Significance
Traditional microscopy suffers from a fixed trade-off between depth-of-field (DOF) and spatial resolution—the higher the desired spatial resolution, the narrower the DOF. We present DeepDOF, a computational microscope that allows us to break free from this constraint and achieve >5× larger DOF while retaining cellular-resolution imaging—obviating the need for z-scanning and significantly reducing the time needed for imaging. The key ingredients that allow this advance are 1) an optimized phase mask placed at the microscope aperture; and 2) a deep-learning-based algorithm that turns sensor data into high-resolution, large-DOF images. DeepDOF offers an inexpensive means for fast and slide-free histology, suited for improving tissue sampling during intraoperative assessment and in resource-constrained settings.
Keywords: deep learning, extended depth-of-field microscopy, end-to-end optimization, phase mask, pathology
Abstract
Microscopic evaluation of resected tissue plays a central role in the surgical management of cancer. Because optical microscopes have a limited depth-of-field (DOF), resected tissue is either frozen or preserved with chemical fixatives, sliced into thin sections placed on microscope slides, stained, and imaged to determine whether surgical margins are free of tumor cells—a costly and time- and labor-intensive procedure. Here, we introduce a deep-learning extended DOF (DeepDOF) microscope to quickly image large areas of freshly resected tissue to provide histologic-quality images of surgical margins without physical sectioning. The DeepDOF microscope consists of a conventional fluorescence microscope with the simple addition of an inexpensive (less than $10) phase mask inserted in the pupil plane to encode the light field and enhance the depth-invariance of the point-spread function. When used with a jointly optimized image-reconstruction algorithm, diffraction-limited optical performance to resolve subcellular features can be maintained while significantly extending the DOF (200 µm). Data from resected oral surgical specimens show that the DeepDOF microscope can consistently visualize nuclear morphology and other important diagnostic features across highly irregular resected tissue surfaces without serial refocusing. With the capability to quickly scan intact samples with subcellular detail, the DeepDOF microscope can improve tissue sampling during intraoperative tumor-margin assessment, while offering an affordable tool to provide histological information from resected tissue specimens in resource-limited settings.
Histopathology, or microscopic examination of thinly sectioned and stained tissue slices on glass slides, is the gold standard to diagnose and guide surgical management of cancer. To prepare histopathology slides, biopsies or surgical specimens are typically formalin-fixed and paraffin-embedded (FFPE), sliced with a microtome to around 5 µm, stained with hematoxylin and eosin (H&E) dyes, and evaluated under a light microscope. For intraoperative assessment, resected surgical specimens will be first cut with a scalpel into 3- to 4-mm-thick slices to access potential tumor margins on cross-sectional surfaces (Fig. 1A); the thick slices can then be quickly frozen to acquire thin (∼5 µm) transverse tissue sections for staining and microscopic examination. While frozen sections can reduce the processing time, a cryostat microtome is required to cut thin sections of frozen tissue, which still must be fixed and stained. Despite the central role of histopathology in cancer diagnosis, the time- and labor-intensive sample-preparation steps require specialized personnel and expensive equipment, while allowing for only limited sampling of resected tissue. In addition, these destructive procedures are susceptible to tissue-processing artifacts (1, 2) and can also interfere with downstream molecular or genetic analysis.
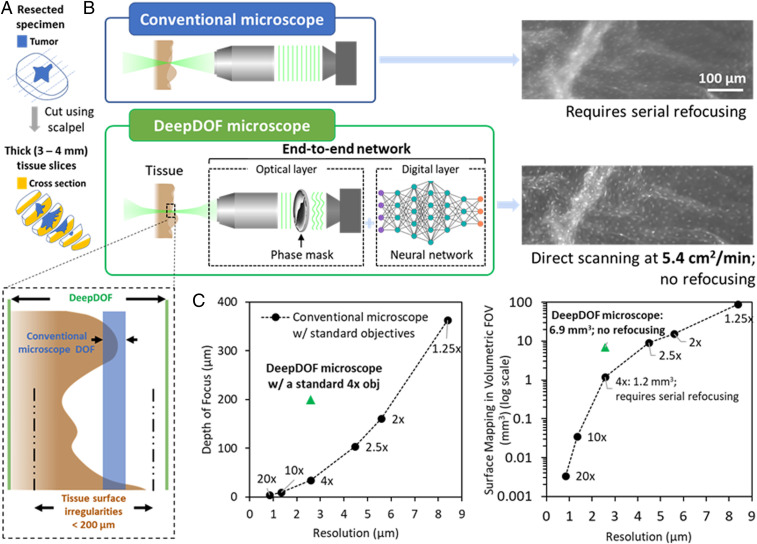
Fig. 1.
DeepDOF microscope schematic and imaging performance in comparison to conventional microscopes for fluorescence imaging of intact tissue specimens. (A) Prior to imaging, the resected specimen is bread-loafed by using a pathology scalpel, and the cross-section surface can be evaluated for tumor-margin assessment. (B) Variations in the surface topology of intact tissue specimens exceed the DOF of a conventional microscope with subcellular resolution. In comparison, with the simple addition of an inexpensive phase mask, the end-to-end optimized DeepDOF microscope allows subcellular imaging of large areas of intact tissue samples at 5.4 cm2/min. (C) Based on a standard 4× objective (obj), the DeepDOF microscope combines wavefront encoding with deep-learning-enabled image reconstruction to significantly improve the DOF and, thus, the volumetric FOV while maintaining subcellular resolution. As a result, the DeepDOF microscope offers fast scanning of the cross-sectional surface of tissue slices without need for refocusing.
In view of the challenges associated with standard histopathology, the ability to image cross-sectional surfaces of thick tissue slices (Fig. 1A) directly and nondestructively is highly desired. Recent studies have demonstrated successful imaging of large areas of intact specimens using fluorescence microscopy, including approaches based on confocal scanning (3, 4), structured illumination (5), and ultraviolet (UV) excitation (6). Clinical application of these techniques, however, is largely hindered by the shallow depth-of-field (DOF). In conventional microscopy, DOF is fundamentally coupled to lateral resolution:
| [1] |
As shown in Fig. 1C, in conventional microscopy with standard objectives, achieving subcellular lateral resolution (∼2 to 3 µm) restricts the DOF to ∼30 µm. This is almost one order of magnitude smaller than that needed to accommodate the variations in surface topography of freshly resected tissue surfaces, which can extend to up to 200 µm (7). As an example, Fig. 1 B, Upper Right shows a fluorescence image of an ex vivo porcine esophageal sample, stained with proflavine to highlight epithelial cell nuclei. In the image acquired with a conventional fluorescence microscope, the resulting defocus blur compromises the ability to visualize detailed cellular structures across a large field of view (FOV) without serial refocusing.
To overcome the intrinsic optical constraints of conventional fluorescence microscopy for extended DOF, different approaches have been employed, such as decoupled illumination and detection in light-sheet microscopy (7), dynamic remote focusing (8, 9), and spatial and spectral multiplexing (10, 11); nonetheless, they usually require customized and expensive optics or complicated geometrical configurations. Alternatively, reflectance-based label-free modalities, including reflectance confocal microscopy and full-field optical coherence tomography, have been demonstrated for cancer-lesion characterization in skin and different types of epithelium (12–15). While initial results are promising, these systems are significantly more expensive (more than $100,000) than conventional microscopes due to their optomechanical complexity (16, 17). Computationally, extended DOF has also been demonstrated by using Fourier ptychographic microscopy (18), but the image reconstruction assumes a thin sample target transilluminated with oblique plane waves and is not suited for clinical fluorescence imaging.
Wavefront encoding, when combined with computational methods, offers a convenient and inexpensive route to enhance imaging performance (19, 20). Wavefront modulating elements, such as cubic phase masks, annular phase masks, and other adaptive optics components, have been employed in photography, microscopy, and optical coherent microscopy to extend the DOF and to correct other forms of aberrations (21–32). Despite their adoption in different modalities, phase masks usually cause image degradation, thus necessitating a separately designed reconstruction algorithm to retrieve original features. Recently, deep learning is emerging as a powerful tool to complement microscopy for analysis of complex microscopic data (33–36). In this work, we integrate a wave-optics model with deep learning to develop a physics-informed, end-to-end optimization framework for extended DOF. In contrast to conventional approaches, the deep-learning framework optimizes the phase-mask design with large realistic data, while codesigning the reconstruction algorithm. Using this data-driven approach, we design, optimize, and experimentally validate the deep-learning extended DOF microscope (DeepDOF microscope), a low-cost (less than $6,000) computational microscope for fast and slide-free histology of surgical specimens. The DeepDOF microscope consists of two key co-optimized components: the phase mask and the image-reconstruction algorithm (Fig. 1 B, Upper Left). As shown in Fig. 1 B, Lower Left, and C, by jointly optimizing the phase mask and reconstruction algorithm, the DOF of the DeepDOF microscope is significantly extended to 200 µm, accommodating for variations in surface topology of thick cross-sectional tissue slices. Thanks to its capability to map irregular surfaces in a high-volumetric FOV (6.9 mm3 in DeepDOF microscope vs. 1.2 mm3 in a conventional microscope) with subcellular resolution, the DeepDOF microscope can image large areas of bread-loafed tissue slices without refocusing. Importantly, this is achieved by using an inexpensive phase-modulating element (less than $10 at production volume of 500 masks) that does not sacrifice optical throughput, making the DeepDOF microscope design readily adaptable to image fluorophores with low brightness.
Here, we describe key components of the end-to-end optimized DeepDOF microscope from initial numerical simulation, to optical design, to subsequent experimental validation. We first present simulated results to jointly design and optimize the DeepDOF microscope optics and algorithm using a deep neural network. We then report characterization of the optimized DeepDOF phase mask, with simulated and experimental data. Furthermore, imaging of resected surgical specimens from the oral cavity is provided to validate clinical performance. We show that, using the current economical sample stage, DeepDOF can scan large specimens at 5.4 cm2/min, offering a fast, easy-to-use, and inexpensive alternative to standard histopathology for assessment of intact biopsies and surgical specimens with cellular detail.
Results
DeepDOF Microscope Design and End-to-End Network Architecture.
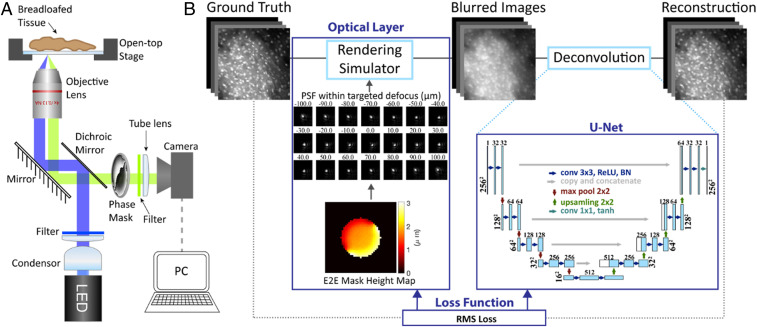
Fig. 2 shows an overview of the DeepDOF microscope design and the deep-learning network for end-to-end optimization; the detailed implementation is described in Materials and Methods. The DeepDOF microscope (Fig. 2A) is built from an epifluorescence microscope with a standard objective (Olympus 4×; numerical aperture [NA] 0.13). A wavefront-encoding phase mask, optimized with image data in the training stage, is placed in the pupil plane to generate a defocus invariant point-spread function (PSF), and a neural-network-based reconstruction algorithm is implemented to perform image deconvolution. To enable automated specimen scanning in the lateral direction, an open-top sample stage is incorporated.
Fig. 2.
DeepDOF microscope setup and the end-to-end network for joint optimization of the phase mask and reconstruction algorithm. (A) The DeepDOF microscope is an epifluorescence microscope with a phase mask in its pupil plane, a co-optimized image-reconstruction algorithm, and an open-top sample stage for fast x–y tissue translation. (B) The end-to-end (E2E) network consists of an optical layer that simulates physics-derived image formation of a fluorescence microscope with a learned phase mask and a sequential digital layer that employs a U-Net to reconstruct in-focus images within the targeted DOF of 200 µm.
The DeepDOF microscope design, including the phase mask and reconstruction algorithm, is modeled and optimized by using the end-to-end network (Fig. 2B). Briefly, the optical layer simulates physically accurate image formation of a fluorescence microscope with the addition of a learned phase mask. In a subsequent digital layer, we employ the U-Net, a widely used neural network (37), to reconstruct in-focus images.
End-to-End Network Is Trained to Resolve Varied Features within a 200-µm DOF.
To leverage the power of deep learning, we trained the neural network to resolve a broad range of imaging features from proflavine-stained resected oral surgical specimens, histopathology images of human tissue of varied types, and natural images with diverse features. To explore the extent to which each component of the deep-learning network affects the imaging performance, we adapted the network to model several microscopy configurations listed in Table 1. Specifically, we trained the network with and without a phase mask. In the masked configurations, we experimented with varied mask learnability, initialization conditions, and loss functions. Using an independent set of test images, the imaging performance of each simulation was evaluated by using the average and SD of image rms error (RMSE) in a layered model of 21 linearly discretized defocus levels within the 200-µm DOF (Table 1 and SI Appendix, Fig. S1).
Table 1.
Testing results in ablation studies
| Network configuration | Learned mask | Initialization | Loss function | Average RMSE (E-02) | RMSE SD (E-02) |
| Conventional microscopy | |||||
| Simulation A | No | NA | NA | 5.59 | 4.37 |
| Conventional microscopy + U-Net reconstruction | |||||
| Simulation B | No | Open aperture | RMS | 3.60 | 2.91 |
| Fixed-phase mask + U-Net reconstruction | |||||
| Simulation C | No | Cubic mask | RMS | 3.79 | 2.29 |
| Learned-phase mask + U-Net reconstruction | |||||
| Simulation D | Yes | Random | RMS | 3.07 | 2.30 |
| Simulation E* | Yes | Cubic mask | RMS | 2.86 | 2.20 |
| Simulation F | Yes | Cubic mask | SSIM | 3.03 | 2.36 |
NA, not applicable.
Used as the optimized DeepDOF microscope design.
First, compared with the baseline conventional microscope (simulation A; RMSE 0.0559), the addition of U-Net reconstruction (simulation B; RMSE 0.0360) improved the reconstruction accuracy; nonetheless, significant variance resulting from defocus was still present (Table 1 and SI Appendix, Fig. S1). As anticipated, the defocus variance was markedly reduced by using the cubic mask as an established method to extend the DOF (simulation C; RMSE 0.0379). However, the average RMSE was also higher using the mathematically derived cubic mask than with U-Net reconstruction, suggesting that its performance can be further improved.
We then employed an end-to-end architecture to jointly optimize the learned phase mask and U-Net reconstruction in simulations D–F. The cubic mask was outperformed by a randomly initialized and end-to-end optimized mask (simulation D; RMSE 0.0307). In fact, the learned, randomly initialized mask achieved lower RMSE at all 21 discrete defocus locations, especially between the −80- and 0-μm defocus range (SI Appendix, Fig. S1). This confirmed that performance of the cubic mask could be improved through a data-driven approach. When initialized with a cubic mask and end-to-end optimized (simulation E; RMSE 0.0286), both the average RMSE and defocus variance were further reduced. Notably, simulation E showed uniform and improved performance across the entire defocus range, while the randomized initialization fell short in the +20- to +80-μm range (SI Appendix, Fig. S1). This is ascribed to the robustness of the cubic mask against defocus. We also tested Structural Similarity Index Measure (SSIM) as a commonly used metric to train the network in simulation F (38, 58), which did not show further enhancement. Based on those results, we chose the optimized DeepDOF design in simulation E as the network operating point; the resulting DeepDOF mask design (SI Appendix, Fig. S2) was subsequently used for experimental fabrication and tissue imaging.
Simulated Performance at the Network Operating Point.
We simulated the performance of the optimized DeepDOF microscope design described above (Fig. 3). In 2,000 image patches from an independent test set, we compared the performance of the DeepDOF microscope (DeepDOF mask + U-Net) with two baseline microscope configurations and a computational microscope based on a cubic mask. The baseline configurations included a conventional microscope using the same Olympus 4×, 0.13-NA objective (baseline I) and a conventional microscope with a reduced aperture, a classical approach to extend DOF (baseline II, Olympus 4× objective + iris, 0.06 NA). In baseline II, we simulated an iris to reduce the pupil size by half and increased the added Gaussian noise accordingly to account for the lower light throughput. The computational microscope was based on a cubic phase mask and corresponding reconstruction U-Net (cubic mask + U-Net).
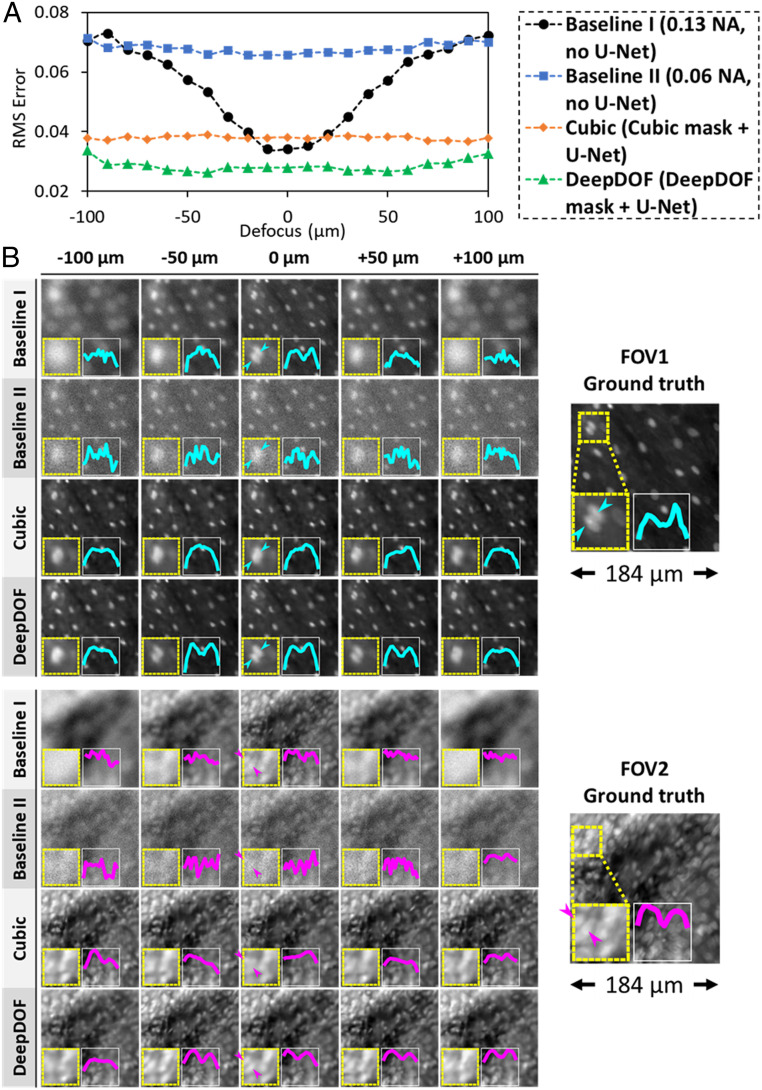
Fig. 3.
Simulated performance of the end-to-end optimized DeepDOF microscope in a test set vs. two conventional baselines and a cubic-mask-enabled computational microscope in a layered model. (A) The DeepDOF microscope design shows superior performance across the entire defocus range. (B) Example reconstructed images from the test set showing sparsely (Upper) and densely (Lower) packed nuclei. In both FOVs, the DeepDOF microscope better resolves adjacent nuclei.
Fig. 3A compares the RMSE vs. defocus for the optimized DeepDOF microscope, the cubic-mask-enabled microscope, and the two baseline configurations. As expected, the DOF was smallest for the standard microscope (baseline I). Despite the increased DOF for baseline II, the associated reduction of light throughput and lateral resolution resulted in an increased average RMSE. In contrast, computational microscopes with either a cubic or DeepDOF mask significantly improved the imaging performance without compromising the light throughput. Importantly, the DeepDOF design achieved superior performance compared to the cubic mask across the entire defocus range. When comparing the modulation-transfer function (SI Appendix, Fig. S3A), the DeepDOF also performed better than the three configurations described in Fig. 3.
Since RMSE is a global measure of imaging performance, we also examined example simulated images at five selected defocus locations. Fig. 3B shows two test images of proflavine-stained oral surgical tissue with sparse and densely packed nuclei, respectively. Insets display a magnified view of two adjacent nuclei and the corresponding intensity profiles. When using a standard microscope (baseline I), the nuclei were only resolved when the sample was at the focal plane. Images obtained with the reduced NA (baseline II) showed degraded resolution and noticeable noise. In contrast, images acquired with the DeepDOF microscope better resolved individual nuclei than the cubic-mask-enabled microscope and both baseline configurations, and the trend was consistent across the targeted 200-µm DOF. Using a test set of 2,000 images, we also compared the DeepDOF reconstruction with conventional deconvolution algorithms, including the Wiener and Richardson–Lucy methods, showing that DeepDOF performed better in the DOF range (SI Appendix, Fig. S3B).
DeepDOF Microscope with Fabricated Mask Achieves Subcellular Resolution within Targeted DOF.
As shown in Fig. 4A, we fabricated the optimized DeepDOF physical mask design (simulation E in Table 1) with photolithography and Reactive Ion Etching (RIE) (39). With the mask inserted in the pupil plane of the DeepDOF microscope, the resulting PSF was measured throughout the DOF (Fig. 4B), and the corresponding U-Net was retrained accordingly.
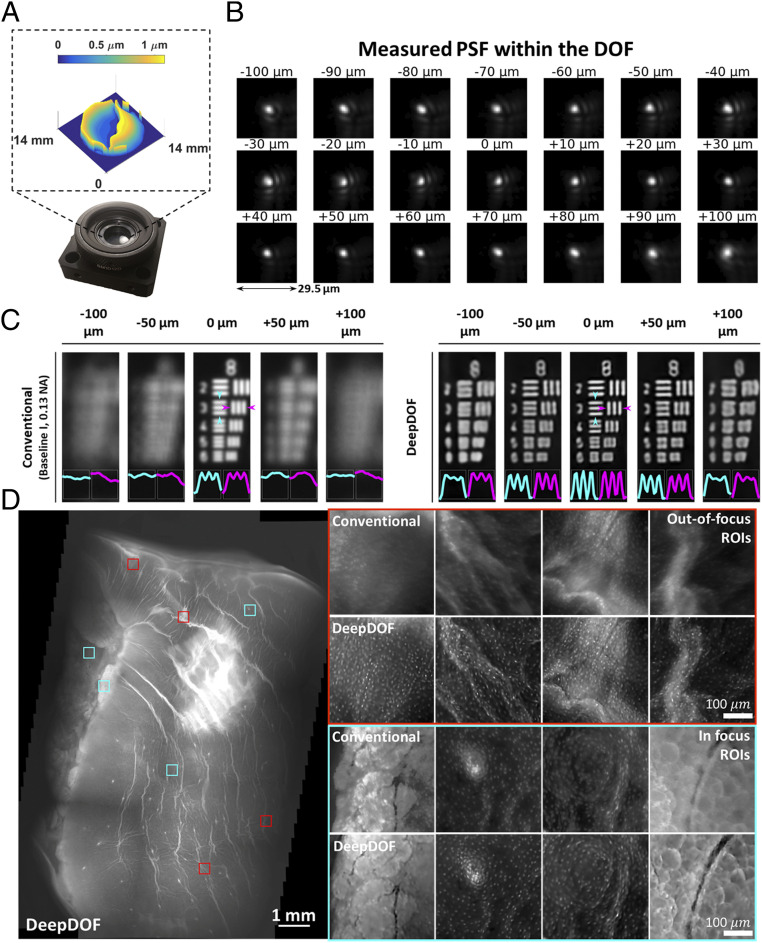
Fig. 4.
Experimental fabrication of the DeepDOF mask and characterization of its imaging performance by imaging a USAF resolution target and an ex vivo porcine esophageal sample. (A) The DeepDOF mask is fabricated based on the end-to-end optimized DeepDOF design and mounted on a plate holder. (B) PSF stacks of the DeepDOF microscope were measured within the 200-µm DOF. (C) Using the fabricated mask and retrained U-Net, the DeepDOF microscope can resolve element 3 of group 8 on the USAF resolution target that corresponds to 323 line pairs per millimeter linewidth. (D) The DeepDOF image clearly visualizes subcellular features, primarily individual nuclei that are evenly distributed on the esophageal wall. Multiple ROIs are shown in regions where the conventional microscope images are out of focus (red) and in focus (cyan).
To further characterize the lateral resolution and DOF, we imaged a US Air Force (USAF) resolution target, comparing performance of the DeepDOF microscope with the standard conventional microscope (baseline I described in Fig. 3; experimental comparison with other configurations is provided in SI Appendix, Fig. S4). For each image, we quantified the image contrast by plotting the intensity across element 3 of group 8 on the USAF resolution target (cyan and magenta in Fig. 4C), which corresponds to 323 line pairs per millimeter linewidth. To facilitate visualization, in each configuration, the image at 0 μm was first normalized to itself; other images in the same configuration were then linearly and equally boosted to ensure that their relative intensities remained unchanged. As expected, the standard conventional microscope (baseline I) could resolve the targeted element at the focal plane; nonetheless, the resolving power degraded quickly as the target moved out of focus. In contrast, using the Sparrow criterion, the DeepDOF was able to resolve the features with significantly improved contrast within the 200-µm DOF. To evaluate the DeepDOF performance for imaging various histology features, we also imaged thin (7 to 10 µm) frozen sections from different types of mouse tissue when they were positioned at varied defocuses across the DOF; in SI Appendix, Fig. S5, our results show that the DeepDOF microscope can consistently resolve varied cellular morphology within the targeted DOF.
We further validated the DeepDOF performance by imaging a large area of squamous epithelium from a resected porcine esophageal specimen in Fig. 4D. Although the ex vivo esophageal sample was not cut cross-sectionally in a manner shown in Fig. 1A, and the imaged superficial epithelium was anticipated to be more uniform than surfaces of resected tumors, significant defocus blur was observed in selected regions of interest (ROIs) (red) from the conventional microscope. In comparison, the DeepDOF microscope clearly revealed subcellular morphology of nuclei and adipose cells in those regions.
DeepDOF Microscope Can Map Intact Oral Surgical Sample without Refocusing.
The ability of the DeepDOF microscope to image tissue with irregular surfaces was evaluated in freshly resected oral specimens from patients undergoing surgery for oral cancer. Specimens were stained topically with proflavine to highlight nuclear morphology. To provide a baseline comparison, each specimen was also imaged by using a maskless conventional microscope with the same objective (Olympus 4×, 0.13 NA).
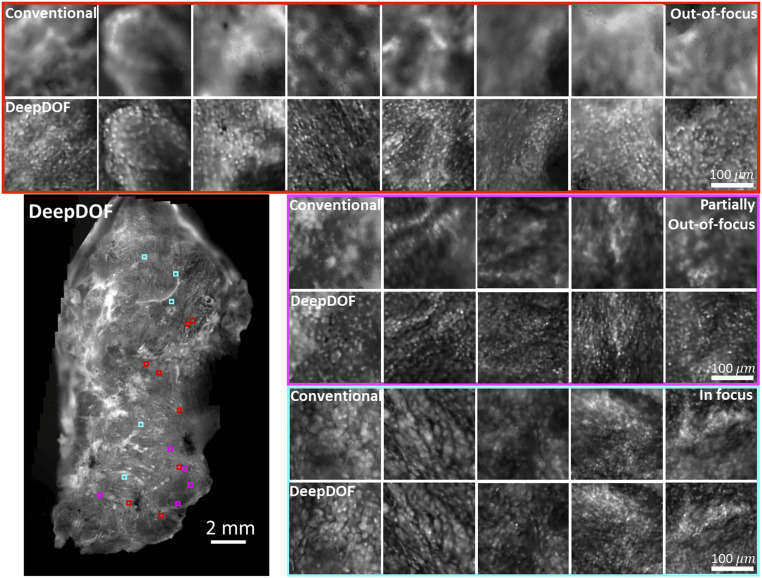
Fig. 5 shows the high-resolution DeepDOF images of a freshly resected oral surgical specimen (∼12 mm × 22 mm in Lower Left) acquired with the DeepDOF microscope and the corresponding conventional microscope. To compare DeepDOF with the conventional microscope, we selected three groups of ROIs across the sample that were significantly out of focus (red), partially out of focus (magenta), and in focus (cyan) in the conventional image. In ROIs highlighted in red, surface irregularity exceeded the DOF of the conventional microscope, and tissue architecture could not be distinguished. In the magenta group, individual nuclei remained difficult to resolve under a conventional microscope. In contrast, DeepDOF was able to resolve the nuclear morphology with subcellular resolution in ROIs in both groups. In the last group (cyan), DeepDOF revealed cellular features in agreement with conventional microscopic images at the focus plane.
Fig. 5.
Images acquired with the DeepDOF microscope and a conventional microscope (Olympus 4×, 0.13 NA) from a large surgically resected specimen from the oral cavity. Three groups of ROIs show regions where the conventional microscope images are significantly out of focus (red), partially out of focus (magenta), and in focus (cyan). The contrast of the conventional images is matched to show the effects of defocus blur. Compared to the conventional microscope, the DeepDOF microscope enables direct imaging of a large area of intact tissue without the need for refocusing.
DeepDOF Microscope Visualizes Important Diagnostic Features of Surgically Resected Oral Tumors with Subcellular Resolution.
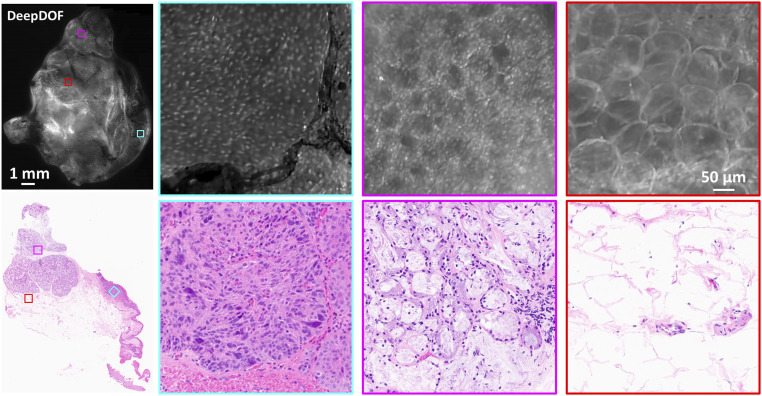
We further assessed the diagnostic potential of the DeepDOF microscope by comparing DeepDOF images acquired from a freshly resected oral tumor to that of gold-standard histopathologic images (Fig. 6). The image of the entire surgical specimen (∼11 mm × 13 mm) acquired with the DeepDOF microscope is shown with the corresponding histopathology section, with three ROIs selected to highlight various histologic features. In the first ROI (cyan), the DeepDOF image revealed architectural disorder and cells exhibiting increased nuclear-to-cytoplasmic area ratio and nuclear pleomorphism, indicating the presence of neoplasia in the squamous epithelial layer. These findings were confirmed by standard histopathology evaluation by the study pathologist (M.D.W.). Interestingly, microvascular features shown in histopathology were also visible in the DeepDOF image, as a result of the strong absorbance of hemoglobin-containing red blood cells. The DeepDOF image of the second ROI (magenta) displayed intact and regularly spaced glandular patterns with discernable individual nuclei in salivary tissue, which is consistent with the histopathology image. In the last ROI (red), both the DeepDOF microscope and histopathology images show adipose cells without prominent nuclear staining. Notably, we observed marked displacement of the epithelium relative to the deep salivary tissue in the histopathology image of the entire specimen, likely resulting from tissue handling and processing. In comparison, by using the DeepDOF microscope, similar diagnostic features with subcellular detail can be directly visualized from an intact tissue sample, circumventing tissue alterations resulting from processing.
Fig. 6.
The DeepDOF microscope visualizes important diagnostic features in large proflavine-stained surgical specimen with subcellular resolution. Three selected ROIs across the DeepDOF image of the specimen reveal a nest of neoplastic cells in the squamous epithelium (cyan), glandular patterns in salivary tissue (magenta), and adipose cells (red), respectively. Image features were confirmed by the gold standard of histopathology.
Discussion
In this work, we develop and validate the DeepDOF microscope for convenient and rapid imaging of intact surgical specimens with an over 5× larger DOF. With the fabricated DeepDOF mask (less than $10), we demonstrate that the DeepDOF microscope can visualize nuclear morphology across highly irregular tissue surfaces without refocusing, a challenging task for conventional microscopes. Furthermore, we show that DeepDOF images reveal cellular features of diagnostic importance, including microvasculature, neoplastic growth, benign salivary tissue, and adipose cells; these findings are consistent with standard histopathology. In future studies, the versatile neural network can also be adapted to provide clinical decision support, including automated tissue annotation and margin segmentation (40–43).
Here, we demonstrated a computational microscope with jointly optimized imaging optics and image-reconstruction algorithm for extended DOF. We show the feasibility and benefits of a data-driven, deep-learning architecture to design and optimize the DeepDOF microscope for fluorescence imaging of irregular tissue surfaces. As shown in Fig. 3 and SI Appendix, Fig. S3, the end-to-end optimized DeepDOF microscope achieves enhanced performance compared to conventional mask design and deconvolution algorithms that are developed separately. Moreover, the DeepDOF microscope is built on a conventional epifluorescence microscope, and its low cost (less than $6,000) allows potential adoption in community and low-resource settings. The robust image reconstruction only takes an average of 0.5 s/mm2 using the TensorFlow package, and its speed can be further optimized. In addition, since wavefront encoding is realized with a simple phase-modulating element, the DeepDOF microscope maintains high light throughput, a key consideration in imaging fluorophores without introducing photobleaching.
The current DeepDOF microscope can be further improved in several ways. First, the topical proflavine dye used in this study preferentially stains nuclei, and we expect that the combined use of nuclear stains and counterstains can contribute to added diagnostic value. In this regard, the DeepDOF architecture can be optimized to image dye combinations for virtual H&E staining (44–46), as well as various fluorescent molecular probes for tumor detection with high specificity (47–50). Second, DeepDOF images were acquired without optical sectioning. While non-optical-sectioned images of proflavine-stained nuclear morphology have been found to show great diagnostic value in a wide range of clinical applications (51–54), image quality can be further improved with confocal scanning or structured illumination (55, 56). This can be particularly important when exploring the use of other nonspecific staining agents, endogenous fluorophores, or label-free modalities. Since wavefront encoding is only performed in the emission optical path of the DeepDOF microscope, we can combine conventional background rejection methods in the context of extended DOF microscopy. In addition, we can also explore the usefulness of UV excitation that offers inherent surface sectioning (6). Finally, future studies are needed to validate the performance in more diverse tissue and label types. Our preliminary results show that different tissue types from frozen mouse sections, as well as ex vivo porcine esophageal tissue, were clearly resolved in the DeepDOF images in Fig. 4D and SI Appendix, Fig. S5. This can be ascribed to the fact that the physics-informed model was trained by using a wide range of complex features from human pathology slides and natural scenes. Moreover, diagnostically important features in oral specimens, including microvasculature, neoplastic squamous epithelium, and salivary and adipose tissue, were visualized with histopathology confirmation in Fig. 6. These results warrant future research to assess the DeepDOF diagnostic value in a blinded study.
In conclusion, we employed an end-to-end deep-learning architecture to significantly enhance conventional microscopy beyond its physical limitations, and we demonstrate the DeepDOF microscope as a fast, easy-to-use, and inexpensive alternative to standard histopathology for assessment of intact tissue specimens with subcellular detail. The capability to provide histologic quality images at a high scanning speed, combined with low cost, warrants further clinical evaluation, especially for improving tissue sampling during intraoperative tumor-margin assessment and in rural settings without access to standard histopathology.
Materials and Methods
DeepDOF Microscope Setup.
As shown in Fig. 2A, the DeepDOF microscope is composed of a conventional fluorescence microscope with simple addition of a learned phase mask. Briefly, Köhler illumination was used to provide uniform fluorescence excitation using a 455-nm blue light-emitting diode (catalog nos. M455L3 and LEDD1B, Thorlabs) and an excitation filter (catalog no. FF01-452/45-25, Semrock). Fluorescence from stained specimens was collected by an Olympus 4× objective (catalog no. RMS4X-PF, 0.13 NA), reflected by a right-angle mirror and imaged by a 20-megapixel complementary metal oxide semiconductor (CMOS) camera (catalog no. BFS-U3-200S6M-C, FLIR Systems) behind a green bandpass filter (catalog no. FF01-550/88-25, Semrock). In this study, we used a 150-mm-focal-length tube lens that provided adequate sampling of the PSF with a pixel size of 0.72 μm on the object plane. During initial DeepDOF microscope testing, we also experimented with a 100-mm-focal-length tube lens.
To allow for convenient placement and mapping of large surgical specimens, we designed an open-top sample stage with a clear imaging window (2” × 3”) during ex vivo imaging. In our initial testing, we also tested mechanically pressing the sample to suppress surface irregularity, but its effectiveness was limited, potentially due to the highly heterogeneous tissue stiffness. Rapid scanning was enabled by two linear motors (catalog no. X-LHM100A, Zaber) in the x and y directions. Using the 20-megapixel CMOS camera, each frame covers a lateral area of 3.9 × 2.6 mm2, while providing sufficient sampling of the PSF. To ensure sufficient overlap for a commercial stitching software (Microsoft Composite Editor), we chose a 2-mm step for motors on the sample stage. With this motor step size, we were able to acquire 135 frames per min. The extended DOF eliminates the need for axial refocusing, allowing intact surgical specimens to be scanned at a speed of 5.4 cm2/min.
DeepDOF Microscope Model: Optical Layer.
The optical layer of the end-to-end network simulates optical imaging of a standard fluorescence microscope from a specimen to the sensor, with an addition of a passive phase-modulating element in the pupil plane. During the simulation in this study, we modeled fluorescence imaging in the green spectrum as incoherent imaging with a discretized wavelength at 550 nm.
Based on Fourier optics, the PSF in the image plane PSF(x2, y2) can be modeled as the squared magnitude of the Fourier transform of the pupil function:
| [2] |
in which F{⋅}denotes the Fourier transform, and the pupil function P(x1, y1) describes the relative amplitude and phase changes of the wavefront at the pupil plane:
| [3] |
When imaging an out-of-focus object with an aberration-free system, the pupil function remains constant across the pupil plane, and the phase term due to defocus is modeled as
| [4] |
in which k is the wavenumber, and z and z0 are the defocused imaging depth and in-focus depth, respectively. The equation above can be further simplified as
| [5] |
with denoting the relative displacement in the pupil plane and being the maximum path-length error at the edge of the pupil due to defocus.
To increase the defocus-invariance of PSF for extended DOF, we modulated the pupil function with a learning phase mask that introduced an additive and depth-independent phase term ΦM(x1, y1). The resulting phase term of the pupil function is
| [6] |
The image formation was thus calculated based on the convolution of a scene Io(x, y; z) and PSF, while signals from varied defocuses are added incoherently to form the final image:
| [7] |
To approximate noise during image capture using a sensor, Gaussian read noise with a SD σ = 0.01 was applied to the normalized blurred image in the range of [0, 1].
While irregular specimen surfaces can be rendered by using Eq. 7, we note that the process requires prior depth information and is computationally expensive. In the context of optimizing the mask design for PSF depth invariance, a layered model can be employed to discretize the DOF, with each layer blurred by the PSF at its corresponding depth. To accommodate for variations in specimen surface topology up to 200 µm, we simulated image formation within a defocus range of −100 to +100 µm, which corresponds to a kWm range of [−9.81, 9.81] in the DeepDOF microscope. As a result, we discretized kWm to [−10:1:10] at 21 imaging depths, and the optical layer output 21 convoluted images at varied focus planes.
Discrete Fourier transform was performed to model the correspondence between the pupil function and the PSF based on Eqs. 2–6 using matrices of 71 × 71 pixels. Specifically, the PSF was discretized based on the sensor pixel size. The pupil function contained a learning phase term modulated by the height map of the phase mask. The heightmap was represented with the first 55 Zernike basis (Zn):
| [8] |
which offers adequate degrees of freedom while ensuring computational efficiency during training and surface smoothness for fabrication. As the first optical layer of the end-to-end network, these coefficients were optimized during training.
DeepDOF Microscope Model: Digital Layer.
The digital layer consisted of a deep neural network trained to reconstruct in-focus images from convoluted outputs of the optical layer in Fig. 2B. In the absence of prior defocus information, this presented a blind deblurring problem best solved using deep neural networks. While there are many network architectures available, we chose the well-known U-Net, as it is widely used in biomedical imaging (37). Briefly, our U-Net adaptation consisted of an encoder and a decoder, with 23 convolution layers and 32 to 512 feature channels (Fig. 2B). During each step of the encoder stage, the input underwent two 3 × 3 convolution layers, each followed by a rectified linear unit (ReLU) and batch normalization (BN). Then, a max pool layer downsampled the input by two. Similarly, during each step of the decoder stage, a 2 × 2 up-convolution upsampled the feature map. It was followed by two 3 × 3 convolutions layers with ReLU and BN. The final layer consisted of a 1 × 1 convolution with tanh activation that mapped the output to the [−1, 1] range. This output was added to the network input to generate the final deblurred reconstruction. For memory efficiency, the U-Net took in blurry images of 256 × 256 pixels and output the corresponding in-focus images at each training iteration. Once the network is trained, it can process images with widths and heights that are multiples of 16.
Network Training: Dataset and Data Augmentation.
To leverage the power of deep learning, a large set of training images presenting a broad range of imaging features is desired. To provide ground truth for training, we acquired microscopic fluorescence images of proflavine-stained oral cancer resections using a 10×/0.25-NA objective (RMS10X) and manual refocusing. The high-frequency features in the ground-truth images will allow physically accurate simulation of image degradation through a 4×/0.13-NA system (with or without a phase mask), primarily due to PSF convolution, defocus blur, and added noise. To further train the end-to-end network for imaging more complex and diverse biological features, we incorporated 600 histopathology images of healthy and cancerous tissue of human brain, lungs, mouth, colon, cervix, and breast from The Cancer Genome Atlas (TCGA) Cancer FFPE slides. As an additional measure to ensure the network generalizability, we also included natural images from the National Institute for Research in Digital Science and Technology (INRIA) Holiday dataset to diversify the scales of features. In total, we assembled a collection of 1,800 images (each 1,000 × 1,000 pixels; gray scale), with 600 images of each type.
The 1,800 images were randomly assigned to training, validation, and testing sets that contained 1,500; 150; and 150 images, respectively (see Data Availability). During training, the images were cropped at random locations and augmented with rotation, flipping, and brightness adjustment. To preclude boundary artifacts when convolving a 256 × 256 image with a 71 × 71 PSF for blurring, a larger 326 × 326 image was cropped initially and then cropped to 256 × 256 after the convolution.
Network Training: Loss Function.
The loss function of the network was the pixel-by-pixel RMSE between the reconstructed-image stack and the ground-truth image set:
| [9] |
where N is the number of pixels, and I* and Î are the ground-truth image and the reconstructed image, respectively. Since the input images were blurred at different defocuses, the network intrinsically updated the mask for defocus-insensitive PSF and required no explicit cost functions to enforce PSF similarity.
In addition to RMSE, we also explored the usefulness of the SSIM, which calculated similarities between windowed regions of two images while disregarding brightness or dynamic-range differences. We chose to minimize the negative SSIM and defined the loss function as follows:
| [10] |
where μ, σ are the mean and cross-correlation, respectively, and c constants to avoid division by 0.
Network Training: Implementation Details.
The network was implemented with the TensorFlow package. We used stochastic gradient descent with the Adam optimizer (57). The learning rate for Adam was chosen empirically at 1e-9 for the optical layer and 1e-4 for the digital layer. To fine tune the U-Net, a learning-rate decay of 0.5 was applied at every 10,000 iterations for the digital layer.
To explore the design space, we carried out a series of ablation studies by varying the mask learnability, initialization conditions, and loss functions. The performance of different models was quantified by calculating the RMSE in 2,000-image patches randomly sampled from the testing set.
In simulation E, we also observed that a two-step optimization improved performance. In this two-step training, we first initialized the network with the cubic mask and then trained the U-Net while fixing the optical layer. Once the network converged, we jointly trained the optical and digital layers to obtain the best performance.
Mask Fabrication.
Once the end-to-end network was trained, we fabricated the optimized phase mask for experimental evaluation. For comparison, we fabricated both the end-to-end trained DeepDOF mask and a cubic mask using photolithography in conjunction with RIE. During simulation, as a trade-off between resolution and computational efficiency, we modeled the phase mask with a pixel size of 485 μm. Once the mask design was optimized during simulation, photolithography allowed us to increase the lateral fabrication resolution to 70 μm/pixel for a smoother profile. Axially, the height maps were phase-wrapped and discretized into 15 steps, with each step being 73 nm thick.
During fabrication, these 15 steps were further multiplexed into four binary patterns. Each of the four patterns was first transferred onto a silicon oxide glass wafer through photolithography (EVG 620 mask aligner) using positive resist. The layer was then etched into the wafer by using RIE (Oxford 100). The remaining resist was stripped away with acetone before the next layer was transferred.
PSF Calibration and Network Retraining.
To account for alignment errors and mask aberration caused by fabrication, we experimentally measured the PSF of the system by imaging fluorescent beads. The fabricated DeepDOF mask was aligned to a 12-mm iris (catalog no. SM1D12D, Thorlabs) that matched the back pupil of the objective lens. The iris was then installed onto the aperture plane of the microscope (Fig. 1B). A monolayer of 1-μm isolated fluorescent beads (catalog nos. 505/515 and F8768, Thermo Fisher Scientific; diluted to 105 per mL) were imaged at 31 depths at 10-μm intervals. At each depth, we averaged temporally over five frames and performed background subtraction to reduce noise.
We selected a target DOF range of 200 µm for network retraining and subsequent imaging based on the invariance of 21 consecutive sections of the PSF. During the retraining phase, we fixed the phase-mask layer and fine-tuned the reconstruction network with measured PSF. The same process was repeated for the cubic mask.
Microscope Calibration.
Once the optimized phase mask was inserted and secured in the pupil plane, the DeepDOF microscope was calibrated by adjusting the right-angle mirror behind the objective to align the sensor with the image plane when imaging a monolayer of 1-µm fluorescent beads (catalog nos. 505/515 and F8768, Thermo Fisher Scientific). Furthermore, we installed a micrometer tilt stage (catalog no. 66-551, Edmund Optics) for sample-stage control. By adjusting the tilt stage when imaging fluorescent beads across a large FOV, uniform focus during sample translation was ensured. Once the DeepDOF microscope was aligned, its imaging performance was assessed by imaging a USAF resolution target (catalog no. 55-622, Edmund Optics) within a 200-µm DOF.
Ex Vivo Imaging of Porcine Esophageal Tissue.
The ex vivo porcine esophageal sample was obtained from an abattoir, freshly resected, and stained with proflavine prior to imaging with the DeepDOF scope. Imaging was then repeated with a conventional microscope using the same standard objective (Olympus 4×, 0.13 NA).
Surgical Sample Imaging and Processing.
We evaluated and validated the imaging performance of the DeepDOF microscope by imaging surgically resected oral cancer specimens. In the ex vivo protocol, consenting patients undergoing surgery for oral cancer resection were enrolled. After surgical resection, the excised specimen was assessed by an expert pathologist and sliced into 3- to 4-mm slices, and selected slices were processed for standard frozen-section pathology. The remaining slices were then stained with proflavine (0.01% weight/volume in phosphate-buffered saline) and imaged by using the DeepDOF microscope. Raw frames from the DeepDOF microscope were processed with U-Net deconvolution and stitched by using Image Composite Editor (Microsoft). After imaging, the specimens were processed by FFPE for permanent histopathology, and histopathology images were acquired with a slide scanner. The study was approved by the Institutional Review Boards at the University of Texas MD Anderson Cancer Center and Rice University.
Supplementary Material
Acknowledgments
This work was supported by NSF Grants 1730574 and 1648451; NSF CAREER Award 1652633; Defense Advanced Research Projects Agency Neural Engineering System Design Grant N66001-17-C-4012; and NIH Award 1RF1NS110501. H&E processing was performed by the MD Anderson Research Histology Core Laboratory, which is supported by National Cancer Institute Grant CA16672. The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013571117/-/DCSupplemental.
Data Availability.
DeepDOF dataset data have been deposited in Zenodo (DOI: 10.5281/zenodo.3922596).
References
- 1.Rastogi V., et al. , Artefacts: A diagnostic dilemma—A review. J. Clin. Diagn. Res. 7, 2408–2413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taxy J. B., Frozen section and the surgical pathologist: A point of view. Arch. Pathol. Lab. Med. 133, 1135–1138 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Gareau D. S., et al. , Confocal mosaicing microscopy in Mohs skin excisions: Feasibility of rapid surgical pathology. J. Biomed. Opt. 13, 054001 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dobbs J., et al. , Confocal fluorescence microscopy for rapid evaluation of invasive tumor cellularity of inflammatory breast carcinoma core needle biopsies. Breast Cancer Res. Treat. 149, 303–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M., et al. , Gigapixel surface imaging of radical prostatectomy specimens for comprehensive detection of cancer-positive surgical margins using structured illumination microscopy. Sci. Rep. 6, 27419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fereidouni F., et al. , Microscopy with ultraviolet surface excitation for rapid slide-free histology. Nat. Biomed. Eng. 1, 957–966 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glaser A. K., et al. , Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat. Biomed. Eng. 1, 0084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shain W. J., Vickers N. A., Goldberg B. B., Bifano T., Mertz J., Extended depth-of-field microscopy with a high-speed deformable mirror. Opt. Lett. 42, 995–998 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Xiao S., Tseng H. A., Gritton H., Han X., Mertz J., Video-rate volumetric neuronal imaging using 3D targeted illumination. Sci. Rep. 8, 7921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrahamsson S., et al. , Fast multicolor 3D imaging using aberration-corrected multifocus microscopy. Nat. Methods 10, 60–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geissbuehler S., et al. , Live-cell multiplane three-dimensional super-resolution optical fluctuation imaging. Nat. Commun. 5, 5830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kose K., et al. , Video-mosaicing of reflectance confocal images for examination of extended areas of skin in vivo. Br. J. Dermatol. 171, 1239–1241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Assayag O., et al. , Large field, high resolution full-field optical coherence tomography: A pre-clinical study of human breast tissue and cancer assessment. Technol. Cancer Res. Treat. 13, 455–468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajadhyaksha M., Marghoob A., Rossi A., Halpern A. C., Nehal K. S., Reflectance confocal microscopy of skin in vivo: From bench to bedside. Lasers Surg. Med. 49, 7–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thouvenin O., Grieve K., Xiao P., Apelian C., Boccara A. C., En face coherence microscopy [Invited]. Biomed. Opt. Express 8, 622–639 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards S. J., et al. , VivaScope® 1500 and 3000 systems for detecting and monitoring skin lesions: A systematic review and economic evaluation. Health Technol. Assess. 20, 1–260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tkaczyk E., Innovations and developments in dermatologic non-invasive optical imaging and potential clinical applications. Acta Derm. Venereol. 176 (suppl. 218), 5–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng G., Horstmeyer R., Yang C., Wide-field, high-resolution Fourier ptychographic microscopy. Nat. Photonics 7, 739–745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sitzmann V., et al. , End-to-end optimization of optics and image processing for achromatic extended depth of field and super-resolution imaging. ACM Trans. Graph. 37, 114 (2018). [Google Scholar]
- 20.Wu Y., Boominathan V., Chen H., Sankaranarayanan A., Veeraraghavan A., “PhaseCam3D—Learning phase masks for passive single view depth estimation” in 2019 IEEE International Conference on Computational Photography, ICCP 2019 (IEEE, Piscataway, NJ, 2019), pp. 1–12. [Google Scholar]
- 21.Zahreddine R. N., Cogswell C. J., Total variation regularized deconvolution for extended depth of field microscopy. Appl. Opt. 54, 2244–2254 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Dowski E. R. Jr, Cathey W. T., Extended depth of field through wave-front coding. Appl. Opt. 34, 1859–1866 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Zhao H., Li Y., Optimized logarithmic phase masks used to generate defocus invariant modulation transfer function for wavefront coding system. Opt. Lett. 35, 2630–2632 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Liu M., Dong L., Zhao Y., Hui M., Jia W., Stationary phase analysis of generalized cubic phase mask wavefront coding. Opt. Commun. 298–299, 67–74 (2013). [Google Scholar]
- 25.Le V. N., Chen S., Fan Z., Optimized asymmetrical tangent phase mask to obtain defocus invariant modulation transfer function in incoherent imaging systems. Opt. Lett. 39, 2171–2174 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Pavani S. R. P., et al. , Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. U.S.A. 106, 2995–2999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnison M. R., Cogswell C. J., Sheppard C. J. R., Török P., “Wavefront coding fluorescence microscopy using high aperture lenses” in Optical Imaging and Microscopy: Techniques and Advanced Systems, P. Török, F.-J. Kao, Eds. (Springer Series in Optical Sciences, Springer, Berlin, 2007), pp. 169–191. [Google Scholar]
- 28.Mo J., de Groot M., de Boer J. F., Focus-extension by depth-encoded synthetic aperture in optical coherence tomography. Opt. Express 21, 10048–10061 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Lorenser D., Yang X., Sampson D. D., Ultrathin fiber probes with extended depth of focus for optical coherence tomography. Opt. Lett. 37, 1616–1618 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Coquoz S., Bouwens A., Marchand P. J., Extermann J., Lasser T., Interferometric synthetic aperture microscopy for extended focus optical coherence microscopy. Opt. Express 25, 30807–30819 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Lee W. M., Yun S. H., Adaptive aberration correction of GRIN lenses for confocal endomicroscopy. Opt. Lett. 36, 4608–4610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C., Ji N., Characterization and improvement of three-dimensional imaging performance of GRIN-lens-based two-photon fluorescence endomicroscopes with adaptive optics. Opt. Express 21, 27142–27154 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Barbastathis G., Ozcan A., Situ G., On the use of deep learning for computational imaging. Optica 6, 921 (2019). [Google Scholar]
- 34.Rivenson Y., et al. , PhaseStain: The digital staining of label-free quantitative phase microscopy images using deep learning. Light Sci. Appl. 8, 23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivenson Y., et al. , Deep learning microscopy. Optica 4, 1437 (2017). [Google Scholar]
- 36.Wang H., et al. , Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronneberger O., Fischer P., Brox T., “U-Net: Convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Navab N., Hornegger J., Wells W., Frangi A., Eds. (Lecture Notes in Computer Science, Springer, Cham, Switzerland, 2015), vol. 9351, pp. 234–241. [Google Scholar]
- 38.Wang Z., Bovik A. C., Sheikh H. R., Simoncelli E. P., Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 13, 600–612 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Wang W., et al. , Generalized method to design phase masks for 3D super-resolution microscopy. Opt. Express 27, 3799–3816 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Barker J., Hoogi A., Depeursinge A., Rubin D. L., Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med. Image Anal. 30, 60–71 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Ehteshami Bejnordi B. et al.; the CAMELYON16 Consortium , Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 318, 2199–2210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma H., Zerbe N., Klempert I., Hellwich O., Hufnagl P., Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput. Med. Imaging Graph. 61, 2–13 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Coudray N., et al. , Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elfer K. N., et al. , DRAQ5 and eosin (‘D&E’) as an analog to hematoxylin and eosin for rapid fluorescence histology of fresh tissues. PLoS One 11, e0165530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cahill L. C., et al. , Rapid virtual hematoxylin and eosin histology of breast tissue specimens using a compact fluorescence nonlinear microscope. Lab. Invest. 98, 150–160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giacomelli M. G., et al. , Virtual hematoxylin and eosin transillumination microscopy using epi-fluorescence imaging. PLoS One 11, e0159337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mizushima T., et al. , Fluorescent imaging of superficial head and neck squamous cell carcinoma using a γ-glutamyltranspeptidase-activated targeting agent: A pilot study. BMC Cancer 16, 411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Behrooz A., et al. , Multispectral open-air intraoperative fluorescence imaging. Opt. Lett. 42, 2964–2967 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Slooter M. D., et al. , Detecting tumour-positive resection margins after oral cancer surgery by spraying a fluorescent tracer activated by gamma-glutamyltranspeptidase. Oral Oncol. 78, 1–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kossatz S., et al. , Validation of the use of a fluorescent PARP1 inhibitor for the detection of oral, oropharyngeal and oesophageal epithelial cancers. Nat. Biomed. Eng. 4, 272–285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Canto M. I. et al.; Confocal Endomicroscopy for Barrett’s Esophagus or Confocal Endomicroscopy for Barrett’s Esophagus (CEBE) Trial Group , In vivo endomicroscopy improves detection of Barrett’s esophagus-related neoplasia: A multicenter international randomized controlled trial (with video). Gastrointest. Endosc. 79, 211–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang S. S., et al. , High resolution microendoscopy for classification of colorectal polyps. Endoscopy 45, 553–559 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Hunt B., et al. , Diagnosing cervical neoplasia in rural Brazil using a mobile van equipped with in vivo microscopy: A cluster-randomized community trial. Cancer Prev. Res. (Phila.) 11, 359–370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quang T., et al. , A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest. Endosc. 84, 834–841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keahey P., Ramalingam P., Schmeler K., Richards-Kortum R. R., Differential structured illumination microendoscopy for in vivo imaging of molecular contrast agents. Proc. Natl. Acad. Sci. U.S.A. 113, 10769–10773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang Y., et al. , In vivo imaging of cervical precancer using a low-cost and easy-to-use confocal microendoscope. Biomed. Opt. Express 11, 269–280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kingma D. P., Ba J., “Adam: A method for stochastic optimization” in 3rd International Conference on Learning Representations: Conference Track Proceedings, Bengio Y., LeCun Y., Eds. (ICLR, San Diego, CA, 2015), pp. 1–15.
- 58.Wang Z., Simoncelli E. P., Bovik A. C., “Multiscale structural similarity for image quality assessment” in The Thirty-Seventh Asilomar Conference on Signals, Systems & Computers, Matthews M. B., Ed. (IEEE, Piscataway, NJ, 2003), pp. 1398–1402. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DeepDOF dataset data have been deposited in Zenodo (DOI: 10.5281/zenodo.3922596).