Significance
Among all cancer types, lung cancer patients exhibit the highest prevalence of brain metastasis, often associated with cognitive impairment, seizures, decline in quality of life, and decreased survival. Limited therapeutic options are currently available to treat brain metastasis. A comprehensive understanding of the signaling pathways and transcriptional networks required for survival and growth of brain-metastatic cancer cells is needed to develop effective strategies to treat this disease. Here, we report that the Heat Shock Transcription Factor 1 (HSF1) is upregulated in brain-metastatic lung cancer cells and is required for brain metastasis in mice. Importantly, we show that the HSF1-dependent expression of E2F target genes implicated in cell cycle progression and survival is decreased by blood–brain barrier-penetrant ABL allosteric inhibitors.
Keywords: brain metastasis, ABL2, HSF1, lung adenocarcinoma, ABL kinases
Abstract
Brain metastases are the most common intracranial tumors in adults and are associated with increased patient morbidity and mortality. Limited therapeutic options are currently available for the treatment of brain metastasis. Here, we report on the discovery of an actionable signaling pathway utilized by metastatic tumor cells whereby the transcriptional regulator Heat Shock Factor 1 (HSF1) drives a transcriptional program, divergent from its canonical role as the master regulator of the heat shock response, leading to enhanced expression of a subset of E2F transcription factor family gene targets. We find that HSF1 is required for survival and outgrowth by metastatic lung cancer cells in the brain parenchyma. Further, we identify the ABL2 tyrosine kinase as an upstream regulator of HSF1 protein expression and show that the Src-homology 3 (SH3) domain of ABL2 directly interacts with HSF1 protein at a noncanonical, proline-independent SH3 interaction motif. Pharmacologic inhibition of the ABL2 kinase using small molecule allosteric inhibitors, but not ATP-competitive inhibitors, disrupts this interaction. Importantly, knockdown as well as pharmacologic inhibition of ABL2 using allosteric inhibitors impairs expression of HSF1 protein and HSF1-E2F transcriptional gene targets. Collectively, these findings reveal a targetable ABL2-HSF1-E2F signaling pathway required for survival by brain-metastatic tumor cells.
Brain metastases stemming from primary tumors of the lung are a major health challenge for patients and often result in devastating neurologic impairments and increased mortality (1–3). Despite numerous studies focused on dissecting genetic and nongenetic molecular mechanisms employed by metastatic tumor cells to survive and colonize the unique microenvironment of the brain (4–8), there is a lack of effective therapies to treat this disease (9). Therefore, there is an urgent need to gain additional insights into the mechanisms employed by metastasizing tumor cells for colonization and survival in the brain, which might lead to new rational approaches for pharmacologic intervention.
The transcriptional activator Heat Shock Transcription Factor 1 (HSF1) is a master regulator of protein homeostasis (10, 11). Under normal physiologic conditions, HSF1 monomers are held inactive in the cytoplasm through intramolecular interactions and a regulatory complex consisting of protein chaperones and the chaperonin TCP1 ring complex (TRiC) (12–14). In response to a diverse array of proteotoxic stresses including heat shock, monomeric HSF1 sheds its inhibitory regulatory complex, translocates to the nucleus, and oligomerizes into transcriptionally active HSF1 trimers (10, 15–17). Active HSF1 then induces transcription of genes encoding molecular chaperones by binding to DNA motifs known as heat shock elements (HSEs) (16). Although the role of HSF1 in the context of protein homeostasis is well established, an emerging body of evidence in recent years has shown that HSF1 also functions to drive transcriptional programs implicated in tumor progression and malignancy independent of the canonical heat shock response (18–25). Previous work identified a divergent cancer-specific transcriptional program driven by HSF1 in highly malignant mammary epithelial cells and tumor cell lines from the lung, breast, and colon (18). Central to this work was the discovery that genome occupancy of HSF1 in the context of tumor malignancy is distinct from that of heat shock and drives expression of target genes implicated in cell cycle, translation, and DNA repair. More recently, HSF1 expression was also identified in T cell acute lymphoblastic leukemia as critical for tumor cell survival by regulating NOTCH1 transcriptional activity (19). Although these studies provide evidence in support of heat shock-independent functions of HSF1, little is known regarding the upstream modulators of HSF1 protein expression as well as the role of HSF1-driven transcriptional programs in the context of tumor metastasis.
In Caenorhabditis elegans, HSF1 was recently shown to function with E2F transcription factors to drive a transcriptional program required during larval stages of development, independent of its role in the cellular response to heat shock and proteotoxic stress (26). In this context, HSF1 binds DNA at “degenerate” heat shock elements adjacent to GC-rich E2F binding domains. We now show that HSF1 protein levels are up-regulated in brain-metastatic lung cancer cells and that HSF1 regulates expression of E2F gene targets in this setting. Genetic inhibition of HSF1 ablates E2F target gene expression and dramatically impairs survival of lung cancer brain metastases both in vitro and in mouse models.
Because therapeutic targeting of transcription factors such as HSF1 and E2F is challenging, we sought to identify actionable upstream regulators of this HSF1-E2F transcription network. We recently characterized a TAZ-AXL-ABL2 feed-forward signaling axis that is activated in brain-metastatic lung cancer cell lines and which is required for successful colonization of these cells in the brain (27). We now show that ABL2 modulates expression of the HSF1 and E2F transcription factors and that knockdown or allosteric inhibition of ABL2 impairs expression of HSF1-dependent and E2F-dependent transcription programs. We report that HSF1 protein is up-regulated in brain-metastatic cancer cells downstream of the ABL2 tyrosine kinase and is required for their survival both in vitro and in vivo. Importantly, we show that HSF1-E2F target gene expression is pharmacologically targetable with ABL kinase allosteric inhibitors (27). These data support a critical role for an actionable ABL2-HSF1-E2F pathway in promoting brain metastasis.
Results
Expression of HSF1 Is Up-Regulated in Brain-Metastatic Lung Adenocarcinoma Cells.
HSF1 expression was previously shown to be up-regulated during progression of normal epithelial cells toward a malignant, tumorigenic state (18). Thus, we questioned whether HSF1 might be differentially expressed in brain-metastatic cells compared to the parental tumor cell lines from which they were derived. Immunoblot analysis of HSF1 protein expression in EGFR mutant (PC9 and HCC4006) and KRAS mutant (H2030) human lung adenocarcinoma cells compared to their respective brain-metastatic variants PC9-BrM3, HCC4006-BrM, and H2030-BrM3, revealed elevated levels of HSF1 protein in the brain-metastatic derivatives (Fig. 1A). Further, immunofluorescence staining of PC9-BrM3 cells revealed high levels of HSF1 in the nucleus, indicative of transcriptionally active HSF1 (Fig. 1B).
Fig. 1.
HSF1 protein is up-regulated in brain-metastatic lung adenocarcinoma cells and is required for cell survival in vitro and colonization of the brain in vivo. (A) Immunoblot analysis of HSF1 protein expression in parental vs. brain-metastatic lung adenocarcinoma (LUAC) cell lines. Protein quantification data for brain-metastatic lines were normalized to actin loading control and corresponding parental cell line expression. (B) Immunofluorescence staining and imaging of HSF1 protein (red) in PC9-BrM3 cells transduced with dox-inducible lentiviral shRNAs against nontarget control (shNTC) or HSF1 (shHSF1 7480). Dapi, nuclear stain (blue). (C) Immunoblot analysis of HSF1 protein expression in PC9-BrM3 cells transduced with distinct dox-inducible shRNA clones targeting HSF1 (7480 or 7481) and treated ± 500 ng/mL dox for the indicated timepoints (H). Protein quantification data were normalized to actin loading control and to corresponding 0 h (untreated) timepoint. (D and E) Cell-Titer Glo assay measuring cell viability in PC9 parental vs. PC9-BrM3 cells transduced with shRNAs against nontarget control or HSF1 clone 7480 (D) or clone 7481 (E) and treated with dox for the indicated timepoints. For all experiments, n = 3 biological replicates per condition. Statistical analysis performed by two-way ANOVA followed by Fisher’s multiple comparison post hoc testing. *P < 0.05; **P < 0.01; ****P < 0.001; ns, not significant. Representative BLI (F) and quantification of brain metastasis burden (G) in mice injected with PC9-BrM3 cells transduced with dox-inducible shRNAs against nontarget control (NTC) or HSF1 (clone 7480). Mice were treated with 3 mg/mL dox water for shRNA induction starting on day 10 postintracardiac injection. After 28 d on dox water, mice were subjected to BLI analysis. Statistical analysis performed by unpaired two-tailed t test. *P < 0.05. (H and I) Individual spider plots (H) and quantification (I) of whole-body metastatic burden in mice as described in Fig. 1 F and G. Statistical analysis was performed using a mixed-effects model in GraphPad Prism 8 software to account for missing values due to premature animal death. **P < 0.01. (J) Kaplan–Meier survival analysis correlating overall survival with mRNA expression of HSF1 in human lung adenocarcinoma patients. Survival groups were separated by tertile based on mRNA expression (n = 720 total patients), and statistical analysis was performed using log-rank Mantel–Cox test.
HSF1 was previously shown to be essential for T cell acute lymphoblastic leukemia cell survival (19). Therefore, we sought to investigate whether the elevated expression of HSF1 in brain-metastatic cells was required for their survival and growth. To examine this possibility, we transduced cells with doxycycline (dox)-inducible lentiviral short hairpin RNAs (shRNAs) against either nontargeting control (shNTC) or HSF1 and measured cell viability across multiple time points after induction of shRNA expression. Immunofluorescence and immunoblot analysis demonstrated the effectiveness of HSF1 knockdown (Fig. 1 B and C). Knockdown of HSF1 with multiple shRNAs resulted in a robust decrease in cell viability starting 48 h post-shRNA induction, which correlated with the loss of HSF1 protein (Fig. 1 C–E and SI Appendix, Fig. S1 A–E). This loss corresponded with an increase in G2/M cell cycle arrest as revealed by FACS analysis (SI Appendix, Fig. S1A) and decreases in Cyclin B1 protein expression (Fig. 1C). Knockdown of HSF1 also resulted in increased levels of cleaved PARP, indicative of apoptotic induction (Fig. 1C). Parental lung cancer cells exhibited decreased survival upon HSF1 knockdown, but to a lesser extent than the brain-metastatic variants (Fig. 1 D and E and SI Appendix, Fig. S1 B–E). These data show that brain-metastatic lung cancer cells are highly dependent on HSF1 for growth and survival.
HSF1 Knockdown Impairs Metastatic Outgrowth and Tumor Cell Survival In Vivo.
Given that loss of HSF1 robustly impairs survival of brain-metastatic lung cancer cells in vitro, we next evaluated whether HSF1 expression might be required during metastatic colonization and outgrowth of lung cancer cells in an in vivo model of brain metastasis. To examine this hypothesis, we performed intracardiac injections of PC9-BrM3 cells transduced with dox-inducible tet-shNTC or tet-shHSF1 shRNAs and coexpressing a luciferase-Tomato (pFuLT) lentiviral reporter. Mice were injected with brain metastatic PC9-BrM3 cells in the left cardiac ventricle and, after assessing colonization of the brain at day 10 postinjection with bioluminescent imaging (BLI), mice were administered 3 mg/mL dox water to induce shRNA expression. The 10-d time point was selected as previous studies have shown that metastasizing tumor cells in circulation complete extravasation into the brain parenchyma within 7 d postinjection (7, 28). We found that the growth rate of metastases in mice injected with PC9-BrM3 cells expressing shRNA against HSF1 was markedly impaired compared to mice injected with nontarget shRNA control cells (Fig. 1F). Metastatic disease burden within the brain was significantly decreased in the HSF1 knockdown group as measured 1 mo after induction with dox water (Fig. 1G). The rate of whole-body metastatic burden was similarly delayed in the HSF1 knockdown cells (Fig. 1 H and I). Together, these results show that HSF1 is necessary for the colonization and outgrowth of metastatic tumor cells in an in vivo model of lung cancer brain metastasis. Notably, analysis of human lung adenocarcinoma patient microarray datasets correlating messenger RNA (mRNA) expression with patient survival showed that high expression of HSF1 correlates with poor overall survival (Fig. 1J).
HSF1 Regulates Transcriptional Expression of E2F Gene Targets in Brain-Metastatic Lung Cancer Cells in a Heat Shock-Independent Manner.
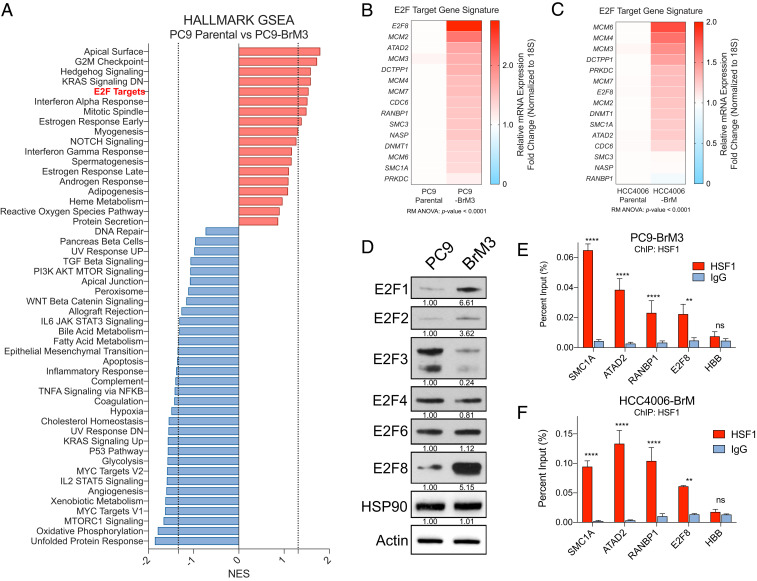
To investigate the mechanism by which HSF1 promotes survival of metastatic lung cancer cells, we employed unbiased transcriptional profiling of these cells with or without HSF1 knockdown in order to gain insights into the transcriptional target gene signatures altered by loss of HSF1. We performed RNA-sequencing (RNA-seq) on PC9-BrM3 cells transduced with dox-inducible nontarget control shRNA or shHSF1 constructs and treated with dox for 48 h, a time point at which HSF1 protein is depleted but prior to loss in cell viability observed at longer time points (Fig. 1 C and D). Gene set enrichment analysis (GSEA) was used on an expanded list of gene signatures from the mSigDB database from The Broad Institute to identify pathways affected by HSF1 loss (29). The signatures most depleted upon HSF1 knockdown corresponded to E2F family gene targets and G2/M cell cycle checkpoint targets, consistent with the observed increase in G2/M cell cycle arrest (SI Appendix, Figs. S1A and S2 A–C and Fig. 2 A–C) and loss of Cyclin B1 expression upon HSF1 knockdown (Fig. 1C). Interestingly, signatures related to the unfolded protein response, while slightly depleted under knockdown conditions, scored relatively poorly despite the known role for HSF1 in this process (Fig. 2A). Similarly, expression of gene targets representative of a canonical heat shock response remained relatively stable despite HSF1 depletion in these cells (Fig. 2D). A list of conserved E2F target genes from a number of E2F gene signatures identified by GSEA, which we confirmed by RT-qPCR to be regulated by E2F family members, was used to generate an HSF1-dependent E2F gene panel (Fig. 2D, upper bracket and SI Appendix, Fig. S2 D and E).
Fig. 2.
Depletion of HSF1 in brain-metastatic lung cancer cells impairs expression of E2F target gene signatures independent of the canonical heat shock response. (A) Waterfall plot of Hallmark GSEA signatures from RNA-seq data ranked by normalized enrichment score (NES) for PC9-BrM3 cells with inducible expression of shNTC or shHSF1. Negative NES, depleted signature; positive NES, enriched signature. Dotted line represents P value cutoff <0.05. (B and C) GSEA plots showing E2F targets (B) or G2/M checkpoint signatures (C) depleted in PC9-BrM3 Tet-shHSF1 knockdown cells. (D) Heatmap depicting relative expression of the E2F target gene panel and representative heat shock gene panel.
We next reasoned that if HSF1 promoted transcription of a subset of E2F target genes, then expression of these target genes might also be increased in brain-metastatic cells expressing high HSF1 levels relative to parental cells. RNA-seq analysis and GSEA comparing PC9-BrM3 cells with PC9 parental cells revealed enrichment for E2F target gene expression as well as G2/M checkpoint targets in the PC9-BrM3 cells (Fig. 3A and SI Appendix, Fig. S3 A–C). RT-qPCR analysis was used to validate increased expression of the HSF1-E2F gene panel in PC9-BrM3 and HCC4006-BrM cells relative to parental PC9 and HCC4006 cells, respectively (Fig. 3 B and C). Notably, RT-qPCR analysis also revealed that E2F family members themselves were differentially expressed in brain-metastatic cells with increased expression of E2F1, E2F2, E2F7, and E2F8, whereas expression of other E2F family members remained unchanged (SI Appendix, Fig. S3D). Further, immunoblot analysis of brain-metastatic cells revealed enrichment of E2F1 and E2F8 relative to the varied expression of other E2F family members, implying a potential role for E2F1 and E2F8 in regulating expression of E2F target genes in this system (Fig. 3D and SI Appendix, Fig. S2 D and E). Of note, analysis of human lung adenocarcinoma patient microarray data revealed that high expression of E2F1 and E2F8 was each individually correlated with poor overall survival (SI Appendix, Fig. S3 E and F). Taken together, these data show that loss of HSF1 in brain-metastatic lung cancer cells results in decreased expression of select E2F target genes with minimal impact on expression of canonical heat shock gene targets.
Fig. 3.
Brain-metastatic lung cancer cells exhibit increased expression of E2F target gene signatures and exhibit HSF1 binding at E2F target genes. (A) Waterfall plot of Hallmark GSEA signatures from RNA-seq data ranked by NES for PC9 parental vs. PC9-BrM3 cells. Negative NES, depleted signature; positive NES, enriched signature. Dotted line represents P value cutoff <0.05. (B and C) Heatmaps of mRNA expression (RT-qPCR) for E2F target genes in PC9 parental vs. PC9-BrM3 cells (B) or HCC4006 parental vs. HCC4006-BrM cells (C). (D) Immunoblot analysis of the indicated proteins expressed in PC9 parental vs. PC9-BrM3 cells. Quantification of protein expression was normalized to actin loading control and parental expression. (E and F) ChIP-qPCR analysis of HSF1 occupancy at degenerate HSEs nearby the indicated E2F target genes in PC9-BrM3 (E) or HCC4006-BrM cells. HBB (Hemoglobin) gene included as negative control. n = 3 biological replicates. For all experiments, *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.001; ns, not significant.
HSF1 was recently shown to function with E2F transcription factors in C. elegans to drive a developmental transcriptional program divergent from the canonical heat shock response (26). Central to this work was the observation that in the context of development, HSF1 occupies genomic loci containing a “degenerate” or partial HSE nearby E2F DNA binding motifs. The canonical DNA element recognized by trimeric HSF1 in response to heat shock consists of triple tandem inverted nGAAn repeats (e.g., nTTCnnGAAnnTTCn or derivations thereof) (11). E2F family members bind DNA sequences at GC-rich motifs consisting of TTTnnCGC (where “n” is either C or G) (SI Appendix, Fig. S4A) (30). Given that loss of HSF1 impairs mRNA expression of E2F family target genes in brain metastatic cells, we hypothesized direct HSF1 binding may occur at degenerate HSEs located near promoter regions of known E2F transcriptional targets. To investigate this possibility, we analyzed the promoter regions of various E2F targets from the E2F target gene panel and found TTTnnCGC motifs upstream of their respective transcriptional start sites (SI Appendix, Fig. S4 B–D). We then searched for partial permutations of the canonical HSE sequence containing varying segments of this DNA motif (SI Appendix, Fig. S4A). We detected a significant number of predicted degenerate HSEs, which were primarily found immediately downstream of the E2F binding motif and upstream of the transcriptional start site, consistent with the report of E2F-HSF1 coregulated DNA binding in C. elegans (SI Appendix, Fig. S4 B–D). In contrast, analysis of the promoter regions of HSP90AB1 and HSPA6, both known target genes of HSF1 during the canonical heat shock response, showed the presence of expected canonical HSEs upstream of the TSS (SI Appendix, Fig. S4 E and F), but no E2F DNA binding motifs were found near the transcriptional start sites of HSP90AB1 and HSPA6. Importantly, chromatin immunoprecipitation (ChIP)-qPCR analysis in PC9-BrM3 and HCC4006-BrM cells revealed empirically that HSF1 occupancy is enriched at predicted degenerate HSEs upstream of E2F target genes (Fig. 3 E and F). Collectively, these findings reveal the presence of degenerate HSF1-binding sequences, occupied by HSF1 and distinct from the canonical HSE motif, near a subset of E2F transcriptional targets.
ABL2-Dependent Regulation of HSF1 Protein Expression in Brain-Metastatic Lung Cancer Cells.
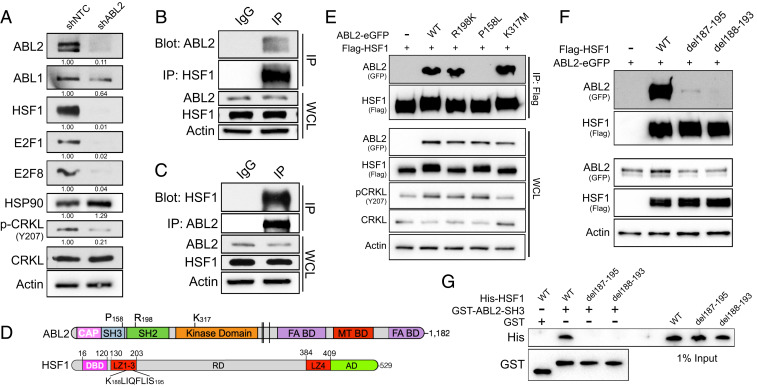
Our data reveal that HSF1 is required for survival of brain-metastatic cancer cells and suggest that targeting of HSF1 could be exploited as a potential therapeutic strategy to treat this disease. The structure of HSF1 is not readily amenable to inhibition by small molecules; however, HSF1 expression and activity might be impaired through inhibition of upstream HSF1 regulators. As our previous work (27) showed that loss of ABL2 in lung cancer cells impaired brain metastasis outgrowth, similar to the phenotype induced by loss of HSF1 in vivo, we evaluated whether ABL2 might also regulate HSF1 expression required for survival and colonization of brain-metastatic cells. Notably, knockdown of ABL2 in PC9-BrM3 cells in vitro resulted in a near complete loss of measurable HSF1, E2F1, and E2F8 proteins (Fig. 4A). Consistent with a heat shock-independent role for HSF1 in brain metastasis, protein expression of HSP90 was not altered by the loss of HSF1 induced by ABL2 knockdown (Fig. 4A). Interestingly, reciprocal coimmunoprecipitation of endogenous HSF1 and ABL2 in PC9-BrM3 cells showed a strong interaction between these two molecules (Fig. 4 B and C). In contrast to HSF1, which shuttles between the cytoplasm and the nucleus, ABL2 is a cytoplasmic protein and lacks a nuclear localization sequence. Coimmunoprecipitation and immunoblot analysis between nuclear and cytoplasmic fractions in cells cotransfected with ABL2-GFP and Flag-tagged HSF1 (Flag-HSF1) revealed that the interaction between these two molecules is restricted to the cytoplasm (SI Appendix, Fig. S5A).
Fig. 4.
ABL2 kinase regulates HSF1 and E2F protein expression. (A) Immunoblots of the indicated proteins in PC9-BrM3 cells transduced with lentiviral shRNAs against nontarget control or ABL2. Quantification was normalized to actin loading control and shNTC, with the exception of phospho-CRKL, which was normalized to total CRKL. (B) Coimmunoprecipitation pulldown assay for endogenous HSF1 protein and immunoblots of indicated proteins in PC9-BrM3 cells. WCL, whole cell lysate. (C) Coimmunoprecipitation pulldown assay for endogenous ABL2 protein and immunoblots of indicated proteins in PC9-BrM3 cells. (D) Linear protein structures for ABL2 and HSF1. (E) Coimmunoprecipitation pulldown assay of HSF1 (Flag) in 293T cells transiently transfected with His-tagged HSF1 and GFP-tagged ABL2 WT, ABL2 R198K, ABL2 K317M, or empty vector control. (F) Coimmunoprecipitation pulldown assay of HSF1 (Flag) in 293T cells transiently transfected with Flag-tagged HSF1 WT or indicated deletion mutants with ABL2-eGFP. (G) GST in vitro pulldown assay of purified HSF1 (His) WT or deletion mutants incubated with purified ABL2 SH3 domain (GST).
To characterize the interaction between ABL2 and HSF1, we employed WT, kinase-inactive (K317M), SH2-inactive (R198K), or SH3-inactive (P158L) ABL2-GFP–tagged proteins (Fig. 4D). Cotransfection of GFP-tagged ABL2 WT or ABL2 mutant proteins with His-tagged HSF1 followed by coimmunoprecipitation assays revealed that expression of the ABL2 P158L mutant resulted in a complete loss of the interaction with HSF1 (Fig. 4E). Further, GST in vitro pulldown assays of purified His-HSF1 and GST-ABL2 SH2 or SH3-SH2 domains revealed a direct interaction between HSF1 and ABL2 that is dependent on the presence of the ABL2 SH3 domain (SI Appendix, Fig. S5B).
Reciprocally, to identify the region of HSF1 responsible for its interaction with ABL2, we generated HSF1 truncation mutants by site-directed mutagenesis (SI Appendix, Fig. S5C). Coimmunoprecipitation of the HSF1-truncated proteins in cells cotransfected with ABL2-GFP revealed a loss in the interaction between HSF1 and ABL2 upon removal of the LZ1-LZ3 domains (amino acids 130–206) in the HSF1 1–129 truncation mutant (SI Appendix, Fig. S5D). Curiously, this region of HSF1 lacks proline-rich sites that normally typify canonical SH3-binding proteins, suggesting that a noncanonical SH3-binding motif might mediate the interaction between HSF1 and ABL2. To test this possibility, we generated 10-amino acid truncation mutants of HSF1 from residues 166–206 to more precisely identify the residues responsible for binding the SH3 domain of ABL2. We found that the loss of residues spanning 187–195 of HSF1 completely ablated the interaction between these two molecules (SI Appendix, Fig. S5E). As recent work revealed that SH3 domains can bind to proline-independent hydrophobic motifs (31), we hypothesized that a hydrophobic six amino acid KLIQFL motif spanning residues 188–193 of HSF1 (Fig. 4D) might be responsible for the SH3-dependent interaction with ABL2. Deletion of this domain in HSF1 del188-193 and HSF1 del187-195 mutants ablated the interaction between HSF1 and ABL2 in cells (Fig. 4F). Furthermore, GST in vitro pulldown assays with purified His-HSF1 WT, His-HSF1 del187-195, or His-HSF1 del188-193 mutants in the presence of purified GST-ABL2 SH3 protein revealed a complete loss in the interaction between the ABL2 SH3 domain and the HSF1 deletion mutants (Fig. 4G). Together these findings uncover an intermolecular interaction between a noncanonical, proline-independent motif of HSF1 with the ABL2 SH3 domain.
Allosteric Inhibition of the ABL Kinases Impairs HSF1-E2F Expression.
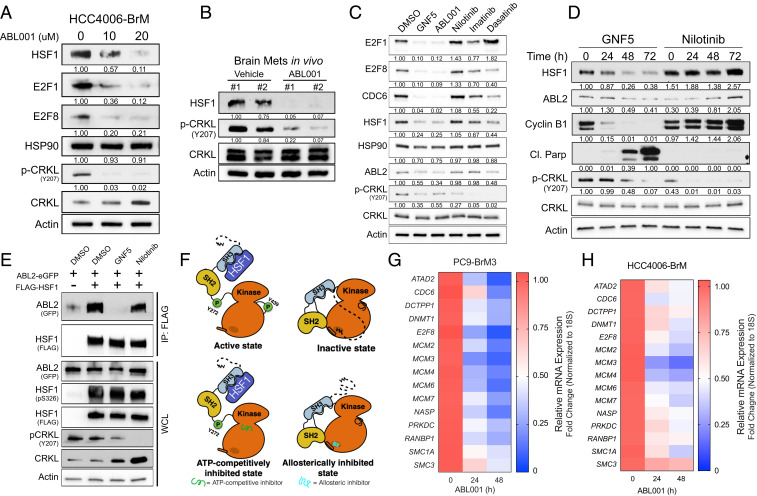
We previously showed that the BBB-penetrable allosteric inhibitor ABL001 (Asciminib) is effective in mouse models of lung cancer brain metastasis (27). Thus, we examined if the loss of HSF1 protein observed in ABL2 knockdown cells was also induced by pharmacologic inhibition of the ABL2 kinase with ABL001. Treatment of HCC4006-BrM cells with ABL001 resulted in a dose-dependent decrease in the expression of HSF1, E2F1, and E2F8 proteins, while expression of HSP90 remained unchanged (Fig. 5A). Notably, inhibition of the ABL kinases with ABL001 via oral gavage in mice harboring established lung cancer brain metastases resulted in near complete loss of HSF1 protein expression and inhibited phosphorylation of the ABL kinase substrate CRKL (Fig. 5B). ABL2-dependent regulation of HSF1 protein expression occurs posttranscriptionally, as RT-qPCR analysis of PC9-BrM3 cells treated with ABL001 showed no measurable change in HSF1 mRNA expression (SI Appendix, Fig. S6A) and it is independent of the proteasome, as proteasomal inhibition with MG132 in cells treated with an ABL allosteric inhibitor did not rescue HSF1 protein expression (SI Appendix, Fig. S6B).
Fig. 5.
ABL allosteric inhibitors impair expression of HSF1 and E2F proteins and downstream transcriptional targets. (A) Immunoblots of indicated proteins in HCC4006-BrM cells treated with indicated concentrations of ABL001 for 72 h. Quantification normalized to actin loading control and DMSO control (left lane). (B) Immunoblots of indicated proteins in established in vivo brain metastases from mice injected with H1975 lung cancer cells and treated with vehicle (n = 2) or 100 mg/kg ABL001 (n = 2) via oral gavage. Mice were treated at 3, 12, and 24 h prior to harvesting brain metastases. Protein quantification data normalized to actin loading control and the leftmost lane (Vehicle #1). (C) Immunoblots of PC9-BrM3 cells treated with DMSO or the ABL allosteric inhibitors GNF5 (10 μM) or ABL001 (10 μM), and ATP-competitive Nilotinib (1 μM), Imatinib (2 μM), or Dasatinib (150 nM) for 48 h. Indicated equipotent doses for each drug were selected after determining corresponding IC50 values. Phospho-CRKL Y207 blot used as surrogate marker for ABL kinase inhibition. (D) Immunoblots of PC9-BrM3 cells treated with 10 μM GNF5 or 1 μM Nilotinib for the indicated timepoints. Protein quantification data normalized to actin loading control and the leftmost lane (DMSO). For all immunoblots of phospho-CRKL, quantification data were normalized to total CRKL levels. (E) Coimmunoprecipitation pulldown assay for Flag-HSF1 in 293T cells cotransfected with ABL2-GFP and treated with the indicated inhibitors for 24 h. (F) Proposed diagram of the SH3-dependent interaction between ABL2 and HSF1 in the absence (Upper) and presence (Lower) of ABL allosteric versus ATP-competitive pharmacologic inhibitors. (G and H) RT-qPCR analysis of E2F target gene mRNA expression in PC9-BrM3 (G) or HCC4006-BrM (H) cells treated with 10 μM ABL001 for indicated timepoints.
ABL kinase inhibitors can be classified into two broad categories: 1) selective allosteric inhibitors that target the unique myristate-binding pocket located within the ABL kinase (SH1) domain and 2) ATP-competitive inhibitors that bind to the catalytic site within the kinase domain and target not only ABL but several other tyrosine kinases (32). Notably, comparison of ABL allosteric inhibitors (GNF5, ABL001) with ATP-competitive inhibitors (Nilotinib, Imatinib, and Dasatinib) revealed that cells treated with the ABL allosteric inhibitors exhibited a profound loss in HSF1, E2F1, and E2F8 proteins relative to ATP-competitive inhibitor treatment (Fig. 5C). Expression of CDC6, an HSF1-E2F target gene, was preferentially decreased by the ABL allosteric inhibitors, whereas expression of HSP90 was not affected by any of the inhibitors tested (Fig. 5C). Similar to HSF1 knockdown, treatment with the ABL allosteric inhibitor GNF5 resulted in loss of Cyclin B1 expression as well as a robust increase in cleaved PARP levels that were not observed by treatment with Nilotinib (Fig. 5D). These findings are consistent with published data showing that in contrast to ABL allosteric inhibitors, the ATP-competitive inhibitors elicit ERK activation in lung cancer cells and other tumors (33). Notably, coimmunoprecipitation and immunoblot analysis of Flag-HSF1 cotransfected with ABL2-GFP in cells treated with either an allosteric or ATP-competitive inhibitor revealed a complete loss in the interaction between HSF1 and ABL2 only under allosteric inhibition (Fig. 5E). Structural studies revealed striking differences in ABL kinase conformation and activity states under ATP-competitive–inhibited vs. allosteric-inhibited conditions (34). In line with these findings, our work suggests that, in contrast to ATP-competitive inhibition, allosteric inhibition of the ABL2 kinase results in an inactive state wherein the ABL2 SH3 domain is sterically inaccessible to binding with SH3-interacting proteins such as HSF1 (Fig. 5F). Collectively, our results show that in contrast to the ATP-competitive inhibitors, the ABL allosteric inhibitors phenocopy the effects of ABL2 knockdown and might be used to disrupt HSF1-E2F transcription in brain-metastatic lung cancer cells. In this regard, PC9-BrM3 and HCC4006-BrM cells treated with ABL001 exhibited profound decreases in mRNA expression of the HSF1-E2F target gene panel (Fig. 5 G and H). Together, these results demonstrate that HSF1-E2F transcription networks can be pharmacologically targeted with ABL kinase allosteric inhibitors.
An HSF1-E2F Transcriptional Signature is Up-Regulated in Lung Adenocarcinoma Patients and Is Predictive of Poor Survival.
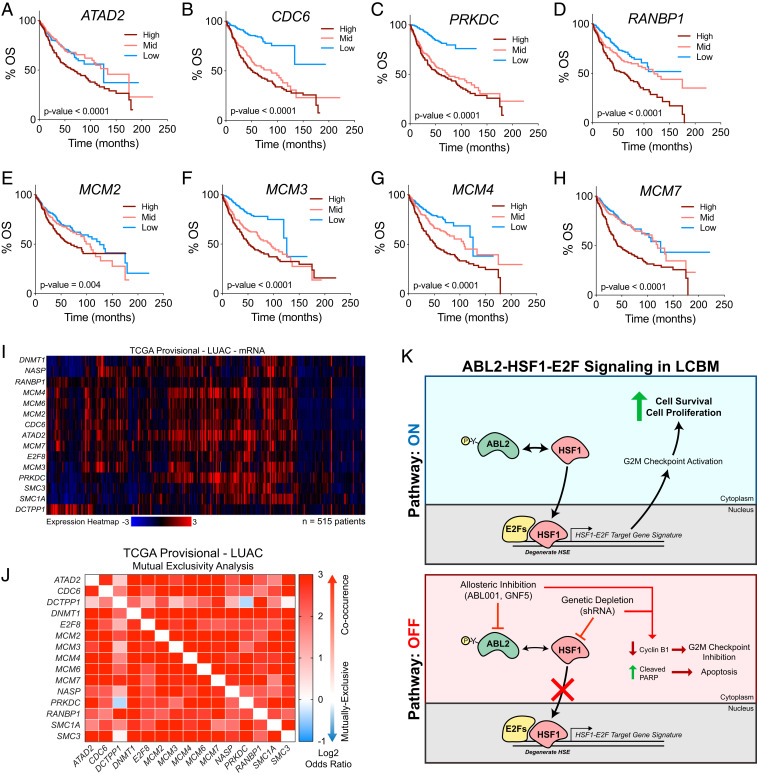
To evaluate whether the HSF1-E2F target gene signature is clinically relevant, we analyzed patient datasets to determine if HSF1-E2F target gene expression is predictive of patient survival outcomes. Analysis of microarray-based mRNA expression correlated to overall patient survival data revealed that high expression of the HSF1-E2F–coregulated target genes ATAD2, CDC6, PRKDC, RANBP1, MCM2, MCM3, MCM4, and MCM7 each individually were predictive of significantly poorer overall survival in lung adenocarcinoma patients (Fig. 6 A–H). In addition, analysis of TCGA datasets for lung adenocarcinoma showed that high expression of the HSF1-E2F target gene signature co-occurred in the same patients, and mutual exclusivity analysis showed high co-occurrence for almost all gene pairs tested from the HSF1-E2F gene panel (Fig. 6 I and J). Collectively, these data provide supporting evidence for an HSF1-E2F–coregulated transcriptional gene signature that may be clinically relevant as a prognostic marker for patient survival in lung adenocarcinoma (Fig. 6K).
Fig. 6.
Expression of HSF1-E2F target genes is predictive of poor survival outcomes and co-occurs in lung adenocarcinoma patients. (A–H) Overall survival (OS) curves derived from human lung adenocarcinoma patient microarray datasets with high versus low expression of HSF1-E2F target genes ATAD2 (A), CDC6 (B), PRKDC (C), RANBP1 (D) MCM2, (E), MCM3 (F), MCM4 (G), and MCM7 (H). Curves were generated using KMPlot analysis tool and patient groups were stratified by tertile (n = 720 total patients). For all survival curves, statistical analysis performed by log-rank Mantel–Cox test. (I) Clustered heatmap depicting mRNA expression for HSF1-E2F target gene panel in human lung adenocarcinoma provisional patient dataset available through TCGA. Data were analyzed using the cBioPortal online analysis software. (J) Mutual exclusivity analysis of HSF1-E2F target gene expression in the same TCGA lung adenocarcinoma dataset to demonstrate high co-occurrence of target gene expression. Heat map scaling based on log2 odds ratio as calculated using cBioPortal analysis tool. (K) Model diagram illustrating ABL2-HSF1-E2F signaling in lung adenocarcinoma brain metastasis.
Discussion
While HSF1 has been shown to support initiation and progression of distinct types of primary tumors, little is known regarding the role of HSF1 during metastasis (18, 20, 22). We now show that brain-colonizing lung cancer cells up-regulate expression of HSF1 and that loss of HSF1 in these cells results in dramatic decreases in cell survival in vitro and metastatic burden in vivo. Additionally, our data show that the transcriptional response driven by HSF1 is strikingly divergent from its canonical role as the master regulator of the heat shock response. Unbiased transcriptomic profiling comparing brain-metastatic and parental cell lines reveals significant enrichment of E2F transcription family gene signatures in brain-metastatic cells whose expression is dependent on HSF1. Previous work in C. elegans revealed an HSF1-E2F transcriptional program is required during development, whereby HSF1 binds DNA at promoter regions of E2F target genes in a heat shock-independent manner (26). Similarly, analysis of promoter regions for predicted HSF1-E2F–coregulated genes in human cells revealed the presence of “degenerate” HSEs downstream of E2F DNA-binding motifs which were confirmed empirically to be occupied by HSF1, thus supporting the hypothesis that HSF1 is a cofactor required for expression of these E2F gene targets in metastatic lung cancer cells. Importantly, analysis of human patient data revealed that high expression of an HSF1-E2F transcriptional signature co-occurs in patient populations with poor survival outcomes, suggesting that an HSF1-E2F target gene panel may hold prognostic value in patients with aggressive metastatic disease. Future studies comparing samples of paired patient primary tumors and corresponding brain metastases should be undertaken to evaluate whether HSF1 protein expression and an HSF1-E2F–coregulated transcription network may be relevant to progression in this disease setting. Furthermore, the importance of HSF1 activation and E2F-coregulated transcription in extracranial metastasis merits further investigation.
The ABL kinases, ABL1 and ABL2, function downstream of known HSF1 activators including EGFR, HER2, and RAS (20, 33, 35–38). Additionally, the ABL kinases regulate expression and activity of multiple factors that function as HSF1 transcriptional coregulators, including E2F1 (39–41). Recent work by our laboratory uncovered a critical role for the ABL2 kinase in the regulation of a TAZ-dependent transcriptional network required for the early colonization of lung cancer brain metastases (27). We now show that the SH3 domain of ABL2 forms a complex with a hydrophobic KLIQFL motif located within the LZ1-3 domains of HSF1, and knockdown of ABL2 results in impaired expression of HSF1, E2F1, and E2F8 proteins in brain-metastatic lung cancer cell lines. Notably, we found that pharmacologic inhibition of the ABL kinases using selective ABL allosteric inhibitors, but not ATP-competitive inhibitors, ablates the physical interaction between ABL2 and HSF1, results in markedly decreased expression of HSF1, E2F1, and E2F8 proteins in brain-metastatic lung cancer cells, and results in depletion of HSF1-E2F transcriptional targets. These findings highlight potential differences affecting intramolecular and intermolecular protein–protein interactions induced by allosteric versus ATP-competitive kinase inhibitors that might have important therapeutic implications. Importantly, the targetable nature of the ABL2-HSF1-E2F signaling network identifies ABL allosteric inhibitors as a potentially effective therapy for the treatment of metastatic lung cancers characterized by high expression of HSF1.
Materials and Methods
Additional experimental details and methods are provided in SI Appendix, including procedures for RNA-seq analysis, real-time PCR analysis, immunoblotting, immunofluorescence, and patient survival analysis. RNA sequencing raw data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession no. GSE149246.
Cell Lines and Cell Culture.
Human non-small cell lung cancer (NSCLC) cell lines HCC4006 and H1975 were purchased from American Type Culture Collection (ATCC). PC9 parental, H2030 parental, and H2030-BrM3 cells were a gift from Joan Massagué (Memorial Sloan Kettering Cancer Center, New York, NY). PC9-BrM3 and HCC4006-BrM cell lines were derived in the A.M.P. laboratory by serial intracardiac injection as described in the following section. Parental and derivative cell line pairs were subjected to short tandem repeat profiling through the Duke University DNA Analysis Facility Human cell line authentication service to confirm their authenticity. NSCLC lines were maintained in RPMI 1640 (Life Technologies) supplemented with 10% tetracycline-screened fetal bovine serum (FBS, HyClone), 10 mM Hepes, 1 mM sodium pyruvate, and 0.2% glucose. H293T cells used for transfection and virus production were purchased from ATCC and were maintained in DMEM (Life Technologies) with 10% FBS (Corning). All cultures were maintained at 37 °C in humidified air containing 5% CO2. In studies employing the use of dox-inducible constructs, the dose of dox (Millipore Sigma) was determined empirically.
For experiments assessing effects of pharmacologic inhibitors in vitro (GNF5, ABL001, Nilotinib, Dasatinib, Imatinib), drugs were dissolved in DMSO with the final concentration of DMSO in culture media not exceeding 0.1% vol/vol. The proteasomal inhibitor MG132 (Sigma) was dissolved in DMSO for use in cell-based in vitro assays. Inhibitors were synthesized by the Duke University Small Molecule Synthesis Facility and were validated by liquid chromatography–mass spectrometry and 1H-NMR techniques in addition to cell-based assays confirming effects on both cell viability (Cell-Titer Glo) and kinase target inhibition (Western blot).
Intracardiac Injection.
All animal studies were performed in accordance with protocols approved by the Duke University Division of Laboratory Animal Resources Institutional Animal Care and Use Committee. PC9-BrM3 cells transduced with a stable pFU-luciferase-Tomato (pFuLT) lentiviral vector in addition to shRNA constructs as described in figure legends were injected intracardially, and mice were subsequently monitored in vivo using an IVIS XR bioluminescent imager to confirm both proper anatomical injection as well as to monitor for metastatic disease progression. Eight- to 12-wk-old age-matched female athymic nu/nu mice were used for all studies (Jackson Laboratory). Mice were anesthetized with 5% isoflurane prior to injections. For all studies, 4 × 105 lung cancer cells suspended in 100 μL of PBS were injected into the left cardiac ventricle with a 30-gauge needle and animals were monitored until full recovery from anesthesia.
ABL001 (Asciminib) was used to pharmacologically inhibit the ABL kinases in tumor-bearing mice in vivo and was prepared as a suspension in sterile 0.5% methylcellulose/0.5% Tween-80 as described previously (42). To evaluate the pharmacologic effects of ABL001 on ABL kinase activity and downstream signaling in brain metastases in vivo, mice were injected with H1975 pFulT lung cancer cells and metastatic burden was allowed to progress over the span of 1 mo. Mice with established brain metastases were administered either vehicle or 100 mg/kg ABL001 via oral gavage at 3, 12, and 24 h prior to euthanasia and tissue harvest. Immediately following euthanasia, mouse brains were freshly dissected and tissue sections with visible brain metastases were excised and lysed in RIPA buffer containing protease-phosphatase inhibitor mixture (Cell Signaling). Harvested samples were digested for 15 min on a rotator at 4 °C followed by centrifugation for 15 min at 4 °C to clear the lysate of myelin and cell debris. Protein lysates were temporarily stored at −20 °C prior to immunoblotting.
Derivation of Brain-Metastatic Lung Cancer Cell Lines.
Derivation of brain-metastatic cell lines was performed as described previously (5). Eight- to 12-week-old athymic nude mice were injected intracardially with either PC9, HCC4006, or H1975 parental cell lines (4 × 105 cells per injection) and were monitored for metastatic progression using an IVIS XR bioluminescent imager. Approximately 30 d postintracardiac injection, subsets of mice presenting with advanced brain metastases were euthanized and whole brains were collected, minced, and digested in RPMI culture medium supplemented with 0.125% collagenase III and 0.1% hyaluronidase. Cells suspended in digestion media were then placed on a rotator for 4–5 h at room temperature. Following incubation, cells were centrifuged and resuspended in 0.25% trypsin for 10 min at 37 °C, followed by resuspension in culture media containing 1× Anti-Anti (Thermo Fisher). Cell lines were expanded to near-confluence in culture prior to sorting for Tomato-positive cells for future experimental use.
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism 8 software. Mouse numbers per group were determined through statistical power calculations (α = 0.05) where 10 mice per group allows for 90% power to detect intergroup differences of 50% and assuming intragroup variability of 25%. For Kaplan–Meier survival analysis, P values were calculated using log-rank (Mantel–Cox) testing. Statistical comparisons of two groups were conducted using Student’s t tests (unpaired, two-tailed). For comparisons involving more than two groups, data were evaluated by ANOVA followed by post hoc testing as described in figure legends. Post hoc testing was performed only when statistical significance was achieved from the ANOVA. For comparisons between groups of unequal size, the mean value was used to allow for statistical analysis by ANOVA. For all tests, we considered a P value less than 0.05 as statistically significant. Data shown represent averages ± SEM unless otherwise indicated in figure legends.
Supplementary Material
Acknowledgments
We thank Dr. Joan Massagué for providing the PC9 parental, H2030 parental, and H2030-BrM3 cell lines, as well as Dr. Anthony Koleske (Yale University, New Haven, CT) for providing DNA plasmids. We thank the Duke University genome sequencing facility for providing assistance with RNA-seq data acquisition, the Duke Flow Cytometry Shared Resource for assistance with cell sorting, and the Duke Light Microscopy Core Facility. We acknowledge Courtney McKernan and Ashley Colemon for technical assistance. This work was supported by NIH National Cancer Institute Grants R01CA195549 (to A.M.P.), F31CA22496001 (to J.P.H.), F99CA245732-01 (to J.P.H.), F31CA243293-01A1 (to B.M.), and 5T32GM007105-44 (to J.P.H. and B.M.), the Lung Cancer Research Foundation Free to Breathe Metastasis Research Grant (to A.M.P.), the Emerson Collective, and Duke SPORE in Brain Cancer Grant P50CA190991.
Footnotes
Competing interest statement: A.M.P. is a consultant and advisory board member for The Pew Charitable Trusts. A.M.P. holds patent no. U.S. 9,931,342 B2, related to this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2007991117/-/DCSupplemental.
Data Availability.
Requests for resources and reagents found in this study should be directed to and will be fulfilled by the corresponding author, A.M.P. (ann.pendergast@duke.edu). All unique and stable reagents generated in this study are available upon completion of a Materials Transfer Agreement. RNA-seq data have been deposited in GEO (accession no. GSE149246).
References
- 1.Schuette W., Treatment of brain metastases from lung cancer: Chemotherapy. Lung Cancer 45, S253–S257 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Stelzer K. J., Epidemiology and prognosis of brain metastases. Surg. Neurol. Int. 4, S192–S202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox B. D., Cheung V. J., Patel A. J., Suki D., Rao G., Epidemiology of metastatic brain tumors. Neurosurg. Clin. N. Am. 22, 1–6 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Zeng Q., et al. , Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 573, 526–531 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valiente M., et al. , Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q., et al. , Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Er E. E., et al. , Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nat. Cell Biol. 20, 966–978 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shih D. J. H., et al. , Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat. Genet. 52, 371–377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fecci P. E., et al. , The evolving modern management of brain metastasis. Clin. Cancer Res. 25, 6570–6580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anckar J., Sistonen L., Regulation of HSF1 function in the heat stress response: Implications in aging and disease. Annu. Rev. Biochem. 80, 1089–1115 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Pastor R., Burchfiel E. T., Thiele D. J., Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neef D. W., et al. , A direct regulatory interaction between chaperonin TRiC and stress-responsive transcription factor HSF1. Cell Rep. 9, 955–966 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi Y., Mosser D. D., Morimoto R. I., Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 12, 654–666 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou J., Guo Y., Guettouche T., Smith D. F., Voellmy R., Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94, 471–480 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Akerfelt M., Morimoto R. I., Sistonen L., Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neudegger T., Verghese J., Hayer-Hartl M., Hartl F. U., Bracher A., Structure of human heat-shock transcription factor 1 in complex with DNA. Nat. Struct. Mol. Biol. 23, 140–146 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Mercier P. A., Winegarden N. A., Westwood J. T., Human heat shock factor 1 is predominantly a nuclear protein before and after heat stress. J. Cell Sci. 112, 2765–2774 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Mendillo M. L., et al. , HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kourtis N., et al. , Oncogenic hijacking of the stress response machinery in T cell acute lymphoblastic leukemia. Nat. Med. 24, 1157–1166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai C., Whitesell L., Rogers A. B., Lindquist S., Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130, 1005–1018 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kourtis N., et al. , FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat. Cell Biol. 17, 322–332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai C., et al. , Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J. Clin. Invest. 122, 3742–3754 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xi C., Hu Y., Buckhaults P., Moskophidis D., Mivechi N. F., Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J. Biol. Chem. 287, 35646–35657 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin X., Moskophidis D., Mivechi N. F., Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab. 14, 91–103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su K. H., Dai S., Tang Z., Xu M., Dai C., Heat shock factor 1 is a direct antagonist of AMP-activated protein kinase. Mol. Cell 76, 546–561.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Chauve L., Phelps G., Brielmann R. M., Morimoto R. I., E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 30, 2062–2075 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoj J. P., Mayro B., Pendergast A. M., A TAZ-AXL-ABL2 feed-forward signaling Axis promotes lung adenocarcinoma brain metastasis. Cell Rep. 29, 3421–3434.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kienast Y., et al. , Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 16, 116–122 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgunova E., et al. , Structural insights into the DNA-binding specificity of E2F family transcription factors. Nat. Commun. 6, 10050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gkourtsa A., van den Burg J., Avula T., Hochstenbach F., Distel B., Binding of a proline-independent hydrophobic motif by the Candida albicans Rvs167-3 SH3 domain. Microbiol. Res. 190, 27–36 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Greuber E. K., Smith-Pearson P., Wang J., Pendergast A. M., Role of ABL family kinases in cancer: From leukaemia to solid tumours. Nat. Rev. Cancer 13, 559–571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu J. J., et al. , Inactivation of ABL kinases suppresses non-small cell lung cancer metastasis. JCI Insight 1, e89647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skora L., Mestan J., Fabbro D., Jahnke W., Grzesiek S., NMR reveals the allosteric opening and closing of Abelson tyrosine kinase by ATP-site and myristoyl pocket inhibitors. Proc. Natl. Acad. Sci. U.S.A. 110, E4437–E4445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srinivasan D., Plattner R., Activation of Abl tyrosine kinases promotes invasion of aggressive breast cancer cells. Cancer Res. 66, 5648–5655 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Mader C. C., et al. , An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 71, 1730–1741 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li D., Yallowitz A., Ozog L., Marchenko N., A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis. 5, e1194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng L., Gabai V. L., Sherman M. Y., Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene 29, 5204–5213 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birchenall-Roberts M. C., et al. , The p120-v-Abl protein interacts with E2F-1 and regulates E2F-1 transcriptional activity. J. Biol. Chem. 272, 8905–8911 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Wong K. K., et al. , v-Abl activates c-myc transcription through the E2F site. Mol. Cell. Biol. 15, 6535–6544 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parada Y., et al. , BCR-ABL and interleukin 3 promote haematopoietic cell proliferation and survival through modulation of cyclin D2 and p27Kip1 expression. J. Biol. Chem. 276, 23572–23580 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Wylie A. A., et al. , The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature 543, 733–737 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for resources and reagents found in this study should be directed to and will be fulfilled by the corresponding author, A.M.P. (ann.pendergast@duke.edu). All unique and stable reagents generated in this study are available upon completion of a Materials Transfer Agreement. RNA-seq data have been deposited in GEO (accession no. GSE149246).