Significance
Information processing in the central nervous system relies on chemical synapses where neurotransmitters such as the inhibitory neurotransmitter γ-aminobutyric acid (GABA) are released at presynaptic nerve endings. GABA is synthesized by glutamic acid decarboxylase (GAD), an enzyme requiring pyridoxal 5'-phosphate (PLP or vitamin B6) as cofactor, the latter being synthesized by pyridoxal kinase (PDXK). Here, we show that the antimalarial drug artemisinin inhibits PDXK and describe the structural basis of this inhibition. The decrease in PLP production reduces the amount of GABA being produced, which, in turn, impacts the efficacy of GABAergic transmission. This study combined with our previous data sheds light on how artemisinins can influence inhibitory synaptic transmissions both presynaptically, as described here, and postsynaptically.
Keywords: artemisinins, pyridoxal kinase, pyridoxal phosphate (PLP), GABA biosynthesis, inhibitory neurotransmission
Abstract
The antimalarial artemisinins have also been implicated in the regulation of various cellular pathways including immunomodulation of cancers and regulation of pancreatic cell signaling in mammals. Despite their widespread application, the cellular specificities and molecular mechanisms of target recognition by artemisinins remain poorly characterized. We recently demonstrated how these drugs modulate inhibitory postsynaptic signaling by direct binding to the postsynaptic scaffolding protein gephyrin. Here, we report the crystal structure of the central metabolic enzyme pyridoxal kinase (PDXK), which catalyzes the production of the active form of vitamin B6 (also known as pyridoxal 5′-phosphate [PLP]), in complex with artesunate at 2.4-Å resolution. Partially overlapping binding of artemisinins with the substrate pyridoxal inhibits PLP biosynthesis as demonstrated by kinetic measurements. Electrophysiological recordings from hippocampal slices and activity measurements of glutamic acid decarboxylase (GAD), a PLP-dependent enzyme synthesizing the neurotransmitter γ-aminobutyric acid (GABA), define how artemisinins also interfere presynaptically with GABAergic signaling. Our data provide a comprehensive picture of artemisinin-induced effects on inhibitory signaling in the brain.
Pyridoxal 5′-phosphate (PLP) is the active form of vitamin B6. In humans, PLP biosynthesis is catalyzed by pyridoxal kinase (PDXK), a member of the ribokinase superfamily. PDXK utilizes inactive forms of vitamin B6 (pyridoxal [PL], pyridoxine, and pyridoxamine) and ATP as substrates, producing PLP along with the byproduct ADP. The corresponding reaction proceeds via a random substrate addition reaction mechanism (1) in which PLP biosynthesis takes place by transferring the γ-phosphate of ATP to the 5′-OH group of the B6 vitamers, in a process assisted by divalent metal ions such as Zn2+ and Mg2+ (2) (Fig. 1A). PLP serves as the essential active site component for more than 160 distinct human enzymatic activities (3) catalyzing crucial cellular processes such as detoxification reactions and multiple metabolic processes including amino acid, carbohydrate, and lipid metabolism. PLP-dependent enzymes also participate in neurotransmitter biosynthesis including the inhibitory neurotransmitters γ-aminobutyric acid (GABA) and glycine (4–6), which are synthesized by glutamic acid decarboxylase (GAD) and serine hydroxymethyl transferase (SHMT), respectively. Vitamin B6 deficiency has been implicated in multiple neurological, psychiatric, and internal disorders possibly including even diabetes, cancer, and autism (3), thus underpinning the importance of a finely tuned PLP biosynthesis.
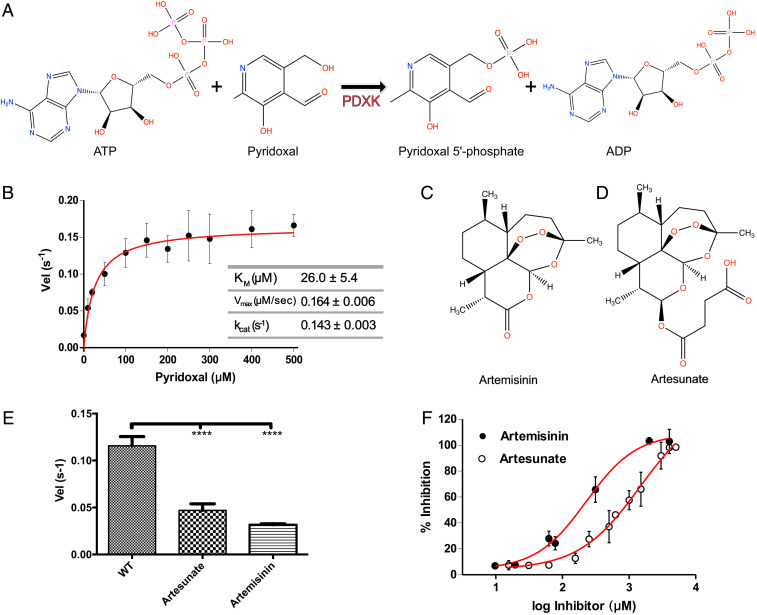
Fig. 1.
Biochemical basis of PDXK inhibition by artemisinins. (A) Schematic representation of the reaction catalyzed by PDXK. (B) Michaelis–Menten curve derived for the enzymatic activity of recombinantly purified PDXK. (C and D) Chemical structures of artemisinin (C) and artesunate (D). (E) Enzymatic activity of wild-type PDXK (WT-PDXK) in the absence and presence of artemisinin derivatives at a concentration of 1.5 mM (artesunate) and 156 µM (artemisinin), respectively. Data are presented as mean ± SEM (P values are *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001) (one-way ANOVA test). (F) Inhibition curves of PDXK by artemisinin and artesunate used to derive the corresponding IC50 values.
Recently, PDXK was identified as one of the mammalian targets of the antimalarial drug artemisinin (7). Artemisinin-containing plant extracts have been used in traditional Chinese medicine for the treatment of malaria (8). Chemically, these small molecules are sesquiterpene lactones with an unusual endoperoxide bridge. Artemisinin and its semisynthetic derivatives artemether and artesunate (collectively referred to as artemisinins), in combination with quinones such as mefloquine and lumefantrine, nowadays represent the standard drug combinations used to treat malaria caused by Plasmodium falciparum (9). In addition to their antiprotozoan activities, these drugs have also been pharmacologically observed to regulate the activities of a variety of mammalian cellular processes, some of which are deregulated in various types of cancer (10, 11). Recently, it was discovered that artemisinins also modulate the differentiation of pancreatic Tα cells by inducing the transdifferentiation of glucagon-producing Tα cells into insulin-secreting Tβ cells, thus suggesting an antidiabetic activity of artemisinins (7). However, two subsequent studies contradicted this observation, thus questioning the potential clinical application of these compounds in the treatment of diabetes (12, 13).
Until recently, in the absence of a single protein crystal structure in complex with artemisinins (neither a plasmodial nor a mammalian protein), the detailed framework describing the target recognition by these small molecules remained enigmatic. The first molecular insights into artemisinin recognition by a target protein were derived by us from crystal structures of the C-terminal domain of the moonlighting protein gephyrin (GephE) in complex with two artemisinin derivatives, artesunate and artemether (14). Gephyrin is the principal scaffolding protein at inhibitory postsynaptic specializations and also catalyzes the final two steps of the evolutionarily conserved molybdenum cofactor (Moco) biosynthesis (15–17). Structures of the GephE–artemisinin complexes demonstrated that artemisinins specifically target the universal receptor binding pocket of this moonlighting protein, without altering its enzymatic activity, thus inhibiting critical interactions of gephyrin with GABA type A receptors (GABAARs) and glycine receptors (GlyRs). As an important functional consequence, artemisinins modulate inhibitory neurotransmission in a gephyrin-dependent manner. In addition to gephyrin, various proteins were identified as putative targets of artemisinins in pancreatic cells, including the central metabolic enzyme PDXK (7), yet the molecular mechanisms underlying the modulation of these targets by artemisinins remained unknown.
Here, we determined the 2.4-Å resolution crystal structure of mouse pyridoxal kinase (mPDXK) in complex with artesunate, a succinate derivative of artemisinin. The artesunate binding site partially overlaps with the substrate (PL)/product (PLP) binding site, thus suggesting a drug-induced inhibitory effect. Enzymatic activity assays in vitro indeed revealed a significant inhibition of PLP production in the presence of artemisinins with Ki values in the high micromolar range. Electrophysiological recordings and measurements of GABA biosynthesis suggest that artemisinins exert their effect by down-regulating the activity of PLP-dependent enzymes such as GAD. Taken together, our data define the molecular basis for the inhibition of PDXK by artemisinins and their consequences at the presynaptic terminals of inhibitory postsynapses and extend our current understanding of the artemisinin-induced modulation of inhibitory neurotransmission beyond gephyrin.
Results
Artemisinins Inhibit PDXK.
To derive the oligomeric state of recombinantly purified mPDXK, we performed multiangle laser light scattering coupled to size exclusion chromatography (SEC-MALLS). The experiments showed that the protein is a dimer in solution (SI Appendix, Fig. S1), as has been reported for the human and also prokaryotic PDXK homologs (18). Next, we measured the enzymatic activity of mPDXK by directly monitoring PLP production in a photometric assay revealing a KM of 26.0 ± 5.4 µM, a Vmax of 0.1640 ± 0.006 µM/s, and a kcat of 0.1436 ± 0.003 s−1 for the substrate PL in the presence of 1 mM of ATP (Fig. 1B), which is in line with reported KM values (3 to 50 μM) for PDXK (19–25).
To understand the effect of artemisinins on the enzyme, we performed the activity assays in the presence of two artemisinins, the parental compound artemisinin and the succinate derivative artesunate (Fig. 1 C and D). The determination of the turnover rates (velocity [Vel]) displayed a highly significant inhibition in mPDXK activity in the presence of artemisinins with observed reductions to 0.032 ± 0.001 and 0.047 ± 0.007 s−1 for artemisinin and artesunate, respectively. Compared to the turnover rate of the enzyme in the absence of these drugs (0.116 ± 0.01 s−1) (Fig. 1E) this corresponds to an approximately threefold decrease. The enzymatic or turnover velocity, Vel, is defined here as the mean number of product molecules generated by a single enzyme per unit time. Statistical analyses revealed a significant reduction in enzymatic activity in the presence of the artemisinins and via a Dixon plot analysis Ki values of 120 ± 2.4 and 1250 ± 4.7 µM were derived for artemisinin and artesunate, respectively (SI Appendix, Fig. S2). To further characterize the inhibitory properties of artemisinins we also determined the IC50 for both compounds. While artesunate displayed an IC50 value of 1,445 ± 1.4 µM, artemisinin was approximately sixfold more potent with an IC50 of 229 ± 1.3 µM (Fig. 1F). These results were in contrast to what we observed earlier for gephyrin, the postsynaptic target of the artemisinins, where the highest affinity and the higher stabilization of the C-terminal E domain were observed with artesunate.
Owing to a superior solubility of artesunate (approaching ∼10 mM at moderate dimethyl sulfoxide (DMSO) concentrations), all subsequent in vitro experiments with PDXK including our crystallization trials were carried out with artesunate.
Structural Basis for the Inhibition of PDXK by Artemisinins.
To gain insights into the mechanism of inhibition at the atomic level we determined three crystal structures of mPDXK. First, we derived the crystal structure of mPDXK in its apo-state and in complex with ATPγS, one of the substrates of the enzyme. These structures were solved by molecular replacement (MR) with the structure of human apo-PDXK as the search model. The apo and the mPDXK-ATPγS structures were refined in space group C2, containing two dimers in the asymmetric unit, to resolutions of 2.45 and 2.9 Å, respectively (Table 1 and SI Appendix, Fig. S3 A and B). The overall architecture of mouse apo-PDXK shares high structural similarity with its human ortholog (26, 27) as reflected in root-mean-square (rms) deviations of 0.84 Å (Protein Data Bank [PDB]: 2YXT; human apo-PDXK) after superposition of all Cα atoms.
Table 1.
Data collection and refinement statistics
| Data collection | PDXK-apo | PDXK-ATPγS | PDXK-ATPγS–artesunate |
| Space group | C2 | ||
| a, b, c (Å) | 279.13, 53.43, 109.37 | 278.60, 53.02, 109.85 | 279.38, 53.04, 110.15 |
| α, β, γ (°) | 90, 90.00, 90 | 90, 91.75, 90 | 90, 91.64, 90 |
| Resolution (Å) | 47.32 to 2.45 (2.53 to 2.45) | 47.16 to 2.9 (3.03 to 2.9) | 47.20 to 2.4 (2.46 to 2.4) |
| Rsym* | 0.098 (0.75) | 0.10 (0.739) | 0.084 (1.069) |
| Rpim† | 0.093 (0.70) | 0.068 (0.488) | 0.053 (0.704) |
| CC1/2 | 0.993 (0.572) | 0.995 (0.600) | 0.996 (0.541) |
| <I/σI>‡ | 9.1 (1.6) | 9.3 (1.6) | 8.1 (1.1) |
| Completeness (%) | 95.6 (99.7) | 99.7 (99.7) | 99.7 (99.6) |
| Redundancy | 3.2 (3.5) | 3.4 (3.2) | 3.4 (3.1) |
| Reflections used in refinement | 57,174 (5,965) | 36,057 (3,532) | 63,558 (6,235) |
| Refinement | |||
| R-work¶ | 0.2162 (0.3042) | 0.2341 (0.3378) | 0.2088 (0.3271) |
| R-free§ | 0.2596 (0.3237) | 0.2548 (0.3366) | 0.2535 (0.3851) |
| No. of nonhydrogen atoms | 9,715 | 9,862 | 9,890 |
| Macromolecules | 9,411 | 9,606 | 9,445 |
| Ligands | 116 | 232 | 304 |
| Solvent | 188 | 24 | 141 |
| RMS (bonds) | 0.002 | 0.005 | 0.002 |
| RMS (angles) | 0.53 | 0.73 | 0.54 |
| Ramachandran favored (%)# | 96.83 | 96.99 | 96.76 |
| Ramachandran allowed (%) | 3.17 | 3.01 | 3.24 |
| Ramachandran outliers (%) | 0.00 | 0.00 | 0.00 |
| Average B-factor (Å2) | 57.79 | 95.87 | 69.33 |
| Protein | 57.62 | 95.89 | 68.94 |
| Ligands | 74.74 | 95.54 | 84.27 |
| Solvent | 55.93 | 91.85 | 63.08 |
Although the β angle of the apo-structure is 90.00, the crystals display monoclinic and not orthorhombic symmetry. Numbers in parentheses refer to the respective highest-resolution data shell in each dataset.
Rsym= ΣhklΣi | Ii − <I> |/ΣhklΣiIi, where Ii is the ith measurement and <I> is the weighted mean of all measurements of I.
Rpim = Σhkl1/(N − 1)1/2 Σi| Ii(hkl) – I (hkl) |/ΣhklΣiI(hkl), where N is the redundancy of the data and I (hkl) the average intensity.
<I/σI> indicates the average of the intensity divided by its SD.
Rwork = Σhkl ||Fo| − |Fc||/Σhkl|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes.
Rfree same as R for 5% of the data randomly omitted from the refinement. The number of reflections includes the Rfree subset.
Ramachandran statistics were calculated with MolProbity.
Closer inspection of the nucleotide binding pocket revealed that ATPγS binding is directly mediated by Val226, which forms a hydrogen bond with the adenine of the nucleotide through its main chain carbonyl oxygen and residues Thr186 and Thr233 as well as Asp118 and Asn150, which coordinate the ATP analog through interactions with the α- and β-phosphates of its triphosphate moiety, respectively (SI Appendix, Fig. S3C). There were no significant conformational changes in the binary complex compared to the mouse apo-structure as reflected in an rms deviation of 0.45 Å for all Cα atoms with minimal structural rearrangements in the ATP binding pocket (SI Appendix, Fig. S3D). A comparison of PDXK sequences derived from organisms representing different evolutionary levels revealed that all residues, which are crucial for the binding of the nucleotide, are strictly conserved (SI Appendix, Fig. S4).
To gain insights into the mechanism of artemisinin inhibition we determined the crystal structure of mPDXK in complex with ATPγS and artesunate (Fig. 2 and Table 1). This structure was obtained by soaking artesunate into preexisting binary mPDXK-ATPγS crystals. After MR with the apo-structure, in addition to the clear density for ATPγS (Fig. 2B), strong difference density in close proximity to the substrate-binding pocket was also observed (Fig. 2D), which allowed us to unambiguously model the bound artesunate. Surprisingly, this density was observed in only one of the four molecules present in the asymmetric unit. The absence of artesunate in the other monomers may be due to the involvement of these protomers in crystal contacts, thus preventing artesunate binding when soaking the compounds into preexisting crystals.
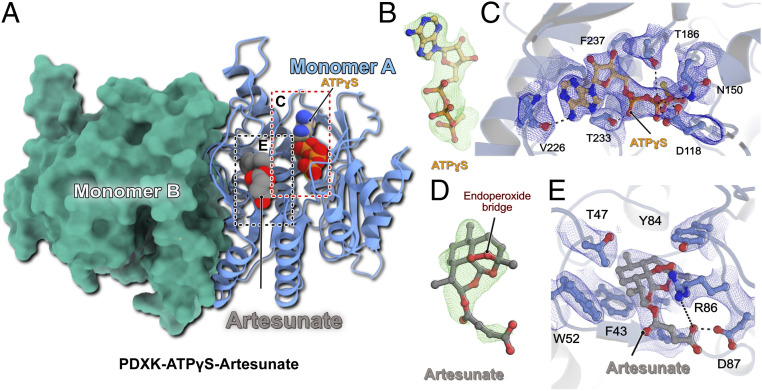
Fig. 2.
Structure of the ternary PDXK-ATPγS–artesunate complex. (A) Overall architecture of the ternary complex. One monomer is shown in cartoon representation with the bound ligands in CPK representation, while the second monomer is shown as a surface in green. (B) Fo-Fc omit electron density for the bound ATPγS contoured at an rms deviation of 3. (C) Enlarged view of the ATPγS binding pocket. The bound ligand and residues, which are crucial for ligand binding, are shown in ball-and-stick representation. The SIGMAA-weighted 2Fo-Fc electron density for the bound ligand and surrounding residues is contoured at an rmsd of 1. Critical protein–ligand interactions are highlighted. (D) Fo-Fc omit electron density for the bound artesunate contoured at an rms deviation of 3. (E) Enlarged view of the artesunate-binding pocket. The bound ligand and residues, which are crucial for ligand binding, are shown in ball-and-stick representation. SIGMAA-weighted 2Fo-Fc electron density for artesunate and interacting residues is contoured at an rmsd of 1. Critical protein–ligand interactions are highlighted.
The fact that all three structures reported here belong to the same space group with similar unit cell parameters and essentially identical crystal packing allowed for a meaningful comparative analysis. The overall architecture of the mPDXK-ATPγS–artesunate structure is identical to that of the apo and binary mPDXK-ATPγS structures; a superposition of the Cα atoms of these two complexes revealed rms deviations of 0.51 and 0.30 Å for the apo and ATPγS-bound structures, respectively. Binding of the substrate analog ATPγS was mediated by the same residues described for the binary mPDXK-ATPγS complex (Fig. 2C and SI Appendix, Fig. S5). A closer inspection of the artesunate-binding pocket revealed that drug binding is mainly mediated by Val41, Thr47, and also Trp52, which generate a hydrophobic pocket that binds artesunate with a buried surface area of 364 Å2 compared to a total surface area of the drug of 538 Å2. In particular, artesunate is sandwiched in between two aromatic residues, Phe43 and Tyr84, which stabilize artesunate through van der Waals interactions. In addition to the hydrophobic interactions, the carboxylate moiety of artesunate comes into proximity of the guanidinium group in the side chain of Arg86, which potentially stabilizes the interactions through electrostatic contacts. Finally, Asp87 favors artesunate binding through a hydrogen bond (2.5 Å) between its side chain and the carboxylate of artesunate, assuming one of these carboxylates is protonated (Fig. 2E).
An analysis of the mPDXK-ATPγS–artesunate structure showed that the ATP binding pocket is in relatively close proximity to the bound artesunate at a distance of ∼21 Å, as measured between the Cα atoms of Phe237 in the ATP binding pocket and Phe43 in the artesunate binding pocket. A comparison of the ternary mPDXK-ATPγS–artesunate and binary mPDXK-ATPγS structures suggested that binding of artesunate neither induced any significant rearrangements in the conformation of the nucleotide nor in the residues mediating its binding, in line with the independent binding of the two ligands (SI Appendix, Figs. S5 and S6). Interestingly, the binding pocket of artesunate, like that of ATP mentioned earlier, is highly conserved (SI Appendix, Fig. S7). To get additional information regarding the mode of inhibition, we also compared our ternary structure with PDXK-PLP structures. Strikingly, when we superimposed the already reported ternary HsPDXK-ATP-PLP (PDB: 3KEU) structure with our ternary complex, a critical partial overlap between the tricyclic ring system of artesunate and the pyridine ring of the product PLP was uncovered (Fig. 3 A and B), which would result in severe van der Waals repulsions for the C2 and C3 atoms of PLP and the C2 and C3 atoms of artesunate, if bound simultaneously. This clearly constitutes a major reason for the inhibition of the PDXK. Moreover, an analysis of the surface properties of the ternary structure revealed a tunnel, which is leading from the protein surface to the distal end of the ATP binding pocket spanning a length of ∼38 Å, which is blocked near its entrance by artesunate. A blockade of this tunnel, in turn, may prevent an efficient turnover of the enzyme. Taken together, these data illustrate the structural basis for the inhibition of mPDXK by artemisinins (Fig. 3 C–E).
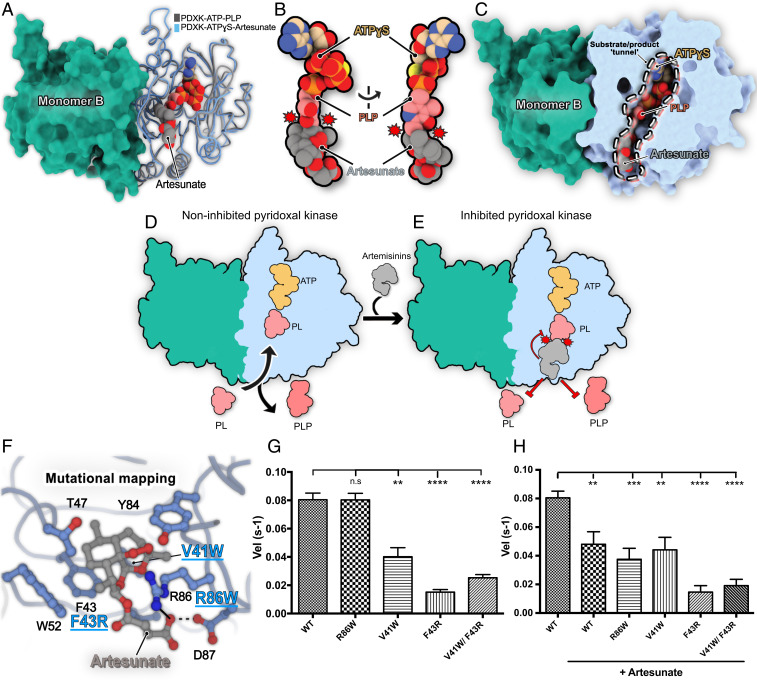
Fig. 3.
Structural basis for PDXK inhibition by artemisinins. (A) Superposition of the crystal structures of the ternary murine PDXK-ATPγS–artesunate (this study) and the human PDXK-ATP-PLP (PDB: 3KEU) complexes. Carbon atoms of artesunate are shown in gray, those of PLP in pink, and those of ATPγS in beige; the other atoms are colored in red (oxygen), blue (nitrogen), orange (phosphorous), and yellow (sulfur). Only ATPγS of the artesunate complex is shown to reduce visual complexity. (B) Enlarged view of the ligand binding pockets of ATP, artesunate, and PLP displaying the partial overlap (indicated by red circles with spikes) between PLP and artesunate. (C) Cutaway view of the superimposed PDXK-ATPγS–artesunate and PDXK-ATPγS–PLP structures displaying how artesunate binding blocks the substrate tunnel, which may also impair enzyme turnover. (D and E) Schematic representation of the structural basis for inhibition of PDXK activity by artemisinins. (F) Mutational mapping of the artesunate binding pocket with the investigated mutants highlighted in blue. (G and H) Comparison of the turnover rates of PDXK variants and the WT in the absence (G) and presence (H) of artesunate (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001) (paired t test).
Mapping of the Artemisinin Binding Pocket.
To validate the observations derived from the crystal structures, we performed site-directed mutagenesis experiments of residues located in the artesunate binding pocket (Fig. 3F) and tested these mutants for PLP production in the presence and absence of artesunate (Fig. 3 G and H). First, we analyzed the mutants through SEC-MALLS, which revealed that all variants retained their dimeric state in solution as observed for the wild-type (WT) protein (SI Appendix, Fig. S8). Among the residues being investigated, we mutated Val41 and Phe43, which are involved in mediating the binding of artesunate through hydrophobic interactions to either introduce steric interference or alter the polar properties of the binding pocket, respectively. The V41W and F43R mutants significantly lowered the turnover rates (0.04 ± 0.006 s−1 for V41W and 0.016 ± 0.001 s−1 for F43R), even in the absence of artesunate in comparison to the WT (0.080 ± 0.004 s−1). This can be easily explained by the fact that these residues also mediate binding of PL and thus play a role in the regular enzymatic turnover of the protein. In contrast, mutation of Arg86, the residue involved in the long-range electrostatic interaction with the carboxylate of artesunate, to the bulky aromatic side chain of Trp did not alter the activity of mPDXK in the absence of artesunate, as demonstrated by its turnover rate of 0.08 ± 0.005 s−1, which is virtually identical to that of the WT (Fig. 3G).
Next, we further analyzed the catalytic activity of the mutants in the presence of artesunate (Fig. 3H and SI Appendix, Fig. S9). The V41W and F43R variants did not result in significant changes in the turnover rates of the enzyme in the presence of artesunate (0.044 ± 0.009 s−1 for V41W and 0.014 ± 0.005 s−1 for F43R compared to 0.040 ± 0.006 and 0.016 ± 0.001 s−1, respectively, in its absence), which is in line with artesunate binding being abolished in both variants (SI Appendix, Fig. S9 C and D). As expected, a similar trend was observed in the case of the V41W/F43R double mutant with turnover rates of 0.024 ± 0.002 s−1 in the presence of artesunate compared to 0.019 ± 0.004 s−1 in its absence (SI Appendix, Fig. S9E). An identical behavior was observed in the case of GephE where mutation of a crucial aromatic residue (Phe330) to Ala completely abolished artemisinin binding (14). In contrast, when we compared the activity of the R86W variant in the absence (0.080 ± 0.005 s−1) and presence (0.038 ± 0.008 s−1) of the drug, a significant reduction in enzymatic activity was observed (SI Appendix, Fig. S9B). Thus, although R86 is involved in an electrostatic interaction as revealed by the crystal structure, the mutational analysis demonstrated that the inhibition potency of artesunate is retained even in the absence of this interaction. This observation is in contrast to the GephE structure where the replacement of Arg (Arg653 in gephyrin) with the bulkier aromatic Trp prevented artesunate binding (14). Thus, our structures help to define the molecular signatures of artemisinin binding pockets, which may aid in the future identification of target sites, especially by in silico approaches.
Artemisinins Inhibit GABA Biosynthesis and Down-Regulate GABAergic Neurotransmission.
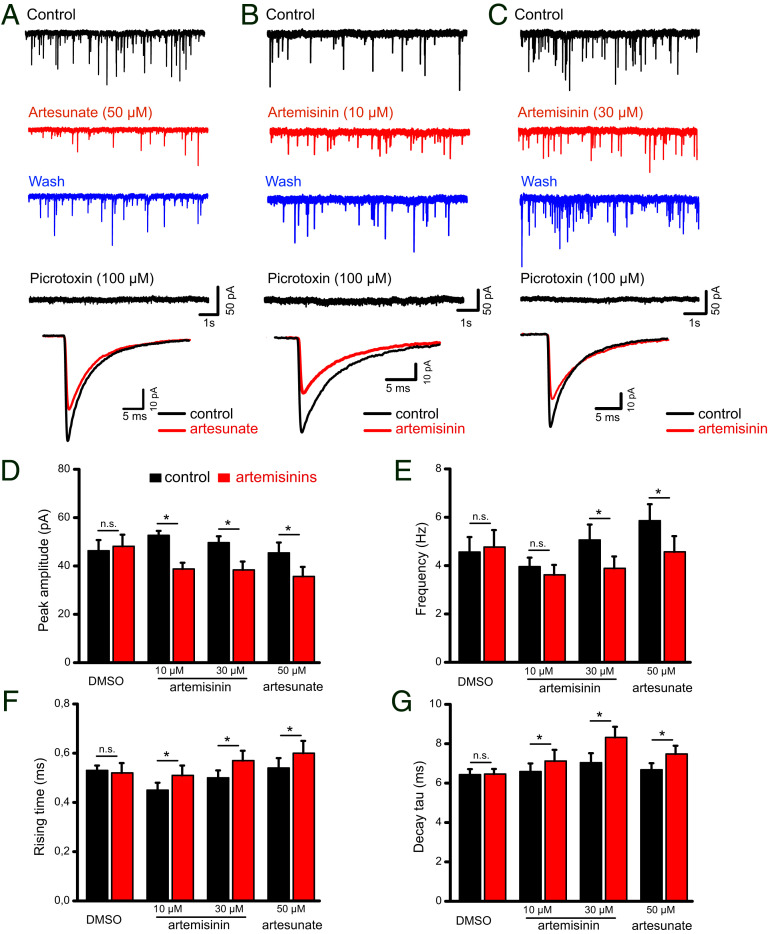
To understand whether the functional consequences of our biochemical and structural analyses correlate with a physiological scenario, whole-cell voltage-clamp recordings (Fig. 4) from CA1 pyramidal cells in hippocampal slices were performed. We determined the properties of GABAergic miniature inhibitory postsynaptic currents (mIPSCs) in the absence and presence of artemisinin (10 and 30 µM, Fig. 4 B and C) as well as artesunate at a concentration of 50 µM (Fig. 4A), which was shown to deregulate the postsynaptic GABAAR–gephyrin clustering. Artesunate treatment induced a significant reduction in the mIPSC amplitudes from 45.4 ± 4.3 to 35.7 ± 3.9 pA (P = 0.0001), while the frequency decreased from 5.9 ± 0.7 to 4.6 ± 0.7 Hz (P = 0.0008, n = 8 from four mice) at the tested concentration (Fig. 4 A, D, and E). In contrast, artemisinin down-regulated mIPSC amplitudes already at 10 µM (from 52.7 ± 1.9 to 38.8 ± 2.6 pA, n = 7 from four mice, P = 0.001, paired t test; Fig. 4 B and D). We attribute the changes in the amplitude to the artemisinin-induced disruption of postsynaptic GABAAR–gephyrin complexes. While a significant reduction in amplitudes was retained at a higher concentration (30 µM), we also observed a concomitant decrease in mIPSC frequency from 5.1 ± 0.6 to 3.9 ± 0.5 Hz (n = 7 from five mice, P = 0.012; Fig. 4 C and E) in the presence of artemisinin. In addition, artemisinin and/or artesunate altered mIPSC kinetics with slower rise and decay times (Fig. 4 F and G). In both cases, a significant decrease in mIPSC frequency is particularly noteworthy in the context of this study as it reflects changes in the presynaptic terminals; e.g., it would be in line with a reduced synthesis of the neurotransmitter GABA (28).
Fig. 4.
Impact of artemisinins on electrophysiological recordings of hippocampal slices. (A–C) Representative voltage-clamp recordings of GABAA receptor-mediated mIPSCs from mouse CA1 pyramidal cells in hippocampal slices were collected before (control), during (10 to 15 min), and after (wash) artesunate (50 µM, A) or artemisinin (10 µM, B and 30 µM, C) application. Picrotoxin was applied to verify the GABAergic origin of these events. Superimposed traces (A–C, Bottom) are averaged events before and during artemisinin treatment. (D–G) Quantifications of how artemisinins affect mIPSC amplitudes (D), frequencies (E), and kinetics (F and G). *P < 0.05, paired t-test.
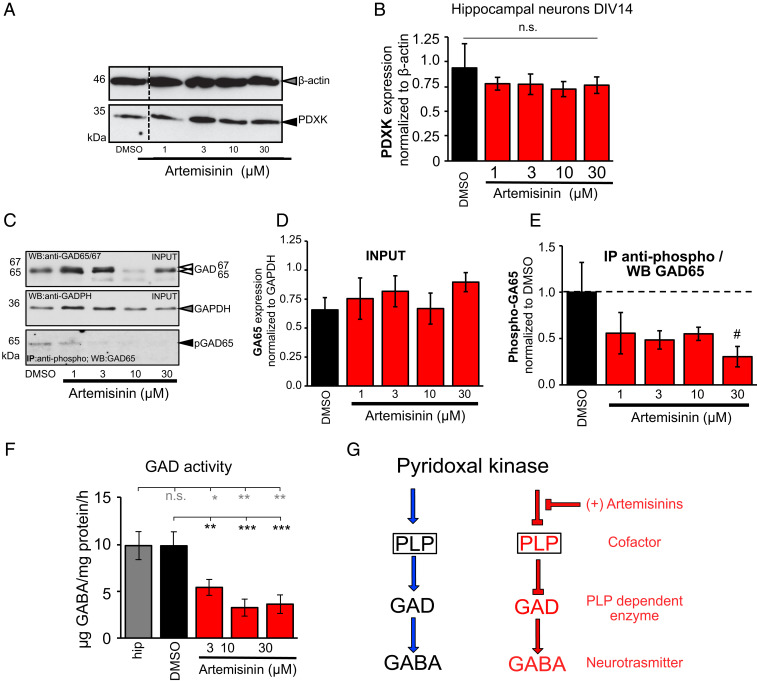
To evaluate whether the changes in the mIPSC frequencies observed were due to changes in the cellular concentration of PDXK, we first analyzed its expression levels in hippocampal neurons at day in vitro (DIV) 14 (Fig. 5 A and B), as PDXK activity is required for the activity of GAD by providing its cofactor PLP. These studies revealed that the PDXK expression level was unaffected by artemisinin treatment. Next, we investigated whether the observed mIPSC frequencies were due to changes in GABA availability upon alterations in the activity of GAD, the GABA synthesizing enzyme. Following treatment with artemisinin at different concentrations, no changes in the overall GAD65 expression level were observed (Fig. 5 C and D). In contrast to GAD67 which is constitutively active, GAD65 is transiently active when there is an increased demand of GABA. Moreover, it was shown that GAD65 is activated by phosphorylation (29, 30). To investigate whether treatment with artemisinin in vitro has an impact on the amount of active phosphorylated GAD65, immunoprecipitation studies were performed. Using hippocampal neurons at DIV14 in culture treated with increasing artemisinin concentrations (1, 3, 10, and 30 µM), a reduction in the levels of phosphorylated GAD65 protein was detected compared to the DMSO-treated control samples (1 µM 55 ± 9% compared to DMSO 100%, n = 4, P = 0.108; 3 µM 48 ± 7%, n = 4, P = 0.059; 10 µM 55 ± 11%, n = 4, P = 0.11; 30 µM 30 ± 6%, n = 2, P = 0.09); however, the data fell just short of reaching statistical significance (Fig. 5 C and E). As a qualitative measure, we also stained hippocampal neurons for PDXK, GAD, and the postsynaptic marker gephyrin, which did not reveal any noticeable differences in the expression of these marker proteins (SI Appendix, Figs. S10 and S11). Finally, to check whether the frequency changes observed in the electrophysiology measurements and the down-regulation observed in the active form of GAD correlate with GABA biosynthesis, we measured the amount of this neurotransmitter in primary hippocampal neurons (DIV14). GABA levels were quantified by the classical ninhydrin reaction (SI Appendix, Fig. S12) by measuring the fluorescence emission of the resulting adduct at 450 nm and calibrating it with a GABA standard. Remarkably, this analysis revealed a significant reduction in the amount of GABA production (5.4 ± 0.8, 3.2 ± 0.9, and 3.6 ± 0.98 µg GABA per milligram protein per hour) in hippocampal neurons treated with artemisinin at concentrations of 3, 10, and 30 µM, respectively (P = 0.0054, 0.0007, and 0.0002 against DMSO measurements and P = 0.018, 0.0087, and 0.0047 against untreated hippocampal measurements for 3, 10, and 30 µM artemisinin concentrations) (Fig. 5F). In comparison, untreated samples resulted in levels of 9.8 ± 1.5 µg GABA per milligram protein per hour. The observed perturbation on the presynaptic side is therefore primarily due to the direct effect of artemisinins on the biosynthesis of PLP, which results in a reduced production of this cofactor, which is required by the GAD enzyme to produce the neurotransmitter GABA (Fig. 5G), while a slight augmentation of this decrease may be due to a reduction in the levels of the phosphorylated form of GAD65.
Fig. 5.
Artemisinins impact GABA biosynthesis by down-regulating GAD activity. (A) Representative image of a Western blot stained for PDXK (35 kDa, black arrowhead) and β-actin (46 kDa, gray arrowhead). DMSO-treated cells served as internal control. (B) PDXK expression analysis after incubation with artemisinins (1, 3, 10, and 30 µM). PDXK expression was normalized to β-actin. (C) Representative images of input probes after artemisinin treatment (1, 3, 10, and 30 µM) used for immunoprecipitation stained for GAD65/67 (65/67 kDa, white arrowheads) and GAPDH (36 kDa, gray arrowhead). Following immunoprecipitation using an anti-phospho antibody, probes were stained with a specific GAD65 antibody (65 kDa, black arrowhead). DMSO-treated cells served as control samples. (D) Quantitative analysis of input probes from lysates of hippocampal neurons DIV14 incubated with increasing concentrations of artemisinin (1, 3, 10, and 30 µM; DMSO served as control) normalized to the GAPDH signal. (E) Precipitated pGAD (phosphorylated GAD65) was normalized to the DMSO control. Note, that the number of experiments was #n = 2 for 30 µM artemisinin; all other data result from n = 4. All values are shown as mean ± SEM. (F) Measurements of GAD activity in hippocampal samples following treatment with different concentrations of artemisinin (3, 10, and 30 µM). Tissues without treatment (gray bar) and treated with DMSO (black bar) served as positive controls. Number of measurements n = 8 to 9 from three independent biological replicates. GAD activity decreased significantly with increasing concentrations of artemisinin. The data were analyzed with a paired t test. **P = 0.0054, ***P = 0.0007, and ***P = 0.0002 against DMSO measurements and *P = 0.018, **P = 0.0087, and **P = 0.0047 against hippocampal measurements for artemisinin concentrations of 3, 10, and 30 µM. (G) Schematic representation of the steps leading to GABA biosynthesis at presynaptic terminals. Left shows how GAD synthesizes GABA by utilizing PLP as a cofactor which is produced by PDXK, while Right shows how artemisinins inhibit the initial step in the biosynthesis by inhibiting PDXK, which, in turn, indirectly impacts downstream biosynthetic processes and eventually down-regulates the amount of neurotransmitter being synthesized.
Discussion
Despite their widespread clinical application as antimalarial drugs, and despite their known effects on various cellular pathways in mammals, the molecular mechanisms of how artemisinins affect cellular pathways are still only poorly understood. Artemisinins can efficiently cross the blood–brain barrier (31) and, strikingly, administrations of high levels of artemisinins are accompanied by severe neurotoxic side effects (32–34). Recently, we were able to derive the first protein–artemisinin structure by X-ray crystallography at 1.5 Å resolution, namely that of the inhibitory postsynaptic scaffolding protein gephyrin in complex with artesunate and artemether (14). Here, we successfully validated and elucidated the mechanism underlying yet another mammalian artemisinin target, the critically important metabolic enzyme PDXK.
Our structural studies demonstrate a competition between the substrate pyridoxal and artemisinins, in line with the observed inhibition of the enzyme derived from kinetic data. As artesunate targets the same binding pocket identified previously for the interaction of (R)-roscovitine with PDXK (35, 36) and for the neurotoxins ginkgotoxin and theophylline (37), our structure suggests that the neurotoxicity induced by artemisinins could be due, at least in part, to their binding to PDXK and the resulting inhibition of its activity.
The presynaptic effect of artemisinin and, at higher concentrations, also for artesunate in our electrophysiological recordings correlates nicely with the down-regulation of PDXK activity and can be extended toward glycine, the other major inhibitory neurotransmitter. This neurotransmitter is synthesized by SHMT, again in a strictly PLP-dependent fashion. Thus, we predict a similar electrophysiological behavior with decreased frequencies at glycinergic synapses as observed for GAD and GABA levels. With respect to the postsynaptic counterparts, we have already demonstrated a decrease in glycinergic currents following artemisinin treatment (14). Structurally, GlyRs, via their β subunit, also depend on the identical binding pocket in GephE for their postsynaptic clustering as do GABAARs. Our GephE–artesunate/artemether structures demonstrated that artemisinins block the N-terminal three-residue stretch from the core binding motifs of the GlyR and GABAARs to mediate the inhibition (398FSI400 of GlyR β subunit vs. 398FNI400 of GABAAR α3 subunit). Therefore, we can extrapolate these results and predict a similar down-regulation of receptor clustering at glycinergic postsynapses.
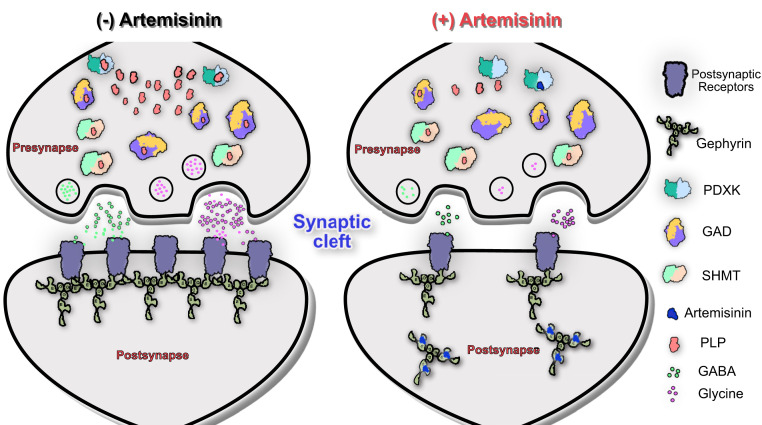
Thus, the data presented here extend our current understanding of how artemisinins act at inhibitory synapses in the central nervous system. The present study shows that artemisinins not only act at the postsynaptic side, but also affect the functionality of the presynaptic terminals via their interaction with PDXK which ultimately leads to a decrease in neurotransmitter biosynthesis (Fig. 6). Although the data presented here and our earlier study (14) define mechanisms underlying the down-regulation of inhibitory neurotransmission by artemisinins, other neurotransmitters such as dopamine, histamine, and serotonin are also synthesized in a PLP-dependent manner (4–6); thus, future studies will be required to comprehensively dissect the molecular details underlying the artemisinin-induced regulation of neurotransmitter levels and the resulting physiological consequences.
Fig. 6.
Schematic representation of inhibitory synapses in the absence and presence of artemisinins. This scheme shows that in the absence of artemisinins (Left) gephyrin clusters the receptors required for inhibitory neurotransmitters at postsynaptic sides, while PDXK contributes to the biosynthesis of neurotransmitters at presynaptic terminals by producing the PLP cofactor for GAD and SHMT enzymes. In contrast, in the presence of artemisinins (Right), gephyrin-mediated clustering of receptors at postsynaptic sites is impaired while neurotransmitter biosynthesis at presynaptic terminals is inhibited.
In addition, a comparison of the artemisinin binding pockets in mPDXK and GephE revealed common denominators of drug recognition. Notably, in both cases, artemisinins engage in crucial van der Waals interactions with aromatic residues. In addition, in GephE as well as PDXK, the side chain of an Arg, contributes to artesunate binding, which stabilizes the drug through an electrostatic interaction with its succinate moiety, thus revealing common signatures of artemisinin binding pockets. Our results thus not only broaden the understanding of target recognition by artemisinins at the structural level, but also provide important insights into how these interactions impair inhibitory synaptic transmission in the brain and how this might account for the neurological side effects of these drugs. Future studies, along with the molecular signatures revealed in our structures, will be required to investigate whether artemisinins indeed directly bind to and also modulate the activities of other mammalian targets such as protein disulfide isomerase and fatty acid synthase which were identified earlier (7).
Materials and Methods
Experimental Model and Subject Details.
For cloning purposes Escherichia coli DH5α was used and the cells were grown on Luria Broth (LB)-agar plates and in LB liquid medium at 37 °C. For recombinant protein expression E. coli SoluBL21 cells were used. The cells were grown at 37 °C initially and were further incubated at 30 °C for 16 to 18 h after induction. Transverse hippocampal slices (350 µm thick) were prepared from sevoflurane-anesthetized adult C57BL/6J mice (2 to 4 months old) of either sex purchased from Charles River. Adult animals (12-week-old male or female mice) were taken from the mouse strain CD1 (strain code 022; Charles River) to isolate the hippocampi. Animals were housed under standard conditions and all procedures were conducted according to the guidelines and with approval of the local government of Lower Franconia. Preparation of brain slices containing the hippocampal formation was performed as previously described (38).
Method Detail.
Cloning, recombinant protein expression, and purification.
The cDNA encoding mPDXK was subcloned into the pETM14 expression vector harboring a 3C precision protease cleavage and BamH1 sites by sequence-independent ligation cloning (SLIC) (39). The proteins (WT and all mutants) were expressed in the E. coli SoluBL21 strain. Cells were grown at 37 °C and expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at an optical density (OD600) of 0.6 to 0.8 and cultures were subsequently incubated at 30 °C for 16 to 18 hours. Following centrifugation at 8,000 × g for 15 min the harvested cells were resuspended in lysis buffer containing 50 mM Tris pH 8, 300 mM NaCl, and 5 mM β-mercaptoethanol (β-ME) and lysis was performed by using a microfluidizer. For purification, a two-step protocol was employed consisting of an initial Ni-affinity chromatography with Ni-iminodiacetic acid (IDA) beads, which was followed by cleavage of the N-terminal His6 tag by incubation with 3C precision protease overnight at 4 °C. Finally, size exclusion chromatography on a Superdex 200 26/60 (GE Healthcare) column was performed in SEC buffer (20 mM Tris pH 8, 150 mM NaCl, and 5 mM β-ME) to purify the protein to apparent homogeneity.
SEC-MALLS.
SEC-MALLS experiments of 100 µM WT and all mutants were carried out by using a Superdex 200 10/300 column (GE Healthcare) in SEC buffer. The experiments were performed at a constant flow rate of 0.5 mL/min at room temperature. The differential refractive index (dRI) and the light-scattering (LS) signals were monitored with a Dawn Helios detector from Wyatt Technologies and molecular masses were derived from the dRI and LS measurements.
Crystallization.
Crystallization of mPDXK was performed in the apo-form and in complex with ATPγS at a protein concentration of 12 mg/mL corresponding to a molar concentration of 0.3 mM. The protein was mixed with 2 mM ATPγS and 5 mM MgCl2 and the complex was incubated on ice for 30 min prior to crystallization. Crystallization was performed with the sitting drop vapor diffusion method by mixing equal volumes of protein and mother liquor at 20 °C. The mPDXK-ATPγS–artesunate structure was determined by soaking mPDXK-ATPγS crystals with different concentrations of artesunate (2 to 10 mM) for 30 to 600 s. Crystals were transferred into mother liquor (0.18 to 0.24 M sodium thiocyanate and 18 to 26% PEG3350) supplemented with different concentrations of artesunate and 25% glycerol as cryoprotectant before flash cooling in liquid nitrogen.
Data collection, structure determination, and refinement.
Data collection for all crystals was performed at the European Synchrotron Radiation Facility (ESRF), Grenoble, France, on beamline ID23A-1 at a wavelength of 0.9724 Å at 100 K. Datasets were indexed and integrated with XDS (40) and subsequently scaled and merged with AIMLESS (41) from the CCP4 suite (42). The apo-structure and the binary mPDXK-ATPγS complex were determined by molecular replacement with PhaserMR (43) using the human PDXK structure (PDB: 2YXT) as search model and the ternary mPDXK-ATPγS–artesunate complex was solved with the apo-mPDXK structure as search model. The protein crystallized in space group C2 with four molecules in the asymmetric unit. Refinement was performed in PHENIX (44) with repeated manual model building in Coot (45). Coordinates and restraints for artesunate were obtained from our gephyrin–artesunate structure (PDB: 6FGC). All figures representing protein structures were generated with PyMOL (Schrodinger LLC), Chimera (46), or ChimeraX (47).
Enzymatic activity assay.
Pyridoxal kinase activity (WT and variants) was measured following a previously described procedure (22). Briefly, the assay was conducted in 10 mM Hepes buffer (pH 7.3) at 37 °C with 100 mM KCl, 1 mM MgCl2, 1 mM Mg-ATP, and 50 µg/mL BSA. The pyridoxal kinase concentration was 20 μg/mL (0.6 µM), and the substrate pyridoxal was added in a range from 10 to 600 μM. The activity was measured following the increase in absorbance at 388 nm due to PLP formation (extinction coefficient of 4,900 M−1⋅cm−1) in a CLARIOstar (BMG LABTECH) microplate reader. All experiments were carried out in triplicate. KM and kcat values were calculated by a Lineweaver–Burk plot (48) with the program Prism (GraphPad Software). For statistical significance of the enzymatic assays, initially, the normality distribution of the data was determined by a D’Agostino and Pearson normality test. After passing the normality test, the statistical significance was determined by a paired t test. For all statistical tests, the P values correspond to *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant. Statistical analyses were performed by using values from four independent experiments.
To derive the Ki values, the assay was performed under the same conditions using pyridoxal at concentrations of 50 and 150 µM. Using both pyridoxal concentrations the assays were performed with twofold serial dilutions of artesunate and artemisinin, starting at concentrations of 2.5 and 0.156 mM, respectively. Ki values for the inhibitors artesunate and artemisinin were estimated by a Dixon plot (49), by using a linear regression fit (P < 0.0001) of the inverted velocity values. The Ki value corresponds to the intersection between the two lines obtained for each individual pyridoxal concentration. For determining the IC50 values, the values of inhibitor concentration were transformed to a logarithmic scale and fitted using a nonlinear regression fit with variable slope. IC50 values were calculated as the concentration of inhibitor that gives a velocity halfway between the minimal and maximal values of the curve. All curve fitting procedures and statistical analyses were performed using Prism (GraphPad Software).
Electrophysiology.
Transverse hippocampal slices (350 µm thick) were prepared from adult C57BL/6J mice. Animals were housed under standard conditions and all procedures were conducted according to the guidelines and with approval of the local government of Lower Franconia. Whole-cell voltage-clamp recordings were obtained from visualized pyramidal cells of the hippocampal CA1 region in a submerged chamber with perfusion solution containing 125 mM NaCl, 3 mM KCl, 2.5 mM CaCl2, 1.5 mM MgCl2, 1.25 mM NaH2PO4, 25 mM NaHCO3, and 10 mM D-glucose at 31 °C, constantly gassed with 95% O2 / 5% CO2 (pH 7.4). mIPSCs were recorded in the presence of the ionotropic glutamate receptor antagonist kynurenic acid (2 mM) and tetrodotoxin (TTX) (0.5 µM) using pipettes filled with solution containing 130 mM CsCl, 5 mM Hepes, 3 mM MgCl2, 5 mM EGTA, 2 mM Na2ATP, 0.3 mM Na3GTP, 4 mM NaCl, and 5 mM QX-314 (pH 7.3). mIPSCs were recorded at a holding potential of −70 mV as downward deflections. Current signals were filtered at 2 kHz and sampled at 20 kHz using a Multiclamp 700B amplifier together with a Digidata 1440A interface and the pClamp10 software (Molecular Devices). Data were analyzed off line with Clampfit 10.6 (Molecular Devices) and were expressed as means ± SEM.
Statistical comparisons of drug effects were performed with OriginPro 2015G (OriginLab Corporation) using a paired t test. Significance was assumed for P values <0.05.
Fluorimetric assay for GAD activity determination.
The basis for the determination of the activity of GAD in brain tissue is the ninhydrin reaction. During this reaction, the substrate ninhydrin reacts with the amino group of GABA, releasing water and forming a Schiff base. Following decarboxylation of GABA and elimination of the amino acid, amino-ninhydrin is formed, which dimerizes with ninhydrin (blue color). The concentration of ninhydrin is proportional to the concentration of the amino acid (law of Lambert–Beer) and was measured with a FluoroMax-4 (HORIBA Scientific) fluorimeter using an excitation wavelength of 375 nm (6 mm slide width) and an emission wavelength of 450 nm (10 mm slide width) (50, 51).
The experimental setup was slightly modified from ref. 51. Hippocampi were transferred into a fresh tube containing sonification buffer (0.5 M KCl, 0.01 M EDTA, 0.5% Triton X-100 in sodium-phosphate buffer, pH 6.4) followed by sonification for 10 s. Protein concentration was measured via a Bradford assay and adjusted to 1 µg/µL. The following conditions were analyzed in triplicates: hippocampi without treatment, DMSO, 3 µM artemisinin, 10 µM artemisinin, and 30 µM artemisinin. For each sample a probe (P) and blank (B) were used with 100 µL homogenized hippocampi and the corresponding DMSO/artemisinin concentrations (3, 10, and 30 µM) diluted in sonification buffer. The B fraction was supplemented with 200 µL 10% trichloroacetic acid (TCA). The P and B fractions were supplemented with 100 µL substrate buffer (100 mM sodium-l-glutamate buffer in 0.4 M sodium phosphate buffer, pH 6.7, 40 µL 50 mM pyridoxal phosphate 5-phosphate buffer) and incubated for 2 hours at 38 °C.
Subsequently the reaction of the P fraction was also stopped by adding 200 µL 10% TCA. Samples were centrifuged at 950 × g for 20 min. A total of 200 µL of all probes was incubated with 400 µL ninhydrin (14 mM in 0.5 M sodium carbonate buffer, pH 9.93) at 60 °C for 30 min. Afterward, probes were incubated with 9 mL copper tartrate (1.6 g sodium carbonate, 329 mg tartaric acid, 300 mg copper-(II) sulfate in 1 L aqua dest) at 22 °C for 20 min and measured (1:10 diluted) in the fluorescence spectrophotometer. A GABA concentration series of 0, 0.5, 1, 3, 5, 20, 50 µM GABA was used as standard.
Preparation of primary hippocampal neurons.
Hippocampal neurons were prepared at embryonic day 17 (E17) from pregnant female wild-type mice. Dissociated cells were grown in neurobasal medium supplemented with 5 mL of l-glutamine (200 mM) and B27 supplement (Life Technologies; A3582801) with an exchange of 50% medium after every 6 d in culture.
Protein lysate preparation.
At DIV14, hippocampal neurons were incubated for 2 hours with different DMSO/artemisinin concentrations (1, 3, 10, and 30 µM). Cells were washed and harvested in phosphate-buffered saline (PBS), pH 7.4, with the help of a cell scraper. After a centrifugation step, the pellet was resuspended in 100 µL of brain homogenate buffer (20 mM Hepes, 100 mM K-acetate, 40 mM KCl, 5 mM EGTA, 5 mM MgCl2, 5 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1% Triton X, protease inhibitor Roche complete, pH 7.2) and sonicated at low power for 5 s. Protein concentration was determined with the Bradford assay. A total of 10 µg per condition was used for Western blot analysis.
Western blot.
For SDS/PAGE, 11% polyacrylamide gels were freshly prepared, followed by Western blot on nitrocellulose membranes (GE Healthcare). Membranes were blocked for 1 hour with 5% BSA in TBS-T (TBS with 1% Tween 20). Primary antibodies were incubated overnight at 4 °C. GAD and PDXK proteins were detected with the GAD67/65-specific antibody (ab11070 1:1,000; abcam) and PDXK-specific antibody (NBP1-88283, 1:1,000; novusBio). β-Actin (GTX26276, 1:5,000; GeneTex/Biozol) served as loading control.
Signals were detected using the ECL plus system (GE Healthcare).
Data analysis of Western blots.
Image quantification was performed using ImageJ (1.51)/Fiji2 (52–54). The data were analyzed using a Student’s t test (analysis of variance) and values below *P < 0.05 were considered significant, **P < 0.01, ***P < 0.001. The values are displayed as mean ± SEM or as otherwise noted.
Immuncytochemical staining.
DIV14 primary hippocampal neurons were incubated for 2 hours with corresponding DMSO/artemisinin concentrations (1, 3, 10, and 30 µM). Neurons were fixed in 4% paraformaldehyde in PBS for 15 min. After washing twice with PBS, 50 mM NH4Cl was added for 10 min followed by blocking with 5% goat serum in PBS (permeabilized with 0.2% Triton X-100) for 30 min at 22 °C. Primary antibodies were incubated for 1 hour in blocking solution without Triton X-100. GAD and PDXK proteins were detected with the GAD67/65-specific antibody (ab11070, 1:500; abcam), gephyrin-specific antibody (147111, 1:500; Synaptic Systems) and PDXK-specific antibody (NBP1-88283, 1:150; novusBio). Secondary antibodies gαmCy3, gαrAlexa488, and gαrCy5 (1:500; Dianova) were applied for 1 hour. Cells were stained with 4′,6 diamino-2-phenylindole (DAPI) and slides were mounted with Mowiol.
Immunoprecipitation.
DIV14 primary hippocampal neurons were incubated for 2 hours with corresponding DMSO/artemisinin concentrations (1, 3, 10, and 30 µM). Cells were lysed in lysis buffer (TBS: 50 mM Tris, 150 mM NaCl, pH 8.0) containing 0.1% Triton X-100, 0.1 mM PMSF, 2 mM EDTA, and complete, EDTA-free protease inhibitor mixture (Roche) and sonicated. The protein concentration of each probe was measured, and 50 µg was used for immunoprecipitation (IP). The protein solution (300 µL) was supplemented with an anti-phospho antibody (1:100, ab17464; abcam) and agitated overnight at 4 °C. A total of 40 µL of protein A-Sepharose beads (REF 11719408001; Roche) was added and agitation continued overnight at 4 °C. Following a short centrifugation step, the supernatant was removed, and beads were washed three times with lysis buffer. Proteins were eluted from beads by incubation with 30 µL of 2× SDS sample buffer at 95 °C for 7 min. Proteins were separated on 11% SDS gels and stained after Western blotting with GAD65 antibody (1:1,000, ab26113; abcam). Input controls were stained with GAPDH antibody (1:1,000, CB1001; Calbiochem) and GAD65/67 antibody (1:1,000, ab11070). All experiments were performed with at least three independent biological replicates (unless otherwise stated). Quantification of input GAD65 protein signals at different artemisinin concentrations (1, 3, 10, and 30 µM) were normalized to GAPDH signals of corresponding input probes. The IP probes were finally normalized to the DMSO signal. Calculated data were compared using an unpaired t test with a probability of error of P < 0.05 considered to be significant.
Supplementary Material
Acknowledgments
We thank Dr. Antje Gohla for providing the cDNA of mPDXK and Dana Wegmann and Christine Schmitt for excellent technical assistance. We also thank the beamline scientists at beamline ID23-A1, ESRF, Grenoble, for technical assistance during data collection. We are grateful to Dr. Kunimichi Suzuki for the critical reading of the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (SCHI425/8-2) and the Rudolf Virchow Center for Integrative and Translational Bioimaging (H.S.). A.P.M. was supported by the Graduate School of Life Sciences at the University of Würzburg. N.S. was supported by funds of the Bavarian State Ministry of Science and the Arts and the Graduate School of Life Sciences. V.B.K. is supported at the Medical Research Council (MRC) Laboratory of Molecular Biology (LMB) by a European Molecular Biology Organization (EMBO) Long-Term Fellowship (ALTF137-2019).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008695117/-/DCSupplemental.
Data Availability.
The coordinates of mPDXK-apo, mPDXK-ATPγS, and mPDXK-ATPγS–artesunate structures have been deposited in the PDB with accession codes 6YJZ, 6YK0, and 6YK1, respectively.
References
- 1.Li M. H., et al. , Conformational changes in the reaction of pyridoxal kinase. J. Biol. Chem. 279, 17459–17465 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Neary J. T., Diven W. F., Purification, properties, and a possible mechanism for pyridoxal kinase from bovine brain. J. Biol. Chem. 245, 5585–5593 (1970). [PubMed] [Google Scholar]
- 3.Merigliano C., Mascolo E., Burla R., Saggio I., Vernì F., The relationship between vitamin B6, diabetes and cancer. Front. Genet. 9, 388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.di Salvo M. L., Safo M. K., Contestabile R., Biomedical aspects of pyridoxal 5′-phosphate availability. Front. Biosci. (Elite Ed.) 4, 897–913 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Percudani R., Peracchi A., A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 4, 850–854 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eliot A. C., Kirsch J. F., Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73, 383–415 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Li J., et al. , Artemisinins target GABAA receptor signaling and impair alpha cell identity. Cell 168, 86–100.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tu Y., Artemisinin-A gift from traditional Chinese medicine to the world (nobel lecture). Angew. Chem. Int. Ed. Engl. 55, 10210–10226 (2016). [DOI] [PubMed] [Google Scholar]
- 9.WHO , “Guidelines for the treatment of malaria” in Guidelines for the Treatment of Malaria (World Health Organization, Geneva, Switzerland, ed. 3, 2015). [Google Scholar]
- 10.Crespo-Ortiz M. P., Wei M. Q., Antitumor activity of artemisinin and its derivatives: From a well-known antimalarial agent to a potential anticancer drug. J. Biomed. Biotechnol. 2012, 247597 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gautam A., Ahmed T., Batra V., Paliwal J., Pharmacokinetics and pharmacodynamics of endoperoxide antimalarials. Curr. Drug Metab. 10, 289–306 (2009). [DOI] [PubMed] [Google Scholar]
- 12.van der Meulen T., et al. , Artemether does not turn alpha cells into beta cells. Cell Metab. 27, 218–225.e214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ackermann A. M., Moss N. G., Kaestner K. H., GABA and artesunate do not induce pancreatic alpha-to-beta cell transdifferentiation in vivo. Cell Metab. 28, 787–792.e783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasaragod V. B., et al. , Elucidating the molecular basis for inhibitory neurotransmission regulation by artemisinins. Neuron 101, 673–689.e611 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Kasaragod V. B., Schindelin H., Structural framework for metal incorporation during molybdenum cofactor biosynthesis. Structure 24, 782–788 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Kuper J., Llamas A., Hecht H. J., Mendel R. R., Schwarz G., Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430, 803–806 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Kasaragod V. B., Schindelin H., Structure-function relationships of glycine and GABAA receptors and their interplay with the scaffolding protein gephyrin. Front. Mol. Neurosci. 11, 317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerry J. A., Rohde M., Kwok F., Brain pyridoxal kinase. Purification and characterization. Eur. J. Biochem. 158, 581–585 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Elsinghorst P. W., di Salvo M. L., Parroni A., Contestabile R., Inhibition of human pyridoxal kinase by 2-acetyl-4-((1R,2S,3R)-1,2,3,4-tetrahydroxybutyl)imidazole (THI). J. Enzyme Inhib. Med. Chem. 30, 336–340 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Hanna M. C., Turner A. J., Kirkness E. F., Human pyridoxal kinase. cDNA cloning, expression, and modulation by ligands of the benzodiazepine receptor. J. Biol. Chem. 272, 10756–10760 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Jones D. C., Alphey M. S., Wyllie S., Fairlamb A. H., Chemical, genetic and structural assessment of pyridoxal kinase as a drug target in the African trypanosome. Mol. Microbiol. 86, 51–64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok F., Churchich J. E., Brain pyridoxal kinase. Purification, substrate specificities, and sensitized photodestruction of an essential histidine. J. Biol. Chem. 254, 6489–6495 (1979). [PubMed] [Google Scholar]
- 23.McCormick D. B., Gregory M. E., Snell E. E., Pyridoxal phosphokinases. I. Assay, distribution, I. Assay, distribution, purification, and properties. J. Biol. Chem. 236, 2076–2084 (1961). [PubMed] [Google Scholar]
- 24.Safo M. K., et al. , Crystal structure of pyridoxal kinase from the Escherichia coli pdxK gene: Implications for the classification of pyridoxal kinases. J. Bacteriol. 188, 4542–4552 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ubbink J. B., Bissbort S., Vermaak W. J., Delport R., Inhibition of pyridoxal kinase by methylxanthines. Enzyme 43, 72–79 (1990). [DOI] [PubMed] [Google Scholar]
- 26.Musayev F. N., et al. , Crystal structure of human pyridoxal kinase: Structural basis of M(+) and M(2+) activation. Protein Sci. 16, 2184–2194 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M. H., et al. , Crystal structure of brain pyridoxal kinase, a novel member of the ribokinase superfamily. J. Biol. Chem. 277, 46385–46390 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Engel D., et al. , Plasticity of rat central inhibitory synapses through GABA metabolism. J. Physiol. 535, 473–482 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei J., Davis K. M., Wu H., Wu J. Y., Protein phosphorylation of human brain glutamic acid decarboxylase (GAD)65 and GAD67 and its physiological implications. Biochemistry 43, 6182–6189 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Chou C. C., et al. , Activation of brain L-glutamate decarboxylase 65 isoform (GAD65) by phosphorylation at threonine 95 (T95). Mol. Neurobiol. 54, 866–873 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Davis T. M. E., et al. , Penetration of dihydroartemisinin into cerebrospinal fluid after administration of intravenous artesunate in severe falciparum malaria. Antimicrob. Agents Chemother. 47, 368–370 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmuck G., Roehrdanz E., Haynes R. K., Kahl R., Neurotoxic mode of action of artemisinin. Antimicrob. Agents Chemother. 46, 821–827 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brewer T. G., et al. , Fatal neurotoxicity of arteether and artemether. Am. J. Trop. Med. Hyg. 51, 251–259 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Wesche D. L., DeCoster M. A., Tortella F. C., Brewer T. G., Neurotoxicity of artemisinin analogs in vitro. Antimicrob. Agents Chemother. 38, 1813–1819 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang L., et al. , Crystal structure of pyridoxal kinase in complex with roscovitine and derivatives. J. Biol. Chem. 280, 31220–31229 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Bach S., et al. , Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 280, 31208–31219 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Gandhi A. K., et al. , Crystal structures of human pyridoxal kinase in complex with the neurotoxins, ginkgotoxin and theophylline: Insights into pyridoxal kinase inhibition. PLoS One 7, e40954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng F., et al. , Activin controls ethanol potentiation of inhibitory synaptic transmission through GABAA receptors and concomitant behavioral sedation. Neuropsychopharmacology 41, 2024–2033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M. Z., Elledge S. J., Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lineweaver H., Burk D., The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56, 658–666 (1934). [Google Scholar]
- 49.Dixon M., The determination of enzyme inhibitor constants. Biochem. J. 55, 170–171 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowe I. P., Robins E., Eyerman G. S., The fluorometric measurement of glutamic decarboxylase and its distribution in brain. J. Neurochem. 3, 8–18 (1958). [DOI] [PubMed] [Google Scholar]
- 51.Holdiness M. R., Justice J. B., Darryl B., Neill B. D., Salamone J. D., Fluorimetric assay for the determination of glutamic acid decarboxylase activity in subregions of rat brain tissue. Anal. Lett. 13, 1333–1344 (1980). [Google Scholar]
- 52.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schindelin J., Rueden C. T., Hiner M. C., Eliceiri K. W., The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of mPDXK-apo, mPDXK-ATPγS, and mPDXK-ATPγS–artesunate structures have been deposited in the PDB with accession codes 6YJZ, 6YK0, and 6YK1, respectively.