Significance
We detail host–pathogen interaction gene-expression profiles of Streptococcus pneumoniae (Spn) and its infected host at disease relevant anatomical sites using mice as experimental models. We identify the shared and organ-specific transcriptomes of Spn, show that bacterial and host gene-expression profiles are highly distinct during asymptomatic colonization versus disease-causing infection, and demonstrate that Spn and host genes with high levels of expression contribute to pathogenesis or host defense, respectively, making them optimal targets for intervention.
Keywords: dual species RNA-seq, in vivo transcriptomics, Streptococcus pneumoniae, mouse models of colonization and invasive disease, host–pathogen interactions
Abstract
Streptococcus pneumoniae (Spn) colonizes the nasopharynx and can cause pneumonia. From the lungs it spreads to the bloodstream and causes organ damage. We characterized the in vivo Spn and mouse transcriptomes within the nasopharynx, lungs, blood, heart, and kidneys using three Spn strains. We identified Spn genes highly expressed at all anatomical sites and in an organ-specific manner; highly expressed genes were shown to have vital roles with knockout mutants. The in vivo bacterial transcriptome during colonization/disease was distinct from previously reported in vitro transcriptomes. Distinct Spn and host gene-expression profiles were observed during colonization and disease states, revealing specific genes/operons whereby Spn adapts to and influences host sites in vivo. We identified and experimentally verified host-defense pathways induced by Spn during invasive disease, including proinflammatory responses and the interferon response. These results shed light on the pathogenesis of Spn and identify therapeutic targets.
The gram-positive microbe Streptococcus pneumoniae (Spn) asymptomatically colonizes the nasopharynx (1). It is also an opportunistic pathogen that commonly infects young children, immunodeficient patients, and the elderly (2). It was estimated that in 2015, 192,000 to 366,000 deaths in children under 5 y of age were the result of pneumococcal infections (3). With the advancement of antibiotics and introduction of the first pneumococcal conjugate vaccine in 2000, and subsequent vaccines, deaths attributable to Spn have declined (3, 4). However, increasing antibiotic resistance and serotype replacement have been observed (5). A study conducted from 2006 to 2016 showed increased resistance to penicillin, amoxicillin, ceftriaxone, and meropenem. Implementation of the PCV13 vaccine in 2010 (6) resulted in increased incidence of nonvaccine Spn serotypes (7). Thus, despite major advances in public health, pneumococcal infections continue to be a significant cause of morbidity and mortality, and further investigations are warranted.
The pneumococcus infects a variety of anatomical sites and causes a range of illnesses, some resulting in acute mortality. Spn is a common cause of sinusitis, otitis media, bronchitis, pneumonia, bacteremia, sepsis, and meningitis (2). Our group has previously shown that during bacteremic episodes, pneumococci in the bloodstream can translocate into the heart and kill cardiomyocytes, resulting in acute heart dysfunction (8). Invasive pneumococcal disease can also lead to renal complications in pediatric patients and hospitalized adults (9).
Current efforts to reduce the impact of pneumococcus on human health include improving the efficacy of vaccines and identifying host-directed therapies to decrease morbidity and mortality. Over the past 20 y, several studies have examined pneumococcal gene expression and virulence determinant requirements in vivo, with consideration for anatomical site-specific differences. One approach was the use of signature tagged mutagenesis to identify mutants that are unable to replicate in vivo in an anatomical site-specific manner (10). Accompanying studies using microarrays to examine in vivo isolated RNA from the bacteria indicated that Spn gene expression varies according to the host site (11). Other RNA-sequencing (RNA-seq) studies have also focused on the host response within the lungs upon pneumococcal infection. These have identified key genes involved in neutrophil recruitment and response to diverse strains and mutants (12, 13). Other forms of sequencing-based studies, such as transposon sequencing (Tn-seq) have also been performed on the pneumococcus in multiple contexts, such as in vivo survival or transmission, in multiple hosts (14–16). These studies, some of which are discussed below in the context of our results, offer an important consideration when identifying vaccine candidates, for which the ideal targets would be universally expressed, essential genes. However, it is important to note that prior studies on the in vivo transcriptome of Spn have employed only a single strain of Spn and used comparisons to in vitro conditions for purposes of normalization, whereas the contrast between in vitro and in vivo transcriptomes reported here suggests a more physiological method of normalization. Finally, we build and expand upon prior studies to provide information on the coordination and regulation of host pathways responsible for restricting pneumococcal infection at different anatomical sites, a process that remains a largely open question.
Here we explored the transcriptome of the pneumococcus and its host using dual species RNA-seq at diverse host anatomical sites in vivo. Blood, kidneys, lungs, and nasopharynx were harvested from mice colonized or infected with three different Spn strains, with uninfected host tissue used as control. These data provide a comprehensive view of dynamic processes impacting bacterial and host biology as the bacterium invades distinct anatomical sites.
Results
Pneumococcal Burden Varies in a Strain- and Site-Specific Manner within the Mouse.
To avoid strain bias, we used three Spn strains each belonging to a distinct capsular serotype and multilocus sequence type: TIGR4 (serotype 4, ST205), D39 (2, ST595), and 6A-10 (6A, ST460). To capture the host and bacterial transcriptomes during infection, mice were challenged with each of these strains and tissue/organs collected from the nasopharynx, lungs, blood, and kidneys (the latter lacking 6A-10 samples). Heart data (TIGR4 only) were included from our previously published study (8).
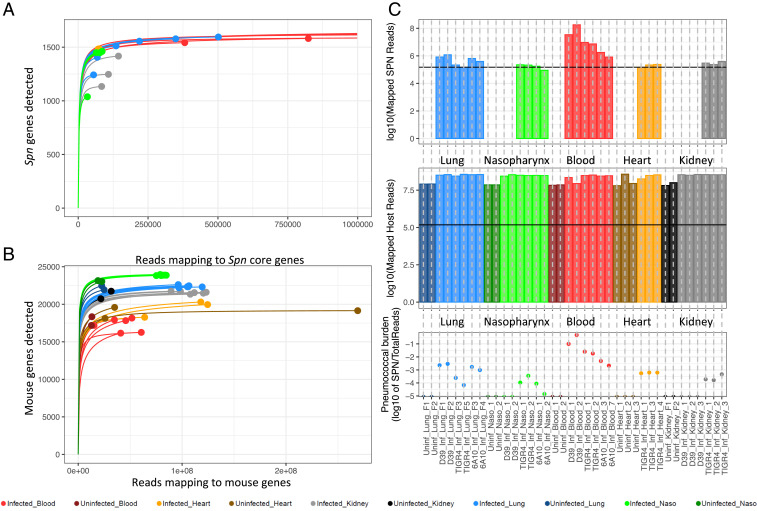
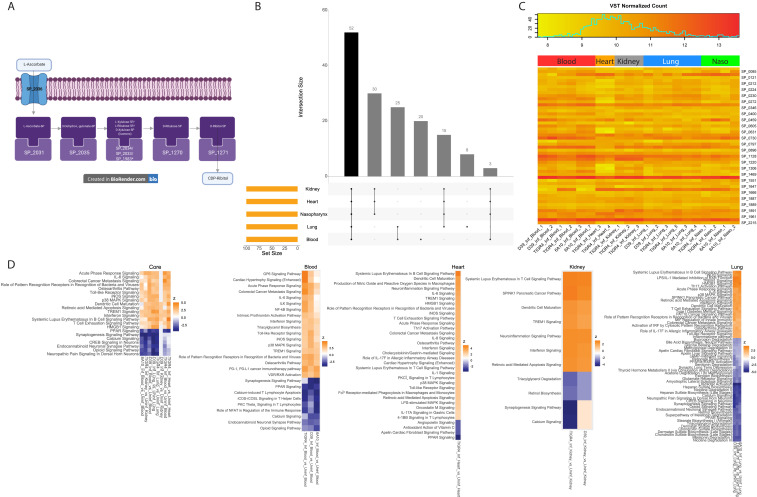
A typical obstacle when performing multispecies RNA-seq is uneven sequence amounts obtained for each species examined. Deeper sequencing is often needed for the minority species, in our case Spn. To attain confidence in the coverage obtained for Spn genes, we generated saturation curves to determine the minimum requirement for reads mapping to the respective Spn and mouse genomes (Fig. 1 A and B). Samples whose curves plateaued or approached plateauing were deemed adequate. The minimum required number of mapped Spn reads per sample was estimated at ∼150,000 reads. Saturation was achieved for all samples except D39-infected kidneys and D39-colonized nasopharynxes that were excluded from all bacterial analyses.
Fig. 1.
Rarefaction curves and estimation of pneumococcal burden. (A) Saturation curves of pneumococcus-infected samples. Samples whose curves plateau harbor enough reads mapped across all 1,682 core Spn genes to achieve saturation. (B) Rarefaction curves of infected and uninfected host samples. (C) Log10 values of reads mapped (Dataset S1) to the pneumococcus-infected samples (Top), host samples (Middle), and relative estimate of pneumococcal burden within each sample. Horizontal black lines (Top and Middle) indicate the minimum number of reads required for samples to plateau.
We estimated the ratios (Fig. 1 C, Bottom) of the number of Spn reads (Fig. 1 C, Top) compared to total sequenced mouse reads (Dataset S1) at each site. Although these ratios should not be directly compared across multiple anatomical sites, and they depend not only on the bacterial burden but also on the transcriptional activity of Spn (and mouse cells) at each site, ratios suggest that Spn infection in the lungs is highest with D39, followed by 6A-10 and TIGR4. On the other hand, TIGR4 appears to dominate over the other strains in the nasopharynx, followed by 6A-10, whereas D39 was more successful in thriving in the blood compared to TIGR4 and 6A-10 during bacteremia. Bacterial read proportions for these strains are consistent with their known virulence phenotypes in mice (17). No conclusions could be drawn within the hearts as our previous study only included the TIGR4 strain. Similarly, for kidneys we lacked 6A-10 samples, but TIGR4 appeared to perform better than D39 at this site. A complete overview of sequenced and mapped reads is provided in Dataset S1. To determine if actual bacterial burdens varied across anatomical sites, we performed colony forming unit (CFU) counts for TIGR4-infected organs; however, no significant differences were found (Dataset S1).
The In Vivo Pneumococcal and Host Transcriptomes Demonstrate Differences in Gene-Expression Profiles during Colonization and Infection.
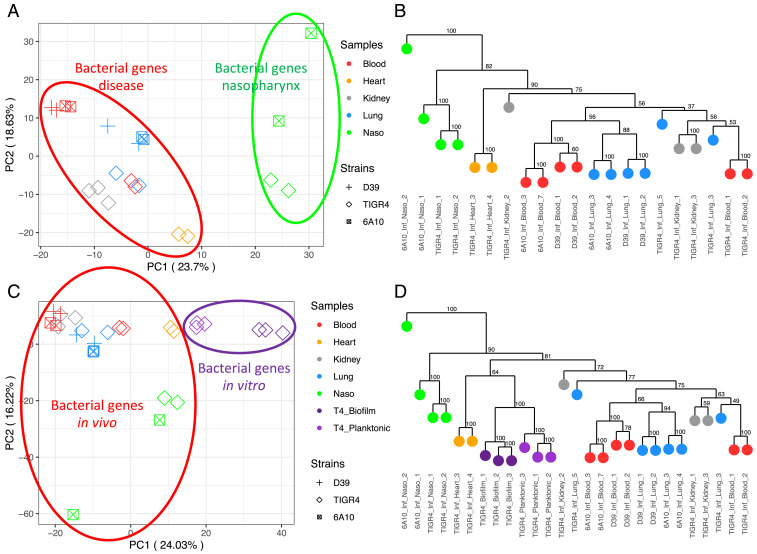
To assess the spectrum of core Spn gene-expression responses at each anatomical site across our samples, and to verify that replicate samples were similar to each other, we performed principal component analyses (PCA) on PanOCT core gene orthologs (18) across the three strains. The bacterial PCA (Fig. 2A) was based on DEseq2’s variance stabilized counts of 1,682 core Spn genes (Dataset S2, which also includes strain-specific Spn and mouse genes). Spn typically colonizes the nasopharynx without harming the host (1). We observed a distinct gene-expression profile in nasopharyngeal samples, which are separated by PC1 away from bacterial profiles seen in the remaining infected organs (Fig. 2A). This pattern revealed that Spn engages in a different expression program during colonization vs. disease. This grouping of samples from infections also indicated that there exist core Spn genes whose expression reflects an infection state at all diseased host anatomical sites.
Fig. 2.
Pneumococcal PCA and dendrograms. (A) PCA of Spn gene-expression profiles (Dataset S2) across in vivo colonized or infected samples. (B) Bootstrapped dendrogram of Spn gene-expression profiles showing replicate clustering of in vivo colonized or infected samples. (C) PCA of A samples with the addition of in vitro grown planktonic and biofilm TIGR4 samples from our previous study (8). (D) Bootstrapped dendrogram with the addition of in vitro samples, again showing replicate clustering.
We observed that PC2 separated the TIGR4-infected samples away from D39 and 6A-10 (Fig. 2A). This indicated that the strains differ in their gene-expression profiles, despite our focus on core genes. There was limited organ-based separation among the infected organs on the PCA because PC2 was more influenced by the TIGR4 variation. A bootstrapped dendrogram of all samples (Fig. 2B) showed this effect. Samples clustered by organ type among the D39 and 6A-10 strains, but TIGR4 samples clustered independently. This finding most likely reflects the fact that D39 and 6A-10 are slightly more genetically similar to each other (SI Appendix, Fig. S1).
To estimate the similarity between in vivo Spn expression profiles and representative in vitro datasets, we performed a PCA on the 1,682 core genes, including Spn planktonic and biofilm in vitro growth datasets (8) (Fig. 2C). Spn grows planktonically within the blood and TIGR4 has been demonstrated to form biofilms within the heart, yet these samples did not cluster closely with in vitro planktonic and biofilm samples, respectively, on the PCA. There was a clear separation of in vivo and in vitro states along PC1. These data suggest that in vitro Spn expression profiles are not representative of in vivo conditions. TIGR4-infected heart samples (8), which include biofilm pneumococci within cardiac microlesions, clustered more closely to the in vitro TIGR4 biofilm samples on a bootstrapped dendrogram (Fig. 2D). However, in vitro samples are all clustered in a clade separated from in vivo samples. On this dendrogram, like on the PCA plot, nasopharyngeal samples still clustered independently from the other states, indicating that nasopharyngeal colonization elicits a unique Spn gene-expression profile significantly different from their infection phenotypes, irrespective of host anatomical site. Many previous studies have assayed the pneumococcal transcriptome in vitro. The D39 transcriptional signature was previously investigated under a panel of in vitro conditions designed to mimic anatomical sites (19). A comparison of the gene expression in the D39 in vitro study to in vivo conditions based on core genes in our study displayed a stark distinction between these datasets (SI Appendix, Fig. S2). The differentially expressed (DE) genes acquired in vitro also showed no significant overlap to those derived from in vivo conditions (SI Appendix, Fig. S3).
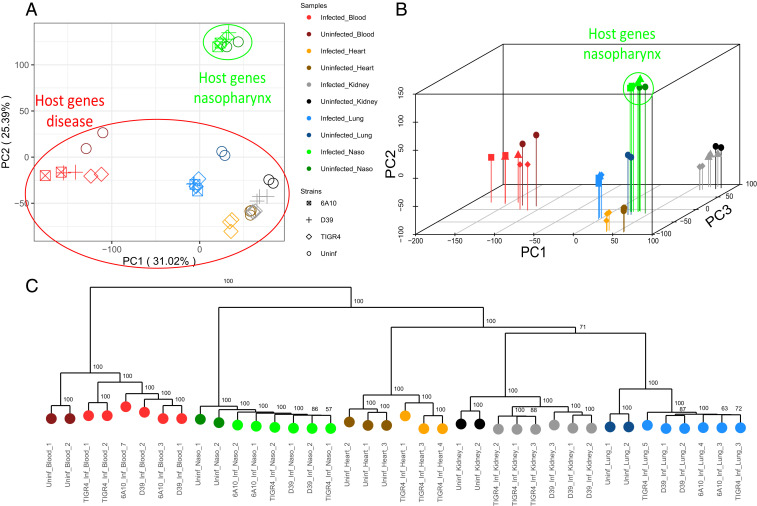
We then generated a PCA for expression profiles of all host genes (Fig. 3A). Host profiles revealed sample clustering based on anatomical site. This was not surprising given that gene programs reflecting specific organ functions would likely supersede genes involved in the response to infection. PC1 separated the blood away from the solid tissue types, and PC2 clearly separated out the nasopharynx as a colonization state away from infected anatomical sites. The lungs, hearts, and kidneys appeared to cluster more closely together. However, the addition of PC3 showed clear separation based on anatomical site (Fig. 3B). Apart from the nasopharynx, each site further formed subclusters of uninfected and infected strain-specific samples. The bootstrapped dendrogram of host samples corroborated the differences between organ types and strains, as well as the differences between infected and uninfected states for nearly all anatomical sites (Fig. 3C). Based on shorter distances between uncolonized and colonized states of the nasopharynx, there appeared to be fewer differences between host gene expression, relative to other anatomical sites, further demonstrating the host’s tolerance to pneumococcal colonization relative to disease states.
Fig. 3.
Host PCA and dendrogram. (A) PCA of host gene-expression profiles (Dataset S2) across samples colonized or infected with Spn, and uninfected. (B) A three-dimensional PCA shows complete separation between anatomical site clusters, with further separation of heart and kidney samples (PC3), as well as infected and uninfected invaded organs. (C) Bootstrapped dendrogram of host gene-expression profiles showing replicate clustering of host samples.
The Pneumococcus and Its Host Differentially Regulate Unique Gene Subsets in a Site-Specific Manner.
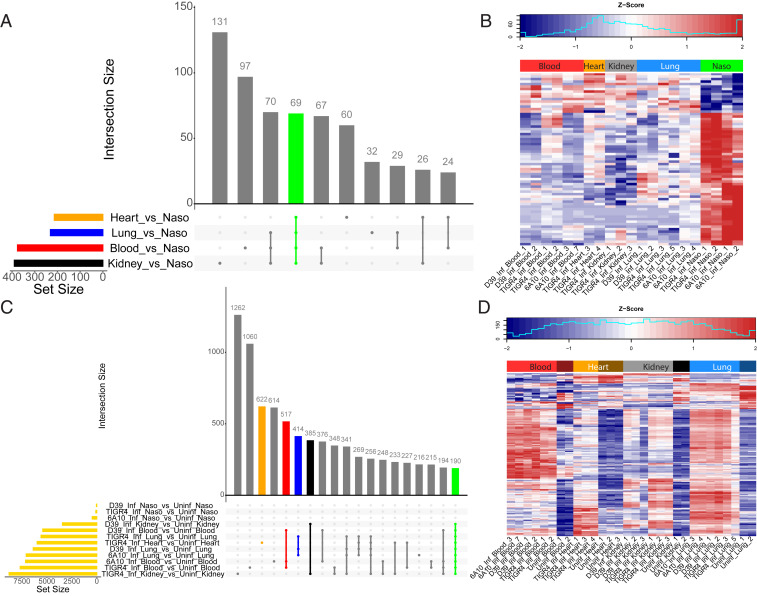
To determine the major Spn responses to each host site, and each host site’s response to Spn, we queried DE genes with their respective control. Statistically significant DE Spn genes, accounting for strain differences, were used to identify key differences between disease states and the colonization state used as baseline. Fig. 4A highlights 69 Spn DE genes shared among all disease states when compared to nasopharyngeal colonization. Of these, all but ribosomal protein L19 (SP_1293) were either up-regulated or down-regulated in the same direction (Fig. 4B). Other comparisons revealed 376 DE genes in the blood versus the nasopharynx, 214 DE genes in the heart, and 231 and 390 DE genes in the lungs and kidneys, respectively (SI Appendix, Fig. S4). Most genes not shared between the three Spn strains did not exhibit differential gene expression in disease-associated organs versus the nasopharynx. Some interesting exceptions include the increased expression of adhesin choline binding protein I in strain TIGR4 (SP_0069) and bacteriocin-containing operons in strain 6A-10 (HKM25_525-HKM25_529). These were up-regulated in the nasopharynx versus disease sites, supporting the requirement for attachment and intraspecies competition, respectively. The comprehensive list of Spn DE genes for each strain-infected organ versus the nasopharynx is available in Dataset S3.
Fig. 4.
DE genes detected in the pneumococcus and host. (A) Upset plot of Spn DE genes (Dataset S4). The individual or connected dots represent the various intersections of genes that were either unique to, or shared among comparisons, similar to a Venn diagram. Vertical bars represent the number of DE genes unique to specific intersections. Horizontal bars represent the number of DE genes for each specific comparison. (B) A z-scored heatmap of expression levels of 69 Spn DE genes shared across all comparisons (green vertical bar from A). (C) Upset plot of host DE genes (Dataset S5). Horizontal gold bars represent the total number of host DE genes for that comparison. Colored vertical bars represent intersections of interest (i.e., specific to the infection of heart, blood, lungs), and an overall infection response in organs susceptible to invasive disease (orange, red, blue, and green vertical bars respectively). (D) A z-scored heatmap of expression levels of 190 host DE genes shared across all comparisons except the nasopharynx (i.e., shared across disease anatomical sites) (green vertical bar from C).
To evaluate the host response to Spn infection, we performed similar host DE gene analyses using respective uninfected tissues as baseline. The use of multiple strains allowed for the identification of a shared host infection response to Spn at each anatomical site (Fig. 4C). The 190 mouse genes that were DE upon invasive infection across four sites followed the same up-regulated or down-regulated trend across all comparisons (Fig. 4D). Other mouse gene sets of interest include 517 DE genes in infected blood, 622 DE genes in infected hearts, 414 DE genes in infected lungs, and 385 DE genes in infected kidneys (SI Appendix, Fig. S4). Nasopharyngeal colonized samples harbored only few DE genes (SI Appendix, Fig. S5). Lists of all Spn and mouse DE genes are provided in Datasets S4 and S5, respectively. To confirm RNA-seq DE gene results, we performed a cross-platform validation of selected DE genes, 10 pneumococcal and 11 mouse genes, using qRT-PCR. A subset of RNA samples that were subjected to RNA-seq, as well as additional infected heart samples, were tested. Results revealed strong correlations with significant P values between the two platforms (SI Appendix, Figs. S6 and S7).
Key Pneumococcal Genes Expressed during Colonization Are Distinct from Those Expressed during Infection.
To identify all potentially relevant DE pathways for our datasets, we used complete DE gene lists (false-discovery rate [FDR] ≤ 0.05) for pathway enrichment analyses. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (20) of Spn DE genes identified several pathways unique to each comparison (Dataset S6), two of which were shared across all disease anatomical sites relative to the nasopharynx: The ascorbate and aldarate metabolism pathway and the pentose and glucuronate interconversions pathway. Unsurprisingly, most of the genes in these two pathways were present in the 69 DE genes subset shared across disease states. Three genes are shared between these two pathways: SP_2033, SP_2034, and SP_2035. These genes are part of the ula operon that metabolizes ascorbate and aldarate (SP_2031–SP_2038). An overview of the uptake and conversion of l-ascorbate (l-ascorbic acid), also known as vitamin C, by Spn is shown in Fig. 5A. l-Ascorbate is transported into the bacteria using a phosphotransferase system (SP_2036, SP_2037, and SP_2038), where it is ultimately converted into cytidine diphosphate-ribitol by other genes within the operon together with genes SP_1983, SP_1270, and SP_1271. Of all of the genes within this pathway, only SP_1983 was not DE above cutoff. Cytidine diphosphate-ribitol is essential for bacteria wall synthesis (21) and, therefore, we hypothesize that interruption of the uptake of l-ascorbate could affect nasopharyngeal colonization.
Fig. 5.
Pneumococcal and host genes of particular interest. (A) Schematic view of two potentially interlinked pneumococcal KEGG pathways (DatasetS6): Ascorbate and aldarate metabolism, and pentose and glucuronate interconversions. *SP_1983 was the only gene not DE relative to the nasopharynx. (B) Upset plot (see description in Fig. 4A) of the 100 most highly expressed Spn genes (Dataset S6) at each anatomical site (on average). (C) Absolute gene-expression level heatmap of the 52 Spn genes (Dataset S6) that are highly expressed genes across all sites (black vertical bar from B). (D) IPA activation z-score heatmaps (average z-score <−3 or >3) of canonical pathways in host DE genes (Dataset S7) present in all diseased anatomical sites. Nasopharyngeal pathways are not shown as too few host DE genes were observed (Fig. 4C).
We also interrogated genes highly expressed by Spn at all anatomical sites, as well as those relevant to disease or colonization. Of the 100 most highly expressed Spn genes at each site (Dataset S6), 52 were shared across all sites (Fig. 5 B and C). While many of these shared genes were housekeeping genes (cell division, translation, and so forth), two were previously studied potential vaccine candidates—lactoferricin-resistance associated PspA and zinc metalloprotease ZmpB (22, 23)—and another was an oligopeptide transporter operon (amiACDEF) shown to have relevance to nasopharyngeal colonization (24). The two vaccine candidates and genes amiA, amiC, and amiD were also shown to be essential for TIGR4 lung survival in vivo in the signature tagged mutagenesis study (10). The identification of these housekeeping genes, virulence factors, and operon among the highest expressed genes suggests that the pneumococcus invests significant resources to produce these proteins at all anatomical sites.
Host Gene Signatures Reveal Induction of the IFN Pathway during Invasive Disease.
For host datasets, we used Ingenuity Pathway Analysis (IPA) to study all DE gene lists (FDR ≤ 0.05, log2 fold-change [LFC] <−2 or >2). Key differences between host responses during colonization compared to invasive disease were observed. Only ∼200 DE genes were observed overall during nasopharyngeal colonization compared to ∼6,000 DE genes on average in invaded tissues (Fig. 4C). A minimal response to strain D39 was observed within the nasopharynx, but a robust response from a modest number of genes was mounted with strains TIGR4 and 6A-10. Both strains induced a similar pattern of induction of proinflammatory cytokines considered part of the acute phase response, including C-reactive protein, tumor necrosis factor, and IL-1β. Their expression signature also included genes indicative of neutrophilic infiltration, and possibly other leukocytes, such as expression of chemoattractant receptors found on myeloid cells, including FPR1, FPR2, LTB4R, and CXCR2, formyl peptide receptors, and LTB4 receptor. This finding is consistent with previous reports showing that nasopharyngeal colonization can lead to recruitment of neutrophils (25, 26). These results indicate that nasopharyngeal colonization can induce subclinical inflammation in the nasopharynx, and that bacterial strains differ in the extent to which they alter host biology during colonization.
As in nasopharyngeal colonization, infection with Spn led to the expected induction of host NF-κB–dependent proinflammatory cytokines typical of the acute-phase response; however, both the magnitude of induction and number of transcripts associated with this response was much greater in infected tissues. In addition, during invasive infection, all tissues—lungs, heart, and kidneys—showed enrichment for genes and pathways associated with leukocyte-specific expression, likely indicative of an inflammatory infiltrate. To obtain an overview of the differences in host response during infection compared to nasopharyngeal colonization common to all infected tissues, we focused on the subset of 190 genes induced during infection in blood and all tissues but not during nasopharyngeal colonization (green bar in Fig. 4C). This list showed significant enrichment of a number of IFN-stimulated genes, genes usually induced following innate immune sensing of viral infection or other abnormal nucleic acids (27), together with typical transcription factors and cytokines associated with the IFN response identified as upstream regulators (Dataset S5). Examination of pathways enriched in the 190 gene set or in the organ-specific DE genes (Dataset S7) showed enrichment of pathways associated with the IFN response: IFN signaling, role of pattern recognition receptors in recognition of bacteria and viruses, and activation of IFN regulatory factors by cytosolic pattern recognition receptors (Fig. 5D). Some organ-specific patterns in expression changes during Spn infection were also observed: Spn infection led to enrichment of coagulation-associated genes in the blood, and pathways associated with cytochrome p450-mediated biosynthesis and degradation were more altered in the lung than in other tissues (Dataset S7).
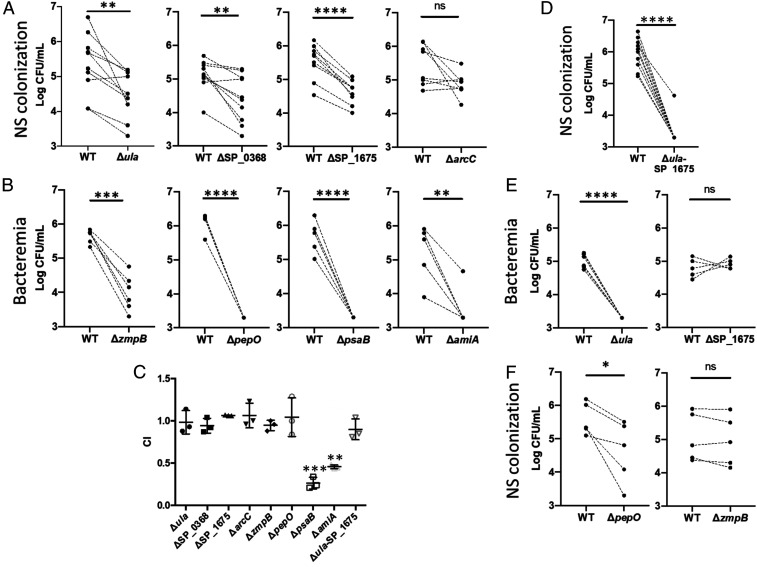
Deletion of Highly Expressed Pneumococcal Genes Lowers Burden at Corresponding Anatomical Site.
To confirm the importance of Spn genes identified as highly expressed (Fig. 5B and Dataset S6) or DE (Fig. 4A and Dataset S4) in an anatomical site-specific manner (Dataset S6), we tested the virulence of their respective isogenic mutants versus WT control using a competitive index assay. Three of four mutants in genes highly expressed only during asymptomatic colonization (ula operon, SP_0368, and SP_1675) were attenuated versus control on day 5 of colonization, each exhibiting an approximate 10-fold reduction in recoverable bacteria from nasal lavage samples. In contrast, mutation of SP_2150 (arcC) had no significant effect, although a general downward trend was observed (Fig. 6A). The ula operon is responsible for the import and degradation of ascorbic acid (28), SP_0368 encodes an O-glycosidase that may contribute to the scavenging of carbohydrates from host glycoconjugates (29), and SP_1675 encodes a kinase (ROK) family member (30). The latter are commonly involved in the regulation of sugar utilization networks (31). arcC is a member of the arginine deaminase operon that, among other functions, allows the pneumococcus to use arginine as a carbon source (32). Mutants deficient in genes identified as highly expressed during pneumonia and invasive disease (SP_0664, SP_1647, SP_1648, and SP_1891) were in turn found to be starkly impaired in their ability to cause bacteremia following pneumonia (Fig. 6B). These genes encode ZmpB, endopeptidase PepO, manganese transporter PsaB, and AmiA. Our results with these mutants agree with studies that have previously linked these proteins to Spn pathogenicity (24, 33–35). Importantly, experiments performed with this panel of mutants in vitro found that only PsaB and AmiA were essential for growth in media, thus the requirement for the remainder of proteins was specific to the in vivo condition (Fig. 6C).
Fig. 6.
In vivo competition experiments between pneumococcal strain TIGR4 and isogenic mutants in highly expressed genes. Mutants were tested using a completive index assay against WT TIGR4. Paired WT and mutant samples are denoted by dots with a connecting dashed line. (A) Bacterial titers in nasal lavage fluid of colonized mice 5 d postinoculation with WT TIGR4 and Spn deficient in genes highly expressed in the nasopharynx. (B) Bacterial titers in the blood of mice 1 d after intratracheal with WT TIGR4 and Spn deficient in genes highly expressed during invasive disease. (C) Mutants tested in A and B were tested for overall fitness in Todd-Hewitt broth with 0.5% yeast extract (THY). Competitive index values were calculated by dividing the numbers of CFU for the isogenic mutants by those for WT. (D) A double mutant, deficient in both ula and SP_1675 was tested versus TIGR4 WT in the nasopharynx model of colonization. (E) Cross infection experiments in which genes highly expressed in nasopharynx (Δula and ΔSP_1675) were tested in the model of lower respiratory tract infection with bacteremia. (F) Genes highly expressed in disease anatomical sites (ΔpepO and ΔzmpB) were tested with the nasopharynx model of colonization. For all panels, mutants were compared to WT using a paired Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant).
Considering that the genes identified as highly expressed during nasopharyngeal colonization are presumably involved in the scavenging of nutrients, we hypothesized that their combined deletion would have a cumulative effect on Spn growth in the nasopharynx. We observed that Spn can rely on the ula operon for growth with ascorbate as a sole carbon source (SI Appendix, Fig. S8), that a double mutant lacking the ula operon and SP_1675 was starkly attenuated during colonization in mice (Fig. 6D), and that the double mutant had normal growth in rich medium in vitro (Fig. 6C). Importantly, cross-infection (i.e., testing of the mutants necessary for colonization in the invasive disease model, and vice versa) revealed that the ula mutant was attenuated for invasive disease, whereas the SP_1675 mutant was not (Fig. 6E). Similarly, the pepO mutant was attenuated for colonization, but the zmpB mutant was not (Fig. 6F). Thus, for most genes tested, a high level of transcription in the nasopharynx or disease site was indicative of their importance under same condition. These findings help prioritize new candidate highly expressed Spn genes as targets for intervention.
IFN Contributes to Host Defense in Pneumococcal Infection.
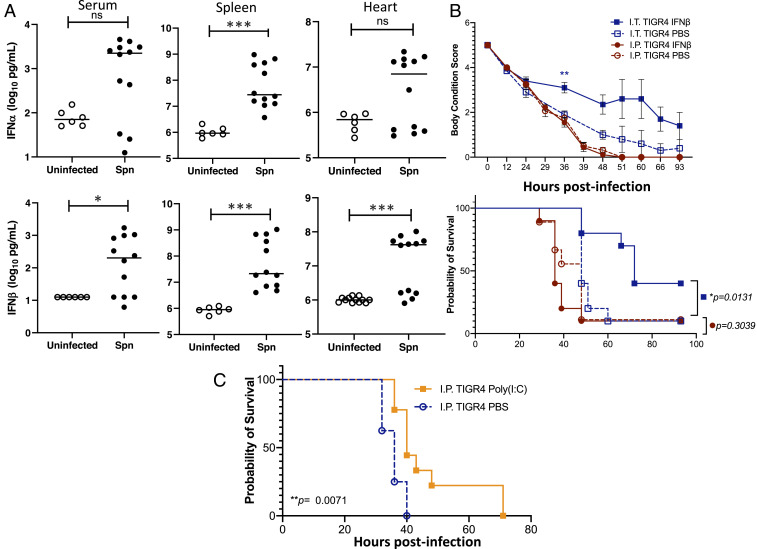
In general, type I IFN is not considered to be a protective mechanism during pneumococcal infection; albeit, a role in alveolar-capillary barrier defense has been described (36, 37). Given the observed enrichment in IFN-stimulated gene expression in infected vs. uninfected organs (Dataset S5), we sought to confirm the production of IFN within tissues during invasive pneumococcal disease and determine its biological consequence. Notably, both IFN-α and IFN-β were detected in the spleen of infected mice at levels higher than uninfected controls (Fig. 7A), with IFN-β also elevated in serum and the heart of infected mice. One-time intranasal pretreatment of mice with IFN-β (isolated from mouse cells, PBL Assay Science) 1 d prior to intratracheal challenge with Spn TIGR4 reduced morbidity and mortality versus the mock-treated controls (Fig. 7B). Next, we probed the role of IFN in protection against intraperitoneal infection. IFN-β pretreatment alone did not protect mice that were pretreated and challenged intraperitoneally with TIGR4 (Fig. 7B). However, pretreatment with Poly(I:C), a potent inducer of both IFN and NF-kB–dependent proinflammatory cytokines, did enhance survival of intraperitoneally infected mice (Fig. 7C) (37). From these results we infer that the IFN response alone primarily confers protection of the airway. However, in combination with other activated host defense pathways, it may also confer protection during systemic infection.
Fig. 7.
Assessment of the IFN response and its contribution to mouse protection. (A) IFN-α and IFN-β levels in the serum, spleen, and hearts of mice experiencing invasive pneumococcal disease. Each data point represents an individual mouse with statistical analysis performed using a two-tailed Student’s t test (*P < 0.05; ***P < 0.001). (B) Mice were administered recombinant IFN-β (30,000 U) in saline or saline alone either intranasally (I.N.; 50 µL volume) or intraperitoneally (I.P.; 100 µL volume) 24 h prior to challenge intratracheally (I.T.) with 5 × 106 CFU or intraperitoneally with 1 × 105 CFU of TIGR4 in 100 µL of saline, respectively. Body condition score and Kaplan–Meier survival of mice treated with IFN-β versus control following challenge with TIGR4. Body condition score significance was tested using a repeated-measures mixed-effects model with Sidak’s multiple comparison posttest of IFN-β versus saline only within each infection group (**P < 0.01); and survival significance was assessed by log-rank Mantel–Cox test (P values indicated on the graph; n = 10 mice per treatment group). (C) Mice were administered 50 µg Poly(I:C) in 100 µL saline intraperitoneally 24 h prior to challenge, intraperitoneally with 1 × 105 CFU of TIGR4. Survival significance was assessed by log-rank Mantel–Cox test (P value indicated on the graph; n = 9 mice per treatment group).
Discussion
We report the shared in vivo pneumococcal and host transcriptome during colonization and invasive disease in relevant anatomical sites in mice. We also identify relevant highly and differentially expressed bacterial and host genes, while exploring the difference between asymptomatic pneumococcal colonization and disease. We also validate the importance of differentially regulated genes for in vivo survival of the bacteria and host, respectively.
We observed major differences between in vitro and in vivo gene-expression results, supporting the use of the in vivo colonization state as the preferred baseline condition for calculating Spn DE genes rather than an in vitro control. Indeed, Spn transcriptional gene signatures investigated under in vitro conditions (8, 19) did not recapitulate our in vivo conditions (SI Appendix, Figs. S2 and S3). It is reasonable to assume that the differences between in vitro and in vivo conditions are in large part due to differences in nutrient availability, the complexity of processes required for sequestering nutrients in vivo, oxygen tension, shear forces, and other physical or physiological aspects of in vivo conditions. It is also likely that the in vitro conditions used in prior studies, like those we used for biofilm formation (8), were optimized for Spn growth, and as such were not accurately replicating the physiological environment of the host. The discordance between in vitro and in vivo studies further demonstrates the potential drawbacks of using results solely from in vitro studies to infer in vivo mechanisms.
Different Spn strains not only have varied transcriptomic signatures within the same organs in vivo (Fig. 2A) but can also result in disparities of Spn activity at distinct sites (Fig. 1C). This is recapitulated in the host responses to each strain as well (Fig. 3). The pneumococcus and the host differentially expressed different numbers and types of genes at each anatomical site, evidenced by our identification of DE gene sets not only commonly expressed across sites but also other sets unique to each, with only few host genes DE in the nasopharynx (Fig. 4 A and C). In both of these cases, the Spn strain effect can be visualized (SI Appendix, Fig. S4). Genes that are DE follow similar trends (i.e., up- or down-regulated), but to varied strain-dependent magnitudes.
We observed that in Spn genes DE only within the blood, multiple metal ion-scavenging genes were up-regulated. Ribosomal proteins were down-regulated in the heart, possibly suggesting a slowdown of translation as the pneumococcus transitions into a biofilm state, as previously described (8, 38). It is tempting to hypothesize that Spn may have evolved mechanisms enabling its transition to a low inflammation-triggering biofilm mode in certain organs. Overall, the pneumococcus is also able to alter its feeding habits, a forte for Spn, respective to each anatomical site. We have shown the up-regulation of l-ascorbate uptake and degradation in the nasopharynx; however, genes involved in galactose and lactose utilization were also observed. It is also interesting to note that mannose transporter genes were up-regulated at all other sites, potentially implicating this feeding mechanism in fitness in disease states.
The identification of targetable highly expressed genes can be particularly useful, especially for the development of vaccines. We detected two previously described pneumococcal antigenic virulence factors, pspA and zmpB, in our highly expressed gene set. Studies have shown that zmpB is conserved across the pneumococcal proteome and is protective in mice (22). However, zmpB has also been shown to be present in closely related commensal streptococcal species colonizing the human host, thereby possibly limiting its vaccine potential (39). PspA is also present in most pneumococcal strains, albeit exhibiting different alleles (40), and this protein has also been demonstrated to be protective against pneumococcal disease (41). Given that these are core proteins, surface-exposed, protective, and now found to be broadly highly expressed by the pneumococcus in vivo, this makes them potentially powerful candidates for the development of a serotype-independent pneumococcal vaccine.
Zafar et al. (42) identified 375 Spn genes necessary for shedding and transmission using a Tn-seq approach. Of these, 72 genes were up-regulated in the nasopharynx, suggesting that these genes could be key factors involved in Spn transmission. Like us, Zafar et al. identified that SP_0368 (O-glycosidase) was up-regulated within the nasopharynx, and we demonstrated its importance for in vivo colonization using knockout mutants. The dlt locus (SP_2173-SP_2176) was also identified by Zafar et al. as one of the major determinants of transmission. Similar Tn-seq studies performed in the context of pneumococcal growth in human saliva have identified and validated that the amiACDEF operon that we found highly expressed at all anatomical sites, was essential for survival (16). This was also essential in vivo in mice within the lungs and nasopharynx (15), correlating well with its high expression at all anatomical sites in our study. Other key genes we identified also correlate with survival genes identified by these Tn-seq studies. For example, three of the eight genes of the vitamin C operon and SP_2150 that were up-regulated in the nasopharynx relative to all other anatomical sites, were specifically essential in the nasopharynx but not in the lung. Similarly, SP_0062, which is part of a phosphotransferase system, was up-regulated in the lung in our dataset and shown to be essential in the lung but not the nasopharynx (15).
Mutants lacking the dltB gene (SP_2175) were shown by Cooper et al. (43) to be selected for during in vivo experimental evolution during colonization, probably because the authors removed the transmission bottleneck by performing multiple experimental passages. Cooper et al. hypothesized that the dlt locus was maintained across the pneumococcal population because it is required for transmission to new hosts despite increased adhesion and colonization when the locus was deleted. Along these lines, our results revealed that the dlt locus was not significantly DE across organs relative to the nasopharynx and all genes within the locus were robustly expressed at all sites. These studies suggest a complex regulatory program of the dlt locus during host–pathogen interactions.
Warrier et al. (44) used data from a variety of RNA-seq techniques focused on Spn strain TIGR4 to characterize the complex operon architecture of the pneumococcus. They identified 474 potential multigene operons with experimental support. They also identified a regulatory element pyrR (SP_1278), essential for virulence and colonization. We found pyrR to be up-regulated within the blood, kidneys, and heart relative to the nasopharynx, validating its role in virulence and infection. Our two operons (ula SP_2031-SP_2038 and SP_1266-SP_1271) up-regulated in the nasopharynx (Fig. 5A) were also detected in the Warrier et al. dataset. A microarray study involving strains of serotype 4 (WCH43) and serotype 6A (WCH16) also identified the ula operon as up-regulated in the nasopharynx relative to the blood, from a total of 62 DE genes. They also showed that a ula knockout mutant had no significant effect on pneumococcal virulence (45). While we also showed that an ula knockout mutant is only mildly attenuated, our double-knockout mutant adding the SP_1675 gene was significantly attenuated in colonization. The other operon consists of lic proteins and phosphorylcholine uptake genes (46) (SP_1266-SP_1271) known to be involved in teichoic acid synthesis (21). The link found between the two operons suggesting activated teichoic acid and bacterial cell wall synthesis, and the resources invested by the pneumococcus for nasopharyngeal colonization, demonstrate the importance of these genes in the initial stage of interactions with the mammalian host.
It has previously been shown that type I IFN decreases expression of platelet-activating factor receptor (PAFr), a pneumococcus binding receptor, and this inhibits pneumococcal transmigration across epithelial and endothelial cell barriers in the airway, preventing invasive disease (36). Our in vivo results revealed a strikingly consistent IFN-stimulated gene-expression signature during acute invasive infection. Enhanced survival of intratracheally IFN-β–pretreated mice during and following the onset of disease provides further evidence for this protective effect. Several previous studies have noted induction of IFN-stimulated genes during pneumococcal infection and have proposed two main models as a cause. First, the STING/cGAS cytosolic DNA-sensing system has been shown to recognize Spn nucleic acids (47). Signaling from well-known bacterial recognition receptors, such as Toll-like receptors (TLRs), also lead to activation of both NF-κB and IFN regulatory factors that mediate the IFN response. Famà et al. (48) demonstrated the effect of nucleic acid-sensing TLRs upon pneumococcal infection in mice. TLR7, TLR9, TLR13, and chaperone protein UNC93B1, responsible for TLR trafficking out of the endoplasmic reticulum, were shown to control pneumococcal infection as knockout mice lacking these genes were moderately to highly susceptible. Examining the levels of these genes within our dataset shows their up-regulation at different sites, mainly the blood and lungs (SI Appendix, Fig. S9), albeit not all passed the DE FDR cutoff of ≤0.05 (Dataset S5).
Second, however, leakage of mitochondrial DNA from damaged mitochondria in stressed or dying cells is also a potential ligand for STING/cGAS (49–51). Since Spn invasion causes cell stress and death, DNA from damaged mitochondria or dying cells would be another potential trigger of the IFN response during invasion. While Skovbjerg et al. (52) observed the up-regulation of IFN-related genes in vitro in human monocytes exposed to intact and autolyzed Spn, they also demonstrated that it was the intact bacteria that were responsible for IFN stimulation, suggesting that exogenous pneumococcal DNA/RNA from autolyzed bacteria may not be the only stimulant to the IFN pathway. A subset of the host IFN-related genes from a different study [identified by pattern B in figure 1 of Skovbjerg et al. (52)] were also up-regulated across all infected tissues in our study. Notably, we observed that pretreatment with IFN-β alone was not protective in mice challenged intraperitoneally (Fig. 7C). This may be due to our use of a single treatment regimen, the fact that the bacteria in the peritoneum do not require invasion to gain access to the bloodstream (and intraperitoneal challenge is therefore considered to be a highly lethal infection model), or that the main target of IFN-β is instead the lungs. Yet we observed that pretreatment with Poly(I:C) conferred protection in this infection model, suggesting that a mixed IFN and NF-κB response has additional effects, most likely occurring at the myeloid cell level, that confers protection during systemic disease. In general, our findings corroborate the prevalence of the IFN responses to Spn infection in mice and its importance in protecting against onset of lethal pneumococcal pneumonia.
Previous studies have shown interactions between the pneumococcus and host cell receptors, including PAFr, polymeric-immunoglobin receptor (pIgR), and laminin receptor (Rpsa), that facilitate pneumococcal invasion (53–55). We observed that PAFr and pIgR were up-regulated in the lungs, blood, and heart as they encounter the pneumococcus. This could suggest that the pneumococcus initiates a feedback invasion mechanism, whereby host receptors hijacked for invasion are being up-regulated, allowing for more invasion. However, for Rpsa, changes were only observed for strain TIGR4, with down-regulation in TIGR4-infected blood, and up-regulation in TIGR4-infected kidneys. Another receptor, found in B cells (CD22), was shown to be important for host resistance to the pneumococcus (56). It was consistently down-regulated in all our infected blood samples but not at other sites, leaving the host susceptible to further infection, and possibly explaining the high titers of Spn we observed in the blood.
In summary, using multiple pneumococcal strains in our in vivo transcriptome dataset, as well as multiple anatomical sites within the host, allowed us to identify shared and strain/organ-specific responses to infection in the pneumococcus and in mice. The focus on core pneumococcal genes shared across the three genetically different strains tested makes it likely that other pneumococcal strains will exhibit similar responses to those identified here when they encounter the host. Exploration of our key shared and organ-specific host genes and pathways, and their human orthologs will provide a robust rationale for designing follow-up studies in other mammalian models and in humans. We demonstrated that the mechanisms of sensing, adaptation, and response of Spn to its host and the host to different Spn isolates are complex, and strikingly different between colonization and disease states.
While we identified pneumococcal and mouse genes playing key roles in host–pathogen interactions, not all of these may be directly relevant in humans, and other important genes have likely been missed. A number of Spn proteins or components have been shown to be strictly restricted to interactions with human cell moieties (57–62) and will not have been interrogated in our mouse models. Similarly, not all mouse responses are directly applicable to human responses and several human-specific responses were likely not observed in this study. Validation of the bacterial and host genes identified here in the human setting, or in models more closely recapitulating human systems will be required. Despite these obvious caveats, we believe that the atlas of transcriptional responses during host–pathogen interactions presented here will constitute an essential resource for the pneumococcal and microbial pathogenesis research communities, and serve as a foundation for identification and validation of key host and pneumococcal therapeutic targets in future studies.
Materials and Methods
All transcriptomics data generated as a part of this study have been deposited at the Gene Expression Omnibus repository under accession no. GSE150788. Additional information about materials and data availability, including genome accession numbers, bacterial strains and constructs, mice, reagents, and software packages, are available in Dataset S8. Details on bacterial growth, mouse housing, and RNA-seq procedures are provided in SI Appendix, SI Appendix 1.
Mice.
Animal experiments were carried out using male and female mice 6 to 12 wk of age. WT C57BL/6 were supplied from The Jackson Laboratory and housed at the University of Alabama at Birmingham Animal Facilities for at least 1 wk prior to their use. All animal experiments were performed using protocols approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee (protocols #21152 and #21231).
Collection of Samples for RNA Isolation.
Mice were anesthetized using vaporized isoflurane at 2.5 to 3% during experimental challenge. For asymptomatic nasopharyngeal colonization, mice were inoculated with 105 CFU of Spn in 10 µL saline with samples collected on day 7 (25). For pneumonia, mice were challenged with 105 CFU of Spn in 100 µL saline by forced aspiration with lungs collected on day 2 (63). For invasive disease, mice were challenged intraperitoneally with 104 CFU of Spn in 100 µL saline with blood, heart, and kidneys collected between 30 and 36 h postchallenge. Data generated from TIGR4-infected blood and heart samples were from a previously published (8) study by our group, with mice infected in the same fashion. Prior to tissue collection, mice were killed by inhalation of vaporized isoflurane (>5% over oxygen) and confirmed by cervical dislocation, bilateral pneumothorax, or cardiac puncture according to approved protocols. For nasopharynx samples, killed mice were decapitated, exterior skin removed, brain and cerebral column excised, and the skull defleshed. The skull and oropharynx were subsequently crushed in RNA Protect Bacteria Reagent (Qiagen) and stored at −80 °C. For blood, samples were obtained by exsanguination via retro-orbital bleed, mixed with RNAprotect Bacteria Reagent, and stored at −80 °C. For lungs, heart, and kidneys, organs were excised, minced down to 1-mm3 fragments with a sterile razorblade, then, and repeated as necessary, rinsed with ice-cold PBS, and gently pressed to the point that blood no longer effused from the tissue. Samples were then placed into RNA Protect Bacteria Reagent, and stored at −80 °C.
RNA Isolation, Library Construction, Sequencing, and Transcriptomics Data Analyses.
These procedures were performed as previously described (8), with minor modifications as detailed in SI Appendix, SI Appendix 1. The numbers of reads generated for each sample are provided in Dataset S1. Common and unique DE genes for both species were determined using gene list intersections and individual heatmaps of DE genes were generated based on z-scores of variance stabilized transformation counts (64) (Figs. 4 and 5 and Dataset S2). Pneumococcal and mouse DE genes are provided in Datasets S3–S5. See Dataset S8 for package source/documentation and Data Availability for additional parameters used in DE gene estimation or heatmap generation.
KEGG Pathway Analysis.
Bacterial DE gene lists were used to determine DE KEGG (20) pathways (with an adjusted P < 0.05) (Dataset S6) using R package KEGGprofile in Rstudio (see Data Availability for additional parameters and Dataset S8 for package source/documentation).
Ingenuity Pathway Analysis.
Mouse DE gene lists were filtered using an LFC cutoff (LFC >2 and <−2) and uploaded into IPA (65) (Qiagen, https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/). Core analyses were conducted on each DE gene list per Spn strain using default parameters. Canonical pathways were determined using the mouse database with default parameters, on mouse organ systems. Analyses were then compared across organ types to estimate core pathways, and across Spn strains for organ-specific responses (Fig. 5). Canonical pathway data were exported (Dataset S7) for use in Rstudio. Heatmaps of canonical pathways were generated for those meeting an average z-score cutoff of >3 and <−3 (nasopharynx samples did not meet this cutoff) in Rstudio (Data Availability).
qRT-PCR Cross-Platform Validation of RNA-Seq.
For qRT-PCR validation, we selected 10 pneumococcal genes and 11 mouse genes that were up- or down-regulated from the 69 Spn shared DE genes at all sites vs. nasopharynx, and 190 host DE genes identified from all diseased sites. Control genes for Spn and host were selected based on high expression at all anatomical sites and low variation in expression (<0.5 SD). Primers for Spn were designed using National Center for Biotechnology Information-Primer BLAST, and those for the mouse were obtained from PrimerBank (66–68). These primer pairs are listed in Dataset S8. We used the Qiagen QuantiTect SYBR Green RT-PCR Kit and generated correlation curves of qRT-PCR ∆∆Ct values (equivalent to LFC) to RNA-seq LFC. These revealed strong correlations with significant P values between the two platforms (SI Appendix, Figs. S6 and S7).
In Vivo Competition Experiments and Assessment of IFN-Mediated Protection.
Experiments were performed using mouse models of asymptomatic nasopharyngeal colonization, lower respiratory tract infection with bacteremia, and intraperitoneal challenge with disseminated disease. For competition assays, logarithmic growth phase Spn TIGR4 strain and its isogenic mutant were mixed at a 1:1 ratio. In all instances, isoflurane-anesthetized C57BL/6 mice were inoculated either intranasally (10 µL in 105 CFU), intratracheally (100 µL in 105 CFU), or intraperitoneally (100 µL in 104 CFU) with physical assessment using a body condition index (25), and nasal lavage and blood collected for enumeration of pneumococci at designated time points. Bacterial titers in the nasopharynx were determined day 5 postchallenge. For competition experiments involving pneumonia with bacteremia, bacterial titers in the blood were determined 2 d postchallenge. For both nasal lavage samples and blood, the number of bacteria present in collected fluids was determined by serial dilution of the sample, plating on blood agar plates, and extrapolation from colony counts the next day. For in vitro experiments testing fitness, the competitive index of each mutant strain was calculated using the following equation: (mutant output CFU/WT output CFU)/(mutant input CFU/WT input CFU). Centrifuged spleen homogenates from mice challenged intraperitoneally with 104 CFU of TIGR4, alternatively uninfected control mice, were used to determine the IFN-α and IFN-β responses to invasive pneumococcal disease. For experiments with IFN-β mice were administered 30,000 U of recombinant mouse IFN-β in 50 µL or 100 µL PBS by intranasal instillation or intraperitoneal injection, respectively, 1 d prior to challenge with TIGR4. For Poly(I:C) experiments, mice were administered 50 µg Poly(I:C) in 100 µL PBS by intraperitoneal injection 1 d prior to intraperitoneal challenge with TIGR4. In all experiments, control mice were administered PBS alone without challenge recombinant protein, drug, or bacteria.
Supplementary Material
Acknowledgments
We thank Mogens Kilian and Matthew Chung for critical reading of this manuscript; and acknowledge essential support provided by members of the Institute for Genome Sciences’ Genomics Resource Center, Genome Informatics Core, and High-Performance Computing Core. This work was supported with funds from Merck, Sharpe, and Dohme, under the Merck Investigator Studies Program Award IISP ID: 57329–Pneumovax entitled: “Dual RNA-seq for characterization of the Streptococcus pneumoniae and host transcriptomes during bacteremia/invasive disease”; and the NIH, National Institute of Allergy and Infectious Diseases under Award R01AI114800, entitled: “Cardiac microlesion formation during invasive pneumococcal disease.” E.F.F. was supported by an Individual Biomedical Research Award from the The Hartwell Foundation.
Footnotes
Competing interest statement: H.T. and C.J.O. received funding from Merck, Sharpe, and Dohme under the Merck Investigator Studies Program; however, no intellectual property is tied to this research.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010428117/-/DCSupplemental.
Data Availability.
R code/scripts used to generate figures and data presented is available on GitHub at https://github.com/admelloGithub/InVivoSpn (69). The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE150788) (70).
References
- 1.Blanchette K. A., et al. , Neuraminidase A-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect. Immun. 84, 2922–2932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engholm D. H., Kilian M., Goodsell D. S., Andersen E. S., Kjærgaard R. S., A visual review of the human pathogen Streptococcus pneumoniae. FEMS Microbiol. Rev. 41, 854–879 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Wahl B., et al. , Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: Global, regional, and national estimates for 2000-15. Lancet Glob. Health 6, e744–e757 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gounder P. P., et al. , Impact of the pneumococcal conjugate vaccine and antibiotic use on nasopharyngeal colonization by antibiotic nonsusceptible Streptococcus pneumoniae, Alaska, 2000–2010. Pediatr. Infect. Dis. J. 34, 1223–1229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldman C., Anderson R., Bacteraemic pneumococcal pneumonia: Current therapeutic options. Drugs 71, 131–153 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Kaur R., Pham M., Yu K. O. A., Pichichero M. E., Rising pneumococcal antibiotic resistance in the post 13-valent pneumococcal conjugate vaccine era in pediatric isolates from a primary care setting. Clin. Infect. Dis., 10.1093/cid/ciaa157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sempere J., de Miguel S., González-Camacho F., Yuste J., Domenech M., Clinical relevance and molecular pathogenesis of the emerging serotypes 22F and 33F of Streptococcus pneumoniae in Spain. Front. Microbiol. 11, 309 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shenoy A. T., et al. , Streptococcus pneumoniae in the heart subvert the host response through biofilm-mediated resident macrophage killing. PLoS Pathog. 13, e1006582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang S. T., et al. , Pneumococcal pneumonia infection is associated with end-stage renal disease in adult hospitalized patients. Kidney Int. 86, 1023–1030 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Hava D. L., Camilli A., Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406 (2002). [PMC free article] [PubMed] [Google Scholar]
- 11.Orihuela C. J., et al. , Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 72, 5582–5596 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minhas V., et al. , In vivo dual RNA-seq reveals that neutrophil recruitment underlies differential tissue tropism of Streptococcus pneumoniae. Commun. Biol. 3, 293 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie N. D., Evans T. J., Dual RNA-seq in Streptococcus pneumoniae infection reveals compartmentalized neutrophil responses in lung and pleural space. mSystems 4, ••• (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe H. M., et al. , Bacterial factors required for transmission of Streptococcus pneumoniae in mammalian hosts. Cell Host Microbe 25, 884–891.e86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Opijnen T., Camilli A., A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 22, 2541–2551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhagen L. M., et al. , Genome-wide identification of genes essential for the survival of Streptococcus pneumoniae in human saliva. PLoS One 9, e89541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shenoy A. T., et al. , Severity and properties of cardiac damage caused by Streptococcus pneumoniae are strain dependent. PLoS One 13, e0204032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouts D. E., Brinkac L., Beck E., Inman J., Sutton G., PanOCT: Automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res. 40, e172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aprianto R., Slager J., Holsappel S., Veening J. W., High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res. 46, 9990–10006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M., KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baur S., Marles-Wright J., Buckenmaier S., Lewis R. J., Vollmer W., Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae. J. Bacteriol. 191, 1200–1210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong Y., et al. , Immunization with a ZmpB-based protein vaccine could protect against pneumococcal diseases in mice. Infect. Immun. 79, 867–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukerji R., et al. , The diversity of the proline-rich domain of pneumococcal surface protein A (PspA): Potential relevance to a broad-spectrum vaccine. Vaccine 36, 6834–6843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruce K. E., Rued B. E., Tsui H. T., Winkler M. E., The Opp (AmiACDEF) oligopeptide transporter mediates resistance of serotype 2 Streptococcus pneumoniae D39 to killing by chemokine CXCL10 and other antimicrobial peptides. J. Bacteriol. 200, e00745–e00717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riegler A. N., Brissac T., Gonzalez-Juarbe N., Orihuela C. J., Necroptotic cell death promotes adaptive immunity against colonizing pneumococci. Front. Immunol. 10, 615 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemon J. K., Miller M. R., Weiser J. N., Sensing of interleukin-1 cytokines during Streptococcus pneumoniae colonization contributes to macrophage recruitment and bacterial clearance. Infect. Immun. 83, 3204–3212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider W. M., Chevillotte M. D., Rice C. M., Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afzal M., Shafeeq S., Kuipers O. P., Ascorbic acid-dependent gene expression in Streptococcus pneumoniae and the activator function of the transcriptional regulator UlaR2. Front. Microbiol. 6, 72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marion C., Burnaugh A. M., Woodiga S. A., King S. J., Sialic acid transport contributes to pneumococcal colonization. Infect. Immun. 79, 1262–1269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Titgemeyer F., Reizer J., Reizer A., Saier M. H. Jr, Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology (Reading) 140, 2349–2354 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Marion C., et al. , Identification of a pneumococcal glycosidase that modifies O-linked glycans. Infect. Immun. 77, 1389–1396 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz C., et al. , Regulation of the arginine deiminase system by ArgR2 interferes with arginine metabolism and fitness of Streptococcus pneumoniae. MBio 5, e01858–e01814 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eijkelkamp B. A., McDevitt C. A., Kitten T., Manganese uptake and streptococcal virulence. Biometals 28, 491–508 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blue C. E., et al. , ZmpB, a novel virulence factor of Streptococcus pneumoniae that induces tumor necrosis factor alpha production in the respiratory tract. Infect. Immun. 71, 4925–4935 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal V., et al. , Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J. Biol. Chem. 288, 6849–6863 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.LeMessurier K. S., Häcker H., Chi L., Tuomanen E., Redecke V., Type I interferon protects against pneumococcal invasive disease by inhibiting bacterial transmigration across the lung. PLoS Pathog. 9, e1003727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mian M. F., et al. , Length of dsRNA (poly I:C) drives distinct innate immune responses, depending on the cell type. J. Leukoc. Biol. 94, 1025–1036 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Morra C. N., Orihuela C. J., Anatomical site-specific immunomodulation by bacterial biofilms. Curr. Opin. Infect. Dis. 33, 238–243 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Kilian M., Tettelin H., Identification of virulence-associated properties by comparative genome analysis of Streptococcus pneumoniae, S. pseudopneumoniae, S. mitis, three S. oralis subspecies, and S. infantis. MBio 10, e01985–e01919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knupp-Pereira P. A., Marques N. T. C., Teixeira L. M., Póvoa H. C. C., Neves F. P. G., Prevalence of PspA families and pilus islets among Streptococcus pneumoniae colonizing children before and after universal use of pneumococcal conjugate vaccines in Brazil. Braz. J. Microbiol. 51, 419–425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akbari E., et al. , Protective responses of an engineered PspA recombinant antigen against Streptococcus pneumoniae. Biotechnol. Rep. (Amst.) 24, e00385 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zafar M. A., et al. , Identification of pneumococcal factors affecting pneumococcal shedding shows that the dlt locus promotes inflammation and transmission. MBio 10, e01032–e01019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper V. S., et al. , Experimental evolution in vivo to identify selective pressures during pneumococcal colonization. mSystems 5, e00352–e00320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warrier I., et al. , The transcriptional landscape of Streptococcus pneumoniae TIGR4 reveals a complex operon architecture and abundant riboregulation critical for growth and virulence. PLoS Pathog. 14, e1007461 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahdi L. K., Van der Hoek M. B., Ebrahimie E., Paton J. C., Ogunniyi A. D., Characterization of pneumococcal genes involved in bloodstream invasion in a mouse model. PLoS One 10, e0141816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadioglu A., Weiser J. N., Paton J. C., Andrew P. W., The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6, 288–301 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Ruiz-Moreno J. S., et al. , The cGAS/STING pathway detects Streptococcus pneumoniae but appears dispensable for antipneumococcal defense in mice and humans. Infect. Immun. 86, e00849–e00817 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Famà A., et al. , Nucleic acid-sensing Toll-like receptors play a dominant role in innate immune recognition of pneumococci. MBio 11, e00415–e00420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X., et al. , Type I IFN expression is stimulated by cytosolic MtDNA released from pneumolysin-damaged mitochondria via the STING signaling pathway in macrophages. FEBS J. 286, 4754–4768 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Gao Y., et al. , Mitochondrial DNA leakage caused by Streptococcus pneumoniae hydrogen peroxide promotes type I IFN expression in lung cells. Front. Microbiol. 10, 630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West A. P., et al. , Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skovbjerg S., et al. , Intact pneumococci trigger transcription of interferon-related genes in human monocytes, while fragmented, autolyzed bacteria subvert this response. Infect. Immun. 85, e00960–e00916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shukla S. D., et al. , An antagonist of the platelet-activating factor receptor inhibits adherence of both nontypeable Haemophilus influenzae and Streptococcus pneumoniae to cultured human bronchial epithelial cells exposed to cigarette smoke. Int. J. Chron. Obstruct. Pulmon. Dis. 11, 1647–1655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iovino F., Molema G., Bijlsma J. J., Streptococcus pneumoniae interacts with pIgR expressed by the brain microvascular endothelium but does not co-localize with PAF receptor. PLoS One 9, e97914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orihuela C. J., et al. , Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J. Clin. Invest. 119, 1638–1646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandes V. E., et al. , The B-cell inhibitory receptor CD22 is a major factor in host resistance to Streptococcus pneumoniae infection. PLoS Pathog. 16, e1008464 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu L., et al. , Species-specific interaction of Streptococcus pneumoniae with human complement factor H. J. Immunol. 181, 7138–7146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J. H., et al. , Monoacyl lipoteichoic acid from pneumococci stimulates human cells but not mouse cells. Infect. Immun. 73, 834–840 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammerschmidt S., Tillig M. P., Wolff S., Vaerman J. P., Chhatwal G. S., Species-specific binding of human secretory component to SpsA protein of Streptococcus pneumoniae via a hexapeptide motif. Mol. Microbiol. 36, 726–736 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Elm C., et al. , Ectodomains 3 and 4 of human polymeric Immunoglobulin receptor (hpIgR) mediate invasion of Streptococcus pneumoniae into the epithelium. J. Biol. Chem. 279, 6296–6304 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Batten M. R., Senior B. W., Kilian M., Woof J. M., Amino acid sequence requirements in the hinge of human immunoglobulin A1 (IgA1) for cleavage by streptococcal IgA1 proteases. Infect. Immun. 71, 1462–1469 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwal V., et al. , Enolase of Streptococcus pneumoniae binds human complement inhibitor C4b-binding protein and contributes to complement evasion. J. Immunol. 189, 3575–3584 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Gonzalez-Juarbe N., et al. , Bacterial pore-forming toxins promote the activation of caspases in parallel to necroptosis to enhance alarmin release and inflammation during pneumonia. Sci. Rep. 8, 5846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krämer A., Green J., Pollard J. Jr, Tugendreich S., Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spandidos A., et al. , A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics 9, 633 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spandidos A., Wang X., Wang H., Seed B., PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X., Seed B., A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 31, e154 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D'Mello A., Tettelin H., InVivoSpn: R code and scripts for custom in vivo RNA-seq analysis. GitHub. https://github.com/admelloGithub/InVivoSpn. Deposited 18 May 2020.
- 70.D'Mello A., et al. , An in vivo atlas of host-pathogen transcriptomes during Streptococcus pneumoniae colonization and disease. Gene Expression Omnibus (GEO) database. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150788. Deposited 18 May 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
R code/scripts used to generate figures and data presented is available on GitHub at https://github.com/admelloGithub/InVivoSpn (69). The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE150788) (70).