Flavobacterium, a class in the Bacteroidetes phylum, where it is predicted to regulate nearby genes homologous to cmpX and oprF but not genes involved in fatty acid metabolism. These regulatory patterns, combined with the atypical tetranucleotide profile of SigX in a majority (84%; Dataset S9) of γ-proteobacterial genomes, suggest that SigX and neighboring genes may have been horizontally acquired by Pseudomonas, possibly from a member of the Flavobacteria. If this is the case, regulation of fatty acid synthesis by SigX in Pseudomonas may reflect regulatory capture over a relatively short time span, raising important questions about the evolutionary pressures that led to this outcome.
Discussion
Predicting gene regulatory interactions from the amino acid sequence of transcriptional regulators is a long-standing goal in biology. To do so requires both the ability to predict the sequence specificity of a novel regulator and the ability to determine significant interactions. Because of the difficulty of meeting both of these challenges, previous work has focused primarily on engineering modular regulatory proteins such as zinc-finger (ZFs) and transcription activator-like (TALs) rather than on de novo prediction of biological targets, a problem that remains largely unsolved (46, 47). In this work, we solve the DNA-specificity code of ECF σs and build a computational pipeline that enables us to use these rules to determine statistically significant putative regulons for ∼67% of bacterial ECF σs.
That the amino acids at a few key conserved positions largely determine promoter specificity is surprising and suggests significant constraints on the evolution of ECF σ DNA binding domains. Such constraints may be imposed by extensive protein–protein interactions of ECF σs with both RNAP and negative regulators (anti-σs), which likely limit the ability of σs to evolve novel DNA specificities through conformational changes. The conservation of DNA determining amino acid positions allows prediction and rational engineering of ECF σ specificity, with exciting implications for tuning bacterial regulation to increase stress resistance or metabolic productivity in industrial settings (48).
To leverage our ability to predict ECF σ promoter specificity, we built a high-throughput, computationally tractable, and statistically rigorous phylogenetic footprinting pipeline using precomputed orthology groups derived from a kingdom-wide analysis of protein evolution (14) and a robust statistical method based on randomized motifs. This pipeline stringently identified real ECF σ promoters in genome sequences, as evidenced by the presence of UP elements, pyrimidine–purine pairs at the TSS, and the 5′UTR length distribution of predicted ECF σ promoters (Fig. 4). The resulting dataset of putative ECF σ regulons is the first of its kind: a comprehensive look at the role of a broadly distributed family of gene regulatory proteins, revealing the breadth of ECF σ regulation, the presence of conserved global regulators and variable local regulators, and opening a window into the evolution of ECF σ regulon membership in different niches.
Although the regulons of most ECF σs were accurately predicted, FecI-like σs were overrepresented among ECF σs without statistically significant predictions. Previous studies have suggested that FecI-like ECF σs may function differently from other ECF σs: FecI is unable to drive transcription without its anti-σ, FecR (5), and mutagenesis of a PvdS (a Pseudomonas aeruginosa FecI-like ECF σ) suggests that the stringency of -35 element recognition is weak (49). Taken together, these data and the lack of statistically significant predictions for FecI-like ECF σs suggest that their promoter recognition properties may substantially differ from those of canonical ECF σs and merit further study.
The approach demonstrated here, which combines covariation analyses with powerful phylogenetic footprinting tools, is not limited to ECF σs but can be broadly applied to study protein–DNA interactions, bridging experimental and computational approaches. By allowing future studies to associate novel genes with known regulators, infer bacterial ecology by the coregulation of disparate stress responses, and reveal the principles of gene regulatory network evolution, this approach will lead to mechanistic and conceptual understanding of bacterial gene regulatory network function and evolution.
Methods
Identifying ECF σs and Their Regulatory Sequences.
Identifying ECF σs.
ECF σs from the proGenomes database (13) were identified by using hmmer (50) to search for protein sequences that contained both a σ2 domain (Pfam:Sigma70_r2, PF004542) and a σ4 domain (Pfam:Sigma70_r4, PF004545 or Pfam:Sigma70_r4–2, PF008281) but lacked a σ3 domain (Pfam:Sigma70_r3, PF004539), as previously described (3). Sequences with more than 65 amino acids between the σ2 domain and the σ4 domain were excluded due to the likely presence of a cryptic σ3 domain (SI Appendix, Fig. S4). The resulting set contained 133,424 ECF σ sequences, which were aligned using hmmalign to custom ECF σ2 and ECF σ4 hmm models. Because alignments frequently contained gaps due to the sequence diversity of ECF σs, alignments were collapsed on either side of conserved residues in domains σ2 and σ4 (positions equivalent to E. coli RpoE Gln51, Phe64, Trp73, and Glu158, respectively) to generate the final alignments (Dataset S2) and manually adjusted where necessary.
Retrieving upstream sequences.
To maximize the chance of finding conserved binding sites, up to three sequences at two lengths were retrieved for each ECF σ. For all ECF σs, sequences were retrieved upstream from the translation start site of the ECF σ (1). If the ECF σ was in an operon (defined as codirectional ORFs < 50 nucleotides apart), the sequence upstream of the first gene in the operon was also retrieved (2). If a divergent gene was located either upstream of the ECF σ or of an operon containing the ECF σ, its upstream sequence was also retrieved (3). Sequences of both 150 and 300 bp were retrieved and analyzed, to increase the statistical power for small clusters (150 bp) and to increase the probability of determining downstream motifs (300 bp).
Duplicate removal.
The genomes of certain organisms (e.g., M. tuberculosis, Streptococcus pneumoniae, and E. coli) are grossly overrepresented in sequence databases such as proGenomes (13). To minimize the influence of duplicate sequences, we only considered one ECF σ from groups in the same phylogenetic clade that were identical in protein sequence, upstream sequence, operon sequence, and divergent gene sequence. The remaining set contained 84,009 ECF σs (Dataset S1).
Clustering.
ECF σs were clustered solely on the basis of their protein sequence. We used two distinct methods to cluster ECF σs. First, we used the eggNOG orthology group of each ECF σ. This clade-specific identifier is based on all-by-all Smith–Waterman alignments of protein sequences from representative genomes (14). Our set contained 1,196 distinct eggNOG orthology groups. Second, we used k-mer distance, as implemented in ClustalW. Besides the advantage of being a distinct clustering method, k-mer distance does not take into account phylogeny, allowing similar ECF σs from distinct bacterial clades to be interrogated together.
Motif discovery and weighting.
For each ECF σ cluster, up to six libraries of upstream regulatory sequences were searched for putative two-block motifs using BioProspector (51). These libraries consisted of 150- and 300-bp sequences from upstream of the ECF σs, upstream of an operon containing the ECF σs, and upstream of a divergently transcribed gene. Motif searches with BioProspector were performed only on the forward strand. Typical searches were of the form W7 w5 G18 g15, where W and w denote the size (bp) of the -35 and -10 motifs, and G and g denote the largest and smallest spacer size. For each cluster in each library, the highest scoring two-block motif was examined and aligned manually. Because an ECF σ could potentially be associated with as many as 12 motifs (150 and 300 bp; k-mer & eggNOG clusters; upstream, operon, and divergent sequences), or as few as 1, motifs were weighted by the reciprocal of the total number of motifs associated with each ECF σ (e.g., the only motif of an ECF σ has a weight of 1, whereas all 12 motifs of an ECF σ with 12 motifs are assigned weights of 1/12). These weights were considered in all downstream analyses.
MI Analysis.
MI analysis was performed on every possible nucleotide position and amino acid position pair. We used the MI formula but accounted for the weights associated with each DNA motif (Motif discovery and weighting) when computing probabilities.
ECF σ Substitution Experiments.
The σ‐promoter GPF measurements were performed as previously described (9). Briefly, assays were performed in E. coli (DH10β) using a three-plasmid system consisting of 1) an IPTG‐inducible T7 RNAP (pN565), 2) a plasmid carrying the σ under the control of an T7 promoter, and 3) a plasmid series carrying a σ promoter driving to sfgfp. Assays were performed using 1 mM IPTG, except those using ECF11_987, which were performed at 10 μM IPTG to avoid toxicity. All assays were performed in a 96‐well format. Overnight liquid transformants were diluted into fresh prewarmed LB +Spec, Amp, Kan, and PTG in a 96‐well cell culture plate and covered with a breathable membrane. Cultures were incubated in a Tecan infinite M1000Pro plate shaker for 37 °C at 582 rpm. OD600 and GFP fluorescence were tracked, and the maximum GFP synthesis rate/OD600 was calculated and used for downstream analysis.
Modeling Studies.
Previously solved ECF σ structures (2MAP and 2H27) were modified using PyMOL and manually assessed for likely interactions.
Predicting ECF σ Motifs.
Motifs were predicted by concatenating the sequence preferences for individual nucleotides based on the alignments described above. Briefly, at each nucleotide position, the log fraction of each DNA base in ECF σs sharing the same amino acids at relevant positions (SI Appendix, Table S1) are taken. Code is available at https://github.com/horiatodor/predictECF.
Phylogenetic Footprinting Pipeline.
A custom pipeline (https://github.com/horiatodor/ECFGenome-Rcpp) was built to perform phylogenetic footprinting based on eggNOG orthology (14). Input consisted of genomes of interest and the predicted -35 and -10 PWMs for each genome. Briefly, each set of PWMs was scored on the upstream sequences (−332 to +2) of each gene in the appropriate genomes. Genes in operons were allowed to take the score of upstream genes, if higher. The score for each orthologous group (OG) was calculated by summing the score of the highest scoring member in each genome, weighted by the relative phylogenetic similarity of all genomes in the set. FDR thresholds were set by randomizing the order of the columns in the PWMs 200× and recomputing ortholog scores.
Using Gene Expression Patterns to Evaluate Regulon Predictions.
To evaluate the accuracy of the predicted regulons of ECF σs, regulons from 13 diverse bacterial species were used: M. tuberculosis H37Rv, S. coelicolor A3 (2), Caulobacter crescentus CB15, Rhizobium etli CFN 42, Rhodobacter sphaeroides 2.4.1, Bacillus subtilis subsp subtilis str. 168, Bacteroides thetaiotaomicron VPI-5482, Porphyromonas gingivalis W83, Bordetella pertussis Tohama I, Burkholderia cenocepacia J2315, E. coli str. K-12 substr W3110, P. aeruginosa, and Shewanella oneidensis MR-1. For each species, the program COnTORT (29) was used to download all publicly available gene expression data from NCBI GEO in order to capture as many unique experimental conditions as possible. Since the precise conditions required to activate each ECF σ are unknown, we used the large dataset of all gene expression data with the idea that the more varied experimental conditions, the more likely the genes in the regulon would have correlated expression patterns. For each predicted regulon with at least two predicted members, the gene expression profiles for all members of the regulon were correlated across all available gene expression datasets. To determine if the correlation was due to random chance, the analysis was performed using the same number of randomly selected genes (repeated 1,000 times). A Wilcoxon rank sum test in the software package R (version 3.6.1) was used to determine statistical significance between the correlation values of the genes within the predicted regulon and the randomly selected genes, with P value of 0.05 being significant.
Tetranucleotide Profiling to Identify HGT Regions.
Tetranucleotide profiling was performed as previously described (39). Regions whose distance from the genomic median was >2.5 MAD higher than normal were considered to have atypical sequence content (and therefore likely horizontally acquired).
Supplementary Material
Acknowledgments
We thank M. Laub, P. Kiley, V. Rhodius, and members of the C.A.G. laboratory for helpful discussions. This work was supported by the NIH (Grants R35 GM118061 to C.A.G. and R01 AG058742 to H.L.), the NSF (Grant 1538946 to C.A.G.), and the US Department of Energy Office of Science (Award DE-SC0018409 to T.J.D.).
Footnotes
Competing interest statement: T.J.D., C.A.G., and M.J.B. are coauthors on a 2019 review article. C.A.G. and M.J.B. are coauthors on a consortium paper [D. Casas-Pastor et al., bioRxiv:2019.12.11.873521 (2019)].
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020204117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Gruber T. M., Gross C. A., Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Paget M. S., Bacterial sigma factors and anti-sigma factors: Structure, function and distribution. Biomolecules 5, 1245–1265 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staroń A., et al. , The third pillar of bacterial signal transduction: Classification of the extracytoplasmic function (ECF) σ factor protein family. Mol. Microbiol. 74, 557–581 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Casas-Pastor D., et al. , Expansion and re-classification of the extracytoplasmic function (ECF) σ factor family. bioRxiv:2019.12.11.873521 (20 December 2019). [DOI] [PMC free article] [PubMed]
- 5.Braun V., Mahren S., Ogierman M., Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6, 173–180 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Mecsas J., Rouviere P. E., Erickson J. W., Donohue T. J., Gross C. A., The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7, 2618–2628 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Donohue T. J., Shedding light on a Group IV (ECF11) alternative σ factor. Mol. Microbiol. 112, 374–384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibb M. J., Domonkos A., Chandra G., Buttner M. J., Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σ(BldN) and a cognate anti-sigma factor, RsbN. Mol. Microbiol. 84, 1033–1049 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Rhodius V. A., et al. , Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 9, 702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urtecho G., Insigne K., Tripp A. D., Brinck M., Lubock N. B., Genome-wide functional characterization of Escherichia coli promoters and regulatory elements responsible for their function. 10.1101/2020.01.04.894907 (6 January 2020). [DOI]
- 11.Rhodius V. A., Suh W. C., Nonaka G., West J., Gross C. A., Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 4, e2 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huerta A. M., Collado-Vides J., Sigma70 promoters in Escherichia coli: Specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 333, 261–278 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Mende D. R., et al. , proGenomes: A resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 45, D529–D534 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huerta-Cepas J., et al. , eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane W. J., Darst S. A., The structural basis for promoter -35 element recognition by the group IV σ factors. PLoS Biol. 4, e269 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campagne S., Marsh M. E., Capitani G., Vorholt J. A., Allain F. H.-T., Structural basis for -10 promoter element melting by environmentally induced sigma factors. Nat. Struct. Mol. Biol. 21, 269–276 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Lin W., et al. , Structural basis of ECF-σ-factor-dependent transcription initiation. Nat. Commun. 10, 710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang C., et al. , Structures and mechanism of transcription initiation by bacterial ECF factors. Nucleic Acids Res. 47, 7094–7104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Fang C., Zhuang N., Wang T., Zhang Y., Structural basis for transcription initiation by bacterial ECF σ factors. Nat. Commun. 10, 1153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim K. Y., Park J. K., Park S., In Streptomyces coelicolor SigR, methionine at the -35 element interacting region 4 confers the -31′-adenine base selectivity. Biochem. Biophys. Res. Commun. 470, 257–262 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Rhodius V. A., Mutalik V. K., Predicting strength and function for promoters of the Escherichia coli alternative sigma factor, sigmaE. Proc. Natl. Acad. Sci. U.S.A. 107, 2854–2859 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodius V. A., Mutalik V. K., Gross C. A., Predicting the strength of UP-elements and full-length E. coli σE promoters. Nucleic Acids Res. 40, 2907–2924 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck H. J., Moll I., Leaderless mRNAs in the spotlight: Ancient but not outdated! Microbiol. Spectr. 6, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendoza-Vargas A., et al. , Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4, e7526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basu R. S., et al. , Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J. Biol. Chem. 289, 24549–24559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Estrem S. T., Gaal T., Ross W., Gourse R. L., Identification of an UP element consensus sequence for bacterial promoters. Proc. Natl. Acad. Sci. U.S.A. 95, 9761–9766 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu T.-P., et al. , DNAshapeR: An R/Bioconductor package for DNA shape prediction and feature encoding. Bioinformatics 32, 1211–1213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubin E. A., Lilic M., Darst S. A., Campbell E. A., Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures. Nat. Commun. 8, 16072 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers K. S., Place M., Noguera D. R., Donohue T. J., COnTORT: Comprehensive Transcriptomic Organizational Tool for simultaneously retrieving and organizing numerous gene expression data sets from the NCBI Gene Expression Omnibus database. Microbiol. Resour. Announc. 9, e00587-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luscombe N. M., et al. , Genomic analysis of regulatory network dynamics reveals large topological changes. Nature 431, 308–312 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Bonneau R., et al. , A predictive model for transcriptional control of physiology in a free living cell. Cell 131, 1354–1365 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Martínez-Antonio A., Collado-Vides J., Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6, 482–489 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Lonetto M. A., Brown K. L., Rudd K. E., Buttner M. J., Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. U.S.A. 91, 7573–7577 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiebig A., Herrou J., Willett J., Crosson S., General stress signaling in the alphaproteobacteria. Annu. Rev. Genet. 49, 603–625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paget M. S., Kang J. G., Roe J. H., Buttner M. J., sigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 17, 5776–5782 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J.-H., Lee J.-H., Roe J.-H., SigR, a hub of multilayered regulation of redox and antibiotic stress responses. Mol. Microbiol. 112, 420–431 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Price M. N., et al. , Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503–509 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Dai J., et al. , An extracytoplasmic function sigma factor-dependent periplasmic glutathione peroxidase is involved in oxidative stress response of Shewanella oneidensis. BMC Microbiol. 15, 34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becq J., Churlaud C., Deschavanne P., A benchmark of parametric methods for horizontal transfers detection. PLoS One 5, e9989 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wecke T., et al. , Extracytoplasmic function σ factors of the widely distributed group ECF41 contain a fused regulatory domain. MicrobiologyOpen 1, 194–213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Q., Pinto D., Mascher T., Characterization of the widely distributed novel ECF42 group of extracytoplasmic function σ factors in Streptomyces venezuelae. J. Bacteriol. 200, e00437-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp J. D., et al. , Comprehensive definition of the SigH regulon of Mycobacterium tuberculosis reveals transcriptional control of diverse stress responses. PLoS One 11, e0152145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rengarajan J., Bloom B. R., Rubin E. J., Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 8327–8332 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanka A., et al. , Identification of the alternative sigma factor SigX regulon and its implications for Pseudomonas aeruginosa pathogenicity. J. Bacteriol. 196, 345–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boechat A. L., Kaihami G. H., Politi M. J., Lépine F., Baldini R. L., A novel role for an ECF sigma factor in fatty acid biosynthesis and membrane fluidity in Pseudomonas aeruginosa. PLoS One 8, e84775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dogan B., Kailasam S., Corchado A. H., Nikpoor N., Najafabadi H. S., A domain-resolution map of in vivo DNA binding reveals the regulatory consequences of somatic mutations in zinc finger transcription factors. bioRxiv:630756 (16 June 2020).
- 47.Zuo Z., et al. , Why do long zinc finger proteins have short motifs? bioRxiv:637298 (15 May 2019).
- 48.Tripathi L., Zhang Y., Lin Z., Bacterial sigma factors as targets for engineered or synthetic transcriptional control. Front. Bioeng. Biotechnol. 2, 33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson M. J., Lamont I. L., Mutational analysis of an extracytoplasmic-function sigma factor to investigate its interactions with RNA polymerase and DNA. J. Bacteriol. 188, 1935–1942 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eddy S. R., Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X., Brutlag D. L., Liu J. S., BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 127–138 (2001). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.