Significance
Many bacteria use the type VI secretion system (T6SS) to compete with other microorganisms in the environment. These systems eject spear-like projectiles that impale neighboring cells to deliver lethal toxins. Here, we report that antibacterial toxins encoded by rearrangement hotspot (rhs) genes are critical for assembly of the T6SS apparatus in Enterobacter cloacae. Rhs proteins fortify the trimeric cell-penetrating spike at the tip of the projectile, and their toxin-encapsulating cage structures are required for full secretion activity. These findings suggest that Rhs proteins mediate a quality control checkpoint to ensure that the spike is loaded with toxic cargo before the cell assembles the T6SS apparatus.
Keywords: bacterial competition, toxin−immunity proteins, self/nonself discrimination
Abstract
Bacteria deploy rearrangement hotspot (Rhs) proteins as toxic effectors against both prokaryotic and eukaryotic target cells. Rhs proteins are characterized by YD-peptide repeats, which fold into a large β-cage structure that encapsulates the C-terminal toxin domain. Here, we show that Rhs effectors are essential for type VI secretion system (T6SS) activity in Enterobacter cloacae (ECL). ECL rhs− mutants do not kill Escherichia coli target bacteria and are defective for T6SS-dependent export of hemolysin-coregulated protein (Hcp). The RhsA and RhsB effectors of ECL both contain Pro−Ala−Ala−Arg (PAAR) repeat domains, which bind the β-spike of trimeric valine−glycine repeat protein G (VgrG) and are important for T6SS activity in other bacteria. Truncated RhsA that retains the PAAR domain is capable of forming higher-order, thermostable complexes with VgrG, yet these assemblies fail to restore secretion activity to ∆rhsA ∆rhsB mutants. Full T6SS-1 activity requires Rhs that contains N-terminal transmembrane helices, the PAAR domain, and an intact β-cage. Although ∆rhsA ∆rhsB mutants do not kill target bacteria, time-lapse microscopy reveals that they assemble and fire T6SS contractile sheaths at ∼6% of the frequency of rhs+ cells. Therefore, Rhs proteins are not strictly required for T6SS assembly, although they greatly increase secretion efficiency. We propose that PAAR and the β-cage provide distinct structures that promote secretion. PAAR is clearly sufficient to stabilize trimeric VgrG, but efficient assembly of T6SS-1 also depends on an intact β-cage. Together, these domains enforce a quality control checkpoint to ensure that VgrG is loaded with toxic cargo before assembling the secretion apparatus.
Bacteria use many strategies to compete against other microorganisms in the environment. Research over the past 15 y has uncovered several distinct mechanisms by which bacteria deliver inhibitory toxins directly into neighboring competitors (1–8). Cell contact-dependent competition systems have been characterized most extensively in Gram-negative bacteria, and the most widespread mechanism is mediated by the type VI secretion system (T6SS) (9). T6SSs are multiprotein complexes related in structure and function to the contractile tails of Myoviridae bacteriophages. T6SS loci vary considerably between bacterial species, but all encode 13 core type VI secretion (Tss) proteins that are required to build a functional apparatus. TssJ, TssL, and TssM form a multimeric complex that spans the cell envelope and serves as the secretion conduit. The phage-like baseplate is composed of TssE, TssF, TssG, and TssK proteins, which form a sixfold symmetrical array surrounding a central “hub” of trimeric valine−glycine repeat protein G (VgrG/TssI). VgrG is structurally homologous to the gp27−gp5 tail spike of phage T4 (10, 11). The T4 tail spike is further acuminated with gp5.4, a small protein that forms a sharpened apex at the tip of the gp5 spike (12). Proline−alanine−alanine−arginine (PAAR) repeat proteins form an orthologous structure on VgrG; and PAAR is thought to facilitate penetration of the target cell outer membrane (13). The T6SS duty cycle begins when the baseplate docks onto TssJLM at the cytoplasmic face of the inner membrane (14). The baseplate then serves as the assembly origin for the contractile sheath and inner tube. The sheath is built from TssB−TssC subunits, and the tube is formed by stacked hexameric rings of hemolysin-coregulated protein (Hcp/TssD). TssA coordinates this assembly process to ensure that the sheath and tube are polymerized at equivalent rates (15). After elongating across the width of the cell, the sheath undergoes rapid contraction to expel the PAAR•VgrG-capped Hcp tube through the transenvelope complex. The ejected tube impales neighboring cells and delivers a variety of toxic effector proteins into the target. After firing, the contracted sheath is disassembled by the ClpV (TssH) ATPase (16), and the recycled TssBC subunits are used to support additional rounds of sheath assembly and contraction.
T6SSs were originally identified through their ability to intoxicate eukaryotic host cells (17), and VgrG proteins were the first effectors to be recognized. VgrG-1 from Vibrio cholerae V52 carries a C-terminal domain that cross-links actin and blocks macrophage phagocytosis (10). Similarly, the VgrG1 protein from Aeromonas hydrophila American Type Culture Collection (ATCC) 7966 carries a C-terminal actin adenosine 5′-diphosphate (ADP) ribosyltransferase domain that disrupts the host cytoskeleton (18). Although the T6SS clearly plays a role in pathogenesis, most of the systems characterized to date deliver toxic effectors into competing bacteria. Because antibacterial effectors are potentially autoinhibitory, these latter toxins are invariably encoded with specific immunity proteins. Antibacterial effectors commonly disrupt the integrity of the bacterial cell envelope. VgrG-3 from V. cholerae carries a lysozyme-like domain that degrades the peptidoglycan cell wall (19, 20). Other peptidoglycan-cleaving amidase toxins are packaged within the lumen of Hcp hexamers for T6SS-mediated delivery (21–24). Phospholipase toxins collaborate with peptidoglycan degrading enzymes to lyse target bacteria (25–27). Other T6SS effectors act in the cytosol to degrade nucleic acids and nicotinamide adenine dinucleotide cofactors (3, 28, 29). Most recently, Whitney and coworkers described a novel T6SS effector that produces the inhibitory nucleotide ppApp (30). These latter toxins are commonly delivered through noncovalent interactions with VgrG. Many effectors contain PAAR domains, which enable direct binding to the C-terminal β-spike of VgrG (13), whereas others are indirectly tethered to VgrG through adaptor proteins (31–33). This combinatorial strategy allows multiple different toxins to be delivered with each firing event.
Rearrangement hotspot (Rhs) proteins are potent effectors deployed by many T6SS+ bacteria (3, 34–37). T6SS-associated Rhs effectors range from ∼150 kDa to 180 kDa in mass and carry highly variable C-terminal toxin domains. The N-terminal region of Rhs proteins often contains two predicted transmembrane (TM) helix regions followed by a PAAR domain. The central region is composed of many Rhs/YD-peptide repeats, which form a β-cage structure that fully encapsulates the toxin domain (38). Genes coding for Rhs were first identified in Escherichia coli K-12 as elements that promote chromosomal duplication (39, 40). This genomic rearrangement was the result of unequal recombination between the rhsA and rhsB loci, which share 99.4% sequence identity over some 3,700 nucleotides. Subsequently, Hill and coworkers recognized that rhs genes are genetic composites (41), and that the variable C-terminal extension domains inhibit cell growth (42). Although E. coli K-12 encodes four full-length Rhs proteins, it lacks a T6SS, and there is no evidence that it deploys Rhs in competition. However, other Rhs/YD-peptide repeat proteins are known to deliver toxins in a T6SS-independent manner. Gram-positive bacteria export antibacterial YD-repeat proteins through the Sec pathway (3), and the tripartite insecticidal toxin complexes released by Photorhabdus and Yersinia species contain subunits with Rhs/YD repeats (38, 43). Thus, the Rhs encapsulation structure has been incorporated into at least three different toxin delivery platforms.
Here, we report that Rhs effectors are critical for the activity of the T6SS-1 locus of Enterobacter cloacae ATCC 13047 (ECL). ECL encodes two Rhs effectors—RhsA and RhsB—which are each exported in a constitutive manner by T6SS-1 (35). Deletion of either rhs gene has little effect on T6SS-1 activity, but mutants lacking both rhsA and rhsB are defective for Hcp1 secretion and no longer inhibit target bacteria. Although ∆rhsA ∆rhsB mutants lose T6SS-1−mediated inhibition activity, they still assemble and fire contractile sheaths at a significantly reduced frequency. We further show that truncated RhsA that retains the PAAR domain still interacts with cognate VgrG2, but the resulting complex does not support Hcp1 secretion or target-cell killing. Full T6SS-1 function requires wild-type Rhs effectors that retain the N-terminal TM helices and PAAR domain together with an intact β-cage. These findings suggest that the Rhs β-cage mediates a quality control checkpoint on T6SS-1 assembly to ensure that VgrG is loaded with a toxic effector prior to export.
Results
ECL T6SS-1 Effectors.
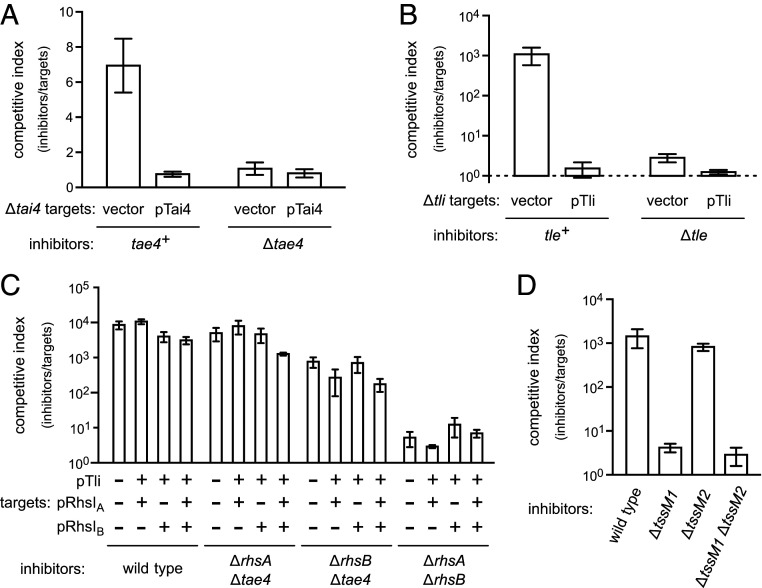
Wild-type ECL cells deploy RhsA (ECL_01567) and RhsB (ECL_03140) effectors through T6SS-1 (35). The T6SS-1 locus also encodes Tae4 (ECL_01542)/Tai4 (ECL_01543) and Tle (ECL_01553)/Tli (ECL_01554) effector/immunity protein pairs (SI Appendix, Fig. S1A). Tae4 is a hydrolytic amidase that cleaves peptidoglycan between D-glutamate and m-diaminopimelate (44, 45). Tle is a predicted phospholipase, and Tli is an ankyrin-repeat protein that presumably neutralizes Tle activity. We tested roles for Tae4 and Tle in competition using ECL ∆tae4 ∆tai4 and ∆tle ∆tli mutant strains as target cells in cocultures with wild-type ECL. Wild-type ECL has a slight growth advantage over ∆tae4 ∆tai4 cells, which is abrogated when targets are provided with plasmid-borne tai4 (Fig. 1A). Tle is much more potent, with wild-type cells outcompeting ∆tle ∆tli mutants almost 103-fold (Fig. 1B). Competitive fitness is restored to these latter targets when complemented with the wild-type tli gene (Fig. 1B). There is an additional PAAR-containing putative effector encoded by ECL_03144 adjacent to the rhsB locus (SI Appendix, Fig. S1B). However, cells lacking ECL_03144 and its presumed immunity gene (ECL_03145) are not inhibited significantly by wild-type ECL (SI Appendix, Fig. S2).
Fig. 1.
ECL T6SS-1 effectors. (A) ECL tae4+ and ∆tae4 inhibitor strains were cultured with ECL ∆tae4 ∆tai4 target cells that harbor an empty vector or a tai4 expression plasmid. (B) ECL tle+ and ∆tle inhibitor cells were cultured with ECL ∆tle ∆tli target cells that harbor an empty vector or tli expression plasmid. (C) The indicated ECL inhibitor strains were cultured with E. coli target cells. Target cells carried immunity protein expression plasmids where indicated (+). (D) The indicated ECL inhibitor strains were cultured with E. coli target cells. All cocultures were seeded at a 1:1 inhibitor to target cell ratio. The competitive index is the ratio of inhibitor to target bacteria at 4 h divided by the initial ratio. Data are the average ± SEM for three independent experiments.
We next examined the contribution of individual T6SS-1 effectors to the inhibition of E. coli cells. E. coli target cells that express rhsIA, rhsIB, and tli—either individually or in combination—are inhibited by wild-type ECL to approximately the same degree as targets with no immunity gene (Fig. 1C). These results are unexpected because the same immunity expression plasmids protect ECL targets that lack T6SS-1 immunity genes (Fig. 1 A and B) (35). To account for Tae4 effector delivery, we also tested ∆rhsA ∆tae4 and ∆rhsB ∆tae4 inhibitor strains and found again that immunity gene expression provided little protection to E. coli targets (Fig. 1C). We note that the combination of rhsIA, rhsIB, and tli immunity genes is expected to completely protect E. coli targets from the latter ECL strains. ECL encodes a second T6SS that could mediate inhibition, but the T6SS-2 locus contains several mutations that should inactivate the system (SI Appendix, Fig. S1C). A large deletion extends from the 3′ end of ompA (ECL_01805) through the first 118 codons of clpV2 (ECL_01806). This lesion probably deleted hcp2, because related Enterobacter T6SS loci encode Hcp proteins between ompA and clpV. The vgrG3 gene (ECL_01807) contains an ochre stop at codon 201. Finally, an IS903 element disrupts the ancestral PAAR-domain gene and ablates the 5′ end of tssF2 (ECL_01817). Nevertheless, we tested T6SS-2 activity and found that deletion of tssM2 has no discernable effect on the competitive fitness of ECL against E. coli targets (Fig. 1D). These results indicate that the T6SS-2 locus is defective and suggest that ECL deploys additional unidentified effectors through T6SS-1.
Rhs Is Required for ECL T6SS-1 Activity.
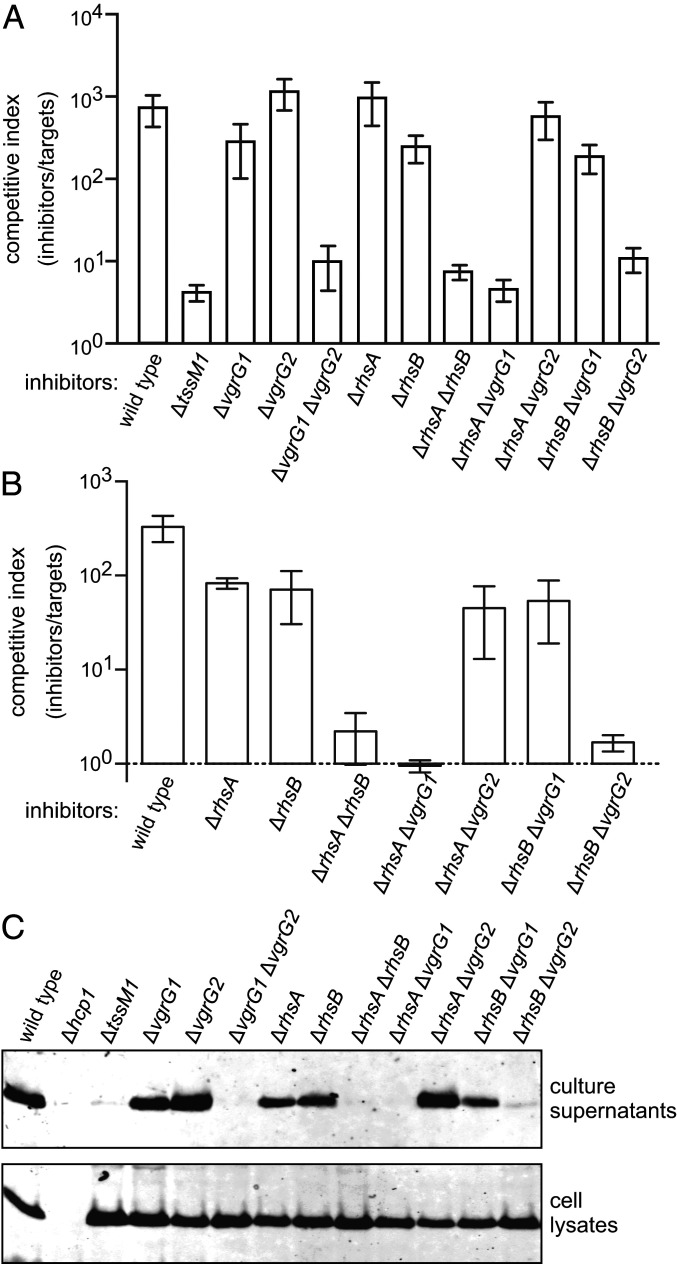
While evaluating T6SS-1 effectors, we found that ECL ∆rhsA ∆rhsB double mutants lose most of their growth advantage against E. coli target bacteria (Fig. 1C). In fact, the competitive fitness of ∆rhsA ∆rhsB cells is similar to that of the T6SS-1 defective ∆tssM1 mutant (Fig. 2A), suggesting that at least one Rhs protein is required to support secretion. Deletion of the vgrG1 and vgrG2 genes also abrogates T6SS-1 activity (Fig. 2A) (35). Furthermore, the deployment of RhsA and RhsB depends on VgrG2 and VgrG1 (respectively) (35), suggesting that ECL builds two distinct T6SS-1 assemblies around cognate RhsA•VgrG2 and RhsB•VgrG1 pairs. Accordingly, ∆rhsA ∆vgrG1 and ∆rhsB ∆vgrG2 mutants, which lack components from each Rhs•VgrG assembly, lose competitive fitness against E. coli (Fig. 2A). By contrast, ∆rhsA ∆vgrG2 and ∆rhsB ∆vgrG1 strains retain about the same growth advantage as the corresponding rhs and vgrG single mutants (Fig. 2A). Given the uncertainty surrounding the number and identity of T6SS-1 effectors deployed against E. coli, we also examined these mutants in growth competitions against ECL ∆tle ∆tli target cells, which are susceptible only to the Tle phospholipase toxin (Fig. 1B). As with the E. coli competitions, individual rhs mutants retain a significant growth advantage against ∆tle ∆tli targets, but the ∆rhsA ∆rhsB strain fails to inhibit target cell growth (Fig. 2B). Similarly, strains lacking components from each Rhs•VgrG assembly (e.g., ∆rhsA ∆vgrG1) also lose their competitive advantage. These inhibition activity defects correlate with the loss of Hcp1 secretion. Hcp1 is detected in the culture supernatant of wild-type ECL, and this export is T6SS-1 dependent because ∆tssM1 mutants accumulate cytoplasmic Hcp1, but fail to secrete the antigen (Fig. 2C). The ∆vgrG1 ∆vgrG2, ∆rhsA ∆rhsB, ∆rhsA ∆vgrG1, and ∆rhsB ∆vgrG2 mutants also exhibit significant defects in Hcp1 secretion (Fig. 2C). Therefore, T6SS-1−mediated secretion appears to require at least one Rhs effector in conjunction with its cognate VgrG protein.
Fig. 2.
Rhs is required for ECL T6SS-1 activity. (A) E. coli target bacteria were cocultured at a 1:1 ratio with the indicated ECL inhibitor strains for 4 h. (B) ECL ∆tle ∆tli target cells were cocultured at a 1:1 ratio with the indicated ECL inhibitor strains for 4 h. The competitive index is the ratio of inhibitor to target bacteria at 4 h divided by the initial ratio. Data are the average ± SEM for three independent experiments. (C) Culture supernatants and cell lysates from the indicated ECL strains were examined by immunoblotting using polyclonal antisera to Hcp1.
Rhs Promotes T6SS-1 Sheath Assembly.
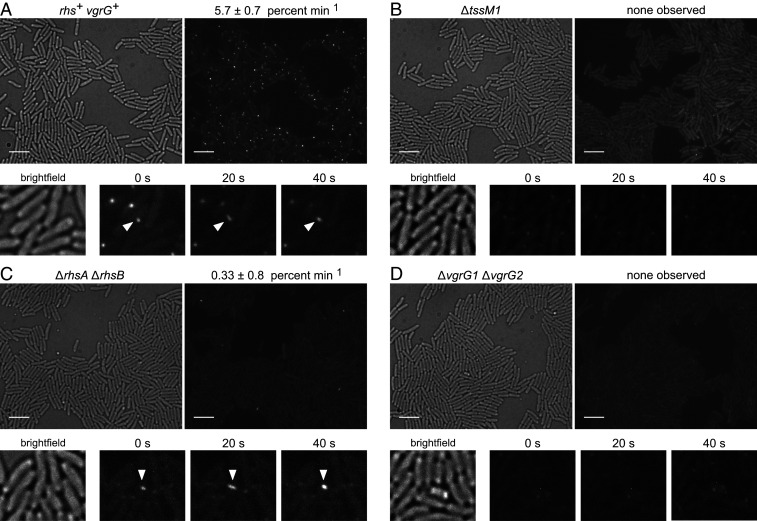
VgrG is a key component around which the T6SS baseplate is assembled (14). To explore whether Rhs is required for baseplate assembly in ECL, we monitored contractile sheath dynamics by time-lapse fluorescence microscopy using a GFP fusion to the TssB sheath protein (46). T6SS-1 firing events were enumerated as the percentage of cells that assemble and contract fluorescent sheaths over a 2.5-min observation period. Wild-type ECL assembles dynamic contractile sheaths at a frequency of 5.7 ± 0.7% min−1 (Fig. 3A and Movie S1). Assembly is completely abrogated in the ∆tssM1 mutant background, with no sheaths detected in 1,000 cells examined (Fig. 3B and Movie S2). Sheath assembly is diminished to 0.33 ± 0.8% min−1 in ∆rhsA ∆rhsB mutants, yet occasional assembly−contraction events were observed in each of five independent experiments (Fig. 3C and Movie S3). Altogether, we detected 28 assembly−contraction events in 2,828 ∆rhsA ∆rhsB cells. Notably, the ∆rhsA ∆rhsB phenotype is distinct from that of ∆vgrG1 ∆vgrG2 mutants, for which no assembly−contraction events were detected in the 877 cells we monitored (Fig. 3D and Movie S4). These results show that Rhs proteins are not strictly required for T6SS-1 assembly and contraction, but they significantly increase the frequency of assembly.
Fig. 3.
Rhs promotes T6SS-1 sheath assembly. Time-lapse fluorescence microscopy of (A) rhs+ vgrG+, (B) ∆tssM, (C) ∆rhsA ∆rhsB, and (D) ∆vgrG1 ∆vgrG strains that express the tssB-gfp fusion. Representative bright-field (Left) and fluorescence (Right) images are presented for each background. (Scale bars, 5 µm.) Time-lapse micrographs are shown below, with white carets indicating coupled assembly−contraction events. T6SS-1 firing frequency is expressed as the number of assembly−contraction events per 100 cells per min. Data are reported as the average ± SEM for at least three independent experiments.
Rhs PAAR Domains Are Not Sufficient to Support T6SS-1 Activity.
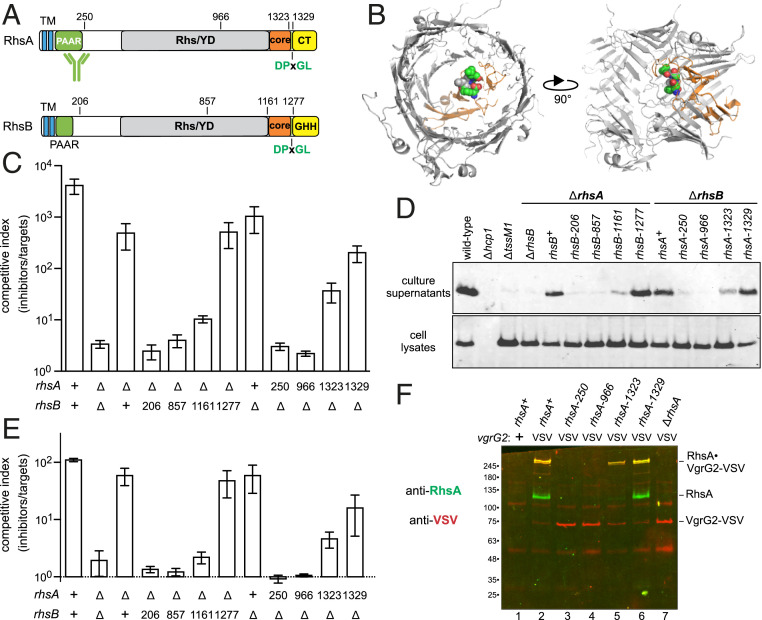
Rhs effectors are considered to be T6SS accessory factors, because many T6SS+ bacteria lack these proteins. However, Rhs proteins often contain PAAR domains (Fig. 4A), which are critical for T6SS activity in Acinetobacter baylyii ADP1 (13). Given that PAAR function is fulfilled by small peptides of ∼95 residues in V. cholerae and A. baylyii, we reasoned that truncated Rhs proteins that retain N-terminal PAAR domains should support T6SS-1 activity. We introduced in-frame stop codons along the chromosomal rhsA and rhsB loci (Fig. 4A) and tested whether the alleles support T6SS-1 activity in ∆rhsB and ∆rhsA backgrounds, respectively. These experiments showed that the Rhs-associated core domain is required for the inhibition of E. coli target bacteria and the secretion of Hcp1 (Fig. 4 A–D). The same results were obtained when the rhs truncation strains were competed against ECL ∆tle ∆tli target cells (Fig. 4E). We also found that contractile sheath dynamics are similar for wild-type rhsA (Movie S5), rhsA-1323 (Movie S6), and rhsA-1329 (Movie S7) when expressed in a ∆rhsB tssB-gfp background. By contrast, strains expressing the rhsA-250 (Movie S8) and rhsA-966 (Movie S9) alleles resemble ∆rhsA deletion mutants (Movie S10).
Fig. 4.
Rhs PAAR domains are not sufficient to support T6SS-1 activity. (A) RhsA and RhsB each contain two predicted N-terminal TM helices. The C-terminal toxin domains are delineated by DPxGL peptide motifs within the Rhs-associated core domain. The activity of RhsA-CT is unknown, and RhsB carries a Tox-GHH DNase domain (Pfam: PF15636). (B) Structural model of the RhsA β-cage. The YD-repeat region, core domain, and DPxGL motif are color coded as in A. (C) ECL inhibitor strains were cocultured with E. coli target bacteria at a 1:1 ratio for 4 h. (D) Culture supernatants and cell lysates from the indicated ECL strains were examined by immunoblotting with polyclonal antisera to Hcp1. (E) ECL inhibitor strains were cocultured with ECL ∆tle ∆tli target cells at a 1:1 ratio for 4 h. (F) VgrG2-VSV was immunoprecipitated from indicated ECL strains with anti-VSV agarose. Immunoprecipitates were analyzed by immunoblotting with antibodies to RhsA (green) and VSV-G (red).
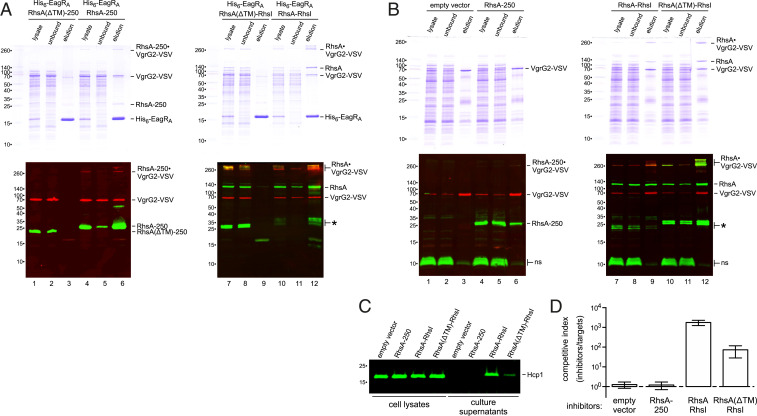
Structural models predict that the RHS core domain closes one end of the β-encapsulation cage (Fig. 4B), suggesting that an intact β-cage is required for T6SS-1 activity. Alternatively, the null phenotypes could be due to insolubility or rapid degradation of truncated Rhs proteins. Endogenous RhsA cannot be detected by immunoblot analysis, so we attempted to enrich truncated RhsA through copurification with the effector-associated gene with Rhs (EagRA) protein encoded upstream of rhsA (SI Appendix, Fig. S1A). Coulthurst and coworkers have shown that EagR adaptor proteins are required to stabilize Rhs for T6SS-dependent delivery in Serratia marcescens (37, 47). EagR dimers are thought to bind the predicted TM helices found at the N terminus of some PAAR domain-containing effectors (29, 48). We first confirmed that eagRA and eagRB are required for the delivery of RhsA and RhsB (respectively) in competition cocultures, showing that each adaptor is specific for its cognate Rhs effector (SI Appendix, Fig. S3A). We then overproduced His6-tagged EagRA in each rhsA truncation strain and isolated His6−EagRA•RhsA complexes by Ni2+-affinity chromatography. RhsA-966 was not isolated by this approach, but truncated RhsA-250, RhsA-1323, and RhsA-1329 were enriched to levels comparable with wild-type RhsA (SI Appendix, Fig. S3B). Thus, soluble RhsA-250 accumulates in the cell, but this truncated form fails to support T6SS-1 activity.
Given that RhsA-250 retains physiologically relevant interactions with EagRA, we next asked whether it also forms a stable complex with VgrG2. We first appended a VSV epitope to the C terminus of VgrG2 and confirmed that the tagged protein supports RhsA delivery (SI Appendix, Fig. S4). The vgrG2-VSV allele was then introduced into rhsA truncation strains to immunoprecipitate RhsA•VgrG2-VSV complexes. Wild-type RhsA and RhsA-1329 were detected in the anti-VSV immunoprecipitates, although there was little monomeric VgrG2-VSV in these samples (Fig. 4F, lanes 2 and 6). Instead, most of the VSV antigen was detected in high-mass species that also react with RhsA antibodies. This higher-order complex is resistant to thermal denaturation in sodium dodecyl sulfate (SDS), and its gel mobility is suggestive of trimeric VgrG2-VSV bound to RhsA. The high-mass complex was also isolated at a lower level from the rhsA-1323 strain (Fig. 4F, lane 5). By contrast, RhsA antigen was not detected in immunoprecipitates from the rhsA-250 and rhsA-966 strains, and all of the isolated VgrG2-VSV migrated as a monomer during SDS/polyacrylamide gel electrophoresis (PAGE) (Fig. 4F, lanes 3 and 4). Together, these results suggest that an intact β-cage is required to form functional RhsA•(VgrG2)3 complexes in ECL.
The Rhs β-Cage Is Required for T6SS-1 Activity.
Although the Rhs β-cage appears to stabilize trimeric VgrG, PAAR•VgrG structural models indicate that RhsA-250 should retain some affinity for VgrG2 (13). Therefore, we tested whether RhsA-250 and VgrG2-VSV interact when overproduced in E. coli cells that lack the T6SS-1 machinery. Under these conditions, RhsA-250 and VgrG2-VSV form a high-mass, thermostable complex that can be isolated by Ni2+-affinity chromatography through interactions with His6-EagRA (Fig. 5A, lanes 4 and 6). Moreover, a large proportion of monomeric VgrG2-VSV copurified with His6-EagRA•RhsA-250, suggesting that the separated PAAR domain retains significant affinity for VgrG2. To ensure that VgrG2-VSV was isolated through specific interactions, we repeated these experiments with RhsA(∆TM)-250, which lacks putative TM residues Ile25 to Leu79. RhsA(∆TM)-250 and VgrG2-VSV accumulated in E. coli cells, but neither copurified with His6-EagRA during Ni2+-affinity chromatography (Fig. 5A, lanes 1 and 3). Similarly, full-length RhsA(∆TM) does not interact with His6-EagRA, although it forms thermostable complexes with VgrG2-VSV (Fig. 5A, lane 7).
Fig. 5.
The Rhs β-cage is required for T6SS-1 activity. (A) His6-EagRA and RhsA variants were overproduced with VgrG2-VSV in E. coli cells for purification by Ni2+-affinity chromatography. Crude lysates and the unbound and elution fractions from Ni2+-nitrilotriacetate (NTA) resin were analyzed by SDS/PAGE on two-layer 7.5%/15% polyacrylamide gels (Top). Samples were also immunoblotted with antibodies to RhsA (green) and VSV-G (red) (Bottom). (B) EagRA and RhsA variants were overproduced with VgrG2-VSV in ECL ∆rhsB ∆rhsA cells for isolation by anti-VSV immunoprecipitation. Samples were analyzed as described for A. Nonspecific (ns) ECL proteins detected by RhsA antisera are indicated. (C) Cell lysates and culture supernatants from the ECL strains in B were examined by immunoblotting with polyclonal antisera to Hcp1. (D) ECL strains from B were cocultured at a 1:1 ratio with E. coli target bacteria. The competitive index is the ratio of inhibitor to target bacteria at 4 h divided by the initial ratio. Data are the average ± SEM for three independent experiments.
Given that overproduced RhsA-250 can stabilize trimeric VgrG-VSV in E. coli, we asked whether this approach restores T6SS-1 activity to ECL ∆rhsB ∆rhsA mutants. Thermostable RhsA-250•VgrG-VSV complexes were not detected in ECL cell lysates (Fig. 5B, lane 4), but the proteins clearly interact, because RhsA-250 coprecipitated with VgrG2-VSV (Fig. 5B, lane 6). The immunoprecipitate also contained a low level of high-mass RhsA-250•VgrG-VSV complex, which is more apparent when the fluorescence intensity is increased (SI Appendix, Fig. S5). Thermostable VrgG2-VSV trimers were not isolated from control cells that do not overproduce RhsA (SI Appendix, Fig. S5). Although overproduced RhsA-250 stabilizes VgrG2-VSV trimers, this ECL strain does not secrete significant amounts of Hcp1 (Fig. 5C), and it fails to inhibit E. coli target bacteria in coculture (Fig. 5D). By contrast, ECL ∆rhsB ∆rhsA cells complemented with wild-type RhsA secrete Hcp1 (Fig. 5C) and outcompete E. coli target bacteria ∼103-fold (Fig. 5D). We also tested RhsA(∆TM) activity in ECL, because it forms thermostable complexes with VgrG2-VSV as efficiently as wild-type RhsA (Fig. 5B, lanes 10 and 12). However, RhsA(∆TM) complementation only partially restored Hcp1 secretion and target-cell killing to ECL ∆rhsB ∆rhsA cells (Fig. 5 C and D). These results show that PAAR-stabilized VgrG trimers are not sufficient for secretion activity and suggest that the Rhs hydrophobic helices and β-cage play important roles in T6SS-1 assembly.
RhsA Is Processed to Release the C-Terminal Toxin Domain.
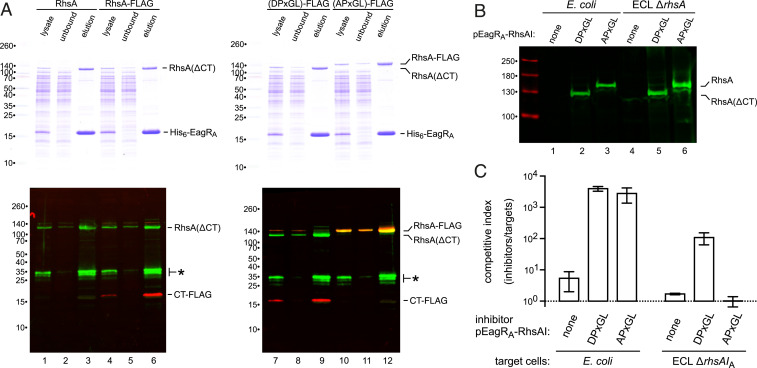
Truncated RhsA-1323 and RhsA-1329 exhibit the same electrophoretic mobility as full-length RhsA, suggesting that the C-terminal toxin domain is processed from the wild-type effector. Indeed, immunoblotting revealed the accumulation of a C-terminal fragment when FLAG epitope-tagged RhsA is produced in E. coli (Fig. 6A, lane 4). This analysis also detected N-terminal RhsA fragments, which may be related to processing recently reported for Rhs in Aeromonas (49). However, the biological significance of these fragments is unclear, because they are not detected when endogenous RhsA is immunoprecipitated from ECL cells with VgrG2-VSV (Fig. 4F, lane 2). Work with tripartite insecticidal proteins has shown that the Rhs-associated core domain acts as an autoprotease to cleave the DPxGL motif and release the toxin domain into the β-cage (38, 43). RhsA is likely processed in the same manner, because mutation of the DPxGL sequence to APxGL abrogates toxin domain processing (Fig. 6A; compare lanes 7 and 10). Moreover, the cleaved C-terminal fragment copurifies with His6-EagRA during Ni2+-affinity chromatography (Fig. 6A, lanes 6 and 9), consistent with retention inside the β-cage. Finally, we tested whether RhsA-CT processing is required for toxin delivery into target bacteria. We introduced the APxGL substitution into an eagRA-rhsAI expression plasmid and confirmed that processing is blocked when expressed in both E. coli and ECL cells (Fig. 6B, lanes 3 and 6). The DPxGL and APxGL constructs were then introduced into ECL ∆rhsA ∆rhsB cells, and the resulting strains tested for inhibition activity. Both constructs restored T6SS-1 function in interspecies competitions against E. coli target bacteria (Fig. 6C), indicating that RhsA-CT processing is not required for association with VgrG2 or assembly of the T6SS-1 apparatus. However, the RhsA APxGL variant did not inhibit ECL ∆rhsA ∆rhsIA target cells (Fig. 6C), which are only susceptible to the RhsA effector. This latter result demonstrates that the C-terminal domain must be cleaved from RhsA to intoxicate target bacteria.
Fig. 6.
RhsA is processed to release the C-terminal toxin domain. (A) His6-EagRA was coproduced with wild-type and FLAG-tagged RhsA variants in E. coli cells for purification by Ni2+-affinity chromatography. The DPxGL and APxGL RhsA constructs carry a SpeI restriction site that introduces an Arg1321Ser substitution. Crude lysates and the unbound and elution fractions from Ni2+-NTA resin were analyzed by SDS/PAGE on two-layer 7.5%/15% polyacrylamide gels (Top). Samples were also immunoblotted (Bottom) with antibodies to RhsA (green) and FLAG (red). Asterisks (*) indicate N-terminal RhsA degradation products. (B) The indicated EagRA-RhsA plasmids were introduced into E. coli and ECL ∆rhsA ∆rhsB cells, and cell lysates analyzed by immunoblotting with anti-RhsA antibodies. (C) ECL ∆rhsA ∆rhsB strains carrying EagRA-RhsA expression plasmids were used as inhibitors against E. coli and ECL ∆rhsA ∆rhsIA target bacteria.
Discussion
Rhs proteins are potent T6SS effectors deployed by many bacterial species (3, 35–37). Although distributed widely, Rhs is considered to be an accessory element of the T6SS, because well-characterized systems from V. cholerae and enteroaggregative E. coli (EAEC) lack these effectors (27, 50). However, we find that ECL requires at least one of its two Rhs proteins to inhibit E. coli target cells. Further, ECL ∆rhsA ∆rhsB mutants are defective for Hcp1 secretion and unable to deploy their phospholipase toxin. These phenotypes suggest that Rhs contributes to the core structure of T6SS-1. Given that RhsA and RhsB appear to be the only functional PAAR-repeat proteins associated with T6SS-1, they likely provide this structure for the apparatus. The PAAR motif folds into a small, conical domain that binds the blunt end of the VgrG spike through specific β-complementation interactions (13). PAAR tapers the spike into an acute apex, raising the possibility that it facilitates penetration through the target cell outer membrane (13). The domain has also been proposed to act as a chaperone that promotes the folding of β-spikes with complex topologies (13, 51). The latter hypothesis predicts that misfolded VgrG trimers could interfere with T6SS assembly, consistent with observed secretion defects in PAAR-deficient A. baylyii and V. cholerae mutants (13). Recent work also shows that the β-spike is critical for T6SS assembly in Acinetobacter (52). By contrast, deletion of the only recognizable PAAR-encoding gene from Edwardsiella tarda has no discernable effect on T6SS effector secretion or virulence in the fish host (53), perhaps indicating that its VgrG is not prone to misfolding. Our findings in ECL are broadly consistent with the chaperone model, with thermostable VgrG2 trimers only detected in cells that also produce RhsA. However, Rhs is not strictly required for secretion, because microscopy reveals occasional T6SS-1 assembly/contraction events in the ∆rhsA ∆rhsB background. Thus, VgrG can presumably fold and form functional baseplates with reduced efficiency in the absence of PAAR/Rhs.
Most T6SS loci encode small PAAR proteins (13), indicating that the free-standing domain is usually sufficient to support secretion activity. We find that the PAAR domain within truncated RhsA-250 retains significant affinity for VgrG2, enabling the proteins to form a thermostable complex when overproduced together. This complex almost certainly contains trimeric VgrG2 with a properly folded β-spike, because the gp5C tail spike trimer from phage T4 also resists dissociation in SDS (54, 55). Although RhsA-250 can stabilize trimeric VgrG2, it does not restore T6SS-1 activity to ECL ∆rhsA ∆rhsB mutants. This failure to complement could reflect the low level of thermostable complex formed with RhsA-250 compared to wild-type RhsA. However, time-lapse microscopy shows that ECL cells typically assemble only one T6SS-1 apparatus at any given time. Therefore, in principle, a single RhsA-250•(VgrG2)3 complex per cell should be sufficient for T6SS-1 assembly. Alternatively, the Rhs β-cage structure may be required to license full T6SS-1 assembly in ECL. This model is supported by recent work from Dong and coworkers, who showed that systematic removal of VgrG-associated effectors from V. cholerae reduces secretion efficiency (56). They propose that the T6SS apparatus discriminates between free VgrG and effector-loaded trimers. Such a checkpoint could be enforced through interactions with the transenvelope subassembly, consistent with electron cryotomography showing that its chamber is filled with density that likely corresponds to VgrG-associated effectors (57). This quality control mechanism would ensure that each VgrG trimer is loaded with toxic cargo before the cell commits to sheath assembly. In ECL, the hydrophobic N terminus of Rhs is also important, because an RhsA variant lacking this region only supports partial secretion activity, despite stabilizing VgrG trimers as efficiently as the wild-type effector. The predicted TM segments could tether Rhs•(VgrG)3 to the membrane to facilitate integration into the T6SS-1 apparatus, or perhaps they enable EagR to play an active role in baseplate assembly.
Finally, these results show that RhsA undergoes the same autoproteolytic processing originally observed with tripartite insecticidal toxin complexes from Photorhabdus and Yersinia (38, 43) and reported more recently for an Rhs effector from Aeromonas (49). Structural studies on the insecticidal complexes revealed that the RHS core domain is an aspartyl protease that releases the C-terminal toxin domain into the β-encapsulation cage (38, 43). This unusual architecture is thought to protect producing cells from autointoxication (38), but it also necessitates a mechanism to open the cage after delivery. Work from the Basler and Mekalanos laboratories indicates that the T6SS of V. cholerae perforates the entire target cell envelope to deliver effectors into the cytosol (58, 59). However, it is not clear that all bacteria transfer VgrG and Hcp directly into the target cell cytoplasm. For example, many Salmonella enterica and E. coli strains deploy “evolved” VgrG (NCBI RefSeq: WP_000371003.1) and Hcp (WP_000502504.1) nuclease effectors that carry pyocin S translocation domains (Pfam: PF06958) (59, 60). All nuclease bacteriocins contain this translocation domain (61), which exploits FtsH to mediate toxin transport from the periplasm to the cytosol (62–64). Thus, many T6SSs probably deposit their effectors into the periplasm, raising the question of how encapsulated Rhs toxin domains reach the cytoplasm. RhsA and RhsB do not contain pyocin S translocation domains, and their processed C-terminal regions lack the “cytoplasm-entry” domains that govern antibacterial CdiA toxin import (65, 66). Work from Quentin et al. (48) suggests that the N-terminal hydrophobic helices of Rhs could mediate this transport. The PAAR-containing Tse6 effector from Pseudomonas aeruginosa uses similar TM helices to dock onto membrane vesicles and transfer its toxic nicotinamide adenine dinucleotide-glycohydrolase domain across the lipid bilayer (48). In the context of Rhs, the hydrophobic segments could perhaps enable the β-cage to fuse with the membrane, thereby releasing toxin into the cytosol. Although this is an attractive model for ECL, there must be other delivery modalities for the many T6SS-associated Rhs nucleases that lack PAAR and N-terminal TM helices (9, 49).
Materials and Methods
Bacterial strains, plasmids, and oligonucleotides are listed in SI Appendix, Tables S1–S3, respectively. The details of all strain and plasmid constructions are provided in SI Appendix. Competition cocultures were performed at a 1:1 inhibitor and target cell ratio on lysogeny broth agar at 37 °C as described in SI Appendix. Viable inhibitor and target cells were enumerated as colony-forming units on antibiotic-selective media, and competitive indices were calculated as the final ratio of inhibitor to target cells divided by the initial ratio. Presented data are the average ± SEM for at least three independent experiments. Hcp1 secretion assays and immunoblot analysis was performed as described in SI Appendix. VgrG2-VSV was immunoprecipitated with anti−VSV-G agarose (Sigma-Aldrich) as described in SI Appendix. ECL strains expressing chromosomal tssB-gfp fusions were visualized by time-lapse fluorescence microscopy as described in SI Appendix. All data are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by Grant GM117930 (C.S.H.) from the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919350117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Aoki S. K., et al. , Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Hood R. D., et al. , A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koskiniemi S., et al. , Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. U.S.A. 110, 7032–7037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamet A., et al. , A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog. 11, e1004592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Souza D. P., et al. , Bacterial killing via a type IV secretion system. Nat. Commun. 6, 6453 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Cao Z., Casabona M. G., Kneuper H., Chalmers J. D., Palmer T., The type VII secretion system of Staphylococcus aureus secretes a nuclease toxin that targets competitor bacteria. Nat. Microbiol. 2, 16183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vassallo C. N., et al. , Infectious polymorphic toxins delivered by outer membrane exchange discriminate kin in myxobacteria. eLife 6, e29397 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruhe Z. C., Low D. A., Hayes C. S., Polymorphic toxin and their immunity proteins: Diversity, evolution, and mechanisms of delivery. Annu. Rev. Microbiol. 74, 497–520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D., de Souza R. F., Anantharaman V., Iyer L. M., Aravind L., Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J., Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spínola-Amilibia M., et al. , The structure of VgrG1 from Pseudomonas aeruginosa, the needle tip of the bacterial type VI secretion system. Acta Crystallogr. D Struct. Biol. 72, 22–33 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Buth S. A., et al. , Structure and biophysical properties of a triple-stranded beta-helix comprising the central spike of bacteriophage T4. Viruses 7, 4676–4706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shneider M. M., et al. , PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunet Y. R., Zoued A., Boyer F., Douzi B., Cascales E., The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 11, e1005545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zoued A., et al. , Priming and polymerization of a bacterial contractile tail structure. Nature 531, 59–63 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A., Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pukatzki S., et al. , Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suarez G., et al. , A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks T. M., Unterweger D., Bachmann V., Kostiuk B., Pukatzki S., Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J. Biol. Chem. 288, 7618–7625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., et al. , Structural basis for recognition of the type VI spike protein VgrG3 by a cognate immunity protein. FEBS Lett. 588, 1891–1898 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Russell A. B., et al. , Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverman J. M., et al. , Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol. Cell 51, 584–593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell A. B., Peterson S. B., Mougous J. D., Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitney J. C., et al. , Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J. Biol. Chem. 288, 26616–26624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell A. B., et al. , Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong T. G., Ho B. T., Yoder-Himes D. R., Mekalanos J. J., Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 2623–2628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flaugnatti N., et al. , A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol. Microbiol. 99, 1099–1118 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Ma L. S., Hachani A., Lin J. S., Filloux A., Lai E. M., Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16, 94–104 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitney J. C., et al. , An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163, 607–619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmad S., et al. , An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unterweger D., et al. , Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 34, 2198–2210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang X., et al. , Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc. Natl. Acad. Sci. U.S.A. 112, 9106–9111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bondage D. D., Lin J. S., Ma L. S., Kuo C. H., Lai E. M., VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koskiniemi S., et al. , Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet. 10, e1004255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitney J. C., et al. , Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92, 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hachani A., Allsopp L. P., Oduko Y., Filloux A., The VgrG proteins are “à la carte” delivery systems for bacterial type VI effectors. J. Biol. Chem. 289, 17872–17884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alcoforado Diniz J., Coulthurst S. J., Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J. Bacteriol. 197, 2350–2360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Busby J. N., Panjikar S., Landsberg M. J., Hurst M. R., Lott J. S., The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 501, 547–550 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Capage M., Hill C. W., Preferential unequal recombination in the glyS region of the Escherichia coli chromosome. J. Mol. Biol. 127, 73–87 (1979). [DOI] [PubMed] [Google Scholar]
- 40.Lin R. J., Capage M., Hill C. W., A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J. Mol. Biol. 177, 1–18 (1984). [DOI] [PubMed] [Google Scholar]
- 41.Hill C. W., Sandt C. H., Vlazny D. A., Rhs elements of Escherichia coli: A family of genetic composites each encoding a large mosaic protein. Mol. Microbiol. 12, 865–871 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Vlazny D. A., Hill C. W., A stationary-phase-dependent viability block governed by two different polypeptides from the RhsA genetic element of Escherichia coli K-12. J. Bacteriol. 177, 2209–2213 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meusch D., et al. , Mechanism of Tc toxin action revealed in molecular detail. Nature 508, 61–65 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Russell A. B., et al. , A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11, 538–549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H., et al. , Structure of the type VI effector-immunity complex (Tae4-Tai4) provides novel insights into the inhibition mechanism of the effector by its immunity protein. J. Biol. Chem. 288, 5928–5939 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J., Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cianfanelli F. R., et al. , VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 12, e1005735 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quentin D., et al. , Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat. Microbiol. 3, 1142–1152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pei T. T., et al. , Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system. Nat. Commun. 11, 1865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng J., Ho B., Mekalanos J. J., Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6, e23876 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchida K., Leiman P. G., Arisaka F., Kanamaru S., Structure and properties of the C-terminal β-helical domain of VgrG protein from Escherichia coli O157. J. Biochem. 155, 173–182 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Lopez J., Ly P. M., Feldman M. F., The tip of the VgrG spike is essential to functional type VI secretion system assembly in Acinetobacter baumannii. mBio 11, e02761-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng J., Leung K. Y., Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Kanamaru S., Gassner N. C., Ye N., Takeda S., Arisaka F., The C-terminal fragment of the precursor tail lysozyme of bacteriophage T4 stays as a structural component of the baseplate after cleavage. J. Bacteriol. 181, 2739–2744 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanamaru S., et al. , Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Liang X., et al. , An onboard checking mechanism ensures effector delivery of the type VI secretion system in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 116, 23292–23298 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang Y. W., Rettberg L. A., Ortega D. R., Jensen G. J., In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep. 18, 1090–1099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vettiger A., Basler M., Type VI secretion system substrates are transferred and reused among sister cells. Cell 167, 99–110.e12 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Ho B. T., Fu Y., Dong T. G., Mekalanos J. J., Vibrio cholerae type 6 secretion system effector trafficking in target bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 114, 9427–9432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma J., et al. , The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence 8, 1189–1202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharp C., Bray J., Housden N. G., Maiden M. C. J., Kleanthous C., Diversity and distribution of nuclease bacteriocins in bacterial genomes revealed using Hidden Markov Models. PLoS Comput. Biol. 13, e1005652 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mora L., Moncoq K., England P., Oberto J., de Zamaroczy M., The stable interaction between signal peptidase LepB of Escherichia coli and nuclease bacteriocins promotes toxin entry into the cytoplasm. J. Biol. Chem. 290, 30783–30796 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang J. W., et al. , Crystal structure of the central and the C-terminal RNase domains of colicin D implicated its translocation pathway through inner membrane of target cell. J. Biochem. 164, 329–339 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Atanaskovic I., et al. , Targeted killing of Pseudomonas aeruginosa by pyocin G occurs via the hemin transporter Hur. J. Mol. Biol. 432, 3869–3880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willett J. L., Gucinski G. C., Fatherree J. P., Low D. A., Hayes C. S., Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc. Natl. Acad. Sci. U.S.A. 112, 11341–11346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bartelli N. L., et al. , The cytoplasm-entry domain of antibacterial CdiA is a dynamic α-helical bundle with disulfide-dependent structural features. J. Mol. Biol. 431, 3203–3216 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.