Significance
Contact dermatitis is an inflammatory skin disorder that arises from direct skin contact with irritants or allergens. Representing over 90% of occupational skin disorders, it has a considerable socioeconomic impact, and patients suffering from contact dermatitis can develop a notable physical handicap. Current diagnostic regimes rely on allergy testing, exposure specification, and follow-up visits. However, distinguishing the clinical phenotype of irritant and allergic contact dermatitis, which is important for appropriate therapeutic strategies, remains challenging. This study identifies and validates biomarkers to distinguish allergic and irritant contact dermatitis in human skin, to be used for the development of novel diagnostic methods and to guide clinical diagnosis.
Keywords: allergic contact dermatitis, irritant contact dermatitis, biomarker, machine learning, artificial intelligence
Abstract
Contact dermatitis tremendously impacts the quality of life of suffering patients. Currently, diagnostic regimes rely on allergy testing, exposure specification, and follow-up visits; however, distinguishing the clinical phenotype of irritant and allergic contact dermatitis remains challenging. Employing integrative transcriptomic analysis and machine-learning approaches, we aimed to decipher disease-related signature genes to find suitable sets of biomarkers. A total of 89 positive patch-test reaction biopsies against four contact allergens and two irritants were analyzed via microarray. Coexpression network analysis and Random Forest classification were used to discover potential biomarkers and selected biomarker models were validated in an independent patient group. Differential gene-expression analysis identified major gene-expression changes depending on the stimulus. Random Forest classification identified CD47, BATF, FASLG, RGS16, SYNPO, SELE, PTPN7, WARS, PRC1, EXO1, RRM2, PBK, RAD54L, KIFC1, SPC25, PKMYT, HISTH1A, TPX2, DLGAP5, TPX2, CH25H, and IL37 as potential biomarkers to distinguish allergic and irritant contact dermatitis in human skin. Validation experiments and prediction performances on external testing datasets demonstrated potential applicability of the identified biomarker models in the clinic. Capitalizing on this knowledge, novel diagnostic tools can be developed to guide clinical diagnosis of contact allergies.
Occupational skin disorders continue to be an important work-related disease, being ranked among the top five occupational diseases in many industrialized countries (1). Contact dermatitis (CD), representing over 90% of occupational skin disorders, causes a significant percentage of work disabilities and a great number of lost workdays, thus it has significant socioeconomic impact (2, 3). Patients suffering from CD can develop a considerable physical handicap leading to reduced quality of life (4).
Two major groups of CD are recognized, namely allergic contact dermatitis (ACD) and irritant contact dermatitis (ICD). ACD is a type IV hypersensitivity reaction that arises from topical exposure to agents with sensitizing potential. In contrast, ICD is caused by direct damage to the skin by exposure to physical or chemical agents, resulting in a nonspecific inflammatory skin reaction (5).
Clinical differentiation of ICD and ACD remains a challenging field in allergology and dermatology. Diagnostic practices include clinical history, physical examination, and diagnostic patch testing. Although patch testing is the current gold standard for ACD diagnosis, subjective clinical interpretation cannot be straightforward and standardized; thus, this approach is prone to errors leading to repeated and additional tests. Therefore, making an appropriate diagnosis and identifying the causative agents is of the greatest importance for proper therapeutic and preventive measures. Currently, available methods for fast and reliable diagnostics are insufficient or even lacking (6).
Several previous studies shed light on pathological processes during CD, identifying a diverse repertoire of biomarkers that can be used to characterize genetic susceptibility and inflammatory conditions in general (7–9). However, the plentitude of contact sensitizers and irritants represents a major challenge in the identification of robust and valid biomarkers. Furthermore, although ACD and ICD are mechanistically dissimilar, irritancy and allergy have multifarious features in common, sharing effector pathways, inflammatory mediators, cell recruitment, as well as cell-death–related phenomena (10). Therefore, simultaneous analysis of a broad range of causative agents may facilitate the identification of valuable biomarkers.
Herein, we performed patch testing using four contact sensitizers and two irritants with widely different physicochemical properties and high relevance to occupational exposures. Employing integrative transcriptome analysis and machine-learning approaches, we aimed to identify composite gene signatures for contact sensitizers and irritants, allowing a distinction between the two highly intertwined disease subtypes to unravel novel molecular biomarkers.
Results
Molecular Profiling of Contact Sensitizers and Irritants.
Positive patch test reactions to the sensitizers methylchloroisothiazolinone/methylisothiazolinone (CM), paraphenylenediamine (PP), epoxy resin, bisphenol A (EP), or nickel (NI), and irritant substances nonanoic acid (NO) or sodium lauryl sulfate (SL), were collected at 48 h postexposure from the patch test areas for analyses according to the scheme in Fig. 1A. The intensities of the allergic test reactions were rated using a scale of +, ++, or +++, conforming to the European guidelines on diagnostic patch testing (11). For the purpose of this study, irritants were rated as IR0 (slight erythema), IR (standard irritant reaction), IR1 (strong irritant reaction), and IR2 (very strong irritant reaction). Representative images and descriptions of clinical grading can be found in SI Appendix (SI Appendix, Fig. S1).
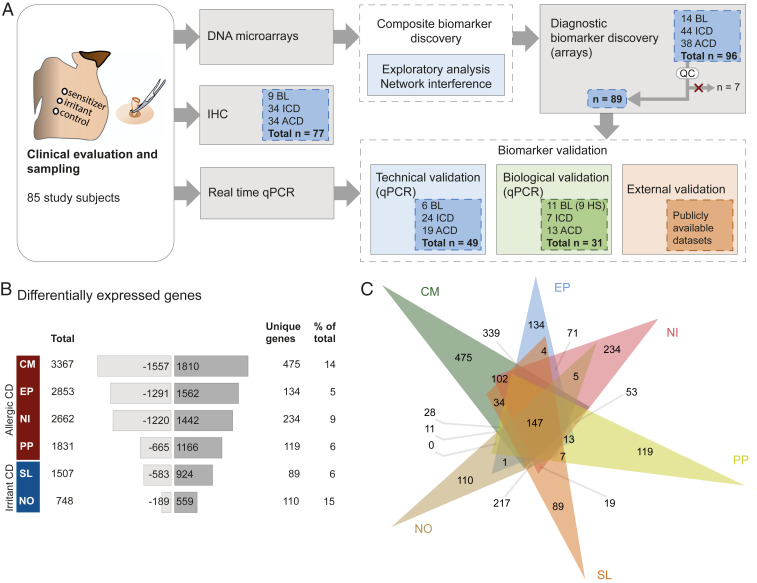
Fig. 1.
Overview of the study scheme and numbers of DEGs in human skin after exposure to allergens and irritants. (A) Schematic illustration of the study design, including total number of study participants (leftmost panel) and number of samples included in each assay (all other panels). Biomarker validation was performed at three levels: Technical (samples used in microarray) and biological validation (independent group of patients) by real-time qPCR, and external validation by testing the performance of the identified biomarkers (external datasets). (B) Numbers of DEGs, including the total number of genes, number of down- and up-regulated genes, and the number and ratio of unique genes. (C) Venn diagram depicting overlaps between exposure groups.
Skin biopsy sample collection was followed by RNA extraction and analysis of global gene-expression levels. Differential gene-expression analysis identified 3,367 differentially expressed genes (DEGs) between CM and baseline (BL), 2,853 DEGs between EP and BL, 2,662 DEGs between NI and BL, 1,831 DEGs between PP and BL, 1,507 DEGs between SL and BL, and 748 DEGs between NO and BL (Fig. 1B). Analysis of the extent of overlap between the groups revealed that the four tested contact sensitizers—PP, NI, EP, and CM—had the largest number of DEGs in common (641), indicating that these substances induce highly similar molecular perturbations during the elicitation phase, regardless of distinct chemical properties. In contrast, the two tested irritants, NO and SL, shared substantially fewer DEGs (339) with one another (Fig. 1C and SI Appendix, Fig. S2), indicating more heterogeneous reaction types. Notably, SL reactions shared 502 DEGs with the contact sensitizers, indicating a higher degree of similarities with contact sensitizers. Moreover, CM induced the largest number of unique DEGs (475), and NI the second largest number (234). All six exposures, including contact sensitizers and irritant substances, induced 152 common DEGs, which could be perceived as a core skin inflammatory response regardless of the type of exposure.
Contact Sensitizers and Irritants Cause Leukocyte Compositional Changes.
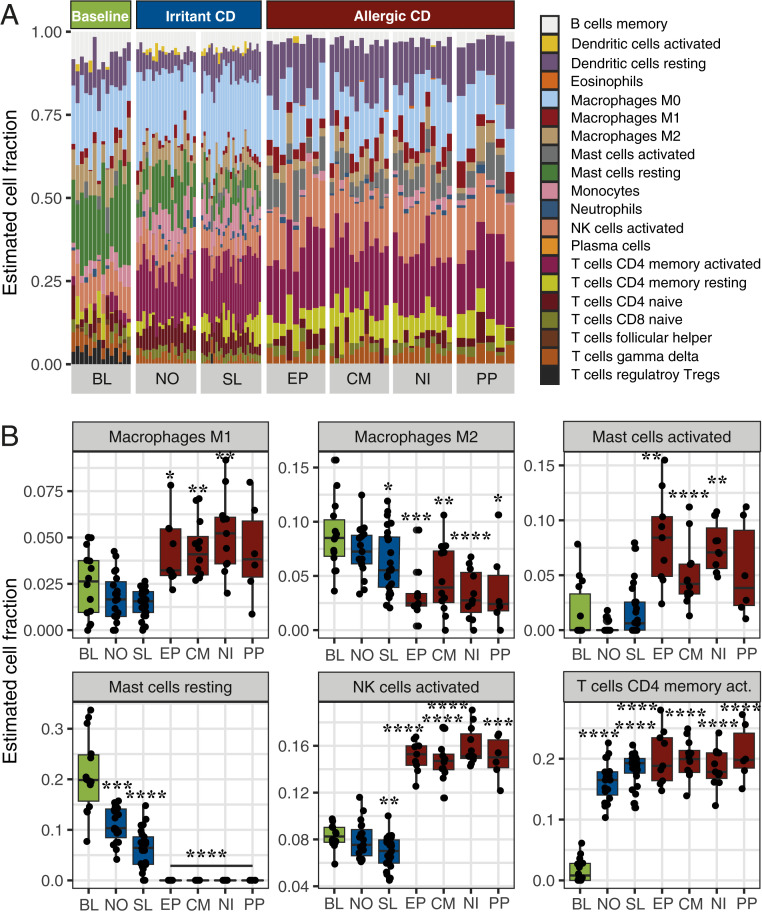
For further analysis of involved cell subsets, we employed a leukocyte deconvolution algorithm, which revealed the accumulation, activation, and polarization of various immune cells in the two investigated types of skin inflammation, allergies or irritant reactions, based on the cutaneous gene-expression profiles. In line with our immunohistochemical analyses, the analysis predicted significant accumulation of T cells in the exposed samples, including CD4+ and CD8+ activated memory and naive T cells. Moreover, the proportion of γδ T cells remained steady across the groups, while T regulatory cells appeared to be absent in all groups except BL. The allergic skin reactions were characterized by a significant accumulation of macrophages, particularly the proinflammatory M1 subset. Notably, there was a significant accumulation of natural killer (NK) cells, as well as activated mast cells, along with the disappearance of resting mast cells. In contrast, irritant reactions displayed a significant decrease in the proportions of resting mast cells and increased proportions of monocytes and T cells (Fig. 2 and SI Appendix, Fig. S3).
Fig. 2.
CIBERSORT analysis of the accumulation of leukocytes in treated skin areas based on the transcriptomes. (A) Overview of estimated cell fractions from gene-expression signatures. (B) Significantly changed estimated cell populations after chemical exposure. Boxplots display the mean and ± SD of estimated cell fractions. P values were generated by CIBERSORT using Monte Carlo sampling and the null hypothesis was tested by Pearson correlation. *P < 0.01; **P < 0.001; ***P < 0.0001; ****P < 0.00001.
Remarkably, our findings are mirrored within the functional enrichment. Pathways such as “Th1 and Th2 signaling,” “Role of macrophages,” and “Natural killer cell signaling” were highly enriched in the gene signatures of each contact sensitizer (SI Appendix, Fig. S4). Moreover, we observed the enrichment of “Aryl Hydrocarbon receptor signaling” in the gene signatures of all exposures. The irritant-induced gene-expression profiles were overrepresented mainly by genes involved in cell cycling (SI Appendix, Figs. S4 and S5A), and to a lesser extent by inflammation-related genes. Similar to the sensitizers, SL stimulated both T cell and dendritic cell activation (SI Appendix, Fig. S5B), however, at a significantly lower level compared to the contact sensitizers. Genes that were most potently stimulated by contact sensitizers included MMP12 and GZMB, while irritants induced S100 protein-coding genes several folds compared with baseline samples (SI Appendix, Fig. S6).
Contact Sensitizers and Irritants Induce Different Transcriptomic Profiles.
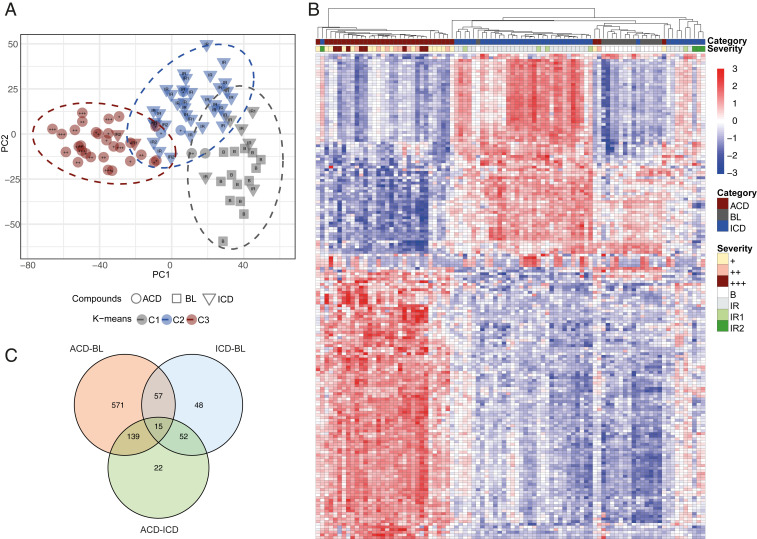
To further examine the extent of similarities and differences between the six exposure groups, we explored the grouping of the transcriptomes, using principal component analysis (PCA) and the k-means algorithm. Using PCA prior to cluster analysis is essential for building robust clustering results (12). Therefore, the PCA procedure was applied first, for projecting high-dimensional gene-expression data into a low-dimensional space, revealing trends in the grouping of ACD and ICD samples. Then, to obtain a clustering result, the k-means algorithm was applied to the first two principal components (PCs), which capture most of the variation in the original dataset. k-means clustering was computed several times by varying k from 1 to 10 clusters (or groups) for different values of k, and the best k was selected based on the maximum average silhouette. Visualization of the results by PCA, revealed three groups clearly separated from each other, including BL, ICD, and ACD, respectively (Fig. 3A). Notably, the reactions clustered further according to severity, with the strongest reactions accumulated to the left in each category (SI Appendix, Table S1). Hierarchical clustering revealed a similar grouping of exposures into three categories (Fig. 3B), and thus both hierarchical and nonhierarchical clustering algorithms demonstrated the presence of three main clusters. Analysis of the enrichment of biological functions based on gene signatures within each category revealed overrepresentation of gene ontology (GO) terms, such as “cytokine-mediated signaling pathway,” “inflammatory response,” and “T cell activation” within ACD, and “peptide cross-linking” and “keratinocyte differentiation” within ICD (SI Appendix, Table S2). Top up- and down-regulated genes within the categories are shown in SI Appendix, Table S3, and a Venn diagram in Fig. 3C illustrates overlaps between DEGs within ICD and ACD relative to BL.
Fig. 3.
The grouping of transcriptomes. The clustering of samples was analyzed by (A) using PCA and k-means algorithms, and by (B) hierarchical clustering, revealing three clearly separated groups; ACD, ICD, and BL. Reaction strength is graded +, ++, or +++ and IR, IR1, and IR2 for ACD and ICD reactions, respectively. In this figure, the reaction strength of BL samples (no reaction) is called “B.” (C) Venn diagram illustrating overlaps of DEGs between ACD and BL, ICD and BL, and ACD and ICD.
Network Analysis Reveals Key Features That Distinguish between Responses to Contact Sensitizers and Irritants.
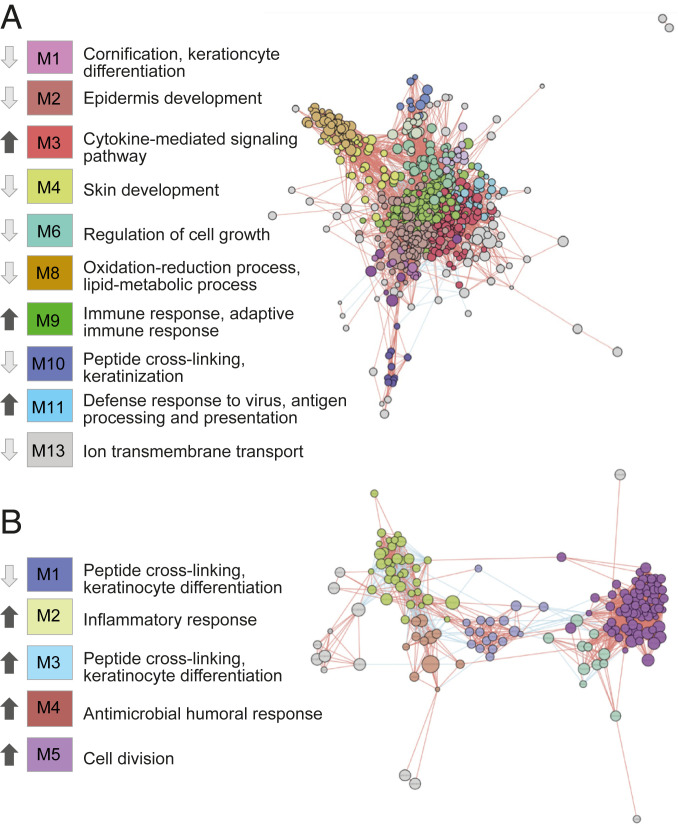
For further analysis of the importance of specific genes and clusters of functions in allergic and irritant inflammatory responses, molecular coexpression networks based on the transcriptomes of the single categories, ACD and ICD, were inferred using the novel tool inference of network response modules (INfORM) (13). The INfORM platform allows data-driven learning of regulatory connections where the algorithm identifies gene modules associated with function, and calculates the importance of genes within each module, based on centrality scores, fold change, and P values. We used all of the genes that were identified as significantly different between ACD and BL samples, and created a gene network (Fig. 4A). Genes were considered as linked in the network if their expression profiles were significantly correlated, and modules were defined based on the similarity of the gene-expression profiles, resulting in 13 modules. Each module of coexpressed genes contributed to specific aspects of ACD pathophysiology, including “cytokine-mediated signaling,” “adaptive immune response,” “antigen processing and presentation,” and “epidermis development.” The analysis revealed that modules enriched for immune signaling contained mostly up-regulated genes, whereas modules that were overrepresented by, for example, skin development, were predominantly down-regulated (Fig. 4A and Dataset S1). The corresponding ICD network (Fig. 4B and Dataset S2) comprised modules enriched for “inflammatory response,” “antimicrobial humoral response,” “cell division,” and “keratinocyte differentiation,” displaying a different pattern of up- and down-regulation compared with the ACD network. INfORM algorithms ranked ADAM8, GPR65, CLEC4A, GPR183, and CD47 as most important genes in the ACD networks, and MELK, CDK1, and RRM2 in the networks of ICD (Datasets S1 and S2). Finally, network inference based on the contrast between ACD and ICD revealed four modules enriched for cornification, cell division, immune signaling, and chemotaxis (SI Appendix, Fig. S7), and ADAM8, BATF, BATF3, and IL13 were ranked as the most important genes (Dataset S3). The analysis revealed up-regulation of immune signaling, and down-regulation of cornification, keratinization, and cell division in ACD compared to ICD (Dataset S3).
Fig. 4.
Network analysis of coexpressed genes associated with ACD or ICD. Networks were inferred based on the transcriptomes identified for positive patch tests against (A) allergens or (B) irritants. The networks were generated and visualized using the INfORM platform. Genes were considered linked in the network, if their expression profiles correlated positively (red edges) or negatively (blue edges). Modules in the network were defined based on the similarity of the gene expression profiles, and the indicated direction of gene expression (black arrow, up-regulated; light grey arrow, down-regulated) is based on average fold-changes of all genes of each module. Top enriched functions were identified by using EnrichR.
Genetic Algorithm-Based Feature Selection Provides Biomarker Models to Distinguish ACD from ICD.
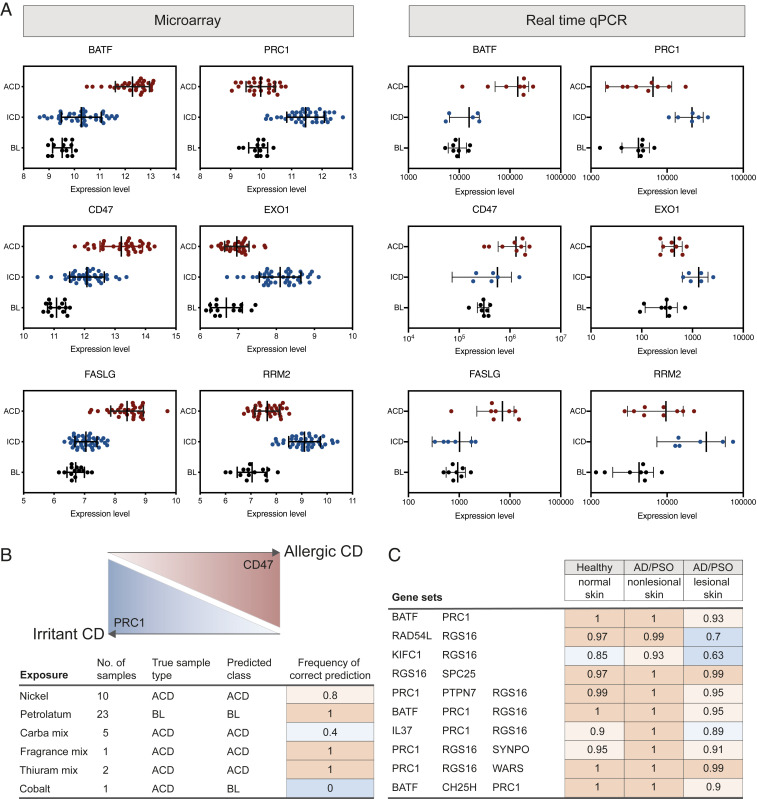
Our observations in the above analyses strongly supported the possibility of discovering biomarkers based on the transcriptomes, with a capacity to discriminate between the three categories: BL, ACD, and ICD. Therefore, we applied a biomarker discovery method, namely GARBO, which utilizes a genetic algorithm (GA) that is coupled with a Random Forest (RF) -based classifier (GA-RF) to optimize the number of features when testing the accuracy of omics-based biomarker panels. If the given classification task can be solved by using only one gene (or molecular feature), then the GARBO process will result in single gene biomarkers. The GA-RF approach was evaluated with fivefold cross-validation in order to estimate more robust classification performances. Using this approach, we discovered 28 novel gene sets, exhibiting high testing accuracy between BL, ACD, and ICD (Table 1). These gene sets employed two to three genes, which classified ACD, ICD, and BL, with an accuracy ranging between 86% and 94%. We also compiled performance metrics of each class, such as precision, recall, and F1-score, in order to investigate the performance of the selected biomarker models with respect to the individual classes: ACD, ICD, and BL (Table 1). We found that the biomarker models selected by GARBO could accurately distinguish ACD from ICD. Indeed, the F1-score reached, on average, 94% for ACD and 92% for ICD. However, the mean F1-score for the BL class was 84%. The biomarkers were validated by real-time qPCR in a small independent group of patients, confirming expression levels as identified by the microarrays (Fig. 5A). Technical validation of all samples by real-time qPCR is shown in SI Appendix, Fig. S8.
Table 1.
Gene sets selected by GARBO
| Accuracy | Precision-ACD | Precision-BL | Precision-ICD | Recall-ACD | Recall-BL | Recall-ICD | F1-score-ACD | F1-score-BL | F1-score-ICD | |
| IL37, SELE | 0.9185 | 0.9412 | 0.9286 | 0.8968 | 0.9412 | 0.8478 | 0.9262 | 0.9412 | 0.8864 | 0.9113 |
| BATF, PRC1 | 0.9222 | 0.9314 | 0.9048 | 0.9206 | 0.9500 | 0.8837 | 0.9.134 | 0.9406 | 0.8941 | 0.9170 |
| BATF, PKMYT1 | 0.9111 | 0.9706 | 0.7857 | 0.9048 | 0.9429 | 0.8462 | 0.9048 | 0.9565 | 0.8148 | 0.9048 |
| BATF, PBK | 0.8926 | 0.9216 | 0.8571 | 0.8810 | 0.9307 | 0.8182 | 0.8880 | 0.9261 | 0.8372 | 0.8845 |
| RAD54L, RGS16 | 0.8926 | 0.9314 | 0.8095 | 0.8889 | 0.9406 | 0.7907 | 0.8889 | 0.9360 | 0.8000 | 0.8889 |
| BATF, EXO1 | 0.9074 | 0.9118 | 0.9524 | 0.8889 | 0.9118 | 0.8511 | 0.9256 | 0.9118 | 0.8989 | 0.9069 |
| CD47, PRC1 | 0.9222 | 0.9118 | 0.9286 | 0.9286 | 0.9394 | 0.9286 | 0.9070 | 0.9254 | 0.9286 | 0.9176 |
| IL37, RGS16 | 0.8926 | 0.9216 | 0.8810 | 0.8730 | 0.9400 | 0.7708 | 0.9016 | 0.9307 | 0.8222 | 0.8871 |
| KIFC1, RGS16 | 0.9259 | 0.9412 | 0.8571 | 0.9365 | 0.9505 | 0.8182 | 0.9440 | 0.9458 | 0.8372 | 0.9402 |
| BATF, IL37 | 0.9111 | 0.9412 | 0.9286 | 0.8810 | 0.9143 | 0.8478 | 0.9328 | 0.9275 | 0.8864 | 0.9061 |
| HIST1H1A, SELE | 0.8926 | 0.9608 | 0.8571 | 0.8492 | 0.9515 | 0.7200 | 0.9145 | 0.9561 | 0.7826 | 0.8807 |
| BATF, TPX2 | 0.9074 | 0.9412 | 0.8810 | 0.8889 | 0.9320 | 0.8409 | 0.9106 | 0.9366 | 0.8605 | 0.8996 |
| PRC1, SELE | 0.9185 | 0.9412 | 0.8810 | 0.9127 | 0.9320 | 0.8222 | 0.9426 | 0.9366 | 0.8506 | 0.9274 |
| RGS16, SPC25 | 0.9148 | 0.9314 | 0.8571 | 0.9206 | 0.9406 | 0.8000 | 0.9355 | 0.9360 | 0.8276 | 0.9280 |
| FASLG, RRM2 | 0.9296 | 0.9118 | 0.8571 | 0.9683 | 0.9688 | 0.8000 | 0.9457 | 0.9394 | 0.8276 | 0.9569 |
| RRM2, SELE | 0.9148 | 0.9412 | 0.8571 | 0.9127 | 0.9412 | 0.7347 | 0.9664 | 0.9412 | 0.7912 | 0.9388 |
| LYPD2, SELE | 0.8963 | 0.9412 | 0.7857 | 0.8968 | 0.9600 | 0.7857 | 0.8828 | 0.9505 | 0.7857 | 0.8898 |
| PRC1, PTPN7, RGS16 | 0.9333 | 0.9510 | 0.8810 | 0.9365 | 0.9604 | 0.8409 | 0.9440 | 0.9557 | 0.8605 | 0.9402 |
| IL37, RRM2, SELE | 0.9370 | 0.9412 | 0.9286 | 0.9365 | 0.9697 | 0.7800 | 0.9752 | 0.9552 | 0.8478 | 0.9555 |
| BATF, PRC1, RGS16 | 0.9259 | 0.9608 | 0.8571 | 0.9206 | 0.9800 | 0.7660 | 0.9431 | 0.9703 | 0.8090 | 0.9317 |
| IL37, PRC1, RGS16 | 0.9296 | 0.9412 | 0.8810 | 0.9365 | 0.9697 | 0.8409 | 0.9291 | 0.9552 | 0.8605 | 0.9328 |
| PRC1, RGS16, SYNPO | 0.9296 | 0.9608 | 0.8571 | 0.9286 | 0.9703 | 0.8000 | 0.9435 | 0.9655 | 0.8276 | 0.9360 |
| PRC1, RGS16, WARS | 0.9222 | 0.9314 | 0.9048 | 0.9206 | 0.9596 | 0.8444 | 0.9206 | 0.9453 | 0.8736 | 0.9206 |
| DLGAP5, IL37, SELE | 0.9259 | 0,9510 | 0.9048 | 0.9127 | 0.9327 | 0.8837 | 0.9350 | 0.9417 | 0.8941 | 0.9237 |
| IL37, SELE, TPX2 | 0.9222 | 0.9412 | 0.8571 | 0.9286 | 0.9412 | 0.9000 | 0.9141 | 0.9412 | 0.8780 | 0.9213 |
| IL37, SELE, THBS2 | 0.9481 | 0.9804 | 0.9286 | 0.9286 | 0.9709 | 0.8667 | 0.9590 | 0.9756 | 0.8966 | 0.9435 |
| LYPD2, PBK, SELE | 0.9148 | 0.9412 | 0.8571 | 0.9127 | 0.9600 | 0.7500 | 0.9426 | 0.9505 | 0.8000 | 0.9274 |
| BATF, CH25H, PRC1 | 0.9185 | 0.8922 | 0.8810 | 0.9524 | 0.9681 | 0.8409 | 0.9091 | 0.9286 | 0.8605 | 0.9302 |
The feature selection process coupled with RF-based classifiers enables the discovery of sets of biomarkers that together can guarantee the classification of the samples in the three classes, BL, ACD, and ICD. The table shows the classification accuracy and its SD calculated on the training and test sets. The stability indicates the occurrence frequency of each model across the five runs.
Fig. 5.
Validation of biomarkers selected by GARBO. (A) Expression levels of selected biomarkers measured by arrays (panels on the Left) and validated by real time qPCR in an independent group of patients (panels on the Right). Error bars correspond to SD. Selected combinations of biomarkers were tested in external datasets of (B) ACD (access code GSE60028) and (C) atopic dermatitis (AD) and psoriasis (PSO) (access code E-MTAB-8149), revealing predictive potential of the biomarkers, reported as the frequency of correct prediction. GSE60028 was used to demonstrate the generalizability of the selected biomarker models on new, external ACD samples; the second external testing dataset (E-MTAB-8149) aimed to prove the uncertainty of the biomarker models when trying to classify lesional and nonlesional skin in psoriasis and atopic dermatitis.
Testing of Identified Gene Sets in Independent Datasets Confirms Predictive Potential of the Biomarkers.
Selected gene sets were tested on microarray gene-expression data derived from two previously published, independent omics studies (8, 14). First, an RF-based classifier with 500 trees was trained for each of the 28 selected biomarker sets by using the entire gene-expression datasets (89 samples) provided in this study. The resulting trained models were then tested in the external microarray datasets. The study by Dhingra et al. (8) (dataset GSE60028, available from Geo DataSets, NCBI) includes gene-expression profiles induced by contact sensitizers (i.e., NI, fragrance, and rubber) compared with petrolatum. The rationale to test this external dataset was to assess the generalizability of the selected biomarker models on new, untested samples. In our analysis, two biomarker models (“CD47, PRC1” and “PRC1, RGS16, SYNPO”) achieved good prediction results (80%). Importantly, we observed that the two biomarker sets correctly classified NI-, thiuram-, and fragrance-exposed samples (Fig. 5B and SI Appendix, Table S4), two of which (thiuram and fragrance mix) were not used by us for biomarker identification.
Furthermore, a study by Fyhrquist et al. (14) (available from EBI ArrayExpress under accession E-MTAB-8149), provided gene-expression profiles from BL samples, lesional and nonlesional skin in psoriasis and atopic dermatitis. The second external testing dataset was particularly useful for assessing the uncertainty of the trained classifiers when they are tested on gene-expression profiles of psoriasis or atopic dermatitis samples. This is important from the point of view that both psoriasis and atopic dermatitis frequently confound ACD and ICD diagnostics, and consequently, biomarkers for ACD or ICD should be disease specific and not recognize other skin inflammatory conditions (7, 11). In our assessment, 10 of the biomarker sets performed as expected in the dataset by Fyhrquist et al. (14), by classifying control samples accurately as BL, while psoriasis and atopic dermatitis nonlesional and lesional skin remained unclassified (Fig. 5C and SI Appendix, Table S5). More technical details on external validation tests are reported in SI Appendix.
The Identified Biomarkers and Immune Cell Infiltration Are Associated with Clinical Severity.
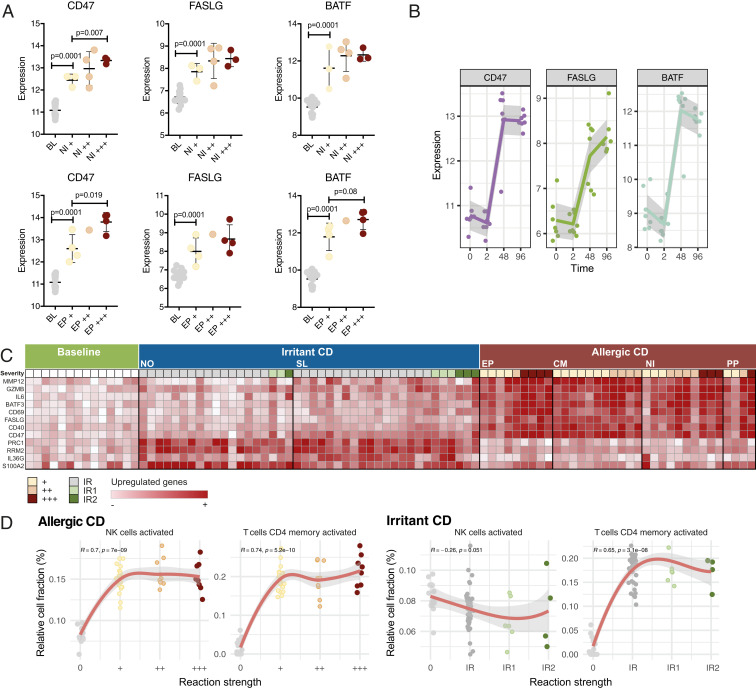
The strength of the reactions to the contact sensitizers, as determined by the dermatologists, associated with the extent of gene expression: That is, the more severe the reaction, the higher the number of dysregulated genes (SI Appendix, Fig. S9A). To ensure that the identified biomarkers are applicable in all degrees of inflammation, ranging from mild to severe reactions, we investigated their expression in each severity category and found that even in the mildest reactions, all identified biomarkers were significantly induced, and their expression levels correlated positively with the strength of the reaction (Fig. 6A and SI Appendix, Fig. S9 B and C). Moreover, we performed a time-course analysis limited to NI+ reactions, observing the kinetics of the expression of three of the identified biomarkers: CD47, BATF, and FASLG. None of them were induced at early time points (2 h), but potently expressed at 48 h, and maintained throughout 96 h postexposure (Fig. 6B). The expression of a selection of genes after allergen or irritant exposure, related to the strength of the reaction, is illustrated in Fig. 6C. Finally, correlating the accumulation of leukocytes (estimated by deconvolution analysis of the transcriptome) in the treated skin areas with reaction strength, revealed associations with activated memory CD4 T cells and activated NK cells, respectively (Fig. 6D). Intriguingly, our analysis shows a clear dependence on NK cells in ACD, but not in ICD, whereas both reactions associate with activated CD4 memory T cells (Fig. 6D), primarily driven by the contrast between the baseline and the positive reactions.
Fig. 6.
Biomarker expression and leukocyte infiltration as a function of severity and time. (A) The expression of selected biomarkers in response to NI (Upper) and EP (Lower) relative to severity. P values were generated by unpaired t test and error bars correspond to SD. (B) The expression of selected biomarkers at time points 2, 48, and 96 h after NI exposure. (C) An overview of ACD and ICD associated key genes in relation to reaction strength (allergic reactions graded +, ++, or +++; irritant reactions graded IR, IR1, or IR2). (D) Spearman’s rank correlation between the proportion of immune cells (estimated by deconvolution analysis of transcriptomes) and reaction strengths reveal dependence of both ACD and ICD on activated CD4 memory T cells, but of ACD only, on activated NK cells.
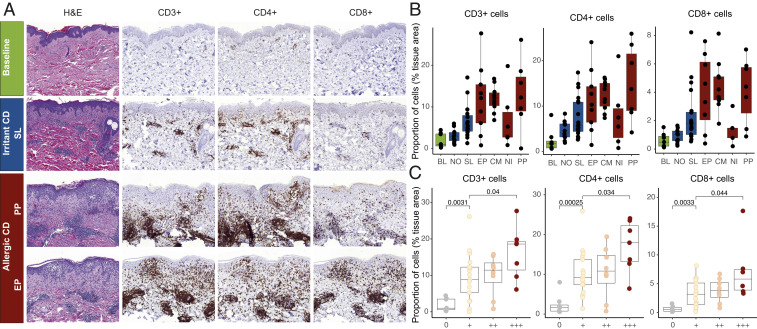
Confirmation of Leukocyte Compositional Changes by Immunohistochemical Analyses.
A second skin biopsy sample was collected from the same skin sites for comparative histological analyses. Chemical-exposed tissues displayed clearly visible perivascular accumulation of lymphocytes and inflammatory involvement of the epidermis (Fig. 7A). Immunohistochemical (IHC) staining and analysis using neural network-based algorithms for recognition of positively stained cells in the tissue, revealed statistically significant accumulation of CD3+, CD4+, and CD8+ cells in the reactions to contact sensitizers (Fig. 7B and SI Appendix, Table S6). Moreover, a significant association was observed between the infiltration of CD3+, CD4+, and CD8+ T lymphocytes and severity in ACD (Fig. 7C), supporting our transcriptomics-based analyses. The association between severity of the response to irritants and the inflammatory infiltrate was nonsignificant, likely due to a low number of samples in the high severity categories (SI Appendix, Fig. S10). SI Appendix, Fig. S11 illustrates the recognition of positively stained cells by deep learning algorithms.
Fig. 7.
IHC staining of chemical exposed human skin. (A) Representative images of BL, SL-, PP-, and EP-exposed samples (original magnification ×10) displaying a larger infiltration of CD3+, CD4+, and CD8+ cells in response to allergen exposure (PP and EP). (B) Positively stained cells identified by deep learning algorithms for each exposure group. Groups were compared using Dunn’s multiple comparison test (results available in SI Appendix, Table S6). (C) Significant association between T lymphocyte infiltration and the severity of the reaction (graded +, ++, and +++). Statistical analysis was performed using Mann–Whitney U test and P values of <0.05 were considered significant. Boxplots display mean, and first and third quartiles.
Discussion
Distinguishing ACD from skin irritation and other dermatoses remains a challenge to dermatologists, and therefore searching for biomarkers that reliably differentiate between the two conditions is essential. Using integrative transcriptome analysis and machine-learning–driven biomarker discovery, we identified robust gene sets for the distinction between the two highly interwoven diseases. We tested these biomarker sets in an independent patient group suffering from CD and demonstrate their potential applicability in reactions induced by agents different from those that were used for biomarker discovery. Thus, we provide high-potential molecular biomarker candidates for further clinical evaluation.
Just a handful of studies have explored the skin for diagnostic markers, and many of them have evaluated only a limited number of markers. A study by Corsini and Galli (15) focused on cytokines and chemokines for the differentiation between ICD and ACD and concluded that while both allergens and irritants induce TNF, GM-CSF, and IFN-γ, instead IL-1A, IL-12 and IL-1B might be useful for the identification of skin allergen-induced reactions. Furthermore, Meller et al. (16) suggested that T cell-driven chemokines, such as CXCL9, CXCL10, and CXCL11 may discriminate between ACD and ICD. Later on, the development of techniques, such as microarray chips and RNA sequencing, allowed for more extensive gene profiling, resulting in studies that deepened our understanding of mechanisms and identified further potential biomarkers. Clemmensen et al. (17) showed that although the irritants NO and SL elicit widely different gene-expression profiles in the skin, they shared 23 biomarkers, which could be used for the identification of cumulative ICD. Moreover, transcriptomics studies in in vitro models developed for chemical safety testing have pinpointed sets of genes that may distinguish between sensitizing and irritant compounds, at least in simplified in vitro models (18). Finally, Dhingra et al. (8) have reported unique gene-activation patterns between a selection of contact sensitizers, concluding that ACD may not be considered as a single entity. Here, we show that although diverse contact sensitizers induce slightly different sets of genes, they also share a remarkable number of common genes, which are distinct from those stimulated by irritants. This allowed for the identification of in vivo biomarkers that distinguished between the two categories—irritants and sensitizers—regardless of the physicochemical properties of the inducing agents.
Unsupervised, data-driven analysis of the clustering of the different exposures gave great promise for biomarker discovery, for the purpose of distinguishing between the two reaction types, ACD and ICD. Each agent was assigned to either category, ACD or ICD, based on the literature. The tested compounds (i.e., EP, CM, NI, and PP) are well-defined agents that commonly cause contact allergies in occupational settings (19–23), and the two irritants, SL and NO, are likewise well-defined skin irritants in the literature (17, 24). Combining the exposures into single categories, either ACD or ICD, revealed gene signatures and functions highly representative of each category, including a prominent inflammatory response in the former and mainly skin development and cell cycling in the latter. Furthermore, network analysis allowed for singling out of important functions and related genes within the two categories, highlighting cytokine signaling and adaptive immunity in ACD, and cell division and inflammatory response in ICD. Genes that were assigned the highest importance within the networks included ADAM8 and CD47 in ACD and RRM2 and SPC25 in ICD. ADAM8 is implicated in various inflammatory diseases by regulating cell recruitment and activation (25, 26), and likely plays a central role in angiogenesis (27). CD47 is a widely expressed transmembrane protein, particularly in NK cells (28), and is important by regulating cell migration and phagocytosis, and by promoting the proliferation of T cells and activation of cytotoxic T lymphocytes (CTLs) (29). Finally, the ICD-specific genes, RRM2 and SPC25, are both involved in functions related to cell cycling (30, 31).
Differential gene expression is important for understanding the biology underlying allergic skin inflammation and skin irritation, but it is not sufficient to optimally address the biomarker discovery problem. Methods that identify significant genes by monovarietal statistical tests (e.g., the empirical Bayes moderated t-statistics), where each gene is considered as independent from the others, are known to perform poorly in external validation tests. Instead, multivariate methods enable the identification of sets of biomarkers with superior diagnostic and prognostic performance compared to single markers in the context of sensitivity, specificity, and robustness, as synergies and antagonisms between potential biomarkers are taken into consideration. Thus, by using GARBO (32), a wrapper approach that couples a GA with an RF-based classifier, we identified an optimal set of biomarkers. The list of biomarkers that this approach generated overlapped our lists of functionally important genes—including CD47, BATF, FASLG, SELE, and IL37—suggesting that in addition to being biomarkers with a diagnostic relevance to distinguish between ACD and ICD, these molecules play key roles in the pathomechanisms of ACD and may therefore also constitute important targets of therapy. The transcription factor BATF, for example, is a key regulator of the differentiation of effector CTLs (33), key players in ACD (34). Moreover, BATF is critical for the differentiation of Th17 cells (35), which have been reported to be involved in the immunopathology of ACD (8, 36). Furthermore, FASLG is a central player in the induction of cell death. By binding to keratinocytes presenting cognate peptides on their MHC I, CTLs induce antigen-specific apoptosis via FAS–FASL interactions. In contrast, NK cells that accumulate in ACD (37), induce target cell death by FASL-dependent cytotoxicity in a nonspecific manner (38). SELE, which encodes E-selectin, in turn, is an important constituent of the beginning of the process of rolling and homing of T cells (39). Finally, IL-37 is a potent inhibitor of innate immune signaling, expressed by effector memory T cells and macrophages, and with an established immunoregulatory role in skin inflammatory disease (40). Thus, the identified biomarkers are part of highly relevant mechanisms in the context of ACD and skin inflammation in general. Finally, our analyses revealed the induction of most biomarkers even in weak reactions to the causing agents, indicating potential as effective diagnostic indicators of allergic or irritant reactions even in the mildest cases. Of note, in this study we accept a potential variability in the grading of severity due to different dermatologists reading the patch tests, and acknowledge that quantitative measurement of erythema (by a colorimeter) is warranted in future studies.
In conclusion, our analysis reveals putative key players in CD, including both common and unique features of ACD and ICD reactions. While T cell-driven immune signaling is a characteristic of the ACD response, T cells also participate in ICD responses, and thus infiltration of T cells per se is a poor differentiator between the two types of skin inflammation. Instead, the presence of innate immune cells, such as macrophages and NK cells, and related signaling makes the difference and is apparently the result of antigen-specific T cell-driven signaling and cross-talk between the two arms of immunity, resulting in the amplification of the response in ACD. Importantly, the identified biomarkers were validated by qPCR in an independent group of patients and distinguished between the two conditions with high accuracy in external datasets (8, 14). The identified sets of genes correctly categorized a wide range of exposing agents, including chemicals that were not part of the biomarker identification process. Moreover, the gene sets did not recognize other types of dermatoses, such as atopic dermatitis or psoriasis, giving promise for a robust, reliable, and unique set of biomarkers for differentiating between ACD and ICD, to be validated further in a prospective clinical study. Finally, the identified biomarkers represent key features of skin inflammation and repair, which might—in addition to serving as biomarkers for diagnostic purposes—prove to be efficient indicators of disease risk or improvement.
Materials and Methods
Patient Recruitment.
Dermatologists at the University of Helsinki and the Finnish Institute of Occupational Health selected patients (n = 85) to be included in the study based on patch test readings and clinical history. Patients over 18 y of age were included in the study, and care was taken to have age- and gender-matched representation. An independent group of patients and samples were collected for validation purposes (Fig. 1A). Exclusion criteria included extensive or disseminated eczema or other skin disorders. Patient characteristics are summarized in Tables 2 and 3 and SI Appendix, Table S7. Skin preparation instructions included avoiding the application of topical emollients for 24 h and topical medication for 2 wk at sampling sites prior to the sampling time point. The experimental protocol followed the Declaration of Helsinki principles and was approved by the Ethics Committee of Helsinki University Central Hospital (approval numbers 43/13/03/00/14 and §214/3.9.2014, dnro 171/13/03/01/2014). All participants signed an informed consent form.
Table 2.
Summary of patient characteristics: Allergic reactions
| Agent | Frequency | Level of reaction | Gender F/M | Age mean ± SD | ||
| + | ++ | +++ | ||||
| CM | 17 | 11 | 6 | 0 | 10/7 | 36 ± 13 |
| EP | 12 | 6 | 1 | 5 | 4/8 | 43 ± 14 |
| NI | 11 | 3 | 4 | 4 | 10/1 | 38 ± 15 |
| PP | 7 | 3 | 1 | 3 | 7/0 | 40 ± 15 |
The severity of allergic reactions is graded +, ++, or +++. F, female; M, male.
Table 3.
Summary of patient characteristics: Irritant reactions
| Agent | Frequency | Level of reaction | Gender F/M | Age mean ± SD | |||
| IR0 | IR | IR1 | IR2 | ||||
| NO | 23 | 3 | 15 | 4 | 1 | 13/10 | 43 ± 14 |
| SL | 26 | 1 | 18 | 4 | 3 | 17/9 | 42 ± 14 |
The severity of irritant reactions is graded IR0, IR, IR1, or IR2. F, female; M, male.
Patient Sample Collection.
Patch testing was performed in accordance with the European Society of Contact Dermatitis guidelines (11) for four allergens (1% EP, 1% PP, 0.02% CM, and 5% NI; Chemotechnique) and two irritants (2% SL, 17.5% NO). A detailed description of the procedure is described in SI Appendix. Two 3-mm punch biopsies were obtained from the test areas when a reaction was seen. From each subject a total of four 3-mm biopsies were obtained, including two from an ACD reaction and two from an ICD reaction. Moreover, biopsies were collected from healthy skin (HS). One biopsy was immersed in RNAlater liquid and stored at −80 °C until further analysis. The second 3-mm biopsy was fixed in formalin and embedded in paraffin for screening and validation of biomarkers by IHC. Negative ACD reactions were collected from part of the patients to represent BL samples, because of a limit to the ethical permit. HS samples were obtained under a different ethical permit. A comparison of the negative reactions and HS showed that gene-expression levels between these two categories were fully comparable (SI Appendix, Fig. S12), and thus, negative reactions and HS were considered as BL samples.
Transcriptomics Data Generation and Analysis.
DNA and RNA were extracted from skin biopsies stored in RNAlater solution using the AllPrep DNA/RNA mini kit (Qiagen) according to the manufacturers’ instructions and only samples with an RNA integrity number > 8 were used for further analysis. SurePrint G3 Human Gene Expression Microarrays (Agilent) were used for transcriptomic analysis according to the manufacturers’ instructions. A detailed description is provided in SI Appendix, Supplementary Methods. Genes were defined as being differentially expressed after fulfilling the requirements of a minimum fold-change of ±1.5 and a maximum, Benjamini–Hochberg-adjusted, P value of 0.05.
Unsupervised Exploratory Analysis.
Samples were grouped according to their expression profiles by using k-mean (with k = 3) and hierarchical clustering algorithms. PCA and heat maps were used to visualize the grouping. PCA was applied to the entire gene-expression datasets, while the heat map was generated using the expression profiles of DEGs across the different comparisons (e.g., EP-BL, SL-BL, and so forth). This analysis was carried out in the R environment.
Leukocyte Deconvolution.
The CIBERSORT algorithm (41) was applied to the gene-expression data to estimate the leukocyte subset proportions of 22 individual immune cells utilizing the “LM22” validated gene-signature matrix as provided from the algorithm. Estimations are based on 1,000 permutations. No significance filter has been applied to the estimated cell fractions to include all samples for further analysis. Correlations between the proportions of leukocytes and reaction strengths were explored by Spearman’s rank correlation (R package ggpubr) (42).
INfORM.
INfORM was applied to generate coexpression networks and to identify gene modules associated with the biological processes and regulatory mechanisms differentiating the expression and regulation of ACD and ICD. First, a robust gene coexpression network was derived by combining multiple networks inferred from the gene-expression profiles. The compendium of gene networks was generated by using different network inference algorithms, including ARACNE (43), MRNET (44), CLR (45), and mutual information-based methods (46). The compiled networks were then merged to define an ensemble gene network that includes only high-confidence edges. A detailed description of the adopted network integration strategy can be found in Marwah et al. (13). Second, gene modules were identified in the network by using community detection algorithms and evaluated based on multiple metrics of node importance. The genes within each module were then ranked based on their centrality scores, differential log2 (fold-change), and differential P value. Moreover, enrichment analysis was performed to find the GO terms overrepresented in each module and compute the similarity between sets of GO terms from different modules.
Biomarker Discovery.
A supervised machine-learning–based approach was used in order to identify sets of genes (or composite biomarkers) whose expression can jointly classify (or distinguish) skin biopsy samples into three groups: ACD, ICD, and BL. Each sample gene-expression profile was first labeled as ACD, ICD, or BL, based on the patch test results. True positive patch testing results against EP, PP, CM, or NI were labeled as ACD, while true positive patch testing results against SL and noncorrosive irritant NO were labeled as ICD. Negative ACD reactions were used to represent BL samples. In order to address the defined ternary ( classes) classification problem, we applied GARBO (32). GARBO implements a wrapper GA coupled with RF-based classifiers in order to search for composite biomarkers that yield the minimum number of genes with maximum accuracy. If one gene is good enough to distinguish the three classes (ACD, ICD, or BL), then GARBO will identify biomarker panels consisting of one feature or gene. However, if GARBO finds biomarker panels including two or more than two genes, then this means that a composite (or multivariate) biomarker model is needed to obtain the highest classification scores. The proposed feature selection strategy is evaluated with fivefold cross-validation in order to simulate different training sets and test the selected gene sets on unseen data. The classification performances are measured using Precision, Recall, F-Measure, and Accuracy rate. Moreover, independent gene-expression datasets are used in order to evaluate the performance of the trained classifiers on independent datasets.
qPCR.
Validation of biomarkers was performed by real-time qPCR in a separate group of patients. mRNA levels of BATF, PRC1, CD47, EXO1, FASLG, and RRM2 (SI Appendix, Table S8) were analyzed by quantitative RT-PCR using Taqman chemistry and the 7500 Fast Real-Time PCR System (Applied Biosystems, Life Technologies). A detailed description is provided in SI Appendix, Supplemental Methods. The results are expressed as relative units (RU), which was calculated by the comparative CT method according to the manufacturer’s instructions (47). Statistical significance was assessed by one-way ANOVA, and P values of <0.05 were considered significant. Statistical analysis and creation of graphs were done in PRISM 8 OS X (GraphPad) and results are presented as ±SD.
Histology and IHC.
Paraffin-embedded, 4-µm-thick skin tissue sections were stained for CD3 (RTU, clone 2GV6, Roche 790-4341), CD4 (1:100 clone SP35, Cell Marque, 104R-16), and CD8 (1:50, clone 4B11, Novocastra, NCL-L-CD8-4B11), using the Ventana Benchmark Ultra instrument (Roche). Tissue sections were pretreated in cell conditioning 1 buffer (pH 8.5) (Roche 950-124) at 98 °C for 64 min, followed by incubation with the antibodies at 37 °C for 44 min. The multimer-based detection kit, Ultraview (Roche 760-700) was used for the detection of CD3, and CD8. Optiview (Roche 760-700) was used to detect CD4. The reactions were visualized with diaminobenzidine (DAB) and counterstained with hematoxylin.
Digitization of Samples.
The anti-CD3, anti-CD8, and anti-CD4 immunostained skin biopsy slides were digitized with a whole-slide scanner (Pannoramic 250 FLASH, 3DHISTECH) equipped with a 20× objective (numerical aperture 0.8) and a 1× adapter, and a global shutter CMOS camera with 4,096 × 3,072 pixels sized 5.5 μm × 5.5 μm (Adimec QUARTZ Q-12A180 camera), resulting in an image in which one pixel represents an area of 0.24 μm × 0.24 μm. Images were stored in a whole slide image format (MRX, 3DHISTECH) and further compressed to a wavelet file format (Enhanced Compressed Wavelet, ECW, ER Mapper, Intergraph) with a target compression ratio of 1:10. The compressed virtual slides were uploaded to a whole-slide image management server (Aiforia Technologies Oy).
Deep Learning-Based Algorithms for Detecting Immunostained Tissue Areas.
The two convolutional neural network-based algorithms (48) were trained using cloud-based software (Aiforia Create, Aiforia Technologies Oy) for image analysis to separately identify tissue areas, as well as the areas stained with DAB, which was used as chromogen for the detection of specific anti-CD3, anti-CD4, and anti-CD8 antigen. The algorithm involves two algorithms in sequence, the first segmenting the tissue areas and the second identifying DAB-stained areas which reflect specific immunostainings. The deep learning network for the detection of tissue areas was trained on anti-CD8 immunostained tissue slides on 35 samples across the expositions as well as the BL. The 500 image annotations used for training the algorithm for the tissue layer corresponded to an area of 1.13-mm2 tissue. The algorithm for detecting anti-CD8 stained areas was trained across 67 samples with 430 image annotations corresponding to an area of 0.053-mm2 tissue. We used a feature size of 100 μm for training the algorithm for detection of the tissue areas and feature size of 20 μm for training the areas with CD8 expression. The augmentation parameters for detection of tissue were 0° to 360° rotation, ±30% scale, ±20% aspect ratio change, ±20% shear distortion, ±20% luminance, and ±20% contrast change and augmentation parameters for the detection of CD8 expression were 0° to 360° rotation, ±10% scale, ±20% aspect ratio change, ±20% shear distortion, ±1% luminance, and ±1% contrast change. All data were flipped both vertically and horizontally. The final algorithms were trained for 10,000 iterations. The same algorithms that were trained for detecting anti-CD8 immunostained tissue areas (SI Appendix, Fig. S11) were used for detecting CD3 and CD4. Prism 8 OS X (GraphPad) and R Studio (v1.2.5001) were used to perform statistical analyses and create graphs (R package ggpubr and ggplot2). Statistical analysis included Dunn’s multiple comparisons test and Mann–Whitney U test, and P values of <0.05 were considered significant.
Supplementary Material
Acknowledgments
This research has received funding from the Finnish Work Environment Fund (project 113314) and from Forskningsrådet för Hälsa, Arbetsliv och Välfärd (FORTE, diarienr. 2018-00601).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. S.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009192117/-/DCSupplemental.
Data Availability.
Original expression microarray data have been deposited in the ArrayExpress, https://www.ebi.ac.uk/arrayexpress/ (accession no. E-MTAB-9501).
References
- 1.Keegel T., Moyle M., Dharmage S., Frowen K., Nixon R., The epidemiology of occupational contact dermatitis (1990-2007): A systematic review. Int. J. Dermatol. 48, 571–578 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Diepgen T. L., et al. , Prevalence of contact allergy in the general population in different European regions. Br. J. Dermatol. 174, 319–329 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Carøe T. K., Ebbehøj N. E., Bonde J. P., Agner T., Occupational hand eczema and/or contact urticaria: Factors associated with change of profession or not remaining in the workforce. Contact Dermat. 78, 55–63 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Moberg C., Alderling M., Meding B., Hand eczema and quality of life: A population-based study. Br. J. Dermatol. 161, 397–403 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Tan C. H., Rasool S., Johnston G. A., Contact dermatitis: Allergic and irritant. Clin. Dermatol. 32, 116–124 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Slodownik D., Lee A., Nixon R., Irritant contact dermatitis: A review. Australas. J. Dermatol. 49, 1–9, quiz 10–11 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Koppes S. A., et al. , Current knowledge on biomarkers for contact sensitization and allergic contact dermatitis. Contact Dermat. 77, 1–16 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Dhingra N., et al. , Molecular profiling of contact dermatitis skin identifies allergen-dependent differences in immune response. J. Allergy Clin. Immunol. 134, 362–372 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Quaranta M., et al. , Intraindividual genome expression analysis reveals a specific molecular signature of psoriasis and eczema. Sci. Transl. Med. 6, 244ra90 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Martin S. F., New concepts in cutaneous allergy. Contact Dermat. 72, 2–10 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Johansen J. D., et al. , European Society of Contact Dermatitis guideline for diagnostic patch testing—Recommendations on best practice. Contact Dermat. 73, 195–221 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Ben-Hur A., Guyon I., Detecting stable clusters using principal component analysis. Methods Mol. Biol. 224, 159–182 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Marwah V. S., et al. , INfORM: Inference of NetwOrk Response Modules. Bioinformatics 34, 2136–2138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fyhrquist N., et al. , Microbe-host interplay in atopic dermatitis and psoriasis. Nat. Commun. 10, 4703 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corsini E., Galli C. L., Epidermal cytokines in experimental contact dermatitis. Toxicology 142, 203–211 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Meller S., et al. , Chemokine responses distinguish chemical-induced allergic from irritant skin inflammation: memory T cells make the difference. J. Allergy Clin. Immunol. 119, 1470–1480 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Clemmensen A., et al. , Genome-wide expression analysis of human in vivo irritated epidermis: Differential profiles induced by sodium lauryl sulfate and nonanoic acid. J. Invest. Dermatol. 130, 2201–2210 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Cottrez F., Boitel E., Auriault C., Aeby P., Groux H., Genes specifically modulated in sensitized skins allow the detection of sensitizers in a reconstructed human skin model. Development of the SENS-IS assay. Toxicol. In Vitro 29, 787–802 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Schalock P. C., et al. , American Contact Dermatitis Society core allergen series: 2017 update. Dermatitis 28, 141–143 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Ahlström M. G., Thyssen J. P., Wennervaldt M., Menné T., Johansen J. D., Nickel allergy and allergic contact dermatitis: A clinical review of immunology, epidemiology, exposure, and treatment. Contact Dermat. 81, 227–241 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Jenkinson C., et al. , Characterization of p-phenylenediamine-albumin binding sites and T-cell responses to hapten-modified protein. J. Invest. Dermatol. 130, 732–742 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Magdaleno-Tapial J., et al. , Contact allergy to isothiazolinones epidemic: Current situation. Contact Dermat. 82, 83–86 (2020). [DOI] [PubMed] [Google Scholar]
- 23.O’Boyle N. M., Delaine T., Luthman K., Natsch A., Karlberg A. T., Analogues of the epoxy resin monomer diglycidyl ether of bisphenol F: Effects on contact allergenic potency and cytotoxicity. Chem. Res. Toxicol. 25, 2469–2478 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Wahlberg J. E., Wrangsjö K., Hietasalo A., Skin irritancy from nonanoic acid. Contact Dermat. 13, 266–269 (1985). [DOI] [PubMed] [Google Scholar]
- 25.Theodorou K., et al. , Whole body and hematopoietic ADAM8 deficiency does not influence advanced atherosclerotic lesion development, despite its association with human plaque progression. Sci. Rep. 7, 11670 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naus S., et al. , The metalloprotease-disintegrin ADAM8 is essential for the development of experimental asthma. Am. J. Respir. Crit. Care Med. 181, 1318–1328 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Mahoney E. T., Benton R. L., Maddie M. A., Whittemore S. R., Hagg T., ADAM8 is selectively up-regulated in endothelial cells and is associated with angiogenesis after spinal cord injury in adult mice. J. Comp. Neurol. 512, 243–255 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nath P. R., et al. , CD47 expression in natural killer cells regulates homeostasis and modulates immune response to lymphocytic choriomeningitis virus. Front. Immunol. 9, 2985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matozaki T., Murata Y., Okazawa H., Ohnishi H., Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol. 19, 72–80 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Grolmusz V. K., et al. , Cell cycle dependent RRM2 may serve as proliferation marker and pharmaceutical target in adrenocortical cancer. Am. J. Cancer Res. 6, 2041–2053 (2016). [PMC free article] [PubMed] [Google Scholar]
- 31.Cui F., Hu J., Fan Y., Tan J., Tang H., Knockdown of spindle pole body component 25 homolog inhibits cell proliferation and cycle progression in prostate cancer. Oncol. Lett. 15, 5712–5720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fortino V., Scala G., Greco D., Feature set optimization in biomarker discovery from genome-scale data. Bioinformatics 36, 3393–3400 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Kurachi M., et al. , The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15, 373–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kehren J., et al. , Cytotoxicity is mandatory for CD8(+) T cell-mediated contact hypersensitivity. J. Exp. Med. 189, 779–786 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schraml B. U., et al. , The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460, 405–409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen J. M., Bonefeld C. M., Poulsen S. S., Geisler C., Skov L., IL-23 and T(H)17-mediated inflammation in human allergic contact dermatitis. J. Allergy Clin. Immunol. 123, 486–492 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Carbone T., et al. , CD56highCD16-CD62L- NK cells accumulate in allergic contact dermatitis and contribute to the expression of allergic responses. J. Immunol. 184, 1102–1110 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Zamai L., et al. , Natural killer (NK) cell-mediated cytotoxicity: Differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J. Exp. Med. 188, 2375–2380 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva M., Videira P. A., Sackstein R., E-selectin ligands in the human mononuclear phagocyte System: Implications for infection, inflammation, and immunotherapy. Front. Immunol. 8, 1878 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teng X., et al. , IL-37 ameliorates the inflammatory process in psoriasis by suppressing proinflammatory cytokine production. J. Immunol. 192, 1815–1823 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Newman A. M., et al. , Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kassambara A., ggpubr:’ggplot2’ Based Publication Ready Plots. R package version 0.3.0.999. https://rpkgs.datanovia.com/ggpubr/. Accessed 15 November 2019.
- 43.Margolin A. A., et al. , ARACNE: An algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7 (suppl. 1), S7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer P. E., Kontos K., Lafitte F., Bontempi G., Information-theoretic inference of large transcriptional regulatory networks. EURASIP J. Bioinform. Syst. Biol. 2007, 79879 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faith J. J., et al. , Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5, e8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer P. E., Lafitte F., Bontempi G., minet: A R/Bioconductor package for inferring large transcriptional networks using mutual information. BMC Bioinformatics 9, 461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmittgen T. D., Livak K. J., Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 48.LeCun Y., Bengio Y., Hinton G., Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original expression microarray data have been deposited in the ArrayExpress, https://www.ebi.ac.uk/arrayexpress/ (accession no. E-MTAB-9501).