Significance
Maize is a global food staple with great economic and cultural importance. Archaeogenomic studies have revealed a process of protracted maize domestication and multiple waves of human-mediated dispersal in the Americas. Maize first arrived in South America as a partial domesticate, where the domestication syndrome became independently fixed and improved varieties developed away from the influence of wild gene flow. We demonstrate that hybrids of some of these improved varieties were likely reintroduced back to Central America. We hypothesize that this backflow of South American genetic material may have contributed to the development of a more productive staple, which was related to the growth and aggregation of human populations, and the formation of more complex social and political structures regionally.
Keywords: maize, archaeogenomics, ancient DNA, agriculture, domestication
Abstract
Maize (Zea mays ssp. mays) domestication began in southwestern Mexico ∼9,000 calendar years before present (cal. BP) and humans dispersed this important grain to South America by at least 7,000 cal. BP as a partial domesticate. South America served as a secondary improvement center where the domestication syndrome became fixed and new lineages emerged in parallel with similar processes in Mesoamerica. Later, Indigenous cultivators carried a second major wave of maize southward from Mesoamerica, but it has been unclear until now whether the deeply divergent maize lineages underwent any subsequent gene flow between these regions. Here we report ancient maize genomes (2,300–1,900 cal. BP) from El Gigante rock shelter, Honduras, that are closely related to ancient and modern maize from South America. Our findings suggest that the second wave of maize brought into South America hybridized with long-established landraces from the first wave, and that some of the resulting newly admixed lineages were then reintroduced to Central America. Direct radiocarbon dates and cob morphological data from the rock shelter suggest that more productive maize varieties developed between 4,300 and 2,500 cal. BP. We hypothesize that the influx of maize from South America into Central America may have been an important source of genetic diversity as maize was becoming a staple grain in Central and Mesoamerica.
Modern maize (Zea mays ssp. mays) is the world’s most productive staple crop (1) with over 1,100 million tons grown in 2018 (https://knoema.com/). Molecular data indicate that maize evolved from the annual grass teosinte (Zea mays ssp. parviglumus, hereafter parviglumus) in southwestern Mexico, and that the first steps toward domestication occurred ∼9,000 calendar years before present (cal. BP) (2, 3). Starch grain and phytolith data from archeological sites in the Balsas region of southwestern Mexico confirm the early use of maize by ∼8,700 cal. BP (4). Archaeobotanical evidence also indicates dispersal of maize out of the Balsas through Central America by ∼7,500 cal. BP (5) and into South America by ∼7,000 cal. BP (6, 7) and ultimately into North America starting around 4,000 cal. BP (8).
Global dispersal of this productive crop beyond the Americas started with European contact with Native Americans in the 15th and 16th centuries (9). In addition to thousands of landraces developed by Indigenous cultivators in the Americas (10–15), experimentation worldwide has led to a staggering array of morphological diversity and adaptations to a wide range of geographic and climatic conditions. This diversity results from human-mediated selection, reproductive isolation from parviglumus (and related Zea mays ssp. mexicana, hereafter mexicana), secondary improvement, and the potential reintroduction of new varieties back to original homelands. A contemporary concern is the backflow of transgenic or commercial hybrid maize varieties to the Mexican heartland because gene flow with extant landraces can result in the loss of genetic diversity and cultural knowledge (10, 16). However, precolonial backflow of divergent maize varieties into Central and Mesoamerica during the last 9,000 y remains understudied, and could have ramifications for the history of maize as a staple in the region.
Morphological evidence from ancient maize found in archaeological sites combined with DNA data confirms a complex and extended domestication history. The earliest maize cobs found in the highlands of Oaxaca, Mexico dating to 6,250 cal. BP are small, and have two nondisarticulating vertical rows of alternating seeds (17, 18) indicating that Indigenous cultivators were controlling plant reproduction. Early four-row cobs from Mexico’s Tehuacán Valley (5,300-4,950 cal. BP) are also small, and show comparable evidence for nondisarticulating seeds consistent with domestication (19, 20). Experimental work has shown that the lower atmospheric CO2 and temperatures in the Late Pleistocene and Early Holocene may have favored phenotypic expression of maizelike inflorescence and seed architecture—nonbranching stalk architecture and naked grains—which could have been promoted and fixed by human-mediated selection (21, 22). Ancient DNA data from Tehuacán cobs (San Marcos Cave) dating between 5,300 and 4,950 cal. BP have alleles comparable to modern maize for inflorescence and seed architecture (td1 – tassel dwarf1; tb1 – teosinte branced1, ba1 – barren stalk1), circadian clock and flowering time (zmg1), and glycogen biosynthesis (bt2 – brittle endosperm2) (23, 24). However, some alleles controlling ear shattering and starch biosynthesis (zag1 – MADS-box gene; su1 – sugary 1, and wx1 – waxy 1) were more teosintelike at that point—four millennia after the onset of domestication. Introgression with mexicana favored adaptation to drier and cooler conditions in the Mexican highlands, and most modern landraces from highland Mexico and Central America carry strong signals of postdomestication mexicana admixture (13, 25). Continued introgression within the natural range of parviglumus and mexicana may also explain the gradual changes in cob size evident in the Tehuacán Valley (19).
The initial first wave of maize dispersal (7,500–7,000 cal. BP) (5) through Central and South America likely occurred when maize was partially domesticated, and before the domestication syndrome—the suite of characters setting a domesticated species apart from its wild counterpart—was fixed (26). Multiple waves of dispersal brought maize out of southwestern Mexico into South America (26, 27), and may have episodically increased admixture and diversity. There is also evidence that deeply structured maize lineages in South America underwent independent fixation of the domestication syndrome and secondary improvement outside the range of parviglumus and mexicana (26). Reduced crop-wild gene flow outside the range of parviglumis and mexicana also enhanced selection for increased cob and seed size (6), and was instrumental in the development of more productive staple grain varieties and greater consumption in Central America starting after 4,700 cal. BP (28, 29). Furthermore, adaptations in the US Southwest—outside the domestication center—resulted in the shorter growing season varieties with earlier flowering times required for dispersal through more temperate parts of North America after 2,000 cal. BP (30). In total, it is clear from the available data that maize diversity and biogeography is complex, and resulted from multiple episodes of human-mediated selection and dispersal throughout the Americas.
Research has focused on the outward dispersal of maize from the original domestication center in southwestern Mexico. However, crop movements were complex, and the archaeological record shows clear evidence of two-way movements of plants and people lasting millennia between Central and South America. It is reasonable to suspect that maize, the most widespread crop species of the precolonial Americas, traveled back toward the domestication center in the hands of skilled farmers as part of this complex history. Here, we sequenced maize genomes from three archaeological samples from El Gigante rock shelter in western Honduras dating to between 2,300 and 1,900 cal. BP and compared these data to published modern landraces of maize and archaeological samples from North, Central, and South America. We use these genomes as a temporal anchor to test the hypotheses that humans moved maize from South America into Central America. In this scenario, the reintroduced germplasm may have been impactful for the development of highly productive varieties. Isotopic evidence from Central America demonstrates substantial maize consumption as a staple grain beginning between 4,700 and 4,000 cal. BP (29). Finally, we use morphological comparisons within the El Gigante maize assemblage to help constrain the timing of this gene flow.
El Gigante and Maize Samples
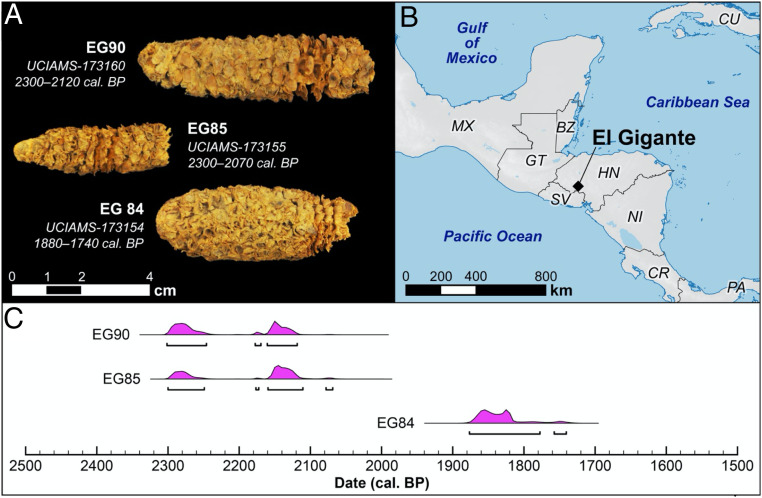
El Gigante rock shelter is located in the highlands of western Honduras (88.06° W, 14.22° N; 1,300 masl; Fig. 1) outside the contemporary range of parviglumus and mexicana (28). The rock shelter is 42 m wide, 17 m deep, 12 m high and was used episodically during the last 11,000 y (31). Dry conditions inside the dripline promoted the preservation of desiccated plant material, including locally available wild plants, tree crops (e.g., avocado [Persea americana Mill.]; hog plum [Spondias sp.]; custard apple [Annona sp.]) and field cultigens (e.g., maize, beans [Phaseolus spp.], and squash [Cucurbita spp.]). Eight phases of occupation were identified based on a Bayesian chronological model of 89 radiocarbon dates through the sequence (28). Wild plant foods dominate the botanical assemblages in the earliest four phases (11,010-7,430 cal. BP; Early and Late Esperanza, Early and Middle Marcala Phases). Squash (Cucurbita spp.) first appears in the Middle Marcala Phase (7,600-7,430 cal. BP) just prior to a long hiatus in rock-shelter use (7,430-4,340 cal. BP). The earliest maize in the sequence dates just after this hiatus in the Late Marcala phase (4,340 and 4,020 cal. BP; Stratum IId-IIa) in a botanical assemblage dominated by wild plant foods. No maize has been directly dated in the subsequent, poorly defined Early Estanzuela Phase deposits (3,390–2,780 cal. BP), but returns as a dominant feature in the Late Estanzuela Phase (2,350 and 1,820 cal. BP; Stratum Id-Ia) found in association with beans and squash.
Fig. 1.
Maize cobs from El Gigante rock shelter (HN) with genomewide data. (A) Photographs of cobs showing morphological characteristics. (B) Map of Central America indicating location of El Gigante rock shelter. (C) Radiocarbon date distributions for the three maize cobs with genomewide data.
Over 10,000 carbonized and uncarbonized maize specimens (cobs, kernels, stalks, leaves) occur in the assemblage. The earliest directly dated maize cobs have 12–14 rows and are relatively slender (conical to lanceolate in shape). There is substantial morphological overlap in cob width, kernel rows, and rachis segment length between the earliest and latest cobs in the assemblage dating to between 2,350 and 1,180 cal. BP. Seed row number is equivalent in the earliest and latest cobs and permutation tests show no statistical differences in cob diameter (P = 0.0693), rachis diameter (P = 0.1006), and cupule length (P = 0.96104) [SI Appendix, Fig. S1; data from ref. (28)]. Temporal differences in cupule width (P = 0.00978) and cupule wing width (P = 0.04228) differ significantly from expectations of random chance. This shift suggests these attributes increased on average between the earlier and later assemblage, suggesting selection for increasing seed size. Our permutation tests are consistent with a robustly domesticated variety of maize developing or arriving in southeastern Mesoamerica by at least 4,300 cal. BP (28) at a time when it was becoming a dietary staple (29).
We attempted to extract DNA from all of the directly dated Late Marcala maize cobs in the assemblage (4,340 and 4,020 cal. BP; n = 20) but preservation was unsuitable for genomic sequencing. We also attempted to extract DNA from 10 Late Estanzuela Phase (2,350–1,900 cal. BP) maize cobs and three specimens produced endogenous DNA sufficient for genomewide sequencing (Table 1).
Table 1.
Maize cobs with endogenous DNA sufficient for genomewide sequencing
| ID | Provenience | Date (cal. BP) | Rows | Diameter (mm) | Shape |
| EG84 | Unit 2, Level 6, Stratum Ib/Ic | 1870–1740 | 16 | 17.55 | Lanceolate |
| EG85 | Unit 2, Level 6, Stratum Ib/Ic | 2300–2070 | 14 | 15.10 | Conical |
| EG90 | Unit 18, Level 14, Stratum Ib2 | 2300–2120 | 10 | 20.50 | Lanceolate |
Genomics
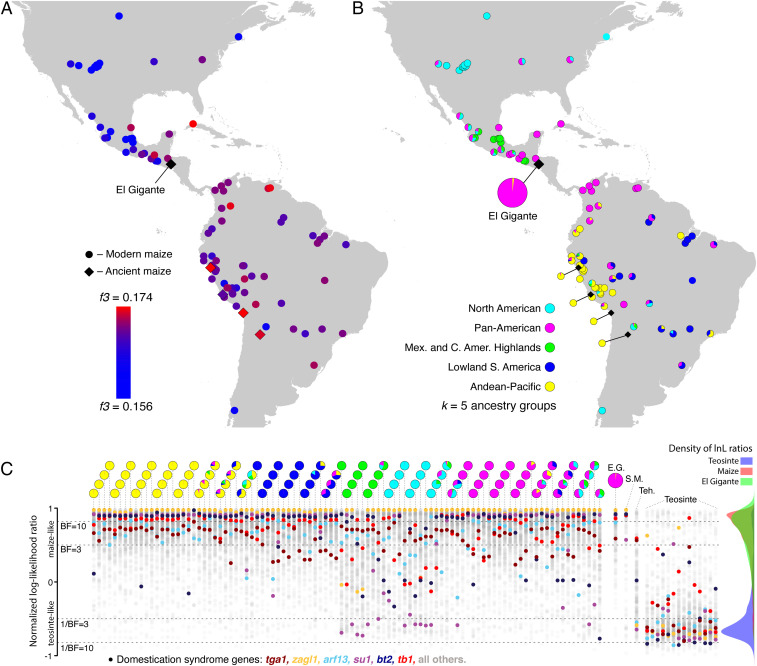
We compared the Late Estanzuela Phase maize genomes with 120 previously published modern (n = 109) and ancient (n = 11) maize genomes (SI Appendix). Model-based clustering revealed that the ∼2,000 cal. BP El Gigante maize is most similar to the “Pan-American” cluster described previously (26) (Fig. 2B). This group is widespread in Mexico, Central America, and South America, with some presence in North America north of Mexico. These genomes carry a surplus of teosinte ancestry compared with other major maize lineages in North and South America, suggesting that this lineage was dispersed away from the domestication center later in time after additional ancient crop–wild gene flow (26). Although model-based clustering primarily places El Gigante maize in the Pan-American cluster, it also identified a small but consistent component of South American ancestry in the El Gigante genomes (Fig. 2B and SI Appendix). We used outgroup-f3 statistics to further interrogate the nearest genetic neighbors to El Gigante maize. We found that its genome most closely resembled modern landraces in South America with predominantly Pan-American ancestry, as well as archaeological genomes from South America (Fig. 2A). One Cuban genome from the maize Hapmap2 project (32) also matched closely. However, this variety (Cuban Flint) was introduced from Argentina in the early 20th century (33). The outgroup-f3 results therefore reinforce a consistent link to South America.
Fig. 2.
Genetic affinities and domestication status of archaeological El Gigante maize. (A) Outgroup-f3 statistics in the form f3(Tripsacum; X, El Gigante) with all other maize samples in position X, showing that maize samples sharing the most drift with El Gigante maize are modern and ancient genomes from South America. The sample in Cuba with a high f3 value is from a HapMap2 landrace with known origins in Argentina. (B) Ancestry proportions of modern and ancient maize estimated via model-based clustering. (C) Estimation of domestication status in El Gigante and other maize via AIMs located near and within domestication syndrome genes. The y axis displays normalized log-likelihood of a gene being drawn from a maizelike (1) vs. teosintelike (−1) reference panel, with significance thresholds marked where Bayes factors (BF) (and 1/BF) ≥3 and ≥10. Each column of dots shows a single genome with up to 278 individual gene lnL ratios, with ancestry proportions corresponding to B above each column. In El Gigante maize (E.G.), domestication genes overlap the modern maize reference panel and deviate strongly from the teosinte panel, and all six specifically analyzed domestication genes are maizelike with at least BF > 3. In contrast, Middle Holocene Tehuacán Valley maize (Teh.) carries a mixture of maizelike and teosintelike variants as previously reported (23). Mid-Holocene San Marcos maize (S.M.) was also previously shown to be a partial domesticate (24), although more maizelike than the Tehuacán specimen (26), a finding reinforced here.
Because maize was well-established in several regions of the Americas by this time, the El Gigante maize at ∼2,000 cal. BP was likely fully domesticated. Therefore, we predicted domestication syndrome genes to be more maizelike than either 1) teosintelike or 2) a combination of maize- and teosintelike signifying partial domestication. Adapting an approach based on ancestry informative markers [AIMs; ref. (23) and SI Appendix], we quantified the proportion of teosintelike character states near domestication genes. The genewise proportion of teosintelike variants in the El Gigante domestication genes overlapped modern maize (Wilcoxon rank-sum test P = 0.75; Welch’s t test P = 0.63), and deviated strongly from modern teosinte (both tests P < 2 × 10−16).
We also used a likelihood-based strategy to assess whether individual genes in El Gigante maize were more likely drawn from a population resembling modern maize or modern teosinte (Fig. 2C). Under this analysis, key domestication genes analyzed in previous aDNA studies—bt2, tga1, tb1, su1, zagl1, and arf13—are all robustly maizelike in the El Gigante genomes (Bayes factors 3.84–78.93, signifying support ranging from “substantial” to “very strong” (34) (SI Appendix). In total, we found strong support that Late Estanzuela El Gigante maize was robustly domesticated.
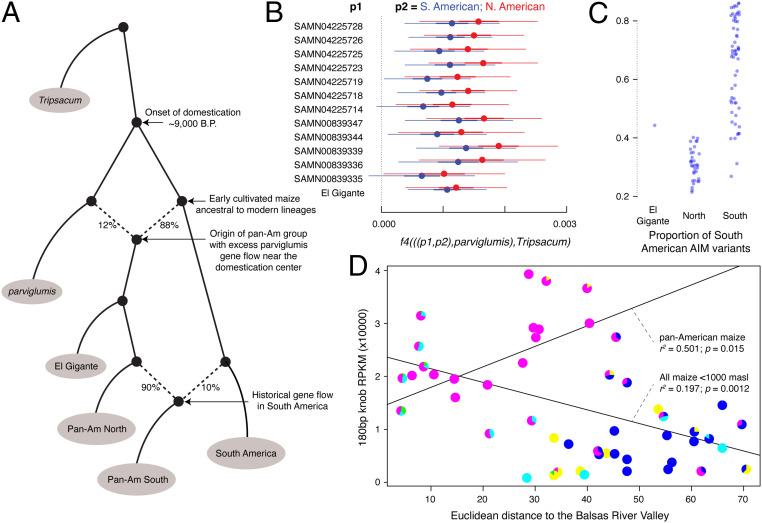
We used admixture graph fitting to refine the relationships between maize populations (Fig. 3A). This method found support (|maxZ| = 2.7) for a model where El Gigante maize falls just outside the range of variation of modern Pan-American maize but shares the historic parviglumis gene flow that characterizes the Pan-American cluster.
Fig. 3.
Population relationships and genome size characteristics. (A) Admixture graph with a good fit to the genomic data, showing El Gigante maize as an early branch of the Pan-American cluster carrying excess parviglumis ancestry. South American members of the Pan-American population carry excess ancestry from earlier-dispersed South American lineages, revealing hybridization in South America. (B) f4-statistics showing that all individual Pan-American genomes and the El Gigante maize carry excess parviglumis ancestry compared with South American and North American lineages. Errors bars at 1 and 3 SEs computed using a block jackknife in 5-Mb blocks. (C) Proportion of characteristic South American lineage alleles at AIMs among geographically southern (samples physically originating in South America) and northern (originating in North and Central America) maize genomes, showing that El Gigante maize is the most South American-like of northern maize. (D) 180 bp heterochromatic knob frequency (RPKM) as a proxy for genome size, compared with distance from the domestication center. The two regression lines show 1) the correlation between distance and genome size in all samples—a general trend to genome contraction with distance—and 2) the reverse trend in samples with ≥90% Pan-American ancestry.
On the basis of f4 statistics, all individual modern Pan-American maize genomes and the El Gigante maize carry this signal for excess teosinte ancestry compared with South American lineages (Fig. 3B and SI Appendix). This reinforces previous findings that Pan-American maize underwent additional crop–wild gene flow in the domestication center after the first wave of maize was carried into South America (26). Although our analysis above demonstrates that El Gigante maize carries a full suite of maizelike alleles at domestication loci, this excess teosinte ancestry is consistently present at low levels on a genomewide basis in El Gigante maize and modern Pan-American genomes.
We also found evidence for introgression from first-wave South American maize into southern members of the Pan-American group, which occurred following the second wave of maize dispersal into South America. This finding suggests hybridization in South American maize fields between established and introduced lineages. This is not surprising in light of millennia of sympatric cultivation. Indigenous farmers apparently maintained the lineage identity of first-wave and second-wave maize to a significant degree in South America, while also integrating admixed varieties into their cropping systems.
We further explored the link between El Gigante maize and South America by identifying a set of AIMs that reliably distinguish between genomes with primarily South American ancestry and all other maize, and quantifying the proportion of SA-like ancestry in each sample (SI Appendix). On this basis, El Gigante maize has the higher proportion of SA-like alleles than any maize originating from north of South America (Fig. 3C; EG maize is 2.53 SD from the mean of the normally distributed southern set; P = 0.0057).
Finally, genome size is extremely variable in maize. We estimated relative genome size among modern maize using a previously established proxy in the heterochromatic knob fraction of the genome (32, 35) (Materials and Methods and Dataset S1). As shown previously (36), we observed that genome size in maize is significantly correlated with elevation in Central and South American highlands (linear model r2 = 0.11, P = 0.0012), and that maize genomes are significantly smaller than teosinte genomes on average (32) (Welch’s t test P = 2.28 × 10−4; Wilcoxon rank-sum test P = 2.24 × 10−4)—albeit with substantially overlapping ranges. Genome size also decreases significantly with distance from the domestication center (linear model r2 = 0.122, P = 6.06 × 10−4), and a multivariate linear model including elevation and distance substantially increases predictive power (multiple r2 = 0.281, P = 3.59 × 10−7). Distance to the domestication center correlates strongly with both a decrease in genomic heterozygosity (25) and an increase in the genomic mutation load (26). Thus, we find that in addition to losing diversity and accumulation of deleterious variants during dispersal, the genome tends to physically contract down the dispersal gradient (Fig. 3D).
The members of the Pan-American group defy this broader pattern. Among low-elevation (<1,000 m) samples with ≥90% Pan-American ancestry on the basis of model-based clustering, genomes near the domestication center are significantly smaller than those in South America (linear model r2 = 0.501, P = 0.014; Fig. 3D). In fact, the largest maize genomes we observe are South American samples with high proportions of Pan-American ancestry. By adding the proportion of Pan-American ancestry as an independent variable alongside distance to the domestication center and elevation, we free up this contradictory relationship to explain additional genome size variance across maize (multivariate linear model multiple r2 = 0.437, P = 5.23 × 10−11).
Discussion
Our genomic analyses establish a clear link between El Gigante maize and modern and ancient maize from South America. On the basis of links to southern Pan-American maize alone, this could be a result of a simple one-way dispersal of the Pan-American lineage where El Gigante maize acts as an outgroup to southern Pan-American genomes. However, this pattern does not explain the AIM-based links between El Gigante and earlier-dispersed South American lineages, nor the outgroup-f3 results tying El Gigante to ancient genomes in South America. Instead, we hypothesize that the South American signal in El Gigante maize provides evidence for Indigenous farmers carrying or dispersing maize northward through the Isthmus of Panama prior to ∼2,000 y ago.
Admixture graphs and f4 statistics reinforce multiple waves of dispersal southward out of the center of domestication. First, by around 7,000 cal. BP, an initial wave of semidomesticated maize was carried southward, where it likely underwent secondary improvement in the southwestern Amazon before diversifying across the continent (26, 37). Later, a second population of maize was transported southward, carrying the signal for excess gene flow with teosinte in the domestication center.
Our analyses here are consistent with this second wave of dispersal reaching into parts of South America where maize was already being grown, resulting in hybridization and experimentation in crop fields. El Gigante maize carries the distinct signal of this South American influence, suggesting that farmers transported hybridized landraces back northward into Central America. In total, the Pan-American lineage may reflect the legacy of the second wave of dispersal southward combined with experimental hybridization with first-wave South American landraces upon arrival.
Genome size dynamics are complex and incompletely understood in maize. Previous work has linked genome size contraction at high elevations with selection on flowering time traits linked with cell dynamics (36), and demonstrated that genome size is greatest in the tropics and lower in temperate zones (32). We extend the latter conclusion to show that genome size has generally decreased with distance from the domestication center, suggesting a contraction of the genome down dispersal gradients during adaptation to new environments. Nonetheless, some of the largest genomes in domesticated maize are observed in members of the Pan-American lineage from northern regions of South America.
The correlation between genome size and distance to the domestication center in the Pan-American group runs counter to the broad pattern in maize. This pattern may reflect a south-to-north dispersal as the ancestors to El Gigante maize—and possibly other landraces in Central America—were dispersed by farmers. This genome size gradient thus may represent the imprint of an ancestral dispersal vector consistent with our other analyses, but more research is necessary to further explore this pattern. Alternatively, it is possible that genome size, or the heterochromatic knob fraction, expanded for unknown reasons during the dispersal history of the Pan-American lineage.
The transit of ancestral Pan-American maize into South America would have conferred reproductive isolation from wild teosinte, perhaps for the first prolonged period in the history of this lineage. The isolation from crop–wild gene flow likely would have encouraged the development of robust domesticated phenotypes and true-breeding landraces. It is also possible that gene flow from first-wave South American lineages boosted Pan-American maize diversity, leading to increased performance through hybrid vigor. In total, ancient interbreeding between ancestral first-wave South American maize and second-wave Pan-American maize could have been responsible for the emergence of robust new forms that were carried northward, as well as spreading with Indigenous farmers throughout South America.
Future work will be required to determine when the backflow of improved South American maize varieties occurred. Ancient DNA is poorly preserved in the early cobs in the El Gigante sequence (4,340–4020 cal. BP), and other cobs dating to the Middle Holocene are rare in this region. However, the morphology of the early cobs from El Gigante are comparable in size and row number to the later cobs [∼2,300–1,900 cal. BP cobs; ref. (28)]. The appearance of robustly domesticated cobs also corresponds well with dietary stable isotope data indicating that maize was becoming a staple grain after ∼4,700 cal. BP (29), and with pollen records revealing maize cultivation in Honduras around the same time (38). The backflow of improved varieties from South America may have occurred by this time and contributed to the development of more productive staple grain varieties. Ultimately, the influx of improved varieties and resulting diversification of maize might have contributed to the growth and aggregation of human populations, helping pave the way for the formation of more complex social and political structures regionally.
Materials and Methods
Complete details of genomic methods and radiocarbon dating used in this study are provided in SI Appendix. Briefly, maize samples were prepared in the dedicated ancient DNA clean laboratory facilities at The Pennsylvania State University Anthropology Department, and the Smithsonian Institution’s Museum Support Center. Standard protocols to prevent and detect contamination were utilized (39), including strict workflow procedures, frequent cleaning with bleach and ethanol, use of complete personal protective equipment, and the preparation and sequencing of negative control reactions. We extracted DNA, prepared sequencing libraries, and screened samples following established protocols for highly degraded ancient DNA (26, 40–42), identifying EG84, EG85, and EG90 as suitable specimens for genomic sequencing. All sequencing was carried out by Admera Health. Genomic data were processed and variants were called exactly as previously described (26), and analytical methods are described fully in SI Appendix.
Each maize cob was subsampled for accelerator mass spectrometer (AMS) 14C dating at in the Human Paleoecology and Isotope Geochemistry Laboratory at The Pennsylvania State University. Full details for AMS dating and calibration are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Instituto Hondureño de Antropología e Historia for granting us permission to conduct this research. We thank Keith Prufer, Nathan Wales, Yoshi Maezumi, and Dolores Piperno for helpful feedback on the manuscript. Funding for this work was provided by grants from the NSF (Archaeology Program, BCS-100343 [K.H.], BCS-1757375 [D.J.K.], BCS-1757383 [A.V], BCS-1757374 [H.B.T.]; Archaeometry Program, BCS-1460367 [D.J.K.], funding from the Pennsylvania State University [D.J.K.], and funding from the Smithsonian Institution [L.K.]). A.B. was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001595), the UK Medical Research Council (FC001595), and the Wellcome Trust (FC001595). We thank Laurie Eccles in the Human Paleoecology and Isotope Geochemistry Laboratory at The Pennsylvania State University for assistance preparing samples for AMS radiocarbon dating. Computations performed for this paper were conducted on the Smithsonian High Performance Cluster, Smithsonian Institution: https://doi.org/10.25572/SIHPC. Portions of the laboratory work were conducted in and with the support of the Laboratories for Analytical Biology (L.A.B.) facilities of the National Museum of Natural History.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.L.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015560117/-/DCSupplemental.
Data Availability.
Raw sequencing data for El Gigante maize specimens have been deposited in the NCBI Sequence Read Archive under BioProject PRJNA636370. Custom scripts, single nucleotide polymorphism (SNP) calls, and SMALT databases are available on Dryad (DOI: 10.5061/dryad.xsj3tx9dc).
References
- 1.Ranum P., Peña-Rosas J. P., Garcia-Casal M. N., Global maize production, utilization, and consumption. Ann. N. Y. Acad. Sci. 1312, 105–112 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Doebley J., The genetics of maize evolution. Annu. Rev. Genet. 38, 37–59 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Matsuoka Y., et al. , A single domestication for maize shown by multilocus microsatellite genotyping. Proc. Natl. Acad. Sci. U.S.A. 99, 6080–6084 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piperno D. R., Ranere A. J., Holst I., Iriarte J., Dickau R., Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc. Natl. Acad. Sci. U.S.A. 106, 5019–5024 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickau R., Ranere A. J., Cooke R. G., Starch grain evidence for the preceramic dispersals of maize and root crops into tropical dry and humid forests of Panama. Proc. Natl. Acad. Sci. U.S.A. 104, 3651–3656 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grobman A., et al. , Preceramic maize from Paredones and Huaca Prieta, Peru. Proc. Natl. Acad. Sci. U.S.A. 109, 1755–1759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombardo U., et al. , Early Holocene crop cultivation and landscape modification in Amazonia. Nature 581, 190–193 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merrill W. L., et al. , The diffusion of maize to the southwestern United States and its impact. Proc. Natl. Acad. Sci. U.S.A. 106, 21019–21026 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebourg C., et al. , Maize introduction into Europe: The history reviewed in the light of molecular data. Theor. Appl. Genet. 106, 895–903 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Perales H. R., Benz B. F., Brush S. B., Maize diversity and ethnolinguistic diversity in Chiapas, Mexico. Proc. Natl. Acad. Sci. U.S.A. 102, 949–954 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruiz Corral J. A., et al. , Climatic adaptation and ecological descriptors of 42 Mexican maize races. Crop Sci. 48, 1502–1512 (2008). [Google Scholar]
- 12.Hufford M. B., Martínez-Meyer E., Gaut B. S., Eguiarte L. E., Tenaillon M. I., Inferences from the historical distribution of wild and domesticated maize provide ecological and evolutionary insight. PLoS One 7, e47659 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Heerwaarden J., et al. , Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl. Acad. Sci. U.S.A. 108, 1088–1092 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Heerwaarden J., Hufford M. B., Ross-Ibarra J., Historical genomics of North American maize. Proc. Natl. Acad. Sci. U.S.A. 109, 12420–12425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa F. M., Silva N. C. de A., Ogliari J. B., Maize diversity in southern Brazil: Indication of a microcenter of Zea mays L. Genet. Resour. Crop Evol. 64, 681–700 (2017). [Google Scholar]
- 16.Abbo S., Rubin B., Transgenic crops: A cautionary tale. Science 287, 1927–1928 (2000). [PubMed] [Google Scholar]
- 17.Benz B. F., Archaeological evidence of teosinte domestication from Guilá Naquitz, Oaxaca. Proc. Natl. Acad. Sci. U.S.A. 98, 2104–2106 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piperno D. R., Flannery K. V., The earliest archaeological maize (Zea mays L.) from highland Mexico: New accelerator mass spectrometry dates and their implications. Proc. Natl. Acad. Sci. U.S.A. 98, 2101–2103 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benz B., Cheng L., Leavitt S. W., Eastoe C., “El Riego and early maize agriculture evolution” in Histories of Maize, Tykot R. H., Benz B., Staller J., Eds. (Elsevier, 2006), pp. 73–80. [Google Scholar]
- 20.Benz B. F., Iltis H. H., Studies in archaeological maize I: The “wild” maize from San Marcos Cave reexamined. Am. Antiq. 55, 500–511 (1990). [Google Scholar]
- 21.Piperno D. R., Assessing elements of an extended evolutionary synthesis for plant domestication and agricultural origin research. Proc. Natl. Acad. Sci. U.S.A. 114, 6429–6437 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piperno D. R., Holst I., Moreno J. E., Winter K., Experimenting with domestication: Understanding macro- and micro-phenotypes and developmental plasticity in teosinte in its ancestral pleistocene and early holocene environments. J. Archaeol. Sci. 108, 104970 (2019). [Google Scholar]
- 23.Ramos-Madrigal J., et al. , Genome sequence of a 5,310-year-old maize cob provides insights into the early stages of maize domestication. Curr. Biol. 26, 3195–3201 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Vallebueno-Estrada M., et al. , The earliest maize from San Marcos Tehuacán is a partial domesticate with genomic evidence of inbreeding. Proc. Natl. Acad. Sci. U.S.A. 113, 14151–14156 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., et al. , The interplay of demography and selection during maize domestication and expansion. Genome Biol. 18, 215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kistler L., et al. , Multiproxy evidence highlights a complex evolutionary legacy of maize in South America. Science 362, 1309–1313 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Freitas F. O., Bendel G., Allaby R. G., Brown T. A., DNA from primitive maize landraces and archaeological remains: Implications for the domestication of maize and its expansion into South America. J. Archaeol. Sci. 30, 901–908 (2003). [Google Scholar]
- 28.Kennett D. J., et al. , High-precision chronology for Central American maize diversification from El Gigante rockshelter, Honduras. Proc. Natl. Acad. Sci. U.S.A. 114, 9026–9031 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennett D. J., et al. , Early isotopic evidence for maize as a staple grain in the Americas. Sci. Adv. 6, eaba3245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swarts K., et al. , Genomic estimation of complex traits reveals ancient maize adaptation to temperate North America. Science 357, 512–515 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Scheffler T. E., Hirth K. G., Hasemann G., The El Gigante rockshelter: Preliminary observations on an early to late Holocene occupation in southern Honduras. Lat. Am. Antiq. 23, 597–610 (2012). [Google Scholar]
- 32.Chia J.-M., et al. , Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 44, 803–807 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Brown W. L., Goodman M. M., “Races of corn” in Corn and Corn Improvement, Sprague G. F., Fuccillo D. A., Perelman L. S., Stelly M., Eds. (American Society of Agronomy, Inc., 1977), pp. 49–88. [Google Scholar]
- 34.Jarosz A. F., Wiley J., What are the odds? A practical guide to computing and reporting Bayes factors. J. Probl. Solving 7, 2–9 (2014). [Google Scholar]
- 35.Jian Y., et al. , Maize (Zea mays L.) genome size indicated by 180-bp knob abundance is associated with flowering time. Sci. Rep. 7, 5954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilinski P., et al. , Parallel altitudinal clines reveal trends in adaptive evolution of genome size in Zea mays. PLoS Genet. 14, e1007162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maezumi S. Y., et al. , The legacy of 4,500 years of polyculture agroforestry in the eastern Amazon. Nat. Plants 4, 540–547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rue D. J., Early agriculture and early Postclassic Maya occupation in western Honduras. Nature 326, 285–286 (1987). [Google Scholar]
- 39.Fulton T. L., Shapiro B., “Setting up an ancient DNA laboratory” in Ancient DNA, Methods in Molecular Biology, Shapiro B., et al., Eds. (Springer, New York, 2019), pp. 1–13. [DOI] [PubMed] [Google Scholar]
- 40.Wales N., Kistler L., “Extraction of ancient DNA from plant remains” in Ancient DNA, Methods in Molecular Biology, Shapiro B., et al., Eds. (Springer, New York, 2019), pp. 45–55. [DOI] [PubMed] [Google Scholar]
- 41.Carøe C., et al. , Single‐tube library preparation for degraded DNA. Methods Ecol. Evol. 9, 410–419 (2018). [Google Scholar]
- 42.Mak S. S. T., et al. , Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience 6, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data for El Gigante maize specimens have been deposited in the NCBI Sequence Read Archive under BioProject PRJNA636370. Custom scripts, single nucleotide polymorphism (SNP) calls, and SMALT databases are available on Dryad (DOI: 10.5061/dryad.xsj3tx9dc).