Significance
The neural crest is a stem cell population that plays a crucial role in the establishment of the vertebrate body plan. Neural crest induction requires a delicate balance of multiple signaling systems, including FGFs. Moderate FGF activity is necessary for neural crest formation, but overactivation of the pathway drives progenitor cells into adopting a neuronal fate. Yet, it remains unclear how levels of FGF are fine tuned to promote neural crest identity. Here, we demonstrate how miRNA-mediated gene silencing modulates the intracellular response to FGF signaling to ensure proper neural crest cell specification. Understanding the function of these miRNAs may broaden our understanding of the neural crest genetic program and shed light on the etiology of neural crest-linked congenital disabilities.
Keywords: neural crest, FGF signaling, miRNAs, signaling systems, ectoderm
Abstract
Ectodermal patterning is required for the establishment of multiple components of the vertebrate body plan. Previous studies have demonstrated that precise combinations of extracellular signals induce distinct ectodermal cell populations, such as the neural crest and the neural plate. Yet, we still lack understanding of how the response to inductive signals is modulated to generate the proper transcriptional output in target cells. Here we show that posttranscriptional attenuation of fibroblast growth factor (FGF) signaling is essential for the establishment of the neural crest territory. We found that neural crest progenitors display elevated expression of DICER, which promotes enhanced maturation of a set of cell-type-specific miRNAs. These miRNAs collectively target components of the FGF signaling pathway, a central player in the process of neural induction in amniotes. Inactivation of this posttranscriptional circuit results in a fate switch, in which neural crest cells are converted into progenitors of the central nervous system. Thus, the posttranscriptional attenuation of signaling systems is a prerequisite for proper segregation of ectodermal cell types. These findings demonstrate how posttranscriptional repression may alter the activity of signaling systems to generate distinct spatial domains of progenitor cells.
Signaling systems play a central role in the patterning of the ectoderm and subsequent segregation of the precursors of the central (CNS) and peripheral (PNS) nervous systems (1–3). In the amniote gastrula, high levels of fibroblast growth factors (FGFs) secreted by the primitive streak induce the formation of the neural plate, the progenitor domain that will give rise to the CNS (4, 5). Concurrently, Wnts and BMPs secreted by the lateral ectoderm drive peripheral cells into adopting an epidermal fate (6, 7). At the border of the neural plate, a precise combination of these signals induces the formation of a cell population known as the neural crest (5, 8, 9). Neural crest cells are stem cells that give rise to the peripheral nervous system, the craniofacial skeleton, and the pigmentation of the skin (10). Despite the requirement of a delicate balance between the levels of FGFs, Wnts, and BMPs for neural crest induction (11), we still have a superficial understanding of how morphogen levels are optimized to properly partition the embryonic ectoderm.
Neural crest formation is orchestrated by a gene regulatory network (GRN) that is initiated by signaling systems during neural plate border induction (10, 12). The existence of a comprehensive network for this molecular program makes the neural crest a tractable model for scrutinizing inductive mechanisms (1, 10). Neural crest cells are formed adjacent to the neural plate, and as a result, the specification of the neural crest is intimately linked to the process of neural induction (13–15). The formation of the neural crest requires high levels of Wnts and low levels of FGFs, while high levels of FGFs drive cells toward a neural fate (5, 9, 16). Although multiple studies have placed FGFs as central players in ectodermal patterning (4, 9, 13, 17), it remains unclear how a gradient of FGFs is converted into a binary response to promote neural vs. neural crest fates.
In recent years, growing evidence has supported a role for microRNA (miRNA)-mediated gene silencing in neural crest development (18–21). In the developing mouse embryo, the tissue-specific deletion of DICER, a key enzyme in miRNA biogenesis, results in a complete loss or malformation of several neural crest-derived structures (22–25). Furthermore, several lines of evidence have underscored a decisive role for specific miRNAs in modulating neural crest stem cell identity (19, 21, 26). In other developmental contexts, miRNAs have been shown to modulate the expression of components of signaling cascades, thus influencing developmental decisions (27–30). These results raise the possibility that posttranscriptional mechanisms may play an important role in the formation of ectodermal derivatives, and in particular, in the development of the neural crest.
Here, we demonstrate that miRNA-mediated gene silencing is required to balance the activity of signaling systems during neural crest induction. We found that neural crest cells express enhanced levels of the miRNA-processing enzyme DICER in chicken embryos. This enrichment of DICER is mediated by a neural crest-specific enhancer that is regulated by components of the neural crest gene regulatory network. High levels of DICER expression are required for the activation of a set of tissue-specific miRNAs. We found that several of these miRNAs act to posttranscriptionally silence components of the FGF signaling pathway. Inhibition of these miRNAs leads to the expansion of the neural plate at the expense of neural crest cells, highlighting their contribution to ensuring proper levels of FGF signaling during neural crest induction. These findings demonstrate how miRNA-mediated gene silencing modulates the intracellular response to signaling systems to ensure proper cell specification.
Results
Enhanced Expression of DICER Is Required for Neural Crest Specification.
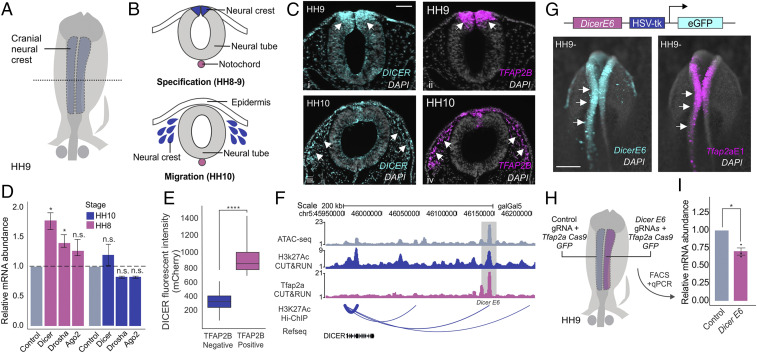
Previous studies indicate a decisive role for DICER in the differentiation and survival of neural crest-derived tissues (22–25, 31). To determine the role of DICER in the early stages of neural crest development (Fig. 1 A and B), we first examined its expression pattern using fluorescence in situ hybridization (FISH) in avian embryos (Fig. 1C). DICER was enriched in the dorsal neural folds (HH9) and displayed strong colocalization with the neural crest mark marker TFAP2B (Fig. 1 C, i and ii) (32, 33). This enrichment was transient, as the DICER signal decreased in migratory neural crest cells (HH10) (Fig. 1 C, iii and iv). Quantitative RT-PCR on neural crest cells isolated with fluorescence-activated cell sorting (FACS) at stages corresponding to specification and migration (HH8 and HH10) (Fig. 1D) confirmed these results. Furthermore, we also observed increased expression of the primary microRNA (pri-miRNA) processing enzyme DROSHA during specification, suggesting a widespread increase in miRNA processing in premigratory neural crest cells (P value <0.05) (Fig. 1D) (34). To test if higher levels of DICER mRNA resulted in an increase in protein expression, we performed immunohistochemistry for DICER and TFAP2B in dissociated embryonic heads. FACS analysis revealed that neural crest cells displayed much higher levels of the DICER protein than TFAP2B-negative cells (P value <0.0001) (Fig. 1E).
Fig. 1.
Neural crest progenitors display high DICER expression during specification. (A) Neural crest cells (blue) are specified on the dorsal folds of the neural tube (gray) during early embryonic development. (B) Transverse section of the neural tube showing the positions of neural crest cells (blue) during specification (HH8 to 9) and migration (HH10). (C) Transverse sections from double fluorescence in situ hybridization for DICER and TFAP2B in chicken embryos at two stages (HH9 and HH10). DICER is enriched in premigratory neural crest cells, having high colocalization with TFAP2B. (D) Quantitative RT-PCR for miRNA biogenesis enzymes DICER, DROSHA, and AGO2 in sorted neural crest cells (Tfap2aE1:GFP+) compared to whole-embryo control cells (Tfap2aE1:GFP−), shows up-regulation of DICER mRNA in specified neural crest cells (n = 6). (E) Immunohistochemistry for DICER reveals high protein expression in TFAP2B+ premigratory neural crest cells (HH8). (F) ATAC, H3k27Ac CUT&RUN, TFAP2A CUT&RUN profiles, and H3k27Ac HiChIP arc plot at the DICER locus. Arc thickness correlates with loop strength in dorsal neural folds. (G) Schematic representation of the enhancer construct. The construct consists of the DicerE6 enhancer region cloned upstream of the HSV-tk minimal promoter driving GFP expression. Transgenic chicken embryos show colocalization of DicerE6:eGFP with the neural crest reporter Tfap2aE1:mCherry in dorsal neural folds. (H) Electroporation scheme for enhancer loss-of-function assay in which control gRNA and Tfap2aE1 Cas9:eGFP (blue) or DicerE6 gRNA and Tfap2aE1 Cas9:eGFP (pink) were injected in different sides of a HH4 chicken embryo (n = 3). (I) Quantitative RT-PCR for DICER in control vs. DicerE6 loss-of-function sides of bilaterally electroporated embryos. Error bars in D and I represent the SD. Arrowheads indicate neural crest cells. HH, Hamburger and Hamilton; n.s., not significant, *P ≤ 0.05, ****P ≤ 0.0001. Scale bar, 50 μM in (C) and 200 μM in (G).
To define why DICER levels are enhanced in neural crest cells, we investigated the cis-regulatory control of the gene in this cell population. By utilizing a high-resolution chromatin interaction map of active enhancers in isolated neural crest cells (H3K27Ac HiChIP) (SI Appendix, Table S1), we identified seven putative enhancers associated with the DICER promoter in neural crest progenitors (Fig. 1F). One of these elements (DicerE6) displayed high levels of H3K27Ac and strong association with the transcription factor TFAP2A (32), a marker of active neural crest enhancers (35) (Fig. 1F). To test if DicerE6 was active in neural crest cells, we cloned it in pTK:eGFP (36) and cotransfected this construct with a neural crest-specific reporter (Tfap2aE1:mCherry) into gastrula-stage chicken embryos (Fig. 1G). We observed that the enhancer was active in premigratory neural crest (HH9-), suggesting it might act to increase in the levels of DICER in this cell population. To test this, we targeted DicerE6 with CRISPR-Cas9 gene editing (Fig. 1 H and I). We transfected the right side of avian embryos with a neural crest-specific Cas9:eGFP vector (Tfap2aE1-CAS9:eGFP) and a double guide RNA vector targeting DicerE6 (37); the left side of the embryos was transfected with nontargeting gRNAs (Fig. 1H). Isolation of transfected cells by FACS, followed by qPCR analysis, revealed that targeting of DicerE6 resulted in a significant decrease in DICER mRNA abundance as compared to control cells (P value <0.05) (Fig. 1I). Furthermore, we transfected several other putative DICER enhancers not bound by TFAP2A, which did not display strong colocalization with the neural crest reporter (SI Appendix, Fig. S1 A and B). These results suggest that TFAP2A, a neural crest-specific transcription factor, can up-regulate DICER levels by interacting with tissue-specific enhancers.
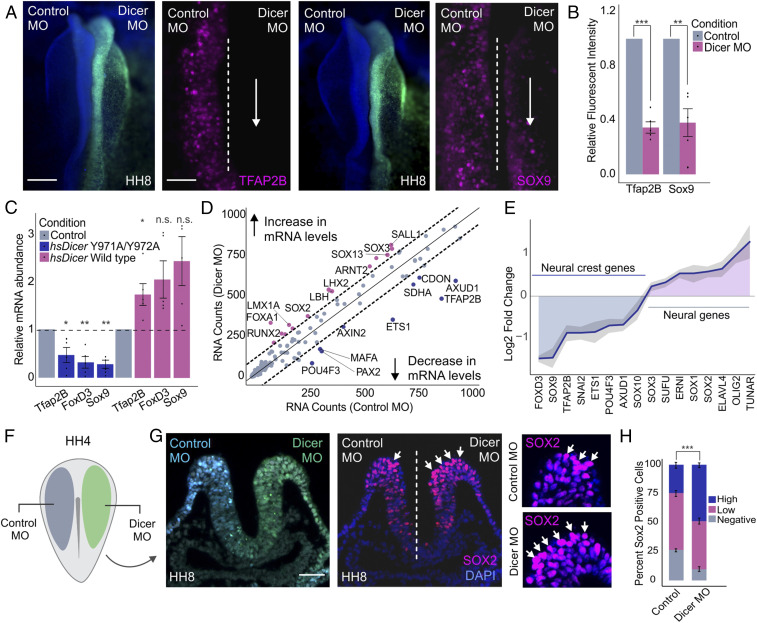
The high levels of DICER in the neural crest suggest posttranscriptional regulation may be particularly important for the specification of these cells. To test this, we knocked down DICER by transfecting embryos with a translation-blocking morpholino (MO) on the right side (green) and a control morpholino (blue) on the left side (Fig. 2F) (38). This treatment resulted in a robust knockdown of DICER in the morpholino-transfected side of the embryo (SI Appendix, Fig. S2A). Loss of DICER led to a significant decrease in the expression of neural crest markers TFAP2B and SOX9, as shown by immunohistochemistry (P value <0.01) (Fig. 2 A and B and SI Appendix, Fig. S2 B and C). To confirm this phenotype was not due to a delay in the specification program, we also analyzed the effect of DICER knockdown in migratory neural crest cells. Immunohistochemistry for markers SOX10 and HNK1 at HH10 demonstrated that a reduction in the number of neural crest cells persists at later developmental stages (SI Appendix, Fig. S2 D–F). To confirm this phenotype was due to the miRNA-processing activity of DICER, and not noncanonical functions of this protein (39–41), we performed rescue experiments using both a control and a mutant form of the human DICER protein. We cotransfected embryos with DICER morpholinos combined with expression constructs encoding either the wild-type (WT) hsDICER or with hsDICERY971A/Y972A, a protein carrying a point mutation that is unable to process miRNAs (42). qPCR analysis for specification genes TFAP2B, FOXD3, and SOX9 showed that the WT construct was able to rescue the knockdown phenotype, whereas hsDICERY971A/Y972A was unable to do so (Fig. 2C).
Fig. 2.
DICER is required for proper neural crest cell specification. (A) Dorsal whole-mount view of HH8 embryo with control MO on the Left and DICER MO on the Right. Immunohistochemistry for neural crest markers AP2B and SOX9 upon DICER knockdown. Dotted line represents embryo midline. (B) Quantification of TFAP2B- and SOX9-positive cells following DICER knockdown, normalized to the control side of the embryo (n = 5). (C) Quantitative RT-PCR for specification genes TFAP2B, FOXD3, and SOX9 in embryos electroporated with DICER morpholino and rescued with hsDICER wild-type or hsDICER Y971A/Y972A expression constructs. Phenotypes were assayed at stage HH8, comparing control to experimental sides of the embryo for each rescue condition (n = 5). (D) Nanostring analysis comparing gene expression profiles of control and DICER knockdown dissected neural folds of bilaterally electroporated embryos. Genes above or below the dotted lines are significantly affected upon DICER knockdown. (E) Nanostring analysis reveals changes in neural crest and neural plate gene expression profiles upon DICER knockdown. The plot represents average log2 fold change across three replicates with SD shaded in gray (n = 3). (F) Electroporation scheme for DICER loss-of-function assays in which control reagent (blue) and targeted reagent (green) were injected in different sides of a HH4 chicken embryo. (G) Immunohistochemistry (transverse section) for neural plate marker SOX2 upon DICER knockdown. Arrows point to high-intensity SOX2-positive cells in control and DICER MO sides of the embryo. (H) Quantification of SOX2-positive (low and high intensity) and -negative cells in the top 1/4 of the dorsal neural tube upon DICER knockdown, as compared to the control. P value represents Student’s t test comparing percentage of high-intensity SOX2-positive cells between control and DICER MO sides of the embryo (n = 5). Error bars in B, C, and H represent the SD. Arrowheads indicate neural crest cells. HH, Hamburger and Hamilton; n.s., not significant; MO, morpholino. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Scale bar, 200 μM in (A). Dorsal whole-mount view of HH8 embryo with control MO on the Left and DICER MO on the Right, 100 μM in (A) immunohistochemistry for neural crest markers, 50 μM in (G).
To globally survey the impact of DICER knockdown on neural crest gene expression, we performed a Nanostring analysis with a set of 200 probes targeting genes involved in neural crest, neural, and placodal development (19). This analysis revealed a robust reduction in expression of neural crest transcription factors, including TFAP2B, AXUD1, and ETS1, as well as an up-regulation of multiple neural factors such as SOX2, SOX3, and SUFU (Fig. 2 D and E). Since we did not detect changes in cell death or proliferation (SI Appendix, Fig. S2 G–I), these results suggested a fate switch upon DICER knockdown, in which presumptive neural crest cells failed to become specified as neural crest and acquired a neural plate identity instead. To examine this phenotype in more detail, we performed immunohistochemistry for SOX2 in DICER morphant embryos. The results revealed a robust increase in SOX2-positive cells in the neural crest territory (P value <0.001) (Fig. 2 G and H and SI Appendix, Fig. S2 K and L), consistent with a neuralization phenotype. This fate switch was confirmed by qPCR analysis of neural folds dissected from morphant embryos, which revealed an up-regulation of various neural plate genes accompanied by a decrease in expression of neural crest specification genes (SI Appendix, Fig. S2J). These results indicate that the processing of miRNAs by DICER is required for proper neural crest induction during early embryonic development.
Small RNA Sequencing Identifies Differentially Expressed miRNAs in Neural Crest and Neural Plate Cells.
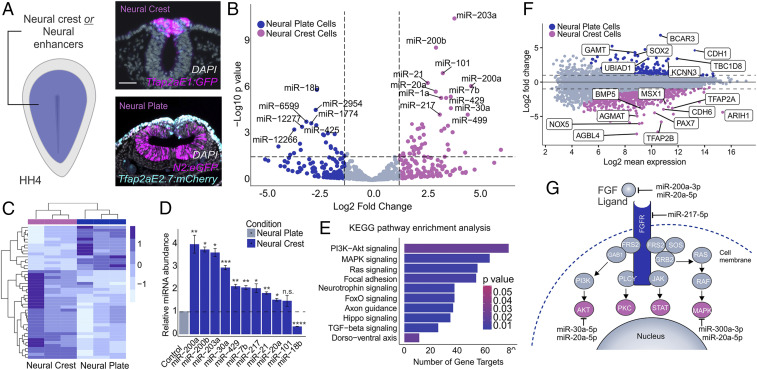
To identify the miRNAs that are involved in the neuralization phenotype observed upon DICER knockdown, we performed small RNA sequencing (RNA-seq) on neural crest and neural plate cells. Pure populations of each cell type were FACS sorted from avian embryos at specification stages (HH8). To label neural crest cells, we utilized the Tfap2aE1:eGFP reporter, whereas for neural plate cells, we employed the neural plate enhancer reporter N2:eGFP (36) and a TFAP2A enhancer active specifically in the nonneural ectoderm (Tfap2aE2.7:mCherry) (Fig. 3A). The latter construct was used to exclude SOX2-positive neural crest cells from the sample (43). Principal component analysis of small RNA-sequencing results demonstrated that the majority of variability was strongly associated with cell identity, as expected (SI Appendix, Fig. S3A). Our datasets revealed several differentially expressed, highly abundant miRNAs between these two cell populations (P value <0.001) (Fig. 3 B and C and SI Appendix, Fig. S3B). Consistent with the enrichment of DICER (Fig. 1), we observed that the neural crest displayed a larger number of enriched miRNAs when compared to the neural plate (Fig. 3B).
Fig. 3.
Small RNA sequencing identifies miRNAs enriched in neural crest progenitors. (A) Electroporation scheme for injection of neural crest or neural plate enhancer reporters in HH4 embryos. Transverse sections of embryos electroporated with the neural crest reporter Tfap2aE1:eGFP or the neural plate reporter N2:eGFP and Tfap2aE2.7:mCherry for exclusion of Sox2-positive neural crest cells. (B) Heatmap displaying the top significantly differentially expressed (P adj. <0.05) miRNAs between neural crest and neural plate cells. Expression levels are z-score normalized. (C) Volcano plot displaying differentially expressed miRNAs between neural crest and neural plate cells. Dotted lines represent log2 fold change and P value thresholds (P value <0.01, log2 fold change greater than or less than 1.5) for differentially expressed miRNAs. miRNAs of interest are labeled based on P value <0.001. (D) Quantitative RT-PCR for top 10 differentially expressed miRNAs in the neural crest compared to the neural plate. miR-18b serves as a negative control that should not be enriched in the neural crest relative to the neural plate. (E) miRPath analysis for top 10 neural crest-enriched miRNAs suggests several gene targets in the FGF signaling cascade (PI3K-Akt and MAPK). The plot shows the top 10 KEGG pathways identified in this analysis. (F) Intensity ratio (MA) plot from differential gene expression analysis between the neural crest and neural plate utilized in miRNA target prediction pipeline. (G) Confidently predicted neural crest miRNA gene targets within the FGF signaling pathway. Error bars in D represent the SD. HH, Hamburger and Hamilton; n.s., not significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Scale bar, 50 μM in (A).
Our datasets included neural crest-enriched miRNAs that have been shown to inhibit epithelial-to-mesenchymal transition in neural crest cells, including miR-200b and miR-203a, which may prevent premature migration prior to delamination (20, 44). We also identified several miRNA candidates that may contribute to neural crest cell formation (Fig. 3C). RT-PCR for the top 10 significantly expressed neural crest miRNAs confirmed up-regulation in this cell type when compared to neural plate cells (Fig. 3D). To elucidate the biological role of the neural crest-enriched miRNAs, we identified their target Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways using DNA Intelligent Analysis (DIANA)-miRPath v3 software (45) (Fig. 3E). miRPath analysis revealed these miRNAs might regulate several signaling systems involved in neural crest development, including FGF-related intracellular signaling pathways MAP kinase (MAPK) and PI3 kinase/Akt (Pi3K/Akt), both of which are important for neural crest development (14, 46). Conversely, miRPath analysis for the top 10 neural plate-enriched miRNAs did not yield FGF-related target pathways (SI Appendix, Fig. S3C).
To determine the role of these miRNAs in the modulation of FGF signaling in neural crest cells, we aimed to identify their target genes within the MAPK and Pi3K/Akt pathways using a multifaceted approach (SI Appendix, Fig. S3D). First, we performed RNA sequencing on FACS-sorted neural crest and neural plate cells, analogous to our small RNA-seq experiments, to identify genes involved in FGF signaling that are differentially expressed between these two cell types (Fig. 3F). KEGG pathway enrichment analysis for neural plate genes revealed enrichment for the Pi3K-Akt and JAK-STAT pathways, therefore suggesting components of these pathways may be subject to posttranscriptional regulation in neural crest cells (SI Appendix, Fig. S3E). Next, we utilized the in silico miRNA target prediction program TargetScanHuman v7.2 to identify confidently predicted genes (context + score < −0.2) that are targeted by the top 10 neural crest-enriched miRNAs (47). Upon merging these datasets, our candidate selection pipeline (SI Appendix, Fig. S3D) identified 4 neural crest miRNAs that may engage in functionally relevant miRNA:mRNA target interactions within the FGF pathway (Fig. 3G and SI Appendix, Fig. S3F). This strategy was able to identify miRNA candidates that may act to attenuate levels of the FGF signaling pathway during neural crest specification (Fig. 3G).
Neural Crest miRNAs Target the FGF Signaling Pathway to Mediate Ectodermal Cell Fate Decisions.
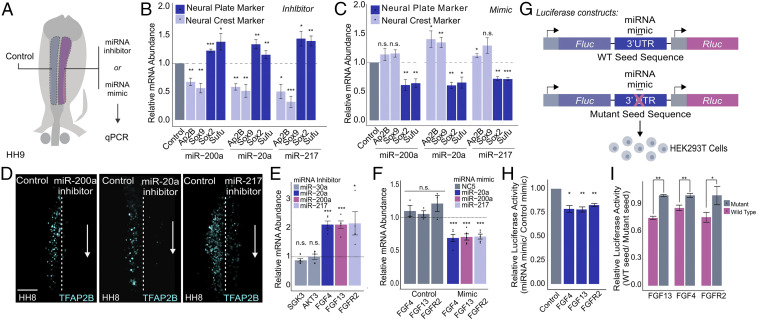
Our small RNA-seq analysis of neural crest cells revealed enrichment of miRNAs that may play essential roles in the formation of this cell type. To test this, we used unilateral transfections in avian embryos (Fig. 4A) to conduct a screen targeting the top 10 enriched miRNAs (Fig. 3D) with IDT miRNA inhibitors (SI Appendix, Fig. S4A). This analysis showed that loss of function of many of these miRNAs resulted in a decreased expression of neural crest markers TFAP2B and SOX9, highlighting a role for posttranscriptional silencing in neural crest formation (the negative control NC5 displayed no effect, SI Appendix, Fig. S4A). We only observed a neuralization phenotype (loss of TFAP2B and SOX9 accompanied by an increase in neural markers SOX2 and SUFU) when targeting the four miRNAs with predicted targets in the FGF signaling pathway (miR-200a, miR-20a, miR-217, and mir-30a). Thus, we pursued miR-200a, miR-20a, and miR-217 for further analysis (Fig. 4B). Immunostaining for TFAP2B in embryos transfected with inhibitors revealed a reduction in the number of neural crest cells (Fig. 4D). Furthermore, the transfection of embryos with miRNA mimic molecules resulted in a decrease of neural plate marker expression, the opposite phenotype to that of transfection with inhibitors (Fig. 4C).
Fig. 4.
Neural crest miRNAs are required for specification and target components of the FGF signaling pathway. (A) Electroporation scheme for miRNA loss- or gain-of-function assays in which control reagent (blue) and targeted reagent (pink) were injected in different sides of a HH4 chicken embryo, which was then developed to HH8 to 9. (B and C) Quantitative RT-PCR for neural crest genes TFAP2B and SOX9 and neural plate genes SOX2 and SUFU upon miRNA loss (B) and gain of function (C) for the top three FGF-targeting neural crest miRNAs (miR-200a, miR-20a, and miR-217) (n = 6). (D) Immunohistochemistry for specification marker TFAP2B upon individually inhibiting FGF targeting miRNAs, miR-200a, miR-20a, and miR-217. Dotted line represents embryo midline. (E and F) Quantitative RT-PCR for target genes upon miRNA loss (E) and gain of function (F). miR-30a targets were excluded from gain-of-function assay due to a lack of observable changes in target gene expression upon loss of function. Gain-of-function assays were also performed using a nontargeting control (NC5) that does not affect target gene expression. (G) Diagram showing the experimental design for 3′ UTR luciferase reporter assays. Wild-type or mutant 3′ UTRs of target genes were cloned into a dual-luciferase reporter (pmiRGlo) and transfected into HEK293T cells with the corresponding miRNA mimic. For mutant 3′ UTRs, the miRNA mimic should not be able to bind the corresponding seed sequence. (H) Luciferase activity of wild-type 3′ UTR reporters. Each construct was transfected with the appropriate targeting miRNA mimic and normalized to a nontargeting miRNA mimic control (n = 3). (I) Luciferase activity of wild-type 3′ UTR luciferase reporters normalized to mutant 3′ UTR luciferase reporters, which should have no regulatory effect (n = 4). Error bars in B, C, E, F, H, and I represent the SD. HH, Hamburger and Hamilton; n.s., not significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Scale bar, 100 μM in (D).
To test if miR-200a, miR-20a, and miR-217 do, in fact, target and repress FGF pathway components, we manipulated miRNA expression in vivo and determined the effect on the mRNA levels of predicted targets. We transfected embryos with either miRNA mimic or inhibitor and performed RT-PCR for the corresponding FGF target gene (Fig. 4 E and F). Inhibition of the three miRNAs resulted in increased levels of FGF4, FGF13, and FGFR2 mRNA in the neural crest territory (Fig. 4E), whereas transfection of mimic molecules had the opposite result (in contrast, inhibition of miR-30a, which was predicted to regulate SGK3 and AKT3, did not affect either gene). Consistent with these results, double fluorescent in situ hybridization found that FGF4, FGF13, and FGFR2 are enriched in the neural plate of neurula-stage embryos (SI Appendix, Fig. S4B). To confirm that such changes in transcript levels of target genes (Fig. 4 E and F) are the result of miRNA-mediated repression, we performed 3′ UTR luciferase reporter assays for our three candidate miRNAs and their gene targets (Fig. 4G). Briefly, the wild type or seed sequence mutant 3′ UTRs of target genes were cloned into the pmiRGlo dual-luciferase reporter and transfected into HEK293T cells. Luciferase expression of UTR reporters was decreased in cells treated with the corresponding miRNA mimic, as compared to a nontargeting control (P value <0.05) (Fig. 4H). Furthermore, the wild-type 3′ UTR reporters for each target gene showed decreased activity in contrast to the mutant reporters, which lack the functional regulatory sequences, confirming miRNA target specificity (P value <0.01) (Fig. 4I).
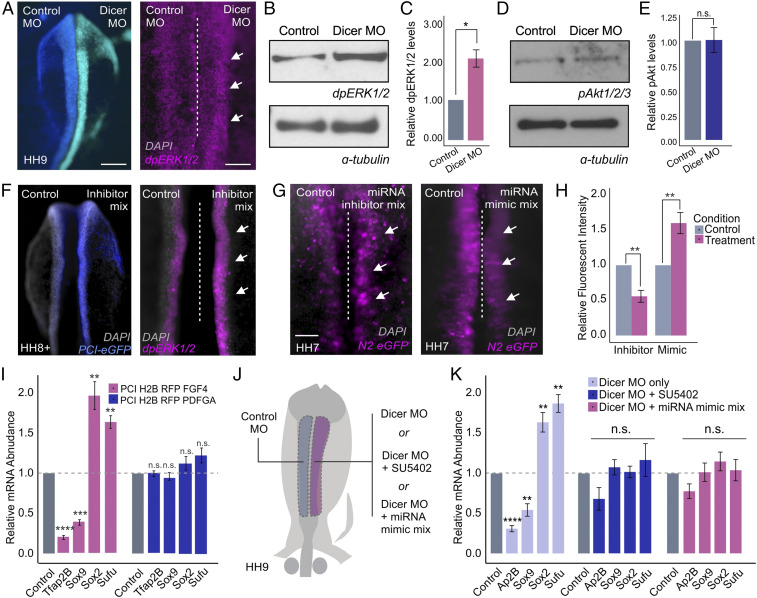
The above results confirm that neural crest-enriched miRNAs, including miR-20a, miR-200a, and miR-217, target and posttranscriptionally repress components of the FGF pathway. To confirm that miRNA-mediated gene silencing affects the activity of this signaling system, we performed stage-specific knockdown of DICER and measured MAPK activity with a dpERK1/2 antibody. Loss of DICER resulted in an increase in ERK1/2 phosphorylation in dorsal neural folds, as shown by both immunohistochemistry (Fig. 5A) and Western blot analysis (P value <0.05) (Fig. 5 B and C). Conversely, we did not observe changes in Akt1/2/3 phosphorylation following DICER knockdown, suggesting that FGF signaling mediated via MAPK is the main target of neural crest-enriched miRNAs (Fig. 5 D and E). To test the requirement of our miRNA candidates for attenuation of FGF signaling, we simultaneously targeted miR-20a, miR-200a, and miR-217 using a mix of inhibitors. This treatment caused an increase in MAPK activity in the dorsal neural folds transfected with the inhibitors (Fig. 5F). Furthermore, manipulation of miRNA levels also affected ectodermal patterning. Cotransfection of either an inhibitor or a mimic mix with the N2:eGFP enhancer (36) (Fig. 5 G and H) revealed an expansion or reduction, respectively, of this neural plate marker in response to modulation of miRNA levels.
Fig. 5.
Posttranscriptional attenuation of FGF signaling is required for neural crest specification. (A) Dorsal whole-mount view of HH9 embryo with control MO on the Left and DICER MO on the Right. Immunohistochemistry for dpERK1/2 upon DICER knockdown. (B) Western blot for endogenous dpERK1/2 levels upon DICER knockdown. (C) Western blot quantification showing relative dpERK1/2 levels in control and DICER knockdown sides of bilaterally electroporated embryos, normalized to alpha-tubulin control (n = 3). (D) Western blot for endogenous pAKT1/2/3 levels upon DICER knockdown. (E) Western blot quantification showing relative pAKT1/2/3 levels in control and DICER knockdown sides of bilaterally electroporated embryos, normalized to alpha-tubulin control (n = 3). (F) Dorsal whole-mount view of HH8+ embryo with control on the Left and miRNA inhibitor mix (miR-200a, miR-20a, and miR-217) and PCI-eGFP electroporation control on the Right. Immunohistochemistry for dpERK1/2 upon miRNA inhibition. (G) Dorsal whole-mount view of HH7 embryos displaying N2:eGFP reporter activity. Embryos were bilaterally electroporated with the N2:eGFP reporter and either miRNA inhibitor or mimic mixes (miR-200a, miR-20a, and miR-217). (H) Quantification of N2:eGFP-positive cells following either miRNA inhibition or overexpression (mimic), normalized to the control side of the embryo (n = 6). (I) Quantitative RT-PCR for neural crest specification genes TFAP2B and SOX9 and neural plate genes SOX2 and SUFU in embryos electroporated with FGF4 or PDGFA overexpression constructs. Each experimental condition was normalized to the control side of unilaterally electroporated embryos for that given condition (n = 5). (J) Electroporation scheme for DICER MO rescue experiments. (K) Quantitative RT-PCR for neural crest specification genes TFAP2B and SOX9 and neural plate genes SOX2 and SUFU in embryos electroporated with DICER MO alone or rescued by incubation with SU5402 chemical inhibitor or coinjection with miRNA mimic mix (miR-200a, miR-20a, and miR-217). Each experimental condition was normalized to the control side of bilaterally electroporated embryos for that given condition (n = 6). Error bars in C, E, H, I, and K represent the SD. Arrowheads indicate neural crest cells. HH, Hamburger and Hamilton; n.s., not significant; MO, morpholino. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Scale bar, 200 μM in (A) and (F). Dorsal whole-mount view of embryo with control on the Left and experimental reagent on the Right, 100 μM in (A) and (F) immunohistochemistry for dpERK1/2, 100 μM in (G).
Several receptor tyrosine kinase (RTK) signaling pathways can activate MAPK and Pi3K/Akt independent of FGF. Namely, platelet-derived growth factor (PDGF) signaling has been shown to be strongly associated with Pi3K/Akt signaling during embryonic development (48, 49). Accordingly, there may be additional miRNA target genes in other signaling systems that are contributing to the DICER knockdown phenotype (46). To confirm that the observed knockdown phenotype is mainly due to posttranscriptional repression of FGF and not PDGF signaling, we used unilateral electroporations to transfect embryos with FGF4 or PDGFA expression vectors. FGF4 overexpression, and not PDGFA, recapitulated the DICER knockdown phenotype as evident by quantitative RT-PCR for neural crest markers TFAP2B and SOX9 and neural plate markers SOX2 and SUFU (Fig. 5I). Additionally, immunohistochemistry for TFAP2B in embryos transfected with the FGF4 overexpression construct revealed a marked reduction in the number of neural crest cells as compared to PDGFA overexpression (SI Appendix, Fig. S5 A and B). Together, these observations support our hypothesis that levels of FGF signaling need to be carefully balanced for neural crest specification to occur.
From these results, we postulated that miR-20a, miR-200a, and miR-217 promote neural crest identity by attenuating levels of FGF signaling. To test this, we employed two strategies for attempting to rescue the neuralization phenotype observed after DICER knockdown (Fig. 5J). First, we treated morphant embryos with the RTK receptor chemical inhibitor SU5402 (200 nm) at the stages of neural crest induction (HH5 to HH9). Second, we coelectroporated embryos with DICER morpholino and a combination of the miR-20a, miR-200a, and miR-217 mimics. Consistent with our model, both conditions were able to rescue the neuralizing effects of DICER knockdown, as shown by quantitative RT-PCR for neural crest markers TFAP2B and SOX9 and neural plate markers SOX2 and SUFU (Fig. 5K). Given that SU5402 is a broad RTK inhibitor also targeting VEGF and PDGF receptors (50), we performed additional rescue experiments with more selective inhibitors of each of these RTKs. These results confirm that inhibition of FGFR1/2/3 (infigratinib), and not VEGFR2 (ZM 323881) or PDGFRα/β (CP-673451) rescues the neuralizing effects of DICER knockdown (SI Appendix, Fig. S5 C and D). These results demonstrate that a set of neural crest-enriched miRNAs, synthesized via DICER, posttranscriptionally attenuates FGF signaling in neural crest territory (Fig. 6).
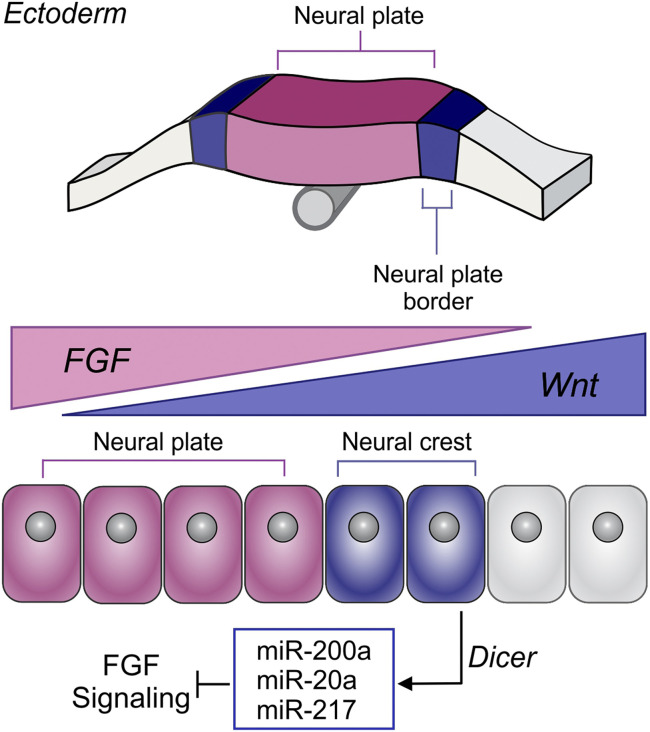
Fig. 6.
A model for the role of DICER and miRNAs in neural crest development. Model summarizing the results. DICER mediates the biogenesis of FGF-targeting miRNAs in neural crest cells, leading to posttranscriptional attenuation of FGF signaling for proper neural crest cell formation.
Discussion
miRNA-mediated gene silencing can function as a regulatory mechanism in critical developmental processes (27, 51). Here, we report that miRNAs influence ectodermal cell fate decisions by modulating the activity of signaling systems. Our results demonstrate that neural crest miRNAs posttranscriptionally attenuate FGF signaling to create conditions for proper cell specification (Fig. 6). These findings are consistent with previous studies that describe a role for miRNAs in lineage specification by regulating signaling networks (29, 52, 53). In human embryonic stem cells (hESCs) and Xenopus embryos, the miR-430/427/302 family promotes mesendoderm formation by targeting TGF-β signaling antagonists (54, 55). Furthermore, in Drosophila embryos, mir-9 targets several components of the FGF signaling pathway to pattern the midbrain–hindbrain boundary (29). Our results establish a decisive role for cell type-specific miRNAs in early developmental patterning events via direct regulation of signaling systems.
We also define why DICER is critical during neural crest formation. In the mouse embryo, Wnt1-Cre DICER knockout mice have severe craniofacial defects, suggesting a pivotal function for miRNAs in neural crest formation (22, 25). Consistent with this, we found that DICER is enriched in neural crest progenitors and that a tissue-specific enhancer mediates this increase in the expression of the enzyme. Although tissue-specific expression of miRNAs is well documented (56–58), we found that expression levels of miRNA processing enzymes may also contribute to specific miRNA function. In neural crest cells, which undergo highly dynamic regulatory state changes and striking morphological transitions, up-regulation of DICER may be necessary for the rapid turnover of miRNAs that facilitate drastic shifts in gene expression. Importantly, DICER is a rate-limiting enzyme in the miRNA biogenesis pathway (59, 60). Enhanced levels of DICER in neural crest cells suggest that distinct cell lineages require variable levels of miRNA processing for proper developmental transitions.
We propose a model (Fig. 6) that describes how the activity of signaling pathways is optimized during neural crest formation. Induction of the neural crest requires a delicate balance of Wnts, FGFs, and BMPs, which cooperate to jumpstart the neural crest gene regulatory program (10, 61). While high levels of FGFs drive ectodermal cells into adopting a CNS fate, complete inhibition of the pathway also abrogates neural crest formation (5, 16, 62). Thus, FGF activity in the neural crest must fall within a specific range for specification to occur. In avian embryos, there is evidence that suggests FGF/MAPK signaling originating from the primitive streak is required for early neural crest induction (5). We propose miRNA-mediated gene silencing modulates MAPK activity to allow for activation of the neural crest gene regulatory network (Fig. 6). This mechanism could also prevent excessive activation of the pathway due to the diffusion of FGFs from other sources, like the paraxial mesoderm (15, 63), and participate in signaling switches observed during neural crest development. For instance, neural crest cells have been shown to shift effector pathway utilization, from MAP kinase to PI3 kinase/Akt, during lineage restriction (14). We postulate posttranscriptional regulation may play an important role not only in spatial patterning, but also in the developmental transitions that take place during differentiation.
Finally, our model underscores a role for posttranscriptional regulation in craniofacial development and disease. Neurocristopathies encompass a wide range of clinical pathologies resulting from the abnormal specification, migration, and differentiation of neural crest cells (64). miRNAs have been implicated in craniofacial development, where they regulate bone and cartilage formation (65–67). Yet, we still know little about the importance of miRNAs in the genesis of craniofacial malformation. Our analysis identified numerous miRNAs that are required for proper neural crest formation and may regulate distinct cellular behaviors. While miRNAs are thought to subtly affect mRNA levels, we found that miRNAs act together to modulate the intracellular response to signaling systems. These sets of tissue-specific miRNAs may be integral components of the gene regulatory networks, acting to balance signal transduction cascades, or the transcriptional outputs of network nodes. Understanding the function of neural crest miRNAs may broaden our understanding of this genetic program and shed light on the etiology of many congenital disabilities.
Materials and Methods
Embryo Collection and Fixation.
Fertilized chicken eggs (Leghorn White) were purchased from the University of Connecticut Department of Animal Science. Eggs were incubated at 37 °C until embryos reached the proper developmental stage. Chicken embryos were collected and cultured according to the Early Chick (EC) protocol (68) and staged based on Hamburger and Hamilton (69). For in situ hybridization, embryos were fixed overnight at 4 °C in phosphate-buffered saline (PBS) containing 4% paraformaldehyde, dehydrated, and stored in methanol. For immunohistochemistry, embryos were fixed with 4% paraformaldehyde in phosphate buffer (PFA-PB) for 20 min at room temperature and processed immediately.
Whole-Mount In Situ Hybridization.
Double fluorescence in situ hybridization was performed using the Tyramide Signal Amplification (TSA) system from PerkinElmer (TSA Plus Cyanine 5 and Fluorescein, NEL754001KT) as previously described (70). Linearized DNA was used to synthesize digoxin and fluorescein-labeled antisense probes with Promega RNA polymerases. RNA probes were purified with Illustra ProbeQuant G-50 Micro-Columns (GE Healthcare, 28-9034-08).
Cryosectioning.
For histological analysis, embryos were incubated in 5% sucrose for 2 h at room temperature, and in 15% sucrose overnight at 4 °C. Embryos were then incubated in 7.5% porcine gelatin for 3 h at 37 °C, embedded in silicone molds, snap frozen in liquid nitrogen, and stored at −80 °C. The 10-μM sections were obtained using the CryoStar NX50 (Thermo Fisher). For imaging, slides were washed in phosphate-buffered saline with 0.1% Tween-20 (PBST) at 42 °C for 15 min for gelatin removal, rinsed twice in PBS, and mounted with PermaFluor Mounting Medium (Thermo Electron Corporation, 434990).
Embryo Electroporation.
Chicken embryos at stages HH4 to 5 were transfected with morpholinos, miRNA reagents, or DNA constructs by ex ovo electroporation, as previously described (38). Morpholinos/miRNA mimics/miRNA inhibitors or DNA expression vectors were injected between the epiblast and vitelline membrane of dissected embryos and electroporated with platinum electrodes (five 50-ms pulses of 5.1 V, with an interval of 100 ms between pulses) (71). In all gene/miRNA knockdown and overexpression experiments, the embryos were injected bilaterally with the control reagent on the left side and the targeted reagent on the right side. Following electroporation, embryos were cultured in albumin at 37 °C until they reached the appropriate developmental stage. All embryos were screened to ensure uniform electroporation and proper embryo morphology prior to further downstream analysis.
Embryo Dissociation and Cell Sorting.
To isolate neural crest cells, embryos were electroporated with 1.5 μg/μL of a neural crest-specific enhancer of the TFAP2A gene (72) (Tfap2aE1:eGFP). To isolate neural plate cells, embryos were electroporated with 1.5 μg/μL of a neural-specific enhancer (Sox2N2:eGFP) and 1.5 μg/μL of a neural crest/nonneural ectoderm enhancer (Tfap2aE2.7:mCherry) to exclude SOX2-positive neural crest cells. Embryos were incubated at 37 °C until the desired stage was reached and screened for robust GFP/mCherry expression. Embryos with weak expression or at incorrect stages of development were discarded. Dissected embryo heads were washed with Dulbecco's phosphate-buffered saline (dPBS) and dissociated with Accumax (Accutase SCR006) cell dissociation solution for 40 min at room temperature (RT). Dissociated cells were passed through a cell strainer (PluriSelect USA, Mini Cell Strainer II, 45-09840-50) and resuspended in Hanks 0.5% bovine serum albumin (BSA). Respective positive and negative cell populations were sorted into the appropriate buffer for downstream analysis using a BD AriaFusion cell sorter.
qRT-PCR.
To determine gene expression levels in sorted neural crest cells, 1,500 GFP+ and GFP− cells from the appropriate stage were sorted directly into 50 μL of lysis buffer from Power SYBR Green Cells-to-CT Kit (Thermo Fisher, 4402953). RNA extraction and cDNA preparation were performed according to the suggested protocol. RT-PCR was performed using the Power SYBR Green PCR master mix (Thermo Fisher, 4368577) in an ABI ViiA7 RT-PCR machine. Cycle threshold (Ct) values of all genes were normalized to reference gene HPRT and presented as a fold change of the control sample (double delta Ct). To quantify changes in gene expression caused by perturbation experiments, single neural folds were dissected from control and targeted sides of the embryo and subsequently lysed in 50 μL lysis buffer from Power SYBR Green Cells-to-CT Kit, following the same protocol as for sorted neural crest cells.
Immunohistochemistry of Cell Suspensions.
Immunohistochemistry of dissociated embryos was performed as previously described (73). In summary, heads of HH8 wild-type embryos were dissected in Ringer’s solution and dissociated in Accumax Cell dissociation solution (Accutase, SCR006) for 20 min at RT under mild agitation. Ten HH8 embryo heads were used for each antibody staining. Following dissociation, cells were washed in PBS and fixed in 0.4% PFA solution for 15 min at RT. Postfixation, cells were permeabilized with PBS + 0.3% Triton solution and blocked with 1% BSA solution (in PBS + 0.1% Tween 20) for 1 h at RT. Cells were incubated overnight with appropriate dilutions of primary antibody in blocking solution. The antibodies mouse anti-DICER (Abcam, ab227518) and rabbit anti-TFAP2B (Abcam, ab221094) were used at a dilution of 1:200. Following primary antibody staining, the cells were washed in PBS + 0.1% Tween 20 solution and incubated with appropriate secondary antibodies diluted to a concentration of 1:1,000 in blocking solution for 1 h at RT. Samples were washed twice in PBS + 0.1% Tween 20 and the antibody staining intensity was measured by FACS analysis using the Attune Nxt flow cytometer.
Enhancer Identification and Analysis.
Identification of enhancer elements using H3K27Ac HiChIP was performed as described by Mumbach and colleagues (74). Briefly, in situ contact libraries were generated according to the HiChIP protocol with modifications for initial low input optimization (74). HiChIP replicates were processed with the HiC-Pro pipeline (75). Raw fastq files (paired-end reads) were aligned to the chicken reference genome (Galgal5) and default settings were used to remove duplicate reads and filter for valid interactions. Preprocessed valid interactions were subsequently submitted to the Hichipper pipeline (76), a platform developed for bias-corrected peak calling, library quality control, and DNA loop calling in HiChIP datasets. Next, the framework Diffloop (77) was employed for identification of differential intrachromosomal chromatin interactions comparing neural crest and whole-embryo datasets. After mango algorithm correction with a false discovery rate (FDR) of 0.05, differential loops (neural crest vs. whole embryo) were called and annotated to promoters and enhancers. Enhancer–promoter loops were filtered using a minimum distance of 1.5 kb, and nonprevalent loops with a minimum of 10 counts in at least two replicates were excluded. Loop log fold change (logFC) and P values <0.05 were used to define neural crest-enriched and -depleted loops. Enhancer-promoter contact positions for the Dicer locus can be found in SI Appendix, Table S1. Putative enhancer elements in the DICER locus were defined by the presence of H3K27Ac chromatin marks (H3K27Ac CUT&RUN), chromatin accessibility via assay for transposase-accessible chromatin-sequencing (ATAC-seq), and enhancer–promoter contacts with logFC >0.5 (H3K27Ac HiChIP). Enhancer elements were amplified from HH8 chicken gDNA and cloned in the pTK-EGFP (36). To assess enhancer activity, HH5 embryos were coelectroporated with Tfap2aE1-mCherry and the pTK-EGFP enhancer constructs. Embryos were incubated at 37 °C to stage HH8 and postfixed overnight in 4% PFA prior to imaging. Whole-mount images were collected using an upright Zeiss Axio Imager fluorescent microscope.
Enhancer Loss-of-Function Assays.
A putative TFAP2A-bound enhancer in the DICER locus was knocked down using a CRISPR-Cas9 system optimized for early chicken embryos (37). Two gRNAs spanning the TFAP2A-associated peak were designed using online resources (http://crispor.tefor.net). gRNAs were cloned downstream of the U6 promoters in the empty cU6.3 vector (a gift from Marianne Bronner, California Institute of Technology, Pasadena, CA) (37). To assay the effects of enhancer knockdown on DICER expression, HH4 embryos were bilaterally electroporated with Tfap2aE1 Cas9:eGFP (modified from pCAGG-nls-Cas9-nls-GFP, a gift from Marianne Bronner) (37) and a control gRNA on the left and Tfap2aE1 Cas9:eGFP and the DicerE6 double gRNA vector on the right. Embryos were incubated at 37 °C to stage HH8 when half heads were dissected for control and targeted sides of the embryo and five heads were pooled per condition. Half heads were dissociated using Accumax as described earlier (embryo dissociation and cell sorting). A total of 300 GFP+ cells for each condition were sorted directly into 50 μL of lysis buffer from the Power SYBR Green Cells-to-CT Kit (Thermo Fisher, 4402953) using a BD AriaFusion cell sorter. DICER expression for control and targeted conditions were determined using quantitative RT-PCR as described above.
miRNA Mimics and Inhibitors.
For miRNA knockdown experiments, steric blocking, antisense miRNA inhibitors that hybridize to the mature miRNA species were utilized (IDT miRNA inhibitors). Each miRNA inhibitor contains a 2′-OMe modification to ribose molecules in the RNA backbone to increase melting temperature and confer resistance to endonucleases (78). Additionally, a terminal N,N-diethyl-4-(4-nitronaphthalen-1-ylazo)-phenylamine (ZEN) modification increases inhibitor binding affinity to the mature miRNA species and confers exonuclease resistance (78). For miRNA overexpression experiments, chemically synthesized, double stranded RNAs which mimic mature endogenous miRNA activity were utilized (miScript miRNA mimics, Qiagen). miRNA mimics and inhibitors were designed based on mature miRNA sequences obtained from miRbase (79). Sequences for all miRNA inhibitors and mimics utilized in this study can be found in SI Appendix, Table S3.
Perturbation Experiments.
DICER knockdown was performed using a fluorescein isothiocyanate (FITC)-labeled translation-blocking morpholino (GeneTools) (SI Appendix, Table S3). Both control and DICER morpholinos were injected at a final concentration of 1.5 mM, supplemented with 1 μg/μL of carrier DNA and 10 mM Tris pH 8.0. For miRNA overexpression experiments, both miRNA mimics (miScript miRNA mimic, Qiagen) and controls (AllStar Negative Control siRNA, Qiagen, 1027280) were electroporated at a concentration of 100 μM, with 1 μg/μL of carrier DNA and 10 mM Tris pH 8.0, to facilitate entry into cells. For miRNA knockdown experiments, both miRNA inhibitors (IDT miRNA inhibitors) and control (NC5 negative control inhibitor, IDT) were electroporated at a concentration of 10 μM, with 1 μg/μL of carrier DNA and 10 mM Tris pH 8.0 miRNA. For DICER knockdown rescue experiments (Fig. 2C), control and DICER morpholinos were injected at a final concentration of 1.5 mM, containing 1 μg/μL of appropriate DICER overexpression vector and 10 mM Tris pH 8.0. For enhancer reporter analysis (Fig. 5 G and H), miRNA mimic or inhibitor mixes were prepared as described above, but supplemented with 1 μg/μL of the N2:eGFP reporter instead of carrier DNA. For DICER knockdown rescue experiments with various RTK receptor inhibitors (Fig. 5 J and K and SI Appendix, Fig. S5 C and D and Table S3), morpholinos were prepared as described above. Electroporated embryos were then placed in albumin supplemented with 200 nm of either SU5402, infigratinib (FGFR1-3 inhibitor), CP-673451 (PDGFRα/β inhibitor), or ZM 323881 (VEGFR2 inhibitor) and developed to the desired stage. Specificity of chemical inhibitors utilized in this study has been previously described (50, 80–82). For DICER knockdown rescue with miRNA mimic mix (Fig. 5 J and K), control and DICER morpholinos were injected at a final concentration of 1.5 mM, supplemented with 1 μg/μL of carrier DNA, miRNA mimics at a final concentration of 100 μM for each and 10 mM Tris pH 8.0.
Immunohistochemistry.
For whole-mount immunohistochemistry, fixed embryos were dissected from the filter paper and washed in Tris-buffered saline (TBS) containing 0.1% Triton X-100 and 1% dimethyl sulfoxide (DMSO) (TBTD). Embryos were blocked for 2 h in TBTD supplemented with 10% donkey serum and incubated in primary antibody diluted in blocking solution, overnight at 4 °C. The following primary antibodies were used: anti-AP2 beta, mouse monoclonal (Santa Cruz Biotechnology, sc-390119); anti-Sox9, rabbit polyclonal (Millipore Sigma, ab5535); anti-Sox2, rabbit polyclonal (Abcam, ab97959); anti-Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), rabbit polyclonal (Cell Signaling Technology, antibody no. 9101); anti-Sox10, goat polyclonal (R&D, AF2864); anti-HNKI, mouse polyclonal (DSHB, AB_2314644); anti-Histone H3 (Phospho S10), rabbit polyclonal (Abcam, ab47297); and anti-Caspase3, rabbit polyclonal (R&D Systems, AF835). Following primary antibody incubation, embryos were washed with TBTD, blocked for 30 min at RT, and stained with appropriate secondary antibodies diluted in blocking solution for 2 h at RT. Secondary antibodies used included donkey anti-mouse/rabbit IgG conjugated with Alexa Fluor 488/568/647 (Molecular Probes, 1:3,000). Following secondary antibody incubation, embryos were washed, stained with DAPI (1:1,000), and postfixed with 4% PFA for 1 h, prior to imaging. Whole-mount images were collected using an upright Zeiss Axio Imager fluorescent microscope. Quantification of fluorescence for phenotype quantification in gain- and loss-of-function studies was performed with ZEISS ZEN imaging software.
Expression Vectors.
DICER wild-type and mutant overexpression constructs (Fig. 2C) were gifts from Phillip Sharp (Addgene plasmid nos. 41590 and 41584; Massachusetts Institute of Technology, Cambridge, MA). Enhancer reporter constructs were assembled as previously described (83). Briefly, enhancer regions were cloned into ptk eGFP (36), such that enhancer activity drives GFP expression. All enhancer sequences were PCR amplified from HH8 chicken gDNA. The FGF4 and PDGFA overexpression constructs (Fig. 5I and SI Appendix, Fig. S5 A and B) were assembled by insertion of the full-length cDNA sequence of avian FGF4 or PDGFA into a pCI-H2B-RFP backbone. All coding sequences were PCR amplified from an HH8 cDNA library and assembled as previously described (32).
Nanostring Analysis.
To assess the global effect of DICER knockdown on neural crest development, expression profiles of control and DICER morpholino-treated neural folds obtained from the same embryo were compared using Nanostring analysis. Stage HH5 chicken embryos were bilaterally electroporated with control morpholino on the left and DICER MO on the right as described above (perturbation experiments). Neural folds from these embryos were dissected and lysed in Ambion lysis buffer (RNAqueous-Micro Kit, AM1931). Control and morpholino-treated neural folds were processed separately. RNA lysates were hybridized for 12 h at 65 °C to a Nanostring probe set containing 200 probes for neural crest, placodal, and neural genes (19). Nanostring data were analyzed using the nSolver software package.
Small RNA Sequencing.
Pure populations of neural crest and neural plate cells were isolated as described above (embryo dissociation and cell sorting). At least 10,000 cells for each cell type were sorted into 1 mL of TRIzol (Thermo Fisher Scientific, 15596026) using a BD AriaFusion cell sorter. Total RNA was isolated according to the suggested manufacturer’s protocol. RNA concentration and integrity were measured using Qubit RNA HS Assay Kit (Thermo Fisher Scientific, Q32852). All six small RNA libraries (three replicates for each cell type) were generated using NEB Next Small RNA Library Prep Set for Illumina (NEB, E7330S), according to manufacturer’s instructions. The PCR amplification was done under the parameters of 100 ng of total RNA input and received 15 cycles. Libraries were sent for quality control using an ABI 3730xl DNA Analyzer for fragment analysis and subsequently size selected on a Pippin Prep instrument for fragments between 105 bp and 155 bp according to manufacturer’s suggestions. Tru-seq barcoded small RNA libraries were sequenced on an Illumina NextSEq. 500 instrument with single-end 75-bp reads.
Small RNA-Sequencing Data Analysis.
Small RNA-sequencing reads were processed and quantified using the miRDeep2 modules mapper and quantifier (84). Briefly, reads were trimmed to remove the 3′ adaptor sequences and aligned to the chicken reference genome Galgal5 using bowtie with no mismatches allowed. Reads consisting of 18 to 35 nucleotides were kept for further analysis. Next, mature and star miRNA sequences were mapped against annotated chicken precursor miRNAs obtained from miRbase (79). Mapped small RNA-sequencing reads were then intersected with the mature miRNA mappings to obtain a read count table. Differential gene expression analysis for mature miRNA reads was performed using DESeq2 (85).
miRNA qRT-PCR.
To measure mature miRNA levels, neural crest and neural plate cells were isolated as described above (embryo dissociation and cell sorting). At least 3,000 cells of each type were sorted directly into 100 μL of RNeasy Plus Micro Kit lysis buffer (Qiagen, 74034). Small RNAs were extracted following the manufacturer’s protocol. Poly(A) tailing and cDNA synthesis were performed using the qScript microRNA cDNA Synthesis Kit (Quanta Biosciences, 95107-025). RT-PCR for individual miRNA was performed as suggested by the kit, with mature miRNA-specific primers, and a universal primer that anneals to the poly-A tail. Ct values were normalized to 18S rRNA and expressed as a fold change as compared to the control sample.
mirPath Functional Analysis.
DIANA-miRPath v3.0 software was utilized to elucidate the functional roles of the top 10 neural crest- or neural plate-enriched miRNAs. The online software utilizes predictive binding algorithms including Target Scan (47) to define a list of potential gene targets for each miRNA. Next, the program identifies miRNA-regulated KEGG pathways ranked by significance level. P values were obtained by enrichment analysis using the Fisher’s exact test and the FDR was estimated using the Benjamini and Hochberg method (86).
RNA Sequencing.
Pure populations of neural crest and neural plate cells were isolated as described above (embryo dissociation and cell sorting). At least 5,000 cells for each cell type were sorted directly into 100 μL of lysis buffer from the RNAqueous-Micro Total RNA Isolation Kit (Thermo Fisher Scientific, AM1931). Total RNA was isolated according to the manufacturer’s protocol. RNA was poly(A) selected using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, E7490). TruSeq-barcoded RNA-seq libraries were generated with the NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs, E7760) and sequenced on an Illumina NextSEq. 500 instrument with single-end 75-bp reads.
RNA-Sequencing Data Analysis.
RNA-sequencing reads were trimmed using Cutadapt (87) to remove adaptor sequences. Reads were then aligned to the chicken reference genome Galgal5 using HiSat2 (88). Read counts were obtained using the feature Counts (89). Differential gene expression analysis was performed using DESeq2 (85).
Identification of miRNA Gene Targets.
Neural plate RNA-sequencing datasets were compared to transcriptome data obtained from neural crest cells at equivalent stages (90). Differentially expressed mRNAs from the RNA-sequencing data between the neural plate and neural crest were determined using DESeq2 (85). Next, the DESeq2 dataset was filtered for genes in the MAPK and PI3/Akt KEGG pathways. Target genes for the top 10 differentially expressed miRNAs in neural crest cells were determined using TargetScanHuman (47), with a context + score threshold of < −0.2. The filtered DESeq2 dataset was then merged with the TargetScanHuman miRNA target predictions, to identify mRNA:miRNA interactions that had an inverse correlation in expression in neural crest cells. Criteria for differentially expressed genes in this analysis included P value <0.01 and log2 fold change < −1.0 (in neural crest as compared to neural plate).
The 3′ UTR Reporter Assays.
Full‐length 3′ UTRs of predicted gene targets were amplified using HH8 chicken gDNA and cloned into the dual-luciferase pmirGLO vector (Promega, E1330) using Gibson assembly. Mutant plasmids for each miRNA seed sequence were generated as previously described (91). For transient transfections, HEK293T cells were plated in a 24-well plate, 24 h prior to transfection. Each well was transfected with 6 ng of dual-luciferase plasmid and 50 nM miRNA mimic using Lipofectamine 2000 (Life Technologies, 1166809). At 48 h posttransfection, cells were lysed using Promega Dual‐Luciferase Kit lysis buffer (Promega, no. E1910). Luciferase activity was detected using the Promega Dual‐Luciferase Kit with a dual-injection luminometer (Turner Biosystems). Normalization of luciferase values for varying conditions was done as previously described (91).
Western Blot Analysis.
Following perturbation experiments (DICER morpholino), neural folds were dissected from control and targeted sides of the embryo and placed directly into modified radioimmunoprecipitation assay (RIPA) buffer. Three neural folds were pooled per condition. Protein extraction and Western blot analysis were performed as previously described (38). Western blot band intensity was quantified using ImageJ and normalized to the alpha-tubulin loading control. The following primary antibodies were used: anti-Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), rabbit polyclonal (Cell Signaling Technology, antibody no. 9101); anti-alpha-tubulin, rabbit polyclonal (Cell Signaling Technology, antibody no. 2144); and Phospho-Akt (Ser473), rabbit polyclonal (Cell Signaling Technology, antibody no. 9271).
Statistical Analysis.
Flow cytometry data analysis.
Flow cytometry data from the cell suspension antibody-staining experiment were analyzed using the FCS Express 6 software package. After assigning appropriate forward scatter and side scatter gates, the neural crest cells were identified as cells withTFAP2B antibody staining intensity above the threshold that was set based on the negative and single-color controls. Next, we obtained the intensity values for DICER in the TFAP2B-positive and -negative cell populations.
Additional statistical analyses.
RT-PCR experiments with sorted cells were performed using three independent biological replicates (each iteration having three technical replicates). RT-PCR performed for perturbation experiments included at least five embryos for each condition. The immunohistochemistry experiments in the embryo were performed with at least five embryos for each antibody and condition. The Nanostring experiment was performed with three replicates of control and DICER morpholino-treated neural folds. The small RNA sequencing for neural crest and neural plate cells was repeated three times, while RNA sequencing was replicated twice. Student’s t test (one tailed) was performed to calculate P values for functional experiments and P < 0.05 was considered to be significant. Wilcoxon signed-rank test was used to calculate P values for flow cytometry data analysis and dual-luciferase 3′ UTR reporter assays.
Supplementary Material
Acknowledgments
We are indebted to Adam Wojno for cell-sorting assistance at the Biotechnology Resource Center Flow Cytometry Cell Sorting Facility at Cornell University. This work was supported by a Cornell Center for Vertebrate Genomics Scholars Award to J.C. and by NIH grant 1DP2HD102043.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009997117/-/DCSupplemental.
Data Availability.
The small RNA-sequencing datasets for sorted neural plate and neural crest cells and RNA-sequencing datasets for sorted neural plate cells have been deposited to the Gene Expression Omnibus under the accession number GSE150007.
References
- 1.Sauka-Spengler T., Bronner-Fraser M., A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557–568 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Stern C. D., Neural induction: Old problem, new findings, yet more questions. Development 132, 2007–2021 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Patthey C., Gunhaga L., Signaling pathways regulating ectodermal cell fate choices. Exp. Cell Res. 321, 11–16 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Streit A., Berliner A. J., Papanayotou C., Sirulnik A., Stern C. D., Initiation of neural induction by FGF signalling before gastrulation. Nature 406, 74–78 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Stuhlmiller T. J., García-Castro M. I., FGF/MAPK signaling is required in the gastrula epiblast for avian neural crest induction. Development 139, 289–300 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García-Castro M. I., Marcelle C., Bronner-Fraser M., Ectodermal Wnt function as a neural crest inducer. Science 297, 848–851 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Wilson P. A., Hemmati-Brivanlou A., Induction of epidermis and inhibition of neural fate by Bmp-4. Nature 376, 331–333 (1995). [DOI] [PubMed] [Google Scholar]
- 8.LaBonne C., Bronner-Fraser M., Neural crest induction in Xenopus: Evidence for a two-signal model. Development 125, 2403–2414 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Yardley N., García-Castro M. I., FGF signaling transforms non-neural ectoderm into neural crest. Dev. Biol. 372, 166–177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simões-Costa M., Bronner M. E., Establishing neural crest identity: A gene regulatory recipe. Development 142, 242–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groves A. K., LaBonne C., Setting appropriate boundaries: Fate, patterning and competence at the neural plate border. Dev. Biol. 389, 2–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meulemans D., Bronner-Fraser M., Gene-regulatory interactions in neural crest evolution and development. Dev. Cell 7, 291–299 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Streit A., Stern C. D., Establishment and maintenance of the border of the neural plate in the chick: Involvement of FGF and BMP activity. Mech. Dev. 82, 51–66 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Geary L., LaBonne C., FGF mediated MAPK and PI3K/Akt Signals make distinct contributions to pluripotency and the establishment of Neural Crest. eLife 7, e33845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pla P., Monsoro-Burq A. H., The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Dev. Biol. 444 (suppl. 1), S36–S46 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Hackland J. O. S., et al. , FGF modulates the axial identity of trunk hPSC-derived neural crest but not the cranial-trunk decision. Stem Cell Reports 12, 920–933 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez I. S., Araujo M., Nieto M. A., Neural induction in whole chick embryo cultures by FGF. Dev. Biol. 199, 42–54 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Sheehy N. T., Cordes K. R., White M. P., Ivey K. N., Srivastava D., The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development 137, 4307–4316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya D., Rothstein M., Azambuja A. P., Simoes-Costa M., Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis. eLife 7, e40556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez-Vásquez E., Bronner M. E., Strobl-Mazzulla P. H., Epigenetic inactivation of miR-203 as a key step in neural crest epithelial-to-mesenchymal transition. Development 146, dev171017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xi J., et al. , Mir-29b mediates the neural tube versus neural crest fate decision during embryonic stem cell neural differentiation. Stem Cell Reports 9, 571–586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang T., Liu Y., Huang M., Zhao X., Cheng L., Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J. Mol. Cell Biol. 2, 152–163 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Huang Z. P., et al. , Loss of microRNAs in neural crest leads to cardiovascular syndromes resembling human congenital heart defects. Arterioscler. Thromb. Vasc. Biol. 30, 2575–2586 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie X., Wang Q., Jiao K., Dicer activity in neural crest cells is essential for craniofacial organogenesis and pharyngeal arch artery morphogenesis. Mech. Dev. 128, 200–207 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zehir A., Hua L. L., Maska E. L., Morikawa Y., Cserjesi P., Dicer is required for survival of differentiating neural crest cells. Dev. Biol. 340, 459–467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gessert S., Bugner V., Tecza A., Pinker M., Kühl M., FMR1/FXR1 and the miRNA pathway are required for eye and neural crest development. Dev. Biol. 341, 222–235 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Alberti C., Cochella L., A framework for understanding the roles of miRNAs in animal development. Development 144, 2548–2559 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Kennell J. A., Gerin I., MacDougald O. A., Cadigan K. M., The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 15417–15422 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leucht C., et al. , MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11, 641–648 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Silver S. J., Hagen J. W., Okamura K., Perrimon N., Lai E. C., Functional screening identifies miR-315 as a potent activator of wingless signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 18151–18156 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiner A. M. J., et al. , Dicer1 is required for pigment cell and craniofacial development in zebrafish. Biochim. Biophys. Acta. Gene Regul. Mech. 1862, 472–485 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Rothstein M., Simoes-Costa M., Heterodimerization of TFAP2 pioneer factors drives epigenomic remodeling during neural crest specification. Genome Res. 30, 35–48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simoes-Costa M., Bronner M. E., Reprogramming of avian neural crest axial identity and cell fate. Science 352, 1570–1573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel D. P., Metazoan microRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rada-Iglesias A., et al. , Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell 11, 633–648 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchikawa M., Ishida Y., Takemoto T., Kamachi Y., Kondoh H., Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4, 509–519 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Gandhi S., Piacentino M. L., Vieceli F. M., Bronner M. E., Optimization of CRISPR/Cas9 genome editing for loss-of-function in the early chick embryo. Dev. Biol. 432, 86–97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simões-Costa M., Stone M., Bronner M. E., Axud1 integrates Wnt signaling and transcriptional inputs to drive neural crest formation. Dev. Cell 34, 544–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cernilogar F. M., et al. , Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480, 391–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coll O., et al. , Dicer-2 promotes mRNA activation through cytoplasmic polyadenylation. RNA 24, 529–539 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neve J., et al. , Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res. 26, 24–35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurtan A. M., Lu V., Bhutkar A., Sharp P. A., In vivo structure-function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA 18, 1116–1122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roellig D., Tan-Cabugao J., Esaian S., Bronner M. E., Dynamic transcriptional signature and cell fate analysis reveals plasticity of individual neural plate border cells. eLife 6, e21620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korpal M., Lee E. S., Hu G., Kang Y., The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 283, 14910–14914 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vlachos I. S., et al. , DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 43, W460–W466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dinsmore C. J., Soriano P., MAPK and PI3K signaling: At the crossroads of neural crest development. Dev. Biol. 444 (suppl. 1), S79–S97 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agarwal V., Bell G. W., Nam J. W., Bartel D. P., Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He F., Soriano P., A critical role for PDGFRα signaling in medial nasal process development. PLoS Genet. 9, e1003851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klinghoffer R. A., Hamilton T. G., Hoch R., Soriano P., An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Dev. Cell 2, 103–113 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Mohammadi M., et al. , Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science 276, 955–960 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Sayed D., Abdellatif M., MicroRNAs in development and disease. Physiol. Rev. 91, 827–887 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Boissart C., Nissan X., Giraud-Triboult K., Peschanski M., Benchoua A., miR-125 potentiates early neural specification of human embryonic stem cells. Development 139, 1247–1257 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Singh B. N., et al. , The etv2-miR-130a network regulates mesodermal specification. Cell Rep. 13, 915–923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi W. Y., Giraldez A. J., Schier A. F., Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318, 271–274 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Rosa A., Spagnoli F. M., Brivanlou A. H., The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 16, 517–527 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Landgraf P., et al. , A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ludwig N., et al. , Distribution of miRNA expression across human tissues. Nucleic Acids Res. 44, 3865–3877 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ro S., Park C., Young D., Sanders K. M., Yan W., Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 35, 5944–5953 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng Y., Zhang X., Graves P., Zeng Y., A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA 18, 2083–2092 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koh H. R., Ghanbariniaki A., Myong S., RNA stem structure governs coupling of dicing and gene silencing in RNA interference. Proc. Natl. Acad. Sci. U.S.A. 114, E10349–E10358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martik M. L., Bronner M. E., Regulatory logic underlying diversification of the neural crest. Trends Genet. 33, 715–727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delaune E., Lemaire P., Kodjabachian L., Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development 132, 299–310 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Monsoro-Burq A. H., Fletcher R. B., Harland R. M., Neural crest induction by paraxial mesoderm in Xenopus embryos requires FGF signals. Development 130, 3111–3124 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Vega-Lopez G. A., Cerrizuela S., Tribulo C., Aybar M. J., Neurocristopathies: New insights 150 years after the neural crest discovery. Dev. Biol. 444 (suppl. 1), S110–S143 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Desvignes T., Contreras A., Postlethwait J. H., Evolution of the miR199-214 cluster and vertebrate skeletal development. RNA Biol. 11, 281–294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ding H. L., et al. , MicroRNA profiling during craniofacial development: Potential roles for Mir23b and Mir133b. Front. Physiol. 7, 281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eberhart J. K., et al. , MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat. Genet. 40, 290–298 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chapman S. C., Collignon J., Schoenwolf G. C., Lumsden A., Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 220, 284–289 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Hamburger V., Hamilton H. L., A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92 (1951). [PubMed] [Google Scholar]
- 70.Denkers N., García-Villalba P., Rodesch C. K., Nielson K. R., Mauch T. J., FISHing for chick genes: Triple-label whole-mount fluorescence in situ hybridization detects simultaneous and overlapping gene expression in avian embryos. Dev. Dyn. 229, 651–657 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Sauka-Spengler T., Barembaum M., Gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 87, 237–256 (2008). [DOI] [PubMed] [Google Scholar]
- 72.Attanasio C., et al. , Fine tuning of craniofacial morphology by distant-acting enhancers. Science 342, 1241006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang C. C., Bajikar S. S., Jamal L., Atkins K. A., Janes K. A., A time- and matrix-dependent TGFBR3-JUND-KRT5 regulatory circuit in single breast epithelial cells and basal-like premalignancies. Nat. Cell Biol. 16, 345–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mumbach M. R., et al. , HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Servant N., et al. , HiC-pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lareau C. A., Aryee M. J., hichipper: A preprocessing pipeline for calling DNA loops from HiChIP data. Nat. Methods 15, 155–156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lareau C. A., Aryee M. J., diffloop: A computational framework for identifying and analyzing differential DNA loops from sequencing data. Bioinformatics 34, 672–674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lennox K. A., Owczarzy R., Thomas D. M., Walder J. A., Behlke M. A., Improved performance of anti-miRNA oligonucleotides using a novel non-nucleotide modifier. Mol. Ther. Nucleic Acids 2, e117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kozomara A., Griffiths-Jones S., miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guagnano V., et al. , Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J. Med. Chem. 54, 7066–7083 (2011). [DOI] [PubMed] [Google Scholar]
- 81.Roberts W. G., et al. , Antiangiogenic and antitumor activity of a selective PDGFR tyrosine kinase inhibitor, CP-673,451. Cancer Res. 65, 957–966 (2005). [PubMed] [Google Scholar]
- 82.Whittles C. E., et al. , ZM323881, a novel inhibitor of vascular endothelial growth factor-receptor-2 tyrosine kinase activity. Microcirculation 9, 513–522 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Simões-Costa M. S., McKeown S. J., Tan-Cabugao J., Sauka-Spengler T., Bronner M. E., Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS Genet. 8, e1003142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Friedländer M. R., Mackowiak S. D., Li N., Chen W., Rajewsky N., miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 87.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 88.Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L., Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Bhattacharya D., Azambuja A. P., Simoes-Costa M., Metabolic reprogramming promotes neural crest migration via yap/tead signaling. Dev. Cell 53, 199–211.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grimson A., et al. , MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 27, 91–105 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The small RNA-sequencing datasets for sorted neural plate and neural crest cells and RNA-sequencing datasets for sorted neural plate cells have been deposited to the Gene Expression Omnibus under the accession number GSE150007.