Significance
Molecular reporters that “remember” the activity history of a cell and give a corresponding signal that can be read out by imaging, sequencing, or other modalities are valuable tools for the study of neural networks. Here, we develop a reporter that drives stable expression of any transgene (such as a fluorescent protein) in response to cellular calcium activity during a user-defined time window. As calcium is a universal proxy for neural activity, our reporter provides a stable “snapshot” of neural activity which can be used for subsequent imaging, analysis, or manipulation of behavior-associated neural networks. A core feature of our reporter is an engineered calcium-activated protease, which cuts a protein sequence to reveal a transcription factor when cellular calcium levels rise.
Keywords: protein engineering, proteases, neuroscience
Abstract
Molecular integrators, in contrast to real-time indicators, convert transient cellular events into stable signals that can be exploited for imaging, selection, molecular characterization, or cellular manipulation. Many integrators, however, are designed as complex multicomponent circuits that have limited robustness, especially at high, low, or nonstoichiometric protein expression levels. Here, we report a simplified design of the calcium and light dual integrator FLARE. Single-chain FLARE (scFLARE) is a single polypeptide chain that incorporates a transcription factor, a LOV domain–caged protease cleavage site, and a calcium-activated TEV protease that we designed through structure-guided mutagenesis and screening. We show that scFLARE has greater dynamic range and robustness than first-generation FLARE and can be used in culture as well as in vivo to record patterns of neuronal activation with 10-min temporal resolution.
A new class of transcriptional integrators with greater specificity and higher dynamic range than previous-generation integrators, such as yeast two hybrid (1), has proven useful in a diverse range of applications, from protein–protein interaction detection (2, 3) and GPCR agonist screens (4) to transsynaptic tracing (5), neurotransmitter detection, and neural circuit mapping (6, 7). These new integrators share a common mechanism, in which the biochemical event of interest (Ca2+ rise, GPCR activation, etc.) triggers intermolecular recruitment or reconstitution of a protease, which in turn releases a membrane-anchored transcription factor (TF) for translocation to the nucleus (Fig. 1A). In some cases, the proteolysis is also gated by light, via fusion of the photosensory LOV domain to the protease recognition sequence. By requiring the coincident arrival of light and the biochemical event for TF activation, these tools provide lower background and can be temporally locked to a user-selected stimulus or experimental time window.
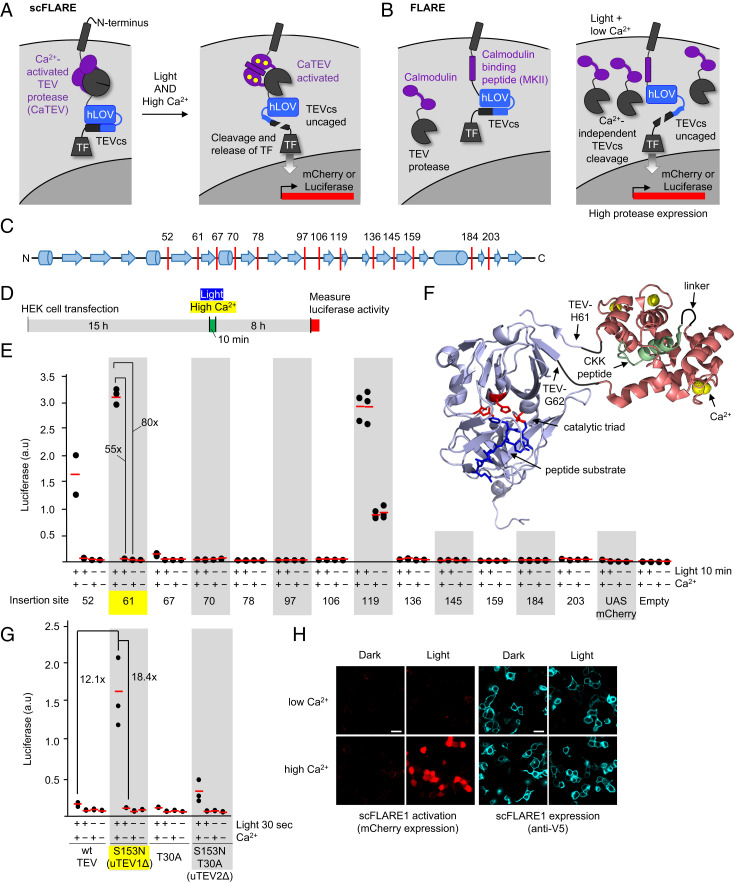
Fig. 1.
Engineering of CaTEV and incorporation into scFLARE. (A) Schematic of scFLARE. From N to C terminus, scFLARE consists of a transmembrane helix, a CaTEV, an LOV domain, a TEV cleavable site (TEVcs), and a TF. High Ca2+ activates the protease, while light alters the conformation of LOV to expose the TEVcs for cleavage. Both light and calcium must arrive coincidentally to enable proteolytic release of the TF, which translocates to the nucleus to drive expression of the reporter gene of choice. (B) The previous two-component FLARE (6, 16) gives background at high protease expression levels. When the FLARE protease copy number is high, TEVcs cleavage can occur under the light + low-Ca2+ condition because of proximity-independent recognition of TEVcs by TEV protease. (C) The domain structure of TEV protease (from Protein Data Bank (PDB) ID: 1LVM) showing loop regions we targeted for insertion (red lines) of Ca2+-sensing modules. (D) The protocol for screening CaTEV variants in the context of scFLARE in HEK 293T cells. Ca2+ was elevated by the addition of 6 mM CaCl2 in the presence of 2 μM ionomycin. Blue light (467 nm) was delivered at 60 mW/cm2 and 33% duty cycle (2 s of light every 6 s). (E) The screening results from D. Calmodulin-CKK was inserted after the indicated residue in TEV, within the context of scFLARE as shown in A. This experiment was performed once with three technical replicates per condition (red lines, mean). (F) Model of the best CaTEV design based on crystal structures of wild-type TEV protease (PDB ID: 1LVM) and calmodulin:CKK complex (PDB ID: 1CKK). The CaM-CKK fusion is inserted between His-61 and Gly-62 of TEV protease. (G) The incorporation of ultraTEV mutations (16) into scFLARE. The same assay as in E, but the light + Ca2+ stimulation time was only 30 s. This experiment was performed once with three technical replicates per condition. (H) Confocal imaging of scFLARE activity in HEK293T cells. scFLARE1 (insertion site 61) was expressed with UAS:mCherry reporter and stimulated for 30 s with blue light and CaCl2 + ionomycin. Cells were fixed, permeabilized, and stained with anti-V5 antibody 8 h later to detect scFLARE expression (scale bars, 20 μm.) This experiment was repeated two times.
In working with these transcriptional integrators, however, we and others have noticed that because of their complex, multicomponent design, their robustness is limited. In particular, performance is highly expression-level dependent because of the intermolecular nature of TF turn on. At high expression levels that exceed the Michaelis constant (KM) of TEV protease–TEVcs (TEV cleavage site) interaction (∼60 μM (8)) or the dissociation constant (Kd) of split TEV reconstitution, for instance, proteolytic release of the TF may occur even if the biochemical event of interest is absent. At low expression levels, the TEVcs may not be significantly cleaved even if the stimulus is present. Only within a narrow band of expression levels do these tools perform as designed.
Here, we undertook a redesign of one of these tools: FLARE, for Fast Light- and Activity-Regulated Expression, which is a calcium- and light-activated TF. We sought to simplify the tool and make its performance much less expression-level dependent. We report single-chain FLARE (scFLARE), in which elements for calcium sensing, light regulation, proteolysis, and transcription are combined in a single polypeptide chain. The core feature is a calcium-activated TEV protease (Ca-TEV) that we engineered via structure-guided screening. We characterize scFLARE and test its performance in living mammalian cells, neuronal culture, and in vivo. The improved design should add to the arsenal of tools available for transcriptional readout of important cellular events.
Results
Engineering a Single-Chain Calcium Integrator.
To develop scFLARE, we needed a mechanism for calcium-triggered proteolysis that does not depend on intermolecular recognition. In FLARE (6), high cytosolic calcium levels drive intermolecular complexation between calmodulin (CaM) and MK2 (a CaM-binding peptide with higher affinity toward CaM in its calcium-bound state), which, in turn, brings low-affinity TEV protease proximal to its cleavage sequence (TEVcs) (Fig. 1B). In Cal-Light (7), high-cytosolic calcium drives the intermolecular reconstitution of split TEV protease.
In nature, calpains are proteases naturally activated by calcium ion. However, they have broad sequence specificity and cleave a wide variety of endogenous proteins, and they are therefore not suitable for use in a tool. The TEV protease used in FLARE and Cal-Light is attractive for its extremely high sequence specificity and orthogonality to mammalian systems. While TEV has previously been split (9), it has not to our knowledge been engineered for analyte or light sensitivity. If we could design a Ca2+-regulated TEV, it could simplify the design of FLARE, overcoming FLARE’s dependence on intermolecular interactions and enabling more specific activation.
Our approach to designing Ca-activated TEV (CaTEV) was similar to that used to design the Ca2+-activated fluorescent proteins (GCaMPs) that have been revolutionary for cell biology and neuroscience (10). We examined the crystal structure of TEV for exposed loops into which we could insert the calcium-sensitive module consisting of CaM split by an effector peptide [NCaM-CKK peptide-CCaM (11)]; this same module when inserted into fluorescent proteins gives them calcium-dependent fluorescence (12). We searched for an insertion site that would produce a minimally active protease in the low-calcium state but restore the active site and give high proteolytic activity in the high-calcium state. We screened 13 different insertion sites in TEV (Fig. 1C), and to read out proteolytic activity, we installed the CaTEV variants into scFLARE itself.
scFLARE, like the original FLARE, features a TF that is tethered to the plasma membrane via a protease-cleavable linker (TEVcs). To give light gating, an optimized hybrid LOV domain [hLOV (2)] was installed N-terminal to TEVcs. The candidate CaTEVs were installed N-terminal to the LOV domain so that intramolecular calcium-dependent cleavage of TEVcs could occur, producing the release of the TF. The opposite configuration, with CaTEV C-terminal to TEVcs, would produce a bulkier cleavage product (CaTEV-TF) that might have impaired translocation to the nucleus.
scFLAREs containing 1 of the 13 candidate CaTEVs were cotransfected with the reporter gene UAS:luciferase into HEK 293T cells. After 10 min of high Ca2+ and light stimulation, we waited 8 h for transcription and translation of the reporter and then quantified luciferase activity using a plate reader (Fig. 1 D and E). Three CaTEV variants, with insertions at amino acids 52, 61, and 119, exhibited significant activity (Fig. 1F). However, CaTEV with insertion at residue 119 was comparably active under both high- and low-Ca2+ conditions. Interestingly, position 119 is also the cut site for split TEV protease (9). CaTEVs with insertions at residues 52 and 61 were highly Ca2+ dependent, but the ±Ca2+ signal ratio was better for the latter (55-fold) than for the former (24-fold). When we repeated the screen by imaging, using UAS:mCherry for readout, we obtained the same results, with insertion site 61 giving the most Ca2+-responsive protease (SI Appendix, Fig. S1).
We also tested alternative CaM-binding peptides (CaMbps) within the insert module in an attempt to further optimize CaTEV (SI Appendix, Fig. S2). In addition to the original “CKK” CaMbp derived from Ca2+/CaM-dependent protein kinase kinase, we screened seven additional variants (13) of the M13 CaMbp (14) derived from skeletal muscle myosin light-chain kinase. In the context of scFLARE, CKK gave the best calcium- and light-dependent turn on.
With the insertion site and CaMbp optimized, we wondered about the absolute activity of CaTEV in the high-Ca2+ state. In general, insertion of domains within internal sites of enzymes will compromise their activity. This is a particular concern for TEV protease, whose natural turnover rate is relatively low [0.19 s−1 (8)] compared to other proteases [for example, trypsin with a kcat of 75 s−1 (15)]. Yet, TEV protease is appealing for its exquisite sequence specificity. We recently used directed evolution to improve the turnover of TEV protease while preserving its high sequence specificity (16). We tested two of the active-site mutations enriched in those selections—S153N and T30A—in the context of CaTEV to see if we could boost its activity in the high-Ca2+ state. Fig. 1G shows that the S153N single mutation does indeed increase luciferase turn on in the live-cell scFLARE assay by 12-fold (when stimulated for just 30 s rather than 10 min) while retaining low activity in the low-Ca2+ state (18-fold difference in luciferase intensity between high- and low-Ca2+ states). The same construct—“scFLARE1” containing CaTEV (insertion site 61 + S153N mutation)—gave similar results when we read out transcriptional activation by microscopy (Fig. 1H).
Characterization of scFLARE1 in HEK Cells.
To characterize scFLARE1 in HEK 239T cells, we first examined its performance across a range of expression levels, using fluorescence-activated cell sorting (FACS) as the readout. HEK cells expressing scFLARE1-p2A-GFP and UAS:mCherry were stimulated with light and Ca2+/ionomycin for 5 min before FACS analysis 8 h later. Fig. 2A shows mCherry turn on across a range of scFLARE1 expression levels (GFP intensities), while mCherry is greatly reduced in the low-calcium negative control. For comparison, we analyzed cells expressing the most recent two-component FLARE variant (FLARE2) (16) in the same manner. To enable a fair comparison, we also installed TEV-accelerating mutations (S153N in the left column; S153N + T30A in the middle column) and the improved hLOV domain into FLARE2. The same FACS analysis showed that these FLARE2 variants gave higher background than scFLARE1 (Fig. 2A, bottom row), especially at high expression levels, and slightly lower signal than scFLARE1 in the high-calcium condition. Quantitation of cell numbers in the red-boxed regions of the FACS plots showed a ±Ca2+ signal ratio of 25.5 for scFLARE compared to just 3.7 for the best FLARE2 variant. In a separate imaging experiment, we showed that scFLARE1 also outperforms first-generation FLARE1 in a side-by-side comparison (SI Appendix, Fig. S3).
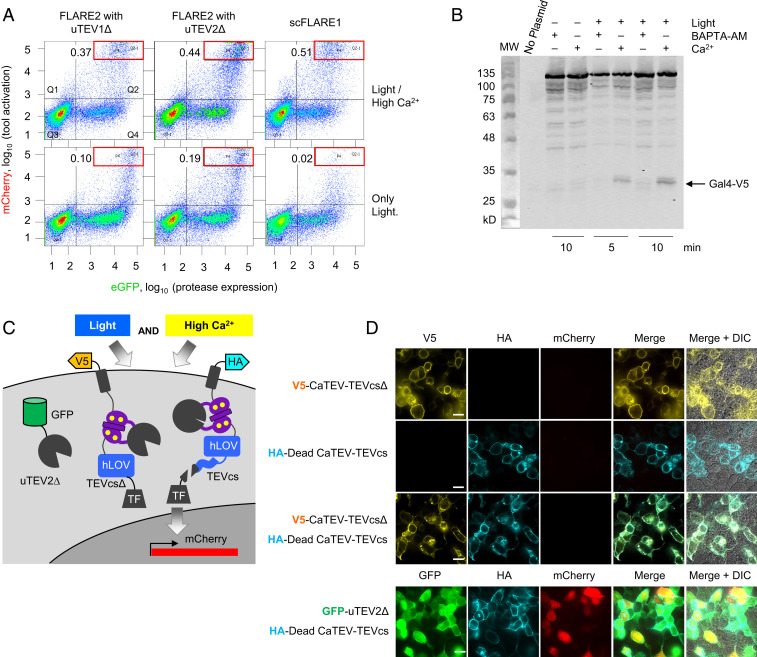
Fig. 2.
Characterization of scFLARE1 in HEK 293T cells. (A) scFLARE1 has lower background than two-component FLARE2 (16). HEK 293T cells expressing the indicated constructs and UAS:mCherry were stimulated for 5 min and analyzed by FACS 8 h later. GFP is fused to TEV protease in FLARE2, and it is expressed via p2a in scFLARE1 (TM-CaTEV(uTEV1Δ)-hLOV-TEVcs-Gal4-p2A-GFP). Ratios in the top left of each FACS plot indicate the number of cells within the red box region divided by the number of cells in Q4. (B) Western blot analysis of scFLARE1 cleavage. HEK cells expressing scFLARE1 were prepared and stimulated (for 5 or 10 min) as in Fig. 1E. Immediately after stimulation, cells were lysed in the presence of 2 mM iodoacetamide (TEV protease inhibitor) and run on 9% SDS/PAGE. scFLARE1 is 145 kDa before cleavage and 32 kDa (V5-containing portion) + 113 kDa after cleavage. BAPTA-AM was added to control cells not receiving Ca2+ stimulation. (C) Constructs used to test scFLARE1 cleavage mechanism. The V5-tagged TF construct lacks a TEVcs. The HA-tagged TF has an inactive protease (C151A mutation (31); DeadCaTEV). (D) The indicated constructs were expressed in HEK 293T cells along with UAS:mCherry. Cells were stimulated for 5 min with light and Ca2+ and then fixed, stained, and imaged 8 h later. The bottom row coexpresses high-affinity TEV protease (GFP tagged) with the HA-tagged DeadCaTEV construct to show that the latter can still be cleaved to generate mCherry reporter signal. Scale bars, 10 μm.
To examine the mechanism, the residues responsible for calcium binding in the four EF hands of CaM within scFLARE1’s CaTEV domain were consecutively mutated. Alanine replacement in each of these Ca2+-binding motifs completely abolished scFLARE1 activation (SI Appendix, Fig. S4 A and B).
Next, we characterized the TF release from scFLARE1 by Western blotting. Fig. 2B shows anti-V5 blotting of whole cell lysate from HEK 293T cells expressing scFLARE1 and subjected to various conditions. Only upon exposure to blue light and Ca2+/ionomycin for 5 min do we observe the appearance of a new band at ∼32 kDa, corresponding to the proteolytically released Gal4 TF. BAPTA-AM addition to the culture media to chelate Ca2+ suppresses TF release. By performing a time course and quantifying the amount of TF released, we could estimate a ∼13-fold difference in cleavage rate for CaTEV under high- versus low-calcium conditions. (SI Appendix, Fig. S4 C and D).
We designed scFLARE1 to carry out intramolecular cleavage, but intermolecular cleavage between two neighboring molecules of scFLARE1 is also theoretically possible. To examine the mechanism of scFLARE1 cleavage, we prepared two variants, one with dead protease (C151A mutation in CaTEV) and one with no TEVcs (Fig. 2C). If intermolecular cleavage is the predominant mechanism, then these two mutants expressed together in the same cell should still be able to produce TF release. Fig. 2D shows HEK cells expressing these scFLARE1 variants along with the UAS-mCherry reporter gene 8 h after light and calcium stimulation. No mCherry expression is observed, consistent with the idea that scFLARE1 requires intramolecular rather than intermolecular proteolysis. A control experiment showed that the dead-protease construct is still capable of being cleaved when high-affinity full-length ultraTEV protease is coexpressed in the same cell (Fig. 2D).
Adapting scFLARE for Use in Neurons.
To adapt scFLARE for use in neurons, we made a number of modifications to the construct, summarized in Fig. 3A. First, we replaced the transmembrane domain (from CD4) with a neurexin-derived transmembrane helix that exhibits improved surface targeting in neurons (6). Second, we replaced the Gal4 TF with the more sensitive TF tTA-VP16, which can drive expression of TRE:mCherry in the nucleus. Third, since physiological Ca2+ rises in neurons are different from ionomycin-based delivery of Ca2+ into the HEK cytosol, we carried out an optimization of Ca2+ affinity and kinetics in scFLARE.
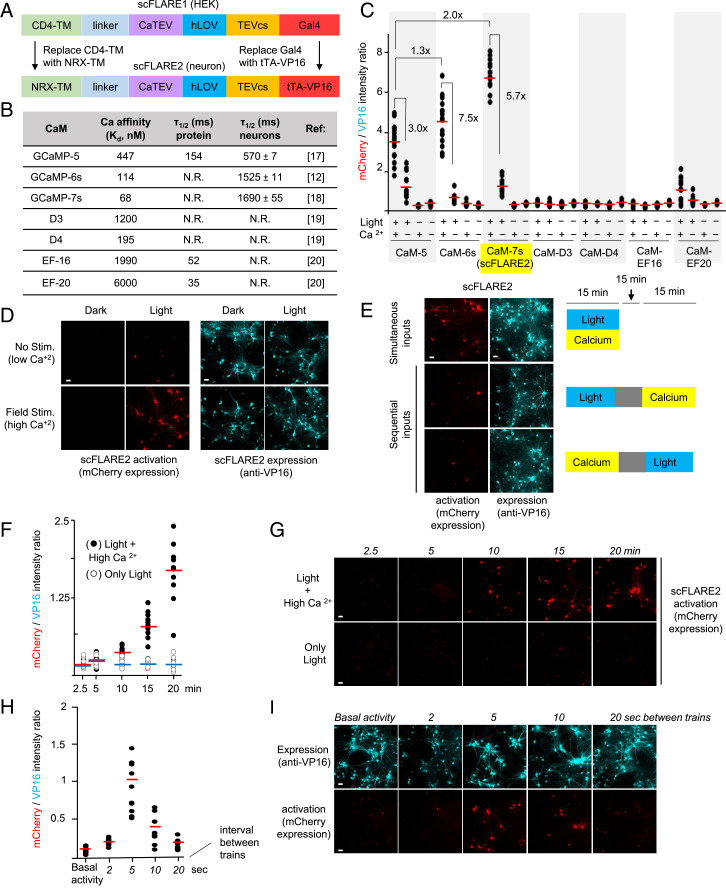
Fig. 3.
scFLARE2 characterization in neurons. (A) Summary of how scFLARE1 was converted to scFLARE2 for use in neurons. (B) Table of CaM variants tested in scFLARE. (C) Screening results for CaM variants in scFLARE. Rat cortical neuron cultures were transduced with scFLARE and TRE:mCherry AAV1/2s. At DIV18, neurons were stimulated with light (467 nm, 60 mW/cm2, 33% duty cycle (2 s every 6 s)) and high Ca2+ (5-s electrical trains consisting of 32 1-ms 50-mA pulses at 20 Hz) for 15 min. Neurons were fixed, immunostained with anti-VP16 antibody, and imaged by confocal microscopy 18 h later. Quantitation of mCherry intensity divided by scFLARE expression (VP16 intensity) was performed across 10 fields of view per condition (red bar, mean). This experiment was performed twice. (D) Confocal images of the best scFLARE variant (scFLARE2) from the experiment in C (scale bars, 10 μm). (E) scFLARE2 is specific for simultaneous (top row) rather than sequential (middle and bottom rows) light and calcium inputs. Ca2+ was elevated by electrical stimulation as in C. In the case of sequential inputs, a 15-min pause separated the two inputs. This experiment was repeated twice. SI Appendix, Fig. S6 shows the same experiment with only a 1-min pause between sequential inputs. (F) scFLARE2 time course. The same conditions as in C were used, but stimulation times varied from 2.5 min to 20 min. Red lines, mean (light + high Ca2+ condition). Blue lines, mean (only light condition). This experiment was performed twice. (G) Representative confocal images from the experiment in F. Additional fields of view are shown in SI Appendix, Fig. S7. (H) scFLARE2 activation is dependent on stimulation frequency. The same conditions as in C were used, but the frequency of electrical stimulation delivered varied from 2 to 20 s over a period of 15 min. Red lines, mean of 10 fields of view per condition. This experiment was performed twice. (I) Representative confocal images from the experiment in H. Additional fields of view are shown in SI Appendix, Fig. S8.
To do so, we drew from the series of real-time Ca2+ indicators (GECIs) that incorporate CaM-based Ca2+-sensing modules. We tested the Ca2+ modules from GCaMP5 (17), GCaMP6s (13), and GCaMP7s (18), in addition to those from the Chameleon-type GECIs D3 (19), D4 (19), EF-16 (20), and EF-20 (20). Fig. 3B summarizes their published affinities and kinetic constants, while Fig. 3C summarizes their performance in the context of scFLARE (SI Appendix, Fig. S5). These experiments were conducted in rat neuron culture, with 15 min of light and electrical stimulation. We found that the GCaMP5-, GCaMP6-, and GCaMP7-derived domains were the best in scFLARE, with GCaMP7s’ Ca2+-sensing module giving the highest signal and good ±Ca2+ signal ratio in neurons.
Fig. 3D shows confocal imaging with our neuron-optimized “scFLARE2” (containing NRX-TM-, tTA-VP16-, and GCaMP7-derived Ca2+-sensing module). Cultures were transduced using Adeno-associated virus (AAV) 1/2s carrying the 3.7-kb scFLARE2 construct and TRE:mCherry. Neurons were exposed to blue light and electrical field stimulation (5-s electrical trains consisting of 32 1-ms 50-mA pulses at 20 Hz) for 15 min and then analyzed by confocal microscopy 18 h later. Anti-VP16 immunostaining shows expression of scFLARE2. mCherry shows scFLARE2 turn on, which is highest in the +light +stimulation condition. Quantitation of >10 fields of view per condition shows a light/dark signal ratio of 27 and a ±Ca2+ signal ratio of 5.7 in neurons (Fig. 3C).
Further Characterization of scFLARE2 in Neurons.
We further characterized the specificity and dynamic range of scFLARE2 in cultured neurons. If scFLARE2 functions as a true “AND” gate for light and calcium, it should not be activated by sequential light and calcium inputs. Fig. 3E shows that this is indeed the case when light and calcium stimuli are staggered by 15 min. When the time window between inputs is reduced to just 1 min, still no scFLARE turn on is observed for the light followed by calcium condition (SI Appendix, Fig. S6), reflecting the fast rate of LOV domain reset. However, if calcium is elevated first, followed by a 1-min pause before blue light, then scFLARE2-driven mCherry expression is observed, likely because CaTEV has not returned to its inactive state. These observations suggest that the reversal time constant for CaTEV (to become inactive again after cytosolic calcium drops) is between 1 and 15 min.
In Fig. 3F, we tested the ability of scFLARE2 to give a transcriptional readout proportional to the extent of integrated calcium activity. Quantification of mCherry imaging data after field stimulation for 0 to 20 min shows that mCherry expression does indeed scale with scFLARE2 stimulation time (Fig. 3G and SI Appendix, Fig. S7). In Fig. 3H, we tested the ability of scFLARE2 to distinguish between different stimulation frequencies. We observed that mCherry expression increased as the interval between stimulation trains decreased from 20 to 5 s. When the interval between trains was further decreased to every 2 s, however, the resulting mCherry expression 18 h later decreased, likely due to neuron desensitization (Fig. 3I and SI Appendix, Fig. S8A). This was confirmed by real-time imaging of the calcium indicator GCaMP5 under matched stimulation conditions (SI Appendix, Fig. S8 B and C and Movies S1–S5). The delivery of electrical trains every 2 s for 15 min indeed desensitized the neurons, while lower-frequency stimulation produced Ca2+ spikes and scFLARE2-driven mCherry expression that was proportional to the Ca2+ firing history (Fig. 3H).
To explore the versatility of scFLARE2 for recording activity in response to different stimuli, we treated neuron cultures with various drugs, in the presence of blue light, for 5 min (SI Appendix, Fig. S9). AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) and (S)-3-5 DHPG (dihydroxyphenylglycine) are agonists of the ionotropic glutamate receptor and group I metabotropic glutamate receptor (mGluR), respectively. Gabazine (SR95531) activates neurons by potently suppressing the activity of inhibitory GABAA channels. Confocal imaging showed that all three drugs induced light-dependent expression of mCherry, especially Gabazine, which up-regulated mCherry expression by 10-fold compared to untreated neurons. By performing GCaMP imaging in parallel, we found that the scFLARE2-driven mCherry expression was correlated with the real-time calcium activity during the drug-treatment window, as expected for a faithful calcium integrator (Movies S6–S8).
Comparison of scFLARE to FLARE1 and FLARE2.
We performed a side-by-side comparison to previous-generation FLAREs to see if scFLARE2 offers a true improvement in performance (Fig. 4A). Rat cortical neuron cultures were transduced with FLARE1 (6), FLARE2 (16), or scFLARE2 AAVs along with the TRE:mCherry reporter gene (Fig. 4B). We observed that scFLARE2 is more sensitive than FLARE1 and able to produce mCherry expression after just 15 min of activation. For the comparison to FLARE2 (16), we examined the tools at both high expression (via transduction with purified and higher titer AAV1/2s) and disproportionate expression (1:0.25 ratio of TF:protease for FLARE2). Fig. 4 C and D and SI Appendix, Fig. S10 show that at high expression levels, FLARE2 gives Ca2+-independent leak, while scFLARE2 continues to show strong Ca2+-dependent turn on. When protease and TF levels are mismatched in FLARE2, barely any mCherry turn on is observed, while scFLARE2 still gives signal. These results suggest that scFLARE2 is a more robust tool than two-component FLARE2 (6), especially under conditions of variable expression.
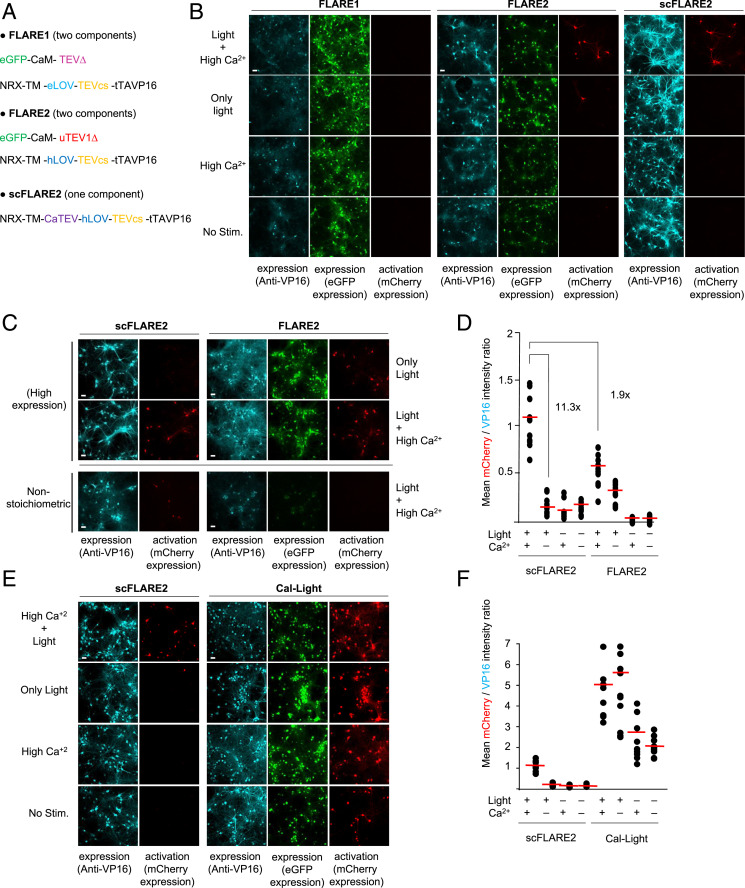
Fig. 4.
scFLARE2 comparison to FLARE1/2 and Cal-Light. (A) The constructs used to test FLARE1, FLARE2, and scFLARE2 in neurons. FLARE2 differs from FLARE1 in its protease and LOV domain (16). (B) A comparison of FLAREs in rat cortical neuron cultures. The neurons were transduced with AAVs encoding the indicated constructs and TRE:mCherry. At DIV18, neurons were stimulated with light (467 nm, 60 mW/cm2, 33% duty cycle [2 s every 6 s]) and electrical field stimulation to raise intracellular Ca2+ (5-s electrical trains consisting of 32 1-ms 50-mA pulses at 20 Hz) for 15 min. Neurons were fixed, immunostained with anti-VP16 antibody, and imaged by confocal microscopy 18 h later. (C) A more detailed comparison of scFLARE2 and FLARE2 at different expression regimes. For high expression, neurons were transduced with fourfold more virus than typically used. For nonstoichiometric expression, we used 0.25:1 ratio of protease:TF for FLARE2 rather than the typical 1:1 ratio. Otherwise conditions were the same as in B. (D) A quantification of the experiment in C (high expression condition). The mCherry/VP16 intensity ratio is plotted for 10 fields of view per condition. Red lines, mean. This experiment was performed twice. Additional fields of view are shown in SI Appendix, Fig. S10. (E) A comparison of scFLARE2 and Cal-Light (7). The rat cortical neuron cultures were transduced with AAV1/2s encoding scFLARE2 or Cal-Light constructs along with TRE:mCherry. The stimulation and imaging conditions were the same as in B. This experiment was performed twice. Additional fields of view are shown in SI Appendix, Fig. S11. (F) A quantification of the experiment in E. Scale bars, 10 μm.
We also compared scFLARE2 to the related calcium integrator Cal-Light (7). Fig. 4 E and F and SI Appendix, Fig. S11 show that Cal-Light gives high background leak in the absence of light or calcium stimulation, whereas scFLARE2 shows clean gating under identical conditions.
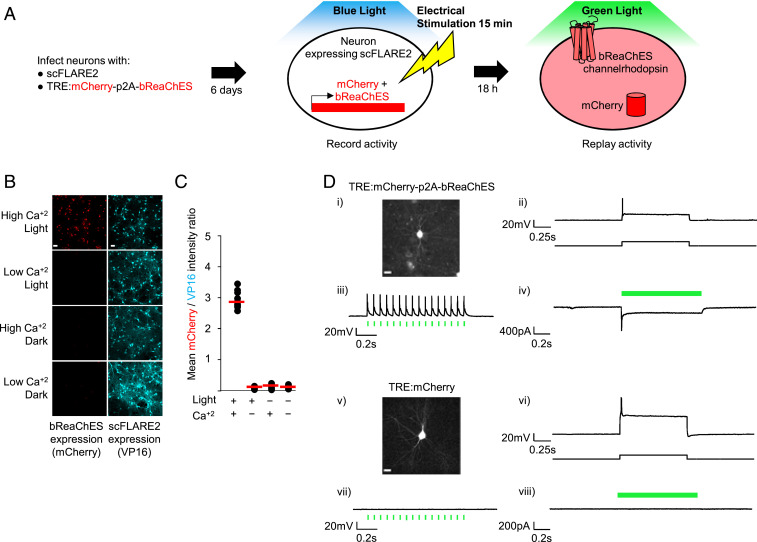
scFLARE2-Driven Channelrhodopsin Expression Enables Neuron Reactivation.
FLARE tools, in contrast to GECIs (10) or CaMPARI (21), allow subsequent manipulation of tagged neuronal subpopulations via activity-dependent expression of effectors such as channelrhodopsin (Fig. 5A). To test this capability with scFLARE2, we transduced cortical neuron cultures with AAV viruses encoding scFLARE2 and TRE:mCherry-p2A-bReaChES [bReaChES is a microbial channelrhodopsin that can be activated by 530 nm light (22)]. Under electrical stimulation coincident with blue light illumination, scFLARE2 drove monocistronic expression of mCherry and bReaChES (Fig. 5 B and C). The omission of light or electrical stimulation prevented expression of mCherry and bReaChES. We performed whole-cell patch clamping of mCherry-positive neurons (coexpressing bReaChES) 18 h after activation of scFLARE2 and were able to use 530 nm illumination to elicit action potentials and inward current (Fig. 5D). As a control, neurons expressing scFLARE2-driven mCherry alone (via TRE:mCherry instead of TRE:mCherry-p2A-bReaChES) did not show any response under 530 nm illumination. In addition to bReaChES, SI Appendix, Fig. S12 shows that scFLARE2 can also drive the expression of an inhibitory opsin, eNpHR3.0-TS. These results highlight the utility of scFLARE2 in driving the expression of effector molecules for subsequent manipulation of previously active neuronal ensembles.
Fig. 5.
scFLARE2 drives channelrhodopsin expression for neuron reactivation. (A) Schematic of reactivation of scFLARE-tagged neurons. bReaChES is a channelrhodopsin that can be activated by green 530-nm light. (B) Light- and activity-dependent expression of mCherry-p2A-bReaChES in scFLARE2-expressing rat cortical neuron cultures. The neurons were transduced with AAVs at DIV12, and stimulation (467 nm light at 60 mW/cm2, 33% duty cycle [2 s every 6 s] + 6-s electrical trains consisting of 32 1-ms 50-mA pulses at 20 Hz for a total of 15 min) was performed at DIV18. Cells were fixed and stained with anti-VP16 antibody 18 h after stimulation (scale bars, 10 μm). Additional fields of view are shown in SI Appendix, Fig. S12. (C) A quantification of the experiment in B, with 10 fields of view per condition. (D) Reactivation of scFLARE2-labeled neurons 18 h after light and electrical stimulation. (i-iv) The neurons expressing mCherry and bReaChES from the experiment in B (high Ca2+, light condition) were analyzed by whole-cell patch clamp electrophysiology. (v-viii) The control neurons expressing mCherry only (scFLARE2-driven expression from TRE promoter) were analyzed in the same manner. Firing could be elicited by a 350-pA depolarizing current injection in both ii and vi. However, optically induced action potentials in response to 15 ms green light (530-nm) stimulation at 16 Hz and an optically induced inward current from 1 s light stimulation were observed only in bReaChES-expressing cells iii and iv (n = 6/6) and not in control cells vii and viii (n = 3/3). Scale bars in i and v, 20 µm.
scFLARE2 for Circuit Labeling In Vivo.
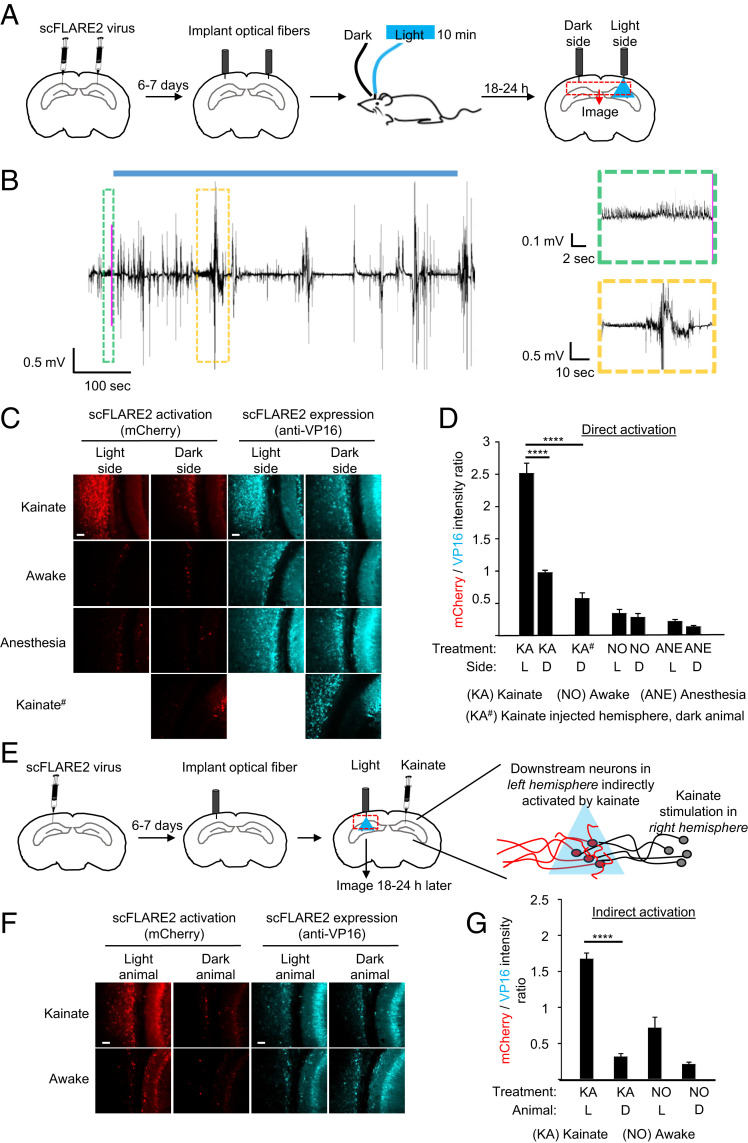
To test whether scFLARE2 could be used for stable tagging of transiently activated neuronal subpopulations in vivo, we utilized a closed-loop system to specifically label neurons active during epileptic seizures induced by intrahippocampal kainate injection, a widely used animal model of temporal lobe epilepsy (23). AAV viruses encoding scFLARE2 and the reporter gene TRE:mCherry were injected bilaterally into the hippocampus and cortex of adult mice (Fig. 6 A and B). Six days later, optical fibers were implanted above the injection sites, and blue light was delivered through the right optical fiber for 10 min while the mice were anesthetized, awake, or experiencing seizures 2 h after kainate injection. After 18 to 24 h to allow time for mCherry transcription and translation, the brains were sectioned and imaged by confocal microscopy (Fig. 6C and SI Appendix, Fig. S13). We observed the strongest mCherry expression (representing scFLARE2 activation) in the +light +kainate condition compared to either the contralateral no-light hemisphere or to kainate-injected animals that received no light at all (Fig. 6D). Some background in the no-light hemisphere of kainate-treated animals may represent mCherry-positive projections extending contralaterally from the illuminated hemisphere. Importantly, little mCherry signal was detected in the awake or anesthetized mice, even with light exposure. These results attest to the strong activity dependence of scFLARE2 in vivo.
Fig. 6.
scFLARE2 labeling in vivo. (A) Concentrated AAV viruses encoding scFLARE2 and TRE:mCherry were injected bilaterally into the hippocampus and cortex of adult mice. After 6 to 7 d of expression, an optical fiber was implanted in both left and right hemispheres, and blue light was delivered to the right hemisphere via the optical fiber (single 10-min session of 473-nm light at 10 mW, 50% duty cycle [2 s light every 4 s]), while mice were subject to either kainate treatment (right hemisphere, dispensed 2 h before light treatment), regular awake conditions (no treatment), or anesthesia. Mice were perfused and immunostained for imaging analysis 18 to 24 h later. (B) An example EEG trace recorded around the time of light stimulation (blue bar). The pink line indicates when the seizure detection system detected a seizure and light delivery was started. The green boxed region shows the EEG signal leading up to the light trigger. The yellow boxed region is an EEG signal showing an example behavioral seizure occurring during the light stimulation. (C) Representative confocal fluorescence images of both hemispheres, following the experiment in A. Activated scFLARE2 drives expression of mCherry (in red), while immunostaining for VP16 (in cyan) shows expression of the tool (scale bars, 10 μm). (D) Quantification of scFLARE2 activation. For each brain hemisphere, we quantified the total mCherry fluorescence intensity divided by scFLARE expression across seven consecutive brain sections around the virus injection site (three to five fields of view for each section). A total of four to six mice per condition were analyzed. See SI Appendix, Fig. S13 for additional fields of view from several animals. The errors bars reflect the SEM; ****P < 0.0001; one-way ANOVA with Tukey post hoc test. (E) To label downstream neurons indirectly activated during seizure, concentrated AAV viruses encoding scFLARE2 and TRE:mCherry were injected into the hippocampus and cortex of adult mice in the left hemisphere contralateral to kainate injection (right hemisphere). After 6 to 7 d of expression, an optical fiber was implanted and blue light was delivered to the left hemisphere (same parameters as in A), while mice were subject to either kainate treatment (dispensed 2 h before light treatment) or regular awake conditions (no treatment). Mice were perfused and immunostained for imaging analysis 18 to 24 h later. (F) Representative confocal fluorescence images of the left hemisphere. Activated scFLARE2 drives expression of mCherry (in red), while immunostaining for VP16 (in cyan) shows expression of the tool (scale bars, 10 μm). See SI Appendix, Fig. S14 for additional fields of view from several animals. Negative controls are from animals not receiving any light. (G) Quantification of scFLARE2 activation. For each animal (2 mice per condition), we quantified the total mCherry fluorescence intensity divided by scFLARE expression across seven consecutive brain sections around the virus injection site (three to five fields of view for each section). Errors bars reflect the SEM; ****P < 0.0001; one-way ANOVA with Tukey post hoc test.
In addition to the identification of neuronal circuits directly activated at a seizure focus, there is growing interest in identifying circuits outside of the focus that are indirectly recruited during seizures. Such neurons could serve as possible targets for therapeutic intervention to ameliorate the psychiatric comorbidities associated with epilepsy (24, 25). Previous studies have shown that optogenetic modulation of neurons in the hippocampus contralateral to the site of kainate injection or even as far away as the cerebellum can help to control seizure activity (23, 26). To test whether scFLARE2 has sufficient sensitivity to detect and tag neuronal subpopulations active outside of the seizure focus, we performed an experiment in which kainate was injected in the right hemisphere to induce seizures, while on-demand, closed-loop scFLARE2 labeling was performed in the contralateral (left) hemisphere (Fig. 6E). Indeed, we observed robust labeling of cells in the hemisphere contralateral to the kainate injection site (Fig. 6F and SI Appendix, Fig. S14), showcasing the efficacy of this tool for identifying critical downstream circuits in vivo (Fig. 6G).
Discussion
Calcium integrators such as FLARE (6), Cal-Light (7), CaMPARI (21), and TRIC (27) represent a new tool class for interrogating activated cell states. In contrast to real-time calcium indicators (10, 28), calcium integrators give stable access to transiently activated cell populations, which facilitates subsequent molecular characterization or functional manipulation. Here, our scFLARE incorporates a Ca-TEV that simplifies the tool design and enhances its robustness across different regimes of expression. We showed that scFLARE can also drive the expression of effectors [in contrast to CaMPARI (21)], is temporally gated by light [in contrast to TRIC (27)], and is fast, requiring only 10 min of stimulation in culture or in vivo [in contrast to Cal-Light (7)].
To develop the Ca-TEV that lies at the heart of scFLARE, we inserted the Ca2+-sensitive module (CaM/CKK) from the well-characterized real-time Ca2+ indicator YC6.1 (12) into various surface-exposed loops in TEV and determined that 61/62 was the best insertion site. Similar approaches, combined with directed evolution as we recently described (29), could potentially be used to generate other analyte-sensitive or conditional variants of TEV. To further enhance Ca-TEV, we also incorporated a catalysis-enhancing mutation discovered in a previous directed evolution study (16). Finally, to optimize its performance in neurons, we replaced the CaM domain in scFLARE with one of the slower off-rate CaM variants from GCaMP7s. With slower Ca2+ dissociation, the Ca-TEV within scFLARE has more time to cleave the TEVcs and release the TF, resulting in higher overall signal.
scFLARE2 was shown to be a robust calcium activity integrator in neuronal culture, giving mCherry expression proportional to the time and frequency of light + calcium stimulation. While sequential inputs (light followed by calcium or the reverse) were unable to activate scFLARE, simultaneous light + calcium for as little as 10 min robustly activated the tool. In vivo, elevated brain activity during seizures in the presence of light led to robust scFLARE2 turn on compared to awake or anesthetized mice in both directly and indirectly activated neuronal populations. Our findings suggest that scFLARE2 could be a valuable tool for the spatiotemporally precise, closed-loop interrogation of activated cellular ensembles in experimental models of complex brain disorders in animals.
Materials and Methods
Cloning.
See the SI Appendix, Plasmid Table for a list of genetic constructs used in this study. The cloning was carried out as described previously (6, 16) (SI Appendix, SI Materials and Methods).
HEK 293T Cell Culture and Transfection.
HEK cell culture and transfection was performed as previously described (6, 16) (for details, see SI Appendix, SI Materials and Methods).
scFLARE Experiments in HEK 293T.
HEK 293T cells expressing scFLARE constructs were processed 15 h posttransfection. To elevate cytosolic calcium, 100 μL of ionomycin and CaCl2 in complete growth media was added gently to the top of the media within a 48-well plate to final concentrations of 2 μM and 6 mM, respectively. For low-Ca2+ conditions, 200 μL complete growth media (with no added Ca2+) was added. For light conditions, cells were irradiated on a blue light-emitting diode light box for the indicated time periods. For the dark condition, HEK 293T cells were wrapped in aluminum foil. Afterward, the media in each well was removed and incubated with 250 μL complete growth media. HEK 293T cells were then incubated in the dark at 37 °C for 8 to 9 h and analyzed by confocal fluorescence microscopy, luciferase activity, or FACS analysis (for details, see SI Appendix, SI Materials and Methods).
Western Blot.
Western blot was carried out as described previously (6, 16) (for details, see SI Appendix, SI Materials and Methods).
Production of AAV Virus Supernatant for Neuron Transduction.
Virus generation was performed as previously described (6, 16) (for details, see SI Appendix, SI Materials and Methods).
scFLARE Experiments in Rat Cortical Neuron Culture.
Cortical neurons were harvested from rat embryos euthanized (we have complied with all relevant ethical regulations) at embryonic day 18 and plated in 24-well plates as previously described (30). A mixture of AAV viruses encoding scFLARE2 and a suitable reporter were added to neurons at DIV 11 to 12. After viral transduction, neurons were grown in the dark and wrapped in aluminum foil, and all subsequent manipulations were performed in a dark room with red-light illumination to prevent unwanted activation of the LOV domain. Neurons were stimulated in the presence or absence of blue light 6 d posttransduction (at DIV 17 to 18). To elevate cytosolic Ca2+, we used electrical stimulation generated by a Master 8 device (AMPI), which induces trains of electric stimuli. For drug stimulation experiments, drugs were diluted in media and added to each well. After stimulation, neurons were incubated in dark conditions at 37 °C before fixation with prewarm paraformaldehyde fixative solution (4% paraformaldehyde, 60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 2 mM MgCl2, 0.12 M sucrose, pH 7.3) for 10 min. Neurons were immunostained using rabbit anti-VP16 antibody and imaged with confocal microscopy (for details, see SI Appendix, SI Materials and Methods).
Electrophysiology on Dissociated Neuron Cultures.
Dissociated neuron cultures plated on glass coverslips were transduced with AAVs encoding scFLARE2 and TRE:mCherry-p2A-bReaChES at DIV (days in vitro) 11. At DIV17, neurons were stimulated with blue light and electrical trains for a total of 15 min. Intracellular recordings were performed 18 h after stimulation in a submerged chamber perfused with oxygenated aCSF (artificial cerebrospinal fluid) (126 mM NaCl, 26 mM NaHCO3, 10 mM glucose, 2.5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 1.25 mM NaH2PO4) at 2 mL/min and maintained at 33 °C by a chamber heater (BadController V, Luigs and Neumann). Neurons were visualized using differential interference contrast (DIC) illumination on an Olympus BX61WI microscope (Olympus Microscopy) and Lambda DG-4 optical illumination system (Sutter Instruments) with a charge-coupled device (CCD) camera (C7500, Hamamatsu). The recording pipettes were pulled from thick-walled borosilicate capillary glass (A-M Systems) using a P97 puller (Sutter Instruments) and were filled with (in millimolar) 126 K-gluconate, 10 Hepes, 4 KCl, 4 ATP-Mg, 0.3 GTP-Na, and 10 phosphocreatine (pH-adjusted to 7.3 with KOH, osmolarity 290 mOsm). Pipettes had a 7-10 megohm tip resistance. Whole-cell recordings were performed on mCherry-expressing neurons. A 350-pA current injection was performed in current-clamp mode to induce action potential firing. A 15-ms 530-nm wavelength light stimulation was delivered at 16 Hz frequency in current-clamp mode to observe optically induced action potentials. A 1-s 530-nm wavelength light stimulation was delivered in voltage-clamp mode at a holding voltage of -65mV to observe optically induced inward current. Data were acquired in pClamp software (Molecular Devices) using a Multiclamp 700B amplifier (Molecular Devices), low-pass filtered at 2 kHz, and digitized at 10 kHz (Digidata 1440A, Molecular Devices). Data analysis was performed using Clampfit (Molecular Devices).
Virus Infusion in Mice.
Adult wild-type male C57BL/6 mice, 12 to 20 wk old (Jackson Laboratory, Bar Harbor, ME), were used for all experiments. All procedures were carried out in accordance with the NIH guidelines for animal care and use and were approved by the Administrative Panel on Laboratory Animal Care of Stanford University. The hippocampus was targeted using the following coordinates from bregma: −2.3 mm AP (anterior-posterior), ±1.5 mm ML (medial-lateral), and −1.35 mm DV (dorsal-ventral) (for details, see SI Appendix, SI Materials and Methods).
Optical Fiber and EEG Implant.
Fiber optic cannula (200 μm core, 0.37 NA; Thorlabs, Newton, NJ) were implanted 0.5 mm above the virus injection site 6 d after virus injection. Mice were stereotaxically injected with kainate (40 nL, 20 mM in saline; Sigma-Aldrich) in the right dorsal hippocampus (−2 mm AP, −1.25 mm ML, and −1.6 mm DV) and implanted with bipolar electroencephalogram (EEG) depth electrodes (PlasticsOne; Roanoke, VA) within the same hemisphere.
scFLARE2 Labeling in Mice.
Light was delivered 6 to 7 d following viral injection. For light delivery, the optical fiber implant from one hemisphere was connected to a 473-nm diode-pumped solid state laser (Shanghai Laser & Optics Century Co., Ltd, China). For anesthetized experiments, mice received light following implant surgery and remained under anesthesia for an additional 30 min following light administration. For awake and kainate experiments, mice were allowed 2 h to recover from implant surgery before light delivery. For kainate experiments, closed-loop seizure detection and light delivery were carried out as previously described (23). Animals in all groups receiving light had one single session of 10-mW 473-nm light delivered at 2-s pulses every 4 s (50% duty cycle) for 10 min total. The light delivery was triggered either by the closed-loop system in kainate-injected animals or manually by the experimenter for animals in the awake or anesthetized group (for additional details, see SI Appendix, SI Materials and Methods).
Brain Slice Preparation and Immunohistochemistry.
Animals were euthanized 18 to 24 h after light administration by being deeply anesthetized with a mixture of ketamine and xylazine (80 to 100 mg/kg ketamine, 5 to 10 mg/kg xylazine; intraperitoneal) and transcardially perfused with 10 mL 0.9% sodium chloride solution followed by 10 mL cold 4% paraformaldehyde (PFA) dissolved in phosphate buffer solution. The excised brains were held in a 4% PFA solution for at least 24 h before being sectioned into 60-μm slices using a vibratome (Leica VT1200S; Leica Biosystems Inc.). Sections were immunostained using rabbit anti-VP16 primary antibody (1:2,000; Abcam) and anti-rabbit Alexa Fluor 647 secondary (1:1,000; Thermo Fisher Scientific) (for details, see SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
We are grateful to Stanford University, the Chan Zuckerberg Biohub, the Beckman Technology Development Seed Grant, and the NIH (R01 MH119353 to A.Y.T., F32 NS106764 to Q.-A.N., and R01 NS94668 to I.S.) for support of this work. M.I.S. was supported by an EMBO long-term postdoctoral fellowship (ALTF 1022-2015). FACS was performed at the Stanford Shared FACS Facility. Dr. Lin Ning (Stanford University) provided rat brain tissue. Gang Liu (MIT) built the light-emitting diode box used for blue-light irradiation of cells. Maja Djuristic (Stanford University) assisted with electrical stimulation of neurons.
Footnotes
Competing interest statement: A.Y.T. and M.I.S. have filed a patent covering some components used in this study (U.S. provisional application 62/906,373; CZB file CZB-123S-P1; Stanford file S19-269; KT file 103182-1132922-002400PR).
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2006521117/-/DCSupplemental.
Data Availability.
DNA plasmids encoding scFLARE1, scFLARE2, and reporter genes have been deposited to Addgene. Addgene ID: 158699 scFLARE1-p2A-eGFP, 158700 scFLARE2, 158701 TRE-mCherry-p2A-bReaches-TS, 158702 TRE-mCherry-p2A-Chrimson-TS, 158703 TRE-mCherry-p2A-eNpHR3.0-TS, 158738 scFLARE1. All other data are included in the article text and supporting information.
References
- 1.Fields S., Song O., A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Kim M. W., et al. , Time-gated detection of protein-protein interactions with transcriptional readout. eLife 6, e30233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kipniss N. H., et al. , Engineering cell sensing and responses using a GPCR-coupled CRISPR-Cas system. Nat. Commun. 8, 2212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim C. K., Cho K. F., Kim M. W., Ting A. Y., Luciferase-LOV BRET enables versatile and specific transcriptional readout of cellular protein-protein interactions. eLife 8, e43826 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talay M., et al. , Transsynaptic mapping of second-order taste neurons in flies by trans-tango. Neuron 96, 783–795.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W., et al. , A light- and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat. Biotechnol. 35, 864–871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee D., Hyun J. H., Jung K., Hannan P., Kwon H. B., A calcium- and light-gated switch to induce gene expression in activated neurons. Nat. Biotechnol. 35, 858–863 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Kapust R. B., et al. , Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Wehr M. C., et al. , Monitoring regulated protein-protein interactions using split TEV. Nat. Methods 3, 985–993 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Lin M. Z., Schnitzer M. J., Genetically encoded indicators of neuronal activity. Nat. Neurosci. 19, 1142–1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osawa M., et al. , A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat. Struct. Biol. 6, 819–824 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Truong K., et al. , FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule. Nat. Struct. Biol. 8, 1069–1073 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Chen T. W., et al. , Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikura M., et al. , Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256, 632–638 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Evnin L. B., Vásquez J. R., Craik C. S., Substrate specificity of trypsin investigated by using a genetic selection. Proc. Natl. Acad. Sci. U.S.A. 87, 6659–6663 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez M. I., Ting A. Y., Directed evolution improves the catalytic efficiency of TEV protease. Nat. Methods 17, 167–174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akerboom J., et al. , Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 32, 13819–13840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dana H., et al. , High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Palmer A. E., et al. , Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem. Biol. 13, 521–530 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Sun X. R., et al. , Fast GCaMPs for improved tracking of neuronal activity. Nat. Commun. 4, 2170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fosque B. F., et al. , Labeling of active neural circuits in vivo with designed calcium integrators. Science 347, 755–760 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Rajasethupathy P., et al. , Projections from neocortex mediate top-down control of memory retrieval. Nature 526, 653–659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krook-Magnuson E., Armstrong C., Oijala M., Soltesz I., On-demand optogenetic control of spontaneous seizures in temporal lobe epilepsy. Nat. Commun. 4, 1376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bui A. D., et al. , Dentate gyrus mossy cells control spontaneous convulsive seizures and spatial memory. Science 359, 787–790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander A., Maroso M., Soltesz I., Organization and control of epileptic circuits in temporal lobe epilepsy. Prog. Brain Res. 226, 127–154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krook-Magnuson E., Szabo G. G., Armstrong C., Oijala M., Soltesz I., Cerebellar directed optogenetic intervention inhibits spontaneous hippocampal seizures in a mouse model of temporal lobe epilepsy. eNeuro 1, ENEURO.0005-14.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X. J., et al. , A transcriptional reporter of intracellular Ca(2+) in Drosophila. Nat. Neurosci. 18, 917–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Broussard G. J., Liang R., Tian L., Monitoring activity in neural circuits with genetically encoded indicators. Front. Mol. Neurosci. 7, 97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y., et al. , Directed evolution of split APEX2 peroxidase. ACS Chem. Biol. 14, 619–635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loh K. H., et al. , Proteomic analysis of unbounded cellular compartments: Synaptic clefts. Cell 166, 1295–1307.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nunn C. M., et al. , Crystal structure of tobacco etch virus protease shows the protein C terminus bound within the active site. J. Mol. Biol. 350, 145–155 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA plasmids encoding scFLARE1, scFLARE2, and reporter genes have been deposited to Addgene. Addgene ID: 158699 scFLARE1-p2A-eGFP, 158700 scFLARE2, 158701 TRE-mCherry-p2A-bReaches-TS, 158702 TRE-mCherry-p2A-Chrimson-TS, 158703 TRE-mCherry-p2A-eNpHR3.0-TS, 158738 scFLARE1. All other data are included in the article text and supporting information.