Significance
Pathological tau accumulation is implicated in a variety of neurodegenerative diseases, including frontotemporal dementia and Alzheimer’s disease. Based on a large-scale forward genetic screen in Drosophila, our study highlights a modifier of tau neurotoxicity, the enzyme biotinidase. Our results suggest that biotin status may represent a druggable metabolic pathway and a potential therapeutic approach to neuroprotection in human tauopathies.
Keywords: tau, mitochondria, biotin, Drosophila, screen
Abstract
Mitochondrial and metabolic dysfunction are often implicated in neurological disease, but effective mechanism-based therapies remain elusive. We performed a genome-scale forward genetic screen in a Drosophila model of tauopathy, a class of neurodegenerative disorders characterized by the accumulation of the protein tau, and identified manipulation of the B-vitamin biotin as a potential therapeutic approach in tauopathy. We show that tau transgenic flies have an innate biotin deficiency due to tau-mediated relaxation of chromatin and consequent aberrant expression of multiple biotin-related genes, disrupting both carboxylase and mitochondrial function. Biotin depletion alone causes mitochondrial pathology and neurodegeneration in both flies and human neurons, implicating mitochondrial dysfunction as a mechanism in biotin deficiency. Finally, carboxylase biotin levels are reduced in mammalian tauopathies, including brains of human Alzheimer’s disease patients. These results provide insight into pathogenic mechanisms of human biotin deficiency, the resulting effects on neuronal health, and a potential therapeutic pathway in the treatment of tau-mediated neurotoxicity.
Regulation of mitochondrial function and cellular metabolism is necessary for cell survival. Mitochondrial dysfunction and concomitant oxidative stress are features of the aging brain. Early stages of neurodegenerative diseases, including Alzheimer’s disease, have shown alterations in mitochondrial function prior to neurodegeneration (1). The reactive oxygen species created from dysfunctional mitochondria can subsequently damage a variety of cellular components including lipids, DNA, and proteins. This oxidative and metabolic stress has the potential to perpetuate a cycle of toxicity that can drive cell death (2–4).
The deposition of the microtubule-associated protein tau into insoluble aggregates is a hallmark pathology of the family of neurodegenerative diseases termed tauopathies, including Alzheimer’s disease and some frontotemporal dementias. Dominant mutations in tau also induce neurodegeneration in human frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17 or FTLD-tau with MAPT mutation), demonstrating that abnormalities in tau alone can cause neuronal loss (5–7). Thus, a greater understanding of the mechanisms underlying the neurotoxicity of tau could highlight critical therapeutic pathways in a variety of neurodegenerative diseases. To identify such mechanisms, we completed a forward genetic screen of over 7,000 genes in the aging adult Drosophila brain and recovered over 350 modifiers of neurotoxicity mediated by the FTDP-17–associated tau mutant, tauR406W. We subsequently focused on the Btnd gene, which encodes biotinidase, an enzyme critical in the homeostasis of the B-vitamin biotin. Biotinidase produces free biotin (vitamin B7) from food for use as a cofactor for carboxylases located primarily in the mitochondria. Importantly, biotin plays a key role in metabolism and has potential for therapeutic manipulation.
We combined Drosophila genetics with a variety of molecular and cellular approaches to explore the role of mitochondrial and metabolic pathways in tauopathy and neuronal survival. We identified dysfunction within the biotin pathway in tauopathy and showed that biotin supplementation both rescues mitochondrial deficits and improves neuronal health in vivo. Furthermore, we demonstrate parallel mechanisms in human Alzheimer’s disease brain. Together, these findings emphasize the importance of biotin handling in mitochondrial and metabolic processes in neurons, suggesting a key role for biotin in both the healthy and diseased brain.
Results
Genetic Screen Implicates Mitochondrial and Metabolic Targets.
We have previously described a Drosophila model of tauopathy based on expression of wild-type or disease-linked forms of human tau (MAPT) in fly neurons (8). Expression of human tau in the Drosophila brain recapitulates key features of human tauopathies, including abnormally phosphorylated tau, age-dependent neurodegeneration, and reduced life span (8–13). We focus here primarily on the FTDP-17–associated mutant form of tau, tauR406W, because expression of tauR406W in aging fly neurons provides a robust, early onset neurodegeneration amenable to large-scale screening. To identify pathways mediating tauopathy pathogenesis, we performed a genome-scale forward genetic screen in flies expressing tauR406W in a pan-neuronal pattern (elav-GAL4 driver). We assayed the effect of 7,204 transgenic RNA interference (RNAi) lines (SI Appendix, Tables S1 and S2) on tauR406W neurotoxicity as evidenced by histological hallmarks of neurodegeneration in the aging adult brain, including vacuole formation and the presence of apoptotic nuclear debris (8). Initial hits were confirmed using a transgenic caspase reporter (UAS-CD8-PARP-Venus) (14, 15). Caspase reporter flies contain a transgenic construct with a fusion of the extracellular and transmembrane domains of mouse CD8 to 40 amino acids from human PARP, including the caspase cleavage site of PARP, and the fluorescent protein Venus (16). Enhancers that exhibited neuropathological evidence of degeneration or increased caspase reporter activity in the absence of transgenic human tauR406W were considered nonspecific and were not pursued further. Candidates not previously identified as modifiers of tauR406W neurotoxicity in Drosophila by our group or other investigators were verified by a second RNAi line, if available. We identified 62 suppressors and 306 specific enhancers of tauR406W neurotoxicity (SI Appendix, Table S1).
Human orthologs of validated hits from the screen were identified using the Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool (DIOPT; http://www.flyrnai.org/diopt) (17). These human orthologs were then entered into the STRING interactome generator to identify protein–protein interaction networks (Fig. 1A) (18). Of the significantly enriched pathways, we noted multiple genetic modifiers of tauR406W-mediated neurotoxicity in metabolic and mitochondrial pathways, supporting previous studies from our laboratory and others defining mechanisms of tau-mediated neurodegeneration (2, 3). In particular, we noted that knockdown of the Btnd gene encoding biotinidase, the enzyme responsible for freeing the B-vitamin biotin, significantly enhanced neurotoxicity. Due to the therapeutic potential of an orally administered B-vitamin supplement, we further pursued the influence of the biotin metabolic pathway in tauR406W-mediated neurodegeneration.
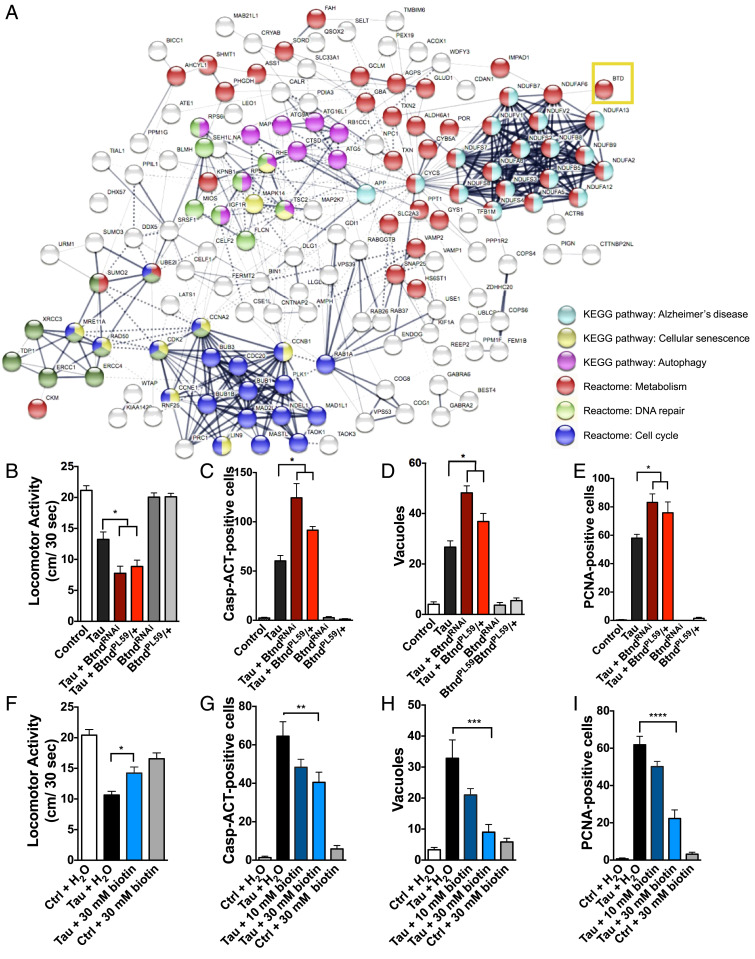
Fig. 1.
Pathway interactome of genetic hits from Drosophila RNAi screen of modifiers of tauR406W-mediated neurotoxicity and validation that biotin status modifies tau-mediated behavioral deficits and toxicity. (A) Predicted interactome from human orthologs of screen hits as shown by STRING analysis, including computationally predicted interactors from public datasets of direct physical and indirect functional interactions. Specific pathways enriched within the interactome are highlighted to the right. Validated screen hits were converted to human orthologs by DIOPT. Biotinidase gene is labeled as the human gene BTD (yellow box). (B) TauR406W-mediated locomotor deficits are enhanced by reduced biotinidase (BtndRNAi or BtndPL59/+) (ANOVA). (C–E) TauR406W-mediated neurotoxicity is enhanced by reduced biotinidase as shown by caspase activation, vacuole formation, and PCNA, an indicator of cell-cycle reentry (ANOVA). (F) TauR406W-mediated locomotor deficits are rescued by biotin feeding. (G–I) TauR406W-mediated neurotoxicity is rescued by biotin feeding as shown by caspase activation, vacuole formation, and PCNA. Biotin was mixed into Drosophila instant food and replaced every third day. All flies were 10 d old at time of experimentation. n = 6 per genotype (two-way ANOVA + Tukey’s post test). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Control genotype B, D, E, F, H, and I: elav-GAL4/+. Control genotype C, G: elav-GAL4/+; UAS-CD8-PARP-Venus/+.
Biotinidase Knockdown Enhances tauR406W-Induced Neurotoxicity.
Reducing biotinidase expression by either RNAi knockdown (BtndRNAi) or heterozygosity for BtndPL59 enhanced tauR406W-mediated locomotor deficits in 10-d-old flies as measured by a previously described Drosophila locomotor assay (Fig. 1B) (19, 20). Reduced biotinidase expression also significantly enhanced tau-mediated neurodegeneration as shown by increased caspase activation, vacuole formation, and cell-cycle reentry, a downstream mechanism of tau-induced neurotoxicity in flies and mammals (Fig. 1 C–E) (9, 21, 22). Reduced biotinidase transcript levels in BtndRNAi and BtndPL59/+ mutant flies were verified by qPCR (SI Appendix, Fig. S1A).
Biotin Feeding Rescues tauR406W-Mediated Neurotoxicity.
Since reducing biotinidase increased neurotoxicity in tauR406W transgenic flies, we next examined if biotin replacement was capable of rescuing toxicity. Chronic biotin feeding significantly improved tauR406W-mediated locomotor deficits in 10-d-old flies (Fig. 1F). TauR406W-mediated neurodegeneration was also significantly reduced with biotin feeding as shown by caspase activation, vacuole formation, and cell-cycle reentry, suggesting that tauR406W-mediated toxicity is modified by the availability of biotin (Fig. 1 G–I). To explore the relevance of our finding to tauopathies beyond those caused by tau mutations, including Alzheimer’s disease which is characterized by deposition of wild-type tau, we assessed the ability of biotin to rescue toxicity in a Drosophila model of human wild-type tauopathy (8, 23). We found that biotin feeding rescued the toxicity of wild-type tau as well as mutant tau (SI Appendix, Fig. S1B). To investigate the specificity of biotin neuroprotection further, we administered biotin to flies modeling spinocerebellar ataxia type 3 (Machado–Joseph disease) through expression of mutant ataxin 3 (24). We observed no alteration in the neuronal toxicity of expanded (Q78) ataxin 3 with biotin administration (SI Appendix, Fig. S1C). Neither genetic manipulation (BtndPL59/+ or BtndRNAi) nor biotin treatment affected tau expression (SI Appendix, Fig. S1D).
Next, we generated Drosophila lines overexpressing biotinidase (BtndOE) using the panneuronal elav-GAL4 driver and verified overexpression of biotinidase by qPCR. Genetic biotinidase overexpression did not significantly rescue tauR406W-mediated neurodegeneration (SI Appendix, Fig. S1 E and F), suggesting that human tauR406W transgenic flies do not have a deficit in biotinidase expression. Because biotinidase overexpression was unable to rescue toxicity yet biotin feeding successfully improved degeneration, we next examined whether human tauR406W transgenic flies had reduced biotin availability.
TauR406W Transgenic Flies Have Reduced Biotin and Decreased Carboxylase Biotinylation and Function.
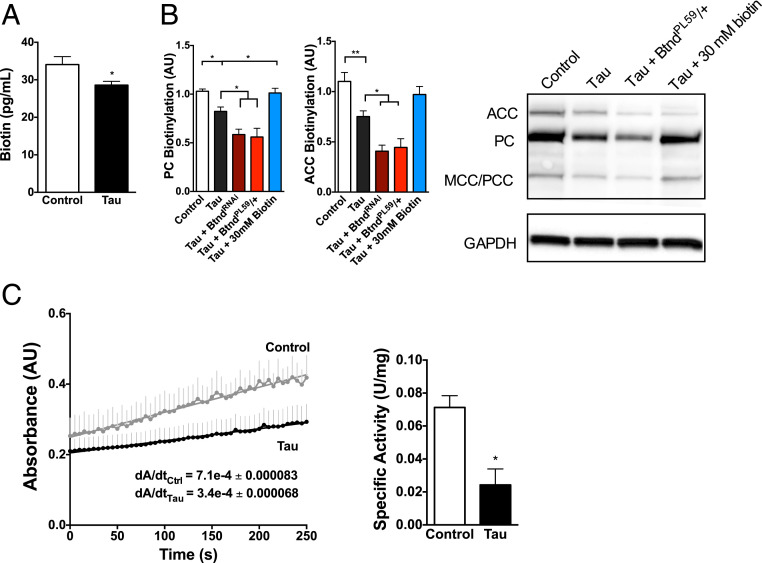
Human tauR406W transgenic flies have a significant reduction in total biotin as shown by biotin enzyme-linked immunosorbent assay (ELISA) on head homogenate (Fig. 2A). While the biological functions of biotin continue to be defined, biotin primarily acts as a cofactor for mitochondrial carboxylases, which are essential catalysts in multiple metabolic reactions. These carboxylases include acetyl-CoA carboxylases 1 and 2 (fatty acid synthesis and oxidation, respectively), pyruvate carboxylase (gluconeogenesis), propionyl-CoA carboxylase (β-oxidation of fatty acids), and methylcrotonyl carboxylase (leucine catabolism) (25). These functions (glucose, amino acid, and fatty acid metabolism) are all critical metabolic processes. Based on the established link between the carboxylases and biotin, we examined the biotinylation of carboxylases in human tauR406W transgenic flies using streptavidin–horseradish peroxidase (HRP) blotting of pooled fly-head homogenate. TauR406W transgenic flies showed a significant decrease in carboxylase biotinylation compared to control flies at 10 d of age (Fig. 2B). TauR406W transgenic flies with reduced biotinidase expression had an even lower level of carboxylase biotinylation, whereas biotin-fed tauR406W transgenic flies had a significantly greater level of carboxylase biotinylation than control-fed animals. These differences in carboxylase biotinylation were not due to changes in overall carboxylase expression levels as verified by qPCR. Next, we showed that human tauR406W transgenic flies also have a significant decrease in carboxylase function compared to control flies using a pyruvate carboxylase enzyme assay, consistent with loss of carboxylase activity due to reduced biotin and loss of carboxylase biotinylation (Fig. 2C).
Fig. 2.
TauR406W transgenic flies have reduced brain biotin and decreases in carboxylase biotinylation and function. (A) Human tauR406W transgenic flies have a significant decrease in biotin levels as shown by biotin ELISA on head homogenate. n = 6 per genotype (t test). (B) TauR406W transgenic flies have reduced carboxylase biotinylation as shown by streptavidin blotting. Reduced biotinidase expression (BtndRNAi or BtndPL59/+) further reduces carboxylase biotinylation, whereas biotin feeding rescues tau-mediated loss of biotin. n = 6 per genotype (ANOVA). ACC: acetyl CoA carboxylase; PC: pyruvate carboxylase; MCC/PCC: methyl crotonyl carboxylase/propionyl coA carboxylase. Carboxylases were identified by molecular weight as previously published. The blot was reprobed with an antibody to GAPDH to illustrate equivalent protein loading. (C) TauR406W transgenic flies have a reduction in carboxylase function as shown by a pyruvate carboxylase enzyme assay on head homogenate. Absorbance of plate fluorescence was quantified on a plate reader, and specific activity was calculated using previously published methods normalized to protein concentration. n = 4 per genotype, where a single sample = 6 pooled heads (t test). *P < 0.05, **P < 0.01. Control genotype: elav-GAL4/+.
Severe Biotin Deficiency Causes Neurodegeneration, Loss of Carboxylase Biotinylation, and Mitochondrial Dysfunction.
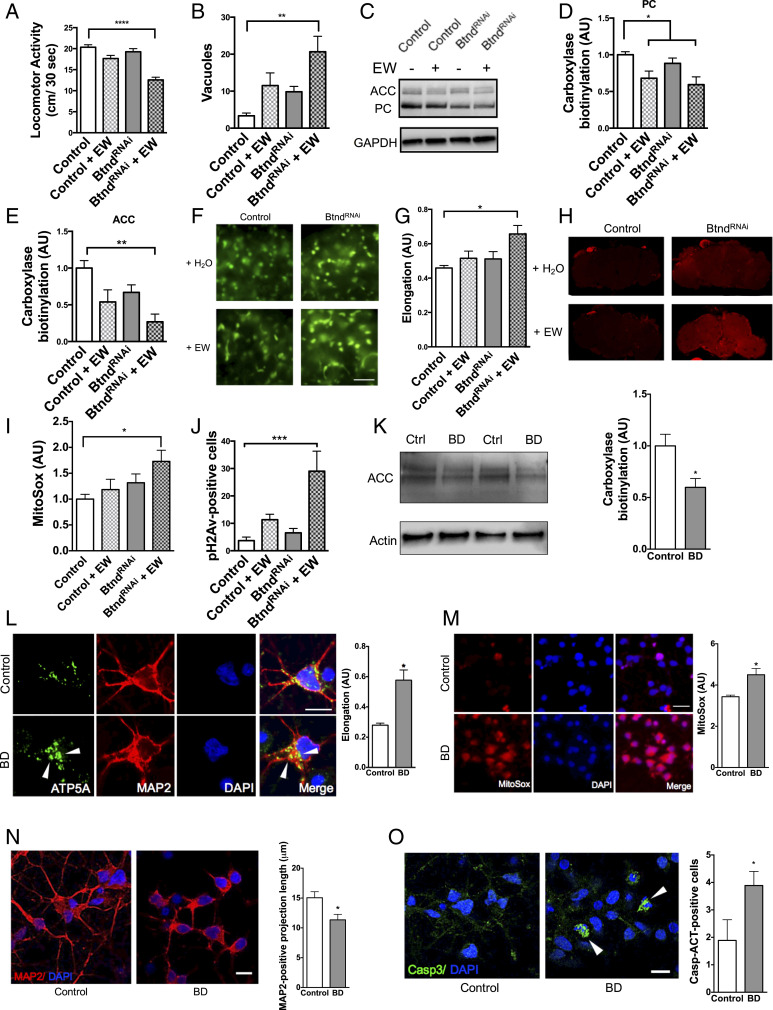
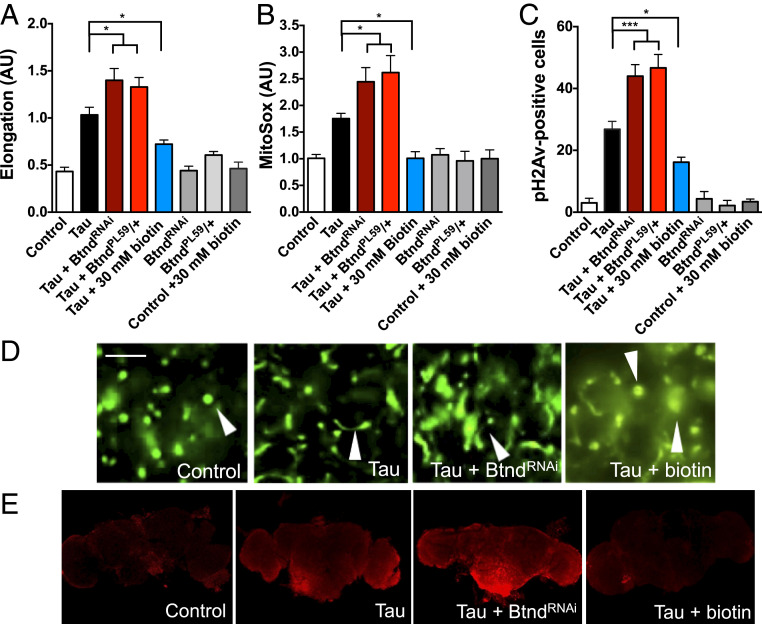
Inborn errors in metabolism leading to marked biotin deficiency preferentially affect the nervous system (26–28). To study the pathways leading to neurotoxicity in biotin-depleted states, we created a Drosophila model of biotin deficiency. Since complete loss of biotinidase function is lethal, we coupled expression of the transgenic BtndRNAi line with chronic egg-white feeding, an avidin-rich diet previously shown to deplete absorption of biotin by high affinity binding to avidin (29). Biotin-deficient flies showed significant locomotor deficits, neurodegeneration as indicated by increased vacuole formation in the brain, and reduced carboxylase biotinylation compared to control flies (Fig. 3 A–E). Due to the known importance of carboxylases to mitochondrial function, we next examined mitochondrial morphology in biotin-deficient flies expressing GFP-tagged mitochondria in neurons. Biotin-deficient flies had significantly elongated mitochondria compared to control flies (Fig. 3 F and G). Biotin-deficient flies also had a significant increase in superoxide levels, an indicator of oxidative stress and mitochondrial function, compared to control flies as shown by the superoxide dye MitoSox (Fig. 3 H and I). Since oxidative stress is a known contributor to DNA damage, we also examined levels of phosphorylated H2Av, the Drosophila ortholog of the human DNA damage marker phospho-H2Ax. Biotin-deficient flies showed significant increases in pH2Av-positive cells, suggesting that mitochondrial dysfunction contributes to DNA damage (Fig. 3J).
Fig. 3.
Biotin deficiency alone is capable of causing neurodegeneration and mitochondrial dysfunction in Drosophila and differentiated human cortical neurons. Biotin-deficient flies were generated by treating BtndRNAi flies with an egg white (avidin)-supplemented diet (+EW). Biotin-deficient flies show behavioral deficits (A), increased markers of neurodegeneration (B), and a loss of carboxylase biotinylation compared to control flies (C–E). (F–I) Biotin-deficient flies show abnormal mitochondrial morphology and function. GFP-tagged mitochondria were analyzed using the Mito ImageJ Plugin (F and G). (Scale bars, 5 μm.) Oxidative stress was indicated using MitoSox dye and quantification from confocal Z-stacks (H and I). (J) Biotin deficiency causes DNA damage as shown by pH2Av-positive foci in the brains of avidin-fed flies. Fly feeding experiments were analyzed by two-way ANOVA with Tukey’s post test; n = 6 per genotype. Control genotype (A–E, H–J) was elav-GAL4/+. Control genotype (F and G) was elav-GAL4/+;UAS-mito-GFP/+. (K–M) Similarly, cortically differentiated iPSCs treated with biotin-deficient (BD) media showed a reduction in carboxylase biotinylation (K), abnormal mitochondrial morphology as shown by ATP5A immunolabeling (L, arrowheads referencing enlarged mitochondria), and an accumulation of superoxide oxidative stress by MitoSox dye (M). Cells treated with biotin-deficient media also showed shortened MAP2-positive projection length (N) and increased caspase activation (O, arrowheads refe1rencing activated caspase-positive cells). (Scale bars = 5 μm.) Cell culture experiments were analyzed by t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Biotin Depletion Causes Mitochondrial Stress in Differentiated Human Cortical Neurons.
Next, we biotin-depleted human induced pluripotent stem cell (iPSC)-derived cortical neurons using biotin-deficient culture media over a 10-d time course. In this mammalian cell model, biotin depletion significantly reduced carboxylase biotinylation (Fig. 3K) and disrupted mitochondrial morphology and function as shown by ATP5A immunofluorescence and the superoxide dye MitoSox, respectively (Fig. 3 L and M). At day 10, biotin depletion also resulted in a reduction in MAP2-positive neuronal process length and an increase in the number of neurons with caspase activation as shown by immunofluorescent analyses (Fig. 3 N and O).
Human tauR406W Transgenic Flies Show Aberrant Expression of Biotin-Related Genes.
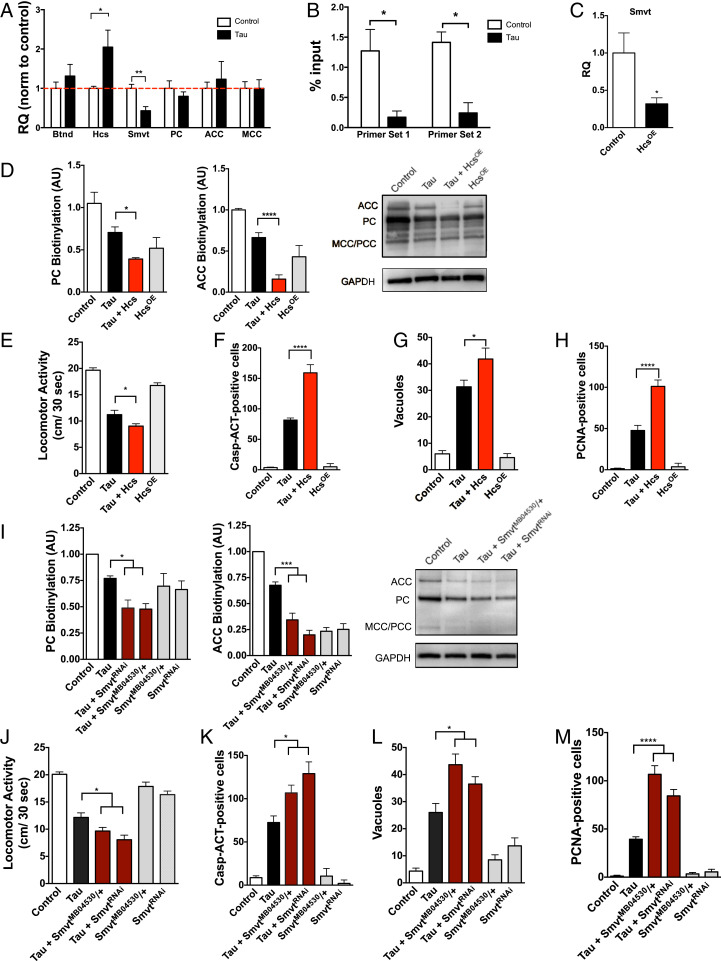
Having found that tauR406W transgenic flies have a biotin deficiency and loss of carboxylase biotinylation, we next examined the underlying mechanism. We thus performed a targeted expression profile to examine other biotin-related genes using qPCR. Targets within this qPCR panel were chosen based on an interactome of protein-protein interaction networks generated by STRING for the human BTD gene (SI Appendix, Fig. S1I). We showed that human tauR406W transgenic flies have both a significant increase in holocarboxylase synthetase (Hcs) expression, an enzyme responsible for linking biotin to carboxylases for their active function, and a significant decrease in expression of the sodium multivitamin transporter (Smvt), which is the transporter that moves biotin across cellular membranes (Fig. 4A) (30–33). The messenger RNA levels of biotinidase and carboxylases remained unchanged. We next explored potential mechanisms for this aberrant expression in the biotin-related genes Hcs and Smvt.
Fig. 4.
Human tauR406W transgenic flies show aberrant changes in expression of biotin-related genes. (A) TauR406W transgenic flies have a significant increase in holocarboxylase synthetase (Hcs) and a decrease in sodium multivitamin transporter (Smvt) level as shown by qPCR. (B) TauR406W transgenic flies have reduced H3K9me2 at the Hcs locus as shown by ChIP-qPCR (C). Overexpression of Hcs reduces Smvt expression in fly heads. Hcs overexpression further reduces carboxylase biotinylation (D), enhances tauR406W-mediated behavioral deficits (E) and neurotoxicity as shown by caspase activation, vacuole formation, and cell-cycle reentry (F–H). Similarly, reductions in Smvt (SmvtRNAi or SmvtMB04530/+) reduce carbozylase biotinylation (I) and enhance tauR406W-mediated behavioral deficits, caspase activation, vacuole formation, and cell-cycle reentry (J–M). n = 6 per genotype. For A–C, t test was used for analysis, and in D–M, ANOVA was used for analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Control genotype (A–E, G–J, L, M): elav-GAL4/+. Control genotype (F and K): elav-GAL4/+;UAS-CD8-PARP-Venus/+.
Tau expression has previously been shown to induce decondensation of constitutive heterochromatin, leading to up-regulated transcription of multiple genes (15). Using predictive tools for heterochromatin sites (annotated tracks in the University of California at Santa Cruz Genome Browser), we determined that the Hcs locus lies within a heterochromatic genomic region in Drosophila. Furthermore, publicly available datasets from Frost et al. (15) in the National Center for Biotechnology Information GeoSets indicate that the Hcs locus is less condensed in human tauR406W transgenic flies than controls based on chromatin immunoprecipitation (ChIP) with an antibody detecting the H3K9me2 histone modification (SI Appendix, Fig. S1G). To further examine heterochromatin markers at this locus, we performed an H3K9me2-based ChIP from human tauR406W transgenic fly heads and performed qPCR using Hcs-targeted primers. Human tauR406W transgenic flies showed a significant reduction in H3K9me2-positive chromatin at the Hcs locus, suggesting heterochromatin decondensation and increased access for transcriptional activity at this site (Fig. 4B).
Hcs not only catalyzes the covalent attachment of biotin to the carboxylases, but also has also been implicated as a general transcriptional repressor (34). More specifically, Hcs decreases the expression of the vitamin transporter Smvt through downstream nuclear signaling pathways (35–37). To examine the effect of Hcs level on Smvt expression, we overexpressed Hcs using a HcsOE Drosophila line and the pan-neuronal elav-GAL4 driver. Hcs overexpression significantly decreased Smvt expression as shown by qPCR (Fig. 4C), resulting in a concomitant decrease in carboxylase biotinylation (Fig. 4D) and an enhancement of tauR406W-mediated behavioral deficits and neurodegeneration (Fig. 4 E–H). Similarly, reduced Smvt expression by either RNAi knockdown (SmvtRNAi) or heterozygosity for SmvtMB04530 also reduced carboxylase biotinylation (Fig. 4I) and enhanced tauR406W-mediated behavioral deficits and neurodegeneration (Fig. 4 J–M).
Biotin Status Alters Mitochondrial Morphology and Function.
Due to the changes to carboxylase biotinylation and function in human tauR406W transgenic flies, we next examined the mitochondrial effects of biotin manipulation in these animals. Using flies with GFP-tagged mitochondria, we showed that reduced biotinidase enhances tau-mediated mitochondrial abnormalities, including significant increases in mitochondrial elongation (Fig. 5A and D). Biotin feeding rescues these measures in tauR406W transgenic flies, showing significant reductions in mitochondrial length. Despite the changes to the morphological characteristics of the mitochondria, there were no significant changes to the total number of mitochondria between the genotypes.
Fig. 5.
Biotin status modifies mitochondrial morphology and function in human tauR406W transgenic flies. (A) Reduced biotinidase enhances tauR406W-mediated mitochondrial elongation, while biotin feeding rescues this abnormal mitochondrial morphology. n = 6 per genotype. (B) Reduced biotinidase enhances tauR406W-mediated superoxide formation, while biotin feeding rescues this abnormal mitochondrial function. n = 10. (C) Reduced biotinidase enhances DNA damage compared to tauR406W transgenic flies, while biotin feeding rescues this damage as shown by immunohistochemical quantification of pH2Av-positive cells. n = 6. (D) Representative images of GFP-tagged mitochondria quantified in A. Arrowheads highlight representative mitochondrial morphology. (Scale bar, 5 μm.) (E) Representative Z-stacks of superoxide dye MitoSox used in B (Imaged at 10× magnification). A–C were analyzed by ANOVA. *P < 0.05, ***P < 0.001. Control genotype (A and D): elav-GAL4/+;UAS-mito-GFP/+. Control genotype (B, C, E): elav-GAL4/+.
Due to the significant changes in mitochondrial morphology following biotin manipulation, we next examined mitochondrial function as indicated by oxidative stress using MitoSox superoxide dye. Reduced biotinidase increases superoxide levels in tauR406W transgenic flies, and, conversely, biotin-fed tauR406W transgenic flies show a significant reduction in superoxide levels, suggesting a rescue of functional mitochondrial deficits (Fig. 5B and E). Finally, reduced biotinidase increased DNA damage compared to tauR406W transgenic flies as measured by the number of pH2Av-positive cells, whereas biotin feeding decreases tauR406W-mediated DNA damage (Fig. 5C).
Brain Carboxylase Biotin Levels Are Altered in Mammalian Tauopathy.
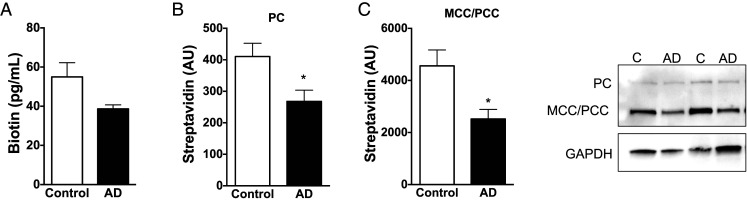
Interestingly, 7-mo-old rTg4510 tau transgenic mice expressing neuronal P301L mutant tau via a CamKII-driven tet on/off system showed decreased carboxylase biotinylation by streptavidin blotting, but no changes in biotin level as shown by biotin ELISA (SI Appendix, Fig. S2D) (38, 39). Recent reports suggest that the rTg4510 mice may have genome-level alterations that could complicate interpretation of these data (40). Thus, we also measured levels of biotin and carboxylase biotinylation in human Alzheimer’s disease frontal cortex. While we saw a nonsignificant trend toward a decrease in biotin in Alzheimer’s disease cortex by ELISA (Fig. 6A), carboxylase biotinylation appears to decrease in at least a subset of Alzheimer’s disease patient brains, raising the possibility that reduced carboxylase biotinylation may be a common pathogenic mechanism in tauopathies (Fig. 6B and C).
Fig. 6.
Reduced carboxylase biotinylation in human Alzheimer’s disease brain. (A) Postmortem cortical tissue from Alzheimer’s disease brain has a nonsignificant trend toward reduced biotin level by ELISA. (B, C) Carboxylase biotinylation decreases in Alzheimer’s disease brain as shown by streptavidin blotting from cortical tissue homogenates. Blots were reprobed with an antibody to GAPDH to illustrate equivalent protein loading. (n = 8). A–C analyzed by t test. *P < 0.05.
Discussion
Here we report a mechanism by which biotin availability modifies the neurodegenerative effects of the microtubule-associated protein tau. Our results suggest that biotin, a covalently bound cofactor required for the activity of the five known biotin-dependent carboxylases, contributes to tauR406W neurotoxicity via changes to carboxylase and mitochondrial function in the Drosophila brain. The importance of mitochondrial function in mechanisms of tau-mediated neurodegeneration reinforces previous work from our laboratory and others (2, 3). Furthermore, these results suggest that neurological effects of biotin deficiency result from changes to carboxylase biotinylation and subsequently altered mitochondrial function.
Innate Biotin Deficiency in Tauopathy.
The current study shows a reduction in biotinylated carboxylase levels in multiple tauopathies, including human tauR406W transgenic flies, tau transgenic mice, and human Alzheimer’s disease brain. While most vitamins are in excess in the body, we define a specific mechanism by which pathological tau promotes a vitamin-deficient state. Pathogenic tau expression alters chromatin structure and genomic stability (15, 41–43). The current study demonstrates that tauR406W-mediated heterochromatin decondensation causes aberrant expression of Hcs, leading to reduced expression of the vitamin transporter Smvt and an innate biotin deficiency within neurons. Hcs is a negative regulator of the Smvt gene, although the exact mechanism of gene regulation is debated (44). Hcs protein has been described as a global transcriptional repressor via two different mechanisms (35, 45). First, Hcs influences soluble guanylate-cyclase–signaling pathways, leading to transcriptional repression (37). While lacking a canonical DNA-binding domain typical of a transcription factor, Hcs has also been implicated in a protein complex that binds regulatory regions to modify transcription of genes such as Smvt (34). Finally, it has been suggested that Hcs mediates histone biotinylation, contributing to epigenetic regulation of the Smvt promoter region (30, 36); however, histone biotinylation remains difficult to assess in vivo (46, 47). Taken together, these results complement existing data on Hcs regulation of Smvt and propose a model for an innate biotin deficiency in tauopathies.
Pathogenesis of Biotin Deficiency.
In patients, severe biotin deficiency due to mutations in the BTD gene typically manifests in infancy or early childhood and displays prominent and severe neurological symptoms, including seizures, ataxia, and hearing loss. Similarly, mouse models of biotin deficiency secondary to either dietary manipulation or biotinidase knock-out show neurological deficits, including demyelination, axonal degeneration, ventriculomegaly, and corpus callosum compression (48). Although loss of carboxylase biotinylation and function has been strongly implicated in the pathogenesis of biotin deficiency, the mechanisms underlying neuronal dysfunction and death remain poorly understood. Prior work has implicated a diverse set of mediators of neuronal toxicity, including brain accumulation of abnormal metabolites, such as lactic acid, ammonia, and other organic acids (49). Here we present approaches to study biotin deficiency, including Drosophila and iPSC-based platforms. Furthermore, through modeling in human tauR406W transgenic flies, biotin-deficient flies, and biotin-depleted human cortical neurons derived from iPSCs, we have shown that reduced biotin results in significant mitochondrial abnormalities, including morphological and functional changes along with oxidative stress. These mechanisms may contribute to neurotoxicity in genetic or acquired biotin deficiency in patients.
Therapeutic Potential of Biotin Supplementation.
Although mechanisms underlying neuronal susceptibility to biotin deficiency are incompletely understood, pharmacological doses of biotin are highly effective and well tolerated in patients with BTD mutations (50–52). Biotin supplementation has been implemented safely as a treatment for a variety of other neurological diseases as well. Biotin reduces the motor symptoms of biotin-thiamine–responsive basal ganglia disease, caused by a mutations in the thiamine transporter-2 (THTR-2) gene (53). Biotin has been also used as a candidate treatment for Leigh’s disease, a childhood metabolic disorder that causes progressive midbrain and brainstem lesions due to mutations in mitochondria-related genes (54). There have also been reports of extreme brain biotin deficiency in severe cases of Leigh’s disease, suggesting that a biotin deficit may contribute to symptom severity and degeneration (28). Finally, biotin is already a successful therapy in adult neurology as a treatment for multiple sclerosis, where a study reported that high doses of biotin (up to 100 to 300 mg/day) were well tolerated and slowed disease progression (55, 56).
While biotin deficiency has yet to be explored in depth in neurodegenerative tauopathies, our data suggest that there may be a similar pathogenic mechanism to human biotin deficiency. The current findings show the presence of biotin-deficient brain carboxylases in postmortem Alzheimer’s disease tissue and rTg4510 tau transgenic mice, suggesting that future studies should focus on the impact on biotin supplementation in vertebrate models of tauopathies (Fig. 6). In addition, our data suggest that biotin may be worth exploring as a biomarker in larger patient populations. These results could be further strengthened by exploring these pathological mechanisms in additional tau isoforms beyond those presented here.
In summary, the Drosophila forward genetic screen at the center of this study led us to identify the biotin pathway as a modifier of tau-mediated neurodegeneration. We further defined a mechanism underlying the neurological effects in chronic biotin deficiency, a set of human disorders with limited mechanistic insight. Finally, these results nominate the biotin pathway as a candidate for future therapeutic and biomarker development in tauopathies.
Materials and Methods
Fly Stocks.
Drosophila crosses and aging were performed at 25 °C. All animals were aged 10 d before experiments were performed. Both male and female flies were used for each study unless otherwise noted. The GAL4-UAS expression system and the pan-neuronal elav-GAL4 driver were used to control transgene expression. Human tau transgenic flies (UAS-tauR406W and UAS-tauWT) and tauopathogy models have been described in detail previously (10, 12, 57). Control animals contained the elav-GAL4 driver in the heterozygous state and other UAS transgenes as appropriate. Control genotypes are provided for each experiment in the figure legends. The Smvt RNAi line (HMJ30134) and SmvtMB04530 were obtained from the Bloomington Drosophila Stock Center. The Btnd RNAi (VDRC16663) line was obtained from the Vienna Drosophila RNAi Center. The following stocks were obtained from the indicated investigators: UAS-CD8-PARP-Venus (D. Williams, King's College London, London, UK); BtndPL59 (A. Vincent and M. Crozatier, Université Paul Sabatier, Toulouse, France); UAS-mito-GFP (T. Schwarz, Boston Children’s Hospital, Harvard Medical School, Boston, MA); and UAS-ataxin 3-Q78 (UAS-MJD-Q78) (N. Bonini, University of Pennsylvania, Philadelphia, PA). The SmvtMB04530 and BtndPL59 mutants were used in the heterozygous state (SmvtMB04530/+ and BtndPL59/+). BtndPL59 is a lethal P-element insertion in the X-chromosome Btnd locus (20).
Genome-Scale Genetic Screen.
To identify pathways and molecules modifying tau-mediated toxicity, we performed an unbiased forward genetic screen using RNAi lines from the Bloomington Drosophila Stock Center and Vienna Drosophila Resource Center. Transgenic RNAi lines were crossed to a tauR406W tester stock (genotype: elav-GAL4; UAS-tauR406W/TM6, GAL80). Histologic sections of the brain at 10 d of age were examined for neuropathological evidence of degeneration. Candidates were then assessed using the UAS-CD8-PARP-Venus transgenic caspase reporter (16). Toxicity was assessed by monitoring the cleavage of the reporter using an antibody (E51, Abcam) specific for cleaved human PARP. Caspase cleavage was assessed at 10 d of age using immunohistochemistry on brain tissue sections. All newly described tauR406W modifiers were validated with two independent transgenic RNAi lines, if available (SI Appendix, Table S1).
Interactome Analysis.
Human orthologs of validated hits from the Drosophila screen were identified using DIOPT (https://www.flyrnai.org/diopt) (17). These orthologs were rank-ordered by weighted DIOPT score for homology as previously described (17), and orthologs with scores above 9 were then entered into the STRING interactome generator (18). This cutoff value was chosen based on the generated score distribution. A total of 145 proteins were entered into STRING, and an additional 30 proteins were included by the generator to create a protein–protein interaction network. Significantly enriched networks previously implicated in tau pathologies were highlighted (P < 0.001).
Transgenic tau Mice rTg4510.
Parental mutant tau and tetracycline-controlled transactivator protein mouse lines were maintained separately and bred to produce rTg4510 mice as described previously (38, 39). Mice used in streptavidin blotting and biotin ELISA were aged to 7 mo prior to being killed. Control genotypes were nontau and nondriver. Mice were on a 12/12 light–dark schedule and had access to food ad libitum. All mouse experiments were performed in keeping with the guidelines of the Institutional Animal Care and Use Committee of Brigham and Women’s Hospital.
Drosophila Biotin Feeding.
Biotin (Fisher) was diluted in ddH2O to 0, 10, and 30 mM concentrations. These biotin solutions (3 mL) were mixed with Drosophila instant food (Formula 4–24, Carolina Biosupply). For biotin depletion studies, 0.8 g/mL of spray-dried egg white was mixed into instant Drosophila food as previously published (29). The flies were fed one of two diets, control or biotin-deficient. Raw egg white contains the protein avidin, which has a high affinity for biotin, and avidin-bound biotin is unavailable for absorption. To eliminate potential confounds of protein content, a small group of tau transgenic animals was fed a protein-matched control diet using 0.56 g/mL of BSA based on previous protocols (29). There was no effect of this treatment. Flies were transferred onto fresh food vials every 3 d and aged to 10 d before experimentation.
qPCR.
RNA was extracted from six Drosophila heads using QIAZOL (Qiagen), and concentrations were measured with a Nanodrop ND-1000 Spectrophotometer. Equal amounts of RNA were reverse-transcribed using a cDNA Reserve Transcription Kit (Applied Biosystems), followed by qPCR. SYBR Green (Life Technologies) qPCR was performed on an Applied Biosystems StepOnePlus Real-Time PCR System with each data point the result of at least three biological replicates, each composed of three technical replicates. Drosophila ribosomal protein RpL32 was used as the internal control gene. Primer sequences were the following: RpL32 5′ GACCATCCGCCCAGCATAC 3′ (forward), 5′ CGGCGACGCACTCTGTT 3′ (reverse); Btnd 5′ CAAGCCGGAAGA-TCCAACCT 3′ (forward), 5′ GTGGCTGTGGCCTGACTATT 3′ (reverse); Hcs 5′ AGTATTGGAATTGGAGAATGC 3′ (forward), 5′ AACTTACATATTGATGGGAAC-C 3′ (reverse); Hcs set 2 5′ CATCAGCAGTACGGCTGGAGG 3′ (forward), 5′ GTTGCACCGACTCAATCCG 3′ (reverse); Smvt 5′ TTTCGGTTTCTTTGCCAAGGGCGA- 3′ (forward), 5′ AGCGAAATGGCGATGGGAATCGTT 3′ (reverse); PC 5′ TCAAAA-ATGGCTACTCCAGCAA 3′ (forward), 5′ GCACGGAACACTCGGATGG 3′ (reverse); MCC 5′ GACATGGGGATTAAGAGCACC 3′ (forward), 5′ CTGCAAACACTCATCTGA-CTGA 3′ (reverse); ACC 5′ CGAGCGGGCCATTAGGTTT 3′ (forward), 5′ GCCATCTTG-ATGTATTCGGCAT 3′ (reverse).
Histological Detection of Mitochondrial Morphology and Markers of Neurodegeneration.
Neurodegenerative outcomes were measured by unbiased counting of brain sections stained for markers of caspase activation, cell-cycle activation, and vacuoles. For tissue sections, adult flies were fixed in formalin and embedded in paraffin. Serial 4-μm frontal sections were prepared through the entire fly brain and placed on a single glass slide. Mouse and human samples were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at a 6-μm thickness. For immunostaining, paraffin slides were processed through xylene, ethanol, and into water. Antigen retrieval was performed by boiling in sodium citrate, pH 6.0, prior to 1 h of blocking in phosphate-buffered saline (PBS) containing 0.3% Triton X-100 and 2% milk. Sections were then incubated in primary antibodies overnight in a humidified chamber (GFP, N86/8, NeuroMab, 1:100; cleaved PARP, E51, Abcam, 1:50,000; proliferating cell nuclear antigen [PCNA], Dako, 1:500). For immunohistochemistry, biotin-conjugated secondary antibodies (1:200, SouthernBiotech) and avidin–biotin–peroxidase complex (Vectastain Elite, Vector Laboratories) staining was performed using 3,3'-diaminobenzidine (Vector Laboratories) as a chromagen. For immunofluorescent studies, appropriate Alexa-Fluor–conjugated secondary antibodies (1:200, Invitrogen) were used. All immunostaining data were replicated with n = 6.
Locomotor Behavior.
The walking assay was performed on 10-d-old flies as previously described (19). Briefly, a single fly was tapped gently to the same initial starting position within its vial, and the vial was placed over a 1-cm gridded surface. The number of centimeters traveled in a 30-s trial was counted four times per fly, and 18 flies of each genotype were assayed.
Pyruvate Carboxylase Assay.
The pyruvate carboxylase assay was performed as previously described with some modifications (58, 59). A total of 95 µL of working solution (0.1 M Tris–HCl, 50 mM NaHCO3, 5 mM MgCl2, 10 mM Acetyl-CoA (CoALA Biosciences), 5 mM ATP, 0.1 M Tris–HCl, pH 8.0) was spiked with 2.5 μL of fresh 3.9 mg/mL DTNB (5,5-dithio-bis-(2-nitrobenzoic acid)) and 0.5 µL 1,000 U/mL citrate synthase (Sigma #C3260). Experimental wells had 5 µL of 0.1 M pyruvate added. A total of 100 µL of working solution (control and experimental) was added to a 96-well plate, and the plate was prewarmed to 30 °C in the plate reader. Ten fresh fly heads were homogenized in 15 µL dH2O, and 5 µL was added to each well. Plates were shaken for 3 s and kinetically measured at 412 nm for 20 min in a plate reader (Molecular Devices Spectra Max M5). Protein concentration of the sample was determined using a BCA (bicinchoninic acid) protein assay with a BSA standard curve (Pierce). Output was plotted, and specific activity was calculated as previously described (Specific activity = Activity/Protein concentration).
Microscopy.
Tissue sections were imaged on a Nikon Eclipse E600 fluorescent microscope with SPOT software for histological quantifications. Images were analyzed with ImageJ. All fluorescent images were taken on a confocal microscope (Olympus Fluoview 100 confocal microscope at the Harvard Medical School Neuroimaging facility, NIH National Institute of Neurological Disorders and Stroke [NINDS] P30NS072030). Control and experimental samples were imaged with the same laser settings.
Western Blotting.
Frozen Drosophila heads were homogenized in Laemmli sample buffer (Sigma), boiled for 10 min at 100 °C, briefly centrifuged, and analyzed by sodium dodecyl sulfate/polyacrylamide gel electrophoresis using 10% gels (Lonza). Proteins were transferred to nitrocellulose membranes (BioRad), blocked in 2% milk in PBS with 0.05% Tween-20, and immunoblotted with primary antibody overnight (actin, JLA20, DSHB, 1:40,000; GAPDH, GA1R, Invitrogen, 1:40,000; tau5, DSHB, 1:100,000), followed by a 2-h incubation with the appropriate HRP-conjugated secondary antibody (SouthernBiotech, 1:20,000). Carboxylase blots were performed by homogenizing six Drosophila heads in PBS with phosphatase and protease inhibitors (Thermo Fisher Scientific). Human and mouse cortical samples (SI Appendix, Table S3) were prepared and homogenized in radioimmunoprecipitation (RIPA) buffer with protease and phosphatase inhibitor mixture using a Kontes 2-mL glass homogenizer. Protein concentration was calculated using the BCA protein assay (Pierce). Following run and transfer as described above, membranes were incubated with streptavidin–HRP (Southern Biotechnology 1:10,000) for 1 h and washed for 2 h prior to imaging. All blots were imaged using SuperSignal West Femto chemiluminescent substrate (Thermo Fisher Scientific) and a Fluorochem HD2 imager (ProteinSimple) and quantified by densitometric analysis in ImageJ. Blots were reprobed for actin or GAPDH to illustrate equivalent protein.
Mitochondrial Morphology and Function.
Mitochondrial morphology was quantified by mitochondrial elongation and interconnectivity measures using a Mito-Morphology Macro in ImageJ (60) using images of fly brain sections with GFP-tagged mitochondria flies (UAS-Mito-GFP). Mitochondrial function was assessed by superoxide accumulation using the commercially available MitoSox dye (Molecular Probes). Briefly, fly brains were dissected and incubated in the dye for 30 min, fixed in 4% paraformaldehyde (PFA) for 10 min, washed in PBS twice for 10 min, and mounted using Fluoromount (Fisher Scientific). Brains were imaged on a confocal microscope as described above. Z-stacks of confocal images through whole fly brains were merged, and the sum of the dye intensity was quantified.
Biotin Depletion in Differentiated Human iPSCs.
Human neurons differentiated from induced pluripotent stem cells (iCell Neurons) were purchased from Cellular Dynamics International. iCell Neurons were generated through a proprietary forebrain differentiation protocol resulting in a neuronal cell population. For immunofluorescent studies, cells were plated onto coated coverslips in 24-well plates with 200,000 cells per well. For MitoSox analysis, cells were plated onto a 96-well plate with 30,000 cells per well. All maintenance was performed as described in the manufacturer’s protocol. A biotin depletion was performed using 5 mL of Pierce Avidin Agarose resin (Fisher Scientific, catalog no. 20219) loaded into a 10-mL column. Biotin depletion of media was confirmed and compared to control media using a standard curve analysis on a biotin ELISA plate as described below. Media was then applied to cultures in a progressive biotin depletion model as follows. On day 3 after plating, 75% of cell media was replaced with either control or 50% biotin-depleted media. Every other day until day 10, 75% of the media was replaced with control or 70% biotin-depleted media. For immunofluorescent studies, cells were fixed in 4% PFA, washed with PBS + 0.1% Trition, blocked with 5% BSA in PBS + 0.1% Trition, and incubated in primary antibodies overnight at 4 °C. The following primary antibody concentrations were used: ATP5A, 15H4C4, Invitrogen, 1:500; MAP2, Millipore: mouse, 1:250, rabbit, 1:500; Caspase 3, Cell Signaling, 1:400. Appropriate fluorescent secondary antibodies (Alexa Fluor, Molecular Probes) were used at 1:1,000.
Biotin ELISA.
Biotin concentrations were calculated using a commercially available biotin ELISA kit (LSBio, catalog no. F10160). For fly tissue, one fly head was homogenized in 30 µL of PBS with protease and phosphatase inhibitors, and 5 µL of this homogenate was used in the assay. Mouse and human brain tissue was homogenized in 100 µL PBS with protease and phosphatase inhibitors and 10 µg of tissue was used in the ELISA, an amount optimized empirically. Protein concentrations were calculated using a Pierce BCA Protein assay. The assay was performed as described in the literature with the following changes: The detection method was modified to use the Quanta Blue Fluorogenic Substrate (Pierce) for detection on a plate reader (fluorescence: 325 nm excitation, 420 nm emission). All samples were run in technical duplicate.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation was performed as previously described (15). Chromatin was prepared and immunoprecipitated with the following modifications: 50 fly heads, representing equal numbers of males and females, were homogenized before formaldehyde cross-linking. Chromatin was sheared to 500 bp by 15- × 30-s bursts of sonication at 100% duty level 5 power with a Branson sonicator. IgG was used as a negative control, and data were normalized to input.
Statistical Analyses.
All sample sizes (n) are biological replicates. Sample sizes were based on previous work with these models and techniques. Statistical analysis was performed using a one-way ANOVA when making multiple comparisons and an unpaired Student’s t test when making comparisons between two samples. Feeding studies were analyzed with a two-way ANOVA with genotype and treatment as factors and Tukey’s post hoc test. Statistical outliers were determined by Grubbs’ Test for Outliers. Each graph is shown as mean ± SEM.
Supplementary Material
Acknowledgments
Fly stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537), the Vienna Drosophila Resource Center, and Drs. D. Williams, A. Vincent, M. Crozatier, T. Schwarz, and N. Bonini were used in this study. We thank the Transgenic RNAi Project at the Harvard Medical School (NIH National Institute of General Medical Sciences R01GM084947) for making transgenic RNAi stocks and the Harvard Medical School Neuroimaging facility (NIH NINDS P30NS072030) for assistance with imaging. Monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development (NICHD of the NIH) and maintained by the Department of Biology, University of Iowa, and the University of California, Davis/NIH NeuroMab Facility. This work was supported by NIH National Institute on Aging (NIA) Grant F32NS100308 (to K.M.L.) and NIH NIA Grants R01 AG044113 and R01 AG057331 (to M.B.F. and C.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922392117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Hirai K., et al. , Mitochondrial abnormalities in Alzheimer’s disease. J. Neurosci. 21, 3017–3023 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dias-Santagata D., Fulga T. A., Duttaroy A., Feany M. B., Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J. Clin. Invest. 117, 236–245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DuBoff B., Götz J., Feany M. B., Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron 75, 618–632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moneim A. E. A., Oxidant/Antioxidant imbalance and the risk of Alzheimer’s disease. Curr. Alzheimer Res. 12, 335–349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutton M., et al. , Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 (1998). [DOI] [PubMed] [Google Scholar]
- 6.Poorkaj P., et al. , Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann. Neurol. 43, 815–825 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Spillantini M. G., Crowther R. A., Kamphorst W., Heutink P., van Swieten J. C., Tau pathology in two Dutch families with mutations in the microtubule-binding region of tau. Am. J. Pathol. 153, 1359–1363 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittmann C. W., et al. , Tauopathy in Drosophila: Neurodegeneration without neurofibrillary tangles. Science 293, 711–714 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Khurana V., Feany M. B., Connecting cell-cycle activation to neurodegeneration in Drosophila. Biochim. Biophys. Acta 1772, 446–456 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khurana V., et al. , A neuroprotective role for the DNA damage checkpoint in tauopathy. Aging Cell 11, 360–362 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Depeint F., Bruce W. R., Shangari N., Mehta R., O’Brien P. J., Mitochondrial function and toxicity: Role of the B vitamin family on mitochondrial energy metabolism. Chem. Biol. Interact. 163, 94–112 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Fulga T. A., et al. , Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat. Cell Biol. 9, 139–148 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Bardai F. H., et al. , A conserved cytoskeletal signaling cascade mediates neurotoxicity of FTDP-17 tau mutations in vivo. J. Neurosci. 38, 108–119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loewen C. A., Feany M. B., The unfolded protein response protects from tau neurotoxicity in vivo. PLoS One 5, e13084 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frost B., Hemberg M., Lewis J., Feany M. B., Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 17, 357–366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams D. W., Kondo S., Krzyzanowska A., Hiromi Y., Truman J. W., Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 9, 1234–1236 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Hu Y., et al. , An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12, 357 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D., et al. , STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merlo P., et al. , p53 prevents neurodegeneration by regulating synaptic genes. Proc. Natl. Acad. Sci. U.S.A. 111, 18055–18060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourbon H.-M., et al. , A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 110, 71–83 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Musi N., et al. , Tau protein aggregation is associated with cellular senescence in the brain. Aging Cell 17, e12840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koseoglu M. M., Norambuena A., Sharlow E. R., Lazo J. S., Bloom G. S., Aberrant neuronal cell cycle re-entry: The pathological confluence of Alzheimer’s disease and brain insulin resistance, and its relation to cancer. J. Alzheimers Dis. 67, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khurana V., et al. , Lysosomal dysfunction promotes cleavage and neurotoxicity of tau in vivo. PLoS Genet. 6, e1001026 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh S., Feany M. B., Comparison of pathways controlling toxicity in the eye and brain in Drosophila models of human neurodegenerative diseases. Hum. Mol. Genet. 13, 2011–2018 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Moss J., Lane M. D., The biotin-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 35, 321–442 (1971). [DOI] [PubMed] [Google Scholar]
- 26.Zempleni J., Hassan Y. I., Wijeratne S. S., Biotin and biotinidase deficiency. Expert Rev. Endocrinol. Metab. 3, 715–724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suchy S. F., McVoy J. S., Wolf B., Neurologic symptoms of biotinidase deficiency: Possible explanation. Neurology 35, 1510–1511 (1985). [DOI] [PubMed] [Google Scholar]
- 28.Baumgartner E. R., et al. , Biotinidase deficiency: A cause of subacute necrotizing encephalomyelopathy (Leigh syndrome). Report of a case with lethal outcome. Pediatr. Res. 26, 260–266 (1989). [DOI] [PubMed] [Google Scholar]
- 29.Smith E. M., et al. , Feeding Drosophila a biotin-deficient diet for multiple generations increases stress resistance and lifespan and alters gene expression and histone biotinylation patterns. J. Nutr. 137, 2006–2012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camporeale G., Giordano E., Rendina R., Zempleni J., Eissenberg J. C., Drosophila melanogaster holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan, and heat tolerance. J. Nutr. 136, 2735–2742 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soló Rzano-Vargas R. S., Pacheco-Alvarez D., León-Del-Río A., Holocarboxylase synthetase is an obligate participant in biotin-mediated regulation of its own expression and of biotin-dependent carboxylases mRNA levels in human cells. Proc. Natl. Acad. Sci. U.S.A. 99, 5325–5330 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mall G. K., Chew Y. C., Zempleni J., Biotin requirements are lower in human Jurkat lymphoid cells but homeostatic mechanisms are similar to those of HepG2 liver cells. J. Nutr. 140, 1086–1092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchida Y., et al. , Major involvement of Na(+)-dependent multivitamin transporter (SLC5A6/SMVT) in uptake of biotin and pantothenic acid by human brain capillary endothelial cells. J. Neurochem. 134, 97–112 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Trujillo-Gonzalez I., et al. , Holocarboxylase synthetase acts as a biotin-independent transcriptional repressor interacting with HDAC1, HDAC2 and HDAC7. Mol. Genet. Metab. 111, 321–330 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Gralla M., Camporeale G., Zempleni J., Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J. Nutr. Biochem. 19, 400–408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zempleni J., Gralla M., Camporeale G., Hassan Y. I., Sodium-dependent multivitamin transporter gene is regulated at the chromatin level by histone biotinylation in human Jurkat lymphoblastoma cells. J. Nutr. 139, 163–166 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pacheco-Alvarez D., et al. , Biotin availability regulates expression of the sodium-dependent multivitamin transporter and the rate of biotin uptake in HepG2 cells. Mol. Genet. Metab. 85, 301–307 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Ramsden M., et al. , Age-dependent neurofibrillary tangle formation, neuron loss, and memory impairment in a mouse model of human tauopathy (P301L). J. Neurosci. 25, 10637–10647 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.SantaCruz K., et al. , Tau suppression in a neurodegenerative mouse model improves memory function. Science 309, 476–481 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gamache J., et al. , Factors other than hTau overexpression that contribute to tauopathy-like phenotype in rTg4510 mice. Nat. Commun. 10, 2479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frost B., Götz J., Feany M. B., Connecting the dots between tau dysfunction and neurodegeneration. Trends Cell Biol. 25, 46–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun W., Samimi H., Gamez M., Zare H., Frost B., Pathogenic tau-induced piRNA depletion promotes neuronal death through transposable element dysregulation in neurodegenerative tauopathies. Nat. Neurosci. 21, 1038–1048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein H.-U., et al. , Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer’s human brains. Nat. Neurosci. 22, 37–46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.León-Del-Río A., Valadez-Graham V., Gravel R. A., Holocarboxylase synthetase: A moonlighting transcriptional coregulator of gene expression and a cytosolic regulator of biotin utilization. Annu. Rev. Nutr. 37, 207–223 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Pacheco-Alvarez D., Solórzano-Vargas R. S., Del Río A. L., Biotin in metabolism and its relationship to human disease. Arch. Med. Res. 33, 439–447 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Healy S., et al. , Biotin is not a natural histone modification. Biochim. Biophys. Acta 1789, 719–733 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Kuroishi T., Rios-Avila L., Pestinger V., Wijeratne S. S. K., Zempleni J., Biotinylation is a natural, albeit rare, modification of human histones. Mol. Genet. Metab. 104, 537–545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pindolia K., et al. , Neurological deficits in mice with profound biotinidase deficiency are associated with demylination and axonal degeneration. Neurobiol. Dis. 47, 428–435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swango K. L., Wolf B., Conservation of biotindase in mammals and identification of the putative biotinidase gene in Drosophila melanogaster. Mol. Genet. Metab. 74, 492–499 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Heard G. S., Secor McVoy J. R., Wolf B., A screening method for biotinidase deficiency in newborns. Clin. Chem. 30, 125–127 (1984). [PubMed] [Google Scholar]
- 51.Wolf B., Heard G. S., Screening for biotinidase deficiency in newborns: Worldwide experience. Pediatrics 85, 512–517 (1990). [PubMed] [Google Scholar]
- 52.Wolf B., Heard G. S., McVoy J. R., Grier R. E., Biotinidase deficiency. Ann. N. Y. Acad. Sci. 447, 252–262 (1985). [DOI] [PubMed] [Google Scholar]
- 53.Tabarki B., et al. , Biotin-responsive basal ganglia disease revisited: Clinical, radiologic, and genetic findings. Neurology 80, 261–267 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Lake N. J., Compton A. G., Rahman S., Thorburn D. R., Leigh syndrome: One disorder, more than 75 monogenic causes. Ann. Neurol. 79, 190–203 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Tourbah A. et al.; MS-SPI study group , MD1003 (high-dose biotin) for the treatment of progressive multiple sclerosis: A randomised, double-blind, placebo-controlled study. Mult. Scler. 22, 1719–1731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sedel F., Bernard D., Mock D. M., Tourbah A., Targeting demyelination and virtual hypoxia with high-dose biotin as a treatment for progressive multiple sclerosis. Neuropharmacology 110, 644–653 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Wittmann C. W., et al. , Tauopathy in Drosophila: Neurodegeneration without neurofibrillary tangles. Science 293, 711–714 (2001). [DOI] [PubMed] [Google Scholar]
- 58.Vemuri G. N., Minning T. A., Altman E., Eiteman M. A., Physiological response of central metabolism in Escherichia coli to deletion of pyruvate oxidase and introduction of heterologous pyruvate carboxylase. Biotechnol. Bioeng. 90, 64–76 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Payne J., Morris J. G., Pyruvate carboxylase in Rhodopseudomonas spheroides. J. Gen. Microbiol. 59, 97–101 (1969). [DOI] [PubMed] [Google Scholar]
- 60.Dagda R. K., et al. , Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 284, 13843–13855 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.