Significance
Gene expression is precisely controlled across human cell types to control cellular functions and phenotypes. Here we present a technology to quantify the level of expression of genes in single cells, which will help to understand how genes are regulated in each cell type in the human body. We demonstrate that this technology improves upon existing tools to enable understanding how DNA regulatory elements control gene expression and mapping the frequencies of different cell types in tissues. This technology will enable new large-scale studies to understand gene control.
Keywords: gene regulation, single cell, enhancers, genomics
Abstract
Single-cell quantification of RNAs is important for understanding cellular heterogeneity and gene regulation, yet current approaches suffer from low sensitivity for individual transcripts, limiting their utility for many applications. Here we present Hybridization of Probes to RNA for sequencing (HyPR-seq), a method to sensitively quantify the expression of hundreds of chosen genes in single cells. HyPR-seq involves hybridizing DNA probes to RNA, distributing cells into nanoliter droplets, amplifying the probes with PCR, and sequencing the amplicons to quantify the expression of chosen genes. HyPR-seq achieves high sensitivity for individual transcripts, detects nonpolyadenylated and low-abundance transcripts, and can profile more than 100,000 single cells. We demonstrate how HyPR-seq can profile the effects of CRISPR perturbations in pooled screens, detect time-resolved changes in gene expression via measurements of gene introns, and detect rare transcripts and quantify cell-type frequencies in tissue using low-abundance marker genes. By directing sequencing power to genes of interest and sensitively quantifying individual transcripts, HyPR-seq reduces costs by up to 100-fold compared to whole-transcriptome single-cell RNA-sequencing, making HyPR-seq a powerful method for targeted RNA profiling in single cells.
Single-cell RNA sequencing (scRNA-seq) has emerged as a powerful and flexible tool for characterizing biological systems (1–5). The ability to measure gene expression in thousands of single cells at once has enabled identifying and characterizing cellular heterogeneity in tissues, defining gene programs in previously uncharacterized cell types, and studying the dynamics of gene expression (6). scRNA-seq has also facilitated high-throughput genetic screens that link CRISPR perturbations to effects on transcriptional programs in single cells (7–9).
However, while scRNA-seq addresses certain biological questions by capturing an unbiased representation of the transcriptome, the utility of this approach is limited for applications that require sensitive detection of specific transcripts of interest. Such applications include profiling the dynamics of low-abundance or nonpolyadenylated transcripts, quantifying rare cell types or states by measuring predefined marker genes, and characterizing the effects of perturbations to cis-regulatory elements on nearby genes. Addressing these questions requires considering the efficiency of detection for individual RNA transcripts [which can range from 5 to 45% for whole-transcriptome scRNA-seq sequenced to saturation (10–12)] and the depth of sequencing required to sample genes of interest (which might require hundreds of thousands of reads per cell for lowly expressed transcripts). To address the latter, previous studies have introduced several strategies to enrich and quantify specific transcripts in single cells, for example using target-specific reverse transcription of RNA during library preparation (13–15) or hybrid selection, PCR, or linear amplification on single-cell cDNA libraries (16–19). However, each of these approaches has certain limitations, such as the number of transcripts that can be measured, the inability to detect nonpolyadenylated transcripts, or the feasibility of profiling very large numbers of cells (SI Appendix, Note S1). New approaches are needed to quantify specific genes of interest in tens of thousands of single cells in a cost-effective and sensitive manner.
We developed an approach called Hybridization of Probes to RNA sequencing (HyPR-seq) that enables targeted quantification of RNAs in thousands of single cells. Our approach builds on the observation that single-molecule fluorescence in situ hybridization (smFISH)—involving hybridization of labeled probes to target RNAs—can detect both polyadenylated and nonpolyadenylated RNAs with very high sensitivity (20–24). We sought to develop a method that quantifies the same hybridization probes via next-generation DNA sequencing, rather than via imaging, to enable simultaneous readouts of hundreds of probes in thousands of single cells.
To do so, HyPR-seq involves hybridizing single-stranded DNA (ssDNA) probes to one or more RNA transcripts of interest, encapsulating single cells in 1-nL droplets, PCR-amplifying the ssDNA probes, and sequencing these amplicons to quantify gene abundance in each cell. This method has greater than 20% sensitivity for individual transcripts, can simultaneously measure hundreds of polyadenylated and nonpolyadenylated transcripts, and can profile more than 100,000 cells in a cost-effective manner. We demonstrate the utility of this approach by measuring the cis-regulatory effects of CRISPR perturbations to noncoding DNA elements, detecting nonpolyadenylated intronic RNA to measure time-resolved changes in gene expression, and quantifying the proportions of cell types in kidney tissue using low-abundance marker genes. By increasing the sensitivity of RNA detection while decreasing both sequencing and reagent costs, HyPR-seq extends the toolkit of single-cell methods to enable new experiments to investigate gene regulation and cellular programs.
Results
We began by adapting probes from the hybridization chain reaction (HCR) smFISH protocol (24, 25) for a sequencing-based readout. In HCR, two “initiator” probes anneal adjacent to each other on a target RNA molecule and are recognized by a metastable “hairpin” oligo, triggering a chain reaction in which fluorescently labeled oligos bind and extend (SI Appendix, Fig. S1). In HyPR-seq, we eliminated the chain reaction and instead hybridize and ligate a single “readout” oligo to one of the initiator probes (Fig. 1A and SI Appendix, Fig. S1). (We retain the cooperative initiator binding and metastable hairpin to increase the specificity of hybridization.) This ligation creates an ssDNA fragment that can be amplified by PCR and quantified by high-throughput sequencing (SI Appendix, Fig. S2). We include a unique molecular identifier (UMI) on the amplified initiator probe to identify sequencing reads that originate from a single hybridization and ligation event.
Fig. 1.
HyPR-seq enables single-cell quantification of selected RNAs. (A) At a probe binding site, initiator probes bind adjacent to one another on an RNA of interest, creating a binding site for a hairpin oligo. A final readout oligo is annealed and ligated, forming a template for PCR amplification containing transcript information, UMI, and handles for PCR and sequencing. (B) Experimental workflow for HyPR-seq. (C) Comparison of three single-cell methods in K562 cells. (Top) HCR measurements of GAPDH and GATA1 using probes targeting the same binding sites used for HyPR-seq. (Middle) Histogram of UMIs per cell for three probes targeting GAPDH and GATA1 from HyPR-seq. (2,148 cells) and (Bottom) 10X Genomics Chromium 3′ scRNA-seq (207,324 cells). (Scale bar, 10 μm.) All experiments performed in K562 cells. (D) HyPR-seq counts per cell for each of the three probes targeting GAPDH (left columns), GATA1 (center columns), or GFP (not expressed, right side). Error bars represent 95% confidence interval (CI) of the mean from two replicates (from 4,605 cells). For more analysis of probe variability, See SI Appendix, Fig. S4D. (E) Counts per cell at a constant depth of 1,000 total UMIs per cell for three genes (GATA1, PGK1, and TUBB) as assayed by 10X Genomics Chromium 3′ scRNA-seq (26) and HyPR-seq. Counts for HyPR-seq are shown for the 22-gene experiment in K562 cells and 2 simulated experiments (based on these counts) measuring different numbers of total genes. Horizontal bars show the caps of the (vertical) error bars, which represent 95% CI of the mean from two replicates. (F) Counts per cell for 19 genes in K562 cells as measured by 10X Genomics Chromium 3′ scRNA-seq (x axis) and HyPR-seq (y axis). Error bars for HyPR-seq measurements show 95% CI of the mean from two replicates (4,605 total cells). (G) In THP1 cells ± LPS, fold changes of 18 genes assayed by bulk RNA-seq (x axis) and HyPR-seq (y axis). Error bars represent 95% CI of the mean from three (RNA-seq) or four (HyPR-seq) replicates (13,957 total cells). (H) To assess mixing, K562 cells were transduced to express two different barcode RNAs, each detectable by three HyPR probes. Scatterplot shows UMI counts for each barcode per droplet (2,727 total droplets). Unassigned cells are in gray.
To use these HyPR-seq probes to detect RNAs in single cells (Fig. 1B), we: 1) Cross-link and permeabilize a population of cells, 2) hybridize initiator probes to hundreds of target RNAs, and 3) hybridize and ligate the hairpin and readout oligos. We then 4) distribute single cells and DNA-barcoded microparticles (“beads”) into an emulsion PCR using a commercially available automated microfluidic droplet-maker. Each bead carries photocleavable primers with a unique clonal barcode. We then 5) cleave primers off of the beads using UV light, 6) PCR-amplify the HyPR-seq probes in emulsion, 7) sequence the resulting amplicons, and 8) quantify the expression of each target gene based on the UMI counts of corresponding initiator probes. (We note that, because we typically include multiple probe pairs per gene, which can bind adjacent to one another on an RNA, some UMI counts for a given gene may derive from the same individual RNA transcript; see SI Appendix, Note S2). During the microfluidic emulsion step, we Poisson-load cells and beads into droplets, expecting 5 to 15% of droplets to contain one cell and at least one bead. Droplets containing more than one bead are computationally identified and merged (Materials and Methods and SI Appendix, Fig. S3). In our implementation, each sample from the droplet-maker yields ∼500 to 2,500 single cells. Our complete HyPR-seq probe design and data analysis pipeline is available at https://github.com/EngreitzLab/hypr-seq.
To test the performance of HyPR-seq, we designed and applied 102 HyPR-seq probes to detect 22 genes (2 to 10 probes per gene) (Dataset S11) in K562 human leukemia cells in two biological replicates. We sequenced the libraries to saturation (17,111 reads per cell) and identified 4,605 total cells (from ∼10,000 loaded cells), with an average of 4,021 UMIs per cell (SI Appendix, Fig. S4A and Dataset S2). UMI counts per probe per cell were highly reproducible between biological replicates (Pearson’s R > 0.99) (SI Appendix, Fig. S4B).
Specificity.
To investigate the specificity of HyPR-seq, we first compared probes targeting two highly expressed genes (GAPDH and GATA1) to control probes that did not target a gene expressed in K562 cells. The GAPDH and GATA1 probes gave 10,000-fold and 500-fold higher signal compared to the control probes: For GAPDH, we measured an average of 303.7 UMIs per cell (101.2 UMIs per probe per cell); for GATA1, an average of 19.1 UMIs per cell (6.4 UMIs per probe per cell); and for the control probes, an average of fewer than 0.03 UMIs per cell (<0.01 UMIs per probe per cell) (Fig. 1 C and D). We detected >100-fold lower signal when we only included one of the initiator probes, confirming the specificity of the split initiator design (SI Appendix, Fig. S4C). As expected, the three independent probes for each gene (targeting different locations on the same mRNA) varied in their UMI counts, presumably due to sequence-specific differences in hybridization efficiency (Fig. 1D). Across all genes in the experiment, ∼75% of probes had UMI counts within 2-fold of the median probe for a given gene (SI Appendix, Fig. S4D) and yielded counts >100-fold above the negative control probes. Based on measurements of a larger panel of genes across a range of expression levels, we estimate that HyPR-seq can specifically detect transcripts expressed above ∼1 transcripts per million (TPM) (Materials and Methods and SI Appendix, Fig. S4E).
Detection Rate and Sequencing Efficiency.
We evaluated the counts per gene observed in HyPR-seq compared to scRNA-seq and smFISH experiments in K562 cells (Fig. 1C). We first counted UMIs in this HyPR-seq experiment (sequenced to ∼70% saturation, ∼4,000 UMIs per cell) compared to a 10X Genomics Chromium 3′ scRNA-seq dataset (26) (sequenced to ∼20% saturation, ∼18,000 UMIs per cell). We observed an average of 36-fold more UMIs per gene in HyPR-seq than in scRNA-seq (Fig. 1C and SI Appendix, Fig. S4F), corresponding to 162-fold more UMIs per cell per 1,000 total UMIs. This enrichment will vary between 30- and 300-fold depending on the number of genes targeted in the HyPR-seq experiment (Fig. 1E). To estimate the sensitivity of our method, we compared HyPR-seq measurements (sequencing UMIs) to smFISH of GATA1 mRNA (counting spots), and found that the best single HyPR-seq probe yielded 20% of the smFISH counts (a lower bound on the per transcript detection efficiency), and together three HyPR-seq probes yielded 31% of the smFISH counts (an upper bound) (SI Appendix, Fig. S4G). In smFISH, 20 to 50 probes per gene are used per RNA to obtain near quantitative detection efficiency (∼90%), assuming that probes bind independently to their target RNAs (27). Under a similar assumption for HyPR-seq, increasing the number of probes per gene should increase detection efficiency for genes expressed above the specificity limit of 1 TPM (SI Appendix, Note S2). The ability to adjust the number of probes per gene will allow users to tune the detection rate and fraction of sequencing reads devoted to different genes.
Quantification Accuracy.
We examined the accuracy of HyPR-seq in quantifying the expression levels of different genes in a single condition (“cross-gene quantification”) and quantifying the fold-change in expression of a given gene across conditions (“cross-condition quantification”).
To assess cross-gene quantification, we compared our HyPR-seq data across 19 genes (not including GFP, blue fluorescent protein [BFP], and the noncoding RNA PVT1) to 10X Genomics Chromium 3′ scRNA-seq data collected for the same cell type (26). UMI counts per cell from HyPR-seq correlated well with scRNA-seq counts for the same genes (Pearson’s R = 0.90) (Fig. 1F). We observed similar concordance for a set of genes across a wider range of expression levels in THP1 cells (Pearson’s R = 0.89) (SI Appendix, Fig. S4E).
To assess cross-condition quantification, we performed two experiments. First, we measured the expression of an RNA encoding BFP under the control of a doxycycline-inducible promoter at 14 timepoints after induction, during which BFP mRNA increased >100-fold. HyPR-seq measurements of BFP mRNA expression correlated well with corresponding measurements by qPCR (Pearson’s R = 0.95) (SI Appendix, Fig. S4H). Second, we examined 18 genes whose expression levels are known to change between 0.5- and 250-fold in THP1 monocytic leukemia cells upon stimulation with bacterial lipopolysaccharide (LPS). The fold-change in gene expression in LPS-stimulated vs. unstimulated cells correlated well between HyPR-seq and bulk RNA-seq (Pearson’s R = 0.98) (Fig. 1G), and these fold-changes were similar across most probes targeting the same gene (SI Appendix, Fig. S4I). These results indicate that HyPR-seq accurately quantifies changes in gene expression across a wide dynamic range.
Single-Cell Purity.
We demonstrated the single-cell purity of HyPR-seq through a single-cell mixing experiment. We engineered two cell lines to each express a unique transcript detectable by HyPR-seq (“detection barcodes”) (Materials and Methods). We applied probes that recognized the two different detection barcodes to both cell lines, then mixed the cells before distributing them into emulsion droplets. We found that 98.5% of droplets contained UMIs from a single detection barcode and 1.5% of the droplets had UMIs from both barcodes. This suggests a cell doublet rate of ∼2.9% (in a 50/50 mix of cells, half of the doublets will involve two cells with the same barcode), compared to a theoretical rate of about 0.9% based on the density of cell loading (Fig. 1H). These data indicate a high level of single-cell purity in the HyPR-seq emulsion PCR (the deviation from the theoretical doublet rate could be due to cell clumping during emulsion generation or low levels of ambient probes leaking from fixed cells).
Having demonstrated the essential technical capabilities of HyPR-seq, we examined its utility in three areas where existing single-cell tools remain limited: 1) Profiling the effects of CRISPR perturbations on individual genes of interest, 2) measuring nonpolyadenylated RNAs such as gene introns, and 3) detecting rare cell types via lowly expressed marker genes in complex tissue.
CRISPR Screens with HyPR-Seq to Map Perturbation Effects on Gene Expression.
We explored the utility of HyPR-seq in generating low-cost readouts of CRISPR perturbations to DNA regulatory elements (26, 28), where sensitive gene detection is critical for obtaining quantitative readouts of the effects of these elements on nearby genes.
We first developed an approach to apply HyPR-seq to pooled CRISPR screens, which requires detecting which guide RNAs (gRNAs) are expressed in each single cell in a population. We designed a lentiviral vector that expresses a gRNA from a U6 promoter as well as a detection barcode from a Pol II promoter (Fig. 2A). We designed HyPR-seq probes to detect these barcodes and thereby determine which gRNA is expressed in a given cell (SI Appendix, Fig. S5A). We note that including three independent and unique probe binding sites in each detection barcode allows us to combinatorially identify a large number of gRNAs with a limited set of probes (Materials and Methods).
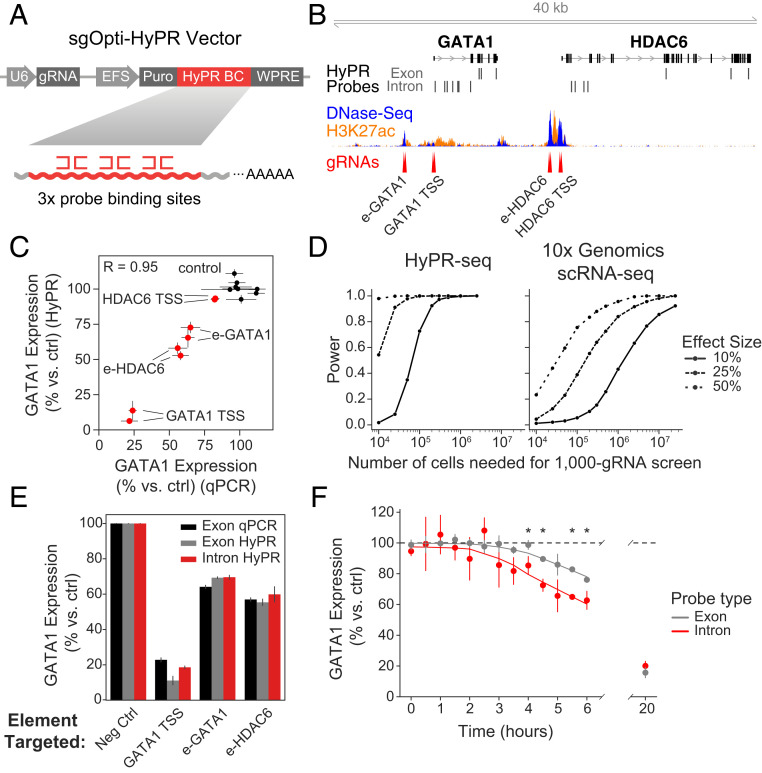
Fig. 2.
Application of HyPR-seq to CRISPR screens for mature mRNAs and introns. (A) Design of sgOpti-HyPR barcoded vector. sgOpti vector was modified to express a detection barcode consisting of three probe binding sites (HyPR BC) from a Pol II promoter along with a sgRNA. (B) Location of CRISPRi gRNAs and HyPR-seq probes in the GATA1 locus, with DNase-seq and H3K27ac ChIP-seq signals from K562 cells. (C) Relative GATA1 expression as measured by qPCR (x axis) and HyPR-seq (y axis) for a small library of 16 gRNAs (8 nontargeting controls [black] and 2 targeting each of 4 elements in the GATA1 locus [red]). qPCR measurements come from cell lines individually infected with each gRNA, while HyPR-seq measurements come from single cells in a pooled screen. Error bars represent 95% CI on the mean of three (qPCR) or two (HyPR-seq) replicates (from 4,605 total cells). (D) Power to detect changes in gene expression of various magnitudes (y axis, lines represent 10%, 25%, and 50% effect sizes) as a function of the total number of cells profiled (x axis). Power is shown for a hypothetical screen of 1,000 CRISPR perturbations and is averaged over 37 genes expressed at >1.5 UMIs per cell in a HyPR-seq dataset (Left) and the same genes in a 10X Genomics Chromium 3′ scRNA-seq dataset (Right) sequenced to ∼18,000 UMIs per cell (26). Power is estimated by testing the indicated number of cells (divided by 1,000) against M total cells (where M = 5,000 for HyPR-seq and 200,000 for 10X Genomics Chromium 3′ scRNA-seq) to mimic a screen with an excess of nontargeting control gRNAs. (E) GATA1 knockdown as measured by qPCR (with primers targeting the mature mRNA, black) or HyPR-seq with probes designed against the exons (gray) or first intron (red) of GATA1. Error bars represent 95% CI on the mean of two replicates (from 4,605 total cells). (F) Direct detection of intronic RNA allows time-resolved analysis of CRISPR knockdown experiments. GATA1 expression in cells with gRNAs targeting GATA1 TSS as measured by both exon- (gray) and intron-targeting (red) probes over a 20-h time course. Mean and 95% CI are shown for three replicates at each of 14 timepoints (from 112,056 total cells), and the curves are fit with lowess regression. Asterisks indicate a significant difference in the knockdown, as measured by introns versus exons.
We tested this approach using a CRISPR interference (CRISPRi) screen in K562 cells to inhibit four elements in the GATA1 locus that we previously found to regulate GATA1 (29, 30) (Fig. 2B). We infected K562 cells expressing KRAB-dCas9 from a doxycycline-inducible promoter with lentiviral constructs encoding 16 gRNAs along with linked detection barcodes (8 control gRNAs and 2 targeting each element). We robustly detected these barcodes (with at least 10 UMIs per cell in 90% of cells) and were able to assign a unique guide to >80% of cells, comparable to existing methods (9, 18) (SI Appendix, Fig. S5 A–C). We quantified the effects of each gRNA on the expression of GATA1 and found that the changes in gene expression detected by HyPR-seq strongly correlated with those measured by qPCR (Pearson’s R = 0.95) (Fig. 2C). We observed good quantification of GATA1 knockdown when down-sampling our data to as low as ∼1,100 reads per cell (>10-fold fewer reads) (SI Appendix, Fig. S5D).
Such an approach could be useful for large-scale screens to measure the effects of enhancers on nearby genes, where existing approaches read out either effects on one gene at a time with very high sensitivity (30) or effects on all genes in the transcriptome with low per gene sensitivity (26, 28). Using data from our pilot experiments, we calculated the power of HyPR-seq to profile the effects of 1,000 CRISPR perturbations on 50 selected genes, the scale needed to systematically connect all putative enhancers in a genomic locus to their target genes (30). For this experiment, HyPR-seq would require profiling ∼25,000 cells at 5,000 reads per cell to achieve 90% power to detect 25% changes in expression for all genes expressed at >1 TPM (Fig. 2 D, Left). In contrast, whole-transcriptome scRNA-seq would require profiling >1,000,000 cells at 20,000 reads per cell to achieve similar power (Fig. 2 D, Right), increasing the total cost by two orders-of-magnitude. Thus, HyPR-seq could provide a powerful and cost-effective approach to profile the effects of CRISPR perturbations on a set of selected genes. See SI Appendix, Note S5 for more details on power calculations.
Detecting Nonpolyadenylated Introns to Measure Perturbation Effects with Increased Temporal Resolution.
Due to its hybridization-based detection of RNA, HyPR-seq can, in theory, quantify nonpolyadenylated transcripts that are difficult to detect with existing droplet-based scRNA-seq approaches. To demonstrate this, we used HyPR-seq to detect gene introns, which are present at lower copy numbers than mature mRNAs due to their short half-lives but whose abundance can be used to estimate gene transcription rates (22). We designed probes targeting the introns of four genes (GATA1, HDAC6, MYC, and PVT1) and performed HyPR-seq in 4,605 K562 cells. We detected absolute signals that correlated with the transcription rates of these four genes as measured by precision nuclear run-on sequencing (Pearson’s R = 0.99) (SI Appendix, Fig. S5E), including an average of 13.1 UMIs per cell for introns of MYC (a highly transcribed gene in K562 cells) and 3.5 UMIs per cell for introns of PVT1 (a less-transcribed gene in K562 cells).
Quantifying intron abundance could enable more temporally precise measurements of the effects of perturbations on gene expression. We examined the effects of promoter or enhancer inhibition with CRISPRi on GATA1 expression over a 20-h time course of KRAB-dCas9 induction in 112,056 K562 cells. We used exon-targeting probes to estimate steady-state mRNA levels and intron-targeting probes to estimate transcription rates. Both intron- and exon-targeted HyPR-seq probes showed equivalent levels of reduction after 20 h, indicating that intron-targeting probes show similar quantitation and specificity (Fig. 2E). However, we detected a decrease in intron signal hours earlier than for exon signal, consistent with the shorter half-life of introns compared to mature mRNAs (Fig. 2F). Thus, HyPR-seq can detect time-resolved changes in gene expression via direct detection of low-abundance gene introns and may enable multiplexed detection of other nonpolyadenylated RNAs that are difficult to capture using existing droplet-based single-cell methods.
Measuring Cell-Type Frequencies and Low-Abundance Genes in Tissue.
HyPR-seq could enable new types of highly multiplexed experiments to measure the expression of genes of interest in complex tissues. For example, determining the frequencies of closely related cell types in a tissue can require the detection of specific marker genes that may not be highly expressed, which is challenging with whole-transcriptome RNA-seq (31).
To test this, we applied HyPR-seq to detect changes in cell type frequency in a mouse model of diabetic kidney disease (DKD) (32). We designed HyPR-seq probes to distinguish 11 cell populations using 25 canonical marker genes. Some of these marker genes were lowly expressed in their corresponding cell types (<2 TPM) and are not robustly detected in existing scRNA-seq datasets (Dataset S7) (33–37). Accordingly, we included up to 20 probe sets per gene to enable sensitive detection (Materials and Methods and SI Appendix, Fig. S6A). We applied HyPR-seq to dissociated single cells from the kidneys of 12-wk-old wild-type (BTBR wt/wt) and diabetic (BTBR ob/ob) mice and detected 29,125 single cells after cell doublet detection and filtering steps (Materials and Methods). UMAP visualization of the HyPR-seq data identified 11 clusters, including all of the targeted cell populations (Fig. 3 A and B, SI Appendix, Fig. S6 B–E, and Dataset S7).
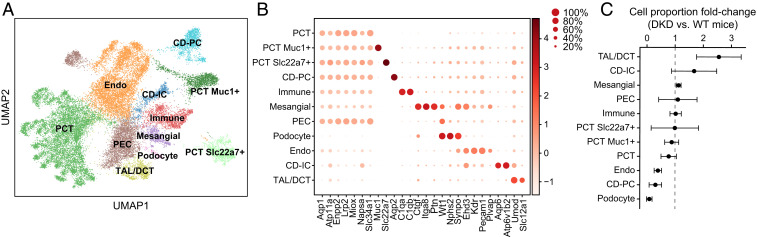
Fig. 3.
HyPR-seq in mouse kidney cells detects changes in cell frequencies in diabetes. (A) UMAP visualization of HyPR-seq data for 31 genes in 14,288 BTBR wt/wt cells and 14,837 BTBR ob/ob cells identifies 11 distinct cell types. Endo, endothelial cell; PCT, proximal-convoluted tubule; PEC, parietal epithelial cell; TAL/DCT, thick ascending limb/distal convoluted tubule cell. (B) Dot plot displays kidney cell types with the fraction of cells in each subtype expressing each gene (circle size) and the relative level of gene expression (color). (C) Change in cell proportions in diabetic versus wild-type mice. Error bars represent the SD of the ratio from two biological replicates (Materials and Methods).
HyPR-seq accurately detected changes in cellular composition in DKD and enabled quantification of specific genes of interest with only an average of 203 UMIs per cell. For example, podocytes represent a rare cell type that plays a key role in the glomerular filtration barrier (38). We observed podocytes—marked by the combined expression of Wt1, Nphs2, and Synpo—at 0.9% frequency in wild-type mice (132 of 14,288 cells). Podocytes decreased in frequency by >10-fold in diabetic mice (to 12 of 14,837 total cells, 0.08%, Cochran–Mantel–Haenszel P < 10−23), consistent with previous reports that podocytes are one of the earliest cell types to be damaged and lost in DKD (Fig. 3C) (39–41). HyPR-seq also detected an increase in thick ascending limb/distal convoluted tubule cells and a decrease in endothelial cells, in agreement with early changes observed in DKD (Fig. 3C) (42). Finally, HyPR-seq detected various subtypes of epithelial cells, including two subclusters of proximal convoluted tubule cells (Slc22a7+ and Muc1+) and collecting duct principal cells (CD-PCs, Aqp2+), and indicated that CD-PCs are reduced in frequency in DKD (Fig. 3 B and C). Together, these observations were possible using only 203 UMIs per cell, ∼10-fold fewer than were used to make similar observations using scRNA-seq (42).
Finally, to determine whether HyPR-seq can scale to measure even more genes simultaneously, we designed an experiment to measure 179 genes expressed in mouse splenocytes with 1,023 HyPR-seq probes (Materials and Methods and SI Appendix, Fig. S8). We compared HyPR-seq data (7,962 single cells) with 10X scRNA-seq data (6,373 single cells), and observed excellent concordance in the abundances of each of the 16 defined cell populations (Pearson’s R = 0.94) and the levels of gene expression within each population (average Pearson’s R between 10X scRNA-seq and HyPR-seq pseudobulk profiles = 0.81) (SI Appendix, Fig. S8 A–C). We conducted a second experiment using a subset of 265 probes against 48 genes (3,269 single cells), and found that using 1,023 probes yielded more total UMIs per cell (493 vs. 197) but was moderately less sensitive for the set of shared genes (average 25% decrease in UMIs per gene per cell) (SI Appendix, Fig. S8 D and E). Finally, we observed that using multiple probes per transcript allowed us to observe lowly expressed marker genes in a larger fraction of cells than would have been possible with a single probe (SI Appendix, Note S2 and Fig. S8F).
Taken together, these experiments show that HyPR-seq will enable high-throughput experiments to measure cellular responses to environmental, chemical, or genetic perturbations in primary cells and complex tissues, including measuring up to hundreds of genes of interest that may be lowly expressed such as drug targets, transcription factors, and signaling molecules.
Discussion
Here, we described HyPR-seq, a microfluidic droplet-based approach for cost-effective and sensitive profiling of a chosen subset of RNA molecules in single cells. HyPR-seq provides a unique combination of capabilities that overcome key limitations of existing single-cell techniques. First, by adapting in situ hybridization probes for a sequencing-based readout, HyPR-seq achieves a per probe RNA detection sensitivity of ∼20% relative to smFISH, while also expanding multiplexing (in this study, we included over 1,000 probes in a single experiment). Second, HyPR-seq can easily be scaled to examine more than 100,000 cells in a single experiment, facilitating large screens (SI Appendix, Note S1). Third, HyPR-seq can detect RNA species that are not targeted by polyA-based scRNA-seq approaches, including introns, enabling time-resolved studies of transcription. Finally, HyPR-seq provides a cost-effective approach for targeted quantification of hundreds of selected transcripts in single cells by reducing reagents costs per cell by 5-fold and sequencing costs per cell by up to 100-fold versus whole-transcriptome scRNA-seq.
HyPR-seq does have several acknowledged limitations. HyPR-seq does not sequence the RNA molecule itself, making it unsuitable for detecting RNA-seq variants or modifications. The current protocol involves multiple rounds of washes for probe hybridization and ligation, which results in some cell loss and requires starting an experiment with 1 million or more cells. Finally, because HyPR-seq involves hybridization of probes, it could be more difficult to detect certain RNA transcripts that have high sequence homology with others.
The unique capabilities of HyPR-seq will enable experiments that were previously impractical using existing tools. For example, HyPR-seq could allow for large-scale CRISPR-based studies to perturb hundreds of regulatory elements in a single locus and profile their effects on all nearby genes. We anticipate that HyPR-seq will be broadly useful for sensitively quantifying RNA expression across a wide range of systems to study gene regulation and cellular heterogeneity (see SI Appendix, Note S3 for design recommendations).
Materials and Methods
Design of HyPR-Seq Probes.
We adapted the HCR (v3) (24) probe design for our droplet-based HyPR-seq method (SI Appendix, Figs. S1 and S2).
Initiator probes.
Two initiator probes, a 5′ probe and a 3′ probe, each contain 25 bp of sequence homologous to the target RNA and other necessary sequences.
The structure of the 5′ probe is: [5′ Initiator] [5′ Spacer] [5′ Homology] [UMI] [Primer Binding Site]
The structure of the 3′ probe is: [3′ Homology] [3′ Spacer] [3′ Initiator]
The constant sequences are:
5′ Initiator:/5Phos/GGAGGGCAGCAAACGG
5′ Spacer: AA
UMI: NNNNNNNNNN
Primer Binding Site: CTCGACCGTTAGCAAAGCTC
3′ Spacer: TA
3′ Initiator: GAAGAGTCTTCCTTTACG.
For probes in this study, we use the “B1” initiator system from HCR (24). Compared to the HCR initiator probes, the modifications we made for HyPR-seq are entirely in the 5′ probe. We added a 5′ phosphate (for ligation in HyPR-seq), attached a 20-bp sequencing adapter (primer binding site) to the 3′ end, and removed 2 bp from the 5′ end of the initiator and added it to the readout oligo, which we found improved the specificity of HyPR-seq.
Following hybridization of the initiator probes, we add the hairpin oligo B1H1 (CGTAAAGGAAGACTCTTCCCGTTTGCTGCCCTCCTCGCATTCTTTCTTGAGGAGGGCAGCAAACGGGAAGAG, Molecular Instruments; note that ordering this component from Molecular Instruments as opposed to IDT is important, likely due to the method of synthesis/purification/quality control).
Finally, we add a “readout” oligo adapted from HCR hairpin B1H2 (CTTACGGATGTTGCACCAGCAAGAAAGAATGCGA, IDT), which is ligated to the 5′ initiator probe (SI Appendix, Fig. S2).
Custom Barcoded Bead Design.
Custom 68mer beads were ordered from Chemgenes on 10-µm Agilent polystyrene beads at an oligo synthesis scale of 10 µmole, using the following sequence: 5′-bead-linker-PC-linker-CAAGCAGAAGACGGCATACGAGATJJJJJJJJJJJJGTTGGCACCAGGCTTACGGATGTTGCACCAGC-3′.
Selecting Target Sequences for HyPR-Seq Probes.
HyPR-seq probes can target any site on an RNA that enables specific binding. We chose the 5′ and 3′ homology sequences on the initiator probes that, similar to HCR, target 25-bp regions on a transcript of interest separated by a 2-bp spacer (52 bp total). We developed a custom design script (https://github.com/EngreitzLab/hypr-seq) to identify 52-bp RNA homology sequences. We tiled candidate sequences across the RNA and excluded those that: 1) Contained homopolymer repeats (n > 5), 2) were predicted to form hairpins or dimers, 3) had GC-content outside of 40 to 65%, 4) contained more than five bases of repetitive sequence [by comparison to RepeatMasker (43)]; or 5) had a match with >25% identity when compared with BLAST to the rest of the transcriptome (RefSeq). From the list of valid homology sequences, we generated the final list by selecting a smaller number of sequences (typically four to six) spaced evenly across the transcript of interest. Then, the 52-bp homology sequence was split to produce the probe homology regions. The reverse complement of bases 1 to 25 (in the 5′-3′ direction on the RNA) form the 3′ homology sequence, and the reverse complement of bases 28 to 52 form the 5′ homology sequence, with bases 26 to 27 acting as an unbound spacer.
Probes targeting the introns of genes were designed similarly, except we confined our search for homology regions to the first 5 kb of the first intron (or first 5 kb of any introns, if the first intron is shorter than 5 kb), in order to detect RNA species whose appearance would most closely correlate with the initiation of transcription.
HyPR-Seq Experimental Protocol.
Cell preparation.
Here, we describe the protocol starting with 5 M cells; all volumes in this section were adjusted according to input cell number. For HyPR-seq, cells are harvested in 1× PBS at 350 × g for 5 min at 4 °C in a swinging bucket rotor. Cells were fixed in 4% formaldehyde solution (4% formaldehyde in 1× PBS and 0.1% Tween 20) for 1 h at room temperature with rocking at a concentration of 1 million cells per milliliter, with up to 10 million cells in a 15-mL conical tube. For all following washes and centrifugations after fixation, cells were spun at 850 × g for 5 min at room temperature. Fixed cells were washed twice with 5 mL 1× PBS containing 0.2% Tween 20 (1× PBST). The washed cells were then permeabilized in 70% ice-cold ethanol 5 mL (at a concentration of 1 M/mL), with up to 10 million cells in a 15-mL conical, and stored at 4 °C for 10 min. The permeabilized cells were harvested and washed twice with 5 mL 1× PBST. Then, the cells were transferred to 2-mL round-bottom tubes with up to 5 million cells per tube. The cells were resuspended in 500 µL (10 M cells/mL) of probe hybridization buffer (5× SSC, 30% formamide, 0.1% Tween 20) and incubated at 37 °C for 5 min. The prehybridized cells were centrifuged, resuspended in 500 µL Probes Mix (prepared by pooling probesets in probe hybridization buffer to a final concentration of 20 nM per probe), and incubated overnight at 37 °C in a hybridization oven (VWR Cat# 10055-006). After hybridization, cells were harvested and washed in 500-µL probe hybridization buffer for 10 min at 37 °C. The wash step was repeated three additional times, for a total of four washes.
In the meantime, snap-cooled hairpin solutions were prepared as follows: In separate tubes, 15 pmol each of the B1H1 hairpin and readout oligo (5 µL each at a concentration of 3 µM) were incubated at 95 °C for 90 s, then allowed to cool to room temperature slowly over 30 min to promote hairpin formation. After the final wash in probe hybridization buffer, cells were resuspended in 500 µL 5× SSCT (5× SSC, 0.1% Tween 20) and incubated at room temperature for 5 min. Cells were centrifuged, then resuspended in 200 µL 75 nM cooled B1H1 hairpin in 5× SSCT. The cells were incubated at 37 °C for 1 h in the hybridization oven. After the B1H1 hairpin hybridization, the cells were washed twice with 200 µL 5× SSCT, then resuspended in 200 µL 75 nM cooled readout oligo in 5× SSCT. Cells were incubated 37 °C for 1 h. After oligo incubation, cells were harvested and washed three times in 500 µL 5× SSCT. After the last spin, cells were washed in 200 µL 1× T4 Ligase reaction buffer (New England Biolabs #B0202S) before being resuspended in 200 µL 1× ligase (1× T4 ligase buffer, 1:100 dilution of T4 DNA ligase New England Biolabs #M0202S) and incubated at room temperature for 1 h. Following ligation, cells were washed three times in 500 µL 1× PBST and filtered through a 20-µm filter (20 µm pluriStrainer cat. 43-0020-01). The cells were then counted using a hemocytometer (Fisher Scientific SKU #DHCF015) and checked for single-cell suspension before proceeding to droplet generation. For long-term storage of the cells, 1 U/µL RNase inhibitor was added to the cell suspension (New England Biolabs #M0314S). After starting with 5 M cells prior to fixation, we typically end with 3 to 4 M cells after probe ligation.
Generation of emulsions.
To generate emulsions, we first combined 12.5 µL 2× EvaGreen Supermix (Bio-Rad Cat #186-4033), 500 nM indexing primer (Dataset S3), 2,000 cells and 20,000 barcoded beads (Chemgenes, as described above) in a 25-µL reaction. Once the PCR mix is made, emulsions were generated using QX200 Digital Droplet Generator (Bio-Rad #1864002) as per the manufacturer’s instructions. Briefly, the droplet generation cartridge (Bio-Rad #186-4007) was inserted into the holder (Bio-Rad #186-3051) and 20 µL of the prepared PCR mix was added to the middle sample well, as per the manufacturer’s instructions. Seventy microliters of the droplet-generating oil (Bio-Rad, #186-4005) was added into the bottom oil well. This was repeated for all wells in a chip, where unused sample wells were filled with 1× PBS. Then, the gasket (Bio-Rad #186-4007) was placed over the filled cartridge and the cartridge was placed in the droplet generator. Once the droplets were formed, the cartridge containing the emulsions was placed under the UV lamp (6.5 J/cm2 at 365 nm) about 3 to 8 cm away from the bulb for 5 min. After UV exposure, ∼50 µL of droplets per well was transferred to 96-well plates (Eppendorf #951020362) and sealed with foil (Bio-Rad #181-4040) using a plate sealer (PX1 Plate Sealer #181-4000) for droplet PCR amplification (Eppendorf Mastercycler Pro #E90030010). The following cycling conditions were used: Denaturation: 94 °C for 30 s; cycling: 30 cycles of 94 °C for 5 s, 64 °C for 30 s, and 72 °C for 30 s; final extension: 72 °C for 5 min. All PCR cycling steps were performed with 50% ramp rate (2 °C/min). To break and clean the emulsions, one to four PCR wells were combined in a tube and 40 µL of 97% 1H,1H,2H,2H-Perfluoro-1-octanol (Sigma-Aldrich #370533) was added. The tubes were vortexed for 5 s to ensure complete emulsion breakage and spun at 1,000 × g for 1 min. The top aqueous layer was carefully removed and cleaned with 1.8× SPRI beads according to manufacturer’s instructions. The amplicon libraries were loaded on a gel to determine size (206 bp) and to ensure no primer dimers remained. The libraries were quantified by Qubit before proceeding to sequencing.
Sequencing.
The libraries were loaded at a concentration of 6 pM on a MiSeq and at 1.8 pM on a NextSeq 550. The sequencing specifications were as follows: Read 1: 35 bp, Index 1: 8 bp and Index 2: 12 bp (SI Appendix, Fig. S2). Sequencing the HyPR-seq libraries on the MiSeq and NextSeq both required custom Read 1 (GACACATGGGCGGAGCTTTGCTAACGGTCGAG, IDT) and custom Index 1 primers (GCTGGTGCAACATCCGTAAGCCTGGTGCCAAC, IDT). The NextSeq additionally required a custom Index 2 primer to read the sample indices. (CTCGACCGTTAGCAAAGCTCCGCCCATGTGTC, IDT). All custom primers were added according to manufacturer’s instructions. The depth of sequencing scaled with the number of genes. For the K562 experiments described (targeting 22 highly expressed genes), we typically aimed to sequence 10,000 reads per cell × 1,000 cells per well = 10 million reads.
HyPR-Seq Computational Pipeline.
We built a custom pipeline to analyze HyPR-seq data by taking raw sequencing reads and constructing a count matrix (counts per probe per cell). Briefly, we filter low-quality reads, identify real bead barcodes, map to our set of probes, remove PCR duplicates using UMIs, and combine data that came from multiple bead barcodes in the same droplet. We have made our analysis pipeline available on GitHub (https://github.com/EngreitzLab/hypr-seq) and detailed the key steps below (standard settings used except where indicated). See SI Appendix, Note S4 for more details.
-
1)
We demultiplex reads by well barcode (Index 2) into separate FASTQs per experiment.
-
2)
We then filter reads, removing short reads in Index 1 (bead barcode) and reads with low quality: fastp –length_required 12.
-
3)
We build a whitelist of bead barcodes to include in downstream analyses. We turn off error correction for sequencing/PCR errors for adjacent bead barcodes and typically rely on the built-in method for finding the UMI threshold to distinguish real cells from background (“knee”): umi_tools whitelist –bc-pattern=NNNNNNNNNN –bc-pattern2=CCCCCCCCCCCC –method=umis –error-correct-threshold 0 –knee-method=distance. In some cases, we found that we needed to manually specify the knee threshold with –set-cell-number=[BB_NUM].
-
4)
We extract bead barcodes and UMIs from the read: umi_tools extract –bc-pattern=NNNNNNNNNN –bc-pattern2=CCCCCCCCCCCC –filter-cell-barcode –whitelist=[WHITELIST].
-
5)
We trim the remaining sequence to 25 bp matching the probe variable sequence: fastx_trimmer -f 1 -l 25 -z -Q33.
-
6)
We map reads to a custom Bowtie index (generated with bowtie-build from a custom FASTA file with one contig per probe): bowtie -v 1.
-
7)
We get sorted and indexed BAM files with SAMTools: View, sort, and index.
-
8)
We construct a table of reads grouped by UMI, transcript, and bead barcode: umi_tools group –per-cell.
-
9)
We feed the resulting (UMI, transcript, bead barcode) tuples into our custom bead barcode clustering algorithm. The purpose of this step (explained in greater detail in SI Appendix, Note S4) is to identify clusters of bead barcodes that share more UMIs than expected by chance, indicating physical coconfinement of the beads carrying these barcodes in the same droplet, and merge them, so as to avoid overcounting the same cell. The output of this algorithm is a new whitelist, this time with bead barcodes from the same droplet grouped together.
-
10)
We repeat steps 4 to 7 above using the new whitelist, this time grouping together all reads that came from any bead barcode in the same droplet: umi_tools extract –error-correct-cell –bc-pattern=NNNNNNNNNN –bc-pattern2=CCCCCCCCCCCC –filter-cell-barcode –whitelist=[NEW_WHITELIST].
-
11)
We generate a count table of UMIs per probe per droplet: umi_tools count –per-gene –per-contig –per-cell.
Cell Culture.
K562.
The immortalized myelogenous leukemia K562 cell line was obtained from ATCC (ATCC, CCL-243). Cells were maintained in RPMI medium 1640 (Corning, 10-040-CM) supplemented with 10% fetal bovine serum (Life Technologies, 16140071) and 1% penicillin-streptomycin (Life Technologies, 15140163). Cell lines were cultured at 37 °C with 5% CO2 and were subcultured twice a week by aspirating off all cell culture medium, except 0.5 mL in a T-75 flask and 1 mL in a T-175 flask, and refreshing the flasks with 13 mL and 49 mL of medium, respectively.
THP1.
The human monocyte THP1 cell line was obtained from ATCC (TIB-202). Cells were maintained in RPMI medium 1640 (Corning, 10-040-CM) supplemented with 10% fetal bovine serum (Life Technologies, 16140071) and 1% penicillin-streptomycin (Life Technologies, 15140163). Cell lines were cultured at 37 °C with 5% CO2 and were subcultured twice a week by aspirating off all cell culture medium, except 0.5 mL in a T-75 flask and 1 mL in a T-175 flask, and refreshing the flasks with 13 mL and 49 mL of medium, respectively. For treatment experiments, cells were stimulated using cell culture medium with LPS at a final concentration of 0.5 µg/mL, diluted from a 5 mg/mL LPS stock (Millipore Sigma, L3024), for 20 h. See SI Appendix, Note S5 for more details on computational analysis of THP1 RNA-seq data.
Hybridization Chain Reaction.
We performed smFISH experiments using HCR. All HCR v3 reagents (probes, hairpins, and buffers) were purchased from Molecular Technologies. Thin sections of tissue (10 µm) were mounted in 24-well glass-bottom plates (VWR, 82050–898) coated with a 1:50 dilution of APTES (Sigma, 440140). Cells were spun at 350 × g for 15 min onto 24-well glass-bottom plates (VWR, 82050-898) coated with WGA (VWR, 80057-710). The following solutions were added to the tissue/cells: 10% formalin (VWR, 100503-120) for 15 min, two washes of 1× PBS (ThermoFisher Scientific, AM9625), ice cold 70% EtOH at −20 2 h to overnight, three washes 5× SSCT (ThermoFisher Scientific, 15557044, with 0.2% Tween-20), Hybridization buffer (Molecular Technologies) for 10 min, probes in Hybridization buffer overnight, four 15-min washes in Wash buffer (Molecular Technologies), three washes 5× SSCT, Amplification buffer (Molecular Technologies) for 10 min, heat denatured hairpins in Amplification buffer overnight, three 15-min washes in 5× SSCT (1:10,000 DAPI, VWR, TCA2412-5MG, in the second wash), and storage/imaging in 5× SSCT. Imaging was performed on a spinning-disk confocal (Yokogawa W1 on Nikon Eclipse Ti) operating NIS-elements AR software. Image analysis and processing was performed on ImageJ Fiji. For K562 cells, StarSearch (https://rajlab.seas.upenn.edu/StarSearch/launch.html) was used to quantify HCR signal in tiff images processed to the same settings using ImageJ Fiji. For HCR quantification in kidney slices, we generated cell masks for parietal epithelial cells and podocytes using Fiji. Cell boundaries were determined by visual inspection. We then calculate the median background-subtracted intensity within each cell mask, where the background is taken as the median fluorescence intensity outside cellular regions.
Determining HyPR-Seq Minimum Specificity.
We computed the relative HyPR-seq signal (counts per cell relative to nontargeting probes) for all probes tested in THP1 cells. (We decided to focus on our experiments in THP1 cells, since we targeted more lowly expressed genes than in K562 cells.) Sixteen of 18 genes tested had at least two probes with >10-fold signal compared to background, including all 15 genes expressed >1 TPM, which we estimate to be our detection threshold (SI Appendix, Fig. S4E).
qPCR.
RNA extraction was performed according to manufacturer’s instructions using Qiagen RNeasy Plus Mini Kit (74136). cDNA was made according to manufacturer’s instructions using Invitrogen SuperScript III First Strand Synthesis (11752-050). Ten percent of undiluted cDNA was loaded into each RT-PCR according to manufacturer’s instructions using SYBR Green I Master (Roche, 04707516001). For qPCR primers see Dataset S4.
Mixing and Single-Cell Purity Experiment.
Two K562 cell lines, each containing a specific detection barcode, were subjected to the standard HyPR-seq protocol. Probes for all 16 possible barcodes were added to the probe mixture during the hybridization step. At the droplet generation step, equal concentrations of each cell line (5,000 cells each) were mixed and loaded into the same well. The droplets containing the cell line mixture were then subjected to the standard PCR and downstream processing for library preparation.
Construction of a gRNA Vector Detectable by HyPR.
We constructed a vector (sgOpti-HyPR) capable of expressing both a gRNA and a “detection barcode” by modifying sgOpti (Addgene 85681) to insert a 400-bp fragment between the puromycin resistance cassette and the Woodchuck hepatitis virus posttranscriptional regulatory element (WPRE). This fragment contains three 52-bp binding sites that, when transcribed into RNA, can be recognized by HyPR-seq probes (probe sequences in Dataset S1 and barcode sequences in Dataset S5). Probe binding sites were selected from a previously validated set of orthogonal 25mer DNA barcode probes (44). Because each detection barcode is composed of three unique probe binding sites, this design allows us to combinatorially encode a large number of gRNAs using a small number of probes. For example, in our study we used 48 probes to recognize each of our 16 detection barcodes; in theory, this allows us to encode gRNAs using all possible combinations of binding sites, facilitating large-scale screens. The binding sites are separated by 50 bp of random sequence and are flanked by primer binding sites. gBlocks containing 16 unique detection barcodes were ordered from IDT, amplified using BrainBar-sgOpti-FWD and BrainBar-sgOpti-REV (Dataset S4), and added to sgOpti digested with MluI (New England Biolabs) using Gibson assembly. Sanger sequencing to confirm the barcode sequences was done using Seq916 (Dataset S4). Knockdown of GATA1 using gRNAs against its transcription start site (TSS) and canonical enhancers (e-GATA1 and e-HDAC6) was identical compared to sgOpti alone, confirming the efficacy of the new plasmid.
Generation of K562 Cell Lines for GATA1 CRISPR Experiment.
gRNAs targeting regulatory elements in the GATA1 locus (GATA1 TSS, HDAC6 TSS, e-GATA1, and e-HDAC6) as well as nontargeting controls were cloned into the sgOpti-HyPR vector, as previously described for sgOpti (29). Each guide was cloned independently into a separately barcoded version of sgOpti-HyPR, so guide–barcode pairings were known in advance (Dataset S6). In these vectors, the gRNA and barcodes are located 1.2 kb away from each other. To minimize viral reassortment, we prepared lentivirus for each of the 16 gRNAs separately, by plating 550 K HEK293T cells on six-well plates (Corning), transfecting 24 h later with 1 μg dVPR, 300 ng VSVG, and 1.2 μg transfer plasmid using XtremeGene9 (Roche Diagnostics), changing media 16 h later, and harvesting viral supernatant 48 h posttransfection. Stable cell lines expressing one gRNA–barcode pair were generated by separate lentiviral transductions in 8 µg/mL polybrene by centrifugation at 1,200 × g for 45 min with 200,000 cells per well in 24 well plates. Twenty-four hours after transduction cells were selected with 1 µg/mL puromycin (Gibco) for 72 h, then maintained in 0.3 µg/mL puromycin. Separately infected cells were counted and pooled after selection with puromycin and KRAB-dCas9 (TRE-KRAB-dCas9-IRES-BFP, Addgene 85449) was induced with 1 µg/mL doxycycline (Millipore Sigma, D3072) for 24 h before experiments. See SI Appendix, Note S5 for more details on computational analysis of GATA1 CRISPR data.
Kidney Single-Cell Dissociation.
All animal work was done according to Broad Institute Institutional Animal Care and Use Committee (IACUC) protocol #0061-07-15-1. Two BTBR wt/wt and two homozygous BTBR ob/ob male mice at 12 wk of age (Jackson Laboratories, 004824) were anesthetized using 4% Isoflurane (029405, Henry Schein Animal Health), then transferred to a nose-cone supply for the duration of the procedure. Performing a combination of blunt dissection and scissor-assisted dissection techniques, the visceral organs were exposed before opening the thoracic cavity to show the thoracic organs. The right atrial chamber was lacerated with scissors before inserting a 27-gauge scalp vein butterfly needle (Excel International, 14-840-38) into the left ventricular chamber and perfusing with 10 to 20 mL of ice-cold 1× PBS (ThermoFisher Scientific, 10010023) using a variable-speed peristaltic pump (VWR, 70730–062) until the heart stops beating and the liver blanches. The kidneys were removed, cut in half, and placed in ice-cold 1× PBS before the renal capsule is removed and the tissue is stored on ice in 1× PBS. Before, we prepared 2.5 mg/mL Liberase TH by diluting 10 mg of Liberase TH (Sigma Aldrich, 5401135001) in 5 mL of DMEM/F12 (Life Technologies, 11320033) and stored at −20 °C. Right before manual dissociation, we thawed and diluted one 100 μL aliquot of 2.5 mg/mL Liberase TH with 0.9 mL of DMEM/F12 per half kidney to make 1× Liberase TH in DMEM/F12. Each half kidney was transferred to a Petri dish and manually dissociated using tweezers and a razor blade, then resuspended well using a 1-mL pipette tip in 1 mL of 1× Liberase TH in an Eppendorf tube. Tubes were incubated in a thermomixer at 600 rpm at 37 °C for 2 h, and were gently and thoroughly pipetted up and down with a 1 mL pipette tip every 10 min. Two kidney halves from the same mouse were combined and homogenized using 40 passes of a dounce homogenizer on ice. This was repeated for the other kidney before combining all samples from the same mouse in a 50-mL conical and adding 40 mL 10% FBS RPMI media to stop the digestion. The media was prepared prior by adding 50 mL FBS (Life Technologies, 16140071) to a 500-mL bottle RPMI (Corning, 10-040-CV) media, then centrifuged at 500 × g for 5 min at room temperature. Next, we aspirated off the supernatant and resuspended pellet in 4 mL of Red Blood Cell Lysing Buffer Hybri-Max (Sigma-Aldrich, R7757-100ML), and centrifuged at 500 × g for 5 min at room temperature. We next aspirated off the supernatant and resuspended pellet in 1 mL Accumax (Stemcell Technologies, 07921) for 3 min at 37 °C, added 20 mL of 10% FBS RPMI to neutralize the Accumax, and centrifuged at 500 × g for 5 min at room temperature. We then aspirated off the supernatant and resuspended the pellet in 4 mL 0.4% BSA/PBS, prepared by dissolving 80 mg of BSA (Sigma-Aldrich, A9418-10G) in 20 mL of PBS the day before. Filtered solution was passed through a 30-µm filter (Corning, 351059), then through a 20-µm pluristrainer (Pluriselect, 43-50020-01). We diluted the sample to assess the cell number and viability using Trypan blue (Sigma-Aldrich, T8154-100ML) and a cellometer (Nexcelom Bioscience, Cellometer Auto T4). Samples were kept on ice before proceeding to fixation in the HyPR-seq protocol. See SI Appendix, Note S5 for more details on computational analysis of kidney data.
Splenocyte Single-Cell Methods for scRNA-Seq and HyPR-Seq.
C57BL/6 mice (strain JR#000664) were acquired from the Jackson Laboratory. Animal procedures were performed under Broad Institute IACUC protocol number 0227-09-18 in accordance with institutional and governmental guidelines.
We followed previously described methods for isolating murine splenocytes (45). Briefly, we dissected mice and harvested the spleens on ice in (1× PBS Gibco, 10010-023). We washed spleens in RPMI media (Gibco, 22400-089) and using a scalpel individually cut the spleens into small pieces. We digested the tissue with a 10× collagenase D (Sigma Aldrich 11088866001)/DNase I (Qiagen, 79524) solution (0.05 g collagenase D and 10 μL DNase I in 1 mL of 1× PBS) diluted to 1× in RPMI by incubation at 37 °C shaking (400 rpm) for 30 min. We transferred the dissociated tissue onto a 70-µm cell strainer (Falcon, 352350) and carefully used a 3-mL syringe plunger (BD, 309578) to compress the tissue through the filter. The filtered solution was centrifuged for 10 min at 300 × g and 4 °C and we resuspended and incubated the subsequent pellet in ACK lysis buffer (Gibco, A1049201) for 10 min at room temperature to lyse the red blood. We quenched the reaction with RPMI media and centrifuged the cells again, then washed twice with cold PBS and used this for final resuspension. We filtered the cells through a 40-µm filter (pluriSelect, 43-50040-01) prior to counting the cells using a disposable hemocytometer (INCYTO, DHC-N01-2). We controlled for cell viability using a live/dead stain (Invitrogen, S34860).
We performed single-cell RNA-seq on splenocytes using the Chromium Single Cell 3′ v3 kit. We performed HyPR-seq as described, in two separate experiments testing different numbers of probes. In one experiment we included all probes designed for 179 genes, whereas for the other we only included only a subset (48 genes) of the genes. See SI Appendix, Note S5 for more details on computational analysis of splenocyte data.
Supplementary Material
Acknowledgments
We thank Jason Buenrostro, Christoph Muus, Aviv Regev, Arjun Raj, Vijay Sankaran, and Tung Nguyen for discussions. This work was supported by the Broad Institute; an NIH Early Independence Award 1DP5OD024583 (to F.C.); the Schmidt Fellows Program at the Broad Institute (F.C.); and NIH Pathway to Independence Awards 1K99HG009917 and R00HG009917 (to J.M.E.). J.M.E. was supported by the Harvard Society of Fellows and the Basic Science and Engineering Research Initiative at the Lucile Packard Children’s Hospital at Stanford University. S.G.R. was supported by the Hertz Graduate Fellowship and the National Science Foundation Graduate Research Fellowship Program (Award 1122374). Q.W. was supported by the Nakajima Foundation Scholarship.
Footnotes
Competing interest statement: J.L.M., V.S., S.G.R., F.C., and J.M.E. are inventors on patent applications filed by the Broad Institute related to this work (62/676,069 and 62/780,889). E.S.L. serves on the Board of Directors for Codiak BioSciences and Neon Therapeutics, and serves on the Scientific Advisory Board of F-Prime Capital Partners and Third Rock Ventures; he is also affiliated with several nonprofit organizations, including serving on the Board of Directors of the Innocence Project, Count Me In, and Biden Cancer Initiative, and the Board of Trustees for the Parker Institute for Cancer Immunotherapy. E.S.L. has served and continues to serve on various federal advisory committees.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010738117/-/DCSupplemental.
Data Availability.
HyPR-seq and scRNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE158002) (46). smFISH images are available at the Center for Open Science OSF (https://osf.io/9acqe/) (47). A detailed protocol is available at protocols.io, https://www.protocols.io/view/hypr-protocol-59rg956 (48).
References
- 1.Ramsköld D., et al. , Full-length mRNA-seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picelli S., et al. , Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Hashimshony T., Wagner F., Sher N., Yanai I., CEL-seq: Single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2, 666–673 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Jaitin D. A., et al. , Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shalek A. K., et al. , Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanay A., Regev A., Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adamson B., et al. , A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixit A., et al. , Perturb-seq: Dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datlinger P., et al. , Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svensson V., et al. , Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14, 381–387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., et al. , Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell 73, 130–142.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Bagnoli J. W., et al. , Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat. Commun. 9, 2937 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saikia M., et al. , Simultaneous multiplexed amplicon sequencing and transcriptome profiling in single cells. Nat. Methods 16, 59–62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mimitou E. P., et al. , Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallejo A. F., et al. Resolving cellular systems by ultra-sensitive and economical single-cell transcriptome filtering. bioRxiv : 10.1101/800631 (6 November 2019). [DOI] [PMC free article] [PubMed]
- 16.Uzbas F., et al. , BART-seq: Cost-effective massively parallelized targeted sequencing for genomics, transcriptomics, and single-cell analysis. Genome Biol. 20, 155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shum E. Y., Walczak E. M., Chang C., Christina Fan H., Quantitation of mRNA transcripts and proteins using the BD Rhapsody™ single-cell analysis system. Adv. Exp. Med. Biol. 1129, 63–79 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Replogle J. M., et al. , Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol. 38, 954–961 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan H. C., Fu G. K., Fodor S. P., Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science 347, 1258367 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Femino A. M., Fay F. S., Fogarty K., Singer R. H., Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Raj A., Peskin C. S., Tranchina D., Vargas D. Y., Tyagi S., Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, e309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levesque M. J., Raj A., Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat. Methods 10, 246–248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi H. M. T., Beck V. A., Pierce N. A., Next-generation in situ hybridization chain reaction: Higher gain, lower cost, greater durability. ACS Nano 8, 4284–4294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi H. M. T., et al. , Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dirks R. M., Pierce N. A., Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. U.S.A. 101, 15275–15278 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasperini M., et al. , A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176, 1516 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S., Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie S., Duan J., Li B., Zhou P., Hon G. C., Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell 66, 285–299.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Fulco C. P., et al. , Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fulco C. P., et al. , Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torre E., et al. , Rare cell detection by single-cell RNA sequencing as guided by single-molecule RNA FISH. Cell Syst. 6, 171–179.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clee S. M., Nadler S. T., Attie A. D., Genetic and genomic studies of the BTBR ob/ob mouse model of type 2 diabetes. Am. J. Ther. 12, 491–498 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Harder J. L. et al.; European Renal cDNA Bank (ERCB); Nephrotic Syndrome Study Network (NEPTUNE) , Organoid single cell profiling identifies a transcriptional signature of glomerular disease. JCI Insight 4, 122697 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czerniecki S. M., et al. , High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22, 929–940.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J., et al. , Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindström N. O., De Sena Brandine G., Ransick A., McMahon A. P., Single-cell RNA sequencing of the adult mouse kidney: From molecular cataloging of cell types to disease-associated predictions. Am. J. Kidney Dis. 73, 140–142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H., Kirita Y., Donnelly E. L., Humphreys B. D., Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 30, 23–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welsh G. I., Saleem M. A., The podocyte cytoskeleton—Key to a functioning glomerulus in health and disease. Nat. Rev. Nephrol. 8, 14–21 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Lin J. S., Susztak K., Podocytes: The weakest link in diabetic kidney disease? Curr. Diab. Rep. 16, 45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alicic R. Z., Rooney M. T., Tuttle K. R., Diabetic kidney disease: Challenges, progress, and possibilities. Clin. J. Am. Soc. Nephrol. 12, 2032–2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu J., et al. , Single-cell RNA profiling of glomerular cells shows dynamic changes in experimental diabetic kidney disease. J. Am. Soc. Nephrol. 30, 533–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson P. C., et al. , The single-cell transcriptomic landscape of early human diabetic nephropathy. Proc. Natl. Acad. Sci. U.S.A. 116, 19619–19625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurka J., et al. , Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Xu Q., Schlabach M. R., Hannon G. J., Elledge S. J., Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl. Acad. Sci. U.S.A. 106, 2289–2294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arora P., Porcelli S. A., An efficient and high yield method for isolation of mouse dendritic cell subsets. J. Vis. Exp. e53824 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doughty B. R., Engreitz J. M., Chen F., HyPR-seq: Single-cell quantification of chosen RNAs via hybridization and sequencing of DNA probes (scRNA-seq dataset). Gene Expression Omnibus (GEO). http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158002. Deposited 16 September 2020. [DOI] [PMC free article] [PubMed]
- 47.Doughty B. R., HyPR-seq. Open Science Framework. https://osf.io/9acqe/. Deposited 2 November 2020.
- 48.Marshall J. L., et al. , HyPR Protocol Protocols.io. https://www.protocols.io/view/hypr-protocol-59rg956. Deposited 6 November 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
HyPR-seq and scRNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE158002) (46). smFISH images are available at the Center for Open Science OSF (https://osf.io/9acqe/) (47). A detailed protocol is available at protocols.io, https://www.protocols.io/view/hypr-protocol-59rg956 (48).