Significance
Approximately 50 human diseases are associated with deposition of abnormally aggregated proteins that propagate in a prion-like manner. The tumor suppressor p53 forms amyloid-like aggregates; however, how p53 aggregates propagate remains unclear. Here, we identified heparan sulfate (HS), the common and major nonprotein component of in vivo deposits of various protein aggregates, as a mediator of prion-like propagation of p53 aggregates in cultured cells. Our immunohistochemical analysis showing codeposition of p53 and highly sulfated domains of HS in ovarian cancer tissues strongly support the role of HS-mediated propagation of p53 aggregates in cancer pathology. Accordingly, our results provide a mechanism of propagation of p53 aggregates that is mediated by HS. Elucidation of detailed biological relevance in cancer is warranted.
Keywords: p53, protein aggregates, heparan sulfate, ovarian cancer, amyloid
Abstract
Sulfated glycosaminoglycans (GAGs) such as heparan sulfate (HS) are heteropolysaccharides implicated in the pathology of protein aggregation diseases including localized and systemic forms of amyloidosis. Among subdomains of sulfated GAGs, highly sulfated domains of HS, called HS S-domains, have been highlighted as being critical for HS function in amyloidoses. Recent studies suggest that the tumor suppressor p53 aggregates to form amyloid fibrils and propagates in a prion-like manner; however, molecules and mechanisms that are involved in the prion-like behavior of p53 aggregates have not been addressed. Here, we identified sulfated GAGs as molecules that mediate prion-like behavior of p53 aggregates. Sulfated GAGs at the cell surface were required for cellular uptake of recombinant and cancer cell-derived p53 aggregates and extracellular release of p53 from cancer cells. We further showed that HS S-domains accumulated within p53 deposits in human ovarian cancer tissues, and enzymatic remodeling of HS S-domains by Sulf-2 extracellular sulfatase down-regulated cellular uptake of p53 aggregates. Finally, sulfated GAG-dependent cellular uptake of p53 aggregates was critical for subsequent extracellular release of the aggregates and gain of oncogenic function in recipient cells. Our work provides a mechanism of prion-like behavior of p53 aggregates and will shed light on sulfated GAGs as a common mediator of prions.
Glycosaminoglycans (GAGs) are linear, unbranched polysaccharides that are attached to a core protein to form proteoglycans. The GAG chain structure is heterogeneous and can vary in net charges. Sulfated GAGs, such as heparan sulfate (HS) and chondroitin sulfate (CS), consist of repeating disaccharide units of a uronic acid and either N-acetylglucosamine or N-acetylgalactosamine. The structural diversity of GAGs confers binding selectivity of GAGs to their ligands and, thereby, plays an essential part in the functions of proteoglycans including organogenesis, signal transduction, and inflammation. Sulfated GAGs have also been implicated in the pathology of several diseases such as cancer and amyloidosis, which is the most common protein aggregation disease (1–3). Since the identification of sulfated GAGs as a common constituent of amyloid deposits in systemic and localized amyloidoses (4), sulfated GAGs have been established to affect the pathogenesis and progression of amyloidoses by promoting the formation and cellular interaction of protein aggregates (1).
The human TP53 gene encodes a nuclear tumor suppressor—p53—that can also respond to several stress conditions to induce cell cycle arrest and apoptosis (5). TP53 gene mutations occur in more than 50% of human cancers, which makes it the most common mutant gene in cancers. The DNA-binding domain of p53 is conformationally unstable (6), and several hotspot mutants are reportedly unfolded (7). p53 was recently reported to form protein aggregates in human cancer tissues and in vitro (8–11), which suggests that p53-mutant cancers may be a new class of protein aggregation diseases. p53 aggregates were proposed to demonstrate prion-like behavior like that of many other protein aggregates (12). We and others reported that the patterns and degrees of GAG’s sulfation modification (13–16), especially the highly sulfated domains of HS (HS S-domains), are essential for the pathology and progression of systemic and localized amyloidoses. However, the involvement of sulfated GAGs in transcellular propagation of p53 aggregates must still be elucidated.
Ovarian cancer is often diagnosed at an advanced stage, and little progress has been achieved in chemotherapy treatment (17). Alterations in the TP53 gene are quite prevalent in ovarian cancer, especially in the most common histological subtype—high-grade serous ovarian carcinoma (96%) (18)—which supports the pathological significance of p53 mutations. In our study here, we investigated the accumulation of HS S-domains in ovarian cancer tissues and the involvement of sulfated GAGs in extracellular release and cellular interaction of p53 aggregates. HS S-domains can be postsynthetically remodeled by Sulf-2 extracellular sulfatase (19). In addition, we studied whether enzymatic remodeling of HS S-domains would affect cellular uptake of extracellularly released p53. Our study supports the common roles of GAGs and their sulfation modifications in transcellular propagation of various protein aggregates.
Results
Cellular Uptake of Recombinant p53 Fibrils Depends on Sulfated GAGs.
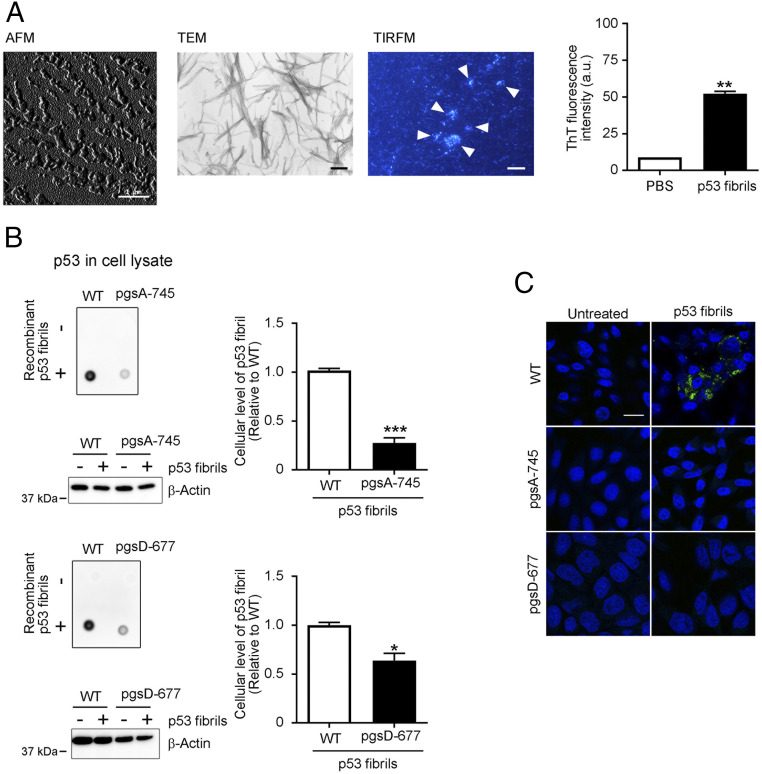
Although several p53 mutations reportedly induced misfolding and aggregation of p53 protein (8, 12, 20), WT p53 protein also formed aggregates in vitro (9) and in cancer cells (11, 21). Here, we successfully prepared fibrils of full-length WT p53 that were expressed in and purified from Escherichia coli, as confirmed by means of atomic force microscopy (AFM), transmission electron microscopy (TEM), total internal reflection fluorescence microscopy (TIRFM) combined with thioflavin T (ThT) fluorescence (22), and ThT fluorescence spectrophotometry (Fig. 1A). Short and thin fibrils (average length of 41.86 nm) were the major population as demonstrated by AFM and TIRFM (Fig. 1A) (SI Appendix, Fig. S1A). To assess the function of sulfated GAGs at the cell surface, we used Chinese hamster ovary (CHO) cells and their variant pgsA-745 cells, which lack sulfated GAGs, including HS and CS (23). In order to distinguish recombinant p53 signals from that of endogenous p53 in dot blots, we used a monoclonal antibody specific to human p53 in detection of p53 fibrils that were taken up by CHO cells. After treatment with recombinant p53 fibrils (25 nM) for 8 h, CHO cells with WT characteristics efficiently took up recombinant p53 fibrils (Fig. 1B), while no signals were observed in nontreated cells, which exclude cross-reaction with endogenous p53 proteins. The fibril content of the p53 fibril preparation was ∼94% (SI Appendix, Fig. S1B). Thus, we concluded that the observed p53 signals represent p53 fibrils that were taken up by cells. Quantitative dot blot analysis indicated that uptake of recombinant p53 fibrils by pgsA-745 cells significantly decreased compared with uptake by CHO WT cells (30%) (Fig. 1B). We also used the pgsD-677 variant, which lacks only HS (23). Again, pgsD-677 cells took up significantly lower amounts of recombinant p53 fibrils (50%, Fig. 1B). Immunocytochemical analysis showed intracellular p53 fibrils that were taken up by CHO WT cells, while no or negligible staining was observed in the GAG-modified variant cells (Fig. 1C). Thus, sulfated GAGs at the cell surface, especially HS, mediated uptake of p53 fibrils, similar to the cellular uptake mechanism of other amyloid fibrils (13–15, 24).
Fig. 1.
Sulfated GAGs at the cell surface mediate cellular uptake of recombinant p53 fibrils. (A) AFM, TEM, and TIRFM images of recombinant p53 fibrils. Arrowheads indicate ThT-positive p53 fibrils. Representative photomicrographs are shown. ThT (10 µM) fluorescent intensity at 485 nm with an excitation wavelength of 445 nm in the absence or presence of recombinant p53 fibrils (5 µM, Right) was assayed with a fluorescence spectrophotometer. Data are means ± SEM of three independent experiments. **P < 0.01. (Scale bars: AFM, 1 µm; TEM, 200 nm; TIRFM, 10 µm.) (B) Recombinant p53 fibrils were taken up by CHO cells with WT characteristics, with uptake being significantly reduced in pgsA-745 sulfated GAG-deficient mutant and pgsD-677 HS-deficient/CS-sufficient mutant CHO cells (23, 40). Cells were treated without (−) or with (+) 25 nM recombinant p53 fibrils for 8 h, after which p53 levels in whole-cell lysates were analyzed by means of dot blotting with the E26 human p53-specific antibody. The dot signals in the blots correspond to recombinant p53 fibrils that were taken up by cells. β-Actin was used as a loading control. The cellular uptake of recombinant p53 fibrils were quantitated by measuring the intensity of dot signals and shown in graphs. Data are means ± SEM of three independent experiments. *P < 0.05, ***P < 0.001. (C) Immunofluorescent staining of CHO WT, pgsA-745, and pgsD-677 variant cells that were treated with 25 nM recombinant p53 fibrils for 8 h. After fixation with 4% paraformaldehyde, cells were stained with the E26 human p53-specific antibody (green). DAPI nuclear counterstaining appears blue. Representative photomicrographs of three independent experiments are shown. (Scale bar: 20 µm.)
CHO Cells Take up Cancer Cell-Derived p53 Aggregates.
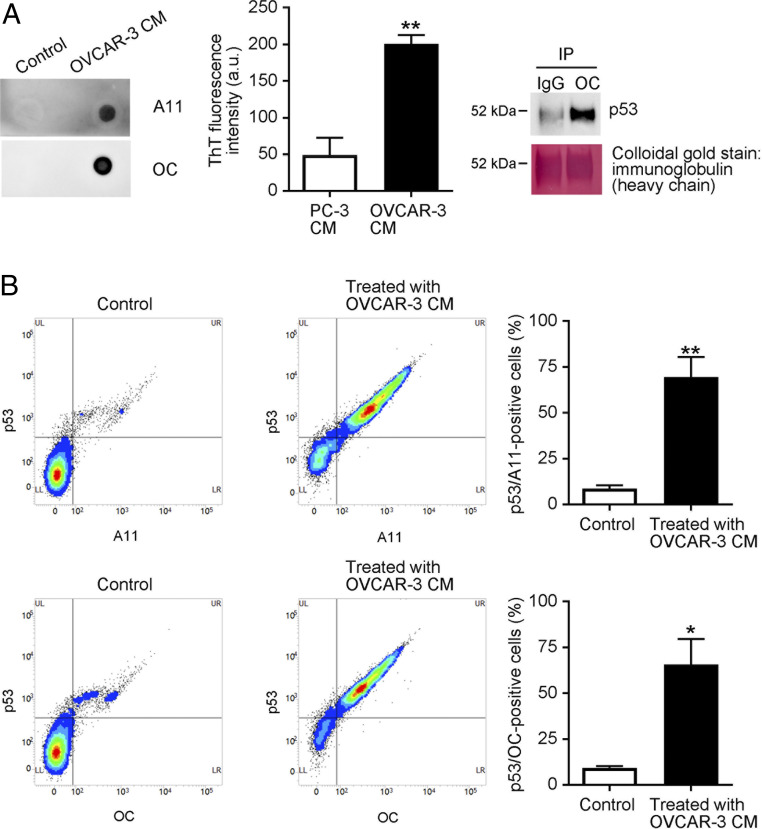
We next addressed whether CHO WT cells take up p53 aggregates that were released from cancer cells. We hypothesized that cancer cells that contain p53 aggregates may extracellularly release them. Because p53 mutations can be found in more than 95% of ovarian cancer patients (18) we used the OVCAR-3 human ovarian carcinoma cells that carry the p53 R248Q mutation and reportedly form p53 aggregates within cells (25). Dot blotting with the A11 anti-oligomer antibody or OC anti-amyloid fibril antibody and fluorescent measurements of an amyloidophilic dye ThT (10 µM) revealed that OVCAR-3 cells released amyloid-like protein aggregates into the culture medium (Fig. 2 A, Left and Center). Released OC-positive protein aggregates included p53 protein (Fig. 2 A, Right). Fluorescence-activated cell sorting (FACS) analysis with the DO-1 anti-p53 monoclonal antibody and the A11 or OC antibody showed that the vast majority of OVCAR-3 conditioned medium (CM)-treated CHO WT cells were positive for both p53 and A11-anti-protein or OC-anti-protein aggregate antibody (75% and 70%, respectively), which confirmed that CHO WT cells efficiently took up cancer cell-derived p53 aggregates (Fig. 2B).
Fig. 2.
CHO cells take up cancer cell-derived p53 aggregates. (A) Human ovarian cancer-derived OVCAR-3 cells released p53 aggregates. OVCAR-3 cells were cultured in serum-free OPTI-MEM for 12 h, after which samples of culture media were collected (OVCAR-3 CM). Protein aggregates in the media were analyzed by means of dot blotting with the A11 anti-oligomer or the OC anti-amyloid fibril antibody (“Control” indicates that fresh RPMI was spotted). ThT fluorescence intensity (10 µM) in CM of PC-3 cells (p53-null cells) and OVCAR-3 cells was recorded at 485 nm with an excitation wavelength of 445 nm (Center) by a multigrating high-speed microplate reader. Amyloid fibrils were immunoprecipitated with the OC anti-amyloid fibril antibody, and the immunoprecipitates were then subjected to Western blotting with the DO-1 anti-p53 monoclonal antibody, which showed that the OC-reactive amyloid fibrils consisted of p53. After examination, the membrane was stained with colloidal gold. (B) CHO WT cells were treated with OVCAR-3 CM for 8 h, after which cells were collected and analyzed by means of FACS with the DO-1 anti-p53 antibody and the A11 and OC anti-protein aggregate antibodies. Detection of cells positive for both p53 and protein aggregates confirmed that CHO cells successfully took up OVCAR-3-derived p53 aggregates. “Control” means that CHO cells were treated with fresh RPMI. The graph shows quantification of p53-positive and A11-positive or OC-positive cells. Data are means ± SEM of three independent experiments. *P < 0.05; **P < 0.01.
Cellular Uptake of OVCAR-3 Cell-Derived p53 Aggregates Depends on Sulfated GAGs at the Cell Surface.
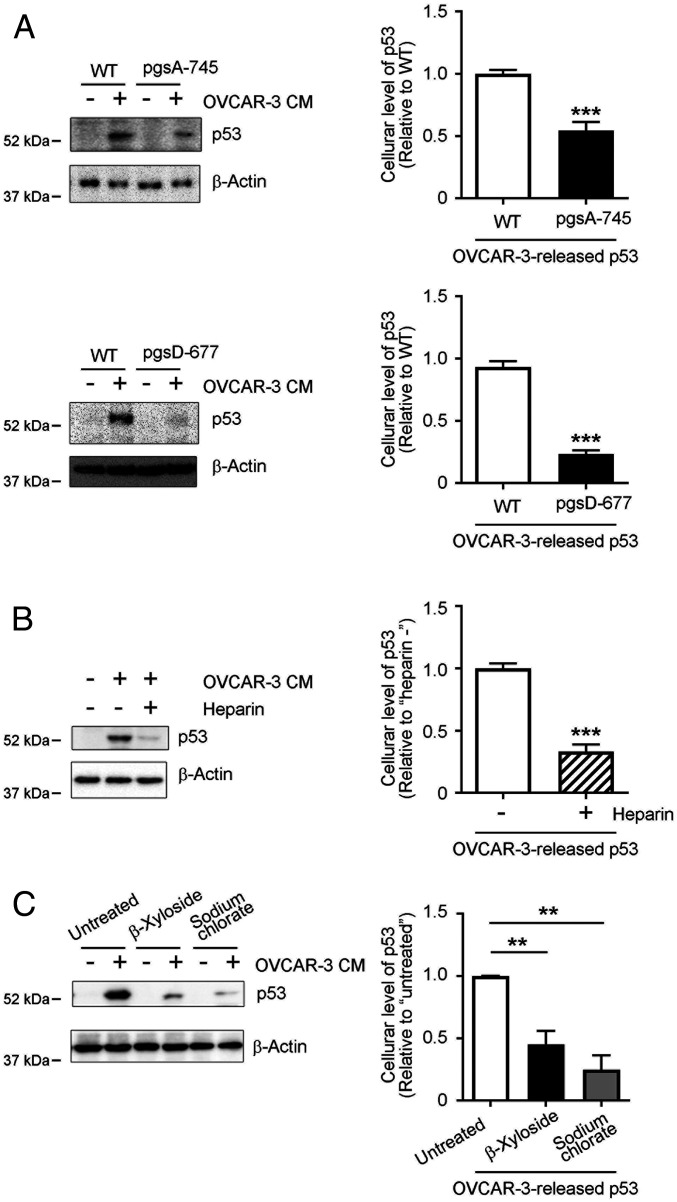
We further investigated whether cellular uptake of p53 aggregates released from cancer cells depends on sulfated GAGs. Cellular uptake of OVCAR-3–released p53 aggregates was significantly down-regulated to ∼30–50% in pgsA-745 and pgsD-677 cells, which lack sulfated GAGs and HS, respectively (Fig. 3A). Heparin is a structural analog of HS S-domains whose disaccharide units are mostly trisulfated (26) and reportedly interfere competitively with interactions of cell surfaces and various protein aggregates such as aggregates of amyloid β (Aβ), apolipoprotein A-I, tau protein, and α-synuclein (14, 15, 24, 27). In our study here, exogenously added heparin significantly suppressed the uptake of OVCAR-3–released p53 aggregates by 65% compared with uptake of control cells (Fig. 3B). To confirm the involvement of cell surface GAGs in the cellular uptake of p53 aggregates, we used β-xyloside and sodium chlorate. Exogenously added xylose and xylose derivatives to cell cultures stimulate xylose-primed GAGs, which are secreted into culture media, and, thus, the proteoglycan form of GAG chains can be eliminated from the cell surface (28). Sodium chlorate is a metabolic inhibitor of the biosynthesis of 3ʹ-phosphoadenosine-5ʹ-phosphosulfate, which is a sulfate donor in sulfate modification, and, thus, it inhibits all cellular sulfation reactions including those of sulfated GAGs (29). β-xyloside (2.5 mM) and sodium chlorate (100 mM) treatment significantly reduced HS (50%) and CS (50%) and HS S-domains (to 30%) in CHO WT cells, respectively (SI Appendix, Fig. S2A). Cellular uptake of OVCAR-3–released p53 aggregates was significantly suppressed by pretreatment with β-xyloside or sodium chlorate by ∼60–70% (Fig. 3C), which suggests that sulfated GAGs at the cell surface mediated the cellular uptake of p53 aggregates that were released from cancer cells.
Fig. 3.
Cellular uptake of OVCAR-3 cell-derived p53 aggregates depends on sulfated GAGs at the cell surface. (A) Significantly reduced uptake of cancer cell-derived p53 by pgsA-745 and pgsD-677 CHO mutant cells. WT, sulfated GAG-deficient pgsA-745, and HS-deficient/CS-sufficient pgsD-677 CHO cells were treated with OVCAR-3 CM for 8 h, after which levels of p53 proteins in whole cell lysates were analyzed by means of Western blotting. (B) Exogenously added heparin, an HS S-domain analog, interfered with cellular uptake of cancer-cell derived p53. CHO cells were treated with OVCAR-3 CM with or without heparin (5 μg/mL), after which whole-cell lysates were prepared and p53 protein levels in the lysates were determined by means of Western blotting. (C) Removal of cell surface sulfated GAGs or inhibition of cellular sulfation modifications significantly inhibited cellular uptake of cancer cell-derived p53. CHO WT cells were pretreated with 2.5 mM β-xyloside or 100 mM sodium chlorate in OPTI-MEM for 24 h, after which cells were treated with OVCAR-3 CM for 8 h. The presence of p53 proteins in whole-cell lysates was determined by means of Western blotting. β-Actin was used as a loading control. Graphs show quantification of cellular uptake of cancer cell-derived p53 aggregates. Data are means ± SEM of three independent experiments. **P < 0.01; ***P < 0.001.
Extracellular Release of p53 Aggregates from Cancer Cells Depends on Sulfated GAGs.
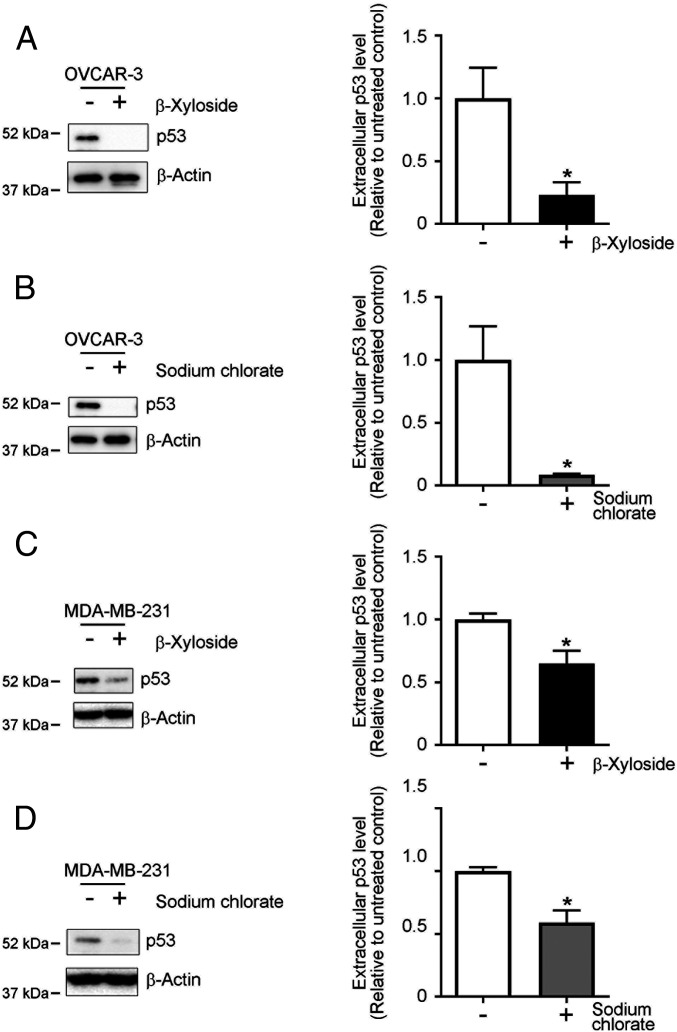
Sulfated GAGs at the cell surface reportedly mediated extracellular release of fibroblast growth factor-2 (FGF-2) via an unconventional protein secretion mechanism (30, 31). Extracellular release and transcellular propagation of the pathological tau protein in Alzheimer’s disease and frontotemporal dementia were also mediated by cellular sulfated GAGs via unconventional protein secretion mechanisms (32–34). Thus, we studied the role of sulfated GAGs in the extracellular release of p53. To modify sulfated GAGs in cells, we pretreated OVCAR-3 cells with β-xyloside or sodium chlorate and then measured the protein levels of p53 protein in culture media by means of Western blotting. Pretreatment with β-xyloside (2.5 mM) or sodium chlorate (100 mM) significantly reduced HS and CS (to 30–50% in OVCAR 3 cells and 40–60% in MDA-MB-231 cells, another cancer cell line), or HS S-domains (to 25% in OVCAR 3 cells and 50% in MDA-MB-231 cells; SI Appendix, Fig. S2 B and C), which confirmed that pretreatment with β-xyloside or sodium chlorate efficiently suppressed synthesis of proteoglycan forms of GAGs or sulfation modification in these cells. Secretion of p53 protein was significantly reduced in OVCAR-3 cells by pretreatment with β-xyloside or sodium chlorate by 75% and 90%, respectively (Fig. 4 A and B). We also confirmed sulfated GAG-dependent extracellular release of p53 in cancer cells by using MDA-MB-231 human breast cancer cells, which carry the p53 R280K mutation (35) and reportedly form intracellular p53 aggregates (25). Again, the levels of p53 protein released from cells decreased by 40% in β-xyloside–treated cells (Fig. 4C) and 50% in sodium chlorate-treated cells (Fig. 4D), which supports the function of sulfated GAGs in the mechanisms of extracellular export of p53 proteins.
Fig. 4.
Extracellular release of p53 from cancer cells depends on sulfated GAGs. Removal of cell surface GAGs or inhibition of cellular sulfation modifications significantly suppressed extracellular release of p53. OVCAR-3 cells (A and B) and MDA-MB-231 cells (C and D) were pretreated with 2.5 mM β-xyloside or 100 mM sodium chlorate in OPTI-MEM for 24 h, after which samples of culture medium were collected. p53 proteins in the samples were analyzed by means of Western blotting. β-Actin was used as a loading control. Graphs show quantification of p53 protein levels in the media. Data are means ± SEM of three independent experiments. *P < 0.05.
HS S-Domains Accumulate with p53 Deposits in Human Ovarian Cancer Tissues and Mediate Cellular Uptake and Subsequent Release of Recombinant or Cancer Cell-Derived p53 Aggregates.
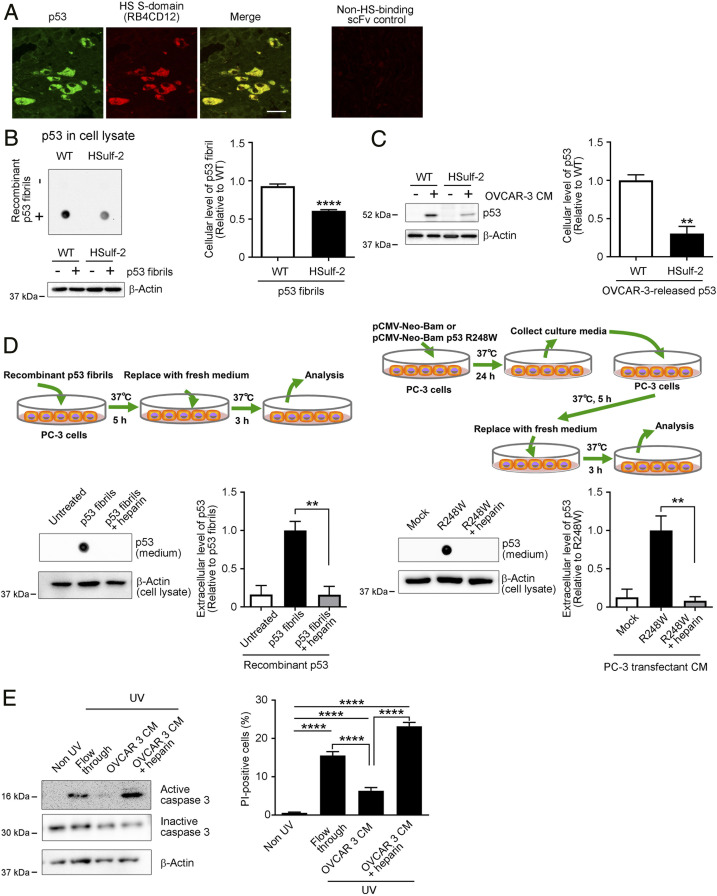
We previously reported the deposition of HS S-domains together with amyloid deposits such as those of Aβ in Alzheimer’s disease (36) and transthyretin (TTR) in familial ATTR amyloidosis (13). We thus investigated whether HS S-domains are a nonprotein component of p53 deposits in ovarian cancer tissues. Of the 11 patients with p53-mutant ovarian cancer included in this study, 6 had high-grade serous carcinoma, 3 had clear cell carcinoma, 1 had mucinous carcinoma, and 1 had colorectal carcinoma that had metastasized to the ovary. Information about the histopathology, p53 mutation status, and p53 deposition for these cases are summarized (Table 1). During the TP53 screening, 12 mutations were detected, including 11 missense mutations and 1 nonsense mutation, and all were located in the DNA-binding domain. In our study here, by using immunohistochemistry with the DO-1 monoclonal anti-p53 antibody and the RB4CD12 anti-HS S-domain scFv antibody (37), we found that HS S-domains localized together with p53 deposits in ovarian cancer tissue with the R248W mutant p53 (Fig. 5A). Additional immunohistochemical images for the RB4CD12 and DO-1 anti-p53 antibody are shown (SI Appendix, Fig. S3). Cytoplasmic colocalization of HS S-domains with p53 deposits was detected in 9 of 11 cases, including not only high-grade serous carcinoma but also clear cell, mucinous, and metastatic colorectal carcinoma cases (SI Appendix, Fig. S3). The staining patterns of p53 were quite similar to those of previous reports on various cancers such as breast cancer (8, 38) and basal cell carcinoma (39). We recently successfully remodeled HS S-domains by stable expression of human Sulf-2 (HSulf-2) in CHO cells (40) and showed that cellular uptake of several protein aggregates such as those of apolipoprotein A-I and TTR depended on HS S-domains (13, 14). Here, we used CHO-HSulf-2 cells whose HS S-domains were reduced by stable expression of HSulf-2 to evaluate whether uptake of p53 aggregates depends on HS S-domains. Amount of recombinant p53 in cell lysates of CHO-HSulf-2 was significantly smaller than that of CHO WT cells (60%), which suggests that CHO-HSulf-2 cells took up lower amounts of recombinant p53 fibrils (Fig. 5B). Similarly, CHO-HSulf-2 cells with reduced HS S-domains took up significantly low amounts of OVCAR-3–released p53 compared with CHO WT cells (30%, Fig. 5C). These results strongly support the role of HS S-domains as receptors that mediate cellular uptake of p53 aggregates.
Table 1.
Histopathology, p53 mutation status, and p53 deposition in ovarian cancer cases
| Sample ID | Histopathology | p53 mutant | p53/HS codeposition |
| OVC-1 | High-grade serous carcinoma | R248W | Positive/positive |
| OVO-11 | High-grade serous carcinoma | R196* | Positive/positive |
| OVO-12 | High-grade serous carcinoma | R273C | Positive/positive |
| OVO-22 | High-grade serous carcinoma | Q136E | Positive/positive |
| OVO-29 | High-grade serous carcinoma | H179Y | Positive/positive |
| OVO-8 | High-grade serous carcinoma | Y205C | Negative/negative |
| OVO-20 | Clear cell carcinoma | E285K | Positive/positive |
| OVO-28 | Clear cell carcinoma | R273C | Positive/positive |
| OVO-19 | Clear cell carcinoma | H179Y, N288D | Negative/negative |
| OVC-2 | Mucinous carcinoma | Y234N | Positive/positive |
| OVO-1 | Metastatic colorectal carcinoma | H193Y | Positive/positive |
*Indicates a nonsense mutation.
Fig. 5.
HS S-domains accumulate with p53 deposits in human ovarian cancer tissues, and mediate cellular uptake and subsequent release of recombinant or cancer cell-derived p53 aggregates. (A) Colocalization of p53 and HS S-domains in human ovarian cancer tissues with R248W mutant p53. Sections obtained from the patient were stained with the RB4CD12 anti-HS S-domain scFv antibody (36, 40) and the DO-1 mouse monoclonal anti-p53 antibody. An additional section was also stained with MPB49 non-HS-binding phage-displayed scFv antibody as a control for RB4CD12, which demonstrated no signals. (Scale bar: 20 μm.) (B and C) Cellular uptake of recombinant or cancer cell-derived p53 aggregates was partly mediated by HS S-domains. CHO cells with WT characteristics and their variant cells that stably express human extracellular sulfatase-2 (HSulf-2) were treated without (−) or with (+) recombinant p53 aggregates (B, 25 nM) or OVCAR-3 CM (C) for 8 h, after which p53 protein levels in whole-cell lysates were analyzed by using dot blotting or Western blotting with the E26 human p53-specific antibody. The dot signals in the blots correspond to recombinant p53 fibrils that were taken up by cells. The intensity of dot signals was measured. Graphs show quantification of p53 protein levels in the whole cell lysates. β-Actin was used as a loading control. (D) Extracellular release of p53 aggregates that were taken up by PC-3 cells was interfered by HS S-domain-analog heparin (5 μg/mL). p53-null PC-3 cells were transfected with pCMV-Neo-Bam (mock) or pCMV-Neo-Bam p53 R248W, and conditioned culture media were collected (PC3 CM-mock and PC3 CM-R248W). Newly prepared PC-3 cells were treated with recombinant p53 aggregates (25 nM, Left), PC3 CM-mock, or PC3 CM-R248W (Right) for 5 h, washed with PBS, and cultured for additional 3 h in fresh medium. p53 protein levels in culture media were analyzed by dot blotting. Graphs show quantification of p53 protein levels in the conditioned culture media. β-Actin was used as a loading control. (E) p53 aggregates-containing OVCAR3 CM reduced caspase 3 activation and UV-induced cell death in p53 WT A549 cells. A549 cells were treated with the p53-free flow through fraction of OVCAR3 CM or unfractionated OVCAR3 CM in the absence or presence of HS S-domain-analog heparin (5 μg/mL) for 12 h, after which cell death was induced by UV irradiation. Activation of caspase-3 was analyzed by means of Western blotting with an anti-cleaved caspase-3 antibody. Data are representative of three independent experiments. β-Actin was used as a loading control. UV-induced cell death was assessed by propidium iodide (PI) uptake and presented as percentages of PI-positive cells. Data are means ± SEM of three independent experiments. Non UV, A549 cells that were cultured without UV irradiation; **P < 0.01; ****P < 0.0001.
We wished to ask whether cultured cells show uptake of p53 aggregates and subsequently release them into culture media. We treated p53-null PC-3 human prostate adenocarcinoma cells (41) with recombinant p53 aggregates in the absence or presence of an excess amount of heparin, a structural analog of HS S-domains, in order to inhibit cellular uptake of p53 aggregates. After the treatment with recombinant p53 aggregates, cells were washed and cultured for an additional 3 h in fresh medium. PC-3 cells released p53 aggregates into the culture medium during the additional cultivation in fresh medium. Release of p53 aggregates was almost completely inhibited by heparin (Fig. 5D). We further confirmed the function of sulfated GAG-mediated cellular uptake of p53 aggregates in propagation of p53 aggregates by using culture medium of p53 R248W-transfected PC-3 cells. p53 R248W-transfected PC-3 cells, but not mock-transfected PC-3 cells, extracellularly released OC-positive p53 aggregates (SI Appendix, Fig. S4). We treated newly prepared PC-3 cells with the culture media prepared from these transfected cells (PC3 CM-mock and PC3 CM-R248W) in the absence or presence of heparin. PC-3 cells released p53 mutant proteins into the culture medium during cultivation in fresh medium for 3 h. Release of p53 mutant proteins was suppressed by the presence of heparin by 80–90% (Fig. 5D). Then, we investigated how p53 aggregates affect recipient cells, because p53 aggregates-containing cells reportedly present a chemoresistant gain-of-function phenotype (11, 42). A549 p53 WT cells (43) treated with OVCAR3 CM showed reduced caspase-3 activation and ultraviolet (UV)-induced cell death (60% reduction) compared to cells that were treated with a p53-free fraction of OVCAR3 CM (Flow through) (Fig. 5E). These effects of OVCAR3 CM were reversed by inhibiting cellular uptake of p53 aggregates by heparin, a HS S-domain analog (Fig. 5E).
Discussion
Growing evidence supports a prion-like behavior of protein aggregates in neurodegenerative diseases, such as aggregates of Aβ, tau, α-synuclein, superoxide dismutase 1, and transactivation-responsive region DNA-binding protein of 43 kDa (TDP-43) (44). In addition, this concept was recently proposed to be expanded to include tumor suppressor p53 in prionoid proteins (12), but the details of p53 propagation, such as which molecules are involved, are not fully understood. We report here a principal mechanism that is involved in the prion-like transcellular propagation of p53 aggregates, that is, sulfated GAG-mediated propagation of p53 aggregates. HS, a major member of the sulfated GAG family, has been established as a major codeposited nonprotein component of tissue amyloid deposits in almost all systemic or localized amyloidoses (1). Our immunohistochemistry study here provides additional evidence that highly sulfated HS subdomains coaccumulated with p53 deposits in human ovarian cancer tissues, which supports the idea that HS is a common nonprotein component in various amyloidoses (4) and that HS S-domains play a role in cancers with p53 deposits. These observations support the role of HS as a mediator of p53 propagation. HS is the first known mediator of the cellular uptake of protein aggregates. For example, HS was shown to be a primary receptor for infectious prion (PrPSc) (45–47). HS also mediated cellular uptake and subsequent cytotoxicity of monomeric or aggregated Aβ at the cell surface (24, 48, 49). We previously showed that HS and especially HS S-domains mediated interactions of cells with amyloid fibrils including those of apolipoprotein A-I and TTR (13, 14), which indicates that HS and HS S-domains are critical for interactions of cells with protein aggregates or cellular uptake of protein aggregates. In accordance with these observations, we showed here that sulfated GAGs, especially HS and HS S-domains, mediated the interaction of p53 aggregates with cells. Extracellular release of p53 from cancer cells also depended on sulfated GAGs, which suggests that p53 may be secreted in an unconventional manner via a pathway that was originally implicated in the secretion of FGF-2 (30) and later in the secretion of hyperphosphorylated tau that may spread to other cells (32, 33) Furthermore, extracellular release of p53 aggregates subsequent to their cellular uptake were significantly suppressed by inhibiting the sulfated GAG-dependent cellular uptake of the aggregates. Propagation of protein aggregates consists of several steps: misfolding and aggregation of proteins, extracellular release of the protein aggregates, cellular uptake of the protein aggregates, and seeding and release of protein aggregates form the recipient cells. Thus, our results provide evidence that HS plays critical roles in p53 propagation at multiple steps by regulating extracellular release and cellular uptake of p53 aggregates.
HS S-domains consist of trisulfated disaccharides [-IdoA(2-OSO3)-GlcNSO3(6-OSO3)-] and play critical roles in signal transduction of growth factors by acting as the site for interaction between the HS chains and ligands (19). We have proposed that HS S-domains are also critical for HS functions in the pathology of amyloidoses, such as acting as scaffolds of amyloidogenesis and receptors for amyloid fibrils (13, 14). Colocalization of HS S-domains and Aβ in amyloid plaques supports the role of HS S-domains in AD (36). In the present study, we further provide evidence that HS S-domains mediate propagation of p53 fibrils. HS S-domains are selectively remodeled by Sulfs, extracellular sulfatases. Here, we showed that remodeling of HS chains significantly suppressed cellular uptake of p53 aggregates, an important step for propagation of p53 aggregates. This raises a notion that HS remodeling by Sulfs might regulate p53 propagation in cancer, which needs to be further clarified. Whether HS S-domains or sulfated GAGs affect the aggregation process of p53 remains to be clarified. Because p53 can translocate into the cytosol, GAG chains may interact with WT p53 or mutant p53 proteins and, thereby, affect the p53 aggregation process, as previously suggested for many other amyloidogenic and aggregate-prone proteins (1). Although our results provide strong evidence of the binding of p53 aggregates and sulfated GAGs at the cell surface, but the precise mechanisms of how p53 protein interacts with sulfated GAGs are still unclear. Additional studies addressing sulfated GAG–p53 interactions and how sulfated GAGs affect p53 aggregation processes present a future challenge. The location of a HS chain is determined by its core protein, and the structure of the HS chain depends on the expression and activity of enzymes that catalyze HS extension and modifications. Because expressions of several HS core proteins and enzymes that are involved in HS metabolism are altered in many cancers (50–54), analysis of GAG structure of p53 deposits and searching for core proteins that are responsible for transcellular p53 aggregate propagation clearly deserves more study.
The pathological consequences of p53 aggregates and their transcellular propagation are still being investigated. The loss of p53 protein function by recruitment of properly folded p53 into aggregated forms can occur even if aggregates are formed in the nucleus (11). This finding is supported by the fact that more than 50% of human cancers, and more than 90% of human ovarian cancers, are associated with p53 mutations (5), which can result in the loss of p53 function. p53 aggregates can also interact with and compromise the function of other proteins such as p63 and p73 (12, 21, 55–57), whose aggregation reportedly underlies the pathology of ankyloblepharon-ectodermal defects-cleft lip/palate syndrome (58). In addition, p53 aggregation can promote tumor activities such as resistance to platinum therapy via interaction with cellular proteins that are involved in cell functions (11). In contrast to the well-established cytotoxic effects of amyloid fibrils or oligomers in neurodegenerative diseases and systemic or localized amyloidoses, whether p53 aggregates are cytotoxic is unclear. A previous report described p53 fibril cytotoxicity (8), whereas several studies suggested that p53 aggregates may contribute to cancer pathology via pathways other than cytotoxicity (9, 21). We provide evidence of gain of oncogenic function of p53 aggregates in recipient cells; however, how p53 aggregates are involved in tumor development and progression requires further elucidation. Nonetheless, that p53 aggregates and their transcellular propagation have critical functions in cancer pathology is now clear, and p53 aggregates may be a promising therapeutic target.
Here, we described a principal pathway that is involved in the transcellular propagation of p53 aggregates: sulfated GAG-dependent internalization of p53 aggregates. We also showed that HS S-domains containing 6-O-sufates, which can be selectively liberated by Sulf-2, mediated interactions of p53 aggregates with cells. We and others have proposed that remodeling of sulfated GAGs including HS in neurodegenerative diseases and in systemic and localized amyloidoses may lead to inhibition of protein aggregation and interaction of these aggregates with cells (15, 59). The current results expand this proposal to include p53 aggregation-associated cancers. Thus, our results suggest that sulfated GAGs, especially HS S-domains, may have a significant role in cancer pathology by mediating transcellular propagation of p53 aggregates, in addition to modulating signal cascades of several growth factors, as previously determined (60). Because enzymatic remodeling of HS S-domains by Sulf-2 resulted in marked suppression of the cellular uptake of p53 aggregates, enzymatic remodeling of HS S-domains (61) may be a promising strategy for targeting transcellular propagation of p53 aggregates as proposed for other amyloidoses.
Materials and Methods
Patients and Samples.
This study included 11 patients with p53-mutant ovarian cancer who underwent surgical resection at Wakayama Medical University Hospital between May 2017 and January 2019. Two or more experienced senior pathologists examined hematoxylin and eosin-stained sections and provided histological diagnoses for all patients. The present study was approved by the Ethics Committee of Wakayama Medical University (authorization no. 2025). All patients in this study provided written informed consent for the use of their tissue samples. All methods were performed according to the Declaration of Helsinki as specified by Wakayama Medical University.
Tumor DNA Extraction and Sequencing.
For TP53 mutation screening, formalin-fixed paraffin-embedded (FFPE) specimens of ovarian cancer were subjected to a histological review, and only those containing sufficient tumor cells (at least 75% tumor cells), as determined by hematoxylin and eosin staining, were used for DNA extraction. Tumor DNA samples (40 ng) were analyzed by using the QIAseq Human Comprehensive Cancer Panel for 275 genes (Qiagen), and libraries were prepared according to the manufacturer’s instructions (62). These purified libraries were pooled and then sequenced by using a NextSeq 500 instrument (Illumina). Reads were aligned to the hg19 human reference genome, and variants were detected according to the manufacturer’s pipeline. Germ-line mutations were excluded on the basis of the Human Genetic Variation Database (https://www.hgvd.genome.med.kyoto-u.ac.jp/) and the Exome Aggregation Consortium database (63).
Materials.
Heparin (average molecular weight 13,000, and <38% sulfur content, catalog no. PH-72210) was purchased from Celsus Laboratories). CHO pgsA-745 and pgsD-677 variant types were from American Type Culture Collection (ATCC) as described previously (40). An anti-β-actin antibody was purchased from Santa Cruz Biotechnology, and the E26 rabbit monoclonal anti-human p53 antibody and DO-1 mouse monoclonal anti-p53 antibody were purchased from Abcam. RB4CD12 anti-HS S-domain antibody (37) was kindly provided by Toin H. van Kuppevelt, Nijmegen Center for Molecular Life Sciences, Radboud University Nijmegen Medical Center, The Netherlands. A mouse monoclonal anti-HS IgM (10E4) was from United States Biological, and a mouse monoclonal anti-chondroitin sulfate IgM (CS-56) was from Abcam. Mouse IgG and rabbit IgG isotype controls were obtained from Cell Signaling Technology. The A11 anti-oligomer antibody (64) and OC anti-amyloid fibrils antibody (65) were obtained from StressMarq Biosciences. ThT was obtained from Sigma. pCMV-Neo-Bam and pCMV-Neo-Bam p53 R248W (Addgene plasmid 16440, http://addgene.org/16440, RRID:Addgene_16436 and Addgene plasmid # 16437, http://addgene.org/16437, RRID: Addgene_16437) were gifts from Bert Vogelstein (Johns Hopkins University School of Medicine, Baltimore, MD) (66).
Cell Culture.
CHO cells that stably expressed the human extracellular endoglucosamine 6-sulfatase HSulf-2 were established as previously described (40). CHO cells with WT characteristics as well as the variants were cultured in a 1:1 mixture of Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F-12 Nutrient Mixture (Sigma) supplemented with 10% heat-inactivated fetal bovine serum (FBS; BioWest), 100 U/mL penicillin, and 100 μg/mL streptomycin (Sigma) at 37 °C in 95% air and 5% CO2. OVCAR-3 human ovarian carcinoma cells were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT/AMED, Japan, and were maintained in RPMI 1640 supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. MDA-MB-231 human breast adenocarcinoma cells and A549 human lung cancer-derived adeno carcinoma from ATCC and p53-null PC-3 human prostate adenocarcinoma cells (41) from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan) were maintained in DMEM and Kaighn’s modification of Ham’s F-12 (F12K) supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin, respectively.
Preparation of p53 Fibrils.
The complementary DNA-encoding WT human p53 with an attached human rhinovirus (HRV) 3C protease recognition sequence was amplified via PCR and was introduced into the pET32a+ expression vector (Novagen). The construct was transformed into an E. coli BL21 Star (DE3) strain (Invitrogen). Expression of WT p53 was induced by adding 500 μM isopropyl-β-d-thiogalactopyranoside and then cultured for 2 h at 37 °C. Bacterial cells were collected by centrifugation at 6,500 × g for 15 min at 4 °C, after which cells were resuspended with 50 mL of Binding Buffer 1 (20 mM Tris/HCl, 500 mM NaCl, 5 mM imidazole, 6 M guanidinium chloride, pH 8.0) and sonicated 4 × 2.5 min on ice (UD-201, TOMY SEIKO), followed by mixing with rotation (10 rpm) at 4 °C overnight. The suspension was centrifuged at 12,000 × g for 30 min at 4 °C, and the supernatant was applied to a nickel column (HisTrap FF 16/25, GE Healthcare) that was preequilibrated with 75 mL of Binding Buffer 1. The column was washed with 100 mL of Binding Buffer 1, and captured p53 proteins conjugated with a His tag at the amino terminus were eluted with a linear gradient of Binding Buffer 1 and Elution Buffer (20 mM Tris/HCl, 500 mM NaCl, 300 mM imidazole, 6 M guanidinium chloride, pH 8.0). The obtained solution was dialyzed against Dialysis Buffer (25 mM phosphate, 150 mM NaCl, 1 mM DTT, pH 7.4) at 4 °C for 4 h followed by an overnight dialysis. His tags were removed from p53-His tag conjugates by treatment with HRV-3C protease (2 U/mL, Takara). Urea (Wako Pure Chemicals) was added to the obtained solution to a final concentration of 8 M, and the solution was incubated with rotation (10 rpm) at 4 °C overnight. The cleaved His tags were removed by using a nickel column (cOmplete His-Tag Purification Column 5 mL, Roche) that was preequilibrated with 50 mL of Binding Buffer 2 (20 mM Tris/HCl, 500 mM NaCl, 5 mM imidazole, 8 M urea, pH 8.0). The p53 protein obtained had two extra amino acids, Gly-Pro, at the amino terminus. The protein preparations were >95% pure as assessed by Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS/PAGE). p53 fibrils were prepared by incubation of 1 mL of recombinant p53 (5 μM in 20 mM sodium phosphate [pH 7.4] containing 150 mM NaCl, 3 M urea, 1 mM Tris (2-carboxyethyl) phosphine hydrochloride, and 5% dimethyl sulfoxide) with rotation in a low protein binding tube (2 mL, Sarstedt) at 10 rpm for 1 wk at 37 °C.
ThT Fluorescence Assay and AFM.
The fibrils obtained were dialyzed against phosphate-buffered saline (PBS, pH 7.4) for ThT fluorescence measurements. Fluorescence intensity of p53 fibrils (5 µM) in PBS in the presence of ThT (10 µM) was recorded 485 nm with an excitation wavelength of 445 nm by using an F-2700 fluorescence spectrophotometer (Hitachi High-Technologies). Fibril formation of the recombinant p53 was also confirmed by using an atomic force microscope as previously described (13). Briefly, 10 μL of fibril solution (30 μM) in 10 mM PBS was diluted with distilled water (45 μL), after which the solution was spotted on freshly cleaved mica (Nilaco Corporation). After washing with 20 μL of distilled water, sample images were acquired at room temperature under ambient conditions on a NanoScope IIIa Tapping Mode AFM (Veeco Instrument) and single-crystal microcantilever OMCLAC160TS-R3 (Olympus) at a scan rate of 0.5 Hz in tapping mode.
TEM and TIRFM Combined with ThT Fluorescence.
For TEM analysis of recombinant p53 fibrils, 2 µL of p53 fibril solution in PBS (5 µM) was spotted on a glow-discharged copper grid (300 mesh) coated with carbon followed by adding 20 µL of 2% phosphotungstic acid solution (adjusted at pH 7.4 with NaOH). The grid was dried in vacuo, and TEM images were obtained on a JEOL JEM-1200EX transmission microscope (JEOL) with an acceleration voltage of 80 kV.
Individual fibrils in the recombinant p53 fibril preparation (5 µM in PBS) were examined by means of TIRFM measurements with a fluorescence microscopic system based on an inverted microscope (IX70; Olympus). ThT was excited by using an argon laser (model 185F02-ADM; Spectra Physics). The fluorescent image was filtered with a bandpass filter (D490/30 Omega Optical) and visualized by using an image intensifier (model VS4–1845; Video Scope International) coupled with a SIT camera (C2400-08; Hamamatsu Photonics).
Analysis of Uptake of Recombinant p53 Fibrils by CHO WT and Variant Cells.
p53 fibrils that were taken up by cells were quantified by using quantitative dot blotting. CHO WT and variant cells were incubated with recombinant p53 fibrils (25 nM) for 8 h at 37 °C, after which whole-cell lysates were prepared by using TCA precipitation (14, 67). Briefly, cells were washed three times with PBS and treated with 10% trichloroacetic acid (wt/vol) in PBS for 30 min on ice, followed by centrifugation at 10,000 × g for 5 min at 4 °C. The resulting precipitates were dissolved in SDS/PAGE sample buffer (0.125 M Tris⋅HCl, 4% [wt/vol] SDS, 20% [vol/vol] glycerol, and 0.01% [wt/vol] bromophenol blue) for preparation of whole-cell lysates. Lysates were spotted on nitrocellulose blotting membranes (Merck Millipore), and membranes were dried completely at room temperature. Recombinant p53 fibrils were probed with the E26 rabbit monoclonal anti-human p53 antibody (1:750) followed by a horseradish peroxidase-labeled anti-rabbit antibody (1:1000, Cell Signaling Technology) and ImmunoStar LD (Wako Pure Chemical Industries). Cell lysate protein contents were normalized to the expression level of β-actin. Signals were visualized by using a LuminoGraph image analyzer (ATTO). To determine the densities of p53-positive dots, image files were analyzed with ImageJ 1.50i (NIH).
Immunocytochemistry.
CHO and their variants were plated on poly-l-lysine–coated glasses and cultured for 12 h, after which they were incubated with recombinant p53 fibrils (25 nM) for 8 h at 37 °C. Cells were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. After the cells were washed three times with PBS, they were blocked and permeabilized with 10% normal goat serum and 0.05% saponin in PBS at room temperature for 20 min. They were then incubated with the E26 rabbit monoclonal anti-human p53 antibody (1:100) followed by an Alexa 488-conjugated secondary antibody (1:500, Thermo Fisher Scientific). Stained specimens were mounted with Vectashield mounting medium containing DAPI (Vector Laboratories) and were examined with an LSM700 (Zeiss).
Analysis of Extracellularly Released p53 and Its Cellular Uptake.
OVCAR-3 cells were used because of their homeostatic synthesis of p53 aggregates (25). OVCAR-3 cells were cultured in serum-free RPMI 1640 for 24 h, after which the culture medium was collected and centrifuged at 2,000 × g for 30 min to remove cells and debris (OVCAR-3 CM). Extracellularly released p53 was measured by means of Western blotting with the E26 rabbit monoclonal anti-human p53 antibody (1:750). OVCAR-3 CM was used to analyze the uptake of extracellularly released p53 aggregates. Protein aggregates in OVCAR-3 CM were confirmed by dot blotting with the OC (1:1000) or A11 (1:200) antibody, and by recording ThT fluorescence intensity (10 µM) at 485 nm with an excitation wavelength of 445 nm with an SH-9000Lab multigrating high-speed microplate reader (Yamato Scientific). We further confirmed that OVCAR-3 CM contained p53 aggregates by means of immunoprecipitation as described below. CHO WT cells and their variants were treated with OVCAR-3 CM for 8 h at 37 °C, and then whole-cell lysates were prepared as described above. To disrupt proteoglycan forms of GAGs or inhibit sulfation modification, CHO WT cells were pretreated with β-xyloside (28) (2.5 mM, Tokyo Kasei) or sodium chlorate (29) (100 mM, Sigma) in OPTI-MEM for 24 h. To analyze GAGs in β-xyloside–treated or sodium chrolate-treated cells, whole-cell lysates were prepared as described above and GAG contents were analyzed by means of dot blotting and Western blotting with 10E4 (1:500), CS-56 (1:1000), or RB4CD12 (1:500). For competition assays with heparin, CHO WT cells were incubated with both OVCAR-3 CM and heparin (5 μg/mL). p53 contents in whole-cell lysates were analyzed by means of Western blotting. Whole-cell lysates were subjected to SDS/PAGE with 5–20% gels (Wako Pure Chemical Industries) and were transferred to polyvinylidene difluoride membranes (Millipore). p53 proteins were probed with the E26 rabbit monoclonal anti-human p53 antibody (1:750) followed by a horseradish peroxidase-labeled anti-rabbit antibody (1:1000) and ImmunoStar LD, after which signals were visualized by using a LuminoGraph image analyzer and image files were analyzed with ImageJ 1.50i. Protein contents were normalized to the expression level of β-actin protein.
In order to see whether extracellular release of p53 by cancer cells depends on cellular sulfated GAGs, we treated cells with β-xyloside or sodium chlorate as described above. In addition to OVCAR-3 cells, we used MDA-MB-231 cells, which also synthesize p53 aggregates (25). Cancer cells were precultured in the presence of 2.5 mM β-xyloside or 100 mM sodium chlorate in OPTI-MEM for 24 h, followed by culture in serum-free RPMI 1640 for 24 h, after which that culture medium was collected and centrifuged at 2,000 × g for 30 min to remove cells and debris. Extracellularly released p53 was measured by means of Western blotting with the E26 rabbit monoclonal anti-p53 antibody (1:750).
For analysis of extracellular release of p53 aggregates that were taken up by cells, p53-null PC-3 cells in 12-well plates were treated with recombinant p53 aggregates (25 nM) for 5 h in the presence or absence of heparin (5 μg/mL), after which cells were washed with PBS three times and cultured for an additional 3 h in fresh F12K. In other experiments, PC-3 cells were transfected with pCMV-Neo-Bam or pCMV-Neo-Bam p53 R248W by using the ViaFect transfection reagent (Promega). After being cultured for 24 h, CM were collected and centrifuged at 2,000 × g for 30 min to remove cells and debris (PC3 CM-mock and PC3 CM-R248W). Newly prepared PC-3 cells in 12-well plates were treated with PC3 CM-mock or PC3 CM-R248W for 5 h, washed with PBS three times, and cultured in fresh F12K for additional 3 h. Culture media were then collected, and proteins in the culture media were precipitated with 10% TCA for 30 min at 4 °C. Precipitates were collected by centrifugation at 10,000 × g for 1 h at 4 °C and lysed with the SDS/PAGE sample buffer. p53 protein levels in samples were analyzed by means of dot blotting as described above.
Flow Cytometric Analysis.
Cells were detached with PBS containing 0.5 mM l-arginine and 4 mM EDTA and were washed twice with 0.5% bovine serum albumin (BSA) and PBS. Cells were then incubated with the DO-1 anti-p53 antibody (1:200) and the A11 anti-oligomer antibody (1:50) or the OC anti-amyloid antibody (1:100) in 0.5% BSA and PBS for 1 h at 4 °C, followed by incubation with an Alexa 647-conjugated polyclonal goat anti-rabbit IgG and an Alexa 488-conjugated polyclonal goat anti-rabbit IgG (1:200, Thermo Fisher Scientific) in 0.5% BSA and PBS for 30 min at 4 °C. After being washed twice, cells were gated by excluding doublets and subjected to flow cytometric analysis by using the BD FACSVerse system (BD Biosciences). Because the generic structural features of conformation-specific antibodies such as OC and A11 are vulnerable to fixation and membrane permeabilization with detergent (68), we carried out the flow cytometric analysis without permeabilization.
Immunohistochemistry.
Paraffin-embedded blocks of human ovarian cancer tissues were cut into 3-μm-thick sections, followed by deparaffinization and rehydration. Antigens were retrieved by using heat treatment: boiling the sections in a pressure cooker in citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 20 min. After sections were blocked in Animal-Free Blocker (Vector Laboratories) for 1 h at room temperature, they were incubated overnight at 4 °C with primary antibodies: a mouse monoclonal anti-p53 antibody (DO-1), 1:200, and the RB4CD12 antibody, 1:100, in Animal-Free Blocker. Samples were then washed in PBS, and primary antibodies were detected with the secondary antibodies Alexa 488-conjugated polyclonal goat anti-rabbit IgG (1:500) and Cy3-conjugated monoclonal anti-vesicular stomatitis virus G glycoprotein (4 μg/mL; Sigma). The sections were mounted with Vectashield mounting medium (Vector Laboratories) and examined with an LSM700 confocal microscope.
Immunoprecipitation.
Samples of culture medium in which OVCAR-3 cells were grown in 6-cm culture dishes were collected and concentrated with Amicon Ultra Filters (Merck Millipore). Protein aggregates in concentrated samples were then immunoprecipitated by using the anti-amyloid OC antibody (1:50) that was conjugated with Dynabeads Protein G (Thermo Fisher Scientific). The precipitates obtained were washed three times with PBS and treated with heat (70 °C) for 10 min in SDS/PAGE sample buffer. The p53 proteins in the supernatants were detected by means of Western blotting with 5–20% gradient gels and the DO-1 anti-p53 monoclonal antibody (1:1000). The signals were visualized and analyzed as described above except that we used a preabsorbed horseradish peroxidase-conjugated anti-mouse IgG (1:10000, Jackson ImmunoResearch Laboratories) in order to minimize the cross-reaction with rabbit IgG.
UV-Induced Cell Death.
A549 nonsmall cell lung cancer cells were chosen for investigating gain of oncogenic function, because A549 cells has a WT p53 status (43). A549 cells were treated with OVCAR3 CM or the p53-free flow through of OVCAR3 CM prepared with 10-kDa cutoff ultrafilter devices (Merck Millipore) at 37 °C for 12 h in order to let A549 cells take up p53 aggregates. Heparin (5 μg/mL) was used to inhibit cellular uptake of p53 aggregates. Cells were then irradiated UV (51 μW/cm2) for 10 min and incubated for additional 24 h. Cell damages were assessed by evaluation of caspase-3 activation and cell death. Activation of caspase-3 was analyzed by means of Western blotting with a rabbit monoclonal anti-cleaved caspase-3 antibody (1:1000, Cell Signaling Technology), and UV-induced cell death was analyzed by using propidium iodide (PI, Thermo Fisher). PI-positive cells were quantified by using the CellInsight image analyzer (Thermo Fisher), and the Cellomics Scan and Cellomics View software (Thermo Fisher).
Statistical Analysis.
Data were analyzed via the unpaired Student’s t test or an ordinary one-way ANOVA with post hoc Dunnett or Tukey’s test by means of Prism software (GraphPad Software, Version 7.04). Results were considered significant when P values were less than 0.05.
Supplementary Material
Acknowledgments
RB4CD12 antibody was kindly provided by Dr. Toin H. van Kuppevelt, Nijmegen Center for Molecular Life Sciences, Radboud University Nijmegen Medical Center, The Netherlands. This work was partly supported by Grants-in-Aid for Young Scientists B-15K19488 and B-17K16123 (to K.N.) and JP18K16776 (to N.I.) from the Japan Society for the Promotion of Science, and Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) (JP15K08265 and JP16KK0202 to K.U.). This study was partially supported by Wakayama Medical University Internal Grant Tokutei-kenkyu-zyosei 2019 and 2020.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009931117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Nishitsuji K., Uchimura K., Sulfated glycosaminoglycans in protein aggregation diseases. Glycoconj. J. 34, 453–466 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Theocharis A. D., Karamanos N. K., Proteoglycans remodeling in cancer: Underlying molecular mechanisms. Matrix Biol. 75-76, 220–259 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Soares da Costa D., Reis R. L., Pashkuleva I., Sulfation of glycosaminoglycans and its implications in human health and disorders. Annu. Rev. Biomed. Eng. 19, 1–26 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Snow A. D., Willmer J., Kisilevsky R., Sulfated glycosaminoglycans: A common constituent of all amyloids? Lab. Invest. 56, 120–123 (1987). [PubMed] [Google Scholar]
- 5.Bykov V. J. N., Eriksson S. E., Bianchi J., Wiman K. G., Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 18, 89–102 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Ang H. C., Joerger A. C., Mayer S., Fersht A. R., Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J. Biol. Chem. 281, 21934–21941 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Gannon J. V., Greaves R., Iggo R., Lane D. P., Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 9, 1595–1602 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ano Bom A. P., et al. , Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J. Biol. Chem. 287, 28152–28162 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S., et al. , p53 amyloid formation leading to its loss of function: Implications in cancer pathogenesis. Cell Death Differ. 24, 1784–1798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S., et al. , Investigating the intrinsic aggregation potential of evolutionarily conserved segments in p53. Biochemistry 53, 5995–6010 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Yang-Hartwich Y., et al. , p53 protein aggregation promotes platinum resistance in ovarian cancer. Oncogene 34, 3605–3616 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Silva J. L., De Moura Gallo C. V., Costa D. C., Rangel L. P., Prion-like aggregation of mutant p53 in cancer. Trends Biochem. Sci. 39, 260–267 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Kameyama H., et al. , The accumulation of heparan sulfate S-domains in kidney transthyretin deposits accelerates fibril formation and promotes cytotoxicity. Am. J. Pathol. 189, 308–319 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Kuwabara K., et al. , Cellular interaction and cytotoxicity of the Iowa mutation of apolipoprotein A-I (ApoA-IIowa) amyloid mediated by sulfate moieties of heparan sulfate. J. Biol. Chem. 290, 24210–24221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes B. B., et al. , Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. U.S.A. 110, E3138–E3147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauch J. N., et al. , Tau internalization is regulated by 6-O sulfation on heparan sulfate proteoglycans (HSPGs). Sci. Rep. 8, 6382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Bois A., Pfisterer J., Future options for first-line therapy of advanced ovarian cancer. Int. J. Gynecol. Cancer 15 (suppl. 1), 42–50 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Network , Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosen S. D., Lemjabbar-Alaoui H., Sulf-2: An extracellular modulator of cell signaling and a cancer target candidate. Expert Opin. Ther. Targets 14, 935–949 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishimaru D., et al. , Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry 42, 9022–9027 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Xu J., et al. , Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 7, 285–295 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Ban T., Hamada D., Hasegawa K., Naiki H., Goto Y., Direct observation of amyloid fibril growth monitored by thioflavin T fluorescence. J. Biol. Chem. 278, 16462–16465 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Esko J. D., Stewart T. E., Taylor W. H., Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 82, 3197–3201 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandwall E., et al. , Heparan sulfate mediates amyloid-beta internalization and cytotoxicity. Glycobiology 20, 533–541 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Rangel L. P., et al. , p53 reactivation with induction of massive apoptosis-1 (PRIMA-1) inhibits amyloid aggregation of mutant p53 in cancer cells. J. Biol. Chem. 294, 3670–3682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallagher J. T., Walker A., Molecular distinctions between heparan sulphate and heparin. Analysis of sulphation patterns indicates that heparan sulphate and heparin are separate families of N-sulphated polysaccharides. Biochem. J. 230, 665–674 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ihse E., et al. , Cellular internalization of alpha-synuclein aggregates by cell surface heparan sulfate depends on aggregate conformation and cell type. Sci. Rep. 7, 9008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson J., Gospodarowicz D., Effect of p-nitrophenyl-beta-D-xyloside on proteoglycan synthesis and extracellular matrix formation by bovine corneal endothelial cell cultures. J. Biol. Chem. 259, 3818–3824 (1984). [PubMed] [Google Scholar]
- 29.Safaiyan F., et al. , Selective effects of sodium chlorate treatment on the sulfation of heparan sulfate. J. Biol. Chem. 274, 36267–36273 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Zehe C., Engling A., Wegehingel S., Schäfer T., Nickel W., Cell-surface heparan sulfate proteoglycans are essential components of the unconventional export machinery of FGF-2. Proc. Natl. Acad. Sci. U.S.A. 103, 15479–15484 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nickel W., The unconventional secretory machinery of fibroblast growth factor 2. Traffic 12, 799–805 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Katsinelos T., et al. , Unconventional secretion mediates the trans-cellular spreading of tau. Cell Rep. 23, 2039–2055 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Merezhko M., et al. , Secretion of tau via an unconventional non-vesicular mechanism. Cell Rep. 25, 2027–2035.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Rabouille C., Pathways of unconventional protein secretion. Trends Cell Biol. 27, 230–240 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Bartek J., Iggo R., Gannon J., Lane D. P., Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene 5, 893–899 (1990). [PubMed] [Google Scholar]
- 36.Hosono-Fukao T., et al. , Heparan sulfate subdomains that are degraded by Sulf accumulate in cerebral amyloid ß plaques of Alzheimer’s disease: Evidence from mouse models and patients. Am. J. Pathol. 180, 2056–2067 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Dennissen M. A., et al. , Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J. Biol. Chem. 277, 10982–10986 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Levy C. B., et al. , Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int. J. Biochem. Cell Biol. 43, 60–64 (2011).Corrected in: Int. J. Biochem. Cell Biol.43, 946 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Lasagna-Reeves C. A., et al. , Dual role of p53 amyloid formation in cancer; loss of function and gain of toxicity. Biochem. Biophys. Res. Commun. 430, 963–968 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Hossain M. M., et al. , Direct detection of HSulf-1 and HSulf-2 activities on extracellular heparan sulfate and their inhibition by PI-88. Glycobiology 20, 175–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isaacs W. B., Carter B. S., Ewing C. M., Wild-type p53 suppresses growth of human prostate cancer cells containing mutant p53 alleles. Cancer Res. 51, 4716–4720 (1991). [PubMed] [Google Scholar]
- 42.Pedrote M. M., et al. , Oncogenic gain of function in glioblastoma is linked to mutant p53 amyloid oligomers. iScience 23, 100820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia L. Q., et al. , Screening the p53 status of human cell lines using a yeast functional assay. Mol. Carcinog. 19, 243–253 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Brettschneider J., Del Tredici K., Lee V. M., Trojanowski J. Q., Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat. Rev. Neurosci. 16, 109–120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horonchik L., et al. , Heparan sulfate is a cellular receptor for purified infectious prions. J. Biol. Chem. 280, 17062–17067 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Hijazi N., Kariv-Inbal Z., Gasset M., Gabizon R., PrPSc incorporation to cells requires endogenous glycosaminoglycan expression. J. Biol. Chem. 280, 17057–17061 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Shyng S. L., Lehmann S., Moulder K. L., Harris D. A., Sulfated glycans stimulate endocytosis of the cellular isoform of the prion protein, PrPC, in cultured cells. J. Biol. Chem. 270, 30221–30229 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Nishitsuji K., Hosono T., Uchimura K., Michikawa M., Lipoprotein lipase is a novel amyloid beta (Abeta)-binding protein that promotes glycosaminoglycan-dependent cellular uptake of Abeta in astrocytes. J. Biol. Chem. 286, 6393–6401 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanekiyo T., et al. , Heparan sulphate proteoglycan and the low-density lipoprotein receptor-related protein 1 constitute major pathways for neuronal amyloid-beta uptake. J. Neurosci. 31, 1644–1651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernfield M., et al. , Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68, 729–777 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Nagarajan A., Malvi P., Wajapeyee N., Heparan sulfate and heparan sulfate proteoglycans in cancer initiation and progression. Front. Endocrinol. (Lausanne) 9, 483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knelson E. H., Nee J. C., Blobe G. C., Heparan sulfate signaling in cancer. Trends Biochem. Sci. 39, 277–288 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maeda D., et al. , Glypican-3 expression in clear cell adenocarcinoma of the ovary. Mod. Pathol. 22, 824–832 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Stadlmann S., et al. , Glypican-3 expression in primary and recurrent ovarian carcinomas. Int. J. Gynecol. Pathol. 26, 341–344 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Kehrloesser S., et al. , Intrinsic aggregation propensity of the p63 and p73 TI domains correlates with p53R175H interaction and suggests further significance of aggregation events in the p53 family. Cell Death Differ. 23, 1952–1960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cino E. A., Soares I. N., Pedrote M. M., de Oliveira G. A., Silva J. L., Aggregation tendencies in the p53 family are modulated by backbone hydrogen bonds. Sci. Rep. 6, 32535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costa D. C., et al. , Aggregation and prion-like properties of misfolded tumor suppressors: Is cancer a prion disease? Cold Spring Harb. Perspect. Biol. 8, a023614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Russo C., et al. , Protein aggregation of the p63 transcription factor underlies severe skin fragility in AEC syndrome. Proc. Natl. Acad. Sci. U.S.A. 115, E906–E915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nishitsuji K., Heparan sulfate S-domains and extracellular sulfatases (Sulfs): Their possible roles in protein aggregation diseases. Glycoconj. J. 35, 387–396 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Vivès R. R., Seffouh A., Lortat-Jacob H., Post-synthetic regulation of HS structure: The Yin and Yang of the Sulfs in cancer. Front. Oncol. 3, 331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishitsuji K., Saito H., Uchimura K., Enzymatic remodeling of heparan sulfate: A therapeutic strategy for systemic and localized amyloidoses? Neural Regen. Res. 11, 408–409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu C., Nezami Ranjbar M. R., Wu Z., DiCarlo J., Wang Y., Detecting very low allele fraction variants using targeted DNA sequencing and a novel molecular barcode-aware variant caller. BMC Genomics 18, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narahara M., et al. , Large-scale East-Asian eQTL mapping reveals novel candidate genes for LD mapping and the genomic landscape of transcriptional effects of sequence variants. PLoS One 9, e100924 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kayed R., et al. , Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Kayed R., et al. , Fibril specific, conformation dependent antibodies recognize a generic epitope common to amyloid fibrils and fibrillar oligomers that is absent in prefibrillar oligomers. Mol. Neurodegener. 2, 18 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baker S. J., Markowitz S., Fearon E. R., Willson J. K., Vogelstein B., Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249, 912–915 (1990). [DOI] [PubMed] [Google Scholar]
- 67.Niikura Y., Nonaka T., Imajoh-Ohmi S., Monitoring of caspase-8/FLICE processing and activation upon Fas stimulation with novel antibodies directed against a cleavage site for caspase-8 and its substrate, FLICE-like inhibitory protein (FLIP). J. Biochem. 132, 53–62 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Kayed R., Glabe C. G., Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 413, 326–344 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.