Significance
Pol ι is a highly conserved Y-family DNA polymerase. Disruptions of the Pol ι gene in cellular and mouse models, however, have so far failed to produce any detectable phenotype. Using unbiased whole genome fitness screens, we find that Pol ι is essential for the genome stability and survival of Fanconi anemia mutant cells. This finding reveals the physiological role for Pol ι as a compensatory factor of Fanconi anemia deficiency. It implies a novel genetic connection between lesion bypass and replication stress response. Furthermore, the identified synthetic lethality between Pol ι and the Fanconi anemia mechanism suggests an enzymatic and therapeutic target for eliminating tumor cells arising from the Fanconi anemia pathway defect.
Keywords: DNA polymerase, Fanconi anemia pathway, whole genome fitness screens, lesion bypass
Abstract
Fanconi anemia (FA) is caused by defects in cellular responses to DNA crosslinking damage and replication stress. Given the constant occurrence of endogenous DNA damage and replication fork stress, it is unclear why complete deletion of FA genes does not have a major impact on cell proliferation and germ-line FA patients are able to progress through development well into their adulthood. To identify potential cellular mechanisms that compensate for the FA deficiency, we performed dropout screens in FA mutant cells with a whole genome guide RNA library. This uncovered a comprehensive genome-wide profile of FA pathway synthetic lethality, including POLI and CDK4. As little is known of the cellular function of DNA polymerase iota (Pol ι), we focused on its role in the loss-of-function FA knockout mutants. Loss of both FA pathway function and Pol ι leads to synthetic defects in cell proliferation and cell survival, and an increase in DNA damage accumulation. Furthermore, FA-deficient cells depend on the function of Pol ι to resume replication upon replication fork stalling. Our results reveal a critical role for Pol ι in DNA repair and replication fork restart and suggest Pol ι as a target for therapeutic intervention in malignancies carrying an FA gene mutation.
Fanconi anemia (FA) is a genomic instability disorder caused by biallelic or x-linked mutations in any of 22 genes. FA patients are characterized by multiple developmental abnormalities, progressive bone marrow failure, and profound cancer susceptibility (1–3). Germ-line FA mutations predispose an individual to breast, ovarian, pancreatic, and hematological malignancies. Somatic FA mutations have been identified in sporadic acute leukemia and breast cancer (4–6).
The FA pathway is the major cellular mechanism responding to DNA crosslinking damage and replication stress. The 22 FA gene products fall into several functional groups. In response to DNA damage, the FANCD2/FANCI complex is monoubiquitinated, signifying the activation of the canonical FA pathway (7, 8). The monoubiquitinated FANCD2/FANCI complex most likely orchestrates the recruitment of nucleolytic factors for the processing of crosslinking DNA damage (9, 10). The FA core complex, consisting of FANCA, -B, -C, -E, -F, -G, and -M, FAAP20, FAAP24, FAAP100, and the RING domain protein FANCL, provides the E3 ligase activity for the damage-induced monoubiquitination of FANCD2/FANCI (11–16). FANCP/XPF and FANCQ/SLX4, the third group of FA gene products, are nucleases or part of the nuclease scaffold, taking part in DNA cleavage for the removal of the crosslinking lesions (8, 17–21). DNA double-strand breaks, as an intermediate structure of ICL (Interstrand CrossLink) repair, depend on the fourth group of FA proteins, required in homologous recombination (FANCD1/BRCA2, FANCO/RAD51C, FANCJ/BARD1, and FANCR/RAD51) (22–26).
In addition to the direct role in crosslinking damage repair, FA pathway components are linked to the protection of replication fork integrity during replication interruption that is not directly caused by damage to the DNA. BRCA1/2 are important in stabilizing stalled forks in an MRE11-dependent manner (27, 28). Similarly, FANCD2 and FANCI have been shown to prevent collapse of stalled replication forks (29, 30). Defects in the FA and recombination mechanisms lead to severe fork erosion and endogenous DNA damage accumulation upon reversible replication block, suggesting that the FA pathway plays a crucial role in DNA replication under both normal and perturbed growth conditions (8, 23, 31–34).
Given the important role of the FA pathway in replication stress, it is perplexing that cells with a completely impaired FA mechanism are capable of sustained proliferation (34, 35). Overt abnormalities are absent in mice with knockout of several key FA genes (36–39). Moreover, individuals can survive without a functional FA pathway for decades (median life expectancy of 30 y for FA patients) (40). More recently, a genome-scale CRISPR-Cas9 guide RNA (gRNA) library screen has defined gene sets essential for proliferation of common model cell lines (41). None of the classic FA genes which participate in the monoubiquitination process appear to be essential in these screens. Cells deficient in classic FA genes can sustain growth despite the accumulation of endogenous DNA damage. Thus, it seems likely that compensatory mechanisms exist in FA mutant cells to support long-term viability.
In this study, we sought to identify cellular mechanisms that are important for the survival of cells deficient in the FA pathway. Comparative genome-scale CRISPR/Cas9 screens were carried out in isogenic FA pathway-proficient and -deficient cells. Genes that exhibit synthetic lethality in FA mutant cells are candidates which compensate for the loss of the FA pathway function. Among the top candidates, we validated and investigated DNA polymerase (Pol) ι as a critical factor for the survival of FA mutant cells. We found that, in FA-deficient cells, Pol ι is crucial in the resumption of stressed replication forks and in suppressing the accumulation of endogenous DNA damage. This reveals a function for Pol ι in relieving DNA damage stress.
Results
Genome-Wide CRISPR Screen Identifies Synthetic Lethal Interactions with the FA Pathway.
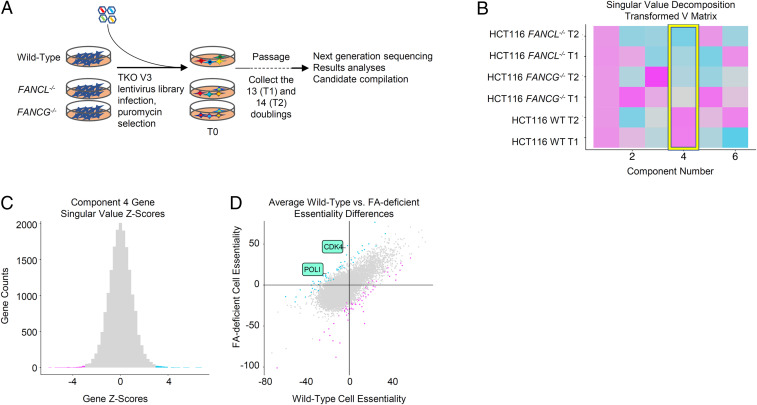
To identify FA synthetic lethal candidates, whole genome dropout screens were carried out to isolate candidates that compromise the survival of FA-deficient cells but not isogenic wild-type controls. Two Fanconi mutant cell lines, FANCL−/− HCT116 and FANCG−/− HCT116, generated via homologous targeting, were used in parallel in the screens (35). This strategy minimizes gene knockout-specific noise and off-target effects from using gRNA-mediated gene inactivation. We screened a whole genome scale gRNA library (42). Wild-type cells and FA-deficient cell lines were separately transduced with the lentivirus-based gRNA library. After infection, the cells were selected with puromycin for 48 h. Cell pellets were collected before (T0) and after passaging these cells for 13 and 14 population doublings (T1 and T2), and each time point had triplicate technical replicates. Genomic DNA from each replicate was extracted independently and labeled with barcodes. The relative change of gRNAs during culture was determined by next-generation sequencing (NGS) of PCR-amplified gRNAs from T0, T1, and T2 cell pellets (Fig. 1A).
Fig. 1.
FA mutant synthetic lethality screen. (A) Schematic diagram depicting the gRNA-mediated whole genome fitness screen. (B) Resulting SVD v matrix of processed quantile-normalized BAGEL essential scores (BF) of FA-deficient and wild-type screens. Color similarity in the matrix represents the direction and amount of signal that is contributing to the individual component. (C) Histogram of Z-transformed component 4 scores coming from matrix u singular values, representing genetic contribution to differences between the FA knockouts and wild-type samples shown in the v matrix above. Blue, Z < −3; pink, Z > 3. (D) Comparison of mean BF scores in the FANCL−/− and FANCG−/− screens versus wild-type screen, colored as in C.
In order to identify target synthetic lethal hits with the corresponding FA-deficient mutant screens, we used a matrix factorization technique, singular value decomposition (SVD). SVD has been used previously for identifying genetic expression differences between backgrounds (43), and analogous techniques have been proposed to identify synthetic lethal targets (44). To our knowledge, however, this method has not been used previously to identify synthetic lethal candidates emerging from genetic fitness screens with single gene knockouts.
For this analysis, the input dataset comprised six separate genome-wide fitness screen samples conducted in HCT116 cells using the TKOv3 library. The six screens were made up of wild-type, FANCL−/−, and FANCG−/− cells at two separate time points/doubling periods. The raw reads generated from these screens were processed with the genetic fitness classifier Bayesian Analysis of Gene Essentiality (BAGEL) (45) software to calculate gene-level fitness scores (Bayes factor [BF]) where positive scores indicate essential genes, and negative scores indicate no knockout phenotype. We compared the screens to previously identified sets of known essential and nonessential fitness genes (41, 46) in order to assess sufficient screen performance (SI Appendix, Fig. S1A). BFs from the six samples were then combined into a single matrix and quantile-normalized, and the resulting matrix was decomposed using SVD. The first component of the SVD v matrix identifies essential genes common to all six screens while the fourth component highlights variation between the FA knockout and wild-type samples (Fig. 1B). This observed variation represents genetic fitness differences that exist between the FA knockouts and wild-type samples (Fig. 1C). We were specifically interested in genes that had high differential fitness scores by SVD that also have a sign change in BF (Fig. 1D and SI Appendix, Fig. S1 B–E): that is, genes that are essential in one background and nonessential in the other. We filtered for genes that were essential in FA knockout cells, nonessential in wild-type isogenic controls, and which showed strong projection on the fourth singular vector (SI Appendix, Fig. S1F). As such, we decided to further validate the top two genes of interest that met both criteria, Pol ι and CDK4 (47, 48).
Pol ι and CDK4 Depletion Leads to Synthetic Growth Suppression in FA Mutant Cells.
The CDK4 kinase drives G1/S cell cycle transition and S phase progression. Its action as a compensatory factor for the FA pathway might emerge if a full complement of CDK4/6 kinase activity is important to overcome cell cycle attenuation arising from accumulation of endogenous lesions in FA mutant cells. It is also possible that CDK function is required in the regulation of lesion bypass activities (49–51). Pol ι was particularly interesting for further study. It is one of 16 known DNA polymerases encoded by mammalian genomes. Although purified Pol ι is known to have translesion polymerase activity for a few examples of DNA adducts, very little is known about its physiological function (52). We hypothesized that its function becomes necessary for cell survival in the absence of an FA-mediated response to replication fork stress.
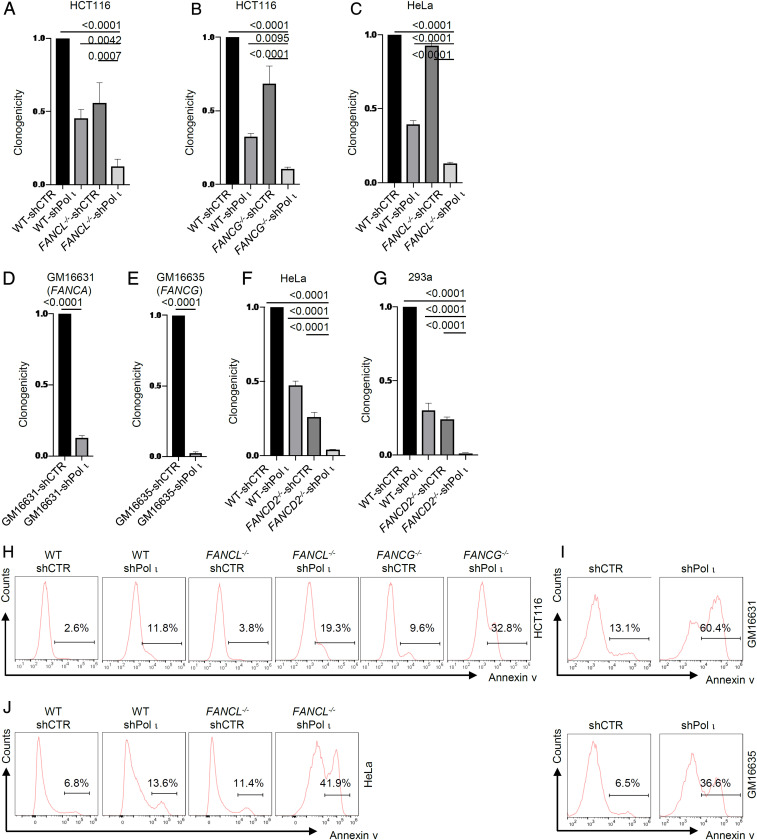
To verify the primary screen results with an independent approach, we used small hairpin RNA (shRNA) to deplete Pol ι or CDK4 in HCT116 wild-type control cells, FANCL-/−, and FANCG−/− and analyzed the effects on cell clonogenicity. Knockdown of Pol ι reduced clonogenicity by 9.4- to 7.9-fold in FANCG−/− or FANCL−/− cells, and by ∼2.7-fold in wild-type HCT116 cells (Fig. 2 A and B and SI Appendix, Fig. S2A). To examine compensatory functions in another cell line, we constructed a FANCL−/− mutant in HeLa cells. We then performed shRNA-mediated knockdown to deplete Pol ι and examined colony-forming ability under unperturbed conditions. Loss of Pol ι in HeLa FANCL−/− cells reduced clonogenicity by 7.6-fold and reduced clonogenicity in wild-type HeLa cells by 2.5-fold (Fig. 2C). Since the primary screens were conducted using FA gene null mutants, we further validated the synthetic phenotype between FA deficiency and Pol ι in fibroblast cells derived from FA patients. Knockdown of Pol ι in both FANCA-deficient and FANCG-deficient cell lines derived from FA patients resulted in 7.8-fold and 40.3-fold reduction of clonogenicity (Fig. 2 D and E). Additionally, the synthetic fitness defect between FA deficiency and Pol ι was observed as nearly abolished cell proliferation in knockouts and patient FA mutant cells devoid of Pol ι (SI Appendix, Fig. S3 A–E).
Fig. 2.
Pol ι loss leads to synthetic lethality with FANCL/FANCG/FANCD2 deficiency. (A–C) Clonogenic efficiency of Pol ι shRNA-knockdown HCT116 FANCL−/− cells, FANCG−/− cells, and HeLa FANCL−/− cells. (D and E) Clonogenic efficiency of Pol ι shRNA-knockdown GM16631 and GM16635 patient cells. (F–G) Clonogenic efficiency of Pol ι shRNA-knockdown HeLa FANCD2−/− and 293A FANCD2−/− cells. (H) Flow cytometry histogram of Annexin V-positive cells in HCT116 wild-type (WT), FANCL−/−, and FANCG−/− cells subjected to shRNA-knockdown of Pol ι. (I) Flow cytometry histogram of Annexin V-positive cells in GM16631 and GM16635 patient cells subjected to shRNA-knockdown of Pol ι. (J) Flow cytometry histogram of Annexin V-positive cells in HeLa WT and FANCL−/− cells subjected to shRNA-knockdown of Pol ι.
To further extend the compensatory function of Pol ι in the FA pathway, we depleted Pol ι from HeLa FANCD2−/− and 293A FANCD2−/− mutants via shRNA-mediated knockdown. Analyses of cell proliferation and clonogenicity showed drastic reduction of both readouts (Fig. 2 F and G and SI Appendix, Fig. S3 F and G). Together, these results show cell fitness is highly dependent on Pol ι in the absence of the FA pathway. It is noteworthy that shRNA-mediated Pol ι depletion had a modest impact on cell proliferation and clonogenicity in FA-proficient cells, which may reflect a transient effect from acute Pol ι loss in wild-type cell culture.
To determine whether the synthetic phenotype of reduced cell proliferation is associated with cell death, we analyzed the apoptotic status of FA-deficient cells lacking Pol ι. Loss of Pol ι led to significant increases in apoptotic cell population in FANCL−/− and FANCG−/− HCT116 cells, FANCL−/− HeLa cells, and FA patient cells, suggesting enhanced genotoxicity under the combined loss of the FA pathway and Pol ι functions (Fig. 2 H–J).
Similarly, we found that loss of CDK4 decreased colony-forming ability and cell proliferation, accompanied by the increase of apoptotic cells, with several fold more pronounced effects in FA-deficient cells than in FA-proficient controls (SI Appendix, Figs. S2B and S4). Given that the function of Pol ι in the DNA damage response mechanism is not clearly understood, we focused on its potential interplay with the FA pathway.
Pol ι Uniquely Sustains the Survival of FA-Deficient Cells among Lesion Bypass Polymerases.
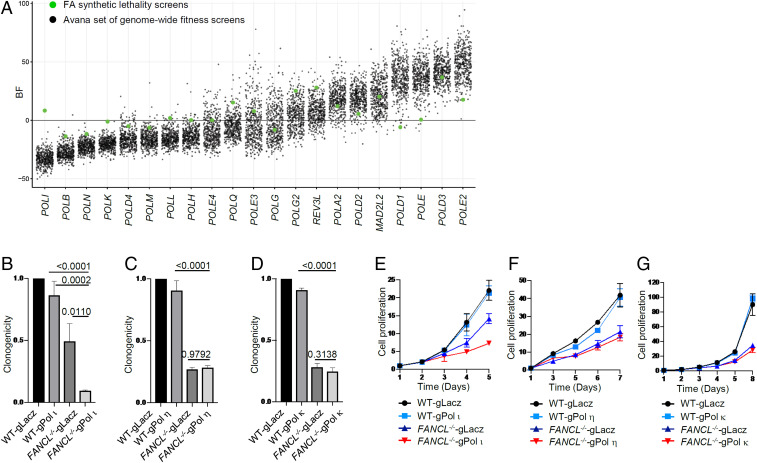
Pol ι is one of the four Y-family DNA polymerases in mammalian cells. Deletion of Pol ι in either mouse or human cells does not produce detectable phenotypes (53–56). To further ascertain the compensatory function of Pol ι in FA-deficient cells, we analyzed 21 replicative and lesion bypass DNA polymerases from 426 CRISPR fitness screens. As shown (Fig. 3A), Pol ι displayed the lowest BF scores from the existing screen data, confirming its less essential role in cell proliferation compared to other DNA polymerases. In FA-deficient cells, however, Pol ι became much more important for cell survival, as reflected by the markedly increased BF score, while the BF scores for other polymerases were similar in FA-proficient and FA-deficient cells. This result suggests that Pol ι is uniquely relevant to the viability of FA mutant cells.
Fig. 3.
Pol ι loss renders unique synthetic lethality with FA deficiency. (A) BF distribution of DNA polymerase subunits from the FA synthetic lethality screens (green dots) and the Avana set (425 screens form Q4 2018) of genome-wide fitness screens (black dots). (B–D) Clonogenicity of Pol ι, η, and κ gRNA knockdown in HCT116 wild-type cells and FANCL−/− cells. (E–G) Cell proliferation of Pol ι, η, and κ gRNA knockdown in HCT116 wild-type cells and FANCL−/− cells.
To test this notion, we used gRNA/CAS9 to deplete Pol ι from FANCL−/− and FANCG−/− cells (SI Appendix, Fig. S5). As shown in Fig. 3B and SI Appendix, Fig. S6A, loss of Pol ι led to significant attenuation (10.5-fold and 5.2-fold, respectively) of clonogenicity in FANCL−/− or FANCG−/− cells compared to knockdown alone (1.2-fold and 1.5-fold, respectively) or the FA mutants alone (2.0-fold and 1.1-fold, respectively). This result is also observed when analyzing cell proliferation of FA wild-type and mutant cells lacking Pol ι (Fig. 3E and SI Appendix, Fig. S6C). These results validate the observation from the primary screen suggesting that Pol ι becomes important in cells devoid of FA pathway function.
To further assess the role of Pol ι in cell fitness acquired as a result of an FA pathway defect, we tested other members of the Y family of DNA polymerases. When polymerase eta (Pol η) was depleted via gRNA/CAS9 treatment from wild-type, FANCL−/−, and FANCG−/− HCT116 cells, no decrease in clonogenic survival was detected in the FA mutant cells compared to the LacZ gRNA control (Fig. 3C and SI Appendix, Fig. S6E). Similarly, depletion of Pol η from FANCL−/− cells and FANCG−/− cells did not affect cell growth/viability (Fig. 3F and SI Appendix, Fig. S6G). These results indicate that cells deficient in the FA pathway do not rely on the function of Pol η for their proliferation. We next tested whether polymerase kappa (Pol κ) is required for the fitness of FANCL−/− and FANCG−/− cells. Both clonogenic and cell growth assays showed that Pol κ knockdown did not render any growth disadvantage to FANCL−/− or FANCG−/− cells (Fig. 3 D and G and SI Appendix, Fig. S6 F and H).
To verify the synthetic phenotype between Pol ι and a defective FA pathway in a different cell background, we examined the effects of depletion of Pol ι, Pol η, and Pol κ in HeLa FANCL−/− and wild-type cells. Consistent with the results from the HCT116 cells, loss of Pol ι distinctly affected clonogenicity and cell proliferation of the HeLa FANCL−/− mutant (SI Appendix, Fig. S6 B and D) while depletion of Pol η or Pol κ produced minimal impact (SI Appendix, Fig. S6 I–L). Taken together, the synthetic fitness phenotype in FA-defective cells is specific to Pol ι and does not extend to Y-family DNA polymerases.
Pol ι Deficiency Exacerbates Endogenous DNA Damage in FA Mutant Cells.
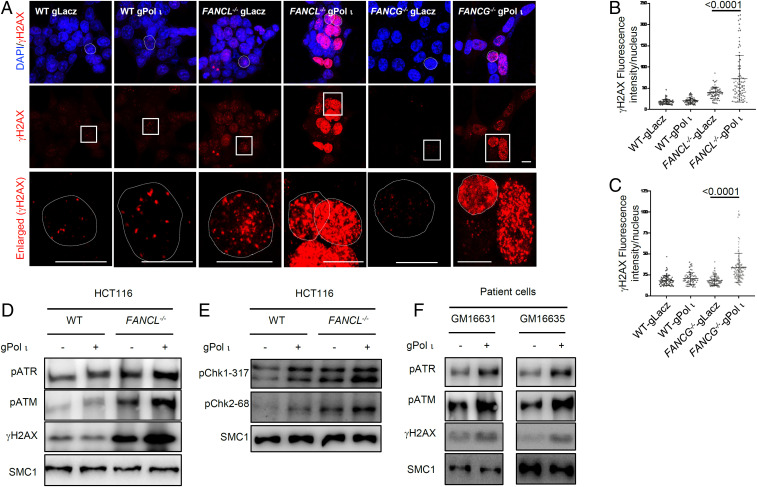
Decreased cell fitness and clonogenicity in FA mutant cells devoid of Pol ι may reflect elevated endogenous genotoxic stress during unperturbed cell proliferation. To test this premise, we analyzed γH2AX focus formation in wild-type, FANCL−/−, and FANCG−/− cells with Pol ι knockdown or control (gLacZ). As shown in Fig. 4 A–C, lack of FANCL alone increased the endogenous γH2AX foci to a detectable level as we have shown previously (34). However, eliminating Pol ι from FA mutant cells resulted in drastically intensified γH2AX staining, suggesting that Pol ι is critical in suppressing the onset of endogenous DNA damage in the absence of the FA mechanism. The intensified γH2AX formation in FA-Pol ι double defective cells likely reflects an escalation of endogenous replication stress and the resulting hyperactivation of ataxia-telangiectasia mutated (ATM)/ATM- and Rad3-related (ATR) signaling. In contrast, knockdown of other Y-family DNA polymerases, Pol κ and Pol η, did not yield any significant increase of γH2AX foci in FA mutant cells (SI Appendix, Fig. S7), again indicating that Pol ι function is specifically important in suppressing endogenous damage in the absence of the FA mechanism.
Fig. 4.
Pol ι loss leads to DNA damage in FANCL/FANCG deficiency cells. (A) γH2AX foci staining of Pol ι gRNA knockdown in HCT116 wild-type cells and FANCL or FANCG deficient cells (72 h post transduction). (Scale bar: 10 µm.) The dotted circles in the first lane indicate the nucleus of the square circled cells in the second lane. The third lane is the enlarged view of the square circled cells. The dotted circles in the third lane indicate the boundary of the nucleus. (B and C) Quantification of γH2AX nuclear fluorescent intensity in experiment shown in A. (D and F) Immunoblotting of γH2AX and phosphorylated ATR and ATM of Pol ι gRNA knockdown in HCT116 wild-type cells and FANCL-deficient cells and mutated patient cells (72 h posttransduction). (E) Immunoblotting of phosphorylated CHK1 and CHK2 of Pol ι gRNA knockdown in HCT116 wild-type cells and FANCL-deficient cells (72 h posttransduction).
We next examined DNA damage signaling markers in FANCL−/− cells lacking Pol ι and observed markedly increased levels of ATR and ATM phosphorylation (Fig. 4D), signifying a strong activation of the replication checkpoint. Consistently, phosphorylation of the ATR and ATM targets Chk1 and Chk2 exhibited a visible increase (Fig. 4E). When Pol ι was depleted from two FA patient cell lines, GM16631 and GM16635, significant increases in ATR and ATM phosphorylation were observed (Fig. 4F). These results suggest that Pol ι plays a critical role in alleviating endogenous replication stress when the FA pathway is comprised.
Pol ι Prevents Replication Fork Stalling and Erosion in FA Mutant Cells.
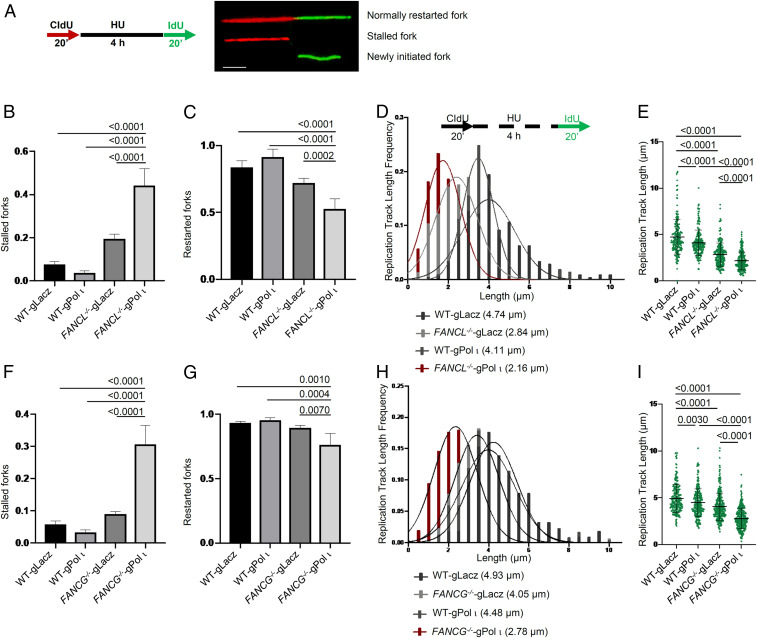
Pol ι loss exacerbated endogenous stress and reduced fitness in cells defective in the FA mechanism. To understand the nature of endogenous replicative stress that gives rise to the loss of cell viability, we examined replication fork dynamics in wild-type and FA mutant cells devoid of Pol ι using the chromatin fiber assay upon transient hydroxyurea (HU) perturbation (Fig. 5A). Analysis of replication fork response to HU blockage showed that depletion of Pol ι from wild-type cells has no detectable impact on replication fork stall and resumption once HU is removed. However, loss of Pol ι caused more than a twofold increase in stalled forks in FANCL−/− cells (Fig. 5B), accompanied by reduced frequency of replication fork restart (Fig. 5C), suggesting an acquired dependency on Pol ι as a result of FANCL deletion.
Fig. 5.
Pol ι loss leads to DNA replication fork collapse in FANCL/FANCG deficiency cells. (A) Schematic of DNA fiber analysis. Red track, CIdU; green track, IdU. (Scale bar: 2 µm.) (B) Proportion of stalled forks in Pol ι gRNA knockdown HCT116 wild-type cells and FANCL−/− cells. (C) Proportion of restarted forks in Pol ι gRNA knockdown HCT116 wild-type cells and FANCL−/− cells. (D) Length frequency of IdU tracts after exposure to HU in Pol ι gRNA knockdown HCT116 wild-type cells and FANCL−/− cells. (E) Distribution of IdU track lengths in Pol ι gRNA knockdown HCT116 wild-type cells and FANCL−/− cells. (F) Proportion of stalled forks in Pol ι gRNA knockdown HCT116 wild-type cells and FANCG−/− cells. (G) Proportion of restarted forks in Pol ι gRNA knockdown HCT116 wild-type cells and FANCG−/− cells. (H) Length frequency of IdU tracts of replication forks after exposure to HU in Pol ι gRNA knockdown HCT116 wild-type cells and FANCG−/− cells. (I) Distribution of IdU track lengths in Pol ι gRNA knockdown HCT116 wild-type cells and FANCG−/− cells. More than two hundred replication forks were analyzed for each sample.
Loss of Pol ι also further reduced replication fork stability in the FANCL−/− mutant (Fig. 5 D and E). The average 5-iodo-2-deoxyuridine (IdU)-labeled replication track lengths in the FANCL−/− cells were significantly shortened from 2.84 ± 1.25 µm to 2.16 ± 0.96 µm (P < 0.0001) as a result of lacking Pol ι, suggesting that Pol ι provides nonepistatic function to the FA mechanism in replication fork stability. Consistently, depletion of Pol ι from the FANCG−/− mutant produced replication fork stress phenotypes similar to that of the FANCL−/− cells (Fig. 5 F–I).
To determine whether Pol ι’s role in replication fork recovery and stability is specific, we depleted Pol η from FANCL−/− and FANCG−/− cells and analyzed their replication fork stability. The results showed that loss of Pol η did not lead to detectable effects in fork stall, recovery, or stability upon HU treatment (SI Appendix, Fig. S8), suggesting that Pol ι function is uniquely involved in compensating for the loss of the FA pathway. Taken together, these results show that Pol ι assumes a more important role in FA-deficient cells to maintain genomic stability.
Discussion
The FA pathway has been firmly established as a key DNA damage response mechanism dealing with strong replication blocking lesions and stabilizing stressed replication forks. Given its crucial function, it is perplexing that complete inactivation of the FA pathway does not lead to organismic or cellular lethality. This is in contrast to the embryonic or cellular lethality caused by the loss of key recombination repair factors, such as Brca1, Brca2, and Rad51 (57–59). We reasoned that the viability of FA mutant cells is likely sustained by a compensatory mechanism(s) and conducted unbiased whole genome screens to identify such a mechanism(s). In this report, we show that Pol ι from the Y-family DNA polymerase plays a critical role in the survival of FA mutant cells, revealing a genetic interaction between the FA pathway and lesion bypass DNA synthesis.
Synthetic lethality against FA deficiency has been sought after by a number of studies, either via compound library screen or via focused small interfering RNA (siRNA) libraries (60, 61). Although these studies produced interpretable candidates, the gRNA/CAS9-based whole genome screen has the advantage of identifying synthetic vulnerability with improved sensitivity and reproducibility in an unbiased manner (41, 45, 48). This is reflected by the consistency of the pair-wide screen data and the fact that our wild-type cell screen data align well with existing gRNA screens (SI Appendix, Fig. S1).
Cell proliferation and clonogenic assays show that Pol ι acquired an indispensable role in the prolonged proliferation and survival of FA mutant cells. This observation was validated with different FA gene knockout mutants derived from different cell backgrounds and with patient-derived FA mutant cells. We found that, in the absence of Pol ι, FA mutant cells, but not wild-type cells, exhibited much-reduced cell growth, augmented endogenous DNA damage, and increased apoptotic cell population. We also tested whether loss of both the FA pathway and Pol ι immediately increase cellular sensitivity to exogenous DNA damage (SI Appendix, Fig. S9 A–D). Using short-term cell growth against mitomycin C (MMC) or cisplatin as a readout, we did not observe increased sensitivity in FA mutants immediately after Pol ι depletion. This result suggests that the synthetic lethality between the FA pathway Pol ι is more reflective of a failure in countering endogenous damage over a prolonged period of cell proliferation.
Our data indicate that FA pathway deficiency generates a dependency on lesion bypass DNA synthesis. Such a dependency is mechanistically congruent with the established function of the FA pathway, which acts to revolve strong replication blocking lesions, such as DNA interstrand crosslinks and DNA-protein crosslinks (62). When FA pathway function falters, DNA replication will inevitably encounter an increased level of endogenous damage and become more dependent on lesion bypass activities to minimize stalled replication forks. The heightened endogenous replication stress in FA mutant cells devoid of Pol ι, reflected by the strong activation of the ATM/ATR checkpoint (Fig. 4 D and F), further confirms that Pol ι provides an indispensable role in reducing replication stress in the absence of the FA pathway. Consistent with this finding, knockdown of FA genes rendered hypersensitivity to the ATM inhibitor M3541 (63).
Previously, FANCD2 has been shown to exhibit synthetic lethality with DNA polymerase θ (Pol θ) in mice (64). This occurs because the Pol θ-mediated end-joining DNA repair pathway is necessary in the absence of the FA pathway and homologous recombinational repair (65–67). These studies point to a diverse spectrum of roles by different nonreplicative DNA polymerases, in addition to or instead of functions in translesion DNA synthesis.
The Y-family DNA polymerases in mammalian cells consist of REV1, Pol ι, Pol κ, and Pol η. Each enzyme has unique fidelity, processivity, and lesion specificity profiles (53). Although Pol ι is a highly conserved enzyme in vertebrates, previous work did not uncover a major biological function. Various members of the Y-family DNA polymerases have been implicated in assisting stressed DNA replication forks (53, 68). Interestingly, the FA pathway has been found important in recruiting or cooperating with several lesion bypass factors, including Rev1, Pol κ, and Pol ζ (69–72). Thus, a defective FA mechanism likely disrupts the effective utilization of these lesion bypass activities. The synthetic phenotype between the FA pathway and Pol ι, on the other hand, predicts that Pol ι recruitment does not rely on an intact FA mechanism. Indeed, chromatin fractionation analysis showed that MMC-induced chromatin enrichment of Pol ι is independent of FANCL and FANCG and enhanced by the absence of these two FA proteins (SI Appendix, Fig. S9E). This result further supports the role of Pol ι as a compensatory factor in FA pathway-deficient cells.
One possible explanation for our finding that Pol ι exhibits synthetic lethality with FA gene mutations is that Pol ι participates in bypass of an endogenous DNA lesion that is normally repaired by an FA-dependent pathway. One distinctive feature of Pol ι is an unusual catalytic pocket that prefers Hoogsteen over Watson–Crick pairing (73), which may accommodate a broader spectrum of lesions. Indeed, Pol ι has been shown to be able to bypass bulky adducts that are unable to be bypassed by other translesion synthesis (TLS) polymerases (74). Furthermore, in the presence of Mn2+, Pol ι is able to bypass a peptide adduct that mimics the DNA-protein crosslink repair intermediate (75, 76). This ability may constitute an effective alternative in countering the accumulation of crosslinking damage from FA pathway loss. The low processivity of Pol ι, compared to other members of the Y-family enzyme, can reduce the nontargeted misincorporations beyond the damaged base (77). These properties might render Pol ι a suitable lesion bypass polymerase to overcome the increased replication stress in FA-deficient cells.
Alternatively, there are hints that Pol ι has a more general role in DNA replication. Kannouche et al. concluded that Pol ι is located in DNA replication foci in undamaged cells and suggested that it may play a role in the maintenance of genome integrity during DNA replication (78). Pol ι has additional mechanisms by which it could be recruited to stalled DNA replication forks, including its interactions with PCNA and REV1 during unperturbed cell proliferation (78, 79). It is plausible that Pol ι is involved in the processing of collapsed replication forks in a manner similar to Pol κ or Pol θ during DNA end rejoining.
Identification of synthetic vulnerability against FA mutant cells can lead to new therapeutic options. Treatment of cancer in FA patients is very challenging because of the increased toxicity of alkylating agent-based systemic chemotherapy (80, 81), and targeted approaches would be more especially desirable. Given that FA gene deficiencies are increasingly identified as somatic mutations (6), the genetic connection between the FA pathway and Pol ι offers a synthetic lethality target for the elimination of tumor cells with FA pathway deficiency.
Methods
Cell Culture, Antibodies, and Plasmids.
HCT116 cells were obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum and 100 units/mL penicillin and 100 μg/mL streptomycin. Patient cells were obtained from the Coriell Institute for Medical Research and cultured in Minimum Essential Media (Eagle) Alpha Modification (GM16631 and GM16635) with 15% fetal bovine serum and 100 units/mL penicillin and 100 μg/mL streptomycin.
Sources of commercial antibodies were as follows: primary antibodies: γH2AX (05-636, Millipore; 613402, BioLegend), α-tubulin (T6199; Sigma-Aldrich), Pol ι (A1942; ABclonal), CDK4 (12790; Cell Signaling Technology), Pol κ (A301-977A; Bethyl Laboratories), Pol η (sc-17770; Santa Cruz Biotechnology), phosphor-ATR (GTX128145; GeneTex), phosphor-ATM (ab81292; Abcam), SMC1 (A300-055A; Bethyl Laboratories), phosphor-CHK1 (2344; Cell Signaling Technology), phosphor-CHK2 (2197; Cell Signaling Technology), 5-bromodeoxyuridine (BrdU) antibody (ldU, 347580; BD Biosciences), and BrdU antibody (CldU, ab6326; Abcam).
For lentiviral RNA interference, annealed shRNAs were cloned into a pLKO-RFP vector as previously reported (82). The shRNA sequences targeting Pol ι and CDK4 were 5′- CCGGTCGGGTCATGTATACAATAATCTCGAGATTATTGTATACATGACCCGATTTTTG -3′ and 5′- CCGGGAGATTACTTTGCTGCCTTAACTCGAGTTAAGGCAGCAAAGTAATCTCTTTTTG-3′. A nontargeting shRNA in mammalians (shCTR) was used as control. Annealed gRNAs were cloned into a LentiCRISPR-EmGFP-sgLacZ vector as previously reported (83). The gRNA sequences targeting Pol ι, Pol κ, and Pol η are 5′- CACCGAGTCATAGTACATGTGGATC-3′, 5′-CACCGGGACTTAATGATAATAAAGC-3′, and 5′-CACCGGTACAGTACAAATCATGGAA-3′.
gRNA Library Screen.
The TKOv3 library contains 70,948 gRNAs targeting 18,053 protein-coding genes (4 gRNAs per gene) with 142 control gRNAs targeting EGFP, LacZ, and luciferase (71,090 total). Library virus production was conducted as described in a previous paper (42). Briefly, 9 million 293T cells were seeded in a T75 flask, and the cells were transfected 24 h later with a mix of 13 μg of lentiviral pLCKO vector containing the library, 13 μg of packaging vector psPAX2, 6.5 μg of envelope vector pMD2.G, 100 μL of TransIT-LT1 Transfection Reagent (Mirus), and 2.0 mL of Opti-MEM medium (Life Technologies). After 24 h, the medium was changed to a serum-free, high-bovine serum albumin (BSA) growth medium (DMEM, 1.1 g/100 mL BSA, 1% penicillin/streptomycin). The virus-containing medium was harvested 48 and 72 h after transfection and centrifuged at 1,500 rpm for 5 min. The supernatant was transferred to a sterile 50-mL tube. One volume of cold PEG-it Virus Precipitation Solution (System Biosciences) was added per 4 volumes of supernatant. The mixture was rotated in a cold room overnight. After incubation, the supernatant/PEG-it mixture was spun down as white pellets at the bottom of the tube at 1,500 × g for 60 min at 4 °C. The pellets were combined together and resuspended in DMEM. The concentrated viruses were stored at −80 °C for long term use.
For gRNA screening, 120 million cells were infected with the TKOv3 library lentiviruses at a targeted multiplicity of infection (MOI) of 0.1 to 0.3. Twenty-four hours after infection, the medium was replaced with fresh growth medium containing 2 μg/mL puromycin. After selection for 2 d, each plate was divided into three replicates containing ∼20 million cells each, passaged every 3 d, and maintained at 200-fold library coverage. At each time point, 25 million cells (>300-fold coverage) were collected for genomic DNA extraction. Genomic DNA was extracted from cell pellets using the QIAamp Blood Maxi Kit (Qiagen), precipitated using ethanol and sodium chloride, and resuspended in Buffer EB (10 mM Tris⋅HCl, pH 7.5). gRNA inserts were amplified via PCR using primers harboring Illumina TruSeq adapters with i5 and i7 barcodes as previously reported (42), and the resulting libraries were sequenced on an Illumina HiSeq 2500 system. The BAGEL algorithm was used to calculate essentiality scores.
Functional Genomic Analysis of Coessential Genes.
SVD analysis.
SVD analysis was conducted using the MASS package version 7.3-50 in R. This method takes an m × n matrix made up of numerical values and breaks it down into three unique matrices: u, v, and d, representing specific features of the input matrix. In this analysis, the input matrix is made up by m representing the number of samples and n the number of genes. The output matrix v can be used to identify differences and consistencies between samples in the input matrix while matrix u can indicate which specific rows are contributing to differences observed in matrix v. Code used for plots can be found in SI Appendix.
Fitness scores and quality control.
Screen quality control and assessment followed procedures demonstrated by BAGEL software found at https://github.com/hart-lab/bagel. All associated code and data used for quality control and screen assessment can be found in SI Appendix.
DRUGZ.
DrugZ data processing followed procedures found at https://github.com/hart-lab/drugz. All associated code and data used can be found in SI Appendix.
Cell Survival Assay.
For cell proliferation assay, cells were seeded at 200 cells per well in 96-well plates. Cells were lysed at selected time points using the CellTiterGlo kit (Promega) following the manufacturer’s instructions, and luminescence was read using the Synergy plate reader (Biotech). The relative luminescence was calculated relative to the value on day 1. For the sensitivity test, cells were continuously exposed to different doses of DNA damage agents cisplatin (232120; Sigma-Aldrich) and MMC (M4287; Sigma-Aldrich) for 4 or 5 d. Cells were lysed and calculated relative to the nontreatment control.
For the clonogenic assay, cells were seeded in triplicate in 6-well plates. After 10 to 14 d of culture, colonies were fixed with methanol and stained with 0.5% (weight/volume [wt/vol]) crystal violet for visualization.
Apoptosis Assay.
The apoptotic cells were quantified using the Annexin V and 7AAD double staining kit (559763; BD Pharmingen). Briefly, FA-deficient cells were infected by shRNA or gRNA/Cas9 expressing lentivirus for 72 h before collection. Harvested cells were washed with phosphate-buffered saline and resuspended in 200 µL of binding buffer containing 5 µL of Annexin V for 15 min. Subsequently, the samples were incubated with 10 µL of 7AAD and immediately analyzed using an Attune NxT Flow Cytometer (Thermo Fisher Scientific). The flow cytometry data were analyzed by FlowJo software (Tree Star).
Western Blotting.
Cell pellets were lysed with 1× sample buffer (50 mM Tris⋅HCl, pH 6.8, 86 mM 2-mercaptoethanol, 2% sodium dodecyl sulfate), boiled for 5 min, and subjected to electrophoresis. Proteins were transferred to nitrocellulose membranes. After blocking with 5% nonfat dried milk in TBS-T (50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, 0.1% Tween 20), the membrane was incubated with the primary antibody and incubated with the appropriate horseradish peroxidase-linked secondary antibody. Chemiluminescence (ChemiDoc imaging system; Bio-Rad) was used to collect digital blot images.
Immunofluorescence Staining.
Cells growing in coverslips were collected 3 d after infection and fixed in ice-cold methanol and incubated sequentially with γH2AX antibody (05-636; Millipore) overnight and a secondary antibody for 1 h, and counterstained with DAPI for 2 min. Images were captured on a Zeiss 880 confocal microscope.
DNA Fiber Assay.
Cells were seeded in 6-well plates. Base-line replication tracts were labeled with 50 mM IdU (I7125; Sigma-Aldrich) followed by exposure to HU (H8627, 4 mM; Sigma-Aldrich) for 4 h after washing off the HU, replication tracts were labeled with 50 mM 5-chlorodeoxyuridine (CIdU) (C6891; Sigma-Aldrich). Cells were prepared as described previously to obtain a single DNA molecule. Fibers were imaged using a Nikon Eclipse 90i, fiber lengths were acquired using ImageJ software, and quantitative data were generated by GraphPad Prism 8 (GraphPad).
Chromatin Fractionation.
The cells were treated with 400 nM MMC for 24 h and harvested for chromatin fractionation. The preparation of soluble fraction and chromatin fraction was described previously (84, 85). Briefly, cells were lysed with NETN buffer [0.5% Nonidet P-40, 1 mM (ethylenedinitrilo)tetraacetic acid, 20 mM Tris⋅HCl, pH 8.0, and 100 mM NaCl]. The supernatant contained soluble fraction and the insoluble pellet contained the chromatin fraction. The insoluble pellets were resuspended in 0.2 M HCl and incubated on ice for 20 min. After centrifuging at 4 °C, the carefully collected supernatant was neutralized with 1 M Tris⋅HCl (pH 8.5) and then used for further analysis.
Statistics.
Statistical analyses were performed using GraphPad Prism 8. Differences between groups were analyzed by the one-way ANOVA. The Student’s t test was used for comparisons of clonogenic survival of patient cells. Error bars indicate SDs. A P value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Drs. Denisse Carvajal-Maldonado, Ying Ye, and Khandan Keyomarsi for discussions and advice. This work is supported in part by P01 CA193124 (project 1 to R.D.W., project 3 to L.L., and project 4 to J.C.); R01 CA190635 (to L.L.), the Cancer Prevention and Research Institute of Texas (RP160667 to J.C. and L.L.), the Olive Stringer endowed professorship (to L.L.), and the Grady F. Saunders Ph.D. Distinguished Research Professorship (to R.D.W.). This research was partly supported by the Fundamental Research Funds for the Central Universities. W.F.L. was supported by Cancer Prevention and Research Institute of Texas (CPRIT) Grant RR160032 and an award from the American Legion Auxiliary. T.H. is a CPRIT Scholar in Cancer Research and is supported by National Institute of General Medical Sciences Grant R35GM130119 and MD Anderson Cancer Center Support Grant P30 CA016672.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2008821117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix. The associated code used for informatics analysis is available upon request.
References
- 1.Alter B. P., Fanconi’s anemia and malignancies. Am. J. Hematol. 53, 99–110 (1996). [DOI] [PubMed] [Google Scholar]
- 2.D’Andrea A. D., Grompe M., The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3, 23–34 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Xie Y., et al. , Aberrant Fanconi anaemia protein profiles in acute myeloid leukaemia cells. Br. J. Haematol. 111, 1057–1064 (2000). [PubMed] [Google Scholar]
- 4.Condie A., et al. , Analysis of the Fanconi anaemia complementation group A gene in acute myeloid leukaemia. Leuk. Lymphoma 43, 1849–1853 (2002). [DOI] [PubMed] [Google Scholar]
- 5.van der Heijden M. S., Yeo C. J., Hruban R. H., Kern S. E., Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 63, 2585–2588 (2003). [PubMed] [Google Scholar]
- 6.Nalepa G., Clapp D. W., Fanconi anaemia and cancer: An intricate relationship. Nat. Rev. Cancer 18, 168–185 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Vandenberg C. J., et al. , BRCA1-independent ubiquitination of FANCD2. Mol. Cell 12, 247–254 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Smogorzewska A., et al. , Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alpi A. F., Pace P. E., Babu M. M., Patel K. J., Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell 32, 767–777 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Meetei A. R., et al. , A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35, 165–170 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Ali A. M., et al. , FAAP20: A novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood 119, 3285–3294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciccia A., et al. , Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell 25, 331–343 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Ling C., et al. , FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 26, 2104–2114 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung J. W., et al. , Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc. Natl. Acad. Sci. U.S.A. 109, 4491–4496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Z., et al. , A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol. Cell 47, 61–75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Z., et al. , A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell 37, 865–878 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogliolo M., et al. , Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 92, 800–806 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhagwat N., et al. , XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol. Cell. Biol. 29, 6427–6437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crossan G. P. et al.; Sanger Mouse Genetics Project , Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 43, 147–152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacKay C., et al. , Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142, 65–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoepker C., et al. , SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 43, 138–141 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Feeney L., et al. , RPA-mediated recruitment of the E3 ligase RFWD3 is vital for interstrand crosslink repair and human health. Mol. Cell 66, 610–621.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howlett N. G., et al. , Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297, 606–609 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Inano S., et al. , RFWD3-mediated ubiquitination promotes timely removal of both RPA and RAD51 from DNA damage sites to facilitate homologous recombination. Mol. Cell 66, 622–634.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Vaz F., et al. , Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 42, 406–409 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Xia B., et al. , Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 39, 159–161 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Schlacher K., Wu H., Jasin M., A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106–116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schlacher K., et al. , Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thongthip S., et al. , Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction. Genes Dev. 30, 645–659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lachaud C., et al. , Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science 351, 846–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan K. L., Palmai-Pallag T., Ying S., Hickson I. D., Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 11, 753–760 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Chen Y. H., et al. , ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol. Cell 58, 323–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lossaint G., et al. , FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol. Cell 51, 678–690 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Tian Y., et al. , Constitutive role of the Fanconi anemia D2 gene in the replication stress response. J. Biol. Chem. 292, 20184–20195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y., et al. , Modularized functions of the Fanconi anemia core complex. Cell Rep. 7, 1849–1857 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houghtaling S., et al. , Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17, 2021–2035 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dubois E. L., et al. , A Fanci knockout mouse model reveals common and distinct functions for FANCI and FANCD2. Nucleic Acids Res. 47, 7532–7547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parmar K., D’Andrea A., Niedernhofer L. J., Mouse models of Fanconi anemia. Mutat. Res. 668, 133–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M., et al. , Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat. Genet. 12, 448–451 (1996). [DOI] [PubMed] [Google Scholar]
- 40.D’Andrea A. D., Susceptibility pathways in Fanconi’s anemia and breast cancer. N. Engl. J. Med. 362, 1909–1919 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hart T., et al. , High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Hart T., et al. , Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 (Bethesda) 7, 2719–2727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alter O., Brown P. O., Botstein D., Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. U.S.A. 97, 10101–10106 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y., Wu M., Liu C., Li X., Zheng J., SL 2 MF: Predicting synthetic lethality in human cancers via logistic matrix factorization. IEEE/ACM Trans. Comput. Biol. Bioinform. 17, 748–757 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Hart T., Moffat J., BAGEL: A computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics 17, 164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hart T., Brown K. R., Sircoulomb F., Rottapel R., Moffat J., Measuring error rates in genomic perturbation screens: Gold standards for human functional genomics. Mol. Syst. Biol. 10, 733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyers R. M., et al. , Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lenoir W. F., Lim T. L., Hart T., PICKLES: The database of pooled in-vitro CRISPR knockout library essentiality screens. Nucleic Acids Res. 46, D776–D780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao L., Washington M. T., Translesion synthesis: Insights into the selection and switching of DNA polymerases. Genes (Basel) 8, 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosal G., Chen J., DNA damage tolerance: A double-edged sword guarding the genome. Transl. Cancer Res. 2, 107–129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prives C., Gottifredi V., The p21 and PCNA partnership: A new twist for an old plot. Cell Cycle 7, 3840–3846 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Goodman M. F., Woodgate R., Translesion DNA polymerases. Cold Spring Harb. Perspect. Biol. 5, a010363 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sale J. E., Lehmann A. R., Woodgate R., Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohkumo T., et al. , UV-B radiation induces epithelial tumors in mice lacking DNA polymerase η and mesenchymal tumors in mice deficient for DNA polymerase ι. Mol. Cell. Biol. 26, 7696–7706 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aoufouchi S., et al. , 129-Derived mouse strains express an unstable but catalytically active DNA polymerase ι variant. Mol. Cell. Biol. 35, 3059–3070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gueranger Q., et al. , Role of DNA polymerases η, ι and ζ in UV resistance and UV-induced mutagenesis in a human cell line. DNA Repair (Amst.) 7, 1551–1562 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Ludwig T., Chapman D. L., Papaioannou V. E., Efstratiadis A., Targeted mutations of breast cancer susceptibility gene homologs in mice: Lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 11, 1226–1241 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Hakem R., et al. , The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell 85, 1009–1023 (1996). [DOI] [PubMed] [Google Scholar]
- 59.Lim D. S., Hasty P., A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16, 7133–7143 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gallmeier E., et al. , High-throughput screening identifies novel agents eliciting hypersensitivity in Fanconi pathway-deficient cancer cells. Cancer Res. 67, 2169–2177 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Kennedy R. D., et al. , Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J. Clin. Invest. 117, 1440–1449 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ceccaldi R., Sarangi P., D’Andrea A. D., The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 17, 337–349 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Cai M. Y., et al. , Cooperation of the ATM and Fanconi Anemia/BRCA pathways in double-strand break end resection. Cell Rep. 30, 2402–2415.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ceccaldi R., et al. , Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carvajal-Garcia J., et al. , Mechanistic basis for microhomology identification and genome scarring by polymerase θ. Proc. Natl. Acad. Sci. U.S.A. 117, 8476–8485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng W., et al. , Genetic determinants of cellular addiction to DNA polymerase θ. Nat. Commun. 10, 4286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wood R. D., Doublié S., DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair (Amst.) 44, 22–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tonzi P., Huang T. T., Role of Y-family translesion DNA polymerases in replication stress: Implications for new cancer therapeutic targets. DNA Repair (Amst.) 78, 20–26 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niedzwiedz W., et al. , The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell 15, 607–620 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Tonzi P., Yin Y., Lee C. W. T., Rothenberg E., Huang T. T., Translesion polymerase κ-dependent DNA synthesis underlies replication fork recovery. eLife 7, e41426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Budzowska M., Graham T. G., Sobeck A., Waga S., Walter J. C., Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link. EMBO J. 34, 1971–1985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim H., Yang K., Dejsuphong D., D’Andrea A. D., Regulation of Rev1 by the Fanconi anemia core complex. Nat. Struct. Mol. Biol. 19, 164–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nair D. T., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K., Human DNA polymerase ι incorporates dCTP opposite template G via a G.C + Hoogsteen base pair. Structure 13, 1569–1577 (2005). [DOI] [PubMed] [Google Scholar]
- 74.McIntyre J., Polymerase ι–An odd sibling among Y family polymerases. DNA Repair (Amst.) 86, 102753 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Naldiga S., et al. , Error-prone replication of a 5-formylcytosine-mediated DNA-peptide cross-link in human cells. J. Biol. Chem. 294, 10619–10627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yeo J. E., et al. , Synthesis of site-specific DNA-protein conjugates and their effects on DNA replication. ACS Chem. Biol. 9, 1860–1868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yoon J. H., Roy Choudhury J., Prakash L., Prakash S., Translesion synthesis DNA polymerases η, ι, and ν promote mutagenic replication through the anticancer nucleoside cytarabine. J. Biol. Chem. 294, 19048–19054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kannouche P., et al. , Localization of DNA polymerases η and ι to the replication machinery is tightly co-ordinated in human cells. EMBO J. 22, 1223–1233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haracska L., et al. , A single domain in human DNA polymerase ι mediates interaction with PCNA: Implications for translesion DNA synthesis. Mol. Cell. Biol. 25, 1183–1190 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Engel N. W., et al. , Fatal myelotoxicity following palliative chemotherapy with cisplatin and gemcitabine in a patient with stage IV cholangiocarcinoma linked to post mortem diagnosis of Fanconi anemia. Front. Oncol. 9, 420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mehta P. A., et al. , Busulfan pharmacokinetics and precision dosing: Are patients with Fanconi anemia different? Biol. Blood Marrow Transplant. 25, 2416–2421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang R., et al. , Tubulin detyrosination promotes human trophoblast syncytium formation. J. Mol. Cell Biol. 11, 967–978 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lu X., et al. , Fine-tuned and cell-cycle-restricted expression of fusogenic protein syncytin-2 maintains functional placental syncytia. Cell Rep. 21, 1150–1159 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Ward I. M., Chen J., Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276, 47759–47762 (2001). [DOI] [PubMed] [Google Scholar]
- 85.Yu X., Harris S. L., Levine A. J., The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res. 66, 4795–4801 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix. The associated code used for informatics analysis is available upon request.