Significance
Bacterial two-component system signaling circuits (TCSs), comprising sensor histidine kinase (HK) and response regulator (RR) proteins, regulate numerous cellular processes in response to environmental or metabolic cues. Phosphorylation, and thereby activation, of RRs is promoted by their cognate HKs but also by low molecular weight phosphodonors. Here, we report that the purine and histidine biosynthetic intermediates ZMP (AICAR) and/or ZTP act as direct phosphodonors to a wide range of RRs in vivo, thereby modulating the activity of many TCS pathways without the necessity of intervention of an HK. The attainment of this reaction confers an additional function for these ancient molecules to serve as global regulators for gene expression.
Keywords: two-component systems, AICAR, phosphorylation, UvrY, response regulator
Abstract
Two-component systems (TCSs) in bacteria are molecular circuits that allow the perception of and response to diverse stimuli. These signaling circuits rely on phosphoryl-group transfers between transmitter and receiver domains of sensor kinase and response regulator proteins, and regulate several cellular processes in response to internal or external cues. Phosphorylation, and thereby activation, of response regulators has been demonstrated to occur by their cognate histidine kinases but also by low molecular weight phosphodonors such as acetyl phosphate and carbamoyl phosphate. Here, we present data indicating that the intermediates of the de novo syntheses of purines and histidine, 5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5′-monophosphate (ZMP) and/or 5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5′-triphosphate (ZTP), activate the response regulator UvrY, by promoting its autophosphorylation at the conserved aspartate at position 54. Moreover, these Z nucleotides are shown to also activate the nonrelated response regulators ArcA, CpxR, RcsB, and PhoQ. We propose that ZMP and/or ZTP act as alarmones for a wide range of response regulators in vivo, providing a novel mechanism by which they could impact gene expression in response to metabolic cues.
Bacterial two-component signal transduction systems (TCSs) are molecular circuits that allow microorganisms to detect, amplify and respond to diverse stimuli. A typical TCS comprises a membrane-bound histidine kinase (HK) and a cytosolic response regulator (RR), that contain a transmitter domain (TD) and a receiver domain (RD), respectively. Signal perception by the HK is thought to trigger conformational changes in the protein that stimulate an adenosine triphosphate (ATP)-dependent autophosphorylation at a conserved histidine residue in its TD. The phosphoryl group (∼P) is, then, transferred to a conserved aspartate residue at the RD of the cognate RR, rendering it active, often as a transcriptional regulator (1). Some TCSs, however, operate by a more elaborate signal transduction pathway, involving sequential phosphoryl group transfers in a His → Asp → His → Asp multistep phosphorelay (2–4). Such TCSs comprise hybrid HKs, which contain, in addition to the TD, an RD and a phosphotransfer domain with a conserved histidine residue. In all cases, in the absence of the stimulus, both the RR and the HK dephosphorylate, resulting in silencing of the system. Phosphorylated RR (RR-P) dephosphorylation occurs either by the inherent lability of the mixed anhydride phosphoaspartyl bond and/or by the RR-P−specific phosphatase activity of the cognate HK (4–8).
RRs can also be phosphorylated by low molecular weight phosphodonors (LMPs), such as acetyl-phosphate (acetyl-P), carbamoyl-phosphate (carbamoyl-P), γ-glutamyl-phosphate, and phosphoramidate (9–12). However, the physiological relevance of such HK-independent RR-phosphorylation events remains unclear, because they have been only observed in vitro, or, in the case of acetyl-P, in vivo by using mutants that lack the cognate HK protein. Nevertheless, it has been suggested that acetyl-P contributes to maintain a basal level of RRs in the phosphorylated state, and that it could act as a global signal for metabolic conditions (13, 14).
The BarA/UvrY TCS of Escherichia coli consists of the membrane-bound hybrid HK BarA and its cognate RR UvrY (15). BarA senses and responds to the presence of acetate and other short-chain carboxylic acids, such as formate or propionate (16). Phosphorylated BarA allows the transphosphorylation of UvrY, a typical RR of the FixJ family (15), which, in turn, activates expression of the noncoding RNAs CsrB and CsrC (17, 18). These regulatory RNAs possess repeated sequence elements that allow the interaction with multiple copies of the RNA-binding protein CsrA, thereby sequestering it and preventing the interaction with its messenger RNA targets (18, 19). CsrA coordinates gene expression by positively or negatively regulating the translation, stability, and/or elongation of its target transcripts (20, 21). Curiously, activation of csrB transcription, which depends directly on UvrY-P, does not take place in a csrA mutant strain (17), suggesting that CsrA has a positive effect on the BarA/UvrY TCS. Recently, it was shown that CsrA is required for proper uvrY expression (22, 23), and is also necessary for activation of the BarA kinase activity (22). Because CsrA is an RNA-binding protein, the effect of CsrA on the activity of BarA was suggested to be indirect. In the present work, on the search of proteins/factors that could affect the activity of the BarA/UvrY TCS, we found that the purine and histidine biosynthetic intermediates ZMP (5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5′-monophosphate), also known as AICAR, and/or ZTP (5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5′-triphosphate) activate UvrY in a BarA-independent manner. Also, we provide evidence that ZMP and/or ZTP activate a wide range of RRs in vivo. We propose that these Z nucleotides act as alarmones that affect two-component signaling, and thereby modulate gene expression in response to metabolic cues.
Results
Disruption of the purA Gene Restores BarA/UvrY Activity in a csrA Mutant Strain.
It has been previously shown that the global translational regulator CsrA is required for proper regulation of the BarA kinase activity in E. coli (22). Therefore, it was suggested that, in addition to acetate, which acts as the physiological stimulus for BarA (16), other factors might be involved in the control of its activity (22). In order to identify such factors, a mini-Tn10 random insertion library of strain IFC5010 (csrA− csrB-lacZ) harboring plasmid pMX543, which expresses uvrY in a CsrA-independent manner (22), was constructed and screened for increased csrB-lacZ expression. In trans uvrY expression was chosen because it has been previously shown that uvrY expression is impaired in csrA mutant strains (22, 23). From a total of ∼40,000 mutants, two positive clones were identified and chosen for further analyses. Localization of the transposon was pursued by constructing plasmid-based genomic libraries of the identified mutants and selecting for the transposon resistance marker, and subsequent sequencing of the plasmids. Surprisingly, transposon insertion in both mutants was found to be located in the purA coding sequence (codons Leu291 and Thr287), the product of which catalyzes the first step in the de novo biosynthesis of adenosine monophosphate (AMP) from inosine monophosphate (IMP) at the expense of l-aspartate and guanosine triphosphate (24). Both purA::Tn10 insertion mutant strains (hereafter referred to as purA−) were unable to grow in M9 minimal medium with glucose as the only carbon source, but they did grow when the media was supplemented with AMP and histidine or in lysogeny broth (LB), in agreement with previous reports (25–27). Because both purA mutant strains showed essentially identical behavior, only results from one of them will be presented.
The expression of the csrB-lacZ reporter, which is located at the λ attachment site in the chromosome and whose expression depends directly on the activity of the BarA/UvrY TCS, in a wild-type (WT) strain (KSB837), and in a csrA mutant strain (IFC5010) and its purA− derivative (IFC6000) both harboring plasmid pMX543, was then compared. As expected, reporter expression was activated in the WT strain at the transition from exponential to stationary phase of growth, whereas no activation of reporter expression was noted in the csrA− strain harboring plasmid pMX543. On the other hand, reporter expression was activated in the csrA− purA− strain harboring plasmid pMX543, reaching ∼60% of WT expression values (Fig. 1A). As expected, the presence of a compatible adenylosuccinate synthase (PurA)-expressing plasmid (pMX558) in the csrA− purA− strain harboring plasmid pMX543 restored reporter expression to csrA− levels (Fig. 1A). Finally, only a slight increase in reporter expression was observed in the csrA− purA− strain in the absence of pMX543 (Fig. 1A), most likely due to the low expression of UvrY caused by the csrA mutation (22). Thus, the purA mutation appears to partially restore the BarA/UvrY activity in the csrA mutant strain.
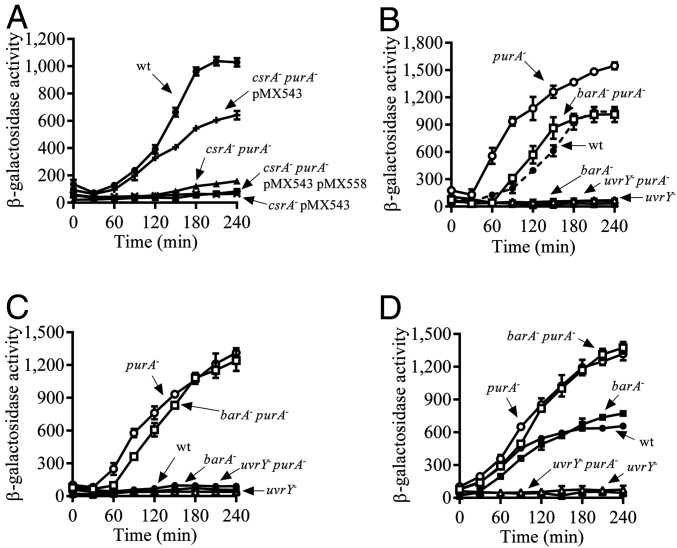
Fig. 1.
Disruption of the purA gene leads to the activation of UvrY. (A) Overnight cultures of the isogenic strains KSB837 (WT) (filled circles), IFC5010 (csrA−) carrying the uvrY-expressing plasmid pMX543 (crosses), IFC6000 (csrA− purA−) harboring plasmid pMX543 (plusses), IFC6000 (csrA− purA−) harboring plasmids pMX543 and pMX558 (filled squares), and IFC6000W (csrA− purA−, without plasmid) (filled triangles) were diluted to an OD600 of ∼0.01 in LB medium, and, after the OD600 reached 0.15, the β-galactosidase activity was followed for 240 min. (B) Cultures of KSB837 (WT) (filled circles), IFC6001 (purA−) (open circles), IFC6002 (barA−) (filled squares), IFC6004 (barA− purA−) (open squares), IFC6003 (uvrY−) (filled triangles), and IFC6005 (uvrY− purA−) (open triangles) were grown in LB medium, and their β-galactosidase activity was followed. (C and D) Cultures of the isogenic strains KSB837 (WT) (filled circles), IFC6001 (purA−) (open circles), IFC6002 (barA−) (filled squares), IFC6004 (purA− barA−) (open squares), IFC6003 (uvrY−) (filled triangles), and IFC6005 (uvrY− purA−) (open triangles) were grown in LB medium, the pH of which had been adjusted and buffered to 5.0 using 0.1 M homopiperazine-N,N′-bis-2-(ethanesulfonic acid). At an OD600 of 0.15, the culture was split in two, and 7 mM acetate was added to one of them (D), whereas the other was used as a control (C), and the β-galactosidase activity was followed every 30 min. The average and SDs from three independent experiments are shown.
Disruption of the purA Gene Leads to UvrY Activation.
We, then, asked whether the purA mutation affects the csrB expression in a WT strain. To this end, the purA mutation was transferred to a WT strain (KSB837), and the expression of csrB-lacZ was examined. It was found that the purA mutant strain expressed csrB-lacZ earlier and reached higher levels of expression than the WT (Fig. 1B), indicating an overstimulation of the BarA/UvrY system.
To ensure that the effect of the purA mutation on csrB expression occurs via the BarA/UvrY TCS, strains IFC6004 (barA− purA−) and IFC6005 (uvrY− purA−) were constructed, and csrB-lacZ expression was monitored. No csrB expression was detected in the uvrY− purA− strain (Fig. 1B), whereas WT levels of csrB expression were obtained in the barA− purA− strain (Fig. 1B). It can, therefore, be concluded that the observed effect of the purA mutation on csrB expression is exerted via the UvrY RR and is independent of the BarA HK.
Interestingly, at pH 5.0, a condition in which the BarA/UvrY system remains silenced (28), unless acetate that acts as a specific stimulus for BarA is added to the growth media (16), activation of csrB-lacZ expression was observed in both the purA− (Fig. 1C) and the purA− barA− strains but not in the WT, barA−, uvrY−, and purA− uvrY− (Fig. 1C) strains. As expected, when acetate was added to the growth medium, reporter expression was activated in the WT and barA− strains, due to acetyl-P−dependent UvrY phosphorylation (16), but not in the uvrY− and purA− uvrY− strains (Fig. 1D). On the other hand, reporter expression remained unchanged in the purA− and the purA− barA− strains (Fig. 1D). Taken together, these results confirm the conclusion that the purA mutation leads to the activation of csrB expression via UvrY, in a BarA-independent manner.
Because overexpression of UvrY has been shown to activate csrB-lacZ expression even in the absence of BarA (22), we tested whether the purA mutation affects UvrY expression. To this end, the amounts of UvrY protein in the WT (KSB837) and the purA− strains (IFC6001) were compared by Western blot analysis, using specific UvrY polyclonal antibodies. Both strains expressed similar amounts of UvrY (Fig. 2A), indicating that expression of UvrY is not affected in the purA mutant strain.
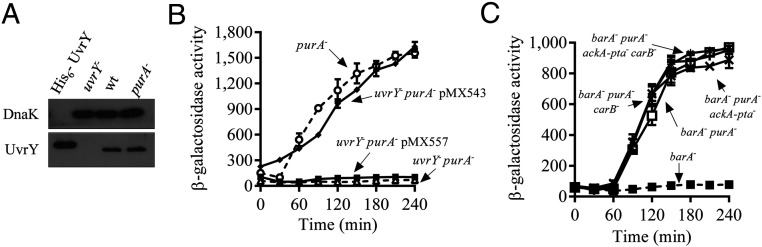
Fig. 2.
(A) The purA disruption does not affect UvrY expression. Levels of UvrY protein (23.890 KDa) in KSB837 (WT) and IFC6001 (purA−) as determined by Western blot analyses using UvrY polyclonal antibodies. Purified His6tagged UvrY (25.380 Da) and an extract from strain IFC6005 (uvrY−) were loaded in lanes 1 and 2, respectively. DnaK (69.115 kDa), detected using DnaK polyclonal antibodies, was used as a loading control. (B) The conserved Asp54 of UvrY is needed for the activation of csrB expression in the purA mutant strain. Cultures of strains IFC6001 (purA−) (open circles), IFC6005 (uvrY− purA−) (open triangles), and IFC6005 (uvrY− purA−) harboring plasmid pMX543 (expressing uvrY) (filled diamonds) or plasmid pMX557 (expressing uvrYD54Q) (filled squares), were grown in LB medium, and β-galactosidase activity was followed for 240 min. The average and SDs from three independent experiments are shown. (C) UvrY autophosphorylation in a purA− strain is not due to acetyl-P or carbamoyl-P. Cultures of strains IFC6002 (barA−) (filled squares), IFC6007 (barA− purA−) (open squares), IFC6006 (barA− purA− ackA-pta−) (crosses), IFC6008 (barA− purA− carB−) (plusses), and IFC6009 (barA− purA− ackA-ptA− carB−) (asterisks) were grown in LB medium, and β-galactosidase activity was measured every 30 min for 240 min. The average and SDs from three independent experiments are shown.
We, then, asked whether the conserved aspartate residue at position 54 of UvrY, the site of phosphorylation, is required for the activation of csrB expression in the purA mutant strain. To test this, plasmids pMX543 and pMX557, carrying the uvrY and uvrYD54Q coding sequences, respectively, were transformed into strain IFC6005 (uvrY− purA−), and csrB-lacZ expression was monitored. It was found that activation of csrB-lacZ expression in strain IFC6005 carrying pMX543 was similar to that observed in the purA− strain, whereas no csrB-lacZ expression was observed in strain IFC6005 carrying pMX557 (Fig. 2B), indicating that the purA− mutation leads to UvrY phosphorylation and subsequent activation of reporter expression.
Neither Acetyl Phosphate nor Carbamoyl Phosphate Is Responsible for UvrY Activation in the purA− Strain.
It is well established that RRs are able to autophosphorylate at the expense of LMP, such as acetyl phosphate or carbamoyl phosphate (9). Therefore, the possibility that the purA mutation results in an increase of such phosphodonors, leading to UvrY phosphorylation and thereby activation of csrB expression, was tested. The acetyl-P or/and carbamoyl-P synthetic pathways were blocked by disruption of the ackA pta genes and/or the carB gene in the barA− purA− (IFC6007) strain, resulting in IFC6006 (barA− purA− ackA-pta−), IFC6008 (barA− purA− carB−), and IFC6009 (barA− purA− ackA-pta− carB−), and csrB-lacZ reporter expression was compared. It was found that nearly identical amounts of csrB-lacZ were expressed in the barA− purA− and its ackA-pta− and carB− derivative strains (Fig. 2C), indicating that neither acetyl phosphate nor carbamoyl phosphate contributes to the activation of UvrY in the purA mutant strain.
Z Nucleotide Accumulation Leads to BarA-Independent UvrY Activation.
Because acetyl-P and carbamoyl-P were found to not be involved in the BarA-independent UvrY activation observed in the purA− strain, the idea that the purA mutation leads to the accumulation of an intermediate in the de novo syntheses of purines and/or histidine pathways (Fig. 3A) that could act as an LMP for UvrY was put forward. To explore this intriguing possibility, deletion mutants that block different steps in the de novo synthesis of purines and in the connection between the purine and histidine pathways were constructed. Firstly, the synthesis of IMP, that is the substrate of PurA to produce adenilosuccinate (S-AMP) (Fig. 3A), was blocked, by deleting the purH gene in strain IFC6007 (barA− purA−). No difference in the levels of reporter expression between the barA− purA− and the barA− purA− purH− strains was observed (Fig. 3B), discarding the possibility that accumulation of IMP is responsible for UvrY activation. Secondly, production of ZMP was obstructed, by deleting the purC− and/or the hisF− genes, generating strains IFC6010 (barA− purA− purC−), IFC6011 (barA− purA− hisF−), and IFC6012 (barA− purA− purC− hisF−). Deletion of the purC gene was decided because of the essentiality of the ZMP-producing enzyme adenylosuccinate lyase (PurB). Deletion of purC in a barA− purA− background strain resulted in a fourfold decrease in csrB-lacZ expression, whereas blocking the histidine pathway, by deletion of the hisF gene in the barA− purA− strain, resulted in twofold lower csrB expression as compared to the one in the barA− purA− strain (Fig. 3B). Finally, when both the purine and the histidine pathways were blocked, by deleting the purC and hisF genes (strain IFC6012), csrB-lacZ expression was practically abolished (Fig. 3B and SI Appendix, Fig. S1A). Thus, the possibility that a factor, other than ZMP and/or ZTP, accumulates, because of the genetic manipulation of the cells, and activates UvrY can be excluded. Considering the fact that the concentration of AMP controls the flow rate of the purine nucleotide synthesis pathway by activating the transcriptional repressor PurR, thereby inhibiting 5-phospho-α-D-ribose 1-diphosphate (PRPP) synthesis (29–31), we reasoned that addition of AMP to a culture of the purA− strain should prevent ZMP/ZTP accumulation and, in consequence, activation of UvrY. It is relevant to mention that, when AMP is added to the medium, it is converted to adenine by UshA and DeoD, taken up by the cells, and reconverted to AMP by Apt and DeoD (32). To this end, strain IFC6007 (barA− purA−) was grown in the presence or absence of 100 μM AMP to an optical density at 600 nm (OD600) of 0.7, and csrB-lacZ reporter expression was compared. It was found that, in the absence of AMP, csrB-lacZ expression was activated (Fig. 3C), in agreement with the above presented results (Fig. 1), whereas, in the presence of AMP, activation of csrB-lacZ expression was completely abolished (Fig. 3C). Moreover, reporter expression in strain IFC6014 (barA− purA− purR−), where the PurR transcriptional repressor was deleted, was significantly higher than in strain IFC6007 (barA− purA−), and the presence of AMP was without effect (Fig. 3C). These results strongly suggest that ZMP/ZTP accumulation is responsible for UvrY activation.
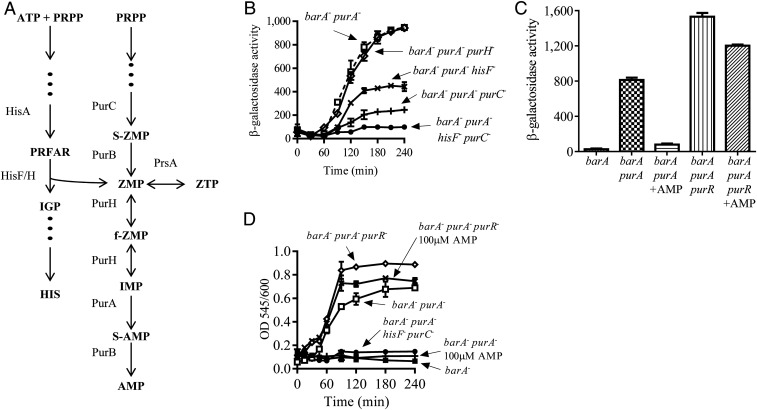
Fig. 3.
Z nucleotide-dependent UvrY autophosphorylation. (A) Representation of the pathways for the de novo synthesis of AMP and histidine [modified from Rohlman and Matthews (47)]. Abbreviations: PurH, IMP cyclohydrolase; HisF/H, complex imidazole glycerol phosphate synthase; HisA, 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino) methylideneamino]imidazole-4-carboxamide isomerase; PurC, phosphoribosylaminoimidazole-succinocarboxamide synthase; PRFAR, phosphoribulosyl-formimino-5-aminoimidazole-4-carboxamide ribotide phosphate; IGP, D-erythro-1-(imidazol-4-yl)-glycerol 3-phosphate; S-ZMP, 5′-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole; F-ZMP, 5-formamido-1-(5-phospho-D-ribosyl)-imidazole-4-carboxamide; HIS, histidine. (B) Cultures of the isogenic strains IFC6007 (barA− purA−) (open squares), IFC6013 (barA− purA− purH−) (open diamonds), IFC6010 (barA− purA− purC−) (plusses), IFC6011 (barA− purA− hisF−) (crosses), and IFC6012 (barA− purA− purC− hisF−) (filled circles) were grown in LB medium, and β-galactosidase activity was followed. The average and SDs from three independent experiments are presented. (C) Strains IFC6002 (barA−), IFC6007 (barA− purA−), and IFC6014 (barA− purA− purR−) were grown in LB medium to an OD600 of ∼0.7, in the presence or the absence of 100 μM of AMP, and β-galactosidase activity was measured. The average from three independent experiments is presented, and SDs are indicated. (D) Strains IFC6001 (purA−) (open circles), IFC6002 (barA−) (filled squares), IFC6007 (barA− purA−) (open squares), IFC6014 (barA− purA− purR−) (open diamonds), and IFC6012 (barA− purA− purC− hisF−) (filled circles) were grown in LB medium, whereas strains IFC6007 (barA− purA−) (plusses) and IFC6014 (barA− purA− purR−) (crosses) were grown in LB medium supplemented with 100 μM of AMP. Samples from cultures were withdrawn every 15 min to 30 min for 240 min, and Z nucleotides were quantified as described in Materials and Methods. Results are expressed as OD545nm/OD600nm to normalize the Z nucleotide concentration (measured as OD545nm) to the cell growth (OD600nm). Data represent the averages from three independent experiments, and the SD values are indicated.
To provide further support to the conclusion that accumulation of ZMP and/or ZTP leads to UvrY activation, the amount of ZMP and ZTP was quantified by the method described by Bratton and Marshall (33) and modified by Nagy et al. (34). This assay, which originally was used for the quantification of antibiotics of the sulfonamide group in blood samples, is based on the diazotization of the primary aromatic amine present in ZMP or ZTP by the addition of sodium nitrite. Only primary aromatic amine groups are diazotized and thereby reactive. Subsequently, coupling the diazonium groups (diazotized amines) with N-(1-naphthyl) ethylenediamine produces an azo compound that can be detected at 545 nm. It is relevant to mention that this method does not differentiate ZMP, ZTP, and associated derivatives, such as AICA (5-aminoimidazole-4-carboxamide) (AICAR without the ribose moiety) or dephosphorylated AICAR. Therefore, hereafter, ZMP and ZTP will be referred as Z nucleotides. Strains IFC6001 (purA−), IFC6002 (barA−), IFC6007 (barA− purA−), IFC6014 (barA− purR− purA−), and IFC6012 (barA− purA− purC− hisF−) were grown in LB supplemented or not with AMP, and the amount of Z nucleotides was determined. It was found that Z nucleotides accumulate in strains IFC6001 (purA−) and IFC6007 (barA− purA−) but not in strain IFC6002 (barA−) or when AMP was added to the culture medium (Fig. 3D and SI Appendix, Fig. S1C). Moreover, accumulation of Z nucleotides was found to be almost completely abolished in strain IFC6012 (barA− purA− purC− hisF−) (Fig. 3D and SI Appendix, Fig. S1C), ruling out the possibility that other nucleotides or aminoimidazole components present in cell extracts interfere with the herein used quantification method. Also, the amount of Z nucleotides was found to be significantly higher in strain IFC6014 (barA− purA− purR−) both in the absence and presence of AMP (Fig. 3D and SI Appendix, Fig. S1C). Finally, Z nucleotide accumulation was observed to occur progressively, and csrB-lacZ expression appears to be activated only when the Z nucleotide concentration reached a threshold of 3 µM (Figs. 3 B and D and SI Appendix, Fig. S1 B and C). Taken together, these results indicate that ZMP and/or ZTP are able to activate UvrY.
UvrY Autophosphorylates at the Expense of the Z Nucleotides.
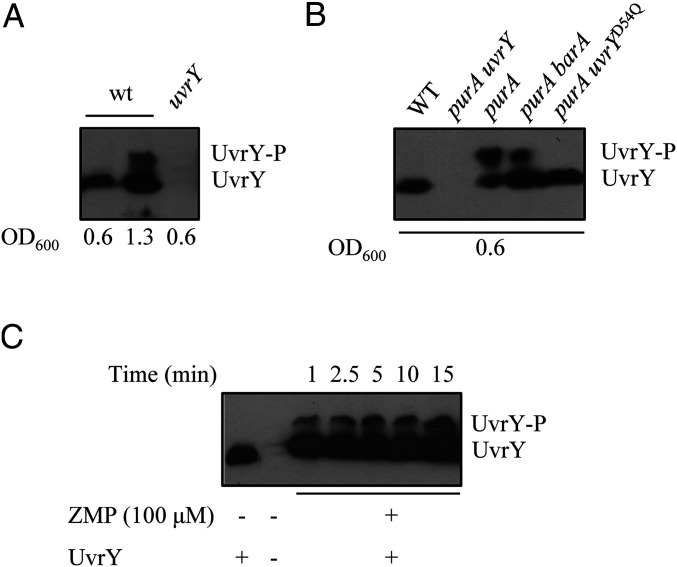
The activation of UvrY by ZMP/ZTP raises the possibility that these nucleotides act as phosphodonors to UvrY, and thereby activate it as a transcriptional regulator. This is supported by the above-presented result demonstrating that the phosphorylatable Asp54 residue of UvrY is essential for csrB expression in the purA− strain (Fig. 2B). To provide more definite evidence, the phosphorylation state of UvrY was evaluated by Phos-tag-acrylamide sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE), which allows the separation of the phosphorylated and the unphosphorylated forms of HKs and RRs (35, 36), followed by Western blot analysis. Protein extracts of strains KSB837 (WT) and IFC6003 (uvrY−) grown at nonstimulatory conditions (OD600 ≈ 0.6), and cell extracts of the WT strain (KSB837) grown at stimulatory conditions (OD600 of 1.3, a growth stage at which the BarA/UvrY system is active, and therefore UvrY is expected to be, at least partly, in its phosphorylated state) were examined by Phos-tag-acrylamide SDS/PAGE and Western blot analysis using UvrY specific antibodies. As anticipated, no UvrY was detected in extracts from IFC6003 (uvrY−), and only one band, corresponding to UvrY, was detected in extracts from the WT strain under nonstimulatory growth conditions (OD600 ≈ 0.6) (Fig. 4A). On the other hand, two bands, corresponding to UvrY and UvrY-P, were detected in extracts from the WT strain at stimulatory growth conditions (OD600 ≈ 1.3) (Fig. 4A). Subsequently, protein extracts from nonstimulatory growth condition (OD600 ≈ 0.6) of strains KSB837 (WT), IFC6004 (barA− purA−), IFC6001 (purA−), and IFC6005 (uvrY− purA−) were assayed. In agreement with the previous result, only UvrY was present in the WT strain, no UvrY was detected in IFC6005 (uvrY− purA−), and both UvrY and UvrY-P were detected in extracts from strains IFC6004 (barA− purA−) and IFC6001 (purA−) (Fig. 4B). Moreover, only UvrY but not UvrY-P was detected in extracts from strain IFC6005 (uvrY− purA−) harboring plasmid pMX557, which carries the uvrYD54Q coding allele (Fig. 4B), demonstrating that Z nucleotide-dependent UvrY phosphorylation occurs at the conserved Asp54 residue. Finally, in order to determine whether Z nucleotides can act as direct phosphodonors for UvrY, purified His6-tagged UvrY was incubated with 100 μM of ZMP, and the kinetics of UvrY phosphorylation were monitored by Phos-tag-acrylamide SDS/PAGE and immunodetection (Fig. 4C). Increasing amounts of UvrY-P were detected in the presence of the Z nucleotide (Fig. 4C). It can, therefore, be concluded that at least ZMP act as a direct phosphodonor to UvrY.
Fig. 4.
UvrY and UvrY-P detection by Phos-Tag SDS/PAGE followed by Western blot analysis. (A) Lanes 1 and 2, KSB837 (WT); lane 3, IFC6003 (uvrY−). In lanes 1 and 3, cell extracts were obtained from cultures at an OD600nm ≈ 0.6 (nonstimulatory conditions), and, in lane 2, cell extracts were obtained from a culture grown to an OD600nm ≈ 1.3 (stimulatory conditions). (B) Lane 1, KSB837 (WT); lane 2, IFC6005 (uvrY− purA−); lane 3, IFC6003 (purA−); lane 4, IFC6004 (barA− purA−); and lane 5, IFC6005 (uvrY− purA−) harboring plasmid pMX577 (expressing UvrYD54Q). Cell extracts were obtained from cultures at an OD600nm ≈ 0.6 (nonstimulatory conditions). (C) Purified His6-UvrY was incubated in a 40-µL reaction mixture with 100 µM ZMP, and, at the indicated time intervals, 5-µL samples were withdrawn and subjected to Phos-Tag SDS/PAGE followed by Western blot analysis.
ZMP and/or ZTP Activate Various Nonrelated RRs.
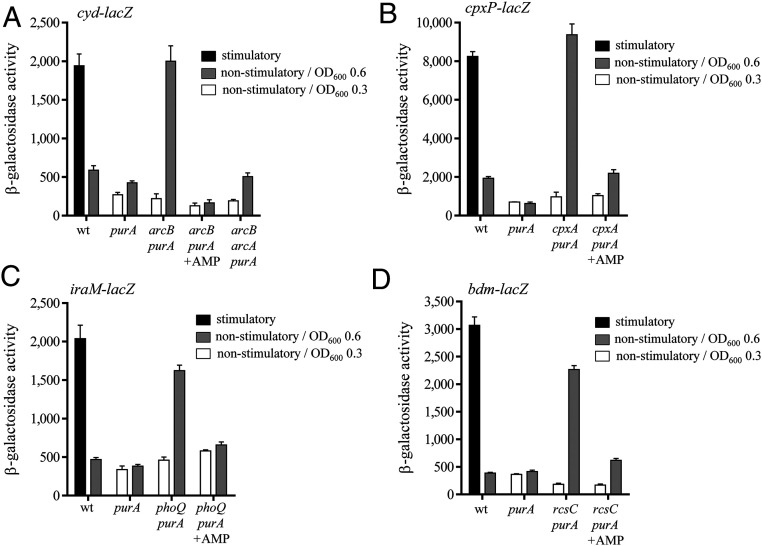
The question of whether the Z nucleotides act as phosphodonors only for UvrY or generally for RRs was, then, addressed. Firstly, the effect of the purA− mutation on the activity of the ArcB/ArcA TCS that is activated under anoxic growth conditions (37) was tested. To this end, the expression of the cyd-lacZ reporter, whose expression is positively regulated by ArcA-P (38), in the purA−, purA− arcB−, and purA− arcB− arcA− strains was monitored under aerobic (nonstimulatory) growth conditions. No activation of reporter expression was observed in any of the tested strains at an OD600 of ∼0.3 (Fig. 5A), a growth stage where the Z nucleotide concentration remains low. On the other hand, at an OD600 of ∼0.6, a growth stage where the concentration of Z nucleotides appears to be higher than the threshold for UvrY activation (Fig. 3C), significant reporter expression was obtained in the purA− arcB− strain but not in the purA− or purA− arcB− arcA− strains (Fig. 5A). As expected, when the culture medium was supplemented with AMP, which prevents Z nucleotide accumulation (Fig. 3D), no activation of cyd-lacZ expression was observed in the purA− arcB− strain (Fig. 5A). Therefore, it can be concluded that ZMP/ZTP activates ArcA, most likely, by triggering its autophosphorylation. This is supported by the fact that no activation of reporter expression was observed in the purA mutant strain, which can be attributed to the ArcA-P−phosphatase activity of ArcB under nonstimulatory conditions.
Fig. 5.
Z nucleotides activate nonrelated RRs under nonstimulatory conditions and in an HK-independent manner. (A) Strains IFC6016 (WT), IFC6017 (purA−), IFC6018 (arcB− purA−), and IFC6019 (arcB− arcA− purA−), all of which carry the ArcA-P−activatable cydA-lacZ reporter, were grown aerobically (nonstimulatory conditions) in LB medium containing 0.1 M Mops (pH 7.4), to an OD600 of 0.3 (white bars) or 0.6 (gray bars). As a control, strain IFC6016 (WT) was grown anaerobically (stimulatory conditions) in the same medium. (B) Strains IFC6020 (WT), IFC6021 (purA−), and IFC6022 (cpxA− purA−), all of which carry the CpxR-P−activatable cpxP-lacZ reporter, were grown in LB medium at pH 5.8 (nonstimulatory conditions) to an OD600 of 0.3 (white bars) or 0.6 (gray bars). To obtain reference values, strain IFC6020 (WT) was grown in LB medium at pH 8.8 (stimulatory conditions). (C) Strains IFC6026 (WT), IFC6027 (purA−), and IFC6028 (phoQ− purA−), all of which carry the PhoP-P−activatable iraM-lacZ reporter, were grown in LB medium (nonstimulatory conditions) to an OD600 of 0.3 (white bars) or 0.6 (gray bars). As a control, strain IFC6026 (WT) was grown in M9 defined minimal medium supplemented with 0.2% (wt/vol) glucose, and, at an OD600 of 0.3, cells were harvested and resuspended in a magnesium-free M9 medium, and incubation continued during 30 min (stimulatory conditions). (D) Cultures of strains IFC6023 (WT), IFC6024 (purA−), and IFC6025 (rcsC− purA−), all of which carry the RcsB-P−activatable bdm-lacZ reporter, were grown in LB medium (nonstimulatory conditions) to an OD600 of 0.3 (white bars) or 0.6 (gray bars). As a control, strain IFC6023 (WT) was grown in M9 defined minimal medium supplemented with 0.2% (wt/vol) glucose, and, at an OD600 of 0.3, 500 mM NaCl were added, and incubation continued for 30 min (stimulatory conditions). For A−D, β-galactosidase activity is measured and expressed in Miller units. Data represent the averages from three independent experiments, and SDs are indicated.
Finally, the effect of the Z nucleotides on the activation of RRs of the nonrelated TCSs CpxA/CpxR, PhoQ/PhoP, and RcsC-RcsD/RcsB, using cpxP-lacZ, iraM-lacZ, and bdm-lacZ chromosomal gene fusions, respectively, was explored (Fig. 5 B–D). First, reference values of the activity of each TCS were obtained by monitoring reporter expression in WT cells under stimulatory and nonstimulatory growth conditions (39–41). Subsequently, reporter expression of purA− strains carrying or lacking the respective HK were analyzed under nonstimulatory growth conditions at OD600 of 0.3 and 0.6. It was found that reporter expression was activated only at OD600 of 0.6 and in the absence of the respective HK (Fig. 5 B–D). Also, when the culture medium was supplemented with AMP, no reporter activation was observed (Fig. 5 B–D), in agreement with the above-presented results. It, thus, appears that the Z nucleotides act as phosphodonors to a wide range of RRs.
Discussion
LMPs, such as acetyl-P, carbamoyl-P, phosphoramidate, and γ-glutamyl-P, have been shown to serve as substrate for RR autophosphorylation, either in vivo or in vitro (9, 11, 12, 42–44). However, with some exceptions, the absence of the cognate HK has been a prerequisite to observe such RR phosphorylation in vivo. Nevertheless, it has been suggested that acetyl-P−dependent RR phosphorylation represents a primitive mechanism of bacteria to sense and respond to changes in the energy status of the cell (14). Also, it has been proposed that acetyl-P may contribute to the maintenance of the steady-state activity of TCSs by providing a baseline level of phosphorylated RRs under nonstimulatory conditions, thus enabling a prompt response upon a shift to stimulatory conditions (9). Here, we report a link between Z nucleotides (ZMP and/or ZTP) and RR-dependent activation of gene expression. Such activation was observed to take place under nonstimulatory conditions, and, in the case of UvrY, it was found that Z nucleotide-dependent activation was mediated by phosphorylation of the conserved aspartate residue that is phosphorylated by the cognate HK BarA. Our in vitro phosphorylation assay suggests that UvrY is able to autophosphorylate, to some extent, at the expense of, at least, ZMP. It is important mentioning that this is a puzzling result if the free energies (ΔG´°) of hydrolysis of ZMP, assumed to be similar to other nucleoside monophosphates, and phosphoaspartate of RRs, assumed to be similar to other acyl phosphates, are considered. However, because experimental ΔG’° values of neither ZMP nor phosphoaspartate linkages in RRs are available, the thermodynamic grounds and plausibility of such phosphoryl transfer reaction cannot yet be assessed. Curiously, we observed that Z nucleotides triggered UvrY phosphorylation both in the absence and in the presence of BarA, and, in the latter case, csrB expression was enhanced under both stimulatory and nonstimulatory conditions. This result suggests that the switch between phosphatase and kinase activities of BarA may be affected by ZMP and/or ZTP, as it was previously shown that BarA possesses, in addition to its kinase activity, a robust phosphatase activity under nonstimulatory conditions (22, 44). However, further studies will be required to explain the specific effect of Z nucleotide accumulation on the regulation of BarA. Finally, as is the case of RR activation by acetyl-P, the presence of the other cognate HKs, namely ArcB, CpxA, PhoQ, and RcsC, impaired Z nucleotide-dependent reporter expression under nonstimulatory conditions, most probably due to their RR-P−specific phosphatase activity.
ZMP is a metabolic intermediate in the de novo synthetic pathway of purines that is subsequently converted to IMP, a precursor of AMP and GMP. ZMP can also be converted to the corresponding triphosphate, ZTP, in a reversible reaction catalyzed by the essential enzyme ribose phosphate diphosphokinase (PrsA), also known as PRPP synthetase (45). Both ZMP and ZTP have been shown to accumulate in bacterial cells during purine or folate depletion, and ZTP has been postulated to act as an alarmone for 10-formyl-THF deficiency in Salmonella typhimurium (46–50). Recently, it was reported that both ZMP and ZTP bind to and activate members of a widespread riboswitch group that regulate genes related to the de novo purine biosynthesis and one-carbon metabolism in bacteria (51). Although no members of this ZTP riboswitch group are present in E. coli, a reporter, based on the plf RNA motif of Clostridium bartlettii, which is a characteristic receptor of this group of riboswitches, was shown to successfully sense ZMP and ZTP accumulation due to folate stress in E. coli (51).
Based on the facts that 1) both the purine and the histidine de novo pathways are widely distributed and remain almost identical among the three domains of life, 2) AICA, which is a precursor of AICAR, can be synthetized under prebiotic conditions, and 3) ZMP and ZTP can both bind to and regulate functional RNA molecules (51–54), it was proposed that these Z nucleotides are ancient molecules (55). On the other hand, it is thought that RRs evolved before the cognate HKs (14), and that primitive TCSs consisted of regulatory proteins that were responsive to available phosphodonors, providing a rudimentary mechanism to sense energy conditions by phosphorylation. Taken together, these suggestions allow us to conceive a scenario in which primitive microorganisms could detect metabolic cues, such as folate deficiency stress, by sensing the Z nucleotide pool through phosphorylatable regulatory proteins, such as RRs.
Summarizing, we provide evidence that ZMP and/or ZTP can act as phosphodonors able to promote the phosphorylation of RRs, thereby modulating gene expression in response to metabolic cues.
Material and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. Strain IFC6000 (csrA::Kanr purA::Tn10 csrB-lacZ) was selected from a mutant library obtained by random insertion of mini-Tn10 transposon present in pBSL181 (56) into strain IFC5010 (csrA::Kanr csrB-lacZ) harboring plasmid pMX543 (22). The purA::Tn10 allele was then transferred to KSB837 (WT, csrB-lacZ), CF7789 (WT), and IFC5010 (csrA::Kanr csrB-lacZ) by P1vir transduction to obtain strains IFC6001, IFC6015, and IFC6000W, respectively. Chromosomal deletions of genes barA, uvrY, carB, hisF, purH, purR, cpxA, rscC, and phoQ were carried out by P1vir transduction of the Kanr-marked mutant allele from the corresponding clone of the Keio collection (57) into suited receptor strains. When needed, the FRT-flanked Kanr cassette was removed using the Flp recombinase encoded in the temperature-sensitive plasmid pCP20 (58). Gene deletion of arcB, arcA, or the ackA-pta was pursued by P1vir transduction of the ΔarcB::Kanr, ΔarcA::Tetr, or ackA::Tetr::pta mutant allele from strain ECL5013 (59), ECL5020 (60), or ECL5336 (61), respectively. Chromosomal deletion of purC was carried out by homologous recombination using the lambda red recombinase system (62). Briefly, a PCR-amplified fragment, using primers del-purC-Fw (5′-AAAGACGTATGTGCGGTATTGTCGGTATCGCCGGTGTTATTGTGTAGGCTGGAGCTGC-3′ and del-purC-Rv (5′-AAGTCAGGGTGCCAGACCGGCACCCTCAGCGAAGGCATCAGAATATCCTCC‐TTAGTTCC-3′) and plasmid pKD4 (62) as the template, was used to replace the purC gene operon with a Kanr cassette. Strains IFC6015 and IFC6019, carrying cydA-lacZ and cpxP-lacZ reporter, respectively, were constructed by P1vir transduction of the λφ(cydA′-lacZ) or λφ(cpxP′-lacZ) alleles from strain ECL5003 (59) or TR50 (39), respectively, into strain CF7789. Strains IFC6022 and IFC6025, carrying chromosomally integrated Ampr-linked bdm-lacZ and iraM-lacZ fusion, respectively, were obtained using the CRIM (conditional-replication, integration, and modular) system (63). Plasmid pAH-bdm, containing a bdm-lacZ operon fusion, was constructed by cloning a 402-bp PCR-amplified fragment containing the transcriptional regulatory region of the bdm gene (using primers bdm-lacZfs-Fw [5′-GCCTGCAGCCACTCTCGGTGGATAC-3′] and bdm-lacZfs-Rv [5′-ATGGATCC-GGTGTCTGCTACGGATC-3′]) into the PstI-BamHI sites of pAH125-bla. Plasmid pAH-iraM, containing an iraM-lacZ operon fusion, was constructed by cloning a 380-bp PCR-amplified DNA fragment containing the transcriptional regulatory region of the iraM gene (using primers iraM-lacZfs-Fw [5′-GCCTGCAGCCACACGTCCGTTTC-3′] and iraM-lacZfs-Rv [5′-GCGGATCCAAATTAATTAAAATGATGGC-3′]) into the PstI-BamHI sites of pAH125-bla (22). Both fusions were integrated into the CF7789 chromosome, as previously described (63), to generate strains IFC6022 and IFC6025.
Table 1.
E. coli strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
| Strain | ||
| CF7789 | MG1655 ΔlacZ (MluI) | Michael Cashel* |
| KSB837 | CF7789 λφ(csrB′-lacZ) | Gudapaty et al. (69) |
| IFC5010 | KSB837 csrA::Kanr | Camacho et al. (22) |
| ECL5336 | MC4100 ackA::Tetr::pta | Liu et al. (61) |
| TR50 | MC4100 λφ(cpxP′-lacZ) | Raivio and Silhavy (39) |
| ECL5003 | MC4100 λφ(cydA′-lacZ) | Kwon et al. (59) |
| ECL5013 | MC4100 ΔarcB::Kanr φ(lldP-lacZ) | Kwon et al. (59) |
| ECL5020 | ΔarcA::Tetr | Georgellis et al. (60) |
| JW2757 | BW25113 ΔbarA::Kanr | Keio collection (57) |
| JW1899 | BW25113 ΔuvrY::Kanr | Keio collection (57) |
| JW2007 | BW25113 ΔhisF::Kanr | Keio collection (57) |
| JW0031 | BW25113 ΔcarB::Kanr | Keio collection (57) |
| JW3970 | BW25113 ΔpurH::Kanr | Keio collection (57) |
| JW1650 | BW25113 ΔpurR::Kanr | Keio collection (57) |
| JW3882 | BW25113 ΔcpxA::Kanr | Keio collection (57) |
| JW5917 | BW25113 ΔrcsC::Kanr | Keio collection (57) |
| JW1115 | BW25113 ΔphoQ::Kanr | Keio collection (57) |
| IFC6000 | IFC5010 purA::Tn10 harboring plasmid pMX543 | This work |
| IFC6000W | IFC5010 purA::Tn10 | This work |
| IFC6001 | KSB837 purA::Tn10 | This work |
| IFC6002 | KSB837 ΔbarA::Kanr | This work |
| IFC6003 | KSB837 ΔuvrY::Kanr | This work |
| IFC6004 | KSB837 purA::Tn10 ΔbarA::Kanr | This work |
| IFC6005 | KSB837 purA::Tn10 ΔuvrY::Kanr | This work |
| IFC6006 | KSB837 purA::Tn10 ΔbarA ackA::Tetr::pta | This work |
| IFC6007 | KSB837 purA::Tn10 ΔbarA | This work |
| IFC6008 | KSB837 purA::Tn10 ΔbarA ΔcarB::Kanr | This work |
| IFC6009 | KSB837 purA::Tn10 ΔbarA ΔcarB::Kanr ackA::Tetr::pta | This work |
| IFC6010 | KSB837 purA::Tn10 ΔbarA ΔpurC::Kanr | This work |
| IFC6011 | KSB837 purA::Tn10 ΔbarA ΔhisF::Kanr | This work |
| IFC6012 | KSB837 purA::Tn10 ΔbarA ΔhisF ΔpurC::Kanr | This work |
| IFC6013 | KSB837 purA::Tn10 ΔbarA ΔpurH::Kanr | This work |
| IFC6014 | KSB837 purA::Tn10 ΔbarA ΔpurR::Kanr | This work |
| IFC6015 | CF7789 purA::Tn10 | This work |
| IFC6016 | CF7789 λφ(cydA′-lacZ) | This work |
| IFC6017 | CF7789 λφ(cydA′-lacZ) purA::Tn10 | This work |
| IFC6018 | CF7789 λφ(cydA′-lacZ) purA::Tn10 ΔarcB::Kanr | This work |
| IFC6019 | CF7789 λφ(cydA′-lacZ) purA::Tn10 ΔarcB::Kanr ΔarcA::Tetr | This work |
| IFC6020 | CF7789 λφ(cpxP′-lacZ) | This work |
| IFC6021 | CF7789 λφ(cpxP′-lacZ) purA::Tn10 | This work |
| IFC6022 | CF7789 λφ(cpxP′-lacZ) purA::Tn10 ΔcpxA::Kanr | This work |
| IFC6023 | CF7789 λφ(bdm’-lacZ) | This work |
| IFC6024 | CF7789 λφ(bdm’-lacZ) purA::Tn10 | This work |
| IFC6025 | CF7789 λφ(bdm’-lacZ) purA::Tn10 ΔrcsC::Kanr | This work |
| IFC6026 | CF7789 λφ(iraM′-lacZ) | This work |
| IFC6027 | CF7789 λφ(iraM′-lacZ) purA::Tn10 | This work |
| IFC6028 | CF7789 λφ(iraM′-lacZ) purA::Tn10 ΔphoQ::Kanr | This work |
| Plasmid | ||
| pBR322 | Cloning vector, Ampr Tetr | Bolivar et al. (64) |
| pAH125-bla | CRIM vector for transcriptional lacZ fusions, Ampr | Camacho et al. (22) |
| pINT-cat | CRIM integration vector, Camr | Camacho et al. (22) |
| pMX543 | uvrY under the control of barA promoter in pEXT21, Spr | Camacho et al. (22) |
| pUY14D54Q | uvrY D54Q in pBR322, Tetr | Tomenius et al. (44) |
| pQE30UvrY | uvrY in pQE30 under an IPTG inducible promoter | Pernestig et al. (15) |
| pMX557 | uvrY D54Q under the control of barA promoter in pEXT21, Spr | This work |
| pMX558 | purA under native promoter in pBR322, Tetr | This work |
| pAH-bdm | bdm-lacZ operon fusion, Ampr | This work |
| pAH-iraM | iraM-lacZ operon fusion, Ampr | This work |
Intramural Research Program, Eunice Kennedy Shriver NICHD, NIH, Bethesda, Maryland.
To construct plasmid pMX557, which carries the uvrYD54Q gene under the control of the barA promoter, a 762-bp DNA fragment was PCR-amplified using primers uvrY (5′-CCCGGATCCCATATGATCAACGTTCTACTTGTTGATGAC‐CACG-3′) and UvrY-Rv-HindIII (5′-CCC AAGCTTCCGTACCACCAGCATCG-3′), and plasmid pUY14D54Q as the template (44). The PCR product, which contains the uvrYD54Q ORF, was used to replace the NdeI-HindIII restriction fragment of pMX543 to generate pMX557. To construct plasmid pMX558, the purA gene and its promoter region were PCR-amplified, using primers purA-Fw-BamHI (5′-CGGATCCTTGGCGGTGGACTTGTGG-3′) and purA-Rv-SacI (5′-CGAGCTCTGGCTTACCCGACACAGC-3′) and chromosomal DNA from strain CF7789 as the template, and cloned into the ScaI site of pBR322 (64).
Unless otherwise stated, cells were grown at 37 °C in LB medium. When necessary, media were supplemented with antibiotics, at the following concentrations: chloramphenicol, 20 µg/mL; kanamycin, 50 µg/mL; ampicillin, 100 µg/mL; spectinomycin, 50 µg/mL; and tetracycline, 10 µg/mL. P1vir transduction was carried out as previously described (65).
Random Mutagenesis.
A random insertion library was generated by inserting mini-Tn10 in the chromosome of strain ICF5010 carrying plasmid pMX543. Cells grown to an OD600 of 0.6 were washed and concentrated for electroporation with the nonreplicative plasmid pBSL181, which carries the mini-Tn10 transposon (56). After electroporation, cells were incubated in 1 mL of LB medium supplemented with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to induce the transposase for 1 h at 37 °C. Mutagenized cells were selected by plating on LB agar containing chloramphenicol, for mini-Tn10 selection, and 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-gal) (40 μg/mL) for detection of increased csrB-lacZ expression. To identify the chromosomal locus of Tn10 insertion in selected mutants, genomic libraries were constructed by Sau3A1 restriction of genomic DNA and cloning of the obtained fragments into BamHI-digested pUC18. Transformed E. coli cells were plated on LB agar containing chloramphenicol, Cm-resistant clones were selected, and plasmids were isolated for DNA sequencing.
β-Galactosidase Activity.
Cells carrying the UvrY-P–activatable csrB′-lacZ reporter were grown aerobically in LB or in LB adjusted to pH 5.0 and buffered with 0.1 M homopiperazine-N,N′-bis(2-ethanesulfonic acid) (HOMOPIPES), at 37 °C. When indicated, acetate was used at a concentration of 7 mM. Cells carrying the ArcA-P–activatable cydA′-lacZ operon fusion were grown aerobically (noninducing condition) in 10 mL to 50 mL of LB medium containing 0.1 M Mops (pH 7.4) in 250-mL baffled flasks at 37 °C with shaking (300 rpm), or were grown anaerobically (inducing condition) in a screw-capped tube filled with buffered LB medium up to the rim at 37 °C and stirred by a magnet. To evaluate the CpxA/CpxR activity, cpxP′-lacZ−bearing strains were grown aerobically in LB medium supplemented with 100 mM sodium phosphate and adjusted at pH 8.0 or 5.5, at 37 °C. Cells harboring the RcsB-P–activatable bdm′-lacZ reporter were grown at 37 °C in LB medium (noninducing condition) or in M9 defined minimal medium (66) supplemented with 0.2% (wt/vol) glucose. At an OD600 of 0.3, 500 mM NaCl were added, and incubation continued for 30 m (inducing condition). Cells bearing the PhoP-P−activatable iraM′-lacZ reporter were grown aerobically at 37 °C in LB (noninducing condition) or in M9 minimal medium supplemented with 0.2% (wt/vol) glucose. At an OD600 of 0.3, cells were harvested and resuspended in a magnesium-free M9 medium, and incubated for 30 m at 37 °C (inducing condition). β-galactosidase activity was assayed and expressed in Miller units as described previously (65).
Western Blotting.
Cells were grown aerobically at 37 °C and harvested by centrifugation during both the midexponential and stationary phase of growth. The cell pellet was resuspended in TE buffer (10 mM Tris⋅HCl, 0.1 mM (ethylenedinitrilo)tetraacetic acid [EDTA], pH 8.0), disrupted by sonication, and cell debris was removed by centrifugation at 10,000 rpm for 10 min. Protein content of cell extracts was measured by the Bradford method, and 5 μg of total protein from each strain were mixed with lysis buffer (50 mM Tris-HCl, 4% SDS, pH 6.8) and separated by SDS/PAGE (15% polyacrylamide gel). Proteins were transferred to a Hybond-ECL filter (Amersham Biosciences). The filter was equilibrated in TTBS (Tween/Tris-buffered saline) buffer (25 mM Tris, 150 mM NaCl, and 0.05% Tween 20) for 10 min and incubated in blocking buffer (1% milk in TTBS) for 1 h at room temperature. UvrY polyclonal antibodies, raised against His6-UvrY (22), or monoclonal antibodies raised against DnaK (Enzo Life Sciences) were added at a dilution of 1:10,000 and incubated for 1 h at room temperature. The bound antibody was detected by using anti-rabbit IgG antibody or anti-mouse IgG antibody conjugated to horseradish peroxidase (1:10,000 dilution) and the Immobilon Western detection system (Millipore).
Analysis of the In Vivo UvrY Phosphorylation State.
UvrY phosphorylation levels were evaluated using Phos-Tag acrylamide gel electrophoresis and subsequent immunoblotting. One-milliliter culture samples were collected by centrifugation at the indicated OD600, and resuspended in 100 μL of 1 M formic acid to stabilize the phospho-Asp residue. Samples were processed as previously reported (35, 67), with slight modifications. Briefly, cell suspensions were vigorously vortexed for 30 s to complete cell lysis. Subsequently, 10 μL of each lysate were transferred to a 1.5-mL microfuge tube, solubilized by addition of 5 μL of loading buffer, and neutralized by addition of 5 μL of 1 M Bis-Tris and 3.1 μL of 2.5 N NaOH. Proteins were separated on a 37.- μM Phos-Tag 10% acrylamide gel. Gel preparation, electrophoresis, and transfer to nitrocellulose membranes were carried out following manufacturer’s instructions (Waco Pure Chemical Industries), including the additional EDTA steps to remove Mn2+ from the gel. Immunoblotting of UvrY was carried out as previous described.
His6-UvrY Purification and In Vitro Phosphorylation Assay.
E. coli M15 cells cotransformed with pREP4 and pQE30UvrY (15) were grown in 1 L of LB medium, supplemented with ampicillin and kanamycin, in a rotary shaker at 37 °C until an OD600 of 0.6. Expression of the His6-tagged UvrY was induced by the addition of 1 mM IPTG. Cells were harvested 5 h after induction, and the cell pellet was resuspended in 10 mL of lysis buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 10 mM imidazole). Finally, protein purification was performed at 4 °C under nondenaturing conditions by Ni-NTA–agarose affinity chromatography, as described previously (15).
For the phosphorylation assay, purified His6-UvrY (10 μg) was incubated with 100 μM ZMP (AICAR phosphate ≥ 95% by high-pressure liquid chromatography, Toronto Research Chemicals) in a 40-μL reaction mixture containing 33 mM Hepes (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, and 10% glycerol, at room temperature. After the indicated times, 5-μL samples were withdrawn and mixed with 1 µL of 1 M formic to stop the reaction and stabilize the phospho-Asp residue. Samples were subjected to analysis by Phos-Tag acrylamide gel electrophoresis separation and immunodetection, as described above.
Z Nucleotide Quantification.
Z nucleotide was quantified as described by Nagy et al. (34), and modified by Bratton and Marshall (33) and Stetten and Fox (68), with slight modifications. Cultures of desired strains were grown in LB, and, at indicated times, 100-μL aliquots were mixed with 900 μL of a solution containing 0.9% (wt/vol) NaCl and 0.001% (wt/vol) SDS, and 15 μL of chloroform was added. Cells were disrupted by vigorous vortexing for 10 s, and 500 μL of 0.4 N HCl and 10 μL of 0.1% (wt/vol) NaNO2 were added. After 3 min, 10 μL of 0.5% (wt/vol) ammonium sulfamate were added, and the mixture was further incubated for 2 min. Finally, 10 μL of 0.1% (wt/vol) N-(1-naphthyl) ethylenediamine were added, and the reactions were incubated for 5 min. The diazotized product was measured at 545 nm. For each sample mixture, the spectrophotometer was blanked using a mixture containing the same components, including the 100 μL of cell culture, except NaNO2. AICAR concentrations were calculated using an ε of 4.5 × 104 M−1⋅cm−1 (34).
Statistics.
All quantitative experiments were performed in triplicate, and experimental results are expressed as mean ± the SD value.
Supplementary Material
Acknowledgments
This work was supported by Grants IN208718 and IN209918 from the Dirección General de Asuntos del Personal Académico-Universidad Nacional Autónoma de México. We are grateful to Claudia Rodríguez Rangel for technical assistance and to Diego Gonzalez Halphen for critically reading the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006209117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Hoch J. A., Silhavy T. J., Two-Component Signal Transduction (American Society for Microbiology, ed. 1, 1995). [Google Scholar]
- 2.Georgellis D., Lynch A. S., Lin E. C. C., In vitro phosphorylation study of the arc two-component signal transduction system of Escherichia coli. J. Bacteriol. 179, 5429–5435 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhl M. A., Miller J. F., Integration of multiple domains in a two-component sensor protein: The Bordetella pertussis BvgAS phosphorelay. EMBO J. 15, 1028–1036 (1996). [PMC free article] [PubMed] [Google Scholar]
- 4.Teran-Melo J. L., et al. , Routes of phosphoryl group transfer during signal transmission and signal decay in the dimeric sensor histidine kinase ArcB. J. Biol. Chem. 293, 13214–13223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stock A. M., Robinson V. L., Goudreau P. N., Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Kenney L. J., How important is the phosphatase activity of sensor kinases? Curr. Opin. Microbiol. 13, 168–176 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez A. F., Barba-Ostria C., Silva-Jiménez H., Georgellis D., Organization and mode of action of two component system signaling circuits from the various kingdoms of life. Environ. Microbiol. 18, 3210–3226 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Georgellis D., Kwon O., De Wulf P., Lin E. C. C., Signal decay through a reverse phosphorelay in the arc two-component signal transduction system. J. Biol. Chem. 273, 32864–32869 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Lukat G. S., McCleary W. R., Stock A. M., Stock J. B., Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U.S.A. 89, 718–722 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng J., et al. , Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J. Bacteriol. 174, 6061–6070 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogura M., Kawata-Mukai M., Itaya M., Takio K., Tanaka T., Multiple copies of the proB gene enhance degS-dependent extracellular protease production in Bacillus subtilis. J. Bacteriol. 176, 5673–5680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe A. J., The acetate switch. Microbiol. Mol. Biol. Rev. 69, 12–50 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCleary W. R., Stock J. B., Ninfa A. J., Is acetyl phosphate a global signal in Escherichia coli? J. Bacteriol. 175, 2793–2798 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolfe A. J., Physiologically relevant small phosphodonors link metabolism to signal transduction. Curr. Opin. Microbiol. 13, 204–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pernestig A. K., Melefors O., Georgellis D., Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J. Biol. Chem. 276, 225–231 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Chavez R. G., Alvarez A. F., Romeo T., Georgellis D., The physiological stimulus for the BarA sensor kinase. J. Bacteriol. 192, 2009–2012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K., et al. , Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184, 5130–5140 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weilbacher T., et al. , A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 48, 657–670 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Liu M. Y., Romeo T., The global regulator CsrA of Escherichia coli is a specific mRNA-binding protein. J. Bacteriol. 179, 4639–4642 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romeo T., Vakulskas C. A., Babitzke P., Post-transcriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ. Microbiol. 15, 313–324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueroa-Bossi N., et al. , RNA remodeling by bacterial global regulator CsrA promotes Rho-dependent transcription termination. Genes Dev. 28, 1239–1251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camacho M. I., et al. , Effects of the global regulator CsrA on the BarA/UvrY two-component signaling system. J. Bacteriol. 197, 983–991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butz H. A., Mey A. R., Ciosek A. L., Payne S. M., Vibrio cholerae CsrA directly regulates varA to increase expression of the three nonredundant Csr small RNAs. MBio 10, e01042-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honzatko R. B., Fromm H. J., Structure-function studies of adenylosuccinate synthetase from Escherichia coli. Arch. Biochem. Biophys. 370, 1–8 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Stepchenkova E. I., Kozmin S. G., Alenin V. V., Pavlov Y. I., Genetic control of metabolism of mutagenic purine base analogs 6-hydroxylaminopurine and 2-amino-6-hydroxylaminopurine in yeast Saccharomyces cerevisiae. Russ. J. Genet. 45, 409–414 (2009). [PMC free article] [PubMed] [Google Scholar]
- 26.He B., Shiau A., Choi K. Y., Zalkin H., Smith J. M., Genes of the Escherichia coli pur regulon are negatively controlled by a repressor-operator interaction. J. Bacteriol. 172, 4555–4562 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He B., Zalkin H., Regulation of Escherichia coli purA by purine repressor, one component of a dual control mechanism. J. Bacteriol. 176, 1009–1013 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondragón V., et al. , pH-dependent activation of the BarA-UvrY two-component system in Escherichia coli. J. Bacteriol. 188, 8303–8306 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi T., et al. , Deregulation of purine pathway in Bacillus subtilis and its use in riboflavin biosynthesis. Microb. Cell Fact. 13, 101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daignan-Fornier B., Pinson B., 5-Aminoimidazole-4-carboxamide-1-beta-D-ribofuranosyl 5′-monophosphate (AICAR), a highly conserved purine intermediate with multiple effects. Metabolites 2, 292–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou G., Smith J. L., Zalkin H., Binding of purine nucleotides to two regulatory sites results in synergistic feedback inhibition of glutamine 5-phosphoribosylpyrophosphate amidotransferase. J. Biol. Chem. 269, 6784–6789 (1994). [PubMed] [Google Scholar]
- 32.Watanabe K., Tomioka S., Tanimura K., Oku H., Isoi K., Uptake of AMP, ADP, and ATP in Escherichia coli W. Biosci. Biotechnol. Biochem. 75, 7–12 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Bratton C., Marshall E. K., A new coupling component for sulfanilamide determination. J. Biol. Chem. 128, 537–550 (1939). [Google Scholar]
- 34.Nagy P. L., Marolewski A., Benkovic S. J., Zalkin H., Formyltetrahydrofolate hydrolase, a regulatory enzyme that functions to balance pools of tetrahydrofolate and one-carbon tetrahydrofolate adducts in Escherichia coli. J. Bacteriol. 177, 1292–1298 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbieri C. M., Stock A. M., Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal. Biochem. 376, 73–82 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinoshita-Kikuta E., Kinoshita E., Eguchi Y., Koike T., Validation of Cis and Trans modes in multistep phosphotransfer signaling of bacterial tripartite sensor kinases by using Phos-Tag SDS-PAGE. PLoS One 11, e0148294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malpica R., Sandoval G. R. P., Rodríguez C., Franco B., Georgellis D., Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8, 781–795 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Lynch A. S., Lin E. C. C., Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: Characterization of DNA binding at target promoters. J. Bacteriol. 178, 6238–6249 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raivio T. L., Silhavy T. J., Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179, 7724–7733 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francez-Charlot A., Castanié-Cornet M.-P., Gutierrez C., Cam K., Osmotic regulation of the Escherichia coli bdm (biofilm-dependent modulation) gene by the RcsCDB His-Asp phosphorelay. J. Bacteriol. 187, 3873–3877 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battesti A., Tsegaye Y. M., Packer D. G., Majdalani N., Gottesman S., H-NS regulation of IraD and IraM antiadaptors for control of RpoS degradation. J. Bacteriol. 194, 2470–2478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schröder I., Wolin C. D., Cavicchioli R., Gunsalus R. P., Phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli. J. Bacteriol. 176, 4985–4992 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattison K., Oropeza R., Byers N., Kenney L. J., A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J. Mol. Biol. 315, 497–511 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Tomenius H., et al. , Genetic and functional characterization of the Escherichia coli BarA-UvrY two-component system: Point mutations in the HAMP linker of the BarA sensor give a dominant-negative phenotype. J. Bacteriol. 187, 7317–7324 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabina R. L., Holmes E. W., Becker M. A., The enzymatic synthesis of 5-amino-4-imidazolecarboxamide riboside triphosphate (ZTP). Science 223, 1193–1195 (1984). [DOI] [PubMed] [Google Scholar]
- 46.Bochner B. R., Ames B. N., ZTP (5-amino 4-imidazole carboxamide riboside 5′-triphosphate): A proposed alarmone deficiency riboside for 10-formyl-tetrahydrofolate deficiency. Cell 29, 929–937 (1982). [DOI] [PubMed] [Google Scholar]
- 47.Rohlman C. E., Matthews R. G., Role of purine biosynthetic intermediates in response to folate stress in Escherichia coli. J. Bacteriol. 172, 7200–7210 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakraborty S., Gruber T., Barry C. E., Boshoff H. I., Rhee K. Y., Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacterium tuberculosis. Science 339, 88–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon Y. K., Higgins M. B., Rabinowitz J. D., Antifolate-induced depletion of intracellular glycine and purines inhibits thymineless death in E. coli. ACS Chem. Biol. 5, 787–795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei J. R., et al. , Depletion of antibiotic targets has widely varying effects on growth. Proc. Natl. Acad. Sci. U.S.A. 108, 4176–4181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim P. B., Nelson J. W., Breaker R. R., An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol. Cell 57, 317–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delaye L., Becerra A., Lazcano A., The last common ancestor: What’s in a name? Orig. Life Evol. Biosph. 35, 537–554 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Joyce G. F., RNA evolution and the origins of life. Nature 338, 217–224 (1989). [DOI] [PubMed] [Google Scholar]
- 54.Ducker G. S., Rabinowitz J. D., ZMP: A master regulator of one-carbon metabolism. Mol. Cell 57, 203–204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vázquez-Salazar A., Becerra A., Lazcano A., Evolutionary convergence in the biosyntheses of the imidazole moieties of histidine and purines. PLoS One 13, e0196349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexeyev M. F., Shokolenko I. N., Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene 160, 59–62 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Baba T., et al. , Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cherepanov P. P., Wackernagel W., Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Kwon O., Georgellis D., Lynch A. S., Boyd D., Lin E. C., The ArcB sensor kinase of Escherichia coli: Genetic exploration of the transmembrane region. J. Bacteriol. 182, 2960–2966 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Georgellis D., Kwon O., Lin E. C. C., Wong S. M., Akerley B. J., Redox signal transduction by the ArcB sensor kinase of Haemophilus influenzae lacking the PAS domain. J. Bacteriol. 183, 7206–7212 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X., et al. , Evidence against the physiological role of acetyl phosphate in the phosphorylation of the ArcA response regulator in Escherichia coli. J. Microbiol. 47, 657–662 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haldimann A., Wanner B. L., Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183, 6384–6393 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolivar F., et al. , Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2, 95–113 (1977). [PubMed] [Google Scholar]
- 65.Miller J. H., Experiments in Molecular Genetics (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, 1972). [Google Scholar]
- 66.Sambrook J., Russell D. W., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory, ed. 3, Cold Spring Harbor, NY, 2001). [Google Scholar]
- 67.Rolfe M. D., et al. , Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J. Biol. Chem. 286, 10147–10154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stetten M. R., Fox C. L. Jr, An amine ed formed by bacteria during sulfonamide bacteriostasis. J. Biol. Chem. 161, 333–349 (1945). [PubMed] [Google Scholar]
- 69.Gudapaty S., Suzuki K., Wang X., Babitzke P., Romeo T., Regulatory interactions of Csr components: The RNA binding protein CsrA activates csrB transcription in Escherichia coli. J. Bacteriol. 183, 6017–6027 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.