Significance
As a genetic malignancy, retinoblastoma (Rb) is caused by RB1 mutations; however, its developmental origin and drug agents for human Rb remain largely unexplored. Here we describe an innovative Rb organoid model derived from human embryonic stem cells with a biallelic mutagenesis of the RB1 gene. We identify tumorigenic growth in the Rb organoids, as well as properties consistent with human primary Rb. We confirm that the Rb cell of origin stemmed from ARR3+ maturing cone precursor cells and SYK inhibitors displaying a significant therapeutic response. Our elegant in-dish Rb organoid model can be used to efficiently and effectively dissect the origin of Rb and mechanisms of Rb tumorigenesis, as well as screen novel therapies.
Keywords: RB1, human embryonic stem cells, retinal organoids, retinoblastoma, cell of origin
Abstract
Retinoblastoma (Rb) is the most prevalent intraocular malignancy in children, with a worldwide survival rate <30%. We have developed a cancerous model of Rb in retinal organoids derived from genetically engineered human embryonic stem cells (hESCs) with a biallelic mutagenesis of the RB1 gene. These organoid Rbs exhibit properties highly consistent with Rb tumorigenesis, transcriptome, and genome-wide methylation. Single-cell sequencing analysis suggests that Rb originated from ARR3-positive maturing cone precursors during development, which was further validated by immunostaining. Notably, we found that the PI3K-Akt pathway was aberrantly deregulated and its activator spleen tyrosine kinase (SYK) was significantly up-regulated. In addition, SYK inhibitors led to remarkable cell apoptosis in cancerous organoids. In conclusion, we have established an organoid Rb model derived from genetically engineered hESCs in a dish that has enabled us to trace the cell of origin and to test novel candidate therapeutic agents for human Rb, shedding light on the development and therapeutics of other malignancies.
Retinoblastoma (Rb) is a life-threatening retinal malignancy in early childhood, with most cases diagnosed before age 5 y. The prognosis of Rb patients is rather dismal, with a global survival rate of <30% (1, 2). Despite the best currently available therapies, significant morbidities, including blindness, remain a substantial challenge (3, 4). One critical issue is the cancerous origin of Rb (5). Over the past several decades, studies using mouse Rb models have identified amacrine, horizontal cells, or Müller glial precursors as originating cells of Rb, depending on the combination of Rb family mutations used (6–8). These mouse Rb models lack human Rb features, however. In humans, cone precursors or retinal progenitors have been proposed as the cells of origin for Rb (9–13). These disparities are due mainly to the lack of a sustainable human Rb model and single-cell resolution to trace cancerous development. Establishing an effective and efficient human Rb model could help unambiguously elucidate the cell of origin of Rb, and to examine the efficacy and toxicities of novel candidate therapeutic drugs for Rb.
Although immortalized Rb cell lines have been widely used for biological and preclinical testing, they lack a self-organized three-dimensional (3D) environment. In vivo models, including genetically engineered mouse models (6, 12) and patient-derived xenografts, are often handicapped by species-specific differences (6, 14) or low engraftment rates and difficulty in genetic manipulations (15).
Human pluripotent stem cells (hPSCs) are capable of generating self-organized 3D retinal organoids (hROs), which could recapitulate retinogenesis in vitro (16–18). This model has provided an extraordinary research path for disease modeling and drug screening of human retinal diseases, especially monogenic disorders (19–21). As a genetic malignancy, Rb is attributed mainly to biallelic inactivation of RB1, a tumor-suppressor gene (2, 4, 22). These advancements prompted us to develop an in-dish model with human Rb organoids (hRBOs) through genetic manipulation and differentiation of hPSCs.
Here we generated gene-edited human embryonic stem cells (hESCs) with a biallelic RB1 mutation (RB1Mut/Mut) or ablation (RB1−/−), which further differentiated into Rb organoids in a stepwise manner. Using this innovative model, we successfully observed tumorigenesis in retinal organoids, revealed high-fidelity genetic and epigenetic signatures, delineated the cancerous cell of origin, and tested potential therapeutic drugs. Our findings showed that this human Rb model can be used efficiently and effectively for dissecting the origination of Rb and mechanisms of Rb tumorigenesis, as well as for screening potential therapies in terms of efficacy and safety.
Results
Mutagenesis of the RB1 Gene in hESCs.
To date, more than 3,000 RB1 mutations have been reported in the online RB1-LOVD database (23). The nonsense mutation p.R320X (c.958C > T) is the mutation most frequently associated with predisposition to Rb (SI Appendix, Fig. S1A). Thus, we generated p.R320X mutation knock-in hESCs using CRISPR/Cas9 genome-editing technology (Fig. 1 A and B and SI Appendix, Fig. S1 B–D). Three clones with homozygous mutation (RB1Mut/Mut) in the RB1 gene were successfully established (SI Appendix, Fig. S1 E and F).
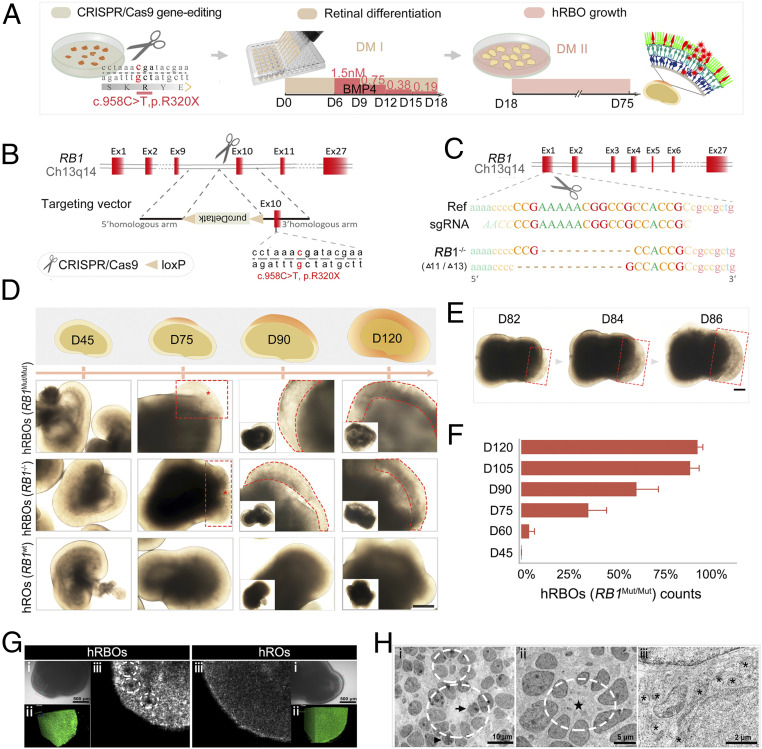
Fig. 1.
Induction of 3D hRBOs from RB1Mut/Mut and RB1−/− hESCs generated by the CRISPR-Cas9 system. (A) Scheme of the stepwise development of hRBOs. (B) Schematic view of the targeting strategy for generation of RB1Mut/Mut hESCs. (C) Targeting strategy for generation of RB1−/− hESCs. (D) Representative images of developing hRBOs in different organoid stages. (Insets) Images of the whole organoids. Wild-type hESC-derived hROs served as controls. Images in the top row show a schematic overview of developing hRBOs. Asterisks indicate the abnormal proliferating “primary foci” of the tumor. Dashed lines denote the tumor-like structures. (Scale bar: 400 µm.) (E) Representative images show the rapid expansion of tumor-like structures (dashed lines) in developing hRBOs. (Scale bar: 2,000 µm.) (F) Quantification of RB1Mut/Mut hESC-derived organoids with tumor-like structures in different organoid stages. Data are mean ± SD. n = 5. (G) Imaging of superior 3D structures of hRBOs and hROs at day 90. (i) Microscope view of screening sites in organoids with extension line. (Scale bar: 500 µm.) (ii) Imaging of screening sites in 3D rotation. (iii) Imaging of screening sites at high x-axis. Dashed lines denote Flexner–Wintersteiner rosettes. See also Movies S2 and S3. (H) TEM image of cells from day 105 hRBOs. Dashed lines denote fleurettes (i) and Flexner–Wintersteiner rosettes (ii); the pentagram marks the rosette cavity. Mitotic figures (arrowheads) represent proliferating cells, which are also represented by invagination in the nucleus (arrow). (iii) Asterisks indicate abundant mitochondria with heterogeneous morphology. (Scale bars: i, 10 µm; ii, 5 µm; iii, 2 µm.)
To further investigate the consequence of RB1 mutagenesis, we created two clones with a biallelic knockout (RB1−/−) by targeting the first exon of the RB1 gene (Fig. 1C and SI Appendix, Fig. S1F). Western blot assays showed significantly reduced pRb expression in all lines (SI Appendix, Fig. S1G), which was validated by immunocytochemistry (SI Appendix, Fig. S1H).
The RB protein family (RB1, p107, and p130) plays a critical role in maintaining hESC self-renewal (24). To investigate the effect of RB1 inactivation on hESCs, we examined the pluripotency and cell cycle properties of hESCs using flow cytometry. In RB1-mutated clones, neither the proportion of OCT3/4+ cells (SI Appendix, Fig. S2A) nor the cell cycle (SI Appendix, Fig. S2 B and C) were altered. Furthermore, RNA sequencing (RNA-Seq) analysis showed that the expression of pluripotency, self-renewal, and cell cycle-associated genes remained unaffected by the mutagenesis of RB1 in hESCs (SI Appendix, Fig. S2D). Karyotyping of these genetically engineered hESCs showed a normal 46,XX karyotype (SI Appendix, Fig. S2E). Taken together, these results demonstrate that the RB1Mut/Mut and RB1−/− hESC lines sustained the primordial state without changing pluripotency and the cell cycle of the hESC lines.
Tumorigenesis in Retinal Organoids Derived from Mutant hESC Lines.
Inspired by the speculation that Rb originates from developing retinal neurons (2, 12, 25), we attempted to investigate Rb tumorigenesis through differentiation of retinal organoids from mutant hESC lines. We generated human retinal organoids using stepwise induction (Fig. 1A and SI Appendix, Fig. S3A) as described previously (16, 17, 19). As expected, the molecular and morphogenetic properties of the organoids recapitulated the developing human retina (SI Appendix, Fig. S3 B–D) (26, 27). There was no difference between hRBOs and hROs before differentiation day 45 (Fig. 1D and SI Appendix, Fig. S3D). Remarkably, at day 75, tumor-like “primary foci” were clearly visible in hRBOs derived from both RB1−/− and RB1Mut/Mut hESCs (Fig. 1D; SI Appendix, Fig. S3F; and Movie S1). In hRBOs, tumor-like structures exhibited obvious uneven interior density, ill-defined edges, and larger size (Fig. 1D and SI Appendix, Fig. S3F). Of note, the tumor-like foci expanded rapidly from the masses thereafter (Fig. 1 E and F and SI Appendix, Fig. S3E).
To further characterize hRBOs, we examined surface topography and internal structure in live organoids with deep optical coherence tomography. As shown in Fig. 1G, ill-defined edges, uneven reflectance, and notable inner cavities resembling Flexner–Wintersteiner rosettes were observed (Movies S2 and S3). Furthermore, transmission electron microscopy (TEM) demonstrated the same ultrastructural features as seen in primary Rb tumors, including a high nucleocytoplasmic ratio, abundant mitochondria with heterogeneous morphology, and tightly packed clusters of cells and rosettes (including Flexner–Wintersteiner rosettes characteristic of Rb and fleurettes characteristic of retinoma), as well as the presence of condensed chromosomes (mitotic figures) (Fig. 1H and SI Appendix, Fig. S3G). Taken together, these results demonstrate that tumorigenesis occurred in hRBOs, indicating that organoid Rbs differentiated from each mutated hESC line could be used for disease modeling.
Rb-Related Molecular Signatures in Induced hRBOs.
To further elucidate the molecular characteristics of hRBOs, we analyzed the gene expression and DNA methylation profiles of hRBOs by RNA-Seq and whole-genome bisulfite sequencing (WGBS). The transcriptomic profiles of hRBOs at different stages clearly demonstrated that proto-oncogenes and Rb signature genes (MKI67, spleen tyrosine kinase [SYK], proliferating cell nuclear antigen [PCNA], DEK, and TOP2A) (28–30) were markedly up-regulated (Fig. 2A). In contrast, the tumor-suppressor genes or low-expressed genes (S100B, VIM, and LATS2) in primary Rb tumors were significantly down-regulated compared with normal hROs (Fig. 2A). Notably, transcriptomic dysregulation was more significant after day 75 (Fig. 2A).
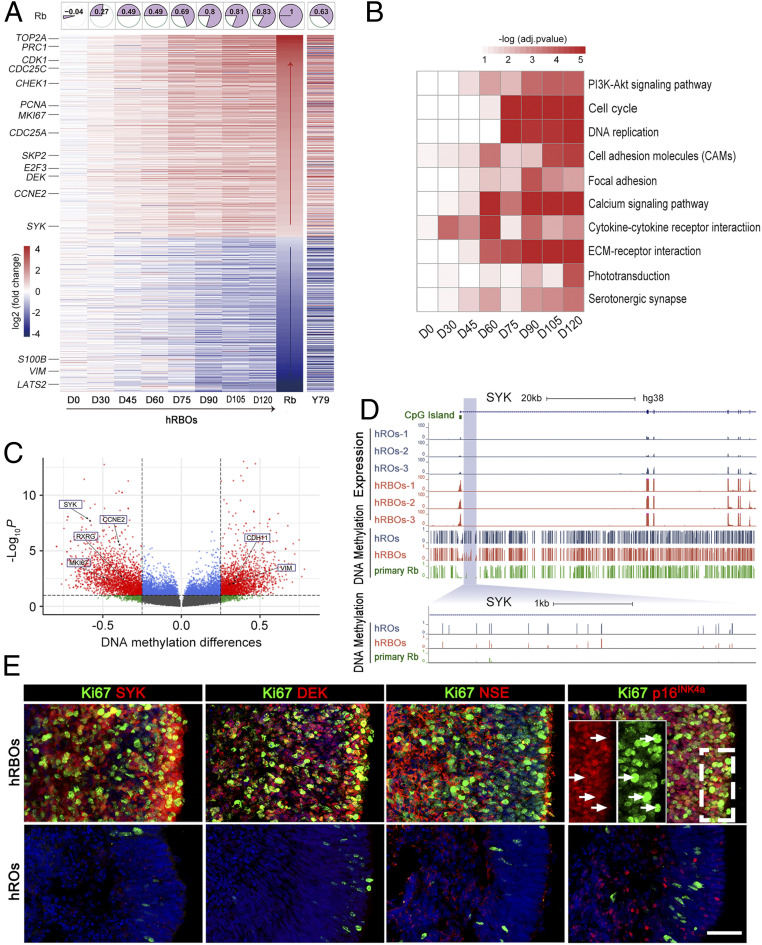
Fig. 2.
Molecular characterization of hRBOs. (A) Heatmaps showing the average log2-fold difference in the indicated genes selected from previously published transcriptome data between the hRBOs and normal hROs at different organoid stages. Representative signature genes (including the known oncogenes or tumor-suppressor genes) of human primary Rbs are shown on the left. Expression data of the indicated genes between the Y79 cell line and normal hROs older than 75 d are also shown in heatmaps. Rb, published transcriptomes of human primary Rbs (28). The top row shows correlation heatmaps of hRBOs at different organoid stages and human primary Rbs based on the expression of indicated genes. The larger the colored fan area, the greater the correlation. (B) Heatmaps of the selected top 10 KEGG pathway enrichments for differentially expressed genes (adjusted log2 fold change >1) between hRBOs and hROs at different organoid stages. Each row represents the enriched pathway terms. (C) Volcano plot visualization of significant differential methylated genes of hRBOs compared to hROs at day 120. Each point represents a distinct gene, and interesting genes are indicated. The lines represent average methylation differences of 0.25 and adjusted P value < 0.05, respectively. (D) Genomic snapshots of RNA-Seq and WGBS for SYK in hRBOs and parental hROs. A representative methylation level of SYK from human primary Rb sample (9) is also shown. An enlarged view of the specific site is shown below. (E) Immunostaining of Rb signature genes, Ki67, SYK, DEK, NSE, and p16INK4a, in hRBOs and hROs at day 105. (Insets) Magnified views of the boxed region. Arrows indicate Ki67+ tumor cells with weak cytoplasmic staining for p16INK4a. (Scale bar: 50 µm.)
When we compared the transcriptomes of hRBOs, primary Rb tumors (28), and the Y79 cell line, we found that 1,014 differentially expressed genes (574 up-regulated and 440 down-regulated) in hRBOs (after differentiation for >75 d) were similar to those reported in primary Rb tumors (Fig. 2A). In the gene expression profile, the hRBOs were more closely correlated with human primary Rb tumors than the Y79 cell line, and this correlation markedly increased across the developmental stages (Fig. 2A), consistent with the results of principal component analysis (SI Appendix, Fig. S4 A and B). A global comparative transcriptomic analysis of all genes between hROs and hRBOs revealed that the number of genes changed by more than twofold and gradually increased with the differentiation stage (SI Appendix, Fig. S4 C and D). Interestingly, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed dysregulation of key cancer-related signaling pathways involved in human Rb tumorigenesis, including the cell cycle and PI3K-Akt pathways (Fig. 2B and SI Appendix, Fig. S4E).
To further understand transcriptional regulation in Rb tumorigenesis, we focused on alterations in DNA methylation, an important epigenetic modification that contributes to transcription dysregulation in tumors. A total of 8,968 differentially methylated regions (DMRs) (4,597 hypermethylation and 4,371 hypomethylation) were identified (Fig. 2C). As expected, the DNA methylation states of key Rb-related genes, such as SYK, RXRG, MKI67, CCNE2, VIM, and CDH11, were significantly changed in hRBOs compared with control hROs (average methylation difference >0.25; adjusted P < 0.05) (Fig. 2C). Of note, SYK in the PI3K-Akt pathway, a pivotal marker gene, was identified as one of the most up-regulated genes during Rb tumorigenesis in hRBOs (Fig. 2A). The methylation level was markedly decreased at the CpG island shore located in the SYK promoter region in hRBOs (average methylation difference = 0.504’ adjusted P = 1.13e-07) and human primary Rb tumors (9) compared with control hROs (Fig. 2D), while the methylation level of VIM, a non–Rb-related gene, was significantly increased (SI Appendix, Fig. S5A), in line with the expression change. Moreover, we found that the DMRs were distributed along the whole genome and enriched in the noncoding regions compared with the coding regions (SI Appendix, Fig. S5 B and C). We also inspected the colocalization between DMR and CGI elements (CGI, CGI shore, and CGI shelf). Most (77%) of the DMRs were distal (>4 kb) to CGI elements, and more than one-half of CGI-proximal DMRs were significantly enriched in the CGI shore regions (χ2 test, P = 2.9e-222) (SI Appendix, Fig. S5D). KEGG pathway analysis identified DMRs closely related to Axon guidance, Pathways in cancer, and the PI3K-Akt signaling pathway (SI Appendix, Fig. S5E).
Immunostaining of organoid sections further confirmed that pRb was readily detected in control hROs, but not in derived hRBOs (SI Appendix, Fig. S5F). In line with the findings from transcriptomic analysis, >70% of cells in hRBOs were highly proliferative (Ki67+), even after week 15 (Fig. 2E and SI Appendix, Fig. S5 F–H), while few cells in normal organoids continued to divide. p16INK4a (CDKN2A), which has been used as a potential marker to distinguish retinoma cells (Rb precursors, Ki67−) from Rb and retina (29), stained strongly positive in hRBOs (Fig. 2E and SI Appendix, Fig. S5H). We also confirmed markedly increased expression of other oncogene markers (SYK, DEK, and NSE) in hRBOs, strongly indicating that Rbs and retinoma-like cells were transformed in the cancerous organoids (Fig. 2E and SI Appendix, Fig. S5H). We did not observe any significant change in the expression of p53 (SI Appendix, Fig. S5F), an important tumor repressor. These results support the previous assumption that loss of RB1 function may initiate retinoma and subsequently lead to malignant transformation and progression (29, 31). Taken together, these results demonstrate that organoid Rb recapitulates human Rb in gene expression, DNA methylation, and protein markers.
Tumorigenicity of hRBOs In Vivo.
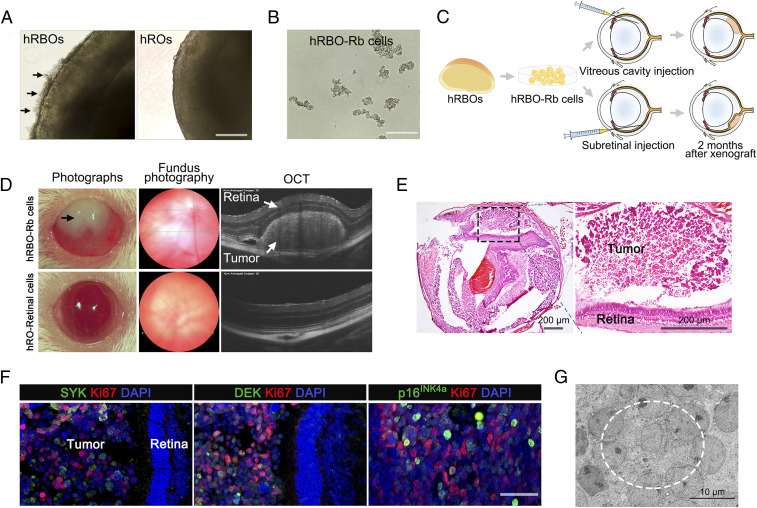
Since hRBOs retained viability and expanded rapidly after 10 wk in vitro, we attempted to assess the tumorigenicity of cancerous cells in vivo. As shown in Fig. 3A, excessively proliferating Rb-like cells in 120-d-old hRBOs could migrate to the medium in suspension conditions (Fig. 3 A and B and SI Appendix, Fig. S6A). We engrafted these cells into the subretinal or vitreous space of immunodeficient NSG mice. Fundography and optical coherence tomography (OCT) imaging were used to monitor the intraocular grafts (Fig. 3C). At 2 mo postinjection, 9 out of 12 of subretinally injected mice and 9 out of 11 vitreous cavity-injected mice with hRBO-derived Rb-like cells showed visible tumors in the intraocular grafts, but an intraocular tumor was not observed in any eyes injected with hRO-derived retinal cells (Fig. 3D and SI Appendix, Fig. S6 B and D), as further confirmed by hematoxylin and eosin (H&E) staining (Fig. 3E and SI Appendix, Fig. S6C). No significant differences in tumor formation were seen between the subretinal and vitreous cavity grafts. Moreover, immunocytochemistry revealed extensive tumor foci positive for Ki67 and oncogene markers (NSE, SYK, and DEK), indicating the tumor characteristics of Rb (Fig. 3F). TEM also demonstrated a high nucleocytoplasmic ratio, abundant mitochondria with heterogeneous morphology, tightly packed clusters of cells and rosettes, and numerous mitotic figures (Fig. 3G and SI Appendix, Fig. S6E), in line with the findings observed in the in-dish hRBOs. These results demonstrate that hRBOs could retain viability and expand after becoming engrafted into immunodeficient mice.
Fig. 3.
Orthotopic engraftment of hRBOs in NSG mice. (A) Representative bright-field image of 120-d-old hRBOs and hROs. Arrows indicate excessive outgrowth of Rb cells, subsequently leading to detachment from the organoids. (Scale bar: 400 µm.) (B) Rb cells derived from hRBOs (hRBO-Rb cells) can be maintained in long-term culture in suspension. (Scale bar: 200 µm.) (C) Schematic view of the procedure for generating tumor engraftments in NSG mice. (D) Ophthalmic examination of mouse eyes after subretinal engraftment. Representative photograph of engrafted eyes at 2 mo after subretinal injection (Left). hRBO-Rb cell-engrafted eye showing leukocoria-like features (black arrow). Representative fundus photography and OCT images of an hRBO-Rb cell-engrafted eye showing an obvious Rb tumor in the retina. Retinal cells derived from hROs (hRO-retinal cells) served as normal controls (Bottom). (E) Representative H&E staining of hRBO-Rb cell-engrafted eyes at 2 mo after subretinal injection. The boxed region is magnified on the right. (Scale bar: 200 µm.) (F) Immunostaining of Rb signature genes Ki67, SYK, DEK, and p16INK4a in hRBO-Rb cell-engrafted eyes at 3 mo after subretinal injection. (Scale bar: 50 µm.) (G) Representative TEM image of Rb engraftment. The dashed line denotes the typical rosette structure. (Scale bar: 10 µm.)
Cellular Identity of hRBOs by Single-Cell Transcriptomes Analysis.
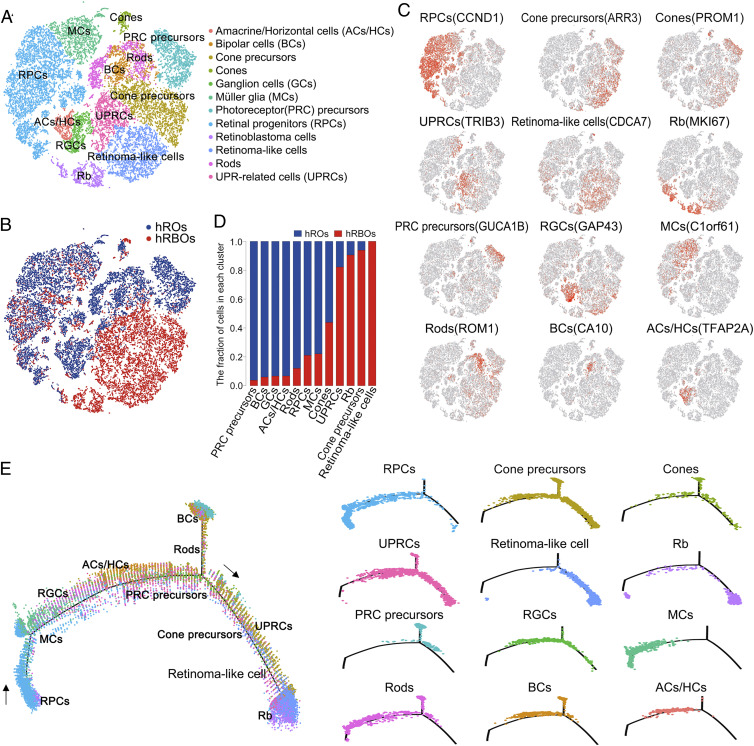
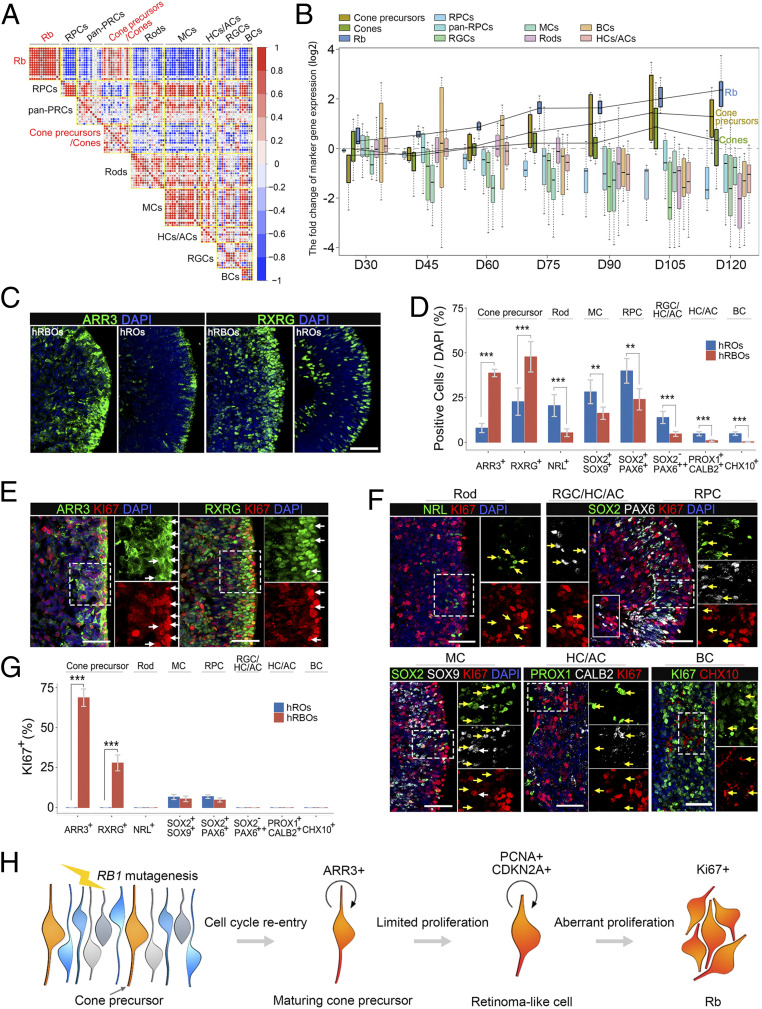
To define heterogeneity, cell types, and tumor-initiating cells (32, 33), single-cell RNA-Seq analysis was performed in both hRBOs and control hROs at day 90. On average, 4,274 hRBO and 3,080 hRO UMIs and 1,784 hRBO and 1,297 hRO genes were detected per cell. As a result, 9,665 and 12,218 single-cell transcriptomes were obtained from hRBOs and control hROs, respectively. As expected, based on cluster-specific marker genes, there were nine distinct retinal cell types, including retinal progenitor, precursor, cone, rod, bipolar, horizontal/amacrine, ganglion and Müller glial cells in control hROs (Fig. 4 A–C). Notably, we found four additional cell clusters—Rb cells, retinoma-like cells, unfolded protein response (UPR)-related cells, and excessive cone precursors—in hRBOs (Fig. 4 A, B, and D). Among these clusters, the Rb cell cluster highly expressed MKI67, CDK1, TOP2A, KIF14, and CDC25C but not VIM, GLUL, CLU, and FRZB (Fig. 4C and SI Appendix, Fig. S7A). The retinoma-like cell cluster strongly coexpressed Rb-enriched genes, including PCNA and DNA replication licensing factors (MCM2-7) and cone precursor marker genes (Fig. 4C and SI Appendix, Fig. S7A). Different from Rbs, these retinoma-like cells have greatly increased expression of CDKN2A and CDCA7 and reduced expression of MKI67 and KLF14 (SI Appendix, Fig. S7B). Thus, we speculated that retinoma-like cells could be an intermediate cell stage between premalignant cone precursors and tumor cells. Interestingly, the number of normal retinal cells was dramatically reduced while the amount of cone precursors was increased in hRBOs (Fig. 4 A, B, and D). In addition, we identified a cluster of abundantly expressed UPR-related genes (Fig. 4 A–C), indicative of endoplasmic reticulum (ER) stress and essential for tumorigenesis and progression in many cancer types (34–36). Notably, we did not detect up-regulation of proapoptotic genes in this cluster, suggesting that these cells were viable and could represent transformed malignant cells. RNA-Seq and immunostaining analysis also confirmed the presence of these UPR-related cells in hRBOs (SI Appendix, Fig. S7 C and D). In addition, an excessive cluster of cone precursors present only in hRBOs expressed abundant cone precursor markers, including ARR3 and RXRG, implicating the cell of origin of Rbs (Fig. 4C and SI Appendix, Fig. S7A).
Fig. 4.
Single-cell RNA-Seq analysis of hRBOs and hROs. (A) t-distributed stochastic neighbor embedding (tSNE) visualization of 21,883 cells in 90-d-old hRBOs and control hROs profiled, with cells color-coded according to cell type annotation. (B) tSNE visualization of 21,883 cells profiled, with cells color-coded according to sample type. (C) Expression of marker genes for the main cell clusters on the tSNE map. (D) Fraction of cells in each cell cluster. (E) Monocle pseudospace trajectory revealing the Rb progression, color-coded according to cell clusters.
To check the effects of cell cycle heterogeneity in the single-cell RNA-Seq data, we reanalyzed the single-cell RNA-Seq data by regressing out cell cycle scores during data scaling. The cell types did not show any obvious changes (SI Appendix, Fig. S7E), suggesting that the cell cycle was not driving cell cluster generation. The violin plots of UMI, mitochondrial, and ribosome counts shown in SI Appendix, Fig. S8 A–C indicate that these counts did not strongly affect clusters. Together, these single-cell transcriptomes reveal distinct cellular diversity and compositions of hRBOs, indicating that the cancerous origin of Rbs may be cone precursors.
Maturing Cone Precursor Is the Cell of Origin of Rbs in hRBOs.
We next sought to confirm the cell of origin of Rbs in the hRBOs using single-cell pseudo-time trajectory analysis and immunocytochemistry. As expected in the retinal developmental trajectory of both the hROs and hRBOs, the population of retinal progenitors was present at the start of the trajectory (Fig. 4E and SI Appendix, Fig. S8 E and F). Notably, across the developmental trajectory specific to hRBOs, ARR3+ maturing cone precursors were present at the branch point, followed sequentially by retinoma-like and Rb cells (Fig. 4E and SI Appendix, Fig. S8E). which was consistent with the results of RNA velocity analysis (SI Appendix, Fig. S8D), a recently developed cell fate tracing and cell lineage reconstructing approach (37). These results suggest that the maturing cone precursor cells might develop into premalignant cone precursors, become converted to retinoma-like cells, and eventually terminate at Rb cells (Fig. 4E and SI Appendix, Fig. S8 D, E, and G).
To further elucidate the cell of origin, we compared the retinal cell type-specific gene expression at different organoid stages based on the RNA-Seq data. As shown in Fig. 5 A and B, genes enriched in retinal progenitor, rod, Müller glial, amacrine, horizontal, ganglion, and bipolar cells were significantly down-regulated in hRBOs after day 75. In contrast, the derived hRBOs exhibited dramatic and consistent up-regulation of cone precursor-enriched genes (Fig. 5 A and B). Consistent with these findings, immunostaining revealed an increasing proportion of cone precursor cells (RXRG+ or ARR3+) and a decreasing number of other retinal neurons in hRBOs with tumorigenesis (Fig. 5 C and D and SI Appendix, Fig. S9A). RNA-Seq data showed no difference between hRBOs and hROs in the expression levels of genes specific for proliferative cells (MKI67), early photoreceptor precursors (THRB and RXRG for cones, NR2E3 for rods, CRX and RORB for cones or rods), or retinal progenitors (OTX2) before day 60 (SI Appendix, Fig. S9B). Remarkably, ARR3 expression was noted at day 60 and showed an abnormal increasing trend thereafter, accompanied by significant up-regulation of MKI67 (SI Appendix, Fig. S9B), suggesting that the abnormal expression of MKI67 may begin with the initial appearance of ARR3+ maturing cone precursors in hRBO. Notably, the developmental time window of postmitotic maturing cone precursors (38) exactly matched the tumorigenesis occurring in derived hRBOs from day 60 to day 90 (SI Appendix, Fig. S3B).
Fig. 5.
Rbs in hRBOs originating from cone precursors. (A) Gene–gene correlation heatmaps of the indicated cell type signature genes from Rbs, cone precursors/cones, and other retinal cells in hRBOs. The signature genes of Rbs in hRBOs had the greatest correlation with most genes specific for cone precursors. Node size and color indicate the degree of correlation. (B) The fold change of cell type-specific marker gene expression in hRBOs and hROs at different organoid stages on days 30, 45, 60, 75, 90, 105, and 120. Most of the genes specific for cone precursors showed elevated expression in hRBOs compared with hROs from day 60. (C) Immunostaining of maturing cone precursor marker ARR3 and pan-cone precursor marker RXRG in day 105 hRBOs and hROs. (Scale bar: 50 µm.) (D) Quantification of ARR3+ or RXRG+ cone precursors, NRL+ rods, SOX2+/SOX9+ Müller cells, SOX2+/PAX6+ retinal progenitors, SOX2−/PAX6++ ganglion/horizontal/amacrine cells, PROX1+ or CALB2+ horizontal/amacrine cells, and CHX10++ bipolar cells in hRBOs and parental hROs. Data are mean ± SD; n = 5. ***P < 0.001; **P < 0.01, unpaired Student’s t test. Markers measured in cells shown in C and SI Appendix, Fig. S9A. (E) Coimmunostaining of ARR3 and RXRG with the proliferative marker Ki67 in day 105 hRBOs. Magnified views of the boxed regions are shown in the corresponding panel on the right. White arrows indicate Ki67+ tumor cells coexpressing ARR3 or RXRG. (Scale bar: 50 µm.) (F) Coimmunostaining of rod, retinal progenitor, ganglion/horizontal/amacrine cell, Müller glial cell, horizontal and amacrine cell, or bipolar cell markers with Ki67 in day 105 hRBOs. Magnified views of the dashed-line boxed regions are shown in the corresponding panels on the right, and the solid-line boxed region is shown in the left panels. Arrows indicate markers specific for different retinal cell types with (white arrows) or without (yellow arrows) Ki67 coexpression. (Scale bar: 50 µm.) (G) Quantification of the percentage of Ki67+ cells expressing the indicated markers specific for cone precursors and other retinal cell types in hRBOs as analyzed by immunostaining. Data are mean ± SD; n = 5. ***P < 0.001, unpaired Student’s t test. (H) Schematic diagram summarizing the cell of origin and development of Rbs in hRBOs.
As postmitotic cells, maturing cone precursors (ARR3+) are nonproliferative and do not express Ki67 (26). Consistently, the few Ki67+ cells detected in the normal hROs after day 75 represented a small number of Müller glial and retinal progenitor cells rather than cone precursor cells (Fig. 5 F and G). In contrast, most ARR3+ maturing cone precursors within hRBOs simultaneously expressed Ki67, indicating the mitotic property of the maturing cone precursors in cancerous organoids (Fig. 5 E and G). We did not detect Ki67 in rod, bipolar, ganglion, amacrine, or horizontal cells in hRBOs (Fig. 5 F and G). In addition, most RXRγ+/ARR3+ cone precursors were positive for Ki67, while RXRγ+ cone precursors lacking ARR3 expression in the hRBOs were negative for Ki67 (SI Appendix, Fig. S9C). Notably, CRX+ early photoreceptor precursors were postmitotic and negative for Ki67 (17). We found that CRX+/ARR3− early photoreceptor precursors were detected in the hRBOs at day 60, and none was positive for Ki67 (SI Appendix, Fig. S9D). In contrast, all ARR3+ cone precursors in the hRBOs at day 60 were strongly Ki67+, indicative of their cell cycle entry. These Ki67+/ARR3+ cone precursors were significantly increased after day 60 (SI Appendix, Fig. S9D). As expected, most ARR3+ maturing cone precursors were also positive for mitosis-associated phosphohistone H3, PCNA, and proto-oncogenic SYK (SI Appendix, Fig. S9 E–G). Only few UPR-related cells (CHOP+) expressed Ki67 in hRBOs (SI Appendix, Fig. S9H). Moreover, in hRBO-derived intraocular tumors, while genes enriched in retinal progenitor, rod, amacrine, horizontal, ganglion, and bipolar cells were barely detectable, ARR3 was detected in the majority of tumor cells and coexpressed with Ki67 (SI Appendix, Fig. S9I). Overall, our results suggest that the ARR3+ maturing cone precursor may serve as the originating cell of tumorigenesis in Rb organoids (Fig. 5H). Thus, we propose that RB1 inactivation induces unusual activation of the cell cycle in maturing cone precursors and drives these postmitotic cells to reenter the cell cycle to induce the Rb precursor (retinoma-like cells, represented by up-regulation of PCNA and CDKN2A), after which the cells undergo undue proliferation (indicated by high expression of Ki67) to eventually transform into Rbs (Fig. 5H).
hRBO-Based Drug Testing Identifies Potential Therapeutic Agents.
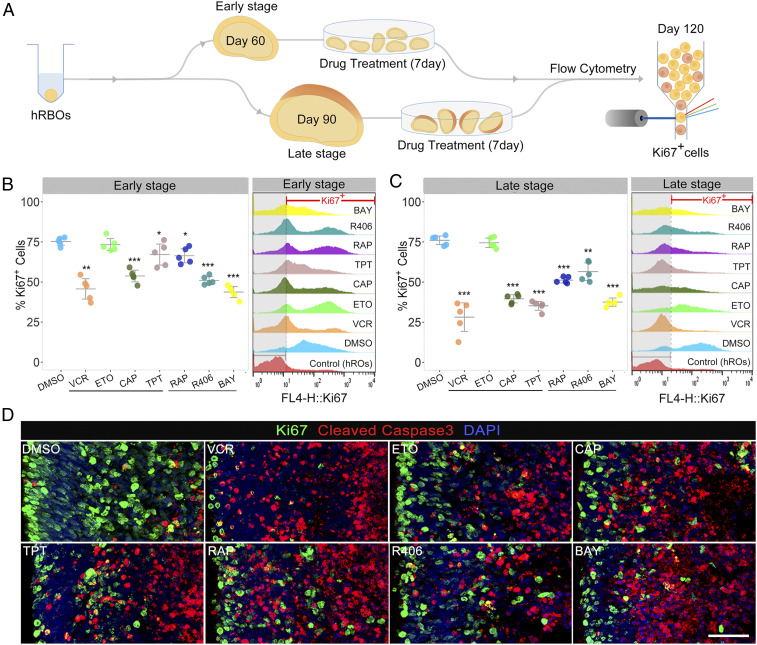
The high-fidelity Rb model enabled us to perform drug testing. We tested four chemotherapeutic drugs currently in clinical use—vincristine (5 nM), etoposide (0.5 µM), carboplatin (10 µM) and topotecan (10 nM)—as a proof of feasibility. As mentioned above, the activator of the PI3K-Akt signaling pathway, SYK, was aberrantly up-regulated in Rb organoids. Thus, we chose three candidate drugs: SYK inhibitors R406 (5 µM) and BAY 61-3606 (5 µM) (39) and the mTOR (downstream of PI3K-Akt signaling) inhibitor rapamycin (10 µM). We investigated the response to these drugs in hRBOs at the early stage (at day 60, referring to hRBOs at the onset of tumorigenesis) and late stage (at day 90, referring to hRBOs with significant tumorigenesis) (Fig. 6A). After 7 d of treatment and continuous culture to day 120, drug responses were determined by flow cytometry and/or immunocytochemistry of Ki67 and cleaved caspase 3-positive cells (Fig. 6A). As expected, flow cytometry showed that all tested drugs except for etoposide caused a significant reduction in the number of Ki67+ cells in hRBOs (Fig. 6 B and C). These results were further confirmed by immunocytochemistry (Fig. 6D and SI Appendix, Fig. S10A). In addition, immunocytochemistry showed a significant increase in cleaved caspase 3 in hRBOs treated with the candidate drugs (Fig. 6D and SI Appendix, Fig. S10A), suggesting that a 7-d administration of these candidate drugs caused remarkable cell apoptosis in cancerous organoids.
Fig. 6.
Chemotherapeutic agents for Rbs tested in hRBOs. (A) Schematic workflow of the chemotherapy treatment and assessment using hRBOs. Timelines of the culture and treatment of hRBOs are shown. (B) Representative fluorescence-activated cell sorting (FACS) profile of Ki67+ cells in day 120 hRBOs after the indicated early-stage treatment with DMSO; four currently used clinical drugs, including vincristine (VCR; final concentration: 5 nM), etoposide (ETO; 0.5 µM), carboplatin (CAB; 10 µM), and topotecan (TOP; 10 nM); and three candidate drugs, including rapamycin (RAP; 10 µM), R406 (5 µM) and BAY 61-3606 (BAY; 5 µM) (Right). Normal day 120 hROs served as a blank control. The dashed line demarcates the Ki67+ gate. (Left) Statistical analysis of the percentage of Ki67+ cells in hRBOs after the indicated treatments as analyzed by FACS. Data are mean ± SD; n = 5. *P < 0.05; **P < 0.01; ***P < 0.001, one-way analysis of variance (ANOVA) with Dunnett’s test. (C) Representative FACS profile of Ki67+ cells in day 120 hRBOs after the indicated late-stage treatment with DMSO, VCR, ETO, CAB, TOP, RAP, R406, and BAY. Normal day 120 hROs served as a blank control (Right). The dashed line demarcates the Ki67+ gate. (Left) Statistical analysis of the percentage of Ki67+ cells in hRBOs after the indicated treatments as analyzed by FACS. Data are mean ± SD; n = 5. **P < 0.01; ***P < 0.001, one-way ANOVA with Dunnett’s test. (D) Immunostaining of Ki67 (green) and apoptosis marker cleaved caspase 3 (red) in day 120 hRBOs after treatment with the indicated drugs. (Scale bar: 50 µm.)
To provide evidence for the specificity of tumor cell killing by these candidate drugs, we also performed parallel toxicity testing with normal hROs. The results suggested that carboplatin, rapamycin, R406, and BAY 61-3606 restored normal cell proliferation, but vincristine and topotecan affected cell viability to some extent in normal hROs (SI Appendix, Fig. S10B). Together, these results suggest that our model can be used to test the effect of potential therapeutic drugs on Rb tumors. The inhibitors of SYK, a key therapeutic target of Rb development, could be considered potential therapeutic drugs for Rb.
Discussion
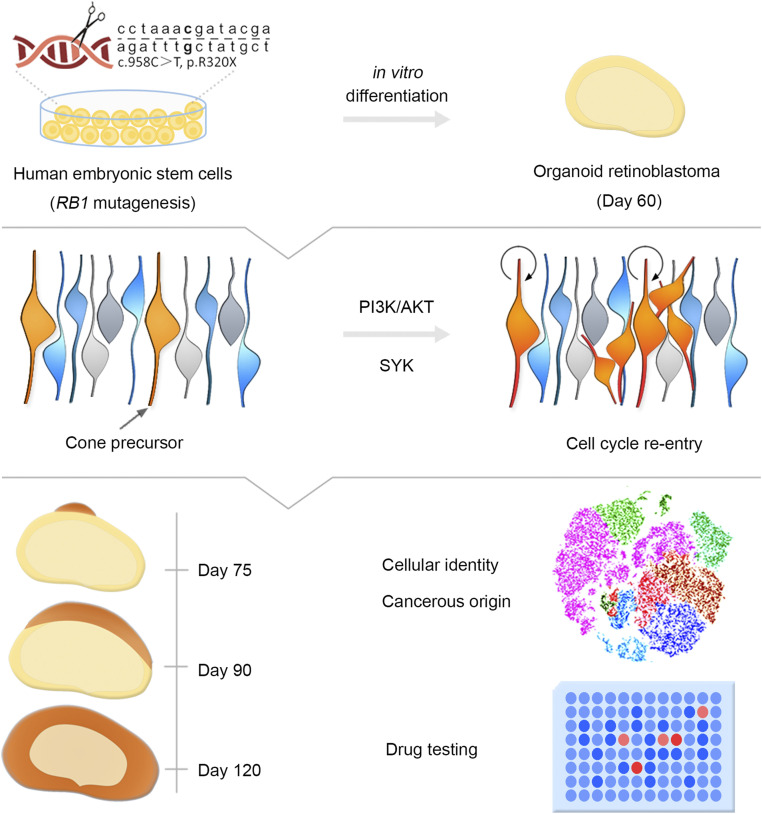
Developing a sustainable human Rb model and further confirming the human cancerous origin are critical for understanding the mechanism of tumorigenesis and progression of Rb. As a monogenic cancer disease caused by mutation of a well-defined tumor suppressor RB1 gene, Rb is suitable for disease modeling to trace its cell of origin (25). In this study, we successfully generated Rb organoids and observed tumorigenesis in a dish by introducing the most frequent RB1 mutation of Rb patients and by fully knocking out RB1 in hESCs using CRISPR/Cas9 technology and stepwise differentiation of 3D retinal organoids. Rb organoids displayed high-fidelity resemblance to primary Rb in molecular, cellular, morphogenetic, and in vivo oncogenic aspects. Consistent with previous studies reported by Xu et al. (13, 31), we demonstrated that the maturing cone precursor is the cell of origin of Rb. We further successfully tested existing chemotherapeutic drugs and candidate therapeutic agents (Fig. 7).
Fig. 7.
Schematic diagram of hRBO development and application. The model hRBOs derived from RB1Mut/Mut or RB1−/− hESCs in a dish exhibit extremely consistent properties of human primary Rbs. The hRBO model is a valuable tool that can be used to study living tumorigenesis, delineate cancerous origin, and test potential drug agents.
Recent studies have reported the ability of hESCs to self-organize stratified retinal organoids under 3D culture conditions (16, 17). Using this technology to develop a human Rb model has several potential advantages. First as a genetically related malignancy, the oncogene of Rb is best known to us (22); it is easy to induce tumorigenic mutation(s) in hESCs with the same genetic background as humans using the gene editing technology. Second, Rb is a primary intraocular tumor that arises from the immature retina, and hESCs can recapitulate the main steps of retinal development from the stratified human retina that contain all major retinal cell types, including precancerous cells, in a 3D in vitro system. Finally, Rb is a rapidly developing malignancy in infants that may occur in utero and is diagnosed at a very young age (8); during this period, the organoids derived from hESCs with inactivation of RB1 allele can mimic the tumorigenesis along with the development of human retina in vitro.
Unlike previous Rb models, the developed hRBOs allow dynamic molecular and cellular analysis of tumorigenesis in a human context during tumor progression, especially for the initial stages of tumorigenesis. These cancerous organoids with the same genetic background as humans are suitable for subsequent pathogenesis research and preclinical investigation. Furthermore, hRBOs, as cancerous retinal organoids that contain multicellular structure composed of tumor and normal retinal cells, can be used to study important properties, such as invasiveness and interaction between tumor and normal tissues.
Over the past several decades, various studies have sought to define the cell of origin of Rb. While a pair of studies on human Rb and retinal explant models suggested that only cone precursors become proliferative subsequent to RB1 loss and are likely the only cells that can transform into Rb (13, 31), McEvoy et al. (12) showed that Rb tumors display similar profiles, with coexpression of signature genes for retinal progenitors and various different mature retinal cell types. Several studies have reported that Rbs are heterogeneous or can be categorized into two distinct subtypes—one originating from retinal progenitor cells and the other from a cone photoreceptor lineage (10, 11). In addition, Rootman et al. (40) identified some tiny early untreated tumors detected by hand-held spectral domain optical coherence tomography and suggested that Rbs might originate from the inner nuclear layer rather than the photoreceptor layer of the retina.
Since precise cellular transcriptomes may dissect the lineage identity and tumor cell of origin in the developed organoids (32, 33, 41), we attempted to establish the cell of origin of Rb in our model and found that Rb tumorigenesis initially occurred in our developed hRBOs from differentiation day 60, the critical period for initial development of cones in derived hROs as well as in fetal retinas (SI Appendix, Fig. S3B) (26, 27, 42, 43). Through dynamic tracking of tumorigenesis and integrative analyses, our data suggest that Rb tumors could stem from ARR3+ maturing cone precursor cells, lending support to previous observations (13, 44). Given that retinal organoids do not generate all retinal cells with normal ratios compared with the retinal tissue in vivo, this system might be biased toward identifying a cell arising from photoreceptor precursors rather than another cell type because photoreceptors predominate, but this is unlikely to result in the very substantial bias in the maturing cone precursor-like state of the majority of proliferating cells. Our findings represent a crucial step toward identifying the cancerous origin, expanding our understanding of the cellular identities of Rb at the single cell level.
Developed hRBOs exhibited the important hallmarks of tumorigenicity and immortality in vivo, which were further confirmed by time-course transcriptomic and epigenetic landscapes. Our results suggested that epigenetic changes occurred in hRBOs after differentiation day 75. However, we are not sure whether these changes are characteristic of tumors or due to the expanded cone precursors that already have these features. Analysis of bulk methylation cannot distinguish these two possibilities. The PI3K-Akt pathway is involved in the regulation of cellular proliferation, differentiation, and survival (45). Irregularities in this pathway in various types of cancer lead to aberrant cell cycle progression and apoptosis inhibition (46, 47). Because the PI3K-Akt signaling pathway was dysregulated and its activator SYK was epigenetically modified and significantly up-regulated in our developed hRBOs, we tested potential agents targeting SYK. Remarkably, our results show that hRBOs, especially at the early stage, exhibited strong responses to R406 and BAY 61-3606 (small-molecule inhibitors of SYK), indicating that SYK inhibitors merit further study in cancerous hRBOs. Moreover, our results suggest the dysregulation of key cancer-related pathways involved in the tumorigenesis of human Rb, including the cell cycle and PI3K-Akt pathways. We also identified a distinct cell cluster with highly expressed genes related to the UPR signaling mediated by ER stress in cancerous organoids. These UPR-related cells can adapt to ER stress and escape apoptosis, and may represent malignantly transformed cells. High-throughput drug screening of small-molecule inhibitors targeting these dysregulated pathways may help identify more effective therapeutic targets based on our human Rb organoid model (4, 36, 48).
Here we have successfully developed a human Rb organoid model differentiated from hESCs with mutagenesis of the RB1 gene. Using this disease-in-a-dish model, we were able to observe tumorigenesis in retinal organoids and unravel its cell of origin and test potential drugs (Fig. 7). Our study provides valuable insights into cancer initiation, modeling, and drug testing for human malignant tumors.
Materials and Methods
Cell Culture.
Undifferentiated H9 hESCs (WiCell) and their derivatives were cultured as described previously (19). More details are provided in SI Appendix.
Mice.
The 4- to 6-wk-old immunodeficient mice (NOD-PrkdcscidIL2rgem2/SMOC, NSG) were purchased from Shanghai Model Organisms (NM-KO-00158). All mice were housed under specific pathogen-free conditions.
CRISPR/Cas9-Mediated Genome Editing.
The RB1Mut/Mut hESCs with the point mutation of c.958C > T; p.R320X in the RB1 gene was generated using CRISPR/Cas9 technology. In brief, 15 single guide RNAs (sgRNAs) were designed using the CRISPR Design Tool (SI Appendix, Table S1). Subsequently, the activity of sgRNAs was evaluated using the Universal Crispr RNA activity assay (Biocytogen), and sgRNA1-C was selected and cloned into pCS-3G to generate the CRISPR/Cas9 guide-carrying plasmids pCS-3G-sgRNA1-C. The LScKO-4G-RB1 targeting vector (LScKO-4G-LR-RR) corresponding to the human RB1 gene containing c.958C > T; p.R320X mutation was designed and constructed by Biocytogen. hESCs were dissociated into single cells using Accutase solution (Stem Cell Technologies; 07920). Approximately 1 × 106 cells were suspended with nucleofector solution prepared according to the manufacturer’s protocol by mixing 82 µL of P3 primary cell solution and 18 µL of supplement 1 (Lonza; V4XP-3032), mixed with 5 μg of plasmid mixture, including 2.5 µg of guide-carrying plasmids and 2.5 µg of RB1 targeting vector, and then transferred into a nucleofection cuvette (Lonza). Cells in the nucleofection cuvette were nucleofected under the program CA-137 using Nucleofector 4D (Lonza). After nucleofection, cells were gently transferred into Matrigel-coated plates containing TeSR-E8 medium supplemented with 10 μM Y-27632 and then cultured in a 37 °C, 5% CO2 incubator. After electroporation, the cells were treated with 2 μg/mL puromycin (Gene Operation; ISY1130-0025MG) for approximately 7 d. Then the puroresistant clones were picked and expanded for genotyping. For detection of positive RB1Mut/Mut clones, the following primers were designed: RB1-L-GT-F, 5′- GTGCTAGTGGGAGAGGCTGACAAAG -3′; RB1-WT-R, 5′- GCCAATGCTACAAAACAGTACCTAA -3′; RB1-WT-F, 5′- CACGCCTGTAATTCTAGTACTTTGG -3′; RB1-R-GT-R, 5′- GAAACGTGAACAAATCTGAAACACT -3′; RB1-R-GT-F1, 5′- TGGGATGTTTGGAAAATCTTGGCAGT -3′. The primers RB1-L-GT-F/RB1-WT-R, RB1-WT-F/RB1-R-GT-R, and RB1-R-GT-F1/RB1-R-GT-R were respectively used to detect HR and non-HR alleles.
A similar procedure was followed to generate RB1−/− hESCs (49). More details are provided in SI Appendix.
Human Retinal and Rb Organoid Differentiation.
Human retinal and Rb organoid differentiation were performed as previously described with minor modifications (16, 19). On day 0, RB1Wt/Wt, RB1Mut/Mut, and RB1−/− hESCs maintained on Matrigel-coated plates were pretreated with 10 μM Y-27632 for 2 h and then carefully dissociated into single-cell suspensions using TrypLE Select (Gibco; 12563011, containing 0.05 mg/mL DNase I and 20 μM Y-27632), plated at a density of 12,000 live cells per well in 96-well low-attachment V-bottom plates in retinal differentiation medium I containing 44% Iscove’s modified Dulbecco’s medium (Gibco; 12440053), 44% Ham’s F-12 (Gibco; 11765054), 10% KnockOut Serum Replacement (Gibco; 10828028), 1% GlutaMAX-I (Gibco; 35050061), 1% penicillin-streptomycin (Gibco; 15140163, 10,000 U/mL), and 20 μM Y-27632. A volume of 1.5 nM (55 ng/mL) recombinant human BMP4 (R&D Systems; 314-BP) was added to the culture on day 6, and its concentration was semireduced with a half-medium change every 3 d. Retinal differentiation medium II, composed of DMEM/F12-Glutamax (Gibco; 10565042) medium containing 10% fetal bovine serum (Biological Industries; 04-002-1A), 1% N2 supplement (Gibco; 17502048), 1% penicillin-streptomycin (10,000 U/mL), 0.5 μM retinoic acid (Sigma-Aldrich; R2625), and 0.1 mM taurine (Sigma-Aldrich; T8691), was exchanged until day 18, when the aggregate bodies were transferred to 9-cm nonadherent dishes (30 bodies per dish) for further long-term culture in a 37 °C, 5% CO2/40% O2 incubator. Half of the volume was changed with fresh retinal differentiation medium II every 5 d. The state and morphology of organoids were monitored in real time using an inverted fluorescence microscope. Detailed methods and protocols are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank all 502 laboratory members for technical assistance and discussion. This study was supported by an institutional grant from BIO (to Z.-B.J.), the National Key R&D Program of China (2017YFA0105300, to Z.-B.J.), the Natural Science Foundation of China for Excellent Young Scholars (81522014, to Z.-B.J.), the Natural Science Foundation of China (81970838, to Z.-B.J.), and the Zhejiang Provincial Natural Science Foundation of China (LQ17H120005, to Z.-B.J.; LQ20C120001, to H.L.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011780117/-/DCSupplemental.
Data Availability.
RNA-Seq, WGBS, and single-cell RNA-Seq data have been deposited in the Gene Expression Omnibus database (accession no. GSE136929).
References
- 1.Dimaras H., et al. , Retinoblastoma. Lancet 379, 1436–1446 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Dimaras H., et al. , Retinoblastoma. Nat. Rev. Dis. Primers 1, 15021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacPherson D., Dyer M. A., Retinoblastoma: From the two-hit hypothesis to targeted chemotherapy. Cancer Res. 67, 7547–7550 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Pascual-Pasto G., et al. , Therapeutic targeting of the RB1 pathway in retinoblastoma with the oncolytic adenovirus VCN-01. Sci. Transl. Med. 11, eaat9321 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Dimaras H., Corson T. W., Retinoblastoma, the visible CNS tumor: A review. J. Neurosci. Res. 97, 29–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D., et al. , Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell 5, 539–551 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Ajioka I., et al. , Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell 131, 378–390 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson D. A., et al. , Neuronal differentiation and synaptogenesis in retinoblastoma. Cancer Res. 67, 2701–2711 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Aldiri I. et al.; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project , The dynamic epigenetic landscape of the retina during development, reprogramming, and tumorigenesis. Neuron 94, 550–568.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapatai G., et al. , Gene expression profiling identifies different sub-types of retinoblastoma. Br. J. Cancer 109, 512–525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kooi I. E., et al. , Loss of photoreceptorness and gain of genomic alterations in retinoblastoma reveal tumor progression. EBioMedicine 2, 660–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McEvoy J., et al. , Coexpression of normally incompatible developmental pathways in retinoblastoma genesis. Cancer Cell 20, 260–275 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X. L., et al. , Rb suppresses human cone precursor-derived retinoblastoma tumours. Nature 514, 385–388 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyer M. A., Lessons from retinoblastoma: Implications for cancer, development, evolution, and regenerative medicine. Trends Mol. Med. 22, 863–876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neal J. T., Kuo C. J., Organoids as models for neoplastic transformation. Annu. Rev. Pathol. 11, 199–220 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Kuwahara A., et al. , Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 6, 6286 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Nakano T., et al. , Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Eldred K. C., et al. , Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 362, eaau6348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng W. L., et al. , Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Reports 10, 1267–1281 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan D., et al. , COCO enhances the efficiency of photoreceptor precursor differentiation in early human embryonic stem cell-derived retinal organoids. Stem Cell Res. Ther. 11, 366 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Z. B., et al. , Stemming retinal regeneration with pluripotent stem cells. Prog. Retin. Eye Res. 69, 38–56 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Friend S. H., et al. , A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323, 643–646 (1986). [DOI] [PubMed] [Google Scholar]
- 23.Fokkema I. F., et al. , LOVD v.2.0: The next generation in gene variant databases. Hum. Mutat. 32, 557–563 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Conklin J. F., Baker J., Sage J., The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 3, 1244 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Dyer M. A., Bremner R., The search for the retinoblastoma cell of origin. Nat. Rev. Cancer 5, 91–101 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Swaroop A., Kim D., Forrest D., Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 11, 563–576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellough C. B., et al. , An integrated transcriptional analysis of the developing human retina. Development 146, dev169474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganguly A., Shields C. L., Differential gene expression profile of retinoblastoma compared to normal retina. Mol. Vis. 16, 1292–1303 (2010). [PMC free article] [PubMed] [Google Scholar]
- 29.Dimaras H., et al. , Loss of RB1 induces non-proliferative retinoma: Increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 17, 1363–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Kivelä T., et al. , Neuronal and glial properties of a murine transgenic retinoblastoma model. Am. J. Pathol. 138, 1135–1148 (1991). [PMC free article] [PubMed] [Google Scholar]
- 31.Xu X. L., et al. , Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell 137, 1018–1031 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young M. D., et al. , Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 361, 594–599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Y., et al. , hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell 24, 487–497.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke H. J., Chambers J. E., Liniker E., Marciniak S. J., Endoplasmic reticulum stress in malignancy. Cancer Cell 25, 563–573 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Wang M., Kaufman R. J., The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 14, 581–597 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Hetz C., Axten J. M., Patterson J. B., Pharmacological targeting of the unfolded protein response for disease intervention. Nat. Chem. Biol. 15, 764–775 (2019). [DOI] [PubMed] [Google Scholar]
- 37.La Manno G., et al. , RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welby E., et al. , Isolation and comparative transcriptome analysis of human fetal and iPSC-derived cone photoreceptor cells. Stem Cell Reports 9, 1898–1915 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., et al. , A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481, 329–334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rootman D. B., et al. , Hand-held high-resolution spectral domain optical coherence tomography in retinoblastoma: Clinical and morphologic considerations. Br. J. Ophthalmol. 97, 59–65 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Zong C. C., Single-cell RNA-seq study determines the ontogeny of macrophages in glioblastomas. Genome Biol. 18, 235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Cordero A., et al. , Recapitulation of human retinal development from human pluripotent stem cells generates transplantable populations of cone photoreceptors. Stem Cell Reports 9, 820–837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaewkhaw R., et al. , Transcriptome dynamics of developing photoreceptors in three-dimensional retina cultures recapitulates temporal sequence of human cone and rod differentiation revealing cell surface markers and gene networks. Stem Cells 33, 3504–3518 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh H. P., et al. , Developmental stage-specific proliferation and retinoblastoma genesis in RB-deficient human but not mouse cone precursors. Proc. Natl. Acad. Sci. U.S.A. 115, E9391–E9400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spangle J. M., et al. , PI3K/AKT signaling regulates H3K4 methylation in breast cancer. Cell Rep. 15, 2692–2704 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., et al. , A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell 31, 820–832.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delaloge S., DeForceville L., Targeting PI3K/AKT pathway in triple-negative breast cancer. Lancet Oncol. 18, 1293–1294 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Alzahrani A. S., PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin. Cancer Biol. 59, 125–132 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Avior Y., Lezmi E., Yanuka D., Benvenisty N., Modeling developmental and tumorigenic aspects of trilateral retinoblastoma via human embryonic stem cells. Stem Cell Reports 8, 1354–1365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq, WGBS, and single-cell RNA-Seq data have been deposited in the Gene Expression Omnibus database (accession no. GSE136929).